Translate this page into:
Comparative antibacterial potential of silver nanoparticles prepared via chemical and biological synthesis
⁎Corresponding author at: Carretera Torreón-Matamoros Km 7.5, Ciudad Universitaria, Torreon, Coah, Mexico. j.marszalek@uadec.edu.mx (Jolanta E. Marszalek)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
New and improved approaches are urgently needed to fight the increasing number of multi-drug resistant bacteria. The antibacterial effect of silver nanoparticles (AgNPs) prepared by standardized chemical and biological syntheses is compered here. Biological systems included extracts of Opuntia ficus-indica mucilage and extracellular growth broth of Aspergillus niger and Bacillus megaterium. The nanoparticles were characterized by infrared spectroscopy, IR, and transmission electron microscopy. All of the AgNPs shared characteristic IR peaks and had an average size of 20–60 nm. The AgNPs were mainly spherical regardless of synthetic path. The synthesis based on the extracellular broth of the fungus, due to the highest biomass and active compounds concentration, resulted in a high yield of nanoparticle formation. These AgNPs also exhibited the highest inhibition zone against Salmonella typhimurium and Staphylococcus aureus. The syntheses reported here have no significant influence on AgNPs physical characteristics, as compared to literature, but represent processes with shorter reaction time. Additionally, the fungal based nanoparticles have superior antibacterial characteristics.
Keywords
Silver nanoparticles
Green nanosynthesis
Antibacterial activity
Characterization
1 Introduction
Nanoparticles, NPs, are units with a size defined between 1 and 100 nm (Ahmed et al., 2016; Mohanraj and Chen, 2007; Negahdary et al., 2015). Their specific properties allow diverse applications in healthcare, cosmetics, food, environmental, electronics, space, and energy industry to name a few. In the biomedical field, they found utilization because of the high surface area, plasmon resonance, chemical stability, and catalytic activity which together allow low concentration dosage (5 mg L−1). At these dilutions, NPs show low toxicity to human cells (Gurunathan et al., 2009; P. Singh et al., 2015; Zhang et al., 2016).
The antibacterial effect of silver species has been known for over 3000 years and it has been demonstrated that at low concentration, silver is noxious to the human body. This is one of the many reasons why silver nanoparticles (AgNPs) have been utilized in biomedical treatments against microbes, in bio-labeling, and in cancer diagnostic and treatment (Panáček et al., 2018; Prabhu and Poulose, 2012; Rai et al., 2009).
Silver nanoparticles’ bactericidal, fungicidal, and insecticidal activity is connected to size, shape, and concentration (El Badawy et al., 2011; Graves et al., 2015; Siddiqi et al., 2018). They have been shown to defeat Escherichia coli, Enterococcus faecalis, Staphylococcus aureus, and many other pathogen microorganisms (Ahmed et al., 2016; Graves et al., 2015; Gurunathan et al., 2009). Interestingly, they have even found an application in the battle against antibiotic-resistant bacteria. AgNPs in combination with antibiotics have a better antibacterial effect than any of these alone (Deng et al., 2017; Kim et al., 2007).
AgNPs have been shown to accumulate inside the cell membrane and can subsequently penetrate the cells causing damage to them. Within the bacterial cells, the silver ions can form complexes with nucleic acids and interact with the nucleosides (Beer et al., 2012; Dakal et al., 2016; Gudikandula and Charya Maringanti, 2016; Siddiqi et al., 2018). Moreover, silver atoms bind to thiol groups (-SH) of enzymes forming stable bonds, this causes the denaturalization and sudden deactivation of enzymes in the cell membrane. Another model has proposed that silver enters the cell and intercalates between the purine and pyrimidine base pairs disrupting the hydrogen bonding between the two anti-parallel strands and thus, denaturing the DNA molecule (Panáček et al., 2018; Prabhu and Poulose, 2012; Zhang et al., 2011).
Because of their wide areas of application nanoparticles are commercially synthesized, commonly using the chemical and physical processes. Unfortunately, these techniques produce a lot of toxic and hard to degrade byproducts (Ahmed et al., 2016). Biological systems are employed in the syntheses of nanoparticles, resulting in environmentally safe processes (Negahdary et al., 2015; Siddiqi et al., 2018; P. Singh et al., 2015).
A typical “bottom-up” aqueous synthesis of silver nanoparticles involves a redox reaction, which requires a reducing agent, metal precursor, and a stabilizing agent (Ahmed et al., 2016). Reducing agents can be of organic or inorganic nature and some can work as reducing and stabilizing agents (Henglein and Giersig, 1999a). If the reducing/stabilization agents come from a biological system the green synthesis generates biodegradable waste products, it is cost-effective, easily scaled up, and there is no requirement for high rates of temperature and pressure (Ahmed et al., 2016; P. Singh et al., 2015). Bacterial and fungal extra/intracellular extracts, as well as flowers, leaves, and fruit extracts, can be employed in the biogenic synthesis of gold and silver nanoparticles (Siddiqi et al., 2018). An extract can be used for nanoparticles synthesis if it contains functional groups that can act as reducing agents for the silver ion to produce elemental silver (Ahmed et al., 2016; Dubey et al., 2010; Gudikandula and Charya Maringanti, 2016; Gurunathan et al., 2009; Henglein and Giersig, 1999b; Siddiqi et al., 2018). In literature, the reduction and stabilization of silver ions by a combination of biomolecules such as proteins, amino acids, enzymes, polysaccharides, alkaloids, tannins, phenolics, saponins, terpenoids, and vitamins is well established (Gade et al., 2010).
