Translate this page into:
Development and molecular modeling of Co(II), Ni(II) and Cu(II) complexes as high acting anti breast cancer agents
⁎Corresponding author. rasayanshg@gmail.com (S.H. Gaikwad)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
A series of cobalt, nickel and copper complexes of bidentate Schiff base derived from the condensation reaction of 4-amino-5-mercapto-3-methyl-1,2,4-triazole with 2-nitrobenzaldehyde had been synthesized. The synthesized Schiff base and their metal complexes have been characterized with the support of more than a few physicochemical techniques, elemental evaluation, magnetic moment measurements, spectroscopic, thermo gravimetric techniques and X-ray powder diffraction. Spectral analysis exhibits square planer geometry for Cu(II) complex while octahedral geometry for Co(II) and Ni(II) complexes. The Schiff base and their complexes have been screened for their anticancer activity using MCF7 cell line. In molecular docking learn exhibits that Ni(II) complex is more active confirmed quantity of interaction in particular hydrogen bond interaction with ASN142 and charge interactions with ASP97 and GLU99.
Keywords
Transition metal complexes
Spectral analysis
Molecular docking
Anticancer
1 Introduction
Cisplatin discovery in 1965 proved a milestone in the drug discovery of metal based anticancer agents. In recent years the metalo-pharmaceuticals attracted attention of the scientific fraternity. Currently various metalo-pharmaceuticals such as bismuth subsalicylate (anti-diarrheal), auranofin (anti-inflammatory for treatment of arthritis), silver sulfadiazine (antibacterial) and zinc pyrithione (antibacterial and antifungal) are in the clinical use. Breast cancer is leading cause of death in women in recent years. Breast cancer is accounting for total 23% of all cancer cases in women, and the death percentage of the patient suffering from this cancer rises to 16% of all cancer deaths. The emergence of the some new types such as triple negative breast cancer, development of new and potent therapeutic agents targeting novel target against breast cancer is need of time. CDK or Cyclin-dependent kinases are the family of the heterodimer kinases which are having critical role in the regulation of cell cycle progression and transcription. Deregulation of these kinases leads to proliferation of cancer cells, and aberrant activity of the number of kinases has been found in variety of the cancers. So inhibition of these kinases will be acting as attractive target for development of new anticancer agents. The chemistry of Schiff bases and their structural analogues have occupied a position of considerable significance (Gornovskii et al., 2009) as they form steady complexes with most transition metal ions (Heshmatpour et al., 2007; Nuria et al., 2005) and exhibit well-established biological properties (Manikshete et al., 2011). 1,2,4-triazole nucleus and their derivatives emerge quickly with the advances of modern heterocyclic chemistry, promising a variety of scientific purposes comparable to antibacterial, antifungal, anticancer, antitumor, anticonvulsant, anti-inflammatory, and analgesic properties (Turan-Zitouni et al., 2005; Walczak et al., 2004; Mavrova et al., 2009; Al-Soud et al., 2003; Almasirad et al., 2004; Amir and Shikha, 2004).
Schiff bases of 1,2,4-triazoles find diverse applications and huge biological activity. The incorporation of the 1,2,4-triazole unit into Schiff base macro cycles is of colossal current curiosity as complexes of 1,2,4-triazoles are being developed for knowledge use in purposes akin to magnetic substances and image chemically pushed molecular instruments (Brandt et al., 2007). Schiff bases derived from 3-substituted 4-amino-5-mercapto-1,2,4-triazoles show analgesic, antimicrobial, anti-inflammatory and antidepressant activities (Bekircan and Bektas, 2006). These motives caused us to carry out gain knowledge of synthesis of Schiff base and its complexes with Co(II), Ni(II) and Cu(II) metal ions.
The intention of present communication is to gain knowledge of bioactivities of 1,2,4-triazole Schiff base and reap the relative derivatives with better curing effect and strengthen bioavailability with the aid of coordinating them with transition metal ions. The synthesized Schiff base and their metal complexes have been characterized with the aid of elemental analysis, magnetic moment measurements, spectroscopic, thermo gravimetric and X-ray powder diffraction approaches. The Current manuscript deals with development of transition metal complexes of 1,2,4-triazoles and their anticancer screening on the MCF 7 cell line. Thus, our study will give new useful insights for designing metallo-pharmaceuticals for anticancer therapy.
2 Experimental
All reagents used, hydrazine hydrate, carbon disulfide, 2-nitrobenzaldehyde, cobalt chloride, nickel chloride and copper chloride were of AR grade. The solvents used were ethanol, ethyl acetate, petroleum ether, dimethyl formamide (DMF) and dimethyl sulfoxide (DMSO). The synthesis of Schiff base ligand and metal complexes is proven in Scheme 1.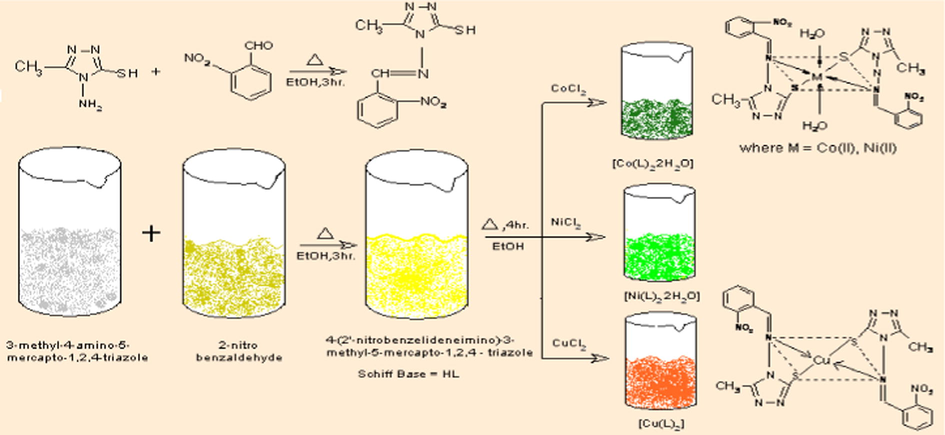
4-Amino-5-mercapto-3-methyl-1,2,4-triazole (AMMT) was prepared by reported literature Method (Bala et al., 1978). The AMMT (1.30 g, 0.01 M) and 2-nitrobenzaldehyde (1.511 g, 0.01 M) were dissolved in ethanol (25 ml) individually in 1:1 molar ratio. The ethanolic solutions were mixed together. The mixture was refluxed on water bath for 3 h. On cooling, a crystalline Schiff base, 4-(2′-nitrobenzylideneimino)-3-methyl-5-mercapto-1,2,4-triazole (HL), used to be separated via filtration and crystals were washed with cold ethanol and recrystallized from ethanol and dried in vacuum over anhydrous CaCl2. The melting point of synthesized compound was found to be 224 °C.
