Translate this page into:
Synthesis and anti-α-glucosidase activity evaluation of betulinic acid derivatives
⁎Corresponding author. xuetaoxu@wyu.edu.cn (Xue-Tao Xu)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
In this article, a series of betulinic acid derivatives (3a ∼ 3u, 4a ∼ 4e) were synthesized through a stepwise structure optimization and evaluated for their anti-α-glucosidase activities. All synthesized derivatives exhibited stronger anti-α-glucosidase activities (IC50: 0.56 ± 0.05 ∼ 3.99 ± 0.23 μM) than betulinic acid (IC50: 7.21 ± 0.58 μM) and acarbose (IC50: 611.45 ± 15.51 μM). Compound 3q presented the outstanding inhibitory activity (IC50: 0.56 ± 0.05 μM), which was ∼ 1100 time stronger than that of acarbose. Compound 3q was revealed as a reversible and noncompetitive α-glucosidase inhibitor by inhibitory mechanism assay. Fluorescence spectra, 3D fluorescence and CD spectra results showed that the interaction of compound 3q with α-glucosidase caused the conformational and secondary structure content change of α-glucosidase. Finally, the molecular docking simulated the interaction between compound 3q with α-glucosidase and the physicochemical parameter was assessed using SwissADME software.
Keywords
Betulinic acid
Synthesis
a-Glucosidase inhibitor
Docking
1 Introduction
Diabetes mellitus is one of chronic metabolic diseases, that requires long-term drug intervention to regulate blood sugar levels (DeFronzo et al., 2015; Kahn et al., 2014). Type 2 diabetes is predominant in diabetes, more than 90 % of all cases (Adeghate et al., 2006; Guariguata et al., 2014). The insufficient insulin secretion and insulin resistance leads to hyperglycemia, that is the hallmark of diabetes (Wang et al., 2020). Thence, glucose control is a key strategy to treat diabetes (Deng et al., 2022). Inhibiting hydrolase enzymes of carbohydrate such as α-glucosidase can obviously control the postprandial hyperglycemia and reduce the risk of diabetes complications (Rafique et al., 2020; Zheng et al., 2021).
α-Glucosidase is an important enzyme in the small intestine, that can catalytic hydrolysis the carbohydrates to produce glucose (Dhameja and Gupta, 2019; Ghani, 2015). α-Glucosidase inhibitors can block the activity of α-glucosidase, resulting in the regulation of the blood glucose and postprandial hyperglycemia (Ali et al., 2017; Hameed et al., 2019). Up to now, only several α-glucosidase inhibitors including voglibose, acarbose, and miglitol have been used clinically to treat type-2 diabetes (Wang et al., 2018; Wang et al., 2017; Wang et al., 2017). However, their long-term use leads to some side effects such as flatulence and diarrhea. Therefore, to find efficient and safer α-glucosidase inhibitors has attracted more and more attention.
Betulinic acid (BA, Fig. 1) is a natural pentacyclic lupane-type triterpenoid, mainly isolated from birch bark (Zhang et al., 2017). In recent years, scientific researches have revealed that BA is a very valuable natural star compound. BA and its derivatives have shown an array of pharmacological activities, including anti-cancer, anti-viral, and anti-inflammatory (Catteau et al., 2018; Kim et al., 2014; Putta et al., 2016). Moreover, BA shows effective antidiabetic effects due to its inhibition activity against α-glucosidase (IC50 = 10.6 μM) and human PTP1B (IC50 = 3.5 μM) (Ding et al., 2018; Ramirez-Espinosa et al., 2011). Besides, some BA derivatives have been reported as potential α-glucosidase agents, such as 2,3-indolobetulinic acid derivatives (Khusnutdinova et al., 2019), methyl ester derivatives of betulinic acid (Khusnutdinova et al., 2017), amide derivatives of betulinic acid (Kazakova et al., 2020), and N-allylated/N-alkylated niacin hybrids derivatives of betulinic acid (Narender et al., 2013) (Fig. 1). Also, combining with our research results on the structural modification of triterpenoid, including oleanolic acid and ursolic acid, we believe that structural modification of BA would be an effective strategy to develop novel α-glucosidase inhibitors.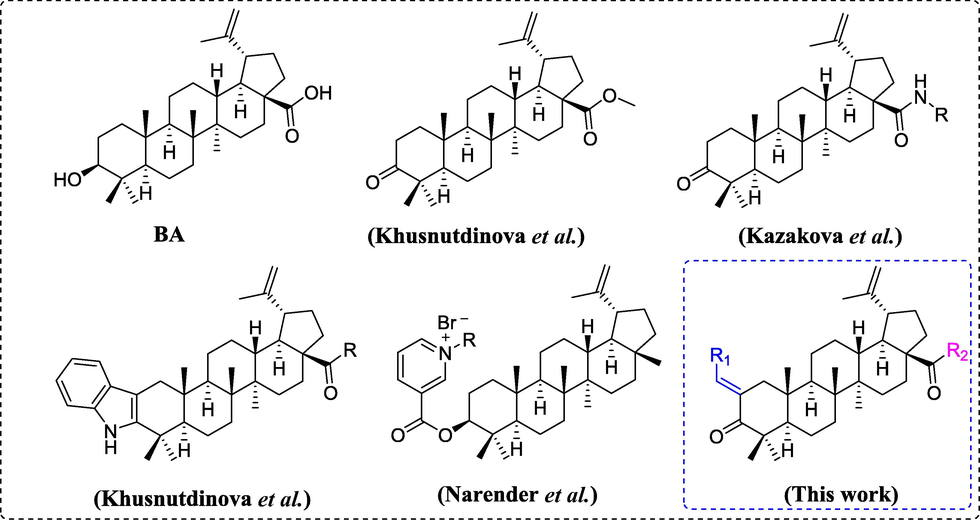
Chemical structures of some reported α-glucosidase inhibitors.
On the other hand, styrene group is an important substituent in many pharmacologically compounds, such as chalcone derivatives (Leong et al., 2018); coumarin derivatives (Saeedi et al., 2017; Zawawi et al., 2015); cinnamic acid derivatives (Xu et al., 2020). Most of them present hypoglycemic and anti-α-glucosidase activity. For developing BA derivatives as potential α-glucosidase inhibitors, bioactive substituent of styrene was incorporated into BA skeleton via the hybridization strategy. In addition, synthesized BA derivatives were assayed for their α-glucosidase.
2 Results and discussion
2.1 Chemistry
The BA derivatives 3a ∼ 3u were firstly synthesized using BA as starting material (Schemes 1). BA (1) was oxidized to produce betulonic acid 2 under 2-lodoxybenzoic acid (IBX). Betulonic acid 2 underwent Claisen-Schmidt condensation reaction with substituted aldehydes to yield BA derivatives 3a ∼ 3u.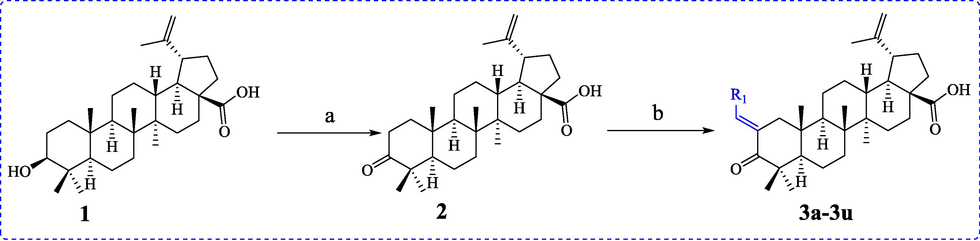
Synthesis of BA derivatives 3a ∼ 3u. Reagents and condition: (a) IBX, DMSO, 0 °C; (b) NaOH, EtOH, rt.
The BA derivatives 4a ∼ 4e were subsequently synthesized using 3q as starting material (Schemes 2). All synthesized BA derivatives were identified by 1H NMR, 13C NMR and HRMS.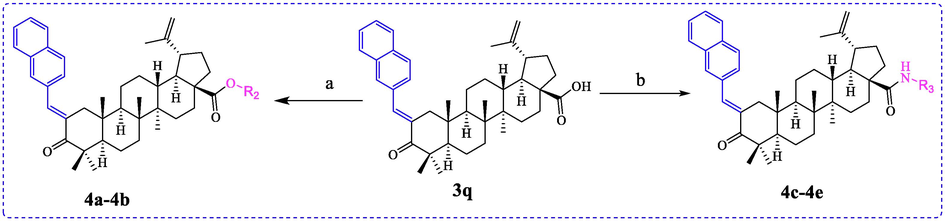
Synthesis of BA derivatives 4a ∼ 4e. Reagents and condition: (a) CH3I or benzyl bromide, K2CO3, DMF, rt; (b) HATU, DIPEA, DMF, 0 °C.
2.2 α-Glucosidase inhibitory activity assay and SAR study
The α-glucosidase inhibitory activities of BA derivatives 3a ∼ 3u were firstly assayed and the results were listed in Table 1. The results revealed that BA derivatives 3a ∼ 3u presented potent inhibitory activity toward α-glucosidase (IC50 from 0.56 ± 0.05 μM to 3.99 ± 0.23 μM), which was stronger than BA (IC50: 7.21 ± 0.58 μM) and standard control acarbose (IC50 611.45 ± 15.51 μM, P < 0.05). The results showed that the incorporation of styrene at C-28 position of BA could effectively strengthen its α-glucosidase inhibitory activity. Among them, derivative 3q showed the strogest inhibitory activity (IC50: 0.56 ± 0.05 μM). Thence, the structural modification of BA might be a benefit method for improving its anti-α-glucosidase activity and obtaining potential inhibitors. The standard error was obtained from four replicates. aP < 0.05, compared to acarbose.
