Translate this page into:
Biotransformation fate and sustainable mitigation of a potentially toxic element of mercury from environmental matrices
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
The deposition of potentially toxic mercury (Hg) in various ecosystems and subsequent entry into the food chain pose serious concerns to the ecosystem, biodiversity, and public health. In terms of toxicity, Hg is considered as a neurotoxin and capable to augment in food chains and bind to the thiol functional entity in living tissue. Moreover, methylated mercury (CH3Hg+) is a highly toxic form of mercury and extremely difficult to remove from living bodies. Mercury methylation is mainly conducted by microbial and/or chemical processes under appropriate conditions. The mechanisms associated with mercury methylation inside the environment, their sources, production/degradation rate, and transport into the living organisms are not well understood. In addition, efficient and sustainable remediation strategies are essential to employ for mercury removal. Therefore, this review signifies a possible mechanism for mercury methylation and its transportation in the environment, including molecular mechanisms and genes associated with microbial-mediated mercury methylation, and identifies the gaps in existing research. The transport of Hg into the human body and associated health risks are given with suitable examples. Moreover, the escalating anthropogenic activities, the rate-limiting factors, and the sustainable remediation strategies implemented for mercury removal from the environment are discussed. This study will provide a scientific base, direction, and progress in future studies.
Keywords
Potentially toxic element
Mercury
Water matrices
Remediation
Nano-constructs
Methylated mercury
Microbial-mediated methylation
1 Introduction
Mercury (Hg) is a global environmental pollutant having seven stable isotopes (204Hg, 202Hg, 200Hg, 201Hg, 199Hg, 198Hg, and 196Hg). Human exposure to Hg is intensively toxic for health through its noxious impacts on embryonic, endocrine, reproductive, immunological, pulmonary, nervous, hematological, renal, and cardiovascular systems (Rice et al., 2014). Due to its toxic nature, it has been ranked 3rd among the most toxic substances by the United States Government Agency for Toxic Substances and Disease Registry (Clifton, 2007; Rice et al., 2014). Accumulation of Hg in the atmosphere, water, soil, and glaciers is rapidly increasing due to escalating anthropogenic activities and easily available to enter into the food web through plants and livestock (Clifton, 2007; Beckers and Rinklebe, 2017). In 2015, the global mercury emission into the atmosphere from anthropogenic activities was estimated approximately, 2220 tons. Among the anthropogenic activities, 24% accounts for stationary combustion of fossil fuels of the estimated emissions, mainly from coal-burning (21%) (UNEP, 2019). Other human sources include direct mercury production, waste disposal (Pehnec et al., 2010), chlor-alkali industry (Busto et al., 2011; Kakareka and Kukharchyk, 2012; Busto et al., 2013), gold production (Wu et al., 2018), steel and iron production (Wang et al., 2016b), and cement production (Wang et al., 2016a). In recent decades, leading emission sources of Hg included fuel combustion (coal) and artisanal and small-scale gold mining (ASGM) (Gworek et al., 2017). According to Gworek et al. (2017), ASGM has the largest share of 32% of Hg emission into the air. Telmer and Veiga (2009) stated that about 100 million people in 55 countries of South America, Africa, and Asia are dependent on AGSM for their incomes. ASGM is one of the main contributors of Hg release in the areas where gold mining and processing are active. In addition, Hg emission has increased threefold with the technical and scientific revolution and up to tenfold in highly industrialized areas (Hylander and Meili 2003). Mercury emission share estimated from metallurgy sector was 13.2% and from cement industry was 10.8% (Gworek et al., 2017).
Similarly, gaseous elemental mercury (GEM) emission contributes 27% from the industrial sector (Zhang et al. 2016). The member states of the European Union (European Union 2016), generated 20% of industrial based Hg emission in 2014 (Gworek et al., 2017). Moreover, factories producing chlorine and sodium hydroxide also contribute to the emission of Hg. The global Hg emission related to the chlor-alkali industry was (18.5–100.8 t/year) in 2010, which is 2.7% of the total emission due to anthropogenic sources Wilson et al. 2012). Several other industries such as electrical and electronic devices, lightening equipment, and batteries production also contribute to Hg emission (Gworek et al., 2017).
Several approaches are adapted to remove toxic heavy metals from the environment. In the past few decades, more sustainable and efficient strategies are developed for metals removal by using microorganisms (Sajjad et al., 2019). Recently, consortia of iron-oxidizing bacteria and bamboo sawdust was used for the removal of metals from low-grade ore that is a potential source of heavy metals released into the environment (Sajjad et al., 2020). Sajjad et al. (2018) reported that the consortia of indigenous microorganisms are more efficient in metals removal. Similarly, Huang et al. (2018) studied biochar-supported nano-chlorapatite (BC-nClAP) for the efficient immobilization of lead-contaminated sediment. In another study, Wang et al. (2020a, 2020b) removed copper and tetracycline from aqueous solution by using steam-activated biochar prepared from bamboo. Recently, Deng et al. (2020) achieved excellent lead remediation performance by using sodium lignin sulfonate stabilized nano-chlorapatite. In addition, enhanced phytoremediation efficiency combined with nanoscale zero-valent iron (nanoparticles) was achieved by Gong et al. (2017) for cadmium removal.
Bioaccumulation of Hg inside the body could occur via the food chain or directly enter through the respiratory system from the vaporization of mercury and/or burning of mercury-containing materials. Although, in living organisms, it can be quickly eliminated from the blood, however, if distributed into different tissues, it will start its adverse effects on human health (Beckers and Rinklebe, 2017). It enters into the environment in the inorganic forms such as mercurous (Hg (I)) or mercuric (Hg (II)) and further methylated into organometallic compounds including methylmercury (CH3Hg+) (Schaefer and Morel, 2009; Regnell and Watras, 2019; Celo et al., 2006; Yuan et al., 2019). Although, all the states of Hg are toxic, however, CH3Hg+ is more toxic compared to inorganic forms, and removal of CH3Hg+ from the body is not so easy. The mechanisms associated with Hg methylation within the environment and their transport into the living organisms are not well understood. Therefore, this paper aimed to provide inclusive details information about the possible mechanism for Hg methylation and its transportation in the environment, including molecular mechanisms associated with microbial-mediated Hg methylation, the sources of transportation in the environment, and transport into the human body. This review explained the activities and efficiency of microbes, responsible microbial genes for Hg methylation, the rate-limiting factors and the remediation strategies implemented for Hg removal from the environment.
2 Risks associated with Hg
Among its three existing forms (elemental, organic, and inorganic), elemental mercury (Hg0) has the higher absorbing potential in the nervous and respiratory systems to induce serious health risks in animals and humans (Clarkson and Magos, 2006; Magos and Clarkson, 2006). Oxidized vapors of Hg can become lipid-soluble and bio-accumulate in the liver, renal cortex, and especially in the brain, thus causing serious toxic effects in the neurological system. When enters into the body, Hg adversely affects human health, but its determination for specific impact is tricky as it is eliminated quickly from the blood, redistributed, and sequestered into various tissues. It shows that an indirect correlation could exist between the blood Hg concentration and severity of the Hg poisoning (Clifton, 2007; Rice et al., 2014; Budnik and Casteleyn, 2019). The entrance of Hg inside the cell and causing toxicity is highly dependent on alterations in cell membrane permeability. At a cellular level, it induces oxidative stress and leads to mitochondrial dysfunction (Rice et al., 2014). The toxicity mechanism caused by is documented in Fig. 1.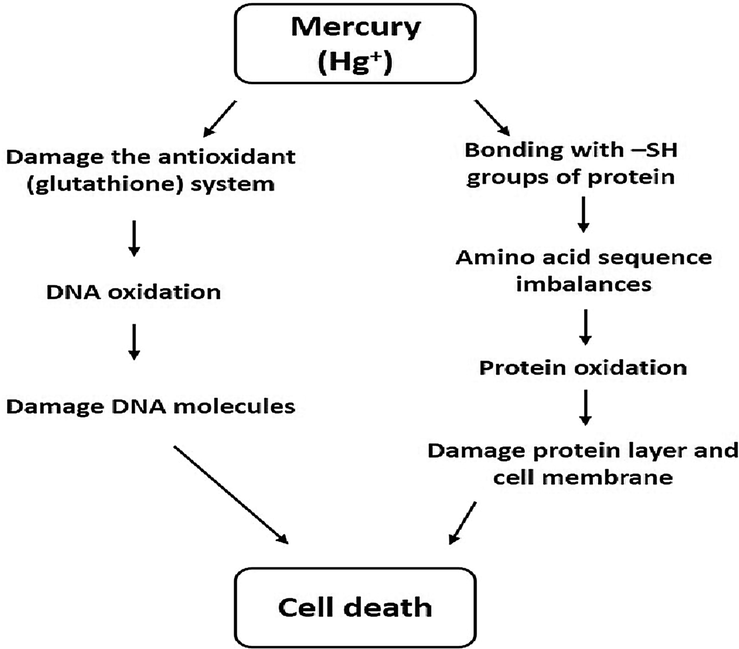
The mechanism of toxicity (apoptosis) caused by Hg in living organisms.
Previously, the association of Hg with cardiomyopathy has been reported. The accumulated Hg is also thought to cause angina after its accumulation. Moreover, Hg has been widely found to cause dental problems, adrenal problems, diabetes, hypertension, renal toxicity, fetal toxicity, and alterations in the genetic, enzyme, and immune system. The entry of CH3Hg+ into the body could inhibit the cardioprotective function of paraoxonase and causes anemia including aplastic and hemolytic anemia as Hg competes with iron for binding to hemoglobin thus, resulting in impaired hemoglobin formation. Furthermore, Hg may also increase the risk of mononucleosis and involved in Hodgkin’s disease and leukemia (Rice et al., 2014; Budnik and Casteleyn, 2019). After Hg ingestion, epithelial cells absorbed Hg causing several digestive disturbances including the inhibition and synthesis of digestive enzymes such as trypsin, pepsin, and chymotrypsin. The gastrointestinal related problems associated with Hg are indigestion, diarrhea, inflammatory bowel disease, and abdominal pain. Moreover, Hg can damage the kidney, can cause necrosis of acute tubules, and renal cancer. It can also cause tubular dysfunction, secondary focal segmental glomerulosclerosis, subacute-onset nephrotic syndrome, nephritic syndrome, and glomerular disease (Clifton, 2007; Rice et al., 2014; Alkaissi et al., 2016; Budnik and Casteleyn, 2019; Mahadappa et al., 2020).
