Translate this page into:
Investigation into the mechanistic action of zhuye shigao decoction in acute pneumonia treatment via liquid chromatography-mass spectrometry-based metabolomics combined with network pharmacology analysis
*Corresponding author: E-mail address: wang.yang1986@hotmail.com (Y. Wang)
-
Received: ,
Accepted: ,
Abstract
The traditional Chinese medicine (TCM) formulation Zhuye Shigao Decoction (ZSD) has demonstrated therapeutic efficacy in the treatment of acute pneumonia. Nevertheless, the precise mechanisms underlying its pharmacological action remain inadequately elucidated. This study aims to assess the therapeutic efficacy of ZSD in managing acute pneumonia and to elucidate the potential mechanisms driving its effects. A lipopolysaccharide (LPS)-induced murine model of acute pneumonia was established. Histopathological examination and serum biochemical assays were conducted to assess the therapeutic efficacy of ZSD for acute pneumonia. An LC-MS-based metabolomics strategy was employed to explore the impact of ZSD on the serum metabolome. To further elucidate the underlying mechanisms of ZSD’s therapeutic effects, a combined metabolomics and network pharmacology strategy was employed. Compared with the model group, ZSD demonstrated the ability to reduce serum levels of Tumor necrosis factor α (TNF-α), Interleukin-1β (IL-1β), and Interleukin-6 (IL-6), while simultaneously enhancing the expression of IgA and IgM. Histological analysis showed that ZSD could significantly improve lung tissue changes caused by LPS exposure. We selected 74 serum metabolites as metabolic biomarkers for acute pneumonia, of which 67 demonstrated significant recovery following treatment with ZSD, thereby highlighting its therapeutic potential in addressing LPS-induced pneumonia. Network pharmacology analysis identified potential targets, including AKT1, IL-6, IL-1β, TNF, Myc proto-oncogene protein (MYC), and Peroxisome proliferator-activated receptor gamma (PPARG). The joint analysis indicated that the potential therapeutic mechanism of ZSD might associated with sphingolipid metabolism, arachidonic acid metabolism, glycerophospholipid metabolism, PPAR signaling pathway, and Janus kinase-signal transducer and activator of transcription ((JAK-STAT) signaling pathway. This study illustrated the comprehensive regulatory role of ZSD in the treatment of acute pneumonia and, for the first time, elucidated the possible mechanisms of ZSD at both the protein and metabolic levels. Furthermore, it offers a new perspective on the pharmacological effects of Chinese materia medica (CMM) in acute pneumonia treatment, with a particular emphasis on metabolic regulation.
Keywords
Acute pneumonia
Liquid chromatography-mass spectrometry
Metabolomics
Network pharmacology
Zhuye Shigao decoction
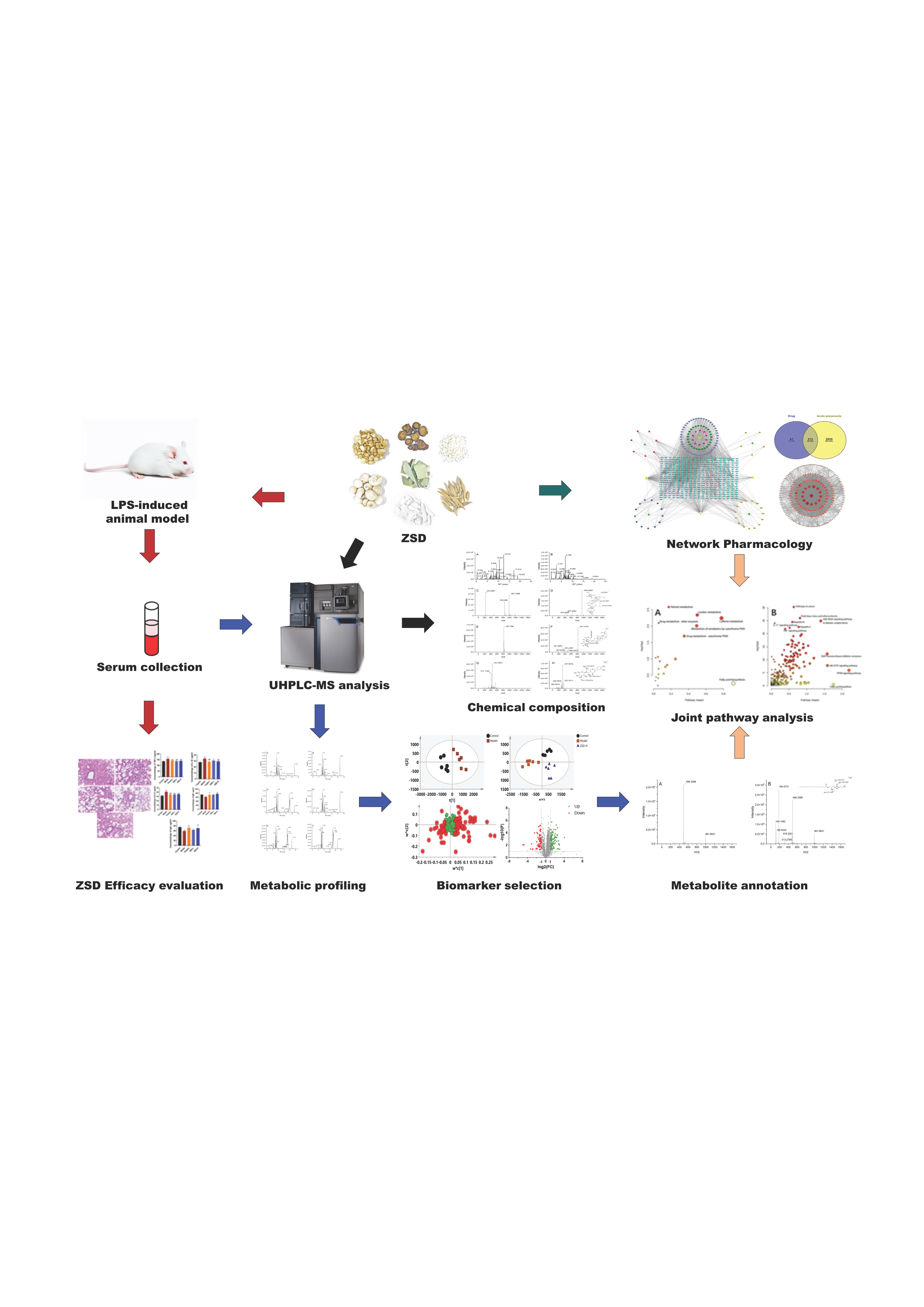
1. Introduction
Acute pneumonia is a clinical condition characterized by impaired respiratory immunity and compromised barrier functions, typically triggered by bacterial, viral, or other pathogenic infections. Clinically, it presents with hallmark symptoms like high fever, cough, chest tightness, dyspnea, and loss of appetite [1]. It shows higher mortality rates in immunocompromised children and elderly individuals [2]. Although antibiotics are primarily used in the clinical treatment of acute pneumonia, their widespread use has contributed to the emergence of bacterial resistance, significantly diminishing the efficacy of monotherapy [3]. Therefore, it is of paramount importance to develop drugs with minimal side effects and robust clinical effects in the prevention and treatment of acute pneumonia.
Compared with the symptomatic treatment approach of modern medicine, traditional Chinese medicine (TCM) pays more attention to the holistic concept, treating pneumonia patients as integrated systems and tailoring therapeutic strategies to specific syndrome types. These strategies include evacuating wind and cold, promoting lung and relieving cough, clearing heat and dissolving phlegm, etc. [4]. A classic prescription Zhuye Shigao decoction (ZSD) comes from the “Shang Han Za Bing Lun” written by Zhang Zhongjing in the Han Dynasty, which includes Folium phyllostachydis henonis, Gypsum fibrosum, Ginseng radix et rhizoma, Ophiopogonis radix, Pinelliae rhizoma, Glycyrrhizae radix et rhizoma, Oryze semen. This formula is traditionally recognized for its ability to clear heat, promote salivation, tonify qi, and regulate the stomach. With a history of over 1000 years, its efficacy on acute pneumonia has also been confirmed in modern clinical practice [5]. Studies have shown that ZSD combined with Yinqiaosan achieved a 91.115 effective rate for severe pneumonia [6]. The combination of ZSD and Maidong decoction showed remarkable efficacy in addressing drug-resistant community-acquired pneumonia, with a reported effective rate of 97.44% [7]. Despite these promising outcomes, its mechanism of action is still unclear. Given the complex composition of ZSD, elucidating its mechanism of action requires the integration of advanced modern techniques, such as omics and comprehensive network pharmacology analysis [8].
Metabolomics, an emerging scientific discipline, focuses on the comprehensive analysis of small molecules within biochemical samples. This strategy provides valuable insights into metabolite fluctuations during metabolic processes and uncovers the interaction and regulatory mechanisms among these metabolites [9]. Changes in the metabolome serve as direct indicators of metabolic network activity and provide foundational information regarding the underlying biological state of the system. As a holistic strategy to understand systemic characteristics, metabolomics aligns seamlessly with the TCM principles of “holistic concept” and “therapy with syndrome differentiation”, which provides new ideas for the modernization of TCM [10, 11]. Network pharmacology, on the other hand, creates biological network models, such as drug-target and gene-disease networks, to investigate the active compounds in Chinese materia medica (CMM), predict their potential targets, and explore their associations with various diseases [12]. This approach enables the identification of multi-target and multi-pathway mechanisms inherent to CMM, providing a theoretical foundation for its research, development, and clinical application. Through the network pharmacology approach, various components in ZSD can be summarized and screened to determine the principal active compounds. Moreover, the mechanism of action of ZSD can be elucidated by mapping the intricate network of interactions between the drug and biomolecules.
In this study, the chemical composition of the ZSD extract was characterized using ultra-high-performance liquid chromatography coupled with high-resolution mass spectrometry (UHPLC-HRMS). The lipopolysaccharide (LPS)-induced acute pneumonia mice model was established, and ZSD was administered via intragastric gavage. The serum metabolic biomarkers were selected by LC-MS-based untargeted metabolomics analysis. The network pharmacology approach was employed to investigate the potential therapeutic targets of ZSD in mice with LPS-induced acute pneumonia. The joint pathway analysis, integrating altered serum metabolites and active targets, was further applied to reveal the potential mechanistic action of ZSD in the treatment of acute pneumonia. This study demonstrated that ZSD could exert therapeutic effects on acute pneumonia by modulating endogenous metabolite levels through a comprehensive regulation of the component-target-pathway network. This finding provides robust scientific evidence supporting the clinical application of ZSD in the treatment of acute pneumonia.
2. Materials and Methods
2.1. Chemicals and reagents
HPLC-grade acetonitrile and methanol were purchased from Fisher Scientific Corporation (Waltham, USA). Formic acid was provided by Aladdin Industrial Corporation (Shanghai, China). The purified water was obtained from Watsons (Guangzhou, China). All traditional herbal medicines listed in Table 1 were provided by Hebei Shenwei Pharmaceutical Co., Ltd. (Shijiazhuang, China). The voucher specimens of these CMMs were deposited at the laboratory of Chinese medicine analysis with the labels ZSD2024001-2024007. Lipopolysaccharide (N28IS220883) was purchased from Shanghai Yuanye Biological Co., Ltd. (Shanghai, China). Dexamethasone acetate tablets (2306152) were purchased from Suicheng Pharmaceutical Co., Ltd. (Zhengzhou, China). The enzyme-linked immunosorbent assay (ELISA) kits were purchased from Shanghai Youxuan Biotechnology Co., Ltd. (Shanghai, China).
| No. | Latin name | English name | Chinese name | Batch No. |
|---|---|---|---|---|
| 1 | Phyllostachys nigra (Lodd.) Munro var. henonis (Mitf.) Stapf ex Rendle. | Folium phyllostachydis henonis | Zhuye | BZ230701 |
| 2 | Gypsum Fibrosum | Gypsum fibrosum | Shigao | 1709271 |
| 3 | Panax ginseng C. A. Mey. | Ginseng radix et rhizoma | Renshen | 23072301 |
| 4 | Ophiopogon japonicus (L.f) Ker-Gawl. | Ophiopogonis radix | Maidong | 20190602 |
| 5 | Pinellia ternata (Thunb.) Breit. | Pinelliae rhizoma | Banxia | XH22082201 |
| 6 | Glycyrrhiza uralensis Fisch. | Glycyrrhizae radix et rhizoma | Gancao | HIQ23041501 |
| 7 | Oryza sativa L. subsp. japonica Kato. | Oryze semen | Jingmi | HL24012101 |
2.2. Preparation of ZSD
The preparation process of ZSD was established based on preliminary experimental research conducted in our laboratory. The CMMs, comprising 27.6 g of Zhuye, 27.7 g of Renshen, 34.5 g of Banxia, 106 g of Maidong, 220 g of Shigao, and 27.6 g of Gancao, were precisely weighed and subsequently decocted in 2 L of boiling water for a duration of 30 mins. After being filtered with a 200 mesh gauze, Jingmi (110 g) was added to the filtrate and decocted for 20 mins. Then, it was filtered by gauze, and the filtrate was concentrated at 800 mL to obtain standard decoction. The decoction was freeze-dried and stored.
