Translate this page into:
Deciphering the mechanism of Ephedra Herba-Armeniacae Semen Amarum herb pairs on COVID-19 by integrated network pharmacology and bioinformatics
⁎Corresponding author. wangzhuoxi0919@163.com (Rui Qie)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background
Effective prevention of COVID-19 is vital for impeding the current pandemic. Our study is devoted to investigating the targets of Ephedra Herba (EH)-Armeniacae Semen Amarum (ASA) herb pairs on COVID-19 and identifying the signature genes.
Methods
Firstly, the active constituents and corresponding targets of EH-ASA were separately obtained through Traditional Chinese Medicine System Pharmacology Database (TCMSP) and UniProt KB database. We searched the Gene Expression Omnibus (GEO) database to identify the differentially expressed genes (DEGs) for COVID-19 and intersected the targets with DEGs to retrieve common genes. Subsequently, the Protein-Protein Interaction (PPI) network and weighted gene coexpression network analysis (WCGNA) were carried out to discover the genes related to COVID-19, which was combined with DEGs performed the GO and KEGG enrichment analysis to investigate the pathogenesis of COVID-19. The receiver operating characteristic curve (ROC) and molecular docking were ultimately exerted to verify the precision and stability of the screened genes.
Results
There were 42 active compounds of the EH-ASA corresponding to 200 targets, including Luteolin, Glycyrol, Glabridin, and Quercetin. Also, 548 DEGs were filtered out, and 74 common genes were obtained after the intersection. In addition, 358 genes associated with COVID-19 were identified through the WGCNA method, mainly involved in Human T-cell leukemia virus one infection and IL-17 signaling pathway, etc. At last, three signature genes were obtained, containing ESR1, IL2, and NFKBIA, and the ROC of these three genes was 1.000, 0.967, and 0.967, respectively, which illuminated the accuracy of the results. Also, molecular docking indicated that the main chemical constituents of the EH-ASA had a good affinity with COVID-19.
Conclusion
EH-ASA herb pairs may play a preventive and therapeutic role in COVID-19 through the three characteristic genes, ESR1, IL2, and NFKBIA.
Keywords
Ephedra Herba
Armeniacae Semen Amarum
Network pharmacology
Bioinformatics
COVID-19
Abbreviations
- EH
-
Ephedra Herba
- ASA
-
Armeniacae Semen Amarum
- TCMSP
-
Traditional Chinese Medicine System Pharmacology Database
- GEO
-
Gene Expression Omnibus
- DEGs
-
differentially expressed genes
- PPI
-
Protein-Protein Interaction
- WGCNA
-
weighted gene coexpression network analysis
- ROC
-
receiver operating characteristic curve
- TCM
-
Traditional Chinese medicine
- OB
-
oral bioavailability
- DL
-
drug-likeness
- GEO
-
Gene Expression Omnibus
- FC
-
fold change
- GO
-
Gene Ontology
- KEGG
-
Kyoto Encyclopedia of Genes and Genomes
- BP
-
biological process
- CC
-
cellular component
- MF
-
molecular function
- GCMs
-
Gene Characteristic Modules
- TOM
-
topological overlap matrix
- GS
-
gene significance
- MM
-
module membership
- GSEA
-
gene set enrichment analysis
- AUC
-
area under the curve
- PLpro
-
apin-like protease
- Mpro
-
main protease
- PDB
-
Protein Data Bank
- TP53
-
tumour protein P53
- AKT1
-
Serine/threonine-protein kinase
- TNF
-
tumour necrosis factor
- CASP3
-
recombinant caspase 3
- Th
-
T helper
- RBD
-
receptor binding domain
- THF
-
hepatocyte growth factor
- NFKBIA
-
Nuclear factor kappa B inhibitor alpha
- IκB
-
NF-κB inhibitory protein
- IKK
-
IKB kinase
- HIV-1.
-
human immunodeficiency virus-1
1 Introduction
A precipitate disease manifested at the end of December 2019: sore throat, cough, and high fever broken out with indistinct pathogenic factors. What was more intractable was the rapid spread of the virus in our country and subsequently around the world, which caused millions of infections in a few months (Wollina, 2020). With the publication of the whole genome sequence of this virus, it was identified and abbreviated as “COVID-19” by the WHO. Nowadays, though the diffusion and destruction of the virus tend to be gentle, four SARS‐CoV‐2 variants elicited by unignorable mutations of spike protein emerging, namely Alpha, Beta, Gamma, and Delta in recent times (Cui et al., 2019; Yakovleva et al., 2021), which might attribute to the weakened effectiveness of neutralizing antibodies and cell‐mediated immunity of the existing vaccines (Hadj Hassine, 2022). Therefore, seeking more safe and economical drugs to promote broadly is imminent.
Traditional Chinese medicine (TCM) exhibits unexpected advantages regarding this disastrous pandemic, contributing to its rich experience in curbing and treating plagues which dates back more than 2500 years. Many ancient monographs and formulas about the epidemic are still in use with remarkable effects (Guo et al., 2023). A systematic review, including 57 randomized controlled trials and cohort studies with 15,520 patients infected with COVID-19, found that TCM could reduce the proportion of progression of severe patients by 55 % and the mortality of critical patients by 49 % (Kang et al., 2022). As an essential part of TCM, Chinese herbs are widely cultivated and relatively safe with varieties of natural active compounds, displaying a prospect in exploring innovative drugs and becoming a complementary remedy. Ephedra Herba (EH) and Armeniacae Semen Amarum (ASA), a pair of drugs often used together in such as Lianhua Qingwen capsules/granules and Qingfei Paidu decoction, etc., which were recommended for the pandemic by authority (Wu et al., 2022), suggesting that the ancient herbs have glowed a new vitality of precaution and anesis on COVID-19. With their higher frequency of utilization, we aimed to clarify the mechanism and predict genes targeting COVID-19.
In the last decades or so, due to the boom of large online public databases and bioinformatics, many study approaches have been invented, such as network pharmacology and molecular docking (Jiashuo et al., 2022). Applying these methods to mine exact anti-viral targets and search for resultful drugs to defend against SARS-CoV-2 at the early stage of the outbreak has saved many losses (Zhang et al., 2021). In the current study, we ingeniously integrated network pharmacology and bioinformatics to decipher the mechanism of EH-ASA herb pairs treat COVID-19 from a molecular level, which has yet to be reported and hopefully will reveal some latent worthy information. The flow chart of this study was shown in Fig. 1.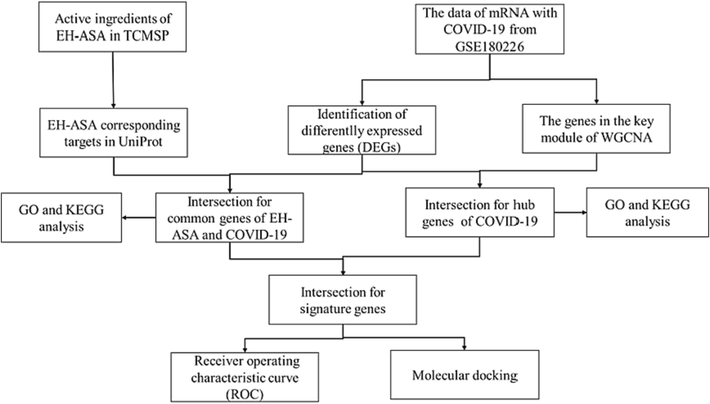
The flow chart of this study.
2 Methods
2.1 Screening of active compounds of EH-ASA herb pairs
The active compounds of EH-ASA were obtained from the Traditional Chinese Medicine Systems Pharmacology (TCMSP) database (Ru et al., 2014) (https://tcmsp-e.com), containing all herbs registered in Chinese Pharmacopoeia, which came into being to promote the integration of modern medicine and traditional medicine in drug research, and further development of TCM. It provides a total of twelve related properties on absorption, distribution, metabolism, and excretion to evaluate herbal medicines with potential biological efficacy, among which oral bioavailability (OB) and drug-likeness (DL) are leading indicators to appraise pharmacokinetics of a particular ingredient and whether it takes effect. We know that the higher the OB and DL scores, the better the component's pharmacological activity is. Thus, we set the screening conditions as OB > 30 % (Xu et al., 2012) and DL > 0.18 (Tao et al., 2013) in our study.
2.2 Predicting of targets of EH-ASA herb pairs
After getting the active ingredients of EH-ASA, we downloaded its corresponding target protein, namely “Target name” in TCMSP, and then the “Target name” was standardized as “Gene name” in the Universal Protein (UniProt) database, and the duplications were removed (https://www.uniprot.org) (UniProt, 2021). Finally, the gene symbol related to EH-ASA was obtained.
2.3 The construction of the Herb-Compound -target network
We used the Cytoscape software (Version.3.6.2) (Shannon et al., 2003) to construct the network of active compounds corresponding target genes from herbs in TCMSP that satisfied the threshold for OB and DL, so that we could observe the core relationship of the compound-target-herb network. The CytoNCA plug-in was used to calculate each node parameter in the network. Deeper color and larger node areas represented more robust connectivity of the node and its neighbors, suggesting the effective critical compound and key candidate target.
