Translate this page into:
Association of genetic variations in Toll-Like receptor 3 with Acute Lymphoblastic leukemia
⁎Corresponding author. Ralonezan@ksu.edu.sa (Rasha Alonaizan),
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Objectives
This study investigates the association between specific SNPs in the Toll-like receptor 3 (TLR3) gene and susceptibility to Acute Lymphoblastic Leukemia (ALL) in the Saudi population, aiming to clarify genetic influences on ALL risk.
Methods
We evaluated four SNPs of TLR3 (rs3775296 C/A, rs5743312 C/T, rs3775291 C/T, and rs3775290 C/T) for their association with ALL. The allelic and genotypic frequencies of these SNPs were compared between 150 patients and 115 control subjects in this case-control study. All participants were genotyped using TaqMan PCR techniques. Additionally, mRNA expression levels were assessed in patients with ALL and matched healthy individuals using Real-Time Quantitative Reverse Transcription PCR (qRT-PCR).
Results
Significant differences in genotype frequencies were observed for the TLR3 SNPs rs5743312 C/T and rs3775290 C/T between the ALL patients and control subjects. No significant differences were detected for rs3775296 C/A and rs3775291 C/T. Notably, the rs5743312 SNP was associated with an increased risk of ALL, while rs3775290 was associated with a decreased risk in the Saudi population.
Conclusions
Our study demonstrates significant associations of the TLR3 SNPs rs5743312 C/T and rs3775290 C/T with ALL in the Saudi population. These polymorphisms could be pivotal in developing genetic screening tools and enhancing risk prediction strategies for ALL.
Keywords
Acute Lymphoblastic Leukemia
Single nucleotide polymorphisms
Toll-like receptor
- SNPs
-
Single Nucleotide Polymorphisms
- TLR3
-
Toll-like receptor 3
- ALL
-
Acute Lymphoblastic Leukemia
- HSCs
-
Hematopoietic Stem Cells
Abbreviations
Data availability
Data will be made available on request.
1 Introduction
Acute Lymphoblastic Leukemia (ALL) represents one of the most prevalent malignant disorders globally, characterized by a complex interplay of genetic and environmental factors. The hallmark of ALL is a rapid proliferation of abnormal leukocytes, which accumulate in the bone marrow and interfere with the production of normal white blood cells (Tebbi, 2021).
Research has consistently shown that ALL involves significant dysregulation of the immune system, particularly evident during early childhood development. Furthermore, polymorphisms in genes related to innate immunity are believed to play critical roles in initiating intrinsic biological changes that predispose individuals to cancer (Monlish et al., 2016).
A key component in understanding the pathogenesis of ALL is the role of Toll-like receptors (TLRs). These receptors, integral to the innate immune response, identify pathogen-associated and damage-associated molecular patterns. TLRs are not only crucial in immune response mechanisms but also in cancer biology. They are expressed across a variety of cells, including hematopoietic stem cells, progenitor cells, and effector immune cells, as well as endothelial cells. Their involvement in cancer development is well-documented, linking aberrant TLR signaling to a spectrum of hematological malignancies, including lymphoproliferative disorders and myelodysplastic syndromes, which possess a high risk of transformation into leukemia (Gao et al., 2019; Sallustio et al., 2019; Monlish et al., 2016).
Toll-like receptor 3 (TLR3) has been targeted as an immunotherapy for different types of cancer. Located in endosomes and plays a vital role in recognizing viral double-stranded RNA, a key step in viral infection responses, and triggers the production of type I interferon (Le Naour et al., 2020). Emerging evidence suggests that TLR3′s expression in tumor cells—or its activation within the tumor microenvironment—may paradoxically either inhibit or promote tumor progression (Zheng et al., 2021). Moreover, research indicates that genetic variations in TLR3 may affect the protein's expression and function, influencing susceptibility to cancer progression (Meng et al., 2020).
In this context, our case-control study has identified potential susceptibility genes that support a polygenic model of cancer predisposition based on genetic and gene expression variations. These insights could prove crucial for understanding tumor aggressiveness and developing prognostic markers for leukemia and other cancers.