2.3. Characterization of ZSD by UHPLC-MS
The freeze-dried ZSD powder (1 g) was precisely weighed and combined with a methanol/water (1:1) solvent mixture (20 mL). Following ultrasonic extraction for 30 mins, the resultant extract was filtered through a 0.22 μm microporous membrane prior to LC-MS analysis. The Mass spectrometry (MS) and MS/MS spectra of ZSD were acquired using full-scan and fast-data-dependent acquisition (DDA) modes, which were utilized to conduct compound annotation and identification through database searching. The detailed LC-MS methods are shown in Supplementary Materials. The data processing was conducted using MS-DIAL software [13], and the parameters were set as follows: Data collection, MS1 tolerance 0.01 Da and MS2 tolerance 0.025Da; Peak detection, Minimum peak height 1000 amplitude and Mass slice width 0.1 Da; Adduct, [M-H]-, [M-H2O-H]-, [M+FA-H]-, [M+H]+, [M+Na]+. Compound annotation was performed by comparing the acquired MS and MS/MS data against database records. The MSP format databases, including MSMS_Public_EXP_NEG_VS17 and MSMS_Public_EXP_Pos_VS17, were obtained from https://systemsomicslab.github.io/compms/msdial/main.html#MSP and subsequently imported into the MS-DIAL software for database searching.
2.4. Animals and treatment
Fifty Institute of Cancer Research (ICR) male mice, weighing 35±2g, were provided by Changchun Yisi Animal Co., Ltd. (Changchun, China). The feeding temperature is maintained at 20-25°C, with relative humidity ranging from 40% to 55%, and a 12-hr light-dark cycle in a climate-controlled environment. The Animal Ethics Committee of the Changchun University of Chinese Medicine approved the whole animal treatment (approval number: 2024313). Following a one-week acclimation period, 50 mice were randomly allocated into five distinct groups, each comprising 10 mice. These groups were designated as control, model, positive, ZSD low-dose (ZSD-L), and ZSD high-dose (ZSD-H).
The mouse model was established according to the literature document with slight modifications [14]. The anesthetized mice were positioned on a 45-degree inclined platform. The mouth of each mouse was gently opened, and the tongue was carefully extended using tweezers. A pipette was used to administer 50 μL of a 1 g/L LPS solution. The control group received an equivalent volume of normal saline, which was slowly instilled into the posterior pharyngeal wall of the mice. Simultaneously, the noses of the mice were pinched for 30 seconds to ensure ingestion, after which the tongue and nose were released, allowing the mice to recover naturally in their cages. Following a two-day modeling period, all groups, with the exception of the control and model groups, received their respective treatments for seven consecutive days: the positive control group was administered dexamethasone (0.6 mg/kg, i.g.), the ZSD-L group received ZSD at a dosage of 5.2 g/kg (i.g.), and the ZSD-H group was given ZSD at a dosage of 10.4 g/kg (i.g.).
The mice were humanely euthanized 12 hrs after final administration, by an intraperitoneal 10% chloral hydrate injection. Blood samples were then collected and divided into two aliquots for subsequent biochemical and metabolomics analyses. The left lobe of the lung was immersed in a 4% formaldehyde solution for histopathological examination.
2.5. Histopathological analysis
The lung tissue was fixed overnight in 4% paraformaldehyde, embedded in paraffin, and stained with hematoxylin and eosin (HE). The pathological state of lung tissue was observed under a microscope.
2.6. Serum inflammatory cytokines and immunoglobulin analysis
The blood samples were left to clot at ambient temperature for 30 mins, followed by centrifugation at 3000 g for 15 mins at 4°C. The resultant supernatant serum was collected for subsequent biochemical analyses. The concentrations of inflammatory cytokines, specifically tumor necrosis factor α (TNF-α), interleukin-1β (IL-1β), and interleukin-6 (IL-6), along with immunoglobulins A (IgA) and M (IgM), were quantified utilizing the appropriate enzyme-linked immunosorbent assay (ELISA) kits.
2.7. Serum metabolic profile
2.7.1. Serum sample preparation and data acquisition
The serum sample was prepared using our previous protocols [15]. Metabolomics focuses on the small molecules in a given biological sample; thus, the organic solvent is always used to remove protein and extract metabolite. In this study, a serum sample of 100 μL was combined with 300 μL of methanol to facilitate protein precipitation. The resulting mixture underwent centrifugation at 15000 g for 15 mins at 4°C. Subsequently, the supernatant was evaporated under a nitrogen stream and reconstituted in 100 μL of ultrapure water. Following a second centrifugation at 15000 g for 15 mins at 4°C, an aliquot of 5 μL from the supernatant was injected into the LC-MS system to acquire the metabolic profile of the serum samples.
LC-MS has high resolution, sensitivity, and throughput merits, so it is considered a major platform for untargeted metabolomics analysis. In this study, an LC-MS-based untargeted metabolomics strategy was applied to analyze the prepared serum samples and to screen the metabolites associated with animal modeling and ZSD intervention. Since a column with sub 2 μm particle size packing could offer excellent chromatographic separation resolution, fast analysis speed, and high detection sensitivity, we chose a C18 column with 1.7 μm to analyze serum samples with the complex matrix. MS detection was conducted on a Q-TOF MS instrument, during which the full scan was applied to obtain the MS information of metabolites acquired for data preprocessing, and the fast-DDA was conducted to acquire the MS/MS spectra of features for compound annotation. Before commencing sample analysis, the precision of the MS instrument was calibrated with standard reagents. The real-time calibration was conducted concurrently with sample analysis to ensure the MS accuracy. For the detailed LC-MS setting, please refer to the Supplementary Material.
2.7.2. Data analysis
Progenesis QI software (Nonlinear Dynamics, Newcastle, UK) was employed for preprocessing raw data and organizing all chromatograms into a comprehensive dataset that included sample groups, peak retention times (RT), mass-to-charge ratios (m/z), and peak intensities for subsequent statistical analyses. Utilizing SIMCA 14.1 software, a partial least squares discriminant analysis (PLS-DA) model was developed based on this dataset. The variable importance in projection (VIP) value of each feature was calculated to assess the significance of variables, with features exhibiting VIP values greater than 1 deemed most critical for sample classification. Metabolic biomarkers associated with LPS-induced acute pneumonia were identified by combining the VIP value with a fold change (FC) >2 and a t-test P value < 0.05. Metabolite annotation was conducted by querying the Human Metabolome Database (HMDB) [16] using the precise m/z and MS/MS data of the extracted features. Subsequently, the HMDB identifiers of the annotated metabolites were input into the MetaboAnalyst 6.0 web tool [17] for metabolic pathway analysis.
2.8. Network pharmacology analysis
2.8.1. Selection of CMM compounds and related targets
The Traditional Chinese Medicine Systems Pharmacology (TCMSP) database [18] was employed to investigate the chemical constituents of CMM, utilizing the keywords “Ginseng,” “Glycyrrhiza uralensis Fisch,” and “Pinellia ternata.” Compounds exhibiting an oral bioavailability (OB) of ≥30% and a drug-likeness (DL) index of ≥0.18 were regarded as active ingredients. Given that TCMSP does not encompass all CMMs within ZSD, the HERB Database [19] was utilized to identify the active compounds present in bamboo leaves and Ophiopogon japonicus. The targets that these active ingredients can regulate were identified and validated using the UniProt Database [20], excluding non-human targets. Consequently, the drug targets of ZSD were ascertained.
2.8.2. Disease target selection and drug-active ingredient-target network construction
The disease target was identified using the GeneCards database [21] with “Acute pneumonia” as the search term. The active ingredients and associated targets of ZSD, along with pneumonia-related targets, were systematically organized in an Excel spreadsheet and subsequently imported into Cytoscape 3.9.1 software [22] to construct a Chinese Materia Medica-active ingredient-target (CMM-AI-T) network. A Venn diagram drawing web tool (https://bioinformatics.psb.ugent.be/webtools/Venn/) was employed to determine the intersection between ZSD targets and disease targets, thereby identifying potential therapeutic targets of ZSD for the treatment of acute pneumonia. The overlapping targets were then imported into the Search Tool for the Retrieval of Interacting Genes (STRING) database [23] to develop a protein-protein interaction (PPI) network with a confidence threshold greater than 0.9. TSV format files of the PPI networks were downloaded and imported into Cytoscape 3.9.1 software to facilitate network visualization, conduct topological analysis, and identify core targets.
3. Results and Discussion
3.1. Characterisation of the chemical composition of ZSD
The chemical composition of ZSD was characterized using UPLC-Q-TOF MS, employing both positive and negative ion modes to ensure a comprehensive analysis of the ZSD extract. The base peak chromatograms (BPC) of ZSD, acquired under different ionization conditions, have been illustrated in Figure 1(a) and 1(b). Following database interrogation, 107 compounds were identified and annotated in ZSD, predominantly terpenoids, flavonoids, and saponins, as detailed in Table 2. To exemplify the annotation results, we used three representative compounds as examples.
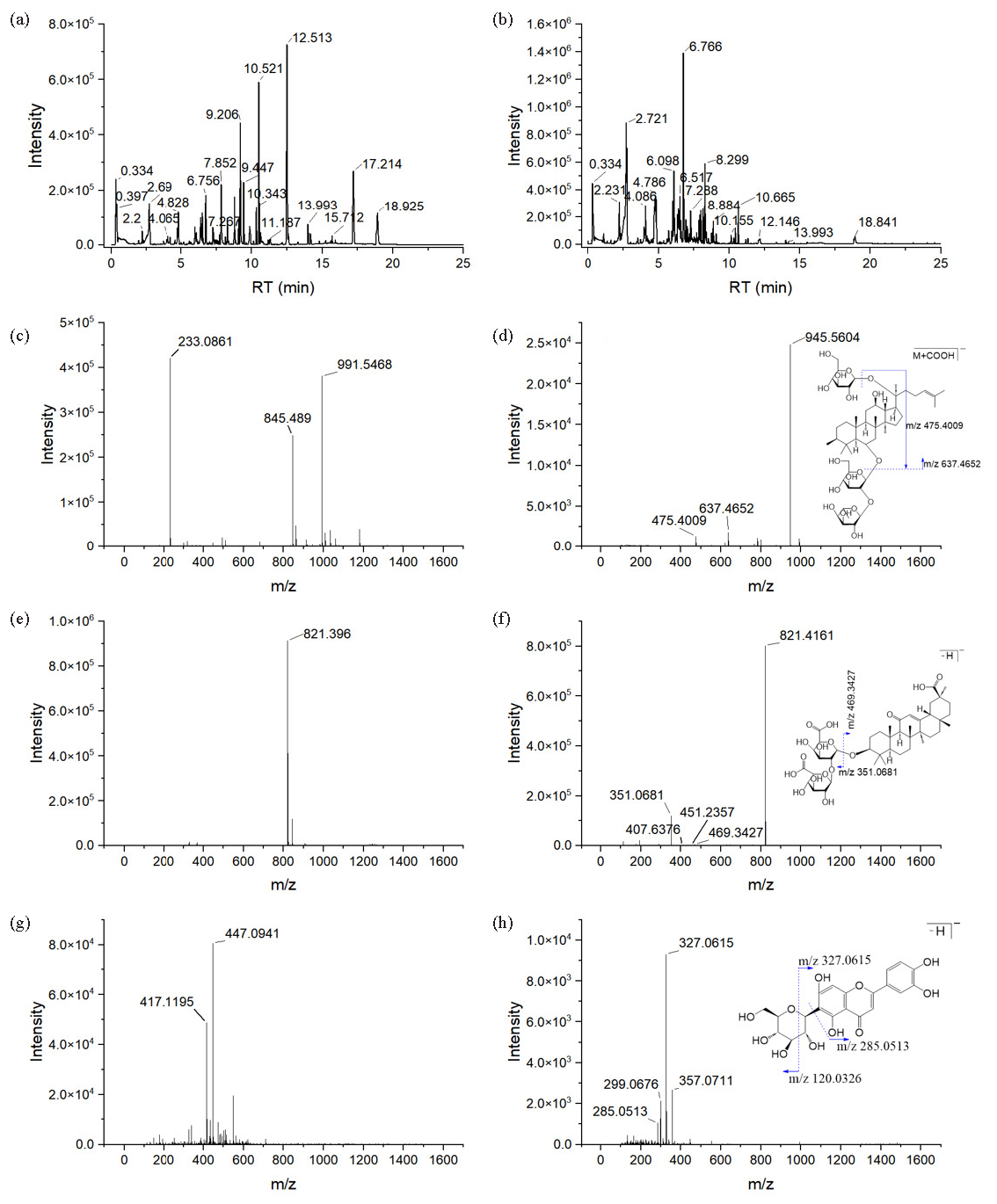
- Characterization of the chemical composition of ZSD. The BPCs of ZSD were obtained from both (a) positive and (b) negative ion modes. MS and MS spectra are presented for (c-d) ginsenoside Re, (e-f) glycopyrrolate, and (g-h) isorhizobin.