2.4 Identification of COVID-19 differentially expressed genes
We browsed the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) (Barrett et al., 2013) with “COVID-19” as the search term and finally selected the dataset of GSE180226 as following subject. The dataset contains 23 cases of lung tissue gene expression, pathology, and plasma protein as clinical sample information, of which 20 samples were from the disease group and others were from the control. The screening of differentially expressed genes (DEGs) between the two groups and the standardization of DEGs were performed using the “limma” package of R language software (Version 3.6.3, https://www.r-project.org/). To identify genes with significant differences in expression, the screening criteria were set as fold change (FC) value (|log2FC|) > 2 and P value < 0.05 (Li et al., 2022).
The standardization results were shown in box plots (Supplementary Fig. 1A–B), while the significantly expressed genes of the GSE180226 dataset were visualized with a heatmap and volcano plot by the “ggplot2” package in R software (Ito and Murphy, 2013). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
2.5 Construction of protein–protein interaction (PPI) network
The Venn online mapping tool (https://bioinformatics.psb.ugent.be/cgi-bin/liste/Venn/) was used to intersect the DEGs and EH-ASA herb pairs targets, namely common genes, which were regarded as potential targets against COVID-19, and were prepared for constructing PPI network. The list of genes obtained above was input into the STRING database (https://string-db.org/), with the species set as “homo sapiens”, the minimum required interaction score set as more than or equal to 0.4 (Szklarczyk et al., 2019), free targets hidden, and the rest of the parameters remained default. The related parameters in the PPI network were saved in “tsv” file format and then imported into Cytoscape software to visualize the relationship between targets of EH-ASA and COVID-19 genes. Subsequently, the nodes revealed critical genes according to the degree value calculated by the “NetworkAnalyzer” plug-in.
2.6 Functional enrichment analysis of common genes
To further illuminate the biological mechanism of active compounds of EH-ASA herb pairs on COVID-19, the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were conducted. First, we imported common genes into the DAVID platform (https://david.ncifcrf.gov/) for GO and KEGG analyses. Then, the functional annotation tables were downloaded and arranged in ascending order according to P value. Notably, the GO analysis examined the following three aspects: biological process (BP), cellular component (CC), and molecular function (MF). Then, we visualized the top 10 common genes of topological analysis and significant terms of GO annotation (top 10 of each category) and KEGG analysis (top 20). Finally, the relationship between hub genes and possible mechanisms was visualized using the function of “SankeyNetwork” of the “Goplot” package of R language software with a Sankey diagram. The P value < 0.05 was considered a threshold for significant terms.
2.7 Weighted gene co-expression network analysis
Weighted gene co-expression network analysis (WGCNA) is a widely used method to discover the Gene Characteristic Modules (GCMs) and key genes by connecting gene networks to clinical traits realized by the scale-free topology criterion. The characteristic module is the first essential element of a GCM, which could be used to indicate the whole expression of the GCM. In this study, we applied the “WGCNA” package of R software (Langfelder and Horvath, 2008) to construct the WGCNA co-expression network of the DEGs from the GSE180226 dataset, with healthy control and COVID-19 as clinical traits. Firstly, the outlier cases were filtered by hierarchical clustering analysis. Secondly, the pickSoft Threshold function was conducted to measure the soft threshold, also called soft power and adjacencies. So far, we have obtained an adjacency matrix, which was subsequently performed into a topological overlap matrix (TOM). The dynamic tree-cutting means was used to determine the modules in the co-expression network. We then appraised the correlation between gene modules and COVID-19 by gene significance (GS) and module membership (MM). The higher the values, the closer the connection between genes and clinical characteristics. At last, we merged the genes of the key module with the most excellent Pearson correlation coefficient and DEGs, namely hub genes, for further investigation.
2.8 Functional enrichment analysis of hub genes
We picked up the hub genes by crossing the genes in the key module obtained by the WGCNA method with DEGs, which were input into the DAVID online analysis platform. The results were visualized as circle plots through the “GOplot” package in R software. In addition, we exerted the gene set enrichment analysis (GSEA) to assess the differences in the stimulation between the COVID-19 samples and phenotypes. GSEA on DEGs was selected with P < 0.05 as a filtration standard to identify the association between hub genes and significant enrichment terms (Subramanian et al., 2005).
2.9 Identification of signature genes
After a series of filtration, we initially acquired common genes via the intersection of EH-ASA herb pairs targets and DEGs of the GSE180226 dataset; next, we also got hub genes of COVID-19 by merging DEGs and genes of the characteristic module with the highest correlation coefficient. Now, we were going to identify the candidate signature genes by combining common genes with hub genes, regarded as signature genes of EH-ASA that could treat COVID-19 with high probability. The diagnostic accuracy of screened signature genes was evaluated by employing the area under the curve (AUC) of the receiver operating characteristic curve (ROC). An AUC was more significant than 0.7, which manifested preferable diagnostic performance.
2.10 Molecular docking
The binding situation and interaction force between the protein target, usually receptor, and small molecule compound, usually ligand, could be predicted and obtained with molecular docking analysis (Pinzi and Rastelli, 2019). In this study, the active compounds from EH-ASA obtained through topological analysis and signature genes acquired by WGCNA were finally picked for molecular docking analysis. Furthermore, we selected another two protein targets, including apin-like protease (PLpro) and main protease (Mpro), from the COVID-19 Docking Server website (https://ncov.schanglab.org.cn/index.php) (Kong et al., 2020), which were considered as promising targets for discovering novel anti-viral drugs. Then, the 2D structure of drug ligands and protein receptors were separately downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) (Wang et al., 2017) and Protein Data Bank (PDB) database (https://www.rcsb.org/) (Wang et al., 2012). Small molecule energy minimization was performed to optimize conformation using PyMol 2.4.0 software. Autodock Vina software was conducted to delete water, add perhydro, calculate charges, detect and choose a torsion tree, an active docking centre via the grid box function, and then processed the molecular docking with default parameters (Trott and Olson, 2010). Finally, the binding energy of docking results was shown in a heatmap. We also chose the best affinity as the final docking conformation and visualized it in Pymol 2.4.0 software (Seeliger and de Groot, 2010) and LigPlot 2.2.8 software.
3 Result
3.1 The screening of active compounds and targets of EH-ASA herb pairs
A total of 42 qualified chemical compounds of EH-ASA were collected in TCMSP, among which Stigmasterol and (+)-catechin were the mutual ingredients of EH and ASA, and the details of these screened constituents are shown in Table 1. Subsequently, we identified the targets corresponding to the above compounds in the UniProt KB database. After discarding the repetition, we finally obtained 200 potentially therapeutic targets of EH-ASA. Whereas compounds MOL001506 from EH and MOL010922, MOL002372 and MOL003410 from ASA could not relate to their targets, so we removed these four chemicals and then built a network about the interaction of active compounds (C) and targets (T) of EH and ASA (drug, D).
NO.
MOL ID
compounds
OB(%)
DL
Herb
MH1
MOL010489
Resivit
30.84
0.27
EH
MH2
MOL002881
Diosmetin
31.14
0.27
EH
MH3
MOL001506
Supraene
33.55
0.42
EH
MH4
MOL002823
Herbacetin
36.07
0.27
EH
MH5
MOL001755
24-Ethylcholest-4-en-3-one
36.08
0.76
EH
MH6
MOL000006
luteolin
36.16
0.25
EH
MH7
MOL001771
poriferast-5-en-3beta-ol
36.91
0.75
EH
MH8
MOL000358
beta-sitosterol
36.91
0.75
EH
MH9
MOL005573
Genkwanin
37.13
0.24
EH
MH10
MOL005043
campest-5-en-3beta-ol
37.58
0.71
EH
MH11
MOL007214
(+)-Leucocyanidin
37.61
0.27
EH
MH12
MOL004798
delphinidin
40.63
0.28
EH
MH13
MOL005842
Pectolinarigenin
41.17
0.3
EH
MH14
MOL000422
kaempferol
41.88
0.24
EH
MH15
MOL001494
Mandenol
42
0.19
EH
MH16
MOL011319
Truflex OBP
43.74
0.24
EH
MH17
MOL000449
Stigmasterol
43.83
0.76
EH
MH18
MOL000098
quercetin
46.43
0.28
EH
MH19
MOL000492
(+)-catechin
54.83
0.24
EH
MH20
MOL004576
taxifolin
57.84
0.27
EH
MH21
MOL010788
leucopelargonidin
57.97
0.24
EH
MH22
MOL004328
naringenin
59.29
0.21
EH
MH23
MOL005190
eriodictyol
71.79
0.24
EH
XR1
MOL010921
estrone
53.56
0.32
ASA
XR2
MOL010922
Diisooctyl succinate
31.62
0.23
ASA
XR3
MOL002211
11,14-eicosadienoic acid
39.99
0.2
ASA
XR4
MOL002372
(6Z,10E,14E,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene
33.55
0.42
ASA
XR5
MOL000359
sitosterol
36.91
0.75
ASA
XR6
MOL000449
Stigmasterol
43.83
0.76
ASA
XR7
MOL005030
gondoic acid
30.7
0.2
ASA
XR8
MOL000953
CLR
37.87
0.68
ASA
XR9
MOL000211
Mairin
55.38
0.78
ASA
XR10
MOL000492
(+)-catechin
54.83
0.24
ASA
XR11
MOL002311
Glycyrol
90.78
0.67
ASA
XR12
MOL003410
Ziziphin_qt
66.95
0.62
ASA
XR13
MOL004355
Spinasterol
42.98
0.76
ASA
XR14
MOL004841
Licochalcone B
76.76
0.19
ASA
XR15
MOL004903
liquiritin
65.69
0.74
ASA
XR16
MOL004908
Glabridin
53.25
0.47
ASA
XR17
MOL005017
Phaseol
78.77
0.58
ASA
XR18
MOL007207
Machiline
79.64
0.24
ASA
XR19
MOL012922
l-SPD
87.35
0.54
ASA
As shown in Fig. 2, it was found that there was a one-to-one correspondence between 42 active constituents and 200 targets of EH-ASA by interconnection in this complex C-T-D network, which contained a total of 238 nodes and 580 edges, demonstrating that EH and ASA could directly or indirectly exert the therapeutic influence on a mass of targets.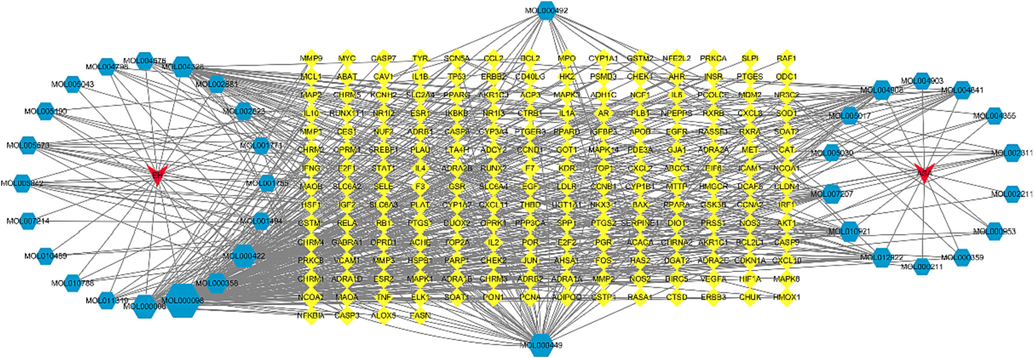
C-T-D network of EH-ASA herb pairs.
