Translate this page into:
Exploring the interaction of calycosin with cyclin D1 protein as a regulator of cell cycle progression in lung cancer cells
⁎Corresponding author at: No. 44 Xiaoheyan Road, Dadong District, Shenyang 110042, Liaoning Province, PR China. w14901@yahoo.com (Wei Wang)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Cyclin D1 has been shown to play a pivotal role in the proliferation of lung cancer cells through regulation of cell cycle progression. Therefore, targeting this protein can be used as a potential strategy in lung cancer treatment. Calycosin has been reported to show potential anticancer effects, however, its possible anticancer mechanisms remain unclear. Therefore, in this study we aimed to explore the interaction of cyclin D1 and calycosin to determine the binding properties and probable structural changes of cyclin D1. We carried out in-depth experimental and computational binding assays of calycosin with cyclin D1 under simulated physiological environment, using intrinsic, extrinsic, synchronous fluorescence, circular dichroism, and differential scanning calorimetry (DSC) analysis. The results showed a spontaneous static mechanism driven from hydrogen bonding and van der Waals forces between hydrophilic residues of cyclin D1 with hydroxyl groups of calycosin. We determined that calycosin led to secondary and tertiary structural changes of cyclin D1 through exposure of hydrophobic residues. Also, it was determined that calycosin resulted in an apparent decrease in the heat capacity changes (ΔCp) and midpoint of unfolding transition (Tm) values of cyclin D1. Cellular studies also indicated that calycosin caused the inhibition of lung cancer cell proliferation through cell cycle arrest at G1 phase, which may be due to denaturation of cyclin D1, although it needs further investigation in the future studies. In general, this study may provide useful preliminary data about the development of calycosin-based anticancer platforms.
Keywords
Cyclin D1
Calycosin
Interaction
Spectroscopy
Lung cancer
1 Introduction
Cancer is one of the leading causes of death in the world. What makes cancer a global health problem today is the growing number of people with the disease (Sung et al., 2021). Among different types of cancers, lung cancer is one of the most common cancers in men, accounting for about 18% of all cancer-related deaths (Doll and Hill, 1956). Among the possible causes of lung cancer, the most worrying factors are smoking, environment pollution, and alcohol consumption (Nichols et al., 2012). In fact, cell proliferation can be one of the chronic inflammatory responses of carcinogens. Cell proliferation is considered to be one of the main symptoms of cancers. Genetic aberrations in the cell regulatory cycle through the G1 phase of the cell cycle are often determined in human cancers (Wikman and Kettunen, 2006). Cyclin D1 as a protein encoded in humans by the CCND1 gene located on the long arm of chromosome 11 (band 11q13) (Wilkerson and Reis-Filho, 2013; Ramos-García et al., 2017).
In fact, overexpression of cyclin D1 as a controller of the G1 phase of the cell cycle has been shown to have a significant portion of tumors and human cancers, mostly due to its induction through oncogenic signals (Ramos-García et al., 2019). Overexpression of this gene may act as an oncogenic stimulus by increasing the transition from G1 to S phase of the cell cycle and increasing the regulatory function of the cell cycle (Montalto and De Amicis, 2020). Overexpression of cyclin D1 is a key molecular change in different cancers and predicts prognostic and clinicopathological significance (Ramos-Garcia et al., 2018).
Indeed, significance of cyclin D1 expression in different cancers including renal cell carcinoma (Li et al., 2020), breast cancer (He et al., 2017), colo-rectal cancer (Li et al., 2014), bladder cancer (Ren et al., 2014); melanoma (González-Ruiz et al., 2021), and lung cancer (Li et al., 2012) has been reported as a potential marker. Cyclin as D1 is a protein that is encoded by a gene belonging to the cyclin family and its function is in the regulation of cell cycle (Diehl, 2002). Cyclins have two domains of an identical all-α helix located at the N- and the C-terminus (Day et al., 2009). Cyclins act as regulators of CDKs (cyclin-dependent kinases) and are involved in mitotic events (Day et al., 2009). Of the three types of cyclin D that each attach to CDK, overexpression of cyclin D1 is mainly associated with tumorigenesis and cell metastasis (Zhao et al., 2017). In addition to regulating CDK in the cell cycle, cyclin D1 is also involved in the regulation of transcription factors, cell cycle transport, tumor regeneration proteins, and cellular metabolism (Huber et al., 2021; Tchakarska and Sola, 2020).
Therefore, any compound that can bind this protein and results in its denaturation and subsequence deactivation can be used as a potential strategy in the development of anticancer platforms against several cancers including lung, prostate, breast, etc.
In fact, some factors contribute to reducing the side effects of carcinogens, including the use of antioxidant and anti-inflammatory supplements which may prevent cell cycle progression and on the other side result in the apoptosis induction (Coccia et al., 2016; Shi et al., 2015).
Calycosin (C16H12O5), 7-hydroxyisoflavones, as the main metabolite in the dry root extract of Radix astragali, has shown potential anticancer effects (Gao et al., 2014; Deng et al., 2021). It has been also shown that calycosin can trigger its anticancer effects through regulation of different signaling pathways (Liu et al., 2021). However, the interaction of small molecules like calycosin and cyclin D1 as one of the main proteins in cancer progression through cell cycle regulation has not been well explored. Indeed, if calycosin can bind this protein and denature its structure, then one of the possible mechanisms by which calycosin can induce its anticancer effect is mediated through denaturation of main proteins involved in the cancer progression. Therefore, in this study, the interaction of calycosin with cyclin D1 was explored by different experimental and theoretical analysis. Also, cell cycle assay on lung cancer cells was performed to assess the cell cycle arrest ability of calycosin.
