Translate this page into:
Comprehensive characterization of narirutin metabolites in vitro and in vivo based on Analogous-Core recursion analysis strategy using UHPLC-Q-Exactive Orbitrap MS/MS
⁎Corresponding authors. wangzhibin@tongrentang.com (Zhibin Wang), zhangjiayu0615@163.com (Jiayu Zhang)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Narirutin, extracted from the dried ripe pericarp of Rutaceae orange and its cultivated varieties, is a dihydroflavone compound with various biological activities and pharmacological effects. So far as we know, the metabolism profiling of narirutin has been insufficient until now. In the present study, an efficient method was employed to perform rapid analysis and identification of narirutin metabolites by using ultra-high performance liquid chromatography quadrupole exactive orbitrap MS/MS (UHPLC-Q-Exactive Orbitrap MS/MS) combined with an analogous core recursion (ACR) analysis strategy. Firstly, according to the basic core structure of the dihydroflavonoid, the cleavage mode and fragmentation ions of narirutin were summarized. Secondly, based on the difference of the substituent groups, the fragment ions of narirutin were preliminarily inferred and verified by the secondary mass spectra of its standard compound. Thirdly, the fragment ions of narirutin in positive and negative ion modes were summarized to provide a basis for the identification of metabolites. Fourthly, candidate metabolites were accurately and tentatively identified according to the cleavage law of mass spectrometry, literature reports, comparison of reference substances, and especially the diagnostic fragment ion clusters (DFICs) and characteristic suggestive ions (CSIs) deduced preliminarily. Finally, a total of 46 metabolites (30 in vivo and 19 in vitro), including prototype drugs were identified based on the ACR analysis strategy, chromatographic retention behavior of metabolites, corresponding ClogP value and accurate molecular weight. Among them, 22 metabolites were discovered in rats plasma, 12 in urine, 4 in liver tissue, 19 in liver microsomes, respectively. Additionally, metabolites relative contents were also studied by extracted ion chromatography (EIC) method. Our result also illustrated that narirutin primarily underwent glucuronidation, sulfation, glutathione binding, deglycosylation, oxidation, reduction, methylation, hydroxylation, ring-opening and their composite reactions. A novel strategy was constructed to comprehensively elucidate the biotransformation pathways of narirutin in vitro and in vivo, and a systematic metabolic profile of narirutin was generated. This study provided a great reference for further understanding of the biotransformation pathways and guiding for clinical application of narirutin.
Keywords
Narirutin
UHPLC-Q-Exactive Orbitrap MS/MS
Metabolites
Analogous-Core Recursion analysis strategy
In vitro and in vivo
1 Introduction
Narirutin (Naringenin 7-O-rutinoside) attributed to dihydroflavone compounds (Duan et al., 2016) has been isolated from the dry and mature peel extract of orange (Citrus reticulata Blanco) as well as its cultivated varieties in Rutaceae. Plenty of studies have shown that Narirutin owns a great many of pharmacological activities, including blood pressure reduction (Fraga et al., 2021), glucose and lipid downregulation (Chiechio et al., 2021), antimicrobial (Shehata et al., 2021), anti-inflammatory (Ha et al., 2012), anti-virus (Yang et al., 2020), anti-tuberculosis (Sahu et al., 2020), anti-cancer (Lim et al., 2015) and anti-allergic (Niu et al., 2021) properties. At the same time, narirutin can improve the level of intracellular cAMP, stimulate endothelial nitric oxide synthase and activate voltage-gated potassium channels in vascular smooth muscle cells, thus playing a role in vascular relaxation (Wong et al., 2021). It also showed protective effects on the immune system and inflammation (Miles and Calder 2021), alcohol-induced liver damage (Park et al., 2013) and gastric Injury (Wu et al., 2021a, 2021b). In recent years, Miscellaneous innovative applications of narirutin was also supported the further study of its pharmacological effects. By using Silico, it was shown that citrus flavone myoglobin interacts significantly with most of the receptors and possesses a good inhibitory activity in vitro, indicating its powerful role in the management of diabetes and its complications (Qurtam et al., 2021). The polymerized nanoparticles were suitable for delivering the flavonoid naringin to the gut, avoiding gastric degradation and improving its bioavailability from oral solid dosage forms, as was the case with other flavonoids such as narirutin (Mohanty et al., 2021).
After oral administration, a series of biological transformation reactions will occur until the drug is eliminated from the body, accompanied by the therapeutic efficacy or toxic side-effect, which provides a pivotal theoretical basis for in vivo mechanisms exploitation of drugs (Wu et al., 2021a, 2021b). Previous studies mostly focused on the pharmacological activity of naringin at present, rather few on narirutin metabolites mining. That is to say, an in-depth study on narirutin metabolic pathways in vivo and in vivo carried out was of great importance. Therefore, it is important to establish an accurate and high-resolution qualitative analysis method for the comprehensive characterization of narirutin metabolites.
Xenobiotic metabolism is a natural reaction to the ubiquity of exotic compounds. However, they are usually metabolized into different forms by different metabolic pathways (Shang et al., 2017). Because of its little amount, the metabolite signal is usually masked by the background noise produced by endogenous substances. In recent years, with the continuous development of liquid chromatography mass spectrometry (LC-MS), its performances of rapid detection speed, high sensitivity and high resolution have already been improved. Therefore, LC-MS has been frequently used to clarify the separation and analysis of drug metabolites (Jiang et al., 2021; Lan et al., 2022). Especially, ultra-high performance liquid chromatography quadrupole exactive Orbitrap MS/MS (UPLC-Q-Exactive Orbitrap MS/MS) method was established to screen and identify the naringin metabolites. In our study, we proposed a strategy named “Analogous Core Recursion” (ACR), which combined with the “Diagnostic Fragment Ion Clusters” (DFICs) and “Characteristic Suggestive Ions” (CSIs) obtained by our recursive analysis, for the inference and recognition of narirutin metabolites to improve the efficiency of metabolite recognition. Finally, based on these detected metabolites, the metabolic pathways of narirutin were also proposed so as to provide the basis for the biological activity study.
2 Material and methods
2.1 Chemicals and reagents
Narirutin (MUST-20111105, its physical and chemical properties were shown in Table 1) was purchased from Chengdu Must Bio-technology Co., Ltd (Sichuan, China). The reference standard with purity higher than 99% was applicable to HPLC analysis. Pooled male Sprague Dawley (SD) rat liver microsomes (20210305, 1 mL, 20 mg/mL), β-nicotinamine adenine dinucleotide phosphate (NADPH), uridine 5′-diphosphoglucuronic acid trisodium salt (UDPGA) and Magnesium chloride (MgCl2) were purchased from NEWGAINBIO Co., Ltd (Wuxi, China). UHPLC grade acetonitrile, methanol and formic acid (FA) were purchased from Thermo Fisher Scientific (Fair Lawn, NJ, USA), and pure water for analysis was bought from Watson Group Co., Ltd (Guangzhou, China). All the other chemicals of analytical grade were available at the work station, Shandong Academy of Chinese Medicine (Jinan, China). Grace Pure™ SPE C18-Low solid phase extraction cartridges (200 mg/3 mL, 59 mm, 70 Å) were purchased from Grace Davison Discovery Science (Deerfield, IL, USA).
Property Name
Property Value
density
1.7 ± 0.1 g/cm3
Boiling point
924.3 ± 65.0 °C at 760 mmHg
melting point
152–190 °C
Molecular Formula
C27H32O14
Molecular Weight
580.5
Exact Mass
580.17920569
Appearance trait
Grey powder
Vapor pressure
0.0 ± 0.3 mmHg at 25 °C
flash point
307.3 ± 27.8 ℃
Refractive index
1.708
XLogP3-AA
−1.1
Hydrogen Bond Donor Count
8
Hydrogen Bond Acceptor Count
14
Rotatable Bond Count
6
Topological Polar Surface Area
225 Å2
Heavy Atom Count
41
2.2 Animals and drug administration
Six male SD rats weighting 200 ± 10 g were obtained from Jinan Pengyue Experimental Animal Breeding Co., Ltd (Shandong, China). The rats were housed in a controlled room at standard temperature (24 ± 2 ℃) and humidity (70 ± 5%) and kept on a 12 h light/12 h dark regime. After a week of acclimation, the rats were randomly divided into two groups: Drug Group (n = 3) and Control Group (n = 3) for testing plasma, urine and faeces individually. They were fasted for 12 h with free access to water prior to the experiment. Narirutin dissolved in normal saline was orally given to the rats in Drug Group were at a dose of 150 mg/kg body weight. Rats in Control Group were given the same volume of normal saline. All the rats were administered for three consecutive days and fed in animal room of Shandong International Biotechnology Park. The animal protocols were approved by the institutional Animal Care and Use Committee at Binzhou Medical University (2021-085). The animal facilities and protocols were complied with the Guide for the Care and Use of Laboratory Animals (USA National Research Council, 1996).
2.3 Sample collection and preparation
2.3.1 Preparation of in vivo samples
Blood samples (0.5 mL) were taken from the suborbital venous plexus of rats at 0.5, 1, 1.5, 2, 4, and 6 h post-administration. Each sample was centrifuged at 3,500 rpm for 10 min to separate plasma samples and preserved in three parts. Urine and faeces samples were collected 0–24 h after oral administration. Rat liver tissues were collected 24 h after the last administration. All the homogeneous biological samples from the same group were merged into a collective sample.
Three copies of plasma samples were treated in three ways. Randomly select any two samples, and methanol or acetonitrile was added to the blood samples at a ratio of 1:3 respectively. Each sample was centrifuged at 3,500 rpm for 10 min to separate supernatant liquid. Another sample of blood plasma (1 mL) were added to solid phase extraction (SPE) cartridges pretreated with methanol (5 mL) and deionized water (5 mL), respectively. Then, the SPE cartridges were successively washed with deionized water (3 mL) and methanol (3 mL).
