Translate this page into:
Dioxomolybdenum(VI) chelates of bioinorganic, catalytic, and medicinal relevance: Studies on some cis-dioxomolybdenum(VI) complexes involving O, N-donor 4-oximino-2-pyrazoline-5-one derivatives
*Corresponding author. Tel.: +91 761 2601303; fax: +91 761 2603752 rcmaurya1@gmail.com (R.C. Maurya)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Available online 18 January 2011
Peer review under responsibility of King Saud University.

Abstract
A new series of five mixed-ligand complexes of dioxomolybdenum(VI) of the general composition [MoO2(L)2(H2O)2], where LH = 4-acetyloxime-3-methyl-1-phenyl-2-pyrazolin-5-one (aomppH), 3-methyl-1-phenyl-4-propionyloxime-2-pyrazolin-5-one (mppopH), 4-butyryloxime-3-methyl-1-phenyl-2-pyrazolin-5-one (buomppH), 4-iso-butyryloxime-3-methyl-1-phenyl-2-pyrazolin-5-one (ibuomppH) or 4-benzoyloxime-3-methyl-1-phenyl-2-pyrazolin-5-one (bomppH) has been synthesized by the interaction of [MoO2(acac)2] with the said ligands in ethanol medium. The complexes so obtained were characterized by elemental analyses, molar conductance, decomposition temperature and magnetic measurements, thermogravimetric analyses, 1H NMR, IR, mass, and electronic spectral studies. The 3D molecular modeling and analysis for bond lengths and bond angles have also been carried out for one of the representative compound, [MoO2(aomppH)2(H2O)2] (1) to substantiate the proposed structure.
Keywords
Dioxomolybdenum(VI) chelates
O, N-donor oximes
3D Molecular modeling
1 Introduction
Oxime/oximate metal complexes have been investigated actively since the beginning of the last century of the last millennium (Kukushkin and Pombeiro, 1999). Dimethylglyoxime (or diacetyldioxime) was the first organic reagent to be used in analytical chemistry for the estimation of nickel (Tschugaeft, 1905, 1908). Since then, the analytical application of a number of bidentate chelate ligands like dioximes and aromatic hydroxyl aldoximes have been extensively studied (Diehl, 1940; Banks, 1963). Apart from analytical applications, studies have been conducted on their stability data with a variety of metal ions in aqueous medium and also on their coordination behavior (Mehrotra et al., 1975; Singh et al., 1974). Although, oximes are widely recognized as versatile ligands and their complexes with various transition metals have been studied in detail (Mehrotra et al., 1975; Singh et al., 1974), much remains to understand the type of structures that are formed. In general the oxime function is known (Chakravorty, 1974) to coordinate in four ways as shown in Fig. 1. Coordination modes (a) and (c) are observed most frequently although the oxime group is a poor donor unless it is a part of the chelate ring. Oximes can react either as such or in the form of a conjugate base. This is what is meant by putting the hydrogen atom in (a) in parenthesis. This atom may or may not be present. In (c) one oxime group is present as such while the second group is present as the conjugate base; the single hydrogen atom is then shared in the O–H⋯O bridge.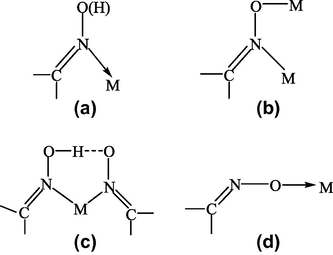
Various coordination modes of oximes.
The chelating behavior of 4-oximino-2-pyrazoline-5-ones (Shah and Shah, 1981, 1982a,b; Patel and Shah, 1985; Maurya et al., 2003d; Masoud et al., 1986) is well established. They coordinate through the cyclic carbonyl oxygen and oximino nitrogen, thus behaving as bidentate O, N donors. Since the increasing use of coordination compounds in analytical chemistry, pigments, medicine and biochemistry, many investigators have studied these topics, especially the important role of oxime complexes in chemistry (Pande, 1966; Schrazer, 1976; Brown, 1973; Gok and Bekaroglu, 1981; Irez and Bekaroglu, 1983; Gul and Bekaroglu, 1982; Kocak and Bekaroglu, 1985; Ozcan and Mirzaoglu, 1988; Ucan and Mirzaoglu, 1990; Sevindir and Mirzaoglu, 1992; Karatas et al., 1991a; Ucan and Karatas, 1991; Pekacar and Ozcan, 1994, 1995a,b; Mercimek et al., 1995; Karatas and Ucan, 1998; Karatas et al., 1991b; Reddy et al., 2000; Alexandrova et al., 2000; Yildim et al., 2003; Zulfikaroglu et al., 2003).
The coordination compounds of molybdenum can catalyze a variety of industrially important chemical reactions such as olefin epoxidation (Abrantes et al., 2003; Li et al., 2010), isomerization of allyl alcohols (Franczek et al., 2002), and olefin metathesis (Schrock, 2004). The useful role of molybdenum is not restricted to artificial catalysis alone, since it is an essential element in diverse biological systems, as nature has made use of the molybdenum center in various redox enzymes (Collision et al., 1996; Hille, 1996; Enemark et al., 2004). Oxidized forms of these molybdoenzymes, e.g., aldehyde and sulfite oxidases are supposed to contain cis-MoX2 units (X = O, S) coordinated to sulfur, nitrogen and oxygen donor atoms of the protein structure.
The presence of the cis-dioxomolybdenum(VI) cation, [MoO2]+2, in the oxidized form of certain molybdoenzymes (Stiefel, 1979a), has stimulated both the search for new structures in which this moiety is coordinated to ligands containing nitrogen, oxygen and/or sulfur donors and also to the study of their chemicals, spectroscopic, and structural properties (Stiefel, 1979b).
Molybdenum bears some resemblance to vanadium. Both vanadate and molybdate can strongly inhibit protein tyrosine phosphatase activity, thus maintaining tyrosine phosphorylation in protein extracts, although molybdate is less strongly bound in the enzyme active site (Elberg et al., 1995). Molybdate was shown to inactivate glycogen synthase and increase the glycolytic flux in rat hepatocytes (Fillat et al., 1992) and to display synergistic stimulation of glucose uptake in rat adipocytes in the presence of H2O2 (Elberg et al., 1995; Goto et al., 1992), again analogous to in vitro effects of vanadate. Sodium molybdate dihydrate (Na2MoO4·2H2O), 0.4–0.5 g L−1 in drinking water and 0.75–1.25 g Kg−1 in food for 8 weeks in STZ-diabetic rats, led to 75% blood glucose lowering and also normalized plasma triglyceride and non-esterified fatty acid levels, without affecting plasma insulin (Ozcelikay et al., 1996), in STZ-diabetic rats. Thus, in view of these findings efforts are being made to synthesize complexes of MoO22+ containing oxygen and/or nitrogen environment, such as, bis(maltolato)dioxomolybdenum(VI) (BMDOM) complex, [MoO2(ma)2] (Shuter, 1995, Fig. 2).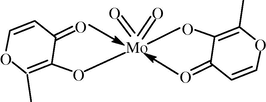
Structure of BMDOM.
Most molybdenum(VI) complexes contain cis-MoO22+ and an octahedral geometry. Although many precursors are available, [MoO2(acac)2] is a convenient source of MoO22+ (Mohanty and Dash, 1990; Syamal and Maurya, 1989).
