Translate this page into:
Antimicrobial activity of sulfur nanoparticles: Effect of preparation methods
⁎Corresponding author. jwrhim@khu.ac.kr (Jong-Whan Rhim)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Sulfur nanoparticles (SNPs) were synthesized using elemental sulfur and sodium sulfide, capped with chitosan as a stabilizer (SNPES), and their properties were compared to SNPs prepared by acidification of sodium thiosulfate (SNPSTS). The SNPs were characterized using UV–visible spectroscopy, EDS, TEM, XRD, and TGA, and their antimicrobial activity was tested using the disk diffusion method and minimum inhibitory concentration (MIC)/minimum bactericidal concentration (MBC) analysis. The SNPES showed a rod-shaped morphology with an average length of 87 nm, while SNPSTS exhibited a spherical shape with an average particle size of 17 nm. The rod-shaped SNPES showed higher thermal stability than the spherical SNPSTS. Both types of SNPs did not show significant antibacterial activity against Gram-negative (E. coli) bacteria but showed significant antibacterial activity against Gram-positive (L. monocytogenes) bacteria. Between the SNPs, SNPES showed higher growth-inhibiting activity against L. monocytogenes than SNPSTS.
Keywords
Sulfur nanoparticles
Rod-shaped
Elemental sulfur
Antibacterial activity
1 Introduction
Sulfur is an abundantly available biologically active element and has long been used for dermatological treatment since ancient times for its efficient antimicrobial properties (Teng et al., 2019). Sulfur and its plethora of chemically diverse organic and inorganic compounds are known to exhibit a diverse spectrum of biological activities such as antimicrobial, antioxidant, anticancer, and radical-scavenging properties (Shankar and Rhim, 2018; Zimmerman et al., 2015). Sulfur has been used as a component of various skin disorders, such as dandruff shampoos and acne ointments (Parcell, 2002). Also, sulfur has been efficiently used as a pesticide or a fungicide for the treatment of a wide range of plant diseases (Abuyeva et al., 2018; Rao and Paria, 2013; Tweedy, 1981). However, its use is now restricted in the agrochemical industries because it needs bulk quantities for application, and it is likely to induce resistance in the target species. Besides, high hydrophobicity has limited the actual use of sulfur. By the way, the advent of nanotechnology opens up a new way of using sulfur. Surface modified nano-sized sulfur nanoparticles (SNPs) has been reported for their excellent antimicrobial and antifungal activities (Abuyeva et al., 2018; Choudhury et al., 2011; Massalimov et al., 2018; Shankar et al., 2018) and used for the production of anticancer and antibacterial agents (Choudhury et al., 2012; Shankar et al., 2018).
SNPs can be synthesized using a variety of physicochemical and biological methods, such as i) acidification of sodium thiosulfate and stabilization of SNPs with surfactants or capping agents (Shankar et al., 2018; Urakaev et al., 1999) ii) acidification of different polysulfide solutions (Massalimov et al., 2018) and iii) the water in oil (W/O) micro-emulsification method, in which chemical reactions between sodium polysulfide and hydrochloric acid by reverse microemulsion using surfactants (Guo et al., 2006; Soleimani et al., 2013), iv) sublimation of sulfur and nucleation in poly(ethylene glycol) (PEG-200) (Xie et al., 2012), v) modification of sulfur surface in surfactant by mechanical and ultrasonic dispersing (Choudhury et al., 2011; Turganbay et al., 2013), vi) biological method by adding acidified plant extract to sodium thiosulfate to obtain monoclinic SNPs (Awwad et al., 2015; Khairan et al., 2019; Paralikar and Rai, 2017). The size and morphology of the SNPs are varied depending on the preparation method and raw materials of SNPs. Various allotropic forms of SNPs (α-sulfur and β-sulfur), can be produced with different sizes (Chodhury et al., 2011; Urakaev et al., 1999). The antimicrobial activity of SNPs is expected to depend not only on the shape and size of SNPs but also on the surfactants or capping agents used to stabilize SNP preparations (Choudhury et al., 2013a; Shankar et al., 2018). In addition, large-scale SNP production requires a cost-effective strategy that uses inexpensive resources. In that sense, production of SNPs using abundantly available elemental sulfur is recommended since sulfur is the most abundant inorganic non-metallic element on the earth's surface and >60 million tons of elemental sulfur (S0) are produced annually as a by-product in the petroleum refining process (Griebel et al., 2016).
To date, reports on the antimicrobial properties of SNPs are limited. In particular, studies on the comparison of antimicrobial properties of SNPs with different morphologies made by different methods are lacking in the literature. Previously, methods for synthesizing rod-shaped SNPs have been reported (Choudhury et al., 2013b; Xie et al., 2012), but these methods require complex microemulsion systems (Choudhury et al., 2013b) and a rather long process time (Xie et al., 2012). Besides, the diameter of the synthesized SNPs is>50 nm. In this study, a convenient and cost-effective method was used for the production of rod-shaped SNPs using elemental sulfur as the primary study for the development of mass-production methods for SNPs. And their properties were compared with spherical SNPs prepared by the chemical precipitation method of acidified sodium thiosulfate. Both types of SNPs were characterized using TEM, EDS, XRD, and UV–vis spectroscopy, and compared their antimicrobial activity.