In males, exposure to Hg can alter reproductive behavior, increase sperm morphologic abnormalities, and reduce sperm count, motility, and daily production (Martinez et al., 2014). Furthermore, it can also decrease luteinizing hormone concentration, antioxidant enzyme activities, and increased lipid peroxidation in testis. It can cause infertility and erectile dysfunction in males, ovarian dysfunction, tipped uterus, premature menopause, and menstruation disorders in females (Rice et al., 2014). Recently, a study revealed higher Hg accumulation in the somatotrophs of anterior pituitary glands and lowered levels of growth hormone (Pamphlett et al., 2019).
3 Sources and transportation of Hg into the environment
The sources of Hg emission can be classified into primary and secondary sources (Beckers and Rinklebe, 2017). Primary sources release Hg from the lithosphere to atmosphere while secondary reservoirs re-emit the deposited Hg from the surface into the atmosphere (Schoch et al., 2019; Raj and Maiti, 2020). Subsequently, this Hg is accumulated in water, soil, glaciers, and sediments (Gosnell et al., 2016; Haynes et al., 2017) (Fig. 2). In these environmental systems, the transformation of deposited Hg occurs regularly from one form into another depending on the availability of suitable conditions. For instance, the methylation process can be reversed by microorganisms (Lu et al., 2016), although the associated mechanism is not clear. The major sources of CH3Hg+ in the aquatic environments are terrestrial and runoff, especially from the wetlands and atmospheric deposition (Celo et al., 2006) (Fig. 2). Moreover, global industrialization increases Hg deposition which will have environmental consequences (Yin et al., 2016). Forest soils are considered among the main sources of CH3Hg+ in aquatic environments (Xu et al., 2019). Atmospheric Hg is dumped onto the cryosphere and emitted back to the atmosphere. This deposition of Hg due to its high mobility in the cryosphere is linked with the process of atmospheric Hg depletion events during the polar spring. Snowmelt is strongly considered to be the main source of Hg release into lakes and water sources (Fig. 2). Ice and melted water containing Hg are indicative of its net deposition onto snow packs. Although, long-range transport of CH3Hg+ is reported in polar regions, Arctic, Antarctic, Tibetan plateau, and the Himalayas (Gionfriddo et al., 2016; Ghimire et al., 2019), more investigations are required to estimate the net Hg deposition during specific periods (Ghimire et al., 2019). In addition, meteorological changes also contributed largely to the Hg reservoirs. Stabile Hg in soil outflows after becoming labile into fluvial systems. Alterations of hydrological and meteorological conditions induce the entrance of Hg into rivers which is then introduced into the marine environment (Gębka et al., 2020). The CH3Hg+ can be biomagnified and bio-accumulated through the aquatic food web (Ghimire et al., 2019; Yuan et al., 2019) and fish species (Bosch et al., 2016; Beckers and Rinklebe, 2017; Graci et al., 2017), enters into the human body (Bradley et al., 2017). Its accumulation in the body increases the concentration of CH3Hg+ as it cannot be easily removed from the living organisms (Wiener, 2013). Therefore, the methylation process of inorganic Hg has been the subject of intensive research over the past few decades (Schaefer and Morel, 2009).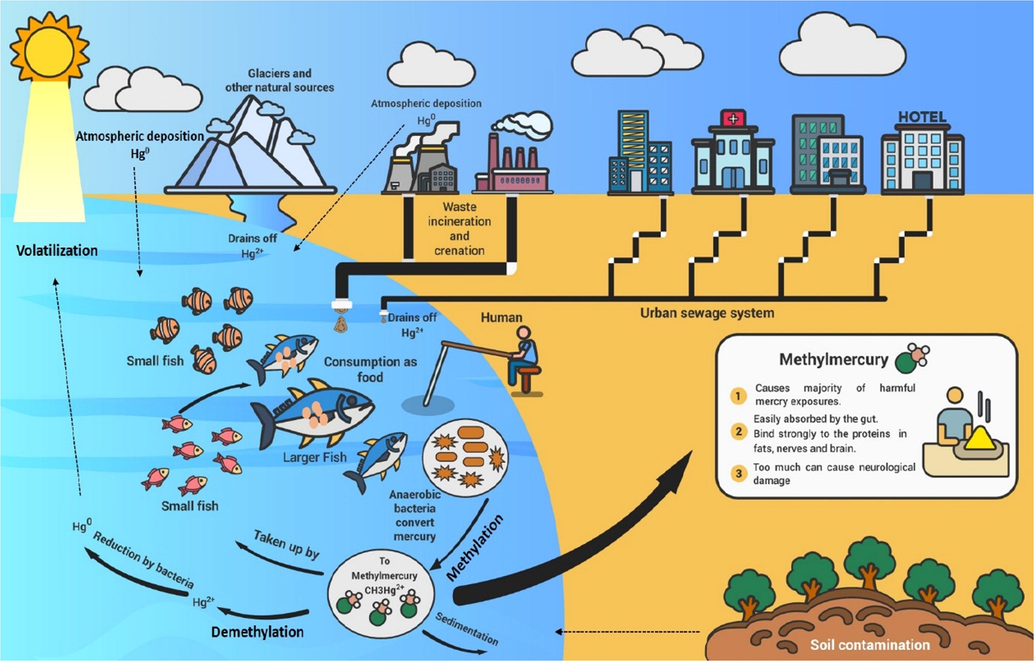
Transformation of Hg (methylation-demethylation) in different resources to the food web and subsequently into humans.
4 Mercury methylation
The presence of methylated Hg co-existing with other forms in the atmosphere indicates that chemical methylation of Hg can occur under natural conditions. Relatively harmless inorganic mercury (Hg(I) and Hg(II)) released by anthropogenic activities such as coal burning and industry undergoes a process of methylation and become lethal monomethyl mercury (Celo et al., 2006; Ghimire et al., 2019). Celo et al. (2006) explained the reaction of Hg with some potential methyl donors based on their laboratory-based investigations of the aqueous environment. Chemical methylation of Hg takes place in the presence of suitable methyl donors, which may be the products of biological processes. Small organic molecules, larger organic components of dissolved organic matter, transmethylation reactions involving organometallic complexes such as methyl cobalamin, and methyl lead or methyl tin compounds are considered as possible pathways for abiotic methylation of Hg in the aquatic environment (Celo et al., 2006).
Furthermore, CH3Hg+ is also produced in the oxygen-limited environmental sources including soils, stratified water columns, and sediments (Gascón Díez et al., 2016). In addition to environmental factors, the content and composition of organic matter modulate biological methylation rates of the inorganic Hg (Drott et al., 2007). Herrero Ortega et al. (2018) investigated the influence of natural organic matter composition on methylation of Hg in boreal beaver ponds across the Sweden. They found that the rates of Hg methylation were fueled by increased production of algal-derived organic matter triggered by enhanced nutrient availability (Herrero Ortega et al., 2018). Algal biomass may enhance the microbial capability of Hg methylation by increasing the abundances of potential microbial methylators in sediments (Bouchet et al., 2018). Algal biomass may promote microbial Hg methylation through increasing the abundances of microbes and microbial methylators into sediments (Lei et al., 2019). Lei et al. (2019) explained that CH3Hg+ levels were increased in sediments with algae settlement and decomposition after algal blooms. This rise in the production of CH3Hg+ in sediments was associated with the algal organic matter but not with other organic components including aromatic proteins.
It is widely known that leaf litter contains labile organic matter and Hg, therefore, during its decomposition, the process of Hg methylation induces by providing oxygen and energy (Britt and Vincent 2004). For instance, Balogh et al. (2005), observed high levels of CH3Hg+ in a stream in Minnesota (USA) having a large mass of litter accumulation. Additionally, further investigation showed that litter if incubated with fresh stream water in a closed container, high levels of CH3Hg+ are produced (Balogh et al., 2005). These investigations were further supported by the study of Tsui et al. (2008), who demonstrated that plant litter can regulate the Hg methylation. Recently, Tang et al. (2019) reported that rice straw amendment could increase CH3Hg+ concentrations in paddy soils. Molecular composition and origin of organic matter are essential parameters in CH3Hg+ formation and accumulation processes in lake sediments and water reservoirs (Bravo et al., 2017).
4.1 Microbes associated with Hg methylation
Recently, iron-reducing bacteria (IRB) and sulfur-reducing bacteria (SRB) have been found as the dominant microbial strains responsible for the Hg methylation in the sediments of the Pearl River, Pearl River Estuary, and the South China Sea (Yuan et al., 2019). Besides the most known anaerobic microbes, sulfate- or iron-reducing Deltaproteobacteria (Gilmour et al., 2011) and Desulfovibrio desulphuricans ND132 (Colombo et al., 2013) are also involved in the Hg methylation process. Since inorganic mercury (Hg0) is oxidized to mercuric ions (Hg2+) in oxic environments, Hu et al. (2013) reported that anaerobic microorganisms can oxidize and methylate the dissolved Hg0. They tested three bacterial strains from Deltaproteobacteria to investigate their ability of oxidation and methylation for Hg under anaerobic conditions. Geobacter sulphurreducens PCA (iron-reducing) and D. desulphuricans ND132 (sulfate-reducing) are Hg methylators while Desulfovibrio alaskensis G20 (sulfate-reducing) is a non-methylator. By comparing the ability of two strains of Desulfovibrio and one strain of Geobacter to oxidize and methylate Hg0 under dark, anaerobic conditions, they found that the rate of methylation was about one-third compared to the methylation rate of oxidized Hg. These investigations indicate that the CH3Hg+ formation could be enhanced by the combined activity of methylating and non-methylating bacteria in anaerobic environments (Hu et al., 2013).