2 Material and methods
2.1 Materials
Cyclin D1 (CAT Nr: SRP5178), Nile red, calycosin ≥ 98% (HPLC) (CAT Nr:20575-57-9), and cell culture reagents were purchased from Sigma. Co. (Shanghai, China). All other materials were obtained from Merck Co. (Shanghai, China) and were of analytical grade.
2.2 Preparation of protein and calycosin samples
The protein was dissolved in 50 mM Tris-Hcl buffer, pH 7.4, 150 mM Nacl, 10 mM glutathione. Calycosin was dissolved in the minimum amount of DMSO solution and prepared to the concentration of 2 mg/mL solution with 50 mM Tris-Hcl buffer, pH 7.4, 150 mM Nacl.
2.3 Fluorescence study
The intrinsic fluorescence spectra of cyclin D1 (5 µM) with or without different concentrations of calycosin in the range of 1–100 nM were read using a fluorescence instrument (Cary-100, Australia). The λex and λem were set to 280 nm and 300–430 nm, respectively at a slit width of 5 nm for both λ. Furthermore, the synchronous fluorescence spectroscopy was carried out to study the microenvironmental changes around tryptophan (Trp) and tyrosine (Tyr) residues at Δλ = 60 nm and Δλ = 15 nm, respectively. The experimental procedure and concentrations were similar as described above. The fluorescence intensity of protein was corrected against buffer, calycosin and inner filter effect. Moreover, the change in the surface hydrophobicity of protein was determined using Nile red dye. The cyclin D1 sample (5 µM) was incubated with Nile red (50 μM) and added by different concentrations of calycosin in the range of 1–100 nM. The Nile fluorescence intensity was then read between 550 and 800 nm while the samples were excited at 550 nm.
2.4 Circular dichroism (CD) spectroscopic analysis
Far-ultraviolet (UV)-CD (195–260 nm) spectra were determined with a JASCO spectropolarimeter (J715, USA). The cyclin D1 concentrations were 10 µM and the protein samples were added by different concentrations of calycosin in the range of 1–100 nM. The CD signals were corrected against buffer and calycosin signals and the experiment was done at room temperature. The secondary structural changes were qualified using the DICHROWEB server with the CONTIN algorithm.
2.5 Differential scanning calorimetry (DSC)
DSC analysis was performed using a calorimeter II (Model 6100, Calorimetry Sciences Corp.). The cyclin D1 sample (50 µM) were firstly degassed and thermograms of calycosin alone or with different concentrations of calycosin ranging from 1 to 100 nM were read at a scan rate of 1 °C per min. The data were analyzed with CpCalc data analysis software (CpCalc 2.1) (Persikov et al., 2004) and heat capacity changes (ΔCp) and midpoint of unfolding transition (Tm) parameters were calculated as reported previously (Jiskoot and Crommelin, 2005).
2.6 Docking study
Molecular docking study was done employing Schrödinger Maestro 9.2 glide module series based on a previous report (Rahman et al., 2021). The cyclin D1 protein crystal structure (PDB ID: 2w96) and calycosin (CID: 5280448) were imported from the PDB (www.rcsb.org) and NCBI, respectively. Protein refinement and optimization of hydrogen bonds was done by the Wizard tool through the atomic force field (Rahman et al., 2021; Prieto-Martínez et al., 2019). Restrained minimization calculation in the optimized potentials for liquid simulations (OPLS) 2005 force-field removed potential steric structural clashes. Structural energy was reduced until the non‑hydrogen atom's root mean square deviation (RMSD) attained a value of 0.3. The calycosin and protein were finally prepared using optimized potentials for liquid simulations (OPLS) 2005 force field. The optimized calycosin structures were imported on the grid for the cyclin D1 and molecular docking was performed. For this analysis, the XP docking system mode was used to dock calycosin with cyclin D1 (Rahman et al., 2021; Patel et al., 2015).
2.7 Cell culture and exposure of calycosin
The human lung adenocarcinoma [A549, National Center for Cell Science (NCCS), India] was used to explore cytotoxicity after incubation with calycosin. The A549 cells were cultured in Dulbecco’s Modified Eagle’s medium (DMEM) supplemented with FBS (10%) and antibiotics (1%) at 5% CO2 and 37 °C. After reaching confluency and cell harvesting, the cells were used for exposure to different concentrations of calycosin with a range of 1–200 µg/mL. Cells incubated with vehicle control were considered as control.
2.8 MTT cell proliferation assay
The MTT (3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide) assay was used to assess the cytotoxicity based on the previous report (Bendale et al., 2017). Briefly, A549 cells (1 × 104 cells/ml) were treated with increasing concentrations (1–200 µg/mL) of calycosin for a period of 24 h, reacted with MTT for 4 h, added by 150 µL DMSO, and finally absorbance at 570 nm was measured spectrophotometrically using a microplate reader.
2.9 Cell cycle analysis
Cell cycle arrest induced by IC50 concentration of calycosin was assessed employing flow cytometry based on the previous report (Bendale et al., 2017). Approximately 2 × 105 cells per well were plated in six-well plates and allowed to attach. Briefly, after treating, the cells were collected, fixed for 4 h, resuspended, centrifuged (2000 rpm for 10 min) washed, and resuspended in 1 ml of PI/RNase A for 30 min. Finally, cell cycle arrest was assessed using a BD FACScan Cell flow Cytometer (Becton Dickinson USA) and data were analyzed with the Cell Quest Pro software.
2.10 Statistical analysis
Statistical comparisons were made using Student’s t-test. All results were expressed as means ± standard deviation (SD) of at least three independent experiments. P-values of less than 0.05 were considered significant. For all assays, the background intensities (buffer, calycosin) were subtracted from the main signal.