| No. | Compound name | RT (min) | Detected m/z | Theoretical m/z | Adducts | Molecular Formula | Mass error (ppm) | Origin |
|---|---|---|---|---|---|---|---|---|
| 1 | Methocarbamol | 0.35 | 242.1045 | 242.1023 | [M+H]+ | C11H15NO5 | 9.09 | - |
| 2 | Xanthotoxin | 0.38 | 215.0329 | 215.0350 | [M-H]- | C12H8O4 | -9.67 | a |
| 3 | Fumaric acid | 0.38 | 115.0044 | 115.0037 | [M-H]- | C4H4O4 | 6.26 | b |
| 4 | Scutellarioside II | 0.38 | 507.1551 | 507.1508 | [M-H]- | C24H28O12 | 8.48 | a, e |
| 5 | Adenosine | 0.42 | 268.1058 | 268.1040 | [M+H]+ | C10H13N5O4 | 6.60 | d |
| 6 | debromohymenialdisine | 0.42 | 246.0996 | 246.0986 | [M+H]+ | C11H11N5O2 | 4.27 | c |
| 7 | Citric acid | 0.45 | 191.0197 | 191.0197 | [M-H]- | C6H8O7 | -0.16 | b |
| 8 | Isoorientin | 0.85 | 447.0915 | 447.0933 | [M-H]- | C21H20O11 | -4.00 | a |
| 9 | Isobavachin | 1.05 | 323.1231 | 323.1289 | [M-H]- | C20H20O4 | -17.89 | a |
| 10 | 3-(3-Hydroxyphenyl)propionic acid | 1.11 | 165.0565 | 165.0557 | [M-H]- | C9H10O3 | 4.73 | a |
| 11 | Epicatechin-3-o-gallate | 1.11 | 441.0791 | 441.0827 | [M-H]- | C22H18O10 | -8.21 | b |
| 12 | verruculotoxin | 1.12 | 262.1928 | 262.1914 | [M+NH4]+ | C15H20N2O | 5.38 | c, e |
| 13 | Kynurenic acid | 1.12 | 190.0507 | 190.0499 | [M+H]+ | C10H7NO3 | 4.37 | a |
| 14 | Shikimic Acid | 1.32 | 173.0457 | 173.0456 | [M-H]- | C7H10O5 | 0.87 | - |
| 15 | Chlorogenic acid | 1.32 | 353.0878 | 353.0878 | [M-H]- | C16H18O9 | -0.03 | a |
| 16 | (3-Formyl-2,4-dihydroxy-6-methylbenzoic acid (2-carboxy-3,5-dihydroxy-4-methylphenyl)methyl ester) Alectorialic acid | 1.32 | 375.0688 | 375.0722 | [M-H]- | C18H16O9 | -8.96 | - |
| 17 | eupomatenoid 5 | 1.35 | 293.116 | 293.1183 | [M-H]- | C19H18O3 | -7.91 | b |
| 18 | Citrinin | 1.39 | 249.0791 | 249.0769 | [M-H]- | C13H14O5 | 9.03 | c, e |
| 19 | Liquiritin | 1.56 | 417.1185 | 417.1191 | [M-H]- | C21H22O9 | -1.46 | e |
| 20 | Naringin | 1.57 | 603.1666 | 603.1684 | [M+Na]+ | C27H32O14 | -3.03 | b, e |
| 21 | Rutin | 1.66 | 609.1465 | 609.1461 | [M-H]- | C27H30O16 | 0.64 | a |
| 22 | 5-Demethylnobiletin | 1.84 | 387.1071 | 387.1085 | [M-H]- | C20H20O8 | -3.72 | c, d |
| 23 | Methyl chlorogenate | 2.02 | 367.1045 | 367.1035 | [M-H]- | C17H20O9 | 2.83 | a |
| 24 | Luteolin 7-O-glucoside | 2.17 | 447.0943 | 447.0933 | [M-H]- | C21H20O11 | 2.26 | a |
| 25 | Orientin | 2.17 | 447.0901 | 447.0922 | [M-H]- | C21H20O11 | -4.67 | a |
| 26 | Apiin | 2.21 | 563.143 | 563.1406 | [M-H]- | C26H28O14 | 4.21 | e |
| 27 | Phenprobamate | 2.25 | 180.1009 | 180.1019 | [M+H]+ | C10H13NO2 | -5.61 | - |
| 28 | Tetrahydropapaveroline | 2.49 | 288.1235 | 288.1230 | [M+H]+ | C16H17NO4 | 1.63 | - |
| 29 | bungeiside C | 2.51 | 453.1364 | 453.1367 | [M+H]+ | C19H26O11 | -0.73 | - |
| 30 | vitexin | 2.64 | 433.1144 | 433.1129 | [M+H]+ | C21H20O10 | 3.42 | a |
| 31 | Convallatoxin | 2.71 | 549.2704 | 549.2705 | [M-H]- | C29H42O10 | -0.22 | a, b, c, e |
| 32 | N-lauroylsarcosine | 2.81 | 272.2244 | 272.2220 | [M+H]+ | C15H29NO3 | 8.74 | d |
| 33 | Agnuside | 3.00 | 467.1538 | 467.1548 | [M+H]+ | C22H26O11 | -2.12 | a |
| 34 | Dihydrohesperetin-7-O-neohesperidoside | 3.06 | 611.1951 | 611.1981 | [M-H]- | C28H36O15 | -4.97 | a |
| 35 | (2-hydroxy-4-methoxy-6-propylbenzoic acid) Divaricatinic acid | 3.11 | 209.0824 | 209.0819 | [M-H]- | C11H14O4 | 2.25 | - |
| 36 | Baicalein | 3.22 | 269.0464 | 269.0456 | [M-H]- | C15H10O5 | 3.16 | d |
| 37 | Diosmetin | 3.32 | 299.0569 | 299.0561 | [M-H]- | C16H12O6 | 2.64 | b, e |
| 38 | Homoeriodictyol | 3.38 | 301.0727 | 301.0718 | [M-H]- | C16H14O6 | 3.12 | a, c, e |
| 39 | Sissotrin | 3.67 | 491.1185 | 491.1195 | [M+HCOO]- | C22H22O10 | -2.04 | e |
| 40 | Jaceosidin | 3.68 | 331.0826 | 331.0812 | [M+H]+ | C17H14O7 | 4.14 | c |
| 41 | Albiflorin | 3.70 | 481.1672 | 481.1704 | [M+H]+ | C23H28O11 | -6.73 | - |
| 42 | Liquiritin apioside | 3.91 | 573.1534 | 573.1579 | [M+Na]+ | C26H30O13 | -7.78 | e |
| 43 | Paprazine | 3.94 | 284.1295 | 284.1281 | [M+H]+ | C17H17NO3 | 4.86 | c |
| 44 | Isoliquiritin | 4.03 | 419.1341 | 419.1337 | [M+H]+ | C21H22O9 | 1.05 | e |
| 45 | Liquiritigenin | 4.03 | 255.0678 | 255.0663 | [M-H]- | C15H12O4 | 5.96 | e |
| 46 | Digitonin | 4.09 | 1273.571 | 1273.5706 | [M+HCOO]- | C56H92O29 | 0.29 | c |
| 47 | licochalcone B | 4.16 | 287.0914 | 287.0914 | [M+H]+ | C16H14O5 | 0.00 | e |
| 48 | 7-O-Methylchrysin | 4.20 | 269.0826 | 269.0808 | [M+H]+ | C16H12O4 | 6.54 | - |
| 49 | Salvianolic acid C | 4.20 | 515.0898 | 515.0949 | [M+Na]+ | C26H20O10 | -9.84 | b, e |
| 50 | Homoharringtonine | 4.37 | 568.249 | 568.2517 | [M+Na]+ | C29H39NO9 | -4.75 | c |
| 51 | Ginsenoside Rf | 4.72 | 823.4821 | 823.4814 | [M+Na]+ | C42H72O14 | 0.81 | b |
| 52 | Rhodojaponin III | 4.98 | 391.2085 | 391.2091 | [M+Na]+ | C20H32O6 | -1.56 | b, e |
| 53 | Tricin | 5.56 | 331.0824 | 331.0812 | [M+H]+ | C17H14O7 | 3.53 | a |
| 54 | Arjungenin | 5.70 | 527.3358 | 527.3343 | [M+Na]+ | C30H48O6 | 2.83 | d |
| 55 | Undecanedioic acid | 5.71 | 215.1296 | 215.1289 | [M-H]- | C11H20O4 | 3.35 | a, b, c, d, e |
| 56 | Hispidulin | 5.82 | 299.0563 | 299.0561 | [M-H]- | C16H12O6 | 0.64 | e |
| 57 | Caperatic acid | 5.84 | 425.2512 | 425.2510 | [M+Na]+ | C21H38O7 | 0.54 | e |
| 58 | Nandrolone | 6.03 | 275.2014 | 275.2006 | [M+H]+ | C18H26O2 | 3.05 | a, b, c |
| 59 | Ginsenoside Rg1 | 6.10 | 845.4884 | 845.4904 | [M-H]- | C42H72O14 | -2.38 | b |
| 60 | Pazopanib | 6.14 | 438.1722 | 438.1707 | [M+H]+ | C21H23N7O2S | 3.49 | - |
| 61 | Formononetin | 6.24 | 267.0673 | 267.0663 | [M-H]- | C16H12O4 | 3.82 | a, b, d, e |
| 62 | Ginsenoside F3 | 6.24 | 815.4769 | 815.4799 | [M+HCOO]- | C41H70O13 | -3.62 | b |
| 63 | Ginsenoside Rb1 | 6.38 | 1153.598 | 1153.6011 | [M+HCOO]- | C54H92O23 | -2.72 | b |
| 64 | Ginsenoside Rh1 | 6.45 | 683.4362 | 683.4376 | [M+HCOO]- | C36H62O9 | -2.03 | b |
| 65 | Licoricesaponin G2 | 6.52 | 837.3901 | 837.3914 | [M-H]- | C42H62O17 | -1.58 | e |
| 66 | Glycyrrhizic acid | 6.72 | 821.396 | 821.3965 | [M-H]- | C42H62O16 | -0.62 | e |
| 67 | Licoricesaponin H2 | 6.76 | 823.4113 | 823.4111 | [M+H]+ | C42H62O16 | 0.29 | e |
| 68 | Ginsenoside Re | 6.79 | 991.5467 | 991.5483 | [M+HCOO]- | C48H82O18 | -1.63 | b |
| 69 | Octyl-methoxycinnamate | 6.89 | 291.1971 | 291.1955 | [M+H-H2O]+ | C18H26O3 | 5.60 | a, d, e |
| 70 | Fenpropimorph | 6.93 | 304.2654 | 304.2635 | [M+H]+ | C20H33NO | 6.28 | a, b, c, d, e |
| 71 | Ginsenoside Ro | 7.09 | 957.5066 | 957.5054 | [M+H]+ | C48H76O19 | 1.30 | b |
| 72 | Lauryldiethanolamine | 7.23 | 274.2754 | 274.2741 | [M+H]+ | C16H35NO2 | 4.89 | a, b, c, d, e |
| 73 | Curcumin | 7.28 | 367.1195 | 367.1187 | [M-H]- | C21H20O6 | 2.15 | a, b, c, d, e |
| 74 | Triacetyl resveratrol | 7.42 | 353.1041 | 353.1031 | [M-H]- | C20H18O6 | 2.95 | a, b, c, d, e |
| 75 | N-Oleoylethanolamine | 7.45 | 326.3077 | 326.3054 | [M+H]+ | C20H39NO2 | 7.17 | d |
| 76 | Alpha-guaiaconic acid | 7.55 | 339.1244 | 339.1238 | [M-H]- | C20H20O5 | 1.77 | b, d, e |
