Translate this page into:
Comparative characterization of the metabolites of phloretin and phlorizin in rats using UHPLC-Q-Exactive Orbitrap mass spectrometer
⁎Corresponding author at: BIN ZHOU Medical University, No. 346 Guanhai Road, Lai’shan District, Yantai 264003, China. upgoping@gmail.com (Jiayu Zhang)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Both phloretin and phlorizin (phloretin 2′-O-glucoside) extracted from the peel of apples are attributed to particular flavonoid dihydrochalcones with multiple pharmacological activities. However, metabolite structural characterization of these two components, which may accumulate to exert their pharmacological effects, remains insufficient. The present study aimed to comparatively clarify the metabolic pathways of phloretin and phlorizin after oral administration individually to Sprague-Dawley (SD) rats. Therefore, a rapid, integrate and systematic analytical strategy based on characteristic fragment ion fishing was proposed for the screening and identification of metabolites coming from phloretin and phlorizin using UHPLC-Q-Exactive Orbitrap mass spectrometry in parallel reaction monitoring mode. As a result, a total of 50 phloretin metabolites and 52 phlorizin metabolites were individually identified from different biological samples including rat plasma, urine, and faeces. Moreover, glucuronidation, sulfation, carbonylation and hydrolyzation were revealed to be the main metabolic pathways of phlorizin, while decarbonylation, glucuronidation and sulfation were regarded as the predominant biotransformation pathways as for phloretin. This is the first systematic study on comparison of metabolic profiles of phlorizin and phloretin in disease states, which was helpful to declare the complicated structure activity relationships between phlorizin and phloretin and shed light to their action mechanism.
Keywords
Analytical strategy
Metabolites
Phloretin
Phlorizin
UHPLC-Q-Exactive Orbitrap mass spectrometer
- DPIs
-
diagnostic product ions
- UHPLC
-
ultra-high performance liquid chromatography
- SPE
-
solid-phase extraction
Abbreviations
1 Introduction
Phloretin and phlorizin (phloretin 2′-O-glucoside) belong to natural flavonoid dihydrochalcone compounds found in apples and have been used in physiological research for more than 50 years (Tsao et al., 2003). In recent years, phloretin and phlorizin have exhibited a variety of biological activities, including anti-tyrosinase (Auner et al., 2005), anti-oxidant (Wei et al., 2017), anti-tumor (Abkin et al., 2016), anti-inflammatory (Huang et al., 2017) and anti-hyperglycemic (Sampath et al., 2017). Pharmacologically, the scientific community has confirmed that phlorizin has a positive effect on colitis, myocardial ischemia and arrhythmia under acute global cerebral ischemia (Feigin et al., 2017; Eberhardt et al., 2000; Malekova et al., 2007). It is worth noting that phlorizin is also effective against kidney disease, eye disease, cardiovascular injury and glycolipid metabolism disorder (Londzin et al., 2018; Han et al., 2017). Phloretin, one of the main metabolites of phlorizin, displays similar biological and pharmacological characteristics with phlorizin. However, phloretin is distinguished from phlorizin by its antiallergic and antithrombotic abilities (Mariadoss et al., 2019). In fact, the hydroxylated metabolite of phloretin (3-OH phloretin) plays a crucial role in the prevention or alleviation of obesity and metabolic diseases by inducing the recovery of adipocyte dysfunction or adipogenesis (Nguyen et al., 2020). Furthermore, previous studies have also demonstrated that the phase II metabolites of phloretin and phlorizin were the primarily detected forms, which also exhibit multiple pharmacological activities, similar to their prototypes (Crespy et al., 2001). For instance, the metabolites of phlorizin showed great anti-inflammatory activities, such as phloretin 4-O-β-D-glucuronide, 6-methoxyl-phloretin-2-O-β-D-glucuronide, and phloretin-2-O-β-D-glucuronide (Zhao et al., 2017).
In the initial stage of drug development, it is essential to study the processes of drug metabolism. Accumulating evidences revealed that prototype drugs could undergo biotransformation reactions in vivo and produced the metabolites with various bioactivities through diverse metabolic pathways (Li et al., 2016; Chen et al., 2014). Consequently, metabolite identification has already been used to discover bioactive constituents and clarify the actional mechanism of drugs. It is well documented that phloretin and phlorizin can be absorbed into the blood circulation, and the absorption rate of phlorizin is faster than that of phlorizin. Further research shows that 24 h after oral administration of phloretin and phlorizin, plasma concentrations almost return to baseline (Wang et al., 2019). This conclusion also suggests that metabolites may be the key bioactive components. However, to date, no one has systematically studied the metabolites of phloretin and phlorizin until now.
Recently, ultra-high-performance liquid chromatography coupled with quadrupole-exactive orbitrap mass spectrometry (UHPLC-Q-Exactive Orbitrap Mass) serves as a rapid analysis platform for complex chemical constituents and has shown excellent performance owing to its high efficiency, sensitivity and selectivity (Lin et al., 2015; Zhang et al., 2017). UHPLC-Q-Exactive Orbitrap mass spectrometry characterized by high trapping capacity and resolution power, MSn scanning, and superior data-mining of extracted ion chromatograms (EICs), plays an important role in metabolite identification (Wang et al., 2018; Xu et al., 2017). In this study, we established a new method for the qualitative analysis of metabolic profiles of phloretin and phlorizin using UHPLC-Q-Exactive Orbitrap MSn coupled with multiple data-processing methods to comparatively characterize the metabolites of phloretin and phlorizin.
2 Materials and methods
2.1 Chemicals and reagents
Phloretin and phlorizin reference standards were purchased from Chengdu Must Bio-technology Co., Ltd. (Chengdu, China) and Chengdu Biopurify Phytochemicals Co., Ltd. (Chengdu, China), respectively. Their purities were all determined to be higher than 98 % according to HPLC-UV analysis, and their structures were fully elucidated by comparing the spectral data (ESI-MS and 1H, 13C NMR spectroscopy) with the literature. LC-MS grade acetonitrile and methanol were manufactured by Fisher Scientific Co., Ltd.(Waltham, USA). Grace Pure SPE C18-Low solid-phase extraction cartridges (200 mg/3 mL, 59 μm, 70 \AA) were purchased from Grace Davison Discovery Science (Deerfield, IL, USA). All the other reagents were of analytical grade.
2.2 Animals and dosing
Twelve male Sprague-Dawley (SD) rats weighing 220 ± 10 g (certification number SCXK (Jing) 2011–0004) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd (Beijing, China) and were housed in SPF level conditions at Beijing University of Chinese Medicine with a temperature of 22–24 °C, humidity of 55 %–65 % and a 12 h light/dark cycle for one week of acclimation. All rats were allowed access to tap water and food ad libitum. The animal experiment was approved by the Animal Care and Use Committee at Binzhou Medical University (2021–085).
Before dosing, twelve SD rats were randomly divided into three groups, the control group (n = 4), phloretin group (n = 4) and phlorizin group (n = 4). Rats in two drug groups were orally administered phloretin and phlorizin dissolved in physiological saline at a dose of 200 mg per kilogram of body weight, respectively,. Equivalent volumes of the physiological saline were administrated to rats in the control group. Rats in the three groups were dosed with physiological saline, phloretin and phlorizin solution by intragastric administration.
2.3 Sample collection and preparation
Before the experiment, all the rats were fasted with permitted to access water for 12 h. Blood samples of each rat were collected into heparinized tubes at 0.5, 1, 2, 4 and 6 h from the orbital vein. Urine and faeces were collected from 0 to 24 h. The collected blood and urine samples were centrifuged at 3,500 rpm for 10 min at 4℃. Fecal samples were respectively dissolved with deionized water with an ultrasonic processing for 60 min, and then the supernatants were collected after centrifuging at 3,500 rpm for 10 min at 4℃. All of the above biological samples were stored at − 80℃ before the subsequent analysis.
All the bioanalyzed samples were prepared using Grace PureTM solid phase extraction (SPE) C18 columns. Prior to sample preparation, SPE columns were activated using 5 mL methanol and 5 mL deionized water. Then, the plasma, urine and fecal supernatants were applied to the SPE columns with a gradient elution program as follows: A volume of 1 mL of biological sample solution was added to a preliminary SPE cartridge. Afterwards, the SPE cartridges were successively washed with 3 mL deionized water and 3 mL methanol. The methanol eluate was evaporated to dryness in a water bath at 70 °C. Then, the residues were redissolved using 100 μL 5 % acetonitrile and vortexed for 3 min. After centrifugation at 14,000 rpm for 20 min, the supernatants were obtained for further LC-MS analysis.
2.4 Analysis condition
Separation was carried out on a Waters ACQUITY UPLC® BEH C18 column (2.1 mm × 50 mm, 1.7 μm) at 35 °C. The mobile phase system was composed of 0.1 % formic acid aqueous solution (A) and 0.1 % formic acid acetonitrile solution (B). Metabolites needed to be eluted using a linear gradient: 0.0–5.0 min, 5 %-50 % B; 5.0–14.0 min, 50 %–95 % B; 14.0–16.0 min, 95 % B; 16.0–16.1 min, 95 %–5% B; 16.1–18.0 min, 5 % B. The flow rate was set at 0.3 mL/min. Two-microliter supernatant samples were injected into a Q-Exactive Orbitrap mass spectrometer system (Thermo Scientific, Bremen, Germany) for analysis.