Urine samples were centrifuged at 12,000 rpm for 15 min to collect supernatant. Rat faeces samples were lyophilized, ground and mixed evenly. Then pure water (10 mL) was added into the excrement powder (2 g) for ultrasonic treatment for 0.5 h and the suspension was centrifuged at 3,500 rpm for 15 min to obtain the supernatant. The rat liver tissues were ground with normal saline and centrifuged at 3,500 rpm for 15 min to obtain the supernatant liquid. The treated urine, faeces and liver tissues samples were passed through SPE cartridges and methanol eluent was collected to use for the further instrumental analysis.
2.3.2 Preparation of in vitro samples
In the incubation system containing MyCl2 (3 mM), liver microsomes (20 mg/mL) and narirutin (0.1 mg/mL), incubated for 5 min at 37 ℃. NADPH solution (25 mg/mL) was added to start the reaction and incubated for 4 h. The total incubation amount in the system was 1 mL. At 5, 10, 15, 30, 45, 60, 120 and 240 min, 100 μL of system solution was taken and 200 μL of cold acetonitrile was added to stop the reaction. The solution at each stage was mixed and centrifuged at 3,500 rpm for 15 min to take the supernatant.
2.3.3 Sample preservation and pre-injection treatment
After all the samples were prepared, the organic solvent was blow-dried with N2 at room temperature. The samples were redissolved with 300 μL methanol, centrifuged at 14,000 rpm for 10 min at high speed. The supernatant was taken in liquid sample bottle for the subsequent instrumental analysis.
2.4 Instruments and analytical conditions
UHPLC analysis was performed on a DIONEX Ultimate 3000 UHPLC system (Thermo Fisher Scientific, MA, USA), which equipped with a binary pump, an automated sampler and a column compartment. The chromatographic separations were performed on Waters ACQUITY BEH C18 column (2.1 × 100 mm, 1.7 μm). HRMS and MS/MS spectra were obtained using a Q-Exactive Orbitrap mass spectrometer.
The mobile phase consisted of 0.1% formic acid aqueous solution (A) and acetonitrile (B) at a flow rate of 0.3 mL/min. The linear gradient procedure was described as follows: 0–5 min, 5%–30%B; 5–10 min, 30%-50%B; 10–27 min, 50%–90%B; 27–30 min, 90%–5%B. The injection volume was 2 μL. An analytical run was performed by mass spectrometry in positive and negative ion modes. The ion source parameters were listed as follows: nitrogen (purity ≥ 99.99%) served as the sheath gas and auxiliary gas. The flow of sheath gas (nitrogen) was 45 arb and auxiliary gas (nitrogen) was 10 arb. A capillary temperature of 320 ℃, probe heater temperature of 320 ℃ and spray voltage of 3,800/3,500 V (+/-) were used. Orbitrap analyzers obtained high-resolution mass spectra with a full scan in m/z 80 ∼ 1,200 mass range at a resolution of 70,000 in MS and a resolution of 17,500 in dd MS/MS.
2.5 Peak selections and data processing
A Thermo Xcalibur 2.1 workstation was used for the data acquisition and processing. To obtain as many ESI-MS/MS fragment ions of narirutin metabolites as possible, the base peak ions detected with an intensity over 40,000 in positive ion mode and 10,000 in negative ion mode were selected for chemical identification. The chemical formulas attributed to the selected peaks were calculated using a formula predictor by setting the parameters as follows: C [5–30], H [5–60], O [1–20], S [0–5], N [0–5] and the ring double bond (RDB) equivalent value [3–20].
3 Results
3.1 Establishment of analytical strategy
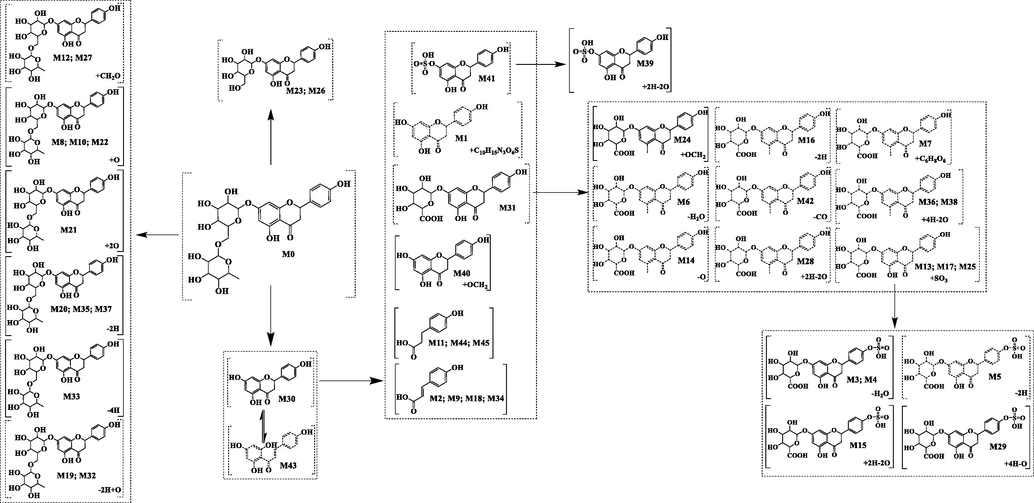
It is important to establish an efficient analytical strategy to capture the signals of narirutin metabolites. Therefore, in this study, an analytical strategy based on ACR was proposed to screen and identify narirutin metabolites using complex mass spectra information. In the analysis strategy, based on the core structure of a large class of substances, a single compound was deduced, and according to the DFICs and CSIs, the metabolites and the possible sites of their metabolic reactions were determined (Fig. 1). Firstly, according to the cleavage mode of the basic core structure of flavonoids, the cleavage rules of narirutin were deduced by mass spectra, and the DFICs of narirutin were identified by combining the secondary mass spectra information of narirutin. Secondly, based on the metabolic characteristics of compounds in vivo, the potential phase II metabolic reactions, such as deglycosylation to aglycone, glucuronidation and sulfation, were predicted. Correspondingly, the DFICs were determined based on the abovementioned metabolites. Thirdly, based on the DFICs obtained in the previous two steps, the metabolites were further speculated. Fourthly, the possible sites of metabolic reactions, such as the A ring, B ring and sugar group, were deduced by indicating the distinction of CSIs. Considering the retention time of metabolites and the corresponding ClogP value, isomers were preliminarily distinguished. Finally, the accuracy and reliability of the analysis strategy were verified by comparison to the fragmentation cleavage behavior of the reference substance and metabolites of narirutin.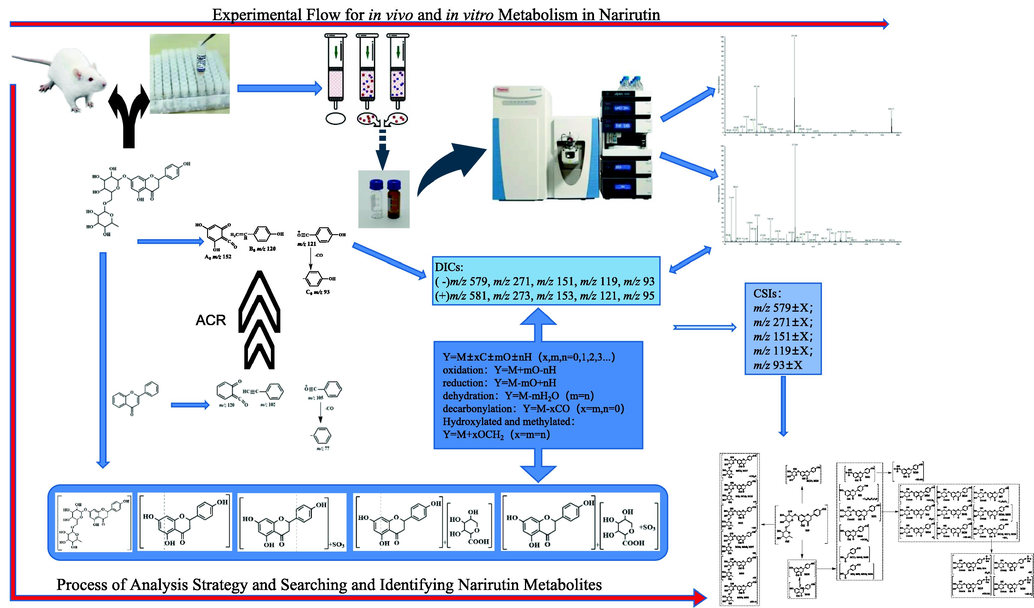
Schematic diagram of analysis strategies and the experimental processes of Narirutin metabolism in vitro and in vivo.