The Schiff base (2.63 g, 0.01 M) was dissolved in 25 ml ethanol and added to metal salt cobalt chloride (1.189 g, 0.005 M), Nickel chloride (1.188 g, 0.005 M) and copper chloride (0.852 g, 0.005 M)]. The mixture was refluxed for 4 h. The products formed were filtered and purified by washing thoroughly with absolute ethanol and finally with acetone and dried in vacuum over anhydrous CaCl2.
The sulphorhodamine B (SRB) evaluates were used for cell density determination, based on the measurement of cellular protein content. The process described right here was once optimized for the toxicity screening of compounds to adherent cells in a 96 well format. After an incubation period, cell monolayers were fixed with 10% (w/v) trichloroacetic acid and stained for 30 min, after which the surplus dye was once removed with the aid of washing repeatedly with 1% (v/v) acetic acid. The protein-bound dye is dissolved in 10 mM tris base solution for OD resolution at 510 nm utilizing a micro plate reader. The results had been linear over a 20-fold variety of cell numbers and the sensitivity was comparable to those of fluorometric approaches.
2.1 Molecular docking
To ascertain the mode of action of the synthesized metal complexes molecular docking calculations are carried out using a biopredicta module of the V life MDS 4.3. Virtual analysis was carried out using the crystal structure of the Human CDK 7 (PDB ID: IUA2) downloaded from Protein Data Bank (www.rcsb.org/pdb) at a resolution of 3.02 Å. Protein structure was refined via deletion of all the heteroatoms including water molecules and addition of the polar hydrogen atoms to get a native conformation. All other bonds were allowed to be rotatable. Structures of the synthesized metal complexes are drawn in the builder module of the V life MDS 4.3 engine. The 2D structures of the molecules were converted into the 3D structures are optimized via application of the MMFF force field. These optimized structures were further utilized for the docking analysis. All calculations were performed on an Intel i3, based machine running windows 7 as operating system. The docked protein ligand complex was further analyzed via docking score of the each of the complex, which is nothing but the binding energy of the complex, for every derivative best 100 binding conformation was analyzed to select the best conformation having the minimum energy of the binding.
3 Results and discussion
3.1 Elemental and physical data
The Schiff base used to be soluble in ethanol, methanol, acetone, DMF and DMSO. The entire metal complexes are colored, non-hygroscopic solid which might be steady in air, insoluble in water and organic solvents but soluble in DMSO and decomposed at higher temperature. The purity of ligands and their metal complexes has been checked with the aid of TLC. Based on elemental and spectral studies the geometry of synthesized compounds has been elucidated. By the elemental analysis, the stoichiometry of ligand and their metal complexes is confirmed. The elemental analyses of ligand and metal complexes are found in agreement with the proposed structure of ligand and the metal complexes. Analytical and physical data of the Schiff base(HL) and Co(II), Ni(II) and Cu(II) complexes are listed in Table 1.
Comp.
Molecular formula
Mol. Wt.
Color
Elemental analysis
C% found (calc.)
H% found (calc.)
N% found (calc.)
M% found (calc.)
HL
C10H9N5SO2
263
Pale yellow
45.20 (45.63)
3.52 (3.42)
26.48 (26.61)
–
HL-Co
C20H20N10S2O6Co [Co(L)22H2O]
618.93
Olive green
38.84 (38.78)
3.16 (3.23)
22.71 (22.62)
9.42 (9.52)
HL-Ni
C20H20N10S2O6Ni [Ni(L)22H2O]
618.91
Green
38.71 (38.78)
3.30 (3.23)
22.58 (22.62)
9.59 (9.52)
HL-Cu
C20H16N10S2O4Cu [Cu(L)2]
587.54
Brick red
40.93 (40.85)
2.81 (2.72)
23.89 (23.83)
10.93 (10.81)
3.2 Electronic spectral analysis and magnetic studies
The electronic spectra and magnetic moments of the compounds are very useful in the evaluation of results acquired by using different ways of structural investigation. The information regarding the geometry of the complexes around the Co(II), Ni(II) and Cu(II) ion was obtained from electronic spectral studies and magnetic moments. The electronic spectra of ligand and their metal complexes were recorded at room temperature utilizing DMSO as a solvent.
The electronic spectra of ligand exhibit band at 29,585 cm−1 which will also be assigned to n → π∗ transition of azomethine group. Within the spectra of complexes, the bands of azomethine chromophore because of n → π∗ transition are shifted to lower frequencies indicate that imine nitrogen is concerned in the coordination of metal ion.