3.2 Identification of different expression genes related to COVID-19
Compared with the healthy samples, there were 548 genes with significant differences in expression of the subjects of COVID-19 total, among which 137 genes were highly up-regulated, while 411 genes were very down-regulated in the GSE180226 dataset. A heat map and a volcano diagram were drawn with the screened DEGs to demonstrate the distribution of these genes in this dataset between the control and COVID-19 group using “limma”, “heatmap”, and “ggplot2” packages in R software (Fig. 3A, B).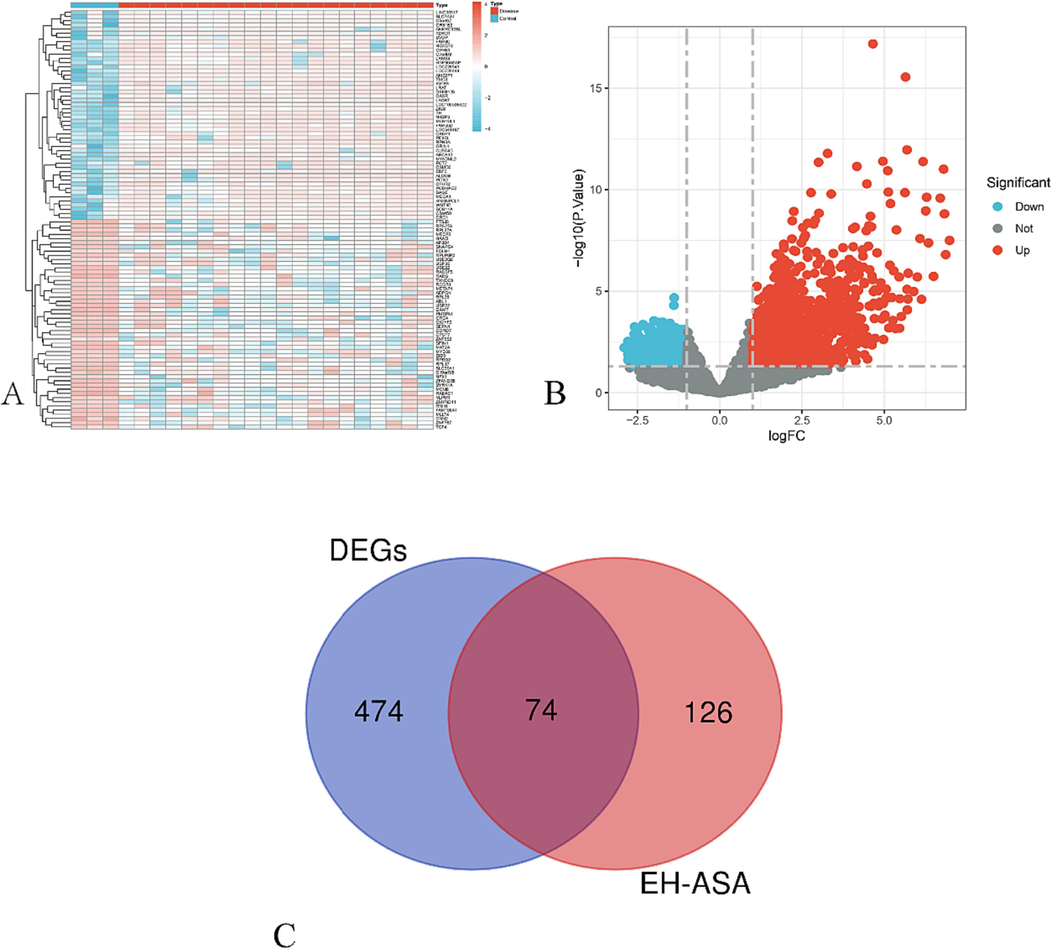
Results of DEGs in GSE180226 related to COVID-19. (A) Heatmap. (B) Volcano plot. The red dots represent upregulated genes, the blue dots represent downregulated genes, and the grey dots represent stable genes. FC, fold change. (C) Venn diagram of the common genes of DEGs with targets of EH-ASA herb pairs. EH, Ephedra Herba; ASA, Armeniacae Semen Amarum.
In addition, the therapeutic effects of EH-ASA on COVID-19 needed to be validated. Thus, As shown in Fig. 3C, after combining the 200 targets corresponding to active constituents of 548 DEGs of the GSE180226 dataset via limma difference analysis, there were a total of 74 overlap genes identified, namely common genes, which were regarded as the potential targets in the treatment of COVID-19 and prepared for the subsequent study.
3.3 Protein-protein interaction (PPI) network construction of common genes
As shown in Fig. 4A, the PPI network was constructed with common genes, including 74 nodes and 1320 edges. To determine the interactive relationships of DEGs with active targets, we applied these 74 common genes to the STRING online platform and formed a PPI network through Cytoscape software. The result showed that multiple targets had strong interactions with each other, such as tumour protein P53 (TP53), Serine/threonine-protein kinase (AKT1), tumour necrosis factor (TNF), and recombinant caspase 3 (CASP3).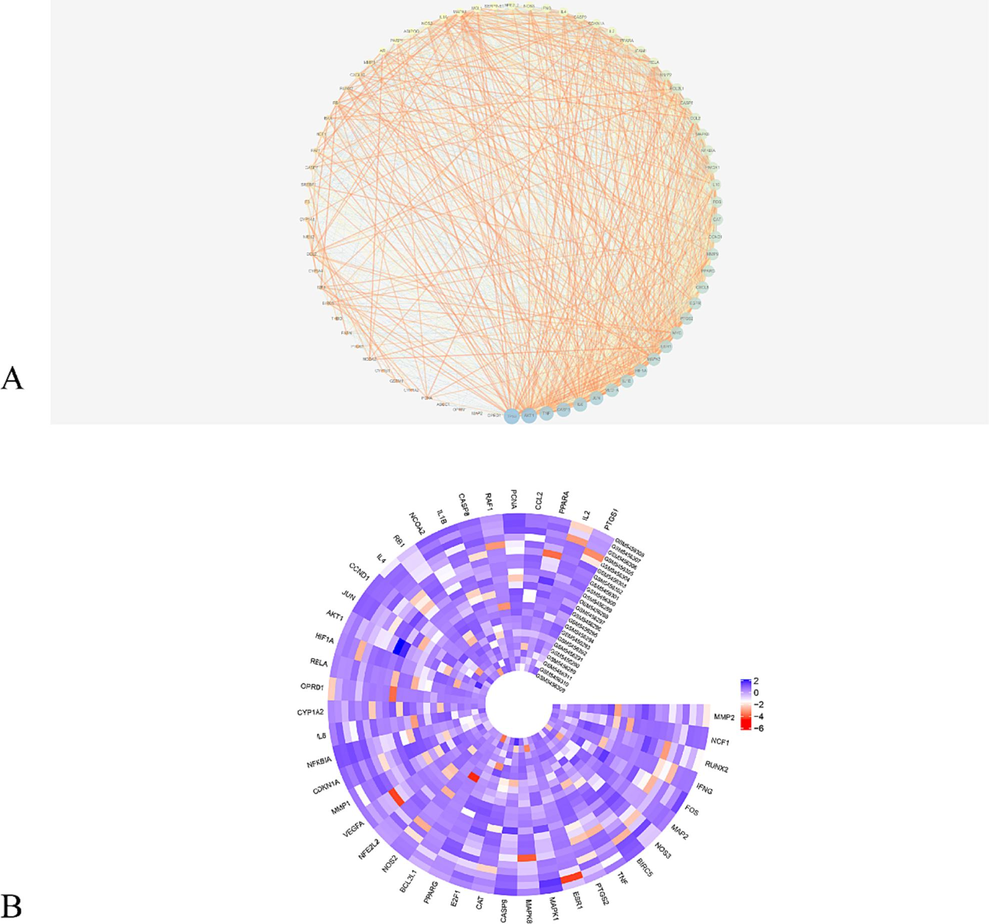
The results of PPI. (A) PPI network of the common genes. The targets were ranked in order based on their degree values, larger dots, and darker colors indicating greater significance. (B) Heatmap of 23 samples of GSE180226 and its corresponding DEGs of common genes.