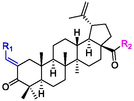
Compound
R1
R2
IC50 (μM)
3a
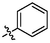
OH
1.06 ± 0.07a
3b
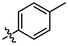
OH
2.63 ± 0.09a
3c
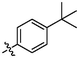
OH
1.11 ± 0.05a
3d
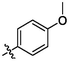
OH
2.91 ± 0.34a
3e
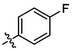
OH
3.06 ± 0.26a
3f
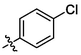
OH
1.89 ± 0.08a
3g
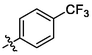
OH
1.39 ± 0.06a
3h
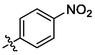
OH
2.58 ± 0.15a
3i
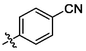
OH
1.90 ± 0.06a
3j
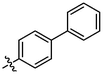
OH
1.40 ± 0.03a
3k

OH
3.43 ± 0.23a
3L
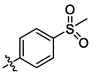
OH
2.10 ± 0.19a
3m
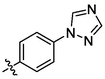
OH
2.23 ± 0.17a
3n
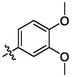
OH
3.69 ± 0.33a
3o
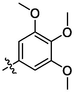
OH
3.25 ± 0.21a
3p
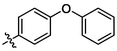
OH
0.86 ± 0.06a
3q
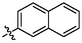
OH
0.56 ± 0.05a
3r
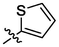
OH
1.67 ± 0.05a
3s
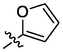
OH
2.03 ± 0.07a
3t
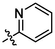
OH
2.12 ± 0.16a
3u
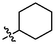
OH
1.37 ± 0.11a
4a
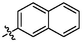
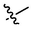
3.39 ± 0.28a
4b
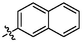
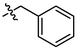
2.71 ± 0.17a
4c
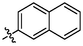
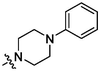
3.98 ± 0.22a
4d
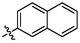
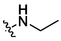
3.92 ± 0.15a
4e
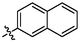
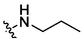
3.99 ± 0.23a
BA
7.21 ± 0.58a
Acarbose
611.45 ± 15.51
For revealing the effect of different substituents on anti-α-glucosidase inhibitory, structure–activity relationship (SAR) was analyzed using derivative 3a as template compound that had no substituent at benzene ring. For BA derivatives 3b ∼ 3q with different substituent at benzene ring, BA derivatives 3b ∼ 3o presented lower inhibitory activities than derivative 3a, suggesting the introduction of CH3, C(CH3)3, OCH3, F, Cl, CF3, NO2, CN, C6H5, N(CH3)2, SO2CH3, (OCH3)2, (OCH3)3, or N3H2 reduced the inhibitory activity. While, BA derivatives 3p (phenyl ether) and 3q (naphthalene) showed stronger inhibitory activities than derivative 3a, indicating that the introduction of phenyl ether or naphthalene enhanced the inhibitory activity. The results showed that the substituents at benzene ring might not be benefit for their anti-α-glucosidase inhibitory, except substituents with conjugate properties, such as phenyl ether and naphthalene. Moreover, the introduction of heterocycle (thiophene (3r), furan (3s), pyridine (3t)) and naphthenic (cyclohexane (3u)) pulled the inhibitory activity lower, compared to derivative 3a. The results indicated that the heterocycle was adverse compared to benzene ring.
In order to obtain more potent inhibitors, the carboxyl moiety of BA was optimized based on compound 3q with naphthalene group, yielding BA derivatives 4a ∼ 4e. Unfortunately, their inhibitory activity assay results showed that the introduction of ester group, amino group, or piperazine group reduced their α-glucosidase inhibitory activities. The results indicated that the carboxyl moiety was critical for keeping the anti-α-glucosidase activity of BA comparing to ester and amide group. All above results gave useful guidance information for the structural modification of BA.
2.3 Inhibitory mechanism assay
Compound 3q (IC50: 0.56 ± 0.05) with strongest inhibitory activity was selected as representative compound to assay their inhibitory mechanism. Fig. 2a showed the plots of initial velocity of the enzymatic reaction via α-glucosidase concentration in the absence and presence of compound 3q at different concentrations. The plots were straight lines with different positive slopes and passed through the ordinate origin, implied that compound 3q inhibited α-glucosidase activity through the reversible inhibition mode (Yang et al., 2021).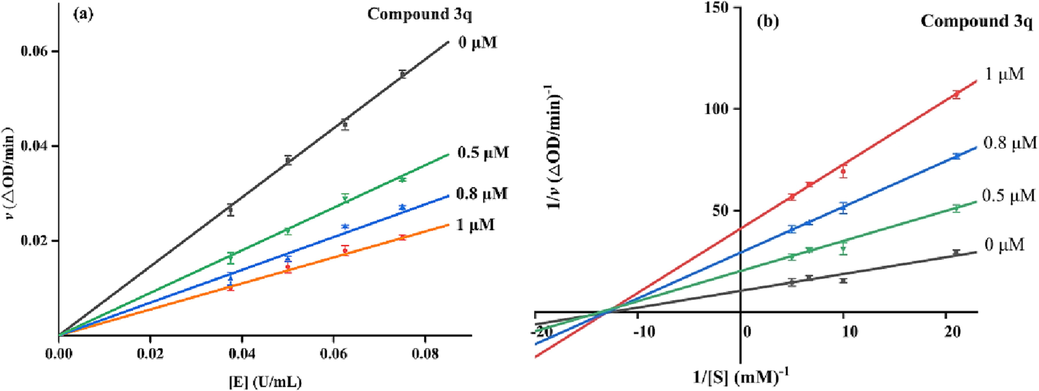
(a) The initial velocity via α-glucosidase concentration in the absence and presence of compound 3q; (b) Lineweaver-Burk plots for the inhibition α-glucosidase in the absence and presence of compound 3q.
The inhibition type of α-glucosidase by compound 3q was assayed by Lineweaver-Burk plots (Fig. 2b). The slope of plot increased with the increase of compound 3q concentration, and all lines intersected at one point on the × axis. These results were coinciding with the traits of noncompetitive inhibition (Yang et al., 2021). In addition, the slops via compound 3q concentrations to give the inhibition constant (KI) was calculated as 0.295 μM.
2.4 Fluorescence quenching analysis
To understand the inhibitory mechanism of compound 3q against α-glucosidase, the effect of compound 3q on the fluorescence of α-glucosidase was analyzed. It could be seen that α-glucosidase appeared an emission peak at 340 nm, which was decreased by the addition of compound 3q (Fig. 3a ∼ d). The results revealed that compound 3q could interact with α-glucosidase, leading to the conformation change of α-glucosidase. Also, the fluorescence quenching degree of α-glucosidase by compound 3q was slightly increased with the increase of temperature.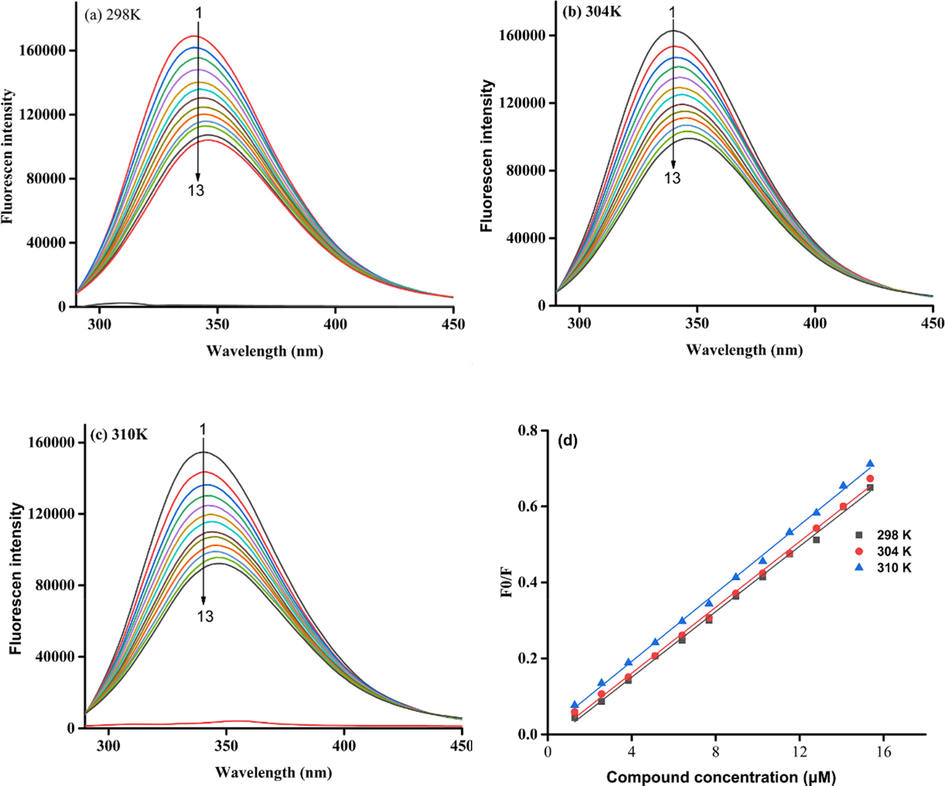
Fluorescence spectra of α-glucosidase in the presence of compound 3q. (a) 289 K, (b) 304 K, (c) 310 K. Compound 3q concentration of curves 1–13 were 0, 1.28, 2.56, 3.84, 5.12, 6.4, 7.68, 8.96, 10.24, 11.52, 12.8, 14.08, and 15.36 μM respectively, (d) the Stern-Volmer plots of α-glucosidase quenched by compound 3q.
The quenching mechanism was analyzed by Stern Volmer equation and the results (Table 2) showed that the quenching constant (Ksv) was increased with the increase of temperature, revealing a static quenching. The binding constants were greater than 105 L mol−1, and the binding bits number was approximately equal to 1. At the same time, the thermodynamic parameters were calculated using the Van’t Hoff and the Gibbs-Helmholtz Eq. The negative values of ΔG suggested the forming of complexes occurring spontaneously, and negative values of ΔH and ΔS implied the mainly interaction being hydrogen band and van der Waals forces.
