Translate this page into:
Exploring the effective components and potential mechanisms of Zukamu granules against acute upper respiratory tract infections by UHPLC-Q-Exactive Orbitrap-MS and network pharmacology analysis
⁎Corresponding authors at: Hunan Province Key Laboratory for Antibody-based Drug and Intelligent Delivery System, School of Pharmaceutical Sciences, Hunan University of Medicine, Huaihua 418000, China (W. Cai), National Institutes for Food and Drug Control, Beijing 100050, China; Xinjiang Uygur Autonomous Region Institute for Drug Control, Urumqi 830054, China (J.B. Yang). yangjianbo@nifdc.org.cn (Jianbo Yang), 20120941161@bucm.edu.cn (Wei Cai)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Acute upper respiratory tract infections (AURTIs) are common diseases of respiratory system, which are caused by adenoviruses and generate the mix of severe clinical presentation. Zukamu granules (ZKMG), a traditional Chinese medicine (TCM) prescription within Health Commission of Xinjiang Uygur Autonomous Region possesses anti-influenza virus, antibacterial and anti-inflammatory effects that exerts therapeutic effects in treatment of AURTIs. However, the main effective chemical components and their corresponding action mechanisms have not been clarified. Therefore, ultra-performance liquid chromatography coupled with quadrupole Exactive orbitrap tandem mass spectrometry (UHPLC-Q-Exactive Orbitrap MS) and network pharmacology were used to detect and identify potentially effective components in ZKMG as well as uncover their pharmacological mechanisms against AURTIs. As a result, a total of 265 components from all 12 composed herbal medicines were characterized based on self-built database and fragmentation patterns, of which 38 compounds were unambiguously confirmed using reference standards. Then, the compound-target-pathway network was constructed that implied potential therapeutic mechanisms of ZKMG on AURTIs. Compounds noscapine, cryptopine, steviol-19-O-glucoside, N-methylnarcotine, allocryptopine, naringenin, boldine, methyl rosmarinate with related targets EGFR, PTGS2, IL2, MMP9, TNF, AKT1, PIK3CA, F3 were considered as the key components and targets. Besides, the results also indicated that PI3K-Akt, AGE-RAGE, PD-L1, HIF-1 signaling pathways contributed significantly to the therapeutic effects of ZKMG on AURTIs. Overall, ZKMG could have an effect on AURTIs based on multicomponent, multitarget, and multichannel mechanisms of action as well as this method provides guiding significance for the further development of TCM treatment.
Keywords
Zukamu granules
Acute upper respiratory tract infections
Effective components
Mechanisms
UHPLC-Q-Exactive Orbitrap-MS
Network pharmacology
1 Introduction
Acute upper respiratory tract infections (AURTIs) are one of the most common respiratory infectious diseases characterized by tonsillitis, pharyngitis, laryngitis, rhinitis, angina and the common cold, which are primarily caused by viruses rather than bacteria, fungi, and helminths (Huang et al., 2019; Xu et al., 2022). It was reported that the global incidence of AURTIs was 17.1 billion worldwide in 2017, which has brought about tremendous socioeconomic burden to public health (James et al., 2018). Although, the majority of AURTIs are slight and self-limited, they could result in undesirable clinical outcomes, as well as their complications might develop into serious diseases and threaten the security of the infected (Huang et al., 2019). The conventional treatment for AURTIs are preliminary medicines that relieve symptoms but do not treat disease infections, such as antipyretic analgesics and antihistamines (Pelzman and Tung, 2021). Antibiotics only for specific AURTIs and validated clinical indications are recommended, but most of antibiotics for common AURTIs are needless and ineffective (Wang et al., 2014; Harris et al., 2016; Zhang et al., 2021). Additionally, the inappropriate utilization of antibiotics could lead to the emergence of resistant bacteria and increase treatment difficulties in clinical practice (Alrafiaah et al., 2017). Therefore, it is necessary that new therapeutic drugs are explored for AURTIs treatment.
Traditional Chinese medicine (TCM) has the advantages of low toxicity and less adverse reactions, reversing multidrug resistance and reducing drug dosage, which has been widely considered to be effective method for various diseases because of its characteristics of multicomponents and multitargets (Chen et al., 2021; Kong et al., 2021). It is by using TCM that AURTIs have been cured for more than 1800 years in China and TCM plays a significant role in disease prevention (Zeng et al., 2022). Zukamu granules (ZKMG) as a famous classical formula was first recorded in the Uyghur medical book karibatin kader over 1500 years ago, which was known as the first choice for the treatment of colds in Uighur (Hou et al., 2019; Zeng et al., 2021). ZKMG is composed of Kaempferia galanga (KG), Pericarpium papaveris (Papaver somniferum) (PP), Matricaria recutita (MR), Ziziphus jujuba (ZJ), Mentha canadensis (MC), Glycyrrhiza uralensis (GU), Althaea rosea (AL), Nymphaea tetragona (NT), Rheum palmatum (RP), Co rdia dicholoma (CRD) (Zeng et al., 2021). Modern pharmacological studies have revealed that ZKMG exhibits preferable anti-inflammatory anti-oxidant stress, regulation of apoptosis, and analgesic activities, as well as it is widely used for treating AURTIs and lung diseases in clinical studies (Li et al., 2022). Alkaloids in PP, flavonoids in ZJ, saponins in GU and quinones in RP have been primarily regarded as the main components of ZKMG prescription herbs. Besides, these components have been found to possess pharmacological activities against AURTIs (Fiore et al., 2008; Song et al., 2014; Liu et al., 2015; Parsaei et al., 2016). However, as a complex TCM prescription, the therapeutic effect of ZKMG is not a function of a single herb as well as active compounds and action mechanism in the treatment of AURTIs remain still unclear because of the multichemical components, multi-pharmacological effects, and multi-action targets of ZKMG, so it is quite necessary to explore the potential pharmacodynamic substances and mechanism, which are related to its therapeutic function.
Apparently, the diversity of the chemical constituents of TCM possibly results in complex interactions between active ingredients and multiple targets in diseases. It is a great challenge that how to comprehensively explore potential effective constituents and mechanisms of action (Huang et al., 2021). Ultra-high performance liquid chromatography coupled with quadrupole-Exactive orbitrap mass spectrometry (UHPLC-Q Exactive Orbitrap-MS) is powerful qualitative analysis technique, which has been developed for rapid characterization of unknown trace components in herbal medicines due to its high sensitivity, high resolution and high selectivity (Wang et al., 2021). Network pharmacology could systematically illustrate complex interactions among drugs, chemical constituents, targets, diseases and pathways, which has been considered to be a reliable approach to predict the pharmacological mechanism of drug treatments for diseases based on the development of systems biology and bioinformatics (Lu et al., 2021).
Therefore, it is a powerful method that UHPLC-MS combined with network pharmacology was used to explore the active ingredients and their potential mechanisms of TCM formula. In this study, a rapid and sensitive UHPLC-Q Exactive Orbitrap-MS method was established to investigate complex chemical compounds of ZKMG and their related mechanisms were explored by network pharmacology analysis. The study has certain guiding significance for clinical use and exploration mechanisms of action of ZKMG in treatment of AURTIs.
2 Materials and methods
2.1 Materials and reagents
Zukamu granules (ZKMG) were provided by Xinjiang Uygur Pharmaceutical Co., Ltd. (220252, Xinjiang, China). The detailed information of reference standards is listed in Supplementary Table S1. Chromatographic grade methanol and acetonitrile were purchased from Merck (New Jersey, USA). Ultrapure water was prepared with a Milli-Q system (Millipore, Milford, MA, USA). LC-MS grade formic acid was obtained from Fisher Scientific (New Jersey, USA). All other reagents were of analytical grade.
2.2 Sample preparation
Zukamu granules (1 g) were extracted in 50 mL 70% (v/v) aqueous methanol with ultrasonic treatment for 1 h at 30 °C and 40 kHz. The extracting solution was filtered and evaporated (50 °C) using the rotary vacuum evaporator to obtain the residue of ZKMG. Then, 100 mg ZKMG extracts were approximately weighted and dissolved in 1 mL methanol. After centrifugation at 12000 rpm for 20 min, an aliquot (2 μL) of supernatant was injected into the UHPLC-MS system for analysis.
Individual stock solutions of standards (1 mg/mL) were respectively accurately weighted and prepared by dissolving in methanol. Then, the solutions of 38 standards were mixed and serially diluted at a concentration of 10 µg/mL stored at −20 °C before UHPLC-MS analysis.
2.3 UHPLC-MS conditions
Each sample was performed on an Ultimate 3000 system (Thermo Fisher Scientific, California, USA) equipped with an Thermo Scientific Syncronis C18 (100 mm × 2.1 mm, 1.7 μm) at a temperature of 45 °C. The mobile phases consisted of 0.1% formic acid in water (A) and acetonitrile (B), delivering at a flow rate of 0.3 mL/min with the following solvent gradient: 0–2 min, 95–92% A; 2–5 min, 92–85% A; 5–10 min, 85–78% A; 10–12 min, 78–50% A; 12–20 min, 50–35% A; 20–25 min, 35–20% A; 25–26 min, 20–95% A; 26–30 min, 95% A.
The UHPLC system was coupled to a Q-Exactive Orbitrap MS (Thermo Fisher Scientific, Bremen, Germany), equipped with a heated electrospray ionization source (HESI) in the negative and positive modes for detection with the mass range of m/z 120–1200. For the spray voltage, the positive ion mode was 3.5 kV and the negative ion mode was 3.0 kV. The MS1 spectra were acquired with full MS mode with a resolution of 35,000, and the MS2 spectra resolution was 17,500 conducted in the data-dependent acquisition (DDA) mode. The sheath gas flow rate of 30 arbitrary units and auxiliary gas flow rate of 10 arbitrary units; and the temperatures of the capillary and auxiliary gas heaters were kept at 320 and 350 °C, respectively. The acquisition mode of the stepped normalized collision energy was set to 30, 40, and 60 %. The mass data were analyzed by Xcalibur 4.2 software (Thermo Fisher Scientific, California, USA).
2.4 The UHPLC-MS data analysis of ZKMG
The approaches for identifying chemical composition of ZKMG were divided into three steps. (1) The self-built database based on searching compounds in domestic and international databases or related natural products research literatures, such as GoogleScholar, PubMed and CNKI, was established for consisted herbs of ZKMG including compounds names, molecular formulars, fragment ions and classifications. Besides, the fragmentation pathways of reference standards obtained were investigated to be beneficial for the characterization. (2) The Xcalibur 4.2 software was applied to acquire extracted ion chromatograms (EICs) of possible compounds, theoretical mass, experimental mass, retention times, mass error and detailed MS/MS fragment ions in both negative and positive modes. (3) The data pre-obtained from (1) was extracted in experimental row data in Xcalibur 4.2 software to identify chemical profiles by matching MS/MS fragments. Furthermore, the fragmentation patterns of reference standards were used to characterize unknown components, which possess similar skeleton structures or fragment ions based on the principle of structural similarity (Fig. 1).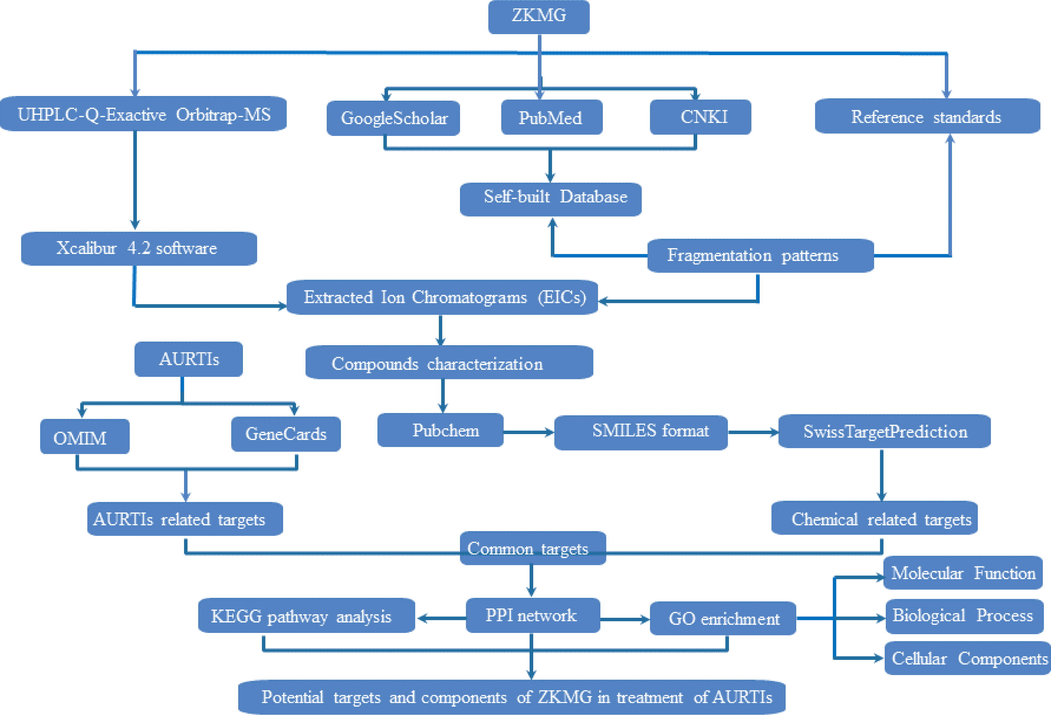
The analytical strategy based on UHPLC-MS and network pharmacology for exploring potential pharmacodynamic substance and mechanisms of action of ZKMG on AURTIs.
