Translate this page into:
Rational design of Zeocin binding protein variants for antibiotic resistance studies
⁎Corresponding author at: Department of Biochemistry, College of Science, Building no 5, P.O. Box 2455, King Saud University, Riyadh 11451, Saudi Arabia. amalik@ksu.edu.sa (Ajamaluddin Malik)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Antibiotic resistance marker was used in this investigation because they are selected;e markers and effective in different hosts. This study used Zeocin binding protein (ZBP) due to its known 3D structure and applicability in prokaryotic and eukaryotic hosts. A library of 22 mutations was developed through a rational design strategy. Subsequently, a selection strategy was used to identify destabilizing mutations in the ZBP. ZBP variants were expressed in E.coli using a leaky expression approach, and minimum inhibitory concentration (MIC) was calculated by cultivating different variants at various Zeocin concentrations. Zeocin resistance was drastically decreased in some ZBP variants. Positive controls (wild-type ZBP) exhibited high resistance, while negative controls (pET vector without ZBP) showed susceptibility. Two variants (ZBP P9E and ZBP R26F) displayed drastic resistance loss. The variants reported in this study may identify molecular chaperones and folding modulators affecting proteostasis. These variants can potentially discover chemical chaperones that improve the stability and solubility of destabilized ZBP variants.
Keywords
Bleomysin
Zeocin
Biosensor
Folding
1 Introduction
The cellular proteostasis network tightly regulates the lifecycle of proteins, from synthesis to termination. The proteostasis network monitors protein activity throughout their functional phase, maintaining proper interactions and cellular balance. The termination phase, marking the end of a protein's lifecycle, involves controlled degradation (Jayaraj et al., 2020). Disturbances in proteostasis are a contributing factor to a range of diseases (Kikis et al., 2010, Sweeney et al., 2017). Alzheimer's, Parkinson's, Huntington's, and Amyotrophic Lateral Sclerosis (ALS) diseases cause neuron degeneration due to protein misfolding (Lamptey et al., 2022). In type II diabetes, amylin misfolding causes pancreatic dysfunction, while CFTR protein misfolding causes cystic fibrosis (Fukuda and Okiyoneda 2020, Hassan et al., 2022). Additionally, misfolded proteins are associated with certain malignancies (Chen et al., 2017, Kawano et al., 2023).
The mammalian proteostasis network comprises over 2000 genes (Klaips et al., 2018, Lualdi et al., 2020). Identification of genes encoding protein folding modulators remains challenging without efficient, widely applicable, and sensitive in vivo protein folding assays. The protein folding quality-control machinery ensures the proper balance between folded and unfolded proteins. All organisms possess an efficient system to minimize the protein misfolding, aided by specialized proteins known as molecular chaperones (Vabulas et al., 2010, Chen et al., 2011). Chaperoning was first observed in 1962 under thermal stress, leading to the term “molecular chaperone” coining in 1978 (Ritossa, 1962, Laskey et al., 1978). During the early 1980s, Heat Shock Proteins (Hsps) were identified as molecular chaperones, later found to be conserved across different species. Traditional methods for identifying molecular chaperones include stress induction, mutation causing proteome instability, homology to known chaperones, binding to other proteins, and in vitro chaperone activity assays (Quan and Bardwell, 2012). While these methodologies have been fruitful, they lack specificity. For instance, heat stress induces the expression of 50–200 genes, with the most strongly induced proteins typically involved in chaperoning, while others are engaged in activities such as regulation, metabolism, nucleic acid binding, and proteolysis (Quan and Bardwell, 2012). Combining multiple approaches and focusing on specific targets can enhance the specificity of chaperone discovery, but traditional methods are time-consuming and labor-intensive.
Given the stochastic nature of chaperone discovery, the current list of molecular chaperones is likely incomplete. Previous studies using beta-lactamase and DsbA folding biosensors identified the periplasmic chaperone Spy (Quan et al., 2011). However, these biosensors are limited to the periplasmic space (Foit et al., 2009, Quan et al., 2011). A partially destabilized ZBP can serve as an effective protein-folding biosensor, facilitating the discovery of novel molecular chaperones and folding modulators. By leveraging the correlation between ZBP expression levels and antibiotic resistance, this biosensor will allow for efficient in vivo screening across diverse hosts, overcoming the limitations of traditional chaperone identification methods.
Zeocin, belonging to the bleomycin/phleomycin antibiotic family, exhibits strong toxicity against a wide range of cells (Drocourt et al., 1990). The zeocin formulation contains the active ingredient phleomycin D1, a basic copper-chelated glycopeptide. Zeocin specifically breaks double-stranded DNA, causing cytotoxicity in both prokaryotic and eukaryotic hosts (Gatignol et al., 1987, Gatignol et al., 1988, Perez et al., 1989, Calmels et al., 1991). The ShBle gene from Streptoalloteichus hindustanus, a bleomycin resistance marker, was employed in this study. ZBP, a 13.6 kDa homodimeric protein, binds stoichiometrically with high affinity to various antibiotics of the bleomycin family (Gatignol et al., 1988). A tight correlation has been established between the antibiotic resistance of the host cell and the level of ZBP gene expression. The development of ZBP as a protein-folding biosensor is motivated by its wide selection range, cost-effectiveness, and powerful selectable phenotype in both prokaryotic and eukaryotic hosts (Dumas et al., 1994).
2 Material and methods
2.1 Materials
The Escherichia coli BL21 (DE3)-RIL competent cell obtained from Agilent Technologies was employed for cloning and Zeocin sensitivity assay. The pET3a plasmid (Novagen) was utilized for the cloning process. The Zeocin was purchased from Invitrogen. Sigma-Aldrich provided LB Broth (Lennox) and LB agar (Lennox agar). Ampicillin was obtained from Biobasic. All the remaining reagents were of analytical grade.
