Amomum tsao-ko essential oil, a novel anti-COVID-19 Omicron spike protein natural products: A computational study
⁎Corresponding author. bluesmar@163.com (Qi Cui) zcmucq@163.com (Qi Cui) zcmucq@163.com (Qi Cui)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Since the outbreak of COVID-19, this virus has been constantly mutating. The latest mutant Omicron has been identified as VOC by WHO. The main reason for its concern is the mutation of 46 amino acids in spike protein, which has brought the global epidemic prevention into another difficulty. Herbal aromatic plant Amomum tsao-ko was excavated from formula 1 and 2 for the treatment of COVID-19 in China, and its active components were extracted and identified. Molecular dynamics simulation and Fpocket were applied to find the key sites on RBDOmicron, and molecular docking was also used to reveal the interaction between A. tsao-ko essential oil (AEO) and RBDOmicron. The AEO components were analyzed and identified by GC/Q-TOF MS. There were 20 kinds of AEO with a relative area percentage of more than 1%, and they were related to the three active centres of RBDOmicron. In this study, virtual screening was used to mine the essential oil components of medicinal plants, and it was found that the components could interact with the spike protein RBD in aerosol to block the interaction of RBD and hACE2, thus cutting off the transmission route and protecting the host. This study has certain guiding significance in the modernization of Traditional Chinese medicine and the prevention of COVID-19.
Keywords
Amomum tsao-ko essential oil
SARS-CoV-2
Omicron
RBD
Molecular dynamics
Molecular docking
1 Introduction
Amomum tsao-ko is a perennial herbaceous plant of Zingiberaceae, which is a kind of unique Chinese medicinal and edible plant that has been used for thousands of years in China (Cui et al., 2017; Guo et al., 2017). In a hot pot, a typical Chinese dish, A. tsao-ko is an indispensable condiment (Jia et al., 2020). It is recorded in the compendium of Materia Medica that A. tsao-ko tastes spicy, warm and belongs to the spleen and stomach meridian (Li et al., 2021; Shi et al., 2021; Wei et al., 2021). A large number of previous studies have shown that A. tsao-ko has multiple pharmacological effects such as sedative, antibacterial and immunity enhancement, and its active substance is mainly volatile essential oil (Qin et al., 2021; Liu et al., 2021; Sabulal and Baby, 2021). In the early stage of the SARS-CoV-2 outbreak in 2019, both formula 1 proposed by Academician Xiaolin Tong and formula 2 proposed by Academician Qi Wang contained A. tsao-ko (Ai et al., 2020). Traditional Chinese Medicine (TCM), the treasure of Chinese civilization, is a complex system, which usually acts synergistically (Khanna et al., 2021; Zhao et al., 2021). It is more suitable to study its mechanism of action by using systematic and holistic research ideas and extracting the complex system from the system for the development of anti-epidemic drugs. Based on the summary of previous studies and the excavation of essential oil components of A. tsao-ko, it can be used as a low-cost drug for the development of COVID-19 prevention (Zhu et al., 2021; Lee et al., 2021).
The COVID-19 pandemic has undergone four major mutations since the outbreak, and the World Health Organization (WHO) began naming in June 2021 after Greek letters: Alpha, Beta, Delta, and the new mutation Omicron that has just been discovered in South Africa (Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern) (https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern; https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/). SARS-CoV-2 is an RNA coronavirus with a simple structure and is prone to mutation. It is mainly composed of four structural proteins, namely spike protein, envelope protein, membrane protein, nucleocapsid protein and sixteen non-structural proteins. The spike protein consists of S1 and S2, of which S1 is composed of N terminal (NTD, 1–217) and receptor-binding domains (RBD, 319–541) and acts directly on hACE2, which is the first step for the SARS-CoV-2 to contact with the host. Blocking the binding of the spike protein and hACE2 can protect the host to a great extent. The RBD part of the Omicron mutant has 15 key amino acid mutations, which is the mutant with the most mutations found at present (Fig. 1). It has been reported that Omicron mutations can enhance immune escape, binding ability with receptors, virulence and so on. Therefore, it is of extreme significance to explore a substance that is easier to combine with spike protein RBD in the air, to play a role in epidemic prevention (Lan et al., 2020; Wang et al., 2021a).
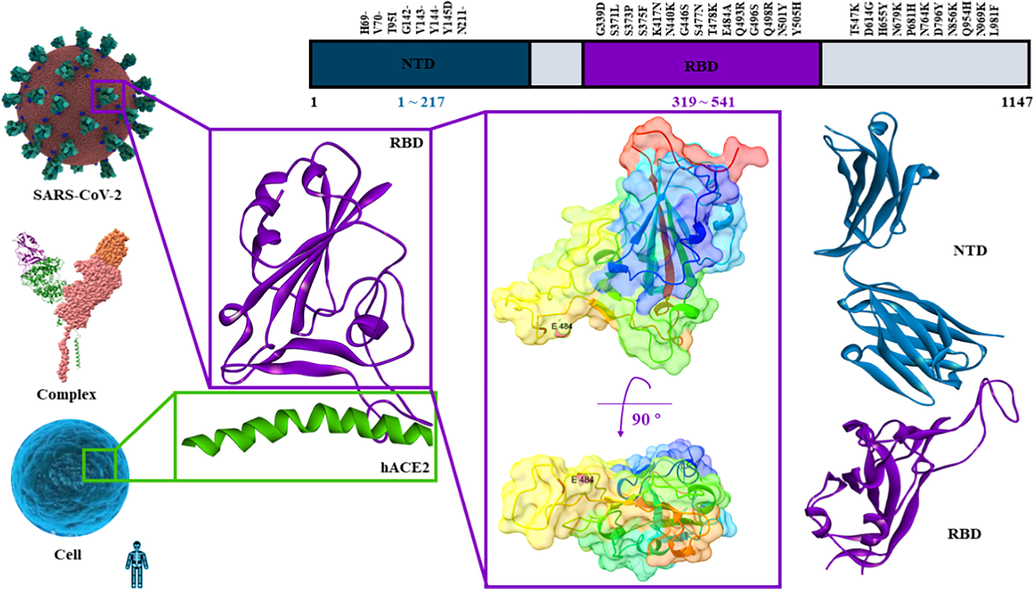
- The structure of spike protein.
In this study, GC/Q-TOF MS was applied to identify and analyze the A. tsao-ko essential oil extracted by hydrodistillation. Meanwhile, homology modeling and molecular dynamics simulation were used to construct and search for key amino acid sites of Omicron’s spike protein RBD. The Omicron RBD was docked with the components with a relative area percentage of more than 1% in A. tsao-ko essential oil by molecular docking to demonstrate the relationship between them. The interaction between RBD and hACE2 was inhibited to some extent through the interaction between essential oil compositions and RBD. Since ancient times, spices have been used for epidemic prevention, not to eliminate viruses, but to inhibit the ability of viruses to bind to receptors floating in the atmosphere in poorly ventilated spaces. The purpose of this study is not to develop a drug to cure COVID-19, but to excavate and discover a new substance from natural medicinal plant essential oil that can reduce the binding ability of SARS-CoV-2 to host for COVID-19 epidemic prevention.