3.2 Interpretation of the narirutin metabolites analysis strategy
There are two basic cleavage methods of flavonoids in mass spectrometry. The first method is the Retro-Diels-Alder (RDA) cleavage that produces 2 ions at m/z 120 and m/z 102, and the second method yields a fragment ion at m/z 105. The three fragment ions produced by these methods are derived from the core structure at m/z 222. CO can be easily removed from the ion at m/z 105 to produce the ion at m/z 77. The aglycone of narirutin is a dihydroflavonoid, which has 3 more Os (2 in the A ring and 1 in the B ring) and 2 more Hs (C2-C3) than the nuclear structure of the basic flavone. Thus, we speculated that the above two cleavage methods were applicable to narirutin. Based on the above cleavage rules, we recursed the DFICs of narirutin as m/z 580, m/z 272, m/z 152, m/z 120 and m/z 94 (Fig. 2). The corresponding fragment ions in different ion modes were as follows: m/z 579, m/z 271, m/z 151, m/z 119 and m/z 93 in the negative ion mode; and m/z 581, m/z 273, m/z 153, m/z 121 and m/z 95 in the positive ion mode. By comparing the information of the narirutin standard in electron spray ionization (ESI)-MS/MS (Fig. 3 and Fig. 4), the correctness of the DFICs obtained by recursive analysis were confirmed, implying a further analysis could be employed.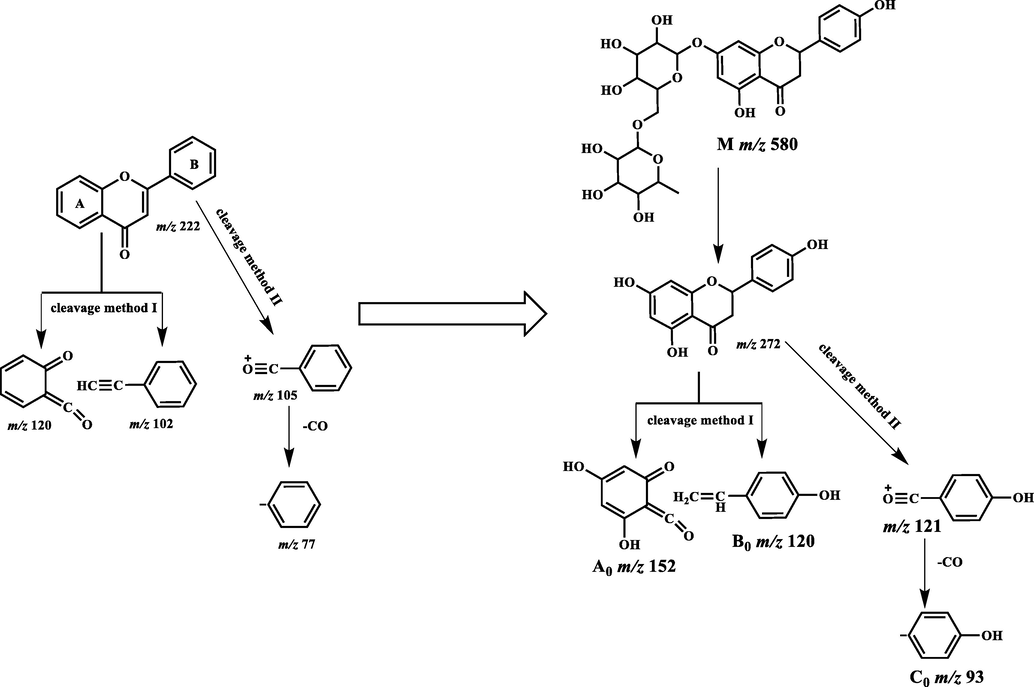
Cleavage of core structure of flavonoids and Narirutin.
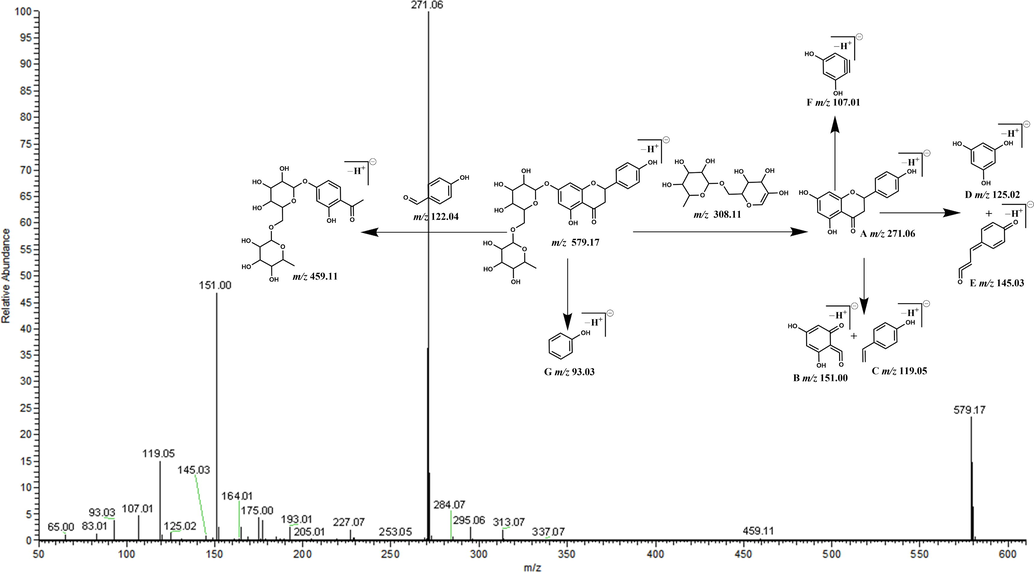
Secondary mass spectrum and cleavage rule in negative mode.
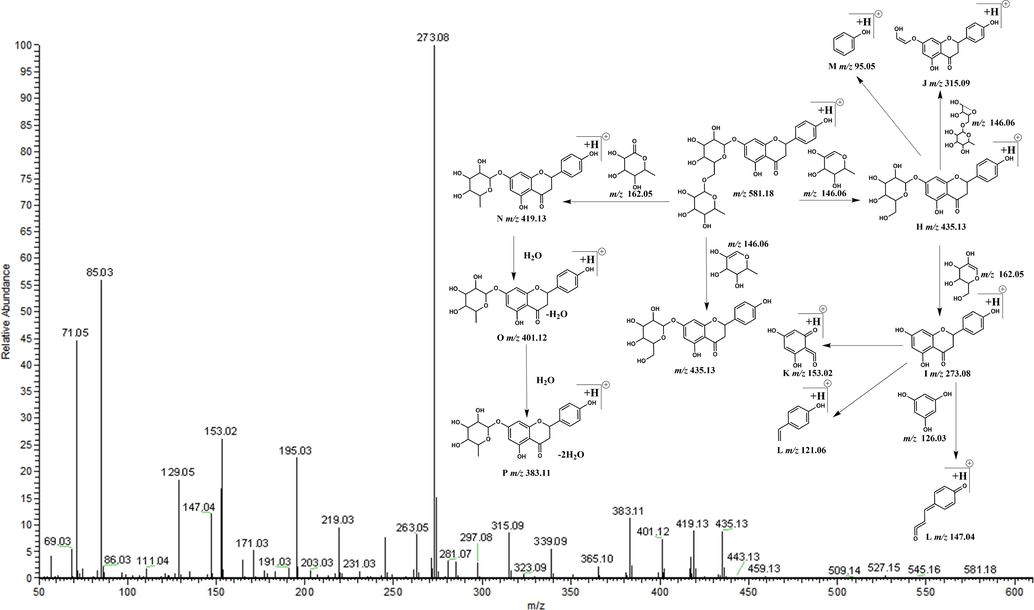
Secondary mass spectrum and cleavage rule in positive mode.
Narirutin is produced by combining the rutinose group at the C7 position on naringenin. The polarity of narirutin is relatively large, and the glucuronidation, sulfation and other binding reactions with narirutin are difficult due to the steric hindrance of the rutinose group. However, the polarity of naringenin produced by metabolism is low, which is not conducive to excretion by the body. Because naringenin has little steric hindrance, it is speculated that glucuronidation, sulfation and other reactions may occur in naringenin aglycone. Fragment ions, such as m/z 176, m/z 114 and m/z 80, can be generated by the glucuronic acid group and sulfate group, thus enriching their DFICs. For example, in the negative ion mode, the DFICs produced by naringenin glucuronic acid were m/z 447, m/z 271, m/z 176, m/z 151, m/z 119, m/z 114 and m/z 93, and the DFICs formed by naringenin sulfate were m/z 351, m/z 271, m/z 151, m/z 119, m/z 93 and m/z 79.
According to the DFICs of the corresponding compounds and the secondary mass spectra information of different samples, the metabolites obtained by oxidation, reduction and dehydration reactions were predicted. In the present study, we proposed the following recursive formula: Y = M ± xC ± mO ± nH; where Y is the exact molecular weight of the metabolite; M is the exact molecular weight of the prototype compound; C, O and H represent the exact molecular weights of carbon, oxygen and hydrogen, respectively; and x, m and n are integers ≥0. According to different types of metabolic reactions, such as oxidation, reduction and dehydration, different x, m and n values were substituted into the formula, and the obtained Y values were searched in the corresponding MS/MS spectra, which allowed a preliminary prediction of the metabolites (Fig. 1).
The regular changes in DFICs indicated where metabolic reactions were likely to occur. Taking narirutin as an example, the following DFICs changes were estimated in negative ion mode: if the metabolic reactions occurred in the A ring, the DFICs changes were 579 ± X, m/z 271 ± X, m/z 151 ± X, m/z 119 and m/z 93; if the reaction occurred in the B ring, the DFICs changes were m/z 579 ± X, m/z 271 ± X, m/z 151, m/z 119 ± X and m/z 93 ± X; and if the reaction occurred at the relatively rare rutinose group, the DFICs changes were m/z 579 ± X, m/z 271, m/z 151, m/z 119 and m/z 93. Furthermore, the retention time of the metabolites and their corresponding ClogP values were combined to preliminarily distinguish and identify the isomers produced by the same reaction at different sites.
3.3 Identification and structural elucidation of narirutin metabolites
A total of 46 narirutin metabolites (narirutin included) were detected and identified in the plasma, urine, faeces, liver tissues and liver microsomes of SD rats using ultra-high performance liquid chromatography quadrupole exactive orbitrap MS/MS (UHPLC-Q-Exactive Orbitrap MS/MS). Among them, 22, 12, 4 and 19 metabolites were identified in plasma, urine, liver and liver microsomes, respectively. However, no metabolites were identified in rat faeces. The relative content of each metabolite was estimated according to the intensity and peak area information of the MS/MS spectra. The LC-MS data of these detected metabolites were summarized in Table 2 and Fig. 5, and the extracted ion chromatogram (EIC) profiles of the metabolites in the different samples are illustrated in Fig. 6. PS: plasma SPE extraction; PM: plasma methanol precipitation; PA: plasma acetonitrile precipitation; U: urine SPE extraction; LS: liver SPE extraction; LM: Prototype drug liver microsomal treatment; detected: + (in vivo), # (in vitro); abundant: ++ (in vivo), ## (in vitro).