Gilmour et al. (2018a, 2018b) demonstrated that the production of CH3Hg+ is mediated by methanogenic archaea. The different strains of methanogens including Methanocorpusculum bavaricum, Methanocella paludicola SANAE, Methanosphaerula palustris E1-9c, Methanofollis liminatans GKZPZ, Methanospirillum hungatei JF-1 (5), Methanolobus tindarius, and Methanomethylovorans hollandica produced CH3Hg+ at different rates. However, Methanococcoides methylutens was found incapable of methylation. Sulfide inhibited the production of CH3Hg+, indicating that methanogens and Deltaproteobacteria respond to Hg complexation in the same way. The Hg methylating bacteria including Desulfovibrio desulfuricans, Desulfobulbus propionicus, Geobacter sulfurreducens, and Desulfovibrio magneticus are majorly affected by the geochemical factors. For instance, the dental wastewater system is generally dominated by the Actinobacteria, Proteobacteria, Chloroflexi, and Bacteroidetes due to the impact of high loads of Hg (Rani et al., 2015).
Fleming et al. (2006) reported that the molybdate mediated inhibition of “microbial sulfate reduction” and sediment sterilization significantly decrease the Hg methylation. Methylation by SRB requires optimum pH values from 4 to 6, however, decreasing the pH enhances the Hg methylation rate and yield of CH3Hg+. Furthermore, microbial-mediated methylation of Hg is highly dependent on sulfate concentration, such that methylation increases at lower sulfate concentration until peak value (0.3 mM). Bravo et al. (2018) also reported similar results while conducting a study regarding the environmental factors controlling the activity of Hg methylating bacteria. Ferruginous geochemical conditions influence the methylation process in the presence of iron and sulfur-transforming bacteria, syntrophs, and methanogens. Furthermore, Geobacteraceae dominated the hgcA carrying communities. Comparatively, sulfate reducers constituted a minor role, though these are generally considered the main Hg methylators (Bravo et al., 2018).
Microorganisms, such as psychrophiles in the cryosphere are considered to be actively involved in the process of Hg methylation (Collins and Margesin, 2019), where the functions of microbial communities are generally thought to be similar (Boetius et al., 2015). The upper layer of the cryosphere (sea ice) facilitates microbial reactions involved in global biogeochemical cycles (Maccario et al., 2014; Ghimire et al., 2019). The activity of microbes including cyanobacteria, eukaryotic Zygnematales, and Zhlamydomonadales is generally higher in the cryosphere due to the organic debris layers (Harding et al., 2011; Hodson et al., 2015). Although microbial-mediated Hg methylation occurs in the cryosphere, there is no adequate data available for biotic and abiotic relations. Algae initiate microbial-mediated methylation of Hg (Louis et al., 2007; Gamberg et al., 2015), thus it could be the source of possible pathway prediction. Cyanobacteria, Firmicutes, Bacillus, Davidiellaceae, Alternaria, and Rhodotorula were detected dumped in ice in the Greenland and Antarctica (Knowlton et al., 2013), which could further be studied for their role in Hg methylation. Moreover, the detection of SRB in the Arctic and sub-Arctic ecosystems indicates that they may play a crucial role in the methylation process in the cryosphere, though it was not related to CH3Hg+ concentrations in the Arctic and sub-Arctic regions (Louis et al., 2007; Ghimire et al., 2019).
In the sea ice, Nitrospina, a marine microaerophilic bacterium has been found to be a potential methylator of Hg (Gionfriddo et al., 2016), indicating the importance of Nitrospina in diverse environments. Accumulation of CH3Hg+ occurs more efficiently compared to inorganic Hg, therefore, Hg transformations into CH3Hg+ control burdens in upper trophic level biota. For the determination of potential CH3Hg+ sources in low-oxygen waters, Munson et al. (2018), measured the rates of methylation process in the oligotrophic central Pacific Ocean. They found that overall rates of methylation over 24 h incubation periods are similar if compared with those of the Arctic and Mediterranean waters. The methylation rate associated with heterotrophic bacteria was found higher in the filtered water than the unfiltered water. Moreover, enhanced demethylation of newly produced methylated Hg was observed, in incubations of unfiltered water in relation to filtered water. Ding et al. (2019) investigated the Hg methylation by Geobacter metallireducens GS-15 under the influence of algae of Skeletonema costatum. This study revealed that the biomass of S. costatum can inhibit the bacterial methylation of Hg. Lehnherr et al. (2011), reported that in seawater samples collected from the Canadian Arctic Archipelago, methylation and demethylation occurs side by side. The investigation further revealed that methylation of inorganic Hg is a significant source of monomethyl mercury in the world’s oceans and pelagic marine food webs in the Arctic.
Studying the levels of CH3Hg+ in the soil using Gas Chromatography Cold Vapor Atomic Fluorescence Spectrometry and Direct Mercury Analyzer, revealed that CH3Hg+ concentration was the highest in seasonal drying and flooding alternating areas. This higher concentration of CH3Hg+ is likely due to soil organic matter abundance and sedimentation from flooding. These observations suggest that water level fluctuation directly affects the production of CH3Hg+ in soils. In these seasonal drying and flooding alternating areas, Deltaproteobacteria and Methanomicrobia were found in abundance, suggesting their association with Hg methylation in soil. However, the low methylation capacity of Methanomicrobia may be the reason for the low total CH3Hg+ levels (Hoy et al., 2018; Xiang et al., 2018). Xu et al. (2019), reported the Hg methylation in boreal forest soils which can be a contributor to CH3Hg+ in aquatic networks. Proteobacteria, Firmicutes, and Methanomicrobia with Deltaproteobacteria (Geobacteraceae) were found dominantly in all soil samples. Various bacteria and archaea involved in Hg methylation under different environmental conditions are well mentioned and discussed in Table 1.
Type of Microorganisms
Class
genus and species
Source habitat
References
Bacteria
Deltaproteobacteria
Pseudodesulfovobrio aespoeensis
Freshwater ecosystem
(Villar et al., 2019)
Desulfomicrobium escambiense
Fresh water
(Villar et al., 2019)
Desulfovibrio africanus
Freshwater
(Gilmour et al., 2013; Gilmour et al., 2018a, 2018b)
Geobacter bemidjiensis
Freshwater Ecosystem
(Gilmour et al., 2013; Gilmour et al., 2018a, 2018b)
Desulfovibrio desulfuricans
Soil, Water, Animal‘s stool, estuarine sediments
(Goldstein et al., 2003; Gilmour et al., 2018a, 2018b; Gilmour et al., 2011; Villar et al., 2019)
Geobacter sulfurreducens PCA
Ditch water, Mud, Wastewater
(Villar et al., 2019; Yu et al., 2018)
Syntrophus aciditrophicus
Soil, Mud, Wastewater
(Gilmour et al., 2013; Bae et al., 2014; Sorokin et al., 2008; Yu et al., 2018)
Desulfobulbus propionicus
Mud ditch, Freshwater, Wastewater, Soil
(Gilmour et al., 2013; Bae et al., 2014; Sorokin et al., 2008; Villar et al., 2019)
Desulfococcus multivorans
Soil, Mud, Wastewater
(Villar et al., 2019)
Geobacter metallireducens
Soil, Mud, Wastewater
(Villar et al., 2019)
Desulfomicrobium baculatum
Soil, Mud, Wastewater
(Villar et al., 2019)
Desulfovibrio sp.X2
Marine, Saline water
(Villar et al., 2019)
Geobacter daltonni
Soil, Mud, Wastewater
(Villar et al., 2019)
Syntrophobacter wolinii
Freshwater ecosystem
(Yu et al., 2018)
Syntrophorhabdus aromaticivorans
Anaerobic ecosystem (Aromatic Compounds)
(Yu et al., 2018)
Geobacter metallireducens
Soil, Mud, Wastewater
(Villar et al., 2019)
Desulphonetronospira thiodismutans
Marine, Saline water
(Villar et al., 2019)
Firmicutes
Dethiobacter alkaliphilus
Marine/Saline water
(Gilmour et al., 2013; Villar et al., 2019)
Syntrophobotulus glycolicus
Freshwater sediments, sea
(Yu et al., 2018)
Acetonema longum
Microbiota
(Villar et al., 2019; Ghimire et al., 2019)
Desulfobacterium dehalogenans
Freshwater, glaciers
(Villar et al., 2019)
Desulfitobaterum metallireducens
Freshwater
(Villar et al., 2019)
Desulfosporosinus youngiae
Freshwater
(Villar et al., 2019)
Desulfosporosinus acidiphilus
Soil, Mud, Wastewater
(Villar et al., 2019; Ghimire et al., 2019)
Ethanoligenens harbinense
Soil, Mud, Wastewater
(Villar et al., 2019)
Archaea
Methanomicrobia
Methanomethylovorans hollandica
Anaerobic sediment, Eutrophic freshwater pond
(Podar et al., 2015; Gilmour et al., 2013; Gilmour et al., 2018a, 2018b; Ghimire et al., 2019)
Methanolobus tindarius
Lake sediment, Freshwater
(Gilmour et al., 2013; Gilmour et al., 2018a, 2018b; Ghimire et al., 2019)
Methanoculleus bourgensis
Tannery byproducts, Sewage sludge Digester
(Ghimire et al., 2019)
Methanobrevibacter smithii
Human feces, Wastewater
(Ghimire et al., 2019)
Methanomassiliicoccus luminyensis
Human feces, Wastewater
(Podar et al., 2015; Gilmour et al., 2018a, 2018b)
Mrthanocorpusculum bavaricum
Bulk sea ice, brine
(Christensen et al., 2016; Ghimire et al., 2019)
Methanococcoides methylutens
Marine sediment, Submarine canyon
(Ghimire et al., 2019)
Methanocella paludicola SANAE
Rice paddy
(Christensen et al., 2016)
Methanosphaerula palustris
Peatlands, Minerotrophic fen
(Gilmour et al., 2018a, 2018b)
Methanofollis liminatans
Wastewater treatment plant
(Christensen et al., 2016)
Methanospirillum hungatei IF1
Sewage sludge, Soil, mud
(Yu et al., 2013; Gilmour et al., 2018a, 2018b; Christensen et al., 2016; Yu et al., 2018)
4.2 Key genes involved in Hg methylation
Production of CH3Hg+ by bacteria involves cellular uptake of Hg2+ by active transport. The methylation process occurs in the cytosol followed by its export from the cell. This methylation is associated with reductive acetyl–CoA pathway and corrinoid proteins of the pathway. However, further molecular and genetic evidence is still needed to confirm and elaborate on the underlying pathways (Parks et al., 2013; Regnell and Watras, 2019). Parks et al. (2013) performed genomic analysis for methylators and non-methylators bacteria to understand the genetic and biochemical basis of Hg methylation on the basis that corrinoid iron-sulfur protein transfers methyl groups to a NiFeS cluster in acetyl-CoA synthase. Hence, similar proteins may also mediate the transferring of the methyl group. They utilized the pre-established basis hgcA and hgcB, which are solely required by D. desulfuricans ND132 and G. sulfurreducens PCA to carry out the Hg methylation process. The deletion of both or any of the genes (hgcA and hgcB) abolished the methylation process. Putative corrinoid protein-encoding gene, hgcA, and a 2(4Fe-4S) ferredoxin, hgcB, were found with consistent roles as that of methyl carrier and an electron donor in corrinoid cofactor reduction, respectively. Moreover, the presence of several genes orthologs in confirmed methylators but not in non-methylators suggest a common Hg methylation pathway (Parks et al., 2013). Recently, Yuan et al. (2019) investigated the microbial communities and associated genes responsible for Hg-methylation in the sediments of the South China Sea, Pearl River Estuary, and Pearl River by using sequencing-based (high-throughput) approaches. The SRB and IRB mediated CH3Hg+ production was found significantly correlated with the abundance of hgcA and hgcB genes (Yuan et al., 2019). The methylation was found significantly correlated with the abundance of hgcA and hgcB genes, indicating a close link between CH3Hg+ and Hg methylating bacteria in the aquatic environment. These observations further suggest that the transformation of inorganic Hg into methylated form is closely associated with microbes carrying hgcA/B genes (Yuan et al., 2019). The copies of Archaea-hgcA methylation genes were found 51 – 397% higher in sediments having algae. Similarly, algal biomass-inhabited methylator microbes contributed to methylation approximately 21% of the total Archaea-hgcA. These results provide evidence that both algal organic biomass mediated enhancement of methylators and algae-associated methylators cause increasing hgcA genes, thereby increasing CH3Hg+ production in sediments (Lei et al., 2019).
