Translate this page into:
Excellent treatment activity of biscoumarins and dihydropyrans against P. aeruginosa pneumonia and reinforcement learning for designing novel inhibitors
⁎Corresponding authors at: Department of Clinical Pharmacy, General Hospital of Western Theater Command, Chengdu, Sichuan, China (D. Qu). yzc627@163.com (Zichen Ye), qvdi@sina.com (Di Qu)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Pseudomonas aeruginosa (P. aeruginosa) is a common clinical pathogen, which can easily cause cystic fibrosis and even bacteremia. In recent years, the antibiotic resistance of P. aeruginosa has been increasing. In an attempt to develop novel antibacterial agents, a series of biscoumarins (1–5) and dihydropyrans (6–10) were successfully prepared. The molecular structures of two representative compounds, that is, 1 and 6 were confirmed by single crystal X-ray diffraction study. The anti-bacterial activity of these synthesized compounds in vitro was evaluated by measuring the MIC values, as well as the P. aeruginosa growth curves. Next, the in vivo treatment activity of these compounds against the P. aeruginosa pneumonia infection was assessed by observing the survival rate of the infected mice and counting the bacterial load with colony plate counting assay. Additionally, the ELISA detection was conducted to evaluate the inflammatory response levels by measuring the IL-1β and TNF-α content released into the plasma, nasal lavage fluid and alveolar lavage fluid. The HE staining was also carried out to detect the protective effect of the compounds on the lung tissue damage. Further, novel anti-bacterial structures that are based on biscoumarin 5 are predicted and evaluated using reinforcement learning technic, and two outstanding pharmaceutical structures with low binding energy and high SA and QED scores are analyzed in detail using molecular docking simulation.
Keywords
Biscoumarins
Dihydropyrans
Pseudomonas aeruginosa
Reinforcement learning
1 Introduction
Pseudomonas aeruginosa (P. aeruginosa) is a clinical common pathogenic strain in hospitals. As an opportunistic pathogen, it can cause serious or even fatal infections in the burn or tumor patients with impaired or reduced immune function (Azam and Khan, 2019; Thi et al., 2020; Miyoshi-Akiyama et al., 2017). In addition, patients are also prone to be infected with P. aeruginosa when undergo surgery or in the certain therapeutic operations. P. aeruginosa can cause sepsis, respiratory tract infections, endocarditis, urinary tract infections, central nervous system infections, bone and joint infections, eye infections, skin and soft tissue infections, digestive tract infections, and so on (Sharma et al., 2014). Among these disease, the lower respiratory tract infections are the leading cause of patients’ death, which has drawn the attention of most researchers in the past decades. However, dur to the rising antimicrobial resistance and the absence of an effective vaccine or novel antimicrobial agents, the treatment of the P. aeruginosa infections is very complicated (Chegini et al., 2020). So, in this present research, new candidates were designed and synthesized for the P. aeruginosa infection treatment.
Dicoumarin and dihydropyran derivatives are widely available in nature. These derivatives exhibit various biological activities, such as anticoagulant, insecticidal, antihelminthic, hypnotic, and antifungal activities, phytoalexin production, and HIV protease inhibition (Li et al., 2015; Sui et al., 2015; Zhou et al., 2016). Moreover, the two kinds of compounds are gaining increasing interest because of their versatile activities through chemical modifications (different substituents on the aromatic ring) (Li et al., 2015). The application values of the dicoumarin and dihydropyran derivatives were needed to be evaluated for the new anti-bacterial compounds development. Recognizing the considerable importance of the compounds, the researchers focused on the synthesis of dicoumarin derivatives and dihydropyran derivatives (Li et al., 2015).
To identify more active compounds, we prepared a series of biscoumarin (Scheme 1) and dihydropyran (Scheme 2) derivatives and then evaluated their antibacterial activities in vitro and in vivo. The results of the compounds’ MIC values and P. aeruginosa growth curves indicated that compound 5 exhibited the most excellent anti-bacterial activity in vivo. At the same, compound 5 could also significantly increase the P. aeruginosa pneumonia mice and reduce the bacterial load. In addition to these, we also revealed the significant inhibitory activity of compound 5 on inflammatory response and lung tissue damage in the infected mice.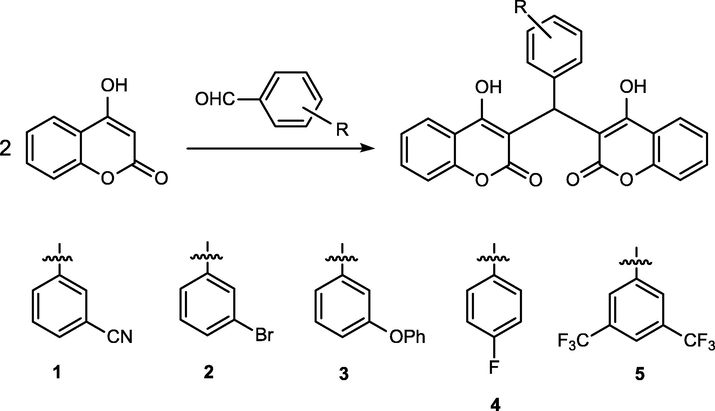
Synthetic route of compounds 1–5.
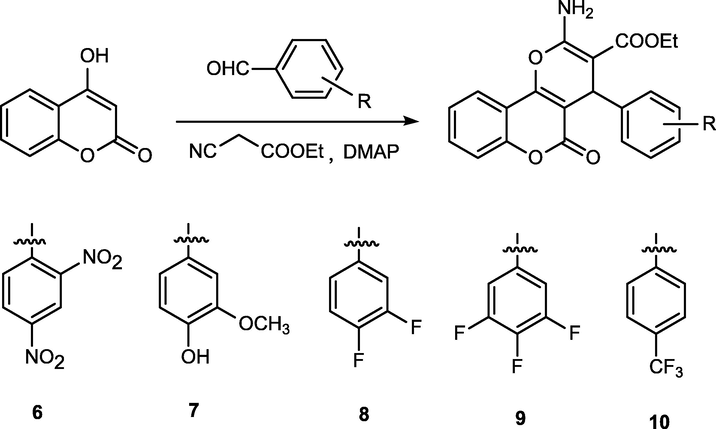
Synthetic route of compounds 6–10.
Besides the experiments, a recently developed autonomous molecule generation method, molecule optimization by reinforcement learning and docking (MORLD), that is a combination of reinforcement learning and molecular docking has been employed for predicting and designing outstanding pharmaceutical structures with low binding energy and high synthetic accessibility and quantitative estimate of drug-likeness scores (Jeon and Kim, 2020). By using compound 5 as the input structure, 6000 structures have been generated by the MORLD model. According to the reinforming learning algorithm, new functional groups could be introduced on to the initial compound 5 automatically, later, molecular docking simulation has been performed on all the generated structures, and potential structures are filtered out which could shed light on the synthesis of anti-bacterial inhibitors.
2 Experimental section
2.1 Apparatus and materials
IR spectra (400–4000 cm−1) were obtained using a Brucker Equinox-55 spectrophotometer (Fremont, CA, USA). 1H NMR spectra were obtained using a Varian Inova-400 spectrometer (at 400 MHz) (Palo Alto, CA, USA). Mass spectra were obtained using a micrOTOF-Q II mass spectrometer (Bruker). The melting points were taken on a XT-4 micro melting apparatus (Taike Company, Beijing, China), and the thermometer was uncorrected.