Peak
Ion Mode
tR/min
Formula
Theoretical
Mass m/zExperimental
Mass m/zRDB
Error
(ppm)MS/MS fragment ions
PS
PM
PA
U
L
LM
Relative content
M0
N
5.92
C27H32O14
579.17193
579.1701
12.5
−1.264
579.17(24.92),459.11(0.39),271.06(100.00),165.02(2.65),151.00(46.95),145.03(0.94),125.02(1.48),119.05(15.62),107.01(4.76),93.03(4.17)
√
√
√
√
√
√
++, ##
P
5.92
C27H32O14
581.18648
581.18591
11.5
−0.984
581.18(0.14),435.13(8.64),419.13(9.16),383.11(10.94),365.10(2.29),339.09(5.77),315.09(8.65),273.08(100.00),219.03(9.55),153.02(26.21),147.04(11.73)
√
√
√
√
√
√
M1
N
3.15
C25H27N3SO11
576.12935
576.13464
14.5
1.081
576.13(100.00),447.09(11.33),400.10(26.57),271.06(35.68),151.00(11.87),119.05(9.11),74.02(2.58),59.01(8.09)
√
+
M2
N
3.73
C9H8O3
163.04007
163.03856
6.5
−2.518
163.04(10.72),119.05(100.00)
√
++
M3
N
3.91
C21H18SO13
509.03953
509.0387
13.5
0.515
509.04(88.50),333.01(35.45),253.05(100.00),113.02(5.24),79.96(6.51)
√
+
M4
N
4.26
C21H18SO13
509.03953
509.03888
13.5
0.869
509.04(70.15),333.01(32.98),253.05(100.00),113.02(5.87),79.96(6.98)
√
+
M5
N
4.64
C21H18SO14
525.03445
525.03381
13.5
0.872
525.03(51.96),445.08(0.87),349.00(18.81),269.04(100.00),113.02(3.45),79.96(1.57)
√
+
M6
N
4.73
C21H18O10
429.08272
429.082
13.5
0.878
429.08(8.48),253.05(100.00),175.02(20.30),113.02(44.19)
√
√
√
+
M7
N
4.79
C27H28O17
623.12537
623.12463
14.5
0.569
623.12(29.33),447.09(100.00),271.06(63.91),175.02(15.05),151.00(27.95),119.05(6.65),113.02(53.95),93.03(2.86)
√
++
M8
N
4.98
C27H32O15
595.16684
595.16571
12.5
−0.061
595.17(55.57),459.11(1.16),287.06(30.79),151.00(5.25),135.04(2.87),125.02(100.00)
√
#
M9
N
5.09
C9H8O3
163.04007
163.03873
6.5
−1.476
163.04(10.38),119.05(100.00)
√
++
M10
N
5.34
C27H32O15
595.16684
595.16602
12.5
0.459
595.17(30.09),459.11(1.26),287.06(62.78),193.01(1.99),151.00(100.00),135.04(34.60),125.02(4.07)
√
##
P
5.35
C27H32O15
597.1814
597.18048
11.5
−1.535
435.13(8.05),399.10(10.55),331.08(8.65),289.07(100.00),219.03(10.68),195.03(24.71),153.02(25.86),129.05(20.59)
√
M11
N
5.40
C9H10O3
165.05572
165.05441
5.5
−1.276
165.05(35.83),147.04(100.00),121.06(17.99),119.05(33.73),93.03(4.53)
√
√
√
++
M12
N
5.45
C28H34O15
609.18249
609.18152
12.5
0.203
609.18(25.63),301.07(14.78),283.06(66.06),271.06(3.27),119.05(29.82),93.03(1.50)
√
#
M13
N
5.51
C21H20SO14
527.0501
527.04956
12.5
1.058
527.05(57.55),447.09(31.97),351.02(53.62),271.06(100.00),175.02(8.07),151.00(16.50),119.05(4.43),113.02(46.33)
√
+
M14
N
5.59
C21H20O10
431.09837
431.09763
12.5
0.828
431.10(100.00),255.06(43.96),175.02(24.69),149.02(36.27),113.02(66.42)
√
+
M15
N
5.63
C21H22SO12
497.07592
497.07568
11.5
1.724
497.08(40.71),321.04(100.00),241.09(23.22),175.02(2.80),135.04(2.90),121.03(12.76),119.05(3.92),113.02(17.38),79.96(8.68)
√
++
M16
P
5.70
C21H18O11
447.09219
447.09164
12.5
−1.225
447.09(9.04),271.06(100.00),153.02(1.38)
√
√
√
+
M17
N
5.74
C21H20SO14
527.0501
527.0495
12.5
0.944
527.05(41.57),447.09(22.43),351.02(61.38),271.06(100.00),175.02(8.35),151.00(36.49),119.05(6.89),113.02(44.47),93.03(3.02)
√
++
M18
N
5.83
C9H8O3
163.04007
163.03879
6.5
−1.108
163.04(26.90),119.05(100.00),93.03(1.39)
√
++
M19
P
5.88
C27H30O15
595.16575
595.16339
12.5
−3.959
595.16(1.87),287.05(100.00),165.02(7.84),129.05(6.04)
√
#
M20
N
5.88
C27H30O14
577.15628
577.15515
13.5
−0.055
577.15(14.10),269.04(100.00),151.00(5.80),119.05(2.08),93.03(0.68)
√
##
P
5.89
C27H30O14
579.17083
579.17004
12.5
−1.367
579.17(3.31), 433.11(27.34), 271.06(100.00), 153.02(0.65),129.05(1.34)
√
M21
N
5.90
C27H32O16
611.16176
611.1601
12.5
−0.918
611.16(100.00),489.12(9.34),303.05(78.48),285.04(78.12),269.04(40.10),151.00(82.42),125.02(58.54),119.05(22.81)
√
#
M22
N
5.91
C27H32O15
595.16684
595.16382
12.5
−3.237
595.17(63.48),287.06(100.00),151.00(73.86),135.04(22.98),125.02(12.72)
√
#
M23
N
6.01
C21H22O10
433.11402
433.11301
11.5
0.2
433.11(2.90),271.06(100.00),151.00(37.10),119.05(12.49),93.03(5.76)
√
#
M24
P
6.05
C22H22O12
479.1184
479.11768
11.5
−1.508
479.11(8.06),303.09(100.00),183.03(44.06),147.04(13.68),136.06(24.43)
√
#
M25
N
6.13
C21H20SO14
527.0501
527.0498
12.5
1.514
527.05(71.79),447.09(35.27),351.02(60.23),271.06(100.00),175.02(8.04),151.00(30.95),119.05(7.35),113.02(52.87)
√
+
M26
N
6.24
C21H22O10
433.11402
433.11319
11.5
0.616
433.11(4.76),271.06(100.00),151.00(32.20),119.05(12.93),93.03(4.14)
√
#
M27
N
6.26
C28H34O15
609.18249
609.1814
12.5
0.006
609.18(21.56),301.07(100.00),271.06(1.54),151.00(5.09),149.06(1.18),125.02(4.11)
√
#
M28
N
6.26
C21H22O9
417.11911
417.11862
11.5
1.466
417.12(71.42),241.09(27.61),175.02(35.26),135.04(13.45),121.03(37.58),119.05(20.25),113.02(100.00),93.03(3.63)
√
√
√
√
++
M29
N
6.27
C21H24SO13
515.08648
515.0838
10.5
−3.083
515.08(4.97),271.06(23.08),151.00(52.48),119.05(6.10),79.96(3.93)
√
√
√
++
M30
P
6.34
C15H12O5
273.07575
273.07535
9.5
−1.465
273.08(82.96),171.03(22.67),153.02(100.00),147.04(47.75),119.05(5.95)
√
√
√
++, #
N
6.35
C15H12O5
271.06065
271.06
10.5
−0.369
271.06(99.44),151.00(100.00),119.05(56.96),93.03(9.99)
√
√
√
M31
N
6.35
C21H20O11
447.09328
447.09244
12.5
0.564
447.09(93.15),271.06(100.00),175.02(52.06),151.00(72.00),119.05(23.58),113.02(61.28),93.03(4.61)
√
√
++, #
M32
P
6.46
C27H30O15
595.16575
595.16473
12.5
−1.708
595.16(1.36),577.16(3.62),435.13(2.36),273.07(100.00),171.03(16.80),153.02(35.66),147.04(10.78)
√
#
M33
N
6.46
C27H28O14
575.14063
575.13977
14.5
0.414
575.14(17.85),433.11(6.95),271.06(100.00),151.00(30.86),119.05(9.39),93.03(3.14)
√
#
M34
N
6.47
C9H8O3
163.04007
163.03859
6.5
−2.334
163.04(3.36),119.05(100.00)
√
++
M35
P
6.50
C27H30O14
579.17083
579.1698
12.5
−1.782
435.13(8.86),273.07(100.00),195.03(4.15),171.03(21.15),153.02(41.65)
√
#
M36
N
6.57
C21H24O9
419.13476
419.13397
10.5
0.743
419.13(100.00),243.10(36.31),175.02(9.64),123.04(6.62),121.03(2.99),119.05(3.35),113.02(40.21),93.03(3.35)
√
√
+
M37
N
6.84
C27H30O14
577.15628
577.15546
13.5
0.482
433.11(12.63),271.06(100.00),151.00(34.73),119.05(14.84),93.03(4.07)
√
#
P
6.84
C27H30O14
579.17083
579.16992
12.5
−1.575
579.17(1.49),417.12(2.76),273.07(100.00),171.03(21.48),153.02(43.29)
√
M38
N
7.03
C21H24O9
419.13476
419.13419
10.5
1.268
419.13(100.00),243.10(46.94),175.02(16.80),123.04(7.29),121.03(3.79),119.05(2.31),113.02(57.02),93.03(2.26)
√
√
+
M39
N
7.06
C15H14SO6
321.04383
321.0433
9.5
1.759
321.04(100.00),241.09(36.73),121.03(52.72),119.05(23.94),93.03(3.41),79.96(23.50)
√
√
√
++
M40
N
7.14
C16H14O6
301.07176
301.07104
10.5
1.247
301.07(100.00),271.06(10.68),181.05(4.07),119.05(48.24),93.03(20.67)
√
++
M41
N
7.40
C15H12SO8
351.01801
351.01727
10.5
1.013
351.02(30.68),271.06(100.00),151.00(37.10),119.05(10.93),93.03(5.37),79.96(7.72)
√
√
+
M42
P
7.83
C20H20O10
421.11292
421.10986
10.5
−7.274
421.11(79.21),403.25(2.22),245.05(100.00),121.03(6.56)
√
++
M43
N
8.49
C15H12O5
271.06065
271.06094
10.5
3.099
271.06(100.00),151.00(83.81),119.05(59.18),93.03(13.77)
√
√
√
++, ##
P
8.5
C15H12O5
273.07575
273.07541
9.5
−1.245
273.08(100.00),171.03(4.27),153.02(99.87),147.04(57.23),119.05(7.63)
√
√
√
M44
N
9.89
C9H10O3
165.05572
165.0544
5.5
−1.337
165.05(48.70),147.04(14.57),121.06(100.00),119.05(10.41),93.03(6.33)
√
√
++
M45
N
10.73
C9H10O3
165.05572
165.05435
5.5
−1.64
165.05(55.67),147.04(26.20),121.06(100.00),119.05(11.15),93.03(8.98)
√
√
++
Metabolic Pathway Profiling and Verification.