2 Materials and methods
2.1 Plant materials
Amomum tsao-ko were purchased from Yunnan Province, China and authenticated by Prof. Chunchun Zhang from Zhejiang Chinese Medical University. The dried samples were powdered (0.30 mm) and deposited at 4 °C until use. Ultrapure water was manufactured by Milli-Q IQ 7000 purification system (Merck, Germany). Chromato-graphic grade n-hexane was purchased from Merck (Merck, Germany).
2.2 Extraction procedure
Hydrodistillation was applied for the extraction of A. tsao-ko essential oil (AEO). with a traditional Clevenger apparatus. An aliquot of 200 g material was introduced into the distillation reaction flask with 2.4 L of water soaking for 30 min before extraction and then extracted for 5 h until the essential oil volume was kept constant. The essential oil was collected in the receiving flask with a circulating water-cooling system and transferred to an amber-colored vial, dehydrated with anhydrous sodium sulphate. Finally, the essential oil was kept at 4 °C with N2 until being used. The experiment was performed in triplicate.
2.3 GC/Q-TOF MS analysis of essential oil
GC/Q-TOF MS analysis was performed on an Agilent quadrupole time of flight mass spectrometer (Agilent Technology Co., LTD, USA) consisting of an Agilent 7890B gas chromatography and an Agilent 7250 time of flight mass spectrometer, using an HP-5MS capillary column (30 m × 250 μm × 0.25 μm film thickness). The operational conditions of GC/Q-TOF MS analysis were preset as follows: helium was used as the carrier gas at 1.0 mL/min flow rate; split ratio 1:20; injection volume 1 μL; injection temperature 230 °C; ion source temperature 230 °C; interface temperature 280 °C; oven temperature progress: 60 °C (holding for 2 min), 60–120 °C (6 °C/min, holding for 2 min), 120–180 °C (5 °C/min), 180–280 °C (30 °C/min); ionization mode used at electronic impact 70 eV; mass range 29–500 m/z; TOF-scan mode at 5 spectra/s; solvent delay 4 min. The mass spectra of essential oil compositions in the NIST14 mass spectral library and the fragmentation patterns of the mass spectra with the data published in the literature were applied for the identification of essential oil compositions.
2.4 Homology modeling of the spike protein
Homology modeling is a highly-accurate method for the prediction and construction of the 3D structure of a protein-based on another protein with a known 3D structure (template). Using the SWISS-MODEL interactive server (Waterhouse et al., 2018), the 3D structures of Omicron mutant spike protein RBD and NTD domains were constructed based on the structures of wild-type SARS-CoV-2 spike protein RBD (PDB ID: 7EKG; 2.63 Å resolution) and NTD (PDB ID: 7BZ5; 1.84 Å resolution) with the sequence identity of 93.30% and 96.31% retrieved from PDB database (https://www.rcsb.org), respectively. The Ramachandran plot and Molprobity scores showed that the built structures were of good quality and suitable for further studies.
2.5 Fpocket analysis
The built structure of RBDOmicron was analyzed by the Fpocket webserver to predict possible active centres (AC) on the protein surfaces. Fpocket is an open-source pocket detection package based on Voronoi tessellation and alpha-spheres (Schmidtke et al., 2010). The PDB file of the RBDOmicron was submitted to Fpocket, and the outputs were analyzed using UCSF Chimera software. Binding sites were the distribution of surrounding residues in AC and acted as the active residues.
2.6 Molecular dynamics simulation
Molecular dynamics (MD) simulations were performed using GROMACS 2018.8 on Intel(R) Core (TM) i7-10870H CPU@ 2.20 GHz, using the OPLS-AA/L all-atom force field to treat the protein (Abraham et al., 2015). RBD was then immersed in a pre-equilibrated octahedral box of TIP3P water and the system was neutralized. The system was then minimized using the energy gradient convergence criterion set to 0.01 kcal/mol Å2 in four steps involving: (i) an initial 5,000 minimization steps (2,500 with the steepest descent and 2,500 with the conjugate gradient) of only hydrogen atoms, (ii) 20,000 minimization steps (10,000 with the steepest descent and 10,000 with the conjugate gradient) of water and hydrogen atoms, keeping the solute restrained, (iii) 50,000 minimization steps (25,000 with the steepest descent and 25,000 with the conjugate gradient) of protein side chains, water and hydrogen atoms, (iv) 100,000 (50,000 with the steepest descent and 50,000 with the conjugate gradient) of complete minimization. Successively, the water, ions and protein side chains were thermally equilibrated in three steps: (i) 5 ns of NVT equilibration with the Langevin thermostat by gradually heating from 0 K to 300 K, while gradually rescaling solute restraints from a force constant of 10 to 1 kcal/mol Å2, (ii) 5 ns of NPT equilibration at 1 atm with the Berendsen thermostat, gradually rescaling restraints from 1.0 to 0.1 kcal/mol Å2, (iii) 5 ns of NPT equilibration with no restraints. Finally, a production run of 200 ns was performed using a timestep of 2 fs. The SHAKE algorithm was used for those bonds containing hydrogen atoms in conjunction with periodic boundary conditions at constant pressure and temperature, particle mesh Ewald for the treatment of long-range electrostatic interactions, and a cutoff of 10 Å for nonbonded interactions.
2.7 Molecular docking
The computational molecular docking of hACE2 (PDB ID: 6 M17.B, receptor) with Omicron mutant spike protein RBD (ligand) was explored by an information-driven flexible docking approach using HADDOCK and Autodock Vina software. Before docking, the ligand and receptor are dehydrated, hydrogenated, and charged. In docking, the receptor was considered semi-rigid while the ligand was flexible, and the grid matrix of the docking area was sized to ensure complete coverage of the protein molecule. The size of the docking range was set to (40,60,60), and the docking centre was (−32,27,17). The best Autodock Vina score with the suitable energetically favored conformations was used for further analysis. The interaction of AEO compositions with amino acid residues of omicron mutant spike protein RBD was analyzed and represented using ChimeraX.
3 Results and discussion
3.1 Chemical compositions of the essential oil
The chemical compositions of AEO were determined and listed in Fig. 2 and Table 1. And twenty components with a relative area percentage of more than 1% were identified in AEO representing 92.82% of the total AEO constituents. The total percentage of monoterpenes was the most abundant compositions in AEO (77.73%), including monoterpene hydrocarbons (14.57%) of α-Phellandrene, β-Pinene, α-Pinene, 3-Carene and oxygenated monoterpenes (63.16%) of 1,8-Cineole, trans-Citral, trans-2-Decenal, α-Terpineol, cis-Citral, trans-Geraniol, Geraniol acetate, (-)-4-Terpineol, Linalool, (-)-cis-Sabinol. The oxygenated monoterpene compounds possessed higher dipolar moments in essential oils and interacted more vigorously with heating, and they can be extracted more easily than monoterpene hydrocarbons.
