Translate this page into:
QSAR study on 4-alkynyldihydrocinnamic acid analogs as free fatty acid receptor 1 agonists and antidiabetic agents: Rationales to improve activity
⁎Corresponding author at: School of Pharmacy, Lloyd Institute of Management and Technology, Greater Noida 201306, UP, India. Tel.: +91 120 2320749. mkgupta5@gmail.com (Manish K. Gupta)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
The free fatty acid receptor 1 (FFAR1) is a class A G-protein coupled receptor and a validated target to develop antidiabetic drugs. The present work describes the quantitative structure–activity relationship (QSAR) study of a series of 4-alkynyldihydrocinnamic acid analogs to rationalize their FFAR1 agonist activity. The various physicochemical and structural descriptors were derived from Molecular Operating Environment (MOE, 2011). The variable selection and model development were carried out using Combinatorial Protocol in Multiple Linear Regressions (CP-MLR) approach. The identified QSAR models highlighted the significance of solvent accessible surface area and associated hydrophobicity to the biological activity. The chemical features to improve hydrophobicity of 4-alkynyldihydrocinnamic acid analogs have been discussed.
Keywords
Free fatty acid receptor 1
GPR40
QSAR
CP-MLR
Physicochemical property

- FFAR1
-
free fatty acid receptor 1
- CP-MLR
-
Combinatorial Protocol in Multiple Linear Regression
- MOE
-
Molecular Operating Environment
- T2DM
-
type 2 diabetes mellitus
- ADCA
-
4-alkynyldihydrocinnamic acid
- QSAR
-
quantitative structure–activity relationship
Abbreviations
1 Introduction
The last decade witnessed a steep rise in the number of patients with type 2 diabetes mellitus (T2DM) especially in young and middle age individuals. The change in life style, irregular eating habits and lack of exercise are contributing factors for the development of T2DM. According to World Health Organization approximately, 347 million people worldwide are affected with diabetes. Here, T2DM alone accounts for more than 90% of total diabetes. It is estimated that the diabetes will be the 7th leading cause of death in 2030 (WHO, 2014). The T2DM is characterized by persistent hyperglycemia due to reduced insulin sensitivity, development of insulin resistance or impaired insulin secretion. If untreated, the hyperglycemia leads to the serious pathological conditions such as coronary heart disease, atherosclerosis, nephropathy, neuropathy and retinopathy. T2DM is a major concern in patients with cardiovascular disease because cardiovascular deaths have been increased up to fourfold in diabetic patients when compared with their nondiabetic counterparts (Inzucchi and Sherwin, 2005). Several classes of drugs such as biguanide (metformin), sulphonylureas (gliclazide), thiazolidinediones (rosiglitazone), dipeptidyl peptidase-4 enzyme inhibitors (sitagliptin) are used as oral hypoglycemic agents, however, use of these drugs is often associated with serious risk factors (Stein et al., 2013). The biguanides and sulphonylureas force the β-cell to secrete insulin regardless of blood glucose levels and thus produce hypoglycemia. The hypoglycemia may lead to the loss of conscious (Tan et al., 2008; Graveling and Frier, 2009). These drugs also cause the loss of islet function (Maedler et al., 2005; Del Guerra et al., 2005). The side effects of mostly used thiazolidinediones also include fluid retention, weight gain, bone fracture and cardiovascular failure (Rizos et al., 2009). These drugs are also reported to be failed in providing desire hypoglycemic effect for the adequate duration of time (Kahn et al., 2006; Grant et al., 2007). Therefore, the development of new and effective antidiabetic drugs with improved patient compliance is of current interest (Waugh et al., 2010).
