Translate this page into:
Anti-inflammatory or anti-SARS-CoV-2 ingredients in Huashi Baidu Decoction and their corresponding targets: Target screening and molecular docking study
⁎Corresponding author at: Prof Hongwei Gao: School of Life Science, Ludong University, Yantai, Shandong, 264025, China. gaohongw369@ldu.edu.cn (Hongwei Gao)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Coronavirus disease 2019 (COVID-19) is a rapidly emerging infectious disease caused by SARS-CoV-2. Inflammatory factors may play essential roles in COVID-19 progression. Huashi Baidu Decoction (HSBD) is a traditional Chinese medicine (TCM) formula that can expel cold, dispel dampness, and reduce inflammation. HSBD has been widely used for the treatment of COVID-19. However, the active ingredients and potential targets for HSBD to exert anti-inflammatory or anti-SARS-CoV-2 effects remain unclear. In this paper, the active ingredients with anti-inflammatory or anti-viral effects in HSBD and their potential targets were screened using the Discovery Studio 2020 software. By overlapping the targets of HSBD and COVID-19, 8 common targets (FYN, SFTPD, P53, RBP4, IL1RN, TTR, SRPK1, and AKT1) were identified. We determined 2 key targets (P53 and AKT1) by network pharmacology. The main active ingredients in HSBD were evaluated using the key targets as receptor proteins for molecular docking. The results suggested that the best active ingredients Kaempferol2 and Kaempferol3 have the potential as supplements for the treatment of COVID-19.
Keywords
COVID-19
Anti-SARS-CoV-2
Anti-inflammatory
Traditional Chinese medicine formula
Target
Active ingredient
1 Introduction
The outbreaks of Coronavirus disease 2019 (COVID-19) caused by acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection have posed a severe threat to global public health (Chan et al., 2020). It is still raging with no signs of vanishment as the more contagious Delta variants spread. For symptomatic COVID-19, the clinical manifestations range from mild influenza-like symptoms to severe respiratory failure. The most common symptoms of COVID‐19 are fever, cough, and shortness of breath (Huang et al., 2020); but the headache, myalgias, diarrhea, cutaneous manifestations, sore throat, neurological involvement, and anosmia have also been reported (Indini et al., 2021; Wang et al., 2021). Although most patients present with mild symptoms, in a subset of infected individuals, patients worsen to pneumonia, multiorgan failure, and eventually die (Maguire et al., 2020).
Available data suggest that a highly deregulated and exuberant inflammatory response is associated with disease severity and lethality in patients with COVID-19 (Pedersen and Ho, 2020; Chen et al., 2020; Kessel et al., 2021). On admission, 83.2 % of COVID-19 patients had lymphopenia, 36.2 % had thrombocytopenia, and 33.7 % had leukopenia (Guan et al., 2020). The abnormal proportion of severe patients (including lymphopenia and leukopenia) was more evident than that of non-severe patients. In order to control inflammation, immune cells in the affected area may be over-recruited and aggregated, causing a decrease in leukocytes and platelets in the blood. When numerous immune cells gather in the diseased area, they stimulate the secretion of a large number of cytokines through positive feedback regulation (Tisoncik et al., 2012). As a result, the immune cells are over-activated, causing tissue injury or a severe immune over-response in the system. This process leads to acute respiratory distress syndrome, multiple organ dysfunction syndromes, and even multiple organ failures (Chousterman et al., 2017; Paules et al., 2020). Therefore, inflammation is a crucial factor in the progression of COVID-19.
Despite the tireless efforts of scientists worldwide, there is still no specific drug for SARS-CoV-2 that can be used for clinical treatment. Traditional Chinese medicine (TCM) formulas are widely used in China against COVID-19 (Ni et al., 2020). Among all recommended TCM formulas for combating COVID-19, Huashi Baidu decoction (HSBD) has attracted much attention (Wang and Qi, 2020). The pharmacological research results and clinical practice of HSBD have demonstrated its effectiveness in the treatment of COVID-19 (Zou et al., 2020; Wang et al., 2022). HSBD is a new TCM formula based on the theory of “epidemic disease” in the clinical diagnosis and therapy of COVID-19 (Zhu et al., 2021). In China’s Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Trial 7th and 8th versions), HSBD is listed as the recommended medicine for patients with severe COVID-19 (Han et al., 2020). HSBD consists of 13 herbs (Han et al., 2020): Chinese Atractylodes (Cangzhu), Fructus Tsaoko (Caoguo), Radix Paeonia Rubra (Chishao), Rhubarb (Dahuang), Poria cocos (Fuling), Liquorice (Gancao), Magnolia officinalis (Houpu), Astragalus menbranaceus (Huangqi), Wrinkled gianthyssop herb (Huoxiang), Chinese ephedra herb (Mahuang), Pepperweed seed (Tinglizi), and Semen armeniacae amarum (Xingren). According to TCM, this formula has the effect of detoxifying dampness, clearing heat and relieving asthma (Cao et al., 2020). However, the active ingredients and potential targets for HSBD to exert anti-inflammatory or anti-SARS-CoV-2 effects have not been fully elucidated.
Computer-aided drug design (CADD) is a commonly used method to develop multi-targeted drugs. It enables the rapid screening of molecules and plays an important role in the drug discovery process (Gentile et al., 2020; Sliwoski et al., 2014). Network pharmacology is an emerging field that integrates systems biology, polypharmacology, computational biology, and network analysis (Hsin et al., 2016). Network pharmacology aims to achieve synergistic effects by combining drugs within the network while targeting individual components in the network (Barabasi and Oltvai, 2004). This approach has been used to decode the TCM mechanisms.
Based on the complicated ingredients of HSBD, it is hard to identify the influence of a single ingredient on COVID-19 accurately. Accordingly, the conventional experimental approaches have some limitations when investigating TCM. However, these problems were dispelled by combining CADD and network pharmacology as a research method. It linked drug components, targets, and disease to establish a “drug-target-disease” interaction network. This approach has displaced the traditional “one drug, one target” model and transformed into a new “multi-component, multi-target” model. Therefore, in the present study, we attempted to use the method of CADD combined with network pharmacology to screen out the main active ingredients and core therapeutic targets of HSBD exerting anti-inflammatory or anti-SARS-CoV-2 effects. Our findings will provide a theoretical basis for future drug research and discovery.