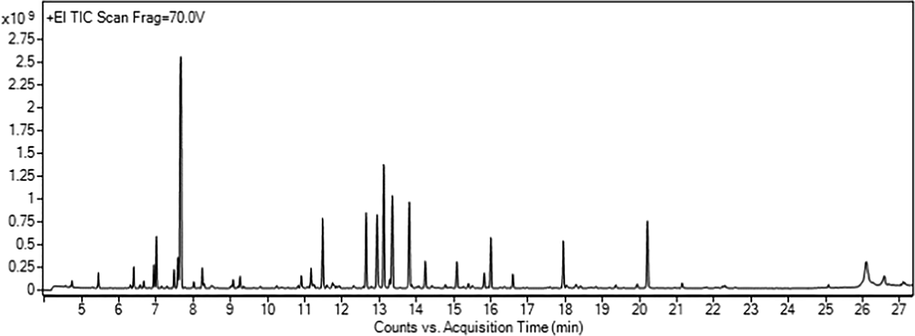
- Total ion flow diagram of AEO.
| No. | Compound | CAS number | Formula | Relative area percentage (%) |
|---|---|---|---|---|
| 1 | Linalool | 78-70-6 | C10H18O | 1.24 |
| 2 | α-Terpineol | 10482-56-1 | C10H18O | 4.02 |
| 3 | α-Phellandrene | 99-3-2 | C10H16 | 7.56 |
| 4 | trans-Geraniol | 106-24-1 | C10H18O | 3.29 |
| 5 | β-Pinene | 2437-95-8 | C10H16 | 3.72 |
| 6 | cis-Nerolidol | 106-25-2 | C15H26O | 2.72 |
| 7 | trans-Citral | 141-27-5 | C10H16O | 4.76 |
| 8 | 1,8-Cineole | 470-82-6 | C10H18O | 36.00 |
| 9 | (−)-4-Terpineol | 562-74-3 | C10H18O | 2.11 |
| 10 | (−)-cis-Sabinol | 3310-02-9 | C10H16O | 1.11 |
| 11 | trans-2-Decenal | 39313-81-3 | C10H18O | 4.45 |
| 12 | cis-Citral | 5392-40-5 | C10H16O | 3.79 |
| 13 | 2-Isopropylbenzaldehyde | 6502-22-3 | C10H12O | 1.91 |
| 14 | trans-Nerolidol | 7212-44-4 | C15H26O | 1.60 |
| 15 | α-Pinene | 80-56-8 | C10H16 | 1.69 |
| 16 | 3-Carene | 13466-78-9 | C10H16 | 1.60 |
| 17 | Geraniol acetate | 105-87-3 | C10H20O2 | 2.39 |
| 18 | 2-Dodecenal | 20407-84-5 | C10H22O | 1.34 |
| 19 | 4-Propylbenzaldehyde | 28785-06-0 | C10H12O | 5.65 |
| 20 | Indane-4-carboxaldehyde | 51932-70-8 | C10H10O | 1.87 |
Due to the rich essential oil components, Chinese herbal aromatic plants not only have a special aroma, but also have the effect of medical care that are widely used in many industries. Essential oil can cause aroma movement and regulate qi2, and have the functions of regulating emotions, coordinating human physiological functions, calming and improving human immunity. Chinese herbal aromatic essential oil can improve the therapeutic outcome of metabolic syndrome by interfering with insulin signal transduction, resisting oxidative stress, reducing the inflammatory response and increasing insulin sensitivity. These results suggest that Chinese herbal aromatic essential oil has the advantages of high efficiency, low toxicity, multi-target and multi-level therapy, and has broad research prospects in the future (Tang et al., 2021). As the predominant component in AEO, 1,8-cineole is an oxygenated monoterpene with a fragrance, camphoraceous features and a spicy flavor. Therapeutic applications of 1,8-cineole for the treatment of respiratory diseases, muscle pain, neurosis, rheumatism and kidney stones have been investigated as well (Cai et al., 2021). As an important part of natural essential oil, traditional Chinese medicine essential oil has a wide variety of sources and has high medicinal, industrial and edible values.
3.2 Mapping of RBDOmicron mutations compared with RBDWT
Compared with RBDWT, RBDOmicron had fifteen additional mutation sites including G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H (Fig. 3C), respectively. Of the fifteen Omicron changes in RBD, nine map to hACE2 binding footprint: K417N, G446S, S477N, E484A, Q493R, G496S, Q498R, N501Y, Y505H, which considerably affects receptor recognition, with N440K and T478K just peripheral (Fig. S1, Dejnirattisai et al., 2022). Additional, mutations occur on G339D, S371L, S373P and S375F, the last three of which are adjacent to a lipid-binding pocket (Toelzer et al., 2020). This pocket has been seen occupied by a lipid similar to linoleic acid in an unusually rigid state of spike protein, where all RBDs are found in a locked-down configuration stabilized by lipid-bridged quaternary interactions between adjacent RBDs, and several were previously shown to modulate ACE2 binding and/or antibody evasion (Harvey et al., 2021). Thus, understanding the underlying molecular basis for the enhanced transmissibility, immune resistance, and virological properties of the Omicron variant may facilitate halting this looming crisis.
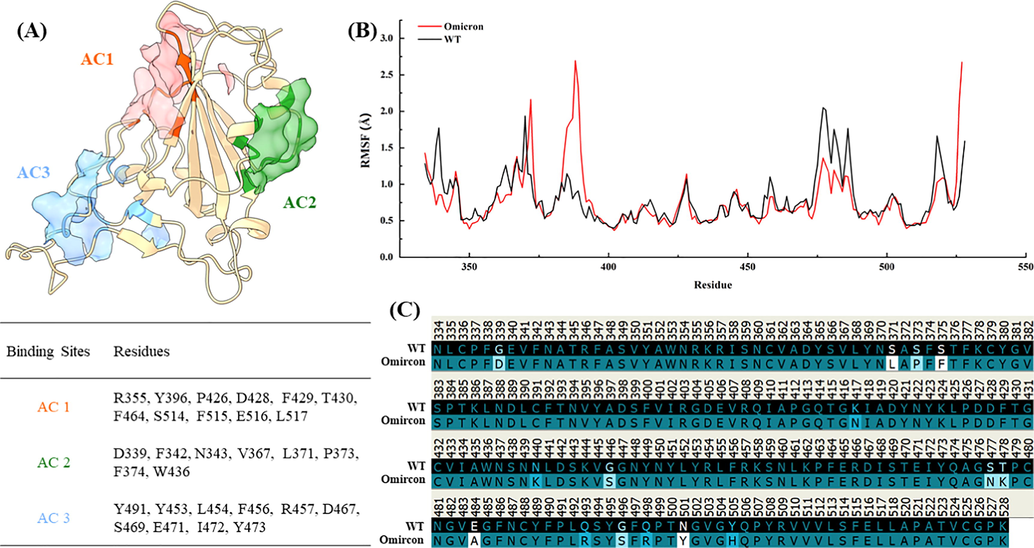
- Active centres discovered on RBD using molecular dynamics simulation: (A) The active centre on RBD, (B) RMSF values of RBD on wild-type (WT) and Omicron mutant, (C) amino acid sequence of RBD on wild-type (WT) and Omicron.
