Translate this page into:
Drug metabolite cluster centers-based strategy for comprehensive profiling of Neomangiferin metabolites in vivo and in vitro and network pharmacology study on anti-inflammatory mechanism
⁎Corresponding author at: School of Pharmacy, Binzhou Medical University, 264003 Yantai, Shandong, China. zhangjiayu0615@163.com (Jiayu Zhang)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Neomangiferin (NMF) is an extremely special xanthone that could be simultaneously attributed to C-glycoside and O-glycoside with a variety of biological activities, such as anti-inflammatory, antitumor, antipyretic, and so on. So far as we know, the metabolism profiling has been insufficient until now. Herein, Drug Metabolite Cluster Centers (DMCCs)-based Strategy has been developed to profile the NMF metabolites in vivo and in vitro. Firstly, the DMCCs was proposed depending on literature-related and preliminary analysis results. Secondly, the specific metabolic rule was implemented to screen the metabolites of candidate DMCCs from the acquired Ultra High Performance Liquid Chromatography Quadrupole Exactive Orbitrap Mass Spectrometry (UHPLC-Q-Exactive Orbitrap MS) data by extracted ion chromatography (EIC) method. Thirdly, candidate metabolites were accurately and tentatively identified according to the pyrolysis law of mass spectrometry, literature reports, comparison of reference substances, and especially the diagnostic product ions (DPIs) deduced preliminarily. Finally, network pharmacology was adopted to elucidate the anti-inflammatory action mechanism of NMF on the basis of DMCCs. As a result, 3 critical metabolites including NMF, Mangiferin (MF) and Norathyriol (NA) were proposed as DMCCs, and a total of 61 NMF metabolites (NMF included) were finally screened and characterized coupled with 3 different biological sample preparation methods including solid phase extraction (SPE), acetonitrile precipitation and methanol precipitation. Among them, 32 metabolites were discovered in rat urine, 30 in rat plasma, 12 in rat liver, 9 metabolites in liver microsomes and 8 in rat faeces, respectively. Our results also illustrated that NMF primarily underwent deglucosylation, glucuronidation, methylation, sulfation, dihydroxylation and their composite reactions in vivo and in vitro. Additionally, network pharmacology analysis based on DMCCs revealed 85 common targets of disease-metabolites, and the key targets were TNF, EGFR, ESR1, PTGS2, HIF1A, IL-2, PRKCA and PRKCB. They exerted anti-inflammatory effects mainly through the pathways of inflammatory response, calcium-dependent protein kinase C activity, nitrogen metabolism, pathways in cancer and so on. In general, our study constructed a novel strategy to comprehensive elucidate the biotransformation pathways of NMF in vivo and in vitro, and provided vital reference for further understanding its anti-inflammatory action mechanism. Moreover, the established strategy could be generalized to the metabolism and action mechanism study of other natural products.
Keywords
Drug Metabolism Cluster Centers-based strategy
Neomangiferin
UHPLC-Q-Exactive Orbitrap MS
Anti-inflammatory
Network pharmacology
- DMCs
-
Drug Metabolite Clusters
- DMCCs
-
Drug Metabolite Cluster Centers
- NMF
-
Neomangiferin
- MF
-
Mangiferin
- NA
-
Norathyriol
- EIC
-
Extracted Ion Chromatography
- DPIs
-
Diagnostic Product Ions
- SPE
-
Solid Phase Extraction
- UHPLC-Q-Exactive Orbitrap MS
-
Ultra High Performance Liquid Chromatography Quadrupole Exactive Orbitrap Mass Spectrometry
- HRMS
-
High Resolution Mass Spectrometry
- GluA
-
glucuronyl
- Glc
-
glucosyl
Abbreviations
1 Introduction
Drug metabolism refers to the process of chemical structural change under the interference of various drug metabolism enzymes in the internal body, also known as biological transformation. To understand the process of biotransformation, we classify it into two phases. The first is called phase Ⅰ that drug molecules are usually modified by reactions such as oxidation, reduction and hydrolysis. The resulting metabolites are prone to further changes, called phase Ⅱ. It involves conjugation of the metabolites with bulky hydrophilic molecules such as glucuronic acid, sulfonic acid and glutathione. The biotransformation process terminates the intended pharmacological action of each drug molecule and proceeds at a drug-specific rate, which is subsequently captured for hepatic or extrahepatic clearance. However, the products of biotransformations may not necessarily be pharmacologically inert. They may be active pharmacologically or highly reactive, disrupting cellular homeostasis sometimes leading to cell death and organ damage (Obach., 2013, Ito., 2014). In recent years, a large number of researches have been carried out on drug metabolism and metabolism-related study (Tekes et al., 2011, Hughes et al., 2020) since they have been increasingly crucial to clarify the action mechanism and to guide clinical medication. However, difficulty remains hard regarding how to screen out the metabolites from the complex ground-substance system.
Consequently, we firstly proposed Drug Metabolite Clusters (DMCs)-based strategy to discover more drug metabolites (Wang et al., 2019). It refers to the primary metabolites produced by specific metabolic enzymes after drug entry into the body, and then these primary metabolites in turn form the primary metabolite cluster centers and drug metabolism primary clusters. The primary metabolites further form secondary metabolites and secondary metabolic clusters, eventually forming DMCs. However, the concentration of drug metabolites in the body is usually too faint low to be further metabolized in fact. Additionally, most of the metabolites identification were tentative rather than accurate, which causing waste of a great deal of efforts to deduce the uncertain results. Thus, it is increasingly essential to develop novel analytical strategy to enhance metabolite identification efficiency and accuracy.
Neomangiferin (NMF) is an extremely special xanthone that could be simultaneously attributed to C-glycoside and O-glucoside (Yang et al., 2016). It is 7-O-glucoside derivative of Mangiferin (MF) that abundant in various herbals and foods, such as rhizome of Anemarrhena anemarrhena (Li et al., 2013, Wang et al., 2021), mangoes and mango leaves (Wu et al., 2020) and Salvia miltiorrhiza (Li et al., 2015). In addition, NMF is an exceedingly essential component associated with anti-inflammatory, antitumor, antipyretic and antioxidant activities (Liu et al., 2012, Gong te al., 2013, Jeong et al., 2014, Han et al., 2015, Wang et al., 2014).
Currently, the study of NMF mainly focuses on its pharmacokinetics (Li et al., 2015, Cai et al., 2010) and quantification in vivo (Yang et al., 2016, Qiu et al., 2016), whereas limited studies were performed to characterize NMF metabolites. For instance, Xie et al (Xie et al., 2016) tentatively identified only 21 potential metabolites of NMF and MF in rat plasma, which indicated the metabolism study of NMF has been relatively insufficient until now.
Thereby, in this study, NMF was taken as an example to further establish Drug Metabolites Clusters Centers (DMCCs)-based analytical strategy coupled with Ultra High Performance Liquid Chromatography Quadrupole Exactive Orbitrap Mass Spectrometry (UHPLC-Q-Exactive Orbitrap MS) to clarify its metabolic transformation process. Moreover, the anti-inflammatory action mechanism of NMF was also explored from the systematic and holistic perspective via network pharmacology.
2 Experiments and methods
2.1 Chemicals and reagents
NMF and MF reference standards were purchased from Chengdu Must Biotechnology Co., ltd. (Sichuan, China). Their structures were fully elucidated by comparing the spectral data (ESI-MS and 1H, 13C NMR). Their purities were acceptable (≥98 %) according to HPLC-UV analysis. UHPLC grade acetonitrile methanol and formic acid (FA) were purchased from Thermo Fisher Scientific (Fair Lawn, NJ, USA). All the other chemicals of analytical grade were available at the work station, Beijing Chemical Works (Beijing, China). The deionized water used throughout the experiment was purchased from Watsons (Guangzhou, China). Nicotinamide adenine dinucleotide phosphate (NADPH) and MgCl2 were provided by Shanghai Macklin Biochemical Co., ltd (Shanghai, China). 6-well plates were obtained from Corning Incorporated-Life Science (Jiangsu, China). Grace Pure™ SPE C18-Low solid phase extraction cartridges (200 mg/3 mL, 59 μm, 70 Å) were produced by Grace Davison Discovery Science (Deerfield, IL, USA). Rat liver microsomes was obtained from Xin Run Biotechnology Co., ltd (Wuxi, China, batch No. 20210305).
2.2 Incubation experiment of liver microsomes
MgCl2 and liver microsomes were dissolved in PBS (pH = 7.4) to obtain the final concentration of MgCl2 of 3 mM and the protein mass concentration of 1 mg/mL. Then, NMF reference substance was dissolved in this solution, and the final drug concentration was reached to 1 mg/mL. After setting the administration group and blank group, the above mixture (900 μL) was added to each well of the 6-well plate. After preheating at 37 °C for 5 min, 100 μL of NADPH (100 μL, 50 mM) was added in them to start the reaction, and the incubation was continued at 37 °C. At 5, 10, 15, 30, 45, 60, 120, 240 and 480 min, the system solution (100 μL) was added to cold acetonitrile (200 μL) to stop the reaction. Finally, the supernatant was centrifuged at 3,500 rpm to obtain the analysis samples.
2.3 Animals experiment
Six male Sprague-Dawley (SD) rats weighting 200 ± 10 g were obtained from Jinan Pengyue Experimental Animals Co., ltd (Jinan, China). The rats were housed in a controlled room at standard temperature (24 °C ± 2 °C) and humidity (70 % ± 5 %), and kept on a 12 h light/12 h dark regime. After 7 days of acclimation, the rats were randomly divided into two groups: Drug Group (n = 3) and Control Group (n = 3) for testing plasma, urine, faeces and liver, respectively. They were fasted for 12 h with free access to water prior to the experiment. NMF was dissolved in normal saline. The rats in the Drug Group were given a dose of 150 mg/kg body weight orally. The rats were given the same volume of normal saline in Control Group. All the rats were administered for 3 consecutive days and fed in the animal room of Shandong International Biotechnology Park. Animal protocols were approved by the institutional Animal Care and Use Committee at Binzhou Medical University (2021–085). Animal facilities and protocols were complied with the Guide for the Care and Use of Laboratory Animals (USA National Research Council, 1996).
2.4 Biological sample processing
Blood samples (0.5 mL) were taken from the suborbital venous plexus of rats at 0.5, 1, 1.5, 2,4 and 6 h after administration. Each sample was centrifuged at 3,500 rpm for 10 min to obtain the supernatant of plasma samples. Urine and faeces samples were collected 0–24 h after administration. 24 h after oral administration, the rats were dissected and their liver tissues were taken off and quenched in liquid nitrogen. All the homogeneous biological samples from the same group were finally merged into a collective sample and stored at −80 °C prior to analysis.