The operating conditions of the mass spectrometer were as follows: electrospray ionization (ESI) source in positive and negative ion mode; sheath gas flow: 40 and 30 arb; auxiliary gas flow: 20 and 10 arb; source voltage: 4.5 and 3.5 kV; capillary temperature: 350 °C; capillary voltage: 25 and − 35 V; and tube lens: 110 and − 110 V. The samples were analyzed using a full scan with a mass range from m/z 100–1,000 and 30,000 of resolution. The three ions with the most intense signals from one-stage mass spectrometry scanning were selected for further analyses. The other key parameters were as follows: 0.25 q of collision-induced dissociation (CID) activation type, 30 ms of activation time, and 35 % of normalized collision energy. All the raw data were processed using Thermo Xcalibur 2.1.
3 RESULTS
3.1 Establishment of the analytical strategy
A rapid and integrate strategy was established for the comprehensive screening and characterization of phloretin and phlorizin metabolites using a UHPLC-Q-Exactive Orbitrap MS coupled with diagnostic fragment ion filtering technique (Fig. 1). Initially, the biosamples, including plasma, urine and faeces, were prepared using the SPE method. These samples were then analyzed by UHPLC-Q-Exactive Orbitrap MS acquired in full mass scanning mode to obtain the full mass raw data. For the subsequent data mining processing, multiple metabolic pathways template was established, which included approximately 76 common metabolic reactions in vivo, such as glucuronidation, methylation, hydrogenation and sulfation. Then, phloretin and phlorizin standards were analyzed to acquire the fragmentation pattern and diagnostic product ions (DPI), and a series of filtering fragment ions (FFI) were obtained by combining multiple metabolic pathways template with DPIs. Simultaneously, an elementary screening of the candidate compounds was performed by using Thermo Xcalibur 2.1 software to obtain comprehensive mass data, such as retention times, accurate molecular weights and secondary fragment ion information, which could be used for the subsequent establishment of a metabolite database. Subsequently, the abovementioned FFIs were used to simultaneously extract and identify metabolite data in the database, and the biotransformation pathways of phloretin and phlorizin were proposed based on these identified metabolites and the corresponding metabolic reactions. Finally, the correctness of the metabolic pathways of phloretin and phlorizin were verified by summarizing the FFIs of the metabolic products and comparing the reference substance (the metabolites produced from certain metabolic pathways) mass spectrometry information.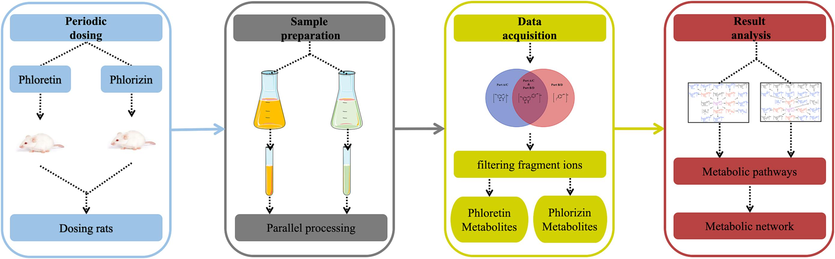
Summary diagram of the developed strategy and methodology.
3.2 DPIs and FFIs determination
To demonstrate the metabolic pattern of phloretin and phlorizin in vivo, we conducted a detailed analysis of their DPIs. The ESI-MS2 spectra of phloretin and phlorizin in negative/positive ion mode were shown in Fig. 2. Owing the same backbone structure, phloretin and phlorizin shared the similar fragmentation pattern in mass spectrometer. Phloretin generated deprotonated [M − H]− and [M + H]+ ions at m/z 273.07681 and m/z 275.09073 with mass errors of 3.882 ppm and −2.436 ppm in ESI-MS spectra, respectively. Due to the existence of carbonyl group at C-3′ position, a series of DPIs could be produced by breaking the bonds in C1-1′, C1′-2′ and C2′-3′ in negative ion mode, such as ions at m/z 167, m/z 107, m/z 151, m/z 121, m/z 119, m/z 93 and so on. Afterwards, the intermediate product ion at m/z 153 (undetected) rapidly produced m/z 123 and m/z 125 by losing CHO and CO groups. In positive ion mode, the cleavage behaviors were similar to those in negative ion mode. These representative DPIs were also detected, such as m/z 169, m/z 153, m/z 123 and m/z 151. The tentative fragmentation pathway of phloretin is shown in Fig. 3A. In the meantime, phlorizin generated [M − H]− and [M + H]+ ions at m/z 435.12967 and m/z 437.14349 with mass errors of 1.670 ppm and −1.678 ppm, respectively. In negative ion mode, phlorizin yielded aglycone ion at m/z 273 by neutral loss of glucose. Then, the characteristic ion at m/z 273 [M − H − Glc]− produced a lot of DPIs same as phloretin at m/z 167, m/z 107, m/z 151, m/z 121, m/z 119 and m/z 93. In positive ion mode, a series of DPIs were detected that were similar to phloretin, such as m/z 169, m/z 153, m/z 123 and m/z 151. The inferred fragmentation behavior of phlorizin is shown in Fig. 3B.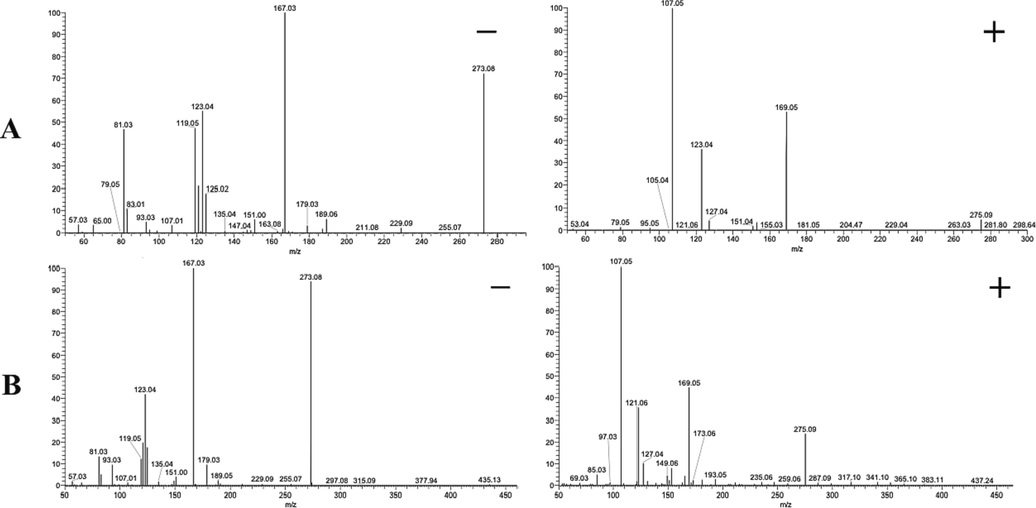
The ESI-MS2 spectra of phloretin (A) and phlorizin (B) in negative/positive ion mode.
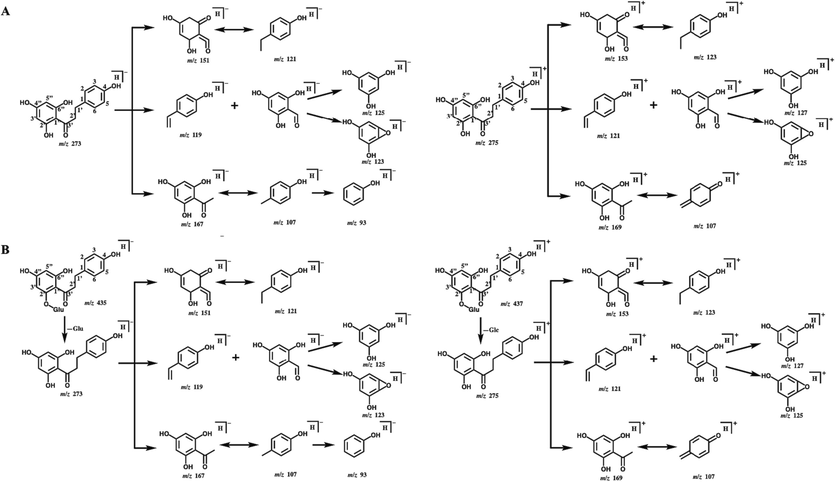
The mass fragmentation behavior of phloretin (A) and phlorizin (B) in negative/ positive ion mode.