2.5 Network pharmacology analysis
Firstly, all the identified compounds based on UHPLC-MS were transformed into canonical SMILES through the PubChem (https://pubchem.ncbi.nlm.nih.gov/), most recently updated in March 2019. However, a number of compounds were not found in the PubChem, then their structures were imported in SwissTargetPrediction (http://www.swisstargetprediction.ch/, updated in 2019) to obtain SMILES, and the canonical SMILES information was uploaded into the SwissTargetPrediction database in “homo sapiens” species to predict all potential targets of compounds with probability ≥ 0.1 as the screening condition (Daina et al., 2019). Then the target genes obtained were verified by the UniProt protein database (https://www.uniprot.org/, updated in 2022) and converted into corresponding standard gene names (UniProt. 2018). Secondly, the keywords about “AURTIs” as well as relevant diseases “acute pharyngitis”, “acute tonsillitis”, “acute rhinitis”, “acute laryngitis” and “acute angina” were searched to obtain AURTIs related gene targets from Online Mendelian Inheritance in Man database (OMIM http://omim.org/, updated in 2018) and GeneCards database (https://www.genecards.org/, version 5.0), and screen genes with “relevance score” of disease ≥ 30, and the repeated targets were deleted (Amberger et al., 2015; Stelzer et al., 2016). Thirdly, the chemical compounds targets and the diseases targets were imported in bioinformatics website (http://bioinformatics.psb.ugent.be/webtools/venn/, updated in 2022) to draw a Venn diagram and obtain their common gene targets. Then, the chemical components of ZKMG and its therapeutic targets in AURTIs were introduced into Cytoscape (https://cytoscape.org/, version 3.9.1) to construct the compound-target network (Shannon et al., 2003). Afterwards, the overlapping targets between the compounds targets and the AURTIs related targets were inputted into STRING database (http://string-db.org/, version 11.0) with a high confidence score ≥ 0.9 to construct the protein–protein interaction (PPI) network. The Cytoscape software (version 3.9.1) was used to visualize and analysis interactions in this network according to the network node topological parameter “degree”, it was used for evaluating the importance in the PPI network of a protein (Szklarczyk et al., 2018; Zhuang et al., 2020). Finally, the Database for Annotation, Visualization and Integrated Discovery (DAVID https://david.ncifcrf.gov/, version 6.8) was used for GO enrichment including molecular function (MF), biological process (BP), cellular components (CC), and KEGG pathway analysis (https://www.bioinformatics.com.cn/, updated in 2022), which provided systematic and comprehensive biological function annotation information. Meanwhile, top 20 enriched pathways of three GO terms (MF, BP, and CC) and KEGG were visualized in a bubble chart as well as three GO terms were drawn together with a bar chart and represented by a box line partition (p < 0.01) (Huang da et al., 2009; Li et al., 2022). Compound-target-pathway networks were constructed by using Cytoscape software (Fig. 1).
3 Results and discussion
3.1 Characterization of chemical components of ZKMG
The chemical ingredients of ZKMG were detected and identified by UHPLC-Q-Exactive Orbitrap MS in both positive and negative modes to obtain as much information as possible. Totally, 265 compounds were identified from ZKMG based on accurate precursor and product ions and comparing their fragmentation patterns with standards or reported in the literatures, including 46 alkaloids, 92 flavonoids, 28 triterpenoid saponins, 27 phenolic acids, 24 phenylpropanoids, 21 quinones, 13 tannins, 4 amino acids, 4 nucleosides, 2 naphthols, 2 phenols, 2 terpenoids in Table 1 and Supplementary Table S2. The extracted ion chromatograms (EICs) of these compounds in both positive mode and negative ion modes were showed in Fig. 2. And 38 compounds in ZKMG were unambiguously identified by comparison with reference standards. * Compared with standard compounds.
peak
tR
Theoretical Mass m/z
Experimental Mass m/z
Error (ppm)
Formula
Identification
peak
tR
Theoretical Mass m/z
Experimental Mass m/z
Error (ppm)
Formula
Identification
1*
0.84
136.0618
136.0617
−0.68
C5H5N5
adenine
134
10.37
463.0882
463.0888
1.31
C21H20O12
quercetin-7-O-glucoside
2*
0.84
191.0561
191.0553
−4.19
C7H12O6
quinic acid
135
10.41
477.0675
477.0680
1.04
C21H18O13
quercetin-3-O-glucuronide
3
0.90
191.0197
191.0190
−3.80
C6H8O7
citric acid isomer
136*
10.47
463.0882
463.0888
1.32
C21H20O12
isoquercitrin
4*
0.92
133.0142
133.0132
−8.24
C4H6O5
malic acid
137
10.52
447.0933
447.0931
−0.45
C21H20O11
luteolin‐5‐O‐glucoside
5
1.05
191.0197
191.0190
−3.80
C6H8O7
isocitric acid
138
10.55
445.0776
445.0781
1.02
C21H18O11
rhein-8-O-β-D-glucoside
6*
1.18
191.0197
191.0190
−3.75
C6H8O7
citric acid
139*
10.66
447.0933
447.0937
0.91
C21H20O11
cynaroside
7
1.28
331.0671
331.0674
1.03
C13H16O10
1-O-galloylglucose
140
10.66
461.0725
461.0730
0.91
C21H18O12
kaempferol-3-O-β-D-glucuronide
8
1.31
268.1040
268.1038
−1.01
C10H13N5O4
adenosine
141*
10.69
623.1981
623.1988
1.04
C29H36O15
acteoside
9
1.32
132.1019
132.1019
−0.19
C6H13NO2
isoleucine
142
10.87
615.0992
615.1002
1.63
C28H24O16
quercetin-O-galloyl-glucopyranoside
10
1.43
284.0989
284.0987
−0.90
C10H13N5O5
guanosine
143
11.08
623.1618
623.1622
0.76
C28H32O16
isorhamnetin-3-O-nehesperidine
11
1.45
132.1019
132.1019
−0.27
C6H13NO2
leucine
144
11.12
370.1649
370.1646
−0.94
C21H23NO5
allocryptopine
12
1.48
331.0671
331.0674
1.03
C13H16O10
gallic acid-4-O-β-D-glucopyranoside
145*
11.26
515.1195
515.1198
0.49
C25H24O12
isochlorogenic acid B
13*
1.49
330.0598
330.0592
−1.72
C10H12N5O6P
adenosine cyclophosphate
146
11.29
340.1543
340.1540
−1.10
C20H21NO4
papaverine
14
1.51
302.1387
302.1383
−1.24
C17H19NO4
morphine N-oxide
147
11.33
579.1719
579.1726
1.07
C27H32O14
naringin
15
1.69
331.0671
331.0674
1.03
C13H16O10
gallic acid-3-O-β-D-glucopyranoside
148*
11.42
593.1512
593.1517
0.91
C27H30O15
kaempferol-3-O-rutinoside
16
1.80
286.1438
286.1433
−1.61
C17H19NO3
morphine
149
11.44
414.1547
414.1541
−1.57
C22H23NO7
noscapine isomer 1
17*
1.98
169.0142
169.0134
−4.95
C7H6O5
gallic acid
150*
11.55
515.1195
515.1201
1.09
C25H24O12
1,5-dicaffeoylquinic acid
18
2.44
166.0863
166.0862
−0.27
C9H11NO2
phenylalanine
151*
11.56
137.0244
137.0235
−7.06
C7H6O3
salicylic acid
19
3.23
179.0350
179.0553
−4.81
C9H8O4
caffeic acid isomer
152
11.63
623.1981
623.1992
1.73
C29H36O15
isoacteoside
20
3.32
197.0455
197.0448
−3.64
C9H10O5
danshensu
153
11.65
400.1391
400.1387
−1.02
C21H21NO7
narcotoline
21
3.51
329.0878
329.0882
1.29
C14H18O9
pseudolaroside B
154*
11.70
515.1195
515.1199
0.72
C25H24O12
isochlorogenic acid A
22
3.75
153.0193
153.0185
−5.76
C7H6O4
protocatechuic acid
155
11.73
491.0831
491.0836
0.97
C22H20O13
isorhamnetin-7-O-glucuronide
23
3.88
153.0557
153.0548
−5.93
C8H10O3
3,4-dihydroxyphenylethanol
156
11.73
519.1872
519.1877
1.07
C26H32O11
pinoresinol 4-O-β-D-glucopyranoside
24
4.15
451.1246
451.1249
0.78
C21H24O11
catechin‐5‐O‐glucoside
157
11.78
577.1563
577.1569
1.11
C27H30O14
apigenin-7-O-rutinoside
25
4.16
205.0972
205.0971
−0.07
C11H12N2O2
tryptophan
158
11.78
623.1618
623.1619
0.18
C28H32O16
isorhamnetin-3-O-rutinoside
26
4.25
483.0780
483.0784
0.83
C20H20O14
gallic acid-O-galloyl-glucoside
159
11.85
414.1547
414.1541
−1.42
C22H23NO7
noscapine isomer 2
27
4.33
448.1966
448.1965
−0.14
C23H29NO8
N-methylnorcoclaurine-7-O-glucoside
160
11.90
565.1563
565.1571
1.45
C26H30O14
hydroxyliquiritin apioside
28
4.33
515.1406
515.1411
0.82
C22H28O14
chlorogenic acid-hexoside
161
11.93
447.0933
447.0936
0.64
C21H20O11
luteolin-4′-O-glucoside
29
4.43
311.0409
311.0412
1.21
C13H12O9
caftaric acid
162
12.11
446.1809
446.1802
−1.71
C23H27NO8
narceine
30
4.47
272.1281
272.1277
−1.47
C16H17NO3
DL-demethylcoclaurine
163
12.17
282.1489
282.1485
−1.26
C18H19NO2
O-nornuciferine
31*
4.51
353.0878
353.0880
0.67
C16H18O9
neochlorogenic acid
164
12.17
433.1140
433.1143
0.55
C21H22O10
naringenin-7-O-glucoside
32
4.52
462.2122
462.2115
−1.70
C24H31NO8
N-methylcoclaurine-7-O-glucoside
165
12.28
431.0984
431.0985
0.19
C21H20O10
emodin-1-O-D-glucoside
33
4.54
300.1594
300.1589
−1.83
C18H21NO3
codeine
166
12.36
445.0776
445.0779
0.62
C21H18O11
apigenin-7-O-glucuronide
34
4.58
285.0616
285.0619