Urine samples were centrifuged at 12,000 rpm for 15 min to collect the supernatant. Faeces samples (2 g) were ultrasonically extracted with water (10 mL) for 30 min (3,500 rpm, 4 °C) and then filtered to obtain the supernatant. Additionally, normal saline (10 mL) was added to liver tissue (1 g) to grind and then the supernatant was centrifuged for 5 min (3,500 rpm, 4 °C) to take the supernatant. And then, the above samples were prepared by solid phase extraction (SPE). The sample processing steps were listed as follows: (a) methanol (3 mL) was added to activate SPE column, and water (3 mL) was added to pretreat the column. (b) urine, faeces and liver tissue samples (1 mL) were added to the pretreated SPE column and subsequently washed with water (3 mL) and methanol (3 mL). (c) the methanol eluate was finally collected into a 5 mL EP tube and evaporated by nitrogen at room temperature.
Then, 3 different methods were utilized to process plasma samples. The first method was used by methanol to precipitate samples. Methanol (3 mL) was added to plasma samples (1 mL) for precipitation, and the supernatant was obtained by centrifuging for 10 min (3,500 rpm, 4 °C). The second method used acetonitrile to precipitate plasma samples. Plasma samples (1 mL) were treated with acetonitrile (3 mL) for precipitation, and then they were centrifuged for 10 min (3,500 rpm, 4 °C) to obtain the supernatants. The third method was performed to prepare samples by SPE method above mentioned.
Finally, all the eluates of various biological samples were blow-dried with nitrogen. Before analysis, all the samples were redissolved in 300 μL methanol, and centrifuged at 14,000 rpm.
2.5 LC-MS conditions
UHPLC analysis was performed using a DIONEX Ultimate 3,000 UHPLC system (Thermo Fisher Scientific, MA, USA) equipped with dual pumps and autosampler. Chromatographic separation using Waters ACQUITY BEH C18 column (2.1 × 100 mm i.d., 1.7 μm; Waters Corporation, Milford, MA, USA) and the column temperature was maintained at 35 °C. Solvent A of mobile phase was 0.1 % FA aqueous solution and solvent B was acetonitrile. The injection volume was 3 μL. The flow rate of mobile phase was set at 0.30 mL/min with linear gradient as follows: 0–10 min, 95 % B; 10–15 min, 95 %-70 % B; 15–32 min, 70 %-50 % B; 32–35 min, 50 %-95 % B.
High resolution mass spectrum (HRMS) spectral analysis was performed on Q-Exactive-Orbitrap MS (Thermo Fisher Scientific, MA, USA). A single analytical run was performed in both positive and negative ion modes, and the tune method set was as follows: nitrogen (purity ≥ 99.99 %) was used as sheath gas and auxiliary gas with a flow rate of 45 and 10 (arbitrary unit), respectively; vaporizer temperature of 320 °C, capillary temperature of 320 °C, and spray voltage of 3,800/3,500 V (+/-) were set. Metabolites were detected using full-scan method at m/z 80–1,200 and resolution of 70,000. The collision energy was set to 30 % of the normalized collision energy to generate adequate fragment ions.
2.6 Data processing
Thermo Xcalibur 2.1 workstation was used for data acquisition and processing. The ESI-MS2 fragment ions of NMF metabolites with intensity over 10,000 were selected for the subsequent structural identification. The parameters were set as follows: C [0–40], H [0–60], O [0–30], S [0–1], the ring double bond (RDB) equivalent value [5–20]. Accurate mass measurements were set within mass error of ± 5 ppm.
2.7 Network pharmacology
2.7.1 Target prediction
The Canonical SMILES of DMCCs were found using Pubchem database (https://pubchem.ncbi.nlm.nih.gov/). They were imported in Swiss Target Prediction database (https://www.swisstargetprediction.ch/), and the species were selected “Homo Sapiens”. The target information related to inflammatory was acquired in GeneCards database (https://www.genecards.org/) with “inflammatory” as the keyword. And then, the potential anti-inflammatory targets of NMF were screened by the intersection of targets of metabolite and inflammatory.
2.7.2 Construction protein-protein interaction (PPI) network and screening of core targets
Potential anti-inflammatory targets of metabolites were imported into String (https://cn.string-db.org/) database, and then “Multiple protein” was selected. Species were selected as “Homo Sapiens” for analysis. Data was imported into Cytoscape 3.8.0 to obtain PPI network diagram and screened out core targets according to the degree value of node connection number.
2.7.3 Analysis of enrichment of gene ontology (GO) pathways and the kyoto encyclopedia of genes and genomes (KEGG) pathways
GO is an international standard classification system for gene function. KEGG database includes the metabolic pathways and functions of gene products in cells. GO and KEGG analyses were performed on the anti-inflammatory corresponding targets of the major metabolites (DMCCs) based on Metascape database (https://metascape.org/) (Zhou et al., 2019). Homo sapiens was set as species, P < 0.01 as threshold.
2.7.4 Construction of disease-target-metabolites network
We expected to ascertain the interaction among targets of metabolites and targets of inflammation and thus a disease-target-metabolite network was constructed. Firstly, we intersected the primary metabolites targets with the genes associated with inflammation and obtained a Venn diagram of the intersected gene symbols. Then, we built a network of complex information based on intersections among the major metabolites, gene symbols, and disease. Finally, we used Cytoscape 3.8.0 to undertake visual analyses of disease-targets-metabolites network.
3 Results
3.1 The establishment of DMCCs-based analytical strategy
Herein, DMCCs-based strategy has been developed to profile the metabolites in vivo and in vitro and reveal the anti-inflammatory mechanism of NMF (Fig. 1). Firstly, NMF, one di-glucosyls xanthone, was preliminarily deduced that it could generate MF and NA during metabolism, where one or two glucosyls were removed, correspondingly (Xie et al., 2016, Liu et al., 2011). Therefore, NMF, MF and NA were regarded as the three most vital DMCCs to produce various secondary metabolites. Secondly, since NMF is a flavonoid compound, the retro-Diels-Alder reaction might stand a good chance during the fragmentation process of NMF. Moreover, diagnostic product ions (DPIs) were extremely critical for the structural identification of NMF metabolites. Since the basic structure of the prototype drug isn’t prone to change much in the biotransformation process, the fragmentation pathways of NMF metabolites could be predicted and elucidated according to the deduced DPIs of NMF standard (Mei et al., 2019). Thirdly, metabolic reactions (Zhang et al., 2008) were applied to DMCCs mentioned above including phase Ⅰ, phase Ⅱ and their composite metabolism pathways, such as oxidation, reduction and hydrolysis, S-glutathionylation, glucuronidation, sulfation, their composite reactions, and so on. If these reactions occurred, the m/z values of metabolites would increase by 16 Da, 2 Da, 18 Da, 307 Da, 176 Da and 80 Da on the basis of prototype drug, respectively. Meanwhile, according to the above m/z values, the extracted ion chromatography (EIC) method and DPIs were adopted to screen and identified the primary candidate metabolites of DMCCs, correspondingly. The above reactions occur further on the basis of the obtained metabolites, and the search for metabolites is not complete until no more EIC signal could be found. Finally, on the basis of DMCCs, we selected 3 key metabolites including NMF, MF and NA to carry out network pharmacology analysis so as to clarify the anti-inflammatory mechanism of NMF. The disease-target-metabolite network and the PPI network were established based on the common targets of metabolites and inflammation, and the core targets were screened out. And then, GO enrichment analysis and KEGG pathway analysis of the core targets were performed.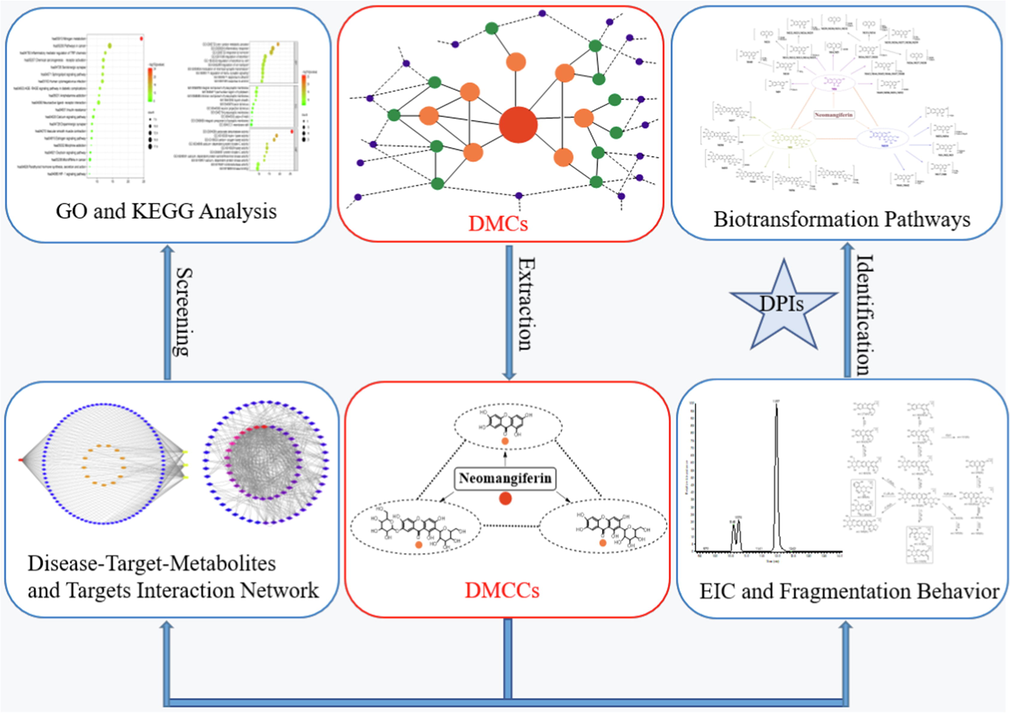
The summary diagram of analytical strategy and methodology. Note: the red node represents prototype drug, the orange nodes represent the primary metabolites, the green nodes represent secondary metabolites and the bule nodes represent tertiary metabolites in DMCs.