In previous research, our research group summarized many preliminary metabolic reactions in vivo, so we combined these reactions with phloretin and phlorizin and then analyzed the possible products and their fragments individually (Wang et al., 2022). Based on the fragmentation pathway of phloretin and phlorizin, we found regular changes in these possible metabolic reaction product fragments. In the ESI-MS2 spectrum of phloretin, we identified the ions of m/z 167 + x and m/z 121 + y (x, y = molecular weight of substituent groups such as SO3 (80 Da), CH3 (15 Da), GluA (176 Da), etc.) as basic ions. And, we also derived a series of characteristic ions as FFIs for screening and identifying metabolites in negative ion mode, such as ions at m/z (167 + 2), m/z (167 + 80), m/z (121–––2), m/z (121 + 14) and so on. As in negative ion mode, we choose the ions of m/z (169 + x) and m/z (123 + y) as basic ions in positive ion mode. Subsequently, a sequence of FFIs were defined such as ions at m/z (169 + 162), m/z (169 + 80), m/z (123–––2), m/z (123 + 14) and so on. For phlorizin, we recognized m/z (167 + m) and m/z (121 + n) as basic ions and inferred a series of FFIs in negative mode, such as ions at m/z (167 + 0), m/z (167 + 2), m/z (121 + 14) and m/z (121–2). Similarly, we chose the ions at m/z (169 + m) and m/z (123 + n) as basic ions in positive mode and inferred a series of FFIs. It is worth noting that the two abovementioned types of FFIs could be divided into Part A/C and Part B/D according to the molecular fracture characteristics of phloretin and phlorizin: m/z (167 + x, 169 + x, 167 + m, 169 + m) belongs to Part A/C and m/z (121 + y, 123 + y, 121 + n, 123 + n) belongs to Part B/D. Since the cleavage directions of Part A/C and Part B/D were opposite, the structure of the metabolites could be directly determined when Part A/C or Part B/D appeared in the metabolite fragments of phloretin and phlorizin. The inferred FFIs of Part A/B/C/D were shown in Fig. 4.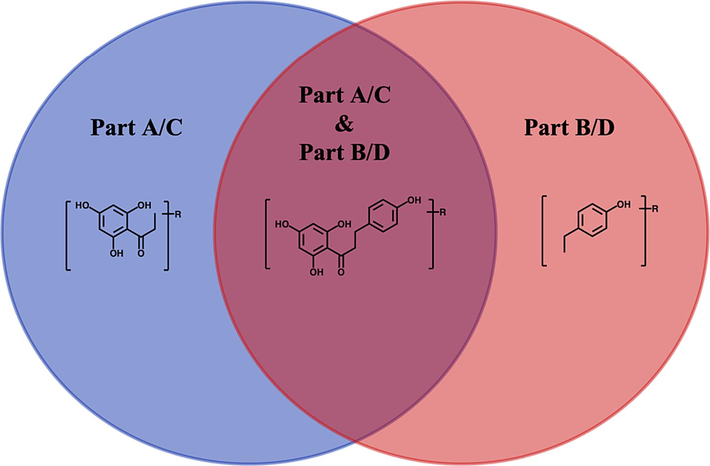
The extraction of metabolites in phloretin and phlorizin using PartA/B/C/D.
3.3 Verification process for the identification of phloretin metabolites in rat urine, plasma and faeces
A total of 50 metabolites were positively or tentatively detected and identified in the plasma, urine and faeces of SD rats using a UHPLC-Q-Exactive Orbitrap mass spectrometer. The chromatographic and MS data of these detected metabolites are summarized in Table 1. Note: tR: retention time; P: plasm; U: urine; F: faeces; “+”: positive mode; “-”: negative mode; “√”: detected.
Peak
tR/min
Formula [M−H]-/[M + H]+
Theoretical Mass (m/z)
Experimental Mass (m/z)
Error (ppm)
MS2 fragment ions
Identification/Reactions
P U F
+
-
+
-
+
-
M1
1.01
C16H15O6
303.08735
303.08447
−1.845
151(100.00),134(2.10),210(1.17),125(1.26),196(0.93)
hydroxylation, methylation
√
M2
1.08
C8H7O5
183.03005
183.02908
1.531
183(33.83),165(5.38),167(3.03)
debenzylation
√
M3
1.86
C8H7O5
183.03005
183.02901
1.148
183(6.31),124(1.07),165(1.16)
debenzylation
√
M4
2.86
C8H7O5
183.03005
183.02913
1.804
124(100.00),183(36.36),165(1.19)
debenzylation
√
M5
3.91
C14H15O5
263.09086
263.09161
0.798
263(100.00),141(10.21),123(5.22)
decarbonylation, hydroxylation
√
M6
4.65
C27H29O17
625.14105
625.14124
2.102
167(100.00),125(99.10),273(85.69),449(65.52),625(4.92),93(3.13)
diglucuronidation
√
M7
4.74
C27H29O17
625.14105
625.14130
2.198
273(100.00),167(98.26),125(96.38),449(80.76),625(9.28),93(2.12)
diglucuronidation
√
M8
4.76
C18H19O5
315.12545
315.12399
4.093
315(100.00),285(10.09),106(9.35), 92(2.92),120(2.56),270(1.65),165(0.87)
trimethylation
√
M9
4.79
C16H17O5
289.10760
289.10678
−0.934
93(100.00),121(94.59),107(45.75),165(16.91),169(7.36),289(7.01)
methylation
√
M10
4.80
C21H19O11
447.09325
447.09344
2.801
271(100.00),119(61.03),93(9.70)
glucuronidation, dehydrogenation
√
M11
4.86
C21H19O12
463.08815
463.08835
2.694
287(100.00),153(93.42),119(7.31),167(5.84),135(2.95),93(1.02),463(0.63)
glucuronidation, carbonylation
√
M12
4.88
C15H11O6
287.05605
287.05600
3.433
153(100.00),287(49.47),167(3.58),135(2.84),93(1.83),125(1.33)
carbonylation
√
M13
4.95
C15H13O5
273.07685
273.07660
0.850
167(100.00),273(67.04),123(54.10),125(22.58),93(8.67),151(6.31)
phloretin
√
M14
4.96
C21H21O11
449.10895
449.10880
2.142
167(100.00),273(93.49),125(19.61),93(6.56),449(5.77)
glucuronidation
√
M15
4.97
C27H29O17
625.14105
625.14117
1.990
167(100.00),273(99.17),449(23.72),125(20.00),625(1.62),93(1.56)
diglucuronidation
√
M16
5.01
C27H29O17
625.14105
625.14142
2.390
167(100.00),273(90.25),449(19.27),125(14.00),93(4.03),625(3.10)
diglucuronidation
√
M17
5.15
C15H15O6
291.08576
291.08643
0.396
291(57.41),93(50.88),107(24.28),124(3.93),169(2.70)
hydroxylation
√
M18
5.20
C21H21O11
449.10895
449.10904
2.677
273(100.00),167(88.58),125(15.42),449(3.69),93(2.37)
glucuronidation
√
M19
5.24
C18H19O5
315.12545
315.12390
3.807
315(100.00),107(35.83),121(18.35),209(1.73)
trimethylation
√
M20
5.76
C15H17O5
277.10710
277.10672
−1.191
277(100.00),93(4.70),107(3.49),157(0.64),171(0.57)
hydrogenation
√
M21
5.88
C15H17O5
277.10710
277.10632
−2.635
93(100.00),107(53.77),121(31.19),157(11.43),171(10.88)
hydrogenation
√
M22
6.09
C15H11O8S
351.01795
351.01801
3.121
271(100.00),119(78.26),351(14.65),125(8.57),93(4.13),147(3.46)
sulfation, hydroxylation, dehydration
√
M23
6.18
C15H11O8S
351.01795
351.01810
3.378
271(100.00),119(55.43),351(8.31),93(7.23),125(2.55)
sulfation, hydroxylation, dehydration
√
M24
6.25
C15H13O9S
369.02855
369.02863
3.119
169(100.00),369(36.36),289(35.85),183(33.69),121(26.71),125(19.58)
hydroxylation, sulfation
√
M25
6.29
C15H11O8S
351.01795
351.01828
3.890
271(100.00),119(42.15),351(11.64),93(5.39)
sulfation, hydroxylation, dehydration
√
M26
6.34
C15H13O9S
369.02855
369.02869
3.281
369(100.00),169(29.92),183(19.94),289(15.52)
hydroxylation, sulfation
√
M27
6.42
C17H17O5
301.10925
301.11130
−1.623
301(100.00),125(1.23),153(0.22),195(0.17),167(0.17),195(0.17),121(0.14)
dimethylation
√
M28
6.77
C15H13O9S
369.02855
369.02759
0.301
369(100.00),289(23.49),183(15.34)
hydroxylation, sulfation
√
M29
6.88
C15H13O9S
369.02855
369.02771
0.626
369(100.00),289(13.76),183(10.38),121(8.62),125(5.42)
hydroxylation, sulfation
√
M30
6.89
C15H11O8S
351.01795
351.01810
3.378
271(100.00),106(71.37),351(7.51),119(6.62),125(2.18)
sulfation, hydroxylation, dehydration
√
M31
6.99
C15H13O9S
369.02855
369.02853
2.848
369(100.00),109(22.72),167(19.16),289(17.10),123(7.04)
hydroxylation, sulfation
√
M32
6.99
C15H11O8S
351.01795
351.01813
3.463
271(100.00),106(26.89),351(20.09),119(13.15)178(12.98)
sulfation, hydroxylation, dehydration
√
M33
7.10
C15H17O5
277.10710
277.10583
−4.403
93(100.00),277(67.91),107(53.29),157(4.39),171(4.18)
hydrogenation
√
M34
7.70
C15H17O5
277.10710
277.10690
−0.542
93(100.00),107(51.09),277(41.13),157(10.23),171(8.68)
hydrogenation
√
M35
8.52
C15H17O5
277.10710
277.10641
−2.310
93(100.00),277(56.50),107(42.92),171(5.40),157(4.29)
hydrogenation
√
M36
8.64
C15H17O5
277.10710
277.10587
−4.259
93(100.00),277(70.06),107(47.65),157(7.66),171(4.16)
hydrogenation
√
M37
8.87
C21H21O14S
529.06575
529.06598
2.509
167(100.00),273(80.90),449(73.64),125(49.07),78(25.88),119(25.78)
glucuronidation, sulfation
√
M38
8.93
C15H13O11S2
432.99045
432.99075
3.166
353(100.00),167(81.33),273(68.84),149(17.76),93(13.93)
disulfation
√
M39
8.93
C15H17O5
277.10710
277.10559
−1.460
93(100.00),277(54.70),107(47.50),157(6.98),171(4.52)
hydrogenation
√
M40
9.12
C15H13O11S2
432.99045
432.99078
3.236
167(100.00),353(97.74),273(90.52),125(16.17)
disulfation