1.16
C12H14O8
uralenneoside isomer1
167*
12.37
609.1825
609.1832
1.21
C28H34O15
hesperidin
35
4.71
163.0401
163.0392
−5.63
C9H8O3
coumaric acid
168*
12.43
515.1195
515.1197
0.37
C25H24O12
isochlorogenic acid C
36
4.80
483.0780
483.0784
0.83
C20H20O14
gallic acid-O-galloyl-glucoside
169
12.43
607.1668
607.1674
0.95
C28H32O15
diosmin
37
4.81
448.1966
448.1962
−0.94
C23H29NO8
N-methylnorcoclaurine-4′-O-glucoside
170*
12.48
359.0772
359.0772
−0.09
C18H16O8
rosmarinic acid
38
4.89
285.0616
285.0619
1.05
C12H14O8
uralenneoside isomer 2
171
12.53
491.0831
491.0834
0.66
C22H20O13
isorhamnetin-3-O-glucuronide
39
5.04
451.1246
451.1249
0.72
C21H24O11
catechin‐7‐O‐glucoside
172
12.56
463.1235
463.1233
−0.49
C22H22O11
tectoridin
40
5.21
339.0722
339.0727
1.58
C15H16O9
esculin
173
12.66
639.1920
639.1920
0.09
C29H34O16
aurantio-obtusin-6-O-rutinoside
41
5.27
137.0244
137.0234
−7.28
C7H6O3
4-hydroxybenzoic acid
174
12.69
463.1235
463.1231
−0.82
C22H22O11
homoplantaginin
42
5.40
483.0780
483.0783
0.58
C20H20O14
gallic acid-O-galloyl-glucoside
175
12.71
463.1246
463.1250
0.90
C22H24O11
hesperetin-7-O-β-D-glucoside
43
5.50
515.1406
515.1412
1.05
C22H28O14
chlorogenic acid-hexoside
176
12.74
549.1614
549.1621
1.36
C26H30O13
licuraside
44
5.63
451.1246
451.1249
0.65
C21H24O11
catechin‐4′‐O‐glucoside
177
12.79
431.0984
431.0986
0.53
C21H20O10
aloe-emodin-3-(hydroxymethyl)-O-β-D-glucopyranoside
45
5.65
483.0780
483.0785
0.96
C20H20O14
gallic acid-O-galloyl-glucoside
178
12.79
591.1719
591.1727
1.25
C28H32O14
liquiritigenin-4′-O-(β-D-3-O-acetyl-apiofuranosyl-(1 → 2)-β-d-glucopyranoside
46
5.71
314.1751
314.1747
−1.15
C19H23NO3
lotusine
179
12.80
463.1235
463.1234
−0.10
C22H22O11
diosmetin-7-O-β-D-glucopyranoside
47
5.74
337.0929
337.0933
1.27
C16H18O8
5-p-coumaroylquinic acid
180
12.85
447.0933
447.0933
0.01
C21H20O11
luteolin-3′-O-glucoside
48
5.86
316.1543
316.1540
−1.00
C18H21NO4
norreticuline
181
12.86
475.0882
475.0885
0.57
C22H20O12
diosmetin-7-O-glucuronide
49
5.92
515.1406
515.1411
0.93
C22H28O14
chlorogenic acid-hexoside
182
12.89
459.1297
459.1302
1.13
C23H24O10
6′-acetylliquiritin
50
6.02
483.0780
483.0786
1.14
C20H20O14
gallic acid-O-galloyl-glucoside
183
12.90
417.1191
417.1195
0.90
C21H22O9
isoliquiritin
51
6.04
451.1246
451.2189
0.85
C21H24O11
catechin-3′-O-glucoside
184
12.92
493.1341
493.1340
−0.17
C23H24O12
aurantio-obtusin-6-O-glucoside
53*
6.14
353.0878
353.0881
0.75
C16H18O9
chlorogenic acid
185
12.96
431.1337
431.1334
−0.69
C22H22O9
ononin
52
6.14
325.0929
325.0931
0.77
C15H18O8
coumaric acid-O-glucoside
186
12.99
417.1191
417.1195
0.97
C21H22O9
neoisoliquiritin
54
6.16
635.0890
635.0895
0.78
C27H24O18
tri-O-galloyl-glucoside
187
13.03
433.0776
433.0779
0.50
C20H18O11
quercetin 3-O-arabinoside
55*
6.37
353.0878
353.0881
0.84
C16H18O9
cryptochlorogenic acid
188
13.04
695.1981
695.2002
−2.09
C35H36O15
licorice-glycoside B
56
6.40
286.1438
286.1436
−0.77
C17H19NO3
coclaurine
189
13.09
725.2087
725.2099
1.64
C36H38O16
licorice-glycoside A
57
6.48
300.1594
300.1592
−0.80
C18H21NO3
N-methylisococlaurine
190
13.09
459.0933
459.0938
1.21
C22H20O11
carboxyl-chrysophanol-O-glucose
58
6.52
291.0146
291.0148
0.55
C13H8O8
brervifolincaboxylic acid
191
13.21
283.0612
283.0607
−1.76
C16H12O5
glycitein
59
6.55
367.1035
367.1041
1.84
C17H20O9
4-feruloylquinic acid
192
13.21
447.0933
447.0935
0.44
C21H20O11
isorhamnetin-3-O-arabinoside
60
6.63
483.0780
483.0785
0.96
C20H20O14
gallic acid-O-galloyl-glucoside
193
13.25
593.1876
593.1881
0.90
C28H34O14
didymin
61
6.63
625.1410
625.1420
1.61
C27H30O17
quercetin-3-O-diglucoside
194
13.27
255.0663
255.0663
0.19
C15H12O4
liquiritigenin
62
6.67
325.0929
325.0933
1.14
C15H18O8
coumaric acid-O-glucoside
195
13.27
373.0929
373.0931
0.59
C19H18O8
methyl rosmarinate
63
6.67
635.0890
635.0898
1.34
C27H24O18
tri-O-galloyl-glucoside
196
13.29
473.1089
473.1090
0.16
C23H22O11
aloe-emodin-8-O-(6-O-acetyl)-glucoside
64
6.67
771.1989
771.2005
2.03
C33H40O21
quercetin-3-O-sophoroside-7-O-rhamnoside
197
13.31
999.4442
999.4453
1.00
C48H72O22
24-hydroxy-licoricesaponin A3
65*
6.71
179.0350
179.0343
−3.87
C9H8O4
caffeic acid
198
13.35
837.3914
837.3927
1.52
C42H62O17
yunganoside K2
66
6.76
635.0890
635.0897
1.06
C27H24O18
tri-O-galloyl-glucoside
199
13.40
489.1038
489.1059
4.25
C23H22O12
luteolin-7-O-6″-ocetylglucoside
67
6.81
328.1543
328.1541
−0.87
C19H21NO4
boldine
200
13.41
407.1348
407.1350
0.48
C20H24O9
torachrysone-8-O-glucoside
68
6.82
344.1856 [M]+
344.1853
−0.88
C20H26NO4+
N-methylreticuline
201*
13.42
285.0405
285.0406
0.45
C15H10O6
luteolin
69
6.85
579.1719
579.1729
1.59
C27H32O14
liquiritigenin-O-diglucuronide
202
13.42
431.0984
431.0985
0.26
C21H20O10
emodin-8-O-D-glucoside
70
7.09
386.1598
386.1597
−0.37
C21H23NO6
glaucamine
203*
13.44
301.0354
301.0356
0.78
C15H10O7
quercetin
71
7.09
711.2141
711.2150
1.10
C32H40O18
glucoliquirtin asioside
204
13.45
415.1035
415.1037
0.66
C21H20O9
chrysophanol-1-O-β-D-glucoside
72*
7.11
342.1699 [M]+
342.1694
−1.62
C20H24NO4+
magnoflorine
205
13.45
253.0506
253.0505
−0.48
C15H10O4
chrysophanol
73
7.15
515.1194
515.1196
0.25
C25H24O12
dicaffeylquinic acid
206
13.46
447.1286
447.1282
−0.77
C22H22O10
calycosin-7-O-β-D-glucoside
74
7.16
314.1750 [M]+
314.1748
−0.86
C19H24NO3+
isolotusine
207
13.46
983.4493
983.4506
1.25
C48H72O21
licoricesaponin A3
75
7.17
337.0928
337.0931
0.56
C16H18O8
3-p-coumaroylquinic acid
208*
13.48
283.0612
283.0611
−0.34
C16H12O5
physcion
76
7.19
325.0928
325.0932
0.95
C15H18O8
coumaric acid-O-glucoside
209
13.50
853.3863
853.3876
1.44
C42H62O18
22-hydroxy-licoricesaponin G2 isomer 1
77
7.24
328.1543
328.1541
−0.59
C19H21NO4
corytuberine
210
13.52
315.0510
315.0513
0.97
C16H12O7
isorhamnetin isomer
78
7.42
153.1273
153.1273
−0.80
C10H16O
camphor
211
13.52
895.3969
895.3982
1.44
C44H64O19
hydroxy acetoxyglycyrrhizin
79
7.42
330.1699
330.1696
−1.29
C19H23NO4
reticuline
212
13.53
415.1035
415.1037
0.59
C21H20O9
chrysophanol-8-O-glucoside
80
7.46
300.1594
300.1592
−0.70
C18H21NO3
N-methylcoclaurine
213
13.62
301.0707
301.0704
−0.88
C16H12O6
tectorigenin
81
7.48
635.0889
635.0896
0.97
C27H24O18
tri-O-galloyl-glucoside
214
13.64
315.0510
315.0515
1.35
C16H12O7
isorhamnetin isomer
82
7.55
337.0928
337.0932
0.92
C16H18O8
4-p-coumaroylquinic acid
215
13.64
853.3863
853.3878
1.74
C42H62O18
22-hydroxy-licoricesaponin G2 isomer 2
83
7.58
593.1511
593.1517
0.91
C27H30O15
vicenin Ⅱ
216
13.70
879.4020
879.4031
1.24
C44H64O18
acetoxy-glycyrrhizic acid
84
7.64
635.0889
635.0895
0.78
C27H24O18
tri-O-galloyl-glucoside
217
13.72
837.3914
837.3924
1.17
C42H62O17
licoricesaponin P2
85
7.67
314.1750
314.1746
−1.43
C19H23NO3
4′-methyl-N-methylcoclaurine
218
13.72
859.3758
859.3742
−1.82
C44H60O17
methyllicorice-saponin Q2 isomer 1
86
7.67
314.1750 [M]+
314.1746
−1.43
C19H24NO3+
magnocurarine
219
13.77
445.1140
445.1145
1.15
C22H22O10
physcion-8-O-β-D-glucoside
87
7.67
639.1567
639.1574
1.06
C28H32O17
isorhamnetin-3,7-O-diglucoside
220
13.78
329.0667
329.0671
1.32
C17H14O7
jaceosidin
88
7.98
328.1543
328.1541
−0.87
C19H21NO4
scoulerine
221*
13.86
271.0612
271.0614
0.75
C15H12O5
naringenin
89
8.02
298.1438
298.1436
−0.74
C18H19NO3
codeinone
222
13.86
835.3758
835.3762
0.46
C42H60O17
yunganoside M
90
8.08
635.0890
635.0898
1.25
C27H24O18
tri-O-galloyl-glucoside
223
13.89
283.0612
283.0613
0.51
C16H12O5
acacetin
91
8.39
356.1492
356.1489
−0.98
C20H21NO5
amurensinine N-oxide A
224
13.89
445.1140
445.1144
0.94
C22H22O10
physcion-1-O-β-D-glucoside
92
8.39
367.1035
367.1045
2.90
C17H20O9
5-feruloylquinic acid
225
13.91
269.0455
269.0457
0.64
C15H10O5
apigenin
93
8.43
635.0890
635.0886
−0.58
C27H24O18
tri-O-galloyl-glucoside
226
13.93
329.0667
329.0669
0.56
C17H14O7
iristectorigenin B
94
8.45
300.1594
300.1591
−1.20
C18H21NO3
6-demethyl-4′-methyl-N-methylcoclaurine
227
13.98
879.4020
879.4040
−1.72
C44H64O18
acetoxy-glycyrrhizic acid
95
8.45
344.1856
344.1852
−1.23
C20H25NO4
codamine
228*
13.99
285.0405
285.0407
0.87
C15H10O6
kaempferol
96
8.60
268.1332
268.1329
−1.03
C17H17NO2
caaverine
229
14.10
301.0707
301.0702
−1.68
C16H12O6
hispidulin
97
8.60
337.0929
337.0935
1.75
C16H18O8
1-p-coumaroylquinic acid
230
14.10
301.0718
301.0721
1.16
C16H14O6
hesperetin
98
8.62
625.1410
625.1417
1.12
C27H30O17
myricetin-3-O-rutinoside
231
14.11
819.3809
819.3823
1.78
C42H60O16
licoricesaponin E2
99*
8.63
163.0401
163.0392
−5.20
C9H8O3
4-coumaric acid