2 Experimental section
2.1 Preparation of active ingredients in HSBD
The active ingredients of all herbs that make up the HSBD were retrieved from the Traditional Chinese Medicines database (TCMdb) and the online public database Traditional Chinese Medicine Systems Pharmacology (TCMSP) using the standard herb names as the search terms. Then, we imported all active ingredients with anti-inflammatory or antiviral effects into Discovery Studio 2020 (DS2020) software for ligands preparation and minimization.
2.2 Screening active ingredients with good druggability
In the early stage of drug development, targeted selection of active ingredients according to the Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) properties is necessary to improve the success rate of drug development and reduce the waste of funds (Arora et al., 2016). We calculated ADMET properties, including blood–brain barrier penetration (BBB) and human intestinal absorption (HIA). Toxicity Prediction by Komputer Assisted Technology (TOPKAT) is a fast and accurate prediction of the toxicity and environmental effects of active ingredients based on the 2D structure of the molecule (Khalaf et al., 2020). The toxicological properties to be considered in this study are Rodent Carcinogenicity, oral LD50 in rats, Mutagenicity, and Aerobic Biodegradability. Active ingredients obtained by ADMET prediction and TOPKAT were then filtered through Lipinski's and Veber's rules.
2.3 Screening the target proteins of HSBD for COVID-19
The reverse finding target process in DS2020 was to match the active ingredients with good druggability against the target proteins. We then converted the target protein names into standard gene names using the UniProtKB search function in the UniProt (https://www.uniprot.org/) database. COVID-19-associated targets were screened from the Human Gene Database (GeneCards; http://www.genecards.org/) using the keywords “SARS-CoV-2” and “COVID-19”. The overlaps between COVID-19 and HSBD targets were supposed to be the potential targets of HSBD for the treatment of COVID-19. Moreover, the efficacy of these potential targets was also identified and distinguished, such as anti-inflammatory targets or anti-SARS-CoV-2 targets.
2.4 Construction of “herb-active ingredient-target” network and protein–protein interaction (PPI) network
A “herb-active ingredient-target” network was constructed using Cytoscape 3.8.0 software to show the active ingredients in HSBD and their corresponding targets in the treatment of COVID-19. The search tool for the Retrieval of Interacting Genes database (STRING, https://string-db.org/) was performed to analyze the potential targets of HSBD for treating COVID-19. The research species was set to “human species” for the PPI network. The results could reflect the core targets among these potential targets.
2.5 Molecular docking verification
The 3D crystal structures of the key target proteins were obtained from the RCSB Protein Data Bank (PDB, https://www.rcsb.org/). Then, we need to prepare the receptor binding site for the target protein with the help of DS2020 software. Molecular docking calculations were performed using CDOCKER, which can simulate the hypotheses before the implementation of specific experiments (Gentile et al., 2020). We applied a consensus scoring process based on different scoring functions (-CDOCKER ENERGY and -CDOCKER INTERATION ENERGY) to determine the best candidate active ingredients for the molecular docking process.
2.6 Molecular dynamics simulation and binding free energy calculation
The ff14SB force field was used for the protein part. The force field for ligands was generated from the generalized Amber force field potential using the antechamber module of Amber 18. All the systems were solvated in a 10 Å truncated octahedral water box using a TIP3P solvent model under periodic boundary conditions. Firstly, a total of 5000 cycles of steepest descent and then 5000 cycles of conjugated gradient minimizations were carried out with restrictions on all solute molecules. Secondly, a total of 5000 cycles of steepest descent and then 5000 cycles of conjugated gradient minimizations were carried out again without any restriction. Thirdly, a 300 ps heating procedure followed by a 500 ps equilibrium simulation was conducted. Finally, a 100 ns MD simulation was performed for each system under NPT conditions to produce a trajectory without any constraints. The temperature was maintained at 310 K by coupling to a Langevin heat bath using a collision frequency of 1 ps-1. The step side was set to 2 fs in all the simulations.
The MMGBSA method was used to calculate the binding free energy for the systems. The calculation formulas are as follows:
In Equation (2), the EMM, Gsol, and TΔS represent the gas-phase molecular mechanics' energy, the solvation free energy, and the vibrational entropy term, respectively. EMM is the calculated as the sum of internal (Eint), van der Waals (Evdw), and electrostatic (Eele) terms. Gsol represents the solvation energy, which is composed of the electrostatic solvation free energy (GGB) and the nonpolar solvation free energy (GSA). The binding free energies were the average values of calculating 4000 snapshots sampling from the last 40 ns trajectory.
3 Results and discussion
3.1 The active ingredients of HSBD
99 active ingredients in HSBD were retrieved from TCMdb and TCMSP, as detailed in Table 1. Since this study required consideration of receptor-ligand interactions, the ligand preparation process is crucial. Different protonated states, isomers and tautomers usually have different 3D geometric structures and binding properties. If the exact binding configuration was unknown, one approach was to enumerate several likely configurations before screening the active substance and running docking protocols. Ligand Preparation helped us to accomplish this task. Therefore, the number of active ingredients reached 298 after ligands preparation and minimization.