2.2 Cloning of ZBP (WT)
The Sh Ble gene from Streptoalloteichus hindustanus has been optimized for expression in E. coli using codon optimization. Following that, a His-tag and a highly specific cleavage site (TEV protease site) were fused at the N-terminus of ZBP. The ZBP fusion protein was inserted into the pET-3a plasmid at the NdeI and BamHI restriction enzyme sites (Genscript). The ZBP (WT) fusion protein was transformed into a chemically competent Escherichia coli BL21 (DE3)-RIL strain and then selected on an LBamp (200 μg/ml) plate.
2.3 Generation of ZBP mutant library
The development of the ZBP mutant library was carried out using a rational design technique. ZBP's primary structure consists of 124 amino acids. An in-depth analysis was conducted on the conserved residues in the primary structure and the 3D crystal structure to assess the residues contributing to the stabilization of ZBP. Furthermore, the crystal structure of ZBP revealed the involvement of 21 residues in the binding pocket (Dumas et al., 1994). A set of 22 mutants (Table 1) was created, omitting the residues directly involved in Zeocin binding or interacting with the Zeocin binding residues. The selection criteria were based on the idea that the mutation should not impact the Zeocin binding affinity. The 22 ZBP variants have been introduced into the Escherichia coli BL21 (DE3)-RIL strain and then selected on an LBamp (200 ug/ml) plate.
Clone No
Clone name
Mutation site on ZBP
Mutation site on ZBP fusion protein
1.
Neg. Control
2.
ZBP (wt)
3.
ZBP P9E
P9E
ZBP P29E
4.
ZBP L11A
L11A
ZBP L31A
5.
ZBP V16A
V16A
ZBP V36A
6.
ZBP V20A
V20A
ZBP V40A
7.
ZBP F22A
F22A
ZBP F42A
8.
ZBP D25G
D25G
ZBP D45G
9.
ZBP D25K
D25K
ZBP D45K
10.
ZBP R26G
R26G
ZBP R46G
11.
ZBP R26F
R26F
ZBP R46F
12.
ZBP L27A
L27A
ZBP L47A
13.
ZBP L27G
L27G
ZBP L47G
14.
ZBP F29A
F29A
ZBP F49A
15.
ZBP R31A
R31A
ZBP R51A
16.
ZBP R43A
R43A
ZBP R63A
17.
ZBP R69A
R69A
ZBP R89A
18.
ZBP L71A
L71A
ZBP L91A
19.
ZBP Y75A
Y75A
ZBP Y95A
20.
ZBP E77A
E77A
ZBP E97A
21.
ZBP W78A
W78A
ZBP W98A
22.
ZBP V81A
V81A
ZBP V101A
23.
ZBP V82A
V82A
ZBP V102A
24.
ZBP L108A
L108A
ZBP L128A
2.4 Screening of unstable ZBP variants
The initial transformation selection was based on the ampicillin resistance present on pET3a. ZBP (wt) clone was used as the positive control, and a pET plasmid lacking the ZBP clone was used as the negative control for Zeocin resistance. At first, all the clones were cultured overnight at 37 °C in Lennox broth supplemented with 200 μg/ml ampicillin. The OD600 of the overnight culture was determined and placed on ice to freeze the growth. The OD600 of each culture was standardized to a concentration of OD1/ml. Afterward, each culture was diluted ten-fold using Lennox broth in a serial manner. Subsequently, a spot-titer experiment was performed using the methodology described in the study by Malik et al (Malik et al., 2014). In summary, a small volume of 2 μl from each dilution (ranging from 10−0 to 10−5) of each clone was placed on Lennox agar plates containing various amounts (0, 25, 100, 250, 500, 1000, 1500, and 2000 μg/ml) of Zeocin. The plates were placed in an incubator and kept overnight at 37 °C. The growth of clones at the highest dilution was quantified and plotted in relation to Zeocin concentration.
3 Results
3.1 Development of a protein-folding biosensor
The principle of ZBP-based folding biosensors is illustrated in Fig. 1. The protein folding biosensor will effectively link the stability of typically unstable, aggregation-prone ZBP variants to the overall Zeocin resistance of the host strain. The construction of a biosensor incorporating a destabilized and partially folded ZBP is anticipated to result in moderate resistance. Destabilized ZBP can attain a folded and soluble state by integrating molecular or chemical chaperoning. Consequently, the host cell will exhibit substantial resistance to Zeocin. However, the destabilized ZBP will be susceptible to aggregation and/or proteolysis under stress or proteases due to its inherent instability. This will significantly diminish or eliminate the biosensor's affinity for Zeocin, leading to the loss of the Zeocin resistance. This approach will enable the identification of molecular chaperones or other folding modulators impacting proteostasis or chemical chaperones that contribute to stabilizing and improving the solubility of the ZBP variants prone to aggregation.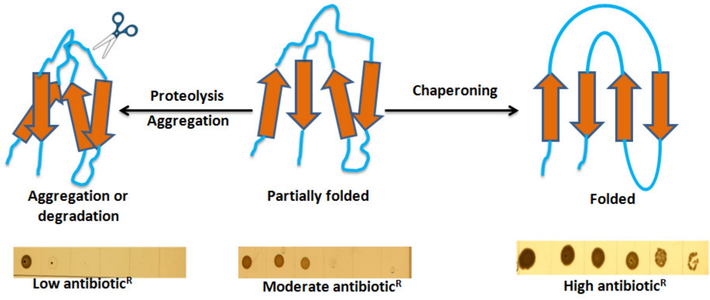
Schematic diagram of biosensor to measure in vivo protein folding.A ZBP-based biosensor (destabilized and partially folded) should result in moderate resistance. In the case of molecular or chemical chaperoning and proper folding, more ZBP biosensors will attain a folded and soluble state and tightly bind zeocin, resulting in high zeocin resistance. Under stress conditions or following protease induction, the unstable or unfolded biosensor will be more prone to aggregation and/or degradation, which will result in little or no binding to zeocin. As a result, zeocin resistance will be lost.