The P. aeruginosa strain ATCC 27,853 was purchased from the ATCC (Florida, USA). The clinical P. aeruginosa strains were isolated from the sputa of cystic fibrosis patients in Zibo Prevention and Treatment Hospital for Occupation Diseases (Zibo, Shandong). All the bacteria were cultured on LB agar plates (Sigma-Aldrich, USA) at 37 °C in an incubator and then transferred into liquid MH broths for strain replication at the condition of 220 rpm, 37 °C (Ahmar et al., 2020).
2.2 Synthesis and characterization of compounds 1–10
Biscoumarins 1–5 were synthesized according to the methods of a previous report (Li et al., 2014). A mixture of 3-cyanobenzaldehyde (3-bromobenzaldehyde, 3-phenoxybenzaldehyde, 4-fluorobenzaldehyde and 3,5-bis(trifluoromethyl)benzaldehyde) (10 mmol) and 4-hydroxycoumarin (20 mmol) was dissolved in 100 mL of EtOH. A few drops of piperidine were added, and the mixture was stirred for 3 h at room temperature. After reaction completion as determined by TLC, water was added until precipitation occurred. The solid was filtered off and then recrystallized from ethanol to give compounds 1–5.
3-(Bis(4-hydroxy-2-oxo-2H-chromen-3-yl)methyl)benzonitrile (1): m.p. 210–211 °C. IR (KBr pellet cm−1): 2910, 1651, 1600, 1560, 1312, 1300, 1024, 760 cm. 1H NMR (DMSO, δ, ppm): 11.564 (s, 1H), 11.353 (s, 1H), 7.996–8.094 (q, 2H), 7.649–7.688 (t, 2H), 7.575–7.593 (t, lH), 7.390–7.507 (m, 7H), 6.069 (s, lH). HRMS (ESI+): m/z: calcd for C26H15NO6: 460.0792 [M + Na]+; found: 460.0743.
3,3′-((3-Bromophenyl)methylene)bis(4-hydroxy-2H-chromen-2-one) (2): m.p. 190–191 °C. IR (KBr pellet cm−1): 2800, 1679, 1610, 1460, 1367, 1300, 1024, 760 cm. 1H NMR (CDCl3, δ, ppm): 11.593 (s, 1H), 11.316 (s, 1H), 8.028–8.109 (q, 2H), 7.649–7.691 (m, 2H), 7.360–7.458 (m, 6H), 7.179–7.245 (m, 2H), 6.086 (s, 1H). HRMS (ESI+): m/z: calcd for C25H15BrO6: 512.9944 [M + Na]+; found: 512.9919.
3,3′-((3-Phenoxyphenyl)methylene)bis(4-hydroxy-2H-chromen-2-one) (3): m.p. 200–201 °C. IR (KBr pellet cm−1): 2970, 1711, 1630, 1493, 1338, 1300, 1192, 761 cm. 1H NMR (CDCl3, δ, ppm): 11.655 (s, 1H), 11.319 (s, 1H), 8.009–8.091 (q, 2H), 7.626–7.668 (m, 2H), 7.412–7.433 (d, 4H), 7.274–7.326 (m, 3H), 6.999–7.055 (m, 4H), 6.900–6.950 (m, 2H), 6.107 (s, 1H). HRMS (ESI+): m/z: calcd for C31H20O7: 527.1101 [M + Na]+; found: 527.1128.
3,3′-((4-Fluorophenyl)methylene)bis(4-hydroxy-2H-chromen-2-one) (4): m.p. 245–246 °C. IR (KBr pellet cm−1): 2848, 1776, 1650, 1590, 1545, 1378, 1124, 768 cm. 1H NMR (CDCl3, δ, ppm): 11.562 (s, 1H), 11.338 (s, 1H), 8.015–8. 108 q, 2H), 7.640–7.683 (m, 2H), 7.432–7.452 (d, 4H), 7.195–7.229 (q, 2H), 7.016–7.059 (t, 2H), 6.079 (s, 1H). HRMS (ESI+): m/z: calcd for C25H15FO6: 453.0745 [M + Na]+; found: 453.0789.
3,3′-((3,5-Bis(trifluoromethyl)phenyl)methylene)bis(4-hydroxy-2H-chromen-2-one) (5): m.p. 197–198 °C. IR (KBr pellet cm−1): 2947, 1710, 1620, 1560, 1423, 1300, 1068, 768 cm. 1H NMR (CDCl3, δ, ppm): 11.527 (s, 1H), 11 0.403 (s, 1H), 7.996–8.101 (q, 2H), 7.814 (s, 1H), 7.644–7.695 (t, 4H), 7.390–7.447 (q, 4H), 6.132 (s, 1H). HRMS (ESI+): m/z: calcd for C27H14F6O6: 571.0587 [M + Na]+; found: 571.0593.
Dihydropyran derivatives (6–10) were also synthesized according to a reported procedure (Li et al., 2016). A mixture of 4-hydroxycoumarin (10 mmol), aromatic aldehydes (10 mmol), Ethyl 2-cyanoacetate (10 mmol) and 4-(dimethylamino)pyridine (DMAP) (1 mmol) in ethanol (100 mL) was refluxed for 2–3 h and then cooled to room temperature. The solid was filtered off and then recrystallized from ethanol to give compounds 6–10.
Ethyl 2-amino-4-(2,4-dinitrophenyl)-5-oxo-4H,5H-pyrano[3,2-c]chromene-3-carboxylate (6): m.p. 265–266 °C. IR (KBr pellet cm−1): 2975, 1710, 1630, 1592, 1332, 1300, 1014, 761 cm. 1H NMR (CDCl3, δ, ppm): 7.95–8.02 (m, 3H), 7.69–7.73 (m, 1H), 7.42–7.51 (m, 3H), 7.19–7.27 (m, 2H), 5.60 (s, 1H), 3.88–3.98 (m, 2H), 0.97–1.00 (t, 3H). HRMS (ESI+): m/z: calcd for C21H15N3O9: 476.0701 [M + Na]+; found: 476.0733.
Ethyl 2-amino-4-(4-hydroxy-3-methoxyphenyl)-5-oxo-4H,5H-pyrano[3,2-c]chromene-3-carboxylate (7):
m.p. 190–191 °C. IR (KBr pellet cm−1): 2887, 1713, 1621, 1593, 1434, 1340, 1024, 760 cm. 1H NMR (CDCl3, δ, ppm): 7.97–8.00 (m, 1H), 7.77 (s, 2H), 7.66–7.71 (m, 1H), 7.42–7.51 (m, 2H), 7.23–7.25 (q, 1H), 7.12–7.15 (t, 1H), 6.83–6.88 (m, 2H), 4.76 (s, 1H), 3.92–3.98 (m, 2H), 3.60 (s, 3H), 1.09–1.12 (t, 3H). HRMS (ESI+): m/z: calcd for C22H19NO7: 432.1054 [M + Na]+; found: 432.1066.
Ethyl 2-amino-4-(3,4-difluorophenyl)-5-oxo-4H,5H-pyrano[3,2-c]chromene-3-carboxylate (8): m.p. 203–204 °C. IR (KBr pellet cm−1): 2989, 1723, 1683, 1590, 1412, 1330, 1014, 760 cm. 1H NMR (CDCl3, δ, ppm): 7.96–7.99 (q, 1H), 7.91 (s, 2H), 7.69–7.74 (m, 1H), 7.45–7.52 (m, 2H), 7.25–7.31 (m, 2H), 7.07–7.09 (m, 1H), 4.69 (s, 1H), 3.97–4.05 (m, 2H), 1.09–1.13 (t, 3H). HRMS (ESI+): m/z: calcd for C21H15F2NO5: 422.0811 [M + Na]+; found: 422.0838.