| 77 | Wulignan A1 | 7.62 | 365.1377 | 365.1359 | [M+H]+ | C20H22O5 | 4.82 | a, c |
| 78 | Demethoxycurcumin | 7.68 | 337.1083 | 337.1082 | [M-H]- | C20H18O5 | 0.44 | b, e |
| 79 | 5-O-methylvisammioside | 7.80 | 453.171 | 453.1755 | [M+H]+ | C22H28O10 | -9.97 | b, c |
| 80 | Octadecanedioic acid | 7.83 | 313.2389 | 313.2384 | [M-H]- | C18H34O4 | 1.50 | b, c, d, e |
| 81 | Ginsenoside Rg2 | 7.92 | 829.4956 | 829.4955 | [M+HCOO]- | C42H72O13 | 0.12 | b |
| 82 | Methylophiopogonanone B | 8.29 | 329.1403 | 329.1384 | [M+H]+ | C19H20O5 | 5.92 | c |
| 83 | y-Linolenic acid | 8.46 | 277.2197 | 277.2173 | [M-H]- | C18H30O2 | 8.66 | d, e |
| 84 | Grayanotoxin I | 8.49 | 413.2535 | 413.2534 | [M+H]+ | C22H36O7 | 0.29 | b, c, d, e |
| 85 | Prosapogenin A | 8.52 | 745.409 | 745.4134 | [M+Na]+ | C39H62O12 | -5.84 | b |
| 86 | Ginsenoside Rg6 | 8.77 | 811.4864 | 811.4849 | [M+HCOO]- | C42H70O12 | 1.81 | b |
| 87 | Kitasamycin | 8.87 | 786.4658 | 786.4634 | [M+H]+ | C40H67NO14 | 3.01 | - |
| 88 | Geranylgeranylhydroquinone | 9.19 | 383.2953 | 383.2945 | [M+H]+ | C26H38O2 | 2.19 | - |
| 89 | Histamine-trifluoromethyltoluide | 9.19 | 383.2057 | 383.2053 | [M+H]+ | C19H25F3N4O | 0.99 | a, b, c, d, e |
| 90 | Ruscogenin | 9.27 | 431.3189 | 431.3156 | [M+H]+ | C27H42O4 | 7.67 | c |
| 91 | Vasicinone | 9.60 | 203.0828 | 203.0815 | [M+H]+ | C11H10N2O2 | 6.40 | e |
| 92 | 5-O-methyllicoricidin | 9.60 | 439.2491 | 439.2479 | [M+H]+ | C27H34O5 | 2.73 | e |
| 93 | Dodecylbenzenesulfonic acid | 9.64 | 325.184 | 325.1843 | [M-H]- | C18H30O3S | -0.89 | a, b, c, d, e |
| 94 | Protolichesterinic acid | 11.18 | 323.2215 | 323.2228 | [M-H]- | C19H32O4 | -3.96 | a, b, c, d, e |
| 95 | Dehydroeburicoic acid monoacetate | 11.32 | 533.3598 | 533.3601 | [M+Na]+ | C33H50O4 | -0.62 | a, b, c, d |
| 96 | Trans-Vaccenic acid | 11.32 | 281.2495 | 281.2486 | [M-H]- | C18H34O2 | 3.20 | a, b |
| 97 | Hederagenin | 11.47 | 471.3484 | 471.3480 | [M-H]- | C30H48O4 | 0.89 | e |
| 98 | Bullatine B | 11.70 | 460.2629 | 460.2670 | [M+Na]+ | C24H39NO6 | -8.82 | a, b, c, d, e |
| 99 | Campesterol | 11.98 | 423.3557 | 423.3597 | [M+Na]+ | C28H48O | -9.54 | b |
| 100 | Panasenoside | 12.13 | 633.1463 | 633.1426 | [M+Na]+ | C27H30O16 | 5.83 | b |
| 101 | Ochrolifuanine A | 12.46 | 439.29 | 439.2856 | [M+H]+ | C29H34N4 | 9.97 | b, c, d, e |
| 102 | Lunarine | 12.46 | 455.2687 | 455.2653 | [M+NH4]+ | C25H31N3O4 | 7.51 | a, b, c, d, e |
| 103 | Cycloprodigiosin | 12.46 | 322.189 | 322.1914 | [M+H]+ | C20H23N3O | -7.42 | - |
| 104 | Valinomycin | 13.24 | 1111.629 | 1111.6384 | [2M+Na]+ | C54H90N6O18 | -8.49 | - |
| 105 | Ginsenoside compound K | 13.31 | 621.4366 | 621.4372 | [M-H]- | C36H62O8 | -0.95 | b |
| 106 | Dehydrotumulosic acid | 14.51 | 485.3601 | 485.3625 | [M+H]+ | C31H48O4 | -5.03 | - |
| 107 | Nonacosane | 15.16 | 409.475 | 409.4768 | [M+H]+ | C29H60 | -4.35 | b |
a, Folium phyllostachydis henonis; b, Ginseng radix et rhizoma; c, Ophiopogonis radix; d, Pinelliae rhizoma; e, Glycyrrhizae radix et rhizome
The mass spectrum of a saponin (#68) at RT 6.79 min_m/z 991.5468 has been shown in Figure 1(c), in which the precursor of m/z 991.5468 coeluted with ions at m/z 233.0861 and m/z 845.5468. The MS/MS spectrum of the ion with m/z 991.5468 has been presented in Figure 1(d). The base peak at m/z 945.5604 exhibits a Δm/z of 46 Da relative to the precursor, suggesting that the precursor ion is [M+HCOO]-, while the ion at m/z 945.5604 represents the deprotonated molecule. Additionally, the fragment ion at m/z 637.4652 results from the loss of a saccharide moiety at the C6 position. The ion at m/z 475.4009 was identified as [M-H-2Glc-Rha]-. Based on the information provided, the compound #68 was assigned to ginsenoside Re.
The MS and MS/MS spectra of compound #66 at RT 6.72 min_m/z 821.3960 have been shown in Figure 1(e) and 1(f). The deprotonated molecule with an m/z value of 821.3960 is identified as the base peak in the MS spectrum. Fragment ions with m/z values of 469.3427 and 351.0681 are attributed to [M-H-gluA-gluA]- and represent two molecules of glucuronic acid, respectively. The ion at m/z 451.2357 is generated by the loss of an H2O molecule from [M-H-gluA-gluA]-, while the ion at m/z 407.6376 corresponds to [M-H-gluA-gluA-H2O-CO2]-. Collectively, this compound was annotated as glycyrrhizic acid.
As illustrated in Figure 1(g), the MS spectrum of compound #8 reveals a base peak at m/z 447.0941, which corresponds to the [M-H]- ion. The MS/MS spectrum, depicted in Figure 1(h), displays ions at m/z 285.0513 and m/z 327.0615, corresponding to the [M-H-glc]- and [M-120.0326]- ions, respectively. Therefore, this compound was assigned to isoorientin, which is a flavonoid.
3.2. Effect of ZSD on acute pneumonia
Histological examination using HE staining was conducted on lung tissue sections to evaluate morphological alterations and determine the therapeutic effects of ZSD. As illustrated in Figure 2, the model group exhibited significant tissue edema, proliferation of inflammatory infiltrates in alveolar epithelial cells, and dilation of the alveolar wall capillaries compared with the control group (Figure 2b). Notably, administration of the drug led to a reduction in symptoms such as tissue edema and inflammatory infiltration (Figure 2c-e). These findings suggest that ZSD confers a protective effect against LPS-induced acute pneumonia.
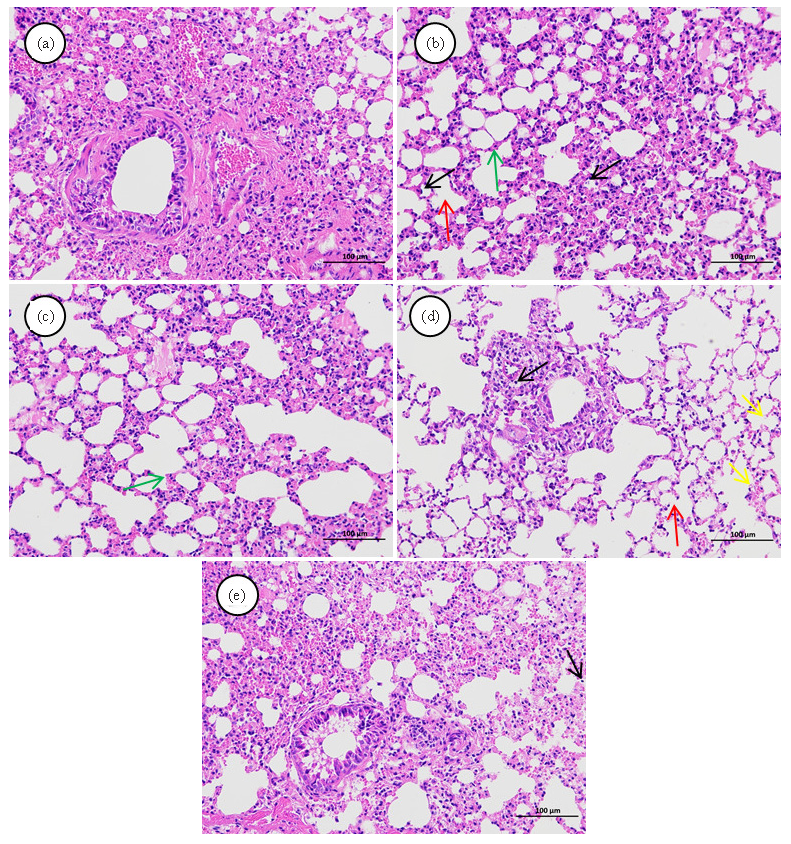
- Hematoxylin and eosin (HE) stained histopathological sections of lung tissue from the (a) control, (b) model, (c) positive, (d) ZSD-L, and (e) ZSD-H groups. The colored arrows illustrate various histopathological alterations in the lungs under different experimental conditions. (magnification, 200×). (The red arrows denote the partial loss of alveolar septa and the coalescence of alveoli into alveolar sacs. Black arrows indicate widespread infiltration of inflammatory cells within the tissue. Green arrows signify hyperplasia of alveolar epithelial cells and thickening of the alveolar septa. Yellow arrows highlight pronounced capillary dilation in the alveolar wall sections.)
The anti-inflammatory properties of ZSD were assessed by measuring the serum levels of the inflammatory cytokines, IgA, and IgM, with results depicted in Figure 3. Relative to the control group, the model group exhibited increased TNF-α, IL-1β, and IL-6 concentration alongside reduced IgA and IgM levels. Following treatment with ZSD, the concentrations of these inflammatory mediators and immune proteins showed a trend towards normalization, aligning more closely with those of the control group.
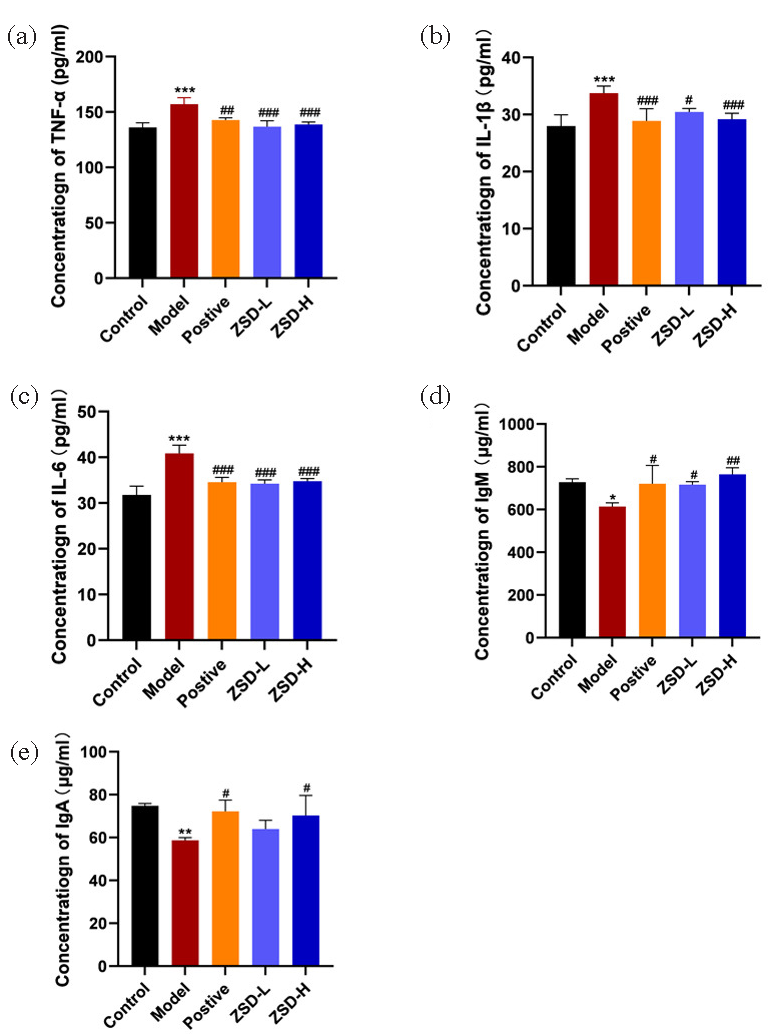
- The expression of inflammatory factors and immune proteins in the serum of each group of mice, including (a) TNF-α, (b) IL-1β, (c) IL-6, (d) IgM, and (e) IgA. Statistical significance is indicated as follows: *P<0.05, **P<0.01, ***P<0.001 when compared to the control group; #P<0.05, ##P<0.01, ###P<0.001 when compared to the model group.