Co(II) complex exhibits two absorption bands at 10,810 cm−1 (γ1) and 21,276 cm−1 (γ3) which might be assigned to 4T1g(F) → 4T2g (F) (γ1); 4T1g(F) → 4T1g (P) (γ3) transitions (Avji et al., 2008; Kulkarni et al., 2011). These are attribute bands of high spin octahedral Co(II) complexes; γ2 is not observed, but it may be calculated by using relation γ2 = γ1 + 10Dq, which may be very nearly (γ3) transition. These transitions propose a octahedral environment around Co(II) ion which was once extra supported by using its magnetic moment value of (μeff) 4.89BM.
Ni(II) complex generally shows three absorption bands in octahedral environment corresponding to 3A2g (F) → 3T2g (F) (γ1), 3A2g (F) → 3T1g (F) (γ2), and 3A2g (F) → 3T1g (P) (γ3) transitions (Cotton et al., 2003). Ni (II) complex exhibits above three transitions at 9891 cm−1 (γ1), 16,129 cm−1 (γ2), and 24,992 cm−1 (γ3) suggesting octahedral geometry which is further proven through its magnetic moment value of (μeff) 3.45 BM.
The electronic spectra of Cu(II) complex show large band at 17,993 cm−1 which could also be cheap be assigned to 2B1g → 2A1g (γ1) transition, consistent with a square planar geometry around Cu(II) (Osman et al., 2004) which is further verified with the aid of its magnetic of moment value (μeff) 1.92 BM. The ligand field parameters like crystal field stabilizing energy (Dq), Racah parameter (B), Nephelauxetic ratio (β) and β% have been calculated for Co(II), Ni(II), Cu(II) complex and observed data are shown in Table 2.
Compd.
γ1 cm−1
γ2 cm−1
γ3 cm−1
Dq cm−1
B cm−1
γ2/γ1 cm−1
β cm−1
β %
μeff. BM
HL-Co
10,810
22,703⁎
21,276
1189.3
770
2.10
0.792
20.70
4.89
HL-Ni
9891
16,129
24,992
989.1
788.16
1.63
0.757
24.29
3.45
HL-Cu
17,993
–
–
1799.3
–
–
–
–
1.92
3.3 IR and 1H NMR spectroscopic studies
The prominent bands observed in IR spectra of Schiff base(HL) and its complexes are represented in Table 3. The IR spectrum of free ligands shows a strong band at 1590 cm−1 assigned to ν(N⚌CH) indicating the formation desired shift base ligand. This used to be further proven by the presence of a new band observed in 483–515 cm−1 region assigned to ν(M—N), which is coordination of metal to azomethine nitrogen. The Schiff base (HL) (Fig. 1) a band at 2760 cm−1 is assigned to ν(SH) vibration (Singh et al., 2006a,b). This band disappeared in the spectra of the metal complexes indicating deprotonation and complexation via sulfur. In the spectra of Schiff base a band due to tautomeric form of C⚌S is appeared at 1114 cm−1, in metal complexes this peak was disappeared.
Sr. no.
Compound
νC⚌N cm−1
νSH cm−1
νC⚌S cm−1
ν(C—-S) cm−1
νH2O cm−1
νM—S cm−1
νM—N cm−1
1
HL
1590
2760
1114
–
–
–
–
2
HL-Co
1525
–
–
718
3395
355
515
3
HL-Ni
1525
–
–
785
3206
379
483
4
HL-Cu
1567
–
–
750
–
350
493
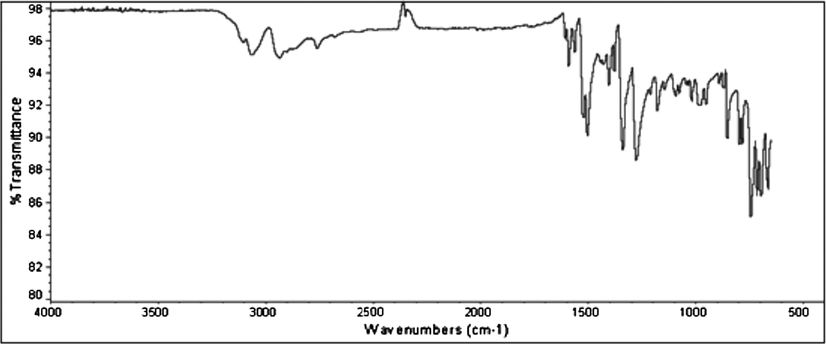
IR spectra of Schiff base (HL).