Ethyl 2-amino-5-oxo-4-(3,4,5-trifluorophenyl)-4H,5H-pyrano[3,2-c]chromene-3-carboxylate (9): m.p. 208–209 °C. IR (KBr pellet cm−1): 2823, 1691, 1702, 1610, 1345, 1300, 1014, 761 cm. 1H NMR (CDCl3, δ, ppm): 7.95–7.98 (m, 3H), 7.70–7.74 (m, 1H), 7.45–7.52 (m, 2H), 7.15–7.19 (q, 2H), 4.69 (s, 1H), 3.97–4.07 (m, 2H), 1.10–1.13 (t, 3H). HRMS (ESI+): m/z: calcd for C21H14F3O5: 440.0716 [M + Na]+; found: 440.0723.
Ethyl 2-amino-5-oxo-4-(4-(trifluoromethyl)phenyl)-4H,5H-pyrano[3,2-c]chromene-3-carboxylate (10): m.p. 221–222 °C. IR (KBr pellet cm−1): 2834, 1671, 1630, 1545, 1412, 1321, 1028, 760 cm. 1H NMR (CDCl3, δ, ppm): 7.98–8.00 (q, 1H), 7.94 (s, 2H), 7.69–7.73 (m, 1H), 7.60–7.62 (d, 2H), 7.45–7.52 (m, 4H), 4.78 (s, 1H), 3.96–4.02 (q, 2H), 1.08–1.11 (t, 3H). HRMS (ESI+): m/z: calcd for C22H16F3NO5: 454.0873 [M + Na]+; found: 454.0812.
2.3 X-ray crystallography
SuperNova diffractometer was utilized in acquiring XRD patterns. CrysAlisPro was employed to analyze intense data and converted the data to HKL files. SHELXS in the light of direct mean and SHELXL-2014 software based on least-squares strategy were employed respectively for the synthesis and refinement of original architectural modes. After the addition of non-H atoms, the anisotropic parameters could be mixed. Eventually, the entire H atoms could be fixed on the C atoms that bridged with AFIX commands in geometry. The as-prepared compounds’ refinement details and crystallography parameters are displayed in Table 1.
Identification code
1
6
Empirical formula
C26H15NO6
C42H30N6O19
Formula weight
437.39
922.72
Temperature/K
293 (2)
293 (2)
Crystal system
Monoclinic
Monoclinic
Space group
P21/c
P21/c
a/Å
9.6579 (9)
10.907 (8)
b/Å
10.2778 (8)
15.214 (12)
c/Å
20.922 (2)
13.911 (10)
α/°
90
90
β/°
96.678 (9)
93.943 (16)
γ/°
90
90
Volume/Å3
2062.6 (3)
2303 (3)
Z
4
2
ρcalcg/cm3
1.408
1.331
μ/mm−1
0.01
0.11
Data/restraints/parameters
3583/0/305
4001/0/307
Goodness-of-fit on F2
0.817
1.081
Final R indexes [I>=2σ (I)]
R1 = 0.0483, ωR2 = 0.0593
R1 = 0.0972, ωR2 = 0.2143
Final R indexes [all data]
R1 = 0.1332, ωR2 = 0.0856
R1 = 0.1773, ωR2 = 0.2398
Largest diff. peak/hole / e Å−3
0.16/-0.16
1.10/-0.33
2.4 MIC determination
After the synthesis of the compounds with novel structure, their minimum inhibitory concentration against the P. aeruginosa was firstly measured. The MIC values of the compounds were measured in this present research strictly in accordance with the instructions (Liao et al., 2019; Liu et al., 2020). Briefly, the P. aeruginosa strain was cultured in MH broth in microtiter plates at the final destiny of 5 × 105 CFU/ml for 12 h. Next, the 100 μL culture medium supplemented with the synthesized compound at several different concentrations (2–256 μg/mL). All the bacteria were incubated with the compounds in an incubatory at the concentration of 37 °C for 24 h, 0.2 % triphenyl tetrazolium chloride was added into the wells for another 2 h’ incubation. The lowest concentration of compounds that showed no red color change were regarded as the MIC values of the compounds.
2.5 Bacterial growth curves
To evaluate the anti-bacterial activity of the synthesized novel compounds against P. aeruginosa, the growth curves of the P. aeruginosa was measured after the incubation of the compounds with indicated concentration. This preformation was carried totally under the guidance of the instructions with some modifications (Nussbaumer-Pröll et al., 2020). In brief, the P. aeruginosa suspension was collected, quantified and added into the bacteria growth plates, then the compounds were added into the wells for treatment at indicated concentrations. The P. aeruginosa strain was cultivated with MH broth medium added with different concentration of the compounds in an automated Bioscreen C system in the automatic bacterial growth curve analyzer (Bioscreen, Finland) at 37 °C, and the absorbance of each well was measured every hour at 600 nm.
2.6 Mice infection model and survival rate
To further assess the treatment effect of the compounds synthesized in this present research, the mouse P. aeruginosa pneumonia infection model was induced firstly strictly in accordance with the protocols previous described (Berube et al., 2016). Before the pneumonia infection model construction, P. aeruginosa strain was cultured in Brain Heart Infusion medium at 37 °C overnight. Then, the bacterial suspension was harvested and centrifuged at 1000 g condition for 10 min, followed by twice 0.9 % NaCl washing. 60 BALB/c mice (5–6 weeks, 20–22 g) used in this present research were purchased from Jiangsu Jicui Yaokang Biotechnology Co., Ltd. (Nanjing, China), then the animals were kept at General Hospital of Western Theater Command central laboratory with the standard condition of 20-25 °C, 12-hour light/dark cycle. All the animals were divided into 6 groups randomly, the control group (n = 10), model group (n = 10), gentamicin treatment group (n = 10), compound 5 treatment groups (5, 10, 20 mg/kg, n = 10). The model and compound treatment mice were anaesthetized with sevoflurane (Abbott, Chicago, IL, USA), and 108 CFU bacteria was dripped into the nasal cavity to induce the mouse P. aeruginosa pneumonia infection model, and the synthesized compound was injected i.p at the concentration of 5, 10, 20 mg/kg for treatment. Survival rates of the mice was assessed once a day for 7 days since infection.
2.7 ELISA assay
To investigate the therapeutic potential of synthesized compound on the P. aeruginosa pneumonia infection, their inhibitory activity on the inflammatory cytokines releasing was assessed with ELISA detection kit in this research. This experiment was carried out according to the manufactures’ protocols with a little change (Bahey-El-Din et al., 2020). In short, the P. aeruginosa pneumonia infection model was induced as previously described, followed by the compounds’ treatment at the concentration of 5, 10, 20 mg/kg. Subsequently, the plasma was collected from all the groups and the content of IL-1β, as well as TNF-α was measured with ELISA assay for at least three times.
2.8 Bacterial load
The bacterial CFU numbers in the plasma, nasal lavage fluid and alveolar lavage fluid was further determined for the evaluation of compounds’ treatment activity on P. aeruginosa pneumonia infection. This conduction was finished in accordance with the instructions with modified changes (Shah et al., 2018). After the construction of the P. aeruginosa pneumonia infection, as well as the indicated compound treatment, the plasma, nasal lavage fluid and alveolar lavage fluid in different groups was harvested and then plated on LB agar plates in triplicate. All the plates were cultured in an incubator at the concentration of 37 °C, 5 %CO2 for 24 h, and the bacterial CFUs was counted. All the results were presented as mean ± SD.