3.3. Validation of LC-MS method for serum metabolomics analysis
Method validation is critical for metabolomics analysis, which ensures that the differences we obtain in our experiments are due to the sample rather than systematic drift or error. The QC samples were pre-injected three times to ensure the system was well-balanced. During objective sample analysis, one QC sample was injected after every six samples, and data were used to validate the established method. After data preprocessing, two datasets were generated from QC injections using the data obtained from different ionization modes. We calculated the relative standard deviation (RSD) of each ion in these two datasets, which showed that more than 80% of features had RSDs of less than 30%, suggesting the excellent reproducibility and stability of the constructed sample analysis method.
3.4. Metabolic difference discovery
LPS administration can provoke pulmonary inflammation in murine models, subsequently altering systemic metabolic pathways. Consequently, serum samples were obtained, and their metabolomic profiles were assessed employing a validated LC-MS technique. The chromatographic peaks of the control, model, and ZSD-H groups exhibit clarity and distinct separation, with notable differences in RT and peak intensities discernible across the chromatograms (Figure 4). Then, a multivariate statistical analysis of serum metabolite data was conducted to archive more precise and intuitive assessments of serum metabolic data. The PLS-DA models were developed utilizing datasets acquired from both positive and negative ionization modes. The PLS-DA score plot for the positive ion mode, illustrated in Figure 5(a), clearly demonstrates a distinct separation between the control and model groups. This separation signifies the occurrence of metabolic alterations in mice with LPS-induced acute pneumonia. This model’s R2Y and Q2 values were 0.982 and 0.872, respectively, suggesting good predictability and fitness. After being treated with ZSD-H, the score plot (Figure 5b) shows that the group separated from the model group to the right side of the y-axis, together with the control group, suggesting the ZSD could modulate the metabolome back to normal to exert the therapeutic effect on acute pneumonia. The permutation test results in Figure 5(c) and (d) show that the calculated R2 and Q2 were lower than the original value, suggesting no overfitness in both PLS-DA models.
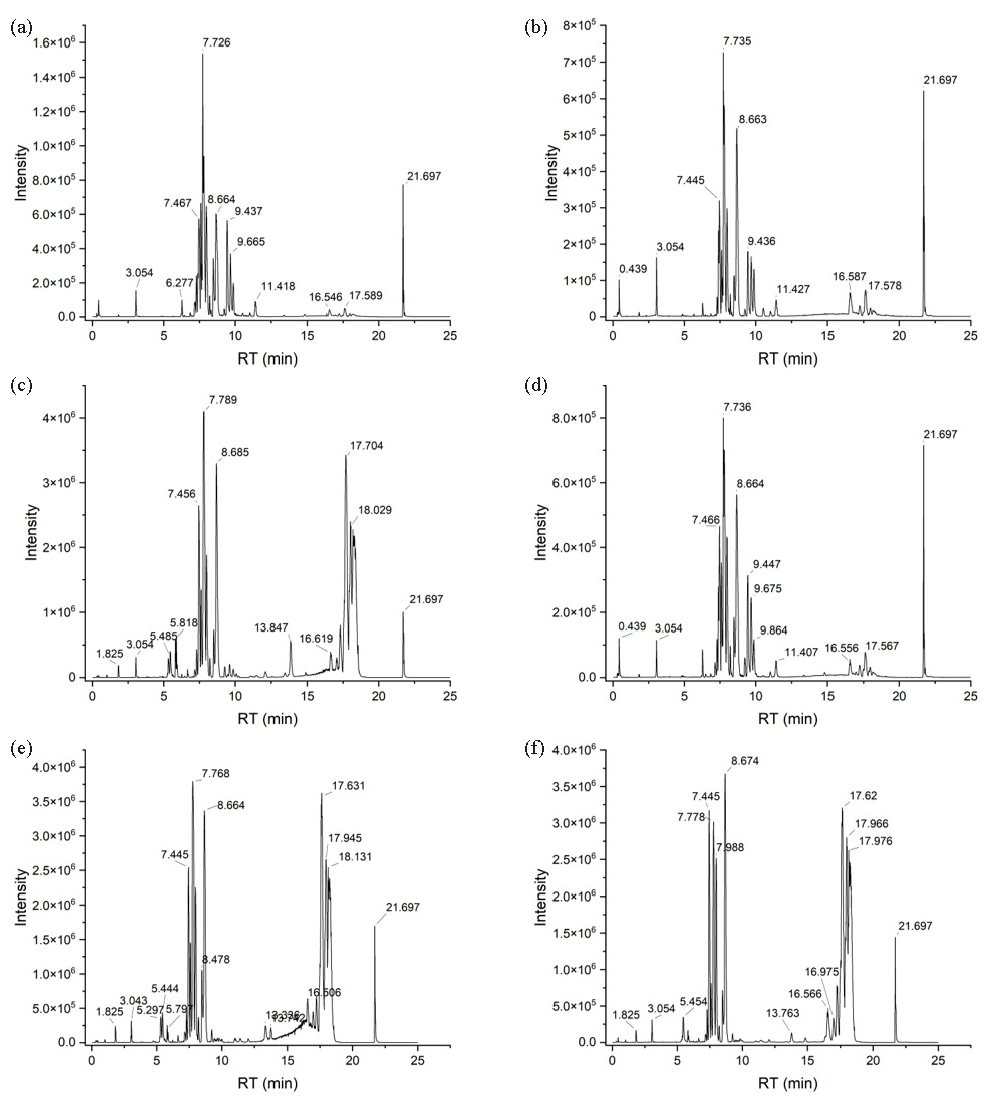
- BPCs for (a-b) the control, (c-d) model, and (e-f) ZSD-H groups, as acquired in (a, c, and e) negative and (b, d, and f) positive ioniza.
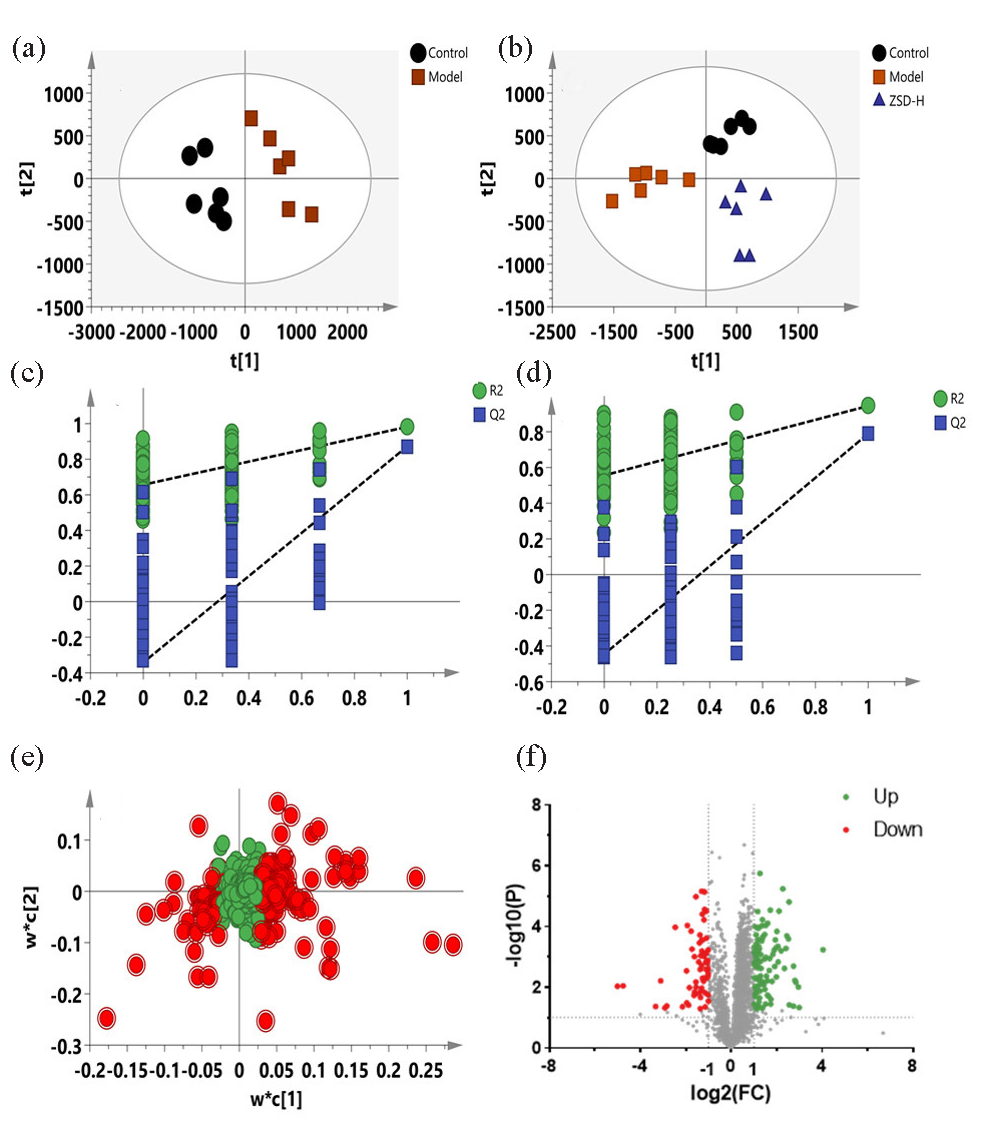
- PLS-DA score plots (a-b), permutation test plots (c-d), PLS-DA loading plots (e), and volcano plots (f) for the control, model, and ZSD-H groups based on the data from negative ion mode.
The VIP value was calculated to show the importance of the feature’s contribution to the sample classifications. The PLS-DA loading plot has been shown in Figure 5(e), in which the point farther away from the origin indicates that the variable contributes more to the classification. The features with VIP >1 are marked in red, and these points are all located far away from the origin, which are selected as the biomarker candidates. Then, the FC and t-test P values were applied to further filter the biomarkers and observe whether their intensities significantly differed between groups. The volcano plot in Figure 5(f) highlights the features that meet the criteria. Metabolites exhibiting a VIP score exceeding 1, a fold change greater than 1.5, and a P value below 0.05 were selected as potential biomarkers associated with acute pneumonia. Similar results have been depicted in Figure 6(a-d), in which the model was constructed using the data obtained from negative ion mode. Biomarkers obtained were highlighted using above procedures (Figure 6e and 6f).