The free fatty acid receptor 1 (FFAR1 or GPR40) is a G-coupled, class A G-protein coupled receptor. FFAR1 is expressed in pancreatic β-cells and its activation by medium and longer chain free fatty acids (FFAs; endogenous ligands for FFAR1) results in enhancement of glucose stimulated insulin secretion (GSIS) (Itoh et al., 2003; Briscoe et al., 2003; Rayasam et al., 2007). The FFAR1 has been suggested as a potential drug target for T2DM. The studies have shown that FFAR1 agonists induce insulin secretion only when needed and thus prevent the patient from severe hypoglycemia (Tan et al., 2008; Feng et al., 2012). The various selective ligands including thiazolidinedione “glitazone” have been identified as FFAR1 agonists and antidiabetic agents (Kotarsky et al., 2003; Bharate et al., 2009).
Christiansen et al. (2011) synthesized a series of 4-alkynyldihydrocinnamic acid (hereafter referred as ADCA analogs) as FFAR1 agonists and antidiabetic agents (Table 1). These ADCA analogs have been prepared by modifying the ring B with substituted aryl and heteroaryls with an aim to optimize their lipophilicity for better activity (Table 1). These compounds have been evaluated for their efficacy to stimulate FFAR1 (Christiansen et al., 2011).
Comp.
Ar
logEC50
Observedb
Predictedc
Obsd.
Eq. (1)
Eq. (2)
Eq. (3)
1
-phenyl
6.70
6.820
7.148
6.898
2a
-pyridin-2-yl
4.88
5.568
5.251
5.607
3
-pyridin-3-yl
5.22
5.637
5.656
5.675
4
-pyridin-4-yl
5.39
5.677
5.914
5.654
5
-6-methylpyridin-2-yl
5.21
5.857
5.441
5.777
6
-3-nitropyridin-2-yl
4.96
4.674
4.614
4.906
7
-2-chloropyridin-3-yl
6.22
6.355
6.005
6.460
8
-6-chloropyridin-3-yl
6.15
6.280
6.241
6.467
9
-2-fluoro-5-methylpyridin-3-yl
6.18
6.759
5.940
6.684
10
-2-chloropyrimidin-4-yl
5.04
5.160
5.487
5.256
11
-2-methoxypyrimidin-4-yl
5.02
4.956
5.085
4.431
12
-2-chloro-5-methylpyrimidin-4-yl
5.37
5.509
5.486
5.350
13
-2-chloro-6-methylpyrimidin-4-yl
5.56
5.437
5.462
5.314
14
-thiazol-5-yl
5.41
5.567
5.877
5.425
15a
-thiophen-2-yl
6.60
6.736
7.139
6.636
16
-3-methylthiophen-2-yl
7.12
7.084
7.263
6.669
17
-o-tolyl
7.34
7.046
7.039
6.901
18a
-2-methyl-pyridin-4-yl
5.95
5.828
5.882
5.763
19
-2-fluoropyridin-4-yl
6.29
6.259
6.816
6.423
20a
-2-chloropyridin-4-yl
6.53
6.353
6.389
6.458
21
-2-methoxypyridin-4-yl
6.28
6.090
5.490
5.770
22
-2-phenoxypyridin-4-yl
6.32
5.522
5.946
6.754
23
-2-phenyl-pyridin-4-yl
6.73
6.194
6.111
6.422
24a
-2-o-tolylpyridin-4-yl
5.97
6.285
6.347
6.672
25a
-2-fluoro-5-methylpyridin-4-yl
6.75
6.606
6.689
6.515
26
-2-fluoro-3-methylpyridin-4-yl
6.92
6.372
6.569
6.285
27
-2-chloro-3-methylpyridin-4-yl
6.87
6.601
6.531
6.552
28
-3-allyl-2-chloropyridin-4-yl
6.50
7.479
6.751
6.930
29a
-7-chloroquinolin-4-yl
7.10
6.955
6.699
6.935
30
-2,6-dichloropyridin-4-yl
7.39
6.874
7.170
7.246
31a
-2,6-dichloro-3-methylpyridin-4-yl
7.36
7.355
7.410
7.410
32d
-2,6-dichloropyridin-4-yl
7.37
7.341
7.501
7.125
The quantitative structure–activity relationship (QSAR) study helps to optimize the desired physicochemical property(s) in a series of compound to improve their potency. The QSAR study assists medicinal chemists to select the optimum structural requirement to improve the activity of a series of compounds under investigation (Hansch et al., 2002). In QSAR study, the parameterization of chemical structure plays an essential role. In enumeration of chemical structures, it is important to note that in isolation, a data point is only a qualified number. A collection of such qualified numbers makes a variable or descriptor. In mathematical models each and every (independent; X) variable communicates with the target (dependent; Y) variable. A meaningful communication between X and Y variables results in the evolution of models with predictive value (Gupta and Prabhakar, 2008).