4.3 Mechanism associated with microbial methylation of Hg
Microbes bind to Hg cations through anionic structures found on the cell surface. The binding of Hg to cellular ligands may disrupt proteins nucleic acids by to sulfhydryl and phosphate/hydroxyl groups, respectively (Ghimire et al., 2019). Despite the damage, Hg transfers into the periplasm (Ayangbenro and Babalola, 2017). The polyelectrolyte nature of the microbial cell wall and the presence of polysaccharide, lipid, and protein in the cell wall are crucial for the binding (Ghimire et al., 2019). Passive diffusion of Hg occurs through the outer layer of microbes, which is then transported to the inner membrane (Ayangbenro and Babalola, 2017), followed by its transport through the inner lipophilic layer. In the case of facilitated diffusion, Hg compounds are transported to the cytoplasm via transmembrane protein pumps. Interestingly, the diffusion-based Hg transport does not require ATP, however, it is dependent on concentration or electrochemical gradient (Ghimire et al., 2019). In the case of active transport, the uptake of Hg depends on thiol compounds (Schaefer and Morel, 2009; Ma et al., 2019). Transport of Hg can also occur through membrane-bound proteins MerP, MerC, and MerT, which facilitate the transfer of Hg from the periplasmic and inner membrane. In this mechanism, Hg attaches to MerP in the periplasm followed by its transport to the cytoplasm through MerT and MerC for further methylation (Ma et al., 2019). This transportation of Hg into cytosol requires high-affinity transport systems; ATP-binding cassette, transporters and P-type ATPase, or couples to cation diffusion facilitator proteins. This transport is further facilitated by P-type ATPase, CBA transporters, and cation diffusion facilitators (Prabhakaran et al., 2016). Furthermore, the methylation of Hg requires the microbes to be resistant to a higher concentration of Hg and involve gene regulation under Hg toxicity stress. To provide a pathway for intracellular information-processing, bacteria contain two-component signaling systems; a bridge between specific adaptive responses and external stimuli with the help of a membrane-bound histidine kinase and a response regulator which regulate gene expression (Prabhakaran et al., 2016; Ghimire et al., 2019). This signal transduction process is further facilitated by sigma factors, methyl-accepting chemotaxis protein, and cyclic-diguanosine monophosphate related proteins and several chemotaxis genes including cheJ, cheB, cheA, cheZ, and cheY. Methyltransferase cheB and methyltransferase cheR are also involved in the processes of signaling and responses to external stimuli (Prabhakaran et al., 2016; Ghimire et al., 2019). Acetyl-CoA pathway is a common pathway for microbial methylation but in the case of D. desulfuricans, the methylation does not require acetyl-CoA, thus suggesting the possibility of an alternate mechanism. DND132_1056 and DND132_1057 genes found in D. desulfuricans ND132 and are thought to encode putative corrinoid protein (Parks et al., 2013). Genes hgcA and hgcB have been found only in Hg methylating microbes, thus may play role in the methylation through encoding corrinoid iron, which is the linkage of carbon monoxide dehydrogenase and acetyl-CoA synthase and sulfur protein (Gionfriddo et al., 2016) which from the reductive acetyl-CoA pathway. MerA and dsrAB are also associated with methylation, which is carbon monoxide dehydrogenase encoding genes. Moreover, organoHg lyase (MerB) is known as a key enzyme related to the methylation process (Gionfriddo et al., 2016). Although the production of CH3Hg+ is generally attributed to the activity of anaerobic bacteria, the formation of CH3Hg+ in the oxic water column negated this idea. Gascón Díez et al., (2016) measured the concentrations of total Hg and CH3Hg+ in settling particles and sediments of Lake Geneva and found that Hg can be methylated in settling particles of oxic environment. It suggests that syntrophs like organisms or SRB are involved in the process (Gascón Díez et al., 2016). The proposed mechanism and pathway used by microbes for methylation of Hg is described in Figs. 3 and 4.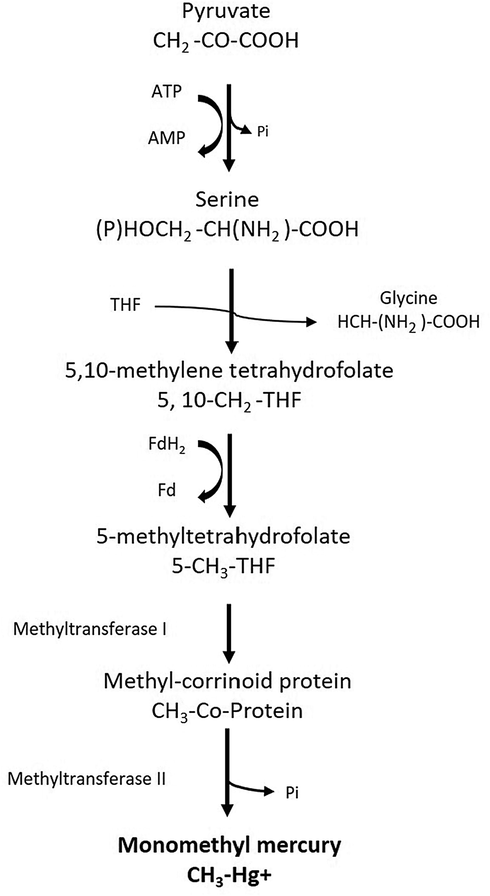
The proposed metabolic pathway involved in Hg methylation by microbes.
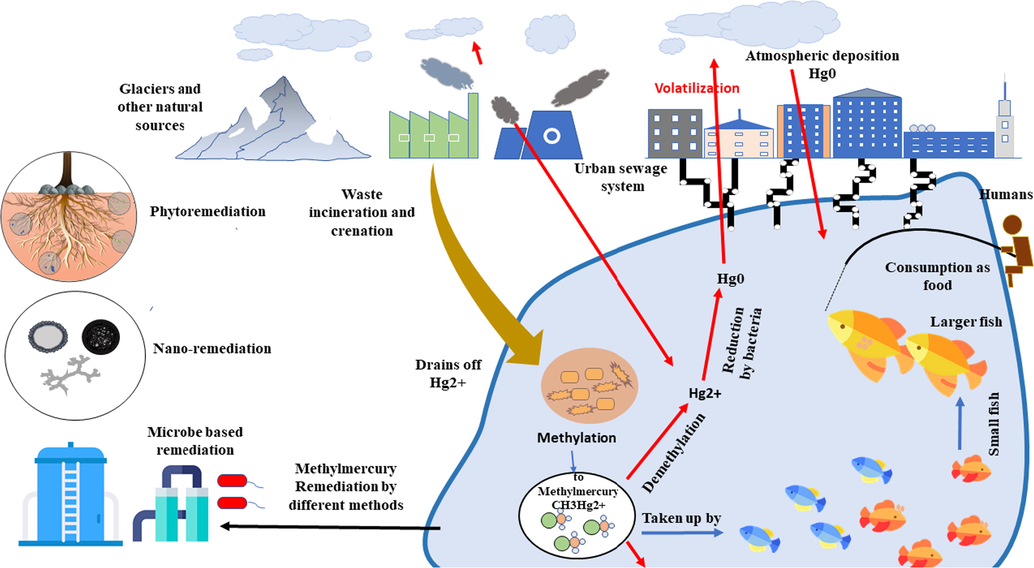
A brief sketch of methyl mercury formation and its remediation.