3.2 DPIs construction based on the mass fragmentation behavior of NMF and MF
DPIs were vital for metabolite identification. NMF and MF were selected as the subjects to ascertain the DPIs based on the comprehensive ESI-MS/MS information of NMF (Fig. 2) and MF standards. In negative ion mode, NMF and MF showed [M−H]- ions at m/z 583.12936 (C25H27O16, −0.451 ppm) and m/z 421.07708 (C19H17O11, 0.722 ppm), respectively. Then, NMF further afforded a series of DPIs at m/z 493, m/z 463, m/z 421, m/z 331, m/z 313, m/z 301, m/z 273 and m/z 259 owing to the respective loss of C3H6O3 (90 Da), C4H8O4 (120 Da), C6H12O6 (162 Da), C6H12O6 + C3H6O3 (252 Da), C12H16O7 (270 Da), C6H12O6 + C4H8O4 (280 Da), C6H12O6 + C4H8O4 + CO (310 Da) and 2 C6H12O6 (324 Da) (Zhang et al., 2011). Since MF only has one less glucose than NMF, their fragmentation pathways might be similar, too. Thus, the products ions of NMF at m/z 493 ± X, m/z 463 ± X, m/z 421 ± X, m/z 331 ± X, m/z 313 ± X, m/z 301 ± X, m/z 273 ± X, m/z 259 ± X [X = molecular weight of substituent groups, such as 14 (CH2), 162 (Glc), 176 (GluA), 80 (SO3) and 16 (O), etc] could also be observed. The methylated metabolite of NMF was taken as an example, which yielded the fragment ions at m/z 507 (m/z 493 + CH2), m/z 477 (m/z 463 + CH2), m/z 435 (m/z 421 + CH2), m/z 345 (m/z 331 + CH2), m/z 327 (m/z 313 + CH2), m/z 315 (m/z 301 + CH2), m/z 287 (m/z 273 + CH2), and m/z 273 (m/z 259 + CH2), which could be applied for the subsequent metabolite identification.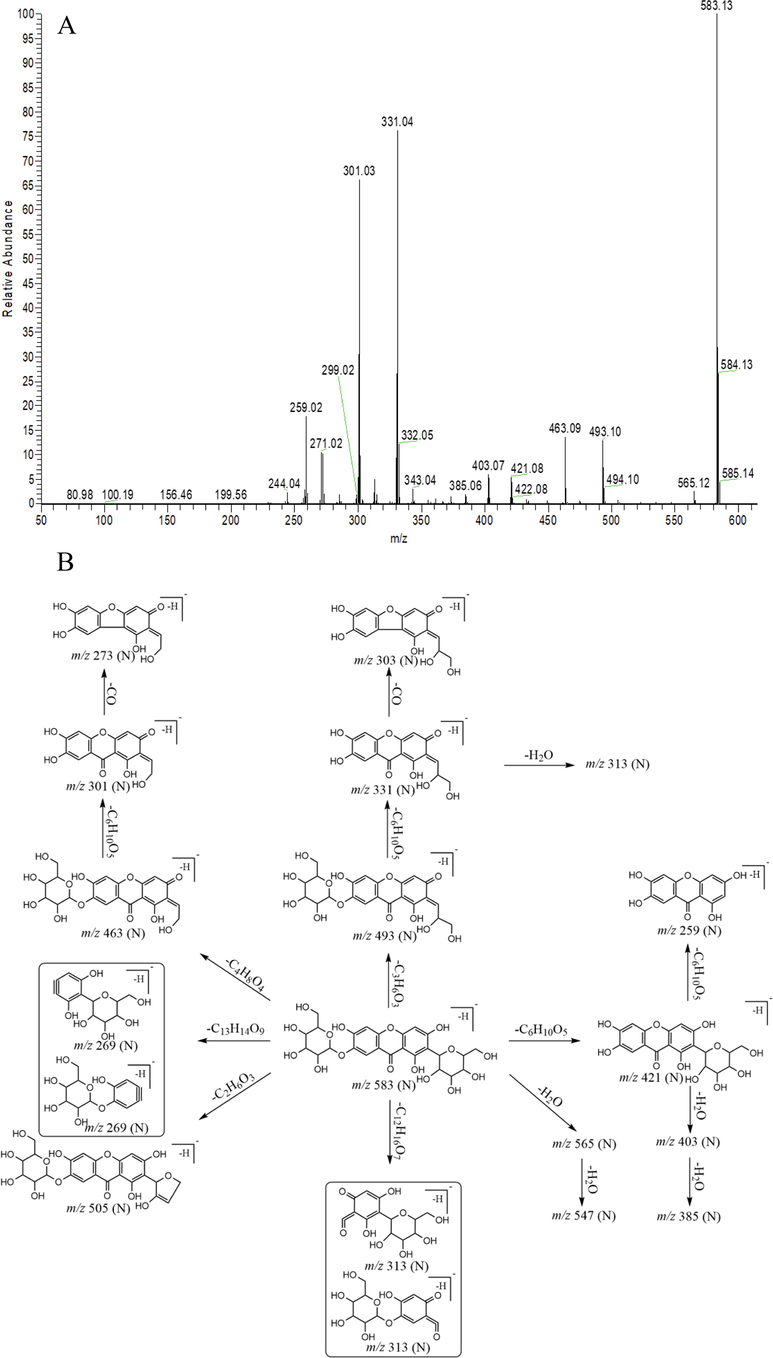
The ESI-MS2 spectrum (A) and mass fragmentation behavior (B) of NMF in negative ion mode.
3.3 Identification of NMF metabolites
A total of 61 NMF metabolites (prototype drug, included) were detected and characterized in SD rat plasma, urine, faeces, liver and liver microsomes (Table 1) based on DMCCs-based analytical strategy coupled with UHPLC-Q-Exactive Orbitrap MS. Among them, 32 metabolites were found in rat urine, 30 in rat plasma, 12 in rat liver, 9 metabolites in liver microsomes and 8 in rat faeces, respectively. High resolution extracted ion chromatography (HREIC) of all the NMF metabolites were illustrated in Fig. 3. Notes: *, Reference; PS, plasma SPE extraction; PA, plasma acetonitrile precipitation; PM, plasma methanol precipitation; US, urine SPE extraction; LC, Prototype drug liver microsomes treatment; FS, faeces SPE extraction; LS, liver SPE extraction.
Peak
tR(min)
Ion Mode
Formula[M-H]-/[M + H]+
Theoretical Mass (m/z)
Experimental Mass (m/z)
RDB
Error (ppm)
MS2 Fragment Ions
Identification/Reactions
Distribution
M0*
8.02
N
C25H27O16
583.12936
583.12891
12.5
−0.451
583(1 0 0), 331(73), 301(62), 259(17), 463(13), 493(13), 421(5), 313(5), 273(2), 505(1)
NMF
PS, PA, LC, LS
8.07
P
C25H29O16
585.14501
585.14392
11.5
−1.091
273(1 0 0), 303(78), 567(39), 585(26), 299(24), 465(17), 285(11), 405(7), 260(6), 271(3), 423(1)
M1
7.90
N
C25H25O17
597.10956
597.10919
13.5
0.027
597(1 0 0), 301(88), 331(79), 598(25), 259(21), 421(16), 271(11), 403(8), 272(8), 275(7), 313(6)
MF glucuronidation/isomers
US, FS
M2
9.27
N
C25H25O17
597.10956
597.10972
13.5
0.647
597(1 0 0), 301(80), 331(74), 259(22), 421(19), 271(15), 243(13), 403(5), 477(4)
MF glucuronidation/isomers
PA
M3
9.35
N
C25H25O17
597.10956
597.10870
13.5
−0.794
259(1 0 0), 435(25), 260(13), 597(10), 436(5), 598(2), 421(1), 331(1), 301(1)
MF glucuronidation/isomers
PS, US, LS
9.35
P
C25H27O17
599.12482
599.12335
12.5
−2.460
261(1 0 0), 437(30), 438(6), 245(3), 423(2), 599(1)
M4
10.60
N
C13H7O6
259.02426
259.02338
10.5
−3.408
259(1 0 0), 221(55), 215(6),175(2), 154(2)
Isomer of NA
PA, FS
M5
10.75
N
C13H7O6
259.02426
259.02380
10.5
−1.787
259(1 0 0), 221(48), 215(4), 175(3), 154(3)
Isomer of NA
PA
M6
11.97
N
C13H7O6
259.02426
259.02420
10.5
−0.242
259(1 0 0), 221(33), 260(14), 219(11), 215(6), 223(5), 231(1)
NA
PA, FS, LS
M7
9.54
N
C20H19O11
435.09273
435.09274
11.5
0.009
435(1 0 0), 287(30), 272(23), 436(22), 315(21), 259(5), 288(4), 273(3), 316(3), 271(2)
MF methylation/isomers
PM, FS
M8
10.12
N
C20H19O11
435.09273
435.09244
11.5
−0.681
272(1 0 0), 435(99), 315(58), 345(26), 259(19), 273(19)
MF methylation/isomers
PA
10.13
P
C20H21O11
437.10838
437.10754
10.5
−1.936
287(1 0 0), 317(82), 341(39), 419(35), 313(33), 353(25), 260(19), 401(19), 371(17), 437(14)
M9
10.23
N
C18H15O11
407.06043
407.06241
11.5
2.393
231(1 0 0), 407(86), 405(29), 113(18), 232(11)
NA decarbonylation and glucuronidation
PA
M10
11.94
N
C13H7O9S
338.98107
338.98099
10.5
−0.258
259(1 0 0), 260(14), 338(13), 321(3), 164(3)
NA sulfation
PS, PA
M11
7.99
N
C25H23O18
601.08843
601.08813
14.5
−0.505
259(1 0 0), 601(45), 260(12), 435(12), 258(3), 175(1)
NA bis-glucuronidation/isomers
US, LS
8.00
P
C25H25O18
603.10408
603.10260
13.5
−2.428
260(1 0 0), 603(22), 437(18), 262(13), 438(4)
M12
9.01
N
C25H23O18
601.08843
601.08801
14.5
−0.701
259(1 0 0), 435(37), 260(13), 601(13), 436(7), 301(1), 175(1)
NA bis-glucuronidation/isomers
US
9.02
P
C25H25O18
603.10408
603.10278
13.5
−2.134
260(1 0 0), 437(19), 262(13), 603(11), 438(4)
M13
9.27
N
C25H23O18
601.08843
601.08856
14.5
0.199
259(1 0 0), 435(42), 601(3), 315(3), 175(1)
NA bis-glucuronidation/isomers
PA, PM
M14
9.42
N
C25H23O18
601.08843
601.08777
14.5
−1.094
259(1 0 0), 435(46), 260(13), 601(2)
NA bis-glucuronidation/isomers
PS, US
9.43
P
C25H25O18
603.10408
603.10254
13.5
−2.526
260(1 0 0), 437(28), 262(13), 603(10), 438(6)
M15
5.33
P
C25H25O18
603.10408
603.10266
13.5
−2.330
260(1 0 0), 437(27), 262(12), 603(8), 438(6)
NA bis-glucuronidation/isomers
US
M16
9.74
N
C19H15O12
435.05635
435.05627
12.5
−0.185
259(1 0 0), 435(31), 260(13), 301(2)
NA glucuronidation/isomers
PA
M17
10.59
N
C19H15O12
435.05635
435.05597
12.5
−0.875
259(1 0 0), 435(29), 260(10), 395(2), 329(2), 302(2)
NA glucuronidation/isomers
PS, PA, PM, LS
M18
10.75
P
C19H17O12
437.07200
437.07135
11.5
−1.489
260(1 0 0), 262(13), 437(8), 243(3), 438(2)
NA glucuronidation/isomers
PA, US
M19*
9.08
N
C19H17O11
421.07708
421.07739
11.5
0.722
301(1 0 0), 421(84), 331(57), 259(25), 273(8), 313(7), 285(5), 260(5), 403(4)
MF
PA, PM, US, LC, FS, LS
9.08
P
C19H19O11
423.09273
423.09183
10.5
−2.142
273(1 0 0), 303(83), 299(31), 405(31), 387(21), 423(20), 260(7)
M20
9.38
P
C19H19O11
423.09273
423.09164
10.5
−2.591
260(1 0 0), 423(8), 299(1), 273(1), 303(1)
MF isomer
US
M21
7.73
P
C25H25O17
597.10917
597.10742
13.5
−2.937
245(1 0 0), 421(15), 246(12), 597(12), 141(5), 422(4)
NA bis-glucuronidation and dehydroxylation/Isomer
US
M22
8.01
N
C26H25O18
625.10408
625.10382
14.5
−0.430
273(1 0 0), 258(64), 449(50), 274(14), 625(10), 450(8)
NA bis-glucuronidation and methylation/isomer
US
8.03
P
C26H27O18
627.11973
627.11823
13.5
−2.406
275(1 0 0), 456(19), 451(18), 286(15), 627(2)