√
M41
9.21
C15H13O11S2
432.99045
432.99057
2.751
167(100.00),273(98.84),353(90.78),119(23.80)
disulfation
√
M42
9.30
C21H21O14S
529.06575
529.06592
2.396
273(100.00),167(91.92),125(65.57),449(51.90),119(25.74),78(4.91),93(3.89)
glucuronidation, sulfation
√
M43
9.35
C15H13O11S2
432.99045
432.99066
2.959
273(100.00),167(69.92),353(64.92),121(16.21)
disulfation
√
M44
9.42
C21H21O14S
529.06575
529.06586
2.283
273(100.00),167(86.86),125(72.89),449(63.19),119(16.84),529(9.77)
glucuronidation, sulfation
√
M45
9.53
C15H13O11S2
432.99045
432.99066
2.959
273(100.00),353(95.71),167(68.04)
disulfation
√
M46
9.67
C15H13O11S2
432.99045
432.99057
2.751
353(100.00),273(91.72),167(80.95),125(17.15)
disulfation
√
M47
9.82
C15H13O11S2
432.99045
432.99072
3.097
167(100.00),273(98.01),353(71.79),121(15.69)
disulfation
√
M48
10.21
C15H13O11S2
432.99045
432.99060
2.820
353(100.00),273(83.47),167(67.23),93(14.45)
disulfation
√
M49
10.34
C21H21O14S
529.06575
529.06586
2.283
167(100.00),273(92.54),78(68.67),125(44.98),449(44.70),119(17.11),529(16.28)
glucuronidation, sulfation
√
M50
14.18
C14H13O4
245.08195
245.08154
2.875
121(100.00),245(60.26),93(15.79),125(6.83)
decarbonylation
√
3.3.1 Verification process for the identification of phloretin metabolites based on Part a (m/z 167/169 + x)
M2, M3 and M4 displayed [M−H]− ions at m/z 183.02908, m/z 183.02901 and m/z 183.02913 (C8H7O5, mass errors within ± 2.00 ppm), with the mass being 90 Da less than that of phloretin in negative ion mode, In the ESI-MS2 spectra, their FFIs at m/z 167 [M−H−O]− suggested that M2, M3 and M4 could be isomeric debenzylation metabolites of phloretin. M13 produced [M−H]− ion at m/z 273.07660 (C15H13O5, mass error 0.850 ppm) with the same molecular formula as phloretin, indicating that it was the prototype of phloretin. The FFIs at m/z 167 and product ions at m/z 273 and m/z 93 confirmed the previous inference. M26, M28 and M31 owned the same theoretical [M−H]− ion at m/z 369.02855 with the predictive molecular formula of C15H13O9S (mass errors within ± 4.00 ppm), respectively. All of them were 96 Da more massive than phloretin, indicating that they could be isomeric hydroxylation and sulfation metabolites of phloretin. In the ESI-MS2 spectra, their FFIs at m/z 167 [M−H−SO3−C7H6O−O]− and product ions at m/z 289 [M−H−SO3]− and m/z 183 [M−H−SO3−C7H6O]− were all observed, which were in accordance with the assumption we mentioned in Fig. 2. Thus, M26, M28 and M31 could be tentatively identified as isomeric hydroxylation and sulfation metabolites of phloretin.
M14 and M18 possessed [M−H]− ions at m/z 449.10880 and m/z 449.10904 (C21H21O11, mass errors 2.142 ppm and 2.677 ppm), respectively. Both of them were 176 Da more than that of phloretin in negative ion mode, which implied that glucuronidation occurred. Product ion at m/z 273 was produced by the neutral loss of glucuronide. The FFIs at m/z 167 [M−H−GluA−C7H6O]− and DPIs at m/z 125 [M−H−GluA−C8H8O−CO]− and m/z 93 [M−H−GluA−C8H6O4−CH2]− confirmed the above-mentioned assumption. Hence, M14 and M18 could be tentatively identified as isomeric glucuronidation metabolites of phloretin. M6, M7, M15 and M16 generated [M−H]− ions at m/z 625.14124, m/z 625.14130, m/z 625.14117 and m/z 625.14142 (C27H29O17, mass errors 2.102 ppm, 2.198 ppm, 1.990 ppm and 2.390 ppm), orderly. Product ions at m/z 449 [M−H−GluA]− and m/z 273 [M−H−2GluA]− were generated by the successive loss of GluA (176 Da). Based on the analysis of M14 and M18, the FFIs at m/z 167 and DPIs at m/z 125 and m/z 93 were all detected again. Therefore, M6, M7, M15 and M16 were all predicted to be isomeric diglucuronidation metabolites of phloretin. M37, M44 and M49 were characterized as sulfation and glucuronidation metabolites of phloretin. The process used to analyze them is provided in the supporting information. In positive ion mode, M35, M36 and M39 were hydrogenation metabolites of phloretin. M17 is a hydroxylation metabolite of phloretin. M38, M40, M41, M45, M46 and M48 were isomeric disulfation metabolites of phloretin. The process by which they were analyzed is provided in the supporting information.
3.3.2 Verification process for identification of phloretin metabolites based on Part B (m/z 121/123 + y)
M1 displayed [M−H]− ion at m/z 303.08447 (C16H15O6, mass error −1.845 ppm) that was 30 Da heavier than phloretin. With one more methoxy than phloretin, FFIs at m/z 151 (121 + OCH2) and DPIs at m/z 210 [M−H−C6H5O]− and m/z 196 [M−H−C6H5O−CH2]− were generated in negative ion mode, which suggested methoxy group attached to C-2′. Therefore, we identified M1 as a hydroxylation and methylation metabolite of phloretin. M50 produced an [M−H]− ion at m/z 245.08154 (C14H13O4, mass error 2.875 ppm) with a mass 28 Da less than that of phloretin, indicating that the occurrence of decarbonization reaction. In the ESI-MS2 spectra, FFIs were characterized at m/z 121 [M−H−C6H4O3]− and DPIs at m/z 125 [M−H−C8H8O]− and m/z 93 [M−H−C6H4O3−C2H4]−, suggesting that it could be a decarbonization metabolite of phloretin. Based on the above analysis, M5 presented [M + H]+ ion at m/z 263.09161 (C14H15O5, mass error 0.798 ppm) in positive ion mode. The complementary ions at m/z 123 [M + H-C7H8O3]+ and m/z 141 [M + H-C7H6O2]+via the C1′-2′ bond breaking pathway were also observed, respectively. In the end, M5 was confirmed as a decarbonization and hydroxylation metabolite of phloretin. M10 is a glucuronidation and dehydrogenation metabolite of phloretin. The process by which they were analyzed is described in the supporting information.
M22, M23, M25, M30 and M32 displayed [M−H]− ions at m/z 351.01801, m/z 351.01828, m/z 351.01810 and m/z 351.01813 (C15H11O8S, mass errors 3.121 ppm, 3.890 ppm, 3.378 ppm and 3.463 ppm), respectively. The product ion at m/z 271 [M−H−SO3]− was attributed to the neutral loss of SO3, FFIs at m/z 119 (121–2) [M−H−SO3−C7H4O4]− was attributed to the RDA cleavage pathway. Thus, M22, M23, M25, M30 and M32 were tentatively identified as isomeric sulfation hydroxylation and dehydration metabolites of phloretin.
3.3.3 Verification process for identification of phloretin metabolites based on Part a (m/z 167/169 + x) with Part B (m/z 121/123 + y)
In negative ion mode, M8 and M19 generated [M−H]− ions at m/z 315.12399 and m/z 315.12390 (C18H19O5, mass errors 4.093 ppm and 3.807 ppm) with retention times of 4.76 min and 5.24 min, respectively. Both of them were 42 Da (3CH2) more than that of phloretin in negative ion mode, which implied that trimethylation occurred. M8 showed FFIs at m/z 165 (167–2) [M−H−3CH3−C7H5O]− and m/z 120 (121–1) [M−H−3CH3−C7H2O4]− and other representative fragment ions at m/z 285 [M−H−2CH3]− and m/z 270 [M−H−3CH3]−. Meanwhile, M19 possessed FFIs at m/z 209 (167 + 42) [M−H−C7H6O]− and m/z 121 [M−H−C10H10O4]−, which were different from those of M8. Therefore, M8 and M19 were all speculated to be isomeric trimethylation metabolites of phloretin with different connection positions of three methyl groups. M12 eluted at 4.88 min and produced [M−H]− ion at m/z 287.05600 (C15H11O6, mass error 3.433 ppm), with a mass 14 Da greater than that of phloretin. The FFIs at m/z 167 [M−H−C7H4O2]− and m/z 135 (121 + 14) [M−H−C7H4O4]− proved that it was a carbonylation metabolite of phloretin. Based on above mentioned analysis, M11 was a glucuronidation and carbonylation metabolite of phloretin. M27 was a dimethylation metabolite of phloretin. M9 was a methylation metabolite of phloretin. The process by which these were analyzed is provided in the supporting information.