In this milieu, we have contemplated a QSAR study on FFAR1 stimulating activity of ADCA analogs. The aim of the study was to identify the specific physicochemical and structural requirement to improve the activity of ADCA analogs. For QSAR study, different physicochemical and structural descriptors have been calculated from the Molecular Operating Environment (MOE, 2011). The Combinatorial Protocol in the Multiple Linear Regression (CP-MLR) approach was used to develop the QSAR models (Prabhakar, 2003).
2 Material and methods
2.1 Dataset
A series of 4-alkynyldihydrocinnamic acid analogs (ADCA analogs) along with their FFAR1 stimulating activity in terms of pEC50 (negative logarithm of effective concentration which showed 50% activity) was selected for the study. The structure database of compounds was developed in MOE. The 3D structures were optimized by MMFF94x force-field as implemented in MOE. The optimized structure dataset was used for the parameterization of compounds. This has resulted in three hundred and seven non-zero descriptors depicting the physicochemical and topological properties of the ADCA analogs. The structure dataset was partitioned into training set (24 compounds) and test set (8 compounds) with the help of single linkage hierarchical clustering of all the descriptors (Table 1). A plot of molecular weights (MW) versus LogP of all ADCA analogs was used to compare the chemical space spanned by the training set and test set compounds (Fig. 1). The plot showed that the test set compounds cover almost entire chemical space corresponding to the training set compounds.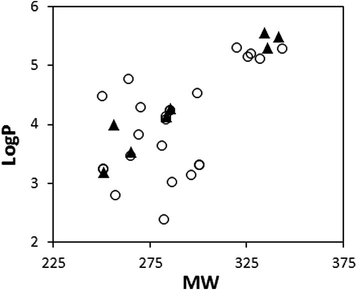
Plot of molecular weights (MW) versus logP of training (circle) and test (triangle) set of ADCA analogs.
2.2 QSAR procedure
The QSAR models were developed using the CP-MLR procedure (Prabhakar, 2003). CP-MLR helps in identifying the information rich descriptors correspond to the biological activity under study from a large descriptor pool. It is a filter based variable selection procedure for QSAR and QSPR studies (Prabhakar, 2003; Gupta and Prabhakar, 2006; Saquib et al., 2007). Its implementation and detailed procedural aspects are given elsewhere (Gupta et al., 2005; Saquib et al., 2007; Prabhakar et al., 2006; Gupta and Prabhakar, 2008; Sharma et al., 2011; Kumar et al., in press). In CP-MLR, the filter-1 seeds the variables by the way of limiting inter-parameter correlations to a predefined level (default acceptable value ⩽0.3); filter-2 controls the variables entry to a regression equation through t-values of coefficients (threshold value ⩾2.0) which confer more than 95% regression coefficients significant level ; filter-3 provides comparability of equations with different numbers of variables in terms of square-root of adjusted multiple correlation coefficient of regression equation “r-bar” and filter-4 estimates the consistency of the equation in terms of cross-validated R2 or Q2. In the present study, the thresholds for filters-1, 2, 3 and 4 were kept 0.3, 2.0, 0.71 and 0.3 ⩽ Q2 ⩽ 1.0, respectively. Reassessment of the identified models was done for chance correlations by performing repeated randomization of the activity. Each identified model was subjected to 100 simulation runs with scrambled activity and the emerging correlations were counted to express the percent chance correlation of the model under examination (Prabhakar et al., 2004). The proposed models were validated through test set. The robustness of models was further evaluated by calculating their overall metric values (Ojha et al., 2011; Roy et al., 2012) and slopes (k and k′) for the least squares regression line (Golbraikh and Tropsha) (Table 2). The metric is represented by the following equation:
Here
and
are correlations between the observed and predicted values with and without intercept, respectively, for the least squares regression lines. The metric
values help to circumvent the overestimation of the quality of prediction due to a wide response range (Y-range) (Ojha et al., 2011; Roy et al., 2012). The slopes k and k′ were calculated by the following equations:
Here, Y and
are observed and predicted activities, respectively. For a good predictive QSAR model the value of k and k′ should be near to 1 (Golbraikh and Tropsha, 2002).