M23
8.38
N
C26H25O18
625.10408
625.10358
14.5
−0.814
273(1 0 0), 258(96), 449(86), 625(32), 450(18)
NA bis-glucuronidation and methylation/isomer
US
8.40
P
C26H27O18
627.11973
627.11890
13.5
−1.337
275(1 0 0), 451(16), 627(14), 276(14), 452(3)
M24
9.52
N
C26H25O18
625.10408
625.10382
14.5
−0.430
449(1 0 0), 258(84), 273(69), 450(22), 625(2)
NA bis-glucuronidation and methylation/isomer
US
M25
9.90
N
C26H25O18
625.10408
625.10382
14.5
−0.430
273(1 0 0), 449(37), 625(29), 113(19), 274(17), 259(9), 175(6), 450(5)
NA bis-glucuronidation and methylation/isomer
PS, US
9.89
P
C26H27O18
627.11973
627.11859
13.5
−1.832
275(1 0 0), 451(22), 276(14), 627(8), 452(5), 260(3)
M26
10.40
N
C19H15O11
419.06043
419.06049
12.5
0.129
243(1 0 0), 113(24), 244(13), 175(11), 418(8), 419(4)
NA glucuronidation and dehydroxylation/isomer
PS, PA, FS, LS
M27
10.68
N
C19H15O11
419.06043
419.06016
12.5
−0.659
243(1 0 0), 113(40), 419(26), 175(17), 244(13)
NA glucuronidation and dehydroxylation/isomer
PS, LS
M28
10.84
N
C19H15O11
419.06043
419.06068
12.5
0.582
243(1 0 0), 419(22), 244(14), 199(8), 113(6), 175(1)
NA glucuronidation and dehydroxylation/isomer
PS, PM
M29
10.46
N
C19H15O10
403.06652
403.06631
12.5
−0.525
227(1 0 0), 113(26), 85(16), 228(13), 175(4), 403(3), 209(2)
Xanthone glucuronidation and bis-hydroxylation /isomer
PS
10.46
P
C19H17O10
405.08217
405.08099
11.5
−2.917
229(1 0 0), 184(29), 104(26), 230(18), 405(12), 387(5)
M30
11.44
N
C19H15O10
403.06652
403.06635
12.5
−0.425
227(1 0 0), 113(30), 403(17), 228(14), 175(11)
Xanthone glucuronidation and bis-hydroxylation /isomer
PM
M31
12.52
N
C19H15O10
403.06652
403.06628
12.5
−0.599
227(1 0 0), 113(59), 175(26), 403(26), 228(11), 359(3)
Xanthone glucuronidation and bis-hydroxylation /isomer
PS, PM
12.51
P
C19H17O10
405.08217
405.08136
11.5
−2.003
229(1 0 0), 230(14), 104(11), 184(8), 141(7), 405(5)
M32
9.95
P
C19H17O10
405.08217
405.08145
11.5
−1.781
229(1 0 0), 230(13), 405(6), 184(3), 104(2), 231(1)
Xanthone glucuronidation and bis-hydroxylation /isomer
PS, LS
M33
9.89
N
C19H15O14S
499.01825
499.01797
12.5
−0.562
243(1 0 0), 499(42), 419(28), 244(17), 420(7), 283(5), 322(5)
NA glucuronidation, dehydroxylation and sulfation/isomer
PS, LS
M34
10.06
N
C19H15O14S
499.01825
499.01846
12.5
0.420
243(1 0 0), 88(43), 499(34), 419(28), 183(15), 440(15)
NA glucuronidation, dehydroxylation and sulfation/isomer
PS
M35
10.06
N
C20H17O12
449.07200
449.07214
12.5
0.310
273(1 0 0), 258(93), 113(25), 274(15), 449(14), 85(12), 259(11), 175(10)
NA glucuronidation and methylation/isomer
PS, LC
M36
10.76
N
C20H17O12
449.07200
449.07199
12.5
−0.024
258(1 0 0), 113(59), 273(49), 449(28), 85(26), 259(21), 175(21), 275(16)
NA glucuronidation and methylation/isomer
PS, US, FS, LS
10.75
P
C20H19O12
451.08765
451.08602
11.5
−3.394
275(1 0 0), 451(36), 260(4), 141(3), 113(3)
M37
11.40
N
C20H17O12
449.07200
449.07187
12.5
−0.291
273(1 0 0), 113(21), 85(11), 259(9), 175(8), 230(5), 449(3)
NA glucuronidation and methylation/isomer
PS, US, FS
11.40
P
C20H19O12
451.08765
451.08608
11.5
−3.260
275(1 0 0), 331(14), 451(11), 261(6), 273(4)
M38
10.52
P
C20H19O12
451.08765
451.08676
11.5
−1.975
275(1 0 0), 331(19), 451(7), 261(4), 243(1)
NA glucuronidation and methylation/isomer
US
M39
11.09
P
C20H19O12
451.08765
451.08621
11.5
−3.194
275(1 0 0), 331(14), 451(14), 261(6)
NA glucuronidation and methylation/isomer
US, LS
M40
12.62
N
C14H9O9S
352.99672
352.99664
10.5
−0.248
273(1 0 0), 258(54), 274(18), 352(14), 259(10)
NA sulfation and methylation
PS
M41
3.15
N
C31H33O23
773.14126
773.14142
15.5
0.205
259(1 0 0), 601(37), 435(15), 260(14), 773(10), 593(4), 436(3), 301(2), 597(2)
MF bis-glucuronidation/isomer
US
M42
2.63
P
C31H35O23
775.15691
775.15485
14.5
−2.660
261(1 0 0), 437(31), 262(13), 438(6), 423(6)
MF bis-glucuronidation/isomer
US
M43
10.45
P
C14H11O6
275.05556
275.05478
9.5
−2.846
275(1 0 0), 260(26), 276(15), 126(6), 233(2), 247(2), 167(1), 123(1)
NA methylation/isomer
US
M44
10.75
P
C14H11O6
275.05556
275.05429
9.5
−4.628
275(1 0 0), 259(18), 276(15), 243(4), 233(2)
NA methylation/isomer
US
M45
11.09
P
C14H11O6
275.05556
275.05435
9.5
−4.410
275(1 0 0), 259(26), 276(15), 232(2), 233(2)
NA methylation/isomer
US
M46
11.40
P
C14H11O6
275.05556
275.05435
9.5
−4.410
275(1 0 0), 259(23), 276(15), 273(4), 232(2), 233(1)
NA methylation/isomer
US
M47
12.97
P
C14H11O6
275.05556
275.05472
9.5
−3.065
275(1 0 0), 259(18), 276(16), 243(4), 233(1)
NA methylation/isomer
US
M48
13.46
P
C14H11O6
275.05556
275.05469
9.5
−3.174
275(1 0 0), 259(26), 276(15), 260(4), 233(1)
NA methylation/isomer
US
M49
9.30
P
C13H9O5
245.04499
245.04375
9.5
−4.547
130(1 0 0), 147(20), 129(19), 116(7), 245(4), 153(1), 109(1)
NA dehydroxylation/isomer
US
M50
10.42
P
C13H9O5
245.04499
245.04407
9.5
−3.788
245(1 0 0), 153(2), 217(1), 109(1), 137(1)
NA dehydroxylation/isomer
US
M51
10.85
P
C13H9O5
245.04499
245.04405
9.5
−3.870
245(1 0 0), 246(14), 217(1), 137(1), 109(1)
NA dehydroxylation/isomer
US
M52
13.17
P
C13H9O5
245.04499
245.04422
9.5
−3.176
245(1 0 0), 217(2), 177(1), 137(1), 109(1)
NA dehydroxylation/isomer
US
M53
9.94
N
C24H23O16
567.09860
567.09882
13.5
0.372
259(1 0 0), 391(54), 260(13), 392(10), 567(9)
MF glucuronidation and de-hydroxymethylation/isomer
US
9.95
P
C24H25O16
569.11425
569.11316
12.5
−1.932
260(1 0 0), 437(39), 262(12), 438(8), 569(1)
M54
10.13
N
C24H23O16
567.09860
567.09851
13.5
−0.175
259(1 0 0), 392(10), 435(2), 567(1), 175(1)
MF glucuronidation and de-hydroxymethylation/isomer
US
M55
8.01
N
C25H27O18
605.11973
605.11908
12.5
−1.071
605(1 0 0), 301(47), 606(33), 331(23), 259(23), 581(17), 495(17), 421(16)
NMF bis-hydroxylation
LC
M56
8.72
N
C26H29O17
603.14047
603.14081
12.5
0.548
603(1 0 0), 301(43), 331(38), 463(11), 302(9), 313(8), 259(7), 343(7), 493(6)
NMF hydroxylation and methylation
LC
M57
9.60
N
C26H29O16
597.14555
597.14557
12.5
0.018
597(1 0 0), 345(53), 315(49), 477(12), 346(11), 507(10), 435(2)
NMF methylation
LC
9.62
P
C26H31O16
599.16020
599.16016
11.5
−1.752
287(1 0 0), 317(82), 581(59), 599(37), 313(28), 353(25), 365(21)
M58
8.43
N
C25H25O16
581.11425
581.11438
13.5
0.208
581(1 0 0), 299(52), 331(26), 259(24), 301(24), 271(16), 401(14), 419(12)
NMF hydroxylation and dehydration
LC
M59
9.19
N
C26H27O16
595.12990
595.13013
13.5
0.371
595(1 0 0), 433(27), 596(27), 313(24), 343(18)
NMF methylation hydroxylation dehydration
LC
M60
7.95
P
C25H27O15
567.13499
567.13422
12.5
−0.226
273(1 0 0), 567(59), 327(52), 299(29), 405(9), 260(6), 435(6), 447(1)
NMF dehydration/isomers
LC
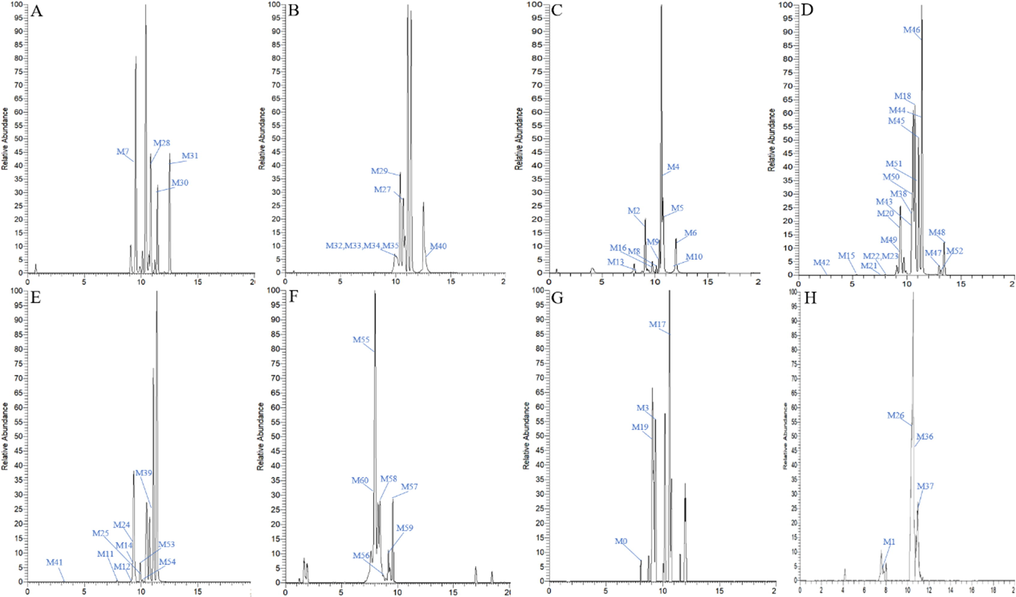
High resolution extracted ion chromatograms for 61 NMF metabolites (A-C for plasma, D-E for urine, F for liver microsomes, G for liver, H for feaces).