The Opuntia ficus generally called “nopal” is a cactus species with the most economical importance in the world. It is grown in America, Africa, Asia, Europe, and Oceania. Opuntia is used as a fodder and is cultivated to harvest fruit and cladodes (Reyes-Agüero et al., 2005). The chemical composition of O. ficus cladode is water (92%), carbohydrates (∼4–6%), proteins (∼1%), vegetable fats (∼0.2%), minerals (∼1%), and vitamins, mainly ascorbic acid. Also, Opuntia ficus is considered a great source of natural antioxidant compounds mostly the high content of polyphenols that is responsible for the antioxidant effects. Moreover, hydroxyl and carboxylic groups are present in the compounds of Opuntia. These types of compounds play an important role during metal ion reduction processes and allow size control and stability of formed NPs (Al-thabaiti et al., 2008; Silva de Hoyos et al., 2012).
The Aspergillus niger is a filamentous fungus, growing aerobically in the organic matter. It is found on decaying organic material and soil. It is a non-pathogenic fungus widely distributed in nature. Such as all microorganisms, Aspergillus family growth depends on temperature, pH, and water activity (Schuster et al., 2002). Some amino acids fabricated by A. niger like aspartic acid, tryptophan, and reductase enzyme are responsible for the reduction of metal ions by this fungus. As a result, A. niger has heavy metal ions biosorption capacity (Sagar, 2012; Vigneshwaran et al., 2007).
Bacillus megaterium grows in both aerobic and anaerobic conditions. It is a Gram-positive bacterium that shows the optimum growth around 30 °C. It is found usually in soil, but it can inhibit rice paddies, seawater, dried food, and even bee honey (Higashihara and Okada, 1974; Vary, 1994; Vary et al., 2007). It has industrial importance because of its unusual, but useful, enzymes and products. The products include penicillin amidase (precursor of semisynthetic antibiotics), β-amylase, steroid hydrolase, and vitamin B12 (Higashihara and Okada, 1974; Vary, 1994; Vary et al., 2007). Other industrial applications of B. megaterium focus on its ability to produce enzymes and products to reduce metal salts (Banu and Balasubramanian, 2015; Saravanan et al., 2011; Vary, 1994).
Here, we compare the antibacterial effect against representative pathogenic bacteria (S. aureus, S. typhimurium) of silver nanoparticles prepared by standardized chemical and biological syntheses with biological extract readily available in the local area, namely Opuntia ficus indica mucilage extract, Aspergillus niger, and Bacillus megaterium broth. Our standardized approach (the same temperature, time, and precursor concentration) allows a more precise comparison between the synthesized nanoparticles and thus more accurate evaluation of the reducing processes.
2 Materials and methods
2.1 Materials
Silver nitrate and sodium citrate were purchased from Jalmek. Tryptic soy broth and nutritive soy broth originated from DIBICO. Müller-Hinton agar was purchased from BD Bioxon. The chemicals were used without additional purification. The strains used were as follows: Bacillus megaterium, ATCC 14581, Aspergillus niger, ATCC 16888, Staphylococcus aureus, ATCC 25923, Salmonella typhimurium, CDBB-B-1251. Opuntia ficus-indica came from a botanic garden in Torreon, Coahuila.
2.2 Preparation of biological extracts
Here, three extracts from different sources (B. megaterium, A. niger, and Opuntia ficus-indica mucilage) were prepared based on the methodology previously reported. Briefly: (1) Bacillus megaterium extract. 50 µL of the B. megaterium pre-inoculum was added to 300 mL of sterilized nutrient broth (composition in g/L: meat extract 3, peptone from gelatin 5) and left to grow for 24 h with stirring. After that time, the growth broth was centrifuged at 10,000 rpm for 15 min at 4 °C and the resulted supernatant was used as the B. megaterium extract (Banu and Balasubramanian, 2015; Saravanan et al., 2011). The bottom pellet was dried in the oven at 100 °C for 24 h and the dried weight was measured to determine the biomass by percent (grams of biomass per 100 mL of broth). (2) Aspergillus niger extract. 50 µL of the pre-inoculum of A. niger was added to 500 mL of sterilized tryptic soy broth (composition in g/L: sodium chloride 5.0, dextrose 2.5, phosphate dipotassium 2.5, casein peptone 17, soy peptone 3) acidified to pH 5 with acetic acid and grown for 115 h with stirring. After the incubation time, the broth was separated from the micelle by vacuum filtration and the resulted solution was named as A. niger extract (Sagar, 2012). After the filtration step, the filtrate paper was dried in the oven at 100 °C for 24 h and then the dried weight was measured to determine the biomass by percent. (3) Opuntia ficus-indica extract (mucilage)(Sepúlveda et al., 2007) The cladodes were rinsed with distillate water, cut into small pieces (average 1 cm2), and immersed in water (1:5 ratio) for 24 h. The aqueous extract was then vacuum filtered, centrifuged at 10,000 rpm for 10 min, and autoclaved at 121 °C for 15 min. After the autoclaved step, 10 mL of the solution was dried in the oven at 100 °C for 24 h and the dry mass was assigned as the biomass (% of mucilage in the solution).
2.3 Determination of antioxidant and reducing activity
Total phenolic content was determined by the 2,2-diphenyl-1-picrylhydrazyl (DPPH) method (Brand-Williams et al., 1995; Choi et al., 2002), total proteins by the Bradford method (Bradford, 1976) and reducing sugars by the Miller method (3,5-dinitro salicylic acid method) (Miller, 1959).