2 Materials and methods
2.1 Materials
Sodium thiosulfate pentahydrate, sodium sulfide, and elemental sulfur were purchased from Sigma-Aldrich (St. Louis, MO, USA). Chitosan (CS-001, molecular weight: 180–200 kDa; viscosity: 110 cp in 1% acetic acid solution at 25 °C; the degree of deacetylation: 90%) was procured from Fine Agar Co., Ltd. (Damyang, Jeonnnam, Korea). Tryptic soy broth (TSB), brain heart infusion broth (BHI), and agar powder were purchased from Duksan Pure Chemicals Co., Ltd (Ansan, Gyeonggi-do, Korea). Yeast malt broth (YMB), and yeast malt agar (YMA) were obtained from KisanBio Co., Ltd. (Seoul, Korea). Potato dextrose broth (PDB), potato dextrose agar (PDA), and peptone water were purchased from Difco (Becton Dickinson, Sparks, MD, USA). YM 3 M Petri film was obtained from 3 M Corporation (St. Paul, MN, USA). Test microbial strains, Staphylococcus aureus (ATCC 13565, enterotoxin A), Listeria monocytogenes ATCC 15313, Escherichia coli O157: H7 (ATCC 11234), Candida albicans (ATCC 18804), and Aspergillus flavus (ATCC 22546), were obtained from the Korean Culture Center of Microorganisms (KCCM, Seoul, Korea).
2.2 Preparation of sulfur nanoparticles
Sulfur nanoparticles (SNPs) were prepared using elemental sulfur (S0) and sodium sulfide following the procedure shown in Fig. 1. For this, 20 mL of 1 M sodium sulfide was heated to 100 °C and followed by the addition of 1.6 g of elemental sulfur reacted for 1 h with vigorous stirring. The yellow color of elemental sulfur turned reddish-orange, indicating the formation of polysulfide and designated as SNPES (Guo et al., 2006; Paralikar and Rai, 2017; Soleimani et al., 2013). Then 40 mL of a 0.5 wt% chitosan solution (dissolved in 1% acetic acid) was added to the polysulfide solution, and an 18 wt% hydrochloric acid solution was added dropwise with stirring until the color of the solution turned into milky white. The formed nanoparticles were collected by precipitation using a centrifuge and washed three times with distilled water until the pH reached 7. The final product was dried at 50 °C and designated SNPES. For comparison, spherical SNPs were prepared using the acidification method of sodium thiosulfate according to the method described previously (Shankar et al., 2018; Chaudhuri and Paria, 2011). Briefly, 2.482 g of sodium thiosulfate pentahydrate was dissolved in 900 mL distilled water and added 100 mL of dilute HCl (0.2 M) with stirring. As described in previous reports (Shankar et al., 2018; Shankar and Rhim, 2018), for coating with chitosan, the precipitated SNPs were washed and dispersed in 50 mL of 0.5 wt% chitosan solution with stirring for 1 h. The SNPs were collected, washed until the pH reached 7, and dried as described above and designated SNPSTS (see Fig. 2).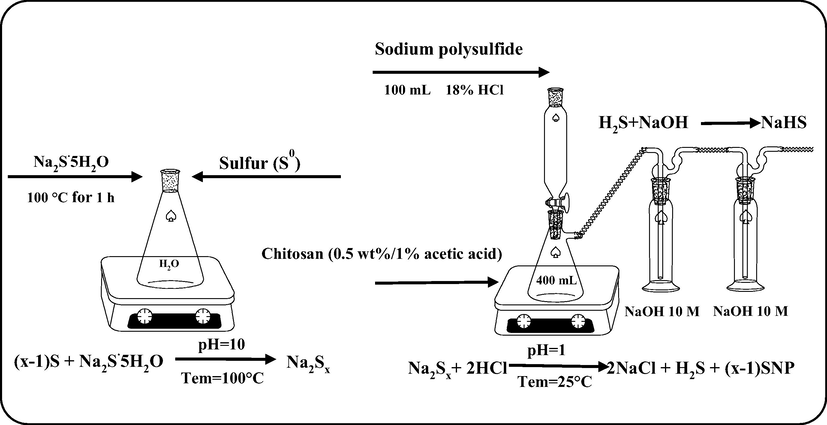
The schematic diagram for the preparation of sulfur nanoparticles from elemental sulfur.
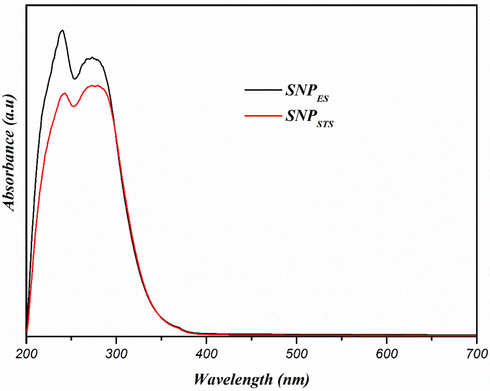
UV–vis spectra of sulfur nanoparticles.