Equations
Metric valuea
Slopeb
Metric
Reverse
Average
k
k′
(1)
0.693
0.602
0.647
0.090
0.997
0.999
(2)
0.742
0.644
0.693
0.098
0.997
0.999
(3)
0.748
0.649
0.698
0.099
1.000
0.996
3 Results and discussion
In the first step of the study, CP-MLR procedure with default parameter settings was used to develop simple one and two descriptor models (base line models). This resulted in identification of three one-parameter and six hundred and sixty-four two-parameter models embedding total one hundred and sixty-five descriptors in them. These one hundred and sixty-five descriptors were selected to discover higher models i.e. four-parameter models to explain the activity of ADCA analogs. The following are selected four-parameter models (Eqs. (1) and (2)) from the pool of equations:
In all regression equations, n is number of compounds, r is correlation coefficient, Q2 is cross-validated R2 from leave-one-out (LOO) procedure, Q2L3O is cross-validated R2 from leave-three-out procedure (where a group of three compounds are randomly kept outside the analysis each time in such a way that all compounds are in the predictive groups for once), s is standard error of the estimate and F is F-ratio between the variances of calculated and observed activities. The ryrand(SD) is the mean correlation coefficient of the regressions in the activity (Y) randomization study with its standard deviation from 100 simulations. In the randomization study, none of the identified models has shown any chance correlation. Additional statistical parameters such as, the “Akaike’s information criterion” (AIC) (Akaike, 1973, 1974), the Kubinyi function ‘FIT’ (Kubinyi, 1994a, 1994b) and the Friedman’s “lack of fit” (LOF) (Friedman, 1990) have also been calculated to further validate the derived QSAR models. The AIC takes into account the statistical goodness of fit and the number of parameters that have to be estimated to achieve that ‘degree of fit’. The Kubinyi function ‘FIT’ closely related to the F-value, proved to be a useful parameter for assessing the quality of the models. The model that produces the lowest AIC value and highest FIT value is considered potentially the most useful. The LOF factor takes into account the number of terms used in the equation and is not biased, as are other indicators, toward large number of parameters (Sharma et al., 2009). The Eqs. (1) and (2) have been validated with a test set of 8 compounds (R2T = R2 of test set compounds). The test set predictions are in agreement with their experimental values (Table 1). The metric values and slopes (k and k′) of developed models (Eqs. (1)–(3)) have been found within their acceptable limits and thus, ensure the good predictability of the models (Table 2).