2.4 Synthesis of nanoparticles
The temperature of all the syntheses was kept at 80 °C and the stirring at 110 rpm. In chemical and biological syntheses, the reaction was heated for 105 min and cooled for 30 min to room temperature.
For the chemical synthesis, the final silver nitrate and sodium citrate concentrations were 0.9 mM and 2.8 mM, respectively (Gudikandula and Charya Maringanti, 2016). These nanoparticles were named CAgNPs.
To 100 mL of the extract (B. megaterium, A. niger, or Opuntia ficus-indica mucilage) AgNO3 solution was added to give the final 0.9 mM concentration of silver (Banu and Balasubramanian, 2015; Sagar, 2012; Silva de Hoyos et al., 2012).
Biosynthesized nanoparticles were labeled as BAgNPs, FAgNPs, and MAgNPs for B. megaterium, A. niger, and Opuntia ficus-indica mucilage extracts, respectively.
Silver nanoparticles purification was done in a centrifuge at 10,000 rpm for 15 min. The nanoparticles synthesized by the chemical path were washed once and the biological reactions were washed 3 times with distilled water (Banu and Balasubramanian, 2015).
2.5 Characterization of nanoparticles
Infrared spectrophotometer (IR) was performed in the Nicolet IR 100 F1-IR Thermo Scientific system with a frequency ranging from 400 to 4000 cm−1 and a resolution of 4 cm−1, using the KBr pellet method to prepare the samples. The NPs were freeze-dried out of the solutions before being mixed with dry KBr.
Transmission Electron Microscopy (TEM) was captured on JEOL JEM-1010. A drop of the diluted samples in deionized water (having a final concentration of <1 µg/mL) were deposited on a carbon-coated copper grid. The samples were completely dried before the analysis, using as a parameter an accelerating voltage in a range 60–80 kV and a resolution of 0.25 nm. The images were taken using the camera Gatan Bioscan 1kx1k and digital program Micrograph 3.1. The analysis of the photos was done with GIMP 2.10.12 software.
2.6 Reaction yield
The PNs samples were freeze-dried in the LABCONCO freeze dry system/Freezone 4.5, for 14 h at 0.058 mBa and −38 °C. Dried samples were weighted to determine the dry mass and then re-suspended to a specific volume of distilled water. The yield of reaction or conversion of silver nitrate into the nanoparticles was calculated in percent by comparing the weight of the freeze-dried nanoparticles compare to the total grams of silver introduced to the reaction.
2.7 Antibacterial assays
All the NPs (CAgNPs, BAgNPs, FAgNPs, and MAgNPs) were analyzed with respect to their antimicrobial activity against Staphylococcus aureus and Salmonella typhimurium (Gram-positive and Gram-negative, respectively). The bioassays were done by well diffusion method on Müller-Hinton agar (MHA) (Gudikandula and Charya Maringanti, 2016). Shortly, wells, 7 mm in diameter, were dug from solidified MHA inoculated with bacterial solutions (50 µL, 1x107 CFU) and were stored for 2 h at 35±°C (Gade et al., 2010). Each plate contained [1] 50 µL of the tested NPs at 500 µg/mL w/v concentrations (BAgNPs, FAgNPs, or MAgNPs); [2] 50 µL amoxicillin 500 ppm (positive control); [3] AgNO3 1 mM (negative control); [4] 50 µL of the extract corresponding to the tested NPs; [5] 50 µL CAgNPs (500 µg/mL w/v). All plates were subjected to subsequent incubation at 37 °C for 24 h at which time the growth inhibition zone in millimeters (GIZ, mm) was recorded. These experiments were done by triplicate.
2.8 Statistical analysis
All experiments of synthesis and yield determination were done in minimum duplicates. The data were evaluated by means of variance analysis (ANOVA) using Minitab 18 software. The significance level was set at p = 0.05 (Tukey HSD).
3 Results and discussion
3.1 Characterization of extracts
The biological extracts, namely Opuntia ficus-indica mucilage extract, extracellular B. megaterium, and A. niger extracts, were evaluated for their dry mass (biomass) content (Table 1). The results indicate that the extract with the higher average biomass concentration (0.20 ± 0.01%) was A. niger but without statistical difference with Opuntia ficus-indica and B. megaterium extracts (0.16 ± 0.05 and 0.09 ± 0.0%, respectively). The biomass amount and composition of the extracts are related to their reducing and stabilizing ability, as reported before (Sepúlveda et al., 2007; Gade et al., 2010; Silva de Hoyos et al., 2012). In the fungus and bacterial extracts, the biomass acts indirectly on the concentration of the reducing agent and at higher biomass concentration the rate of reaction should increase (Banu and Balasubramanian, 2015; Sagar, 2012; Saravanan et al., 2011). Also, the active compound concentrations for the same extracts were determined (Fig. 1). The A. niger extract shows the highest concentration of all the tested compounds as per antioxidants (438 ± 66 ppm), reducing sugars (103 ± 2 ppm), and proteins (7.9 ± 1 ppm). These levels indicate a higher bioactivity potential of A. niger extract in comparison with other extracts used. The antioxidants are most likely represented by phenolic compounds (ascorbic and galacturonic acid), also reducing sugars and catalytic proteins, among other not determined compounds. Previous reports on this fungus indicated that while its biomass increased the reducing sugars also were on the rise, thus possibly aiding in the reduction of the silver ions (Sagar, 2012). In respect to B. megaterium extract, the concentrations of antioxidants (306 ± 16 ppm) and proteins (7.2 ± 0.2 ppm) are not significantly different from the fungus extract levels, however, the concentration of the reducing sugar is much lower (4 ± 0.7 ppm). On the other hand, regardless of rather high biomass, the Opuntia ficus-indica mucilage extract has rather low phenolic compounds (54 ± 8 ppm), reducing sugars (5 ± 0.1 ppm), and protein concentrations (2 ± 0.3 ppm). It has been reported that mucilage is principally composed of sugars and phenolic compounds, which is in accordance to our findings. Its pectins account for various proportions of D-galactose, L-arabinose (pyranose and furanose forms), D-xylose, and L-rhamnose arranged in the core chain with branching (Goycoolea and Cárdenas, 2003). a = No significant difference (p = 0.05). Values are the average of three replicates ± SD. (p < 0.05).