T(K)
KSV
(×105 L mol−1)Ka
(×105 L mol−1)n
△H
(KJ/moL)△G
(KJ/moL)△S
(J/(mol·K)
298
4.31
1.59
1.09
−78.17
−29.67
−147.67
304
4.33
6.80
0.99
–33.28
310
4.48
8.24
0.90
–33.75
2.5 3D fluorescence spectra assay
3D fluorescence spectra assay was carried out to analyze the effect of compound 3q toward the conformational change of α-glucosidase. As shown in the 3D fluorescence spectra of α-glucosidase (Fig. 4a), Tyr with Trp residues appeared spectral properties at peak 1 (λex = 280 nm, λem = 340 nm), protein structure transition from the n → π* caused the fluorescence feature at peak 2 (λex = 225 nm, λem = 340 nm). However, the fluorescence intensity of peak 1 reduced by 11.93 %, and Peak 2 reduced by 34.64 %, after treated with compound 3q (Fig. 4b). The results showed that the interaction of compound 3q with α-glucosidase would cause the conformational change of the enzyme.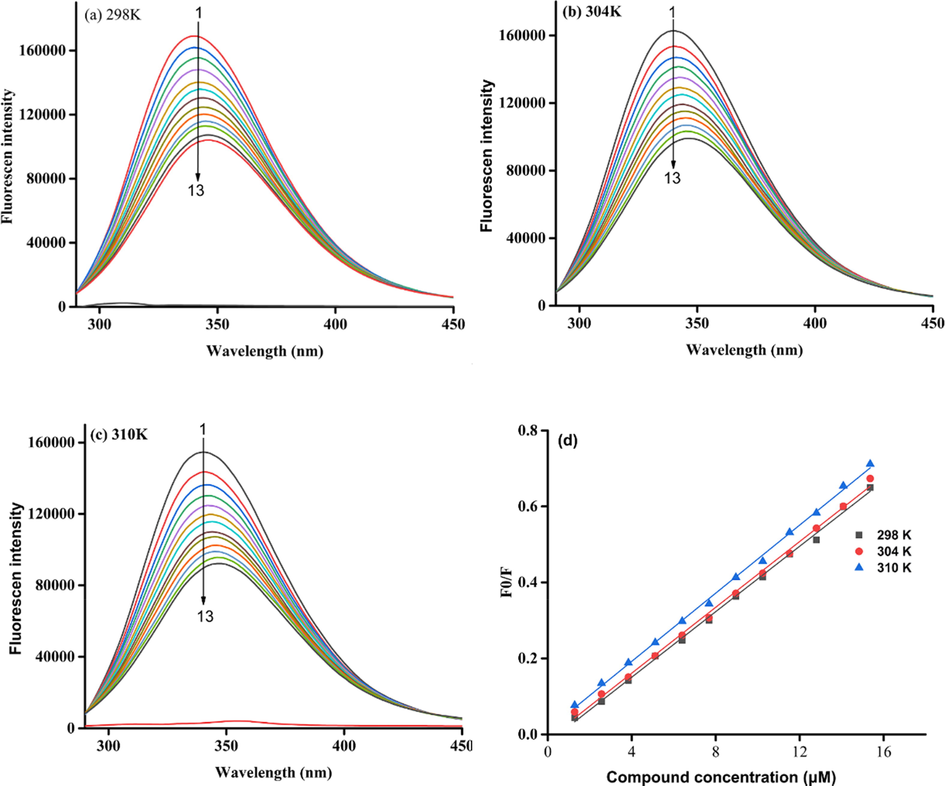
The 3D fluorescence spectra of α-glucosidase (a) and α-glucosidase with compound 3q system (b).
2.6 CD spectra assay
The CD spectra of protein could reflect its secondary structure content. The CD spectra of α-glucosidase appeared two negative bands at 210 and 222 nm, that were the typical features of α-helixes (Fig. 5). Treatment with compound 3q lead to the obviously reduction of negative bands intensity (Fig. 5). The secondary structure content change of α-glucosidase caused by compound 3q was also obtained (Table 3). Compound 3q (molar ratios: 3:1) caused a decrease in the content of the α-helix (from 11.00 to 9.40 %), β-turn (from 20.20 to 19.50 %), and random coil (from 33.30 to 32.50 %), while an increase in β-Sheet (from 30.20 to 34.40 %), respectively. These results suggested that the binding of compound 3q to α-glucosidase altered the conformation followed by the active center transfer or the shutdown.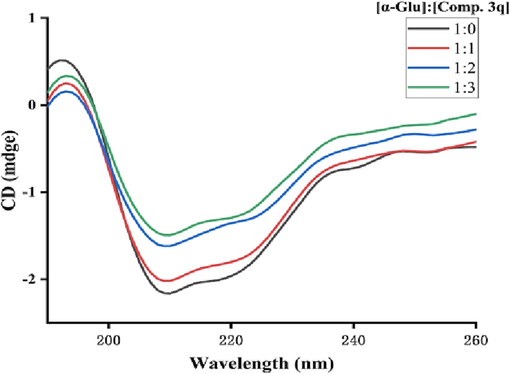
CD spectra of α-glucosidase and compound 3q system.
Molar ratio
[α-Glu]:[Comp.20]α-Helix
(%)β-Sheet
(%)β-Turn
(%)Rndm Coil
(%)
1:0
11.00
30.20
20.20
33.30
1:1
10.40
30.30
20.20
33.70
1:2
9.50
33.20
19.7
32.90
1:3
9.40
34.40
19.50
32.50
2.7 Molecular docking
Finally, the binding mode of compound 3q with α-glucosidase was simulated using molecular docking software and the docking results was presented in Fig. 6. Compound 3q was anchored into the middle side of active site of α-glucosidase with naphthalene ring part locating outside of the active site and carboxyl moiety of betulinic acid nesting inside (Fig. 6a). The detailed interaction was investigated and found that the carboxyl moiety formed hydrogen bonds with Asp349 (bond length: 2.6 Å), which was reported as the key interaction between compound and protein. At the same time, compound 3q made hydrophobic interactions with Phe157, Phe158, Phe177, Als278, and Phe300. Mentioned above interactions helped compound 3q to bind into the active site of the α-glucosidase.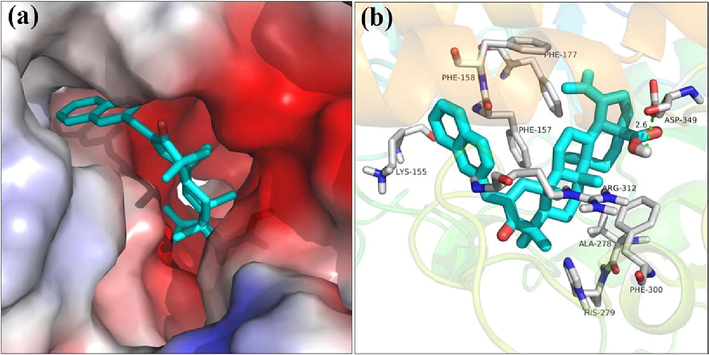
The molecular docking of α-glucosidase with compound 3q.
2.8 Physicochemical parameters assay
The physicochemical parameters of betulinic acid derivatives 3a, 3c, 3f, 3g, 3i, 3j, 3p, 3q, 3r, and 3u were further assessed using SwissADME software (https://www.swissadme.ch/index.php) to assess the drug-like profile and the results were listed in Table 4. Overall, the studied betulinic acid derivatives presented favourable drug-likeness profile, in spite of the molecular weight of each derivative was higher than 500, due to the high molecular weight of betulinic acid (456.7). In particular, RB, HBA, and HBD values (RB < 10, HBA < 5, HBD < 10) were compatible with good TPSA permeability (TPSA < 90 Å2). The physicochemical parameters assay results would contribute to the further structural modification of betulinic acid and in-depth pharmacology research. MW (Molecular Weight), Na (Num. heavy atoms), RB (Rotatable bonds), HBA (H-Bond acceptor atoms), HBD (H-Bond donor atoms), TPSA (Topology polar surface area), WS (Water solubility).
Compound
MW (g/mol)
Na
RB
HBA
HBD
TPSA (Å2)
LogPo/w
WS
3a
542.79
40
3
3
1
54.37
7.63
Poorly soluble
3c
598.90
44
4
3
2
54.37
8.77
Poorly soluble
3f
577.24
41
3
3
1
54.37
8.11
Poorly soluble
3g
610.79
44
4
6
1
54.37
8.65
Poorly soluble
3i
567.80
42
3
4
1
78.16
7.40
Poorly soluble
3j
618.89
46
4
3
1
54.37
8.76
Insoluble
3p
634.89
47
5
4
1
63.60
8.78
Insoluble
3q
592.85
44
3
3
1
54.37
8.47
Insoluble
3r
548.82
39
3
3
1
82.61
7.62
Poorly soluble
3u
548.84
40
3
3
1
54.37
7.98
Poorly soluble
3 Conclusion
In conclusion, a series of betulinic acid derivatives (3a ∼ 3u, 4a ∼ 4e) were synthesized and assayed their α-glucosidase inhibitory activity. All synthesized derivatives exhibited stronger anti-α-glucosidase activities than betulinic acid and acarbose. Compound 3q presented the outstanding inhibitory activity (IC50: 0.56 ± 0.05 μM). Compound 3q was revealed as a reversible and noncompetitive α-glucosidase inhibitor. 3D fluorescence and CD spectra were used to assay the effect of compound 3q on the conformational and secondary structure content change of α-glucosidase. Molecular docking was also used to simulate the interaction between compound 3q and α-glucosidase. Overall, betulinic acid derivatives could be used as the leading compound in the management of Type 2 diabetes.
4 Experimental
4.1 General procedure for preparation of BA derivatives
4.1.1 General procedure for preparation of BA derivatives 3a ∼ 3u
2-lodoxybenzoic acid (2.46 g 4.4 mol) was added slowly to a solution of BA (2 g, 2.2 mmol) in dry DMSO (25 mL) at 0 °C. The mixture was stirred until TLC indicated completed consumption of BA. The reaction was quenched with water and extracted with ethyl acetate, followed by evaporation under reduced pressure to give betulonic acid 2. To a solution of betulonic acid 2 (1 mmol) and NaOH (6 mmol) in ethanol (10 mL) at 0 °C was added substituted aldehydes (3 mmol). After reaction completed, the mixture was quenched by addition of dilute hydrochloric acid (pH = 4 ∼ 5), the resulting solution was extracted with ethyl acetate. The combined organic extract was washed with saturated NaCl, dried Na2SO4, and concentrated in vacuo. Final purification of the residue by column chromatography afforded BA derivatives 3a ∼ 3u.