As the Omicron variant showed a considerable number of positively charged amino acids at sites of mutation, the electrostatic potential at the RBD-ACE2 interface and isoelectric point (pI) change of mutation sites on RBD were analyzed. It can be found that the increased pronouncedly electro positivity in the Omicron variant (T478K, Q493R, Q498R and Y505H), and also lost a positively charged amino acid at the interaction interface of K417N (Fig. S2). Combined with existing studies, the largest difference between RBDWT and RBDOmicron was found at positions 417 and 493 (Socher et al., 2021). For the Omicron variant that exchanged K417 to N417, a strongly reduced interaction with D30, while Q493 mutated to R493 reduced the interaction with K31, which may be due to electrostatic repulsion between the positively charged R and K. The pI of protein is the pH value at which its surface is completely charged but its net charge is zero. Also, the amino acid pI of a protein can determine the local electric charge of the protein, which is an important basis for finding targets in drug design (Ren et al., 2020a; Xia 2021). A pI value of more than 7 indicates that the protein or amino acid is alkaline, whereas a value less than 7 indicates that it is acidic. At present, there are few studies on pI of RBD mutation of SARS-CoV-2 spike protein (Krebs et al., 2021). In this study, it is considered that the change of pI has a great influence on the mutation of SARS-CoV-2, which is a problem neglected by the scientific community at present. The theoretical pI of the RBDWT and RBDOmicron variant were 8.38 and 8.73, indicating that the Omicron variant was expected to have more alkaline pI. As shown in Table 2, the pI of RBDOmicron mutation sites greater than 7 included N440K, T478K, Q493R, Q498R and Y505H. In addition to steric hindrance, the number of basic amino acids also affects the binding affinity between RBD and hACE2. Studies have found that in the omicron virus multiplication process, supplying excessive basic amino acids is required (Hanai, 2022). Hence, the transmissible strength of COVID-19 variants depended on the mutated basic amino acids that can enhance the binding affinity with ACE-2.
| Mutation site | pI | Mutation site | pI | Mutation site | pI |
|---|---|---|---|---|---|
| N440K | 5.41–9.60 | K417N | 9.60–5.41 | G339D | 6.06–2.85 |
| T478K | 5.60–9.60 | S477N | 5.68–5.41 | G446S | 6.06–5.68 |
| Q493R | 5.65–10.76 | S371L | 5.68–6.01 | G496S | 6.06–5.68 |
| Q498R | 5.65–10.76 | S373P | 5.68–6.30 | N501Y | 5.41–5.64 |
| Y505H | 5.64–7.60 | S375F | 5.68–5.49 | E484A | 3.15–6.11 |
The instability index of less than 40 indicates that the protein structure is stable. In our study, the range remained 20.43–28.51, indicating the great stability of RBDWT and RBDOmicron. Aliphatic indexes of 70.31 and 73.21 for RBDWT and RBDOmicron indicated that the protein was temperature stable across a wide temperature range. The greater a protein’s aliphatic index, the more thermostable it was. A protein with a low grand average of hydropathicity (GRAVY) value is nonpolar and has a stronger affinity for water, indicating that it is intrinsically hydrophilic. The higher amino acid composition of F, L, I and P in RBDOmicron, when compared with RBDWT, suggested that RBDOmicron included more hydrophobic amino acids. The amino acid composition of RBDOmicron was low in polar amino acids such as Q, S and E, while was high in nonpolar amino acids such as L, F and P. These results indicated that the Omicron variant was more hydrophobic and more easily combined with hACE2, which played an important role in increasing its transmissibility.
3.3 Molecular dynamics simulation results
The final predicted 3D structure model of RBDOmicron was shown in Fig. 3A. Although there were many active pocket mining tools, there was no statistically significant difference in structure type and tool performance (Clark et al., 2020). These tools could provide relatively accurate prediction results for protein active pockets regardless of the presence of ligands. Some researchers used Fpocket as an important tool to analyze the protein-ligand binding pocket, screened the active pocket of methionyl-tRNA synthetase, and proved it to be a natural inhibitor of the lead compound with inhibitory ability in subsequent docking experiments (Rowaiye et al., 2022). Similar studies have also reported that Fpocket can be used as a well-known tool to mine active pockets and predict specific action targets (active pockets) in both the structure and protein of COVID-19, accurately predict active pockets and provide the necessary basis for the accurate positioning of amino acids (Gervasoni et al., 2020). All of these are inseparable from the accurate prediction ability of Fpocket. Therefore, Fpocket was applied to detect the pockets on RBDOmicron surface that were suitable to bind small molecule ligands. It can be found that there were three active centres (AC) on RBDOmicron surface by Fpocket analysis, considered as ligand binding sites in this protein. Important amino acid residues presented in these three AC were determined as AC1 (orange red) including R355, Y396, P426, D428, F429, T430, F464, S514, F515, E516, L517, AC2 (forest green) including D339, F342, N343, V367, L371, P373, F374, W436, AC3 (cornflower blue) including Y453, L454, F456, R457, D467, S469, E471, I472, Y473, Y491. Since AC1 was the part connected with NTD, the remaining ACs were mainly used for molecular docking with small molecule ligands in subsequent experiments. It is worth noting that among the fifteen mutations of RBDOmicron, four mutations, namely G339D, S371L, S373P and S375F, were located at AC2, while the remaining eleven mutations were located at or near AC3, which was just on the interaction interface with hACE2.
Topological domain-specific average backbone root mean square deviation (RMSD) value of RBDOmicron was less than that of RBDWT as shown in Fig. S3. For RBDWT, the structure ensembled from simulation (100 ns) deviated from the backbone of RBDWT by 0.30 Å on average, while the RBDOmicron possessed averaged RMSD value of 0.29 Å, indicating that the mutations only slightly altered the structure of RBDOmicron from the RBDWT. Hence, the mutations in the Omicron variant did not significantly reduce the RBD stability, instead, the hACE2-RBDOmicron complex was even slightly more stable than hACE2-RBDWT (Lupala et al., 2022). Molecular dynamics was used to calculate the root mean square fluctuations (RMSF) value of amino acids. RMSF means the degree of freedom of each amino acid, which can prove to some extent which amino acid residues are more active in space. The residue fluctuations were analyzed by calculating the RMSF of RBDOmicron and RBDWT (Fig. 3B). According to the average values of RMSF, the RBDOmicron (0.75 Å) was more rigid than RBDWT (0.80 Å). The reduction of RMSF was more pronounced at the interfacing residues of RBD, also known as the receptor-binding motif (Yi et al., 2020).