M20, M21, M33 and M34 generated [M + H]+ at m/z 277.10672, m/z 277.10632, m/z 277.10583 and m/z 277.10690 (C15H17O5, mass errors −1.191 ppm, −2.635 ppm, −4.403 ppm and −0.542 ppm) that were 2 Da more massive than phloretin, proving the occurrence of a hydrogenation reaction. Due to the discovery of the FFIs at m/z 171 (169 + 2) [M + H-C7H6O]+ and m/z 123 [M + H-C7H6O4]+ in their ESI-MS2 spectra, they were tentatively identified as isomeric hydrogenation metabolites of phloretin. M24 and M29 showed [M−H]− ions at m/z 369.02863 and m/z 369.02771 (C15H13O9S, mass errors 3.119 ppm and 0.626 ppm), respectively. Then, the product ions at m/z 289 was produced by loss of SO3 group. The FFIs at m/z 183 (167 + 16) [M−H−SO3−C7H6O]− and m/z 121 [M−H−SO3−C7H4O5]− were also generated in negative ion mode, which gave us a clue that hydroxylation occurred in the Part A. Consequently, M24 and M29 were all assigned as isomeric hydroxylation and sulfation metabolites of phloretin. M43 and M47 were disulfation metabolites of phloretin. M42 was a glucuronidation and sulfation metabolite of phloretin. The process by which they were analyzed is provided in the supporting information.
3.4 Identification of phlorizin metabolites in rat urine, plasma and faeces
A total of 52 metabolites were positively or tentatively detected and identified in the plasma, urine and faeces of SD rats using a UHPLC-Q-Exactive Orbitrap mass spectrometer. The chromatographic and MS data of these detected metabolites are summarized in Table 2. Note: tR: retention time; P: plasm; U: urine; F: faeces; “+”: positive mode; “-”: negative mode; “√”: detected.
Peak
tR/min
Formula [M−H]-/[M + H]+
Theoretical Mass (m/z)
Experimental Mass (m/z)
Error (ppm)
MS2 fragment ions
Identification/Reactions
P
U
F
P1
1.05
C16H15O6
303.08735
303.08463
−0.925
151(100.00),93(17.67),125(10.57),303(7.09),120,(5.22)
hydrolyzation, hydroxylation, methylation
√
P2
1.97
C8H7O5
183.03005
183.02911
1.695
123(100.00),167(10.22),183(3.04),125(1.69)
hydrolyzation, debenzylation
√
P3
2.80
C27H31O16
611.16177
611.16180
1.863
167(100.00),273(90.40),125(30.17),93(8.26),435(4.73),149(4.60),611(3.49),107(1.10),121(0.37)
glucuronidation
√
P4
3.99
C8H15O11
287.05605
287.05890
−1.988
287(100.00),151(7.32),153(3.34),93(2.1),125(1.96),167(1.44),135(1.01)
hydrolyzation, carbonylation
√
P5
4.29
C27H31O16
611.16177
611.16199
2.174
167(100.00),273(93.36),119(13.84),611(9.60),435(7.11),93(5.87),149(1.99)
glucuronidation
√
P6
4.58
C27H29O17
625.14097
625.14136
2.294
167(100.00),125(61.57),449(47.30),625(4.18),315(1.88)
glucuronidation, carbonylation
√
P7
4.65
C27H29O17
625.14105
625.14136
2.294
167(1 0 0),273(97.65),125(61.54),449(47.30)123(35.21)
hydrolyzation, diglucuronidation
√
P8
4.70
C27H29O17
625.14105
625.14130
2.198
167(1 0 0),273(96.72),123(33.96),449(22.11),125(21.62)
hydrolyzation, diglucuronidation
√
P9
4.70
C27H29O17
625.14097
625.14130
2.198
167(100.00),449(22.11),125(21.62),119(16.20),93(3.62),315(2.79),625(2.32)
glucuronidation, carbonylation
√
P10
4.74
C21H23O14S
531.08137
531.08209
3.366
289(100.00),531(54.49),451(40.26),167(11.13),137(9.35),93(8.76)
hydroxylation, sulfation
√
P11
4.76
C18H19O5
315.12545
315.12393
3.903
315(100.00),92(3.26),120(2.52),135(1.03),165(1.01)
hydrolyzation, trimethylation
√
P12
4.76
C27H29O17
625.14105
625.14117
1.990
167(100.00),273(92.74),123(33.15),449(21.30),125(19.67)
hydrolyzation, diglucuronidation
√
P13
4.76
C15H13O6
289.07060
289.07010
−1.953
93(100.00),135(23.26),125(17.93),169(16.41),137(5.00)
hydrolyzation, carbonylation
√
P14
4.78
C21H19O11
447.09325
447.09378
3.561
271(1 0 0),151(81.54),119(59.06),93(6.69),117(4.72),447(3.85),125(1.24)
hydrolyzation, glucuronidation, dehydrogenation
√
P15
4.79
C27H29O17
625.14097
625.14117
1.990
167(100.00),449(21.30),125(19.67),119(16.84),93(3.89),315(2.64),625(1.85)
glucuronidation, carbonylation
√
P16
4.81
C21H23O13S
515.08647
515.08691
2.955
167(100.00),273(92.95),515(26.39),125(20.61),119(15.21)93(9.84),435(8.01)
sulfation
√
P17
4.87
C22H23O12
479.11937
479.11996
3.251
303(100.00),288(87.10),153(13.16),479(3.98)
carbonylation, hydroxylation, methylation
√
P18
4.88
C27H29O17
625.14097
625.14124
2.102
167(100.00),449(23.75),125(19.23),93(3.87),315(2.04),625(1.84)
glucuronidation, carbonylation
√
P19
4.89
C21H23O10
435.12967
435.12994
3.141
167(100.00),273(96.95),123(40.78),81(15.34),119(13.94),93(10.95),179(10.52)
phlorizin
√
P20
4.91
C21H23O10
435.13021
435.13018
−0.983
167(100.00),273(96.95),123(40.78),91(9.87),435(0.84),121(0.48)
carbonylation, dehydroxylation, hydrogenation
√
P21
4.92
C15H13O5
273.07685
273.07687
4.101
167(1 0 0),273(58.15),125(17.76),93(9.72),107(3.13),121(0.51)
phloretin
√
P22
4.93
C21H21O11
449.10887
449.10870
1.920
167(100.00),125(19.79),93(6.52),449(5.16),287(4.13)
carbonylation
√
P23
4.96
C21H23O11
451.12350
451.12250
−2.190
169(100.00),275(6.81)
hydrolyzation, glucuronidation
√
P24
4.99
C27H29O17
625.14097
625.14117
1.990
167(100.00),273(97.90),449(22.78),93(4.54),315(2.95),625(2.33)
glucuronidation, carbonylation
√
P25
5.07
C21H23O11
451.12342
451.12283
−1.458
107(100.00),169(10.20),275(6.95),149(3.85),151(3.05),127(2.93),131(2.56)
carbonylation
√
P26
5.08
C21H23O13S
515.08647
515.08630
1.771
167(100.00),273(91.78),515(24.16),125(20.21),119(9.35),93(7.71),435(6.08)
sulfation
√
P27
5.16
C15H13O5
273.07685
273.07684
3.992
167(1 0 0),273(71.60),125(18.46), 93(15.72),107(3.39),121(0.20)
phloretin
√
P28
5.18
C21H23O14S
531.08137
531.08185
2.914
289(100.00),451(44.84),531(35.46),167(33.37),93(11.59),123(7.44)
hydroxylation, sulfation
√
P29
5.20
C21H21O11
449.10895
449.10892
2.410
167(100.00),273(1 0 0),125(16.70),449(3.26),123(32.56),93(2.50)
hydrolyzation, glucuronidation
√
P30
5.20
C21H23O11
451.12350
451.12244
−2.323
107(100.00),169(12.78),275(8.75),151(3.30),123(0.52),121(0.48)
hydrolyzation, glucuronidation
√
P31
5.36
C22H23O11
463.12507
463.12524
3.783
152(100.00),287(52.69),272(36.08),463(21.41),124(18.27)
carbonylation, methylation
√
P32
5.38
C21H23O14S
531.08137
531.08197
3.140
289(100.00),451(70.15),531(31.20),167(13.58)
hydroxylation, sulfation
√
P33
5.41
C22H23O12
479.11937
479.11993
3.188
303(100.00),288(21.69),168(7.12),151(4.52),479(28.49),153(11.95),167(7.20)
carbonylation, hydroxylation, methylation
√
P34
5.65
C22H27O10
451.16042
451.15808
−3.975
107(100.00),169(12.35),275(6.50),151(3.51),349(1.74),183(1.64)
methylation
√
P35
5.78
C21H21O11
449.10887
449.10931
3.278
167(100.00),125(18.15),449(4.81),287(3.75),93(1.27)
carbonylation
√
P36
5.83
C15H11O8S
351.01795
351.01843
4.318
271(100.00),151(51.61),119(42.91),351(13.09),93(8.70)
hydrolyzation, sulfation, hydroxylation, dehydration
√
P37
5.88
C15H13O8S
353.03365
353.03394
3.896
167(100.00),273(95.36),123(40.08),353(19.35),93(9.67),151(4.24),121(0.42)
hydrolyzation, sulfation
√
P38
5.98
C15H13O5
273.07685
273.07709
4.907
167(100.00),273(66.42),125(17.56), 93(11.62),107(2.35),121(1.69)
phloretin
√
P39
6.00
C20H23O9
407.13477
407.13757
3.911
407(100.00),125(4.12),121(0.96),245(0.67)
decarbonylation
√
P40
6.09
C20H23O9
407.13477
407.13184
−1.819
407(100.00),245(6.04),125(2.71),121(0.54)
decarbonylation
√
P41
6.14
C15H13O8S
353.03365
353.03381
3.528
167(100.00),273(81.61),123(41.45),353(33.35),93(13.53),125(11.26),151(3.29),107(1.46)
hydrolyzation, sulfation
√
P42
6.29
C15H13O9S
369.02855
369.02881
3.607
289(100.00),369(58.38),137(16.13),167(9.69),125(4.16),151(3.92)
hydrolyzation, hydroxylation, sulfation
√
P43
6.32
C15H17O5
277.10710
277.10587
−4.259