3 primary metabolites including NMF, MF and NA were regarded as DMCCs in the following analysis were preliminarily observed. M0 produced [M−H]- ion at m/z 583.13045 with the retention time of 8.02 min (C25H27O16, mass error of −0.451 ppm). It afforded the characteristic ions at m/z 493 [M−H−C3H6O3]-, m/z 463 [M−H−C4H8O4]-, m/z 421 [M−H−Glc]-, m/z 331 [M−H−Glc−C3H6O3]-, m/z 313 [M−H−C12H16O7]-, m/z 301 [M−H−Glc−C4H8O4]- and m/z 259 [M−H−2Glc]-, which were consisted with those of NMF reference standard. Consequently, M0 was accurately identified as NMF. M19 possessed the fragment ions at m/z 331 [M−H−Glc−C3H6O3]-, m/z 313 [M−H−C12H16O7]-, m/z 301 [M−H−Glc−C4H8O4]- and m/z 259 [M−H−2Glc]-. Consequently, it was unambiguously characterized as MF. Additionally, M6 (C13H7O6, mass error of −0.242 ppm) possessed [M−H]- ion at m/z 259.02420 with the retention time of 11.95 min, which was 162 Da less than that of MF. The characteristic product ions at m/z 231 [M−H−CO]- and m/z 215 [M−H−CO−O]- were detected in its ESI-MS2 spectra, indicating M6 could be judged as NA (Liu et al., 2018).
With the retention time of 7.90 min, 9.27 min and 9.35 min, M1-M3 displayed the deprotonated molecular ions (C25H25O17) at m/z 597.10919, m/z 597.10972 and m/z 597.10870 with mass errors of 0.027, 0.647 and −0.794 ppm, respectively. Additionally, they were 176 Da more than that of MF, and the fragment ions at m/z 421 [M−H−GluA]-, m/z 331 [M−H−GluA−C3H6O3]-, m/z 303 [M−H−GluA−C4H8O4]- and m/z 259 [M−H−GluA−Glc]- have been all observed in their ESI-MS2 spectra, which indicated glucuronidation reaction might occur (Fig. 4). Consequently, M1-M3 were all speculated to be glucuronidation products of MF (Day 2000).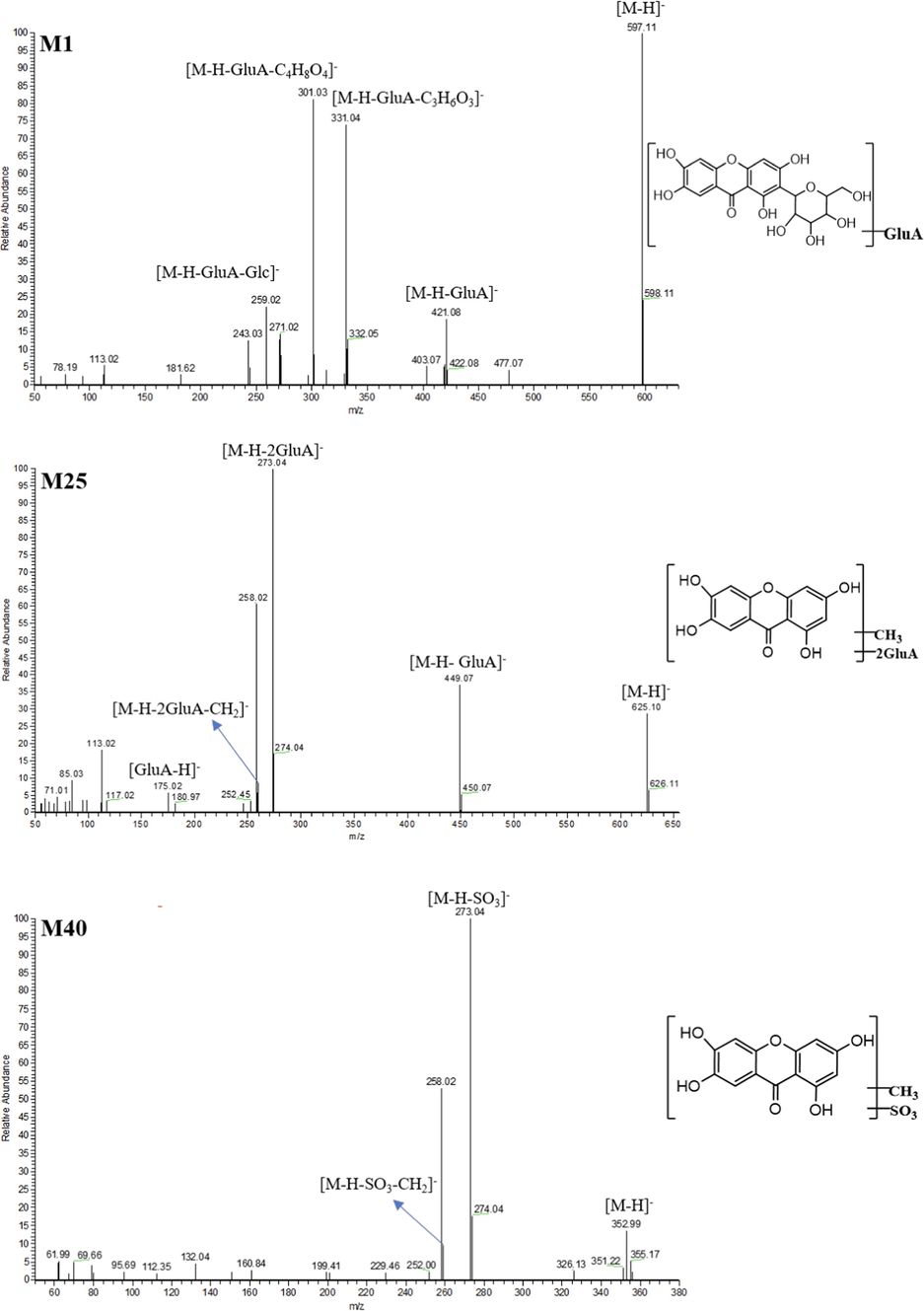
The ESI-MS2 spectra and chemical structures of M1, M25 and M40.
Both M7 and M8 yielded their significant [M−H]- ions at m/z 435.09274 and m/z 435.09244 (C20H19O11, mass errors of 0.009 and −0.681 ppm), which were eluted at 9.54 min and 10.12 min, respectively. The methylation reaction could be deduced to occur because they were 14 Da more than that of MF. In their ESI-MS2 spectra, the characteristic fragment ions at m/z 345, m/z 315, m/z 273 and m/z 259 were generated by the subsequent loss of C3H6O3, C4H8O4, Glc and (Glc + CH2). Thus, both M7 and M8 were tentatively characterized as methylation products of MF.
M41 and M42 were respectively eluted at 3.15 min and 2.63 min, and showed the deprotonated molecular ions at m/z 773.14142 and m/z 775.15485 (C31H33O23 and C31H35O23, mass errors of 0.205 and −2.660 ppm). M41 produced the fragment ions at m/z 601 [M−H−Glc]-, m/z 597 [M−H−GluA]-, m/z 435 [M−H−Glc−GluA]- and m/z 259 [M−H−Glc−2GluA]- in its ESI-MS2 spectrum. Meanwhile, M42 possessed the essential fragment ions at m/z 423 [M + H-2GluA]+ and m/z 261 [M + H-2GluA-Glc]+. Therefore, M41 and M42 could be deduced as bis-glucuronidation products of MF.
M9 generated the deprotonated molecular [M−H]- ion at m/z 407.06241 (C18H15O11, mass error of 2.393 ppm) with the retention time at 10.23 min. It was 148 Da than that of NA, manifesting that decarbonylation and glucuronidation reactions in vivo might have occurred. In its ESI-MS2 spectrum, the fragment ion at m/z 231 generated from the loss of GluA was observed. Therefore, M9 was tentatively identified as decarbonylation and glucuronidation product of NA.
M10 was 79 Da more than that of NA, which gave rise to [M−H]- ion at m/z 338.98099 with mass error of 0.206 ppm and the retention time of 11.94 min. It generated the fragment ion at m/z 259 [M−H−SO3]- in its ESI-MS2 spectrum. Based upon this, M10 was tentatively identified as sulfation product of NA.