3.4 In silico molecular docking analysis
HADDOCK (high ambiguity-driven protein-protein docking) algorithm was applied to perform the restraint docking of WT, Delta and Omicron variants with hACE2 receptor by defining the interface residues as reported previously (Koukos et al., 2021; Khandia et al., 2022). The docking scores for RBDWT-hACE2 and RBDDelta-hACE2 complex as −49.4 kcal/mol and −64.3 kcal/mol, while the docking score for RBDOmicron-hACE2 complex was −141.4 kcal/mol (Table 3). The data showed that the reported mutations have increased the binding ability of RBDOmicron to hACE2 than RBDWT and RBDDelta, which was similar to the results previously reported (Khan et al., 2021). Furthermore, a comparison of van der waals, electrostatic and desolvation energy values also revealed considerable differences among WT, Delta and Omicron variants. For RBDWT and RBDDelta, the van der waals energies were −18.1 kcal/mol and −24.0 kcal/mol, while RBDOmicron had a van der waals energy of −55.2 kcal/mol. The electrostatic energies for RBDWT, RBDDelta and RBDOmicron were −114.9 kcal/mol, −213.6 kcal/mol and −343.6 kcal/mol, respectively. Both the van der waals energy and electrostatic energy of RBDOmicron were increased, and higher electrostatic energy was considered the primary reason for enhanced binding (Khan et al., 2022).
| WT | Delta | Omicron | |
|---|---|---|---|
| HADDOCK score | −49.4 ± 4.5 | −64.3 ± 13.0 | −141.4 ± 5.5 |
| Van der Waals energy (kcal/mol) | −18.1 ± 2.6 | −24.0 ± 6.6 | −55.2 ± 4.9 |
| Electrostatic energy (kcal/mol) | −114.9 ± 16.9 | −213.6 ± 37.8 | −343.6 ± 49.8 |
| Desolvation energy (kcal/mol) | −8.3 ± 1.1 | −0.6 ± 2.7 | −20.1 ± 2.8 |
| Z-Score | −0.2 | −1.8 | −2.0 |
The interactions at the interface of the ACE2-RBD complex for WT have been previously reported in the perspectives of both static crystal structures and dynamical conformations (Wang et al., 2020; Lupala et al., 2021). Cryo-EM showed that eleven of the fifteen mutations in RBDOmicron (K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H) were located at ACE2-RBD interface, consequently changing the electrostatics surface changes at the interface. As can be seen from Fig. 4 the salt bridge E484-K31 and K417-D30 of RBDWT-ACE2 were lost in the Omicron variant, while hydrogen bonds Q493-E35, Q498-K353 and Y505-E37 were enhanced by the Omicron mutations, forming new interactions, such as R498-Y41, R498-Q42 and H505-K353. Also, an additional π-π stacking interaction Y501-Y41 was introduced. The key interactions Y449-D38, Y453-H34, A475-S19, N487-Y83, T500-N330 and T500-D355 in RBDWT-ACE2 were preserved in the Omicron variant. Cryo-EM structural analysis of the RBDOmicron-hACE2 complex showed that new salt bridges and hydrogen bonds were formed at R493, S496 and R498 mutations, which may compensate for other Omicron mutations, such as K417N which is known to reduce the affinity between ACE2 and S protein (Mannar et al., 2022). The interactions R493-E35 have previously been shown to be responsible for the anchor locking and correct orientation of the RBDOmicron when binding to hACE2. Based on docking studies, the Q493R, N501Y, S371L, S373P, S375F, Q498R and T478K mutations contributed significantly to high binding affinity with hACE2. Analysis revealed that mutations at N440K, T478K and N501Y sites may impart ten times more infectivity to WT (Thakur and Ratho, 2022). The differences in binding pattern between the WT and the Omicron variant complexes revealed that the key mutations are responsible for enhanced binding and the consequent transmissibility and infectivity. Dejnirattisai et al., 2022 found Omicron variant caused widespread escape from neutralization by serum obtained following infection by a range of SARS-CoV-2 variants, meaning that previously infected individuals will have little protection from infection with Omicron.
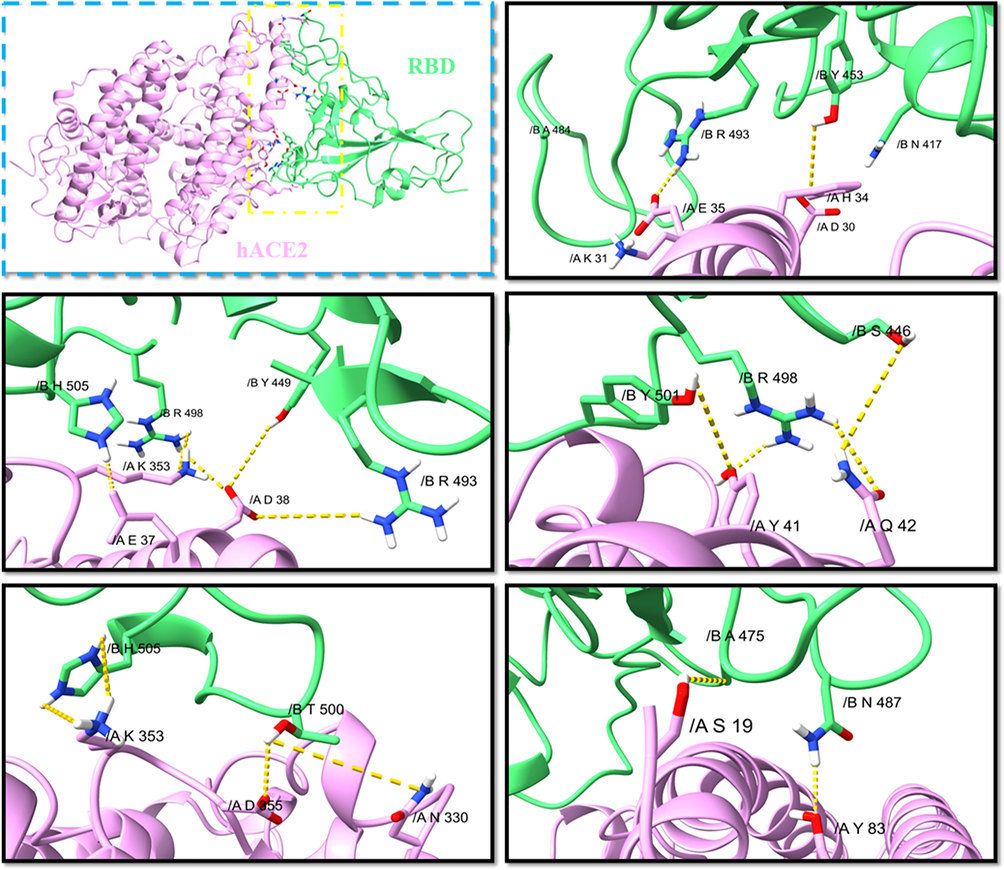
- The interactions between RBD-hACE2 of the Omicron variant.