(3a, C37H50O3). White sold; Yield 66 %; m. p. 182 – 185 °C; 1H NMR (500 MHz, Chloroform - d) δ 7.51–7.46 (m, 1H), 7.41 (d, J = 5.8 Hz, 4H), 7.33 (tt, J = 5.5, 2.6 Hz, 1H), 4.76 (d, J = 2.2 Hz, 1H), 4.65 (t, J = 1.9 Hz, 1H), 3.03 (ddd, J = 22.3, 13.6, 3.2 Hz, 2H), 2.34–2.18 (m, 3H), 1.99 (q, J = 8.3, 7.9 Hz, 2H), 1.73 (s, 3H), 1.67 (t, J = 11.4 Hz, 1H), 1.55–1.38 (m, 12H), 1.33–1.22 (m, 2H), 1.13 (d, J = 10.8 Hz, 7H), 1.03 (s, 6H), 0.97 (s, 3H), 0.79 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 207.45, 181.41, 149.70, 136.54, 135.09, 133.31, 129.48, 127.61, 127.58, 108.80, 55.58, 51.91, 48.28, 47.55, 45.96, 44.31, 43.58, 41.63, 39.64, 37.58, 36.16, 35.61, 32.18, 31.18, 29.74, 28.80, 28.61, 24.73, 21.47, 20.77, 19.48, 18.64, 14.95, 14.60, 13.75.
(3b, C38H52O3). White sold; Yield 70 %; m. p. 289.7–290.5 °C; 1H NMR (500 MHz, Chloroform-d) δ 7.47 (d, J = 2.6 Hz, 1H), 7.33 (d, J = 8.0 Hz, 2H), 7.22 (d, J = 7.9 Hz, 2H), 4.76 (d, J = 2.2 Hz, 1H), 4.65 (t, J = 1.9 Hz, 1H), 3.09–2.98 (m, 2H), 2.38 (s, 3H), 2.33–2.17 (m, 3H), 2.06–1.94 (m, 2H), 1.73 (s, 3H), 1.67 (t, J = 11.4 Hz, 1H), 1.57–1.38 (m, 11H), 1.33–1.20 (m, 4H), 1.14 (s, 3H), 1.11 (s, 3H), 1.03 (s, 3H), 0.97 (s, 3H), 0.90–0.81 (m, 1H), 0.78 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 208.38, 182.45, 150.69, 138.79, 137.66, 133.39, 133.26, 130.57, 129.35, 109.78, 56.59, 52.86, 49.28, 48.58, 46.97, 45.23, 44.66, 42.62, 40.63, 38.58, 37.16, 36.57, 33.18, 32.19, 30.74, 29.81, 29.65, 25.74, 22.46, 21.77, 21.53, 20.48, 19.64, 15.95, 15.60, 14.75.
(3c, C41H58O3). White sold; Yield 84 %; m. p. 261 – 263 °C; 1H NMR (500 MHz, Chloroform-d) δ 7.48–7.43 (m, 3H), 7.40 (d, J = 8.5 Hz, 2H), 4.78 (d, J = 2.2 Hz, 1H), 4.67 (dd, J = 3.3, 1.7 Hz, 1H), 3.11–3.00 (m, 2H), 2.33–2.21 (m, 3H), 2.06–1.94 (m, 2H), 1.74 (s, 3H), 1.68 (t, J = 11.4 Hz, 1H), 1.59–1.49 (m, 3H), 1.49–1.40 (m, 7H), 1.35 (s, 9H), 1.33 (s, 1H), 1.30–1.25 (m, 3H), 1.14 (s, 3H), 1.11 (s, 3H), 1.04 (s, 3H), 0.98 (s, 3H), 0.88 (t, J = 6.9 Hz, 2H), 0.79 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 208.46, 182.40, 151.94, 150.85, 137.47, 133.40, 133.24, 130.54, 125.66, 109.69, 56.62, 52.79, 49.28, 48.59, 46.96, 45.17, 44.82, 42.63, 40.64, 38.59, 37.16, 36.54, 34.93, 33.17, 32.19, 31.73, 31.48, 31.35, 30.79, 29.82, 29.73, 25.79, 22.80, 22.42, 21.81, 20.51, 19.70, 16.02, 15.60, 14.75, 14.27.
(3d, C38H52O4). White sold; Yield 73 %; m. p. 271.9–274.2 °C; 1H NMR (500 MHz, Chloroform-d) δ 7.47 (t, J = 2.0 Hz, 1H), 7.41 (d, J = 2.1 Hz, 1H), 7.40 (d, J = 2.3 Hz, 1H), 6.95 (d, J = 2.0 Hz, 1H), 6.94 (d, J = 2.1 Hz, 1H), 4.77 (d, J = 2.3 Hz, 1H), 4.69–4.64 (m, 1H), 3.85 (s, 3H), 3.03 (ddd, J = 15.5, 6.3, 4.4 Hz, 2H), 2.35–2.16 (m, 3H), 2.06–1.96 (m, 2H), 1.74 (s, 4H), 1.68 (t, J = 11.4 Hz, 1H), 1.58–1.48 (m, 4H), 1.48–1.40 (m, 6H), 1.34–1.22 (m, 2H), 1.14 (s, 4H), 1.11 (s, 3H), 1.03 (s, 3H), 0.97 (s, 3H), 0.79 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 208.30, 182.19, 159.94, 150.78, 137.45, 132.37, 132.07, 128.79, 114.13, 109.75, 56.59, 55.45, 52.76, 49.30, 48.63, 46.97, 45.13, 44.80, 42.63, 40.64, 38.59, 37.16, 36.52, 33.18, 32.19, 30.76, 29.81, 29.75, 25.78, 22.45, 21.81, 20.51, 19.67, 15.99, 15.60, 14.76.
(3e, C37H49FO3). White sold; Yield 72 %; m. p. 297.3–300.7 °C; 1H NMR (500 MHz, Chloroform-d) δ 7.44 (t, J = 2.0 Hz, 1H), 7.42–7.37 (m, 2H), 7.13–7.07 (m, 2H), 4.76 (d, J = 2.2 Hz, 1H), 4.66 (dd, J = 2.3, 1.4 Hz, 1H), 3.05–2.96 (m, 2H), 2.33–2.21 (m, 2H), 2.18 (dd, J = 16.7, 2.9 Hz, 1H), 2.04–1.94 (m, 2H), 1.73 (s, 4H), 1.67 (t, J = 11.4 Hz, 1H), 1.56–1.40 (m, 12H), 1.34–1.22 (m, 2H), 1.13 (d, J = 12.2 Hz, 7H), 1.03 (s, 3H), 0.97 (s, 3H), 0.78 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 208.25, 182.41, 163.64, 161.65, 150.66, 136.36, 133.93, 132.36, 132.29, 132.22, 132.20, 115.80, 115.63, 109.82, 56.58, 52.86, 49.27, 48.55, 46.96, 45.27, 44.52, 42.62, 40.63, 38.57, 37.15, 36.59, 33.15, 32.17, 30.72, 29.80, 29.61, 25.72, 22.45, 21.78, 20.46, 19.63, 15.94, 15.60, 14.74.
(3f, C37H49ClO3). White sold; Yield 59 %; m. p. 306.5–308.9 °C; 1H NMR (500 MHz, Chloroform-d) δ 7.43 (dd, J = 3.0, 1.5 Hz, 1H), 7.39 (d, J = 2.1 Hz, 1H), 7.38 (d, J = 2.3 Hz, 1H), 7.34 (d, J = 8.7 Hz, 2H), 4.76 (d, J = 2.2 Hz, 1H), 4.67–4.64 (m, 1H), 3.06–2.95 (m, 2H), 2.34–2.21 (m, 2H), 2.20–2.15 (m, 1H), 2.02–1.96 (m, 2H), 1.73 (s, 4H), 1.67 (t, J = 11.4 Hz, 1H), 1.54–1.43 (m, 12H), 1.27 (dq, J = 16.5, 4.7 Hz, 4H), 1.14 (s, 3H), 1.12 (s, 3H), 1.03 (s, 3H), 0.97 (s, 3H), 0.78 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 208.20, 150.60, 136.16, 134.83, 134.50, 134.48, 131.63, 128.86, 109.87, 77.41, 77.36, 77.16, 76.91, 56.57, 52.90, 49.27, 48.55, 46.96, 45.33, 44.52, 42.63, 40.64, 38.57, 37.16, 36.62, 33.16, 32.18, 30.72, 29.84, 29.80, 29.57, 25.70, 22.47, 21.78, 20.45, 19.62, 15.94, 15.60, 14.75.
(3 g, C38H49F3O3). White sold; Yield 89 %; m. p. 283.0–283.4 °C; 1H NMR (500 MHz, Chloroform-d) δ 7.67 (s, 1H), 7.65 (s, 1H), 7.50 (s, 1H), 7.49 (s, 1H), 7.48–7.45 (m, 1H), 4.76 (d, J = 2.3 Hz, 1H), 4.65 (t, J = 1.8 Hz, 1H), 3.01 (ddd, J = 16.3, 6.6, 3.1 Hz, 2H), 2.33–2.17 (m, 3H), 2.06–1.94 (m, 2H), 1.72 (s, 4H), 1.66 (t, J = 11.4 Hz, 1H), 1.52–1.41 (m, 11H), 1.30–1.24 (m, 2H), 1.14 (s, 3H), 1.13 (s, 3H), 1.02 (s, 3H), 0.97 (s, 3H), 0.88 (t, J = 6.9 Hz, 1H), 0.79 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 208.14, 182.33, 150.58, 139.57, 136.41, 135.66, 130.36, 125.56, 125.53, 125.50, 125.47, 109.87, 56.57, 52.98, 49.26, 48.51, 46.96, 45.44, 44.44, 42.64, 40.65, 38.57, 37.15, 36.69, 33.15, 32.17, 31.73, 30.72, 29.79, 29.50, 25.68, 22.80, 22.49, 21.78, 20.44, 19.62, 15.95, 15.61, 14.74, 14.27.
(3 h, C37H49NO5). White sold; Yield 53 %; m. p. 214.5–216.7 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.26 (d, J = 8.8 Hz, 2H), 7.53 (d, J = 8.8 Hz, 2H), 7.47 (q, J = 1.5 Hz, 1H), 4.75 (d, J = 2.2 Hz, 1H), 4.65 (t, J = 1.8 Hz, 1H), 3.05–2.95 (m, 2H), 2.33–2.18 (m, 3H), 1.98 (q, J = 7.7, 6.9 Hz, 2H), 1.71 (s, 3H), 1.78–1.61 (m, 2H), 1.54–1.38 (m, 8H), 1.34–1.21 (m, 4H), 1.14 (d, J = 7.0 Hz, 7H), 1.02 (s, 3H), 0.97 (s, 3H), 0.78 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 207.91, 182.41, 150.47, 147.20, 142.60, 137.88, 134.62, 130.77, 123.82, 109.94, 56.54, 52.97, 49.23, 48.48, 46.95, 45.51, 44.52, 42.64, 40.64, 38.54, 37.14, 36.73, 33.12, 32.14, 30.67, 29.77, 29.46, 25.63, 22.49, 21.79, 20.41, 19.59, 15.98, 15.59, 14.72.