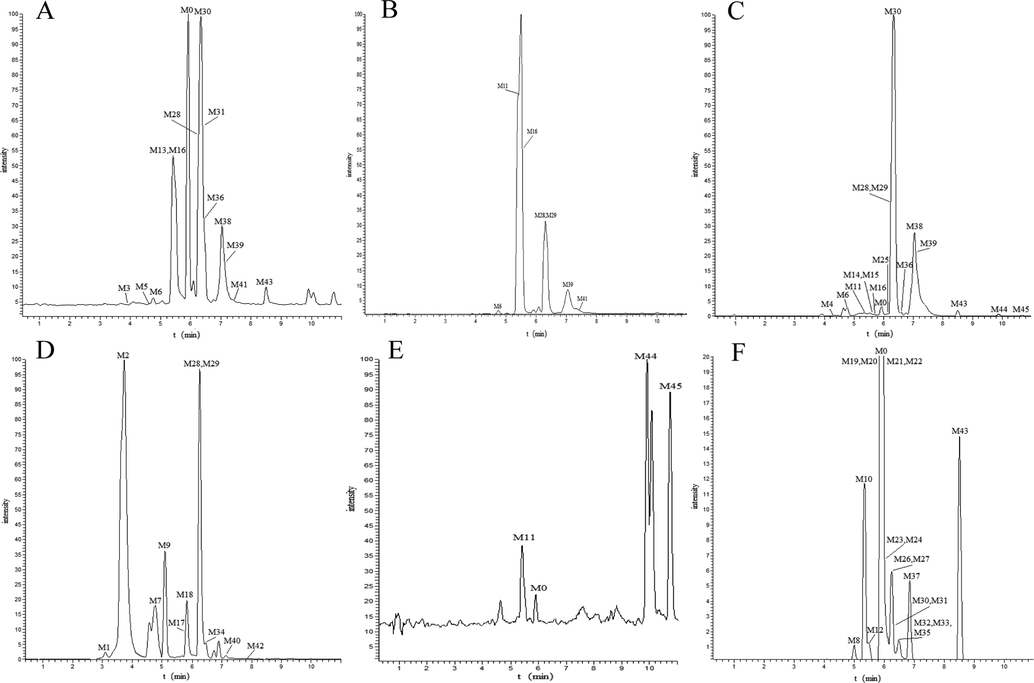
The EIC profiles of the metabolites in the different samples (A: PS, plasma SPE extraction; B: PM, plasma methanol precipitation; C: PA, plasma acetonitrile precipitation; D: U, urine SPE extraction; E: LS, liver SPE extraction; F: LM, Prototype drug liver microsomal treatment).
3.3.1 Identification of narirutin metabolites
The primary metabolite of naringin was mainly obtained from liver microsomes in vitro. Only the metabolites of narirutin deglycosylation and aglycone glucuronic acid were identified in vitro and in vivo, and there was an obvious difference of in vitro and in vivo metabolites. In addition to using the changes of CSIs, the metabolites were identified using DFICs at m/z 579, m/z 271, m/z 151, m/z 119 and m/z 93 in negative ion mode as well as at m/z 581, m/z 273, m/z 153, m/z 121 and m/z 95 in positive ion mode.
M30 and M43 were 308 Da less massive than narirutin, suggesting that they may be products of narirutin lacking the rutinoside group. Based on the DFICs, we found that M30 and M43 possessed the same theoretical [M + H]+/[M−H]- ions at m/z 273.07575 (C15H11O5, mass error ≤3 ppm) in the positive ion mode as well as at m/z 271.06065 (C15H13O5, mass error ≤3 ppm) in the negative ion mode. In the ESI-MS/MS spectra of M30 and M43, a series of diagnostic ions for naringin were observed, including m/z 151.00, m/z 119.05 and m/z 93.03 in the negative ion mode as well as m/z 153.02, m/z 121.06 and m/z 95.05 in the positive ion mode, which provided evidence that M30 and M43 were naringin and its isomer. Dihydroflavone and its corresponding chalcone were isomers of each other. Chalcone is a planar molecule, and its hydrophobicity is stronger than the corresponding dihydroflavone, resulting in a larger ClogP value. In reversed-phase separation mode, compounds with a higher ClogP value generally exhibit longer retention times. Thus, M30 was identified as naringenin, and M43 was identified as naringenin chalcone.
In the negative ion mode, the same base peak ions at m/z 433.11402 ([M−H−C6H12O4]-) and some other DFICs at m/z 151.00, m/z 119.05 and m/z 93.03 were present in the M23 and M26 metabolites. Glucose-binding sites were determined by comparing the retention time and ClogP value of glucose-binding hydroxyl groups at different positions of naringenin. The retention times of M23 and M26 were relatively similar and were between those of naringenin aglycone and narirutin. Therefore, M23 and M26 were deduced as 7-glucose-naringin and 4′-glucose-naringin, respectively.
Through the DFICs and the changes of CSIs, we speculated that some metabolites, such as M8, M10, M12, M19, M20, M21, M22, M27, M32, M33, M35 and M37, were obtained by oxidation, hydroxylation and other reactions. The oxidation reaction is a type of chemical reaction that easily occurs in the body via many mechanisms, including increased oxygen oxidation, increased dehydrogenation and increased coexistence of oxygen and dehydrogenation.
M8, M10 and M22 gave rise to the [M−H]- ion at m/z 595.16571, 595.16602 and 595.16382 (C27H33O15, error < 4 ppm), respectively. These metabolites were 16 Da more massive than narirutin, which indicated that an oxidation reaction may have occurred. At the same time, DFICs at m/z 287.06 ([M−C12H20O9 + O]-, m/z 271.06 + 15.99), m/z 151.00, m/z 135.04 (m/z 119.05 + 15.99) were observed, which suggested that the oxidation site occurred in the B ring or at C-3. M8, M10 and M22 were identified as single increased oxygen oxidation products.
M21 generated the [M−H]- ion at m/z 611.16010 (C27H31O15, error = -0.918 ppm) with a retention time of 5.90 min, and it was 32 Da more massive than narirutin, which suggested that M21 was a double increased oxygen oxidation product of narirutin. According to the DFICs at m/z 611.16 (m/z 579.17 + 31.99), m/z 303.05 (m/z 271.06 + 31.99) and m/z 151.00, the oxidation reaction occurred on aglycone and not on the A ring. Based on the unique cracking law (Yang et al., 2015) of flavonoid compounds, the CSIs at m/z 285.04 ([M−H−C6H12O4−H2O]-) and m/z 269.04 ([M−H−C6H12O4−H2O−O]-) were compared. Thus, M21 was deduced as 3, 6′-OH-narirutin.
M20 generated its [M−H]- ion at m/z 577.15515 with a mass error of −0.055 ppm and its [M + H]+ ion at 579.17004 with a mass error of −1.367 ppm. At the same time, DFICs at m/z 151.00 (-), m/z 93.03 (-) and m/z 153.02 (+) were observed. The CSIs at m/z 269.04 in the negative ion mode and m/z 271.06 in the positive ion mode were 2 Da smaller than narirutin, which indicated that dehydrogenation occurred on aglycones. M20 was tentatively inferred as 7-rutinoside-apigenin. However, due to the lack of sufficient fragment ion information, we were unable to confirm that its aglycone was apigenin, indicating that the specific structure of M20 requires further experimental investigation. M37 was 2 Da less massive than narirutin. However, the DFICs at m/z 271.06, m/z 151.00 and m/z 119.05 in the negative ion mode as well as m/z 273.07 and m/z 153.02 in the positive ion mode were the same as those of narirutin, which indicated that the dehydrogenation reaction occurred at the rutinoside group. Thus, M20 and M37 were metabolites produced by the dehydrogenation of rutinoside groups.
M19 and M32 with the same theoretical [M + H]+ ions at m/z 595.16575 (C27H30O15, mass error within 4 ppm) were observed at 5.88 min and 6.46 min, respectively. In the ESI-MS/MS spectrum of M19, the CSIs at m/z 287.05 (m/z 273.07 + 13.98) could indicate that the oxidization has occurred in the aglycone group. The DFICs of M32 such as those at m/z 435.13 ([M + H-C6H12O4]+), m/z 273.07 ([M + H-C27H30O14]+), m/z 153.02 and 147.04 provided evidences to help us determined where the oxidation occurred in the rutinose site. Thus, M19 and M32 were deduced as the dehydrogenation and oxygenation metabolites of narirutin.
M12 and M27 possessed identical [M−H]- ions at m/z 609.18249 (C28H34O15, error < 4 ppm) with retention times of 5.45 min and 6.26 min, respectively. Presumably they were from the oxidation of narirutin. They were 30 Da more massive than Narirutin. Presumably, they were hydroxylation and methylation products of narirutin. In the ESI-MS/MS spectrum, M12 showed its DFICs at m/z 271.06 ([M−H−C6H12O4]-), m/z 119 and m/z 93.03 and CSIs at m/z 301.07 ([M−H−C6H12O4 + OCH2]-), m/z 283.06 ([M−H−C6H12O4 + OCH2-H2O]-), It was proved that the hydroxylation and methylation reaction took place in the A ring. The DFICs and CSIs at m/z 301.07 ([M−H−C6H12O4]-), m/z 271.06 ([M−H−C6H12O4−OCH2]-), m/z 151.00 and m/z 149.06 (m/z 119.05 + 30.01) could be determined that the hydroxylation and methylation reaction of M27 occurred in the B ring. Thus, M12 and M27 were tentatively identified as the hydroxylation and methylation products of Narirutin.