Biological extract
Biomass (%)
Opuntia ficus-indica
0.16 ± 0.05a
Bacillus megaterium
0.09 ± 0.07a
Aspergillus niger
0.20 ± 0.01a
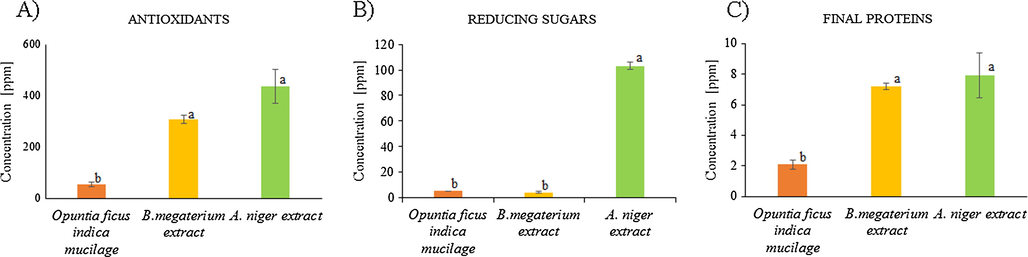
Concentration (in parts per million) of phenolic compounds or antioxidants (A), final proteins (B), and reducing sugars (C) in the biological extracts: Opuntia ficus-indica (orange), B. megaterium (yellow), and A. niger (green). The same letter following the values indicated no significant difference, different letters indicated there is a significant difference. (P = 0.05, Tukey HSD).
The actual compounds involved in ion reduction in biological syntheses are not clearly known, however, it has been discussed in the literature that after the microorganism growth, the enzymes and other primary metabolites, like antioxidants, can act as the reducing agents (Brand-Williams et al., 1995; Sagar, 2012; Siddiqi et al., 2018). In the case of fungal-supported reduction, the electron transfer via enzymatic metal reduction process is most likely involved (Verma et al., 2010). It has also been reported that alpha-nicotinamide adenine dinucleotide phosphate reduced form (NADPH)-dependent nitrate reductase enzymes could play a key role in the biosynthesis of metal NPs (Ahmad et al., 2003). Similarly, some of the reducing enzymes (nitrogenase, hydrogenase) as well as silver-binding peptides can be found in the extracellular bacterial extracts and could be responsible for the silver ion reduction (R. Singh et al., 2015).
Therefore, based on biomass and active compounds concentrations, A. niger extract should show the highest metal-reducing activity.
3.2 The yield of AgNPs
The syntheses of nanoparticles were successfully carried out at the same temperature, time, and silver precursor concentrations (80 °C, 135 min, 0.9 mM). After the purification of the reactions, 5 mL of the nanoparticles solution was freeze-dried under standard conditions (14 h at 0.058 mBa, and −38 °C). Thus, the yield of the nanoparticles formation was evaluated from the dry mass of the nanoparticles. At the same time, the conversion of silver ions into silver nanoparticles was determined (Table 2). The initial silver content was based on the presence of AgNO3 in the reaction. The final silver amount was taken as the weight of the freeze-dried samples where the coating by sodium citrate or any other stabilizers was ignored. Our extracts show the presence of antioxidants, reducing sugars, and residual proteins, thus a mixture of these compounds could be responsible for the reduction reaction of silver ion. The A. niger extract showed the highest contents of these compounds so it was expected to show the highest metal-reducing activity. Indeed, statistical analysis of the reaction yields indicates that synthesis based on Aspergillus niger extract results in the highest amount of AgNPs. Statistically similar results were seen for mucilage synthesis. This is surprising since the concentrations of the active compounds for this extract were rather low. Silver nanoparticles synthesized with mucilage extract, MAgNPs, must have a well-developed coating separating them and leading to higher than expected yields. It is also possible that that coating, composed of high molecular sugars, added weight to the dry mass of the MAgNPs changing their yield calculation. However, the presence of such coating is not fully confirmed by IR analysis discussed in the next section. Values are the average of three replicates ± SD. The same letter following the values indicated no significant difference. (P = 0.05, Tukey HSD).
NPs Samples
Concentration (µg/mL)
Conversion of Ag + to NPs (%)
CAgNPs
33.33 ± 13b
34.4 ± 0.7b
BAgNPs
20.4 ± 16.4b
21 ± 2.0b
MAgNPs
53.2 ± 16.4a
54.8 ± 2.0a
FAgNPs
67.2 ± 29.4a
69.2 ± 3.6a
The capping action of the stabilizing agents, as in CAgNPs and FAgNPs, is also believed to stop the agglomeration in the nanoparticles and allows higher yields (Ahmed et al., 2016; Sagar, 2012; Gudikandula and Charya Maringanti, 2016; Siddiqi et al., 2018). However, in our experimental data, the CAgNPs' silver ion conversion represented about 50% of the FAgNP conversion. Taking into consideration the levels of biological compounds (Fig. 1), the silver ion conversion into metal silver is related to higher concentrations of reducing sugars and antioxidants. The higher biomass occurring during the preparation of fungus extract also plays a role in the reduction process, increasing the conversion rate, thus producing a larger number of nanoparticles.