Now in terms of physical meaning descriptors, a_nN and vsurf_CP (Pearson correlation r = −0.691 and 0.687, respectively) are among the highest correlating descriptors with the activity (Table 3). The a_nN refers to the number of nitrogen atoms in ADCA analogs. Negative correlation of a_nN with activity indicated that presence of nitrogen atom(s) in aryl rings is detrimental for the activity. Closure look into the structures of ADCA analogs showed that phenyl/thiophenyl group containing analogs (compounds 1 and 15, Table 1) exhibited better activity than nitrogen containing pyridyl (2, 3 and 4), pyrimidyl (11) and thiazolyl (14) analogs. The descriptors a_nCl (number of chlorine atoms) and a_nF (number of fluorine atoms) show positive correlation with the activity. Here, chloro (20, 30) or fluoro (19) substituted pyridyl derivatives appeared superior to the unsubstituted one (2, 3, 4). This may be suggested that the increase in activity of chloro or fluoro substituted pyridyl or pyrimidyl derivatives may be due their positive effect on the hydrophobicity of these analogs. The descriptor std_dim2 (Eq. (1)) and vsurf_CP (Eq. (2)) belongs to the descriptor class representing surface area, volume and shape of the compounds. These descriptors are favorable for activity. The std_dim2 depend on the structure connectivity and conformation of compounds. The vsurf_CP is a critical packing descriptor which considers surface and shape of molecules. The higher surface area of molecules is associated with either hydrophobicity or important for dispersion interactions. Its positive correlation with activity indicates that the descriptor is conducive for the activity. SlogP_VSA2 and SlogP_VSA6 (Eq. (2)) are indicative of accessible van der Waals surface area with lag 2 and 6 respectively of the compounds associated with SlogP. Their positive correlations with the activity indicate that the compounds with high accessible van der Waals surface area would have higher activity. The chloro function in compound 20 has higher accessible van der Waals surface area than fluoro containing compound 19 and exhibited better activity. However, compounds 26 (fluoro) and 27 (chloro) showed comparable activity (Table 1).
Des. no.
Descriptorsa
1
2
3
4
5
6
7
8
9
1
a_nCl
1.000
2
a_nF
−0.260
1.000
3
a_nN
0.236
−0.055
1.000
4
dipoleX
0.291
0.084
0.235
1.000
5
SlogP_VSA2
−0.269
1.000
−0.055
0.084
1.000
6
SlogP_VSA6
0.946
−0.232
0.141
0.161
−0.232
1.000
7
std_dim2
−0.069
−0.007
0.105
0.123
−0.007
0.105
1.000
8
vsurf_CP
−0.038
−0.277
−0.896
−0.292
−0.277
0.041
−0.003
1.000
9
logP(o/w)
0.349
0.005
−0.554
−0.053
0.005
0.391
0.257
0.633
1.000
−logEC50b
0.294
0.150
−0.691
0.114
0.150
0.367
0.249
0.687
0.857
The series of ADCA analogs were designed by Christiansen et al. to study the effect of hydrophobicity. They assumed that compounds with less hydrophobicity or high hydrophilicity would be favorable for activity. To study the influence of hydrophobicity on the activity of ADCA analogs, the Eq. (3) was developed by replacing parameter std_dim2 in Eq. (1) with logP(o/w).
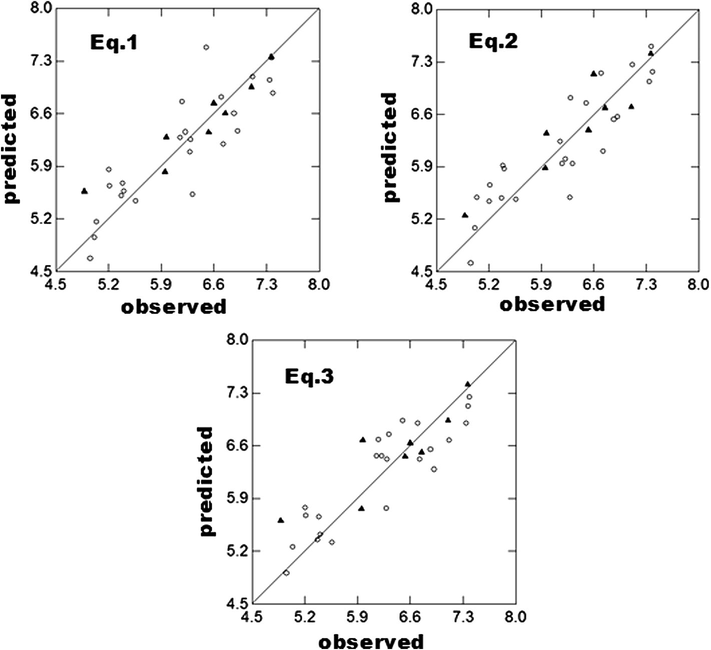
Plots for observed versus predicted activity of training (circle) and test (triangle) set of ADCA analogs.