Co(II), Ni(II) and Cu(II) complexes (Fig. 2a–c) show band at 350–379 cm−1 have been assigned to ν(M—S) (Liver, 1968). Within the spectra of all complexes, bands appeared at 718–785 cm−1 and 350–379 cm−1 were assigned to ν(C—S) and ν(M—S), respectively (Osman et al., 2004; Singh et al., 2006a,b). In the spectra of Co(II) and Ni(II) complexes, a broad band in the region 3206–3395 cm−1 indicated the presence of coordinated water molecules. The presence of water molecule was also confirmed by thermal analysis.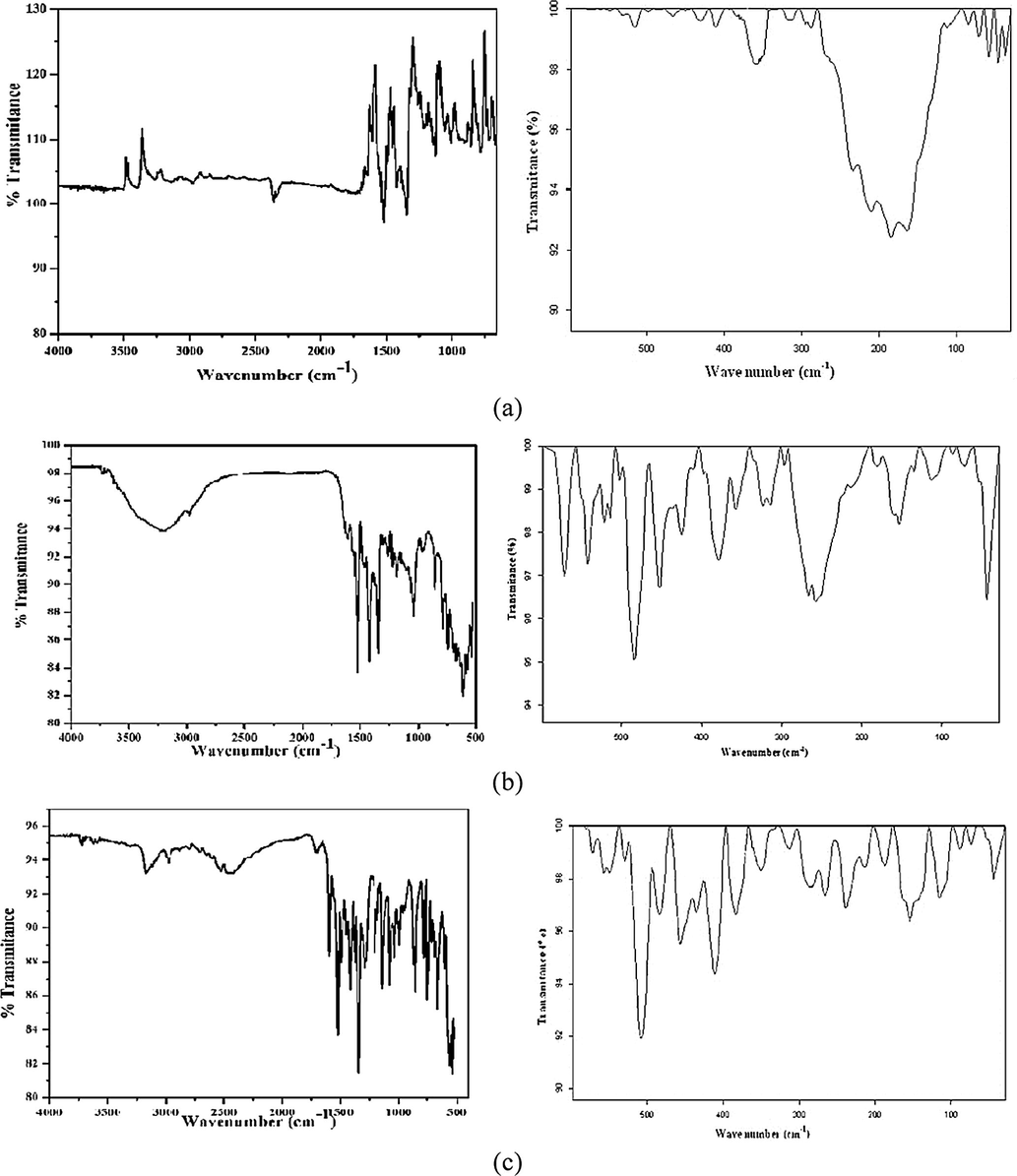
(a–c): IR spectra of Co(II), Ni(II) and Cu(II) complexes.
The 1H NMR spectrum of Schiff base was recorded in DMSO, using Tetramethylsilane (TMS) as an internal standard. The 1H NMR spectrum of ligand shows characteristic azomethine proton singlet at δ 11.07 ppm. The signal at δ 13.27 ppm is ascribed to SH proton. The aromatic proton of Schiff base appeared as multiplate at δ 7.27–7.99 ppm and singlet at δ 2.28 ppm is due to methyl group. Since Co(II), Ni(II) and Cu(II) complexes are paramagnetic, their 1H spectra could not be obtained (Canpolat and Kaya, 2005). 1H NMR spectrum of Schiff base(HL) is indicated in Fig. 3.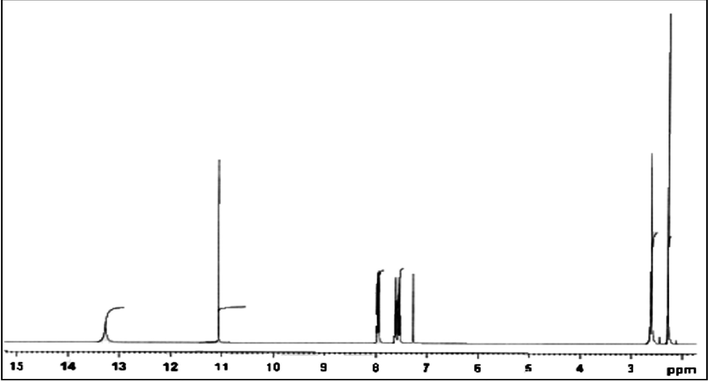
1H NMR spectra of Schiff base(HL).
3.4 Thermal analysis
Thermal decomposition of Co(II), Ni(II) and Cu(II) complexes has been studied as a function of temperature by means of TGA-DTA method. The decomposition temperatures, paralyzed products, percentage mass losses of the complexes and ash percentage are listed in Table 4. The Co(II) and Ni(II) complexes exhibit three steps of decomposition. In first step, decomposition takes place from 50 to 180 °C due to total cleavage of the base metal complex along with hydroxide to oxide transformation followed by concomitant release of water molecules corresponding to loss of coordinated water molecules. The second decomposition takes place from 175 to 425 °C attributed to the release of organic moiety. Third decomposition around 400–550 °C can be attributed to the release of triazole moiety. The decomposition of both the complexes ended with oxide formation above 550 °C. For Cu(II) complex, first step of decomposition at 50–425 °C corresponds to loss of organic moiety while second step corresponds to removal of triazole molecules at 425–750 °C. After 750 °C metal oxide is formed as a residue. Thermal curve analysis of Co(II) complex is shown in Fig. 4.