2.9 Hematoxylin and eosin (H&E) staining
In the MRSA infection model, parts of lung and liver tissues were fixed in 10 % neutral buffered formalin for 24 h, dehydrated and processed into paraffin sections totally in accordance with a standard procedure. The paraffin sections were then stained with hematoxylin and eosin (H&E) staining for histopathology analysis, and the morphologies of the lung tissues were observed with microscope.
2.10 Ethics statement
All the preformation in vivo was approved by the Committee of Animal Ethics, which was finished totally under guidance of the institutional policies and the guidelines formulated by the animal welfare committee.
2.11 tatistical analysis
All the experiments performed in this present study were repeated at least three times and the results were presented as mean ± SD. Unpaired student’s t-test was used to analyze the statistical difference between two groups, and the one-way ANOVA analysis was used for statistical analysis among three or more groups. P < 0.05 was denoted as significant statistical significance. GraphPad Prism software (San Diego, USA) was recommended for Statistical analysis.
2.12 Simulation methods
The reinforcement learning and docking simulation for designing novel anti-bacterial inhibitors that were based on biscoumarin 5 were performed by using the molecule optimization by reinforcement learning and docking (MORLD) model, which is an online server for predicting the molecules having high predicted binding affinities against the target protein (Zhou et al., 2019). To perform the designing and filtering procedure, the SMILES code of biscoumarin 5 has been provided as the input of the initial structure. From our previous studies we could know that biscoumarins have excellent potentials in the applications anti-bacterial field, however, there are too many sites on the biscoumarins that can be modified for the pharmaceutical development, the validation of all those potential structures is extremely challenging. The machine learning technic provides a new efficient solution for the predicting and designing of new pharmaceutical molecules. In the current study, the Thymidylate kinase (TMPK) has been employed as the receptor protein for probing the binding energy of the newly designed molecular structures, since it is an attractive therapeutic target for the development of novel antibacterial agents. The crystal structure of TMPK was obtained from protein data bank, and the PDB ID is 4QGG (https://www.rcsb.org). Its protonated structure has been uploaded to the server as the receptor. The elements that are allowed during the reinforcement learning procedure are C, O, N and F. The maximum allowed number of molecular structures is 6000, and for each of the structure the maximum allowed number of modifications/attempts is 16. The weight of both the synthetic accessibility score (SA) and quantitative estimate of drug-likeness (QED) score is set to 1.0. For the scoring of the binding poses between the newly generated molecular structure and the TMPK receptor, the docking pocket has been placed at the coordinates of × = 11.666, y = 1.721 and z = 5.140 Å, the number of grid points on each direction is 50.
3 Results and discussion
3.1 Molecular structure
X-ray single crystal diffraction analysis show that 1 crystallize in a monoclinic crystal system with P21/c space group. The molecular structure of 1 is made up of three parts two 4-hydroxy-2H-chromen-2-one groups [(C15, C21, C19, C29, C31, C33, C16, C9, C22, O4, O5 and O7) and (C10, C12, C20, C28, C39, C14, C18, C30, C23, O1, O2 and O3)] and one benzene ring (C1-C6). Three parts were further connected by the C8 atom, which show the twisted tetrahedral structure (Fig. 1). There exists one cyano group C7-N1 on the benzene ring via the bond of C5-C7. And further, the adjacent molecules are connected by the intermolecular interaction to give a supramolecular structure (Fig. 2).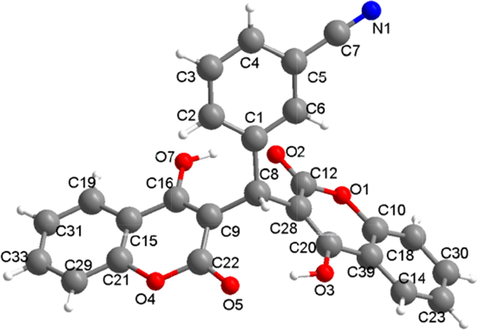
The molecule structure for 1.
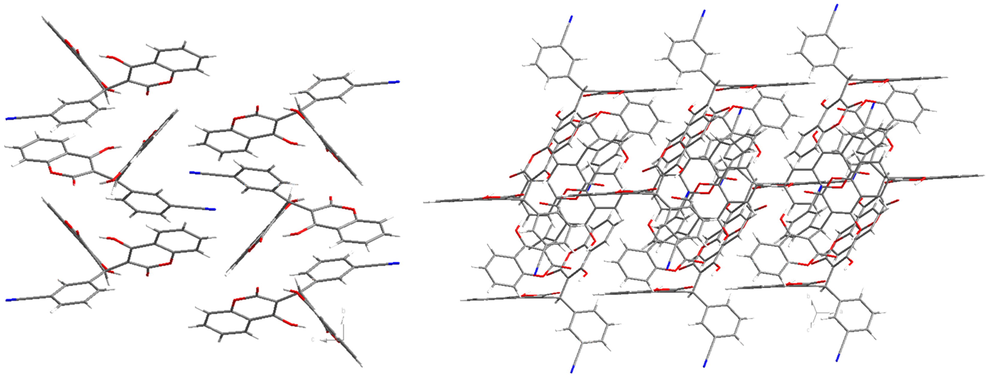
The crystal packing structure of 1.
X-ray single crystal diffraction data analysis reveal that 6 crystallizes in monoclinic crystal system with the P21/c space group. As shown in the Fig. 3, the molecule structure consists of two benzene rings [(C10-C15) and (C16-C21)] and two six-membered rings [(C6, C15, C10, O4, C9, C7) and (C4-C8, O3)]. The benzene ring (C10-C15) connects the six-membered ring (C6, C15, C10, O4, C9, C7) via the C15-C10, the two six-membered rings connect each other via the bond of C6-C7. The benzene ring (C16-C21) connects the six-membered ring (C4-C8, O3) via the bond of C8-C16. There exist two nitro groups on the benzene ring (C16-C21) via the N3-C19 and N2-C21, respectively. The ethyl formate group (C1H3C2H2-O2C3 = O1-) and amino group (N1H2) connect C4 and C5 atoms of the six-membered ring. The O5 atom connects the six-membered ring via the double ring C9 = O5. And further there exist on rigid water molecule in 6. The adjacent molecules were further connected to give a supramolecuar structure (Fig. 4).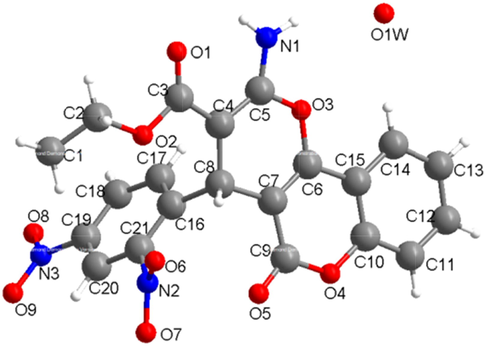
The molecule structure for 6.
The crystal packing structure of compound 6.
3.2 Anti-bacterial activity evaluation of compounds against P. aeruginosa strain in vitro
For the treatment of P. aeruginosa pneumonia infection, serial small-molecular chemical compounds with novel structure were designed and synthesized in this present research, as described above. For further evaluating the inhibitory activity of the chemical compounds, the MICs of the compounds were determined. As the results showed in Table 2, these two different kinds of derivatives significantly different anti-bacterial activities. The biscoumarin derivatives showed much stronger anti-bacterial effect than the dihydropyran derivatives, reflected as the lower MIC values. And among the biscoumarin derivatives, compound 5 showed the most excellent anti-bacterial effect with the lowest MIC value of 2–4 μg/ml. In addition to this, the influence of these compounds on the P. aeruginosa strain survival was also measured. In consistence with the above results, the inhibitory effect of the biscoumarin derivatives was much stronger than the dihydropyran derivatives, and compound 5 exhibited the strongest suppression activity on the P. aeruginosa growth. These results showed that the growth of P. aeruginosa was significantly inhibited by compound 5. However, different from compound 5, other compounds exhibited weaker or even no inhibitory activity on the P. aeruginosa growth (Fig. 5).