4 Concluding remarks
The QSAR study on ADCA analogs as FFAR1 agonist indicated that hydrophobicity is an important physicochemical property for their activity. Hydrophobicity of organic molecules depends upon their solvent accessible surface area and thus, substituted aryl functions (phenyl or thiophenyl) with high solvent accessible surface area would lead to better activity. In aliphatic substituents, linear one possesses more van der Waals surface area than corresponding branched substituents. Therefore, aryl function in ADCA analogs may be substituted with open chain alkyl groups to increase their hydrophobicity and activity. In addition to this, aryl groups substituted with chloro or fluoro functions at ortho, meta or para positions would show superior FFAR1 agonistic activity. Presence of nitrogen atom decreases the hydrophobicity and thus nitrogen containing heterocyclic ring should be avoided for better FFAR1 agonistic activity.
Acknowledgments
Authors thank Dr. Y.S. Prabhakar, Scientist, Central Drug Research Institute, Lucknow (UP) India for providing software for the QSAR study. The support and facilities provided by Chairman, ISF College of Pharmacy, Moga (PB) India is greatly acknowledged.
References
- Akaike, H., 1973. Information Theory and an Extension of the Minimum Likelihood Principle. AkademiaiKiado, Budapest, pp. 267–281.
- A new look at the statistical identification model. IEEE Trans. Autom. Control.. 1974;AC-19:716-723.
- [Google Scholar]
- Progress in the discovery and development of small-molecule modulators of G-protein-coupled receptor 40 (GPR40/FFA1/FFAR1): an emerging target for type 2 diabetes. Expert Opin. Ther. Pat.. 2009;19:237-264.
- [Google Scholar]
- The orphan G protein-coupled receptor GPR40 is activated by medium and long chain fatty acids. J. Biol. Chem.. 2003;278:11303-11311.
- [Google Scholar]
- Identification of a potent and selective free fatty acid receptor 1 (FFA1/GPR40) agonist with favorable physicochemical and in vitro ADME properties. J. Med. Chem.. 2011;54:6691-6703.
- [Google Scholar]
- Effects of prolonged in vitro exposure to sulphonylureas on the function and survival of human islets. J. Diabetes Complications. 2005;19:60-64.
- [Google Scholar]
- GPR40: a therapeutic target for mediating insulin secretion. Int. J. Mol. Med.. 2012;30:1261-1266.
- [Google Scholar]
- Friedman, J., 1990. Technical Report No. 102 Laboratory for Computational Statistics. Stanford University, Stanford.
- How doctors choose medications to treat type 2 diabetes: a national survey of specialists and academic generalists. Diabetes Care. 2007;30:1448-1453.
- [Google Scholar]
- Topological descriptors in modeling the antimalarial activity of 4-(3′,5′-disubstituted anilino)quinolines. J. Chem. Inf. Model.. 2006;46:93-102.
- [Google Scholar]
- QSAR study on tetrahydroquinoline analogues as plasmodium protein farnesyltransferase inhibitors: a comparison of rationales of malarial and mammalian enzyme inhibitory activities for selectivity. Eur. J. Med. Chem.. 2008;43:2751-2767.
- [Google Scholar]
- CP-MLR directed QSAR studies on the antimycobacterial activity of functionalised alkenols – topological descriptors in modeling the activity. Bioorg. Med. Chem.. 2005;13:343-351.
- [Google Scholar]
- Chem-bioinformatics: comparative QSAR at the interface between chemistry and biology. Chem. Rev.. 2002;102:783-812. <http://www.who.int/mediacentre/factsheets/fs312/en/index.html> (accessed on 18.03.13)
- [Google Scholar]
- The prevention of type 2 diabetes mellitus. Endocrinol. Metab. Clin. North Am.. 2005;34:199-219.
- [Google Scholar]
- Free fatty acids regulate insulin secretion from pancreatic beta cells through GPR40. Nature. 2003;422:173-176.