Since the outbreak of COVID-19, several large-scale mutations have all been carried out around spike proteins, among which RBD mutation is the most common. The evolution direction of this virus is dependent on transmission from human to human or from animal to human, through the host immune system to complete the screening (Kupferschmidt, 2021). In short, the pandemic’s weakness has led to the evolution of the SARS-CoV-2, which is centred on spike proteins, particularly the RBD part (Sanjuán and Domingo-Calap, 2016). Traditional Chinese medicine has been playing an important role in the prevention and treatment of COVID-19 from the early stage to the present. Studies have shown that Chinese medicine components have good inhibitory capacity on both structural and non-structural proteins of SARS-CoV-2. The study of Chinese medicine components by network pharmacology is one of the most widely used methods to explore the inhibition of SARS-CoV-2 binding, infection and replication from Chinese medicine at present (Ren et al., 2020b). Although this research method has some limitations, it can also provide the best binding skeleton in a specific active centre and provide a good research strategy for drug design and drug screening (Sahlan et al., 2021; Dotolo et al., 2021). Traditional idea of Traditional Chinese medicine is to prevent and control the spread of diseases before they get sick through systematic regulation, which is highly consistent with modern epidemiological views (Zhang et al., 2021).
Based on the theory of Traditional Chinese medicine, the research idea of blocking the binding of viruses to receptors is more valuable than the discovery of inhibitors of key enzymes of viruses. Therefore, this study focuses on the study of the herbal aromatic plant Amomum tsao-ko. It can be found that all components in AEO can interact with RBD and NTD by molecular docking the components with a relative area percentage of more than 1% with RBDWT, RBDDelta, RBDOmicron, NTDWT and NTDOmicron. The binding energy between AEO components and RBDOmicron varied from −7.0 kJ/mol to −4.5 kJ/mol. A more negative value of binding energy implies stronger binding efficacy (Kar et al., 2020). And the binding sites of most AEO components were located at AC2 and AC3 or near the interface. Among them, EO02, 03, 08 and 17 with high relative content and strong binding energy with RBD (Fig. 5 and Table S1). Meanwhile, RBD is the earliest binding region of COVID-19 with the receptor. It was imperative to research spike protein, especially on how to reduce its binding ability with hACE2 in the epidemic prevention of COVID-19. The binding energy of RBD and hACE2 was between −9.8 and −14.0 kcal/mol (COVID-19 spike-host cell receptor GRP78 binding site prediction), which was a relatively strong binding ability. In current studies, the binding energy between no matter the known drug molecules or the drug molecules obtained by virtual screening cannot reach less than −9.8 kcal/mol, that is, the minimum binding energy of RBD and hACE2 (Shah et al., 2020; Cavasotto and Filippo, 2021), indicating that it was difficult to design the inhibition of spike protein to inhibit viral infection. Therefore, this study focused on reducing the ability of the virus to spread in the air and contact with the host, which was consistent with the results of relevant literature and could provide proof for this study to a certain extent (Abdelli et al., 2021; Kumar et al., 2020; Yu et al., 2020; Naeem et al., 2020; Xiong et al., 2020). Virtual screening showed that most components of AEO could interact with AC2 and AC3 of RBDOmicron, indicating that the interaction between AEO and RBD was a synergistic effect of multiple targets (Fig. 5, Fig. 6 and Fig. S4-12). Spike protein RBD is the most externally exposed part of the SARS-CoV-2 structure. If RBD contact with AEO molecules in the air, the two can interact directly and form a relatively stable complex, thus protecting the host to a certain extent (Wang et al., 2021b). The essential oil is volatile itself and can interact with SARS-CoV-2 spike protein RBD in the air to inhibit the binding ability of RBD and hACE2, thus achieving epidemic prevention effect.
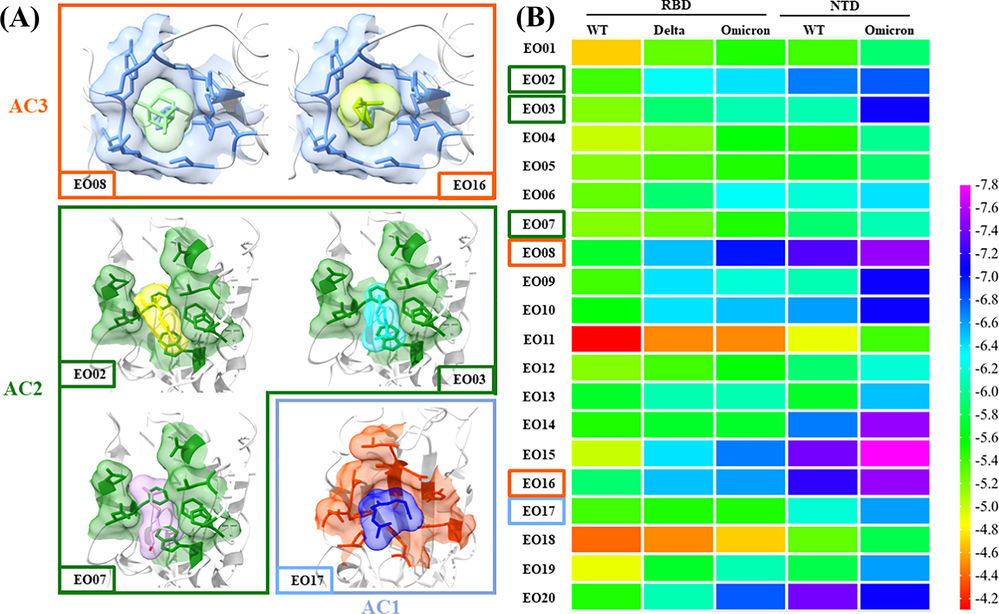
- Molecular interactions of AEO components against RBDOmicron (A) and the heatmap of the binding energy of AEO to RBD and NTD (B).