232
14.11
837.3914
837.3922
0.94
C42H62O17
licoricesaponin Q2
100
8.67
579.1719
579.1727
1.28
C27H32O14
liquiritigenin-O-diglucuronide
233
14.11
859.3758
859.3737
−2.39
C44H60O17
methyllicorice-saponin Q2 isomer 2
101
8.68
563.1406
563.1412
0.96
C26H28O14
schaftoside
234
14.11
967.4544
967.4556
1.19
C48H72O20
haoglycyrrhizin isomer 1
102
8.83
459.0933
459.0937
0.94
C22H20O11
carboxyl-chrysophanol-O-glucose
235*
14.14
315.0510
315.0514
1.16
C16H12O7
isorhamnetin
103
8.84
625.1410
625.1417
1.12
C27H30O17
quercetin-7-O-diglucoside
236
14.22
301.0707
301.0705
−0.65
C16H12O6
diosmetin
104
8.87
344.1856
344.1853
−0.97
C20H25NO4
laudanine
237
14.22
837.3914
837.3923
1.08
C42H62O17
uralsaponin N
105
8.89
312.1594
312.1589
−1.54
C19H21NO3
thebaine
238
14.24
863.4071
863.4081
1.17
C44H64O17
acetoxyglycyrrhaldehyde
106
8.91
326.1387
326.1382
−1.45
C19H19NO4
pacodine (7-O-demethylpapaverine)
239
14.27
449.1453
449.1456
0.69
C22H26O10
torachrysone-O-acetylglucoside
107
8.96
367.1035
367.1043
2.24
C17H20O9
3-feruloylquinic acid
240
14.35
255.0663
255.0661
−0.64
C15H12O4
isoliquiritigenin
108
9.08
463.0882
463.0887
0.97
C21H20O12
quercetin-5-O-glucoside
242*
14.36
821.3965
821.3973
0.94
C42H62O16
glycyrrhizic acid
109
9.36
433.1140
433.1143
0.55
C21H22O10
naringenin-4′-O-glucoside
241
14.36
329.0667
329.0668
0.47
C17H14O7
aurantio-obtusin
110
9.37
314.1751
314.1749
−0.45
C19H23NO3
armepavine
243
14.38
837.3914
837.3920
0.72
C42H62O17
hydroxyglycyrrhizin
111
9.40
417.1180
417.1178
−0.43
C21H20O9
daidzin
244
14.48
457.1140
457.1146
1.18
C23H22O10
chrysophanol-O-acetylglucoside
112
9.41
342.1700
342.1698
−0.45
C20H23NO4
tetrahydrocolumbamine
245
14.55
354.1336
354.1335
−0.17
C20H19NO5
pseudoprotopine
113
9.59
579.1719
579.1726
1.07
C27H32O14
liquiritigenin-O-diglucuronide
246
14.55
967.4544
967.4561
1.76
C48H72O20
haoglycyrrhizin isomer 2
114
9.59
621.1097
621.1104
1.01
C27H26O17
apigenin-7-O-diglucuronide
247
14.60
837.3914
837.3924
1.17
C42H62O17
hydroxyglycyrrhizin
115
9.63
326.1387
326.1384
−0.87
C19H19NO4
palaudine (3′-O-demethylpapaverine)
248
14.73
821.3965
821.3975
1.23
C42H62O16
licoricesaponin H2
116
9.67
549.1614
549.1619
1.03
C26H30O13
liquiritin apioside
249
14.76
807.4172
807.4185
1.51
C42H64O15
licoricesaponin B2
117
9.69
595.1668
595.1679
1.79
C27H32O15
eriocitrin
250
14.80
955.4908
955.4923
1.58
C48H76O19
yunganoside C1
118
9.72
417.1191
417.1195
0.83
C21H22O9
neoliquiritin
251
14.82
369.1333
369.1329
−1.07
C21H20O6
gancaonin N
119
9.81
433.1140
433.1144
0.97
C21H22O10
naringenin-5-O-glucoside
252
14.94
955.4908
955.4924
1.70
C48H76O19
yunganoside A1
120
9.94
428.1704
428.1699
−1.21
C23H25NO7
N-methylnarcotine
253
15.03
479.2650
479.2654
0.83
C26H40O8
steviol-19-O-glucoside
121
9.95
549.1614
549.1617
0.69
C26H30O13
isoliquiritin apioside
254
15.10
297.0405
297.0406
0.33
C16H10O6
6-methyl-rhein
122
9.97
517.0412
517.0438
4.86
C26H14O12
1,1,3,4,5,6,8,8′-octahydroxy-9H,9′H-2,2′-bixanthene-9,9′-dione
255
15.17
953.4752
953.4742
−0.96
C48H74O19
yunganoside D1
123
9.98
417.1191
417.1193
0.51
C21H22O9
liquiritin
256*
15.44
283.0248
283.0249
0.31
C15H8O6
rheic acid
124
10.00
370.1649
370.1643
−1.75
C21H23NO5
cryptopine
257
15.54
343.0823
343.0825
0.57
C18H16O7
eupatilin
125*
10.01
609.1461
609.1467
1.02
C27H30O16
rutin
258
15.67
283.0612
283.0613
0.40
C16H12O5
biochanin A
126
10.03
537.1038
537.1040
0.28
C27H22O12
lithospermic acid isomer
259
15.72
369.1333
369.1329
−1.07
C21H20O6
glicoricone
127
10.06
354.1336
354.1336
−0.05
C20H19NO5
protopine
260
15.72
807.4172
807.4184
1.43
C42H64O15
22-dehydrouralsaponin C
128*
10.09
300.9990
300.9991
0.40
C14H6O8
ellagic acid
261
15.83
355.1187
355.1193
1.63
C20H20O6
uralenin
129
10.10
431.0984
431.0985
0.33
C21H20O10
apigenin-7-O-β-D-glucoside
262
15.98
589.1351
589.1359
1.21
C31H26O12
1-methyl-8-hydroxy-9,10-anthraquinone-3-O-(6′-O-cinnamoyl)-glucoside
130
10.12
358.2013
358.2008
−1.24
C21H27NO4
laudanosine
263*
16.27
593.1301
593.1309
1.34
C30H26O13
procyanidin
131*
10.23
463.0882
463.0888
1.32
C21H20O12
hyperoside
264*
17.72
269.0455
269.0458
0.87
C15H10O5
emodin
132
10.29
593.1512
593.1520
1.43
C27H30O15
luteolin-7-O-rutinoside
265*
23.15
469.3323
469.3333
1.95
C30H46O4
18 β-glycyrrhetintic acid
133
10.34
344.1856
344.1855
−0.51
C20H25NO4
tetrahydropapaverine
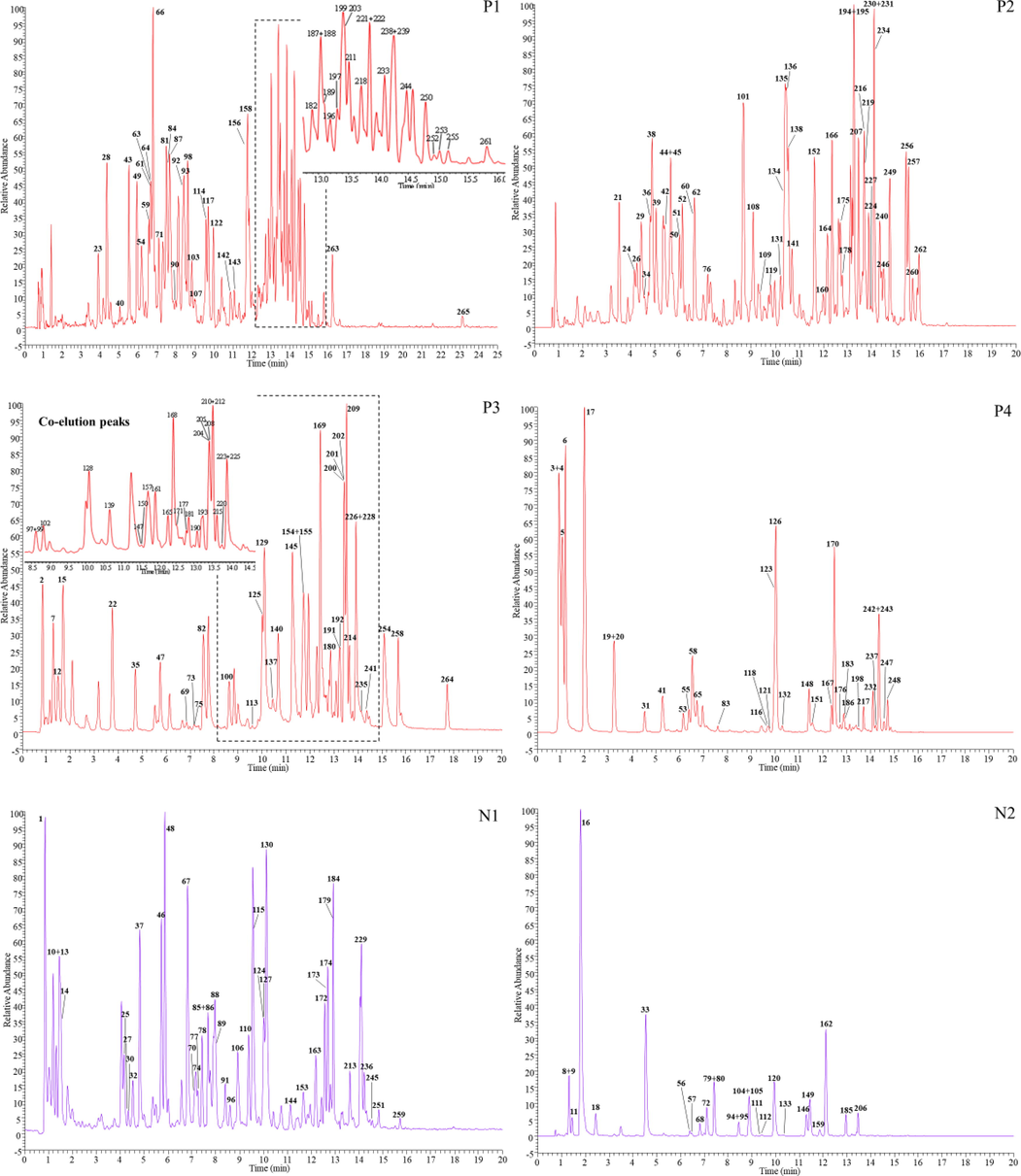
The high resolution extracted ion chromatograms (EICs) of ZKMG in the positive (P) and negative ion mode (N). P1. m/z 153.0557, 271.0611, 301.0353, 339.0721, 355.1187, 367.1034, 433.0776, 449.1453, 457.1140, 459.1296, 469.3323, 473.1089, 479.2650, 489.1038, 515.1406, 517.0412, 519.1871, 593.1300, 595.1668, 615.0991, 621.1097, 623.1617, 625.1410, 635.0889, 639.1566, 695.1981, 711.2141, 725.2087, 771.1989, 835.3757, 859.3757, 863.4070, 895.3969, 953.4751, 955.4908, 999.4442; P2. m/z 255.0662, 283.0248, 285.0615, 301.0717, 311.0408, 325.0928, 329.0878, 343.0823, 373.0928, 433.1140, 445.0776, 445.1140, 451.1245, 463.0881, 463.1245, 477.0674, 483.0780, 563.1406, 565.1562, 589.1351, 591.1719, 623.1981, 807.4172, 819.3808, 879.4019, 967.4544, 983.4493; P3. m/z 153.0193, 163.0400, 191.0561, 253.0506, 269.0455, 283.0611, 285.0404, 297.0404, 300.9989, 315.0510, 329.0666, 331.0670, 337.0928, 407.1347, 415.1034, 431.0983, 447.0932, 459.0932, 461.0725, 475.0881, 491.0831, 515.1194, 577.1562, 579.1719, 593.1875, 607.1668, 609.1461, 853.3863; P4. m/z 133.0142, 137.0244, 169.0142, 179.0349, 191.0197, 197.0455, 291.0146, 353.0878, 359.0772, 417.1191, 537.1038, 549.1613, 593.1511, 609.1824, 821.3965, 837.3914; N1. m/z136.0617, 153.1273, 205.0971, 268.1332, 272.1281, 282.1488, 284.0989, 298.1437, 301.0706, 302.1386, 314.1750, 314.1761, 316.1543, 326.1386, 328.1543, 330.0597, 354.1335, 356.1492, 358.2012, 369.1332, 370.1648, 386.1598, 400.1390, 448.1965, 462.2122, 463.1234, 493.1340, 639.1919; N2. m/z 132.1019, 166.0862, 268.1040, 286.1437, 300.1594, 312.1594, 330.1699, 340.1543, 342.1699, 344.1856, 414.1547, 417.1180, 428.1703, 431.1336, 446.1809, 447.1285.
3.1.1 Identification of alkaloids
In this study, a total of 46 alkaloids mainly derived from PP were identified, which could be further divided into different alkaloids, including 2 tetrahydroprotoberberines, 5 aporphines, 20 benzyltetrahydroisoquinolines, 5 phthalideisoquinolines, 6 protopines, 5 morphinans, 3 benzylisoquinolines. The proposed fragmentation pathways of each-type representative alkaloids (peaks 88, 72, 56, 153, 144, 16, 146) were observed in Supplementary Figure S3.
3.1.1.1 Identification of tetrahydroprotoberberine alkaloids
Tetrahydroprotoberberine is a four-ring structure, which derived from two isoquinoline rings connected by sharing one nitrogen atom, and its C2, C3, C9, C10 contain oxygen groups (Yang et al., 2016). The peaks 88 and 112 were deemed to be tetrahydroprotoberberine alkaloids because of their MS/MS spectral patterns, which produced characteristic ions by Retro Diels-Alder (RDA) cleavage. They were tentatively identified as scoulerine and tetrahydrocolumbamine based on MS/MS fragmentation pattern of tetrahydroprotoberberine alkaloids. The fragmentation ions of scoulerine and tetrahydrocolumbamine were observed at m/z 178.086 [C10H12O2N]+ and 151.075 [C9H11O2]+ due to RDA fragmentation and cleavage of the B-ring, respectively. Another characteristic ion at m/z 163.060 was indicated by the loss of the methyl radical from the ion at m/z 178.086 (Jeong et al., 2012).
3.1.1.2 Identification of aporphine alkaloids
For aporphine alkaloids, the elimination of CH3NR group as well as lose CH3OH moiety and CO were the characteristic fragmentation patterns. Peak 72 showed precursor ion [M]+ at m/z 342.1699, which produced characteristic ions at m/z 297.111 [M-(CH3)2NH]+, 282.088 [M-(CH3)2NH-CH3]+, 265.085 [M-(CH3)2NH-CH3OH]+. Comparing with reference standards, peak 72 was accurately identified as magnoflorine. Similarly, peaks 67, 77 were pairs of isomers with [M + H]+ ion at m/z 328.1543 and exhibited three major diagnostic fragment ions at m/z 297.111 [M + H-CH3NH2]+, 265.085 [M + H-CH3NH2-CH3OH]+, 237.090 [M + H-CH3NH2-CH3OH-CO]+. Therefore, peaks 67, 77 were respectively determined as boldine and corytuberine according to their retention behavior on the chromatographic column (Conceição et al., 2020). Similarly, peaks 96 and 163 were considered as caaverine and O-nornuciferine.