3.3 Infrared spectroscopy of AgNPs
The nanoparticles were characterized by infrared spectroscopy to assess the efficiency of the purification procedure and to evaluate the presence of the coating compounds. AgNO3 spectrum was compared to a sample of CAgNP to determine the presence of the ionic salt residue in the samples. The two IR spectra are very different and do not share any of the major peaks proving that there is no silver nitrate left in the purified AgNP.
Fig. 2 displays a series of IR spectra, each of them represents AgNP synthesized using the different reducing agents. All the nanoparticles regardless of the preparation have identical peaks with some differences in intensity. This proves an efficient purification procedure of all the samples and also that they share the characteristics of the binding compound (Gade et al., 2010; Siddiqi et al., 2018). The maximum peaks that appeared on the graph were 3415, 2850, 2852, 1724, 1639, 1446, and 1074 cm−1 corresponding to various functional groups. The peak near 3415 cm−1 is related to the presence of O-H stretching originating from sodium citrate or polysaccharides. The peak at 2850 cm−1 corresponds to C-H stretching of saturated hydrocarbons. The peak at 1724 cm−1 is attributed to C = O stretching and at 1639 cm−1 corresponds to the presence of the amide bonds due to the vibrations of N-H related to native proteins, 1446 cm−1 represents the symmetric stretching vibrations of —CO—O— groups of amino acids residues, and 1074 cm−1 can be assigned to C—O stretching of residual polysaccharides (Banu and Balasubramanian, 2015; Das et al., 2017; Jain and Mehata, 2017; Saravanan et al., 2011; Tripathy et al., 2010; Vigneshwaran et al., 2007). These peaks provide an inside to the nature of the reducing and stabilizing agents that were participating in the reduction of the silver ion into nano-silver. That is to say, some peaks can be related to the presence of amino acids and polysaccharides residues. During growth, the microorganisms used here produce polysaccharides. The gender Aspergillus spp. can produce extracellular polysaccharide and other polymers formed by L-alanine, L-asparagine, L-glutamine, L-leucine, L phenylalanine, and several others (Kumaran et al., 2014). Bacillus megaterium also can produce extracellular polysaccharides formed principally of fucose, arabinose, mannose, and glucose (Roy Chowdhury et al., 2011). In the case of Opuntia ficus-indica, the polysaccharides are found in the mucilage and are composed of pectin-like polysaccharides (Sepúlveda et al., 2007; Gade et al., 2010; Silva de Hoyos et al., 2012).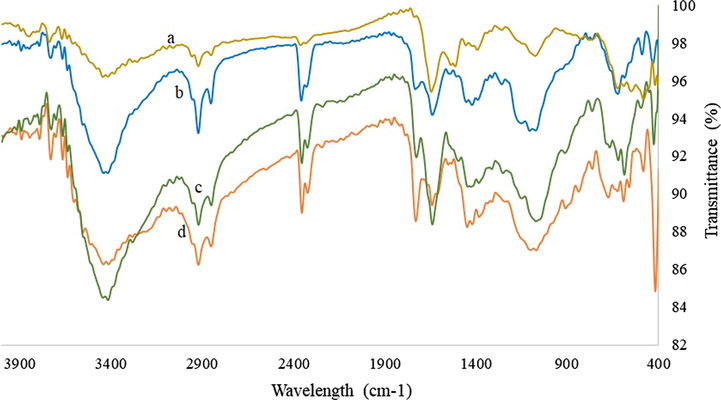
Infrared spectroscopy of silver nanoparticles synthesized with (a) Aspergillus niger extract, (b) Bacillus megaterium extract, (c) chemical synthesis, (d) with nopal mucilage.
Interestingly, the peaks that represent sugars are also present in CAgNPs, which only had sodium citrate in the solution. On the other hand, there are the characteristic peaks only visible in citrate reduced nanoparticles, specifically at 1520 cm−1 (C⚌O stretching asymmetric in COO—) and a weak signal at 1390 cm−1 (C⚌O stretching symmetric in COO—) (Ranoszek-Soliwoda et al., 2017).
Our results and literature reports are not enough to conclude that the peaks in IR spectra of the silver nanoparticles correspond to specific compounds from the extracts. It could be, however, hypothesized that the polysaccharides, amino acids, or their residues promote the reduction of the silver ions and induced the formation of a capping coating around the nanoparticles since these were present in the extracts. Based on the composition of the initial material (plant cladode, initial growth media) and tested extract composition we could draw assumptions that in the case of nopal Opuntia ficus the reduction is driven by hydrocarbons presence, wherein the case of microorganisms after-growth broth is the combination of hydrocarbons, antioxidants, and proteins, thus the reduction is handled by the combination of these. It is evident, that more research is needed to provide stronger evidence to the nature of the reducing and/or capping compounds in these nanoparticles.