(3i, C38H49NO3). White sold; Yield 58 %; m. p. 292.2–295.5 °C; 1H NMR (500 MHz, Chloroform-d) δ 7.72–7.66 (m, 2H), 7.48 (d, J = 8.3 Hz, 2H), 7.45–7.37 (m, 1H), 4.76 (d, J = 2.3 Hz, 1H), 4.65 (t, J = 1.8 Hz, 1H), 3.05–2.93 (m, 2H), 2.33–2.16 (m, 3H), 2.06–1.94 (m, 2H), 1.72 (s, 3H), 1.79–1.62 (m, 2H), 1.55–1.40 (m, 10H), 1.35–1.19 (m, 4H), 1.14 (s, 6H), 1.12 (s, 3H), 1.02 (s, 4H), 0.97 (s, 3H), 0.78 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 207.98, 182.29, 150.57, 140.61, 137.34, 135.06, 132.32, 130.61, 118.78, 111.73, 109.87, 56.54, 52.95, 49.24, 48.49, 46.94, 45.46, 44.51, 42.64, 40.64, 38.53, 37.14, 36.70, 33.12, 32.14, 30.69, 29.77, 29.48, 25.66, 22.47, 21.79, 20.41, 19.61, 15.96, 15.58, 14.72.
(3j, C43H54O3). White sold; Yield 66 %; m. p. 283.9–285.6 °C; 1H NMR (500 MHz, Chloroform-d) δ 7.67–7.63 (m, 4H), 7.52 (t, J = 6.1 Hz, 3H), 7.46 (dd, J = 8.4, 6.9 Hz, 2H), 7.39–7.34 (m, 1H), 4.76 (d, J = 2.2 Hz, 1H), 4.67–4.64 (m, 1H), 3.11 (dd, J = 16.5, 1.6 Hz, 1H), 3.02 (td, J = 10.8, 4.8 Hz, 1H), 2.27 (dtd, J = 15.8, 9.0, 7.7, 3.4 Hz, 3H), 2.01 (dq, J = 15.7, 7.4 Hz, 2H), 1.73 (s, 4H), 1.67 (t, J = 11.4 Hz, 1H), 1.56–1.43 (m, 12H), 1.29–1.24 (m, 2H), 1.16 (s, 3H), 1.13 (s, 3H), 1.04 (s, 3H), 0.98 (s, 3H), 0.81 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 208.37, 150.70, 141.23, 140.43, 137.14, 135.06, 134.30, 131.07, 129.01, 127.80, 127.23, 127.17, 109.80, 56.59, 56.56, 52.88, 49.29, 48.60, 46.96, 45.29, 44.80, 42.64, 40.65, 38.59, 37.16, 36.62, 33.19, 32.19, 30.74, 29.81, 29.68, 25.75, 22.48, 21.82, 20.50, 19.65, 16.01, 15.61, 14.77.
(3 k, C39H55NO3). White sold; Yield 54 %; m. p. 292.2–294.8 °C; 1H NMR (500 MHz, Chloroform-d) δ 10.62 (s, 1H), 7.48 (s, 1H), 7.41 (d, J = 8.4 Hz, 2H), 6.79 (s, 2H), 4.78 (d, J = 2.3 Hz, 1H), 4.66 (t, J = 1.9 Hz, 1H), 3.03 (s, 8H), 2.33–2.18 (m, 3H), 1.99 (q, J = 8.4 Hz, 2H), 1.77 (q, J = 7.9, 5.7 Hz, 1H), 1.74 (s, 3H), 1.68 (t, J = 11.4 Hz, 1H), 1.61–1.38 (m, 9H), 1.35–1.22 (m, 2H), 1.12 (d, J = 18.1 Hz, 7H), 1.03 (s, 3H), 0.97 (s, 3H), 0.79 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 208.03, 182.22, 171.37, 150.91, 150.49, 138.62, 132.66, 129.41, 124.09, 111.91, 109.64, 60.56, 56.60, 52.62, 49.30, 48.71, 46.95, 45.08, 44.92, 42.61, 40.63, 40.27, 38.59, 37.16, 36.40, 33.20, 32.21, 30.79, 29.90, 29.82, 25.83, 22.44, 21.80, 21.19, 20.53, 19.70, 16.04, 15.59, 14.75, 14.32.
(3 L, C38H52O5S). Yellow sold; Yield 71 %; m. p. 287.7–289.3 °C; 1H NMR (500 MHz, Chloroform-d) δ 7.98–7.94 (m, 2H), 7.57–7.53 (m, 2H), 7.48–7.46 (m, 1H), 4.75 (d, J = 2.3 Hz, 1H), 4.65 (t, J = 1.8 Hz, 1H), 3.09 (s, 3H), 3.05–2.93 (m, 2H), 2.32–2.16 (m, 3H), 1.98 (q, J = 7.8, 7.1 Hz, 2H), 1.71 (s, 4H), 1.66 (t, J = 11.4 Hz, 1H), 1.55–1.38 (m, 11H), 1.25 (ddt, J = 12.9, 9.6, 3.4 Hz, 3H), 1.14 (d, J = 5.0 Hz, 6H), 1.02 (s, 3H), 0.96 (s, 3H), 0.78 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 207.97, 182.32, 150.48, 141.62, 139.75, 137.48, 134.95, 130.75, 127.60, 109.96, 56.52, 53.00, 49.23, 48.47, 46.95, 45.50, 44.58, 44.38, 42.63, 40.64, 38.54, 37.14, 36.73, 33.14, 32.15, 30.67, 29.76, 29.45, 25.64, 22.50, 21.78, 20.40, 19.59, 15.95, 15.60, 14.73.
(3 m, C39H51N3O3). White sold; Yield 75 %; m. p. 213.1–215.6 °C;1H NMR (500 MHz, Chloroform-d) δ 8.66 (s, 1H), 8.14 (s, 1H), 7.75 (d, J = 8.1 Hz, 2H), 7.55 (d, J = 8.2 Hz, 2H), 7.49 (s, 1H), 4.76 (d, J = 2.3 Hz, 1H), 4.64 (t, J = 1.8 Hz, 1H), 3.07–2.97 (m, 2H), 2.34–2.20 (m, 3H), 2.05–1.95 (m, 2H), 1.72 (s, 4H), 1.66 (t, J = 11.4 Hz, 1H), 1.58–1.39 (m, 12H), 1.35–1.22 (m, 3H), 1.15 (s, 3H), 1.13 (s, 3H), 1.03 (s, 3H), 0.97 (s, 3H), 0.79 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 208.09, 181.79, 152.53, 150.64, 136.41, 136.19, 135.70, 135.54, 131.74, 119.97, 109.84, 77.37, 56.51, 52.93, 49.25, 48.57, 46.97, 45.39, 44.57, 42.64, 40.64, 38.55, 37.16, 36.66, 33.17, 32.20, 30.70, 29.79, 29.55, 25.71, 22.50, 21.80, 20.44, 19.61, 15.96, 15.61, 14.74.
(3n, C39H54O5). White sold; Yield 61 %; m. p. 200.7–202.2 °C; 1H NMR (500 MHz, Chloroform-d) δ 7.44 (t, J = 2.0 Hz, 1H), 7.07 (dd, J = 8.4, 1.8 Hz, 1H), 6.98–6.90 (m, 2H), 4.64 (q, J = 1.8 Hz, 1H), 3.92 (d, J = 2.5 Hz, 3H), 3.88 (d, J = 1.7 Hz, 3H), 3.11–2.96 (m, 2H), 2.33–2.16 (m, 3H), 2.05–1.94 (m, 2H), 1.58–1.37 (m, 12H), 1.34–1.21 (m, 12H), 1.12 (dd, J = 12.5, 1.9 Hz, 7H), 1.03 (d, J = 1.6 Hz, 3H), 0.97 (d, J = 3.3 Hz, 3H), 0.91–0.84 (m, 5H), 0.79 (d, J = 2.1 Hz, 3H); 13C NMR (126 MHz, CDCl3) δ 208.23, 150.67, 149.54, 148.72, 137.56, 132.60, 129.03, 123.12, 114.50, 111.07, 109.76, 76.91, 76.91, 56.60, 56.58, 56.03, 56.00, 52.79, 49.27, 48.70, 46.95, 45.19, 44.83, 42.62, 40.63, 38.61, 37.15, 36.51, 33.19, 32.17, 30.73, 29.82, 29.69, 25.73, 22.45, 21.85, 20.47, 19.64, 16.02, 15.61, 14.77.
(3o, C40H56O6). White sold; Yield 72 %; m. p. 233.6–235.9 °C; 1H NMR (500 MHz, Chloroform-d) δ 7.48–7.35 (m, 1H), 6.64 (s, 2H), 4.73 (d, J = 2.2 Hz, 1H), 4.62 (t, J = 1.7 Hz, 1H), 3.88 (d, J = 14.7 Hz, 9H), 3.09 (dd, J = 16.3, 1.6 Hz, 1H), 3.00 (td, J = 10.7, 5.0 Hz, 1H), 2.33–2.22 (m, 2H), 2.17 (dt, J = 16.1, 2.2 Hz, 1H), 2.03–1.95 (m, 2H), 1.75 (d, J = 13.4 Hz, 1H), 1.70 (s, 3H), 1.64 (t, J = 11.4 Hz, 1H), 1.55–1.38 (m, 12H), 1.38–1.20 (m, 2H), 1.13 (d, J = 4.1 Hz, 6H), 1.01 (s, 3H), 0.97 (s, 3H), 0.82 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 208.26, 182.24, 153.03, 150.29, 138.58, 137.50, 133.91, 131.61, 109.92, 107.78, 61.09, 56.54, 56.31, 52.96, 49.20, 48.76, 46.93, 45.41, 44.54, 42.60, 40.64, 38.58, 37.11, 36.62, 33.20, 32.14, 30.65, 29.82, 29.48, 25.58, 22.48, 21.88, 20.40, 19.55, 16.04, 15.63, 14.78.