3.1.1.3 Identification of benzyltetrahydroisoquinoline alkaloids
For benzyltetrahydroisoquinoline alkaloids, they were easy to observe the elimination of NRH2, CH3OH groups and other characteristic ions at m/z 175.075 [C11H11O2]+, 161.059 [C10H9O2]+, 121.064 [C8H9O]+, 107.049 [C7H7O]+. Peak 30 with the parent ion [M + H]+ at m/z 272.12811, was easy to produce fragment ions at m/z 255.101 [M + H-NH3]+, 161.059 [M + H-NH3-C6H5O]+, and 107.049 [M + H-C9H11N2O]+. Consequently, it was tentatively determined to be DL-demethylcoclaurine based on the related literature (Oh et al., 2018). Peak 56 showed the [M + H]+ ion at m/z 286.1437, which yielded characteristic ions at m/z 269.117 [M + H-NH3]+, 237.090 [M + H-NH3-CH3OH]+, 175.075 [M + H-NH3-C6H6O]+, so peak 56 was tentatively named coclaurine (Menéndez-Perdomo et al., 2021). Similarly, peaks 27, 32, 37, 46, 57, 74, 80, 85, 86, 94, 110 were successfully identified as N-methylnorcoclaurine-7-O-glucoside, N-methylcoclaurine-7-O-glucoside, N-methylnorcoclaurine-4′-O-glucoside, lotusine, N-methylisococlaurine, isolotusine, N-methylcoclaurine, 4′-methyl-N-methylcoclaurine, magnocurarine, 6-demethyl-4′-methyl-N-methylcoclaurine, armepavine. Peak 130 had a retention time of 10.12 min and the [M]+ ion at m/z 358.2012. The fragmentation ions at m/z 327.158 [M + H-CH3NH2]+, 206.117 [C12H16NO2]+, 189.090 [C12H16NO2-NH3]+, and 151.075[C9H11O2]+ suggested that peak 130 was tentatively inferred to be laudanosine. On the basis of this method, peaks 48, 79, 68, 95, 104, 133 were deduced to be norreticuline, reticuline, N-methylreticuline, codamine, laudanine, tetrahydropapaverine.
3.1.1.4 Identification of phthalideisoquinoline alkaloids
For phthalideisoquinolines, the fragment ions were obtained by the isoquinone after bond cleavage with the phthalide ring as well as the elimination of H2O and OCH3 groups (Menéndez-Perdomo et al., 2021). Peak 153 showed a protonated adduct ion at m/z 400.1390 [M + H]+, which fragmented to [M + H-C10H10O4]+ at m/z 206.081, [M + H-H2O]+ at m/z 382.127 and [M + H-H2O-OCH3]+ at m/z 351.110. Therefore, peak 153 could be considered as narcotoline (Menéndez-Perdomo et al., 2021). Similarly, peaks 120, 149, 159, 162 were identified as N-methylnarcotine, noscapine isomer 1, noscapine isomer 2, narceine.
3.1.1.5 Identification of protopine alkaloids
For protopines, it produced characteristic ions by RDA fragmentation, and subsequent loss of water from the isoquinoline fragment (Jeong et al., 2012). Peak 144 showed the parent ion at m/z 370.1648 [M + H]+, and fragment ions at m/z 352.154 [M + H-H2O]+, 206.081 [C11H12NO3]+, and 188.070 [C11H12NO3-H2O]+. Then, peak 144 was characterized as allocryptopine based on the Orbitrap Traditional Chinese Medicine Library (OTCML). Likewise, peaks 70, 91, 124, 127, 245 were assigned as glaucamine, amurensinine N-oxide A, cryptopine, protopine, pseudoprotopine according to the OTCML database and relevant literature (Oh et al., 2018).
3.1.1.6 Identification of morphinan alkaloids
Peak 16 exhibited a protonated molecular ion at m/z 286.1437 [M + H]+, which fragmented to [M + H-CH2CHNHCH3-CO]+ at m/z 201.090 and [M + H-CH2CHNHCH3-H2O]+ at m/z 211.075. Therefore, this information led to the tentative conclusion that peak 16 was identified as morphine (Menéndez-Perdomo et al., 2021). Peak 33 with [M + H]+ ion at m/z 300.1594, was 14.015 Da (CH2) higher than the mass of the [M + H]+ ion of peak 16 and generated fragment ions at m/z 215.106 [M + H-CH2CHNHCH3-CO]+, 225.090 [M + H-CH2CHNHCH3-H2O]+. Peak 33 was characterized as codeine (Menéndez-Perdomo et al., 2021). Moreover, peaks 14, 89, 105 were respectively considered as morphine N-oxide, codeinone and thebaine (Oh et al., 2018; Menéndez-Perdomo et al., 2021).
3.1.1.7 Identification of benzylisoquinoline alkaloids
The precursor ion of peak 146 at m/z 340.1543 and the fragment ions at m/z 202.085 [C12H12O2N]+ and 324.122 [M + H-CH4]+ were formed by rearrangement of the C3′ and C4′ methoxy groups to a methylenedioxy bridge. Hence, peak 146 was identified as papaverine (Menéndez-Perdomo et al., 2021). Similarly, peaks 106, 115 were considered as pacodine and palaudine, respectively.
3.1.2 Identification of flavonoids
Flavonoids usually consist of the framework C6-C3-C6 that are formed when two phenyl rings (A and B) bind with C3, most of them undergo RDA cleavage (Luo et al., 2019). Totally, 92 flavonoids were characterized including 7 chalcones, 5 flavan-3-ols, 24 flavones, 25 flavonols, 21 flavonones, and 10 isoflavones in ZKMG. The proposed fragmentation pathways of each-type representative flavonoids (peaks 183, 24, 129, 235, 221, 213) were observed in Supplementary Figure S2.
3.1.2.1 Identification of chalcone flavonoids
The chalcones (1,3-diaryl-2-propen-1-ones) mainly derived from GU, which are open chain flavonoids. Peaks 183 and 186 showed the same [M−H]- ion at m/z 417.1191, which further yielded fragment ions at m/z 255.066 [M−H−C6H10O5]- by the elimination of glucosyl group, 135.007 [C7H3O3]- and 119.048 [C8H7O]- obtained due to RDA cleavage of the C-ring. Based on literature, peaks 183 and 186 were respectively identified as isoliquiritin and neoisoliquiritin (Xue et al., 2021). Peak 240 exhibited [M−H]- ion at m/z 255.0662, which generated characteristic ions at m/z 135.007 [C7H3O3]- and 91.017 [C7H3O3-CO2]-, so it was deduced as isoliquiritigenin based on OTCML database. Similarly, peaks 121, 176, 188, 189 were identified as lsoliquiritin apioside, licuraside, licorice-glycoside B, licorice-glycoside A according to the OTCML database and literature (Xue et al., 2021).
3.1.2.2 Identification of flavan-3-ol flavonoids
Peaks 24, 39, 44, 51 eluted at different time with the same precursor ion [M−H]- at m/z 451.1245, and the MS2 spectrum showed fragment ions at m/z 289.072 [M−H−C6H10O5]- by a loss of glucose, 245.081 [M−H−C6H10O5−CO2]-, 137.023 [M−H−C6H10O5−C8H8O3]-. Thus, peaks 24, 39, 44, 51 were tentatively identified as catechin-5-O-glucoside, catechin-7-O-glucoside, catechin-4′-O-glucoside, catechin-3′-O-glucoside based on ClogP values. Peak 263 was unambiguously identified as procyanidin in accordance with the reference standard by comparing with retention time and MS/MS fragmentations.
3.1.2.3 Identification of flavone flavonoids
Peak 225 displayed the protonated molecule ion [M−H]- at m/z 269.0455, which subsequent fragment ions at m/z 227.034 [M−H−C2H2O]-, 225.055 [M−H−CO2]-, 201.055 [M−H−C3O2]-, and 151.002 [C7H3O4]- due to RDA cleavage, so peak 225 was assigned as apigenin based on OTCML database. Peak 129 exhibited molecular ion [M−H]- at m/z 431.0983 and the main fragment ion at m/z 269.045 [M−H−C6H10O5]-, which indicated peak 225 as aglycone of peak 129. Therefore, peak 129 was tentatively identified as apigenin-7-O-β-D-glucoside. Likewise, peaks 114, 157, 166 were tentatively characterized as apigenin-7-O-diglucuronide, apigenin 7-O-rutinoside, apigenin-7-O-glucuronide. Peak 201 showed [M−H]- ion at m/z 285.0404, and it could form the fragment ions m/z 241.049 [M−H−CO2]-, 217.049 [M−H−C3O2]-, 199.039 [M−H−C2H2O−CO2]-, 243.029 [M−H−C2H2O]-, 175.039 [M−H−C2H2O−C3O2]-, 133.028 [C8H5O2]- and 151.002 [C7H3O4]- due to RDA cleavage. Thus, peak 201 was accurately identified as luteolin by comparing with the reference standard. Peaks 137, 139, 161, 180 displayed the same precursor ion [M−H]- at m/z 447.0932, which produced the fragment ion at m/z 285.040 [M−H−C6H10O5]-. Based on the standard and literature, peaks 137, 139, 161, 180 were respectively assigned as luteolin‐5‐O‐glucoside, cymaroside, luteolin-4′-O-glucoside, luteolin-3′-O-glucoside (Zhao et al., 2019). Similarly, peaks 83, 199, 132, 101, 261, 236, 181, 179, 169, 257, 229, 174, 220, 223 were tentatively characterized as vicenin II, luteolin-7-O-6″-ocetylglucoside, luteolin-7-O-rutinoside, schaftoside, uralenin, diosmetin, diosmetin-7-O-glucuronide, diosmetin-7-O-β-D-glucopyranoside, diosmin, eupatilin, hispidulin, homoplantaginin, jaceosidin, acacetin based on OTCML database.
3.1.2.4 Identification of flavonol flavonoids
Peak 235 exhibited [M−H]- ion at m/z 315.0510, yielded fragment ions at m/z 300.027 [M−H−CH3]-, 271.024 [M−H−CH3−CHO]- and 151.002 [C7H3O4]- by RDA cleavage. Therefore, peak 235 was identified as isorhamnetin according to the retention time of isorhamnetin reference standard and the comparison with MS2 fragment ions. The protonated molecular ion [M−H]- of peaks 155 and 171 was m/z 491.0831, which could form MS2 fragments at m/z 315.051 [M−H−C6H8O6]- by loss of glucuronide unit and subsequent loss of CH3 at m/z 300.027 [M−H−C6H8O6−CH3]-, so they were respectively assigned as isorhamnetin-7-O-glucuronide and isorhamnetin-3-O-glucuronide by comparing with their structures and chromatographic elution order (Nakamura et al., 2018). Correspondingly, peaks 87, 143, 158, 192, 210, 214 were respectively characterized as isorhamnetin-3,7-O-diglucoside, isorhamnetin-3-O-nehesperidine, isorhamnetin-3-O-rutinoside, isorhamnetin-3-O-arabinoside, isorhamnetin isomer, isorhamnetin isomer. Peak 203 displayed deprotonated molecular ion [M−H]- at m/z 301.0353, which produced characteristic ions at m/z 151.002 [C7H3O4]-, 178.997 [C8H3O5]- due to RDA cleavage. Based on reference standard and MS2 data, peak 203 was identified as quercetin. Likewise, peaks 61, 64, 98, 103, 108, 125, 131, 134, 135, 136, 140, 142, 148, 187, 228 were respectively identified as quercetin-3-O-diglucoside, quercetin-3-O-sophoroside-7-O-rhamnoside, myricetin-3-O-rutinoside, quercetin-7-O-diglucoside, quercetin-5-O-glucoside, rutin, hyperoside, quercetin-7-O-glucoside, quercetin-3-O-glucuronide, isoquercitrin, kaempferol-3-O-β-D-glucuronide, quercetin-O-galloyl-glucopyranoside, kaempferol-3-O-rutinoside, quercetin 3-O-arabinoside and kaempferol.
3.1.2.5 Identification of flavonone flavonoids
Peak 194 exhibited the precursor [M−H]- ion at m/z 255.0662 and yielded characteristic ions at m/z 135.007 [C7H3O3]-, 91.017 [C7H3O3-CO2]-, and 119.048 [C8H7O]- obtained due to RDA cleavage, suggesting that it was characterized as liquiritigenin by comparison with OTCML database. Moreover, peaks 69, 71, 100, 113, 116, 118, 122, 123, 160, 178, 182 were considered as liquiritigenin-O-diglucuronide, glucoliquirtin asioside, liquiritigenin-O-diglucuronide, liquiritigenin-O-diglucuronide, liquiritin apioside, neoliquiritin, 1,1,3,4,5,6,8,8′-Octahydroxy-9H,9′H-2,2′-bixanthene-9,9′-dione, liquiritin, hydroxyliquiritin apioside, liquiritigenin-4′-O-(β-D-3-O-acetyl-apiofuranosyl-(1 → 2)-β-D-glucopyranoside, 6′-acetylliquiritin in accordance with the reference literatures (Wang et al., 2020; Xue et al., 2021). Peak 221 showed protonated molecular ion at m/z 271.0611, which produced characteristic fragment ions at m/z 151.002 [C7H3O4]-, 119.049 [C8H7O]- due to RDA cleavage. Therefore, peak 221 was identified as naringenin by comparison with reference standard along with the retention and the characteristic product ions. Meanwhile, based on similar fragmentation patterns, peaks 109, 117, 119, 147, 164, 167, 175, 230 were identified as naringenin-4′-O-glucoside, eriocitrin, naringenin-5-O-glucoside, naringin, naringenin-7-O-glucoside, hesperidin, hesperetin-7-O-β-D-glucosidehesperetin.
3.1.2.6 Identification of isoflavone flavonoids
Peak 213 generated the [M + H]+ ion at m/z 301.0706, and the MS2 spectrum showed fragment ions at m/z 286.046 [M + H-CH3]+, 258.054 [M + H-CH3-CO]+, 168.005 [C7H4O5]+, which allowed its identification as tectorigenin by comparison with the OTCML database. Based on similar fragmentation patterns, peaks 111, 172, 185, 191, 206, 226, 251, 258, 259 were respectively deduced as daidzin, tectoridin, ononin, glycitein, calycosin-7-O-β-D-glucoside, iristectorigenin B, Gancaonin N, biochanin A and glicoricone according to OTCML database.