Moreover, we elucidated the interaction between targets of EH-ASA and DEGs of the GSE180226 dataset related to COVID-19. Therefore, the expression of 74 genes that were necessary from these 23 COVID-19 samples was selected, and a heatmap was then created using “circlize” and “ComplexHeatmap” packages in R software (Fig. 4B).
3.4 Functional enrichment analysis of common genes
We also applied the common genes to GO and KEGG enrichment analysis to clarify the active mechanism. As shown in Fig. 5A-B, the BP category extensively focused on cellular response to reactive oxygen species, ageing and process of apoptosis, and positive regulation of transcription from the RNA polymerase II promoter. CC was mainly involved in the cytoplasm, mitochondria, and RNA polymerase II transcription factor complex. Moreover, the targets were widely aggregated in transcription factor binding and protein binding in MF. When referred to the KEGG analysis, the most common targets were enriched in the TNF signaling pathway, Influenza A, and Toll-like receptor signaling pathway. The complex overlapping relationship between genes and pathways was also shown in the Sankey diagram, in which MAPK3 is related to most KEGG pathways, suggesting a crucial status in associating the various cross-talk with different pathways.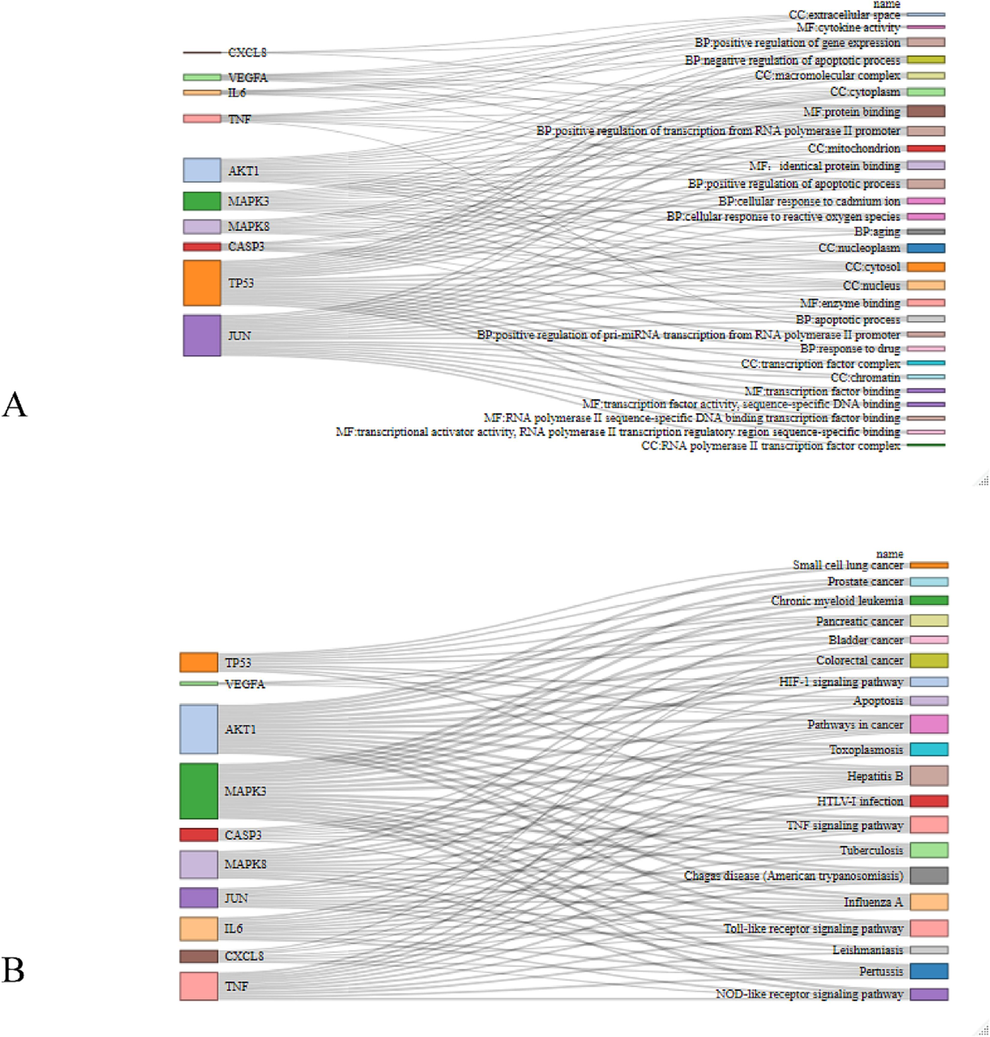
Sankey diagram of functional enrichment analysis of common genes. (A) GO; (B) KEGG. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
3.5 Construction of co-expression network and selection of key module
The WGCNA of COVID-19 subjects and healthy controls in the GSE180226 dataset was conducted. We finally established a scale-free co-expression network through hierarchical clustering and dynamic tree cut. A scale-free index of 0.85 and relatively good mean connectivity identified the soft threshold power as β = 19 (Fig. 6A, B). The cluster gene dendrogram is shown in Fig. 6C-E. Finally, a total of 17 differently coloured modules were obtained, among which the MEsinna3 module, including 1226 genes, was considered to be the most significantly associated with COVID-19 (r = 0.9, P < 1e-200) (Fig. 6F, G). As shown in Fig. 5H, a total of 189 mapping genes between DEGs and genes in the Mesinna3 module were shown in a Venn diagram.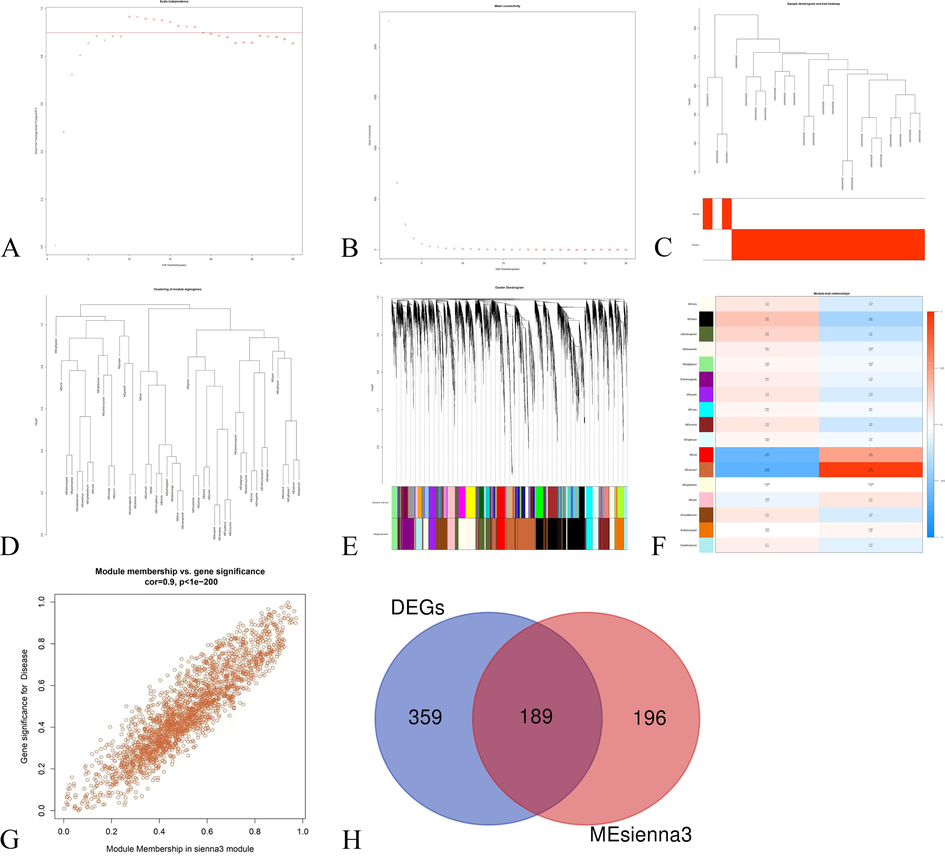
The results of WGCNA of GSE180226. (A) The soft thresholding power of scale independence. (B) The soft thresholding power of mean connectivity values. (C) Heatmap of the sample cluster. (D) Clustering of module eigengenes of correlation diagram of modules. (E) Merged dynamic tree cut diagram of all DEGs according to the cluster. The different color stripes show the results of the monolithic analysis in automation. (F) The heatmap of the correlation between module characteristic genes and phenotypes. The vertical coordinate is the correlation coefficient of feature modules, and the darker color indicates a more significant correlation. (G) The scatter diagram of module characteristic genes and phenotypes. (H) Venn diagram of DEGs with genes in the key module.