Compounds
1
2
3
4
5
6
7
8
9
10
MIC (µg·ml−1)
32
16
16
64
2–4
>256
>256
>256
>256
>256
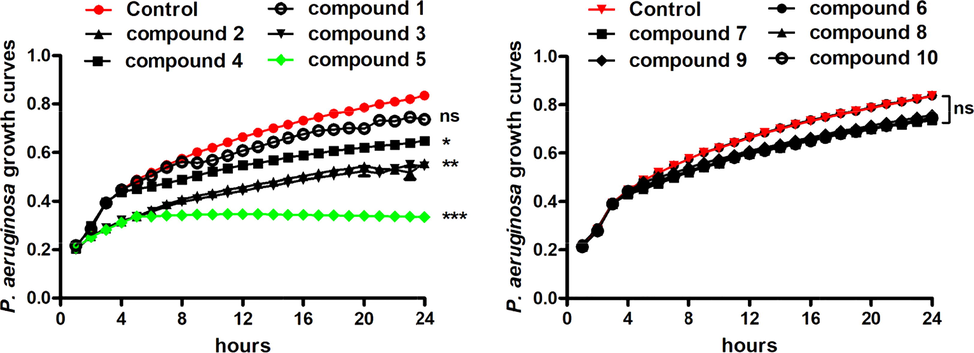
In vitro anti-bacterial activity evaluation of the synthesized compounds against P. aeruginosa. The P. aeruginosa strain was seeded into plates and incubated with synthesized compounds at serial concentrations. MIC values of the compounds were determined by broth microdilution. The growth curves of the P. aeruginosa were measured with automatic bacterial growth curve analyzer after compounds treatment.
3.3 Protective activity of the compound against P. aeruginosa pneumonia by reducing the bacterial loading
As reported above, compound 5 exhibited the strongest inhibitory activity on P. aeruginosa growth in vitro. However, whether compound 5 could still exert the excellent treatment activity on the P. aeruginosa pneumonia infection mice was still need to be explored. After the construction of the bacterial model and the indicated compound treatment, the survival of the mice was observed once a day for 7 days since infection. The survival curves of the P. aeruginosa pneumonia mice showed that compared with the control group, there was a significantly reduced survival rate of the infected mice, with p < 0.05. As expected, after compound 5 treatment, the survival curves of the P. aeruginosa pneumonia mice were up-regulated obviously. This protective activity of compound 5 showed a dose dependent relationship, which increases combined with the dosage increases (Fig. 6A). Furthermore, the P. aeruginosa CFU numbers was also counted and the date suggested that the bacterial load in the plasma, nasal lavage fluid and alveolar lavage fluid was reduced dose dependently under compound 5 exposure (Fig. 6B). All the data above indicated that compound 5 also showed excellent protective activity on the P. aeruginosa pneumonia infection mice in vivo.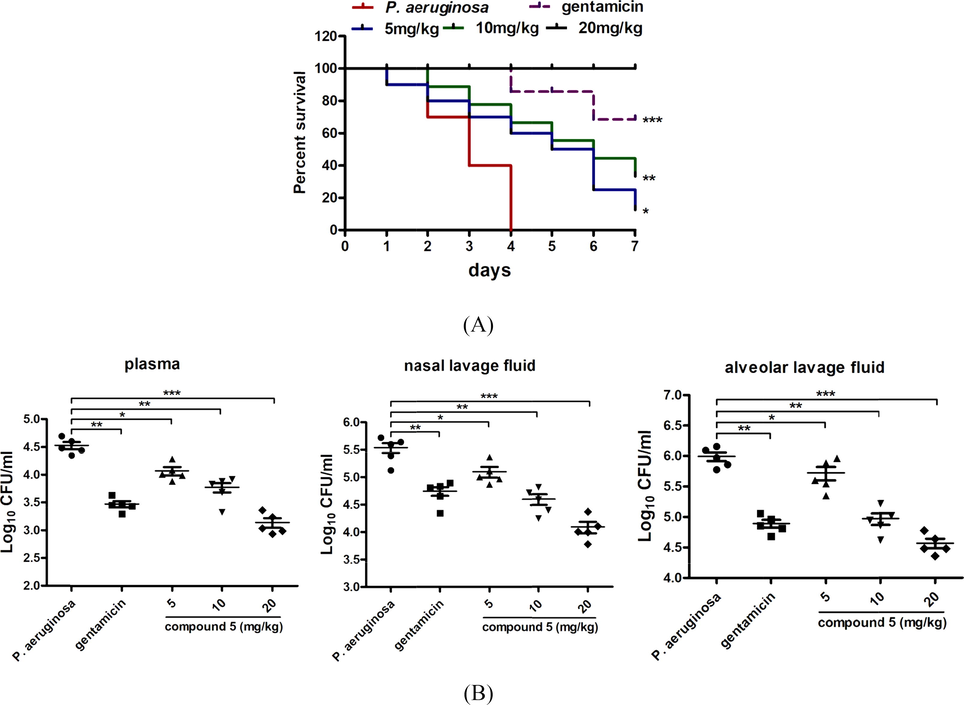
Compound 5 increased the survival rate of the P. aeruginosa pneumonia infection mice and reduced the P. aeruginosa CFU numbers. 60 BALB/c mice were divided into control group (n = 10), model group (n = 10), gentamicin treatment group (n = 10), compound 5 treatment groups (5, 10, 20 mg/kg, n = 10). After the indicted treatment, the survival rate of the infected mice was measured (A) and the bacterial burden in the plasma, nasal lavage fluid and alveolar lavage fluid was determined with colony plate counting assay (B).