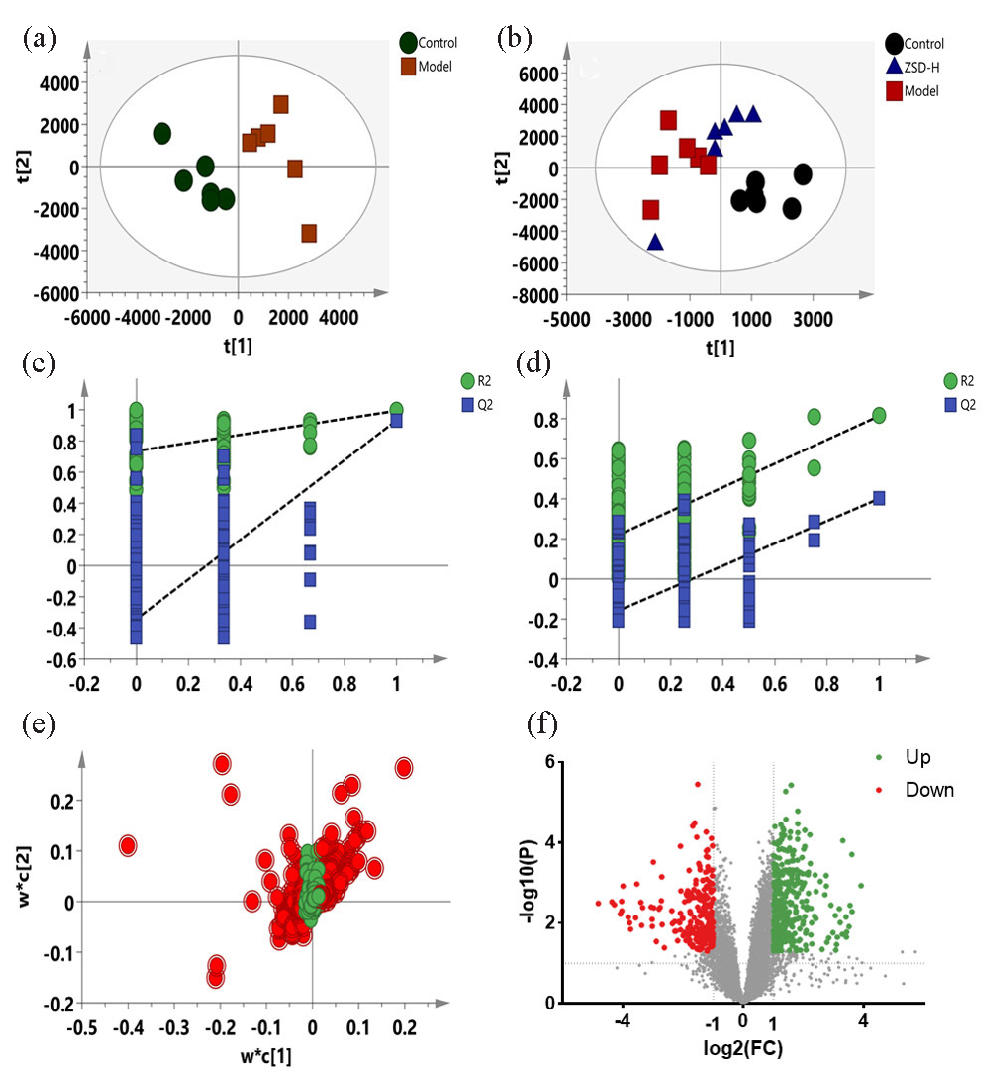
- PLS-DA score plots (a-b), permutation test plots (c-d), PLS-DA loading plots (e), and volcano plots (f) for the control, model, and ZSD-H groups based on the data from positive ion mode.
Chemical structures of the above-selected biomarkers were annotated by database searching in HMDB using its accurate m/z value MS/MS information. The annotation process of LysoPC (16:0/0:0) at RT 17.62 min_m/z 496.3398 was described as follows. The MS spectrum of this metabolite has been presented in Figure 7(a). The peak observed at m/z 496.3398 corresponds to the protonated molecule, while the ion at m/z 991.6641 is associated with the [2M+H]+ species. In Figure 7(b), the fragment ion at m/z 478.3301 is generated through the loss of an H2O molecule from the precursor ion, whereas the ion at m/z 313.2746 results from the detachment of the headgroup unit at m/z 184.0731. The fragment ions detected at m/z 104.1082 and m/z 125.0032 are attributed to the [(CH3)3N(CH2)2O+H]+ moiety from the headgroup and the [head group-(CH3)3N+H]+ ion, respectively. Based on this spectral data, the compound was tentatively identified as LysoPC (16:0/0:0). A total of 74 serum metabolites were finally identified, and the results have been listed in Table S1. The impact of ZSD on these metabolites was then assessed by comparing the intensities between the ZSD-H group and the model group, and the results showed that 67 of the 74 changed metabolites were reversed-regulated, suggesting the ZSD exerted the therapeutic effect on acute pneumonia through modulating these metabolisms of these compounds.
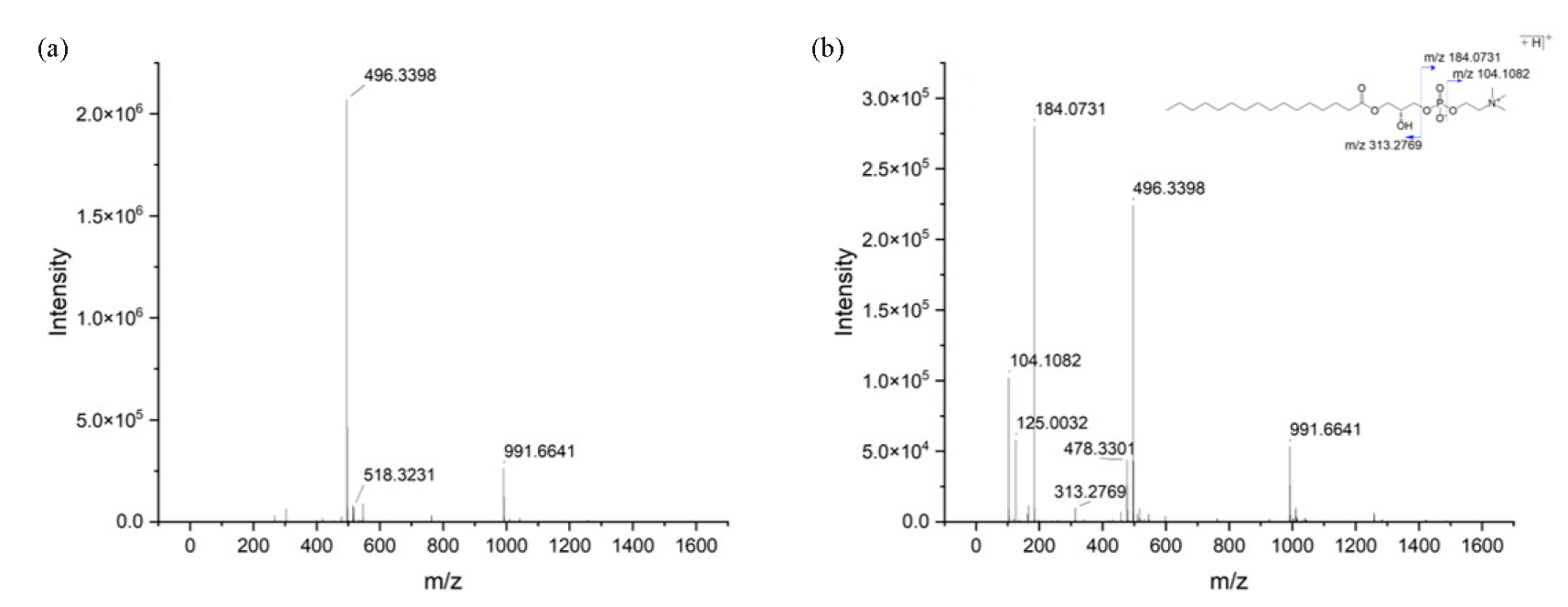
- (a) MS and (b) MS/MS spectra of LysoPC (16:0/0:0) obtained in positive ion mode.
3.5. Network construction and target selection
The CMM-AI-T network was constructed to exhibit the relationship network among the active ingredient and their targets and highlight the core bioactive components of ZSD based on connectivity degree. As shown in Figure 8(a), this CMM-AI-T network contains 511 nodes and 11786 edges. A Venn diagram was employed to intersect the targets linked to the active ingredients of ZSD and those implicated in acute pneumonia (Figure 8b). This analysis revealed a total of 213 common targets, indicating that ZSD has the potential to modulate multiple targets, thereby exerting therapeutic effects on acute pneumonia.
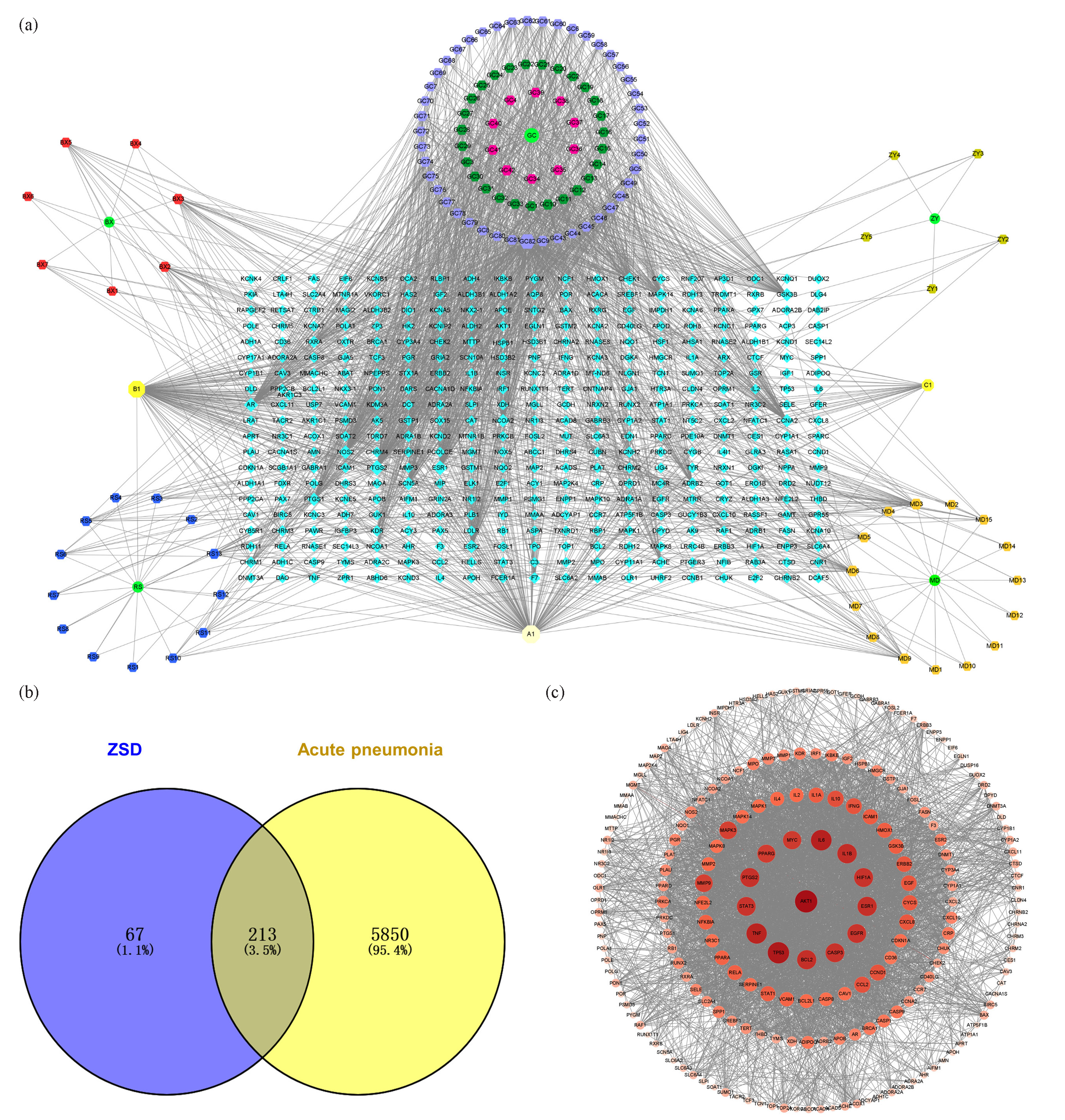
- (a) The CMM-active ingredient-target (CMM-AI-T) network, CMM (•), active ingredient (⬣), target (⬥), common ingredients of CMMs (⬢). (b) The Venn diagram overlap highlighted the potential target of ZSD for treating acute pneumonia. (c) The PPI network was constructed using the selected potential targets.
The PPI network of overlapping targets associated with ZSD and acute pneumonia was constructed utilizing the STRING database. As illustrated in Figure 8(c), the PPI network comprises 212 nodes and 3924 edges. The degree of each target was computed, where a higher degree signifies a more pivotal role of the node within the PPI network. The findings revealed that proteins such as AKT1, IL-6, IL-1β, TNF, MYC, PPARG, PTGS2, STAT3, TP53, BCL2, CASP3, EGFR, ESR1, and HIF1A exhibited degree values exceeding 90, underscoring their significant involvement in the network and their potential as therapeutic targets for ZSD in the treatment of acute pneumonia. The detailed information on the nodes in network pharmacology analysis has been listed in Table S2.
3.6. Joint pathway analysis
Changes in biomarkers reflect altered metabolic phenotypes and provide us with information to understand the metabolic regulation action of ZSD. Key targets were obtained from network pharmacology analysis to show the potential action of the mechanism of ZSD. The joint pathway analysis was performed by integrating these 74 biomarkers and key targets into the MetaboAnalyst 6.0 software for enrichment analysis, with the results presented in Figure 9. The findings indicate that multiple pathways, such as arachidonic acid metabolism, sphingolipid metabolism, glycerophospholipid metabolism, the PPAR signaling pathway, and the JAK-STAT signaling pathway, were affected during the LPS modeling and subsequent ZSD treatment.