M16 and M17 possessed their [M−H]- ions at m/z 435.05627 and m/z 435.05597 (C19H15O12, mass errors of −0.185 and −0.875 ppm) in negative ion mode, respectively. They were 176 Da more than that of NA, indicating that glucuronidation reaction has occurred in vivo. The fragment ion at m/z 259 [M−H−GluA]- was observed in their ESI-MS2 spectra. In addition, M18, possessing the experimental [M + H]+ ion at m/z 437.07135 (C19H17O12, mass error of −1.489 ppm), was detected at 10.75 min in positive ion mode. The fragment ion at m/z 259 suggested that glucuronic acid has been lost during the fragmentation process, and thus M16-M18 could be attributed to be glucuronidation products of NA.
M11, M12 and M14 afforded the same theoretical [M + H]+/[M−H]- ions at m/z 603.10408 (C25H25O18, mass error < ±3 ppm)/m/z 601.08843 (C25H23O18, mass error < ±3 ppm) in positive and negative ion modes, respectively. These 3 metabolites were 352 Da more massive than that of apigenin, implying that they might be bis-glucuronidation products of NA. In their ESI-MS2 spectra, the base peak ion at m/z 259 [M−H−2GluA]-, the fragment ions at m/z 435 [M−H−GluA]- and m/z 175 [GluA-H]- in negative ion mode, m/z 260 [M + H-2GluA]+ and m/z 177 [M + H-GluA]+ in positive ion mode were respectively observed. Furthermore, M13 and M15 showed the same fragmentation behaviors, and thus M11-M15 were speculated to be bis-glucuronidation products of NA.
Six isomeric metabolites, M43-M48, displayed [M + H]+ ions at m/z 275.05556 (C14H11O6, mass error < ± 5 ppm) These metabolites were 14 Da more than that of NA. Moreover, the characteristic fragment ions at m/z 260 [M + H-CH2]+, m/z 247 [M + H-CO]+, m/z 233 [M + H-CH2-CO]+, m/z 167 [C8H6O4 + H]+ and m/z 123 [C7H6O2 + H]+ indicated RDA reaction have occurred. Therefore, M43-M48 were all deduced as isomeric methylation products of NA.
M49-M52 were respectively eluted at 9.30 min, 10.42 min, 10.85 min and 13.17 min, and showed the protonated molecular ions (C13H9O5) at m/z 245.04375, m/z 245.04407, m/z 245.04405 and m/z 245.04422 with the mass errors within ± 5 ppm. In their ESI-MS2 spectrum, a battery of characteristic fragment ions at m/z 217 [M + H-CO]+, m/z 137 [C7H4O3 + H]+ and m/z 109 [C6H4O2 + H]+ were observed. Additionally, their [M + H]+ ions were 16 Da less massive than that of NA, which suggested dehydroxylation reaction have occurred in vivo. Therefore, M49-M52 were all characterized as the dehydroxylation products of NA.
M40 generated [M−H]- ion at m/z 352.99664 (C14H9O9S, mass error of −0.248 ppm) with the retention time of 12.62 min. It was 93 (14 + 79) Da more massive than that of NA, implying that it might be the sulfated and methylated product of NA. Fragment ions at m/z 273 [M−H−SO3]- and m/z 259 [M−H−SO3−CH2]- were exhibited in its ESI-MS2 spectra (Fig. 4). Hence, M40 was characterized as sulfation and methylation product of NA.
In negative ion mode, M57 produced [M−H]- ion at m/z 597.14557 (C26H29O16) with mass error of 0.018 ppm. In its ESI-MS2 spectrum, the fragment ions were assigned as m/z 507 [M−H−C3H6O3]-, m/z 477 [M−H−C4H8O4]-, m/z 435 [M−H−Glc]-, m/z 345 [M−H−Glc−C3H6O3]-, m/z 315 [M−H−Glc−C4H8O4]- and m/z 273 [M−H−2Glc]-. In addition, it was 14 Da massive than that of NMF, demonstrating the methylation reaction might occur. Based on this, M57 was deduced as methylation product of NMF (van der Woude et al., 2004). M59 produced the deprotonated molecular ion at m/z 595.13013 (C26H27O16, mass error of 0.371 ppm) in negative ion mode, which was 2 Da more massive than that of M57. Characteristic fragment ions at m/z 433 [M−H−Glc]-, m/z 343 [M−H−Glc−C3H6O3]-, m/z 313 [M−H−Glc−C4H8O4]- and m/z 285 [M−H−Glc−C4H8O4−CO]- were all observed correspondingly. Hence, M59 was identified as methylation + hydroxylation + dehydration products of NMF. M58 was also 2 Da less massive than NMF, and its fragment ions at m/z 419 [M−H−Glc]-, m/z 299 [M−H−Glc- C4H8O4]- and m/z 259 [C13H8O6-H]- in negative ion mode were detected. And thus, M58 was tentatively identified as hydroxylation + dehydration product of NMF (Morand et al., 2000).
M21 was eluted at 7.73 min and showed [M + H]+ ion at m/z 597.10742. Its formula was speculated as C25H25O17 with mass error of −2.937 ppm. It was 16 Da less massive than that of M15, suggesting that dehydroxylation reaction occurred. In its ESI-MS2 spectrum, the DPIs were assigned as m/z 421 [M + H-GluA]+ and m/z 245 [M + H-2GluA]+. Thus, M21 was tentatively identified as the bis-glucuronidation and dehydroxylation product of NA. M22-M25 were 14 Da more than that of M15, and the DPIs at m/z 449 [M−H−GluA]-, m/z 273 [M−H−2GluA]- and m/z 259 [M−H−2GluA−CH2]- also demonstrated that methylation reaction might have occurred (Fig. 4). Therefore, M22-M25 were all characterized as bis-glucuronidation and methylation products of NA.
3.4 Proposed biotransformation pathways of NMF in rats
In our study, a total of 61 NMF metabolites (NMF included) were detected and characterized in vivo and in vitro. Moreover, we proposed its biotransformation pathways depicted in Fig. 5. The results demonstrated that the prototype drug first generated 2 the primary metabolites including MF and NA, which could be regarded as DMCCs (NMF included) for the subsequent analysis. And then, a large number of metabolites were mined on the basis of DMCCs analytical strategy. NMF mainly underwent deglucosylation, glucuronidation, methylation, sulfation, dihydroxylation and their composite reactions. Among them, the phase Ⅱ reactions and their composite reactions were predominated the biotransformation pathways, which indicated that the increased water solubility of drugs was beneficial to excrete after the biotransformation in vivo.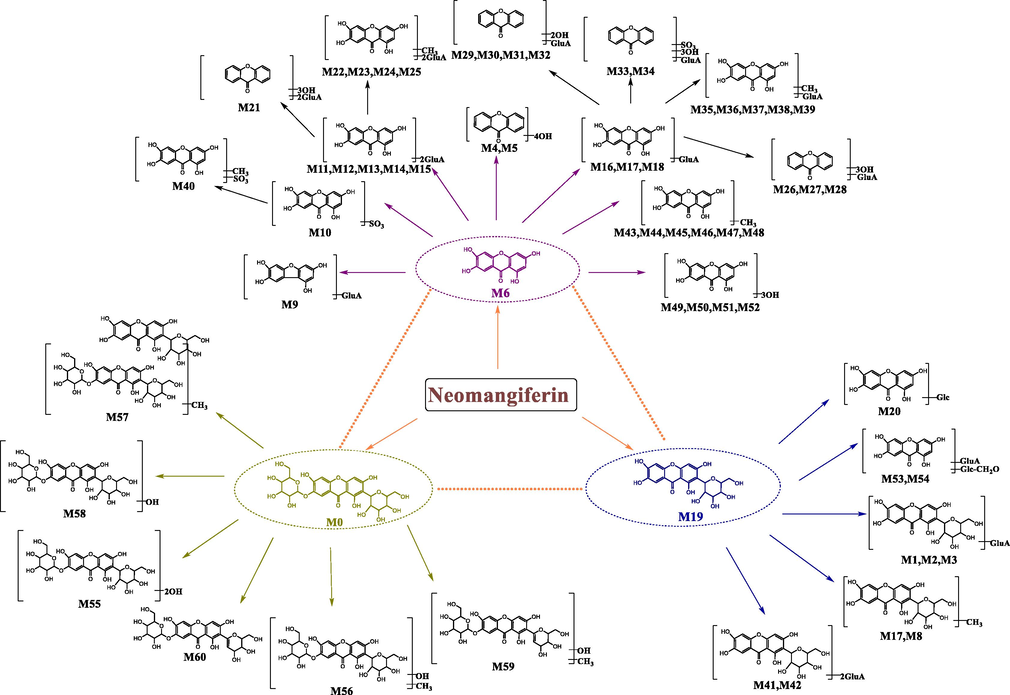
The proposed NMF biotransformation pathways of NMF in vivo and in vitro.
3.5 Network pharmacology analysis
DMCCs have been analyzed by network pharmacology to explore the action mechanism of NMF. We respectively searched 60, 73 and 72 targets of DMCCs by using Swiss Target Prediction database, and 98 were obtained after removing the repeated values. And then, 13,045 targets of inflammatory have been found by using GeneCards as well. 85 common targets were obtained by interaction analysis later (Fig. 6A). Integration between the metabolites and disease targets were imported into Cytoscape 3.8.0 software to construct the disease-target-metabolites network. There were 102 nodes and 288 edges, among which the red node represented disease and the yellow nodes represented 3 main metabolites. Besides, those 85 blue nodes represented the overlapping gene symbols between the disease and metabolites while the orange nodes represented other genes (Fig. 6B).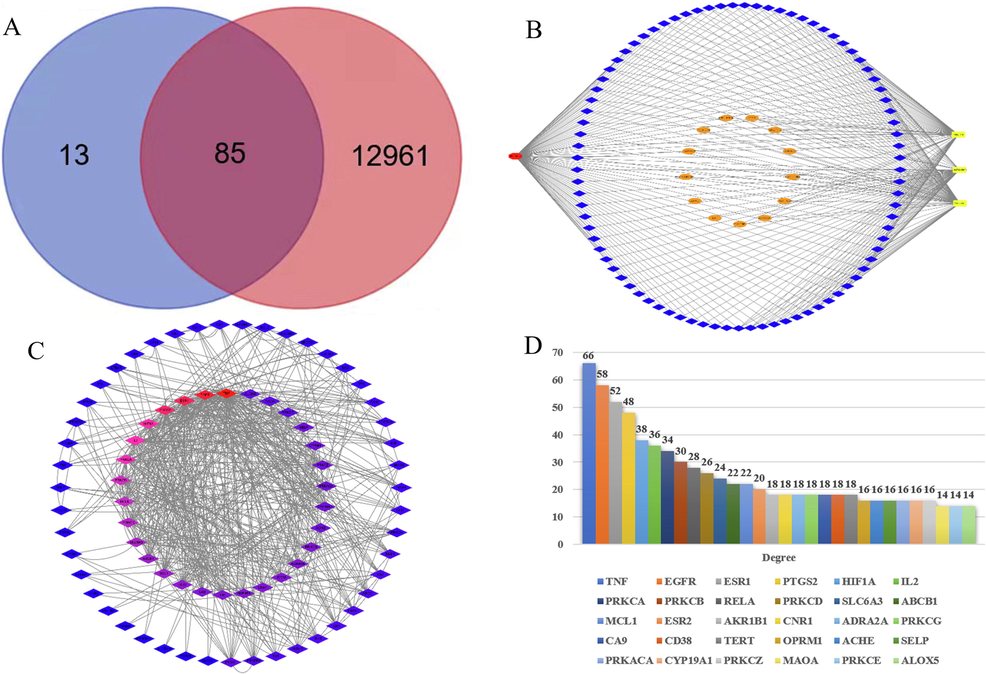
85 overlapping gene symbols between the 3 main metabolites (A), disease-target-metabolites network (B), Interaction network between targets (C) and 3 major metabolites core targets for anti-inflammatory (top 30) (D).