3.1.3 Identification of triterpenoid saponins
In this study, a total of 28 saponins primarily derived from GU were characterized in ZKMG. Saponins are composed of a sapogenin of 3α-hydroxy oleanolic acid and sugar residues, such as glucose (Glc), glucuronic acid (GluA), rhamnose (Rha), and xylose (Xyl), which is mainly regarded as structure of 11-oxo-12-ene, 12-ene skeleton (Cheng et al., 2021). The proposed fragmentation pathway of glycyrrhizic acid (peaks 242) was observed in Supplementary Figure S3.
Peak 242 showed precursor ion [M−H]- at m/z 821.3965, and its primary characteristic ions appeared at m/z 351.057 [2GluA-H]-, 193.034 [GluA-H]- and 175.024 [GluA-H2O-H]-. Therefore, it was exactly identified as glycyrrhizic acid by comparing the retention time and fragmentation patterns with reference standard. Peaks 198, 217, 232, 237, 243, 247, which eluting at retention time of 13.35, 13.72, 14.11, 14.22, 14.38, 14.60 min respectively, exhibited the same molecular ion at m/z 837.39142 and the main fragment ions at m/z 351.057 [2GluA-H]-, 193.034 [GluA-H]- and 175.024 [GluA-H2O-H]-. Thus, they were respectively deduced as yunganoside K2, licoricesaponin P2, licoricesaponin Q2, uralsaponin N, hydroxyglycyrrhizin, hydroxyglycyrrhizin based on their MS2 fragmentation behavior, chromatographic retention time, and comparison with the similar known compounds and other reference evidence (Wang et al., 2020). Likewise, peaks 197, 207, 209, 211, 215, 216, 218, 222, 227, 231, 233, 238, 242, 248, 249, 260, 265 were respectively as 24-hydroxy-licoricesaponin A3, licoricesaponin A3, 22-hydroxy-licoricesaponin G2 isomer 1, hydroxy acetoxyglycyrrhizin, 22-hydroxy-licoricesaponin G2 isomer 2, acetoxy-glycyrrhizic acid, methyllicorice-saponin Q2 isomer 1, yunganoside M, acetoxy-glycyrrhizic acid, licoricesaponin E2, methyllicorice-saponin Q2 isomer 2, acetoxyglycyrrhaldehyde, glycyrrhizic acid, licoricesaponin H2, licoricesaponin B2, 22-dehydro uralsaponin C, 18 β-glycyrrhetintic acid according to their similar fragmentation patterns, standard, OTCML database. Peak 255 gave a precursor ion [M−H]- at m/z 953.4751, which yielded characteristic fragment ions at m/z 497.115 [2GluA + Rha-H]-, 435.114 [2GluA + Rha-H2O-CO2-H]-, 339.093 [GluA + Rha + H2O-H]-, 321.082 [GluA + Rha-H]-. Compared with the data in literatures, peak 255 was assigned as yunganoside D1 (Ji et al., 2014). Similarly, peak 234, 246, 250, 252 were respectively characterized as haoglycyrrhizin isomer 1, haoglycyrrhizin isomer 2, yunganoside C1, yunganoside A1 based on their analogous fragmentation pathway and published data (Ji et al., 2014; Xue et al., 2021).
3.1.4 Identification of phenolic acids
In this research, a total of 27 phenolic acids were characterized. The proposed fragmentation pathway of ellagic acid (peaks 128) was observed in Supplementary Figure S3. Peak 2, 4, 6, 17, 65, 99, 128, 151 were exactly and respectively identified as quinic acid, malic acid, citric acid, gallic acid, caffeic acid, 4-coumaric acid, ellagic acid and salicylic acid with the reference standards. Peaks 3, 5, 6 indicated the same parent [M−H]- ion at m/z 191.0197, could give main product ions at m/z 173.008 [M−H−H2O]-, 129.018 [M−H−H2O−CO2]-, 111.007 [M−H−2H2O−CO2]-. Based on their chromatographic elution orders, further MS2 fragmentation patterns and reported literature (Al Kadhi et al., 2017). Peaks 7, 12, 15 exhibited the same precursor ion [M−H]- at m/z 331.0670, which produced daughter ions at m/z 271.046 [M−H−C2H4O2]-, 211.024 [M−H−2C2H4O2]- and 169.013 [M−H−C6H10O5]- by loss of a glucose moiety. Based on similar fragmentation patterns and chromatographic elution orders, they were respectively identified as 1-O-galloylglucose, gallic acid-4-O-β-D-glucopyranoside, gallic acid-3-O-β-D-glucopyranoside (Jin et al., 2007). Peak 19 showed the similar protonated molecular ion at m/z 179.0349 with peak 65, and yielded characteristic ion at m/z 135.044 [M−H−CO2]-, so it was presumed to be caffeic acid isomer. Peaks 20, 22, 35, 41 were tentatively identified as danshensu, protocatechuic acid, coumaric acid, 4-hydroxybenzoic acid, respectively according to the OTCML database. Peaks 52, 62, 76 with the same protonated molecular ion [M−H]- at m/z 325.0928, were glucose moiety more than peak 35, which produced characteristic ions at m/z 163.039 [M−H−C6H10O5]-, 145.028 [M−H−C6H10O5−H2O]- and 119.049 [M−H−C6H10O5−CO2]-. Their fragment patterns were similar with peak 35, so they were tentatively assigned as coumaric acid-O-glucoside. Peak 126 exhibited [M−H]- ion at m/z 537.1038 and fragment behaviors are similar with lithospermic acid while retention time could not bring into correspondence with standard. Thus, it was tentatively assigned to lithospermic acid isomer. Peak 156 showed precursor ion [M−H]- at m/z 519.1871 and yielded characteristic ions at m/z 357.134 [M−H−C6H10O5]-, 151.039 [C8H7O3]-, indicating that peak 156 was inferred as pinoresinol 4-O-β-D-glucopyranoside. Peak 193 generated the quasi-molecular ion [M−H]- at m/z 593.1875 and fragment ions at m/z 309.077 and 285.076. Thus, it was tentatively characterized as didymin. Peak 21 showed protonated molecular ion at m/z 329.0878, and produced fragment ions at m/z 167.034 [M−H−C6H10O5]-, 152.010 [M−H−C6H10O5−CH3]- and 123.044 [M−H−C6H10O5−CO2]-, suggesting that it was pseudolaroside B. Peaks 34 and 38 exhibited the same precursor ion [M−H]- at m/z 285.0615, and it could form the fragment ions m/z 153.018 [M−H−C5H8O4]- by loss of an arabinose group and 109.028 [M−H−C5H8O4−CO2]-, suggesting that they were uralenneoside isomers.
3.1.5 Identification of phenylpropanoids
A total of 24 phenylpropanoids comprising of 4 coumaroylquinic acids, 3 feruloylquinic acids, 11 caffeylquinic acids and 6 other type acids were identified in ZKMG extract. The proposed fragmentation pathway of rosmarinic acid (peaks 170) was observed in Supplementary Figure S3. Peaks 31, 53, 55, 141, 145, 150, 154, 168, 170 were exactly identified as neochlorogenic acid, chlorogenic acid, cryptochlorogenic acid, acteoside, isochlorogenic acid B, 1,5-dicaffeoylquinic acid, isochlorogenic acid A, isochlorogenic acid C, rosmarinic acid, respectively. Likewise, peak 73 gave precursor ion [M−H]- at m/z 515.1194, with the fragment ions at m/z 353.088 [M−H−caffeoyl]-, 191.055 [quinic acid-H]-, 179.034 [caffeic acid-H]-, 135.044 [caffeic acid-H-CO2]-, which were consistent with the corresponding ions of dicaffeoylquinic acids. Therefore, peak 73 was inferred as dicaffeylquinic acid. Peaks 28, 43, 49 with the same parent ion [M−H]- at m/z 515.1406 were a glucose group (C6H10O5 162.052 Da) more than peaks 31, 53, 55 and possessed similar characteristic fragment ions at m/z 173.044 [quinic acid-H-H2O]-, 191.055 [quinic acid-H]-, 179.034 [caffeic acid-H]- and 135.044 [caffeic acid-H-CO2]-. Thus, they were tentatively identified as chlorogenic acid-hexosides. Peaks 47, 75, 82, 97 were found to elute at 5.74, 7.17, 7.55, 8.60 min, with [M−H]- ion at m/z 337.0928, and they could yield characteristic fragment ions at m/z 163.039 [coumaric acid-H]-, 119.048 [coumaric acid-H-CO2]-, 191.055 [quinic acid-H]-, 173.044 [quinic acid-H-H2O]-, suggesting that these compounds might be coumarylquinic acid. Therefore, Peaks 47, 75, 82, 97 were respectively identified as 5-p-coumaroylquinic acid, 3-p-coumaroylquinic acid, 4-p-coumaroylquinic acid, 1-p-coumaroylquinic acid according to their chromatographic elution behavior (Zhao et al., 2014). Peaks 59, 92, 107 were respectively eluted at 6.55, 8.39, 8.96 min with the parent ion [M−H]- at m/z 367.1034 and displayed characteristic secondary fragments at m/z 193.050 [ferulic acid-H]-, 149.059 [ferulic acid-H-CO2]-, 134.036 [ferulic acid-H-CO2-CH3]-, 173.044 [quinic acid-H-H2O]-, suggesting that these compounds might be feruloylquinic acid. Hence, they were tentatively identified as 4-feruloylquinic acid, 5-feruloylquinic acid, 3-feruloylquinic acid based on their chromatographic elution orders, further MS2 fragmentation patterns and reported literature (Zheleva-Dimitrova et al., 2017). Peak 29 with the precursor ion [M−H]- at m/z 311.0408, gave characteristic product ions at m/z 149.008 [tartaric acid-H]-, 179.034 [caffeic acid-H]-, 135.044 [caffeic acid-H-CO2]-, suggesting that it was tentatively assigned to caftaric acid according to the OTCML database. Peak 40 was tentatively identified as esculin based on the OTCML database. Peak 195 with the parent ion [M−H]- at m/z 373.0928, was methyl (CH2 14.015 Da) more than peak 170, and produced characteristic ions at m/z 197.045 [M−H−C9H7O3]-, 179.034 [M−H−C9H9O4]-, 161.023 [M−H−C9H9O4−H2O]-, 135.044 [M−H−C9H9O4−CO2]-, which were the same fragment patterns as peak 170. Therefore, it was identified as methyl rosmarinate based on the OTCML database. Peaks 141, 152 both gave precursor ion [M−H]- at m/z 623.1981, and yielded fragment ions at m/z 461.166 [M−H−C9H6O3]-, 179.034 [C9H7O4]-, 161.023 [C9H7O4-H2O]-, 135.043 [C9H7O4-CO2]-, suggesting that they were a group of isomers. Peak 141 was confirmed as acteoside by standard, so peak 152 was identified as isoacteoside based on their chromatographic retention behavior.
3.1.6 Identification of anthraquinones
A total of 21 quinones were identified in ZKMG, including 3 rheic acid-types, 3 physcion-types, 3 emodin-types, 6 chrysophanol-types, 3 aurantio-obtusin-types, 2 aloe-emodin-types and 1other compound, which are all derived from RP. The proposed fragmentation pathway of rhein (peak 256) was observed in Supplementary Figure S3.
Peaks 208, 256, 264 were unambiguously attributed to physcion, rheic acid, emodin by comparison with the authentic standards. Rhein as the main anthraquinone in ZKMG, was used to characterize the fragmentation pathways. It exhibited a parent ion [M−H]- at m/z 283.0248, and yielded characteristic product ions at m/z 255.030 [M−H−CO]-, 239.034 [M−H−CO2]-, 211.039 [M−H−CO2−CO]-, 183.044 [M−H−CO2−2CO]-. Based on these fragmentation patterns, peaks 138, 254 were identified as rhein-8-O-β-D-glucoside and 6-methyl-rhein. Physcion showed a precursor ion [M−H]- at m/z 283.0611, which produced characteristic ions at m/z 268.037 [M−H−CH3]-, 240.042 [M−H−CH3−CO]-. Therefore, peaks 219, 224 were respectively identified as physcion-8-O-β-D-glucoside and physcion-1-O-β-D-glucoside according to the chromatographic elution orders, similar fragment patterns. Emodin indicated the parent ion [M−H]- at m/z 269.0455, and produced product ions at m/z 241.050 [M−H−CO]-, 225.054 [M−H−CO2]-. Based on these fragmentation patterns, peaks 165, 202 were identified as emodin-1-O-D-glucoside and emodin-8-O-D-glucoside. Likewise, peaks 102, 173, 177, 184, 190, 196, 204, 205, 212, 241, 244, 262 were assigned to carboxyl-chrysophanol-O-glucose, aurantio-obtusin-6-O-rutinoside, aloe-emodin-3-(hydroxymethyl)-O-β-D-glucopyranoside, aurantio-obtusin-6-O-glucoside, carboxyl-chrysophanol-O-glucose, aloe-emodin-8-O-(6-O-acetyl)-glucoside, chrysophanol-1-O-β-D-glucoside, chrysophanol, chrysophanol-8-O-glucoside, aurantio-obtusin, chrysophanol-O-acetylglucoside, 1-Methyl-8-hydroxy-9,10-anthraquinone-3-O-(6′-O-cinnamoyl)-glucoside, respectively, according to the similar fragment patterns.