5 Remediation strategies against Hg
Advance technologies like oxidation, reduction, adsorption, and desorption are used to remove Hg contamination or to reduce highly toxic forms into less toxic (Lewis et al., 2016). Containment and stabilization techniques immobilize the Hg relocation by physical trapping and chemical complexation, respectively (He et al., 2015). In situ capping is an effective remediation option to isolate and immobilize the contaminated sediments by completely sealing contaminants to decreasing the risk of release into the upper aquatic environment. Chemically capped isolation of dissolved metals (activated carbon) is also an active sorption option (Randall and Chattopadhyay, 2013). To stabilize the Hg level with chemicals in contaminants, nitrate (like calcium nitrate) is widely used (Matthews et al., 2013). Nitrate inhibits heavily-reduced conditions in water bodies, while sulfate and iron encourage microbe’s metabolic activity. The prevention of methylmercury synthesis by nitrate has been reported for aquatic systems (Matthews et al., 2013). Hypolimnion and benthic sediments oxygenation can prevent methylmercury production by SRB in many ways. As like, SRB reduces sulfate for a living but does not thrive in oxygenated waters. If more oxygen is provided to the water, less will be the SRB and CH3 zone drop into benthic sediments where oxygenated hypolimnion does not diffuse methylmercury upward and become bioavailable (Munson et al., 2018).
Novel approaches have been examined in recent studies to completely remediate mercury, including techniques and synthetic materials that possess high surface area, active sites, and large porosity for adsorption (Budnik and Casteleyn, 2019; Ghimire et al., 2019; Xu et al., 2019). Activated carbon adsorption or thermal desorption conventional techniques for Hg remediation are not environmentally friendly and less cost-effective, compared to novel innovative techniques. Innovative techniques for soil, water, and air Hg contaminant treatment are based on materials and organisms like bacteria, algae, and plant metabolism. The remediation strategies against Hg and its various forms in different mediums and conditions are mentioned in Table 2. By introducing remediation and/or Hg emission reduction measures significant benefits could be achieved globally. Reduction of mercury emission will reduce the exposure of human and wildlife to methyl mercury. According to Sundseth et al. (2010), the reduction of mercury emission from coal power plants will subsequently reduce particulate matter and sulfur dioxide emission. Particulate matter causes cardiovascular and lung problems while sulfur dioxide induces acidification and corrosion. Substantial potential benefits and uncertainties associated with the reduction of mercury emission are discussed by Sundseth et al. (2010) and Pacyna et al. (2010).
Strategy Used
Type of media
Remarks
References
Nano-material
Fe3O4
Water containing Hg2+
Absorbed 181.8 mg/g Hg
(Wang et al., 2020a, 2020b)
Single-walled carbon nanotubes
Absorbed 1666 mg/g Hg
Multi-walled carbon nanotubes
Absorbed 112 mg/g Hg
Silica-coated Fe3O4 nanoparticles
Absorbed 82 mg/g Hg
Gold
99% Hg absorbance
CeO2
Flue Gas containing Hg0
86.5% Hg absorbance
Mesoporous silica
Water containing Hg2+
Absorbed 42.8 mg/g Hg
ZnS
Flue Gas containing Hg0
Absorbed 0.50 mg/g Hg
FeOOH
Water containing Hg2+
Absorbed 217 mg/g Hg
Graphene oxide
Absorbed 829.27 mg/g Hg
MoS2
Absorbed 425.5 mg/g Hg
Nano-cellulose
Absorbed 1989 mg/g
Carbon nanofiber
Flue Gas containing Hg0
90% Hg absorbance
CuS
Absorbed 122.4 mg/g Hg
Polymers
Sulfur
Flue Gas containing Hg0
Absorbed 0.151 mg/g Hg
(Wang et al., 2020a, 2020b)
Polypyrrole
Absorbed 2042.7 mg/g Hg
Zr(IV) phosphoborate
Absorbed 153.85 mg/g Hg
Terpolymer hydrogel
Absorbed 1172.97 mg/g Hg
Sulfur
84% Hg absorbance
Chitosan
Absorbed 1594 mg/g Hg
Graphene
Graphene oxide
Water containing Hg2+
68.8 mg/g adsorption potential
(Wang et al., 2020a, 2020b)
Magnetic Graphene oxide
71.3 mg/g adsorption potential
Silver Graphene oxide
280.8 mg/g adsorption potential
Dithiocarbamate Magnetic Graphene oxide
181.82 mg/g adsorption potential
Thymine oligonucleotide Graphene oxide
180.18 mg/g adsorption potential
Other material
Manganese oxides
Hg0 in flue gas
Efficiently removed Hg0
(Scala and Cimino, 2015)
Birnessite
in situ Hg contaminated sediments
Significantly removed
(Leven et al., 2018)
Pyrolusite
Significantly removed
(Vlassopoulos et al., 2018)
Chelating resin
Water containing Hg2+
526.9 mg/g removal capacity
(Xiong et al., 2015)
Phytoremediation
Cyrtomium macrophyllum
Residues of the mining area
Accumulated 36 mg/kg Hg
(Xun et al., 2017)
Festuca rubra
Field study/mining area
Accumulated 84 mg/kg Hg
(Fernández et al., 2017)
Leontodon taraxacoides
Accumulated 78 mg/kg Hg
Equisetum telmateya
Accumulated 77 mg/kg Hg
Datura stramonium
Gold mining area
Significantly accumulated Hg
(Mbanga et al., 2019)
Phragmites australis
Hyperaccumulation of Hg in roots and leaves
Persicaria lapathifolia
Melilotus alba
Accumulated Hg at a minimal level
Panicum coloratum
Accumulated Hg
Cyperus eragrostis
Waste derived material/Biochar/Bio-sorbents
Pine cone
Hg contaminated soil
(Beckers et al., 2019)
Rice husk
73% adsorptive potential
(Wang et al., 2020a, 2020b)
Pine sawdust
Hg contaminated water
Cotton straw
Hg contaminated flue gas
Seaweed
(Liu et al., 2018)
Waste tea
(Wang et al., 2020a, 2020b)
Tobacco straw
Seaweed
Wheat straw
Bioelastomeric foam
Significantly adsorbed and removed Hg
Waste tire
Coal fly ash
Flue Gas containing Hg0
Algae and fungi-based remediation
Ulva lactuca
Wastewater containing Hg2+
Due to several functional groups, significantly accumulated Hg
(Henriques et al., 2015)
Gracilaria gracilis
Fucus vesiculosus
Aspergillus sp. A31
In vitro study
Removed 100% Hg in Culture medium
(Pietro-Souza et al., 2020)
Curvularia geniculata P1
Lindgomycetaceae P87
Westerdykella sp. P71
Aspergillus niger
Potato dextrose agar (PDA) supplemented with HgCl2
Adsorbed 25 mg/kg of Hg
(Hindersah et al., 2018)
Aspergillus flavus
Bacterial remediation of Mercury
Enterobacter cloacae
Plate count agar with CH3Hg and CH3HgCl
Significant capacity of CH3Hg degradation
(Adelaja and Keenan, 2012)
Alkaligenes faecalis
Pseudomonas fluorescens
Citrobacter braakii
Pseudomonas putida SP-1
Bioremediation of HgCl2
89% removal of Hg, volatilized 100% of total Hg
(Zhang et al., 2012)
5.1 Phytoremediation
Phytoremediation, an essential remediation process to degrade, immobilize, volatize or extract soil and certain plant contaminants by using plants (Fig. 5), also called hyper-accumulators. (Liu et al., 2018; Mbanga et al. 2019). Plant's ability to accumulate high concentrations of toxic metals above the ground by natural owing or microorganism’s assessment knows as hyper-accumulation. Many plant species have been reported as Hg hyper-accumulators (Chamba et al., 2017). Xun et al. (2017). The ability of Cyrtomium macrophyllum plant examined by pot experiment in a Hg-contaminated mining area to extract mercury and observed that the mercury level reached 36 mg kg−1 with 2.62 translocation factor, and also a high resistance to mercury stress was shown by cyrtomium macrophyllum leaf tissues. A study using native plant species (like Equisetum telmateya; Leontodon taraxacoides, and Festuca rubra), conducted by Fernández et al. (2017) in mercury and arsenic (Hg-As) mining area to extract mercury showed that the Hg accumulated in plant leaves with 77, 78 and 84 mg kg-1, as the highest concentration reached, respectively. Mercury concentrations may vary in different tissues of the plants and mostly accumulated by roots, followed by leaves and stems, because of the transport function of stems it can accumulate very less Hg, compared to leaves which showed 2-fold higher accumulation of Hg (Marrugo-Negrete et al., 2015, Marrugo-Negrete et al. 2016). Other plant species including Jatropha curcas, Spartina alterniflora, Phragmites australis, Typha latifolia, Rhus lancea, Eucalyptus camaldulensis, and P. australis have been utilized in the phytoremediation of Hg. Mbanga et al. (2019) studied the remediation abilities of some plant species, such as Datura stramonium, Phragmites australis, Panicum coloratum, Cyperus eragrostis, Persicaria lapathifolia, and Melilotus alba, in wetland contaminants in a gold mine. Phragmites australis was found with the highest remediation ability during the dry season, whereas Melilotus alba with lowest remediation ability during both dry and wet seasons. In situ detoxification strategy can be induced by developing genetically transformed plants. Such as transforming the plants with bacterial enzyme organomercurial lyase genes (merA/B) for organic Hg detoxification, enhances CH3Hg+ concentrations than wild-type plants. Plants having genes (merA/B) showed 10-fold higher concentrations of Hg, compared to plants with merB alone. The above discussion indicates the value of these genes for a plant to convert Hg to volatile and much less toxic Hg0, thus detoxify organic Hg (Bizily et al., 2000).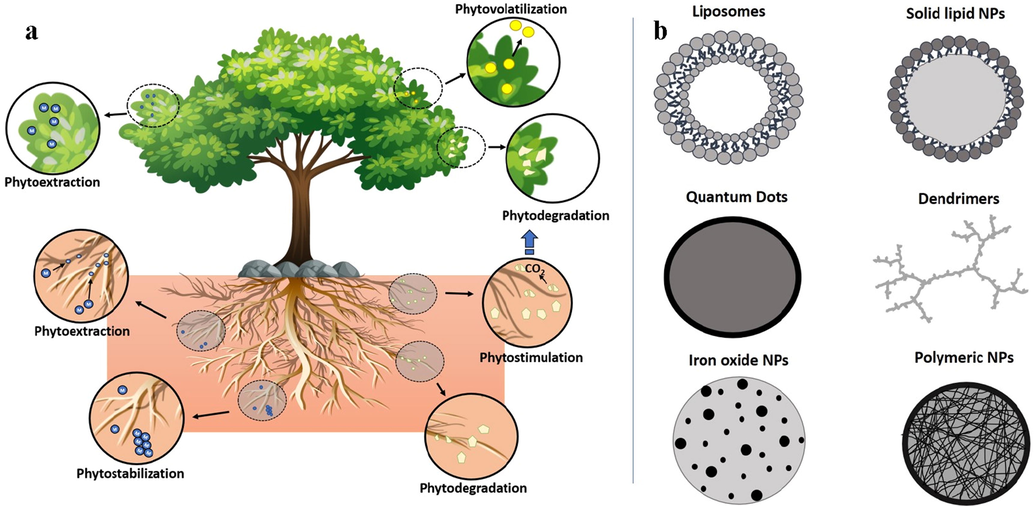
(a) Phytoremediation and (b) Nano polymers for remediation of CH3Hg+contaminants.