2 Materials and methods
2.1 Criteria for sample selection
Human whole blood samples from a total of 265 individuals were collected for the study. Gender and age-matched controls and cases were selected. The study included 150 patients diagnosed with ALL without any other known pathologies, hematological disorders, or previous cancer. Additionally, 115 unrelated healthy individuals (both female and male) without any clinical signs of cancer or other diseases served as controls. The characteristics of the ALL patients and control subjects are shown in Table 1.
Patient
Control
P-value
Number of subjects
mean ± St.Dev
Min
Max
Number of subjects
Mean ± St.Dev
Min
Max
150
22.45 ± 20.27
1
80
115
18.68 ± 15.53
2
85
P=0.10
2.2 Ethics approval
The study was approved by the medical ethics committee at King Khalid University Hospital and the ethics committee of King Saud University, Riyadh, Saudi Arabia (Ref. No. 20/0525/IRB).
2.3 Blood sample collection
Blood samples were collected by venipuncture in the presence of an anticoagulant, ethylenediaminetetraacetic acid (EDTA) from all subjects. Genomic DNA was isolated from whole blood using the commercial kit, DNeasy Blood & Tissue Kit (QIAGEN).
2.4 Single nucleotide polymorphisms and genotyping
In this research, we identified Four SNPs in TLR3 gene (rs3775296 C/A, rs5743312 C/T, rs3775291 C/T, and rs3775290 C/T) in all subjects by the TaqMan allelic discrimination through the dbSNP databases (https://www.ncbi.nlm. nih.gov/snp/). SNP were selected according to their minor allele frequency (MAF) ≥ 5 %.; Hardy-Weinberg equilibrium (HWE) P value cut-off > 0.005. TLR3 genotyping of the Four SNPs was performed by VIC- and FAM-labelled allelic discrimination method, using assay-on-demand TaqMan assays ordered from Applied Biosystems according to the manufacturer's instructions using ViiA™7, v.1.1. real-time PCR (Applied Biosystems, USA).
Real-time PCR was implemented in 10 μl a reaction system containing 0.26 μl 2x SNP Genotyping Assay, 5.5 μl 2xPower Taq Master Mix, 2.24 μl Nuclease-Free Water, and 2 μl DNA template (100 ng/μl). The PCR conditions for all SNPs were 1 cycle at 95 ˚C for 10 min followed by 40 cycles (95 ˚C for 15 s, 55 ˚C for 30 s, and 72 ˚C for 30 s) and a final extension at 72 ˚C for 5 min. For confirmation, about 5 % of the samples were randomly chosen for repeat genotyping.
2.5 mRNA relative quantification by quantitative PCR
A Reverse Transcription System Kit (Promega, Madison, USA) was used for cDNA synthesis and genomic DNA elimination. Quantitative PCR (qPCR) reactions were performed using TaqMan® gene expression assay (ThermoFisher) specific probes for TLR3 gene (Catalog # 4,331,182 and assay ID: Hs00152933_m1). For the reference gene, GAPDH was used as the normalizing endogenous control and its relative expression was performed using TaqMan® gene expression assay (Catalog #4331182 and Assay ID: Hs02758991_g1) (ThermoFisher). The reactions were performed in triplicate, using 25 ng of cDNA in Quantastudio flex ViiA™7 Real-Time PCR (Applied Biosystems, USA). The program of amplification was an initial denaturation at 95 ˚C for 5 min followed by 40 cycles of denaturation at 95 ˚C for 15 s and annealing at 60 ˚C for 60 s and 72 ˚C for 30 s, and a final extension for 10 min at 72 ˚C. The relative quantitation of the gene expression was calculated as the fold-change relative to the average expression of the controls using the comparative CT method (the 2−ΔΔCt method). Statistical analysis was performed using GraphPad Prism 9.
2.6 Statistical analysis
The control data was assessed for The Hardy-Weinberg equilibrium test to detect any deviation in the control samples. The chi-square analysis was used to compare Genotype distribution and allele frequency differences across groups. SNP genotypes were coded into three groups: homozygous (ancestral allele), heterozygous, and homozygous (minor allele) to assess the minor allele frequency (MAF), Odds ratios (OR) with 95 % confidence intervals (CI) were calculated to estimate the strength of the associations. The cutoff for significance was a two-tailed p-value ≤ 0.05. All statistical calculations were performed using SPSS version 22 (SPSS Inc., Chicago, IL, USA).