Herb
Active ingredients name
Effect
Cangzhu
Atractylenolide I
anti-inflammatory
Atractylone
anti-inflammatory
(+)-Eudesma-4(15),7(11)-dien-8-one
anti-inflammatory
Syringin
anti-inflammatory
Limonin
antiviral
Caoguo
quercetin-3-o-galactopyranoside
anti-inflammatory
citral
anti-inflammatory
isoquercetin_qt
anti-inflammatory
(4E,6E)-1,7-bis(4-hydroxyphenyl)hepta-4,6-dien-3-one
anti-inflammatory
protocatechuic acid
anti-inflammatory
quercetin 3-o-rhamnopyranosyl
anti-inflammatory
vanillic acid
anti-inflammatory
(-)-nopinene
anti-inflammatory
EIC
anti-inflammatory
ent-Epicatechin
anti-inflammatory
Aldrich
anti-inflammatory
6,7-dihydroxy-indan-4-carbaldehyde
anti-inflammatory
(-)-catechin
anti-inflammatory
quercertin,3-o-beta-d-glucopyranoside
antiviral
quercetin,3-o-glucoside
antiviral
Hirsutrin
antiviral
quercetin
anti-inflammatory, antiviral
quercetin,3-o-rutinoside
anti-inflammatory, antiviral
(L)-alpha-Terpineol
anti-inflammatory, antiviral
Hyperin
anti-inflammatory, antiviral
Chishao
Paeoniflorin
anti-inflammatory
beta-Sitosterol
anti-inflammatory
Eugeniin
antiviral
Dahuang
Alizarin
anti-inflammatory
Gallic acid
anti-inflammatory
Piceid
anti-inflammatory
(+)-Catechin
antiviral
Procyanidin B2 3,3′-di-O-gallate
antiviral
Xingren
Dihydroquercetin
anti-inflammatory
Eriodictyol
anti-inflammatory
Linalool
antiviral
Fuling
Dihydroquercetin
anti-inflammatory
Astilbin
anti-inflammatory
Gancao
6,8-Bis(C-beta-glucosyl)-apigenin
anti-inflammatory
Glycyrrhetinic acid
anti-inflammatory
Isoliquiritin
anti-inflammatory
Isoquercitrin
anti-inflammatory
Licochalcone A
anti-inflammatory
Liquiritic acid
anti-inflammatory
Pinocembrin
anti-inflammatory
beta-Sitosterol
anti-inflammatory
3,3′-Dimethylquercetin
antiviral
Glycycoumarin
antiviral
Glycyrrhisoflavone
antiviral
Glepidotin D
antiviral
Isolicoflavonol
antiviral
Licopyranocoumarin
antiviral
Glycyrrhizic acid
anti-inflammatory, antiviral
Rutin
anti-inflammatory, antiviral
Houpu
Terpinen-4-ol
anti-inflammatory
Linalool
antiviral
Huangqi
Robinin
anti-inflammatory
beta-Sitosterol
anti-inflammatory
Quercetin
antiviral
Kaempferol
anti-inflammatory, antiviral
Huoxiang
Acacetin
anti-inflammatory
Friedelan-3-one
anti-inflammatory
beta-Pinene
anti-inflammatory
beta-Sitosterol
anti-inflammatory
Linalool
antiviral
Pachypodol
antiviral
Cinnamaldehyde
antiviral
Mahuang
(4S,5R) Ephedroxane
anti-inflammatory
Isoquercitrin
anti-inflammatory
N-Methylephedrine
anti-inflammatory
Kaempferol
anti-inflammatory, antiviral
Banxia
beta-Sitosterol
anti-inflammatory
Tinglizi
Arachic acid
anti-inflammatory
DBP
anti-inflammatory
palmitic acid
anti-inflammatory
methyl palmitate
anti-inflammatory
beta-sitosterol
anti-inflammatory
Isorhamnetin
anti-inflammatory
Erucic acid
anti-inflammatory
myristic acid
anti-inflammatory
Syringaldehyde
anti-inflammatory
3-(4-Hydroxyphenyl)propionitrile
anti-inflammatory
Sinapinic acid
anti-inflammatory
EIC
anti-inflammatory
Isovanillic acid
anti-inflammatory
Exceparl M-OL
anti-inflammatory
hederagenin
anti-inflammatory
beta-sitosterol
anti-inflammatory
Tropeolin
anti-inflammatory
Dihomolinolenic acid
anti-inflammatory
4-O-Methylgallic acid
anti-inflammatory
nicotinic acid
anti-inflammatory
linolenic acid
anti-inflammatory
oleic acid
anti-inflammatory
HMF
anti-inflammatory
stearic acid
anti-inflammatory
alexandrin
antiviral
Kaempferol
anti-inflammatory, antiviral
quercetin
anti-inflammatory, antiviral
3.2 Screening active ingredients with good druggability
We used DS2020 to evaluate the ADMET properties of 298 active ingredients. The results are shown in Fig. 1. The blue ellipse represents the 99 % confidence interval of the absorption, and the green ellipse represents the 99 % confidence interval of the BBB. An important factor that promotes the efficacy of drugs in the body depends on the absorption of drugs in the intestinal tract (Song et al., 2014). The absorption outside the 95 % ellipse tends to drop pretty quickly. The BBB is a significant barrier that blocks the physical diffusion of most proteins or compounds from the blood to the brain (Quaegebeur et al., 2011). Notably; the screened compounds must have blood–brain barrier permeability to ensure their activity during systemic administration (Banks, 2016; Albarracin et al., 2012). If a molecule is outside the 99 % confidence interval of the BBB model; the prediction of the molecule is considered unreliable. 95 active ingredients in the intersection of blue and green ellipses were retained for further screening.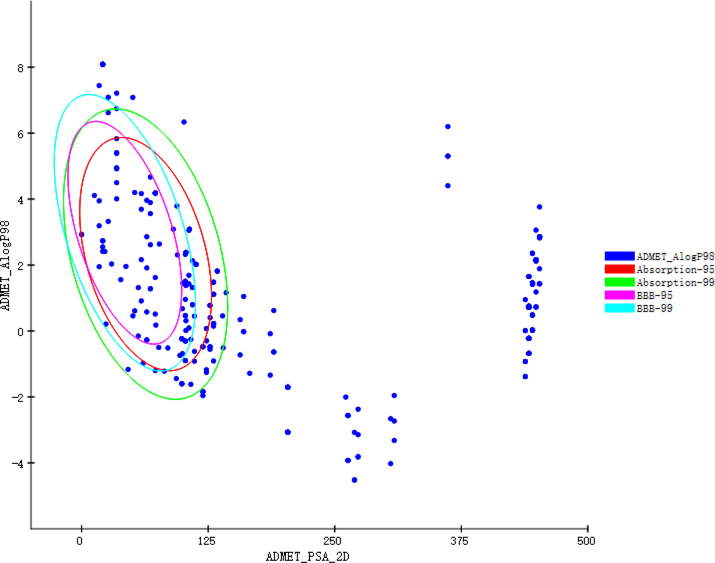
ADMET properties of active ingredients.