2.3 Characterization
The UV–vis light absorption patterns of the SNPs were examined in the range of 200–700 nm using a UV–visible spectrophotometer (Mecasys Optizen POP Series UV/Vis, Seoul, Korea) to confirm the formation of SNPs.
The morphology such size and shape of the SNPs were evaluated using a transmission electron microscopy (TEM). For this, 10 μL of SNPs suspension in ethanol was dropped on the carbon-coated copper grid and allowed to dry. The morphological image was observed using the JEOL-1010 TEM instrument (Tokyo, Japan) operated at an accelerating voltage of 200 kV. The particle size distribution of the SNPs was performed using Image J software (ImageJ 1.46r, National Institute of Health, USA).
Elemental analysis of the SNPs was performed using energy-dispersive X-ray spectroscopy (EDX) equipped on SEM instrument (FE-SEM, S-4800, Hitachi Co., Ltd., Matsuda, Japan).
X-ray diffraction patterns of SNPs were investigated using an XRD diffractometer (PANalytical X’pert Pro MRD Diffractometer, Amsterdam, Netherlands). The spectra were recorded using Cu Kα radiation (wavelength of 0.1541 nm) and a nickel monochromator operated at the voltage of 40 kV and a current of 40 mA at diffraction angles in the range of 2θ = 5-80° with a scanning speed of 0.4°/min.
The thermal stability of the SNPs was evaluated using a thermogravimetric analyzer (Hi-Res TGA 2950, TA Instrument, New Castle, DE, USA). About 10 mg of each sample in a standard aluminum pan was heated from room temperature to 600 °C at a heating rate of 10 °C/min under a nitrogen flow rate of 50 cm3/min. Derivatives of TGA (DTG) were calculated using the central finite difference method.
The surface charge of NPs was evaluated by zeta potential and particle size using the dynamic light scattering (DLS) method using a Zeta PALS-zeta potential analyzer (Brookhaven Instruments Corporation, NY, USA).
2.4 Antibacterial activity of SNPs
The antibacterial activity of the SNPs for E. coli, S. aureus, L. monocytogenes, and C. albicans was investigated using a disk diffusion method. For this, 100 µL of microbial suspensions (∼106 CFU/mL) were inoculated by spreading on agar plates. Then, added 30 μL of each SNPs suspension (512 µg/mL) onto a sterile paper disc surface (Advantec 8 mm, Toyo Roshi Kaisha, Ltd., Tokyo, Japan), placed on the agar surface and incubated at 36 °C for 24 h. The zone of inhibition around the paper disc was measured using a caliper. For A. flavus, agar disc diffusion assay was performed by the same procedure as described above except for incubation temperature and time, which was incubated at 25 °C for 4 days to determine the zone of inhibition.
In addition, the minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of the SNPs for E. coli and L. monocytogenes were determined using a modified broth microdilution method (Shankar et al., 2018). For this, 100 μL of two-fold serially diluted SNPs were prepared in a 96-well microtiter plate to obtain final concentration ranged from 2 to 1024 μg/mL and added 50 μL of TSB and BHI media for E. coli and L. monocytogenes, respectively. 100 μL of the bacterial culture broth (∼106 CFU/mL) was added to each well and incubated the bacteria at 37 °C for 15 h, then added 10 µL of resazurin (0.4 mg/mL solution) and incubated for 3 h. The growth was assessed using a microtiter plate reader by measuring the optical density (OD) at 595 nm. The MIC was determined as the lowest concentration of the SNPs to inhibit the bacterial growth completely. The MBC was determined with the SNPs that gave MIC by subculturing on fresh TSB and BHI agar plates. The MIC and minimum fungicidal concentrations (MFC) of A. flavus were determined using the same procedure after incubation at 25 °C for 4 days.
3 Results and discussion
3.1 Characterization of SNPs
In the process of preparing SNPs, the yellow of elemental sulfur turned reddish-orange as polysulfide was formed, and then turned milky white as SNPs were formed. The formation of SNPs was also confirmed using a UV–visible spectrophotometer. The UV–visible spectra of both types of SNPs (SNPES and SNPSTS) are shown in Fig. 1. Both SNPs exhibited similar absorption patterns showing two maximum absorption peaks around 240 and 280 nm without light absorption of visible light above 400 nm. The absorption of UV light was due to the n → σ* transition of nonbonding electrons, and this property can be used as preliminary detection of the SNPs (Xie et al., 2012).
Shankar and Rhim (2018) also found two absorbance peaks of SNPs in chitosan-based nanocomposite films. Xie et al. (2012) observed two light absorption peaks of nano-sulfur sol prepared by dissolving sublimed sulfur in a solvent of PEG-200. They found a sharp peak centered at 218 nm and a weak shoulder at 260 nm. Shankar et al. (2018) reported a clear light absorption peaks of SNPs around 280 nm, but they missed the other absorbance peak at a lower wavelength since they scanned from 250 nm. The light absorption peaks around 240 and 280 nm confirms the formation of sulfur nanoparticle (Paralikar & Rai, 2017; Xie et al., 2012).