5.2 Nanotechnology-based remediation
Remediation techniques based on nanotechnology gained attention on heavy metals removal from water, air, and soil, because of low cost, nonhazardous to the environment, and high efficacy (Fig. 5). For Hg removal, different techniques like, immobilization, adsorption, and stabilization by nanoparticles are used (Almaroai et al., 2014). Series of innovative catalysts having combine advantages of pillared-clay (PILCs) and Ce-Mn were evaluated for the removal of elemental mercury, and more than 90% removal efficiency was reported (He et al., 2014). Jampaiah et al. (2019) used TiO2 nanorods-supported MnOx-FeOx-CrOx catalyst and carried out Hg removal (∼80–83%) at low temperature in the presence of oxygen. Similarly, Chalkidis et al. (2019) carried out maximum elemental mercury removal with the help of α-MnO2 nanotubes. Adsorptive materials remediate Hg by adsorption in internal structures and on the surface (Wei et al., 2013). Adsorption is an effective technique for the removal of heavy metals in soil contaminants.
During the remediation processes, toxic contaminants are rapidly converted into significantly less toxic products (Wei et al., 2013). Different effective nanomaterials can be utilized to remove heavy metals from the wastewater (Kumar et al., 2014) or aquifer (Rabbani et al., 2016). For the removal of Hg, either calorimetric based or fluorescence-based nanoparticles are used, like gold nanoparticles with 11- mercaptoundecanoic acid and chitosan. Zero-Valent Iron Nanoparticles (ZVIN) are small in size, nontoxic (Sugunan et al., 2004), and having significant reducing and adsorption properties that make them to react with heavy metals such as Hg (Hg2+) (Cundy et al., 2008), Chromium (Cr6+), Cadmium (Cd2+), Nickel (Ni2+), and Lead (Pb2+) from soil contaminants (Caliman et al., 2011). The main mechanisms of Hg remediation are adsorption and reduction that convert heavy metals to less toxic species, immobilized, or limited available (Cundy et al., 2008). Nanoparticles like, stabilized iron sulfide (FeS) with carboxymethyl cellulose is used in fields and sediments to immobilize Hg (Gong et al., 2012). For immobilization of sedimented mercury, FeS is a good option. Xiong et al. (2015) reported that FeS nanoparticles treated mercury contaminants at 26.5 M ratio (FeS-to-Hg), reduced 97% mercury concentration in the water body with 99% TCLP leachability (Rabbani et al., 2016).
5.3 Microbial remediation
Bacteria resistant to Hg contain organomercurial lyase (merB) enzyme, that demethylate toxic (organic) methylmercury (CH3Hg+) to less toxic Hg (inorganic) (Ralston et al., 2007) and mercuric ion reductase enzyme (merA) that catalyzes IHg reduction to Hg0, which is less harmful e.g. Pseudomonas aeruginosa and Proteus species (Ayangbenro and Babalola, 2017). Applying microbial action to remediation involves the transformation of the genes (hgcA and hgcB) from microorganisms that convert CH3Hg+ or other Hg compounds into Hg0. Microorganisms also play a critical role in the process of Hg remediation by plants, such as rhizosphere bacteria (mostly highlighted) (Liu et al., 2018). Rhizosphere bacteria increase the bioavailability of metal in different ways, such as alteration of the soil pH, redox reactions, and releasing chelators (Chamba et al. 2017). Franchi et al. (2017), reported that plant-associated growth-promoting bacteria (PGPB) increased 45% of phytoextraction efficacy for mercury remediation. Metal mobility and bioavailability alteration encourage the phytoremediation process by PGPB.
6 Conclusion and future prospective
Environmental contamination by Hg is one of the most vital issues of the contemporary world. In the past few decades, industrialization, transportations, human travels, animals’ migrations, and erosion of mercury-containing soil are increased that enhanced the Hg released in the environment and subsequent availability to the food chain. Methylated Hg is a highly toxic specie that already occupied space in the environment could be more alarming for humans and other terrestrial and aquatic life shortly. Therefore, the concerned authorities must adopt efficient and sustainable strategies to cope with the problem of Hg entry into the environment and food chain and minimize the associated risks. One of the promising strategies for effective remediation of Hg can be adsorption which can be made cost-effective by applying waste-derived substances for Hg adsorption and reusing of the adsorption materials. However, further research is crucial to develop methods for the regeneration of adsorbents and identify materials having higher reuse abilities. Moreover, further studies should focus on exploration, characterization, and developing mercury resistant microorganisms and plant species that could efficiently uptake methyl mercury from soil and water sources and convert them into the non-toxic usable form.
In addition, Hg methylation processes based on biotic and abiotic strategies are equally important, however, biotic processes are more dominant sources of methylmercury in aquatic and soil environments. Therefore, it is deemed necessary that biological entities related to methylation and demethylation should be investigated on wide scales. These studies could help in the identification of the most efficient species to further control the mercury methylation processes. There is a complex relationship between environmental stimulators and microbial communities’ regulations that are required for methylmercury formation and degradation. Changes in both environmental factors and gene regulations affect the shifts in abiotic and biotic processes, respectively. Therefore, understanding these complex relationships can help to determine the rates, efficiencies, and impact of different methylation processes. Notably, environmental factors can affect both biotic and abiotic methylation processes at the same time. These relationships are often very complex to understand. It is, therefore, necessary to estimate the levels of mercury in aquatic environments, soils, and cryosphere reservoirs worldwide and identify the key factors associated with their depositions. In addition, the identification of conditions most favorable to methylation and demethylation, associated microbial communities and genes, and forms of mercury (or its compounds) that could be non-toxic, can help to develop strategies to protect the living organisms from the adverse effects of toxic mercury.
Funding
The current project was funded by the Postdoctoral research grant from Chinese Postdoctoral Science foundation, China for Suliman Khan and National Natural Science Foundation of China (grant number 31501260).
Acknowledgments
We acknowledge Postdoctoral research grant from Second Affiliated Hospital of Zhengzhou University (SK), Region of Mid-line of South-to-North Diversion Project, Collaborative Innovation Center of Water Security for Water Source Region of Mid-line of South-to-North Diversion Project of Henan Province (PD), PIFI Fellowship from the Chinese Academy of Sciences (2020PC0052) (WS).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Environmental mercury and its toxic effects. J. Prev. Med. Public. Heal. 2014;47(2):74.
- [Google Scholar]
- Cycling of mercury in the environment: Sources, fate, and human health implications: A review. Crit. Rev. Env. Sci. Technol.. 2017;47:693-794.
- [Google Scholar]
- Mercury methylation-related microbes and genes in the sediments of the Pearl River Estuary and the South China Sea. Ecotoxicol. Environ. Saf.. 2019;185:109722
- [Google Scholar]
- High methylation rates of mercury bound to cysteine by Geobacter sulfurreducens. Nat. Geosci.. 2009;2:123-126.
- [Google Scholar]
- Microbial mercury methylation in aquatic environments: A critical review of published field and laboratory studies. Environ. Sci. Technol.. 2019;53:4-19.
- [Google Scholar]
- Abiotic methylation of mercury in the aquatic environment. Sci. Total Environ.. 2006;368:126-137.
- [Google Scholar]
- The toxicology of mercury and its chemical compounds. Crit. Rev. Toxicol.. 2006;36:609-662.
- [Google Scholar]
- Overview of the clinical toxicity of mercury. Ann. Clin. Biochem.. 2006;43:257-268.
- [Google Scholar]
- Mercury pollution in modern times and its socio-medical consequences. Sci. Total Environ.. 2019;654:720-734.
- [Google Scholar]
- Genome-wide association study to identify genes related to renal mercury concentrations in mice. Environ. Health Perspect.. 2016;124:920-926.
- [Google Scholar]
- Effect of plastic foreign body impaction on rumen function and heavy metal concentrations in various body fluids and tissues of buffaloes. Ecotoxicol. Environ. Saf.. 2020;189:109972
- [CrossRef] [Google Scholar]
- Spatial patterns and temporal trends in mercury concentrations in common loons (Gavia immer) from 1998 to 2016 in New York’s Adirondack Park: has this top predator benefitted from mercury emission controls? Ecotoxicology. 2019;6:1-12.