Adopting the TOPKAT process saves time to market, reduces animal testing, and assesses health risks (Wang et al., 2021). Based on the TOPKAT results, we removed all active ingredients from our list that exceeded the four properties' (Rodent Carcinogenicity, oral LD50 in rats, Mutagenicity, and Aerobic Biodegradability) optimal prediction spaces (OPS). The number of active ingredients was down to 60. Lipinski's and Veber's rules can determine whether the compounds have good absorption, penetration, and oral bioavailability (Lipinski et al., 1997). Finally, the 60 active ingredients were further filtered by applying reverse finding target, reducing the number of active ingredients to 48.
3.3 The target proteins of HSBD for COVID-19
We found that the anti-inflammatory or anti-viral target proteins of the 48 active ingredients in HSBD were Antigen peptide transporter 1 (TAP1), Retinol-binding protein 4 (RBP4), Interleukin 1 receptor antagonist (IL1RN), Centromere-associated protein E (CENPE), Beta-secretase 1 (BACE1), Transthyretin (TTR), Tyrosine-protein kinase Fyn (FYN), Thyroid hormone receptor alpha (THRA), Pulmonary surfactant-associated protein D (SFTPD), Nuclear receptor subfamily 0 group B member 2 (NR0B2), Cellular tumor antigen p53 (P53), SRSF protein kinase 1 (SRPK1), RAC-alpha serine/threonine-protein kinase (AKT1) and Protein Mdm4 (MDM4). Next, we used “COVID-19” and “SARS-COV-2” as keywords to search for targets related to COVID-19 in the GeneCards database. Table 2 showed 8 overlaps between COVID-19 and HSBD targets, namely RBP4, IL1RN, TTR, FYN, SFTPD, P53, SRPK1, and AKT1.
PDB ID (Target protein)
Gene name
Effect
1RBP (Retinol-binding protein 4)
RBP4
anti-inflammatory
1SRI (Interleukin 1 receptor antagonist)
IL1RN
anti-inflammatory
1TYR (Transthyretin)
TTR
anti-inflammatory
3BEG (SRSF protein kinase 1)
SRPK1
anti-SARS-CoV-2
3CQU (RAC-alpha serine/threonine-protein kinase)
AKT1
anti-inflammatory, anti-SARS-CoV-2
3CQW (RAC-alpha serine/threonine-protein kinase)
AKT1
anti-inflammatory, anti-SARS-CoV-2
2DQ7 (Tyrosine-protein kinase Fyn)
FYN
anti-inflammatory, anti-SARS-CoV-2
2ORJ (Pulmonary surfactant-associated protein D)
SFTPD
anti-inflammatory, anti-SARS-CoV-2
2X0U (Cellular tumor antigen p53)
P53
anti-inflammatory, anti-SARS-CoV-2
2X0V (Cellular tumor antigen p53)
P53
anti-inflammatory, anti-SARS-CoV-2
3.4 Analysis of the “herb-active ingredient-target” network and PPI network
The 8 potential targets and related herbs and their active ingredients in HSBD were used to construct the “herb-active ingredient-target” network by Cytoscape-3.8.0 software. 9 essential herbs were Huangqi, Tinglizi, Xingren, Mahuang, Huoxiang, Gancao, Dahuang, Caoguo, and Mahuang. As shown in Fig. 2, 9 herbs of HSBD were marked in green; 48 active ingredients screened from 9 herbs with anti-inflammatory or anti-SARS-CoV-2 effects were marked in pink; 8 potential targets of HSBD are marked in yellow, and the effects of the targets are marked in red. The black line represented that a specific active ingredient comes from a specific herb; the gray line represented the interaction between the active ingredient and the target, and the red line represented a particular target against inflammation or SARS-CoV-2. Some active ingredients were common to several herbs. For example, Kaempferol1, Kaempferol2, and Kaempferol3 were common ingredients of Huangqi, Tinglizi, and Mahuang. In addition, each target is bound to two or more active ingredients, indicating that these targets could be affected by multiple active ingredients simultaneously to exert different effects. The result revealed the mystery of the “multi-component and multi-target” of TCM formulas.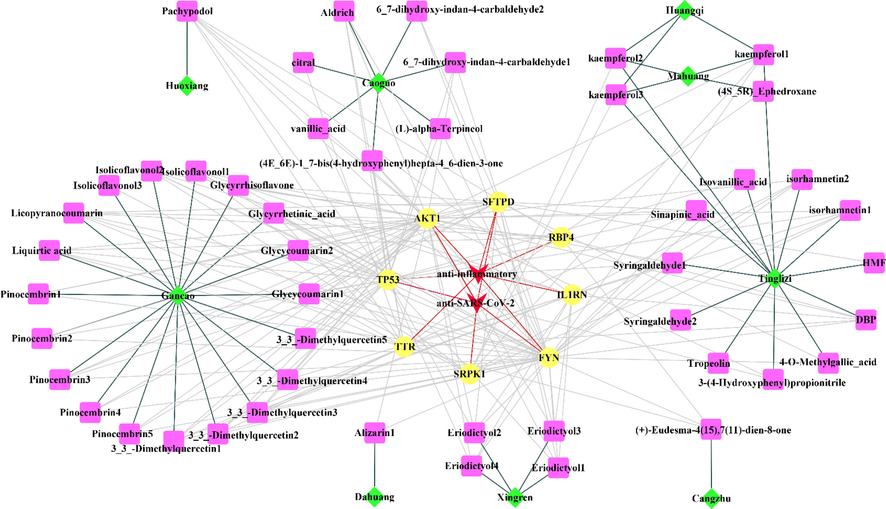
“herb-active ingredient-target“ network.