The morphology of SNPs prepared with different sulfur compound sources was observed using TEM, and the TEM image and size distribution of SNPs are shown in Fig. 3. The shape and size of the SNPs depended on the sulfur source and manufacturing method. The SNPs prepared by the acidification of sodium thiosulfate (SNPSTS) have a spherical shape with an average particle size of 17 nm, while the SNPs made with elemental sulfur (SNPES) have a rod-shape with an average length and diameter of 87 and 22 nm, respectively.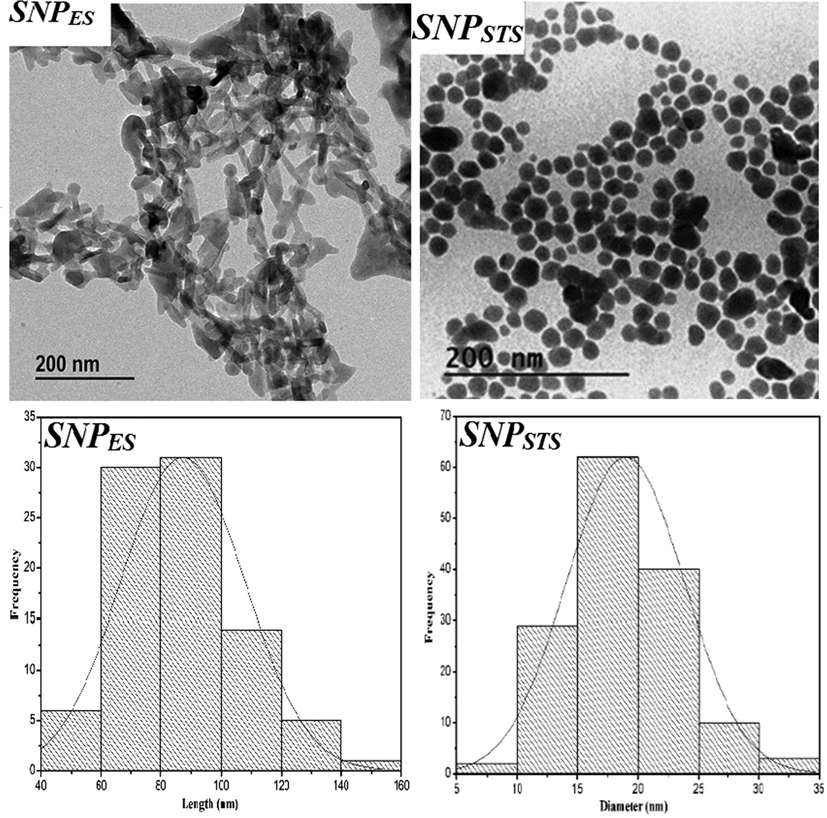
TEM images and size distribution of sulfur nanoparticles.
Also, the average particle size of both SNPs was determined by the dynamic light scattering (DLS), and the results are presented in Table 1. These values are significantly higher than the actual size determined by TEM. This is due to the fact that the DLS method determines the hydrodynamic radius of particles in a solution (Shankar et al., 2018). It has been reported that the shape of the SNPs varies depending on the manufacturing method. Spherical SNPs were obtained when the SNPs were prepared using sodium polysulfide in a microemulsion system (Guo et al., 2006), using sodium polysulfide in the presence of leaf extract (Paralikar & Rai, 2017), and using sodium polysulfide and ammonium polysulfide with polyethylene glycol-400 (PEG-400) as a surface stabilizing agents (Choudhury et al., 2011). On the contrary, Xie et al. (2012) obtained rod-shaped SNPs having a typical diameter of about 80 nm and an average aspect ratio of 6–8 by dissolving sublimed sulfur in a PEG-200 solvent. Choudhury et al. (2013b) also obtained rod-shaped SNPs having a diameter of around 50 nm using a water-in-oil microemulsion technique. In contrast, Massalimov et al. (2014) reported that the size of the SNPs obtained by acidification of polysulfide and sodium thiosulfate increased with increasing acid/precursor concentration ratio. The zeta potential values of SNPs are presented in Table 1. The positive value of SNP is due to the chitosan used for capping, which is cationic at pH = 7. They also reported that the type of acid affects the size of synthesized SNPs, and the size of the SNPs prepared using calcium polysulfide was>100 nm. Chaudhuri et al. (2011) reported that the presence of a surfactant decreased the size of SNPs effectively.