107(100.00),171(6.91),123(9.89)
hydrolyzation, hydrogenation
√
P44
6.46
C15H13O9S
369.02855
369.02878
3.525
289(100.00),369(54.39),137(14.54),167(10.79),125(6.00),151(3.83)
hydrolyzation, hydroxylation, sulfation
√
P45
6.65
C15H13O9S
369.02855
369.02863
3.119
289(100.00),183(85.14),369(43.79),287(13.53),93(10.92),125(9.04),167(7.33)
hydrolyzation, hydroxylation, sulfation
√
P46
6.79
C15H17O5
277.10710
277.10660
−1.624
107(100.00),277(47.99),121(27.07),189(23.67),109(15.57),151(12.59),171(9.08)
hydrolyzation, hydrogenation
√
P47
7.04
C15H11O8S
351.01795
351.01819
3.634
271(100.00),351(52.21),119(9.94)
hydrolyzation, sulfation, hydroxylation, hydrolyzation, sulfation, hydroxylation, dehydration
√
P48
7.65
C15H17O5
277.10710
277.10687
−0.650
107(100.00),189(40.86),277(40.44),121(29.29),151(19.77),189(15.17),171(5.02)
hydrolyzation, hydrogenation
√
P49
7.90
C15H17O4
261.11160
261.11274
−2.051
261(100.00),121(51.40),107(46.85),109(20.61),161(17.33),117(14.22),123(4.79),111,155
hydrolyzation, dehydroxylation, hydrogenation
√
P50
8.23
C15H17O5
277.10710
277.10568
−4.944
277(100.00),107(51.98),121(27.27),119(23.76),151(9.60),171(3.97)
hydrolyzation, hydrogenation
√
P51
8.42
C15H17O5
277.10710
277.10803
0.980
277(100.00),107(41.42),119(26.09),121(24.78),151(7.83),171(5.41)
hydrolyzation, hydrogenation
√
P52
8.49
C15H17O5
277.10710
277.10645
−2.166
277(100.00),107(45.36),119(25.38),121(23.49),151(7.03),171(5.09)
hydrolyzation, hydrogenation
√
3.4.1 Verification process for the identification of phlorizin metabolites based on Part C (m/z 167/169 + m)
P2 showed [M−H]− ion at m/z 183.03005 (C8H7O5, mass error 1.695 ppm). The FFIs at m/z 167 [M−H−O]− and product ions at m/z 125 [M−H−O−C2H2O]− and m/z 123 [M−H−O−C2H4O]− proved that phenol hydroxyl group of phloretin was transferred to C-2′ position and then loss benzyl group. Thus, P2 could be tentatively identified as a debenzylation metabolite of phloretin. P5 eluted at 4.29 min and afforded [M−H]− ion at m/z 611.16199 (C27H31O16, mass error 2.174 ppm) with a mass 176 Da more massive than that of phlorizin. Due to the neutral loss of GluA and Glc groups, product ions at m/z 435 [M−H−GluA]− and m/z 273 [M−H−GluA−Glc]− were generated in its ESI-MS2 spectrum, respectively. In addition, the FFIs at m/z 167 [M−H−GluA−Glc−C7H6O]− and other fragment ions at m/z 119 and m/z 93 were also detected. Thus, P5 could be deduced a glucuronidation metabolite of phlorizin. P19 showed [M−H]− ion at m/z 435.12994 (C21H23O10, mass error 3.141 ppm) with the same molecular formula as phlorizin. The FFIs at m/z 167 and character ions at m/z 273, m/z 119, m/z 123 and m/z 93 confirmed that P19 was the prototype of phlorizin. P7, P8, and P12 were isomeric diglucuronidation metabolites of phloretin. P29 was glucuronidation metabolites of phloretin. The process by which they were analyzed is provided in the supporting information.
With retention times of 4.93 min and 5.78 min, P22 and P35 generated [M−H]− ions at m/z 449.10870 and m/z 449.10931 (C21H21O11, mass errors 1.920 ppm and 3.278 ppm), respectively. In their ESI-MS2 spectrum, FFIs at m/z 167 [M−H−Glc−C7H4O2]− was generated and product ions at m/z 287 and m/z 125, implying that carbonylation reaction did not occur on Part C. Furthermore, product ion at m/z 93 was produced by losing CO group, which suggested the presence of a carbonyl group. Therefore, P22 and P35 were characterized as isomeric carbonylated metabolites of phlorizin. In positive ion mode, P25 was an isomeric carbonylated product. P17 and P33 were isomeric carbonylation hydroxylation and methylation metabolites of phlorizin. P6, P9, P15, P18 and P24 were isomeric glucuronidation and carbonylation metabolites of phlorizin. The process by which they were analyzed is provided in the supporting information.
P16 and P26 gave rise to [M−H]− ions at m/z 515.08691 and m/z 515.08630 (C21H23O13S, mass errors 2.955 ppm and 1.771 ppm) with masses 80 Da more than that of phlorizin, indicating that they could be isomeric sulfation metabolites of phlorizin. In their ESI-MS2 spectra, a series of product ions at m/z 435 [M−H−SO3]−, m/z 273 [M−H−SO3−Glc]−, m/z 125 [M−H−SO3−Glc−C8H8O−CO]−, m/z 119 [M−H−SO3−Glc−C7H6O4]− and m/z 93 [M−H−SO3−Glc−C8H6O4−CH2]− and FFIs at m/z 167 [M−H−SO3−Glc−C7H6O]− were evidence that confirmed our deduction. According to the above indication, P10, P28 and P32 possessed the same theoretical [M−H]− ions at m/z 531.08137 (C21H23O14S, mass errors within ± 4 ppm) in negative ion mode, which showed the FFIs at m/z 167 [M−H−SO3−Glc−C7H6O2]− and product ions at m/z 451, m/z 289, m/z 123 and m/z 93. Consequently, P10, P28 and P32 could be identified as isomeric hydroxylation and sulfation metabolites of phlorizin. P34 eluted at 5.65 min and afforded [M + H]+ ion at m/z 451.15808 (C22H27O10, mass error −3.975 ppm) with a mass 14 Da greater than that of phlorizin. The fragment ions at m/z 275 [M + H-Glc-CH2]+, m/z 183 [M + H-Glc-CH2-C6H4O]+ and m/z 107 [M + H-Glc-CH2-C8H8O4]+and FFIs at m/z 169 [M + H-Glc-CH2-C7H6O]+ provided substantial evidence that it was a methylated metabolite of phlorizin. P42 and P44 were isomeric hydroxylation and sulfation metabolites of phloretin. P41 was sulfation metabolite of phloretin. P43 was hydrogenation metabolite of phloretin. P45 is a hydroxylation and sulfation metabolite of phloretin. The process by which they were analyzed is provided in the supporting information.
3.4.2 Verification process for the identification of phlorizin metabolites based on Part D (m/z 121/123 + n)
P1 possessed [M−H]− ion at m/z 303.08463 (C16H15O6, mass error −0.925 ppm), with a mass 30 Da greater than that of phloretin, which yielded product ions at m/z 125 and m/z 93. The FFIs at m/z 151 (121 + 30) [M−H−C7H4O4]− and m/z 120 [M−H−C7H4O4−OCH3]− were also observed. Hence, P1 was characterized as a hydroxylated and methylated metabolite of phloretin. P39 and P40 generated [M−H]− ions at m/z 407.13757 and m/z 407.13184 (C20H23O9, mass errors 3.911 ppm and −1.819 ppm) in negative ion mode. They weighed 28 Da less than phlorizin, indicating that the occurrence of decarbonization reaction. The product ion at m/z 245 was generated by neutral loss of Glc group, and FFIs at m/z 121 [M−H−Glc−C6H4O3]− proving occurrence of decarbonization. Therefore, P39 and P40 were tentatively deduced as isomeric decarbonization metabolites of phlorizin. P31 showed [M−H]− ion at m/z 463.12524 (C22H23O11, mass error 3.783 ppm). The product ions at m/z 448 [M−H−CH3]− and m/z 287 [M−H−CH3−C6H9O5 (Glc-H, 161)]− suggested that methyl was located in Glc group. In the ESI-MS2 spectrum, the FFIs at m/z 135 [M−H−CH3−C6H9O5 (Glc-H, 161)-C7H4O4]− was also detected. On the basis of these data, P31 was characterized as an isomeric carbonylation and methylation metabolite of phlorizin. P14 eluted at 4.78 and generated [M−H]− ion at m/z 447.09378 (C21H19O11, mass error 3.561 ppm), with a mass 174 Da more massive than that of phloretin. The [M−H]− ion afforded fragment ions at m/z 271 [M−H−GluA]− by neutral loss of GluA, and the FFIs at m/z 119 (121–2) [M−H−GluA−C7H4O4]− indicating that dehydrogenation reaction occurred in Part D. Moreover, a series of DPIs at m/z 125, m/z 117 (119–2) and m/z 93 were also observed. Therefore, P14 could be a glucuronidation and dehydrogenation metabolite of phloretin. P36 and P47 are isomeric sulfation and dehydration metabolites of phloretin. The process by which they were analyzed is provided in the supporting information.