Most proteins do not function in isolation, and their interactions with other proteins define their cellular functions (Liu et al., 2018). Understanding PPI will help to identify pharmacological targets and design drugs (Chen et al., 2010). The PPI analysis of 8 target proteins was carried out using the STRING plug-in of Cytoscape 3.8.0 (Fig. 3). There were 6 nodes and 12 edges in the PPI network. 2 disconnected targets (IL1RN and SFTPD) were excluded from the network. The nodes represent targets, and the size of the nodes represents the degree value. The larger the shape of the nodes was, the larger the degree value and the more important the targets were. Each edge represented an interaction between targets; the thicker the edge, the higher the combined score. The minimum score was set as the highest confidence of 0.9 to ensure high cross-confidence information. The targets within the highest confidence are P53, AKT1, TTR, and RBP4. Based on Fig. 3, P53 and AKT1 with higher degree value and confidence scores played more important roles in the network, which were likely to be the core therapeutic targets of HSBD on COVID-19.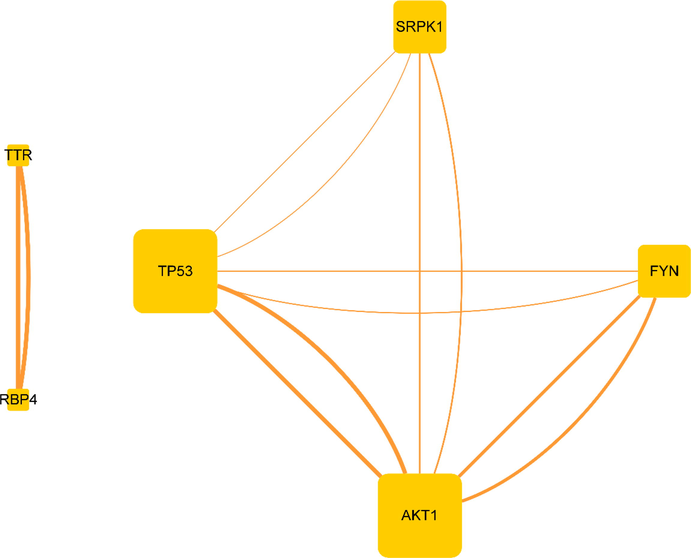
PPI network.
3.5 Analysis of molecular docking
Molecular docking is a calculation method used to predict the binding affinity of two molecules, usually a small molecule ligand and a target protein (Banchi et al., 2020). It can be seen from Table 2 that the PDB IDs corresponding to P53 and AKT1 were 2X0U, 2XOV, 3CQU and 3CQW. We used 2X0U and 3CQW with good resolution and few mutations as target receptors for docking studies. There are 48 active ingredients that were docked into the protein active site in this study. The molecular docking results (top 5) evaluated by 3 different scoring functions were shown in Table 3, including -CDOCKER_ENERGY, -CDOCKER_INTERATION_ENERGY, and Consensus score. The structures of the top 5 active ingredients for 2X0U and 3CQW are presented in Fig. 4.
Target protein
Rank
Active ingredients name
-CDOCKER_ENERGY
-CDOCKER-INTERATION_ENERGY
Consensus
2X0U
1
2-amino substituted benzothiazole scaffold
14.02
25.82
2
2
Kaempferol2
35.58
36.11
2
3
Eriodictyol3
34.22
37.30
2
4
(4E_6E)-1_7-bis(4-Hydroxyphenyl)hepta-4_6-dien-3-one
31.80
39.58
2
5
Kaempferol1
26.86
36.34
2
6
Alizarin1
26.55
31.11
2
3CQW
1
Spiroindoline 13j
21.23
29.60
2
2
Kaempferol3
49.69
57.62
2
3
Isorhamnetin2
46.70
62.21
2
4
Eriodictyol3
42.89
43.01
1
5
Kaempferol2
42.53
41.47
1
6
Kaempferol1
42.37
43.05
2
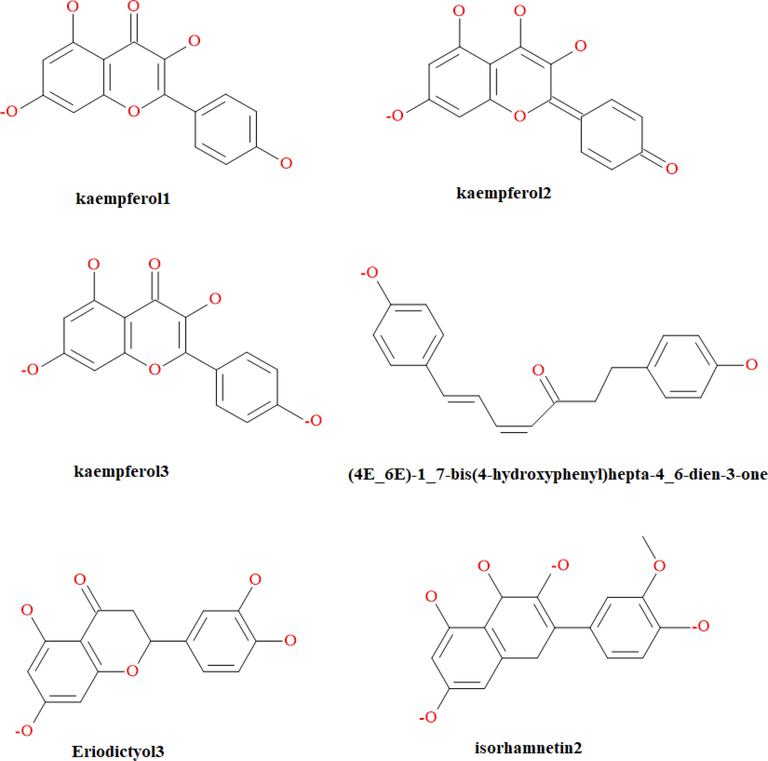
Structures of the docking compounds for 2X0U and 3CQW.
The CDOCKER score was reported as the negative value (i.e., -CDOCKER_ENERGY), which was calculated based on the internal ligand strain energy and receptor-ligand interaction energy (Rampogu and Rampogu Lemuel, 2016). In addition, the -CDOCKER_INTERATION_ENERGY represented the energy of non-bonding interactions between proteins and ligands. Higher -CDOCKER energy and -CDOCKER_INTERATION_ENERGY value indicated more favorable binding. From Table 4, the -CDOCKER ENERGY (35.58) and -CDOCKER_INTERATION_ENERGY (36.11) of Kaempferol2 were all higher than -CDOCKER ENERGY (14.02) and -CDOCKER_INTERATION_ENERGY (25.82) of the control compound (2-amino substituted benzothiazole scaffold) (Basse et al., 2010). In addition, the -CDOCKER ENERGY (49.69) and -CDOCKER_INTERATION_ENERGY (57.62) of Kaempferol3 were all higher than -CDOCKER ENERGY (21.23) and -CDOCKER_INTERATION_ENERGY (29.60) of the control compound (Spiroindoline 13j) (Lippa et al., 2008). Moreover, some experiments have shown that kaempferol is able to protect cells from virus-induced cell death and may be a promising antiviral drug for SARS-CoV-2 (Khan et al., 2021). Taken together, Kaempferol2 and Kaempferol3 can show better effects in the treatment of COVID-19, both in computer simulation screening and in vitro inhibition experiments.