Compound.
Step
Temp. (°C)
TG mass%
No. of moles
Assignment
Calc.
Found
[Co(L)22H2O]
1
50–175
5.81
6.74
2
—H4O2(water molecules)
2
175–425
48.14
46.37
2
—C14H10N4O4(organic moiety)
3
425–540
36.51
34.34
2
—C6H6N6S2(triazole ring)
12.10
12.57
—CoO(residue)
[Ni(L)22H2O]
1
50–180
5.81
6.92
2
—H4O2(water molecules)
2
180–400
48.14
46.11
2
—C14H10N4O4(organic moiety)
3
400–550
36.51
34.09
2
—C6H6N6S2(triazole ring)
12.10
12.94
—NiO(residue)
[Cu(L)2]
1
50–425
45.95
44.46
2
—C14H10N4O4(organic moiety)
2
425–750
43.22
42.84
2
—C6H6N6S2(triazole ring)
12.10
12.69
—CuO(residue)
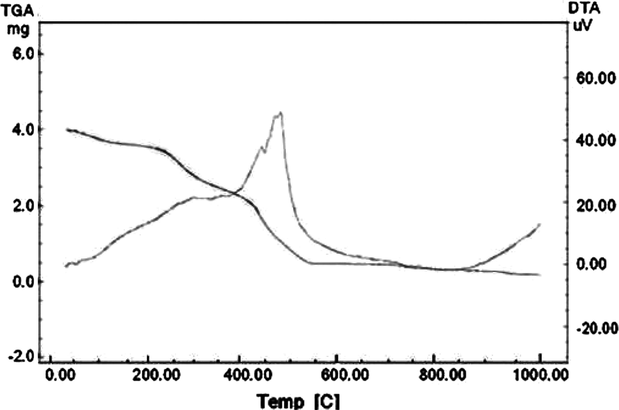
TGA-DTA curve of Co(II) complex.
3.5 Mass spectroscopic studies
The mass spectrum of ligand reveals parent peak due to molecular ion (M+). The proposed molecular formulation of this compound used to be proven through comparing their molecular formula weight with m/z value. The molecular ion peak used to be acquired at m/z 264. This worth is in excellent agreement with the proposed molecular components of compound. A mass spectrum of Schiff base (HL) is exhibited in Fig. 5.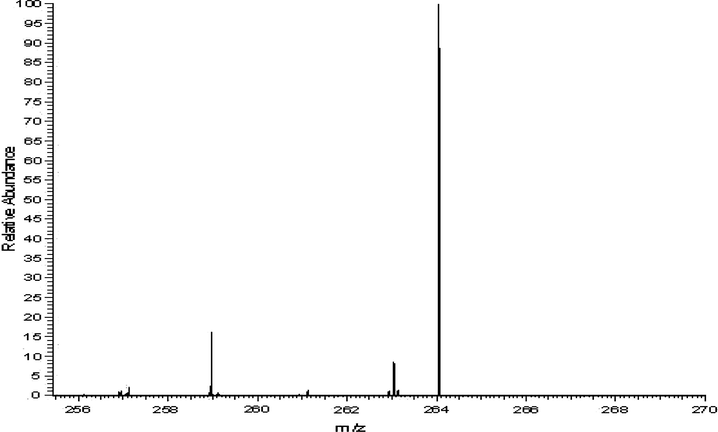
Mass spectrum of Schiff base (HL).
3.6 X-ray diffraction analysis
The X-ray diffraction measurements were obtained using a X-ray diffractometer (Rigaku Ultima IV, Japan made). The Cu Kα radiation tube with line focus was operated at 40 kV and 40 mA. The X-ray powder diffraction patterns were obtained in the range of 20–80° in steps of 0.0990. X-ray powder diffraction analysis of the ligand and its metal complexes was carried out to determine the type of crystal system, lattice parameters and the cell volume. As shown in Fig. 6(a) and (b) the XRD patterns indicate a crystalline nature for the ligand and Cu(II) complex, while Ni(II) and Co(II) complexes have amorphous nature. Indexing of the diffraction patterns was performed using High Score Plus software. For the ligand and Cu(II) complex d values, FWHM and relative intensities are given in Tables 5 and 6. From the indexed data the unit cell parameters, volume, space group and average crystallite size were also calculated and are listed in Table 7. The powder XRD patterns of the Cu(II) complex are completely different from those of ligand. XRD studies reveal that ligand has an orthorhombic structure, while Cu(II) complex has tetragonal structure. The full width at the half-maximum (FWHM) of the diffraction peaks obtained from the refinement was used to calculate the particle size. The X-ray diffraction data were determined by using Cu Kα radiation (λ = 1.54060 Å). The average size of the samples was calculated with the help of the Debye-Scherrer equation,
where λ is the wavelength (Cu Kα), β is the full width at the half maximum (FWHM) and θ is the diffraction angle. The diffraction peaks indicate that the synthesized materials are in the nanometer range.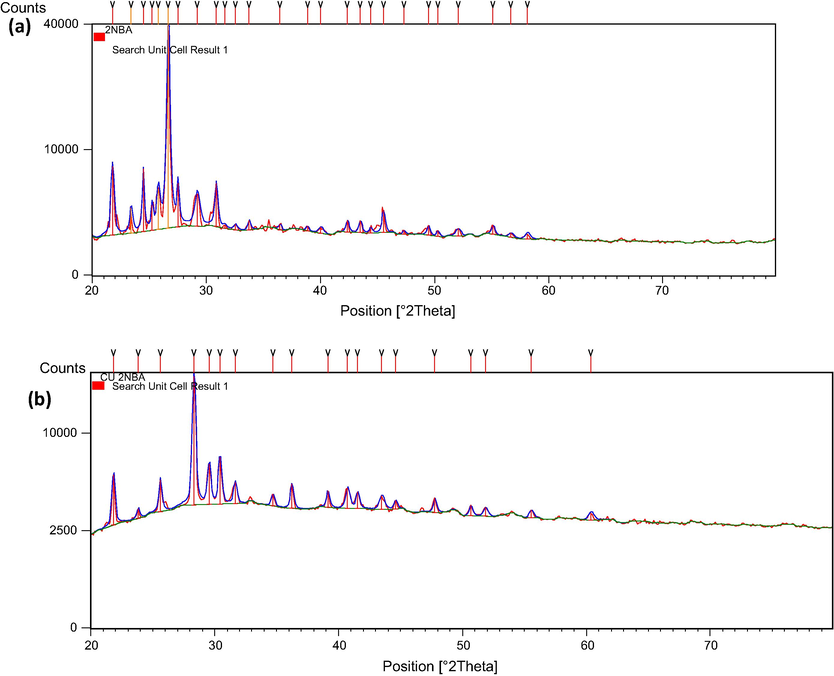
XRD pattern of (a) ligand and (b) Cu(II) complexes.