3 Results
3.1 Association between TLR3 (rs3775296 C/A) gene variant in patients with ALL and control group
Frequencies of the TLR3 rs3775296 CC (wild type), CA (heterozygous), and AA (polymorphic homozygous) genotypes were 85.3 %, 12 %, and 2.6 % in ALL patients, and 86.9 %, 10.4 %, and 2.6 % in the healthy control group, respectively. Moreover, the frequency of the TLR3 rs3775296 C allele was 0.91 % and 0.92 % in the ALL patient and control groups, respectively. The genotype and allele frequencies of the rs3775296 in TLR3 gene in ALL patients and the control group are shown in Table 2. However, a statistically significant difference in the distribution of TLR3 rs3775296 genotypes, as well as in allele frequencies between the patient and control groups was not detected. The genotype and allele frequency did not differ between the ALL patients and the control group for all genetic models (p ˃.05). Abbreviations; OR, odds ratio; CI, confidence interval, n, number of individuals; Boldfaced values indicate a significant difference at the P≤0.05 level.
Locus
Model
Genotype
ALL %
n = 150
Control%
n = 115
OR (95 % CI)
P-value
AIC
rs3775296 C/A
Alleles
C
0.91
0.92
1
0.75
A
0.09
0.08
1.1176 (0.59––2.09)
Codominant
CC
128 (85.3 %)
100 (86.9 %)
1
534.8
CA
18 (12 %)
12 (10.4 %)
0.853 (0.39–1.85)
0.68
AA
4 (2.6 %)
3 (2.6 %)
0.960 (0.21–4.38)
0.95
Dominant
CC
128 (95.3 %)
100 (87 %)
1
0.38
535.8
CA-AA
22 (14.7 %)
15 (13 %)
2.55 (0.30–21.70)
Recessive
CC-CA
146 (97.3 %)
112 (97.4 %)
1
0.17
534.6
AA
4 (2.7 %)
3 (2.6 %)
0.00 (0.00-NA)
Over-dominant
CC-AA
132 (88 %)
103 (89.6 %)
1
0.17
534.7
CA
18 (12 %)
12 (10.4 %)
5.15 (0.45–59.07)
Log-additive
−--
−--
−--
1.42 (0.25–8.20)
0.69
536.4
rs5743312 C/T
Alleles
C
0.84
0.91
1
0.02
T
0.16
0.09
1.896 (1.10–3.26)
Codominant
CC
109 (72.7 %)
96 (83.5 %)
1
536.5
CT
34 (22.7 %)
17 (14.8 %)
1.761 (0.92–3.35)
0.08
TT
7 (4.7 %)
2 (1.7 %)
3.083 (0.62–15.19)
0.14
Dominant
CC
109 (72.7 %)
96 (83.5 %)
1
0.84
536.5
CT-TT
41 (27.3 %)
19 (16.5 %)
0.76 (0.05–11.26)
Recessive
CC-CT
143 (95.3 %)
113 (98.3 %)
1
0.17
534.6
TT
7 (4.7 %)
2 (1.7 %)
0.00 (0.00-NA)
Over-dominant
CC-TT
116 (77.3 %)
98 (85.2 %)
1
0.72
536.4
CT
34 (22.7 %)
17 (14.8 %)
1.71 (0.09–33.64)
Log-additive
−--
−--
−--
0.53 (0.06–4.94)
0.57
536.2
rs3775291 C/T
Alleles
C
0.79
0.75
1
0.34
T
0.21
0.25
0.823 (0.55–1.24)
Codominant
CC
95 (63.3 %)
66 (57.4 %)
1
CT
46 (30.7 %)
41 (35.7 %)
0.779 (0.46–1.31)
0.35
533.2
TT
9 (6 %)
8 (7 %)
0.782 (0.28–2.13)
0.62
Dominant
CC
95 (63.3 %)
66 (57.4 %)
1
0.23
535.1
CT-TT
55 (36.7 %)
49 (42.6 %)
4.81 (0.32–72.06)
Recessive
CC-CT
141 (94 %)
107 (93 %)
1
0.16
534.6
TT
9 (6 %)
8 (7 %)
0.00 (0.00-NA)
Over-dominant
CC-TT
104 (69.3 %)
74 (64.3 %)
1
0.056
532.9
CT
46 (30.7 %)
41 (35.6 %)
20.45 (0.58–725.43)
Log-additive
−--
−--
−--
1.75 (0.24–12.76)
0.58
536.2
rs3775290 C/T
Alleles
C
0.75
0.69
1
T
0.25
0.31
0.733 (0.50–1.07)
0.11
Codominant
CC
89 (59.3 %)
58 (50.4 %)
1
CT
48 (32 %)
43 (37.4 %)
0.727 (0.43–1.23)
0.23