3.4 Size and polydispersity of AgNPs
The samples of AgNPs were analyzed by transmission electron microscopy (TEM). Factors like size, shape, and polydispersity related to the AgNPs bactericidal activity were evaluated. Fig. 3 shows representative micrographs of the silver nanoparticles synthesized at 80 °C during 135 min of reaction using biological and chemical reducing/stabilizing agents. It is apparent to see that the represented nanoparticles are smaller than 100 nm, but they show a dispersion of sizes, which is most evident in MAgNPs and CAgNPs. The size of the NPs was determined using the Image Manipulation software GIMP 2.10.12 (Marszalek et al., 2013). The shape of the nanoparticles is generally spherical with a few larger particles showing ellipsoidal shape. This characteristic is present in all the samples. Fig. 4 compares the particle sizes of each synthetic method and shows that the AgNPs synthesized here are between 20 and 60 nm. The statistical analysis established no significant difference between the samples. Besides, all the samples are characterized by a large polydispersity in their sizes, evident in large standard deviation values.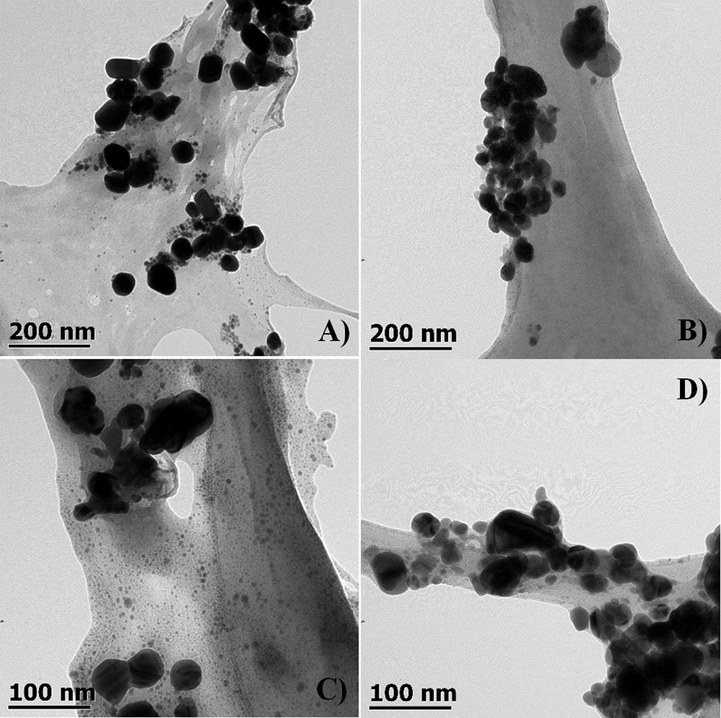
Transmission electron microscope (TEM) micrographs of silver nanoparticles synthesized at 80 °C during 135 min of reaction using: (A) A. niger extract (B) B. megateriums extract, (C) Opuntia ficus-indica mucilage extract, (D) sodium citrate at 2.8 mM as reducing and stabilizing agent.
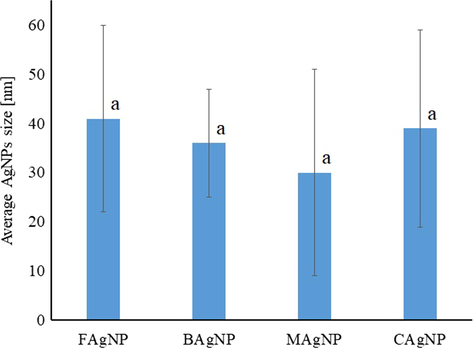
The size of AgNPs synthesized using different reagents (chemical – CAgNPs, bacterial – BAgNPs, mucilage – MAgNPs, and fungal – FAgNPs) estimated based on TEM micrographs. Values are the average of three replicates ± SD. (p < 0.05).
In previous studies, the particles synthesized at 100 °C using sodium citrate as a reducing agent at 2 mM, had 40–85 nm diameter (Asta et al., 2009). Others using AgNO3:Na3C6H5O7 (1:7) obtained AgNPs of 15–50 nm (Ranoszek-Soliwoda et al., 2017). Hence, the chemical synthesis conditions do not allow control over the growth of the NPs and lead to large size polydispersity.
The biological synthesis also leads to polydispersed NPs. AgNPs prepared with whole cladodes blend of the Opuntia ficus-indica, had sizes between 12 and 23 nm and ellipsoidal and spherical shapes (Sagar, 2012; Gade et al., 2010; Silva de Hoyos et al., 2012). Syntheses using the extracellular extract of B. megaterium resulted in spherical, 46–80 nm diameter NPs (Banu and Balasubramanian, 2015; Saravanan et al., 2011). Finally, the NPs synthesized with A. niger extract were about 1–20 nm in size, with a spherical shape (Sagar, 2012; Gade et al., 2008). It is important to highlight that all those reactions were carried out at room temperature and they lasted at least 24 h.
Comparing our results with those previously reported, it can be deduced that the conditions of the reaction do not influence the size and size distribution of the silver nanoparticles. Thus, it could be inferred that the new parameters used in this work and the parameters used by other investigations are not related to the size and size distribution of the nanoparticles, however, it is necessary to gather more information to determine if there is no direct relation between them.
3.5 Antibacterial activity of AgNPs
The freeze-dried nanoparticles were resuspended in water (500 ppm) and used to perform the antibacterial assays. After the incubation time of 24 h, all the AgNPs samples and positive control of antibiotics showed antibacterial activity. The negative control (biological extracts) did not show antibacterial activity. Fig. 5 shows the results of the growth inhibition zone (GIZ) for each NPs type: chemically synthesized (CAgNPs), from bacterial extract (BAgNPs), mucilage supported synthesis (MAgNPs), and fungal extract-based synthesis (FAgNPs). In general, the nanoparticles synthesized via biological path show higher GIZ than the chemically synthesized CAgNPs for both bacteria tasted. Against Salmonella typhimurium the values are 12 ± 1.4, 13 ± 0.0, 13 ± 0.0 mm for BAgNPs, MAgNPS, and FAgNPs, respectfully. Similar diameters were recorded for the Staphylococcus aureus with values 12.5 ± 0.7, 13.5 ± 0.7, and 16 ± 1.4 mm for BAgNPs, MAgNPS, and FAgNPs, respectfully.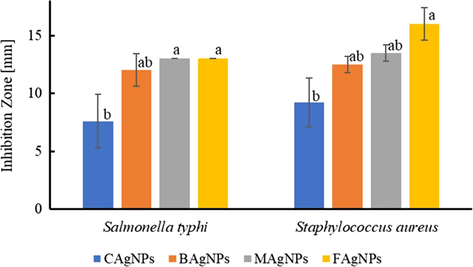
Growth inhibition zone (mm) of AgNPs (chemical – CAgNPs, bacterial – BAgNPs, mucilage – MAgNPs, and fungal – xFAgNPs) against pathogenic bacteria (S. aureus, S, typhi). Values are the average of three replicates ± SD. (p < 0.05).