Pos. [°2Th.]
Height [cts]
FWHM [°2Th.]
d-spacing [Å]
Rel. Int. [%]
21.7677
6477.85
0.2922
4.08292
16.87
23.4049
1732.92
0.2922
3.80089
4.51
24.4578
5927.24
0.1948
3.63961
15.44
25.2393
1805.91
0.1948
3.52865
4.70
25.7877
3311.21
0.2922
3.45484
8.62
26.6576
38391.45
0.2922
3.34404
100.00
27.5059
3839.91
0.1948
3.24280
10.00
29.1995
2666.54
0.4871
3.05847
6.95
30.8622
3803.80
0.2922
2.89737
9.91
31.6022
186.68
0.2922
2.83120
0.49
32.5568
302.31
0.3897
2.75034
0.79
33.7386
576.15
0.2922
2.65665
1.50
36.4505
341.38
0.2922
2.46498
0.89
38.8494
258.24
0.2922
2.31812
0.67
40.0260
335.37
0.4871
2.25265
0.87
42.3468
700.98
0.2922
2.13441
1.83
43.4795
733.64
0.2922
2.08140
1.91
44.3787
282.75
0.2922
2.04129
0.74
45.5015
1526.86
0.2922
1.99350
3.98
47.2986
207.64
0.2922
1.92186
0.54
49.4470
535.30
0.2922
1.84328
1.39
50.2646
286.13
0.2922
1.81520
0.75
52.0517
385.04
0.4871
1.75700
1.00
55.1115
524.00
0.2922
1.66648
1.36
56.6904
200.42
0.4871
1.62376
0.52
58.1296
234.93
0.4752
1.58564
0.61
Pos. [°2Th.]
Height [cts]
FWHM [°2Th.]
d-spacing [Å]
Rel. Int. [%]
21.8271
3375.11
0.2922
4.07195
26.67
23.8238
577.05
0.2922
3.73499
4.56
25.6116
2163.59
0.2922
3.47820
17.09
28.3234
12657.35
0.2922
3.15104
100.00
29.5388
3022.92
0.2922
3.02410
23.88
30.4227
3626.43
0.2922
2.93823
28.65
31.6520
1500.20
0.2922
2.82686
11.85
34.6912
762.07
0.2922
2.58585
6.02
36.2253
1666.66
0.2922
2.47979
13.17
39.1207
1117.85
0.2922
2.30266
8.83
40.7282
1366.82
0.3897
2.21542
10.80
41.5311
1083.03
0.2922
2.17443
8.56
43.4820
876.44
0.4871
2.08128
6.92
44.5875
551.90
0.2922
2.03222
4.36
47.7587
940.21
0.2922
1.90442
7.43
50.6635
654.54
0.2922
1.80184
5.17
51.8466
532.77
0.3897
1.76347
4.21
55.5373
465.59
0.3897
1.65471
3.68
60.3664
362.06
0.4752
1.53212
2.86
Compound
Lattice constants (Å)
Volume, (Å)3
Space group
Crystallite size, (Å)
HL
a = 16.61, b = 8.16, c = 3.91
529.96
I m a m
337
HL-Cu
a = 4.90, b = 4.90, c = 21.1,
506.90
I 41/a c d
182
3.7 Biological activity studies
All the newly synthesized compounds were screened for their anticancer activity using SRB assay on the MCF 7 breast cancer cell line. In the current protocol each cell line is inoculated on a pre-incubated microtiter plate. The test agents are added at a single concentration and the culture is incubated for 48 h. End point determinations are made with Sulforhodamine B, a protein binding dye. The results for each test agents are reported as the percentage growth of the tested cells. The compounds that reduce the growth of any one of the cell lines to 32% or less (negative numbers indicates cell kill) are passed on for evolution over a 5-log dose range. In the present screening program all the compounds possessed growth to less than 32% are regarded as active compounds (Holla et al., 2003, 2006). Adriamycin was served as positive control compound in the cytotoxic assay. The observed results are listed in Table 8.
Sr. no.
Molecular code
GI50
1
HL
64.1
2
HL-Co
35.7
3
HL-Ni
59.9
4
HL-Cu
54.9
3.8 Structure activity relationship (SAR studies)
In Fig. 7 of Co(II), Ni(II) and Cu(II) complexes are found to be active against MCF 7 cell line up to molar dose of 10−4 M which indicates their anticancer potential. SAR analysis of the synthesized complexes indicated following points,
-
1,2,4-Triazole shows moderate activity against MCF −7 cell line. HL shows GI50 value, 64.1 which indicates the substitution of the good electron withdrawing group —NO2 sufficiently affecting the polarity of the molecules.