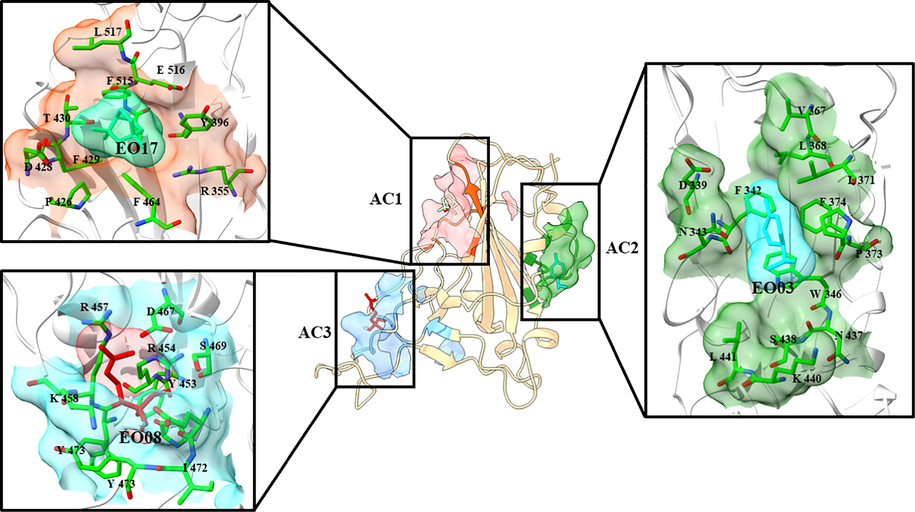
- Surface and maps of AEO components docked into binding sites of RBDOmicrion different active centres.
4 Conclusions
This study focused on the Epidemic prevention formula 1 and 2 in China, combined with the previous research on the medicinal aromatic plant, Amomum tsao-ko. It was found that there were twenty kinds of AEO with a relative area percentage of more than 1% extracted by hydrodistillation and identified by GC/Q-TOF MS. Meanwhile, AEO possessed a good diffusion effect in the air because of its volatility. Given the newly emerged Omicron mutation, RBDOmicron was established by homology modeling in this study. Molecular dynamics simulation and the Fpocket method were used to mine its active centres. RBDOmicron was found to bind hACE2 more strongly than RBDWT and may be a potentially highly pathogenic mutant using HADDOCK. The molecular docking of 20 components in AEO and RBDOmicron showed that they all had a good interaction, and were distributed in three RBDOmicron active centres, and each AEO component and RBDOmicron had a variety of different active centres with similar binding energy. In the absence of direct evidence at present, this study proves that Omicron is indeed a more pathogenic mutant of COVID-19, excavates essential oils from Traditional Chinese medicine that can inhibit its binding with receptors, and proposes a prevention method for preventing transmission of SARS-CoV-2 prevention.
5 Limitations, research gap, future prospectives
Although the world is fighting with the virus, the ever-pervasive COVID-19 continues to evolve, posing a challenge to SARS-CoV-2 epidemic prevention. This study aims to prove from the perspective of simulation that the material basis of the medicinal plant Amomum tsao-ko, which has been reported to have SARS-CoV-2 prevention and control ability, namely its natural essential oil components, has the potential inhibitory ability on COVID-19 especially the Omricon mutants. Starting with the spike protein of the Omricon variant, molecular dynamics and Fpocket were used to dig out the active centres and identify the key amino acid residues. The amino acids in key parts of RBD were described from two aspects of polarity and isoelectric point. Molecular docking proved that various AEO components possessed a good binding relationship with RBD, and maybe in a competitive relationship with the binding of RBD and hACE2, thus achieving the ability to prevent COVID-19. This study provides a valuable reference for the prevention and treatment of COVID-19 with Amomum tsao-ko and aromatic Chinese medicinal materials. Aromatic medicinal plants are usually based on volatile components, which may have certain research value in the prevention and treatment of airborne viruses such as COVID-19. In recent years, it is rare to use molecular dynamics to investigate key residues of proteins in COVID-19 studies, but there have been a large number of studies on the artificial evolution of proteins and the trend is on the rise. Several important mutations in COVID-19 that gradually enhance its infectivity or immune escape, can be regarded as an evolution. Molecular dynamics studies of amino acids can prove the evolutionary direction of the key amino acids. With further research, it would be possible to predict future mutations and develop antibodies or inhibitors in advance to stop outbreaks.
CRediT authorship contribution statement
Ju-Zhao Liu: Formal analysis, Visualization, Writing – original draft, Writing – review & editing. Hong-Chang Lyu: Formal analysis, Methodology. Yu-Jie Fu: Investigation, Writing – review & editing. Qi Cui: Conceptualization, Methodology, Data curation, Writing – original draft, Visualization, Writing – review & editing.
Acknowledgements
This research was funded by China Postdoctoral Science Foundation (2021M692893, 2021M702927), Natural Science Foundation of Zhejiang Province (LQ22H280007) and Traditional Chinese Medicine Modernization Special Project of Zhejiang Traditional Chinese Medicine Science and Technology Plan (2021ZX008). We appreciate the great experimental support from the Public Platform of Medical Research Center, Academy of Chinese Medical Science, Zhejiang Chinese Medical University.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- In silico study the inhibition of angiotensin converting enzyme 2 receptor of COVID-19 by Ammoides verticillata components harvested from Western Algeria. J. Biomol. Struct. Dyn.. 2021;39:3263-3276.
- [Google Scholar]
- GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19-25.
- [Google Scholar]
- “Fei Yan No. 1” as a combined treatment for COVID-19: An efficacy and potential mechanistic study. Front. Pharharmacol.. 2020;11:581277.
- [Google Scholar]
- 1,8-Cineole: a review of source, biological activities, and application. J. Asian Nat. Prod. Res.. 2021;23:938-954.
- [Google Scholar]
- In silico drug repurposing for COVID-19: targeting SARS-CoV-2 proteins through docking and consensus ranking. Mol. Inform.. 2021;40:2000115.
- [Google Scholar]
- Predicting binding sites from unbound versus bound protein structures. Sci. Rep.. 2020;10:15856.
- [Google Scholar]
- Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. Available online: https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern.
- Rapid extraction of Amomum tsao-ko essential oil and determination of its chemical composition, antioxidant and antimicrobial activities. J. Chromatogr. B. 2017;1061–1062:364-371.
- [Google Scholar]
- SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185:467-484.
- [Google Scholar]
- A review on drug repurposing applicable to COVID-19. Brief Bioinform.. 2021;22:726-741.
- [Google Scholar]
- A comprehensive mapping of the druggable cavities within the SARS-CoV-2 therapeutically relevant proteins by combining pocket and docking searches as implemented in Pockets 2.0. Int. J. Mol. Sci.. 2020;21(15):5152.
- [Google Scholar]
- The preservative potential of Amomum tsaoko essential oil against E. coil, its antibacterial property and mode of action. Food Control. 2017;75:236-245.
- [Google Scholar]
- Quantitative in silico analysis of SARS-CoV-2 S-RBD omicron mutant transmissibility. Talanta. 2022;240:123206.
- [Google Scholar]
- COVID-19 Genomics UK (COG-UK) Consortium. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol.. 2021;19:409-424.
- [Google Scholar]
- Records of medicinal and edible plants in the herb section of compendium of materia medica and their revelation. Mod. Chin. Med.. 2020;22:1769-1777.
- [Google Scholar]
- Anisotine and amarogentin as promising inhibitory candidates against SARS-CoV-2 proteins: a computational investigation. J. Biomol. Struct. Dyn.. 2020;11:1-11.
- [Google Scholar]
- Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: an insight from structural data. J. Cell. Physiol.. 2021;236(10):7045-7057.