3.3.2 Identification of naringenin and naringenin Chalcone’s metabolites
M1 was eluted at 3.15 min and showed its [M−H]- ion at m/z 576.13464 (C25H27O11N3S, error = 1.081). It was 305 Da more than M30 and M43, indicating that one of them were glutathione (GSH) binding product. In the ESI-MS/MS spectrum, the DFICs at m/z 271.06, m/z 151.00 and m/z 119.05 from narirutin and at m/z 447.09 ([M−H−C5H7O3N]-), m/z 400.10 ([M−H−C5H7O3N−CH4S]-), m/z 74.02 ([C2H4NO2]-), m/z 59.01 ([C2H3O2]-) from Glutathione group provided evidence that M1 was glutathionated metabolites.
In the negative ion mode, metabolite M31 possessed [M−H]- ion at m/z 447.09328 (C21H20O11, error = 0.564) with the retention time of 6.35 min. It was 176 Da more than naringenin. The DFICs at m/z 271.06 ([M−H−GluA]-), m/z 175.02 ([GluA-H]-), m/z 151.00, m/z 119.05, m/z 113.02 ([GluA-H-CH2O3]-) and m/z 93.03, suggesting the present of glucuronidation. By comparing the ClogP value and retention time of metabolites M30 and M43, it can be determined that M31 is the glucoaldehyde acid product of naringenin chalcone (M43).
M40 (C16H14O6, error = 1.247) was 30 Da more massive than M30 and M43 and eluted at 7.14 min. The DFICs such as m/z 301.07 ([M−H]-), m/z 271.06 ([M−H−OCH3]-) and m/z 119.05 and the CSIs at m/z 181.05 (m/z 151.00 + 30.05) suggested that M40 was identified as the methoxylation product of M30 and M43 and the OCH2 group may be introduced into the A ring. According to the retention time and ClogP value, M40 was tentatively deduced as the methoxylation product of naringenin (M30).
M41 generated the ion at m/z 351.01727 (C15H12SO8, error = 1.013) with the retention time of 7.40 min. It was 80 Da more massive than M30 and M43, indicating that it might be the sulfated product of them. The DFICs such as m/z 271.06 ([M−H−SO3]-), m/z 151.00, 119.05, m/z 93.03 and m/z 79.96 ([SO3]-) could prove that M41 was the sulfated product of M30 and M43. The introduction of sulfate group will increase the polarity of the prototype compound. The final identification will be based on the comparison between the polarity and ClogP value of the compound. Therefore, M41 was identified as the sulfated product of naringenin chalcone (M43).
3.3.3 Identification of naringenin glucuronidation metabolites
M7 was eluted at 4.79 min and was 176 Da more massive than M31. The deprotonation ion at m/z 623.12537 (C27H28O17, error = 0.569) was generated. In addition, the DFICs involving m/z 447.09 ([M−H−GluA]-), m/z 271.06 ([M−H−2GluA]-), m/z 175.02 ([GluA-H]-), m/z 151.00, m/z 119.05, m/z 113.02 ([GluA-H-CH2O3]-) and m/z 93.03 were identical to those of M31. Thus, M7 was identified as the glucuronidation product of M31 [double glucuronidation product of naringenin chalcone (M43)].
M13, M17 and M25 were 80 Da more than M31. They were presumed to be sulfated products of M31. They showed the theoretical [M−H]- ion at 527.05010 (C21H20O14S, error < 4). The DFICs at m/z 271.06 ([M−H−SO3−GluA]-), m/z 175.02, m/z 151.00, m/z 119.05, m/z 113.02 and the CSIs at m/z 447.09 ([M−H−SO3]-), m/z 351.02 ([M−H−GluA]-) indicated that glucuronic acid and sulfate were combined at different locations of M31, respectively. It could be inferred that M13, M17 and M25 were products of glucuronidation and sulfate of M31.
Reduction reactions also occur in living organisms. M28 generated the [M−H]- ion at m/z 417.11911 (C21H22O9, error = 1.466) with the retention time of 6.26 min. The DFICs such as m/z 175.02, m/z 113.02, m/z 119.05 and m/z 93.03, could infer that the reduction reaction did not happen in the glucuronic group and the B ring. But, according to the CSIs at m/z 241.09, m/z 135.04 and m/z 121.03, we could finally deduce that M28 was the hydrogenation and double-deoxidized product of M31 and the reduction reaction occurred in the A ring. M36 and M38 with theoretical [M−H]- ions at 419.134 (C21H24O9, mass error within 4 ppm) were observed at 6.57 min and 7.03 min, respectively. Their reaction site and the DFICs were similar to that of M28 and one step more hydrogenation reduction reaction occurred than that of M28. Finally, they were identified as the double deoxygenation and hydrogenation metabolites of M31. In a similar way, M14 eluted at 5.59 min and gave rise to a [M−H]- ion at m/z 431.09219 (C21H20O10, error = 0.828). It was 16 Da less massive than M31. In the ESI-MS/MS spectrum, the DFICs such as these at m/z 255.06 ([M−H−GluA−O]-), m/z 175.02 ([GluA-H]-) and m/z 113.02 ([GluA-H-CH2O3]-), supported our to identify it as the deoxygenation product of M31.
M16 with the [M + H]+ ion at m/z 447.09219 (C21H18O11, the mass error −1.225 ppm) was observed at 5.70 min. It was 2 Da less massive than M31. The DFICs and CSIs at m/z 271.06 and m/z 153.02, could support us to deduce that M16 was the dehydrogenation product of M31.
M6 was 18 Da less massive than M31. It eluted at 4.73 min and its formula was speculated as C21H18O10, with the mass error of 0.878. According to the DFICs at m/z 429.08 ([M−H]-), m/z 253.05 (m/z 271.06–18.01), m/z 175.02 and m/z 113.02, indicating that M6 was the dehydration product of M31.
M42 with the ion at m/z 421.11292 (C20H20O10, error = -1.274) was observed at 7.83 min. It was 28 less massive than M31 and presumed to be a metabolite formed by M31 took off the carbonyl group. The DFICs at m/z 245.05 and m/z 121.03 further indicated that M42 was the decarbonyl product of M31.
M24 was eluted at 6.05 min and was 30 Da more massive than M31. It showed its [M + H]+ ion at m/z 479.11840 (C22H22O12, error = -1.508). In its ESI-MS/MS spectrum, the DFICs and CSIs at m/z 303.09 ([M + H + OCH2]+), 183.03 (m/z 153.02 + 30.01), m/z 147.04 and m/z 136.06, could indicate that OCH2 groups were introduced into the A ring. M24 was finally identified as the methoxylation product of M31.
3.3.4 Identification of naringenin sulfate metabolites
M39 generated the ion at m/z 321.04330 (C15H14SO6, error = 1.759) with the retention time of 7.06 min. It was 30 Da less massive than M41. Because the M39 did not have a –OCH3 group. The DFICs such as m/z 241.09 (m/z 271.06–29.97), m/z 121.03 (m/z 151.00–29.97), m/z 119.05 and m/z 93.03 were deduced that M39 was the hydrogenation and double-deoxidized metabolite of M41 and the reaction happened in the A ring.
3.3.5 Identification of naringenin glucuronidation and sulfate metabolites
M3 and M4 possessed the same theoretical ion at m/z 509.03953 (C21H18SO13, error < 4) in the negative ion mode. These metabolites were 18 Da less massive than M13, M17 and M25, indicating that they might be the dehydration product. The dehydration reaction occurred on the basis of glucuronidation and sulfate according to the DFICs at m/z 333.01 ([M−H−GluA]-), m/z 253.05 ([M−H−GluA−SO3]-), m/z 113.02 ([GluA-H-CH2O3]-) and m/z 79.96 ([SO3]-). Binding the CSIs at m/z 253.05 (m/z 271.06–18.01), M3 and M4 were tentatively identified as the dehydration product of M13, M17 and M25.
M5 eluted at 4.64 min and was 2 Da less than M13, M17 and M25. Combined with the DFICs at m/z 445.08 ([M−H−SO3]-), m/z 349.00 ([M−H−GluA]-) and m/z 269.04 ([M−H−GluA−SO3]-), M5 was finally deduced as the dehydrogenation product of M13, M17 and M25.
M15 generated the ion at m/z 497.07592 (C21H22O12S, error = 1.759) with the retention time of 5.63 min. It was 30 Da less massive than M13, M17 and M25. The DFICs such as these m/z 321.04 ([M−H−GluA]-), m/z 241.09 ([M−H−GluA−SO3]-), m/z 175.02, m/z 119.05, m/z 113.02 and m/z 79.96 provided evidence to infer that M15 was the metabolites of M13, M17 and M25. And according to the CSIs at m/z 241.09 (m/z 271.06–29.97), m/z 135.04 and m/z 121.03 (m/z 151.00–29.97), it was proved that the reduction reaction occurred in the A ring. Finally, M15 was identified as the hydrogenation and double-deoxidized metabolite of M13, M17 and M25.
M29 eluted at 6.27 min and showed its [M−H]- ion at m/z 515.08 (C21H24SO13, error = -3.083). Observations of its DFICs at m/z 271.06, m/z 151.00, m/z 119.05 and m/z 79.96 were found that reduction reaction did not occur on aglycone and Sulfate group. Therefore, it was preliminarily speculated that the reduction reaction took place at the glucuronidation group. Thus, M29 was characterized as the deoxygenation and double-hydrogenation product of M13, M17 and M25.