-
Cobalt metal complex showing profound activity than the corresponding nickel and copper complexes. Chemical properties of copper make it to take part in number of biological process such as electron transfer and catalysis. Cobalt complex has ability to kill cancer cells are correlated with the induction of the oxidative stress. Cobalt is an important trace elements for humans in the form of vitamin B12 (Cobalamin), and this metal plays a vital role in Protein synthesis.
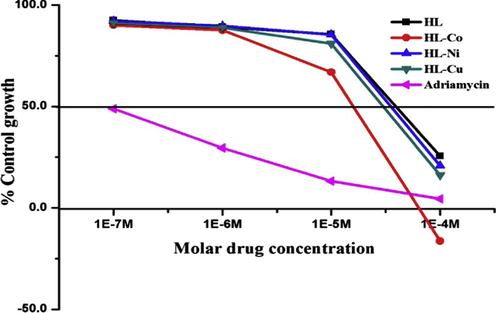
- Growth curve: human breast cancer cell line MCF-7.
3.9 Molecular docking studies
In order to understand the possible mode of action of the synthesized metal complex the molecular docking analysis was carried out using crystal structure of the Human CDK 7. Optimized structure of the metal complexes was utilized for the grip based docking simulation was protein structure is kept rigid and ligand structure is kept flexible. All the molecules were docked into the similar binding site of CDK 7 with binding energy ranging from −33.57 kcal/mol to 96.36 kcal/mol. Nickel complex is most active molecule having binding score of −84.54 kcal/mol having critical interaction with amino acids such as ASN142 (Hydrogen Bond interaction), GLU99 and ASP97 (Charge Interaction) and hydrophobic interaction with GLY21 and GLU99. Hydrogen Bond Interactions and Charge Interactions of Schiff base (HL) and Co(II) Complex are shown in Fig. 8(a) and (b) respectively. Binding energy and critical interaction of all the compounds are given in Table 9. Correlation of binding energy and GI50 of derivatives of Schiff base and metal complexes are shown in Fig. 9.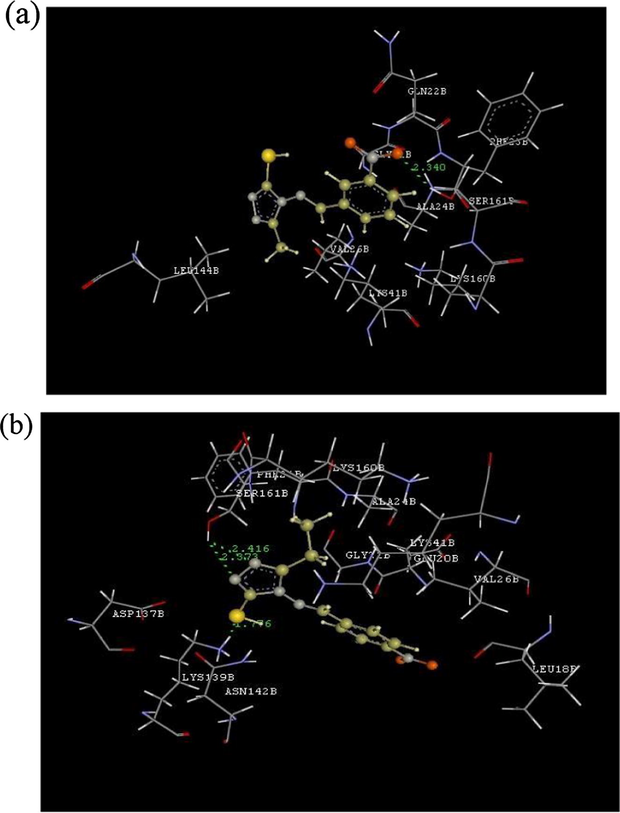
(a): Hydrogen bond interactions and charge interactions of HL; (b): hydrogen bond interactions and charge interactions of HL-Co.
Sr. no.
Molecule code
Binding energy kcal/mol
Interaction with amino acid
H-Bond
Charge
Hydrophobic
1
HL
−40.53
GLN22
–
LYS41 VAL26
2
HL-Co
−48.98
–
3
HL-Ni
−84.54
ASN142
GLU99, ASP97
GLY21, GLU99
4
HL-Cu
−69.36
ASN142
–
LYS41 ALA39
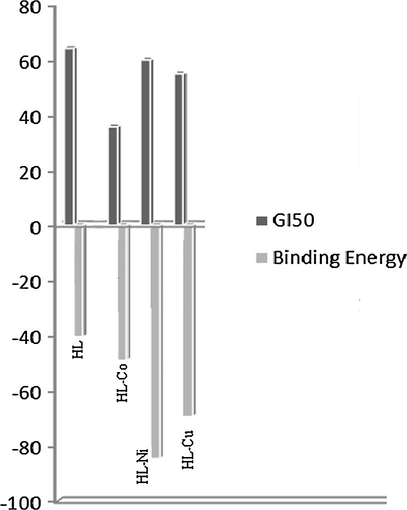
Correlation of binding energy and GI50 of HL and its complexes.
4 Conclusion
The synthesized Schiff base acts as bidentate ligands and is coordinated with the metal ion via nitrogen and sulfur. The bonding of ligand to metal ion is validated through elemental evaluation, spectral reviews (electronic, IR, 1H NMR, mass), TGA and magnetic measurement. The Co(II) and Ni(II) complexes exhibit octahedral and Cu(II) tricky displays square planar geometry. The entire compounds show off anticancer endeavor against MCF-7 cell line. Metal complexes are more potent in comparison with free ligand. The Co(II) complex indicates quality anticancer exercise.