532.5
TT
13 (8.7 %)
14 (12.2 %)
0.01 (0.00–0.70)
0.04
Dominant
CC
89 (59.3 %)
58 (50.4 %)
1
0.04
532.4
CT-TT
61 (40.7 %)
57 (49.6 %)
0.07 (0.00–1.20)
Recessive
CC-CT
137 (91.3 %)
101 (87.8 %)
1
0.81
536.5
TT
13 (8.7 %)
14 (12.2 %)
0.64 (0.02–23.62)
Over-dominant
CC-TT
102 (68 %)
72 (62.6 %)
1
0.01
530.6
CT
48 (32 %)
43 (37.4 %)
0.01 (0.00–0.69)
Log-additive
−--
−--
−--
0.28 (0.04–1.75)
0.14
534.4
3.2 Association between TLR3 (rs5743312 C/T) gene variant in patients with ALL and control group
Frequencies of the TLR3 rs5743312 CC (wild type), CT (heterozygous), and TT (polymorphic homozygous) genotypes were 72.7 %, 22.7 %, and 4.7 % in ALL patients, and 83.5 %, 14.8 %, and 1.7 % in the healthy control group, respectively. Moreover, the frequency of the TLR3 rs5743312 C allele was 84 % and 91 % in the ALL patient and control groups, respectively. Moreover, a statistically significant difference in the distribution of TLR3 rs3775291 genotypes of the five different genetic models between the patient and control groups was not detected (p ˃ 0.05). However, carriers of the minor allele T were at significantly increased risk of ALL compared with carriers of the C allele (OR: 1.896; χ2 = 5.43; CI: 1.100–3.268; p = 0.02). Table 2 shows the obtained genotype and allele frequencies and the significance of the genotype and allele distribution of the tested SNP.
3.3 Association between TLR3 (rs3775291 C/T) gene variant in patients with ALL and control group
Frequencies of the TLR3 rs3775291 CC (wild type), CT (heterozygous), and AA (polymorphic homozygous) genotypes were 63.3 %, 30.7 %, and 6 % in ALL patients, and 57.4 %, 35.7 %, and 7 % in the healthy control group, respectively. Moreover, the frequency of the TLR3 rs3775291 C allele was 79 % and 75 % in the ALL patient and control groups, respectively. The genotype and allele frequencies of the rs3775291 in TLR3 gene in ALL patients and the control group are shown in Table 2.
However, statistically significant differences in the distribution of TLR3 rs3775291 genotypes and allele frequencies between the patient and control groups for all genetic models were not detected. The genotype and allele frequency did not differ between the ALL patients and the control group. Except for the Over-dominant model was borderline significantly associated with increased risk of ALL (P=0.056) but did not reach significant.
3.4 Association between TLR3 (rs3775290 C/T) gene variant in patients with ALL and control group
Frequencies of the TLR3 rs3775290 CC (wild type), CT (heterozygous), and TT (polymorphic homozygous) genotypes were 59.3 %, 32 %, and 8.7 % in ALL patients, and 50.4 %, 37.4 %, and 12.2 % in the healthy control group, respectively. Moreover, the frequency of the TLR3 rs3775290 C allele was 0.75 % and 0.69 % in the ALL patient and control groups, respectively. The genotype and allele frequencies of the rs3775290 in TLR3 gene in ALL patients and the control group are shown in Table 2.