3.4 Compound exhibited the protective activity in vivo by reducing the inflammatory response and tissue damage
To further the mechanism of compound 5 on the P. aeruginosa pneumonia infection treatment, more biological experiments were conducted in this research. As previous reported, the P. aeruginosa could induce the inflammatory response significantly in the infected mice. Thus, in this research, the content of the IL-1β and TNF-α released into the plasma was measured with ELISA detection kit. From the results in Fig. 7A, we can see that the levels of the IL-1β and TNF-α in the P. aeruginosa pneumonia infection model mice was significantly higher than the control group, which could be significantly reduced under compound 5 treatment. The anti-inflammatory activity of compound 5 showed a dose dependent manner, which is consistence with the above results (Fig. 3A). Besides, the tissue damage of the infected mice was also detected with HE staining. The results also suggested that in the P. aeruginosa pneumonia infection model mice, there was a phenomenon of abnormal alveolar structure, necrosis, congestion, as well as the neutrophils infiltration and accumulation in the lung tissue. While, after compound 5 treatment (5, 10, 20 mg/kg, i.p), the levels of the tissue damage were reduced and the neutrophils infiltration was also relieved significantly (Fig. 7B).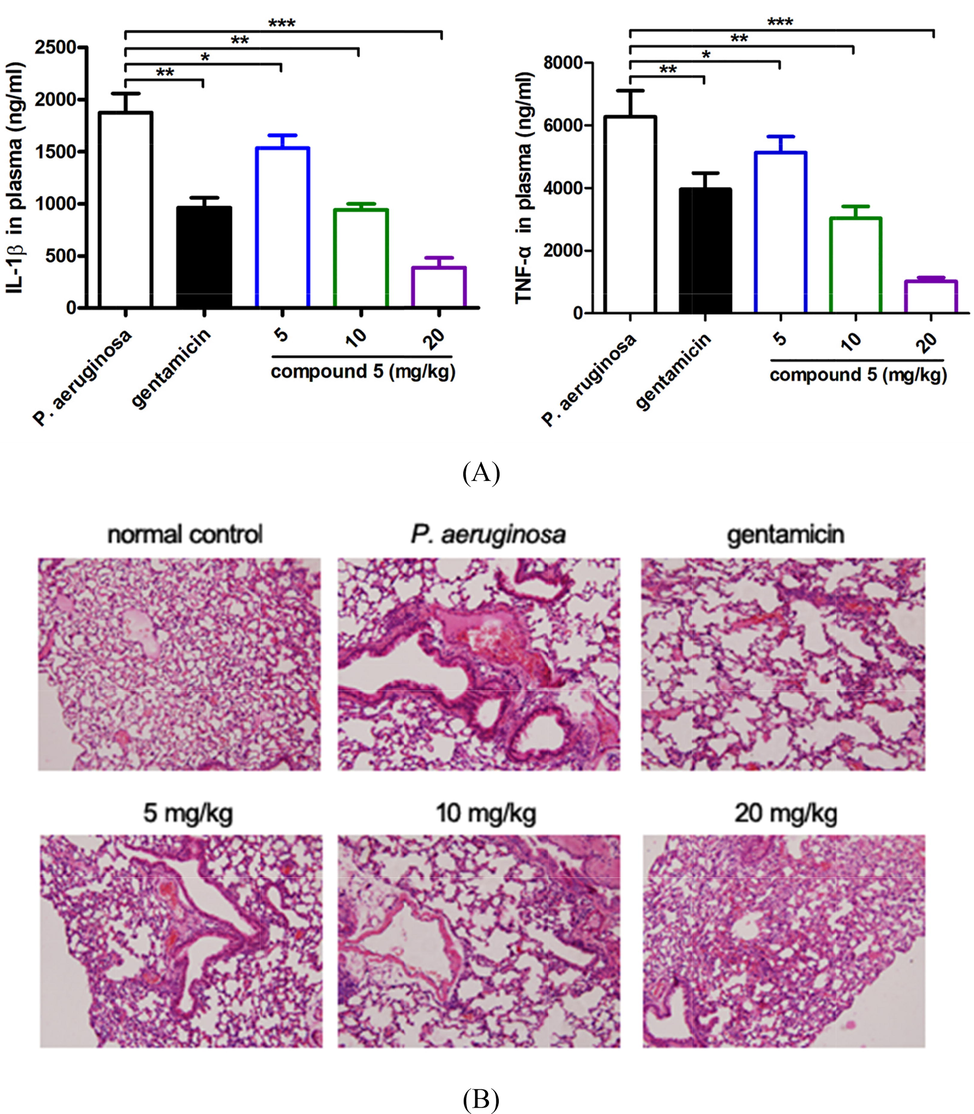
Compound 5 obviously reduce the IL-1β and TNF-α releasing and relief the lung tissue damage. After the construction of the P. aeruginosa pneumonia infection animal model, as well as the compound treatment, the plasma IL-1β and TNF-α levels determination (A). The lung tissue damage was evaluated with HE staining (B).
3.5 Reinforcement learning predictions and molecular docking simulations
In the molecule optimization by reinforcement learning and docking (MORLD) model, for the generating of every new structure, the same input (the SMILES code of the biscoumarin structure) will be used, up to 16 attempts of modifications are allowed with respect to the biscoumarin structure, those attempts are based on the MolDQN framework (Bahey-El-Din et al., 2020), explicitly, for each of the modification attempt, it contains multiple actions where a new atom (bond) will be inserted or deleted on a certain site on the biscoumarin structure via a chemical valid manner, the valence constrains must be satisfied after the attempt of modification, the validation of such constrain on every attempt was performed using software package RDKit (https://www.rdkit.org/). Such reinforcement learning procedure on each attempt will continue until the final attempt is achieved as the maximum number of allowed modifications, which will be further evaluated with molecular docking procedure with QuickVina 2. A more details determination of the optimization and the criterion for every attempt can be found in the flowchart in Kim’s work (Jeon and Kim, 2020).
The intention of the MORLD model is to obtain new structure which has high SA and QED scores and low binding energy, in order to validate the reinforcement learning progress for the newly generated structures, the averaged binding energy over every 100 structures are summarized in Fig. 8. As we can see from Fig. 8, the overall trend of the binding energy decreases as the reinforcement learning progress proceeds further, indicating the MORLD model could predict new structures that are based on the initial biscoumarin with lower and lower binding energies.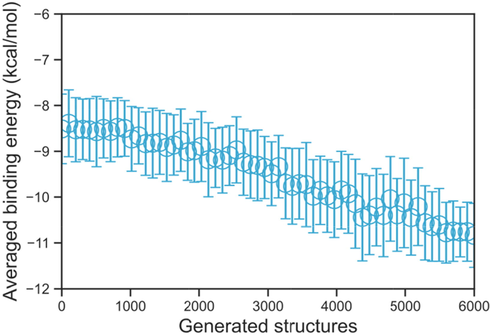
The averaged binding energies over every 100 generated structures, as one can see, as the autonomous reinforcement learning process keep proceeding, the binding energy of the newly generated structure has a decreasing trend.
The initial biscoumarin has already been a well-known pharmaceutical molecular that has excellent anti-bacterial activity, thus, we want to examine the qualities and performances of the newly designed structures towards the anti-bacterial application. In Fig. 9A the binding energy of all 6000 generated structures are displayed, the pink dotted line indicates the binding energy of the biscoumarin structure (−9.3 kcal/mol). It can be seen that not all the structures would have better performance than the default initial structure, this is attributed by the fact that at the earlier stage of the reinforcement learning progress, the choices of the action (add or delete new atom/bond) are basically random, due to the decaying ∊-greedy method, the ∊ for the random choice will gradually decrease from 1.0 to 0.0, meanwhile, the weight for the deep Q-Networks in MolDQN increases, consequently, new structure with relatively lower binding energy will be gradually generated as the reinforcement learning progress keep going. In Fig. 9B the binding energy distribution probability has been displayed, where we can see that the probability of structures with high binding energies is not dominant, instead, the summation of probabilities of structures with lower binding energies is about 55.4 % (using the binding energy of initial structure as the benchmark). The above results indicate that more and more structures with lower binding energies will be generated in comparison to the initial structure, suggesting an effectively and successfully implemented reinforcement learning procedure.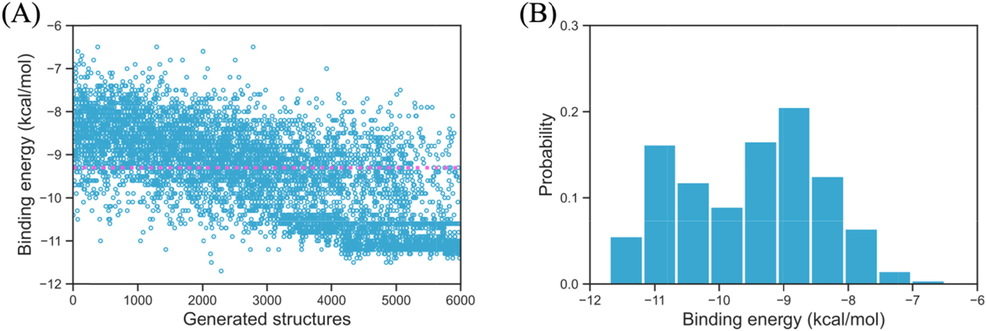
The binging energy of all 6000 generated structures (A) and their binding energy probability (B), the pink dotted line indicates the binding energy of the input biscoumarin structure.