3.2 Construction of ZBP (WT) fusion protein
ZBP is a protein composed of two identical subunits (homodimer) consisting of 124 amino acids (Fig. 2). To optimize the expression of ZBP in E. coli, the codon was optimized. In order to facilitate purification, a His-tag was attached to the N-terminus of ZBP. A specific cleavage site was included between the his-tag and ZBP to eliminate the his-tag from ZBP after purification. Each construct contained an identical sequence of the N-terminus fusion tag, specifically MGSHHHHHHSGSENLYFQ↓SG. In order to facilitate leaky expression in E. coli, suitable cloning and experiment parameters were chosen for this work. The cloning was done on pET3a, which lacks the lacI gene. Thus, lac repressor was absent. The ZBP (WT) clone was transformed in an E. coli strain, which was also lacking repressor plasmid such as pLysS (Escherichia coli BL21 (DE3)-RIL strain). The presence of the bla gene in the pET 3a plasmid facilitated the selection on an ampicillin plate.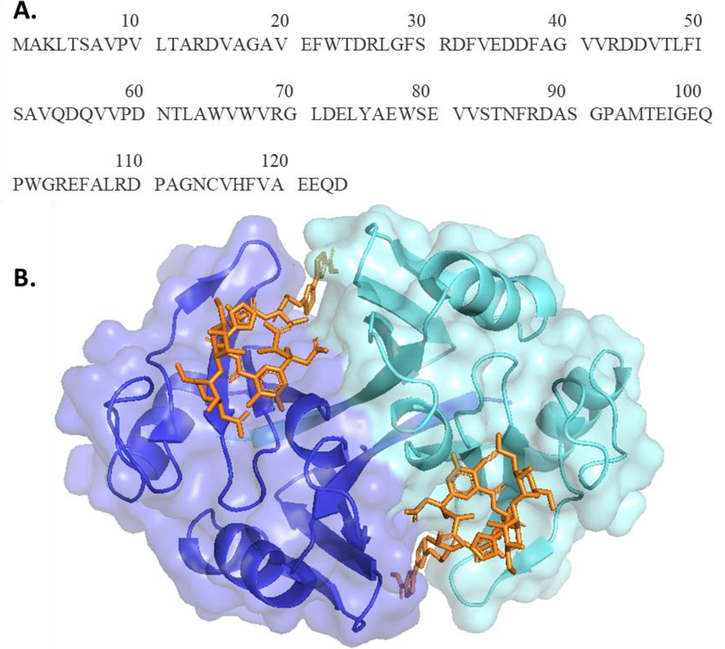
(A) Sequence of Zeocin binding protein (ZBP) of Streptoalloteichus hindustanus. Sh bleomycin gene encodes 124 amino acids long protein product. (B) Symmetry-related homodimeric crystal structure of ZBP (PDB ID: 1XRK), shown in cyan and blue. Zeocin binding in a long crevice at the dimer interface. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3 Generation of ZBP variants
The structure of ZBP is characterized by two identical halves that are folded in the same folds. Each subunit's half comprises four beta-sheets and one alpha-helix. When ZBP forms a dimer, there is a 2:2 ratio between ZBP and Zeocin, meaning two Zeocin molecules bind to each ZBP dimer. A significant portion of ZBP, approximately one-sixth (21 out of 124 residues), was either directly involved in binding with Zeocin or was in close proximity to the Zeocin molecule (Dumas et al., 1994). Consequently, careful considerations were necessary in designing variants to destabilize ZBP without compromising its binding affinity for Zeocin. The binding pocket for Zeocin traverses the subunit interface, as illustrated in Fig. 3, where the 21 residues crucial for Zeocin binding are highlighted in blue. This binding pocket comprises residues from both subunits. This study introduced 22 mutations (Table 2) at locations far from the binding site, as indicated in red in Fig. 4. These mutation sites on alpha-helix and beta-sheets were strategically chosen to disrupt the electrostatic and hydrophobic interactions, aiming to destabilize ZBP.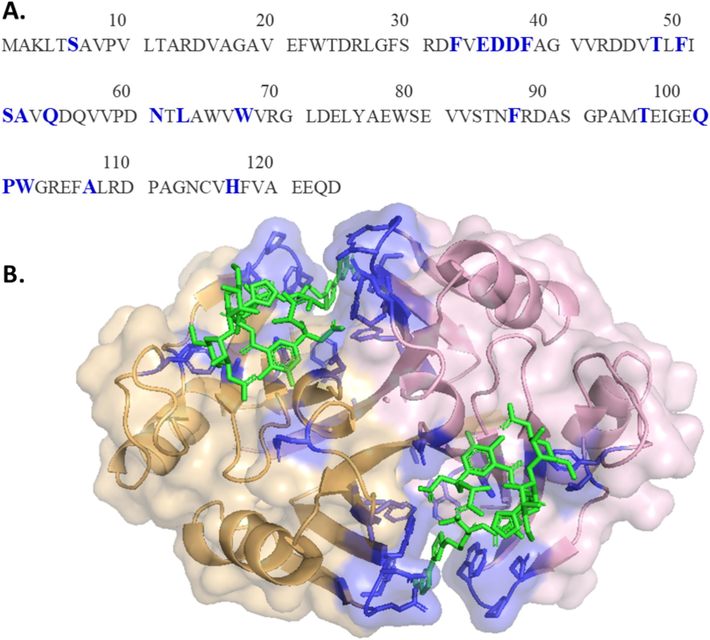
(A) Binding pocket residues were highlighted in blue on the ZBP sequence. (B) Binding pocket residues were shown in blue on the ZBP structure. Each binding pocket consists of 21 residues of both subunits. The Zeocin molecule was shown in green. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
ZBP variants
Sequence
ZBP (WT)
MGSHHHHHHSGSENLYFQSGMAKLTSAVPVLTARDVAGAVEFWTDRLGFSRDFVEDDFAGVVRDDVTLFISAVQDQVVPDNTLAWVWVRGLDELYAEWSEVVSTNFRDASGPAMTEIGEQPWGREFALRDPAGNCVHFVAEEQD
ZBP P29E
MGSHHHHHHSGSENLYFQSGMAKLTSAVEVLTARDVAGAVEFWTDRLGFSRDFVEDDFAGVVRDDVTLFISAVQDQVVPDNTLAWVWVRGLDELYAEWSEVVSTNFRDASGPAMTEIGEQPWGREFALRDPAGNCVHFVAEEQD
ZBP L31A
MGSHHHHHHSGSENLYFQSGMAKLTSAVPVATARDVAGAVEFWTDRLGFSRDFVEDDFAGVVRDDVTLFISAVQDQVVPDNTLAWVWVRGLDELYAEWSEVVSTNFRDASGPAMTEIGEQPWGREFALRDPAGNCVHFVAEEQD
ZBP V36A
MGSHHHHHHSGSENLYFQSGMAKLTSAVPVLTARDAAGAVEFWTDRLGFSRDFVEDDFAGVVRDDVTLFISAVQDQVVPDNTLAWVWVRGLDELYAEWSEVVSTNFRDASGPAMTEIGEQPWGREFALRDPAGNCVHFVAEEQD
ZBP V40A
MGSHHHHHHSGSENLYFQSGMAKLTSAVPVLTARDVAGAAEFWTDRLGFSRDFVEDDFAGVVRDDVTLFISAVQDQVVPDNTLAWVWVRGLDELYAEWSEVVSTNFRDASGPAMTEIGEQPWGREFALRDPAGNCVHFVAEEQD
ZBP F42A
MGSHHHHHHSGSENLYFQSGMAKLTSAVPVLTARDVAGAVEAWTDRLGFSRDFVEDDFAGVVRDDVTLFISAVQDQVVPDNTLAWVWVRGLDELYAEWSEVVSTNFRDASGPAMTEIGEQPWGREFALRDPAGNCVHFVAEEQD
ZBP D45G
MGSHHHHHHSGSENLYFQSGMAKLTSAVPVLTARDVAGAVEFWTGRLGFSRDFVEDDFAGVVRDDVTLFISAVQDQVVPDNTLAWVWVRGLDELYAEWSEVVSTNFRDASGPAMTEIGEQPWGREFALRDPAGNCVHFVAEEQD
ZBP D45K
MGSHHHHHHSGSENLYFQSGMAKLTSAVPVLTARDVAGAVEFWTKRLGFSRDFVEDDFAGVVRDDVTLFISAVQDQVVPDNTLAWVWVRGLDELYAEWSEVVSTNFRDASGPAMTEIGEQPWGREFALRDPAGNCVHFVAEEQD
ZBP R46G
MGSHHHHHHSGSENLYFQSGMAKLTSAVPVLTARDVAGAVEFWTDGLGFSRDFVEDDFAGVVRDDVTLFISAVQDQVVPDNTLAWVWVRGLDELYAEWSEVVSTNFRDASGPAMTEIGEQPWGREFALRDPAGNCVHFVAEEQD
ZBP R46F
MGSHHHHHHSGSENLYFQSGMAKLTSAVPVLTARDVAGAVEFWTDFLGFSRDFVEDDFAGVVRDDVTLFISAVQDQVVPDNTLAWVWVRGLDELYAEWSEVVSTNFRDASGPAMTEIGEQPWGREFALRDPAGNCVHFVAEEQD
ZBP L47A
MGSHHHHHHSGSENLYFQSGMAKLTSAVPVLTARDVAGAVEFWTDRAGFSRDFVEDDFAGVVRDDVTLFISAVQDQVVPDNTLAWVWVRGLDELYAEWSEVVSTNFRDASGPAMTEIGEQPWGREFALRDPAGNCVHFVAEEQD
ZBP L47G
MGSHHHHHHSGSENLYFQSGMAKLTSAVPVLTARDVAGAVEFWTDRGGFSRDFVEDDFAGVVRDDVTLFISAVQDQVVPDNTLAWVWVRGLDELYAEWSEVVSTNFRDASGPAMTEIGEQPWGREFALRDPAGNCVHFVAEEQD
ZBP F49A
MGSHHHHHHSGSENLYFQSGMAKLTSAVPVLTARDVAGAVEFWTDRLGASRDFVEDDFAGVVRDDVTLFISAVQDQVVPDNTLAWVWVRGLDELYAEWSEVVSTNFRDASGPAMTEIGEQPWGREFALRDPAGNCVHFVAEEQD
ZBP R51A
MGSHHHHHHSGSENLYFQSGMAKLTSAVPVLTARDVAGAVEFWTDRLGFSADFVEDDFAGVVRDDVTLFISAVQDQVVPDNTLAWVWVRGLDELYAEWSEVVSTNFRDASGPAMTEIGEQPWGREFALRDPAGNCVHFVAEEQD
ZBP R63A
MGSHHHHHHSGSENLYFQSGMAKLTSAVPVLTARDVAGAVEFWTDRLGFSRDFVEDDFAGVVADDVTLFISAVQDQVVPDNTLAWVWVRGLDELYAEWSEVVSTNFRDASGPAMTEIGEQPWGREFALRDPAGNCVHFVAEEQD
ZBP R89A
MGSHHHHHHSGSENLYFQSGMAKLTSAVPVLTARDVAGAVEFWTDRLGFSRDFVEDDFAGVVRDDVTLFISAVQDQVVPDNTLAWVWVAGLDELYAEWSEVVSTNFRDASGPAMTEIGEQPWGREFALRDPAGNCVHFVAEEQD
ZBP L91A
MGSHHHHHHSGSENLYFQSGMAKLTSAVPVLTARDVAGAVEFWTDRLGFSRDFVEDDFAGVVRDDVTLFISAVQDQVVPDNTLAWVWVRGADELYAEWSEVVSTNFRDASGPAMTEIGEQPWGREFALRDPAGNCVHFVAEEQD
ZBP Y95A
MGSHHHHHHSGSENLYFQSGMAKLTSAVPVLTARDVAGAVEFWTDRLGFSRDFVEDDFAGVVRDDVTLFISAVQDQVVPDNTLAWVWVRGLDELAAEWSEVVSTNFRDASGPAMTEIGEQPWGREFALRDPAGNCVHFVAEEQD
ZBP E97A
MGSHHHHHHSGSENLYFQSGMAKLTSAVPVLTARDVAGAVEFWTDRLGFSRDFVEDDFAGVVRDDVTLFISAVQDQVVPDNTLAWVWVRGLDELYAAWSEVVSTNFRDASGPAMTEIGEQPWGREFALRDPAGNCVHFVAEEQD
ZBP W98A
MGSHHHHHHSGSENLYFQSGMAKLTSAVPVLTARDVAGAVEFWTDRLGFSRDFVEDDFAGVVRDDVTLFISAVQDQVVPDNTLAWVWVRGLDELYAEASEVVSTNFRDASGPAMTEIGEQPWGREFALRDPAGNCVHFVAEEQD
ZBP V101A
MGSHHHHHHSGSENLYFQSGMAKLTSAVPVLTARDVAGAVEFWTDRLGFSRDFVEDDFAGVVRDDVTLFISAVQDQVVPDNTLAWVWVRGLDELYAEWSEAVSTNFRDASGPAMTEIGEQPWGREFALRDPAGNCVHFVAEEQD
ZBP V102A
MGSHHHHHHSGSENLYFQSGMAKLTSAVPVLTARDVAGAVEFWTDRLGFSRDFVEDDFAGVVRDDVTLFISAVQDQVVPDNTLAWVWVRGLDELYAEWSEVASTNFRDASGPAMTEIGEQPWGREFALRDPAGNCVHFVAEEQD
ZBP L128A
MGSHHHHHHSGSENLYFQSGMAKLTSAVPVLTARDVAGAVEFWTDRLGFSRDFVEDDFAGVVRDDVTLFISAVQDQVVPDNTLAWVWVRGLDELYAEWSEVVSTNFRDASGPAMTEIGEQPWGREFAARDPAGNCVHFVAEEQD
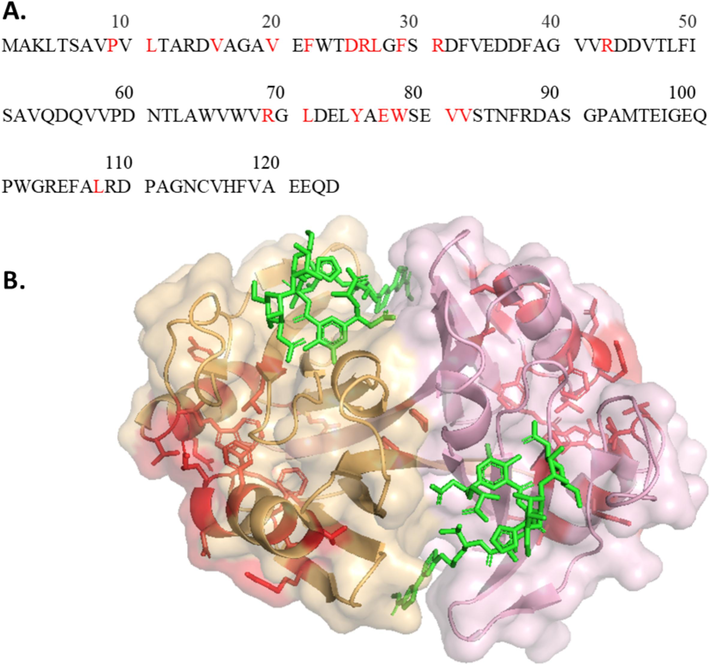
(A) Mutation sites highlighted in red on the ZBP sequence (B) Mutation sites selected away from binding site residues shown on ZBP structure in red. Mutation site residues neither interact with Zeocin nor with the binding site residues. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4 Screening of Zeocin resistance of ZBP variants
The resistance of ZBP variants to Zeocin was assessed by determining the minimum inhibitory concentration (MIC), which represents the lowest Zeocin concentration inhibiting the growth of E. coli expressing ZBP variants. The MIC experiments were conducted following the methodology by Malik et al. (Malik et al., 2014) with slight modifications. Initially, cultures were prepared by inoculating a single colony into 10 ml of Lennox media supplemented with 200 μg/ml ampicillin, followed by overnight incubation at 37 °C. The optical density at 600 nm (OD600) of the overnight culture was adjusted to 1.0. Subsequently, the E. coli culture was serially diluted, and dilutions ranging from 10-0 to 10-5 were spotted on LB plates containing increasing concentrations of Zeocin. After overnight incubation at 37 °C, the growth or no growth thereof for each dilution at each Zeocin concentration was observed and used to calculate MIC values, following the approach described by Foit et al. (Foit et al., 2009). Interestingly, our results aligned with the hypothesis, revealing significant Zeocin resistance loss in certain ZBP variants (see Fig. 4).
The positive control (wild-type ZBP) exhibited growth at the highest dilutions (10-5 dilutions), even in the presence of 2000 μg/ml Zeocin, due to the high stability and solubility of ZBP. Conversely, the negative control (pET vector without ZBP) failed to survive even in the presence of 25 μg/ml Zeocin. Notably, two variants, namely ZBP P9E (Fig. 4A) and ZBP R26F (Fig. 4C), displayed a pronounced loss of resistance, potentially attributed to destabilization. Thirteen other variants (L11A, V16A, V20A, R26G, L27A, L27G, F29A, R31A, R33A, L71A, E77A, W78A, L108A) exhibited a moderate level of loss in Zeocin resistance, suggesting varying degrees of destabilization in these variants.
4 Discussion
Two factors influence protein production levels: the folding capacity of the host and the intrinsic stability or aggregation tendency of the protein of interest (Mayer et al., 2007, Espargaro et al., 2008, Foit et al., 2009). This study presents the process of creating a system that establishes a relationship between the biosensor's stability and antibiotic resistance. Zeocin resistance markers align with our objectives due to their potent cell-killing and broad effectiveness across different species. Antimicrobial resistance can be easily identified by spot titer methods (Foit et al., 2009). Furthermore, it is feasible to assess the influence of different conditions or mutations on protein stability or their propensity to fold. We opted to use the Zeocin-based markers because its 3D structure was known and applicable in both prokaryotic as well as eukaryotic hosts, thus allowing the discovery of novel protein folding modulators even in mammalian cell lines (Dumas et al., 1994).
Our objective was to identify destabilizing mutation sites within ZBP. To accomplish this aim, we developed a library of 22 mutations through a rational design strategy. With positive and negative controls, the total number of samples would be 24 and can fit in the microtiter plate format. The ZBP fusion protein variants were cloned on a strong T7 promoter on a plasmid with high copy numbers, and we opted for leaky expression instead of induced expression. Leaky expression in E. coli refers to protein expression without specific inducing conditions and offers several advantages. By choosing leaky expression, researchers can simplify the experimental process and establish selection conditions across a range of Zeocin concentrations without additional inducers like IPTG. This approach provides continuous, albeit low, protein expression, which can be particularly useful in maintaining selective pressure and enabling the screening of multiple conditions simultaneously. The use of leaky expression allows for a more streamlined and cost-effective protocol. Eliminating the need for IPTG reduces the methods complexity and the potential variability introduced by inducer concentration and timing. Additionally, the continuous low-level expression ensures that selection pressures are consistently applied, facilitating the identification of optimal Zeocin concentrations for selecting desired variants (Francis and Page 2010, Rosano and Ceccarelli 2014, Kato 2020).
Resistance testing using MIC experiments revealed significant Zeocin resistance loss in certain ZBP variants, validating the hypothesis. Positive controls (wild-type ZBP) exhibited high resistance, while negative controls (pET vector without ZBP) showed susceptibility (Fig. 5). Two variants (ZBP P9E and ZBP R26F) displayed pronounced resistance loss, while thirteen others exhibited a moderate loss, indicating variable degrees of destabilization. A significant correlation has been established between host cell Zeocin resistance and ZBP variants. The destabilized variants will identify molecular chaperones and folding modulators affecting proteostasis or chemical chaperones that enhance the stability and solubility of aggregation-prone ZBP variants.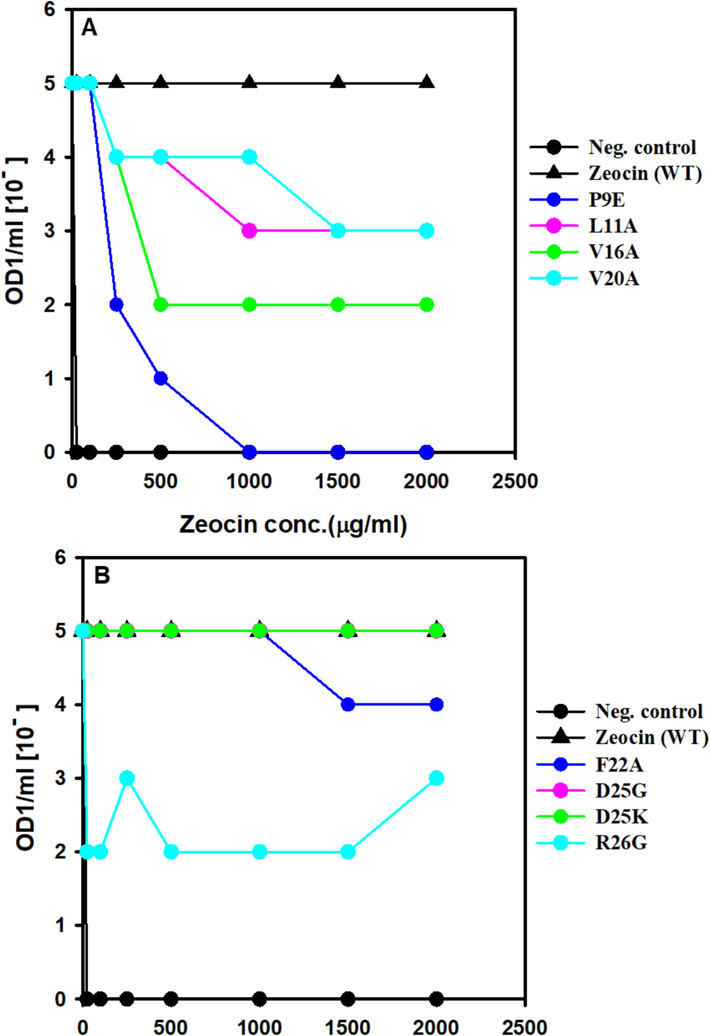
Screening of ZBP variants for MIC. E. coli expressing ZBP variations that were cultivated overnight were adjusted to a concentration of OD1/ml. Cultures were diluted in a series from 10 to 0 to 10 − 5 and then applied as spots on Lennox agar plates that contained varying doses of zeocin. Following an incubation period of 24 h at 37 °C, the presence or absence of growth for each dilution at various zeocin concentrations was recorded and utilized to determine the minimum inhibitory concentration (MIC) values.
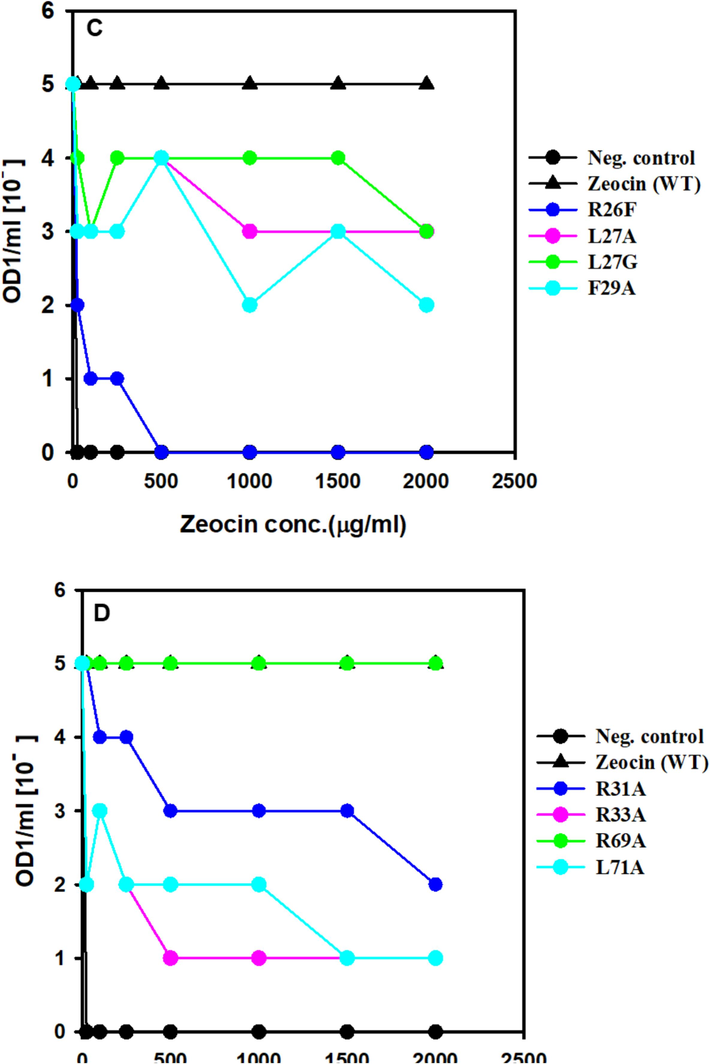
Screening of ZBP variants for MIC. E. coli expressing ZBP variations that were cultivated overnight were adjusted to a concentration of OD1/ml. Cultures were diluted in a series from 10 to 0 to 10 − 5 and then applied as spots on Lennox agar plates that contained varying doses of zeocin. Following an incubation period of 24 h at 37 °C, the presence or absence of growth for each dilution at various zeocin concentrations was recorded and utilized to determine the minimum inhibitory concentration (MIC) values.
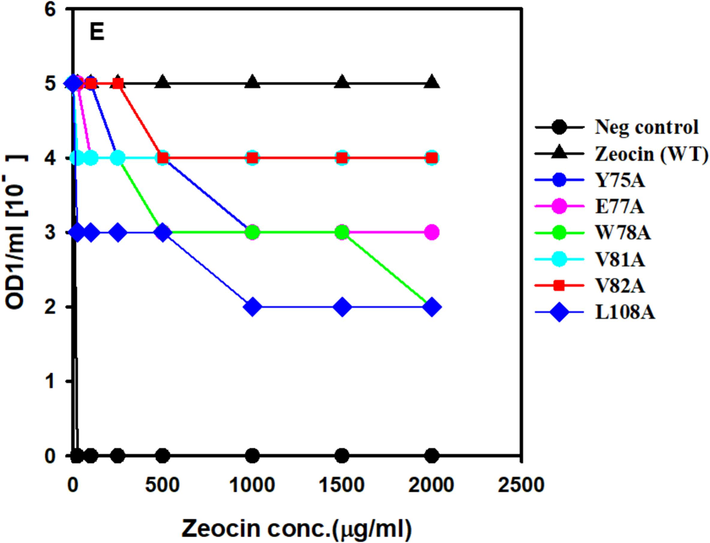
Screening of ZBP variants for MIC. E. coli expressing ZBP variations that were cultivated overnight were adjusted to a concentration of OD1/ml. Cultures were diluted in a series from 10 to 0 to 10 − 5 and then applied as spots on Lennox agar plates that contained varying doses of zeocin. Following an incubation period of 24 h at 37 °C, the presence or absence of growth for each dilution at various zeocin concentrations was recorded and utilized to determine the minimum inhibitory concentration (MIC) values.
The results of this study reveal several promising possibilities. The strong link observed between the host cell resistance to Zeocin antibiotic and ZBP variants highlights the crucial role of protein stability in antibiotic resistance. The significant and controlled loss of resistance found in several ZBP variants suggests that changes in amino acids can greatly destabilize the protein, impacting the survival of the host organism under selection pressure. Discovering molecular chaperones and folding modulators interacting with the destabilized ZBP variants could offer valuable insights into the physiological mechanisms responsible for maintaining proteostasis. Exploring chemical chaperones that can stabilize and improve the solubility of aggregation-prone ZBP variants presents an exciting opportunity. These chemicals can potentially enhance the productivity and effectiveness of recombinant proteins in biotechnological applications.
5 Conclusion
The study focuses on developing a selection system based on the ZBP to identify destabilizing mutations and their impact on antibiotic resistance. The choice of ZBP-based markers was advantageous due to their known 3D structure, applicability in both prokaryotic and eukaryotic hosts, and potential for discovering protein folding modulators, even in mammalian cell lines. The study introduces a rational design strategy to create a library of mutations in ZBP, using the leaky expression in E. coli for selection conditions at different Zeocin concentrations. Although leaky expression can be beneficial, it may not completely mimic the conditions of induced high-level protein expression. In addition, the library consisting of 22 mutations do not cover every possible destabilizing mutation in ZBP. In the future, the study should broaden the mutation library to investigate various destabilizing sites. Furthermore, studying the reported destabilized variants with molecular and chemical chaperones could provide more information about proteostasis mechanisms. Creating high-throughput in vivo assays for a more complete screening of chaperones and folding modulators would improve our understanding of their potential applications in biotechnology and pharmaceutical development.
CRediT authorship contribution statement
Sara Alharbi: Data curation, Writing – original draft. Ajamaluddin Malik: Conceptualization, Formal analysis, Supervision, Writing – original draft, Writing – review & editing. Abdulaziz Alamri: Formal analysis, Investigation, Project administration, Funding acquisition. Javed Masood Khan: Formal analysis, Investigation, Methodology, Writing – original draft. Mohd. Shahnawaz Khan: Funding acquisition, Investigation, Methodology, Project administration. Abdullah Alhomida: Project administration, Resources, Supervision. Tauseef Ahmad: Data curation, Methodology.
Acknowledgments
This study was funded by the National Plan for Science, Technology and Innovation (MAARIFAH), King Abdulaziz City for Science and Technology, Kingdom of Saudi Arabia, Award Number (13-BIO843-02).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- High efficiency transformation of Tolypocladium geodes conidiospores to phleomycin resistance. Curr. Genet.. 1991;20(4):309-314.
- [Google Scholar]
- Enhanced degradation of misfolded proteins promotes tumorigenesis. Cell Rep.. 2017;18(13):3143-3154.
- [Google Scholar]
- Cellular strategies of protein quality control. Cold Spring Harb. Perspect. Biol.. 2011;3(8):a004374
- [Google Scholar]
- Cassettes of the Streptoalloteichus hindustanus ble gene for transformation of lower and higher eukaryotes to phleomycin resistance. Nucl. Acids Res.. 1990;18(13):4009.
- [Google Scholar]
- Crystal structure and site-directed mutagenesis of a bleomycin resistance protein and their significance for drug sequestering. EMBO J.. 1994;13(11):2483-2492.
- [Google Scholar]
- The in vivo and in vitro aggregation properties of globular proteins correlate with their conformational stability: the SH3 case. J. Mol. Biol.. 2008;378(5):1116-1131.
- [Google Scholar]
- Francis, D. M. and R. Page, 2010. Strategies to optimize protein expression in E. coli. Curr Protoc Protein Sci. Chapter 5 (1) 5 24 21-25 24 29.
- Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) ubiquitylation as a novel pharmaceutical target for cystic fibrosis. Pharmaceuticals (Basel).. 2020;13(4)
- [Google Scholar]
- Phleomycin resistance encoded by the ble gene from transposon Tn 5 as a dominant selectable marker in Saccharomyces cerevisiae. Mol Gen Genet. 1987;207(2–3):342-348.
- [Google Scholar]
- Bleomycin resistance conferred by a drug-binding protein. FEBS Lett.. 1988;230(1–2):171-175.
- [Google Scholar]
- Linking hIAPP misfolding and aggregation with type 2 diabetes mellitus: a structural perspective. Biosci. Rep.. 2022;42(5)
- [Google Scholar]
- Functional modules of the proteostasis network. Cold Spring Harb. Perspect. Biol.. 2020;12(1)
- [Google Scholar]
- Extremely low leakage expression systems using dual transcriptional-translational control for toxic protein production. Int. J. Mol. Sci.. 2020;21(3)
- [Google Scholar]
- The anti-oncogenic effect of 17-DMAG via the inactivation of HSP90 and MET pathway in osteosarcoma cells. Oncol. Res.. 2023;31(5):631-643.
- [Google Scholar]
- Protein homeostasis in models of aging and age-related conformational disease. Adv. Exp. Med. Biol.. 2010;694:138-159.
- [Google Scholar]
- Pathways of cellular proteostasis in aging and disease. J. Cell Biol.. 2018;217(1):51-63.
- [Google Scholar]
- A review of the common neurodegenerative disorders: current therapeutic approaches and the potential role of nanotherapeutics. Int. J. Mol. Sci.. 2022;23(3)
- [Google Scholar]
- Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978;275(5679):416-420.
- [Google Scholar]
- Proteostasis and proteotoxicity in the network medicine era. Int. J. Mol. Sci.. 2020;21(17)
- [Google Scholar]
- Cytosolic selection systems to study protein stability. J. Bacteriol.. 2014;196(24):4333-4343.
- [Google Scholar]
- Correlation of levels of folded recombinant p53 in escherichia coli with thermodynamic stability in vitro. J. Mol. Biol.. 2007;372(1):268-276.
- [Google Scholar]
- Phleomycin resistance as a dominant selectable marker for plant cell transformation. Plant Mol. Biol.. 1989;13(4):365-373.
- [Google Scholar]
- Genetic selection designed to stabilize proteins uncovers a chaperone called Spy. Nat. Struct. Mol. Biol.. 2011;18(3):262-269.
- [Google Scholar]
- A new puffing pattern induced by temperature shock and DNP in drosophila. Experimentia.. 1962;18:571-573.
- [Google Scholar]
- Recombinant protein expression in Escherichia coli: advances and challenges. Front. Microbiol.. 2014;5:172.
- [Google Scholar]
- Protein misfolding in neurodegenerative diseases: implications and strategies. Transl. Neurodegener.. 2017;6:6.
- [Google Scholar]
- Protein folding in the cytoplasm and the heat shock response. Cold Spring Harb. Perspect. Biol.. 2010;2(12):a004390
- [Google Scholar]







