Translate this page into:
Fluorescent DNA biosensors for Pb(II) detection based on G-quadruplex formation
⁎Corresponding author. qiujieqiongqjq@163.com (Jieqiong Qiu) qiujieqiong@zstu.edu.cn (Jieqiong Qiu)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University. Production and hosting by Elsevier.
Abstract
Pb(II) ions pose a grave concern due to their ability to inflict irreparable harm upon humans, even at exceedingly low concentrations. Conventional techniques have limitations due to the need for complex sample pre-treatment, specialized operation, and expensive equipment for widespread Pb(II) ion detection. Consequently, there is a burgeoning interest in developing a DNA biosensor that employs fluorescence methodology to detect Pb(II) ions. It offers heightened sensitivity, an impressive lower detection limit, and user-friendly operation, thus rendering it a focal point of contemporary scientific inquiry. One noteworthy observation is the Pb(II) ion’s capacity to form remarkably stable G-quadruplex structures when paired with G-base-rich oligonucleotide sequences (GOS), surpassing the stability achieved with K(I) ions. This observation opens up exciting avenues and possibilities for detecting Pb(II) ions. In this review, we undertake a comprehensive classification and summarization of diverse fluorescent biosensors for Pb(II) ion detection, relying on the formation of G-quadruplex structures through both label-free and labeled methodologies. The categorization encompasses signal on–off/off–on models and enzymatic-assisted approaches within the label-free methods. In the labeling method, our scrutiny delves into various classes of quenching groups, including conventional quenchers, continuous G bases, and nanomaterials, offering a comprehensive discussion of their respective merits and applications. Moreover, this review endeavors to explicate the formation of G-quadruplex structures triggered by Pb(II) ions, thereby fostering a more explicit understanding among researchers of these intricate structures.
Keywords
Pb(II) ions detection
G-quadruplex
Fluorescence
Label-free methods
Labeled methods
1 Introduction
Lead is frequently encountered on the Earth's surface and is commonly found within ores. In the wake of burgeoning heavy industries, the extensive global application of Pb(II) has resulted in a significant release of Pb(II) ions waste into the natural environment (Du et al., 2015). Upon investigation, it has come to light that contaminated soil, food, and drinking water harbor discernible Pb(II) ions (Liu et al., 2016). Of all the pernicious heavy metals, Pb(II) ions rank among the most detrimental, characterized by their resistance to degradation, prolonged half-life, and potent bioaccumulation (Chen et al., 2021). Even minute quantities of Pb(II) ions present in food or potable water can inflict considerable damage upon vital human organs and systems (Fu et al., 2020), including the kidneys, liver (Du et al., 2015), immune system (Chen et al., 2021), as well as the neurological and urinary systems (Ozdemir et al., 2017). This dire threat gravely jeopardizes human life and health. In recognition of this critical issue, the U.S. Environmental Protection Agency (EPA) and the World Health Organization (WHO) have set strict safety thresholds for Pb(II) in drinking water at 53.6 nM and 35.7 nM, respectively (Chen et al., 2020). Consequently, it is necessary to investigate and develop a facile and highly sensitive methodology capable of detecting low concentrations of Pb(II) ions, applicable to real-life scenarios.
Numerous conventional techniques have been devised for Pb(II) ion detection, comprising atomic absorption spectrometry (Bechlin et al., 2014, Chahid et al., 2014), atomic emission spectrometry (Yu et al., 2017), inductively coupled plasma mass spectrometry (ICP-MS) (Ozdemir et al., 2017, Jia et al., 2018), and anodic stripping voltammetry (Jayadevimanoranjitham and Narayanan, 2019). Regrettably, these methods suffer significant drawbacks that hinder their applicability in on-site and real-time analyses. Chief among these limitations is the considerable expense of the requisite instrumentation, coupled with the intricacy and tedium of the pre-processing steps, necessitating specialized technicians for operation. Moreover, the detection sensitivity of these traditional approaches is fraught with uncertainty, influenced by the sample's inherent nature, the method employed for sample preparation, instrumentation nuances, and the specific operating conditions (Dil et al., 2020, Dil et al., 2021, Mehrabi et al., 2023). As a result, these unstable outcomes are prone to manifest, severely constraining the daily expeditious detection of Pb(II) ions.