In continuation of interest in the synthesis and characterization of coordination compounds of bioinorganic and medicinal relevance, a study was undertaken of the coordination chemistry of dioxomolybdenum(VI) complexes involving 4-oximino-2-pyrazoline-5-ones, such as, 4-acetyloxime-3-methyl-1-phenyl-2-pyrazoline-5-one (aomppH, I), 3-methyl-1-phenyl-4-propionyloxime-2-pyrazoline-5-one (mppopH, II), 4-butyryloxime-3-methyl-1-phenyl-2-pyrazoline-5-one (buomppH, III), 4-isobutyryloxime-3-methyl-1-phenyl-2-pyrazoline-5-one (ibuomppH, IV), 4-benzoyloxime-3-methyl-1-phenyl-2-pyrazoline-5-one (bomppH, V). The structure of the oxime derivatives used in the present study is shown in Fig. 3.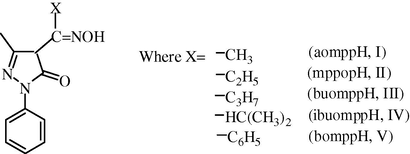
Structures of various oximes.
2 Experimental
2.1 Materials
3-Methyl-1-phenyl-2-pyrazolin-5-one- (Lancaster, UK), benzoyl chloride, acetyl chloride and propionyl chloride, hydroxylamine hydrochloride, acetylacetone, calcium hydroxide (Thomas Baker Chemicals Limited, Mumbai), butyryl chloride, iso-butyryl chloride (Merck, Germany) and Sodium hydroxide (E. Merck India Limited, Mumbai), ammonium molybdate tetrahydrate (Sisco Chem. Industries, Mumbai) were used as supplied. Alcohol was purchased from Bengal Chemicals and Pharmaceuticals Limited, Kolkata. All other chemicals used were of analytical reagent grade.
2.2 Preparation of parent compound
The parent compound bis(acetylacetonato)dioxomolybdenum(VI), [MoO2(acac)2] was prepared by the method of Chen et al. (Chen et al., 1976).
2.3 Preparation of different 4-acyl-3-methyl-1-phenyl-2-pyrazolin-5-one
They were prepared by the Jensen’s (Jensen, 1959) method with slight modification. Into a one liter 3-necked quick fit flask containing DMF (100 mL) and carrying a dropping funnel, a mechanical stirrer, and a reflux condenser, was placed 3-methyl-1-phenyl-2-pyrazolin-5-one (17 g, 0.098 M). A solution was obtained by gentle heating, and benzoyl chloride (12 mL)/acetyl chloride (5.4 mL)/propionyl chloride 8.94 mL)/butyryl chloride (10.33 mL)/isobutyryl chloride (11 mL) was added drop wise within 2–3 min. The reaction was exothermic and the reaction mixtures became a paste. The mixture was allowed to cool and then refluxed with stirring for 1 h on a sand bath, during which period the bright yellow complex formed initially turned yellowish brown. The complex was decomposed by pouring the reaction mixture into chilled dilute hydrochloric acid solution (500 mL, 3 N). A yellowish brown solid settled, which was filtered in a sintered glass crucible, washed with distilled water until the washings were colorless, and dried in air and recrystallized from n-heptane. Some important physical properties are given in the Table 1. bmppH = 4-benzoyl-3-methyl-1-phenyl-2-pyrazolin-5-one. bumppH = 4-butyryl-3-methyl-1-phenyl-2-pyrazolin-5-one. amppH = 4-acetyl-3-methyl-1-phenyl-2-pyrazolin-5-one. pmppH = 3-methyl-1-phenyl-4-propionyl-2-pyrazolin-5-one. Iso-bumppH = 4-iso-butyryl-3-methyl-1-phenyl-2-pyrazolin-5-one.
Abbreviation
Empirical formula
m.p.
Color
Yield (%)
bmppH
(C17H14N2O2) (278)
90
Yellow
75
bumppH
(C14H16N2O2) (244)
65
Yellow
70
amppH
(C12H12N2O2) (216)
60
Yellow
50
pmppH
(C13H14N2O2) (230)
68
Yellow
60
iso-bumppH
(C14H16N2O2) (244)
57
Yellow
50
The reaction scheme related to the synthesis of 4-acyl-3-methyl-1-phenyl-2-pyrazolin-5-one is as follows (Scheme 1.).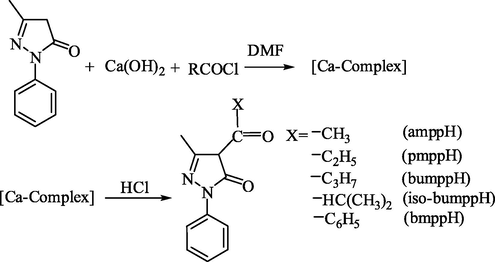
Reaction scheme for the synthesis of 4-acyl-3-mthyl-1-phenyl-2-pyrazolin-5-one derivatives.
2.4 Preparation of oxime derivatives of bmppH, bumppH, amppH, pmppH, iso-bumppH
These oximes were prepared by following the general method given below: A solution of hydroxylamine hydrochloride (0.347 g, 5 mmol) and sodium hydroxide (0.20 g, 5 mmol) in 40 mL aqueous/ethanol (1:1) was added to a solution of 4-benzoyl-3-methyl-1-phenyl-2-pyrazolin-5-one (1.39 g, 5 mmol), 4-acetyl-3-methyl-1-phenyl-2-pyrazolin-5-one (1.08 g, 5 mmol), 3-methyl-1-phenyl-4-propionyl-2-pyrazolin-5-one (1.15 g, 5 mmol), 4-butyryl-3-methyl-1-phenyl-2-pyrazoline-5-one (1.22 g, 5 mmol), or 4-iso-butyryl-3-methyl-1-phenyl-2-pyrazolin-5-one (1.22 g, 5 mmol) in 20 mL ethanol. The mixture was refluxed for two hours. On cooling a crystalline product was obtained, which was filtered by suction, washed several times with water and finally with ethanol. The oxime of 4-benzoyl-3-methyl-1-phenyl-2-pyrazolin-5-one was precipitated by pouring the reaction mixture in excess (150 mL) of distilled water. The characterization data of the oxime derivatives are given in Table 2.
S. No.
Ligand(empirical formula) (F.W.)
Analyses, found/(calcd.), %
Color
Decomposition temp. (°C)
C
H
N
I
aomppH(C12H13N3O2) (231)
62.22 (62.34)
5.52 (5.63)
18.09 (18.18)
Golden brown
170
II
mppopH(C13H15N3O2) (245)
63.47 (63.67)
6.05 (6.12)
17.10 (17.14)
Pale cream
160
III
buomppH(C14H17N3O2) (259)
64.75 (64.86)
6.48 (6.56)
16.18 (16.22)
Daffodil
165
IV
ibuomppH(C14H17N3O2) (259)
64.70 (64.86)
6.48 (6.56)
16.15 (16.22)
Volcano
140
V
bomppH(C17H15N3O2) (293)
69.50 (69.62)
5.06 (5.12)
14.29 (14.33)
Sugar cane color
155
2.5 Preparation of complexes
The oximes derivative, bomppH (0.586 g, 2 mmol), aomppH (0.462 g, 2 mmol), mppopH (0.490 g, 2 mmol), buomppH (0.518 g, 2 mmol) or ibuomppH (0.518 g, 2 mmol) was dissolved by heating in 20 mL of ethanol. To this solution, an ethanolic solution (10 mL) of bis(acetylacetonato)dioxomolybdenum(VI) (0.326 g, 1 mmol) was added. The resulting solution was refluxed for 10–12 h and then concentrated to half of its volume. The resulting precipitate was suction filtered and washed several times using 1:1 ethanol–water and dried in vacuo. The analytical data of the complexes are given in Table 3.