There were 77 nodes and 560 edges in the PPI network diagram, and the average degree value was 14.55. Node represented the target and color represented the degree value of target. A target possessed a higher degree of red and a lower degree of blue (Fig. 6C). The top 30 targets of node degree value were selected to draw a histogram. Apparently, TNF, EGFR, ESR1, PTGS2, HIF1A, IL-2, PRKCA and PRKCB were the core targets, which were twice greater than the average of degree value (Fig. 6D).
85 protein targets in the protein interaction network were imported into Metascape database for GO and KEGG enrichment analysis, and P < 0.01 pathways were selected. The top 10 biological processes, cellular components and molecular functions of GO were selected for the diagram. During the GO biological process, it mainly affected the one-carbon metabolic process, inflammatory response and response to hormone. As for the GO cellular component, it critically affected the integral component of presynaptic membrane, perinuclear region of cytoplasm and intrinsic component of presynaptic membrane. At last, it vitally affected the carbonate dehydratase activity, hydro-lyase activity, carbon–oxygen lyase activity and calcium-dependent protein kinase C activity at molecular function. KEGG pathway analysis demonstrated that nitrogen metabolism, pathways in cancer, inflammatory mediator regulation of TRP channels and chemical carcinogenesis-receptor activation were mostly related to anti-inflammation (Fig. 7).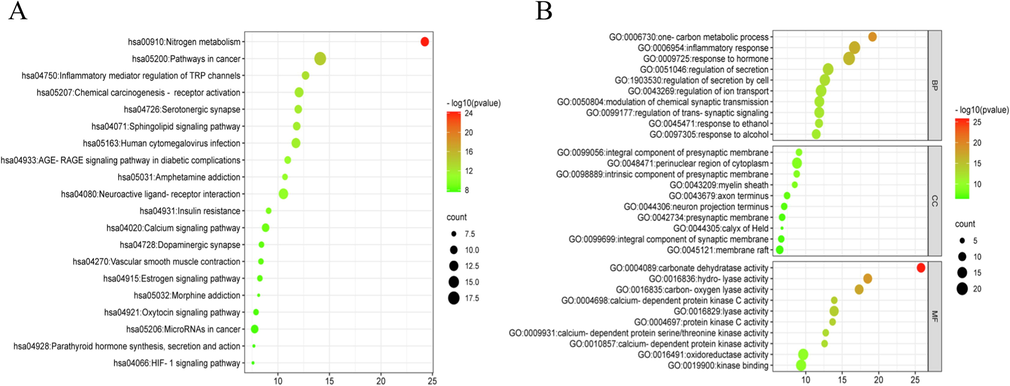
Bubble diagrams of KEGG (A) and GO (B) analysis, Note: Biological processes (BP), Cellular components (CC) and Molecular functions (MF).
4 Discussion
To clarify the metabolic pathways of drug has been playing an increasingly important role in exploring the action mechanism of drug efficacy. Nevertheless, few strategies were universally applied to clarify metabolic pathways of drug. Therefore, developing a systematic strategy of drug metabolism has become a research hots pot until now. Based on the DMCs analytical strategy we ever established, we further developed DMCCs-based strategy in the present study. We screened and assigned NMF, MF and NA as DMCCs. It is worth noting that DMCCs weren’t isolated and there might be some transformation relationships between them (Liu et al., 2020). For instance, NMF could generate MF and NA, while MF and NA could also be transmuted into each other.
In this study, a total of 61 metabolites were screened and identified in biological samples after the oral administration of NMF. By comparing metabolites, there were major 2 steps in the metabolic process of NMF. Firstly, NMF generated MF or NA by loss of 1 or 2 glucosyl groups, correspondingly. Afterwards, MF or NA underwent further reactions such as glucuronidation, sulfation, methylation, hydroxylation and their composite reactions. Among them, phase Ⅱ metabolism and its complicated reactions were predominant, which could produce more water-soluble metabolites so as to be beneficial to the drug excretion. However, the immediate binding reaction products of NMF in vivo were barely detected in the present study. This might be due to the presence of two glucosyls in NMF, which made the steric hindrance of the 3 hydroxyl binding sites, contributing to Ⅱ phase reaction difficult to occur. Study have shown that given the C2-OH and C7-OH position occupied by two glucosyl molecules, and thus NMF has a low exposure in vivo, and it mainly transformed into MF and NA (Xie et al., 2018).
The results also demonstrated that the 18 metabolites in rat plasma obtained by SPE cartridge, which were 10 and 5 more than those obtained by methanol or acetonitrile precipitation, respectively. It indicated that the SPE cartridge was the most suitable method to acquire NMF metabolites in plasma compared to the other two methods. Acetonitrile and methanol could only be served as a dilution and extraction during sample precipitation. Additionally, a total of 32 and 30 metabolites were found in rat urine and plasma, respectively. Apparently, most metabolites of NMF were detected in rat urine and plasma. Moreover, few metabolites were detected in rat faeces and liver, implying that the metabolites of NMF seldom excreted in faeces and liver. Therefore, urine and plasma samples would be more appropriate for investigation of NMF metabolites in rats after oral administration.
Furthermore, network pharmacology combined with systems biology research methods were preliminarily used to reveal the anti-inflammatory mechanism of NMF. It demonstrated that the effective form of drugs isn’t necessarily the natural phytochemical, but may be the metabolites produced by the biotransformation in the body. In the present study, a total of 8 core targets, TNF, EGFR, ESR1, PTGS2, HIF1A, IL-2, PRKCA and PRKBC, were screened out according to the target protein interaction network. Among them, TNF was primitively regarded as a circulating factor which could lead to tumor necrosis, while it was later identified as a critical regulator of the inflammatory response. TNF interacts with two distinct receptors, TNFR1 and TNFR2, which were differentially expressed on cells and tissues and initiate distinct and overlapping signaling pathways. These diverse signaling cascades caused a series of cellular responses, including cell survival, death, differentiation, proliferation and migration (Bradley., 2008). EGFR signaling pathway was regulated by multiple feedback regulators including mitogen-inducible gene 6 protein, which regulated the production of inflammatory mediators (TNF-α, il-1β) by inhibiting the over-activation of EGFR, thereby inhibiting the MAPKs signaling pathway (ERK1/2, p-38) (Chen et al., 2018).
The KEGG metabolic pathway analysis showed that the DMCCs ameliorated inflammation by regulating many pathways, and their regulation of nitrogen metabolism, pathways in cancer, inflammatory mediator regulation of TRP channels and chemical carcinogenesis-receptor activation were particularly important. In 1997, some scholars proposed a link between the process of nitrogen metabolism and the early development of inflammatory respiratory diseases in the body (Janson et al., 1997). Branched-chain amino acids (BCAAs, namely valine, leucine and isoleucine) could produce macromolecules such as nucleotides, which were nitrogen donors. It is well known that nucleotides are indispensable for the growth of human cancer cells. BCAAs metabolism, also known as nitrogen metabolism, is closely related to cancer development (Peng et al., 2020). However, inflammation is a key component of tumor progression, and many cancers occur at sites of infection, chronic irritation, and inflammation. Thereby, there may be a close relationship between nitrogen metabolism and inflammation. Tumor cells often overexpress pro-inflammatory mediators such as macrophage migration inhibitory factor (MIF), TNF-α, IL-6, IL-17, IL-12, IL-23, IL-10 and TGF-β, which can promote or inhibit tumor development. The tumor microenvironment is primarily coordinated by inflammatory cells and is an essential participant in the tumor process, promoting proliferation, survival and migration (Mantovani et al., 2008, Singh et al., 2019, Diakos et al., 2014). Therefore, the occurrence of cancer is inseparable from the appearance of inflammation.
GO enrichment analysis demonstrated that 3 vital metabolites had a great influence on the biological processes such as the one-carbon metabolic process and inflammatory response, the cellular components afforded the integral component of presynaptic membrane, perinuclear region of cytoplasm, intrinsic component of presynaptic membrane and the molecular functions like the carbonate dehydratase activity, calcium-dependent protein kinase C activity and oxidoreductase activity. Cell growth and proliferation require the construction of new cellular components, including proteins, lipids, and nucleic acids. Amino acid metabolism, including serine, glycine and threonine, and the carbon units they provide, can meet the basic requirements of building new cellular components for cell growth and proliferation. One-carbon metabolism uses serine to produce active one-carbon donors, such as 5, 10-methylene tetrahydrofolate, for many fundamental processes, including nucleotide and lipid synthesis (Kory et al., 2018). Modern cancer treatment stems in part from the assumption that folate antagonists can reduce the proliferation of malignant blood cells. The antagonism of folate metabolism and its downstream effectors, such as nucleotide metabolism, have been used in chemotherapy for more than 60 years (Locasale 2013). As mentioned above, there is a close relationship between cancer and inflammation. Consequently, one-carbon metabolism can affect the inflammatory process. Calcium-dependent protein kinase C can activate high-mobility group box 1 protein (HMGB1), and thus HMGB1 has been studied as a key mediator of inflammatory diseases, including sepsis (Oh et al., 2009). And some studies have already revealed found that calcium-dependent protein kinase C related to inflammatory reaction (Li et al., 1998, Boneh et al., 2015). Additionally, Lidocaine enhances AMPK phosphorylation by calcium- dependent protein kinase β and down-regulates inflammatory cytokines, IL-1β, and TNF-α, thereby reducing morphine tolerance (Zhang et al., 2017). Oxidoreductase activity, such as xanthine oxidoreductase, inhibition of xanthine oxidoreductase (XOR) has proven beneficial in a plethora of inflammatory disease processes thanks to a net reduction in pro-inflammatory oxidants and secondary nitrating species. Electrophilic nitrated fatty acid derivatives, such as nitro-oleic acid (OA-NO2) are also noted to display a broad spectrum of anti-inflammatory effects via interaction with critical signaling pathways. An alternative process in which nitrated fatty acids may extend anti-inflammatory actions is via inactivation of XOR, a process that is more effective than allo/oxypurinol-mediated inhibition (Kelley 2019). In general, our results indicated that the screened biological processes and pathways were closely related to the anti-inflammatory mechanism of the main of NMF metabolites.