The newly generated structures should not only have good performance in the docking simulation with the given receptor, they also have to prove that they are realistic in the synthesis procedure. Thus, the synthetic accessibility scores (SA) of all 6000 generated structures are evaluated and are shown in Fig. 10A. In the current study, the extensively studied biscoumarin (Li et al., 2015; Sui et al., 2015; Zhou et al., 2016) seems to be the easiest structure to synthesize with its SA score of 0.79. Further, in consistent with the binding energy, as the reinforcement learning process keep going, the newly generated structures are seen to have higher SA scores. In Fig. 10B the distribution of the SA scores is shown, where we can see that 76.6 % of the structures have a SA score>0.5. And the most probable SA scores are ranging from 0.55 to 0.68.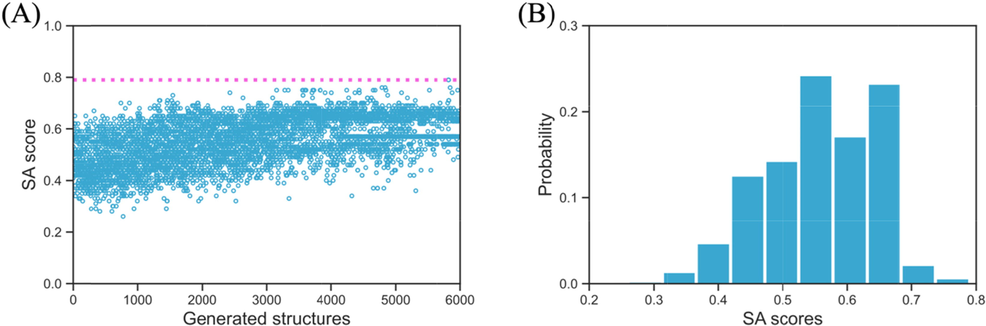
The synthetic accessibility score (SA) of all 6000 generated structures (A) and their SA score probability (B), the pink dotted line indicates the SA score of the input biscoumarin structure.
Besides the SA score, another quantity has to be taken into consideration that measures the similarity between the newly generated molecules and the known drugs in terms of various physicochemical properties and structural features, that’s the quantitative estimate of drug-likeness (QED) score. As can be seen in Fig. 11A, the initial biscoumarin structure has a QED score of 0.2, and similar to the trends in both the binding energy and SA scores, as more and more structures that have been generated by the reinforcement learning procedure, the structures tend to have higher QED scores. And about 56.8 % structures have higher QED scores than the initial biscoumarin structure. The distribution probability of QED score is shown in Fig. 11B, where we can see the most probable QED scores are within the range of 0.30–0.35.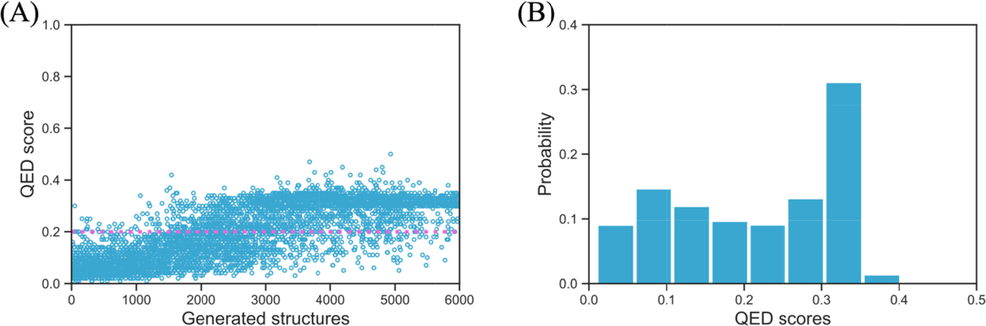
The quantitative estimate of drug-likeness (QED) score of all 6000 generated structures (A) and their QED score probability (B), the pink dotted line indicates the QED score of the input biscoumarin structure.
From the above results that summarize the binding energy, SA and QED scores one could understand that not all generated structures are suitable as the anti-bacterial molecule. A typical structure that has the realistic anti-bacterial activity should have low binding energy and high SA and QED scores. In Scheme 3, we listed the initial biscoumarin structure and two carefully selected structures among the 6000 structure which are seen to satisfy all three criterions. As the benchmark, the initial biscoumarin structure has a binding energy of −9.3 kcal/mol, the SA and QED scores are 0.79 and 0.2, respectively. For the two selected structures, the binding energies are −10.6 and −11.3 kcal/mol, the SA scores are 0.65 and 0.55, and both structures have the same QED score of 0.39. We can see all these values are within the most probable range in the aforementioned distribution probability results.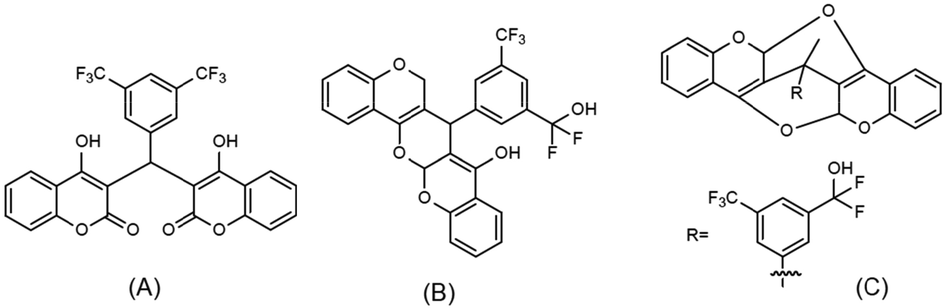
(A): The initial biscoumarin structure that was used as the input for the reinforcement learning; (B) and (C): the generated potential inhibitors with relatively low binding energy and high SA and QED scores, the binding energies are −10.6 and −11.3 kcal/mol, the SA scores are 0.65 and 0.55, and the QED scores are 0.39 and 0.39, respectively.
After qualifying multiple criterions of the two carefully selected structures, the detailed interactions are analyzed in detail to see whether they have anti-bacterial activities with the given receptor TMPK protein. The binding poses are shown in Fig. 12, in Fig. 12A, we can see that the initial biscoumarin structure has two binding interactions with the active residue ARG-48, explicitly, the F atoms on the -CF3 functional groups are interacting with the –NH2 group, the binding lengths of the corresponding hydrogen bonds are 2.9 and 3.0 Å, respectively. In Fig. 12B we can see that the first new structure also formed two binding interactions with the receptor protein, the newly added –OH group interacts with the carboxyl group on active residue GLU-37 with the distance of hydrogen bond equal 2.8 Å, and the F atom on the -CF3 functional group interacts with the –NH2 on active residue ARG-70, the formed hydrogen bond is 3.2 Å long. In Fig. 12C, we can see that one binding interaction has been formed between the second new structure and the receptor protein, the newly added –OH group interacts the carbonyl functional group on active residue GLN-101, the bond length of the hydrogen bonding interaction is 2.7 Å. The above results indicate that the newly generated structure indeed have good anti-bacterial activities, and also have the realistic potential to be synthesized and used as novel pharmaceutical molecules.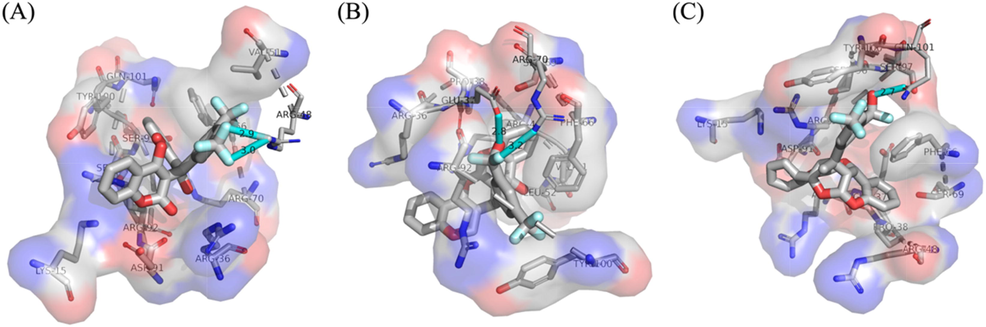
The binding poses of the biscoumarin and two related derivatives that are corresponding to the structures in Scheme 1.