S. No.
Ligands
ν(C⚌O)(pyrazolone)
ν(C⚌N)(oxime)
ν(C⚌N)(pyrazolone)
ν(OH)
ν(N–O)
I
aomppH
1660
1620
1590
3200
1020
II
mppopH
1664
1656
1602
3451
1044
III
bomppH
1685
1620
1590
3040
1020
IV
ibuomppH
1685
1620
1590
3040
1020
V
bomppH
1675
1610
1590
3080
1010
2.6 Analyses
Carbon, hydrogen, and nitrogen were determined micro-analytically at the Central Drug Research Institute, SAIF, Lucknow. The metal content in each chelate was determined (Maurya et al., 1993) as follows: A weighed amount (∼200 mg) of the chelate was first decomposed by heating with concentrated nitric acid and then strongly heating the residue over 800 °C for 1 h until a constant weight was obtained. The residue was weighed as MoO3.
2.7 Physical methods
Electronic spectra of the complexes were recorded in dimethylformamide on an ATI Unicam UV-2–100 UV/visible spectrophotometer in our Department. 1H NMR spectra of compounds were recorded in DMSO-d6 at Central Drug. Research Institute, SAIF, Lucknow. The solid-state infrared spectra were recorded in KBr pellets using Perkin–Elmer model 1620 FT-IR spectrophotometer at the Indian Institute of Technology, Roorkee and also on FT-IR 8400S SHIMADZU at our own Department. Conductance measurements were done at room temperature in dimethylformamide using a Toshniwal Conductivity Bridge and dip type cell with a smooth platinum electrode of cell constant 1.02. Decomposition temperatures of the oxime derivatives and chelates were recorded using an electrothermal apparatus having the capacity to record temperature up to 360 °C. Thermogravimetric analyses of the samples were done at SAIF, Indian Institute of Technology, Mumbai. Mass spectra were recorded at Sophisticated Analytical Instrumentation Facility, CDRI. Lucknow. Magnetic measurement was performed by the Vibrating Sample Magnetometer method.
2.8 Molecular modeling studies
The 3D molecular modeling of a representative compound was carried out on a CS Chem 3D Ultra Molecular Modeling and Analysis Program (www.cambridgesoft.com). It an interactive graphics program that allows rapid structure building, geometry optimization with minimum energy and molecular display. It has ability to handle transition metal compounds.
3 Results and discussion
3.1 Composition and characterization of the oxime derivatives
The oxime derivatives used in the present investigation were prepared as shown in reaction scheme given in Scheme 2.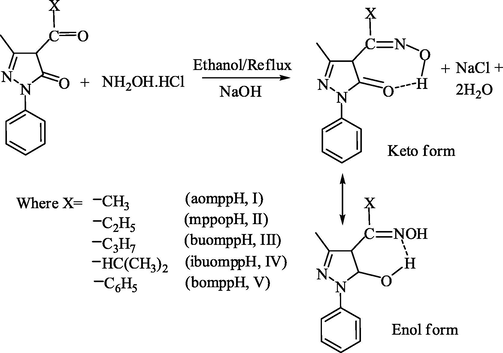
Synthesis of oxime derivatives.
The compositions of the ligands are consistent with the micro-analytical data. The IR spectra of these oximino derivatives have a band at 1660–1685 cm−1, which is assigned to ν(C⚌O) (cyclic) of the pyrazolone skeleton. A strong band at 1610–1656 cm−1 in these derivatives is assigned to v(C⚌N) (oxime). Another strong band observed in all the oxime derivatives at 1590–1602 cm−1 is most probably due to ν(C⚌N) (cyclic) of the pyrazolone skeleton. All the oxime derivatives show medium/broad bands with fine structure in the range 3040–3451 cm−1, which may be due to v(OH) of the oxime group. The observed low value of v(OH) suggests the presence of intramolecular hydrogen bonding in the derivatives. A new band observed at 1010–1044 cm−1 in the oxime derivatives, unlike the parent carbonyl compound, may be assigned to v(N–O). The overall IR results conclude that all the oxime derivatives exist in ketonic form in the solid state as shown in the scheme above.
The proton NMR spectra of two representative ligands namely mppopH (II) (Fig. 4) and buomppH (III) (Fig. 5) were recorded in DMSO-d6. The proton signals due to aromatic protons of the phenyl ring in the two ligands displayed as multiplets in the region δ 7.194–7.913 and δ 7.173–7.939 ppm, respectively. The other proton signals in case of ligand (II) at δ 2.740–2.789 (quartet), δ 2.342 (singlet) and at δ 1.051–1.140 (triplet) ppm are most probably due to proton groups, such as –CH2 (b), –CH3 (a), and –CH3 (c), respectively. In case of the ligand (III) proton signals at δ 2.642–2.858 (multiplets), δ 2.317 (singlet), δ 1.576–1.623 (triplet) and δ 0.902–0.927 (triplet) ppm are due to proton groups likely to be –CH2 (c), –CH3 (a), –CH2 (b) and –CH3 (d), respectively. Both the ligands displayed two singlet proton signals at δ 10.240–10.422 and δ 10.986–11.101 ppm most probably due to enolic (OH) (cyclic) and –NOH (oxime), respectively. The indexing of various protons is given in Fig. 6.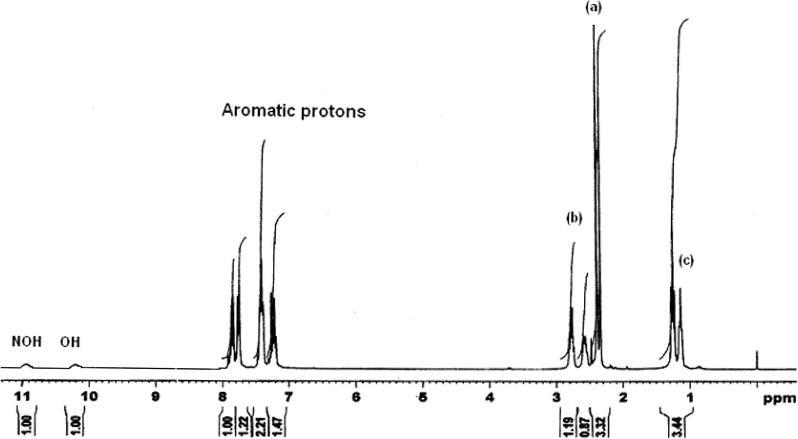
1H NMR spectrum of mppopH (II).
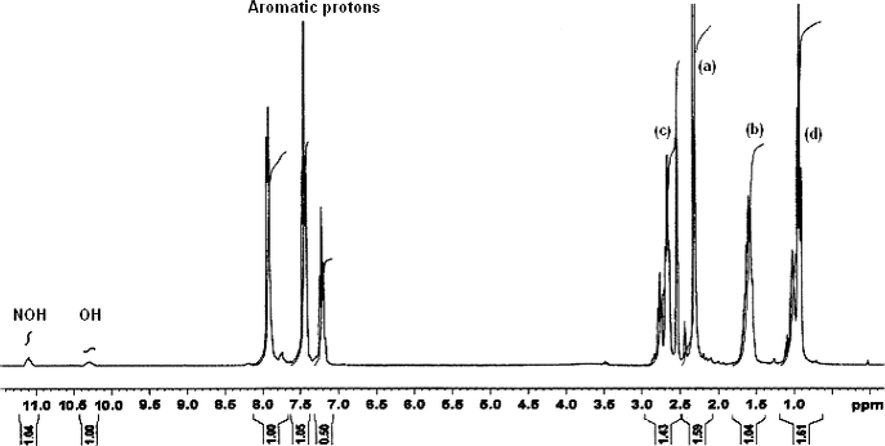
1H NMR spectrum of buomppH (III).
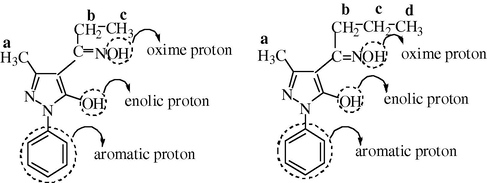
Indexing of various protons in ligands II and III.