5 Conclusion
In this study, a total of 61 metabolites (NMF, included) were screened and identified in rat plasma, urine, faeces, liver and liver microsomes using UHPLC-Q-Exactive Orbitrap MS coupled with DMCCs-based analytical strategy. Among them, 32 metabolites were accurately or tentatively characterized in rat urine, 30 in rat plasma,11 rat in liver, 10 in rat faeces and 9 in rat liver microsomes were, respectively. And the main metabolic reactions included deglucosylation, glucuronidation, methylation, sulfation, dihydroxylation and their composite reactions. Additionally, network pharmacology analysis revealed 85 common targets of disease-metabolites, and the key targets were TNF, EGFR, ESR1, PTGS2, HIF1A, IL-2, PRKCA and PRKCB. They exerted anti-inflammatory effects mainly through the pathways of inflammatory response, calcium-dependent protein kinase C activity, nitrogen metabolism, pathways in cancer, and so on. In general, our study developed a novel strategy to comprehensive elucidate the biotransformation pathways processes of NMF in vivo and in vitro, and also could provide vital reference for further understanding its anti-inflammatory mechanism. Moreover, the established strategy could be also generalized to the metabolism and action mechanism study of other natural products.
CRediT authorship contribution statement
Xianming Lan: Methodology, Writing – original draft. Yanan Li: Methodology. Haoran Li: Methodology. Shuyi Song: Methodology. Xiaoqing Yuan: . Hongyan Zhou: . Qimei Chen: . Jiayu Zhang: Conceptualization, Funding acquisition, Project administration, Methodology, Supervision.
Acknowledgments
This work has been financially supported by Taishan Young Scholar Program of Shandong (TSQN202103110), Young and Creative Team for Talent Introduction of Shandong Province (10073004), Shandong Province Chinese Herbal Medicine and Decoction Piece Standard Research Topic (2020-246) and Major Scientific and Technological Innovation Projects in Shandong Province (2021CXGC010511).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Signal transduction in inherited metabolic disorders: a model for a possible pathogenetic mechanism. J. Inherit. Metab. Dis.. 2015;38:729-740.
- [CrossRef] [Google Scholar]
- Simultaneous determination of active xanthone glycosides, timosaponins and alkaloids in rat plasma after oral administration of Zi-Shen Pill extract for the pharmacokinetic study by liquid chromatography-tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci.. 2010;878:1845-1854.
- [CrossRef] [Google Scholar]
- Mig6 reduces inflammatory mediators production by regulating the activation of EGFR in LPS-induced endotoxemia. J. Cell Physiol.. 2018;233:6975-6983.
- [CrossRef] [Google Scholar]
- Conjugantion position of quercetion glucuronides and effect on biological activity. Free Rad. Biol. Med.. 2000;29:1234-1243.
- [Google Scholar]
- Cancer-related inflammation and treatment effectiveness. Lancet Oncol.. 2014;15:e493-e503.
- [CrossRef] [Google Scholar]
- Anti-inflammatory effects of mangiferin on sepsis-induced lung injury in mice via up-regulation of heme oxygenase-1. J. Nutr. Biochem.. 2013;24:1173-1181.
- [CrossRef] [Google Scholar]
- Rhizoma Anemarrhenae extract ameliorates hyperglycemia and insulin resistance via activation of AMP-activated protein kinase in diabetic rodents. J. Ethnopharmacol.. 2015;172:368-376.
- [CrossRef] [Google Scholar]
- Metabolic forest: predicting the diverse structures of drug metabolites. J. Chem. Inf. Model.. 2020;60:4702-4716.
- [CrossRef] [Google Scholar]
- Nitric oxide, nitrogen metabolism and inflammatory respiratory disease. an hypothesis. Ups J. Med. Sci.. 1997;102:21-33.
- [CrossRef] [Google Scholar]
- The rhizome mixture of anemarrhena asphodeloides and coptidis chinensis ameliorates acute and chronic colitis in mice by inhibiting the binding of lipopolysaccharide to TLR4 and IRAK1 phosphorylation. Evid. Based Complement. Alternat. Med.. 2014;2014:809083
- [CrossRef] [Google Scholar]
- Diminishing inflammation by reducing oxidant generation: nitrated fatty acid-mediated inactivation of xanthine oxidoreductase. Adv. Exp. Med. Biol.. 2019;1127:59-65.
- [CrossRef] [Google Scholar]
- SFXN1 is a mitochondrial serine transporter required for one-carbon metabolism. Science (New York, N.Y.). 2018;362
- [CrossRef] [Google Scholar]
- The mixture of salvianolic acids from salvia miltiorrhiza and total flavonoids from Anemarrhena asphodeloides attenuate sulfur mustard-induced injury. Int. J. Mol. Sci.. 2015;16:24555-24573.
- [CrossRef] [Google Scholar]
- A calcium-dependent tyrosine kinase splice variant in human monocytes. activation by a two-stage process involving adherence and a subsequent intracellular signal. J. Biol. Chem.. 1998;273:9361-9364.
- [CrossRef] [Google Scholar]
- Rhizome of Anemarrhena asphodeloides counteracts diabetic ophthalmopathy progression in streptozotocin-induced diabetic rats. Phytother. Res.. 2013;27:1243-1250.
- [CrossRef] [Google Scholar]
- Simultaneous determination of five components in rat plasma by UPLC-MS/MS and its application to a comparative pharmacokinetic study in Baihe Zhimu Tang and Zhimu extract. Molecules. 2015;20:6700-6714.
- [CrossRef] [Google Scholar]
- Structure elucidation of in vivo and in vitro metabolites of mangiferin. J. Pharm. Biomed. Anal.. 2011;55:1075-1082.
- [CrossRef] [Google Scholar]
- Absorption, metabolism, and pharmacokinetics profiles of Norathyriol, an Aglycone of Mangiferin, in rats by HPLC-MS/MS. J. Agric. Food Chem.. 2018;66:12227-12235.
- [CrossRef] [Google Scholar]
- Comprehensive analysis of resveratrol metabolites in rats using ultra high performance liquid chromatography coupled with high resolution mass spectrometry. Arab. J. Chem.. 2020;13:7055-7065.
- [CrossRef] [Google Scholar]
- Total saponins from Rhizoma Anemarrhenae ameliorate diabetes-associated cognitive decline in rats: involvement of amyloid-beta decrease in brain. J. Ethnopharmacol.. 2012;139:194-200.
- [CrossRef] [Google Scholar]
- Serine, glycine and one-carbon units: cancer metabolism in full circle. nature reviews. Cancer. 2013;13:572-583.
- [CrossRef] [Google Scholar]
- Comprehensive metabolism study of polydatin in rat plasma and urine using ultra-high performance liquid chromatography coupled with high-resolution mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci.. 2019;1117:22-35.
- [CrossRef] [Google Scholar]
- Respective bioavailability of quercetin aglycone and its glycosides in a rat model. BioFactors (Oxford, England). 2000;12:169-174.
- [CrossRef] [Google Scholar]
- Pharmacologically active drug metabolites: impact on drug discovery and pharmacotherapy. Pharmacol. Rev.. 2013;65:578-640.
- [CrossRef] [Google Scholar]
- HMGB1 is phosphorylated by classical protein kinase C and is secreted by a calcium-dependent mechanism. J. Immunol.. 2009;182:5800-5809.
- [CrossRef] [Google Scholar]
- Multifaceted role of branched-chain amino acid metabolism in cancer. Oncogene. 2020;39:6747-6756.
- [CrossRef] [Google Scholar]
- Simultaneous determination of mangiferin and neomangiferin in rat plasma by UPLC-MS/MS and its application for pharmacokinetic study. J. Pharm. Biomed. Anal.. 2016;124:138-142.
- [CrossRef] [Google Scholar]
- Aliphatic and aromatic oxidations, epoxidation and S-oxidation of prodrugs that yield active drug metabolites. Curr. Med. Chem.. 2011;18:4885-4900.
- [CrossRef] [Google Scholar]
- Identification of 14 quercetin phase II mono- and mixed conjugates and their formation by rat and human phase II in vitro model systems. Chem. Res. Toxicol.. 2004;17:1520-1530.
- [CrossRef] [Google Scholar]
- The genus Anemarrhena Bunge: a review on ethnopharmacology, phytochemistry and pharmacology. J. Ethnopharmacol.. 2014;153:42-60.
- [CrossRef] [Google Scholar]
- Identification and pharmacokinetics of saponins in Rhizoma Anemarrhenae after oral administration to rats by HPLC-Q-TOF/MS and HPLC-MS/MS. Acta Pharm.. 2021;71:567-585.
- [CrossRef] [Google Scholar]
- Drug metabolite cluster-based data-mining method for comprehensive metabolism study of 5-hydroxy-6,7,3',4'-tetramethoxyflavone in rats. Molecules. 2019;24:3278.
- [CrossRef] [Google Scholar]
- HPLC fingerprinting-based multivariate analysis of phenolic compounds in mango leaves varieties: correlation to their antioxidant activity and in silico α-glucoidase inhibitory ability. J. Pharm. Biomed. Anal.. 2020;191:113616
- [CrossRef] [Google Scholar]
- Metabolism and pharmacokinetics of major polyphenol components in rat plasma after oral administration of total flavonoid tablet from Anemarrhenae Rhizoma. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci.. 2016;1026:134-144.
- [CrossRef] [Google Scholar]
- Metabolism, pharmacokinetics, and hepatic disposition of xanthones and saponins on Zhimu treatments for exploratively interpreting the discrepancy between the herbal safety and timosaponin A3-induced hepatotoxicity. Acta Pharmacol. Sin.. 2018;39:1923-1934.
- [CrossRef] [Google Scholar]
- Quantification of neomangiferin in rat plasma by liquid chromatography-tandem mass spectrometry and its application to bioavailability study. J. Pharm. Anal.. 2016;6:335-340.
- [CrossRef] [Google Scholar]
- Lidocaine alleviates morphine tolerance via AMPK-SOCS3-dependent neuroinflammation suppression in the spinal cord. J. Neuroinflamm.. 2017;14:211.
- [CrossRef] [Google Scholar]
- Characterization and determination of the major constituents in Belamcandae Rhizoma by HPLC-DAD-ESI-MS(n) J. Pharm. Biomed. Anal.. 2011;56:304-314.
- [CrossRef] [Google Scholar]
- Drug Metabolism in Drug Design and Development: Basic Concepts and Practice (first ed.). Hoboken: John Wiley & Sons Inc; 2008.
- Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun.. 2019;10:1523.
- [CrossRef] [Google Scholar]