(3p, C43H54O4). White sold; Yield 68 %; m. p. 269.2–269.9 °C; 1H NMR (500 MHz, Chloroform-d) δ 7.49–7.45 (m, 1H), 7.44–7.35 (m, 4H), 7.19–7.14 (m, 1H), 7.09–7.05 (m, 2H), 7.04–6.99 (m, 2H), 4.77 (d, J = 2.3 Hz, 1H), 4.65 (t, J = 1.8 Hz, 1H), 3.03 (ddd, J = 15.4, 6.6, 3.1 Hz, 2H), 2.34–2.17 (m, 3H), 2.07–1.95 (m, 2H), 1.72 (s, 4H), 1.67 (t, J = 11.4 Hz, 1H), 1.58–1.38 (m, 11H), 1.37–1.22 (m, 2H), 1.15 (s, 3H), 1.11 (s, 3H), 1.03 (s, 3H), 0.98 (s, 3H), 0.90–0.83 (m, 1H), 0.80 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 208.28, 182.46, 158.08, 156.35, 150.65, 136.95, 132.99, 132.34, 130.80, 130.05, 124.15, 119.89, 118.16, 109.82, 56.58, 52.78, 49.27, 48.59, 46.97, 45.18, 44.77, 42.62, 40.63, 38.58, 37.16, 36.53, 33.16, 32.18, 30.74, 29.80, 29.72, 25.75, 22.44, 21.80, 20.50, 19.64, 15.99, 15.60, 14.74.
(3q, C41H52O3). White sold; Yield 86 %; m. p. 265.8–266.9 °C; 1H NMR (500 MHz, Chloroform-d) δ 7.93–7.84 (m, 4H), 7.69 (d, J = 2.7 Hz, 1H), 7.59–7.51 (m, 3H), 4.78 (d, J = 2.2 Hz, 1H), 4.67 (t, J = 1.9 Hz, 1H), 3.18 (dd, J = 16.2, 1.6 Hz, 1H), 3.04 (td, J = 10.7, 4.7 Hz, 1H), 2.36–2.22 (m, 3H), 2.09–1.97 (m, 2H), 1.75 (s, 4H), 1.69 (t, J = 11.4 Hz, 1H), 1.62–1.40 (m, 11H), 1.29 (ddd, J = 13.2, 9.2, 3.4 Hz, 2H), 1.19 (d, J = 3.5 Hz, 6H), 1.06 (s, 3H), 0.99 (s, 3H), 0.83 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 208.44, 182.46, 150.53, 137.74, 134.57, 133.70, 133.25, 133.07, 130.51, 128.60, 128.10, 127.76, 127.37, 126.90, 126.49, 109.85, 56.57, 52.99, 49.28, 48.53, 46.95, 45.38, 44.48, 42.62, 40.64, 38.54, 37.14, 36.74, 33.17, 32.18, 30.73, 29.80, 29.58, 25.66, 22.53, 21.74, 20.47, 19.62, 15.95, 15.61, 14.76.
(3r, C35H48O3S). White sold; Yield 56 %; m. p. 243.5–246.2 °C; 1H NMR (500 MHz, Chloroform-d) δ 7.71 (t, J = 2.2 Hz, 1H), 7.52 (d, J = 5.0 Hz, 1H), 7.31 (d, J = 3.6 Hz, 1H), 7.13 (dd, J = 5.1, 3.7 Hz, 1H), 4.79 (d, J = 2.2 Hz, 1H), 4.67 (d, J = 2.0 Hz, 1H), 3.04 (ddd, J = 16.9, 9.5, 3.3 Hz, 2H), 2.34–2.25 (m, 2H), 2.18–2.11 (m, 1H), 2.07–1.95 (m, 2H), 1.81 (dq, J = 12.9, 3.5 Hz, 1H), 1.74 (s, 3H), 1.71–1.63 (m, 2H), 1.60–1.33 (m, 11H), 1.27 (dt, J = 12.2, 2.7 Hz, 2H), 1.15 (s, 3H), 1.08 (s, 3H), 1.04 (s, 3H), 0.99 (s, 3H), 0.83 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 207.77, 182.52, 150.64, 139.58, 132.75, 131.18, 130.43, 129.83, 127.70, 109.84, 56.61, 52.54, 49.28, 48.75, 47.00, 45.20, 44.99, 42.61, 40.59, 38.61, 37.15, 36.49, 33.09, 32.18, 30.76, 29.84, 25.77, 22.32, 21.90, 20.54, 19.64, 16.46, 15.54, 14.79.
(3 s, C35H48O4). White sold; Yield 73 %; m. p. 280.7–283.2 °C; 1H NMR (500 MHz, Chloroform-d) δ 7.57 (d, J = 1.7 Hz, 1H), 7.30 (d, J = 1.2 Hz, 1H), 6.59 (d, J = 3.5 Hz, 1H), 6.50 (dd, J = 3.5, 1.8 Hz, 1H), 4.79 (d, J = 2.3 Hz, 1H), 4.67 (t, J = 1.9 Hz, 1H), 3.13 (dd, J = 17.5, 1.8 Hz, 1H), 3.05 (td, J = 10.7, 4.7 Hz, 1H), 2.34–2.23 (m, 2H), 2.22–2.14 (m, 1H), 2.08–1.95 (m, 2H), 1.74 (s, 4H), 1.69 (t, J = 11.4 Hz, 1H), 1.58–1.32 (m, 9H), 1.27 (dt, J = 11.8, 2.5 Hz, 3H), 1.14 (s, 4H), 1.08 (s, 3H), 1.03 (s, 3H), 0.99 (s, 3H), 0.82 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 207.68, 182.32, 152.74, 150.77, 144.55, 131.34, 124.34, 115.57, 112.34, 109.78, 56.61, 52.65, 49.29, 48.58, 46.99, 44.98, 44.87, 42.63, 40.60, 38.64, 37.17, 36.07, 33.16, 32.19, 30.76, 29.83, 29.80, 25.81, 22.35, 21.85, 20.52, 19.65, 16.36, 15.58, 14.76.
(3 t, C36H49NO3). White sold; Yield 87 %; m. p. 239.9–242.6 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.73 (dd, J = 4.9, 1.8 Hz, 1H), 7.72 (td, J = 7.7, 1.9 Hz, 1H), 7.43–7.35 (m, 2H), 7.23–7.17 (m, 1H), 4.76 (d, J = 2.6 Hz, 1H), 4.64 (t, J = 1.9 Hz, 1H), 3.46 (dd, J = 17.8, 1.9 Hz, 1H), 3.03 (td, J = 10.7, 4.7 Hz, 1H), 2.43–2.35 (m, 1H), 2.32–2.22 (m, 2H), 1.99 (q, J = 8.4, 8.0 Hz, 2H), 1.72 (s, 4H), 1.66 (t, J = 11.3 Hz, 1H), 1.61–1.36 (m, 11H), 1.34–1.21 (m, 3H), 1.15 (s, 3H), 1.11 (s, 3H), 1.02 (s, 3H), 0.97 (s, 3H), 0.79 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 209.01, 182.18, 155.42, 150.76, 149.46, 138.96, 136.56, 134.23, 126.72, 122.47, 109.73, 56.57, 52.94, 49.29, 48.31, 46.98, 45.44, 44.90, 42.64, 40.59, 38.59, 37.16, 36.42, 33.16, 32.21, 30.76, 29.82, 29.50, 25.72, 22.38, 21.79, 20.49, 19.62, 16.08, 15.59, 14.75.
(3u, C37H56O3). White sold; Yield 95 %; m. p. 284.7–285.8 °C; 1H NMR (500 MHz, Chloroform-d) δ 6.43 (dq, J = 9.8, 1.4 Hz, 1H), 4.76 (d, J = 2.2 Hz, 1H), 4.64–4.62 (m, 1H), 3.03 (td, J = 10.8, 4.9 Hz, 1H), 2.70 (dd, J = 15.9, 1.5 Hz, 1H), 2.35–2.23 (m, 3H), 2.14 (dt, J = 10.3, 3.7 Hz, 1H), 2.07–1.94 (m, 2H), 1.85 (dd, J = 15.8, 2.6 Hz, 1H), 1.81–1.73 (m, 1H), 1.71 (s, 4H), 1.67 (s, 1H), 1.54 (dt, J = 23.7, 9.8 Hz, 4H), 1.43 (d, J = 4.0 Hz, 8H), 1.34 (dd, J = 11.9, 3.9 Hz, 1H), 1.32–1.08 (m, 5H), 1.05 (d, J = 2.6 Hz, 7H), 1.01 (s, 3H), 0.98 (s, 4H), 0.77 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 208.28, 182.42, 150.61, 146.67, 131.86, 109.89, 77.37, 56.56, 53.19, 49.28, 48.51, 47.01, 45.26, 42.63, 42.24, 40.62, 38.65, 37.19, 37.03, 36.27, 33.32, 32.20, 31.82, 31.73, 30.66, 29.80, 29.31, 26.02, 25.76, 25.70, 22.41, 21.73, 20.42, 19.53, 15.75, 15.67, 14.74.
4.1.2 General procedure for preparation of BA derivatives 4a ∼ 4b
To a solution of dry DMF (15 mL) containing derivative 3q (0.4 mmol) and K2CO3 (0.8 mmol), CH3I or benzyl bromide (1.2 mmol) was added. The mixture was stirred at room temperature until the reaction completed. After quenched by water, the solution was extracted with ethyl acetate, followed by the wash with saturated NaCl, dry with Na2SO4, concentration in vacuo, and subsequently purification with column chromatography to yield BA derivatives 4a ∼ 4b.