3.4.3 Verification process for identification of phloretin metabolites based on Part C (m/z 167/169 + m) with Part D (m/z 121/123 + n)
P49 afforded [M + H]+ ion at m/z 261.11274 (C15H17O4, mass error −2.051 ppm) in positive ion mode. It yielded the FFIs at m/z 123 [M + H-C7H6O3]+ and m/z 155 (169–16 + 2) [M + H-C7H6O]+. Then, DPIs at m/z 121 [M + H-C7H8O3]+, m/z 111 [M + H-C8H8O-OCH2]+ and m/z 107 [M + H-C8H10O3]+ were also observed. Thus, P49 was tentatively deduced as a dehydroxylation and hydrogenation metabolite of phloretin. P21, P27 and P38 produced [M−H]− ions at m/z 273.07687, m/z 273.07684 and m/z 273.07709 (C15H13O5, mass errors 4.101 ppm, 3.992 ppm and 4.907 ppm) with a mass 162 Da less than phlorizin, suggesting that they were isomers of phloretin. The FFIs at m/z 167 [M−H−C7H6O]− and m/z 121 [M−H−C7H4O4]− and DPIs at m/z 125, m/z 107 and m/z 93 supported the previous deduction. Therefore, they were determined to be isomers of phloretin. P4 possessed [M−H]− ion at m/z 287.05890 (C8H15O11, mass error −1.988 ppm) with 14 Da more than phloretin. The FFIs at m/z 167 [M−H−C7H4O2]− and m/z 135 (121 + 14) [M−H−C7H4O4]− proved that carbonyl was located at C-1′, and the DPIs at m/z 125, m/z 123 and m/z 93 were also observed. Through the above inference, P4 could be deduced as a carbonylation metabolite of phloretin. Based on the above analysis of P4, P13 was also a carbonylation metabolite of phloretin with the FFIs at m/z 169 and m/z 137 (123 + 14) in positive ion mode. P23 and P30 were glucuronidation metabolites of phloretin. P37 was sulfation metabolite of phloretin. The process by which they were analyzed is provided in the supporting information.
Five isomeric metabolites of P46, P48, P50, P51 and P52 yielded the same theoretical [M + H]+ ion at m/z 277.10710 (C15H17O5, mass errors within ± 5 ppm) in positive ion mode, which were two hydrogens more than that of phloretin. They shared FFIs at m/z 171 (169 + 2) [M + H-C7H6O]+ and m/z 123 [M + H-C7H6O4]+, proving hydrogenation reaction occurred at the carbonyl, and the DPIs at m/z 107 [M + H-C8H10O4]+ and m/z 121 [M + H-C7H8O4]+ were clearly detected. Hence, P46, P48, P50, P51 and P52 were determined to be hydrogenation metabolites of phloretin. P11 produced [M−H]− ion at m/z 315.12393 (C18H19O5, mass error 3.903 ppm). The FFIs at m/z 165 (167–2) [M−H−C8H8O−2CH3]− and m/z 120 (121–1) [M−H−C9H8O4−CH3]− and representative ion at m/z 92 proved that two methyl groups were attached to Part C and one on Part D. Therefore, we identified P11 as a trimethylation metabolite of phloretin. P3 was 176 Da more than phlorizin, which yielded [M−H]− ion at m/z 611.1618 (C27H31O16, mass error 1.863 ppm). The typical ions at m/z 435 [M−H−GluA]−, m/z 273 [M−H−GluA−Glc]−, m/z 125 [M−H−GluA−Glc−C8H8O−CO]−, m/z 107 [M−H−GluA−Glc−C8H6O4]−, m/z 93 [M−H−GluA−Glc−C8H6O4−CH2]− and FFIs at m/z 167 [M−H−GluA−Glc−C7H6O]− and m/z 121 [M−H−GluA−Glc−C7H4O4]− showed that P3 could be confirmed as a glucuronidation metabolite of phlorizin. P20 generated [M−H]− ion at m/z 435.12994 (C21H23O10, mass error 3.141 ppm). The product ion at m/z 91 [M−H−Glc−C8H6O4−O]− indicated a hydroxy was attached to C-1′, and the FFIs at m/z 167 [M−H−Glc−C7H6O]− and m/z 121 [M−H−Glc−C7H4O4]− were also detected. Thus, P20 could be deduced as a carbonylation, dehydroxylation and hydrogenation metabolite of phlorizin.
4 Discussion
The identification of metabolites of biologically active substances with potential drug development value can help elucidate their efficacy, mechanism and toxicity. The establishment of a metabolic profile was helpful to the study of drug pharmacokinetics and conversion in vivo. Almost all drugs undergo metabolic processes in vivo that producing biologically active or inert metabolites. Due to the structural similarity of metabolites to their prototypes, bioactive metabolites may preserve the intrinsic activity of the prototypes for a range of receptors. In some instances, biologically active metabolites can exhibit distinct activities that may produce adverse effects or lead to new discoveries in biology and medicine. There is evidence that some metabolites possess superior efficacy, pharmacokinetic and safety profiles than their prototype and have been developed as new drugs (López-Muñoz and Alamo, 2013; Cerny et al., 2020). The complexity of the biological matrix makes it more difficult to characterize the metabolic status of the drug in the body. Glycosides generally have aglycones that are simpler in structure and easier to analyze for metabolic pathways. Therefore, the metabolic profile of phloretin was used to supplement the metabolic profile of phlorizin. In this study, a new method of drug metabolism analysis in vivo was established by using UHPLC-Q-Extractive MSn and characteristic ion screening. A total of 50 phloretin metabolites were identified, including 23 metabolites in plasma, 13 metabolites in urine and 14 metabolites in faeces, based on the further reaction of the prototype, glucuronidated phloretin, sulfated phloretin and decarbonylated phloretin. Additionally, 52 phlorizin metabolites were identified, including 22 metabolites in plasma, 16 metabolites in urine and 14 metabolites in faeces; these metabolites indicated the further biotransformation of prototype, phloretin, glucuronidated phlorizin, sulfated phlorizin and carbonylated phlorizin. The proposed metabolic pathways of phloretin and phlorizin in rats are illustrated in Fig. 5. Therefore, most metabolites of phloretin and phlorizin were in the systemic circulation along with wide enterohepatic circulation, which was consistent with previous research results (Mei et al., 2016). Compared with the traditional analysis method, the characteristic ion screening method established in this paper is more efficient, which greatly saves time during the identification process and retains the accuracy as much as possible. In our research, we obtained more than 100 metabolites of phlorizin and phloretin through the use of new analysis methods, and the results were far superior to those involving the dozens of metabolites analyzed by previous researchers (Li et al., 2014). In addition, biological sample preparation technology and the powerful separation ability and high resolution of UHPLC allow us to eliminate endogenous material interference and screen low-level metabolites as much as possible.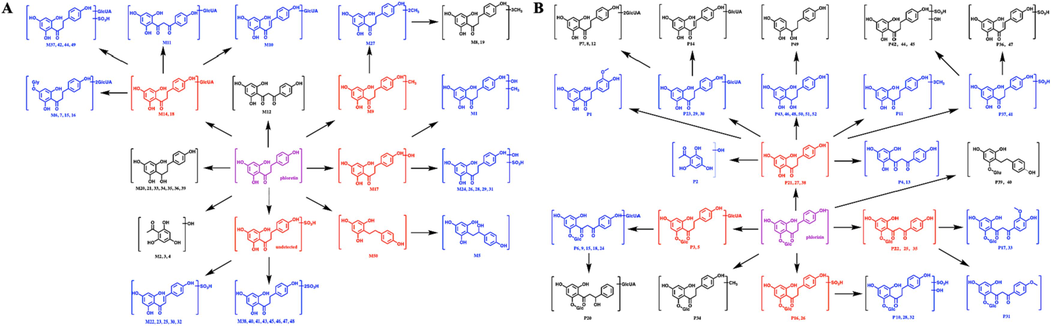
The proposed phloretin (A) and phlorizin (B) metabolic patterns in vivo. Red: Major phase I metabolites; Blue: Major phase II metabolites. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The in vivo metabolic pathways of phloretin and phlorizin all shared common metabolites, such as hydroxylation-methylation, hydroxylation-sulfation, glucuronidation, sulfation and hydrogenation metabolites of phloretin. In addition, a variety of unique metabolites of phloretin were detected in bio-samples, such as hydroxylation, hydroxylation-sulfation, disulfation, decarbonylation, decarbonylation-hydroxylation, methylation, dimethylation, glucuronidation- sulfation and glucuronidation-carbonylation metabolites. Similarly, glucuronidation, glucuronidation-carbonylation, glucuronidation-carbonylation-hydrogenation, carbonylation, carbonylation-methylation, carbonylation-hydroxylation-methylation, sulfation, sulfation-hydroxylation, methylation, decarbonization, hydrolyzation-sulfation, hydrolyzation-sulfation-hydroxylation and hydrolyzation-dehydroxylation-hydrogenation metabolites were identified as exclusive metabolites of phlorizin. Both phloretin and phlorizin were detected in the biological samples, and the former was found as a metabolite of the latter, proving that some phlorizin was metabolized to phloretin in vivo, followed by phase I and II metabolism to be quickly transformed into other metabolites, especially the glucuronidation, sulfation and hydrogenation conjugates observed in this paper. Phloretin and phlorizin from Lithocarpus polystachyus Rehd could engage in hydrolyzation, sulfation, sulfation-glucuronidation, hydroxylation, hydroxylation-sulfation and hydroxylation-methylation reaction to produce metabolites in rat plasma, urine and faeces (Li et al., 2014). In contrast, our results showed that phloretin and phlorizin not only included these above reactions but also involved in many other reactions, which was responsible for the identification of abundant phloretin and phlorizin metabolites. Phlorizin has potential in anti-diabetes and stress hyperglycemia clinical applications, but few related mechanisms have been studied (Zhang et al., 2020). OH-phloretin (M17) was proved to be one of the important metabolites of phloretin in this study. Woo et al. (Woo et al., 2023) suggested that 3-OH phloretin could inhibits obesity by preventing macrophage infiltration into adipose tissue and alleviating insulin resistance. Another study showed that a series of methylated derivatives of phloretin exhibited moderately strong cytotoxicity toward cancer cell lines, such as CH3-phloretin (M9), 2CH3-phloretin (M27) and 3CH3-phloretin (M8, M19 and P11) (Wang et al., 2014). There was also evidence that phlorizin and its metabolites were potential therapeutic agents for the treatment of osteoporosis in the elderly (Antika et al., 2017). Thus, all these insights demonstrated that identification of phloretin and phlorizin metabolites contributed to clarifying their pharmacodynamic material basis and actional mechanism. But the fly in the ointment was that the content of the identified metabolites of phloretin and phlorizin could not be determined due to the lack of standards for the corresponding metabolites. Our goal will be to synthesize the identified metabolites as the protocol in quantitatively measuring the distribution of phloretin and phlorizin and their metabolites for a better understanding of toxicity and efficacy.