3.1.7 Identification of tannins
Tannins might exist in ZKMG as they are important compounds found in the crude drug RP. Totally, 13 tannins were identified in ZKMG extract. The proposed fragmentation pathway of tri-O-galloyl-glucoside (peaks 54) was observed in Supplementary Figure S3. Peaks 54, 63, 66, 81, 84, 90, 93 gave the precursor [M−H]- ion at m/z 635.0889, and produced characteristic ions at m/z 483.077 [M−H−C7H4O4]-, 465.067 [M−H−C7H4O4−H2O]-, 313.057 [M−H−2C7H4O4−H2O]-, 169.013 [C7H5O5]-, and 125.023 [C7H5O5-CO2]-, thus they were assigned as tri-O-galloyl-glucoside isomers. According to this method, peaks 26, 36, 42, 45, 50, 60 were identified as gallic acid-O-galloyl-glucoside isomers.
3.1.8 Identification of other compounds
The other compounds including 4 amino acids, 2 naphthols, 2 phenols, 2 terpenoids were detected in ZKMG. The proposed fragmentation pathway of brervifolincaboxylic acid (peak 58) was observed in Supplementary Figure S3. Peaks 1 and 13 were confirmed as adenine and adenosine cyclophosphate, respectively, compared with known reference compounds. Peaks 9 and 11 both showed [M + H]+ at m/z 132.1019, and gave the same MS2 fragmentation ion at m/z 86.096 [M + H-CO-H2O]+, indicating that they were isomers. Consequently, peaks 9 and 11 were respectively characterized as isoleucine, leucine based on chromatographic elution orders. Peak 200 exhibited a precursor ion [M−H]- at m/z 407.1347, and showed the product ions at m/z 245.081[M-H-C6H10O5]- by loss of a glucosyl group, 230.058 [M−H−C6H10O5−CH3]- by elimination of a CH3 radical. Hence, it was identified as torachrysone-8-O-glucoside. Peak 239 with the parent ion [M−H]- at m/z 449.1453, was acetyl (C2H2O 42.010 Da) more than peak 200, and yielded the same fragment ions. Thus, it was tentatively characterized as torachrysone-O-acetylglucoside. Peak 58 showed [M−H]- ion at m/z 291.0146, and gave MS2 fragmentation ions at m/z 247.024 [M−H−CO2]-, 219.029 [M−H−CO2−CO]-, 191.034 [M−H−CO2−2CO]-, 173.023 [M−H−CO2−2CO−H2O]-, suggesting that it was identified as brervifolincaboxylic acid (Chen et al., 2022). Besides, peaks 8, 10, 18, 23, 25, 78, 253 were respectively characterized as adenosine, guanosine, phenylalanine, 3,4-dihydroxyphenylethanol, tryptophan, camphor, steviol-19-O-glucoside based on the OTCML database.
3.2 Network pharmacology analysis
3.2.1 Potential bioactive compounds and targets of ZKMG in the treatment of AURTIs
In this study, UHPLC-MS was used to detect a total of 265 chemical components of ZKMG. By searching the Swiss Target Prediction, 836 targets were obtained from identified compounds, and 1317 AURTIs related targets based on OMIM and GeneCards database (Supplementary Table S3). Finally, 120 overlapping targets were obtained by precisely matching the potential targets of the above two steps through the online tool Venny 2.1, suggested that ZKMG would play a role in treating AURTIs associated with these 120 common targets (Supplementary Figure S4).
3.2.2 Compound‑target network analysis
The active ingredient potential target network of ZKMG in (Fig. 3). There are 271 nodes and 1117 edges in the network, among which the 155 pink nodes represent the main components of ZKMG, the 116 purple nodes represent the targets of AURTIs, and 1117 edges represent the interactions between the components and the targets of AURTIs. The size of the compounds in the network increases with the number of edges (degree of targets). The fact that the same active ingredient can act on multiple targets and the same target also corresponds to different chemical components were observed from the network, which fully reflect the multicomponent and multitarget characteristics of ZKMG in the treatment of AURTIs. The compounds were screened with a degree and betweenness centrality greater than the mean, such as 18 β-Glycyrrhetintic acid, noscapine, n-methylnarcotine, adenosine, methyl rosmarinate, thebaine, 6-methyl-rhein, which were possibly potential active ingredients of ZKMG in the treatment of AURTIs.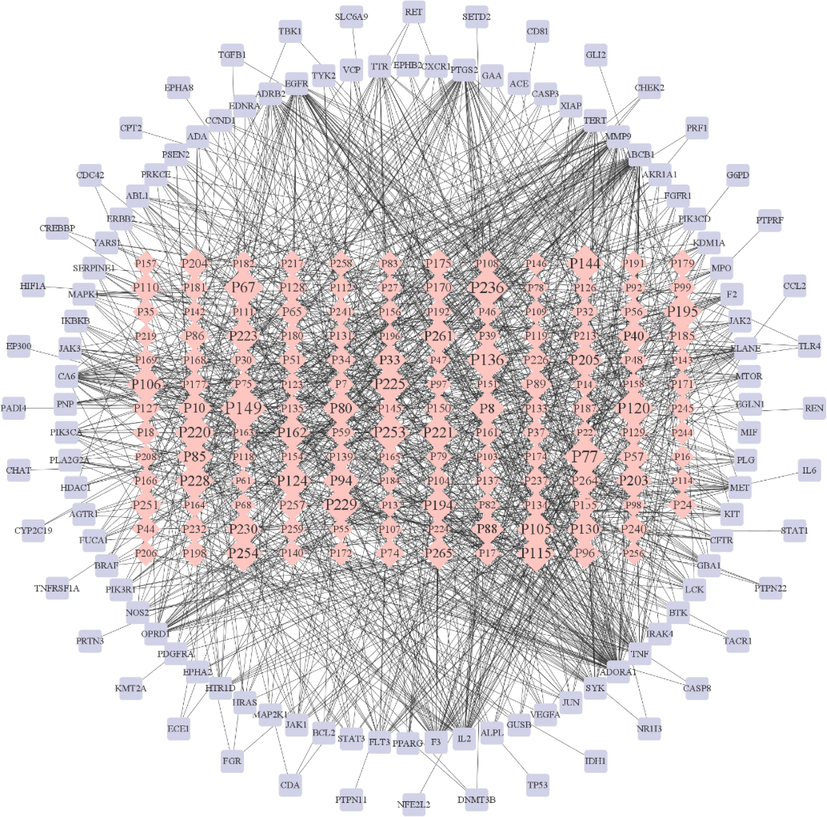
Compound-target network. pink rhombus nodes represent compounds, and purple rectangular nodes represent targets.
3.2.3 PPI network analysis
STRING analysis was used to compare 120 overlapping targets and produce a PPI network, as well as the visualization was realized by Cytoscape software (Fig. 4). There were 117 nodes and 1562 edges were observed with a combined score of greater than 0.4 (Supplementary Table S4). The size and color of the node reflected the importance of the degree. The larger the degree, the more important the node is in the network, suggesting that it may be a key target of ZKMG in the treatment of AURTIs. The top 10 nodes were selected as the major genes, including TNF, TP53, IL6, AKT1, EGFR, VEGFA, STAT3, HRAS, JUN, ERBB2, which were likely to be the critical genes in the development of AURTIs.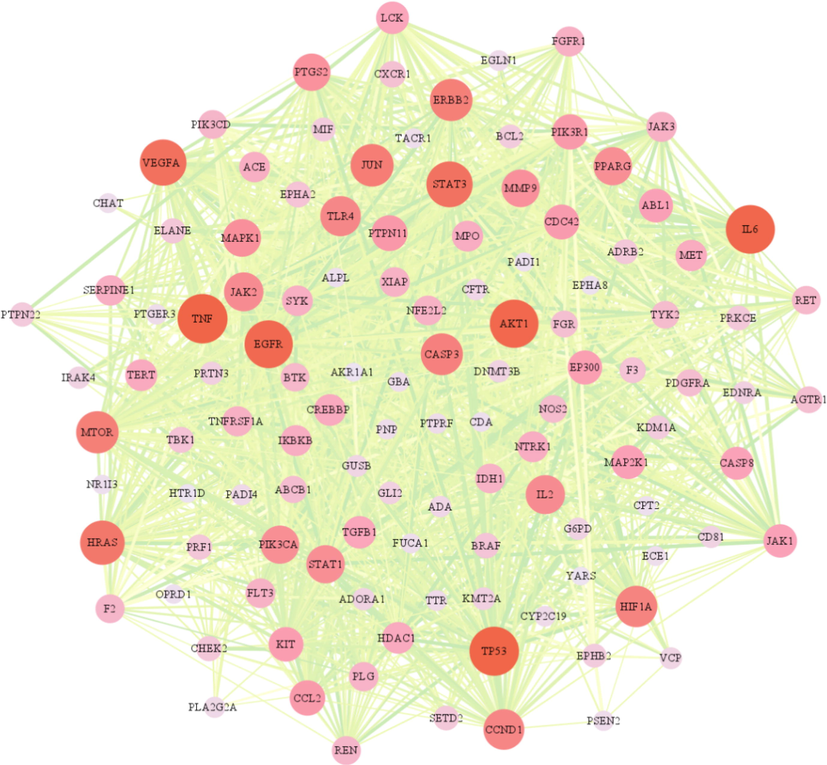
Protein-protein interaction (PPI) network.
3.2.4 GO analysis and KEGG pathway analysis
GO function and KEGG pathway analysis of the 120 candidate target genes were posted on the DAVID database to explore the molecular mechanism of ZKMG in treating AURTIs. GO evaluations were illustrated by using biological process (BP), cell component (CC), and molecular function (MF) terms. The results of GO analysis showed that potential target genes were enriched, which involved with 638 pathways, including 505 BPs, 52 CCs and 81 MFs (P<0.05). The top 20 pathways of BP, CC, MF with the highest number of genes involved were shown in the Fig. 5. In BP, the targets mainly involved peptidyl-tyrosine phosphorylation, protein phosphorylation, inflammatory response. In CC, the targets mainly involved plasma membrane, receptor complex, macromolecular complex. In MF, the targets mainly involved transmembrane receptor protein tyrosine kinase activity, protein tyrosine kinase activity, identical protein binding, protein kinase activity (Supplementary Table S5). KEGG pathway annotation indicated that potential target genes were involved in 148 pathways (P<0.05). The top 20 KEGG pathways with the highest number of genes were shown in the Fig. 6, including PI3K-Akt signaling pathway, AGE-RAGE signaling pathway in diabetic complications, PD-L1 expression and PD-1 checkpoint pathway in cancer, HIF-1 signaling pathway (Supplementary Table S6).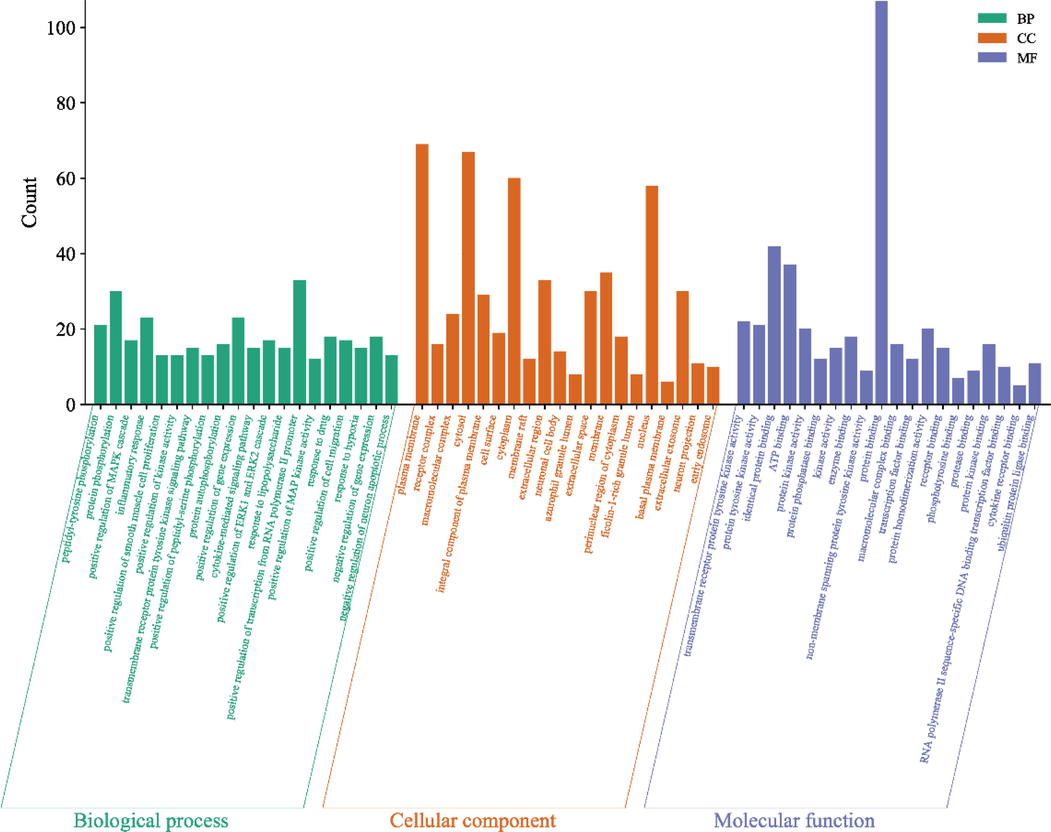
GO term histograms.