A statistically significant difference in the distribution of TLR3 rs3775290 genotypes under co-dominant, dominant as well and over-dominant models between the patient and control groups was detected, CT vs. CC (OR: 0.01; CI: 0.00–0.70; p = 0.04), CT/TT vs. CC (OR: 0.07; CI: 0.00–1.20; p = 0.04), CT vs. CT/TT (OR: 0.01; CI: 0.00–0.69; p = 0.01) respectively. For the significant genetic models, the Over-dominant model was the best fit (AIC=530.6). However, the allele frequency did not differ between the ALL patients and the control group (p = 0.352).
3.5 Association of TLR3 relative mRNA expression and ALL
TLR3 mRNA expression was assessed in healthy and diseased subjects. In comparison to the control group, TLR3 mRNA expression was higher in ALL patients than in healthy individuals. In summary, the Δ Ct mean of TLR3 expression in the ALL patients were 8.541 ± 0.348, and for the healthy donors was 11.444 ± 0.482, P- value = 0.001 (Table 3). That was also confirmed by the 2^-ΔΔ Ct method (fold change: 8.9029 ± 1.7049, p-value: 0.0001) (Fig. 1). Mood median test for TLR3, Data are shown as mean ± SE (standard Error).
Control
Patient
P- value
Δ Ct TLR3
11.444 ± 0.482
8.541 ± 0.348
17.3
0.001
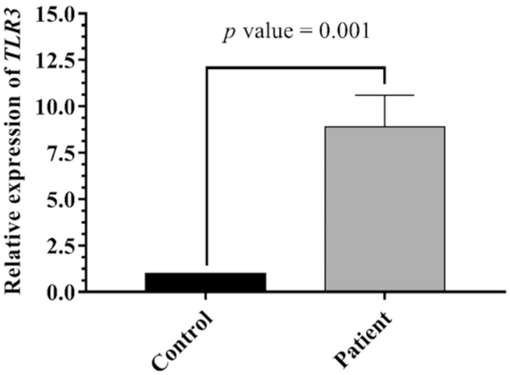
TLR3 expression level in ALL and control. TLR3 level varied significantly between patients and controls (p = 0.001).
4 Discussion
To date, no studies have explored the potential impact of TLR3 variants on the risk of blood cancers in the Saudi population. This study is the first to investigate several SNPs within TLR3 that have not been previously assessed in the context of leukemia. Initially, we analyzed the genotypic distribution of four TLR3 SNPs (rs3775296, rs5743312, rs3775291, and rs3775290) in Saudis and compared these distributions to those reported in other populations.
The minor allele (A) of rs3775296 in healthy Saudi individuals had a frequency of 0.08, the lowest recorded when compared to the global populations reported in the 1000 Genomes Project. In contrast, the major allele (C) had a frequency of 0.92, the highest among the surveyed groups. Notably, populations such as African Caribbeans in Barbados, Esan in Nigeria, and Japanese in Tokyo exhibited higher frequencies of the A allele.
Significantly, TLR3 rs3775296 displayed a distinct allele distribution between ALL patients and healthy controls, particularly in individuals diagnosed after the age of 18, indicating a more than ninefold increase in ALL risk associated with this allele in the Saudi population. This SNP's association with various diseases has been sporadically reported. For instance, a study by Fan et al. (2019) linked rs3775296 to an increased risk of breast cancer initiation and progression, while Wicherska-Pawłowska et al. (2022) found no significant differences in allele or genotype frequencies of this SNP in acute myeloid leukemia patients.
We concluded that rs3775296, located in the 5′-untranslated region of TLR3, could potentially affect amino acid residues and impact the gene's role in the innate immune system. These genetic variations might influence gene expression in response to inflammatory cytokines, leading to transcriptional modifications of TLR3 that could reduce it signaling efficiency and contribute to the development of cancer. Additionally, the minor allele (T) of rs5743312 in healthy Saudis was observed at a frequency of 0.09, comparable to frequencies in African, Mexican, and Peruvian populations but higher than those in Chinese and Japanese populations. Variant genotypes of the intronic TLR3 rs5743312 were significantly associated with an increased risk of developing ALL, particularly in males and adult patients. This finding aligns with research by Fan et al. (2019), which also noted an association between rs5743312 and breast cancer risk. A meta-analysis by Wang et al. (2015) further supported a link between rs5743312 and a heightened cancer risk overall.