The presence of a proton signal at δ 10.240–10.422 ppm suggests that the ligand exist in enolic form in DMSO-d6 (solvent used for NMR). Contrary to this, all the ligands exist in ketonic form in the solid state as concluded from the IR spectral studies discussed above.
3.2 Characterization of complexes
The formation of metal chelates can be represented by the following general equation: The driving force for the reaction in order to form these chelates along with the elimination of two molecules of acetylacetone (acacH) is the better donating (Maurya et al., 2003c) capability of the oxime derivatives compared to acetylacetone anion.
The synthesized complexes are colored, non-hygroscopic and air stable solids. They are thermally stable and their decomposition temperatures are given in the Table 4. These are soluble in dimethylformamide and dimethylsulfoxide, and insoluble in all other common organic solvents. Some physical properties of the complexes are also given in Table 4. The formulations of these complexes are based on their elemental analyses, infrared spectra, magnetic measurements, NMR, mass, thermogravimetry, and electronic spectral studies.
S. No.
Complex* (empirical formula) (F.W.)
Analyses, found/(calcD.), %
Color
Decomp. temp. (°C)
Yield (%)
ΛM (Ohm−1 cm2 mol−1)
C
H
N
Mo
1
[MoO2(aomppH)2(H2O)2](C24H28N6O8Mo) (623.94)
46.04 (46.16)
4.29 (4.49)
13.25 (13.46)
15.28 (15.38)
Middle buff
210
50
11.5
2
[MoO2(mppopH)2(H2O)2](C26N32N6O8Mo) (651.94)
47.67 (47.86)
4.82 (4.91)
12.68 (12.88)
14.60 (14.72)
Daffodil
220
60
15.2
3
[MoO2(buomppH)2(H2O)2](C28H36N6O8Mo) (679.94)
49.29 (49.42)
5.15 (5.29)
12.15 (12.35)
14.05 (14.11)
Yellow
240
52
14.1
4
[MoO2(ibuomppH)2(H2O)2](C28H36N6O8Mo) (679.94)
49.30 (49.42)
5.12 (5.29)
12.20 (12.35)
14.02 (14.11)
Deep green
220
50
12.4
5
[MoO2(bomppH)2(H2O)2](C34H32N6O8Mo) (747.94)
54.42 (54.55)
4.20 (4.28)
11.13 (11.23)
12.67 (12.83)
Leaf brown
200
52
10.1
3.3 IR spectral studies
The MoO22+ moiety prefers to form a cis-dioxo grouping due to the maximum utilization of the d-orbital for bonding. The dioxo-configuration is characterized by two infrared active modes of νs(O⚌Mo⚌O) and νas(O⚌Mo⚌O) in C2V symmetry. The trans-MoO22+ moiety would exhibit a single infrared active stretching band of νas(O⚌Mo⚌O). The presence of two infrared bands in the 910–913 and 941–991 cm−1 regions due to νas(O⚌Mo⚌O) and νs(O⚌Mo⚌O), respectively, in the present complexes is strongly indicative of the cis-MoO22+ structures (Syamal and Maurya, 1989; Maurya et al., 1995a).
The coordination of oxime nitrogen is inferred by a shift to lower frequencies57 (Maurya et al., 2003d) in the ν(C⚌N) (Oxime) (1567–1578 cm−1) in all the complexes as compared to that of the free oxime derivatives at 1610–1656 cm−1. This is further supported by shifting of the ν(N–O) to a higher wave number (1073–1091 cm−1) (Yildim et al., 2003; Maurya et al., 2003d; Karatas et al., 1991c) in the complexes as compared to that of the free oxime derivatives appearing at 1010–1044 cm−1. All the oxime derivatives display a band in the region of 1590–1602 cm−1, which may be due to ν(C⚌N2) (pyrazoline ring). This band remains almost unchanged in all the metal chelates. This indicates that the ring nitrogen N2 does not take part in coordination, which is in agreement to our previous observation (Maurya et al., 2003b). The absence of a strong band at 1660–1685 cm−1 due to ν(C⚌O) and the appearance of a new band in the region of 1126–1227 cm−1 assignable to ν(C–O) (enolic), may be taken as diagnostic of coordination of cyclic oxygen to the metal center in the enol form (Kharodawala and Rana, 2003; Maurya et al., 2007) after deprotonation. The coordination of oxime nitrogen and cyclic oxygen is further supported by the appearance of two non-ligand bands at 508–517 and 430–497 cm−1 assignable to ν(M–O) and ν(M–N), respectively (Maurya et al., 2003a).
The IR spectra of all the complexes show a broad band centered at 3375–3437 cm−1. This suggests the presence of coordinated water molecules. A weak band at 1658–1724 cm−1 in the IR spectra of all the complexes can be ascribed to the inter-molecular hydrogen bonded O–H⋯O (between oxime OH group and coordinated water oxygen) bending vibration (Chakravorty, 1974). The important infrared spectral bands of the oxime derivatives and complexes in the present investigation are given in Tables 3 and 5, respectively. The IR spectra of the ligand, mppopH (II), and its complex [MoO2(mppopH)2(H2O)2] (2) are given in Figs. 7 and 8, respectively.
S. No.
Complex
νs(MoO2)
νas(MoO2)
ν(C–O) (Enolic)
ν(C⚌N) (oxime)
ν(C⚌N) (pyrazolone)
ν(OH)
δ(O–H⋯O)
ν(Mo–O)
ν(Mo–N)
ν(N–O)
1
[MoO2(aomppH)2(H2O)2]
950
910
1126
1578
1612
3401
1724
508
430
1091
2
[MoO2(mppopH)2(H2O)2]
941
913
1167
1578
1605
3437
1659
517
497
1079
3
[MoO2(buomppH)2(H2O)2]
941
913
1227
1567
1605
3430
1658
512
468
1079
4
[MoO2(ibuomppH)2(H2O)2]
991
912
1221
1571
1608
3432
1708
510
456
1091
5
[MoO2(bomppH)2(H2O)2]
951
912
1164
1569
1599
3375
1686
510
450
1073
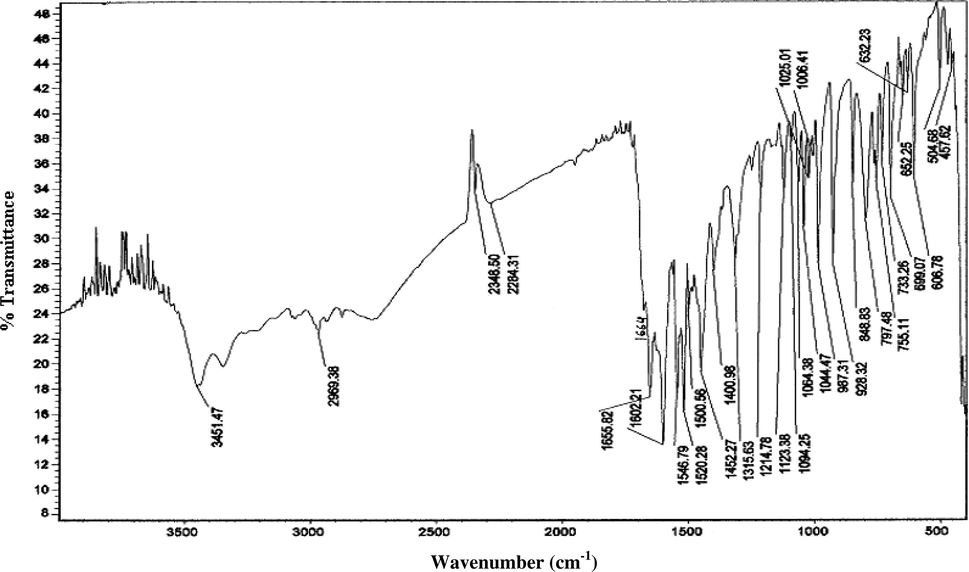
IR spectrum of mppopH (II).