- [Google Scholar]
- The Omicron (B.1.1.529) variant of SARS-CoV-2 binds to the hACE2 receptor more strongly and escapes the antibody response: Insights from structural and simulation data. Int. J. Biol. Macromol.. 2022;200:438-448.
- [Google Scholar]
- Emergence of SARS-CoV-2 Omicron (B.1.1.529) variant, salient features, high global health concerns and strategies to counter it amid ongoing COVID-19 pandemic. Environ. Res.. 2022;209:112816.
- [Google Scholar]
- Herbal immune-boosters: Substantial warriors of pandemic Covid-19 battle. Phytomedicine. 2021;85:153361.
- [Google Scholar]
- Shape-restrained modelling of protein-small molecule complexes with HADDOCK. J. Chem. Inf. Model.. 2021;61(9):4807-4818.
- [Google Scholar]
- Isoelectric point determination by imaged CIEF of commercially available SARS-CoV-2 proteins and the hACE2 receptor. Electrophoresis. 2021;42:687-692.
- [Google Scholar]
- Geranium and lemon essential oils and their active compounds downregulate angiotensin-converting enzyme 2 (ACE2), a SARS-CoV-2 spike receptor-binding domain, in epithelial cells. Plants-Basel. 2020;9:770.
- [Google Scholar]
- Viral mutations may cause another ‘very, very bad’ COVID-19 wave, scientists warn. Science 2021
- [Google Scholar]
- Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215-220.
- [Google Scholar]
- Traditional Chinese herbal medicine at the forefront battle against COVID-19: Clinical experience and scientific basis. Phytomedicine. 2021;80:153337.
- [Google Scholar]
- Correlation analysis of compounds in essential oil of Amomum tsaoko seed and fruit morphological characteristics, geographical conditions, locality of growth. Agronomy. 2021;11:744.
- [Google Scholar]
- Polyphenol extract and essential oil of Amomum tsao-ko equally alleviate hypercholesterolemia and modulate gut microbiota. Food Funct.. 2021;12:12008-12021.
- [Google Scholar]
- Computational simulations reveal the binding dynamics between human ACE2 and the receptor binding domain of SARS-CoV-2 spike protein. Quant Biol.. 2021;9:61-72.
- [Google Scholar]
- Mutations on RBD of SARS-CoV-2 Omicron variant result in stronger binding to human ACE2 receptor. Biochem. Bioph. Res. Co.. 2022;590:34-41.
- [Google Scholar]
- SARS-CoV-2 Omicron variant: antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science. 2022;375(6582):760-764.
- [Google Scholar]
- Herbs, immunity and nCOVID-19: Old performers in new pandemic. Pak. J. Pharm. Sci.. 2020;33:1747-1753.
- [Google Scholar]
- Effects on volatile oil and volatile compounds of Amomum tsao-ko with different pre-drying and drying methods. Ind. Crop. Prod.. 2021;174:114168.
- [Google Scholar]
- A coordinative dendrimer achieves excellent efficiency in cytosolic protein and peptide delivery. Angew. Chem. Int. Ed.. 2020;59:4711-4719.
- [Google Scholar]
- Identifying potential treatments of COVID-19 from Traditional Chinese Medicine (TCM) by using a data-driven approach. J. Ethnopharmacol.. 2020;258:112932.
- [Google Scholar]
- Identifying potential natural inhibitors of Brucella melitensis Methionyl-tRNA synthetase through an in-silico approach. PLOS Neglect Trop D.. 2022;16(3):e0009799.
- [Google Scholar]
- Molecular interaction analysis of Sulawesi propolis compounds with SARS-CoV-2 main protease as preliminary study for COVID-19 drug discovery. J. King Saud. Univ. Sci.. 2021;33:101234.
- [Google Scholar]
- fpocket: online tools for protein ensemble pocket detection and tracking. Nucleic Acids Res.. 2010;38:W582-W589.
- [Google Scholar]
- In silico studies on therapeutic agents for COVID-19: Drug repurposing approach. Life Sci.. 2020;252:117652.
- [Google Scholar]
- Identification of in vitro-in vivo components of Caoguo using accelerated solvent extraction combined with gas chromatography-mass spectrometry integrated with network pharmacology on indigestion. Ann. Transl. Med.. 2021;9:1247.
- [Google Scholar]
- Computational decomposition reveals reshaping of the SARS-CoV-2-ACE2 interface among viral variants expressing the N501Y mutation. J. Cell. Biochem.. 2021;122:1863-1872.
- [Google Scholar]
- Research progress on aromatic traditional Chinese medicine essential oil intervening metabolic syndrome. Chin. Herb. Med.. 2021;52:6088-6095.
- [Google Scholar]
- OMICRON (B.1.1.529): A new SARS-CoV-2 variant of concern mounting worldwide fear. J. Med. Virol.. 2022;94:1821-1824.
- [Google Scholar]
- Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein. Science. 2020;370:725-730.
- [Google Scholar]
- Tracking SARS-CoV-2 variants. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/.
- Specific epitopes form extensive hydrogen-bonding networks to ensure efficient anti-body binding of SARS-CoV-2: Implications for advanced antibody design. Comput. Struct. Biotec.. 2021;19:1661-1671.
- [Google Scholar]
- Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130-135.
- [Google Scholar]
- Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894-904.
- [Google Scholar]
- SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res.. 2018;W1:W296-W303.
- [Google Scholar]
- Chemical profiling of Huashi Baidu prescription, an effective anti-COVID-19 TCM formula, by UPLC-Q-TOF/MS. Chin. J. Nat. Med.. 2021;19:473-480.
- [Google Scholar]
- Domains and functions of spike protein in SARS-Cov-2 in the context of vaccine design. Viruses. 2021;13:109.
- [Google Scholar]
- Chinese herbal medicine for coronavirus disease 2019: A systematic review and meta-analysis. Pharmacol. Res.. 2020;160:105056.
- [Google Scholar]
- Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies. Cell. Mol. Immunol.. 2020;17:621-630.
- [Google Scholar]
- Computational screening of antagonists against the SARS-CoV-2 (COVID-19) coronavirus by molecular docking. Int. J. Antimicrob. Ag.. 2020;56:106012.
- [Google Scholar]
- TCM prevention and treatment of epidemic disease based on “Preventing before disease, disease prevention before change, prevention and relapse after recovery. Western J. Tradit. Chin. Med.. 2021;34:1-4.
- [Google Scholar]
- Prevention and treatment of COVID-19 using Traditional Chinese Medicine: A review. Phytomedicine. 2021;85:153308.
- [Google Scholar]
- Analyzing the potential therapeutic mechanism of Huashi Baidu Decoction on severe COVID-19 through integrating network pharmacological methods. J. Tradit. Complemnt. Med.. 2021;11:180-187.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2022.103916.
Appendix A
Supplementary material
The following are the Supplementary data to this article:







