A Multi-Target mechanism of Withania somnifera bioactive compounds in autism spectrum disorder (ASD) Treatment: Network pharmacology, molecular docking, and molecular dynamics simulations studies
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Autism spectrum disorder (ASD) is a developmental disorder resulting from variations in brain structure and function. Individuals with ASD often experience challenges in communication and social interaction, along with engaging in repetitive or restricted patterns of behavior and interests. The lack of documented data and the wide range of pathophysiological processes associated with ASD make it challenging to work which causes a major financial burden on health care management. Withania somnifera, often referred to as ashwagandha is the tropical winter cherry in the Solanaceae family that can be used to treat several ailments such as asthma, stress, hypertension, diabetes, cancer, arthritic, and neural disorders including ASD. In this investigation, we examined the active compound-target-pathway network and discovered that Withanolide J, Withanone, and Withaferin A have a great role in the onset of ASD by influencing the IL6 gene. Later, the molecular docking method was applied for confirmation of the active compound's effective action against the prospective target. The molecular dynamics simulation exhibited that the complexes of Withanolide J and Withanone had stable intermolecular binding conformation and unveiled very stable dynamics during the simulation time. A combined network pharmacology and molecular docking approach demonstrated that W. somnifera exhibits a promising preventive impact on ASD by targeting relevant signaling pathways associated with the disorder. This establishes a foundation for comprehending the underlying mechanism of the anti-ASD activity of W. somnifera.
Keywords
Bioactive compounds
Autism spectrum disorder
Withania somnifera
Network pharmacology
Bioinformatics, molecular modeling
1 Introduction
Autism Spectrum Disorder (ASD) is a complicated and long-term neurodevelopmental disorder that manifests during childhood and is characterized by persistent deficits that include behavioral impairments, social difficulties, language communication challenges as well and repetitive and stereotypical actions (Bhandari et al., 2020). ASD includes anxiety, intellectual impairment, epilepsy, motor abnormalities, attention deficit hyperactivity disorder, difficulty in sleep, dysregulation of the immune system, and digestive issues (Sandler et al., 2001). It is a chronic and lifelong condition that is irreversible (Chaplin et al., 2018). The overall prevalence of autism is slightly below 1 %, although estimates in high-income countries tend to be higher (Lord et al., 2020). It has not yet been determined what causes ASD. On the other hand, pharmacological intervention, rehabilitation training, sensory integration, comprehensive education, and nutrition treatment are by far the most often employed therapeutic techniques (Linlin et al., 2022). The target treatments for this condition are in the early development stages, needing a longer disease course to gain therapeutic impact while growing evidence shows that traditional therapies can be effective (Feng et al., 2022).
Withania somnifera, widely referred to as “Ashwagandha” falls within the Solanaceae family and is a popular herb in Ayurvedic medicine. The roots of W. somnifera have powerful medicinal properties. It has been utilized as a natural remedy and nutritious diet to treat a variety of ailments and human maladies (Gurav et al., 2020). The pharmacological benefits of W. somnifera include immunostimulatory, hepatoprotective, antidepressant, anxiolytic, nootropic, antioxidant, anti-inflammatory, stress-reducing, anti-tumor, anticonvulsant, and genoprotective characteristics (Mishra et al., 2000). It is utilized in the treatment of a variety of illnesses which include cancer, diabetes, hypertension, stress, arthritic, and neurodegenerative disorders (Singh et al., 2010).
Because Chinese herbal medicine contains a wide range of components and therapeutic purposes, it requires efforts to adequately investigate its pharmacological action using established pharmacological approaches (Ma et al., 2015). Network pharmacology is a novel strategy for discovering new drugs or studying pharmacological processes based on a variety of fields including pharmacology and computer science (Hopkins, 2007). This strategy in contrast to the “single-component-single-target” method of drug development, employs a “multicomponent-multitarget” complicated network structure. Currently, numerous drugs work on multiple targets. This method allows a more comprehensive and extensive investigation of drug-disorder interactions at the system level (Li et al., 2021). Gai et al. have used the approach of network pharmacology along with molecular docking to treat ASD with the bioactive chemical Naringenin (Gai et al., 2022). Chen et al. explored the active components of Baohewan Heshiwei Wen Dan Tang in order to treat ASD using a network pharmacology-based method (Chen et al., 2022).
Using complicated network and visualization technologies, network pharmacology may illustrate numerous targets, multiple pathways, and the combined effects of Traditional Chinese Medicine (TCM) in the treatment of various illnesses, potentially providing novel concepts and efficient approaches for exploring the mechanism of TCM. Additionally, exploration across multiple databases could make it easier to understand mechanisms and repurpose drugs (Tanoli et al., 2021). The objective of this research is to provide the foundation for future research by identifying the useful elements, targets, and processes of W. somnifera in addressing ASD through the use of network pharmacology, molecular docking, and molecular dynamic simulations approaches.
2 Materials and methods
2.1 Identification and ADME evaluation of potential traditional Chinese medicine compounds
Information about the active compounds of W. somnifera was retrieved from IMPPAT (https://cb.imsc.res.in/imppat/) (Mohanraj et al., 2018), ChEBI (https://www.ebi.ac.uk/chebi/) (Hastings et al., 2016), KNApSAcK Core System database (http://www.knapsackfamily.com/KNApSAcK/) (Y. Nakamura et al., 2014); PCIDB (https://www.genome.jp/db/pcidb/), Dr. Duke's Phytochemical and Ethnobotanical Databases (https://phytochem.nal.usda.gov/phytochem/search) (Duke and Bogenschutz, 1994), and wide-scale review of the literature. The compounds were tested utilizing the ADME evaluation approach, with oral bioavailability (OB) and drug-likeness (DL) being the most essential parameters. For further investigation, bioactive components having an OB of ≥ 30 % and DL ≥ 0.18 were chosen (Xu et al., 2012); (Sadaqat et al., 2023). PubChem (https://pubchem.ncbi.nlm.nih.gov/) (Kim et al., 2016) along with ChemSpider (http://www.chemspider.com/) (Pence & Williams, 2010) were also utilized for information collection on molecular weight and chemical ID (Kaur et al., 2023).
2.2 Prediction and screening of target genes associated with compounds and disease
The STITCH (http://stitch.embl.de/) (Kuhn et al., 2007) and Swiss Target Prediction (http://www.swisstargetprediction.ch/) (Daina et al., 2019) databases were utilized to determine the targets associated with each active chemical. Both databases used “Homo sapiens” as a reference organism. STITCH and Swiss Target Prediction databases were employed to choose targets having a combined score/probability score of ≥ 0.7 (Fatima et al., 2024). A compound-target network was constructed in Cytoscape using these compounds and corresponding targets as input.
The keyword “autism spectrum disorder” was searched to retrieve disease-related targets through the GeneCards database (https://www.genecards.org/) (Safran et al., 2010). Subsequently, a Venn diagram was created using the online tool Evenn (http://www.ehbio.com/test/venn/#/) (Chen et al., 2021) to illustrate the overlapping targets between compound and disease, facilitating the identification of common targets. The resultant common targets were assumed as key targets and processed for further analysis.
2.3 GO enrichment and KEGG Pathway analysis
The DAVID (https://david.ncifcrf.gov/) database was used for gene ontology (GO) analysis that comprised three categories (Dennis et al., 2003). These categories are biological process (BP), cellular component (CC), and molecular function (MF) (Fatima et al., 2024). By lowering the likelihood score to less than 0.05, the top ten GO terms and KEGG pathways associated with ASD were chosen for more in-depth analysis. A bubble chart was constructed using the online program SRplot (https://www.bioinformatics.com.cn/en) to analyze the related functional pathways of the targeted genes.
2.4 Protein-Protein interaction (PPI) analysis
The STRING database (https://string-db.org/) was utilized for the PPI analysis and “Homo sapiens” was taken as the reference (Mering et al., 2003). The output network in TSV format was downloaded and a parameter of combined score ≥ 0.5 was employed (Alshehri & Alshehri, 2023). The network was then loaded into Cytoscape v.3.9.1 where the CytoHubba plugin (Chin et al., 2014) was used. CytoHubba's MCC, MNC, degree, betweenness, and closeness scoring algorithms were utilized to predict hub genes. Common genes from the above algorithms were considered as main genes and processed for further analysis.
2.5 Network construction
To comprehend the mechanism of W. somnifera in ASD, network analysis was carried out. The network diagram was created and displayed using the Cytoscape 3.9.1 program (Kohl et al., 2011). The network's nodes represented bioactive components and target genes, on the other hand edges were used to show the association between the compounds as well as targets. The Cytoscape Network Analyzer (Saito et al., 2012) was then utilized for the calculation of the degree, a topological measure that shows the relevance of a component/target/pathway in the network.
2.6 Analysis of molecular docking
The interaction of the hub gene with bioactive substances was predicted using molecular docking (A. Fatima, Arora, et al., 2023); (A. Fatima, Khanum, et al., 2023). The PDB database (https://www.rcsb.org/) was employed to retrieve and download the 3D structure of the hub gene in PDB format (Kouranov et al., 2006). The PubChem database (https://pubchem.ncbi.nlm.nih.gov/) was utilized to collect the 3D structure of bioactive chemicals in SDF format (Kim et al., 2019); (Jindal et al., 2023). The SDF format was then transferred to the PDBQT format through Open Babel software (O’Boyle et al., 2011). To remove the water molecules and natural ligand groups from the protein structure UCSF Chimera (Pettersen et al., 2004) was employed. The CASTp tool (https://sts.bioe.uic.edu/castp/) was used to forecast binding pockets (Tian et al., 2018). Additional molecular docking investigations were performed via the PyRx software interface using AutoDock Vina (Trott & Olson, 2010). In PyRx the docking grid for IL6 was set at coordinates X: 12.1066, Y: –32.4441, and Z: 0.4771 with dimensions X: 21.1750, Y: 22.1155, and Z: 18.1749 Å from the center, and an exhaustiveness level set to 8. The best-docked complex was chosen with the best postures, higher binding energy, and lowest root mean square deviation (RMSD). The interactions between the top three docking complexes with the higher binding energy were displayed in 2D by utilizing Discovery Studio Visualizer (Headquarters, 2008) and in 3D using UCSF ChimeraX (Pettersen et al., 2021).
2.7 Analysis of molecular dynamic simulation
The AMBER22 program was used to carry out a molecular dynamic simulation (Harris et al., 2022); (Ahmad, 2023). A molecular dynamic simulation (MDs) is an in-silico simulation technique mostly utilized for macromolecule and docked atom analysis (Wani et al., 2021). A macromolecule that is allowed to show dynamic behavior for a specific amount of time in a molecular dynamic simulation pipeline and the atom and molecular trajectories are originated by solving Newton's equations of motion (Gupta, 1992). The AMBER software was used in this research to determine the dynamic behavior of the compounds in complex with the receptor (Mortier et al., 2015). The purpose of this was to read how the compound affinity for the receptor enzyme with time. The complexes were processed via the Antechamber program. In the case of receptor processing the FF19Sb force field was applied while for ligand processing the GAFF2 force field was used. An appropriate quantity of counter ions was introduced into the system to achieve charge neutrality (Guterres et al., 2022). A cubic box size of eight angstroms was supposed to be enough to solvate the complexes and the steepest descent and conjugate gradients algorithms were applied (Nicholls & Honig, 1991). The heating of the complexes was achieved gradually till 310 K followed by system equilibration and a production run of 100 ns. The trajectory analyses were performed using CCPTRAJ (Ahmad et al., 2020).
2.8 MMPB/GBSA analysis
The binding free energies of docked ligands with the enzyme were calculated using MMPB/GBSA analysis (Zaman et al., 2021). This was achieved using a script called MMPBSA.py which is part of the AMBER v22 program (Miller et al., 2012). The script assumed 5000 frames from the trajectories selected at regular intervals. The formula for MMPB/GBSA energy:
The terms in the equation are ΔGligand for free ligand energy, ΔGreceptor for the free energy of the receptor protein, ΔGcomplex for complex free energy, and ΔGbind for total binding free energy. The following formula will be used to find the unique free energies of a complex, protein, and ligand. It is found that the performance of the MM/PBSA and MM/GBSA approaches is similar. The MM/PBSA uses the Poisson-Boltzmann equation to determine the electrostatic energy contribution to the free energy, while the MM/GBSA uses the Generalized Born equation which is believed to be a faster solution for the earlier equation.
3 Results
3.1 Identification and selection of bioactive compounds of w. Somnifera
Collectively 242 bioactive constituents of W. somnifera have been collected and retrieved from various databases and a review of the literature (Supplementary Table S1). After the screening and removal of duplicates, 13 putative compounds ((+)-Catechin, 24-Methyldesmosterol, Beta-Sitosterol, Campesterol, Fucosterol, Kaempferol, Oleanolic acid, Quercetin, Stigmasterol, Stigmasterone, Withaferin A, Withanolide J, and Withanone) which retained the criteria of DL ≥ 0.18 and OB ≥ 30 % were selected. The details of compounds, their PubChem ID (CID), molecular formula (MF), molecular weight (MW), OB, DL, and 2D structure were enlisted in Table 1.
| Compound Name | MF | MW | DL | OB | 2D Structure | CID |
|---|---|---|---|---|---|---|
| (+)-Catechin | C15H14O6 | 290.27 | 0.64 | 0.55 |
|
9064 |
| 24-Methyldesmosterol | C28H46O | 398.7 | 0.76 | 0.55 |
|
193,567 |
| Beta-Sitosterol | C29H50O | 414.7 | 0.78 | 0.55 |
|
222,284 |
| Campesterol | C28H48O | 400.7 | 0.59 | 0.55 |
|
173,183 |
| Fucosterol | C29H48O | 412.7 | 0.85 | 0.55 |
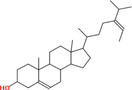
|
5,281,328 |
| Kaempferol | C15H10O6 | 286.24 | 0.5 | 0.55 |
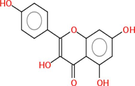
|
5,280,863 |
| Oleanolic acid | C30H48O3 | 456.7 | 0.37 | 0.85 |
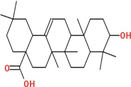
|
10,494 |
| Quercetin | C15H10O7 | 302.23 | 0.52 | 0.55 |
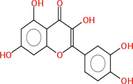
|
5,280,343 |
| Stigmasterol | C29H48O | 412.7 | 0.62 | 0.55 |
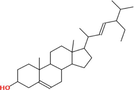
|
5,280,794 |
| Stigmasterone | C29H46O | 410.7 | 0.5 | 0.55 |
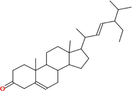
|
14,807,783 |
| Withaferin A | C28H38O6 | 470.6 | 0.37 | 0.55 |
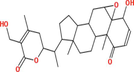
|
265,237 |
| Withanolide J | C28H38O6 | 470.6 | 0.46 | 0.55 |
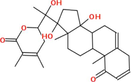
|
21,679,022 |
| Withanone | C28H38O6 | 470.6 | 0.45 | 0.55 |
|
21,679,027 |
3.2 Identification and screening of potential targets
From 13 active compounds, a total of 134 putative potential targets were retrieved through STITCH and Swiss Target Prediction databases (Fig. 1A; Supplementary Table S2). On the other hand, a total of 8172 targets linked with autism spectrum disorder were obtained through the GeneCards database (Supplementary Table S3). A Venn diagram was generated to determine the shared targets from compound-related along ASD-related targets (Fig. 1B). As a result, 70 potential anti-ASD genes from W. somnifera were chosen and regarded as key targets (Supplementary Table S4).
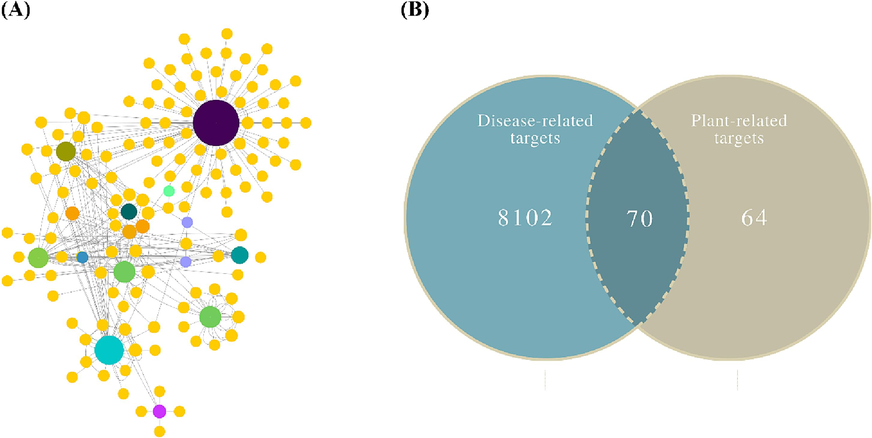
-
(A) Compound-target network, (B) A Venn diagram drawn between compound and disease-related targets to find common target genes.
3.3 GO and KEGG signaling Pathway analyses
To comprehend the possible biological function of W. somnifera targets, gene ontology (GO) at three biological levels: biological processes (BP), cellular components (CC), and molecular function (MF) have been done and a total of 234 BP, 34 CC, 68 MF, and 106 KEGG pathways terms were significantly found to be enriched. Additionally, the analysis of KEGG pathways was also carried out. According to BP, most of the genes were involved in the apoptotic process of negative regulation, peptidyl-serine phosphorylation, xenobiotic stimulus–response, and protein autophosphorylation (Fig. 2A). CC indicates that these genes are predominantly present in cytosol, membrane raft, extracellular exosome, and cell membrane (Fig. 2B). MF revealed that these genes are associated with the binding of ATP, DNA, and protein, protein serine/threonine kinase activity, and protein tyrosine kinase activity (Fig. 2C). Furthermore, KEGG pathways analysis pointed toward significant involvement in HIF-1 signaling, MAPK signaling as well and TNF signaling pathways (Fig. 2D).
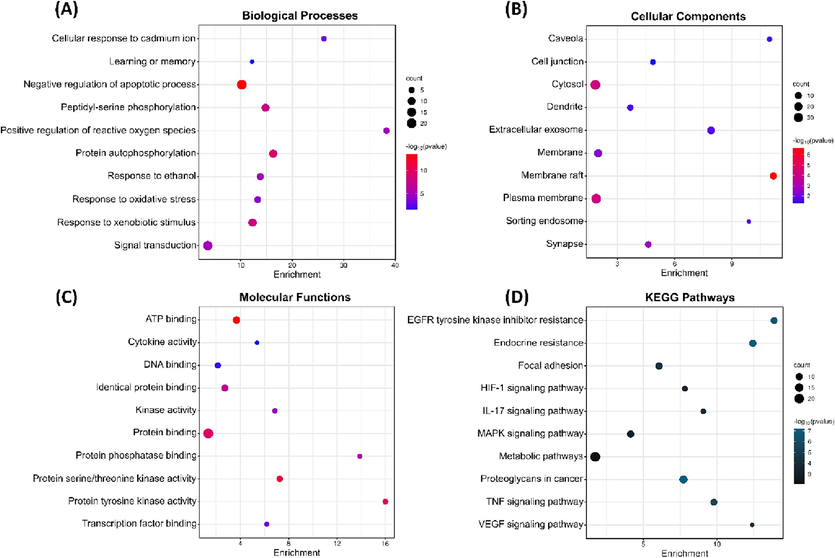
- GO functional enrichment and KEGG pathways analysis, (A) Biological processes, (B) Cellular components, (C) Molecular functions, and (D) KEGG pathways.
3.4 Protein-Protein interaction (PPI)
Interactions of proteins with other proteins were determined through the STING database. MCC, MNC, degree, betweenness, and closeness centralities were used. A Venn diagram was generated to predict common genes from all these five methods (Supplementary Table S5; Fig. 3A). As a result, 6 common genes were analyzed. These are IL6, CASP3, AKT1, TNF, SRC, and EGFR (Fig. 3B). In comparison with KEGG pathways one target gene IL6 was utilized for the analysis of molecular docking.
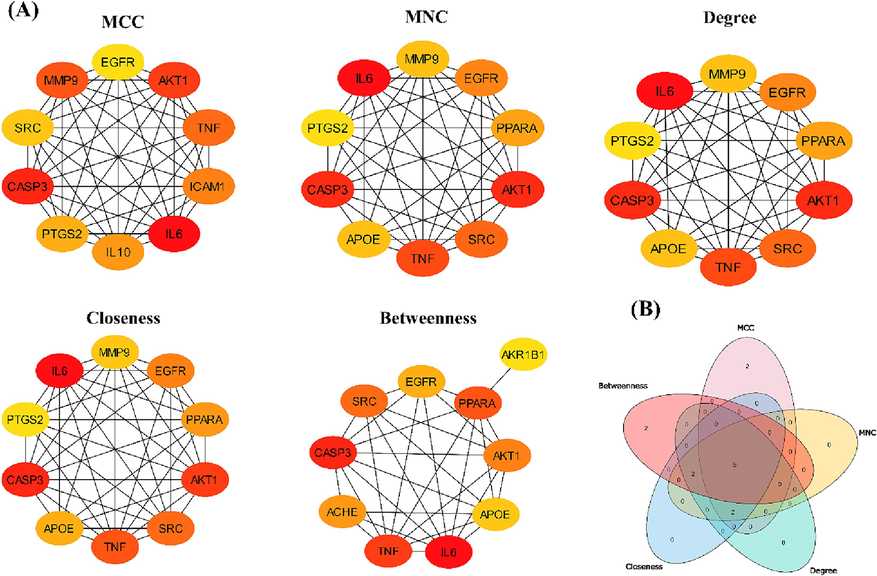
-
(A) Calculation of hub genes by MCC, MNC, Degree, Closeness, and Betweenness scoring algorithms of CytoHubba, (B) Venn diagram showing the common genes of 5 different scoring algorithms.
3.5 Compound-target-pathways network
A network of compound-target pathways was developed with the help of Cytoscape v.3.9.1. Active compounds, hub genes, and pathways were represented by nodes. These nodes are connected through edges (Fig. 4). There were 25 nodes and 58 edges in the network. QSPR models utilizing graph theoretical approaches are vital for predicting molecular properties based on topological indices, offering insights into structure–property relationships efficiently and comprehensively.
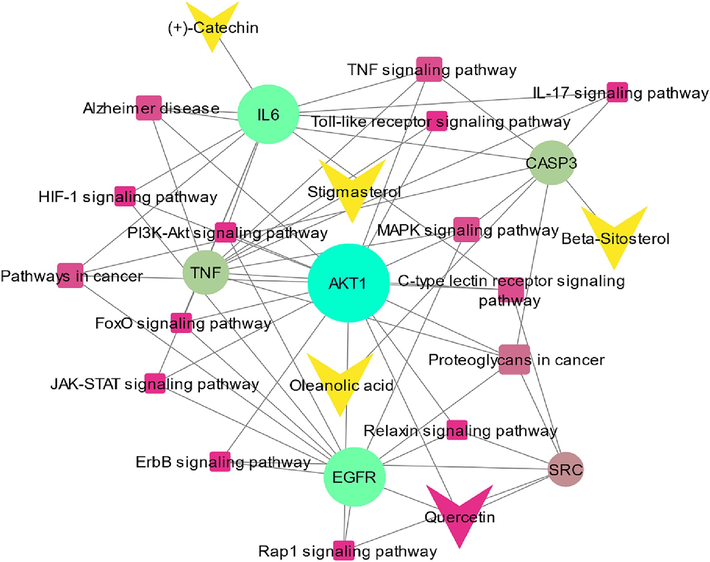
- A network of compound-target-pathways. Arrow symbols represent active constituents, circle symbols represent hub genes, and square symbols represent the pathways related to hub genes.
According to the network ASD is mediated by MAPK, PI3K-Akt, TNF, ErbB, C-type lectin receptor, FoxO, Relaxin, Rap1, IL-17, Toll-like receptor, HIF-1, and JAK-STAT signaling pathways (Table 2).
| Rank | Name | Compounds | Score | Pathways |
|---|---|---|---|---|
| 1 | IL6 | (+)-Catechin | 32 | Pathways in cancer, Alzheimer's disease, Pathway of IL-17 Signaling |
| 2 | CASP3 | Beta-Sitosterol / Oleanolic acid | 29 | MAPK signaling pathway, Proteoglycans in cancer |
| 3 | AKT1 | Quercetin | 29 | Pathway of JAK-STAT signaling, HIF-1 signaling pathway, Pathway of Rap1 signaling, and ErbB signaling |
| 4 | TNF | Stigmasterol | 28 | Pathways of TNF signaling, Toll-like receptor signaling, and C-type lectin receptor signaling |
| 5 | SRC | Quercetin | 27 | Relaxin signaling, Rap1 signaling, and C-type lectin receptor signaling pathways |
| 6 | EGFR | Quercetin | 24 | Pathway of MAPK signaling, Relaxin signaling, and FoxO signaling |
3.6 Molecular docking
All 13 active constituents were utilized for molecular docking analysis in which some compounds were plant-specific compounds (Withaferin A, Withanolide J, and Withanone). Based on PPI and KEGG pathways analysis one hub gene named IL6 was employed for the analysis of molecular docking. Protein structure (IL6 (PDB ID: 1ALU)) was gained through the Protein Data Bank (PDB). UCSF Chimera v1.16 was used to improve structure, remove non-standard amino acids, and minimize energy use. The possible targets of substances that might reduce the occurrence of ASD were screened out utilizing molecular docking. The binding energy and docking scores were employed as major criteria for the compound selection (Table 3). Withanolide J was stably bound with 1ALU protein with a binding affinity of −6.9 kcal/mol and RMSD of 1.03 Å. Withanolide J side chains formed hydrogen bonds with LEU A:92, VAL A:96, LYS A:120, PRO A:141, ALA A:145, and LEU A:148. Withanone and Withaferin A have the highest binding affinity −6.8 kcal/mol and −6 kcal/mol with RMSD of 0.157 Å and 0.536 Å respectively (Fig. 5). Withanone and Withaferin A made strong bonding with LYS A:120, ILE A:123, PRO A:141, ASN A:144 and GLN A:127, PRO A:141, ASN A:144 residues. All active compounds show strong hydrogen bonding with PRO A:141 except Oleanolic acid, Stigmasterone, and Quercetin. These results suggest that the IL6 target protein is efficiently bound by active components of W. somnifera, which then interact as ASD repressors.
| Complex | Binding Affinity (kcal/mol) | RMSD | Interacting Residues |
|---|---|---|---|
| IL6_Withanolide J | -6.9 | 1.03 | LEU A:92, VAL A:96, LYS A:120, PRO A:141, ALA A:145, LEU A:148 |
| IL6_Withanone | -6.8 | 0.157 | LYS A:120, ILE A:123, PRO A:141, ASN A:144 |
| IL6_Withaferin A | -6 | 0.536 | GLN A:127, PRO A:141, ASN A:144 |
| IL6_Fucosterol | -6 | 1.13 | LYS A:120, ILE A:123, PRO A:139, PRO A:141, ALA A:145 |
| IL6_Stigmasterol | -6 | 1.667 | LEU A:92, GLU A: 99, ILE A:123, PRO A:139, PRO A:141, ALA A:145 |
| IL6_(+)-Catechin | -5.9 | 2.237 | GLU A:95, GLU A:99, LYS A:120, THR A:138, PRO A:141 |
| IL6_Oleanolic acid | -5.9 | 2.407 | LEU A:92, GLU A:95, LYS A:120, ILE A:123, LYS A:128 |
| IL6_Stigmasterone | -5.9 | 2.423 | LEU A:92, LYS A:120, ILE A:123, ASN A:144 |
| IL6_Kaempferol | -5.6 | 0.895 | GLU A:95, GLN A:116. LYS A:120, PRO A:139, PRO A:141 |
| IL6_Quercetin | -5.6 | 1.282 | ASN A:63, GLU A:93, VAL A:96, ASN A:144 |
| IL6_Campesterol | -5.3 | 0.961 | LEU A:92, LYS A:120, ILE A:123,PRO A:139, PRO A:141 |
| IL6_24-Methyldesmosterol | -5.1 | 0.68 | LYS A:120, ILE A:123, PRO A:141 |
| IL6_Beta-Sitosterol | -4.8 | 0.675 | LYS A:120, ILE A:123, PRO A:139, PRO A:141 |
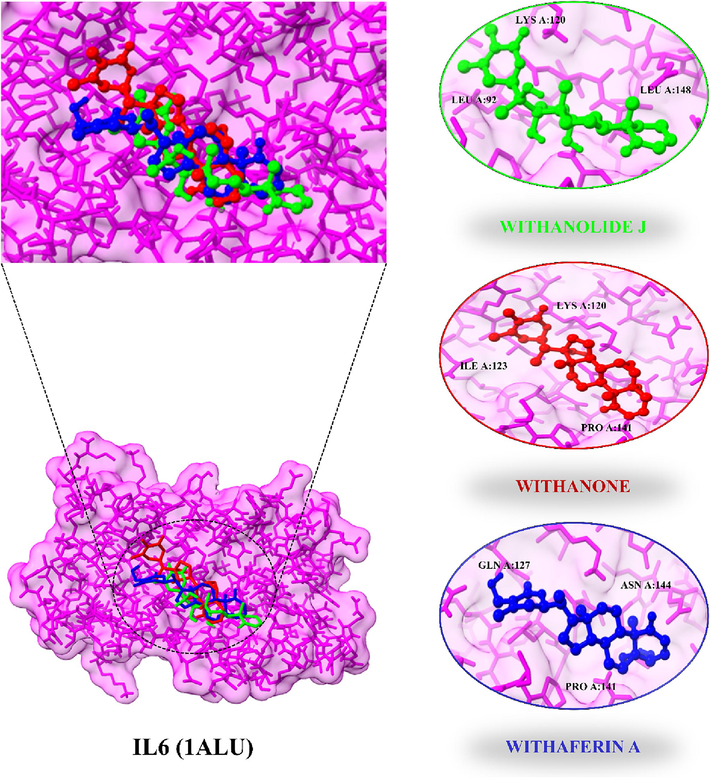
- IL6 docking complexes with their three highest binding affinity compounds.
3.7 Molecular dynamic simulation
Essentially, the dynamic behavior of macromolecules is confirmed by a molecular dynamic simulation study. The simulations analysis includes root mean square deviation (RMSD) (Sargsyan et al., 2017), radius of gyration (RoG) (Lobanov et al., 2008), and root mean square fluctuation (RMSF) (Pitera, 2014). The foundation for all of these studies was the complexes' carbon alpha atom. The purpose of these investigations was to ascertain whether the ligand binding to the receptors was stable and whether the interactions continued throughout the simulation time. Stable receptor-ligand contact will guarantee that the ligand is correctly presented to the IL6 target protein. The RMSD plot of the systems was uniform at first indicating no discernible structural changes. In Fig. 6A, the mean values of IL6_Withanolide J, and IL6_Withanone were found to be (2.00 Å) and (2.21 Å) respectively, whereas the highest values of the systems' root mean square deviation (RMSD) ranged up to 3 Å. Second, the RMSF was calculated to reveal information on the flexibility of the receptor residues in the presence of the ligand molecule (Fig. 6B). Nearly all system residues were within the region of average stability (<5 Å). The RMSF of the systems for IL6_Withanolide J, and IL6_Withanone showed maximum values of (3.69 Å and 7.25 Å), mean values of (1.01 Å and 1.06 Å), and minimum values of (0.45 Å and 0.44 Å). The higher degree of flexibility seen in some of the residues was shown to be a result of the loop pressure inside the system, the compactness of the system was analyzed against time using the RoG analysis. The way ligands attached to the receptors was unaffected by these variations, though. With IL6_Withanolide J and IL6_Withanone having RoG maximum values of (16.40 Å and 16.85 Å), mean values of (16.11 Å, and 16.34 Å), and minimum values of (15.86 Å and 15.88 Å) (Fig. 6C). RMSD demonstrated that all the systems were compact and did not experience any notable alterations after the simulation. The Beta Factor study yielded the following results: IL6_Withanolide J and IL6_Withanone had mean values of (39.77 Å and 50.09 Å), lowest values of (5.46 Å and 5.13 Å), and maximum values of (358.89 Å and 1385.31 Å) (Fig. 6D).
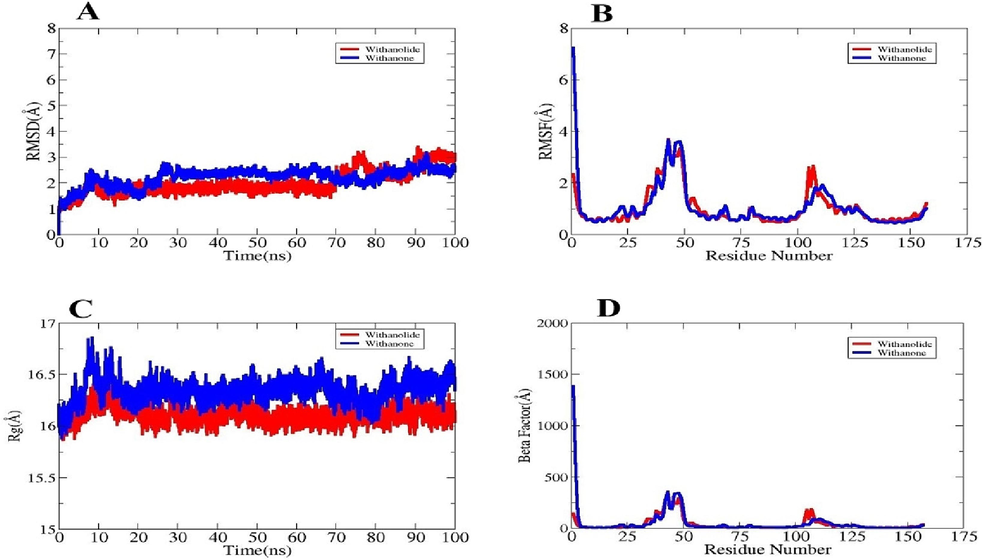
- Different simulation based statistical assays. (A) RMSD, (B) RMSF, (C) RoG, and (D) Beta Factor plots.
To learn more about the surface area of the IL6 that interacts with the solvent molecules, solvent-accessible accessible surface area (area) analysis was performed for the ligands. IL6_Withanolide J (9526.49 nm2), and IL6_Withanone (9776.63 nm2) are the average values for the systems. According to (Fig. 7), the lowest values of SASA for IL6_Withanolide J, and IL6_Withanone were (8364.2 nm2 and 8891.45 nm2). The maximum values were (10730.5 nm2 and 10795.4 nm2), respectively. Plots show the significant variations observed upon ligand binding.
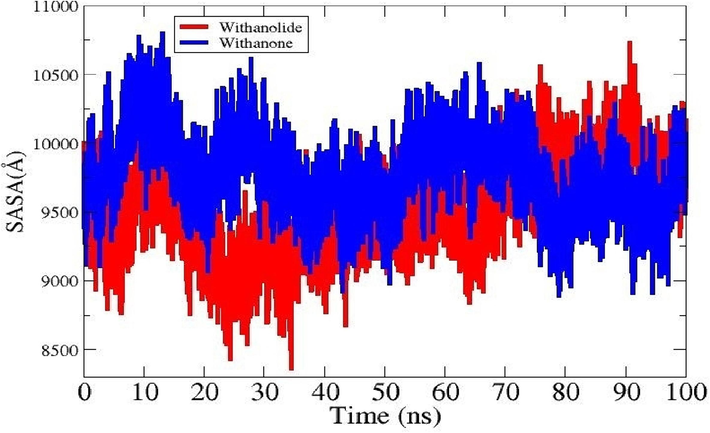
- showing SASA plot for IL6_Withanolide and IL6_Withanone.
3.8 MMPB/GBSA analysis
The MMPB/GBSA analysis was completed for the selected docked complexes. These methods are believed to be more effective in determining the docked ligand binding affinity with the receptor protein. The very negative net binding energies of all docked and control complexes demonstrated the formation of strong intermolecular systems and stable complexes. It was discovered that the van der Waals force, which keeps ligand docking at the docked site and stabilizes systems, is the most potent force produced by the complexes. The net van der Waals energies of the IL6_Withanolide J, and IL6_Withanone were −36.15 kcal/mol and −13.64 kcal/mol, respectively. Furthermore, it was discovered that the electrostatic energy of every docked complex was incredibly constant. The solvation energy was the least favorable of the calculated energies and contributed negatively to the net energy. In MM-GBSA, the net solvation energy of the IL6_Withanolide J, and IL6_Withanone was 8.20 kcal/mol and 7.68 kcal/mol, respectively. In contrast, in MM/PBSA, the net solvation energies of the IL6_Withanolide J, and IL6_Withanone were 7.54 kcal/mol, and 8.01 kcal/mol, respectively. Additional details on the energy terms and values are provided in Table 4.
| Parameter | IL6_Withanolide J | IL6_Withanone |
|---|---|---|
| MM/GBSA | ||
| Energy van der Waals | −36.15 | −48.21 |
| Energy Electrostatic | −12.01 | −13.64 |
| Total Gas Phase Energy | −48.16 | −61.85 |
| Total Solvation Energy | 8.20 | 7.68 |
| Net Energy | −39.96 | −54.17 |
| MM/PBSA | ||
| Energy van der Waals | −36.15 | −48.21 |
| Energy Electrostatic | −12.01 | −13.64 |
| Total Gas Phase Energy | −48.16 | −61.85 |
| Total Solvation Energy | 7.54 | 8.01 |
| Net Energy | −40.62 | −53.84 |
4 Discussion
In recent times, there has been an increasing interest in analyzing the genetic as well as cellular foundations of ASD. There is a lot of evidence to back up the assumption that immunological, environmental, and genetic variables all play a role in its development. A notable risk factor for the disease is strong genetic variability, which includes copy number variation, single nucleotide polymorphism, exon deletions, aberrant methylation of genes, and disturbance of messenger ribonucleic acid (mRNA) transcription or protein exon deletions (Manoli & State, 2021). ASD is now being treated by altering neurotrophic factor levels, cell synapse levels, gene expression, oxidative stress responses, and protein homeostasis (Basilico et al., 2020).
W. somnifera has previously been found as an immunostimulant (Muralikrishnan et al., 2010). In addition it is utilized to treat a variety of infectious and non-infectious disorders. The herb is known to boost mitochondrial and endothelial function while also lowering oxidative stress and inflammation. Due to these activities, W. somnifera holds the potential to serve as a therapeutic alternative for various disorders. W. somnifera plant extracts have no recorded severe toxicity or negative effects, making them suitable for use in humans (Dar, 2020).
In this investigation, a comprehensive set of 70 mutual targets of W. somnifera to treat ASD was identified from IMPPAT, KNApSAcK, GeneCards, and various relevant databases, and 6 important targets were discovered after filtering and screening. IL6 exhibited a strong binding affinity to Withanolide J, Withanone, and Withaferin A, according to molecular docking studies. Autism-like behaviors can be modulated by IL6 elevation by impairing synapse formation, development of dendritic spine development, and neural circuit balance (Xu et al., 2015). Elevated levels of IL6 have been found in the cerebellum of individuals with ASD and it has been associated with repetitive and restricted behaviors in children with ASD (Hughes et al., 2022). Additionally, research has indicated that IL6 can alter neural cell adhesion, migration, and synaptic formation, potentially contributing to the onset of autism (Wei et al., 2011).
Analysis of GO enrichment was carried out to gain biological information about target genes. GO functional study revealed that anti-ASD targets of W. somnifera were involved in apoptotic process negative regulation, response to xenobiotic stimulus, protein autophosphorylation, peptidyl-serine phosphorylation, protein serine/threonine kinase activity, binding of protein, and protein tyrosine kinase activity. The findings of the KEGG pathways indicated that the candidate targets were considerably enriched in several pathways including the MAPK, PI3K-Akt, TNF, ErbB, C-type lectin receptor, Relaxin, pathways of FoxO signaling, Rap1, IL-17, HIF-1, Toll-like receptor, and JAK-STAT signaling pathways.
Animal models of ASD have highlighted the significant role of IL6 in the pathophysiology of ASD. Elevated levels of IL6 have been associated with a pro-inflammatory state in ASD, indicating its involvement in immune dysregulation and neuroinflammation (Kutuk et al., 2020). Studies show that systemic maternal inflammation leading to increased IL6 levels, can promote abnormal behaviors in mice, emphasizing the impact of IL6 in ASD (Majerczyk et al., 2022). Moreover, inhibition of IL6 trans-signaling in the brain has been linked to increased sociability in the BTBR mouse model of autism, suggesting a potential therapeutic target for ASD (Wei et al., 2016). These findings underscore the complex interplay between immune factors like IL6 and behavioral manifestations in ASD animal models, providing valuable insights for potential therapeutic interventions.
As per the topological features of the compound-target-pathway network, IL6 was determined as the primary target. Furthermore, molecular docking was used to validate the IL6 key target, which demonstrated that Withanolide J, Withanone, and Withaferin A bonded securely to the core target. Due to their capacity to interact securely with core targets, the docking study results showed that these compounds may be employed to treat ASD. Further in the molecular dynamic simulation study, the stability of the IL6 receptor-ligand complexes was rigorously examined through RMSD, RMSF, RoG, and Beta Factor analyses. The RMSD plots revealed consistent structural stability throughout the simulation, with mean values for IL6_Withanolide J and IL6_Withanone indicating stable ligand binding. The RMSF analysis illustrated the flexibility of receptor residues, mostly within an average stability range, influenced by internal loop pressure. RoG analysis confirmed the unaffected attachment of ligands to receptors despite system compactness variations. The Beta Factor study indicated notable stability, particularly with IL6_Withanolide J.
Solvent-accessible surface area (SASA) analysis unveiled variations in IL6 interactions with solvent molecules upon ligand binding. MMPB/GBSA analysis showcased strong intermolecular systems, dominated by van der Waals forces. The net binding energies, notably negative, affirmed stable complexes, with van der Waals forces being the most influential. Solvation energy, the least favorable, contributed negatively to the net energy. MM/GBSA and MM/PBSA results consistently demonstrated the effectiveness of IL6_Withanolide J in forming stable complexes with the IL6 receptor, showcasing the promising potential for further exploration in drug design. It also establishes a foundation for future investigations to explore the mechanism behind the fundamental therapeutic effects of diverse Chinese herbal medicines. Future research is required to identify safe drugs investigate the way they work, and ultimately enhance the standard of living for families impacted by ASD because of the variety of phenotypes of ASD and the molecular complexity in pathways.
5 Conclusion
This research identifies new therapeutic targets for ASD and lays a scientific basis for the drug regimen efficacy with several components and targets. By employing network pharmacology along with molecular docking, the study revealed the molecular mechanism of W. somnifera to treat ASD. According to network analysis, W. somnifera contains compounds that target multiple pathways associated with ASD simultaneously. Additionally, the molecular docking results suggest that IL6 is a viable and effective target for preventing and reducing ASD, potentially enhancing its efficacy. The comprehensive molecular dynamic simulation and MMPB/GBSA analyses offered valuable insights into the stability and binding affinity of IL6 receptor-ligand complexes. The consistently negative net binding energies, particularly driven by potent van der Waals forces, highlight the robust nature of the interactions, emphasizing the potential therapeutic relevance of IL6_Withanolide J in targeting the IL6 receptor for drug development. Nevertheless, the study is constrained by certain limitations, and more phytochemical as well as pharmacological research is necessary to substantiate these findings.
Acknowledgment
The authors extend their appreciation to the King Salman Center for Disability Research for funding this work through Research Group no KSRG-2023-553.
Declaration of competing interest
The author declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- DFT, molecular docking, molecular dynamics simulation, and hirshfeld surface analysis of 2-phenylthioaniline. Polycycl. Aromat. Compd. 2023:1-23.
- [Google Scholar]
- Molecular dynamics simulation of zika virus NS5 RNA dependent RNA polymerase with selected novel non-nucleoside inhibitors. J. Mol. Struct.. 2020;1203:127428
- [Google Scholar]
- Network pharmacology-based screening of active constituents of Avicennia marina and their clinical biochemistry related mechanism against breast cancer. J. Biomol. Struct. Dyn. 2023:1-16.
- [Google Scholar]
- Molecular mechanisms for targeted ASD treatments. Curr. Opin. Genet. Dev.. 2020;65:126-137.
- [Google Scholar]
- Neuropsychopathology of autism spectrum disorder: complex interplay of genetic, epigenetic, and environmental factors. Pers. Food Interv. Ther. Autism Spectr. Disord. Manag. 2020:97-141.
- [Google Scholar]
- Noncontiguous finished genome sequence of Megasphaera sp. ASD88, isolated from faeces of a child with autism spectrum disorder. New Microbes New Infect.. 2018;22:13-16.
- [Google Scholar]
- Network Pharmacology integrated molecular docking to reveal the autism and mechanism of baohewan heshiwei wen dan tang. Curr. Pharm. Des.. 2022;28(39):3231-3241.
- [Google Scholar]
- EVenn: Easy to create repeatable and editable Venn diagrams and Venn networks online. J. Genet. Genomics= Yi Chuan Xue Bao. 2021;48(9):863-866.
- [Google Scholar]
- cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol.. 2014;8(4):1-7.
- [Google Scholar]
- SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res.. 2019;47(W1):W357-W364.
- [Google Scholar]
- Neurodegenerative diseases and Withania somnifera (L.): An update. J. Ethnopharmacol.. 2020;256:112769
- [Google Scholar]
- DAVID: database for annotation, visualization, and integrated discovery. Genome Biol.. 2003;4(9):1-11.
- [Google Scholar]
- J. Duke and M. J. Bogenschutz, Dr. Duke’s phytochemical and ethnobotanical databases. USDA, Agricultural Research Service Washington, DC, 1994.
- Experimental spectroscopic, computational, hirshfeld surface, molecular docking investigations on 1H-Indole-3-Carbaldehyde. Polycycl. Aromat. Compd.. 2023;43(2):1263-1287.
- [Google Scholar]
- DFT, molecular docking, molecular dynamics simulation, MMGBSA calculation and hirshfeld surface analysis of 5-sulfosalicylic acid. J. Mol. Struct.. 2023;1273:134242
- [Google Scholar]
- South African Journal of Botany Advanced network pharmacology and molecular docking-based mechanism study to explore the multi-target pharmacological mechanism of Cymbopogon citratus against Alzheimer ’ s disease. South African J. Bot.. 2024;165:466-477.
- [CrossRef] [Google Scholar]
- “Traditional Chinese medicine intervention for autism spectrum disorders: Retraction: A protocol for systematic review and network meta-analysis,”. Medicine (baltimore).. 2022;101(9)
- [Google Scholar]
- J. Gai et al., “Exploration of potential targets and mechanisms of Naringenin in treating autism spectrum disorder via network pharmacology and molecular docking,” Medicine (Baltimore)., vol. 101(46), 2022.
- Computing aspects of molecular dynamics simulation. Comput. Phys. Commun.. 1992;70(2):243-270.
- [Google Scholar]
- Studies on Ashwagandha Ghrita with reference to murcchana process and storage conditions. J. Ayurveda Integr. Med.. 2020;11(3):243-249.
- [Google Scholar]
- CHARMM-GUI high-throughput simulator for efficient evaluation of protein–ligand interactions with different force fields. Protein Sci.. 2022;vol. 31(9):e4413.
- [Google Scholar]
- GPU-accelerated all-atom particle-mesh Ewald continuous constant pH molecular dynamics in Amber. J. Chem. Theory Comput.. 2022;18(12):7510-7527.
- [Google Scholar]
- ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res.. 2016;44(D1):D1214-D1219.