![IR spectrum of [MoO2(mppopH)2(H2O)2] (2).](/content/184/2015/8/3/img/10.1016_j.arabjc.2011.01.010-fig10.png)
IR spectrum of [MoO2(mppopH)2(H2O)2] (2).
3.4 1H NMR spectral studies
The 1H NMR spectrum of a representative compound [MoO2(buomppH)2(H2O)2] (3) (Fig. 9) was recorded in DMSO-d6 to compare its result with the respective ligand. The absence of an enolic proton (cyclic) signal at δ 10.422 in the ligand was found to be absent in the spectrum of this complex. This shows the coordination of enolic oxygen after deprotonation to the metal center in this complex. This is in agreement with the IR spectral results related to the coordination of ligands under discussion. Other proton signals in the complex are δ 2.424 (singlet) –CH3 (a), δ 1.577–1.625 (triplet) –CH2 (b), δ 2.595–2.912 (multiplets) –CH2 (c), δ 0.916–0.969 (triplet) –CH3 (d) and δ 7.268–8.144 (multiples) (aromatic protons).![1HNMR spectrum of [MoO2(buomppH)2(H2O)2] (3).](/content/184/2015/8/3/img/10.1016_j.arabjc.2011.01.010-fig11.png)
1HNMR spectrum of [MoO2(buomppH)2(H2O)2] (3).
Two more proton signals were observed at δ 12.501 and δ 13.173 ppm in this complex, which are most probably due to protons of hydrogen bonded coordinated water molecules, and hydrogen bonded oxime proton (Zulfikaroglu et al., 2003), respectively.
3.5 Electronic spectral studies
Electronic spectra of all the complexes were recorded in 10−3 M in dimethylformamide solutions. The electronic spectral peaks observed in each of the complexes along with their molar extinction coefficient are given in Table 6. The high intensity spectral peak in UV-region in each of the complexes is due to intra-ligand transition. A medium to weak intensity peak near 443–444 nm in complexes (2) and (4) may be due to the ligand to metal charge transfer transitions (LMCT). The nonappearance of the LMCT band in the other three complexes is most probably due to its poor intensity. The electronic spectrum of [MoO2(ibuomppH)2(H2O)2] (4) is given in Fig. 10.
S. No.
Complex
λmax (nm)
ε (l mol−1 cm−1)
Peak assignments
1
[MoO2(aomppH)2(H2O)2]
287
3444
Intra-ligand transition
328
3703
337
3777
348
3851
2
[MoO2(mppopH)2(H2O)2]
282
3461
Intra-ligand transition
307
3500
332
3576
347
3884
355
3730
444
1269
LMCT
3
[MoO2(buomppH)2(H2O)2]
296
3513
Intra-ligand transition
335
4054
357
4432
4
[MoO2(ibuomppH)2(H2O)2]
269
1105
Intra-ligand transition
443
194
LMCT
5
[MoO2(bomppH)2(H2O)2]
293
3642
Intra-ligand transition
340
3607
352
3714
![Electronic spectrum of [MoO2(ibuomppH)2(H2O)2] (4).](/content/184/2015/8/3/img/10.1016_j.arabjc.2011.01.010-fig12.png)
Electronic spectrum of [MoO2(ibuomppH)2(H2O)2] (4).
3.6 Thermogravimetric studies
The thermogravimetric curves of two representative compounds [MoO2(mppopH)2(H2O)2] (2) (Fig. 11) [MoO2(buomppH)2(H2O)2] (3) (Fig. 12) were recorded in the temperature range of 25–1000 °C at the heating rate of 15 °C per minute. The compound (2) shows a weight loss of 5.3% at 300 °C with start of weight loss at 240 °C, corresponding to the elimination of two moles of coordinated water (calcd. weight loss for two mole H2O = 5.52%). It shows a second weight loss of 42.2% at 587 °C corresponding to the elimination of one coordinated oxime ligand group (calcd. weight loss of one oxime ligand group = 42.94%). The final weight loss (obs. = 80.0%) at 778.12 °C corresponds to the elimination of one mole oxime ligand group (calcd. weight loss = 80.37%). The final residue at ∼780 °C (obs. = 20.0%) corresponds to MoO3 (calcd. = 22.07%).![TG curve of [MoO2(mppopH)2(H2O)2] (2).](/content/184/2015/8/3/img/10.1016_j.arabjc.2011.01.010-fig13.png)
TG curve of [MoO2(mppopH)2(H2O)2] (2).
![TG curve of [MoO2(buomppH)2(H2O)2] (3).](/content/184/2015/8/3/img/10.1016_j.arabjc.2011.01.010-fig14.png)
TG curve of [MoO2(buomppH)2(H2O)2] (3).
Similar to compound (2), the compound (3) displayed the following three weight losses:
S. No.
% wt. loss (obs.)
% wt. loss (calcd.)
Elimination of
at (°C)
1
5.3
5.29
Two coordinated water molecules
300
2
43.5
43.23
One ligand group
520
3
80.0
81.16
One ligand group
725
The final residue at ∼780 °C (obs. = 21.0%) corresponds to MoO3 (calcd. = 21.03%). As the MoO3 is known to volatilize above 800 °C (melting point 795 °C (Greenwood and Earnshaw, 1984) and loose the weight, the TG plots will never be flat if these were reordered up to 1000 °C. Hence, both the TG plots appeared to be effectively recorded up to 800 °C, and so these are flat after ca. 750 °C.
3.7 Mass spectral studies
The FAB mass spectrum of a representative compound [MoO2(buomppH)2(H2O)] (3) (Fig. 13) is recorded on a JEOL SX 102/DA-6000 Mass Spectrometer/Data system using argon/xenon (6 KV, 10 mA) as the FAB gas. The accelerating voltage was 10 KV and the spectrum was recorded at room temperature. m-Nitrobenzyl alcohol (NBA) was used as the matrix. The matrix peaks were supposed to appear at m/z 136, 137, 154, 289, 307 in the absence of any metal ion. If metal ions are present, these peaks may be shifted accordingly.![Mass spectrum of [MoO2(buomppH)2(H2O)2] (3).](/content/184/2015/8/3/img/10.1016_j.arabjc.2011.01.010-fig15.png)
Mass spectrum of [MoO2(buomppH)2(H2O)2] (3).
The mass spectral peaks observed at 136 and 154 m/z are matrix peaks. The spectral peak observed at 257, 513, 370, 387, 600, and 542 m/z are most probably due to the following types of ion associations:
-
[buomppH]+ (258) − H+ = 257
-
[buomppH]+ (258) + (Mo = O)+ (111.94) = 370
-
[buomppH]+ (258) + [O⚌Mo⚌O]+ (127.94) + H+ = 387
-
2[buomppH]+ (516) − 3H+ = 513
-
[Base peak]+ (600) − ∗[C3H7]+ (43) − ∗[CH3]+ (15) = 542
-
[Molecular ion]+ (643.94) − ∗[C3H7]+ (43) = 600 (Base peak)
∗From butyryl oxime
These results are consistent with the proposed molecular composition of the complex (3).
3.8 Conductance measurements
The molar conductivities of all the complexes in 10−3 M DMF solution are in the range 10.1–15.2 ohm−1 cm2 mol−1 (Table 4) as expected for non-electrolytes (Geary, 1971). Such a non-zero molar conductance value for each of the complexes in the present study is most probably due to the strong donor capacity of DMF, which may lead to the displacement of the anionic ligand and change of electrolyte type.