3 Results
3.1 Fluorescence quenching analysis
Proteins show intrinsic fluorescence intensity derived from aromatic residues, mainly Trp and Tyr amino acids (Esfandfar et al., 2016; Zeinabad et al., 2016). After interaction of proteins with ligands, the quenching fluorescence intensity occurs through a concentration-dependent manner. Hence, fluorescence spectroscopy analysis can be employed as a crucial technique for revealing the mechanism of protein interaction with different ligands (Zeinabad et al., 2016). In this assay, cyclin D1 concentration was remained constant and was added by different concentrations of calycosin ranging from 1 to 100 nM. Afterwards, the fluorescence intensity of protein sample either alone or with different concentrations of calycosin was read at three temperatures of 298 K (Fig. 1A), 310 K (Fig. 1B), and 315 K (Fig. 1C). It was shown that addition of calycosin resulted in a fluorescence quenching of cyclin D1 in a concentration-dependent manner (Fig. 1A-C). Also, it was seen that the rate of fluorescence quenching was reduced as the temperature of the system was increased (Fig. 1A-C). Hence, we then aimed to further analyze the quenching mechanism as well as the calculation of the binding and thermodynamic parameters.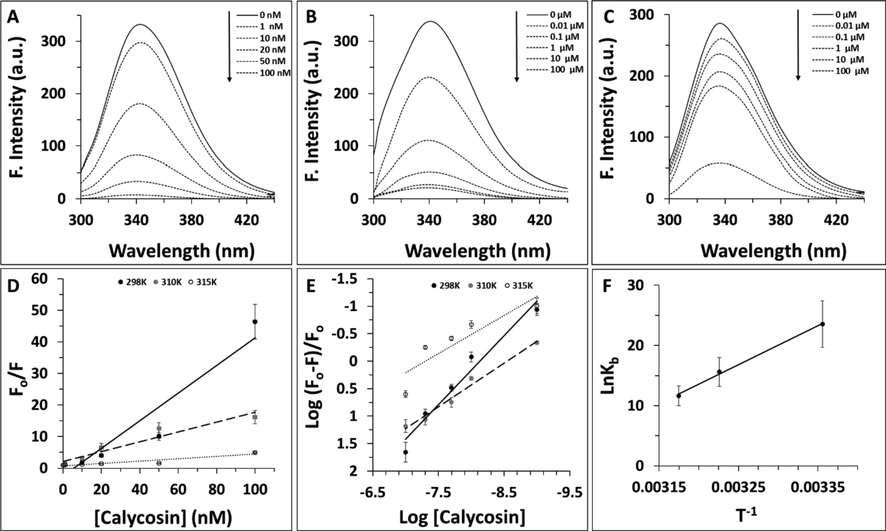
Fluorescence quenching study of cyclin D1 in the presence of increasing concentration of calycosin at (A) 29 K, (B) 310 K, (C) 315 K. (D) Stern-volmer plot of cyclin D1 in the presence of increasing concentration of calycosin. (E) Modified Hill plot of cyclin D1 in the presence of increasing concentration of calycosin. (F) van't Hoff plot of cyclin D1 in the presence of increasing concentration of calycosin.
3.1.1 Quenching mechanism analysis
To further analyze the quenching mechanism of cyclin D1 induced by calycosin, the Stern–Volmer equation (1) was used as follows (Zeinabad et al., 2016).
Fig. 1D exhibits the plot of F0/F for cyclin D1 versus different concentrations of calycosin at three different temperatures. Linear coloration of outcomes was used for estimation of KSV and kq values and the resultant data were tabulated in Table 1. It was reported that the kq at 298 K, 310 K, and 315 K were 0.43 ± 0.07 × 1017 L/mol s, 0.15 ± 0.02 × 1017 L/mol s, and 0.03 ± 0.001 × 1017 L/mol s, respectively, which are much higher than 2.0 × 1010 L/mol s, as the indicator of dynamic quenching mechanism (Rahmani et al., 2018; Liu et al., 2018). Also, based on the inverse coloration between Ksv and kq with temperature, it can be indicated the fluorescence quenching mechanism of cyclin D1 by calycosin was static.
Temperature (K)
Ksv (10 9, L/mol)
kq (1017, L/mol s)
R
298
0.43 ± 0.07
0.43 ± 0.07
0.92
310
0.15 ± 0.02
0.15 ± 0.02
0.94
315
0.03 ± 0.001
0.03 ± 0.001
0.90
3.1.2 Determination of binding constant and binding site
The study of cyclin D1 interaction with different concentrations of calycosin was further analyzed by modified Hill equation (2) to determine the binding constant (Kb) and number of binding sites (n) as follows (Zeinabad et al., 2016):
Then, the values of Kb and n at three temperatures of 298 K, 310 K, and 315 K for calycosin–cyclin D1 interaction were calculated based on Fig. 1E and the relevant data are gathered in Table 2.
Temperature (K)
logKb (L/mol)
n
R
298
10.23 ± 1.19
1.25 ± 0.08
0.96
310
6.79 ± 0.81
0.79 ± 0.05
0.98
315
5.07 ± 0.43
0.69 ± 0.04
0.90
It was seen the values of logKb and n at three different temperatures of 298 K, 310 K, and 315 K for calycosin–cyclin D1 interaction were 10.23 ± 1.19, 6.79 ± 0.81, 5.07 ± 0.43 L/mol and 1.25 ± 0.08, 0.79 ± 0.05, and 0.69 ± 0.04, respectively, suggesting that there is a strong binding interaction and about one binding site on cyclin D1 for calycosin, which the levels of binding affinity and binding sites were reduced upon increase of the solution temperature from 298 K to 315 K. This data indicates the partial unfolding of cyclin D1 at higher temperatures relative to lower ones may result in the partial unfavorable interaction of protein with calycosin. In other words, the increase of temperature to 315 K may result in partial structural changes of protein and corresponding displacement of some amino acid residues on the protein surface which may influence the resultant protein–ligand interaction.