3.3.6 Identification of naringenin other metabolites
According to the data, naringin can generate aglyen naringin under the action of intestinal flora. And naringenin can generate p-hydroxyphenylpropionic acid, p-hydroxycinnamic acid and their isomers through ring-opening reaction under the microorganism (Liu et al., 2012; Zeng et al., 2020a, 2020b). It is speculated that narirutin can also have similar reaction. M11, M44 and M45 were possessed the same theoretical [M−H]- ions at m/z 165.05572 (C9H10O3, error < 4 ppm) in the negative ion mode, respectively. The fragment ions such as m/z 147.04 ([M−H−H2O]-), m/z 121.06 ([M−H−CO2]-) and 119.05 were exhibited in the ESI-MS/MS spectra. Hence, M11, M44 and M45 were identified as p-Hydroxyphenylpropionic acid and their isomers. M2, M9, M18 and M34 were 2 Da less massive than M11, M44 and M45. Therefore, it was speculated that they were metabolites formed by dehydrogenation of M11, M44 and M45 in the liver. Identify them according to the fragment ions at m/z 163.04007 ([M−H]-), m/z 119.05 and m/z 93.03, M2, M9, M18 and M34 were eventually identified as p-hydroxycinnamic acid and their isomers.
4 Discussion
In previous literatures, we found that many researches primarily focus on pharmacokinetics of narirutin. Bioavailability and route of excretion were studied in healthy humans after taking fruit juice (Brett et al., 2009; Silveira et al., 2014; Aschoff et al., 2016) or rats oral drugs containing narirutin (Zhang et al., 2018; Li et al., 2019; Fu et al., 2020). Zhang et al. studied after human consumption of grapefruit juice (the main flavonoids were naringin and narirutin), screened flavonoid metabolites from urine, and found that 9 metabolites were naringenin glucuronides, naringenin sulfates, naringenin glucuronide sulfates and naringenin diglucuronide and their isomers (Zhang and Brodbelt 2004). The above studies indicated that it was not enough for studying the metabolites of narirutin.
In the present study, the in vitro and in vivo metabolism of narirutin was studied in SD rats, including in rat plasma (three different treatments (Jordan et al., 2009)), urine (Nema et al., 2010), faeces, liver tissues and liver microsomes (Knights et al., 2016), to obtain as many narirutin metabolites as possible. The relevant information of each metabolite was captured in positive and negative ion modes by UPLC-Q-Exactive Orbitrap MS/MS after oral administration of narirutin. Combined with the chromatographic retention behavior, accurate molecular weight, positive and negative ion cleavage modes and the cleavage rules of the reference product, a total of 46 metabolites (including prototype drugs) were identified. A total of 22 metabolites were identified in rat plasma, including 16, 13 and 8 metabolites identified from solid phase extraction (SPE), acetonitrile treatment and methanol treatment, respectively. In addition, the metabolites treated with methanol were found in the products of the other two sample treatments, which indicated that the results of plasma samples treated with methanol were not ideal. Therefore, the combination of SPE and acetonitrile treatment may provide a more comprehensive characterization of plasma metabolites.
Based on the above results, we found that the metabolic pathways of narirutin in vitro and in vivo were different. Only three metabolites (M30, M31 and M43) in vivo had similar metabolic pathways in vitro. Narirutin was decomposed to its aglycones, namely, naringenin and naringenin chalcone, in vivo (M30 and M43, the relative content was abundant). Naringenin and naringenin chalcone produced a series of metabolites, such as glucuronidation (M31), sulfation (M41) and glutathione binding (M1). On the basis of these metabolites, further oxidation, reduction, dehydration, decarbonylation and other reactions occurred. Another pathway occurred, in which naringenin and naringenin chalcone were cleaved by the microbiota-mediated ring fission of unabsorbed flavonoids to produce p-hydroxyphenylpropionic acid, p-hydroxycinnamic acid and their isomers, as well as the metabolites excreted by enterohepatic circulation (Zeng et al., 2020a, 2020b). This was precisely why we found them only in metabolites in vivo, but not in the metabolism of liver microsomes, because of the lack of the ring-opening effect from intestine microorganism. Therefore, we found p-hydroxyphenylpropionic acid and its isomers (M11, M44 and M45) in plasma and liver tissues, and we found p-hydroxycinnamic acid and its isomers (M2, M9, M18 and M34) in urine. Therefore, these findings suggested that p-hydroxyphenylpropionic acid and its isomers were further transformed to p-hydroxycinnamic acid and its isomers, which were excreted from the body. In addition, the relative contents of these compounds in the body were high, suggesting that this metabolic pathway is one of the main metabolic pathways of narirutin in vivo. Regarding the in vitro metabolites, most of the metabolites in liver microsomes were based on the metabolic reactions of narirutin, including oxidation, reduction, hydroxylation and methylation reactions. In liver microsomes, we found that the M23 and M26 metabolites were compounds on naringenin that could bind only one glucose. Therefore, we speculated that there may be two generation paths as follows: one was formed by the removal of rhamnose from narirutin, and the other was that narirutin was broken down into naringenin, which was then combined with glucose. In the in vitro and in vivo metabolic pathways, a prototype drug was found. The relative content of narirutin was the highest in plasma followed by urine, and the relative content of narirutin in liver microsomes was also relatively abundant. Therefore, the prototype drug was metabolized and eliminated from the body, which was also one of the main metabolic forms of narirutin. Finally, we concluded that the main difference between in vitro and in vivo metabolism of narirutin lies in the time and degree of its metabolism. In vivo experiments, after three days of intragastric administration, a large number of drugs entered the body of rats, and metabolic reactions fully occurred. Therefore, we not only found primary metabolites of narirutin, but also secondary and tertiary metabolites were also observed. Meanwhile, under the action of intestinal flora, the ring-opening reaction of nariutin and its metabolites were promoted, which enriched the metabolic spectrum of narirutin in vivo. However, in vitro, the liver microsomes experiment was administered only once and conducted for 240 min, which did not allow sufficient time for deeper reactions. Therefore, most of the reactions were based on the primary metabolites of narirutin, and few secondary metabolites were produced.
In the present study, an analysis strategy based on ACR, DFICs and CSIs was proposed to rapidly identify narirutin metabolites. This strategy can be applied not only to the identification of narirutin metabolites in the present study but also to the identification of other flavonoid metabolites, even saponins and alkaloids. For these compounds with a basic core structure, this strategy can be used to quickly identify metabolites from LC-MS information. The fragmentation law of the core structure of the compound is summarized to obtain the corresponding fragment ions. According to the functional groups bound to the compound, the fragmentation ion clusters of the compound are deduced, which are called DFICs. The accuracy of DFICs are tested by the cracking information of the reference substance. Accurate DFICs information facilitates the rapid identification of metabolites. According to the structural information and metabolic characteristics of compounds in vivo, the possible main metabolites are deduced. Based on this node, the possible metabolites are deduced by the recurrence formula. The metabolites are identified by combining the DFICs and secondary fragment information. Further, suggestive fragment ions, called CSIs, are identified from the DFICs to preliminarily infer the sites where metabolic reactions occur, and the metabolites are then identified.
Several major metabolites produced during metabolism may be part of the related pharmacological effects of narirutin. Narirutin and the metabolized forms of naringenin, including naringenin chalcone and naringenin chalcone glucuronidation, were detected in vivo and in vitro. The relative content of these compounds in the metabolites in vivo were abundant, indicating that they are the key metabolites in the metabolic process of narirutin. Recent studies have reported that naringenin has a variety of pharmacological effects similar to narirutin, such as anti-inflammatory, antiviral (Tutunchi et al., 2020) and antidiabetic (Den Hartogh and Tsiani 2019) effects. Thus, narirutin possesses pharmacological effects, in part, due to its metabolism in the body to produce naringenin. Naringenin also has potential immunomodulatory effects (Zeng et al., 2018) and regulatory effects in nonalcoholic fatty liver disease (Wang et al., 2020; Hua et al., 2021). Escribano-ferrer et al. reported that naringenin chalcone may be a useful alternative for topical treatment of inflammatory and allergic skin diseases. The present study linked the metabolism and pharmacological effects of narirutin in vivo, suggesting that studies on narirutin are warranted (Escribano-Ferrer et al., 2019). Citrus peel extract has a positive effect on fat production and breakdown (Chou et al., 2018), and one of the main components of citrus peel extract is narirutin. Horiba et al. reported that narirutin improves adipocyte metabolic functions and exerts insulin-sensitizing effects by activating an adiponectin-related pathway. Therefore, these findings suggest that naringenin chalcone may be the key metabolite of narirutin in improving adipocyte function (Horiba et al., 2010). Kolot et al. reported that naringenin chalcone is bioavailable in humans from a dietary source but that its availability is poor. The intramolecular cyclisation and extended metabolism of naringenin chalcone likely contribute to the inactivation of the α,β-unsaturated reactive center and the excretion of the biologically active molecule, respectively, which may explain the paucity of studies on naringenin chalcone (Kolot et al., 2020).
5 Conclusion
The present study comprehensively characterized the metabolites of narirutin in vitro and in vivo, which provided a basis to study the pharmacological activity mechanism of narirutin. In addition, the present study proposed a new analysis strategy for the rapid identification of metabolites, providing support for the study of compound metabolism.
The annotations of some abbreviations:
ACR: analogous core recursion, DFICs: diagnostic fragment ion clusters, CSIs: characteristic suggestive ions, EIC: Extracted Ion Chromatography, SPE: Solid Phase Extraction, UHPLC: ultra-high-performance Liquid Chromatography, MS: Mass Spectrum, GluA: glucuronyl, Glc: glucosyl.
CRediT authorship contribution statement
Shuyi Song: Methodology, Writing – original draft. Hongyan Zhou: Methodology. Xianming Lan: Methodology. Xiaoqing Yuan: Methodology. Yanan Li: . Shuteng Huang: . Zhibin Wang: Conceptualization, Funding acquisition, Project administration, Methodology, Supervision. Jiayu Zhang: Conceptualization, Funding acquisition, Project administration, Methodology, Supervision.