3.9 3D Molecular modeling and analysis
Based on the proposed structures (Fig. 15), the 3D molecular modeling of one of the representative compounds, viz., [MoO2(aomppH)2(H2O)2] (1), was carried out with the CS Chem 3D Ultra Molecular Modeling and Analysis Programme. The details of bond lengths, bond angles as per the 3D structure (Fig. 14) are given in Tables 7 and 8, respectively. For convenience of looking over the different bond lengths and bond angles, the various atoms in the compound in question are numbered in Arabic numerals. In all, 243 measurements of the bond lengths (83 in number), plus the bond angles (160 in number) are listed in the Tables 7 and 8. Except a few cases, optimal values of both the bond lengths and the bond angles are given in the Tables along with the actual ones. The observed bond lengths/bond angles given in the Tables are calculated values as a result of energy optimization in CHEM 3D Ultra [44], while the optimal bond length/optimal bond angle values are the most desirable/favorable (standard) bond lengths/bond angles established by the builder unit of the CHEM 3D. The missing of some values of standard bond lengths/bond angles may be due to the limitations of the software, which we had already noticed in the modeling of other systems (Maurya and Rajput, 2006; Maurya et al., 2008, 2010). In most of the cases, the observed bond lengths and bond angles are close to the optimal values, and thus the proposed structure of compound (1) (and also others) is acceptable (Maurya and Rajput, 2006; Maurya et al., 2008, 2010).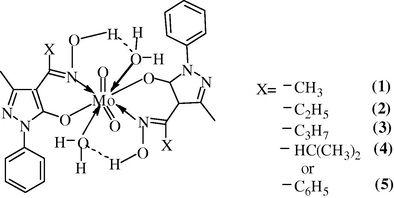
Proposed structure of complexes.
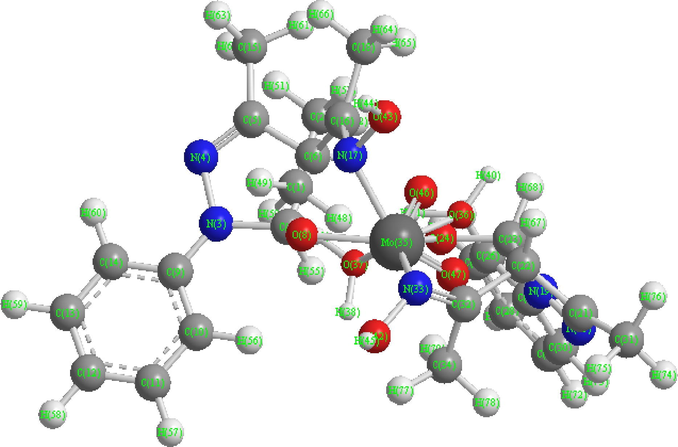
3D structure of compound (1).
S. No.
Atoms
Actual bond length
Optimal bond length
S. No.
Atoms
Actual bond length
Optimal bond length
1
C(2)–H(53)
1.113
1.113
43
C(22)–C(32)
1.497
1.497
2
C(2)–H(52)
1.113
1.113
44
C(22)–C(23)
1.523
1.514
3
C(2)–H(51)
1.113
1.113
45
C(21)–C(31)
1.497
1.497
4
C(1)–H(50)
1.113
1.113
45
C(21)–C(22)
1.497
1.497
5
C(1)–H(49)
1.113
1.113
47
N(20)–C(21)
1.4988
1.26
6
C(1)–H(48)
1.113
1.113
48
N(19)–C(25)
1.266
1.462
7
C(1)–C(2)
1.523
1.523
49
N(19)–C(23)
1.47
1.47
8
O(43)–H(44)
0.942
0.942
50
N(19)–N(20)
1.23
1.426
9
O(42)–H(45)
0.942
0.942
51
C(18)–H(66)
1.113
1.113
10
O(37)–H(39)
0.986
–
52
C(18)–H(65)
1.113
1.113
11
O(37)–H(38)
0.986
–
53
C(18)–H(64)
1.113
1.113
12
O(36)–H(41)
0.986
–
54
N(17)–Mo(35)
1.9741
–
13
O(36)–H(40)
0.986
–
55
N(17)–O(43)
1.316
–
13
Mo(35)–O(47)
1.5353
–
56
C(16)–C(18)
1.497
1.497
15
Mo(35)–O(46)
1.8428
–
57
C(16)–N(17)
1.377
1.377
16
O(36)–Mo(35)
1.8488
–
58
C(15)––H(63)
1.113
1.113
17
Mo(35)–O(37)
1.5202
–
59
C(15)–H(62)
1.113
1.113
18
C(34)–H(79)
1.113
1.113
60
C(15)–H(61)
1.113
1.113
19
C(34)–H(78)
1.113
1.113
61
C(14)–H(60)
1.1
1.1
20
C(34)–H(77)
1.113
1.113
62
C(13)–H(59)
1.1
1.1
21
N(33)–O(42)
1.316
–
63
C(13)–C(14)
1.337
1.42
22
N(33)–Mo(35)
1.976
–
64
C(12)–H(58)
1.1
1.1
23
C(32)–C(34)
1.497
1.497
65
C(12)–C(13)
1.337
1.42
24
C(32)–N(33)
1.6947
1.377
66
C(11)–H(57)
1.1
1.1
25
C(31)–H(76)
1.113
1.113
67
C(11)–C(12)
1.337
1.42
26
C(31)–H(75)
1.113
1.113
68
C(10)–H(56)
1.1
1.1
27
C(31)–H(74)
1.113
1.113
69
C(10)–C(11)
1.337
1.42
28
C(30)–H(73)
1.1
1.1
70
C(9)–C(14)
1.337
1.42
29
C(29)–H(72)
1.1
1.1
71
C(9)–C(10)
1.337
1.42
30
C(29)–C(30)
1.337
1.42
72
O(8)–Mo(35)
1.94
–
31
C(28)–H(71)
1.1
1.1
73
C(7)–H(55)
1.113
1.111
32
C(28)–C(29)
1.337
1.42
74
C(7)–O(8)
1.402
1.391
33
C(27)–H(70)
1.1
1.1
75
C(6)–H(54)
1.113
1.113
34
C(27)–C(28)
1.337
1.42
76
C(6)–C(16)
1.497
1.497
35
C(26)–H(69)
1.1
1.1
77
C(6)–C(7)
1.523
1.514
36
C(26)–C(27)
1.337
1.42
78
C(5)–C(15)
1.497
1.497
37
C(25)–C(30)
1.337
1.42
79
C(5)–C(6)
1.497
1.497
38
C(25)–C(26)
1.337
1.42
80
N(4)–C(5)
1.26
1.26
39
O(24)–Mo(35)
1.94
–
81
N(3)–C(9)
1.266
1.462
40
C(23)–H(68)
1.113
1.111
82
N(3)–C(7)
1.47
1.47
41
C(23)–O(24)
1.402
1.391
83
N(3)–N(4)
1.4219
1.426
42
C(22)–H(67)