4 Conclusions
In this present research, after the synthesis of these two different kinds of derivatives, their anti-bacterial activity was evaluated against the P. aeruginosa firstly in vitro. The data of the MIC values and P. aeruginosa growth curves suggested that the anti-bacterial activity of biscoumarin derivatives was much stronger than dihydropyran derivatives, showed as the lower MIC values and inhibited bacterial growth curves. The difference between these two kinds of derivatives may be due to intramolecular hydrogen bonds made the compounds enter the bacterial cells more easily compared with the dihydropyran derivatives. Next, in the five biscoumarin derivatives, we found that compound 5 exhibited the most excellent inhibitory activity on the P. aeruginosa. According to this phenomenon, we speculate that the substituent of compound 5 could increase the polarity of the compound, which then increase the concentration of the compound in the plasma. After the in vitro anti-bacterial evaluation of the serial compounds, compound 5 was selected for the following in vivo P. aeruginosa pneumonia assessment. The compound 5 was given to the P. aeruginosa pneumonia mice for treatment at the concentration of 5, 10, 20 mg/kg. The results suggested that it could increase the P. aeruginosa pneumonia mice survival rate and reduce the bacterial load in the plasma, nasal lavage fluid and alveolar lavage fluid. All the biological effect of compound 5 exhibited a dose dependent relationship. After the above exploration, the detail mechanism of compound 5 was next discussed, the ELISA data indicated the strong inhibitory activity of compound 5 on the inflammatory response in of the plasma, nasal lavage fluid and alveolar lavage fluid of the mice, showed as the reduced content of IL-1β and TNF-α. As the over releasing of the inflammatory cytokines could lead the tissue damage, the HE staining was further conducted and the results was consistence with the ELISA results. The lung tissue damage in the infected mice was obviously relieved by compound 5 application. All the data in this this research showed that compound 5 may be an excellent candidate for the P. aeruginosa pneumonia treatment.
Through the reinforcement learning technic, two new structures are carefully selected from 6000 tested structure with the consideration of low binding energy and high synthetic accessibility (SA) and quantitative estimate of drug-likeness (QED) scores. Both of the structures exhibit values of all three criterions within the most probable range, suggesting that they should have anti-bacterial activities, which have been further demonstrated in their docking poses. The reinforcement learning technic shows excellent potential in predicting new pharmaceutical molecules when using a well-known structure as input and benchmark, which sheds light on the novel design of pharmaceutical molecule that targets the anti-bacterial application.
Author contributions
Di Qu and Zichen Ye designed the project and wrote the manuscript. Jing Li, Jiangtao Li and Hongjiang Ren synthesized and characterized the compounds. Yingwei Qu provided the P. aeruginosa strains and constructed the animal model. Huiqing Shi and Yan Wu finished the in vitro and in vivo antibacterial evaluation. Zichen Ye performed the reinforcement learning predictions and molecular docking simulations.
Data Availability
The data used to support the findings of this study are included within the article.
Funding
The research was supported by the project of Natural Science Basic Research Plan in Shaanxi Province of China (2022JM-075); General Hospital of Western Theater Command (2021-XZYG-B14); Special Funding of China Postdoctoral Science Foundation (Di Qu).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Culture of Small Colony Variant of Pseudomonas aeruginosa and Quantitation of its Alginate. J. Vis. Exp. 2020
- [CrossRef] [Google Scholar]
- Updates on the pathogenicity status of Pseudomonas aeruginosa. Drug Discov. Today. 2019;24:350-359.
- [Google Scholar]
- Recombinant N-terminal outer membrane porin (OprF) of Pseudomonas aeruginosa is a promising vaccine candidate against both P. aeruginosa and some strains of Acinetobacter baumannii. Int. J. Med. Microbiol.. 2020;310:151415
- [Google Scholar]
- Bacteriophage therapy against Pseudomonas aeruginosa biofilms: a review. Ann. Clin. Microbiol. Antimicrob.. 2020;19:45.
- [Google Scholar]
- Autonomous molecule generation using reinforcement learning and docking to develop potential novel inhibitors. Sci. Rep.. 2020;10:22104.
- [Google Scholar]
- Synthesis, photoluminescent, antibacterial activities and theoretical studies of 4-hydroxycoumarin derivatives. J. Mol. Struct.. 2014;1075:509-514.
- [Google Scholar]
- New biscoumarin and dihydropyran derivatives as antimicrobials. Res. Chem. Intermed. 2015
- [CrossRef] [Google Scholar]
- Synthesis of biscoumarin and dihydropyran derivatives with promising antitumor and antibacterial activities. Bioorg. Med. Chem. Lett.. 2015;25:5520-5523.
- [Google Scholar]
- Synthesis of Biscoumarin and Dihydropyran Derivatives and Evaluation of Their Antibacterial Activity. Molecules. 2015;20:17469-17482.
- [Google Scholar]
- Synthesis of biscoumarin and dihydropyran derivatives as two novel classes of potential anti-bacterial derivatives. Arch. Pharm. Res.. 2016;39:1349-1355.
- [Google Scholar]
- Antibacterial activity and mechanism of silver nanoparticles against multidrug-resistant Pseudomonas aeruginosa. Int. J. Nanomedicine. 2019;14:1469-1487.
- [Google Scholar]
- Antibacterial activity and mechanism of linalool against Pseudomonas aeruginosa. Microb. Pathog.. 2020;141:103980
- [Google Scholar]
- Emergence and Spread of Epidemic Multidrug-Resistant Pseudomonas aeruginosa. Genome Biol. Evol.. 2017;9:3238-3245.
- [Google Scholar]
- Impact of thrombocytes, on bacterial growth and antimicrobial activity of selected antibiotics. Eur. J. Clin. Microbiol. Infect. Dis.. 2020;39:593-597.
- [Google Scholar]
- Antimicrobial Activity of Ibuprofen against Cystic Fibrosis-Associated Gram-Negative Pathogens. Antimicrob Agents Chemother.. 2018;62 e01574-17
- [Google Scholar]
- Pseudomonas aeruginosa biofilm: potential therapeutic targets. Biologicals. 2014;42:1-7.
- [Google Scholar]
- Antibacterial and Antitumor Activities of Biscoumarin and Dihydropyran Derivatives. Molecules. 2015;20:17614-17626.
- [Google Scholar]
- Antitumor activities of biscoumarin and dihydropyran derivatives. Bioorg. Med. Chem. Lett.. 2016;26:3876-3880.
- [Google Scholar]