3.6 Functional enrichment analysis of hub genes
To further discover the pathogenesis of the GSE180226 dataset related to COVID-19, we performed GO and KEGG enrichment analysis with 189 hub genes (Fig. 7A-D). The BP annotation implied that positive regulation of regulatory T cell differentiation, response to epidermal growth factor, and UMP biosynthetic process were mainly enriched. Also, lateral element, brush border, and terminal bouton were primarily collected in CC. In MF, transcription coactivator binding, amino acid binding, and transcription cofactor binding were mainly involved. As for KEGG analysis, the above hub genes were primarily focused on the IL-17 signaling pathway, Human T-cell leukaemia virus one infection, Type I diabetes mellitus, and so on. In addition, the GSEA results suggested that terms of fat digestion and absorption, glycosaminoglycan degradation, linoleic acid metabolism, maturity onset diabetes of the young and mucin type O-glycan biosynthesis were significantly up-regulated in COVID-19 samples; meanwhile, autophagy, cysteine and methionine metabolism, legionellosis, proteasome and ribosome were activated in control samples (Fig. 7E, F).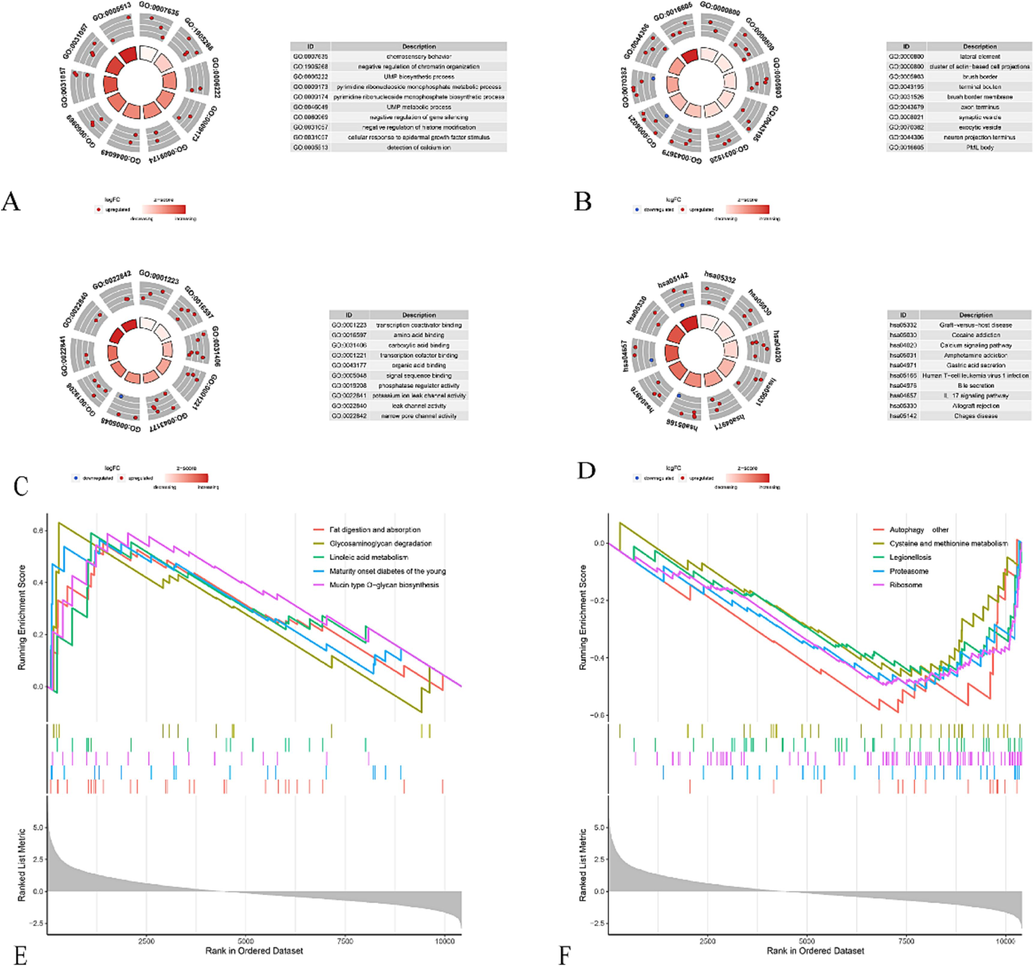
Circle plot of functional enrichment analysis of hub genes. (A) BP; (B) CC; (C) MF; (D) KEGG. (E) GSEA in COVID-19 group; (F) GSEA in control group. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes. UMP, uridine monophosphate.
3.7 Diagnostic accuracy of signature genes in COVID-19
We further performed difference analysis to determine the diagnostic efficacy of signature genes screened through the PPI and co-expression networks of WGCNA. The result showed that these genes significantly distinguished the subjects infected with COVID-19 from those who were healthy, indicating that these genes from EH-ASA might perform a good remedy for COVID-19 (Fig. 8A-C). Among them, ESR1 and IL2 were increased in COVID-19 samples (P < 0.01), while NFKBIA increased in control samples significantly (P < 0.01). Furthermore, the AUC of the signature gene was 1.00 of ESR1, 0.967 of IL2, and 0.967 of NFKBIA, respectively (Fig. 8D-F), suggesting that these three signature genes might have good diagnostic accuracy in detecting COVID-19 and might be against the virus.
The performance of signature genes. The boxplot of gene expression of ESR1(A), IL2(B) and NFKBIA(C). The ROC of diagnostic accuracy of ESR1(D), IL2(E) and NFKBIA(F). Con, Control; Treat, COVID-19.
3.8 Molecular docking
The scores of molecular docking between the main active compound of EH-ASA and signature genes were shown in Fig. 8. It is known that the lower the binding energy, the stronger the docking affinity is, indicating that the compounds were closer to the targets and were more effective (Pantsar and Poso, 2018). The corresponding PDB IDs were ESR1 (1L2I), IL2 (4NEJ), NFKBIA (1NFI), Mpro (6LU7), and PLpro (6WUU), respectively, and the details of main active compounds for molecular docking were shown in Table 2.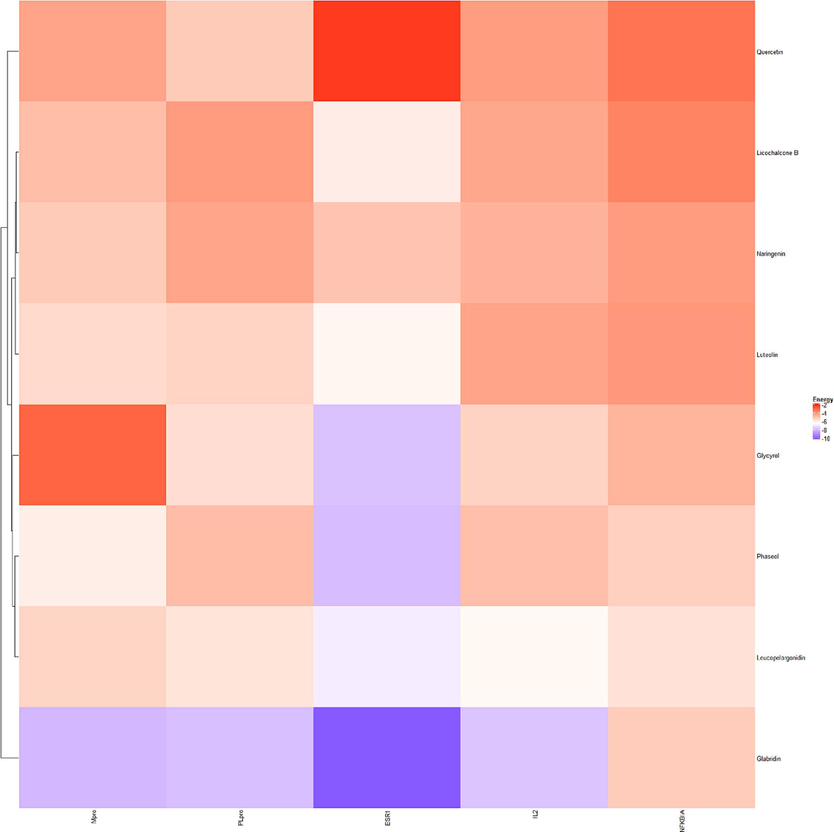
Heatmap of Molecular docking.