In agreement with previous reports about the benefits of biological methods for AgNPs synthesis (Gudikandula and Charya Maringanti, 2016), the NPs obtained in the presence of biological extracts display a higher antibacterial effect than the ones synthesized by chemical reduction. Our statistical analysis shows a significant difference in GIZ between the silver nanoparticles synthesized using sodium citrate and A. niger extract as reducing agents.
The results reported here are similar to previously published studies, where GIZ of 10 ± 0.5 mm for S. aureus using 10 µL of biologically synthesized NPs (32 ppm) were used (Das et al., 2017). In the study of Logeswari et al. (2015), silver nanoparticles bio-synthesized using plant extracts such as Ocimum tenuiflorum, Citrus sinensis, Syzygium cumini had GIZ between 12 and 17 mm using 50 µL of the AgNPs solution (at an unknown concentration) against S. aureus. Other research (Saravanan et al., 2011) demonstrated a GIZ of 14.5 mm using 10 µL of AgNPs (at an unknown concentration) synthesized with B. megaterium extract against S. typhi. Bio-synthesized nanoparticles using A. niger extract against S. aureus produced a GIZ of 16 ± 1 mm using a concentrated solution of AgNPs (Sagar, 2012). Finally, the silver nanoparticles bio-synthesized using Opuntia ficus-indica (the whole cladode) extract resulted in a GIZ between 7 and 9 mm against S. aureus using 10 µL of a high concentration of AgNPs solution (Gade et al., 2010).
CAgNPs had lower inhibition zones compared to the biosynthesized AgNPs, which can be directly related to the stability of bio-nanoparticles. It has been reported that biomolecules acting as stabilizing agents give better antibacterial activity than chemical synthesized AgNPs (Gudikandula and Charya Maringanti, 2016; Siddiqi et al., 2018). It is established that during the synthesis in the presence of fungal extracellular extract the silver nanoparticles are surrounded by fungal protein (Durán et al., 2011). Silver nanoparticles from biosynthesis using Aspergillus niger were cytotoxic to E. coli showing a complete disruption of the bacterial membrane after a few minutes of contact (Gade et al., 2008).
It is necessary to point out that even though results from this work are similar to previously published inhibition rates using green nanoparticles, all previous works used the temperature around room temperature and lasted between 24 and 72 h of the reaction for biobased syntheses (Das et al., 2017; Sagar, 2012; Gade et al., 2010; Gudikandula and Charya Maringanti, 2016; Logeswari et al., 2015; Saravanan et al., 2011). In our case, the reactions were carried out at 80 °C and the time of reaction was only 2 h.
The FAgNPs showed the highest inhibition zones thus it is reasonable to assume that these AgNPs will have a supreme bactericidal effect against other human pathogens. The A. niger extract used in the synthesis comes from the fungus line that is able to excrete a high concentration of metabolites during growth thus produced a high yield of nanoparticles (Sagar, 2012; Gudikandula and Charya Maringanti, 2016; Siddiqi et al., 2018). Also, it is believed that FAgNPs have a fungal protein coating that helps in their suspension and must enhance their antibacterial characteristics (Durán et al., 2011).
The FAgNPs were used to determine if their antibacterial characteristics are dependent on the line of bacteria, that is if these bio-nanoparticles act differently against Gram-positive or Gram-negative bacteria. The results with the statistical analyses are reported in Table 3. There is no significant difference in the GIZ of the FAgNPs on different bacterial types producing inhibition zones 13 ± 0.00 and 16 ± 1.40 mm diameters for Salmonella typhimurium and Staphylococcus aureus, respectfully. This is unlike the previously reported findings where the Gram-negative bacteria were more susceptible to the bactericidal action of silver nanoparticles (Ahmed et al., 2016; Ankanna et al., 2010). The reason is understood in the different structures of the Gram-negative and Gram-positive cell walls. Gram-positive bacteria have a thicker layer of peptidoglycan at the surface than Gram-negative bacteria. Peptidoglycan is negatively charged, and silver ions are positively charged; creating a strong attraction resulting in an irreversible linking. However, a theory explains that silver ions are attached stronger to the cell wall of the Gram-negative bacteria. Even though there is a lower concentration of negatively charged peptidoglycan there are other cell wall components that could attract AgNPs (Ahmed et al., 2016; Pazos-Ortiz et al., 2017). Sondi and Salopek-Sondi (2004) described the formation of holes in E. coli bacterial cell wall upon exposure to AgNPs solution. These holes allow AgNPs penetration and lead to cell destruction. Nevertheless, in the case of the result presented here, there is not enough information to detect a significant difference between the two types of bacteria hence further research is needed. Values are the average of three replicates ± SD. a = No significant difference (p < 0.05).