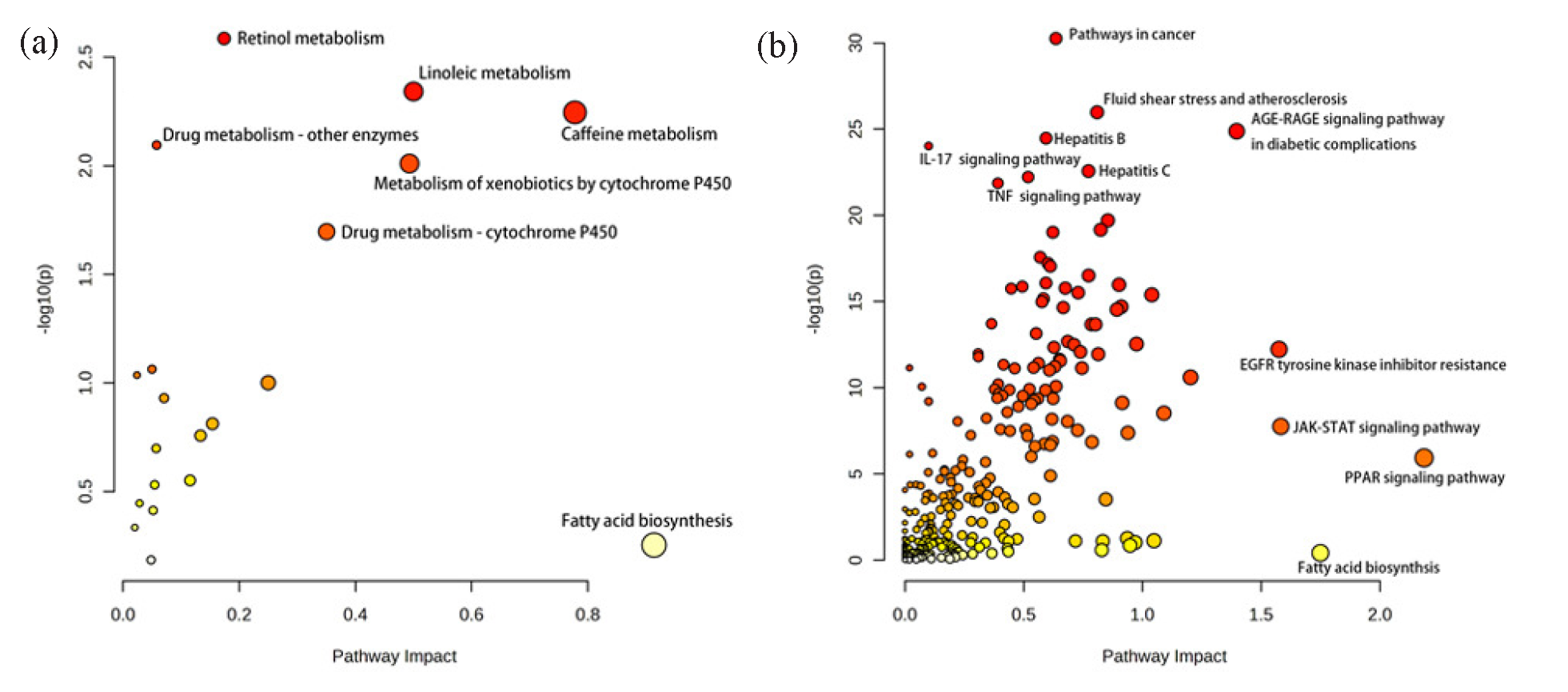
- Bubble diagram of joint pathway analysis using the data obtained from (a) negative and (b) positive ion modes.
3.7. Discussion
Acute pneumonia is predominantly attributable to infections, with its onset primarily associated with the overwhelming proliferation and heightened virulence of pathogenic microorganisms. The common symptoms are high fever, coughing, and difficulty in breathing. ZSD is a typical TCM formula with the medicinal effects of removing heat to promote salivation, tonifying qi, and regulating the stomach. Based on the principles of TCM diagnosis and treatment, ZSD demonstrates therapeutic potential for patients with acute pneumonia during the recovery phase. Specifically, it is beneficial for individuals experiencing high fever, those whose fever has resolved but exhibits lingering symptoms of residual fever, or those suffering from a dual deficiency of qi and yin, characterized by restlessness, thirst, and general debility. However, the precise mechanism underlying its action has yet to be fully elucidated. In this study, we established a mouse model of LPS-induced acute pneumonia and gave ZSD for treatment. ZSD treatment has been shown to mitigate tissue inflammatory cell infiltration, suppress alveolar epithelial cell hyperplasia, and alleviate capillary dilation within the alveolar walls, all of which are characteristic features observed in the model group. Proinflammatory cytokine aggregation has been reported as a mechanism for the pathogenesis of pneumonia [24]. Studies have demonstrated that TNF-α, IL-1β, and IL-6 can influence the central thermoregulatory system, leading to persistent hyperthermia. Among these, IL-1β is particularly notable for its strong proinflammatory activity, as it induces the production of a wide array of proinflammatory mediators, including cytokines and chemokines [25]. IgA plays a pivotal role in the mucosal defense system of the human respiratory tract, while IgM is essential for the body’s humoral immune response. In this study, the ZSD treatment groups, in comparison to the model group, exhibited a significant decrease in the expression levels of pro-inflammatory cytokines TNF-α, IL-1β, and IL-6, alongside a notable increase in the expression levels of IgA and IgM. These findings suggested that ZSD possessed therapeutic potential for acute pneumonia by modulating the expression of inflammatory cytokines, mitigating inflammatory responses, and enhancing immune function.
In this study, a total of 74 serum metabolites were identified as potential biomarkers through an LC-MS-based metabolomics approach. Among these, 67 metabolites, primarily comprising lipids and fatty acids, were reversely modulated by ZSD treatment. Compared with the model group, the intensities of lysophosphatidylcholine (LysoPC), sphingomyelin (SM), phosphatidylcholine (PC), and phosphatidylserine (PS) were significantly elevated in the ZSD-H group, while oxidized phosphatidic acid (PA), phosphatidylglycerol (PG), and cardiolipin (CL) were significantly reduced. These metabolites are mainly involved in arachidonic acid metabolism, sphingolipid metabolism, and glycerophospholipid metabolism. PA is the simplest phospholipid that can bond to several organic molecules, such as the combination with phosphate groups to produce LysoPC, PC, PS, PG, etc. [9].
LysoPC is a monoglycerophospholipid involved in various biological functions, including promoting inflammation, oxidative stress, apoptosis induction, and anti-infection effects [26]. LysoPC is primarily generated through the hydrolysis of phosphatidylcholine by phospholipase A2 (PLA2) at the sn-2 position. It undergoes further metabolism and catabolism via various enzymatic pathways, producing free fatty acids like arachidonic acid (AA) [27, 28]. These fatty acids are subsequently converted into active proinflammatory mediators, including prostaglandins, thromboxanes, leukotrienes, and lipoxins [29]. LysoPC can be converted into lysophosphatidic acid (LysoPA) by autotaxin (ATX), an ectonucleotide pyrophosphatase/phosphodiesterase 2 enzyme with lysophospholipase D activity. As a proinflammatory mediator, ATX represents a promising therapeutic target for potential future treatments of inflammatory diseases [30, 31]. Besides, LysoPC can enhance ATX production by promoting the synthesis of cyclooxygenase-2 and various inflammatory cytokines, which in turn activate multiple signaling pathways involved in oxidative stress and inflammation [32]. In addition, LysoPC can participate in AA metabolism, thereby increasing the production of proinflammatory cytokines in macrophages [33, 34] and promoting the transition of macrophages to proinflammatory M1 phenotype through a G2A-mediated pathway [35].
The result of integrative analysis of metabolomics and network pharmacology showed that the peroxisome proliferator-activated receptor (PPAR) signaling pathway was modulated during ZSD treatment and was intricately associated with inflammatory processes. It was reported that the LPS stimulation increased Th1 cytokines (e.g., TNF-α, IL-1β, IL-6, IL-12) and decreased the Th2 cytokine IL-10 in macrophages of PPARγ-deficient mice [36]. In research on fatty acid-induced inflammation, LysoPC significantly reduced IL-6, ATF3, and CXCL3 expression triggered by palmitic acid. Studies suggest that LysoPC activates PPARδ, a critical pathway through which PPARδ exerts its anti-inflammatory effects by reducing cytokine and chemokine expression and release in muscle cells, adipocytes, and other cell types. Thus, PPARδ activation protected skeletal muscle from fatty acid-induced inflammatory damage and modulated the anti-inflammatory activity of adipocytes and macrophages, preventing leukocyte recruitment to endothelial cells [37]. Therefore, ZSD may exert a therapeutic effect on acute through the PPAR signaling pathway by increasing the serum concentration of LysoPC.
Sphingolipid metabolism plays a crucial role in regulating inflammatory signaling pathways [38]. The first evidence for the involvement of sphingolipids in lung inflammation is the production of autoantibodies against sphingolipids by responsive Mycoplasma pneumonia infection of the lungs [39]. A study demonstrated that sphingolipids were implicated in a range of pathological conditions, such as pulmonary edema, the modulation of inflammatory mediators, and the decline in lung function [40]. Sphingolipid metabolites, particularly sphingolipids, ceramides, and sphingosine-1-phosphate (S1P), were associated with lung inflammation. In the present study, serum SM levels increased significantly after treatment with ZSD, which is consistent with the finding that SM levels significantly decreased in asthmatic patients [41]. Therefore, ZSD may intervene in lung inflammation by affecting SM metabolism.
SM is synthesized by transferring phosphocholine from phosphatidylcholine to ceramide in a reaction catalyzed by SM synthase (SMS). Ceramide is produced by hydrolysis of SM via the sphingomyelinase (SMases), neuroide de novo, and recycling pathways. The biosynthetic pathway of SM and ceramide is known as the “SM cycle” [42]. Ceramides are bioactive lipids that play a role in numerous physiological and pathological processes, like inflammation. Ceramide is converted to sphingosine through the action of ceramidases, including human basic phytoceramidase, human basic ceramidase 1, and human basic ceramidase 2. These enzymes use ceramide as a substrate, thereby regulating intracellular levels of sphingosine and S1P [43]. Inhibition of de novo ceramide synthesis can mitigate oxidative and nitrosative stress, reduce epithelial cell apoptosis, and alleviate airway inflammation, consequently enhancing respiratory function and ameliorating histopathological abnormalities [44]. Moreover, elevated ceramide production in airway epithelial cells has been observed in experimental asthma models, and alterations in the ceramide content of these cells play a pivotal role in the pathogenesis of inflammatory lung diseases.
The joint pathway analysis highlighted that the JAK-STAT signaling pathway involved disease occurrence and treatment using ZSD. JAK-STAT signaling pathway has been reported to play an essential role in inflammatory and autoimmune diseases. The JAK-STAT signaling pathway serves as a critical mediator of cytokine signaling, activated by various proinflammatory molecules to transduce downstream effects and initiate gene transcription [45]. A study on endogenous ceramide production in cultured fibroblasts treated with SMase demonstrated that ceramide activated STAT1 and STAT3 via tyrosine phosphorylation initiated by JAK2 activation [46]. Research also demonstrated that ceramide stimulated matrix metalloproteinase 9 (MMP-9) production by activating the JAK2-STAT3 pathway, identifying it as a potent inducer of MMP-9 in airway epithelial cells [47]. MMP-9 is a pivotal enzyme involved in the remodeling of the extracellular matrix, responsible for the degradation of elastin, collagen, fibronectin, and other extracellular proteins. It plays a critical role in facilitating the infiltration of inflammatory cells into the airways during inflammatory responses [48]. Furthermore, up-regulated levels of MMP-9 were detected in exhaled breath condensate, bronchoalveolar lavage fluid (BALF), and sputum of patients suffering from asthma and chronic obstructive pulmonary disease (COPD) [49]. Consequently, sphingolipid metabolites have the potential to activate the JAK-STAT signaling pathway, thereby modulating inflammatory responses. This mechanism may represent an additional pathway through which ZSD exerted its therapeutic effects in the management of acute pneumonia.
ZSD could regulate biosynthetic metabolic pathways, such as arachidonic acid and sphingolipid metabolism, to influence the concentrations of related metabolites. The changed metabolites could further exert activities through modulating the signaling pathways, like PPAR and JAK-STAT signaling pathways, to exert a therapeutic effect on acute pneumonia. This study exhibited the comprehensive regulatory role of ZSD in disease treatment, which reflected the characteristics of TCM in terms of multiple components, targets, and pathways.