Acknowledgments
This work has been financially supported by Shandong Taishan Scholars Young Expert Project (tsqn202103110), Shandong Province Youth Talents Introducing and Cultivating Program-Innovative Research Team (10073004), Supporting plan for leading talents above provincial level in Yantai (10073801).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Urinary excretion of Citrus flavanones and their major catabolites after consumption of fresh oranges and pasteurized orange juice: A randomized cross-over study. Mol. Nutr. Food Res.. 2016;60:2602-2610.
- [CrossRef] [Google Scholar]
- Absorption, metabolism and excretion of flavanones from single portions of orange fruit and juice and effects of anthropometric variables and contraceptive pill use on flavanone excretion. Br. J. Nutr.. 2009;101:664-675.
- [CrossRef] [Google Scholar]
- A Standardized Extract Prepared from Red Orange and Lemon Wastes Blocks High-Fat Diet-Induced Hyperglycemia and Hyperlipidemia in Mice. Molecules (Basel, Switzerland).. 2021;26
- [CrossRef] [Google Scholar]
- Immature Citrus reticulata Extract Promotes Browning of Beige Adipocytes in High-Fat Diet-Induced C57BL/6 Mice. J. Agric. Food Chem.. 2018;66:9697-9703.
- [CrossRef] [Google Scholar]
- Antidiabetic Properties of Naringenin: A Citrus Fruit Polyphenol. Biomolecules. 2019;9
- [CrossRef] [Google Scholar]
- Discrimination of Citrus reticulata Blanco and Citrus reticulata 'Chachi' by gas chromatograph-mass spectrometry based metabolomics approach. Food Chem.. 2016;212:123-127.
- [CrossRef] [Google Scholar]
- In Vivo Anti-inflammatory and Antiallergic Activity of Pure Naringenin, Naringenin Chalcone, and Quercetin in Mice. J. Nat. Prod.. 2019;82:177-182.
- [CrossRef] [Google Scholar]
- Blood pressure and body fat % reduction is mainly related to flavanone phase II conjugates and minor extension by phenolic acid after long-term intake of orange juice. Food Funct.. 2021;12:11278-11289.
- [CrossRef] [Google Scholar]
- Development of a sensitive and rapid UHPLC-MS/MS method for simultaneous quantification of nine compounds in rat plasma and application in a comparative pharmacokinetic study after oral administration of Xuefu Zhuyu Decoction and nimodipine. Biomedical chromatography : BMC.. 2020;34:e4872.
- [Google Scholar]
- Narirutin fraction from citrus peels attenuates LPS-stimulated inflammatory response through inhibition of NF-κB and MAPKs activation. Food Chem. Toxicol.. 2012;50:3498-3504.
- [CrossRef] [Google Scholar]
- Naringenin chalcone improves adipocyte functions by enhancing adiponectin production. Mol. Cell. Endocrinol.. 2010;323:208-214.
- [CrossRef] [Google Scholar]
- Naringenin alleviates nonalcoholic steatohepatitis in middle-aged Apoe(-/-)mice: role of SIRT1. Phytomedicine : international journal of phytotherapy and phytopharmacology.. 2021;81:153412
- [CrossRef] [Google Scholar]
- A comprehensive profiling and identification of liquiritin metabolites in rats using ultra-high-performance liquid chromatography coupled with linear ion trap-orbitrap mass spectrometer. Xenobiotica; the fate of foreign compounds in biological systems.. 2021;51:564-581.
- [CrossRef] [Google Scholar]
- Selection of SPE cartridge for automated solid-phase extraction of pesticides from water followed by liquid chromatography-tandem mass spectrometry. Anal. Bioanal. Chem.. 2009;394:2257-2266.
- [CrossRef] [Google Scholar]
- In Vitro Drug Metabolism Using Liver Microsomes. Curr. Protocols Pharmacol.. 2016;74:7.8.1-7.8.24.
- [CrossRef] [Google Scholar]
- Bioavailability of naringenin chalcone in humans after ingestion of cherry tomatoes. International journal for vitamin and nutrition research. Internationale Zeitschrift fur Vitamin- und Ernahrungsforschung. Journal international de vitaminologie et de nutrition.. 2020;90:411-416.
- [CrossRef] [Google Scholar]
- Drug metabolite cluster centers-based strategy for comprehensive profiling of Neomangiferin metabolites in vivo and in vitro and network pharmacology study on anti-inflammatory mechanism. Arab. J. Chem.. 2022;15
- [CrossRef] [Google Scholar]
- Simultaneous Quantification of Five Flavanone Glycosides in Rat Plasma by Ultra-Performance Liquid Chromatography-Tandem Mass Spectrometry: Application to a Comparative Pharmacokinetic Study of Aurantii Fructus Immaturus and Aurantii Fructus Extracts. J. AOAC Int.. 2019;102:781-787.
- [CrossRef] [Google Scholar]
- Bioconversion of Citrus unshiu peel extracts with cytolase suppresses adipogenic activity in 3T3-L1 cells. Nutr. Res. Pract.. 2015;9:599-605.
- [CrossRef] [Google Scholar]
- Metabolism and excretion studies of oral administered naringin, a putative antitussive, in rats and dogs. Biopharm. Drug Dispos.. 2012;33:123-134.
- [CrossRef] [Google Scholar]
- Effects of Citrus Fruit Juices and Their Bioactive Components on Inflammation and Immunity: A Narrative Review. Front. Immunol.. 2021;12:712608
- [CrossRef] [Google Scholar]
- Naringin as Sustained Delivery Nanoparticles Ameliorates the Anti-inflammatory Activity in a Freund's Complete Adjuvant-Induced Arthritis Model. ACS Omega. 2021;6:28630-28641.
- [CrossRef] [Google Scholar]
- Application of silica-based monolith as solid phase extraction cartridge for extracting polar compounds from urine. Talanta. 2010;82:488-494.
- [CrossRef] [Google Scholar]
- Inhibitory activity of narirutin on RBL-2H3 cells degranulation. Immunopharmacol. Immunotoxicol.. 2021;43:68-76.
- [CrossRef] [Google Scholar]
- Narirutin fraction from citrus peels attenuates alcoholic liver disease in mice. Food Chem. Toxicol.. 2013;55:637-644.
- [CrossRef] [Google Scholar]
- Citrus Flavanone Narirutin, In Vitro and In Silico Mechanistic Antidiabetic Potential. Pharmaceutics.. 2021;13
- [CrossRef] [Google Scholar]
- Structure-based Discovery of Narirutin as a Shikimate kinase Inhibitor with Anti-tubercular Potency. Curr. Comput. Aided Drug Des.. 2020;16:523-529.
- [CrossRef] [Google Scholar]
- Rapid profiling and identification of puerarin metabolites in rat urine and plasma after oral administration by UHPLC-LTQ-Orbitrap mass spectrometer. Journal of chromatography. B, Analytical technologies in the biomedical and life sciences.. 2017;1068–1069:180-192.
- [CrossRef] [Google Scholar]
- Antioxidant and antimicrobial activities and UPLC-ESI-MS/MS polyphenolic profile of sweet orange peel extracts. Current research in food science.. 2021;4:326-335.
- [CrossRef] [Google Scholar]
- Pharmacokinetics of flavanone glycosides after ingestion of single doses of fresh-squeezed orange juice versus commercially processed orange juice in healthy humans. J. Agric. Food Chem.. 2014;62:12576-12584.
- [CrossRef] [Google Scholar]
- Naringenin, a flavanone with antiviral and anti-inflammatory effects: A promising treatment strategy against COVID-19. Phytotherapy research : PTR.. 2020;34:3137-3147.
- [CrossRef] [Google Scholar]
- Naringenin attenuates non-alcoholic fatty liver disease by down-regulating the NLRP3/NF-κB pathway in mice. Br. J. Pharmacol.. 2020;177:1806-1821.
- [CrossRef] [Google Scholar]
- Relaxation effect of narirutin on rat mesenteric arteries via nitric oxide release and activation of voltage-gated potassium channels. Eur. J. Pharmacol.. 2021;905:174190
- [CrossRef] [Google Scholar]
- Preventive Effect of Gonggan (Citrus Reticulata Blanco Var. Gonggan) Peel Extract on Ethanol/HCl-Induced Gastric Injury in Mice via an Anti-oxidative Mechanism. Front. Pharmacol.. 2021;12:715306
- [CrossRef] [Google Scholar]
- Metabolite Identification in the Preclinical and Clinical Phase of Drug Development. Curr. Drug Metab.. 2021;22:838-857.
- [CrossRef] [Google Scholar]
- Chemical composition and pharmacological mechanism of Qingfei Paidu Decoction and Ma Xing Shi Gan Decoction against Coronavirus Disease 2019 (COVID-19): In silico and experimental study. Pharmacol. Res.. 2020;157:104820
- [CrossRef] [Google Scholar]
- Elucidation of the fragmentation pathways of a complex 3,7-O-glycosyl flavonol by CID, HCD, and PQD on an LTQ-Orbitrap Velos Pro hybrid mass spectrometer. Chin. J. Nat. Med.. 2015;13:867-872.
- [CrossRef] [Google Scholar]
- Naringenin as a potential immunomodulator in therapeutics. Pharmacol. Res.. 2018;135:122-126.
- [CrossRef] [Google Scholar]
- Metabolite Profiling of Naringin in Rat Urine and Feces Using Stable Isotope-Labeling-Based Liquid Chromatography-Mass Spectrometry. J. Agric. Food Chem.. 2020;68:409-417.
- [CrossRef] [Google Scholar]
- Tissue distribution of naringin and derived metabolites in rats after a single oral administration. J. Chromatogr. B, Anal. Technol. Biomed. Life Sci.. 2020;1136
- [CrossRef] [Google Scholar]
- Screening flavonoid metabolites of naringin and narirutin in urine after human consumption of grapefruit juice by LC-MS and LC-MS/MS. Analyst. 2004;129:1227-1233.
- [CrossRef] [Google Scholar]
- Pharmacokinetic Study of 7 Compounds Following Oral Administration of Fructus Aurantii to Depressive Rats. Front. Pharmacol.. 2018;9:131.
- [CrossRef] [Google Scholar]