Sample
Particle size by TEM (nm)
Particle size by DLS (nm)
Zeta Potential (mV)
SNPES
87 nm
267.34 ± 24.3
34.3 ± 0.3
SNPSTS
17 nm
129.6 ± 15.2
32.6 ± 0.7
However, the SNPs synthesized without chitosan capping were very unstable and precipitated immediately, while the SNPs capped with chitosan formed stable suspension of SNPs. The chitosan adsorbed on the surface of SNPs contributes to the creation of a stabilized suspension of SNPs by inducing a net positive charge on the surface of SNPs and inducing electrostatic repulsion between synthesized particles. Thus, chitosan coating results in smaller and more stable SNP nanoparticles. It has been reported that spherical SNPs were prepared when sodium polysulfide and other stabilizers were used (Choudhury et al., 2011; Guo et al., 2006; Paralikar and Rai, 2017), but in this study, rod-shaped SNPs were obtained using chitosan as a stabilizing agent. Though the mechanism for the formation of rod-shaped SNPs in the presence of chitosan is not clear, capping the synthesized SNPs using chitosan formed smaller particle sizes with narrower size distribution (Shankar et al., 2018).
With regard to the various molecular structures and allotropes of sulfur, the allotropic form of the synthesized SNPs should be determined. XRD analysis was performed to determine the allotropic form of the SNPs, and the XRD patterns of elemental sulfur and the synthesized SNPs are shown in Fig. 4. The position and intensity of the diffraction peaks of the SNPs are in good agreement with the XRD patterns of SNPs having an orthorhombic (α) S8 structure of sulfur (Choudhury et al., 2013c; Xie et al., 2012). However, the allotropic forms of SNPs may vary depending on the sulfur source and preparation method of SNPs. Choudhury et al. (2013c) synthesized SNPs with different allotropic forms of sulfur. They reported that PEG-stabilized SNPs had a rhombic (α), whereas Span-80/Tween-80 coated SNPs showed a monoclinic (β) allotrope. SNPs from line broadening of the most intense diffraction peaks corroborates their TEM size distribution. The crystallite size of elemental sulfur, SNPSTS, and SNPES from line broadening of the most intense diffraction peak was determined using the Scherrer equation were 11.64, 12.88, and 17.33 nm, respectively. Soleimani et al. (2013) reported a simple preparation of SNPs with a crystallite size of 22 nm, while Deshpande et al. (2008) reported the preparation of SNPs with the crystallite size of 10 nm. Khairan et al. (2019) reported that the presence of garlic extract decreased the crystallite size of synthesized SNPs. The crystallite size of SNPSTS and SNPES determined by the Scherrer equation is in good agreement with the average diameter of those determined from the TEM results. The difference in the intensity of diffraction peaks at 2θ = 23.7° results from the difference in (2 2 0) crystal plane of sulfur (Falentin-Daudré et al., 2017).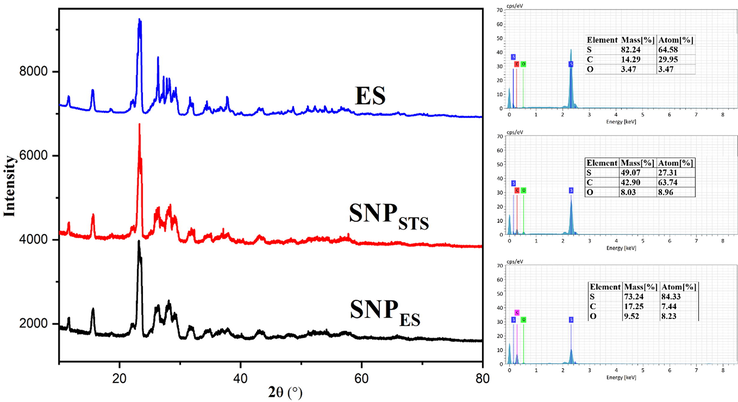
XRD patterns and EDX spectra of elemental sulfur and sulfur and sulfur nanoparticles.
The results of the elemental analysis of the SNPs and elemental sulfur determined using EDS are also shown in Fig. 4. Elemental sulfur (ES) showed the typical peak of sulfur at around 2.4 keV. In addition to the sulfur peak, both SNPs showed additional oxygen peaks due to the presence of chitosan on the surface of the prepared SNPs (Shankar et al., 2018). Also, an intense carbon peak in the EDS spectrum of SNPES implies that a higher amount of chitosan was bound at the SNP surface. The higher amount of chitosan on the SNPES surface was due to the presence of the chitosan molecule used as a stabilizer during SNPES formation. Chitosan not only prevents aggregation by stabilizing SNPs but also provides hydrophilicity to SNPs, helping to create a more stable SNP suspension.