- [CrossRef] [Google Scholar]
- Sources, bioaccumulation, health risks and remediation of potentially toxic metal(loid)s (As, Cd, Cr, Pb and Hg): an epitomised review. Environ. Monit. Assess.. 2020;192:108.
- [Google Scholar]
- Seasonal cycling and transport of mercury and methylmercury in the turbidity maximum of the Delaware estuary. Aquat. Geochem.. 2016;22:313-336.
- [Google Scholar]
- Mobility and transport of mercury and methylmercury in peat as a function of changes in water table regime and plant functional groups. Global Biogeochem. Cycles. 2017;31:233-244.
- [Google Scholar]
- Anaerobic mercury methylation and demethylation by Geobacter bemidjiensis Bem. Environ. Sci. Technol.. 2016;50:4366-4373.
- [Google Scholar]
- Historical Records of Mercury Stable Isotopes in Sediments of Tibetan Lakes. Sci. Rep.. 2016;6:1-10.
- [Google Scholar]
- Mercury methylating microbial communities of boreal forest soils. Sci. Rep.. 2019;9:1-13.
- [Google Scholar]
- Role of settling particles on mercury methylation in the oxic water column of freshwater systems. Environ. Sci. Technol.. 2016;50:11672-11679.
- [Google Scholar]
- Importance of dissolved neutral mercury sulfides for methyl mercury production in contaminated sediments. Environ. Sci. Technol.. 2007;41:2270-2276.
- [Google Scholar]
- High methylmercury formation in ponds fueled by fresh humic and algal derived organic matter. Limnol. Oceanogr.. 2018;63:S44-53.
- [Google Scholar]
- Linking microbial activities and low-molecular-weight thiols to Hg methylation in biofilms and periphyton from high-altitude tropical lakes in the Bolivian altiplano. Environ. Sci. Technol.. 2018;52:9758-9767.
- [Google Scholar]
- Mechanisms of algal biomass input enhanced microbial Hg methylation in lake sediments. Environ. Int.. 2019;126:279-288.
- [Google Scholar]
- A comparison of total mercury and methylmercury export from various Minnesota watersheds. Sci. Total Environ.. 2005;340:261-270.
- [Google Scholar]
- Effects of stream water chemistry and tree species on release and methylation of mercury during litter decomposition. Environ. Sci. Technol.. 2008;42:8692-8697.
- [Google Scholar]
- Molecular composition of organic matter controls methylmercury formation in boreal lakes. Nat. Commun.. 2017;8:1-9.
- [Google Scholar]
- Geobacteraceae are important members of mercury-methylating microbial communities of sediments impacted by waste water releases. ISME J.. 2018;12:802-812.
- [Google Scholar]
- Potential of thermal treatment for decontamination of mercury containing wastes from chlor-alkali industry. J. Hazard. Mater.. 2011;186(1):114-118.
- [Google Scholar]
- Oxidation and methylation of dissolved elemental mercury by anaerobic bacteria. Nat. Geosci.. 2013;6:751-754.
- [Google Scholar]
- A New Strategy for Heavy Metal Polluted Environments : A Review of Microbial Biosorbents. Int. J. Environ. Res. Public Health. 2017;14(1):94.
- [CrossRef] [Google Scholar]
- Phytodetoxification of hazardous organomercurials by genetically engineered plants. Nat. Biotechnol.. 2000;18:213-217.
- [Google Scholar]
- Dynamic mercury methylation and demethylation in oligotrophic marine water. Biogeosciences. 2018;15:6451-6460.
- [Google Scholar]
- The genetic basis for bacterial mercury methylation. Science. 2013;80(339):1332-1335.
- [Google Scholar]
- RSC Advances Microbial stress response to heavy metals in the. RSC Adv.. 2016;6:109862-109877.
- [Google Scholar]
- Technology Mercury methylation by anaerobic microorganisms : A review. Crit. Rev. Env. Sci. Technol. 2019:1-44.
- [Google Scholar]
- Water level fluctuations influence microbial communities and mercury methylation in soils in the Three Gorges Reservoir, China. J. Environ. Sci. (China). 2018;68:206-217.
- [Google Scholar]
- Microbial methylation of mercury in the water-level fluctuation zone of the Three Gorges Reservoir, China. J. Environ. Sci. (China). 2018;68:218-220.
- [Google Scholar]
- Methylation of inorganic mercury in polar marine waters. Nat. Geosci.. 2011;4:298-302.
- [Google Scholar]
- Mercury methylation by Geobacter metallireducens GS-15 in the presence of Skeletonema costatum. Sci. Total Environ.. 2019;671:208-214.
- [Google Scholar]
- Microbial analyses of ancient ice core sections from Greenland and Antarctica. Biology (Basel). 2013;2:206-232.
- [Google Scholar]
- Methylated mercury species in Canadian high Arctic marine surface waters and snowpacks. Environ. Sci. Technol.. 2007;41:6433-6441.
- [Google Scholar]
- Mercury in the Canadian Arctic Terrestrial Environment: An Update. Sci. Total Environ.. 2015;509–510:28-40.
- [Google Scholar]
- Mercury contaminated sediment sites—An evaluation of remedial options. Environ. Res.. 2013;125:131-149.
- [Google Scholar]
- Microbes in high arctic snow and implications for the cold biosphere. Appl. Environ. Microbiol.. 2011;77:3234-3243.
- [Google Scholar]
- Cryospheric ecosystems: a synthesis of snowpack and glacial research. Environ. Res. Lett.. 2015;10:110201
- [Google Scholar]
- Potential drivers of microbial community structure and function in Arctic spring snow. Front. Microbiol.. 2014;5:413.
- [Google Scholar]
- Microbial ecology of the cryosphere : sea ice and glacial habitats. Nat Publ Gr 2015:1-14.
- [Google Scholar]
- Psychrophilic lifestyles: mechanisms of adaptation and biotechnological tools. Appl. Microbiol. Biotechnol.. 2019;103:2857-2871.
- [Google Scholar]
- Geochemical influences and mercury methylation of a dental wastewater microbiome. Sci. Rep.. 2015;5:1-20.
- [Google Scholar]
- Anaerobic oxidation of Hg (0) and methylmercury formation by Desulfovibrio desulfuricans ND132. Geochim. Cosmochim. Acta. 2013;112:166-177.
- [Google Scholar]
- Mercury exposed: advances in environmental analysis and ecotoxicology of a highly toxic metal. Environ. Toxicol. Chem.. 2013;32:2175-2178.
- [Google Scholar]
- Mercury accumulation in Mediterranean Fish and Cephalopods Species of Sicilian coasts: correlation between pollution and the presence of Anisakis parasites. Nat. Prod. Res.. 2017;31:1156-1162.
- [Google Scholar]
- Trends of mercury emissions from the Chlor-Alkali industry in EECCA countries. International J. Environ. Sci.. 2012;2(3):1585-1595.
- [Google Scholar]
- A review of mercury bioavailability in humans and fish. Int. J. Environ. Res. Public Health. 2017;14:169.
- [Google Scholar]
- Mercury accumulation in Yellowfin tuna (Thunnus albacares) with regards to muscle type, muscle position and fish size. Food Chem.. 2016;190:351-356.
- [Google Scholar]
- Mobility of mercury in soil and its transport into the sea. Environ. Sci. Pollut. Res.. 2020;27:8492-8506.
- [Google Scholar]
- UNEP, 2019. United Nation Environment Protection.
- Emission-dominated gas exchange of elemental mercury vapor over natural surfaces in China. Atmos. Chem. Phys.. 2016;16(17):11125-11143.
- [Google Scholar]
- Mercury mass flow in iron and steel production process and its implications for mercury emission control. J. Environ. Sci.. 2016;43:293-301.
- [Google Scholar]
- Mercury flows in large-scale gold production and implications for Hg pollution control. J. Environ. Sci.. 2018;68:91-99.
- [Google Scholar]
- Leaching behaviour of mercury from hazardous solid waste generated by chlor-alkali industry. EDP Sciences; 2013.
- 60-Day chronic exposure to low concentrations of HgCl2 impairs sperm quality: hormonal imbalance and oxidative stress as potential routes for reproductive dysfunction in rats. PLoS ONE. 2014;9(11)
- [Google Scholar]
- Elemental analysis of aging human pituitary glands implicates mercury as a contributor to the somatopause. Front. Endocrinol.. 2019;10:419.
- [Google Scholar]
- Mercury remediation in wetland sediment using zero-valent iron and granular activated carbon. Environ. Pollut.. 2016;212:366-373.
- [Google Scholar]
- Phytoremediation of mercury-contaminated soils by Jatropha curcas. Chemosphere. 2015;127:58-63.
- [Google Scholar]
- Erato polymnioides–A novel Hg hyperaccumulator plant in ecuadorian rainforest acid soils with potential of microbe-associated phytoremediation. Chemosphere. 2017;188:633-641.
- [Google Scholar]
- Tolerance of TBT-resistant bacteria isolates to methylmercury. Res. J. Environ. Sci.. 2012;6:1-13.
- [Google Scholar]
- Natural and synthesised iron-rich amendments for As and Pb immobilisation in agricultural soil. Chem. Ecol.. 2014;30:267-279.
- [Google Scholar]
- Syntrophs dominate sequences associated with the mercury methylation-related gene hgcA in the water conservation areas of the Florida Everglades. Appl. Environ. Microbiol.. 2014;80:6517-6526.
- [Google Scholar]
- Impact of biochar on mobilization, methylation, and ethylation of mercury under dynamic redox conditions in a contaminated floodplain soil. Environ. Int.. 2019;127:276-290.
- [Google Scholar]
- Soil and groundwater cleanup: benefits and limits of emerging technologies. Clean Technol. Environ. Policy. 2011;13:241-268.
- [Google Scholar]
- Development and validation of broad-range qualitative and clade-specific quantitative molecular probes for assessing mercury methylation in the environment. Appl. Environ. Microbiol.. 2016;82:6068-6078.
- [Google Scholar]
- Use of iron-based technologies in contaminated land and groundwater remediation: A review. Sci. Total Environ.. 2008;400:42-51.