3.1.3 Thermodynamic parameters
The interaction forces between a ligand and proteins are mostly derived from hydrophobic, hydrogen bonding, van der Waals, and electrostatic forces. It has been shown that the signs of the thermodynamic parameters [enthalpy changes (ΔH) and entropy changes (ΔS)] can be used to determine the kind of interaction between proteins and ligands (Liu et al., 2018). That is, if both ΔH and ΔS are positive, then the main interaction is hydrophobic forces. If both ΔH and ΔS are negative, then van der Waals and hydrogen-bonding interactions play key roles in the ligand-protein interaction. Electrostatic interactions are dominant when ΔH is negative and ΔS is positive (Zeinabad et al., 2016).
To determine such data, the application of the present outcomes have been further explored through thermodynamic characteristics calculated for cyclin D1–calycosin interaction using the van't Hoff equation (3) (Zeinabad et al., 2016);
Where, T and R are temperature and universal gas constant, respectively. The free energy change (ΔG) was then determined from the following equation (4) (Zeinabad et al., 2016):
According to the Kb at the three different temperatures of 298 K, 310 K and 320 K, the thermodynamic parameters were calculated based on Fig. 1F using equations (3) and (4) and resultant data were tabulated in Table 3. According to obtained data, it was concluded that the binding of calycosin to cyclin D1 occurs through a spontaneous process (ΔG < 0), accompanied by a negative ΔS value. This binding then can be suggested that it involves an exothermic interaction as deduced by the negative ΔH that is consistent with involvement of hydrogen bonds and van der Waals interaction (Zeinabad et al., 2016).
Temperature (K)
ΔH (kj/mol)
TΔS (kj/mol)
ΔG (kj/mol)
298
−536.421 ± 49.37
−477.96 ± 38.12
−58.4609 ± 4.11
310
−536.421 ± 49.37
−497.207 ± 29.19
−39.2142 ± 3.61
315
−536.421 ± 49.37
−505.226 ± 37.91
−31.1947 ± 2.91
3.2 Synchronous and extrinsic fluorescence spectroscopic studies
Synchronous fluorescence spectroscopy as a simple and sensitive method was used to explore the structural changes around aromatic residues through reading the probable shift in position of λmax (Zeinabad et al., 2016; Ahmad et al., 2020). Therefore, synchronous fluorescence spectra of cyclin D1 at the wavelength interval (Δλ) of 60 nm and 15 nm were read to monitor the structural changes around Trp and Tyr residues, respectively. Fig. 2A and Fig. 2B shows the synchronous fluorescence spectra cyclin D1 in the presence of varying concentrations of calycosin at Δλ of 60 nm and 15 nm, respectively. It can be deduced from Fig. 2A and B, that the emission λmax of cyclin D1 at both Δλ of 60 nm and 15 nm showed an apparent red shift upon addition of varying concentrations of calycosin in the range of 1–100 nM. It can be indicated that the polarity around both aromatic amino acid residues increased based on the displacement of these residues in a more hydrophilic environment. This data implies that the interaction of calycosin with cyclin D1 may lead to a substantial conformational alteration of Trp and Tyr residue microenvironment.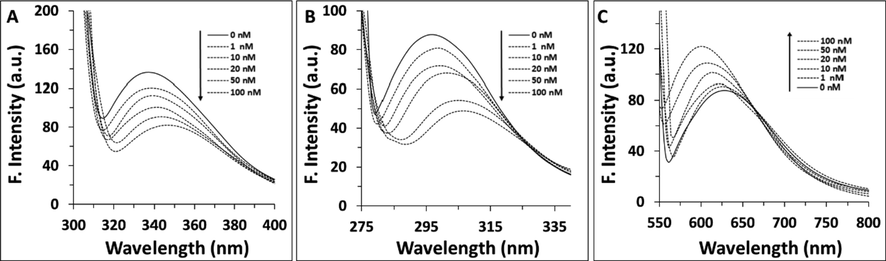
Synchronous fluorescence study of cyclin D1 in the presence of increasing concentration of calycosin at (A) Δλ = 60 nm, (B) Δλ = 15 nm. (C) Nile red fluorescence study of cyclin D1 in the presence of increasing concentration of calycosin.
Nile red fluorescence assay as a sensitive method for the formation of hydrophobic residues was also done to further explore the effect of calycosin on the structure of cyclin D1 (Chabok et al., 2019). It was seen that calycosin resulted in an apparent increase in the Nile red fluorescence intensity accompanied by a blue shift, indicating the exposure of hydrophobic residues of cyclin D1 upon infarction with calycosin (Fig. 2C). This data in good agreement with synchronous fluorescence outcome indicated that calycosin can disrupt the tertiary structure of cyclin D1.
3.3 Circular dichroism (CD) study
To ascertain the probable effect of calycosin interaction on the secondary structure of cyclin D1, CD measurement was also performed in the absence and presence of calycosin. The data read in the range 260–190 nm depicts the presence of two minima around 210 and 222 nm, indicating the presence of α-helix in cyclin D1 structure (Fig. 3) (Zeinabad et al., 2016). Furthermore, it was shown that for the calycosin-cyclin D1 complex, the appearance of the CD spectrum is not exactly similar to that of cyclin D1 alone, and this difference was more significant in the higher concentrations of calycosin than those of lower concentrations. This data further reveals that although the structure of cyclin D1 after addition of calycosin was also predominantly α-helix, the presence of calycosin could increase the distribution of random coil structure (Table 4). Therefore, the different CD spectra of the cyclin D1 with and without calycosin reveal that, at least in this experimental setup, there is a detectable secondary structural change of the cyclin D1 upon binding with the calycosin. Thus, calycosin can influence the secondary structure of cyclin D1 and disrupt its structure (Fig. 3) (Table 4).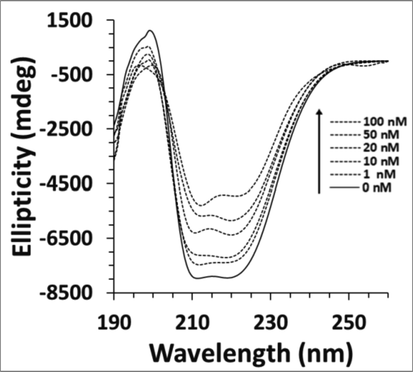
Far-UV CD study of cyclin D1 in the presence of increasing concentration of calycosin.
[Calycosin] (nM)
α-Helix (%)
β-Sheets (%)
Random coil (%)
0
57.32 ± 2.44
12.74 ± 0.93
29.94 ± 1.97
1
56.52 ± 2.19
12.29 ± 0.89
31.19 ± 1.75
10
55.39 ± 2.13
11.88 ± 0.75
32.73 ± 2.08
20
54.29 ± 2.02
11.09 ± 0.63
34.62 ± 2.11
50
53.51 ± 1.98
10.23 ± 0.65
36.26 ± 2.24
100
52.09 ± 1.92
9.97 ± 0.68
37.94 ± 2.18
Therefore, CD and fluorescence spectroscopy indicated that the presence of calycosin can affect the secondary structure of cyclin D1.
3.4 DSC analysis
To explore further detail about denaturation of cyclin D1 upon interaction with calycosin, Tm and ΔCp were determined using DSC analysis. As exhibited in Fig. 4, ΔCp values of cyclin D1 were plotted against temperature and the relevant Tm and ΔΔCp were calculated as described in Material and method section and the outcomes are summarized in Table 4.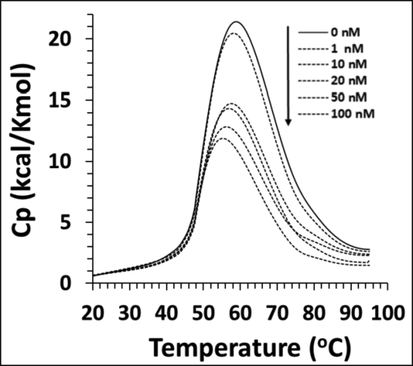
Differential scanning calorimetry (DSC) profile of cyclin D1 in the presence of increasing concentration of calycosin.
The ΔCp data for the cyclin in the presence of various concentrations of calycosin, i.e. 0, 1, 10, 20,50, and 100 nM were 2.17 ± 0.11, 1.97 ± 0.09, 1.74 ± 0.09, 1.63 ± 0.08, 1.26 ± 0.06, 0.82 ± 0.04 kcal/Kmol, respectively (Table 5). ΔCp as a thermodynamic parameter describing the involvement of the hydrophobic forces in the protein structure and corresponding stability (Precupas et al., 2021; Durowoju et al., 2017). The ΔCp value of cyclin D1 was seen to be reduced after addition of calycosin in a concentration-dependent manner, indicating that hydrophobic forces in the protein structure are gradually weakened upon addition of calycosin. Furthermore, the Tm data for cyclin D1 samples based on the above order were 58.21 ± 1.08, 57.98 ± 1.05, 57.19 ± 0.88, 56.33 ± 0.73, 55.92 ± 0.56, 55.04 ± 0.47 °C, respectively (Table 5). In general, ΔCp data in combination with Tm results indicated that calycosin led to a significant decrease in the contribution of hydrophobic forces and thermal stability of cyclin D1.
[Calycosin] (nM)
Tm (oC)
ΔCp (kcal/Kmol)
0
58.21 ± 1.08
2.17 ± 0.11
1
57.98 ± 1.05
1.97 ± 0.09
10
57.19 ± 0.88
1.74 ± 0.09
20
56.33 ± 0.73
1.63 ± 0.08
50
55.92 ± 0.56
1.26 ± 0.06
100
55.04 ± 0.47
0.82 ± 0.04
3.5 Docking studies
Molecular docking analysis was explored to reveal how the calycosin (Fig. 5A) binds cyclin D1. The docked calycosin-cyclin D1 complex is shown in Fig. 5B. The binding energy was found to be −5.93 ± 0.42 kcal/mol (Table 6) which reveals a strong binding affinity between the receptor and ligand. Visualization of the docked site was done by using CHIMERA (www.cgl.ucsf.edu/chimera) and PyMOL (http://pymol.sourceforge.net/) tools (Fig. 5C). It was seen that interacting residues within 4 Å were Asn-198, Pro-199, Ser-201, Glu-35, Leu-32, Lys-238, Ile-237 (Table 6). These residues reveal the presence of hydrophobic interactions, hydrogen bonding, and electrostatic interactions upon interaction of cyclin D1 with calycosin. It seems that the OH groups, aromatic rings and oxygen moiety in the calycosin structure establish hydrogen bonding, hydrophobic interactions, and electrostatic interaction with cyclin D1 residues, respectively. Indeed, hydrogen bonding, hydrophobic and electrostatic interactions can play an important role in the binding of a small molecule with a receptor. The docking score also indicated that the calycosin attaches strongly to the cyclin D1. Spectroscopy data indicated that the hydrogen bonding and van der Waals interactions result in the formation of calycosin-cyclin D1 complex. Docking study also indicated that hydrogen bonding was one of the main forces involved in the interaction observed between calyosin and cyclin D1.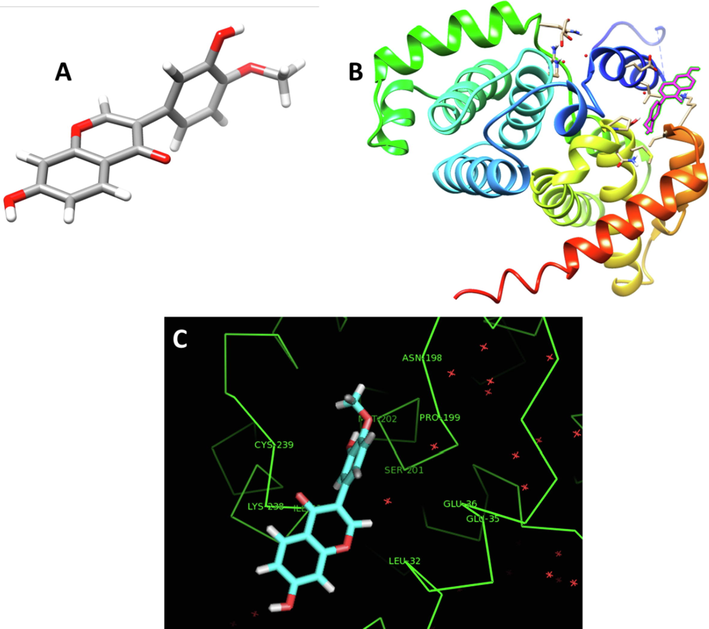
(A) Calycosin structure, (B) Docking complex of calycosin-cyclin D1, (C) Interactive residues involved in the interaction of calycosin and cyclin D1.
Docking score (kcal/mol)
Interacting residues within 4 Å
−5.93 ± 0.42
Hydrophobic; Pro-199, Leu-32, Ile-237
Electrostatic; Glu 35, Lys-238,
Hydrogen bonding; Asn-198, Ser-201
3.6 Cytotoxicity and cell cycle arrest of calycosin on lung cancer cells
Lung cancer cells were treated with increasing concentration of calycosin for 24 h and cytotoxicity was assessed by MTT assay. It was observed that calycosin mitigated cell growth (Fig. 6A) in a concentration-dependent manner. In fact, calycosin induced the highest cytotoxic effects on lung cancer cells at 200 µg/mL. It was then determined that the IC50 concentration of calycosin against A549 cells was about 170.81 ± 19.54 µg/mL, which was used for the cell cycle assay.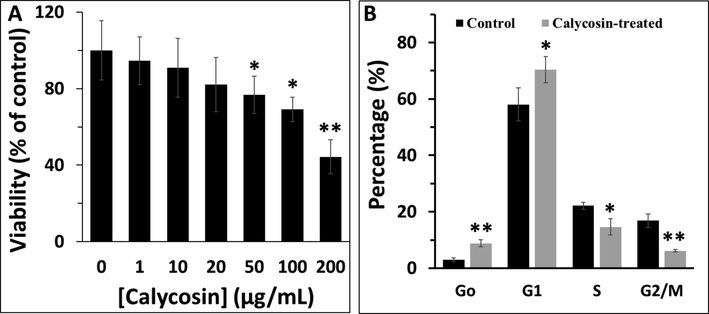
(A) MTT assay of A549 cells in the presence of increasing concentration of calycosin after 24 h. (B) Cell cycle assay of A549 cells in the presence of IC50 concentration of calycosin after 24 h.
Flow cytometry analysis was further used to explore the effect of calycosin on the cell cycle arrest especially at G1 phase as cyclin D1 controls the transition of cells from G1 to S phase. A significant increase in the percentage of cells in G1 phase was reported in A549 cells after incubation with IC50 concentration of calycosin, relative to control cells (Fig. 6B). In fact, as shown in Fig. 6B, when cells were incubated with IC50 concentration of calycosin for 24 h, the percentage of sub-G1 (as a marker of apoptosis) and G1 population showed a marked increase, whereas the percentage of S and G2/M had a significant decrease compared with those in control group. All these data indicated that inhibition of cell growth at G1 phase led to decreased cells entering in S and G2/M phases, subsequently triggering cell apoptosis (Zeinabad et al., 2016).
One of the possible reasons for the cell cycle arrest in the G1 phase is the probable denaturation of cyclin D1 as a key controller for S phase entry, which needs further in vitro and in vivo experiments in the future.
4 Discussion
Cyclin D1 is a protein responsible for transition of cells from G1 to S phase in cell cycle and can regulate the proliferation of cancer cells. Therefore, development of some therapeutic approaches targeting the structure cyclin D1 by some compounds can be used as a therapeutic potential against different kinds of cancers. In this study, the interaction of calycosin as a promising anticancer bioactive small molecule with cyclin D1 was explored by different in vitro experimental and theoretical analyses. In this experimental study, the thermodynamic parameters and side effect of the calycosin upon interaction with cyclin D1 was explored It was shown that the addition of calycosin to cyclin D1 reduces the emission of intrinsic fluorescence of the calycosin D1 through the static mechanism and changes in the secondary and tertiary structures of protein. Indeed, it was seen that calycosin changes the secondary structure of the protein by reducing the regular structure of the α-helix and β-sheets and increasing the structures of the random coils, which indicates a decrease in the stability of the secondary structure of the protein. This data was also verified by DSC analysis which indicated that Tm and ΔCp values of cyclin D1 as the marker of stability of protein through contribution of hydrophobic forces were decreased in the presence of calycosin. The above results showed that calycosin can bind to cyclin D1 and alter its structure.
In another study, it has been indicated that calycosin can increase the stability of interferon gamma as an anticancer agent (Yang et al., 2021). Also, it has been shown that calycosin did not induce any effect on the stability of tau protein, however due to its unique structure can mitigate the tau protein aggregation (Zhenxia et al., 2021). These differences between the effect of calycosin on protein stability may be derived from the type of protein and the contribution of different forces (Zeinabad et al., 2016). At the time of writing this paper, no other studies were available on the interaction of calycosin with proteins by spectroscopic techniques. In agreement with our data, it has been shown that polyphenols can affect the protein structure based on the kind of interaction derived from the unique structure of polyphenol (Xu et al., 2019; Shamsi et al., 2020).
Regarding the binding parameters, it has been shown that the n value is close to unity, revealing a 1:1 complexation with a wide number of polyphenols and the protein (Bose, 2016). Also, the structural alteration in the polyphenols has been realized to change the Kb value and the different moieties attached to the rings of the polyphenols have been shown to play an important role in this aspect (Bose, 2016). It has been found that any changes in polyphenol moiety alters the binding of these small molecules with proteins (Bose, 2016). It seems calycosin due to the presence of different groups such as hydroxy, methoxyphenyl, and chromen can establish different forces with proteins as was investigated in this paper. Therefore, it should be noted that calycosin can bind cyclin D1 and induce some apparent structural changes on this protein which can be considered as a helpful strategy for cell cycle arrest in cancer cells.
A drug’s affinity to bind to regulatory protein in cancer progression can provide useful information in drug development, because drugs mainly affect protein structure and corresponding activity. Cyclin D1 is a regulatory protein in the G1 phase of cell cycle which plays a key role in cancer cell proliferation and calycosin as a small biomolecule with potential therapeutic activities against human cancer can result in denaturation of this protein.
Therefore, this study provides new biochemical and structural detail on the calycosin’s binding to cyclin D1 that is interesting in the exploration of its therapeutic effects in future medicine through inhibiting the proliferation of lung cancer cells.
Hence, in the future some complementary data should be gathered in vitro and in vivo to cover some limitations of this paper based on the expression of cyclin D1 in the presence of calycosin in cancer cells and exploring the structural changes of cyclin D1 in vivo.
It is hoped that the outcomes of this study will hold a great promise for the development of some potential compounds with fewer side effects in cancer therapy.
5 Conclusion
In the present paper, we have systematically investigated the structural and thermodynamic basis of interactions of calycosin and cyclin D1 with multiple experimental and modeling analyses. The steady-state fluorescence analysis revealed that the formation of cyclin D1–calycosin resulted in an apparent reduction in the fluorescence of cyclin D1 in a concentration-dependent fashion. The related Stern-Volmer parameters indicated that the formation of stable cyclin D1–calycosin complexes via a static mechanism.
The calculation of binding parameters for the cyclin D1–calycosin complex formation revealed a strong association constant and one binding site. The negative thermodynamic parameters indicated that the overall cyclin D1–calycosin interaction was spontaneous and the complex was formed mainly due to the presence of hydrogen binding and van der Waals forces. To further support the formation of stable cyclin D1–calycosin complexes, molecular docking study was run. It was found that calycosin showed a strong docking score after interaction with cyclin D1 through different forces and Asn-198, Ser-201 contributed to the formation of hydrogen binding with calycosin. The collective experimental outcomes from synchronous/ Nile red fluorescence and CD spectroscopy demonstrated that calycosin binding induced an apparent conformational change in cyclin D1 structure. Also, DSC analysis indicated that calycosin decreased the Tm and ΔCp values of cyclin D1, revealing the probable denaturation of cyclin D1 through perturbation of hydrophobic forces. Also, cellular studies showed that calycosin mitigated the proliferation of lung cancer cells through cell cycle arrest in the G1 phase regulated by cyclin D1.
Acknowledgement
This study was supported by the PhD Start-up Fund of Natural Science Foundation of Liaoning Province (NO. 2019-BS-296), and the Natural Science Foundation of Liaoning Province (NO. 2018011172-301).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Deciphering the mechanism of hafnium oxide nanoparticles perturbation in the bio-physiological microenvironment of catalase. Nano Express.. 2020;1(3):030006
- [Google Scholar]
- Evaluation of cytotoxic activity of platinum nanoparticles against normal and cancer cells and its anticancer potential through induction of apoptosis. Integrative Med. Res.. 2017;6(2):141-148.
- [Google Scholar]
- Interaction of tea polyphenols with serum albumins: A fluorescence spectroscopic analysis. J. Lumin.. 2016;169:220-226.
- [Google Scholar]
- Nile red-doped fluorescent semiconducting polymer dots as a highly sensitive hydrophobicity probe: protein conformational changes detection and plasma membrane imaging. J. Iran. Chem. Soc.. 2019;16(3):535-543.
- [Google Scholar]
- Extra-virgin olive oil phenols block cell cycle progression and modulate chemotherapeutic toxicity in bladder cancer cells. Oncol. Rep.. 2016;36(6):3095-3104.
- [Google Scholar]
- Crystal structure of human CDK4 in complex with a D-type cyclin. Proc. Natl. Acad. Sci.. 2009;106(11):4166-4170.
- [Google Scholar]
- Calycosin: a review of its pharmacological effects and application prospects. Expert Rev. Anti-Infective Therapy.. 2021;19(7):911-925.
- [Google Scholar]
- Lung cancer and other causes of death in relation to smoking. Br. Med. J.. 1956;2(5001):1071.
- [Google Scholar]
- Differential scanning calorimetry—a method for assessing the thermal stability and conformation of protein antigen. JoVE (J. Visualized Exp.). 2017;121:e55262
- [Google Scholar]
- Spectroscopic studies of interaction between CuO nanoparticles and bovine serum albumin. J. Biomol. Struct. Dyn.. 2016;34(9):1962-1968.
- [Google Scholar]
- Pharmaceutical properties of calycosin, the major bioactive isoflavonoid in the dry root extract of Radix astragali. Pharm. Biol.. 2014;52(9):1217-1222.
- [Google Scholar]
- Prognostic and Clinicopathological Significance of CCND1/Cyclin D1 Upregulation in Melanomas: A Systematic Review and Comprehensive Meta-Analysis. Cancers (Basel).. 2021;13(6):1314.
- [CrossRef] [Google Scholar]
- Clinicopathological and prognostic significance of cyclin D1 amplification in patients with breast cancer: a meta-analysis. J. Buon.. 2017;22(5):1209-1216.
- [Google Scholar]
- The multifaceted role of cell cycle regulators in the coordination of growth and metabolism. The FEBS Journal.. 2021;288(12):3813-3833.
- [Google Scholar]
- Methods for Structural Analysis of Protein Pharmaceuticals. Arlington: VA: AAPS Press; 2005. p. :573-589.
- Prognostic significance of cyclin D1 expression in renal cell carcinoma: a systematic review and meta-analysis. Pathol. Oncol. Res.. 2020;26(3):1401-1409.
- [Google Scholar]
- Effects of the cyclin D1 polymorphism on lung cancer risk-a meta-analysis. Asian Pac. J. Cancer Prev.. 2012;13(5):2325-2328.
- [Google Scholar]
- Prognostic significance of cyclin D1 expression in colorectal cancer: a meta-analysis of observational studies. PLoS ONE. 2014;9(4):e94508
- [Google Scholar]
- Calycosin induces mitochondrial-dependent apoptosis and cell cycle arrest, and inhibits cell migration through a ROS-mediated signaling pathway in HepG2 hepatocellular carcinoma cells. Toxicol. In Vitro. 2021;70:105052
- [Google Scholar]
- Identification differential behavior of Gd@ C82 (OH) 22 upon interaction with serum albumin using spectroscopic analysis. Spectrochim. Acta Part A Mol. Biomol. Spectrosc.. 2018;203:383-396.
- [Google Scholar]
- Cyclin D1 in cancer: a molecular connection for cell cycle control, adhesion and invasion in tumor and stroma. Cells.. 2020;9(12):2648.
- [Google Scholar]
- Causes of death of patients with lung cancer. Arch. Pathol. Lab. Med.. 2012;136(12):1552-1557.
- [Google Scholar]
- Identification of chebulinic acid as potent natural inhibitor of M. tuberculosis DNA gyrase and molecular insights into its binding mode of action. Comput. Biol. Chem.. 2015;59:37-47.
- [Google Scholar]
- Equilibrium thermal transitions of collagen model peptides. Protein Sci.. 2004;13(4):893-902.
- [Google Scholar]
- Calorimetric, spectroscopic and computational investigation of morin binding effect on bovine serum albumin stability. J. Mol. Liq.. 2021;333:115953
- [Google Scholar]
- Acoplamiento molecular: avances recientes y retos. TIP Revista Especializada en Ciencias Químico-Biológicas.. 2019;21(S1):65-87.
- [Google Scholar]
- Spectroscopic and molecular modelling study of binding mechanism of bovine serum albumin with phosmet. Spectrochim. Acta Part A Mol. Biomol. Spectrosc.. 2021;244:118803
- [Google Scholar]
- Probing the interaction of silver nanoparticles with tau protein and neuroblastoma cell line as nervous system models. J. Biomol. Struct. Dyn.. 2018;36(15):4057-4071.
- [Google Scholar]
- Prognostic and clinicopathological significance of cyclin D1 expression in oral squamous cell carcinoma: A systematic review and meta-analysis. Oral Oncol.. 2018;83:96-106.
- [Google Scholar]
- Relevance of chromosomal band 11q13 in oral carcinogenesis: An update of current knowledge. Oral Oncol.. 2017;72:7-16.
- [Google Scholar]
- Predictive value of CCND1/cyclin D1 alterations in the malignant transformation of potentially malignant head and neck disorders: Systematic review and meta-analysis. Head Neck. 2019;41(9):3395-3407.
- [Google Scholar]
- The impact of cyclin D1 overexpression on the prognosis of bladder cancer: a meta-analysis. World J. Surg. Oncol.. 2014;12(1):1-8.
- [Google Scholar]
- Evaluation of Binding of Rosmarinic Acid with Human Transferrin and Its Impact on the Protein Structure: Targeting Polyphenolic Acid-Induced Protection of Neurodegenerative Disorders. Oxid. Med. Cell. Longevity. 2020;5:2020.
- [Google Scholar]
- Apigenin, a dietary flavonoid, inhibits proliferation of human bladder cancer T-24 cells via blocking cell cycle progression and inducing apoptosis. Cancer Cell Int.. 2015;15(1):1-2.
- [Google Scholar]
- Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J. Clin.. 2021;71(3):209-249.
- [Google Scholar]
- Regulation of the G1/S phase of the cell cycle and alterations in the RB pathway in human lung cancer. Expert Rev. Anticancer Ther.. 2006;6(4):515-530.
- [Google Scholar]
- The 11q13-q14 amplicon: clinicopathological correlations and potential drivers. Genes Chromosom. Cancer. 2013;52(4):333-355.
- [Google Scholar]
- Comparative studies of interaction of β-lactoglobulin with three polyphenols. Int. J. Biol. Macromol.. 2019;136:804-812.
- [Google Scholar]
- Enhancement of interferon gamma stability as an anticancer therapeutic protein against hepatocellular carcinoma upon interaction with calycosin. Int. J. Biol. Macromol.. 2021;185:813-820.
- [Google Scholar]
- Thermodynamic and conformational changes of protein toward interaction with nanoparticles: a spectroscopic overview. RSC Adv.. 2016;6(107):105903-105919.
- [Google Scholar]
- miR-149 suppresses human non-small cell lung cancer growth and metastasis by inhibiting the FOXM1/cyclin D1/MMP2 axis. Oncol. Rep.. 2017;38(6):3522-3530.
- [Google Scholar]
- Inhibition of tau aggregation and associated cytotoxicity on neuron-like cells by calycosin. Int. J. Biol. Macromol.. 2021;171:74-81.
- [Google Scholar]