1.113
1.113
S. No.
Atoms
Actual bond angles
Optimal bond angles
S. No.
Atoms
Actual bond angles
Optimal bond angles
1
H(53)–C(2)–H(52)
109.5199
109
81
O(47)–Mo(35)–O(24)
36.6947
–
2
H(53)–C(2)–H(51)
109.4619
109
82
O(47)–Mo(35)–N(17)
142.7106
–
3
H(53)–C(2)–C(1)
109.4615
110
83
O(47)–Mo(35)–O(8)
133.1707
–
4
H(52)–C(2)–H(51)
109.4419
109
84
O(46)–Mo(35)–O(37)
70.4451
–
5
H(52)–C(2)–C(1)
109.4416
110
85
O(46)–Mo(35)–O(36)
29.6984
–
6
H(51)–C(2)–C(1)
109.5005
110
86
O(46)–Mo(35)–N(33)
153.3882
–
7
H(50)–C(1)–H(49)
109.5198
109
87
O(46)–Mo(35)–O(24)
30.6141
–
8
H(50)–C(1)–H(48)
109.4619
109
88
O(46)–Mo(35)–N(17)
79.0453
–
9
H(50)–C(1)–C(2)
109.4617
110
89
O(46)–Mo(35)–O(8)
96.5432
–
10
H(49)–C(1)–H(48)
109.442
109
90
O(37)–Mo(35)–O(36)
87.443
–
11
H(49)–C(1)–C(2)
109.4416
110
91
O(37)–Mo(35)–N(33)
131.1412
–
12
H(48)–C(1)–C(2)
109.5003
110
92
O(37)–Mo(35)–O(24)
65.2425
–
13
H(72)–C(29)–C(30)
119.9998
120
93
O(37)–Mo(35)–N(17)
95.5127
–
13
H(72)–C(29)–C(28)
119.9997
120
94
O(37)–Mo(35)–O(8)
50.2039
–
15
C(30)–C(29)–C(28)
120.0004
–
95
O(36)–Mo(35)–N(33)
125.2943
–
16
H(71)–C(28)–C(29)
120.0002
120
96
O(36)–Mo(35)–O(24)
23.4196
–
17
H(71)–C(28)–C(27)
120.0002
120
97
O(36)–Mo(35)–N(17)
101.4075
–
18
C(29)–C(28)–C(27)
119.9996
–
98
O(36)–Mo(35)–O(8)
125.1198
–
19
H(70)–C(27)–C(28)
119.9996
120
99
N(33)–Mo(35)–O(24)
134.9523
–
20
H(70)–C(27)–C(26)
120.0003
120
100
N(33)–Mo(35)–N(17)
109.4999
–
21
C(28)–C(27)–C(26)
120.0001
–
101
N(33)–Mo(35)–O(8)
109.4999
–
22
H(73)–C(30)–C(29)
120
120
102
O(24)–Mo(35)–N(17)
109.5001
–
23
H(73)–C(30)–C(25)
120.0005
120
103
O(24)–Mo(35)–O(8)
109.4999
–
24
C(29)–C(30)–C(25)
119.9995
–
104
N(17)–Mo(35)–O(8)
58.7561
–
25
H(69)–C(26)–C(27)
120.0001
120
105
H(66)–C(18)–H(65)
109.5196
109
26
H(69)–C(26)–C(25)
119.9997
120
106
H(66)–C(18)–H(64)
109.4624
109
27
C(27)–C(26)–C(25)
120.0003
–
107
H(66)–C(18)–C(16)
109.4617
110
28
C(30)–C(25)–C(26)
120.0001
120
108
H(65)–C(18)–H(64)
109.4421
109
29
C(30)–C(25)–N(19)
119.9998
120
109
H(65)–C(18)–C(16)
109.4418
110
30
C(26)–C(25)–N(19)
120.0001
120
110
H(64)–C(18)–C(16)
109.4998
110
31
H(76)–C(31)–H(75)
109.5202
109
111
O(43)–N(17)–Mo(35)
109.5273
–
32
H(76)–C(31)–H(74)
109.4624
109
112
O(43)–N(17)–C(16)
109.5273
–
33
H(76)–C(31)–C(21)
109.4617
110
113
Mo(35)–N(17)–C(16)
109.1575
–
34
H(75)–C(31)–H(74)
109.4417
109
114
H(59)–C(13)–C(14)
120.0003
120
35
H(75)–C(31)–C(21)
109.4415
110
115
H(59)–C(13)–C(12)
119.9996
120
36
H(74)–C(31)–C(21)
109.4999
110
116
C(14)–C(13)–C(12)
120
–
37
C(21)–N(20)–N(19)
114.4919
115
117
H(58)–C(12)–C(13)
120.0002
120
38
C(25)–N(19)–C(23)
124.5
108
118
H(58)–C(12)–C(11)
120.0003
120
39
C(25)–N(19)–N(20)
124.4998
124
119
C(13)–C(12)–C(11)
119.9995
40
C(23)–N(19)–N(20)
111.0002
–
120
H(57)–C(11)–C(12)
119.9995
120
41
H(68)–C(23)–O(24)
108.4689
106.7
121
H(57)–C(11)–C(10)
120
120
42
H(68)–C(23)–C(22)
114.5222
109.39
122
C(12)–C(11)–C(10)
120.0005
–
43
H(68)–C(23)–N(19)
111.7232
107.5
123
H(60)–C(14)–C(13)
119.9999
120
44
O(24)–C(23)–C(22)
107.5002
107.7
124
H(60)–C(14)–C(9)
119.9996
120
45
O(24)–C(23)–N(19)
110.5003
–
125
C(13)–C(14)–C(9)
120.0005
–
46
C(22)–C(23)–N(19)
104
–
126
H(56)–C(10)–C(11)
120.0003
120
47
C(31)–C(21)–C(22)
128.9413
117.2
127
H(56)–C(10)–C(9)
119.9997
120
48
C(31)–C(21)–N(20)
128.941
115.1
128
C(11)–C(10)–C(9)
120
–
49
C(22)–C(21)–N(20)
102.1176
115.1
129
C(18)–C(16)–N(17)
120.0001
120
50
H(79)–C(34)–H(78)
109.5202
109
130
C(18)–C(16)–C(6)
120.0002
117.2
51
H(79)–C(34)–H(77)
109.4617
109
131
N(17)–C(16)–C(6)
119.9997
120
52
H(79)–C(34)–C(32)
109.4617
110
132
Mo(35)–O(8)–C(7)
109.4999
–
53
H(78)–C(34)–H(77)
109.442
109
133
H(55)–C(7)–O(8)
108.4688
106.7
54
H(78)–C(34)–C(32)
109.4417
110
134
H(55)–C(7)–C(6)
114.5224
109.39
55
H(77)–C(34)–C(32)
109.5
110
135
H(55)–C(7)–N(3)
111.7237
107.5
56
H(67)–C(22)–C(32)
107.8496
109.39
136
O(8)–C(7)–C(6)
107.5
107.7
57
H(67)–C(22)–C(23)
112.9886
109.39
137
O(8)–C(7)–N(3)
110.4999
–
58
H(67)–C(22)–C(21)
112.9885
109.39
137
C(6)–C(7)–N(3)
103.9999
–
59
C(32)–C(22)–C(23)
109.47
109.51
139
C(14)–C(9)–C(10)
119.9995
120
60
C(32)–C(22)–C(21)
109.4699
109.51
140
C(14)–C(9)–N(3)
120.0006
120
61
C(23)–C(22)–C(21)
104.0001
109.51
141
C(10)–C(9)–N(3)
120
120
62
H(45)–O(42)–N(33)
120.0002
142
H(63)–C(15)–H(62)
109.5198
109
63
C(34)–C(32)–N(33)
115.4565
120
143
H(63)–C(15)–H(61)
109.4621
109
64
C(34)–C(32)–C(22)
115.4567
117.2
144
H(63)–C(15)–C(5)
109.4618
110
65
N(33)–C(32)–C(22)
129.0868
120
145
H(62)–C(15)–H(61)
109.4423
109
66
H(39)–O(37)–H(38)
120.0001
–
146
H(62)–C(15)–C(5)
109.4415
110
67
H(39)–O(37)–Mo(35)
121.5008
–
147
H(61)–C(15)–C(5)
109.4999
110
68
H(38)–O(37)–Mo(35)
95.5876
–
148
H(54)–C(6)–C(16)
107.8498
109.39
69
H(41)–O(36)–H(40)
120.0004
–
149
H(54)–C(6)–C(7)
112.9885
109.39
70
H(41)–O(36)–Mo(35)
55.2093
–
150
H(54)–C(6)–C(5)
112.9885
109.39
71
H(40)–O(36)–Mo(35)
139.3508
–
151
C(16)–C(6)–C(7)
109.4701
109.51
72
O(42)–N(33)–Mo(35)
120.0284
–
152
C(16)–C(6)–C(5)
109.4698
109.51
73
O(42)–N(33)–C(32)
120.0282
–
153
C(7)–C(6)–C(5)
104.0001
109.51
74
Mo(35)–N(33)–C(32)
59.6129
–
154
C(9)–N(3)–C(7)
126.1086
108
75
Mo(35)–O(24)–C(23)
109.5
–
155
C(9)–N(3)–N(4)
126.1079
124
76
H(44)–O(43)–N(17)
120.0002
–
156
C(7)–N(3)–N(4)
107.7834
–
77
O(47)–Mo(35)–O(46)
65.3054
–
157
C(15)–C(5)–C(6)
124.4999
117.2
78
O(47)–Mo(35)–O(37)
83.0583
–
158
C(15)–C(5)–N(4)
124.5001
115.1
79
O(47)–Mo(35)–O(36)
41.3612
–
159
C(6)–C(5)–N(4)
111
115.1
80
O(47)–Mo(35)–N(33)
98.7039
–
160
C(5)–N(4)–N(3)
113.2158
115
4 Conclusions
The satisfactory analytical data and all the physico-chemical studies presented above suggest that the complexes under this investigation may be formulated as [MoO2(L)2(H2O)2], where LH = 4-acetyloxime-3-methyl-1-phenyl-2-pyrazoline-5-one (aomppH), 3-methyl-1-phenyl-4-propionyloxime-2-pyrazoline-5-one (mppopH), 4-butyryloxime-3-methyl-1-phenyl-2-pyrazoline-5-one (buomppH), 4-isobutyryloxime-3-methyl-1-phenyl-2-pyrazoline-5-one (ibuomppH), 4-benzoyloxime-3-methyl-1-phenyl-2-pyrazoline-5-one (bomppH). Considering the octa-coordination (Maurya et al., 1995b), tentative structures proposed for these complexes are shown in the Fig. 15.