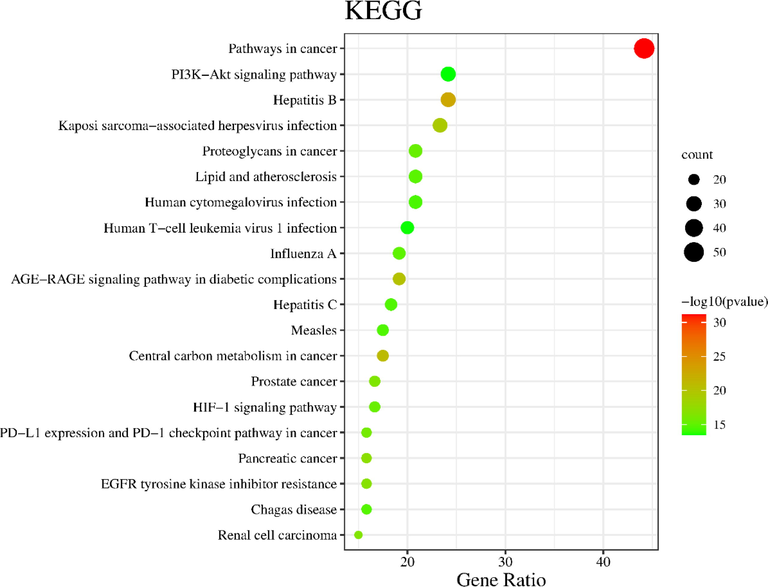
Bubble map of KEGG pathway analysis.
3.2.5 Compound‑target‑pathway network analysis
In order to explore the key compounds of ZKMG in treating AURTIs, the top 20 KEGG pathways, corresponding targets and compounds were constructed to the component-target-pathways network as shown in Fig. 7. The network contained 227 nodes with 134 representative components, 73 representative targets, 20 representative pathways, and 1193 edges. The results indicated that alkaloids mainly derived from PP played an important role in the treatment of ZKMG in treating AURTIs due to their higher degrees, such as noscapine (degree = 24), cryptopine (degree = 17), N-methylnarcotine (degree = 16), allocryptopine (degree = 15). In addition, the targets and active components were distributed in different pathways and played a key role in the treatment of AURTIs, which profoundly reflected the multicomponent, multitarget, and multipathway features of TCM.
Compound-target-pathway network. green circular nodes represent chemical compounds, red V-shaped nodes represent targets, and blue rectangular nodes represent pathways.
4 Conclusion
In this study, a rapid and sensitive UHPLC-Q-Exactive Orbitrap-MS method combined with network pharmacology was established to characterize pharmacodynamic Substance and predict potential molecular mechanisms of ZKMG in treating AURTIs. Finally, a total of 265 were identified or tentatively characterized including 46 alkaloids, 92 flavonoids, 28 triterpenoid saponins, 27 phenolic acids, 24 phenylpropanoids, 21 quinones, 13 tannins, 4 amino acids, 4 nucleosides, 2 naphthols, 2 phenols and 2 terpenoids. Based on the pharmacological network analysis of compound‑target‑pathway, EGFR, PTGS2, IL2, MMP9, TNF, AKT1, PIK3CA and F3 were regarded as key targets for ZKMG to exhibit its effects against AURTIs. Alkaloids, flavonoids, phenylpropanoids and terpenoids such as noscapine, cryptopine, steviol-19-O-glucoside, N-methylnarcotine, methyl rosmarinate, allocryptopine, naringenin and boldine indicated that multiple compounds possessed comprehensive activities by interacting with the above targets against AURTIs. In conclusion, the result revealed that the relationship between compounds and effects of ZKMG treated the AURTIs as well as could lay the foundation of quality control research and clinical application of ZKMG in the future. However, the effective constituents, active targets, and signaling pathways obtained based on UHPLC-MS and network pharmacology need to be confirmed and validated by animal experiment in further studies.
Funding
This study was funded by the Science and Technology Innovation Program of Hunan Province (no. 2022RC1228), Xinjiang Uygur Autonomous Region Institute for Drug Control, and Hunan Province Social Science Innovation Research Base (Ethnic medicine and ethnic culture research base).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Development of a LC-MS/MS method for the simultaneous detection of tricarboxylic acid cycle intermediates in a range of biological matrices. J. Anal. Methods Chem. 2017:5391832.
- [Google Scholar]
- Are the Saudi parents aware of antibiotic role in upper respiratory tract infections in children? J. Infect. Public Heal.. 2017;10(5):579-585.
- [Google Scholar]
- Amberger, J.S., Bocchini, C.A., Schiettecatte, F., Scott, A.F., and Hamosh, A., 2015. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 43 (Database issue), D789-798
- Identification of active compounds and mechanism of Huangtu Decoction for the treatment of ulcerative colitis by network pharmacology combined with experimental verification. Drug Des. Devel. Ther.. 2021;15:4125-4140.
- [Google Scholar]
- Chen, L.M., Talaiti, Zulipiya., Aili, Yusufujiang., Li, J.M., Huo, S.X., Gao, H.M. Establishment of a quality control method for simultaneous determination of multiple active components in Uygur medicine of nymphaeae flos. Chin J Exp Tradit Med Formulae. 1-10.
- A strategy for practical authentication of medicinal plants in traditional Chinese medicine prescription, paeony root in ShaoYao-GanCao decoction as a case study. J. Sep. Sci.. 2021;44(12):2427-2437.
- [Google Scholar]
- Rapid structural characterisation of benzylisoquinoline and aporphine alkaloids from Ocotea spixiana acaricide extract by HPTLC-DESI-MS(n) Phytochem. Anal.. 2020;31(6):711-721.
- [Google Scholar]
- SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res.. 2019;47(W1):W357-W364.
- [Google Scholar]
- Appropriate antibiotic use for acute respiratory tract infection in adults: advice for High-Value care from the American college of physicians and the centers for disease control and prevention. Ann. Intern. Med.. 2016;164(6):425-434.
- [Google Scholar]
- Analysis of anti-inflammation mechanism of Zukamu granules based on network pharmacology and HS-SPME-GC-MS. J. Int. Pharm. Res.. 2019;46(06):441-455.
- [Google Scholar]
- Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc.. 2009;4(1):44-57.
- [Google Scholar]
- Huang, Z., Pan, X., Zhou, J., Leung, W.T., Li, C., and Wang, L., 2019. Chinese herbal medicine for acute upper respiratory tract infections and reproductive safety: A systematic review. BioScience Trends. advpub
- Systematic investigation of the pharmacological mechanism for renal protection by the leaves of Eucommia ulmoides Oliver using UPLC-Q-TOF/MS combined with network pharmacology analysis. Biomed. Pharmacother.. 2021;140:111735
- [Google Scholar]
- Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. The Lancet. 2018;392(10159):1789-1858.
- [Google Scholar]
- Identification of structurally diverse alkaloids in Corydalis species by liquid chromatography/electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom.. 2012;26(15):1661-1674.
- [Google Scholar]
- New triterpene saponins from the roots of Glycyrrhiza yunnanensis and their rapid screening by LC/MS/MS. J. Pharm. Biomed. Anal.. 2014;90:15-26.
- [Google Scholar]
- Simultaneous analysis of multiple bioactive constituents in Rheum tanguticum Maxim. ex Balf. by high-performance liquid chromatography coupled to tandem mass spectrometry. Rapid Commun. Mass Spectrom.. 2007;21(14):2351-2360.
- [Google Scholar]
- Combination of UPLC-Q-TOF/MS and network pharmacology to reveal the mechanism of Qizhen Decoction in the treatment of colon cancer. ACS Omega. 2021;6(22):14341-14360.
- [Google Scholar]
- A novel therapeutic approach for IPF: based on the “Autophagy-Apoptosis” balance regulation of Zukamu Granules in alveolar macrophages. J. Ethnopharmacol.. 2022;297:115568
- [Google Scholar]
- Antiviral effect of emodin from Rheum palmatum against coxsakievirus B5 and human respiratory syncytial virus in vitro. J. Huazhong Univ. Sci. Technol. Med. Sci.. 2015;35(6):916-922.
- [Google Scholar]
- Study of the active ingredients and mechanism of Sparganii rhizoma in gastric cancer based on HPLC-Q-TOF-MS/MS and network pharmacology. Sci. Rep.. 2021;11(1):1905.
- [Google Scholar]
- Study on the material basis of Houpo Wenzhong decoction by HPLC fingerprint, UHPLC-ESI-LTQ-Orbitrap-MS, and network pharmacology. Molecules. 2019;24(14)
- [Google Scholar]
- Benzylisoquinoline alkaloid analysis using high-resolution Orbitrap LC-MS(n) J. Mass Spectrom.. 2021;56(2):e4683.
- [Google Scholar]
- Lymphatic metabolites of quercetin after intestinal administration of quercetin-3-glucoside and its aglycone in rats. Arch. Biochem. Biophys.. 2018;645:126-136.
- [Google Scholar]
- Identification and metabolite profiling of alkaloids in aerial parts of Papaver rhoeas by liquid chromatography coupled with quadrupole time-of-flight tandem mass spectrometry. J. Sep. Sci.. 2018;41(12):2517-2527.
- [Google Scholar]
- Identification of medicinal plants effective on common cold: an ethnobotanical study of Shiraz. South Iran.. 2016;8:90-97.
- [Google Scholar]
- A symptom-directed paradigm for the evaluation and management of upper respiratory tract infections. Med. Clin. North Am.. 2021;105(1):199-212.
- [Google Scholar]
- Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res.. 2003;13(11):2498-2504.
- [Google Scholar]
- Differential pharmacokinetics and the brain distribution of morphine and ephedrine constitutional isomers in rats after oral administration with Keke capsule using rapid-resolution LC-MS/MS. J. Sep. Sci.. 2014;37(4):352-359.
- [Google Scholar]
- Stelzer, G., Rosen, N., Plaschkes, I., Zimmerman, S., Twik, M., Fishilevich, S., 2016. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr Protoc Bioinformatics. 54, 1.30.31-31.30.33.
- STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res.. 2018;47(D1):D607-D613.
- [Google Scholar]
- UniProt Consortium, T., 2018. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 46 (5), 2699
- Use and prescription of antibiotics in primary health care settings in China. JAMA Intern. Med.. 2014;174(12):1914-1920.
- [Google Scholar]
- Chemical profiling and tissue distribution study of Jingyin Granules in rats using UHPLC-Q-Exactive Orbitrap HR-MS. China J. Chin. Mater. Med.. 2020;45(22):5537-5554.
- [Google Scholar]
- A novel and comprehensive strategy for quality control in complex Chinese medicine formula using UHPLC-Q-Orbitrap HRMS and UHPLC-MS/MS combined with network pharmacology analysis: take Tangshen formula as an example. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci.. 2021;1183:122889
- [Google Scholar]
- The effectiveness and safety of Chaiqin Qingning Capsule in upper respiratory tract infections with fever: a prospective, double-blinded, randomized, multicenter controlled trial. Complement. Ther. Med.. 2022;68:102840
- [Google Scholar]
- The combination of UHPLC-HRMS and molecular networking improving discovery efficiency of chemical components in Chinese Classical Formula. Chin. Med.. 2021;16(1):50.
- [Google Scholar]
- Recent research progress in the pharmacological effects of tetrahydroprotoberberines. Prog. Pharm. Sci.. 2016;40(10):739-744.
- [Google Scholar]
- Effectiveness and safety of Danmu extract syrup for acute upper respiratory tract infection in children: a real-world, prospective cohort study. J. Evid. Based Med.. 2022;15(1):19-29.
- [Google Scholar]
- Network pharmacology-based analysis of Zukamu granules for the treatment of COVID-19. Eur. J. Integr. Med.. 2021;42:101282
- [Google Scholar]
- Chinese patent herbal medicine (Shufeng Jiedu capsule) for acute upper respiratory tract infections: a systematic review and meta-analysis. Integr. Med. Res.. 2021;10(3):100726
- [Google Scholar]
- Rapid detection and characterization of major phenolic compounds in Radix Actinidia chinensis Planch by ultra-performance liquid chromatography tandem mass spectrometry. J. Pharm. Biomed. Anal.. 2014;98:311-320.
- [Google Scholar]
- Chemical fingerprint analysis and metabolic profiling of 50% ethanol fraction of Lomatogonium rotatum by ultra-performance liquid chromatography/quadrupole-time of flight mass spectrometry. Biomed. Chromatogr.. 2019;33(11):e4651.
- [Google Scholar]
- HPLC-UV and LC-MS Analyses of acylquinic acids in Geigeria alata (DC) Oliv. & Hiern. and their contribution to antioxidant and antimicrobial capacity. Phytochem. Anal.. 2017;28(3):176-184.
- [Google Scholar]
- Can network pharmacology identify the anti-virus and anti- inflammatory activities of Shuanghuanglian oral liquid used in Chinese medicine for respiratory tract infection? Eur. J. Integr. Med.. 2020;37:101139
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2023.104875.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary data 1
Supplementary data 1