Target protein
Active ingredients
Interaction amino acids
2X0U
Kaempferol2
Thr150, Pro223, Pro222, Glu221, Asp228, Trp146, Val147, Pro151, Cys229, Leu145, Tyr220, Thr230, Phe109, Leu257
Eriodictyol3
Gly154, Tyr220, Pro153, Thr155, Glu221, Pro222, Thr230, Leu145, Pro223, Cys229, Val147, Thr150, Pro151
(4E_6E)-1_7-bis(4-Hydroxyphenyl)hepta-4_6-dien-3-one
Trp146, Leu145, Leu257, CYS229, Thr230, Thr155, Thr150, Pro152, Pro219, Gly154, GRG202, Pro223, Pro151, Val147, Pro222, Tyr220, Glu221, Pro153
Kaempferol1
Thr150, Pro223, Pro222, Glu221, Leu257, Leu145, Phe109, Asp228, Trp146, Val147, Pro151, Cys229, Tyr220, Thr230
Alizarin1
Asp228, Pro223, Thr230, Trp146, Cys229, Leu145, Leu257, Tyr220, Glu221, Thr155, Pro151, Pro222, Val147, Thr150
3CQW
Kaempferol3
Lys276, Asn279, Asp292, Thr160, Gly159, Phe161, Gly162, Leu181, Lys179
Isorhamnetin2
Glu198, Gly294, Asn279, Lys179, Asp292, Lys276, Leu181, Lys163, Val164, Gly162, Phe161, Gly159, Thr160
Eriodictyol3
Glu278, Asn279, Asp292, Thr291, Lys179, Met227, Thr211, Met281, Glu228, Val164, Tyr229, Ala230, Phe438, Ala177, Leu156, Gly157, Lys158, Glu234
Kaempferol2
Lys179, Asp292, Met227, Thr291, Thr211, Glu228, Tyr229, Ala230, Leu156, Ala177, Met281, Phe438, Val164, Gly157, Lys,158, Gly159
Kaempferol1
Lys276, Asn279, Asp292, Thr160, Gly159, Phe161, Gly162, Leu181, Lys179, Glu198, Gly294
Selecting a high activity 2-amino substituted benzothiazole scaffold as a control, it can be found that the active binding site of 2X0U is close to the amino acid residues Thr155, Pro151, Thr230, Val147, Cys220, Pro222, Leu145, and Pro223. Table 4 showed the interaction of amino acids in the ligand–protein for the top 5 docking active ingredients. By comparing the interacting amino acids in the top 5 docking compounds, it was found that Val147, Leu145, Pro222, Pro223, and Thr230 are the crucial amino acid residues for 2X0U. Selecting a high activity Spiroindoline 13j as a control, it can be found that the active binding site of 3CQW is close to the amino acid residues Ala230, Ala177, Leu156, Met227, Val164, Asp292, Thr291, Asn279, Thr291, Glu234, Met281and Pro223. By comparing the interacting amino acids in the top 5 docking active ingredients, it was found that Asp292 and Asn279 are the crucial amino acid residues for 3CQW.
The docking pose of Kaempferol2 was shown in Fig. 5 (A), which displayed 6 key residues around Kaempferol2, consisting of Pro151, Pro222, Pro223, Thr150, Val147, and Tyr220. Val147, Pro222, and Pro223 are the crucial residues for Kaempferol2 based on the result of Table 4. The 2D docking interaction plots of Kaempferol2 with the 2X0U active sites are shown in Fig. 5 (B). The docking pose of Kaempferol2 showed 3 Pi-Alkyl interactions binding with Pro151, Val147, and Pro223; 1 Pi-Antion interaction binding with Tyr220; and 1 conventional hydrogen bond interaction binding with Thr150. The docking pose of Kaempferol3 was shown in Fig. 6 (A), which displayed 4 key residues around Kaempferol3, consisting of Lys179, Phe161, The160, and LYS276. The 2D docking interaction plots of Kaempferol3 with the 3CQW active sites are shown in Fig. 6 (B). The docking pose of Kaempferol3 showed 2 salt bridges keying with Lys276 and Lys179; 2 conventional hydrogen bond interactions binding with Thr160 and Phe161; and 1 Pi-Pi T-shaped interaction binding with Phe161.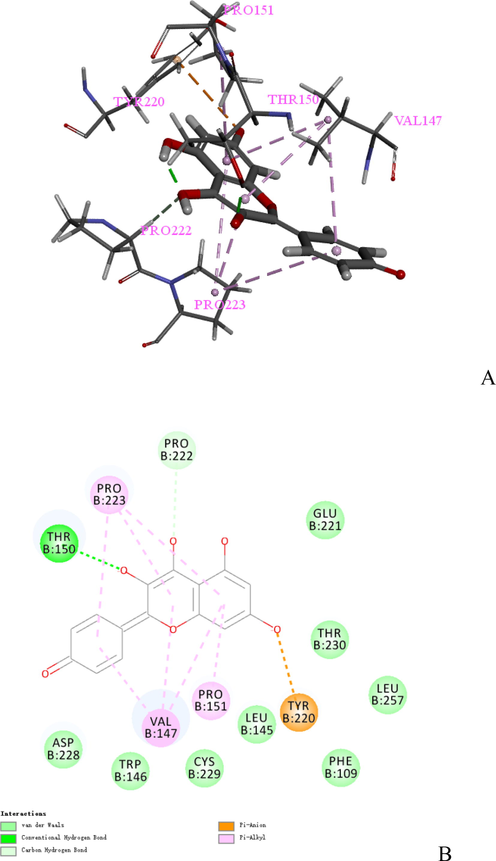
(A) The docking pose of Kaempferol2. (B) 2D docking Interaction plots of Kaempferol2 at the 2X0U active site.