Acknowledgments
The authors are thankful to Professor R.R. Mishra, Vice-Chancellor, Rani Durgavati University, Jabalpur, MP, India, for the encouragement. Analytical facilities provided by the Central Drug Research Institute, Lucknow, India, and the Sophisticated Instrumentation Centre, Indian Institute of Technology, Roorkee and Mumbai are gratefully acknowledged.
References
- Organometallics. 2003;22:2112.
- J. Am. Chem. Soc.. 2000;21:5189.
- Analytical Chemistry. Amsterdam: Elsevier; 1963.
- Prog. Inorg. Chem.. 1973;18:177.
- Coord. Chem. Rev.. 1974;13:1.
- Inorg. Chem.. 1976;15:2612.
- Chem. Soc. Rev. 1996:25.
- Diehl, H. 1940. The Application of the Dioxime to Analytical Chemistry (G. Fredrick Smith Chemical Co., Columbus, Ohio). Van Nostrand, New York.
- Biochemistry. 1995;34:6218.
- Chem. Rev.. 2004;104:1175.
- Biochem. J.. 1992;282:659.
- Inorg. Chem. Commun.. 2002;5:384.
- Coord. Chem. Rev.. 1971;7:81.
- Synth. React. Inorg. Met.-Org. Chem.. 1981;11:611.
- Biochem. Pharmacol. 1992;44:177.
- Chemistry of the Elements. Pergamon Press; 1984. p. 1173
- Synth. React. Inorg. Met.-Org. Chem.. 1982;12:889.
- Chem. Rev.. 1996;96:2757.
- Synth. React. Inorg. Met.-Org. Chem.. 1983;13:781.
- Acta Chem. Scand.. 1959;13:1670.
- Synth. React. Inorg. Met.-Org. Chem.. 1998;28:383.
- Synth. React. Inorg. Met.-Org. Chem.. 1991;21:1031.
- Synth. React. Inorg. Met.-Org. Chem.. 1991;21:1031.
- Synth. React. Inorg. Met.-Org. Chem.. 1991;21:1031.
- Synth. React. Inorg. Met.-Org. Chem.. 2003;33:1483.
- Synth. React. Inorg. Met.-Org. Chem.. 1985;14:89.
- Coord. Chem. Rev.. 1999;181:147.
- J. Mol. Cat. A: Chem.. 2010;322:55.
- Pol. J. Chem.. 1986;60:731-740.
- J. Mol. Struct.. 2006;794:24.
- Polyhedron. 1993;12:2045.
- Synth. React. Inorg. Met.-Org. Chem.. 1995;25:437.
- Bull. Chem. Soc., Jpn.. 1995;68:1589.
- Synth. React. Inorg. Met.-Org. Chem.. 2003;33:817.
- Synth. React. Inorg. Met.-Org. Chem.. 2003;33:699.
- Synth. React. Inorg. Met.-Org. Chem.. 2003;33:309.
- Synth. React. Inorg. Met.-Org. Chem.. 2003;33:435.
- Maurya, R.C., Chourasia, J., Martin, M.H., Jhamb, S. 2007. Proceed. Nuclear and Radiochemistry Symposium (NUCAR-2007), Vadodara, India, CA-2, p. 187.
- Indian J. Chem.. 2008;47A:517.
- Int. J. Curr. Chem.. 2010;1:309.
- Inorg. Chim. Acta. 1975;13:91.
- Synth. React. Inorg. Met.-Org. Chem.. 1995;25:1571.
- Polyhedron. 1990;9:1011.
- Synth. React. Inorg. Met.-Org. Chem.. 1988;18:1559.
- Ozcelikay, A.T., Baker, D.J., Ingemba, L.N., Pottier, A.M., Henquin, J.C., Brichard, S.M. 1996. Am. J. Physiol., 270 (Endocrinol, Metab. 33), E 344.
- Pande, K.C., Patent, U.S., 1966. 3, 352, 672; Chem. Abstr. 1967, 66, 28891C.
- Indian J. Chem.. 1985;24:800.
- Macromol. Rep.. 1994;31:651.
- Macromol. Rep.. 1995;32:1161.
- Synth. React. Inorg. Met.-Org. Chem.. 1995;15:859.
- Indian J. Chem. Sec. A.. 2000;39:557.
- Angew. Chem., Int. Ed.. 1976;15:417.
- J. Mol. Catal. A: Chem.. 2004;213:21.
- Synth. React. Inorg. Met.-Org. Chem.. 1992;22:851.
- J. Indian Chem. Soc.. 1981;58:851.
- Indian J. Chem.. 1982;21A:176.
- Indian J. Chem.. 1982;21A:312.
- Shuter, E. 1995. M. Sc. Thesis University of British Columbia.
- J. Organomet. Chem. Rev.. 1974;64:145.
- Prog. Inorg. Chem.. 1979;22:1.
- Coughltan M.P., ed. Molybdenum and Molybdenum-Containing Enzymes. Oxford: Pergamen Press; 1979. p. :43.
- Coord. Chem. Rev.. 1989;95:183.
- Z. Anorg. Allg. Chem.. 1905;46:144;.
- Ber.. 1908;41:2219.
- Synth. React. Inorg. Met.-Org. Chem.. 1991;21:1083.
- Synth. React. Inorg. Met.-Org. Chem.. 1990;20:437.
- Synth. React. Inorg. Met.-Org. Chem.. 2003;33:1253.
- Synth. React. Inorg. Met.-Org. Chem.. 2003;33:625.