The thermal stability of the SNP was evaluated using thermogravimetric analysis (TGA), and the results of the TGA and DTG thermograms are shown in Fig. 5. Previously, Shankar et al. (2018) found that the thermal stability of SNPs was not affected by the chitosan coating, so the TGA test of the uncoated SNPSTS was not performed. Both SNPs (SNPSTS and SNPES) showed similar thermal degradation behavior of thermal degradation, but the thermal degradation temperature was slightly different. The initial weight loss of SNPSTS occurred at 80–100 °C due to the evaporation of moisture (Shankar et al., 2018). However, SNPES did not show weight loss below 100 °C, possibly due to the complete dehydration of moisture during the drying process. Perhaps the tubular form of SNPES promoted the easier removal of water vapor during the drying process. The main thermal degradation of SNPSTS took place in the range of 210–302 °C with the maximum thermal degradation at 282 °C, and that of SNPES occurred in the range of 210–324 °C with the maximum thermal degradation at 306 °C. The main thermal degradation of the SNPs is presumably due to the partial degradation of sulfur (Shankar et al., 2018). The TGA results indicated that SNPES was slightly more thermostable than SNPSTS.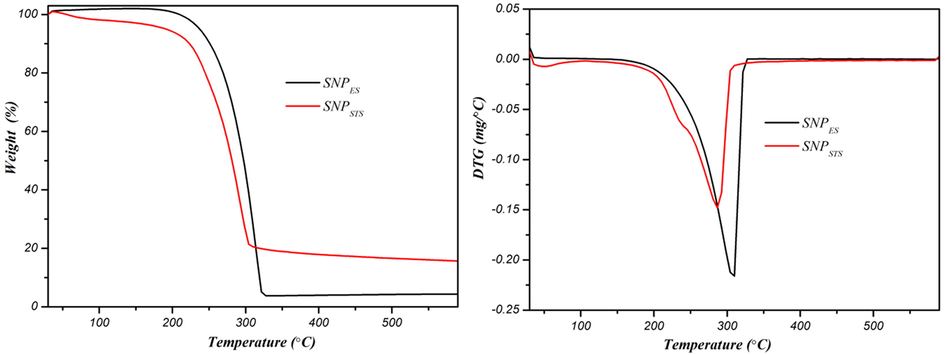
TGA and DTG thermograms of sulfur nanoparticles.
3.2 Antimicrobial activity
The results of the antimicrobial activity of the SNPs against E. coli, L. monocytogenes, S. aureus, A. flavus, and C. albicans determined by the disk diffusion method and their MIC/MBC(MFC) values are shown in Table 2. The disk diffusion test results for E. coli and L. monocytogenes showed that both types of SNPs did not show growth inhibition for E. coli, but showed significant growth inhibition of L. monocytogenes (Fig. 6). Shankar et al. (2018) compared the antibacterial activity of chitosan-capped SNPs (SNPSTS) with the SNPs without chitosan capping, and they found that the SNPSTS showed more potent antibacterial activity than the SNPs without capping. So, the antibacterial activity of chitosan-capped SNPs was compared with the SNPES in this study. Between the types of SNPs, SNPES showed a more significant inhibition zone of growth of L. monocytogenes. Consistently, the MIC/MBC test results showed similar results of the antibacterial activity of the SNPs as the disk diffusion test. All the test results of the disk diffusion and MIC/MBC analysis indicated that both types of SNPs did not show a significant antibacterial effect against the Gram-negative (E. coli) bacteria.
Zone of inhibition (mm)
MIC/MBC (MFC) (μg/mL)
E. coli
L. monocytogenes
S. aureus
A. flavus
C. albicans
E. coli
L. monocytogenes
S. aureus
A. flavus
C. albicans
SNPES
0.0 ± 0.0
18.8 ± 0.4
13.0 ± 0.0
15.3 ± 0.7
22.6 ± 0.6
>1024/>1024
64/128
64/128
256/1024
256/1024
SNPSTS
0.0 ± 0.0
15.2 ± 0.7
14.7 ± 0.6
13.0 ± 2.0
17.3 ± 0.6
>1024/>1024
256/512
64/128
256/1024
256/1024
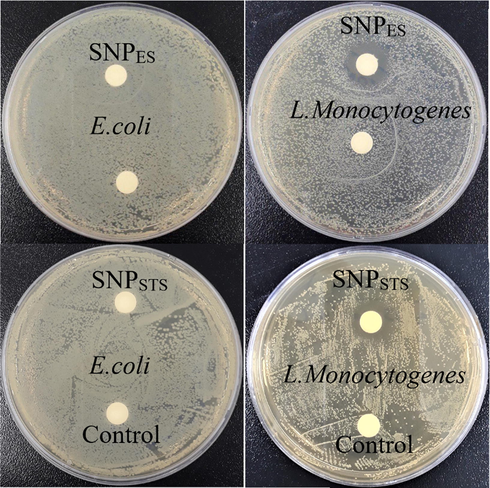
Antibacterial activity of SNPs.
On the contrary, they showed a substantial effect on the Gram-positive bacteria. They also indicated that SNPES exhibited more potent antibacterial activity against L. monocytogenes than SNPSTS. It has been reported so far that there is controversy over the antibacterial effect of SNPs against Gram-positive and Gram-negative bacteria.