Acknowledgment
The authors gratefully acknowledge the ACTREC, Tata memorial centre, Kharghar, New Mumbai, for providing anticancer activity. We also acknowledge Dr. P.P. Wadgaonkar, NCL, Pune, India, for their support and suggestions. We are thankful to Dr, H. N. More Principal Bharati Vidyapeeth College of Pharmacy for computational work.
References
- Synthesis and anticonvulsant activity of new 2- substituted-5-[2-(2-fluorophenoxy) phenyl]-1,3,4-oxadiazoles and 1,2,4-Triazoles. Bioinorg. Med. Chem. Lett.. 2004;14(24):6057-6059.
- [Google Scholar]
- Synthesis and properties of new substituted 1,2,4-triazoles: potent antitumor agents. Bioinorg. Med. Chem.. 2003;11(8):1701-1708.
- [Google Scholar]
- Synthesis and anti-inflammatory, analgesic, ulcerogenic and lipid peroxydation activities of some new 2-[(2,6-dichloroanilino) phenyl] acetic acid derivatives. Eur. J. Med. Chem.. 2004;39(6):535-545.
- [Google Scholar]
- Synthesis, spectral, thermal, solid state d.c. electrical conductivity and biological studies of Co (II), Ni (II) and Cu (II) complexes with 3-substituted-4-amino (indole-3-aldehydo)-5-mercapto-1,2,4-triazole Schiff bases. J. Coord. Chem.. 2008;61(12):1884-1896.
- [Google Scholar]
- Heterocyclic systems containing bridgehead nitrogen atom: part XXXIII – synthesis of s-Triazolo[3,4-b][1,3,4] thiadiazine, s-triazolo-[3,4-b][1,3,4] thiadiazino[6,7-b] quinoxaline and as- triazino-[3,4-b][1,3,4] thiadiazines. Ind. J. Chem.. 1978;16:481-483.
- [Google Scholar]
- Synthesis and structure of 3, 5-disubstituted 1,2,4-triazol head units and incorporation of 3,5-dibenzoyl-1,2,4-triazolate into new [2+2] Schiff base macrocyclic complexes. Supramolecular Chem.. 2007;19(1–2):17-27.
- [Google Scholar]
- Studies on mononuclear chelates derived from substituted schiff base ligands (Part 4): Synthesis and characterization of a new 5-hydroxysalicyliden-P-aminoacetophenoneoxime and its complexes with Co(II), Ni(II), Cu(II) and Zn(II) Turk. J. Chem.. 2005;29:409-415.
- [Google Scholar]
- Advanced Inorganic Chemistry (sixth ed.). New York: Wiley; 2003.
- Molecular design of mononuclear complexes of acyclic Schiff base ligands. J. Coord. Chem.. 2009;62(2):151-204.
- [Google Scholar]
- Synthesis, characterization and molecular structure of a new tetrameric palladium(II) complex containing Schiff-bases derived from AMTTO (AMTTO=4-amino-6-methyl-1,2,4-triazinethione-5-one) Z. Anorg. Allg. Chem.. 2007;633(8):1178-1182.
- [Google Scholar]
- Synthesis, characterization and anticancer activity studies on some Mannich bases derived from 1,2,4-triazoles. Eur. J. Med. Chem.. 2003;38:759-767.
- [Google Scholar]
- Synthesis and studies on some new fluorine containing triazolothiadiazines as possible antibacterial, antifungal and anticancer agents. Eur. J. Med. Chem.. 2006;41:657-663.
- [Google Scholar]
- DNA clevage and antimicrobial investigation of Co (II), Ni (II) and Cu (II) complexes with triazole Schiff base: synthesis and spectral characterization. Med. Chem. Res.. 2011;20(3):346-354.
- [Google Scholar]
- Inorganic Spectroscopy. Amsterdam: Elsevier; 1968.
- Synthesis, characterization and antimicrobial activity of new cobalt (II), nickel (II) and copper (II) complexes with 2-(2-hydroxy-1,2- phenylethylideneamino) benzoic acid. Inorg. Chem. Comm.. 2011;14:618-622.
- [Google Scholar]
- Synthesis, cytotoxicity and effects of some 1,2,4-triazole and 1,3,4 thiadiazole derivatives on immunocompetent cells. Eur. J. Med. Chem.. 2009;44(1):63-69.
- [Google Scholar]
- Synthesis, structure and biological activity of new and efficient Cd (II)-uracil derivative complex system for cleavage of DNA. J. Biological Inorg. Chem.. 2005;10(8):924-934.
- [Google Scholar]
- Synthesis, characterization and photochemical studies of some of Schiff bases derived from 3-hydrazino-6-methyl[1,2,4]triazin-5(4H) one. Synth. React. Inorg. Met. – Org. Chem.. 2004;34(6):1069-1085.
- [Google Scholar]
- Antibacterial Co(II), Ni(II), Cu(II) and Zn(II) complexes with of Schiff bases derived from flurobenzaldehyde and triazoles. J. Enzyme Inhibit. Med. Chem.. 2006;21(5):557-562.
- [Google Scholar]
- Some bivalent metal complexes of Schiff bases containing N and S donor atom. J. Enzyme Inhibit. Med. Chem.. 2006;21(6):749-755.
- [Google Scholar]
- Synthesis and antimicrobial activity of 4-phenyl/cyclohexyl-5-(1-phenoxyethyl)-3-[N-(2-thiazolyl)acetamido]thio-4H-1,2,4-triazole derivatives. Eur. J. Med. Chem.. 2005;40(6):607-613.
- [Google Scholar]
- Synthesis and anti-tuberculosis activity of N-aryl-C-nitroazoles. Eur. J. Med. Chem.. 2004;39(10):849-853.
- [Google Scholar]