MOL ID
Compounds
Pubchem ID
Molecular formula
Source
2D Structure
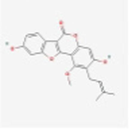
MOL002311
Glycyrol
5320083
C21H18O6
Armeniacae Semen Amarum
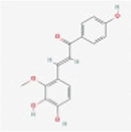
MOL004841
Licochalcone B
5318999
C16H14O5
Armeniacae Semen Amarum
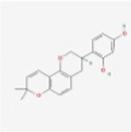
MOL004908
Glabridin
124052
C20H20O4
Armeniacae Semen Amarum
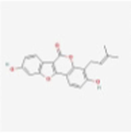
MOL005017
Phaseol
44257530
C20H16O5
Armeniacae Semen Amarum
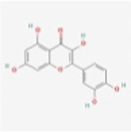
MOL000006
Luteolin
5280445
C15H10O6
Ephedra Herba
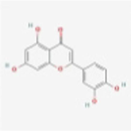
MOL000098
Quercetin
5280343
C15H10O7
Ephedra Herba
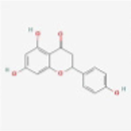
MOL004328
Naringenin
932
C15H12O5
Ephedra Herba
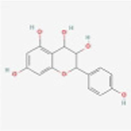
MOL010788
Leucopelargonidin
3286789
C15H14O6
Ephedra Herba
The combined scores of the top six conformations were in the rankings, Glycyrol-ESR1, Glabridin-ESR1, Glabridin-PLpro, Phaseol-ESR1, Luteolin-ESR1, Glabridin-Mpro, suggesting that the above-mentioned active compounds of EH-ASA might possess an excellent affinity with COVID-19 related genes.
In addition, hydrogen bonds and hydrophobic contacts were recorded to meticulously observe the interactions between protein and ligand, which were considered the most common type of interactions (Ferreira de Freitas and Schapira, 2017). As shown in Fig. 10A-L, Glycyrol and Glabridin are bound to ESR1 with the amino acid residues GLU-353 and ARG-394, forming three hydrogen bonds and two hydrophobic contacts. Glabridin bound to PLpro with the amino acid residues LYS-306, forming three hydrogen bonds and two hydrophobic contacts. Phaseol is tied to ESR1 with the amino acid residues ARG-394 and GLU-353, forming three hydrogen bonds and two hydrophobic contacts. Luteolin bound to ESR1 with the amino acid residues ARG-394, GLU-353, LEU-387, LEU-346, and GLY-521, forming three hydrogen bonds and nine hydrophobic contacts. Glabridin bound to Mpro with the amino acid residues GLU-71 and MET-17, forming three hydrogen bonds and four hydrophobic contacts.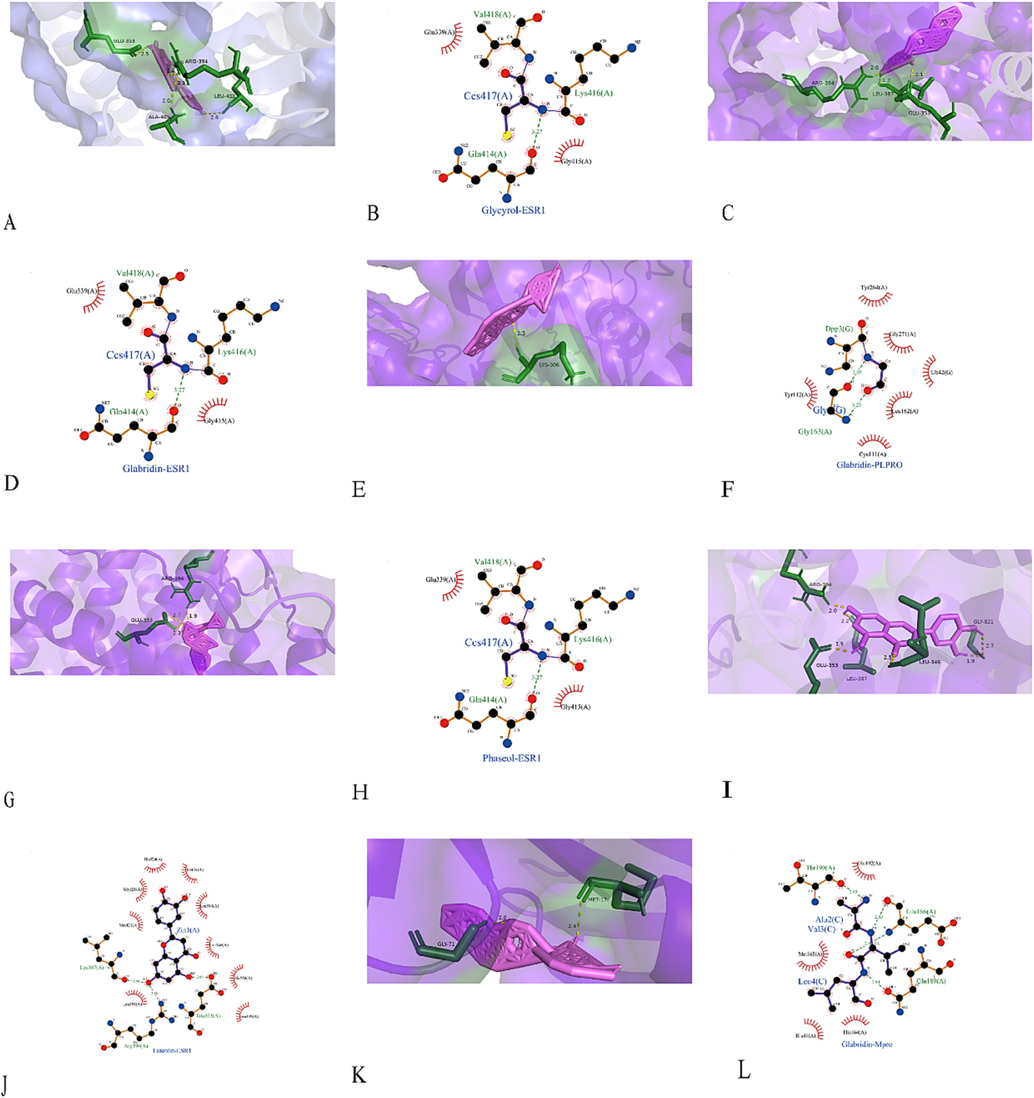
Results of Molecular docking. Molecular docking models(3D and 2D) with the lower binding energy of sites of Glycyrol with ESR1(A, B); Site of Glabridin with ESR1(C, D); Site of Glabridin with PLpro(E, F); Site of Phaseol with ESR1(G, H); Site of Luteolin with ESR1(I, J); Glabridin with Mpro(K, L). GLU, Glutamate; ARG, Arginine; LYS, Lysine; LEU, Leucine; GLY, Glycine; MET, Methionine; PLpro, Papain-Like protease; Mpro, Main protease; Green dotted lines, hydrogen bonds; Red combs, hydrophobic contacts.
4 Discussion
Since the outbreak of the epidemic, it has caused a significant threat to the lives and property of global citizens. Therefore, effective preventive and curative methods are urgently needed to save the colossal loss. Although antivirus agents and neutralizing antibody work rapidly, they may have no effects on mutations (Wang et al., 2022). Also, the high prices of these monoclonal antibodies and immunomodulatory drugs limit their utilization (Najjar-Debbiny et al., 2023).
In this study, we revealed potential targets corresponding to active ingredients of EH-ASA herb pairs on COVID-19 and investigated pathological mechanisms based on network pharmacology and molecular docking binding to bioinformatics. There were the following findings after a series of screenings: (1) We obtained active compounds of EH-ASA through the TCMSP database, among which were mainly flavonoids known as anticancer, antiviral, and anti-inflammatory. We also got 548 DEGs of COVID-19 and further explored their mechanisms by functional enrichment analysis. (2) We then collected 74 common genes by intersecting the targets of EH-ASA and DEGs, which are considered potential substances for treating COVID-19. (3) We constructed a complicated and biologically active PPI network, in which all genes mainly collectively affected Human T-cell leukaemia virus type 1 infection, Influenza A and cancer pathways, etc. Concluded the above-described 1–3 findings, the results suggested that EH-ASA might exert therapeutic effects on this virus through multiple targets cooperating. Furthermore, the pathological mechanism of COVID-19 might be associated with excessive anti-inflammatory response and abnormal tumour regulation. (4) We then collected core COVID-19 genes in significant modules through WGCNA, and three signature genes (ESR1, IL2, NFKBIA) were finally identified after combining common genes with core genes. Then, we performed molecular docking with these genes and primary active compounds, suggesting that EH-ASA may primarily treat this virus through these three signature genes and may be biomarkers for COVID-19.
The following implications deserve more attention. Firstly, we found three signature genes, of which IL2 was significantly upregulated in CIVID-19. IL-2 is known as a cytokine with vigorous T cell growth factor activity that can counteract dysregulated immunity via regulatory T cells when autoimmunity and inflammatory disorder were applied to treat a variety of autoimmune diseases (Sharfe et al., 1997; Gregersen and Olsson, 2009; Hernandez et al., 2022; Vanderniet et al., 2022). Our result was in line with other studies; for example, COVID-19 was characterized by the cytokine profile with chronically elevated IL2 and IL17 in A Brazilian (Queiroz et al., 2022). Similarly, Li detected high-expressed IL2, TNFα, and INFγ in sars-cov-2-specific CD4 and CD8 T cells response with 1096 rehabilitative cases based on a cohort study.