Shankar et al. (2018) found that the SNPs produced by acidification of sodium thiosulfate and capping with chitosan exhibited strong antibacterial activity against E. coli (Gram-negative) and Staphylococcus aureus (Gram-positive) with MIC/MBC values of 16/64 and 16/32 μg/mL, respectively. Paralikar et al. (2017) also found that SNPs synthesized using leaf extracts from medicinal plants showed a similar degree of antibacterial activity against E. coli and S. aureus with a MIC of 200 µg/mL. Choudhury et al. (2013b) reported that rod-shaped monoclinic sulfur nanoparticles (β-SNPs) inhibited the growth of S. aureus at 576 µg/mL, while inhibited the growth of E. coli and K. pneumoniae at 2304 µg/mL. On the other hand, Libenson et al. (1953) reported that the micro-sized elemental sulfur did not show any antibacterial activity against E.ocli. Suleiman et al. (2015) reported that SNPs showed significant antibacterial activity against Gram-positive bacteria (S. aureus), but they did not show any activity against Gram-negative bacteria (E. coli and P. aeruginosa) even at high concentration of 800 µg/mL. They explained that Gram-negative bacteria are less vulnerable to SNPs due to the protection by the outer membrane of Gram-negative bacteria. The change in morphology and membrane integrity induced by the SNPES was observed using FE-SEM, as shown in Fig. 7. By treatment with SNPES, changes in the morphological structure of the microorganisms appeared in different ways. After treatment with SNPES, the cell wall of S. aureus was destroyed entirely, but the cell wall of E. coli was shrinking without destruction. This may explain why Gram-negative bacteria (E. coli) are more resistant to SNPs. The SNP treatment destroyed spores and mycelium of A. flavus and intact spherical cell buds of C. albicans.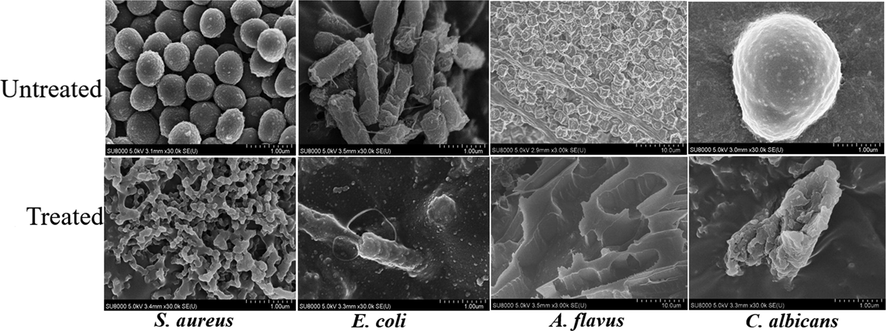
FE-SEM images of microorganisms before and after treatment with SNPES.
Though the exact mechanism of antibacterial action of SNPs has not been yet elucidated, several probable antimicrobial mechanisms of SNPs have been proposed, such as interactions with biological molecules on the surface of bacteria resulting in the membrane rupture and lysis of the cell membrane, the production of toxic H2S gas that reacts with thiol groups of proteins and lipids, and interaction with DNA resulting in denaturation of DNA and cell death (Rai et al., 2016). Previously, Libenson et al. (1953) explained that the antimicrobial activity of elemental sulfur in micro-size was due to the binding of elemental sulfur to an enzyme containing a -SH functional group, but they did not observe the micro-sized elemental sulfur attached to the enzyme. However, Shankar et al. (2018) demonstrated that the stabilized SNPs capped with chitosan (SNPSTS) exhibited more potent antibacterial activity than the non-stabilized SNPs. In this study, a rod-shaped orthorhombic α-SNP (SNPES) showed stronger antibacterial activity than the spherical orthorhombic α-SNP one (SNPSTS). On the other hand, Choudhury et al. (2013c) reported that a spherical α-SNPs hadd stronger antimicrobial activity than tetrapod-like β-SNPs. They reported that the MIC values of α-SNPs were lower than those of β-SNPs. In the case of fungi, the interaction of SNPs with cellular lipids may reduce the total lipid contents to alter the cellular structure, leading to the destruction of the fungal cell (Choudhury et al., 2012; Roy Choudhury et al., 2012). Since the antimicrobial activity of SNPs varies depending not only on the properties of the SNPs such as size, shape, source of sulfur, and preparation method but also on the type of target microorganisms and solvent for dissolving SNPs, further research is needed to draw conclusions on this issue.
4 Conclusions
Rod-shaped orthrombic sulfur nanoparticles (α-SNPs) were synthesized using elemental sulfur and sodium sulfide for the large-scale production of sulfur nanoparticles (SNPES), and their properties were compared with SNPs prepared by the acidification of sodium thiosulfate (SNPSTS). Except for shape and size, the SNPES showed similar characteristics to the SNPSTS. However, the SNPES showed higher thermal stability and antimicrobial activity against L. monocytogenes than the SNPSTS. The SNPES manufacturing method can be applied to large-scale production of functional SNPs using an abundantly available sulfur source.
Acknowledgement
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2019R1A2C2084221).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Preparation of ointment materials based on sulfur nanoparticles in water-soluble polymers. Mater. Today Proc.. 2018;5:22894-22899.
- [Google Scholar]
- Novel approach for synthesis sulfur (S-NPs) nanoparticles using Albizia julibrissin fruits extract. Adv. Mat. Lett. 2015;6:432-435.
- [Google Scholar]
- Growth kinetics of sulfur nanoparticles in aqueous surfactant solutions. J. Colloid Interface Sci.. 2011;354:563-569.
- [Google Scholar]
- Colloidal stability and coagulation kinetics study of different sized sulphur nanoparticles. J. Exp. Nanosci.. 2013;8:267-272.