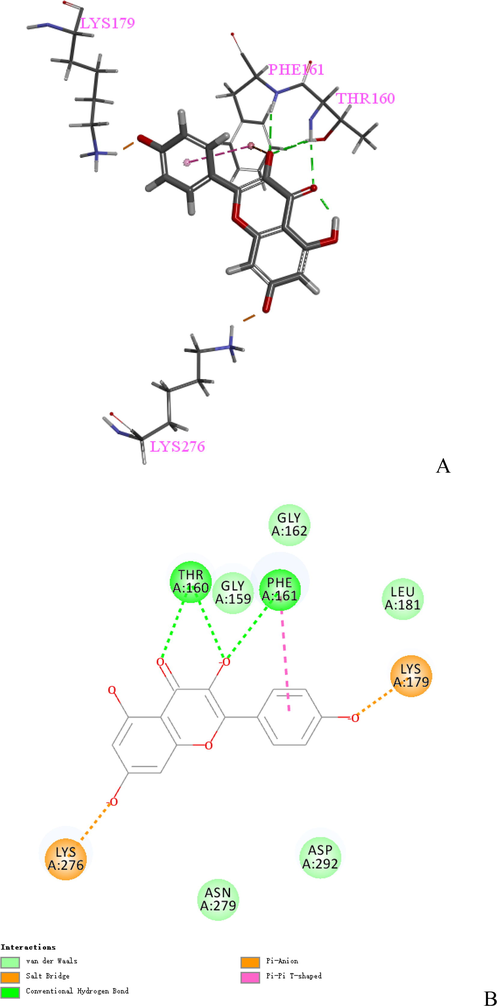
(A) The docking pose of Kaempferol3. (B) 2D docking Interaction plots of Kaempferol3 at the 3CQW active site.
3.6 Global fluctuation of each system and binding free energy analysis
In order to explore the rationality of conformational samplings, the time evolutions of root-mean-square deviation (RMSD) for heavy atoms referenced to the corresponding energy-minimized initial structure were calculated on each system. As shown in Fig. 7, all systems reached equilibrium at about 60 ns. Thus, the last 40 ns trajectories were used to calculate the binding free energy.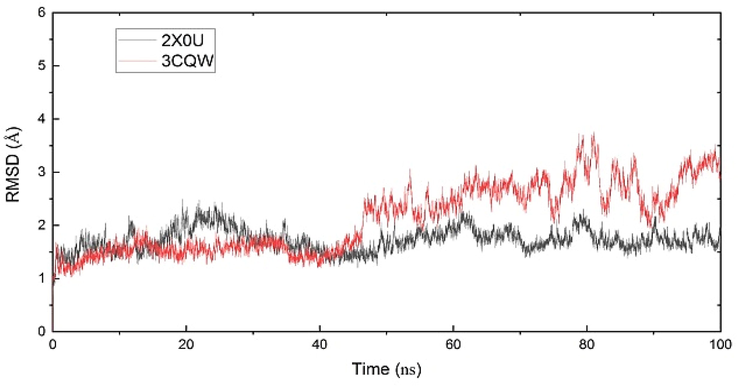
The RMSD curves for the complexes.
In our work, MMGBSA was applied to calculate the binding free energy for each complex. As listed in Table 5, the binding free energy of 2X0U-Kaempferol2 was −12.9 kcal mol−1, and that of 3CQW-Kaempferol3 was –22.5 kcal mol−1. These values are all less than zero, implying that the binding of both complexes is energetically feasible.
ΔEele
ΔEvdw
ΔGGB
ΔGSA
ΔGpol
ΔGnonpol
ΔGMMGBSA
2X0U
−14.1
−20.6
24.1
−2.3
−34.7
21.8
−12.9
3CQW
−40.5
−23.5
45.6
−4.1
−64.1
41.5
−22.5
4 Conclusion
In the present study, we used network pharmacology combined with CADD approach to proving that HSBD affects COVID-19 through interaction between multiple components and multiple targets. The specific process is shown in Fig. 8. We discovered 8 COVID-19-associated targets (RBP4, IL1RN, TTR, FYN, SFTPD, P53, SRPK1, and AKT1) of 48 active ingredients in QFPD. The PPI network revealed that AKT1 and P53 are expected to be the most probably core targets for COVID-19 intervention. Through molecular docking, we found that Kaempferol2 and Kaempferol3 may be the main active ingredients of HSBD for COVID-19. The possibility of receptor protein binding to inhibitor molecules was verified by molecular dynamics simulations and free energy calculations. The results of the study provided a theoretical foundation for clinical application and represented a significant advance in the drug discovery program for the treatment of COVID-19.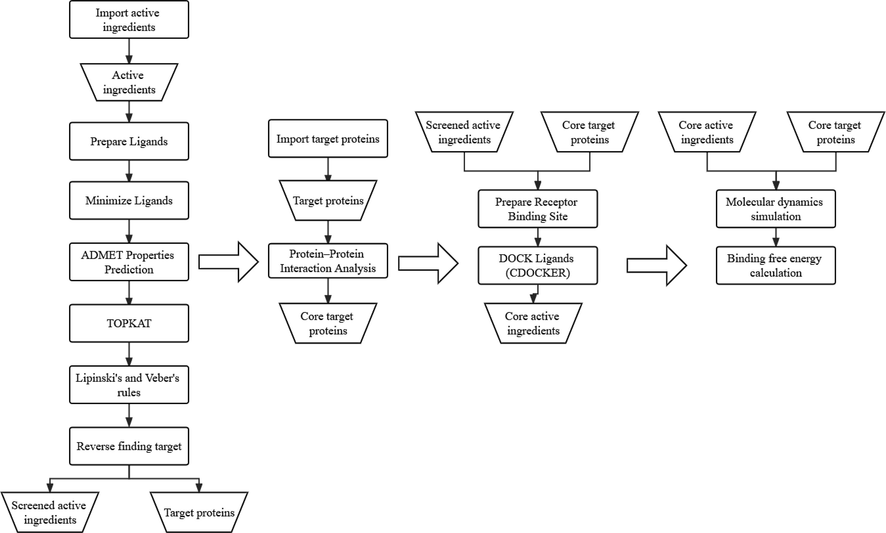
Calculation flow chart.