Conversely, our results indicated no significant association between TLR3 rs3775291 polymorphisms and ALL risk, although a marginal correlation with increased susceptibility to ALL was noted. Consistent with earlier studies, this SNP did not alter TLR3 expression levels or its intracellular localization, though it negatively impacted dsRNA recognition and binding (Ma et al., 2016).
In TLR3 rs3775290, the presence of one copy of the mutant allele T provides resistance against the development of ALL. The frequency of the variant allele T for the investigated SNPs in healthy Saudis was 0.31, similar to that reported for the 1000 Genomes Project in various populations, such as Vietnamese, British (in England and Scotland), and Finnish (in Finland) (1000 Genomes Project Consortium, 2015). Conversely, many previous studies have reported a significant association between the nonsynonymous rs3775290 polymorphism and the increased risk of other types of cancer or diseases. A meta-analysis conducted by Cheng et al. (2014) revealed that the rs3775290 polymorphism, present in exon 4 of the TLR3 gene, might increase the risk of cancer. Additionally, individuals heterozygous at rs3775290 exhibited an increased relative risk of cervical cancer in Tunisian women (Zidi et al., 2018), although this relationship was not detected in India (Pandey et al., 2011). In a study by Hamdy et al. (2018), TLR3 rs3775290 was genotyped in a group of Egyptians infected with chronic hepatitis C virus; this study found that the CC genotype was a risk genotype for chronic hepatitis C virus persistence, a result also observed in the Tunisian population (Sghaier et al., 2017). However, Etokebe et al. (2009) reported no association between TLR3 rs3775290 and rs5743312 with breast cancer, prostate cancer risk (Mandal et al., 2012), and bladder cancer (Singh et al., 2013). Similarly, variant genotypes of TLR3 rs3775290 have not been correlated with susceptibility to multiple sclerosis in various ethnic populations (Deeba et al., 2019).
This present study demonstrated a statistically significant difference between the mean levels of TLR3/GAPDH mRNA expression in the blood of patients with ALL compared with normal cases. Although TLR3 is located in endocytic compartments inside the cell, increasing evidence has shown that TLR3 is also expressed in tumor cells. TLR3 pattern recognition is associated with dsRNA, which has been proposed as an endogenous ligand for its activation. dsRNA released from injured cells could initiate paracrine activation of TLR3, thus inducing tumor cell growth and facilitating evasion from immune surveillance, suggesting that TLR3 activation could play an antagonistic role in tumor progression (Zheng et al., 2021). Importantly, the expression and functionality of TLR3 receptors in ALL have rarely been investigated. Our results follow other data published in 2008 by Jiang and his colleagues; (Jiang et al., 2008) they screened various human and murine tumor cell lines and found that all tested tumor cell lines expressed TLR3 at different levels, suggesting that the ligation of TLR3 can initiate signaling cascades and result in the activation of downstream effector molecules. These molecules include NF-κB, p38, JNK, and IFN regulatory factors (IRFs), which may be involved in tumor growth and apoptosis. The results shown here are also in line with previous findings in other types of cancer. Oblak & Jerala (2011) and González-Reyes et al. (2011), indicated that TLR3, TLR4, and TLR9 expression levels were significantly and positively associated with tumor size, stage, metastasis, and tumoral aggressiveness in breast cancer. Therefore, TLR3 may be applied as a promising therapeutic target for breast cancer. Additionally, González-Reyes et al. (2011) conducted the same experiment in prostate carcinoma and found high expression of TLR3, TLR4, and TLR9 in comparison with benign prostate. Furthermore, Fernandez-Garcia et al. (2014) revealed that gastric carcinomas show an increased level of TLR3, TLR4, and TLR9 expression, and this high expression of TLR3 was found to be significantly correlated with poor outcomes in gastric cancer patients.
In contrast, Sánchez-Cuaxospa et al. (2016) found that low expression of TLR3 in lymphocytes (T, B, and NK cells), monocytes, and dendritic cells from pediatric patients with ALL compared to the healthy group. However, Abdi et al. (2013) reported that primary cells from multiple myeloma patients showed low expression of TLR2, TLR3, and TLR5, while all human myeloma cell lines exhibited robust expression of TLR3. Thus, this may be due to the influence of the tissue origin, which may have a potential impact on determining the TLR expression pattern. Furthermore, Eiró and his colleagues (2012) observed that TLR2, TLR3, TLR4, and TLR5 expression was absent in colorectal carcinoma patients. Therefore, TLRs overexpression can have diverse biological significance in the context of the complex relationship between inflammation and cancer development.