- [Google Scholar]
- Inhibitory effects of sulfur nanoparticles on membrane lipids of Aspergillus niger: a novel route of fungistasis. Curr. Microbiol.. 2012;65:91-97.
- [Google Scholar]
- Surface-modified sulfur nanoparticles: an effective antifungal agent against Aspergillus niger and Fusarium oxysporum. Appl. Microbiol. Biotechnol.. 2011;90:733-743.
- [Google Scholar]
- Evaluation of physicochemical properties, and antimicrobial efficacy of monoclinic sulfur-nanocolloid. J. Nanoparticle Res.. 2013;15:1491.
- [Google Scholar]
- Investigation of antimicrobial physiology of orthorhombic and monoclinic nanoallotropes of sulfur at the interface of transcriptome and metabolome. Appl. Microbiol. Biotechnol.. 2013;97:5965-5978.
- [Google Scholar]
- Sulfur nanoparticles synthesis and characterization from H2S gas, using novel biodegradable iron chelates in aqueous surfactant systems. Arch: MRS Online Proc. Libr; 2008. p. :1103.
- Highly crystalline sphere and rod-shaped TiO2 nanoparticles: A facile route to bio-polymer grafting. Front. Lab. Med.. 2017;1:217-223.
- [Google Scholar]
- Polymerizations with elemental sulfur: A novel route to high sulfur content polymers for sustainability, energy and defense. Prog. Polym. Sci.. 2016;58:90-125.
- [Google Scholar]
- Preparation and characterization of monoclinic sulfur nanoparticles by water-in-oil microemulsions technique. Powder Technol.. 2006;162:83-86.
- [Google Scholar]
- Khairan, K., Zahraturriaz, Jalil, Z., 2019. Green synthesis of sulphur nanoparticles using aqueous garlic extract (Allium sativum). Rasayan J. Chem. 12, 50–57. https://doi.org/10.31788/RJC.2019.1214073
- Antibacterial Effect of Elemental Sulfur. J. Infect. Dis.. 1953;93:28-35.
- [CrossRef] [Google Scholar]
- Chemical precipitation of sulfur nanoparticles from aqueous solutions. Russ. J. Appl. Chem.. 2014;87:700-708.
- [Google Scholar]
- Coprecipitation of nanocomposites based on colloidal particles of sulfur and carbonates of alkaline-earth metals from polysulfide solutions. Colloid J.. 2018;80:407-417.
- [Google Scholar]
- Bio-inspired synthesis of sulphur nanoparticles using leaf extract of four medicinal plants with special reference to their antibacterial activity. IET Nanobiotechnology. 2017;12:25-31.
- [Google Scholar]
- Sulfur in human nutrition and applications in medicine. Altern. Med. Rev.. 2002;7:22-44.
- [Google Scholar]
- Sulfur and sulfur nanoparticles as potential antimicrobials: from traditional medicine to nanomedicine. Expert Rev. Anti. Infect. Ther.. 2016;14:969-978.
- [Google Scholar]
- Use of sulfur nanoparticles as a green pesticide on Fusarium solani and Venturia inaequalis phytopathogens. RSC Adv.. 2013;3:10471-10478.
- [Google Scholar]
- Polyethylene glycol-stabilized sulphur nanoparticles: an effective antimicrobial agent against multidrug-resistant bacteria. J. Antimicrob. Chemother.. 2012;67:1134-1137.
- [Google Scholar]
- Preparation of sulfur nanoparticles and their antibacterial activity and cytotoxic effect. Mater. Sci. Eng. C. 2018;92:508-517.
- [Google Scholar]
- Preparation of sulfur nanoparticle-incorporated antimicrobial chitosan films. Food Hydrocoll.. 2018;82:116-123.
- [Google Scholar]
- A new and simple method for sulfur nanoparticles synthesis. Colloid J.. 2013;75:112-116.
- [Google Scholar]
- Synthesis of nano-sized sulfur nanoparticles and their antibacterial activities. J. Mater. Environ. Sci.. 2015;6:513-518.
- [Google Scholar]
- Applications and challenges of elemental sulfur, nanosulfur, polymeric sulfur, sulfur composites, and plasmonic nanostructures. Crit. Rev. Environ. Sci. Technol.. 2019;49:2314-2358.
- [Google Scholar]
- Surface-modification of sulfur nanoparticles with surfactants and application in agriculture. Advanced Materials Research. Trans Tech Publ 2013:475-479.
- [Google Scholar]
- Kinetics of homogeneous nucleation of monodisperse spherical sulphur and anatase particles in water–acid systems. J. Cryst. Growth. 1999;205:223-232.
- [Google Scholar]
- Facile synthesis, spectral properties and formation mechanism of sulfur nanorods in PEG-200. Mater. Res. Bull.. 2012;47:3665-3669.
- [Google Scholar]
- Sulfur and selenium antioxidants: Challenging radical scavenging mechanisms and developing structure–activity relationships based on metal binding. J. Inorg. Biochem.. 2015;145:30-40.
- [Google Scholar]