(4a, C42H54O3). White sold; Yield 75 %; m. p. 147.9–149.6 °C; 1H NMR (500 MHz, Chloroform-d) δ 7.91–7.82 (m, 4H), 7.67–7.64 (m, 1H), 7.53 (ddd, J = 12.4, 7.4, 2.5 Hz, 3H), 4.75 (d, J = 2.2 Hz, 1H), 4.64 (t, J = 1.9 Hz, 1H), 3.67 (s, 3H), 3.15 (dd, J = 16.2, 1.6 Hz, 1H), 3.00 (td, J = 10.8, 4.4 Hz, 1H), 2.31 (d, J = 2.9 Hz, 1H), 2.29–2.22 (m, 3H), 1.95–1.85 (m, 2H), 1.72 (s, 4H), 1.63 (t, J = 11.4 Hz, 1H), 1.56–1.32 (m, 10H), 1.29–1.20 (m, 2H), 1.17 (s, 3H), 1.15 (s, 3H), 1.10 (dd, J = 13.1, 4.1 Hz, 1H), 1.02 (s, 3H), 0.96 (s, 3H), 0.81 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 208.43, 176.76, 150.74, 137.66, 134.66, 133.72, 133.25, 133.07, 130.50, 128.61, 128.11, 127.77, 127.38, 126.89, 126.49, 109.74, 56.71, 53.03, 51.48, 49.49, 48.61, 47.00, 45.39, 44.49, 42.60, 40.65, 38.40, 37.06, 36.75, 33.21, 32.19, 30.78, 29.78, 29.59, 25.71, 22.59, 21.80, 20.53, 19.65, 15.97, 15.56, 14.78.
(4b, C48H58O3). White sold; Yield 79 %; m. p. 154.6–157.5 °C; 1H NMR (500 MHz, Chloroform-d) δ 7.91–7.82 (m, 4H), 7.65 (d, J = 2.6 Hz, 1H), 7.52 (ddd, J = 9.7, 7.2, 2.4 Hz, 3H), 7.40–7.29 (m, 5H), 5.15 (d, J = 12.3 Hz, 1H), 5.09 (d, J = 12.3 Hz, 1H), 4.74 (d, J = 2.3 Hz, 1H), 4.63 (t, J = 2.0 Hz, 1H), 3.14 (d, J = 16.1 Hz, 1H), 3.02 (td, J = 11.0, 4.7 Hz, 1H), 2.34–2.19 (m, 3H), 1.96–1.83 (m, 2H), 1.71 (s, 4H), 1.63 (t, J = 11.3 Hz, 1H), 1.53–1.34 (m, 9H), 1.27 (dtd, J = 31.6, 13.4, 3.8 Hz, 2H), 1.17 (s, 4H), 1.15 (s, 3H), 1.08 (dd, J = 13.0, 4.1 Hz, 1H), 1.00 (s, 3H), 0.88 (t, J = 6.8 Hz, 1H), 0.79 (s, 6H); 13C NMR (126 MHz, CDCl3) δ 208.44, 175.90, 150.74, 137.66, 136.56, 134.67, 133.72, 133.25, 133.06, 130.49, 128.63, 128.61, 128.42, 128.23, 128.11, 127.77, 127.38, 126.89, 126.49, 109.73, 65.91, 56.68, 53.03, 49.48, 48.61, 46.96, 45.39, 44.48, 42.59, 40.62, 38.34, 37.03, 36.73, 33.20, 32.13, 30.75, 29.66, 29.58, 25.74, 22.59, 21.80, 20.52, 19.65, 15.97, 15.42, 14.74.
4.1.3 General procedure for preparation of BA derivatives 4c ∼ 4e
To a solution of dry DMF (6 mL) containing derivative 3q (0.4 mmol), HATU (0.4 mmol), and DIPEA (0.8 mmol), piperazines or amines (0.8 mmol) are added. After the reaction was completed, the mixture was quenched by water and extracted with ethyl acetate, followed by the wash with saturated NaCl, dry with Na2SO4, concentration in vacuo, and subsequently purification with column chromatography to yield BA derivatives 4c ∼ 4e.
(4c, C51H64N2O2). White sold; Yield 80 %; m. p. 277.2–279.5 °C; 1H NMR (500 MHz, Chloroform-d) δ 7.91–7.82 (m, 4H), 7.64 (s, 1H), 7.56–7.48 (m, 3H), 7.29 (t, J = 7.8 Hz, 2H), 6.93 (dd, J = 14.7, 7.6 Hz, 3H), 4.75 (d, J = 2.3 Hz, 1H), 4.62 (s, 1H), 3.77 (s, 4H), 3.51–3.42 (m, 1H), 3.15 (t, J = 11.3 Hz, 5H), 3.04–2.91 (m, 2H), 2.33–2.24 (m, 1H), 2.17 (dt, J = 13.7, 3.5 Hz, 1H), 2.03 (dd, J = 11.2, 7.1 Hz, 1H), 1.89 (dt, J = 19.1, 8.1 Hz, 1H), 1.72 (s, 4H), 1.67–1.56 (m, 2H), 1.55–1.36 (m, 9H), 1.26 (ddt, J = 28.1, 13.5, 3.8 Hz, 2H), 1.16 (d, J = 9.7 Hz, 6H), 1.03 (s, 3H), 0.98 (s, 3H), 0.82 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 208.53, 173.62, 151.48, 151.11, 137.63, 134.78, 133.74, 133.26, 133.07, 130.50, 129.55, 129.37, 128.62, 128.11, 127.77, 127.37, 126.87, 126.47, 120.54, 117.44, 116.46, 109.33, 54.75, 53.12, 52.73, 49.73, 48.94, 45.71, 45.40, 44.56, 42.12, 40.65, 37.08, 36.80, 36.11, 33.29, 32.63, 31.52, 29.91, 29.57, 25.87, 22.56, 22.08, 20.52, 19.98, 16.04, 15.73, 14.77, 0.14.
(4d, C43H57NO2). White sold; Yield 72 %; m. p. 161.5–165.3 °C; 1H NMR (500 MHz, Chloroform-d) δ 7.89–7.82 (m, 4H), 7.66–7.63 (m, 1H), 7.52 (ddd, J = 13.1, 7.4, 2.5 Hz, 3H), 5.61 (t, J = 5.7 Hz, 1H), 4.75 (d, J = 2.2 Hz, 1H), 4.62 (t, J = 1.8 Hz, 1H), 3.33 (dp, J = 13.6, 6.9 Hz, 1H), 3.25–3.19 (m, 1H), 3.19–3.12 (m, 2H), 2.53 (td, J = 12.3, 3.5 Hz, 1H), 2.28 (dd, J = 16.6, 2.9 Hz, 1H), 2.04–1.90 (m, 2H), 1.76–1.73 (m, 1H), 1.71 (s, 3H), 1.61 (d, J = 11.3 Hz, 1H), 1.59–1.55 (m, 1H), 1.55–1.51 (m, 1H), 1.49 (d, J = 7.2 Hz, 5H), 1.44 (ddd, J = 11.6, 7.5, 4.1 Hz, 2H), 1.40–1.31 (m, 1H), 1.30–1.23 (m, 1H), 1.21 (d, J = 3.4 Hz, 1H), 1.17 (s, 3H), 1.15 (s, 3H), 1.12 (t, J = 7.2 Hz, 3H), 1.07 (dd, J = 13.0, 4.1 Hz, 1H), 1.02 (s, 3H), 0.98 (s, 3H), 0.81 (s, 3H). 13C NMR (126 MHz, CDCl3) δ 208.47, 175.98, 151.23, 137.65, 134.70, 133.69, 133.24, 133.06, 130.49, 128.60, 128.11, 127.76, 127.34, 126.87, 126.47, 109.40, 55.60, 53.04, 50.20, 48.70, 46.82, 45.38, 44.49, 42.66, 40.73, 38.50, 37.86, 36.75, 34.20, 33.86, 33.29, 31.07, 29.57, 29.49, 25.82, 22.56, 21.85, 20.51, 19.78, 15.98, 15.74, 15.20, 14.71.
(4e, C44H59NO2). White sold; Yield 70 %; m. p. 164.6–168.9 °C; 1H NMR (500 MHz, Chloroform-d) δ 7.90–7.81 (m, 4H), 7.66–7.62 (m, 1H), 7.52 (ddd, J = 11.8, 7.4, 2.5 Hz, 3H), 5.61 (t, J = 5.9 Hz, 1H), 4.75 (d, J = 2.3 Hz, 1H), 4.62 (t, J = 1.8 Hz, 1H), 3.26 (dq, J = 13.3, 6.6 Hz, 1H), 3.19–3.08 (m, 3H), 2.53 (td, J = 12.4, 3.5 Hz, 1H), 2.28 (dd, J = 16.1, 2.9 Hz, 1H), 2.01–1.89 (m, 2H), 1.77–1.64 (m, 8H), 1.63–1.56 (m, 2H), 1.48 (tdd, J = 23.6, 7.7, 3.6 Hz, 10H), 1.30–1.18 (m, 2H), 1.16 (s, 3H), 1.15 (s, 3H), 1.02 (s, 3H), 0.98 (s, 3H), 0.92 (t, J = 7.4 Hz, 3H), 0.81 (s, 3H).13C NMR (126 MHz, CDCl3) δ 208.27, 175.84, 150.99, 137.42, 134.47, 133.46, 133.00, 132.82, 130.26, 128.36, 127.87, 127.52, 127.10, 126.63, 126.23, 109.16, 55.49, 52.81, 49.97, 48.48, 46.55, 45.14, 44.26, 42.43, 40.83, 40.49, 38.35, 37.62, 36.53, 33.69, 33.07, 30.84, 29.33, 29.28, 25.59, 22.98, 22.31, 21.63, 20.28, 19.55, 15.75, 15.53, 14.47, 11.37.
4.2 a-Glucosidase inhibition and kinetics assay
The α-glucosidase (from Saccharomyces cerevisiae, Sigma-Aldrich) inhibitory activity of BA derivatives was determined with acarbose and BA as control (Yang et al., 2021; Yang et al., 2021). After α-glucosidase incubated with BA derivatives for 10 min in phosphate buffered saline (0.1 M, pH 6.8), pNPG solution was added into the mixture, and the absorbance change at 405 nm was recorded. All samples were performed in parallel.
The inhibition kinetic of compound 3q against α-glucosidase was assayed with the same procedure above-mentioned. For the inhibitory mechanism, the inhibitory activity of compound 3q was assayed with different α-glucosidase concentration (Taha et al., 2017). For the inhibition type, the inhibitory activity of compound 3q was assayed with different pNPG concentration (Khusnutdinova et al., 2017). The obtained curves revealed the kinetics of compound 3q.