4. Conclusions
In this study, we investigated the efficacy of ZSD in the treatment of acute pneumonia using an LPS-induced acute pneumonia mouse model. Its potential mechanisms were explored using LC-MS-based metabolomics combined with network pharmacology analysis. Serum biochemical analyses and HE staining showed that ZSD reduced inflammatory factors and improved lung tissue changes. A total of 74 metabolites were selected in serum as metabolic biomarkers that changed significantly during modeling, 67 of which were reversed-regulated after being treated with ZSD. Network pharmacology analysis highlighted seven proteins that may associated with the efficacy of ZSD on acute pneumonia. Joint pathway analyses suggest that the medicinal efficacy of ZSD may be related to sphingolipid metabolism, AA metabolism, and PPAR and JAK-STAT signaling pathways. Additional research is necessary to validate these findings, which will advance our comprehension of the regulatory mechanisms of ZSD in the context of acute pneumonia. This study lays the groundwork for the prospective application of ZSD in the treatment of acute pneumonia.
Acknowledgment
This study was supported by the China National Traditional Chinese Medicine Standardization Project (Grant No. ZYBZH-Y-JL-25) and the Changchun University of Chinese Medicine (Grant No. 2023JQ05).
CRediT authorship contribution statement
Zhiliang Sun and Haipeng Tang: Methodology, Software; Zhiliang Sun: Writing - Original draft preparation; Guangzhi Cai and Kejin Xu: Investigation, Data curation; Jiyu Gong: Supervision; Yang Wang: Supervision and Writing- Reviewing and Editing.
Declaration of competing interest
The authors declare that there is no conflict of interest.
Declaration of Generative AI and AI-assisted technologies in the writing process
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript and no images were manipulated using AI.
Supplementary data
Supplementary material to this article can be found online at https://dx.doi.org/10.25259/AJC_192_2024.
References
- Application of metabolomics in viral pneumonia treatment with traditional Chinese medicine. Chinese Medicine. 2019;14:8. https://doi.org/10.1186/s13020-019-0229-x
- [Google Scholar]
- Anti-inflammatory Effect of Symplocos prunifolia Extract in an In Vitro Model of Acute Pneumonia. Plant Foods for Human Nutrition. 2024;79:893-900. https://doi.org/10.1007/s11130-024-01231-5
- [Google Scholar]
- Clinical Effect of Tanreqing Injection Combined With Ceftriaxone Sodium on Acute Pneumonia. China Continuing Medical Education. 2017;13:176-7. https://doi.org/10.3969/j.issn.1674-9308.2017.13.097
- [Google Scholar]
- The Holistic Philosophy of Traditional Chinese Medicine and Conflicts With Modern Medicine. Holistic Nursing Practice. 2023;37:153-160. https://doi.org/10.1097/HNP.0000000000000508
- [Google Scholar]
- Textual Research on Key Information of Classic Prescription Zhuye Shigaotang. Chinese Journal of Experimental Traditional Medical Formulae. 2022;13:176-183. https://doi.org/10.13422/j.cnki.syfjx.20230413
- [Google Scholar]
- Effect of Yinqiao San Combined with Lophatherum and Gypsum Decoction and Antibiotic De-escalation Therapy in the Treatment of Severe Pneumonia. Medical Journal of Liaoning. 2023;37:88-90.
- [Google Scholar]
- Clinical Study of Using Zhuye Shigao Decoction Combined with Maimendong Decoction in the Treatment of Drugresistant Community-acquired Pneumonia. Journal of Sichuan of Traditional Chinese. 2017;35:81-3. https://doi.org/CNKI:SUN:SCZY.0.2017-02-032
- [Google Scholar]
- Technological advances in current metabolomics and its application in tradition Chinese medicine. RSC Advances. 2017;7:53516-53524. https://doi.org/10.1039/c7ra02056b
- [Google Scholar]
- Sera and lungs metabonomics reveals key metabolites of resveratrol protecting against PAH in rats. Biomedicine & Pharmacotherapy. 2021;133:110910. https://doi.org/10.1016/j.biopha.2020.110910
- [Google Scholar]
- Multi-omics analysis of lung tissue metabolome and proteome reveals the therapeutic effect of Shegan Mahuang Decoction against asthma in rats. Journal of Ethnopharmacology. 2024;322:117650. https://doi.org/10.1016/j.jep.2023.117650
- [Google Scholar]
- Integrated metabolomics and network pharmacology to reveal the lipid-lowering mechanisms of Qizha Shuangye granules in hyperlipidemic rats. Journal of the Science of Food and Agriculture. 2024;104:3265-3274. https://doi.org/10.1002/jsfa.13213
- [Google Scholar]
- Network pharmacology: towards the artificial intelligence-based precision traditional Chinese medicine. Briefings in Bioinformatics. 2024;25:bbad518. https://doi.org/10.1093/bib/bbad518
- [Google Scholar]
- MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nature Methods. 2015;12:523-6. https://doi.org/10.1038/nmeth.3393
- [Google Scholar]
- Exploring the Mechanisms of the Huangqin Qingfei Decoction in Pneumonia Treatment for Lipopolysaccharide-Induced Mouse Model: A Network Pharmacology Analysis and Experimental Validation. Natural Product Communications. 2024;19:1-21. https://doi.org/10.1177/1934578X241239843.
- [Google Scholar]
- LC-MS-based metabolomics reveals the in vivo effect of Shegan Mahuang Decoction in an OVA-induced rat model of airway hyperresponsiveness. Molecular Omics. 2022;18:957-966. https://doi.org/10.1039/d2mo00216g
- [Google Scholar]
- HMDB 5.0: the Human Metabolome Database for 2022. Nucleic Acids Research. 2022;50:D622-D631. https://doi.org/10.1093/nar/gkab1062
- [Google Scholar]
- MetaboAnalyst 6.0: towards a unified platform for metabolomics data processing, analysis and interpretation. Nucleic Acids Research. 2024;52:W398-W406. https://doi.org/10.1093/nar/gkae253
- [Google Scholar]
- TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics. 2014;6:13. https://doi.org/10.1186/1758-2946-6-13
- [Google Scholar]
- HERB: a high-throughput experiment- and reference-guided database of traditional Chinese medicine. Nucleic Acids Research. 2021;49:D1197-D1206. https://doi.org/10.1093/nar/gkaa1063
- [Google Scholar]
- UniProt: a hub for protein information. Nucleic Acids Research. 2015;43:D204-D212. https://doi.org/10.1093/nar/gku989
- [Google Scholar]
- GeneCards Version 3: the human gene integrator. Database. 2010;2010:baq020. https://doi.org/10.1093/database/baq020
- [Google Scholar]
- Cytoscape: Software for Visualization and Analysis of Biological Networks. In: Hamacher M., Eisenacher M., Stephan C., eds. Data Mining in Proteomics: From Standards to Applications. Totowa, NJ: Humana Press; 2011. p. :291-303.
- [Google Scholar]
- The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Research. 2021;49:D605-D612. https://doi.org/10.1093/nar/gkaa1074
- [Google Scholar]
- Anti-inflammatory mechanisms of the novel cytokine interleukin-38 in allergic asthma. Cellular & Molecular Immunology. 2020;17:631-646. https://doi.org/10.1038/s41423-019-0300-7
- [Google Scholar]
- The role of IL-1β and TNF-α in intervertebral disc degeneration. Biomedicine & Pharmacotherapy. 2020;131:110660. https://doi.org/10.1016/j.biopha.2020.110660
- [Google Scholar]
- Adipocyte-derived Lysophosphatidylcholine Activates Adipocyte and Adipose Tissue Macrophage Nod-Like Receptor Protein 3 Inflammasomes Mediating Homocysteine-Induced Insulin Resistance. EBioMedicine. 2018;31:202-216. https://doi.org/10.1016/j.ebiom.2018.04.022
- [Google Scholar]
- Lysophosphatidylcholine: Potential Target for the Treatment of Chronic Pain. Journal. 2022:23. https://doi.org/10.3390/ijms23158274
- [Google Scholar]
- Lowering of lysophosphatidylcholines in ovariectomized rats by Curcuma comosa. PLoS One. 2022;17:e0268179. https://doi.org/10.1371/journal.pone.0268179
- [Google Scholar]
- Synopsis of arachidonic acid metabolism: A review. Journal of Advanced Research. 2018;11:23-32. https://doi.org/10.1016/j.jare.2018.03.005
- [Google Scholar]
- Lysophosphatidic acids, cyclic phosphatidic acids and autotaxin as promising targets in therapies of cancer and other diseases. Acta biochimica Polonica. 2008;55:227-240.
- [Google Scholar]
- Autotaxin (ATX) inhibits autophagy leading to exaggerated disruption of intestinal epithelial barrier in colitis. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2023;1869:166647. https://doi.org/10.1016/j.bbadis.2023.166647
- [Google Scholar]
- Cyclooxygenase-2 induction by lysophosphatidylcholine in cultured rat vascular smooth muscle cells: involvement of the p38MAPK pathway. Biomedical research. 2008;29:1-8. https://doi.org/10.2220/biomedres.29.1
- [Google Scholar]
- LysoPC and PAF Trigger Arachidonic Acid Release by Divergent Signaling Mechanisms in Monocytes. Journal of Lipids. 2011;2011:532145. https://doi.org/10.1155/2011/532145;
- [Google Scholar]
- Nanomolar concentrations of lysophosphatidylcholine recruit monocytes and induce pro-inflammatory cytokine production in macrophages. Biochemical and Biophysical Research Communications. 2008;370:348-352. https://doi.org/10.1016/j.bbrc.2008.03.087
- [Google Scholar]
- Lysophosphatidylcholine perpetuates macrophage polarization toward classically activated phenotype in inflammation. Cellular Immunology. 2014;289:185-190. https://doi.org/10.1016/j.cellimm.2014.04.010
- [Google Scholar]
- Parsing the Role of PPARs in Macrophage Processes. Frontiers in immunology. 2021;12:783780. https://doi.org/10.3389/fimmu.2021.783780
- [Google Scholar]
- Lysophosphatidylcholines activate PPARδ and protect human skeletal muscle cells from lipotoxicity. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2016;1861:1980-1992. https://doi.org/10.1016/j.bbalip.2016.09.020
- [Google Scholar]
- Dietary and Endogenous Sphingolipid Metabolism in Chronic Inflammation. Nutrients. 2017;9:1180. https://doi.org/10.3390/nu9111180
- [Google Scholar]
- Role of Sphingolipids in the Pathobiology of Lung Inflammation. Mediators of Inflammation. 2015;2015:487508. https://doi.org/10.1155/2015/487508
- [Google Scholar]
- The role of sphingolipid metabolism disruption on lipopolysaccharide-induced lung injury in mice. Pulmonary Pharmacology & Therapeutics. 2018;50:100-110. https://doi.org/10.1016/j.pupt.2018.04.008
- [Google Scholar]
- Serum sphingolipid profile in asthma. Journal of Leukocyte Biology. 2021;110:53-9. https://doi.org/10.1002/JLB.3MA1120-719R
- [Google Scholar]
- Role of ceramide/sphingomyelin (SM) balance regulated through “SM cycle” in cancer. Cellular Signalling. 2021;87:110119. https://doi.org/10.1016/j.cellsig.2021.110119
- [Google Scholar]
- Sphingolipids in Lung Pathology in the Coronavirus Disease Era: A Review of Sphingolipid Involvement in the Pathogenesis of Lung Damage. Frontiers in Physiology. 2021;12:760638. https://doi.org/10.3389/fphys.2021.760638
- [Google Scholar]
- Ceramide: a key signaling molecule in a Guinea pig model of allergic asthmatic response and airway inflammation. The Journal of Pharmacology and Experimental Therapeutics. 2008;324:548-557. https://doi.org/10.1124/jpet.107.131565
- [Google Scholar]
- JAK-STAT Pathway: A Novel Target to Tackle Viral Infections. Viruses. 2021;13:2379. https://doi.org/10.3390/v13122379
- [Google Scholar]
- Activation of the JAK/STAT pathway by ceramide in cultured human fibroblasts. FEBS letters. 2001;507:163-8. https://doi.org/10.1016/s0014-5793(01)02977-5
- [Google Scholar]
- Ceramide induces MMP-9 expression through JAK2/STAT3 pathway in airway epithelium. Lipids in Health and Disease. 2020;19:196. https://doi.org/10.1186/s12944-020-01373-w
- [Google Scholar]
- Expression of matrix metalloproteinase-9 and -12 in porcine lung infections. Journal of comparative pathology. 2012;146:253-7. https://doi.org/10.1016/j.jcpa.2011.05.005
- [Google Scholar]
- The Variants in the 3’ Untranslated Region of the Matrix Metalloproteinase 9 Gene as Modulators of Treatment Outcome in Children with Asthma. Lung. 2018;196:297-303. https://doi.org/10.1007/s00408-018-0113-y
- [Google Scholar]