Moreover, the T cell response could be maintained 12 months after the initial infection and not be disturbed by the variant strains, especially the delta variant (Guo et al., 2022). Further, Tappe (Tappe et al., 2022) found the increased exhaustion of T helper (Th) cells with signs of weakened Th activity and cytokine response in COVID-19 patients, which exacerbated alveolar oedema and hypoxia ascribed to the aggrandized vascular permeability and integrity of blood/air barrier damaged (Ahmadpoor and Rostaing, 2020). Thus, it may be related to the ability of IL2 to suppress the immune response and to enhance this immune suppression through Tregs and Th cells. The regulation is manifested as driving Th1 and Th2 fate decisions but suppressing Th17 differentiation (Ross and Cantrell, 2018). One of the known pathogenesis of COVID-19 is related to the “hyperinflammation” caused by these chemokines, namely “Cytokines Strom” (Ahmadpoor and Rostaing, 2020). Finally, the immune microenvironment may be out of balance due to a prolonged state of emergency (Mei et al., 2016).
In TCM theory, EH is to lung meridian and is commonly applied to treat fever, chills, cough, wheezing, and edema caused by influenza. Studies have found that various components of EH, such as ephedrine (Mei et al., 2016), Ephedra polysaccharides (Zhang et al., 2022), and Ephedra aqueous extract (Tang et al., 2016), could relieve asthma by restraining Th17, IL13, and other inflammatory factors, and rectify immune disorder of Th1/Th2 and Th17/Treg. The latest research suggested that Quinoline-2-carboxylic acid in EH could directly intercept the invading virus by blocking the bond of ACE2 to the SARS-CoV-2 Spike protein receptor binding domain (RBD) (Mei et al., 2021). At present, a trial vaccine acting on the RBD site could heighten high nAb titers and Th1/Th2 associated cytokines, and maintain the neutralization against β and γ variants, namely ZF2001, is being explored in Anhui China (Yang et al., 2021; Zhao et al., 2021). Besides, the anticancer effect of EH has also been demonstrated (Hyuga et al., 2013); Yukio found that EH could inhibit the process of Non-small cell lung cancer by accelerating the endocytosis of hepatocyte growth factor (HGF)-stimulated MET and phosphorylated (p)-MET (Nishimura et al., 2016). Through enrichment analysis, we detected several cancer-associated pathways and identified higher IL2 mRNA profiling in COVID-19 patients, so we speculate that EH might target IL2 against COVID-19. From the abovementioned argumentation, we assumed that IL2 was inclined to irritate the immune response, which impaired SARS-CoV-2 and defended the subsequent invasion. Therefore, EH could play an auxiliary role in antiviral.
Nuclear factor kappa B inhibitor alpha (NFKBIA), another signature gene we screened, will interfere with NF-κB activation when specific point mutations cause post-translational modification changes (Chear et al., 2022). Generally, NF-κB is inactivated when combined with NF-κB inhibitory protein (IκB) regulated by IKB kinase (IKK) in the cytoplasm. Once IKK phosphorylates and ubiquitinates IκB, it is proteinized, and then it could activate NF-κB and nuclear translocation, where it exerts a role as a transcription factor targeting genes associated with immunity, inflammation, apoptosis, and proliferation (Kanarek and Ben-Neriah, 2012; Zinatizadeh et al., 2021). A Spanish study has shown that patients with severe COVID-19 had significantly lower NFKBIA rs696 GG genotypes compared with healthy controls, indicating that NFKBIA might be activated by SARS-CoV-2, which was consistent with our findings (Camblor et al., 2022). Given the above analysis, we expected that EH and ASA could keep NF-κB in silence or maintain NFKBIA stable. Fortunately, Scientists in Japan have demonstrated that Ephedra extract could induce nuclear translocation of NF-κB to eliminate latent viral reservoirs in patients with human immunodeficiency virus-1 (HIV-1) (Murakami et al., 2008). Moreover, our scholars also proved that Mahuang decoction resisted the influenza virus by inhibiting the TLR4/MyD88/TRAF6 signaling pathway with lower NF-κB and higher IL2 (Peng et al., 2016). The activated cytokines are important to initiate and coordinate immune responses, but they also might be a source of exacerbations and complications in COVID-19 patients; thus, striving to restore the balance of immunity is urgent.
Furthermore, researchers have affirmed that Glabridin, the main flavonoid active compound screened in our study, could exert a pivotal function of anti-inflammation, anti-oxidation, and anti-tumor (Modarresi et al., 2019; Parlar et al., 2020; Goel et al., 2021), by negatively degrading IκBs, restraining nuclear translocation of p65, and impairing DNA binding and transcription of NF-κB/Rel (Kang et al., 2005). Jen found that glabridin binding to tamoxifen could regulate ESR1 and Bcl-2 to reduce cell growth in Ishikawa cells, performing as estrogenic (Jen et al., 2015). Additionally, Glabridin and other chemical ingredients have been proven safe and possess ideal docking activity with potential targets for drug development.
This work has several strengths. Our study is the first to investigate the pathological mechanisms of COVID-19 based on the technical strategy of network pharmacology and molecular docking combined with bioinformatics from the molecular level. Moreover, given that the components of EH and ASA with biological activity could enrich pathways such as cancer, excessive inflammation and immune response, they were considered potential therapeutic agents for COVID-19, which offered an initial direction for developing novel drugs. We hope that this work will foster more research about TCM in the treatment of COVID-19 to be carried out. Nevertheless, we must honestly acknowledge that this study has certain limitations. Firstly, although we have already verified the potential therapeutic targets by molecular docking, the findings are concluded based on network pharmacology. Hence, its reliability needs further confirmed through vivo- or/and in vitro experiments in the real world. Secondly, the results might be biased because the dataset employed in this study was picked from the public database and included small samples. Thus, we performed a bioactive function analysis by the most widely used GSEA approach to explore the mechanism of these genes. Yet, single-cell sequencing is still indispensable for obtaining more accurate information. Last but not least, except for the mechanism needs to be verified, the clinical efficacy and symptom improvement rate of EH and ASA resisting the SARS-CoV-2 of COVID-19 also require confirmation by follow-up studies, which are multicenter, multiracial, different clinical stages as well as extensive clinical samples.
Since last century, thanks to the great convenience of transportation, human communication worldwide has never been closer, immensely facilitating the transmission of epidemics, and nobody can stay away. The world has passed this pandemic, or the emergency phase caused by the omicron variant. However, the infection morbidity is still high (7.9–18.9 %) (Chemaitelly et al., 2022) despite the citizens having been vaccinated or being infected naturally. Moreover, the virus seems to be characterised as a weaker pathogenicity but with a higher contamination rate, leading to extensive infection (Meng et al., 2023). In consideration of the ability of SARS-CoV-2 to evolve rapidly, we realized that the virus might never be eradicated and we would coexist with it for a while. Therefore, exploring economical and safe anti-coronavirus vaccines is imperative, and TCM may provide a favourable development space in this post-pandemic era.
5 Conclusion
In summary, we found that EH-ASA herb pairs might play a role in treating COVID-19, potentially associated with cancer-related pathways, excessive inflammatory and immune response, and abnormal apoptosis. Besides, three signature genes were screened out through this study, namely ESR1, IL2, and NFKBIA, providing a perspective on the prevention and treatment of COVID-19.
CRediT authorship contribution statement
Zhuoxi Wang: Conceptualization, Writing – original draft. Jifang Ban: Methodology, Software. He Wang: Validation, Project administration. Rui Qie: Visualization. Yabin Zhou: Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Why the immune system fails to mount an adaptive immune response to a COVID-19 infection. Transpl. Int.. 2020;33(7):824-825.
- [CrossRef] [Google Scholar]
- NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41(Database issue), D991–995
- [CrossRef] [Google Scholar]
- Genetic variants in the NF-kappaB signaling pathway (NFKB1, NFKBIA, NFKBIZ) and risk of critical outcome among COVID-19 patients. Hum Immunol. 2022;83(8–9):613-617.
- [CrossRef] [Google Scholar]
- A Novel De Novo NFKBIA Missense Mutation Associated to Ectodermal Dysplasia with Dysgammaglobulinemia. Genes (Basel). 2022;13(10)
- [CrossRef] [Google Scholar]
- Protection from previous natural infection compared with mRNA vaccination against SARS-CoV-2 infection and severe COVID-19 in Qatar: a retrospective cohort study. Lancet Microbe. 2022;3(12):e944-e955.
- [CrossRef] [Google Scholar]
- Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181-192.
- [CrossRef] [Google Scholar]
- A systematic analysis of atomic protein-ligand interactions in the PDB. Medchemcomm. 2017;8(10):1970-1981.
- [CrossRef] [Google Scholar]
- In-vitro antitumor activity of compounds from Glycyrrhiza glabra against C6 glioma cancer cells: identification of natural lead for further evaluation. Nat Prod Res. 2021;35(23):5489-5492.
- [CrossRef] [Google Scholar]
- Recent advances in the genetics of autoimmune disease. Annu Rev Immunol. 2009;27:363-391.