5 Conclusion
In this paper, we created a new method for the analysis of linear polyphenol metabolites in vivo. Compared to the traditional method, our method involved the analysis of a series of characteristic fragment ions by analyzing the cracking laws and metabolic reactions of standard products. Then, the metabolic profiles of phloretin and phlorizin in rat plasma, urine and faeces were systematically and comprehensively investigated. A total of 50 phloretin metabolites and 52 phlorizin metabolites were identified and summarized through rapid, sensitive and accurate UHPLC-Q-Extractive MSn coupled with characteristic ion screening and multiple metabolic pathways template. The results of previous studies have shown that glucuronidation and sulfation were the main metabolic pathways of phloretin and phlorizin, while our analysis results proved that they also have other metabolic pathways, such as decarbonylation, carbonylation and hydrogenation. These results directly confirm the efficiency and accuracy of the new method used in this study and provide a solid foundation for future in vivo pharmacodynamic and pharmacokinetic studies of phloretin and phlorizin.
Human and Animal rights
No humans were involved in this study. The reported experiments on animals were in accord with the standards set forth in the 8th Edition of the Guide for the Care and Use of Laboratory Animals (https://grants.nih.gov/grants/olaw/Guide-for-thecare-and-use-of-laboratory-animals.pdf) published by the National Academy of Sciences.
CRediT authorship contribution statement
Haoran Li: Methodology. Hong Wang: Methodology. Pingping Dong: Methodology. Huajian Li: . Shaoping Wang: . Jiayu Zhang: Conceptualization, Funding acquisition, Project administration, Methodology.
Acknowledgement
This work was supported by the National Natural Science Foundation of China (No. 82174039), Natural Science Foundation of Shandong Province (No. ZR2020MH371), Taishan Young Scholar Program of Shandong (No. TSQN202103110), Schoollocality Combination Integrated Development Project in Yantai City, and Matching Support Program for Provincial and Above Leading Talents in Yantai City (No. 10073801).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Phloretin increases the anti-tumor efficacy of intratumorally delivered heat-shock protein 70 kDa (HSP70) in a murine model of melanoma. Cancer Immunol Immunother.. 2016;65:83-92.
- [CrossRef] [Google Scholar]
- Dietary phlorizin enhances osteoblastogenic bone formation through enhancing β-catenin activity via GSK-3β inhibition in a model of senile osteoporosis. J Nutr Biochem.. 2017;49:42-52.
- [CrossRef] [Google Scholar]
- Interaction of phloretin and 6-ketocholestanol with DPPC-liposomes as phospholipid model membranes. Int J Pharm.. 2005;294:149-155.
- [CrossRef] [Google Scholar]
- Effective application of metabolite profiling in drug design and discovery. J Med Chem.. 2020;63:6387-6406.
- [CrossRef] [Google Scholar]
- Metabolism of flavonoids in human: A comprehensive review. Curr. Drug Metab.. 2014;15:48-61.
- [CrossRef] [Google Scholar]
- Bioavailability of phloretin and phloridzin in rats. J Nutr.. 2001;131:3227-3230.
- [CrossRef] [Google Scholar]
- Inhibitory effect of phloretin on α-glucosidase: Kinetics, interaction mechanism and molecular docking. Int J Biol Macromol.. 2017;95:520-527.
- [CrossRef] [Google Scholar]
- Phloretin attenuates allergic airway inflammation and oxidative stress in asthmatic mice. Front. Immunol.. 2017;8:134.
- [CrossRef] [Google Scholar]
- Comprehensive characterization of the in vitro and in vivo metabolites of geniposide in rats using ultra-high-performance liquid chromatography coupled with linear ion trap-Orbitrap mass spectrometer. Xenobiotica.. 2016;46:357-368.
- [CrossRef] [Google Scholar]
- Identification of the bioactive components of orally administered Lithocarpus polystachyus Rehd and their metabolites in rats by liquid chromatography coupled to LTQ Orbitrap mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci.. 2014;962:37-43.
- [CrossRef] [Google Scholar]
- Types, principle, and characteristics of tandem high-resolution mass spectrometry and its applications. RSC Adv.. 2015;5:107623-107636.
- [CrossRef] [Google Scholar]
- Phloridzin, an Apple Polyphenol, Exerted Unfavorable Effects on Bone and Muscle in an Experimental Model of Type 2 Diabetes in Rats. Nutrients.. 2018;10:1701.
- [CrossRef] [Google Scholar]
- Active metabolites as antidepressant drugs: the role of norquetiapine in the mechanism of action of quetiapine in the treatment of mood disorders. Front Psychiatry.. 2013;4:102.
- [CrossRef] [Google Scholar]
- Inhibitory effect of DIDS, NPPB, and phloretin on intracellular chloride channels. Pflugers Arch.. 2007;455:349-357.
- [CrossRef] [Google Scholar]
- Pharmacological Aspects and Potential Use of Phloretin: A Systemic Review. Mini Rev Med Chem.. 2019;19:1060-1067.
- [CrossRef] [Google Scholar]
- Insulin Sensitivity-Enhancing Activity of Phlorizin Is Associated with Lipopolysaccharide Decrease and Gut Microbiota Changes in Obese and Type 2 Diabetes (db/db) Mice. J Agric Food Chem.. 2016;64:7502-7511.
- [CrossRef] [Google Scholar]
- Biocatalytic Production of a Potent Inhibitor of Adipocyte Differentiation from Phloretin Using Engineered CYP102A1. J Agric Food Chem.. 2020;68:6683-6691.
- [CrossRef] [Google Scholar]
- Specific bioactive compounds in ginger and apple alleviate hyperglycemia in mice with high fat diet-induced obesity via Nrf2 mediated pathway. Food Chem.. 2017;226:79-88.
- [CrossRef] [Google Scholar]
- Polyphenolic profiles in eight apple cultivars using high-performance liquid chromatography (HPLC) Agric. Food Chem.. 2003;51:6347-6353.
- [CrossRef] [Google Scholar]
- Comparative oral and intravenous pharmacokinetics of phlorizin in rats having type 2 diabetes and in normal rats based on phase II metabolism. Food Funct.. 2019;10:1582-1594.
- [CrossRef] [Google Scholar]
- Synthesis, Crystal Structure, and Biological Evaluation of a Series of Phloretin Derivatives. Molecules.. 2014;19:16447-16457.
- [CrossRef] [Google Scholar]
- Profiling and identification of chlorogenic acid metabolites in rats by ultra-high-performance liquid chromatography coupled with linear ion trap-Orbitrap mass spectrometer. Xenobiotica.. 2018;48:605-617.
- [CrossRef] [Google Scholar]
- Comprehensive Analysis of Pterostilbene Metabolites In Vivo and In Vitro Using a UHPLC-Q-Exactive Plus Mass Spectrometer with Multiple Data-Mining Methods. ACS Omega.. 2022;7:38561-38575.
- [CrossRef] [Google Scholar]
- Molecular model and in vitro antioxidant activity of a water-soluble and stable phloretin/hydroxypropyl-β-cyclodextrin inclusion complex. J. Mol. Liq.. 2017;236:68-75.
- [CrossRef] [Google Scholar]
- 3-OH Phloretin Inhibits High-Fat Diet-Induced Obesity and Obesity-Induced Inflammation by Reducing Macrophage Infiltration into White Adipose Tissue. Molecules.. 2023;28:1851.
- [CrossRef] [Google Scholar]
- Rapid screening and identification of diterpenoids in Tinospora sinensis based on high-performance liquid chromatography coupled with linear ion trap-Orbitrap mass spectrometry. Molecules.. 2017;22:912.
- [CrossRef] [Google Scholar]
- Phlorizin ameliorates obesity-associated endotoxemia and insulin resistance in high-fat diet-fed mice by targeting the gut microbiota and intestinal barrier integrity. Gut Microbes.. 2020;12:1-18.
- [CrossRef] [Google Scholar]
- Development of a comprehensive flavonoid analysis computational tool for ultrahigh-performance liquid chromatography-diode array detection-high-resolution accurate mass-mass spectrometry data. Anal. Chem.. 2017;89:7388-7397.
- [CrossRef] [Google Scholar]
- Immunomodulatory activities of phlorizin metabolites in lipopolysaccharide stimulated RAW264.7 cells. Biomed Pharmacother.. 2017;91:49-53.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2023.105597.
Appendix A
Supplementary data
The following are the Supplementary data to this article:Supplementary Data 1
Supplementary Data 1