- [Google Scholar]
- Discovery Studio Life Science Modeling and Simulations. Researchgate. Net 2008:1-8.
- [Google Scholar]
- Increased Monocyte Production of IL-6 after Toll-like Receptor Activation in Children with Autism Spectrum Disorder (ASD) Is Associated with Repetitive and Restricted Behaviors. Brain Sci.. 2022;vol. 12(2):220.
- [Google Scholar]
- Copper (II) Monomer Bearing Phenolate-Based Ligand: Theoretical and Experimental Visions. Polycycl. Aromat. Compd.. 2023;43(4):3489-3501.
- [Google Scholar]
- Dimeric ZnII complex of carboxylate-appended (2-pyridyl) alkylamine ligand and exploration of experimental, theoretical, molecular docking and electronic excitation studies of ligand. J. Mol. Struct.. 2023;1276:134715
- [Google Scholar]
- PubChem substance and compound databases. Nucleic Acids Res.. 2016;44(D1):D1202-D1213.
- [Google Scholar]
- PubChem 2019 update: improved access to chemical data. Nucleic Acids Res.. 2019;47(D1):D1102-D1109.
- [Google Scholar]
- “Cytoscape: software for visualization and analysis of biological networks”. In: Data Mining in Proteomics. Springer; 2011. p. :291-303.
- [Google Scholar]
- The RCSB PDB information portal for structural genomics. Nucleic Acids Res.. 2006;vol. 34(suppl_1):D302-D305.
- [Google Scholar]
- STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res.. 2007;vol. 36(suppl_1):D684-D688.
- [Google Scholar]
- Cytokine expression profiles in Autism spectrum disorder: A multi-center study from Turkey. Cytokine. 2020;133:155152
- [Google Scholar]
- Systematic analysis of the molecular mechanisms mediated by coffee in Parkinson’s disease based on network pharmacology approach. J. Funct. Foods. 2021;87:104764
- [Google Scholar]
- A multi-target and multi-channel mechanism of action for Jiawei Yinhuo Tang in the treatment of social communication disorders in autism: network pharmacology and molecular docking studies. Evidence-Based Complement. Altern. Med.. 2022;vol:2022.
- [Google Scholar]
- Radius of gyration as an indicator of protein structure compactness. Mol. Biol.. 2008;42:623-628.
- [Google Scholar]
- Insight into the molecular mechanism of a herbal injection by integrating network pharmacology and in vitro. J. Ethnopharmacol.. 2015;173:91-99.
- [Google Scholar]
- Systemic maternal inflammation promotes ASD via IL-6 and IFN-γ. Biosci. Rep.. 2022;vol. 42(11)
- [Google Scholar]
- Autism spectrum disorder genetics and the search for pathological mechanisms. Am. J. Psychiatry. 2021;178(1):30-38.
- [Google Scholar]
- MMPBSA. py: an efficient program for end-state free energy calculations. J. Chem. Theory Comput.. 2012;8(9):3314-3321.
- [Google Scholar]
- Scientific basis for the therapeutic use of Withania somnifera (ashwagandha): a review. Altern. Med. Rev.. 2000;5(4):334-346.
- [Google Scholar]
- IMPPAT: A curated database of I ndian M edicinal P lants, P hytochemistry A nd T herapeutics. Sci. Rep.. 2018;vol. 8(1):4329.
- [Google Scholar]
- The impact of molecular dynamics on drug design: applications for the characterization of ligand–macromolecule complexes. Drug Discov. Today. 2015;20(6):686-702.
- [Google Scholar]
- Immunomodulatory effects of Withania somnifera on azoxymethane induced experimental colon cancer in mice. Immunol. Invest.. 2010;39(7):688-698.
- [Google Scholar]
- KNApSAcK metabolite activity database for retrieving the relationships between metabolites and biological activities. Plant Cell Physiol.. 2014;55(1):e7-e.
- [Google Scholar]
- A rapid finite difference algorithm, utilizing successive over-relaxation to solve the Poisson-Boltzmann equation. J. Comput. Chem.. 1991;12(4):435-445.
- [Google Scholar]
- UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem.. 2004;25(13):1605-1612.
- [Google Scholar]
- UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci.. 2021;30(1):70-82.
- [Google Scholar]
- Expected distributions of root-mean-square positional deviations in proteins. J. Phys. Chem. B. 2014;118(24):6526-6530.
- [Google Scholar]
- Advanced network pharmacology study reveals multi-pathway and multi-gene regulatory molecular mechanism of Bacopa monnieri in liver cancer based on data mining, molecular modeling, and microarray data analysis. Comput. Biol. Med.. 2023;161:107059
- [Google Scholar]
- The pediatrician’s role in the diagnosis and management of autistic spectrum disorder in children. Pediatrics. 2001;107(5):1221-1226.
- [Google Scholar]
- How molecular size impacts RMSD applications in molecular dynamics simulations. J. Chem. Theory Comput.. 2017;13(4):1518-1524.
- [Google Scholar]
- Exploration of databases and methods supporting drug repurposing: a comprehensive survey. Brief. Bioinform.. 2021;22(2):1656-1678.
- [Google Scholar]
- CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res.. 2018;46(W1):W363-W367.
- [Google Scholar]
- AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem.. 2010;31(2):455-461.
- [Google Scholar]
- STRING: a database of predicted functional associations between proteins. Nucleic Acids Res.. 2003;31(1):258-261.
- [Google Scholar]
- A potential anticancer dihydropyrimidine derivative and its protein binding mechanism by multispectroscopic, molecular docking and molecular dynamic simulation along with its in-silico toxicity and metabolic profile. Eur. J. Pharm. Sci.. 2021;158:105686
- [Google Scholar]
- IL-6 is increased in the cerebellum of autistic brain and alters neural cell adhesion, migration and synaptic formation. J. Neuroinflammation. 2011;8(1):1-10.
- [Google Scholar]
- Inhibition of IL-6 trans-signaling in the brain increases sociability in the BTBR mouse model of autism. Biochim. Biophys. Acta (BBA)-Molecular Basis Dis.. 2016;1862(10):1918-1925.
- [Google Scholar]
- A novel chemometric method for the prediction of human oral bioavailability. Int. J. Mol. Sci.. 2012;13(6):6964-6982.
- [Google Scholar]
- Inflammatory cytokines: potential biomarkers of immunologic dysfunction in autism spectrum disorders. Mediators Inflamm.. 2015;2015
- [Google Scholar]
- Targeting protein tyrosine phosphatase to unravel possible inhibitors for Streptococcus pneumoniae using molecular docking, molecular dynamics simulations coupled with free energy calculations. Life Sci.. 2021;264:118621
- [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2024.105772.
Appendix A
Supplementary data
The following are the Supplementary data to this article:
Supplementary data 1
Supplementary data 1
Supplementary data 2
Supplementary data 2
Supplementary data 3
Supplementary data 3
Supplementary data 4
Supplementary data 4
Supplementary data 5
Supplementary data 5