Bacterium
FAgNPs
Salmonella typhimurium
13 ± 0.00a
Staphylococcus aureus
16 ± 1.41a
4 Conclusions
In conclusion, there is no significant difference in the sizes of the nanoparticles regardless of the biological or chemical synthetic paths. Our bio-based syntheses resulted in sizes comparable to literature where the synthesis conditions were very different from ours; specifically, room temperature, darkness, and 24–72 h of reaction. Thus, the conditions of reaction applied here significantly decreased the time of reaction and the use of the temperature of 80 °C should not be considered an extreme condition.
Silver nanoparticles synthesized by biological routes showed better antibacterial activity against pathogen bacteria than the nanoparticles synthesized with sodium citrate. Based on the results, the stabilization potential of the biological extract is the most likely reason for this higher activity. As such, the silver nanoparticles synthesized using A. niger extract as reducing and stabilizing agents were produced at the highest conversion rate and thus higher concentration was obtained. These FAgNPs, when applied at the same concentration as others, exhibited the highest inhibition zone against pathogen bacteria. Thus keeping the same reaction conditions in different chemical and biological paths of synthesis gives us better-understood systems and results in a clear determination of superior nanoparticles, which are the nanoparticles synthesized with the fungal extract.
CRediT authorship contribution statement
Dafne P. Ramírez Aguirre: Investigation, Writing - original draft. Erika Flores Loyola: Conceptualization, Writing - review & editing. Norma M. De la Fuente Salcido: Methodology, Writing - review & editing. Lucio Rodríguez Sifuentes: Methodology, Writing - review & editing. Agustina Ramírez Moreno: Conceptualization, Methodology. Jolanta E. Marszalek Jola: Conceptualization, Investigation, Methodology, Project administration, Writing - review & editing.
Acknowledgment
We would like to extend our thanks to J. Mota Morales and M. Castillo from Centro de Física Aplicada y Tecnología Avanzada of the Universidad Nacional Autónoma de México, Campus Querétaro for the use of transmission electron microscopy.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Colloids Surfaces B Biointerfaces. 2003;28:313-318.
- J. Adv. Res.. 2016;7:17-28.
- Colloids Surfaces B Biointerfaces. 2008;67:230-237.
- Dig. J. Nanomater. Biostructures. 2010;5:369-372.
- Mater. Sci.. 2009;15:1-7.
- Environ. Sci. Technol.. 2011;45:283-287.
- Parasitol. Res.. 2015;114:4069-4079.
- Toxicol. Lett.. 2012;208:286-292.
- Anal. Biochem.. 1976;72:248-254.
- LWT - Food Sci. Technol.. 1995;28:25-30.
- Plant Sci.. 2002;163:1161-1168.
- Front. Microbiol.. 2016;7:1-17.
- Arab. J. Chem.. 2017;10:862-876.
- Colloids Surfaces A Physicochem. Eng. Asp.. 2010;364:34-41.
- Appl. Microbiol. Biotechnol.. 2011;90:1609-1624.
- J. Arid Environ.. 2007;68:534-545.
- Eur. J. Exp. Biol.. 2012;2(5):1654-1658.
- Curr. Nanosci.. 2010;6:370-375.
- J. Biobased Mater. Bioenergy. 2008;2:243-247.
- J. Prof. Assoc. Cactus Dev.. 2003;5:17-29.
- Front. Genet.. 2015;5:1-13.
- J. Exp. Nanosci.. 2016;11:714-721.
- Colloids Surfaces B Biointerfaces. 2009;74:328-335.
- Am. Chem. Soc.. 1999;46556:9533-9539.
- J. Phys. Chem. B. 1999;103:9533-9539.
- Agric. Biol. Chem.. 1974;38:1023-1029.
- Hua Deng, Danielle McShan, Ying Zhang, Sudarson S. Sinha, Zikri Arslan, P.C.R., H.Y., 2017. Environ. Sci. Technol. 50, 8840–8848.
- Sci. Rep.. 2017;7:1-13.
- African J. Microbiol. Res.. 2014;8:2155-2161.
- J. Saudi Chem. Soc.. 2015;19:311-317.
- J. Biomed. Mater. Res. Part A. 2013;101A:1502-1510.
- Anal. Chem.. 1959;31:426-428.
- Trop. J. Pharm. Res.. 2007;5:561-573.
- Biomed. Res.. 2015;26:794-799.
- Nat. Nanotechnol.. 2018;13:65-71.
- J. Nanomater.. 2017;2017:1-9.
- Int. Nano Lett.. 2012;2:32.
- Biotechnol. Adv.. 2009;27:76-83.
- J. Nanoparticle Res.. 2017;19:273.
- Agrociencia. 2005;39:395-408.
- Bioresour. Technol.. 2011;102:6629-6632.
- Colloids Surfaces B Biointerfaces. 2011;88:325-331.
- Appl. Microbiol. Biotechnol.. 2002;59:426-435.
- J. Nanobiotechnology. 2018;16
- Int. J. Nanomedicine. 2015;10:2567-2577.
- Appl. Microbiol. Biotechnol.. 2015;99:4579-4593.
- J. Colloid Interface Sci.. 2004;275:177-182.
- J. Nanoparticle Res.. 2010;12:237-246.
- Microbiology. 1994;140:1001-1013.
- Appl. Microbiol. Biotechnol.. 2007;76:957-967.
- Nanomedicine. 2010;5:33-40.
- Mater. Lett.. 2007;61:1413-1418.
- Environ. Sci. Technol.. 2011;45:4422-4428.
- Int. J. Mol. Sci.. 2016;17:1-34.