- [CrossRef] [Google Scholar]
- SARS-CoV-2-specific antibody and T-cell responses 1 year after infection in people recovered from COVID-19: a longitudinal cohort study. Lancet Microbe. 2022;3(5):e348-e356.
- [CrossRef] [Google Scholar]
- Da-Yuan-Yin decoction polyphenol fraction attenuates acute lung injury induced by lipopolysaccharide. Pharm Biol. 2023;61(1):228-240.
- [CrossRef] [Google Scholar]
- Covid-19 vaccines and variants of concern: A review. Rev Med Virol. 2022;32(4):e2313.
- [Google Scholar]
- Engineering IL-2 for immunotherapy of autoimmunity and cancer. Nat Rev Immunol. 2022;22(10):614-628.
- [CrossRef] [Google Scholar]
- Herbacetin, a constituent of ephedrae herba, suppresses the HGF-induced motility of human breast cancer MDA-MB-231 cells by inhibiting c-Met and Akt phosphorylation. Planta Med. 2013;79(16):1525-1530.
- [CrossRef] [Google Scholar]
- Application of ggplot2 to Pharmacometric Graphics. CPT Pharmacometrics Syst Pharmacol. 2013;2(10):e79.
- [Google Scholar]
- The Combinatory Effects of Glabridin and Tamoxifen on Ishikawa and MCF-7 Cell Lines. Nat Prod Commun. 2015;10(9):1573-1576.
- [Google Scholar]
- Integration strategy of network pharmacology in Traditional Chinese Medicine: a narrative review. J Tradit Chin Med. 2022;42(3):479-486.
- [CrossRef] [Google Scholar]
- Regulation of NF-kappaB by ubiquitination and degradation of the IkappaBs. Immunol Rev. 2012;246(1):77-94.
- [CrossRef] [Google Scholar]
- Efficacy and mechanisms of traditional Chinese medicine for COVID-19: a systematic review. Chin Med. 2022;17(1):30.
- [CrossRef] [Google Scholar]
- Glabridin, an isoflavan from licorice root, inhibits inducible nitric-oxide synthase expression and improves survival of mice in experimental model of septic shock. J Pharmacol Exp Ther. 2005;312(3):1187-1194.
- [CrossRef] [Google Scholar]
- COVID-19 Docking Server: a meta server for docking small molecules, peptides and antibodies against potential targets of COVID-19. Bioinformatics. 2020;36(20):5109-5111.
- [CrossRef] [Google Scholar]
- WGCNA: an R package for weighted correlation network analysis. BMC Bioinf.. 2008;9(1471–2105 (Electronic)), 559
- [CrossRef] [Google Scholar]
- Li, G., Sun, J., Zhang, J., Lv, Y., Liu, D., Zhu, X., et al. (2022). Identification of Inflammation-Related Biomarkers in Diabetes of the Exocrine Pancreas With the Use of Weighted Gene Co-Expression Network Analysis. Front Endocrinol (Lausanne) 13(1664-2392 (Print)), 839865. doi: doi: 10.3389/fendo.2022.839865.
- Antipyretic and anti-asthmatic activities of traditional Chinese herb-pairs, Ephedra and Gypsum. Chin J Integr Med. 2016;22(6):445-450.
- [CrossRef] [Google Scholar]
- Active components in Ephedra sinica stapf disrupt the interaction between ACE2 and SARS-CoV-2 RBD: Potent COVID-19 therapeutic agents. J Ethnopharmacol. 2021;278(114303):5.
- [Google Scholar]
- Evaluation of the Cytotoxic and Apoptogenic Effects of Glabridin and Its Effect on Cytotoxicity and Apoptosis Induced by Doxorubicin Toward Cancerous Cells. Adv Pharm Bull. 2019;9(3):481-489.
- [CrossRef] [Google Scholar]
- Ephedrae herba, a component of Japanese herbal medicine Mao-to, efficiently activates the replication of latent human immunodeficiency virus type 1 (HIV-1) in a monocytic cell line. Biol Pharm Bull. 2008;31(12):2334-2337.
- [CrossRef] [Google Scholar]
- Effectiveness of Paxlovid in Reducing Severe Coronavirus Disease 2019 and Mortality in High-Risk Patients. Clin Infect Dis. 2023;76(3):e342-e349.
- [CrossRef] [Google Scholar]
- Ephedrae herba stimulates hepatocyte growth factor-induced MET endocytosis and downregulation via early/late endocytic pathways in gefitinib-resistant human lung cancer cells. Int J Oncol. 2016;48(5):1895-1906.
- [CrossRef] [Google Scholar]
- Glabridin Alleviates Inflammation and Nociception in Rodents by Activating BK(Ca) Channels and Reducing NO Levels. Biol Pharm Bull. 2020;43(5):884-897.
- [CrossRef] [Google Scholar]
- Antiviral effects of Yinhuapinggan granule against influenza virus infection in the ICR mice model. J Nat Med. 2016;70(1):75-88.
- [CrossRef] [Google Scholar]
- Molecular Docking: Shifting Paradigms in Drug Discovery. Int J Mol Sci. 2019;20(18)
- [CrossRef] [Google Scholar]
- Cytokine Profiles Associated With Acute COVID-19 and Long COVID-19 Syndrome. Front Cell Infect Microbiol. 2022;12:922422
- [CrossRef] [Google Scholar]
- Signaling and Function of Interleukin-2 in T Lymphocytes. Annu Rev Immunol. 2018;36:411-433.
- [CrossRef] [Google Scholar]
- TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13.
- [CrossRef] [Google Scholar]
- Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des. 2010;24(5):417-422.
- [CrossRef] [Google Scholar]
- Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498-2504.
- [CrossRef] [Google Scholar]
- Human immune disorder arising from mutation of the alpha chain of the interleukin-2 receptor. Proc Natl Acad Sci U S A. 1997;94(7):3168-3171.
- [CrossRef] [Google Scholar]
- Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545-15550.
- [CrossRef] [Google Scholar]
- STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607-D613.
- [CrossRef] [Google Scholar]
- Ephedra water decoction and cough tablets containing ephedra and liquorice induce CYP1A2 but not CYP2E1 hepatic enzymes in rats. Xenobiotica. 2016;46(2):141-146.
- [CrossRef] [Google Scholar]
- Tao, W., Xu X Fau - Wang, X., Wang X Fau - Li, B., Li B Fau - Wang, Y., Wang Y Fau - Li, Y., Li Y Fau - Yang, L., et al. (2013). Network pharmacology-based prediction of the active ingredients and potential targets of Chinese herbal Radix Curcumae formula for application to cardiovascular disease. J Ethnopharmacol 145(1), 1-10. doi: doi: 10.1016/j.jep.2012.09.051.
- COVID-19 patients share common, corticosteroid-independent features of impaired host immunity to pathogenic molds. Front Immunol. 2022;13:954985
- [CrossRef] [Google Scholar]
- AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455-461.
- [CrossRef] [Google Scholar]
- UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49(D1):D480-D489.
- [CrossRef] [Google Scholar]
- Epidemiology of Type 1 Diabetes. Curr Cardiol Rep. 2022;24(10):1455-1465.
- [CrossRef] [Google Scholar]
- PubChem BioAssay: 2017 update. Nucleic Acids Res. 2017;45(D1):D955-D963.
- [CrossRef] [Google Scholar]
- Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature. 2022;608(7923):603-608.
- [CrossRef] [Google Scholar]
- PubChem's BioAssay Database. Nucleic Acids Res. 2012;40(Database issue):D400-D412.
- [CrossRef] [Google Scholar]
- Effect and Mechanism of Qingfei Paidu Decoction in the Management of Pulmonary Fibrosis and COVID-19. Am J Chin Med. 2022;50(1):33-51.
- [CrossRef] [Google Scholar]
- A novel chemometric method for the prediction of human oral bioavailability. Int J Mol Sci. 2012;13(6):6964-6982.
- [CrossRef] [Google Scholar]
- Tracking SARS-COV-2 Variants Using Nanopore Sequencing in Ukraine in Summer 2021. Res Sq 2021
- [CrossRef] [Google Scholar]
- Safety and immunogenicity of a recombinant tandem-repeat dimeric RBD-based protein subunit vaccine (ZF2001) against COVID-19 in adults: two randomised, double-blind, placebo-controlled, phase 1 and 2 trials. Lancet Infect Dis. 2021;21(8):1107-1119.
- [CrossRef] [Google Scholar]
- A network pharmacology based approach for predicting active ingredients and potential mechanism of Lianhuaqingwen capsule in treating COVID-19. Int J Med Sci. 2021;18(8):1866-1876.
- [CrossRef] [Google Scholar]
- Ephedrae Herba polysaccharides inhibit the inflammation of ovalbumin induced asthma by regulating Th1/Th2 and Th17/Treg cell immune imbalance. Mol Immunol. 2022;152:14-26.
- [CrossRef] [Google Scholar]
- Neutralisation of ZF2001-elicited antisera to SARS-CoV-2 variants. Lancet Microbe. 2021;2(10):e494.
- [Google Scholar]
- The Nuclear Factor Kappa B (NF-kB) signaling in cancer development and immune diseases. Genes Dis. 2021;8(3):287-297.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2023.105540.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary data 1
Supplementary data 1