While this study primarily employs traditional statistical methods to analyze the association between TLR3 polymorphisms and ALL susceptibility, incorporating machine learning techniques such as artificial neural networks (ANNs) represent a promising avenue for future research. ANNs could potentially enhance the predictive accuracy of genetic susceptibility to ALL by learning complex patterns and interactions within large genomic datasets.
ANNs have been effectively used in various genetic studies to model nonlinear relationships and interactions that are not easily captured by conventional statistical approaches. For instance, studies like Sekban et al. (2024) and Garg et al. (2022a,b) have demonstrated the utility of ANN and other machine learning methods in predicting material properties and biological responses, suggesting a similar approach could be applied to genetic data in leukemia research.
Future studies could explore using ANNs to integrate genetic markers with clinical data (e.g., age, gender, and medical history) to develop a more comprehensive predictive model of ALL risk. This approach might uncover new insights into the genetic architecture of ALL and improve the precision of risk stratification in clinical settings.
5 Conclusion
In conclusion, our study identifies and quantifies the association of specific SNPs in the TLR3 gene with ALL in the Saudi population. We found that the rs5743312 C/T polymorphism is associated with an increased risk of ALL, while the rs3775290 C/T polymorphism is inversely associated, suggesting a protective effect against the development of ALL. These findings enhance our understanding of the genetic basis of ALL and could guide future research into more effective, personalized treatment approaches.
CRediT authorship contribution statement
Rasha Alonaizan: Writing – review & editing, Project administration, Funding acquisition. Fadwa M Alkhulaifi: Writing – original draft, Methodology, Formal analysis, Data curation. Ahmed rady: Validation, Data curation. Suliman Alomar: Writing – review & editing, Supervision, Conceptualization.
Acknowledgment
The authors extend their appreciation to the Deputyship for Research & Innovation, “Ministry of education” in Saudi Arabia for funding this research work through the project no. (IFKSUDR_H222).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Characterization of the toll-like receptor expression profile in human multiple myeloma cells. PLoS One. 2013;8(4):1-9.
- [CrossRef] [Google Scholar]
- Association between Toll-like receptor 3 polymorphisms and cancer risk: a meta-analysis. Tumor Biol.. 2014;35(8):7837-7846.
- [CrossRef] [Google Scholar]
- Complete sequence analysis of human toll-like receptor 3 gene in natural killer cells of multiple sclerosis patients. Mult. Scler. Relat. Disord.. 2019;33(January):100-106.
- [CrossRef] [Google Scholar]
- Study of the expression of toll-like receptors in different histological types of colorectal polyps and their relationship with colorectal cancer. J. Clin. Immunol.. 2012;32(4):848-854.
- [CrossRef] [Google Scholar]
- Single-nucleotide polymorphisms in genes encoding toll-like receptor-2,-3,-4, and-9 in case–control study with breast cancer. Genet. Test. Mol. Biomarkers. 2009;13(6):729-734.
- [CrossRef] [Google Scholar]
- Toll-like receptor 3 acts as a suppressor gene in breast cancer initiation and progression: a two-stage association study and functional investigation. OncoImmunology. 2019;8(6)
- [CrossRef] [Google Scholar]
- Clinical significance of toll-like receptor 3, 4, and 9 in gastric cancer. J. Immunother.. 2014;37(2):77-83.
- [CrossRef] [Google Scholar]
- 196 to -174del, Rs4696480, Rs3804099 Polymorphisms of Toll-like Receptor 2 Gene Impact the Susceptibility of Cancers: Evidence from 37053 subjects. Biosci. Rep.. 2019;39(12)
- [CrossRef] [Google Scholar]
- Machine learning models for predicting the compressive strength of concrete containing nano silica. Comput. Concr.. 2022;30(1):33.
- [CrossRef] [Google Scholar]
- Predicting elemental stiffness matrix of FG nanoplates using Gaussian Process Regression based surrogate model in framework of layerwise model. Eng. Anal. Bound. Elem.. 2022;143:779-795.