4.3 Fluorescence spectra
Fluorescence spectra of α-glucosidase with sample were recorded at 298, 304 and 310 K. Compound 3q was titrimetrically added into α-glucosidase solution. The excitation wavelength was 280 nm and the emission wavelength was from 290 to 450 nm. The quenching mechanism was analyzed by Stern Volmer equation and the thermodynamic parameters were calculated using the Van’t Hoff and the Gibbs-Helmholtz Eq (Jia et al., 2020; Jia et al., 2021).
4.4 3D fluorescence spectra assay
To a PBS solution of α-glucosidase, compound 3q was added, followed by the fluorescence spectra scanning at excitation and emission wavelengths of 200–600 nm and compared to that of α-glucosidase alone (Taha et al., 2016). The data is imported into Matlab for processing.
4.5 CD spectroscopy
To a solution of α-glucosidase, compound 3q was added, followed by the CD spectrum scanning and compared to that of α-glucosidase alone (Taha et al., 2017). The CDNN was used to analyze the proportion of secondary conformation of protein.
4.6 Molecular docking
Molecular docking between compound 3q and α-glucosidase was completed with SYBYL software using our previous method (Li et al., 2021). Compound 3q was constructed, treated with energy minimization program, and charged with Gasteiger-Hückle model. The homology α-glucosidase protein was built according to previous works (Yu et al., 2018; Hu et al., 2021; Zhang et al., 2022), followed by the optimization of removing water molecules, adding hydrogen atoms, adding charge, and repairing end residues. After the active pocket was generated, the docking was operated. The protomol frid box dimensions data (x, y, z) was 27 × 27 × 25. The results were visualized by Pymol and Discover studio software.
4.7 Statistical analysis
All data were presented as mean ± SD. One-way ANOVA was performed to evaluate the difference between groups. P < 0.05 was considered significant.
Acknowledgements
This work was financially supported by the Fundamental and Applied Basic Research Fund of Guangdong Province (No. 2022A1515011657), Department of Education of Guangdong Province (Nos. 2019KZDXM035, 2021KTSCX135, 2021KCXTD044), and Jiangmen Science and Technology Plan Project (2021030103150006664). Special Funds for the Cultivation of Guangdong College Students’ Scientific and Technological Innovation (“Climbing Program” Special Funds, pdjh2021a0504 and pdjh2022b0532).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- An update on the etiology and epidemiology of diabetes mellitus. Ann. N. Y. Acad. Sci.. 2006;1084:1-29.
- [Google Scholar]
- Hydrazinyl arylthiazole based pyridine scaffolds: Synthesis, structural characterization, in vitro alpha-glucosidase inhibitory activity, and in silico studies. Eur. J. Med. Chem.. 2017;138:255-272.
- [Google Scholar]
- Natural and hemi-synthetic pentacyclic triterpenes as antimicrobials and resistance modifying agents against Staphylococcus aureus: a review. Phytochem. Rev.. 2018;17(5):1129-1163.
- [Google Scholar]
- Synthesis and bioactivities evaluation of oleanolic acid oxime ester derivatives as alpha-glucosidase and alpha-amylase inhibitors. J. Enzyme Inhib. Med. Chem.. 2022;37(1):451-461.
- [Google Scholar]
- Synthetic heterocyclic candidates as promising alpha-glucosidase inhibitors: An overview. Eur. J. Med. Chem.. 2019;176:343-377.
- [Google Scholar]
- New Insights into the Inhibition Mechanism of Betulinic Acid on alpha-Glucosidase. J. Agric. Food Chem.. 2018;66(27):7065-7075.
- [Google Scholar]
- Re-exploring promising alpha-glucosidase inhibitors for potential development into oral anti-diabetic drugs: Finding needle in the haystack. Eur. J. Med. Chem.. 2015;103:133-162.
- [Google Scholar]
- Global estimates of diabetes prevalence for 2013 and projections for 2035. Diabetes Res. Clin. Pract.. 2014;103(2):137-149.
- [Google Scholar]
- Synthesis of benzotriazoles derivatives and their dual potential as alpha-amylase and alpha-glucosidase inhibitors in vitro: Structure-activity relationship, molecular docking, and kinetic studies. Eur. J. Med. Chem.. 2019;183:111677
- [Google Scholar]
- Novel cinnamic acid magnolol derivatives as potent alpha-glucosidase and alpha-amylase inhibitors: Synthesis, in vitro and in silico studies. Bioorg. Chem.. 2021;116:105291
- [Google Scholar]
- Characterization, antioxidant activities, and inhibition on α-glucosidase activity of corn silk polysaccharides obtained by different extraction methods. Int. J. Biol. Macromol.. 2020;163:1640-1648.
- [Google Scholar]
- Chemical structure and inhibition on α-glucosidase of polysaccharides from corn silk by fractional precipitation. Carbohyd. Polym.. 2021;252:117185
- [Google Scholar]
- Pathophysiology and treatment of type 2 diabetes: perspectives on the past, present, and future. The Lancet. 2014;383(9922):1068-1083.
- [Google Scholar]
- Evaluation of cytotoxicity and alpha-glucosidase inhibitory activity of amide and polyamino-derivatives of lupane triterpenoids. Molecules. 2020;25(20)
- [Google Scholar]
- Synthesis and evaluation of 2,3-indolotriterpenoids as new α-glucosidase inhibitors. Med. Chem. Res.. 2017;26(11):2737-2742.
- [Google Scholar]
- Structural modifications of 2,3-indolobetulinic acid: Design and synthesis of highly potent alpha-glucosidase inhibitors. Bioorg. Chem.. 2019;88:102957
- [Google Scholar]
- Beneficial effect of betulinic acid on hyperglycemia via suppression of hepatic glucose production. J. Agric. Food Chem.. 2014;62(2):434-442.
- [Google Scholar]
- In vitro and in silico evaluations of diarylpentanoid series as alpha-glucosidase inhibitor. Bioorg. Med. Chem. Lett.. 2018;28(3):302-309.
- [Google Scholar]
- Insight into interaction mechanism between theaflavin-3-gallate and alpha-glucosidase using spectroscopy and molecular docking analysis. J. Food Biochem.. 2021;45(1):e13550.
- [Google Scholar]
- Synthesis of novel triterpene and N-allylated/N-alkylated niacin hybrids as alpha-glucosidase inhibitors. Eur. J. Med. Chem.. 2013;63:162-169.
- [Google Scholar]
- Therapeutic potentials of triterpenes in diabetes and its associated complications. Curr. Top. Med. Chem.. 2016;16(23):2532-2542.
- [Google Scholar]
- Synthesis of new indazole based dual inhibitors of alpha-glucosidase and alpha-amylase enzymes, their in vitro, in silico and kinetics studies. Bioorg. Chem.. 2020;94:103195
- [Google Scholar]
- Antidiabetic activity of some pentacyclic acid triterpenoids, role of PTP-1B: in vitro, in silico, and in vivo approaches. Eur. J. Med. Chem.. 2011;46(6):2243-2251.
- [Google Scholar]
- Heterocyclic compounds: effective alpha-amylase and alpha-glucosidase inhibitors. Curr. Top. Med. Chem.. 2017;17:428-440.
- [Google Scholar]
- Synthesis, alpha-glucosidase inhibitory, cytotoxicity and docking studies of 2-aryl-7-methylbenzimidazoles. Bioorg. Chem.. 2016;65:100-109.
- [Google Scholar]
- Synthesis and study of the alpha-amylase inhibitory potential of thiadiazole quinoline derivatives. Bioorg. Chem.. 2017;74:179-186.
- [Google Scholar]
- Biology-oriented drug synthesis (BIODS) of 2-(2-methyl-5-nitro-1H-imidazol-1-yl)ethyl aryl ether derivatives, in vitro alpha-amylase inhibitory activity and in silico studies. Bioorg. Chem.. 2017;74:1-9.
- [Google Scholar]
- Design, synthesis and biological evaluation of vincamine derivatives as potential pancreatic beta-cells protective agents for the treatment of type 2 diabetes mellitus. Eur. J. Med. Chem.. 2020;188:111976
- [Google Scholar]
- Discovery of 3,3-di(indolyl)indolin-2-one as a novel scaffold for alpha-glucosidase inhibitors: In silico studies and SAR predictions. Bioorg. Chem.. 2017;72:228-233.
- [Google Scholar]
- Synthesis, in vitro evaluation and molecular docking studies of novel triazine-triazole derivatives as potential alpha-glucosidase inhibitors. Eur. J. Med. Chem.. 2017;125:423-429.
- [Google Scholar]
- Synthesis, biological evaluation, and docking studies of novel 5,6-diaryl-1,2,4-triazine thiazole derivatives as a new class of alpha-glucosidase inhibitors. Bioorg. Chem.. 2018;78:195-200.
- [Google Scholar]
- Synthesis and biological evaluation of coumarin derivatives as alpha-glucosidase inhibitors. Eur. J. Med. Chem.. 2020;189:112013
- [Google Scholar]
- Inhibition mechanism of α-amylase/α-glucosidase by silibinin, its synergism with acarbose, and the effect of milk proteins. J. Agric. Food Chem.. 2021;69:10515-10526.
- [Google Scholar]
- Comparative study of inhibition mechanisms of structurally different flavonoid compounds on α-glucosidase and synergistic effect with acarbose. Food Chem.. 2021;347:129056
- [Google Scholar]
- Comparison of two docking methods for peptide-protein interactions. J. Sci. Food Agric.. 2018;98(10):3722-3727.
- [Google Scholar]
- Synthesis, in vitro evaluation and molecular docking studies of biscoumarin thiourea as a new inhibitor of alpha-glucosidases. Bioorg. Chem.. 2015;63:36-44.
- [Google Scholar]
- Pentacyclic triterpenes as alpha-glucosidase and alpha-amylase inhibitors: Structure-activity relationships and the synergism with acarbose. Bioorg. Med. Chem. Lett.. 2017;27(22):5065-5070.
- [Google Scholar]
- Synthesis and biological evaluation of coumarin derivatives containing oxime ester as a-glucosidase inhibitors. Arab. J. Chem.. 2022;15:104072
- [Google Scholar]
- In vitro and in silico studies of bis (indol-3-yl) methane derivatives as potential alpha-glucosidase and alpha-amylase inhibitors. J. Enzyme Inhib. Med. Chem.. 2021;36(1):1938-1951.
- [Google Scholar]