- [Google Scholar]
- Glycemic durability of rosiglitazone, metformin, or glyburide monotherapy. N. Engl. J. Med.. 2006;355:2427-2443.
- [Google Scholar]
- A human cell surface receptor activated by free fatty acids and thiazolidinedione drugs. Biochem. Biophys. Res. Commun.. 2003;301:406-410.
- [Google Scholar]
- Variable selection in QSAR studies. I. An evolutionary algorithm. Quant. Struct. Act. Relat.. 1994;13:285-294.
- [Google Scholar]
- Variable selection in QSAR studies. II. A highly efficient combination of systematic search and evolution. Quant. Struct. Act. Relat.. 1994;13:393-401.
- [Google Scholar]
- Kumar, V., Gupta, M.K., Singh, G., Prabhakar, Y.S., 2012. CP-MLR/PLS directed QSAR study on the glutaminylcyclase inhibitory activity of imidazoles: rationales to advance the understanding of activity profile. J. Enzyme Inhib. Med. Chem. (in press).
- Sulfonylurea induced beta-cell apoptosis in cultured human islets. J. Clin. Endocrinol. Metab.. 2005;90:501-506.
- [Google Scholar]
- MOE, 2011. The Molecular Operating Environment from Chemical Computing Group Inc., 1255 University Street, Suite 1600, Montreal, Quebec, Canada H3B 3X3. <www.chemcomp.com>.
- Further exploring metrics for validation of QSPR models. Chemom. Intell. Lab. Syst.. 2011;107:194-205.
- [Google Scholar]
- A combinatorial approach to the variable selection in multiple linear regression analysis of Selwood et al. data set – a case study. QSAR Comb. Sci.. 2003;22:583-595.
- [Google Scholar]
- CP-MLR/PLS directed structure–activity modeling of the HIV-1RT inhibitory activity of 2,3-diaryl-1,3-thiazolidin-4-ones. QSAR Comb. Sci.. 2004;23:234-244.
- [Google Scholar]
- A high dimensional QSAR study on the aldose reductase inhibitory activity of some flavones: topological descriptors in modeling the activity. J. Chem. Inf. Model.. 2006;46:86-92.
- [Google Scholar]
- Fatty acid receptors as new therapeutic targets for diabetes. Expert Opin. Ther. Targets. 2007;11:661-671.
- [Google Scholar]
- How safe is the use of thiazolidinediones in clinical practice? Expert Opin. Drug Saf.. 2009;8:15-32.
- [Google Scholar]
- Comparative studies on some metrics for external validation of QSPR models. J. Chem. Inf. Model.. 2012;52:396-408.
- [Google Scholar]
- C-3 alkyl/arylalkyl-2,3-dideoxy hex-2-enopyranosides as antitubercular agents: synthesis, biological evaluation, and QSAR study. J. Med. Chem.. 2007;50:2942-2950.
- [Google Scholar]
- Modeling of cyclooxygenase-2 and 5-lipooxygenase inhibitory activity of apoptosis-inducing agents potentially useful in prostate cancer chemotherapy: derivatives of diarylpyrazole. J. Enzyme Inhib. Med. Chem.. 2009;24:607-615.
- [Google Scholar]
- CP-MLR/PLS directed QSAR study on apical sodium-codependent bile acid transporter inhibition activity of benzothiepines. Mol. Divers. 2011;15:135-147.
- [Google Scholar]
- A review of the efficacy and safety of oral antidiabetic drugs. Expert Opin. Drug Saf.. 2013;12:153-175.
- [Google Scholar]
- Selective small-molecule agonists of G protein-coupled receptor 40 promote glucose-dependent insulin secretion and reduce blood glucose in mice. Diabetes. 2008;57:2211-2219.
- [Google Scholar]
- Newer agents for blood glucose control in type 2 diabetes: systematic review and economic evaluation. Health Technol. Assess.. 2010;36:1-248.
- [Google Scholar]
- WHO, 2014. <http://www.who.int/mediacentre/factsheets/fs312/en/> (accessed on 12.12.14).