- [Google Scholar]
- Phytoremediation capability of native plant species living on Pb-Zn and Hg-As mining wastes in the Cantabrian range, north of Spain. J. Geochem. Explor.. 2017;174:10-20.
- [Google Scholar]
- Microbial mercury methylation in the cryosphere: Progress and prospects. Sci. Total Environ.. 2019;697:134150
- [CrossRef] [Google Scholar]
- Robust mercury methylation across diverse methanogenic Archaea. MBio. 2018;9:e02403-02417.
- [Google Scholar]
- Sulfate-reducing bacterium Desulfovibrio desulfuricans ND132 as a model for understanding bacterial mercury methylation. Appl. Environ. Microbiol.. 2011;77:3938-3951.
- [Google Scholar]
- Mercury methylation by novel microorganisms from new environments. Environ. Sci. Technol.. 2013;47:11810-11820.
- [Google Scholar]
- Desulfovibrio desulfuricans bacteremia and review of human Desulfovibrio infections. J. Cli. Microbiol. 2003;41:2752-2754.
- [Google Scholar]
- Immobilization of mercury in field soil and sediment using carboxymethyl cellulose stabilized iron sulfide nanoparticles. Nanotechnology. 2012;23:294007
- [Google Scholar]
- Study on bioaccumulation and biosorption of mercury by living marine macroalgae: prospecting for a new remediation biotechnology applied to saline waters. Chem. Eng.. 2015;281:759-770.
- [Google Scholar]
- Isolation of mercury-resistant fungi from mercury-contaminated agricultural soil. Agriculture. 2018;8:33.
- [Google Scholar]
- Application of carbon nanotubes in heavy metals remediation. Crit. Rev. Env. Sci. Tec. 2014;44:1000-1035.
- [Google Scholar]
- Characterization of manganese oxide amendments for in situ remediation of mercury-contaminated sediments. Environ Sci-Proc Imp. 2018;20:1761-1773.
- [Google Scholar]
- Removal of elemental mercury by bio-chars derived from seaweed impregnated with potassium iodine. Chem. Eng.. 2018;339:468-478.
- [Google Scholar]
- Mercury accumulation and biotransportation in wetland biota affected by gold mining. Environ. Monit. Assess.. 2019;191:186.
- [Google Scholar]
- Mercury resistance and bioremediation mediated by endophytic fungi. Chemosphere. 2020;240:124874
- [Google Scholar]
- Global prevalence and distribution of genes and microorganisms involved in mercury methylation. Sci. Adv.. 2015;1:e1500675
- [Google Scholar]
- Application of nanotechnology to remediate contaminated soils, Environmental Remediation Technologies for Metal-Contaminated Soils. Springer 2016:219-229.
- [Google Scholar]
- Elemental mercury capture and oxidation by a regenerable manganese-based sorbent: The effect of gas composition. Chem. Eng.. 2015;278:134-139.
- [Google Scholar]
- Dethiobacter alkaliphilus gen. nov. sp. nov., and Desulfurivibrio alkaliphilus gen. nov. sp. nov.: two novel representatives of reductive sulfur cycle from soda lakes. Extremophiles. 2008;12:431-439.
- [Google Scholar]
- Sugunan, A., Juilland, P., Thanachayanont, C., Dutta, J., Hilborn, J.G., 2004. Synthesis of bio-compatible gold nanoparticles. In: Proceedings of International Conference on Smart Materials (SMARTMAT-04), Chiang Mai, Thailand.
- Villar, E., Cabrol, L., Heimbürger-Boavida, L.E., 2019. Widespread microbial mercury methylation genes in the global ocean. bioRxiv, 648329.
- Manganese (iv) oxide amendments reduce methylmercury concentrations in sediment porewater. Environ Sci-Proc Imp. 2018;20:1746-1760.
- [Google Scholar]
- Remediation of mercury contaminated soil, water, and air: A review of emerging materials and innovative technologies. Environ. Int.. 2020;134:105281
- [Google Scholar]
- Influence of waste dump remediation on the levels of mercury in the air. Bull. Environ. Contam. Toxicol.. 2010;84(5):623-627.
- [Google Scholar]
- Applications of nanomaterial-based membranes in pollution control. Crit. Rev. Env. Sci. Tec. 2013;43:2389-2438.
- [Google Scholar]
- Selective removal of Hg (II) with polyacrylonitrile-2-amino-1, 3, 4-thiadiazole chelating resin: batch and column study. Chem. Eng.. 2015;259:257-265.
- [Google Scholar]
- Mercury accumulation plant Cyrtomium macrophyllum and its potential for phytoremediation of mercury polluted sites. Chemosphere. 2017;189:161-170.
- [Google Scholar]
- Mercury methylation by the methanogen Methanospirillum hungatei. Appl. Environ. Microbiol.. 2013;79:6325-6330.
- [Google Scholar]
- Characterization of a marine-isolated mercury-resistant Pseudomonas putida strain SP1 and its potential application in marine mercury reduction. Appl. Microbiol. Biotechnol.. 2012;93:1305-1314.
- [Google Scholar]
- In situ remediation technologies for mercury-contaminated soil. Environ. Sci. Pollut. Res.. 2015;22(11):8124-8147.
- [Google Scholar]
- Whole-lake nitrate addition for control of methylmercury in mercury-contaminated Onondaga Lake, NY. Environ. Res.. 2013;125:52-60.
- [Google Scholar]
- Screening of native plant species for phytoremediation potential at a Hg-contaminated mining site. Sci. Total Environ.. 2016;542:809-816.
- [Google Scholar]
- Importance of molar ratios in selenium-dependent protection against methylmercury toxicity. Biol. Trace Elem. Res.. 2007;119(3):255-268.
- [Google Scholar]
- Phytoremediation of a multi contaminated soil: mercury and arsenic phytoextraction assisted by mobilizing agent and plant growth promoting bacteria. J Soil Sediment. 2017;17(5):1224-1236.
- [Google Scholar]
- Air Contamination by mercury, emissions, and transformations—a review. Water Air Soil Pollut.. 2017;228:123.
- [Google Scholar]
- World emissions of mercury from small scale and artisanal gold mining. In: Pirrone N., Mason R., eds. Mercury fate and transport in the global atmosphere: emissions, measurements and models. New York: Springer; 2009. p. :131-172.
- [Google Scholar]
- 500 years of mercury production: global annual inventory by region until 2000 and associated emissions. Sci. Total Environ.. 2003;304(1):13-27.
- [Google Scholar]
- Observed decrease in atmospheric mercury explained by global decline in anthropogenic emissions. PNAS. 2016;113(3):526-531.
- [Google Scholar]
- Wilson, S., Kindbom, K., Yaramenka, K., Steenhuisen, F., Telmer, K., 2012. Part A: global emissions of mercury to the atmosphere. UNEP/AMAP 2012, Technical Report. www.unep.org/hazardoussubstances/GlobalAtmosphericM.
- Low-temperature elemental mercury removal over TiO2 nanorods-supported MnOx-FeOx-CrOx. Catal. Today. 2019;324:174-182.
- [Google Scholar]
- Regenerable α-MnO2 nanotubes for elemental mercury removal from natural gas. Fuel Process. Technol.. 2019;193:317-327.
- [Google Scholar]
- Adsorption and Oxidation of Elemental Mercury over Ce-MnOx/TiPILCs. Environ. Sci. Technol.. 2014;48(14):7891-7898.
- [Google Scholar]
- Metals Extraction from Sulfide Ores with Microorganisms: The Bioleaching Technology and Recent Developments. Trans. Indian Inst. Met.. 2019;72(3):559-579.
- [CrossRef] [Google Scholar]
- Dissolution of Cu and Zn-bearing ore by indigenous iron-oxidizing bacterial consortia supplemented with dried bamboo sawdust and variations in bacterial structural dynamics: A new concept in bioleaching. Sci. Total Environ.. 2020;709(2020):136136
- [CrossRef] [Google Scholar]
- Bioleaching of copper- and zinc-bearing ore using consortia of indigenous iron-oxidizing bacteria. Extremophiles. 2018;22:851-863.
- [CrossRef] [Google Scholar]
- Remediation of lead-contaminated sediment by biochar-supported nano-chlorapatite: accompanied with the change of available phosphorus and organic matters. J. Hazard. Mater.. 2018;348:109-116.
- [Google Scholar]
- Synergistic removal of copper and tetracycline from aqueous solution by steam-activated bamboo-derived biochar. J. Hazard. Mater.. 2020;384 121470–121470
- [CrossRef] [Google Scholar]
- Eco-friendly remediation for lead-contaminated riverine sediment by sodium lignin sulfonate stabilized nano-chlorapatite. Chem. Eng.. 2020;397:125396
- [Google Scholar]
- Stabilized nanoscale Zerovalent iron mediated cadmium accumulation and oxidative damage of Boehmeria nivea (L.) Gaudich Cultivated in Cadmium Contaminated Sediments. Environ. Sci. Technol.. 2017;51(19):11308-11316.
- [Google Scholar]
- Economic benefits from decreased mercury emissions: Projections for 2020. J Clea Prod. 2010;18(4):386-394.
- [Google Scholar]
- An assessment of costs and benefits associated with mercury emission reductions from major anthropogenic sources. J. Air Waste Manag. Assoc.. 2010;60(3):302-315.
- [CrossRef] [Google Scholar]
- Increased methylmercury accumulation in rice after straw amendment. Environ. Sci. Technol.. 2019;53(11):6144-6153.
- [CrossRef] [Google Scholar]
- Britt, DHall., Vincent L.S.L., 2004. Methylmercury and total mercury in plant litter decomposing in upland forests and flooded landscapes. Environ. Sci. Technol. 38(19), 5010–5021.
- Mercury Methylation from Unexpected Sources: Molybdate-Inhibited Freshwater Sediments and an Iron-Reducing Bacterium. Appl. Environ. Microbiol.. 2006;72(1):457-464.
- [Google Scholar]