- [CrossRef] [Google Scholar]
- Study of TLR3, TLR4, and TLR9 in prostate carcinomas and their association with biochemical recurrence. Cancer Immunol. Immunother.. 2011;60(2):217-226.
- [CrossRef] [Google Scholar]
- Association of Toll-like receptor 3 and Toll-like receptor 9 single-nucleotide polymorphisms with hepatitis C virus persistence among Egyptians. Arch. Virol. 2018;163(9):2433-2442.
- [CrossRef] [Google Scholar]
- Poly I: C enhances cycloheximide-induced apoptosis of tumor cells through TLR3 pathway. BMC Cancer. 2008;8:1-8.
- [CrossRef] [Google Scholar]
- Trial watch: TLR3 agonists in cancer therapy. OncoImmunology. 2020;9(1):1-13.
- [CrossRef] [Google Scholar]
- Association of toll-like receptor 3 polymorphism rs3775291 with age-related macular degeneration: a systematic review and meta-analysis. Sci. Rep.. 2016;6(1):19718.
- [CrossRef] [Google Scholar]
- Association of Toll-like receptor (TLR) 2, 3 and 9 genes polymorphism with prostate cancer risk in North Indian population. Mol. Biol. Rep.. 2012;39(7):7263-7269.
- [CrossRef] [Google Scholar]
- Effect of TLR2 on the proliferation of inflammation-related colorectal cancer and sporadic colorectal cancer. Cancer Cell Int.. 2020;20(1):1-13.
- [CrossRef] [Google Scholar]
- The role of toll-like receptors in hematopoietic malignancies. Front. Immunol.. 2016;7:390.
- [CrossRef] [Google Scholar]
- Toll-like receptor 4 activation in cancer progression and therapy. Clin. Dev. Immunol.. 2011;2011
- [CrossRef] [Google Scholar]
- Pandey, S., Mittal, B., Srivastava, M., Singh, S., Srivastava, K., Lal, P., & Mittal, R. D. (2011). Evaluation of Toll-like receptors 3 (c. 1377C/T) and 9 (G2848A) gene polymorphisms in cervical cancer susceptibility. Mol. Biol. Rep. 38(7), 4715-4721. doi: 10.1007/s11033-010-0607-z.
- Role of toll-like receptors in actuating stem/progenitor cell repair mechanisms: different functions in different cells. Stem Cells Int.. 2019;2019
- [CrossRef] [Google Scholar]
- Low expression of Toll-like receptors in peripheral blood mononuclear cells of pediatric patients with acute lymphoblastic leukemia. Int. J. Oncol.. 2016;49(2):675-681.
- [CrossRef] [Google Scholar]
- Investigating formability behavior of friction stir-welded high-strength shipbuilding steel using experimental, finite element, and artificial neural network methods. J. Mater. Eng. Perform.. 2024;1–9
- [CrossRef] [Google Scholar]
- Role of TLRs and IL-6 in the outcome of chronic hepatitis C treatment in Tunisian population. Cytokine. 2017;99:297-304.
- [CrossRef] [Google Scholar]
- Single-nucleotide polymorphisms in genes encoding toll-like receptor -2, -3, -4, and -9 in a case-control study with bladder cancer susceptibility in a north indian population. Arch. Med. Res.. 2013;44(1):54-61.
- [CrossRef] [Google Scholar]
- TLR3 gene polymorphisms in cancer: a systematic review and meta-analysis. Chin. J. Cancer. 2015;34(6)
- [CrossRef] [Google Scholar]
- Polymorphisms in the genes coding for TLRs, NLRs and RLRs are associated with clinical parameters of patients with acute myeloid leukemia. Int. J. Mol. Sci.. 2022;23(17):9593.
- [CrossRef] [Google Scholar]
- Roles of toll-like receptor 3 in human tumors. Front. Immunol.. 2021;12(April):1-9.
- [CrossRef] [Google Scholar]
- RETRACTED ARTICLE: impact of toll-like receptors 2/3/4/9, IL-1-α/β and TNF-α polymorphisms in cervical cancer susceptibility in tunisia: genetic polymorphisms implicated in the occurence of cervical cancer. Pathol. Oncol. Res.. 2018;24(2):197.
- [CrossRef] [Google Scholar]