CRediT authorship contribution statement
Zixuan Wang: Software, Writing – original draft, Writing – review & editing. Hongwei Gao: Resources, Project administration, Supervision.
Acknowledgements
This work was financially supported by the High-end Talent Team Construction Foundation [Grant No. 108-10000318], the Cooperation Project of University and Local Enterprise in Yantai of Shandong Province (2021XDRHXMXK23).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Effects of natural antioxidants in neurodegenerative disease. Nutr. Neurosci.. 2012;15(1):1-9.
- [Google Scholar]
- Esculetin induces antiproliferative and apoptotic response in pancreatic cancer cells by directly binding to KEAP1. Mol. Cancer. 2016;15(1):1-15.
- [Google Scholar]
- From blood–brain barrier to blood–brain interface: new opportunities for CNS drug delivery. Nat. Rev. Drug Discov.. 2016;15(4):275-292.
- [Google Scholar]
- Network biology: understanding the cell's functional organization. Nat. Rev. Genet.. 2004;5(2):101-113.
- [Google Scholar]
- Toward the rational design of p53-stabilizing drugs: probing the surface of the oncogenic Y220C mutant. Chem. Biol.. 2010;17(1):46-56.
- [Google Scholar]
- The important role of polysaccharides from a traditional Chinese medicine-Lung Cleansing and Detoxifying Decoction against the COVID-19 pandemic. Carbohydr. Polym.. 2020;240(1):116346.
- [Google Scholar]
- A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514-523.
- [Google Scholar]
- KUPS: constructing datasets of interacting and non-interacting protein pairs with associated attributions. Nucleic Acids Res.. 2010;39(suppl_1):D750-D754.
- [Google Scholar]
- Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest.. 2020;130(5):2620-2629.
- [Google Scholar]
- Cytokine Storm and Sepsis Disease Pathogenesis, Seminars in Immunopathology. Springer; 2017. p. :517-528.
- Deep docking: a deep learning platform for augmentation of structure based drug discovery. ACS Cent. Sci.. 2020;6(6):939-949.
- [Google Scholar]
- Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med.. 2020;382(18):1708-1720.
- [Google Scholar]
- The therapeutic efficacy of Huashi Baidu Formula combined with antiviral drugs in the treatment of COVID-19: A protocol for systematic review and meta-analysis. Medicine. 2020;99(42):1-3.
- [Google Scholar]
- systemsDock: a web server for network pharmacology-based prediction and analysis. Nucleic Acids Res.. 2016;44(W1):W507-W513.
- [Google Scholar]
- Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The lancet. 2020;395(10223):497-506.
- [Google Scholar]
- Triage process for the assessment of coronavirus disease 2019-positive patients with cancer: The ONCOVID prospective study. Cancer. 2021;127(7):1091-1101.
- [Google Scholar]
- Discrimination of COVID-19 From Inflammation-Induced Cytokine Storm Syndromes Using Disease-Related Blood Biomarkers. Arthritis Rheumatol.. 2021;73(10):1791-1799.
- [Google Scholar]
- Synthesis, docking, computational studies, and antimicrobial evaluations of new dipeptide derivatives based on nicotinoylglycylglycine hydrazide. Molecules. 2020;25(16):3589.
- [Google Scholar]
- In silico and in vitro evaluation of kaempferol as a potential inhibitor of the SARS-CoV-2 main protease (3CLpro) Phytother. Res.. 2021;35(6):2841-2845.
- [Google Scholar]
- Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev.. 1997;23(1–3):3-25.
- [Google Scholar]
- Synthesis and structure based optimization of novel Akt inhibitors. Bioorg. Med. Chem. Lett.. 2008;18(11):3359-3363.
- [Google Scholar]
- An AP-MS-and BioID-compatible MAC-tag enables comprehensive mapping of protein interactions and subcellular localizations. Nat. Commun.. 2018;9(1):1-16.
- [Google Scholar]
- Prognostic factors in patients admitted to an urban teaching hospital with COVID-19 infection. J. Transl. Med.. 2020;18(1):1-10.
- [Google Scholar]
- Combating COVID-19 with integrated traditional Chinese and Western medicine in China. Acta Pharm. Sin. B. 2020;10(7):1149-1162.
- [Google Scholar]
- The neurovascular link in health and disease: molecular mechanisms and therapeutic implications. Neuron. 2011;71(3):406-424.
- [Google Scholar]
- Network based approach in the establishment of the relationship between type 2 diabetes mellitus and its complications at the molecular level coupled with molecular docking mechanism. Biomed Res. Int.. 2016;2016(1):1-6.
- [Google Scholar]
- Inhibitory effect of ent-Sauchinone on amyloidogenesis via inhibition of STAT3-mediated NF-κB activation in cultured astrocytes and microglial BV-2 cells. J. Neuroinflammation. 2014;11(1):1-14.
- [Google Scholar]
- Deciphering the active compounds and mechanisms of HSBDF for treating ALI via integrating chemical bioinformatics analysis. Front. Pharmacol.. 2022;13(1):879268-1876268.
- [Google Scholar]
- Traditional Chinese medicine to treat COVID-19: the importance of evidence-based research. Drug Discoveries Therapeut.. 2020;14(3):149-150.
- [Google Scholar]
- Screening out anti-inflammatory or anti-viral targets in Xuanfei Baidu Tang through a new technique of reverse finding target. Bioorg. Chem.. 2021;116(1):105274.
- [Google Scholar]
- Next-generation sequencing and proteomics of cerebrospinal fluid from COVID-19 patients with neurological manifestations. Front. Immunol.. 2021;12(1):5323-12325.
- [Google Scholar]
- Analyzing the potential therapeutic mechanism of Huashi Baidu Decoction on severe COVID-19 through integrating network pharmacological methods. J. Tradit. Complement. Med.. 2021;11(2):180-187.
- [Google Scholar]
- Experience summary and suggestion on diagnosis and treatment program of traditional Chinese medicine treating severe type coronavirus disease 2019. J. Tradit. Chin. Med.. 2020;61(15):1289-1293.
- [Google Scholar]