DNA-based Pb(II) ion detection represents an innovative and recently developed methodology. Its effectiveness hinges on deploying oligonucleotide aptamers, lauded for their remarkable binding capacity, facile adaptability, exceptional stability, and uncomplicated synthesis procedures (Krishnamoorthy, 2018, Amalraj et al., 2021, Arunjegan et al., 2021). A pivotal component of this approach is the formation of G-quadruplex (G-4) structures, a nucleic acid secondary conformation that arises from the folding of sequences rich in consecutive guanine (G) bases (Sun et al., 2019). The interaction between Pb(II) and G-rich sequences is of particular significance, giving rise to highly stable tetrameric structures (Sun et al., 2015). It is noteworthy that Pb(II) exhibits higher stability to G-4 compared to K(I) (Sun et al., 2013), an essential property ingeniously harnessed by scientists to devise an array of ingenious techniques and concepts for Pb(II) ions detection. The fluorescence method represents a powerful and innovative approach for detecting and tracking individual target molecules, such as Pb(II), with exceptional sensitivity (Amalraj and Perumal, 2022, Mehta et al., 2022, Zhang et al., 2023). Its capacity to monitor fluorescence signals allows for high precision and a broader linear range of detection, enabling the identification of targets across a broad concentration spectrum (Krishnamoorthy, 2018). The fluorescence-based approach boasts several distinct advantages (Ravikumar et al., 2017, Sivakumar and Lee, 2021). For example, operational simplicity, lower cost, and allow for real-time analysis. Thus, integrating fluorescence and aptamer technology presents a promising avenue to address the pressing challenges posed by Pb(II) detection, propelling scientific inquiry and advancements in this critical domain.
This paper expatiates a synthetical review of contemporary investigations concerning forming Pb/G-4 complexes by employing Pb(II)-specific oligonucleotides. A novel fluorescence-based technique is elucidated with this context, specifically designed to detect low concentrations of Pb(II) ions in aqueous environments. Emphasis is placed on analyzing two fluorescence methods, namely label-free and labeled methods, each boasting unique attributes and mechanisms for recognizing fluorescent signals. The following essays are specifically discussed: signal on–off/off–on (Li et al., 2010, Xu et al., 2015, Huang et al., 2016) models, enzyme-assisted (Wen et al., 2016, Rajaji and Panneerselvam, 2020) for label-free methods. Moreover, the different quenchers of the labeled probes are classified as common quenchers (Liu et al., 2009, Lu et al., 2019), G bases (Wang et al., 2013, Zhang et al., 2021) and nanomaterials (Li et al., 2013, Li et al., 2013, Khoshbin et al., 2019).
2 Label-free methods
Label-free methods significantly advance in detecting specific structures, such as G-4 or Pb(II) binding G-4 structures. Notably, various intercalator dyes have proven effective in discerning G-4/G-4-Pb(II) structures, including N-Methylmesoporphyrin IX (NMM) (Guo et al., 2012), SYBR Green I (Zhan et al., 2014, Huang et al., 2016), Thiazole orange (TO) (Jacobi et al., 2012), Malachite green (MG) (Li et al., 2018), AMT (Geng et al., 2020), and GeneFinderTM (Zhan et al., 2015). These fluorochromes are capable of inducing fluorescence changes, and their interactions with G-4/G-4-Pb(II) structures can manifest in different fluorescence models, characterized as either “on–off” or “off–on”.
2.1 “On-off” model
In the “on–off” model, the fluorescence is enhanced by binding to the target DNA structures, while the fluorescence is quenched upon interaction with the G-4-Pb(II) structures. In 2013, Zhang et al. devised a DNA sensor (Fig. 1a) predicated on the exchange between double-stranded DNA (dsDNA) and G-4 structures to detect Pb(II) ions (Zhang et al., 2013). SYBR Green I was employed, which could be embedded into dsDNA. GOS-1 (Table 1) and its complementary strand could form a dsDNA configuration that exhibited robust fluorescence. However, upon introducing Pb(II) ions, GOS-1 underwent a conformational change, adopting a compact G-4 structure. As a result, this structural transition induced the detachment of the fluorophore SYBR Green I from dsDNA, resulting in a substantial decline in fluorescence intensity. The sensor demonstrated a consistent decrease in fluorescence as the concentration of Pb(II) ions increased from 10 to 100 nM and 100 to 1000 nM, displaying a reliable linear relationship with concentration (Table 2). However, preparation of dsDNA takes up to 3 h and 2 h to monitor the Pb(II) detection, making the detection process time-consuming and laborious. Despite these limitations, the pioneering work in designing the DNA sensor contributes to Pb(II) ion detection, offering valuable insights for future developments and improvements in DNA-based sensing strategies.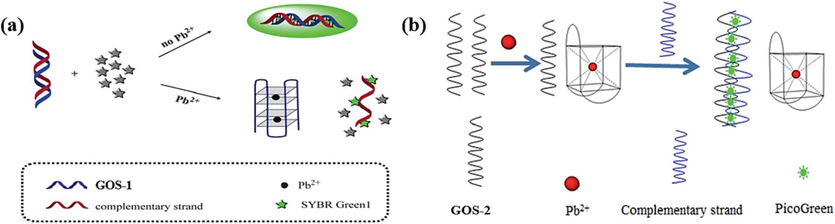
Based on the interplay between dsDNA and G-4 structural conversions: (a) SYBR Green I as the fluorescence indicator (Zhang et al., 2013); (b) PG as the fluorescence indicator to monitor Pb(II) (Huang et al., 2016).
Probe
Nucleotide sequence
GOS-1
GTGGGTAGGGCGGGTTGG
GOS-2
GGTTGGTGTGGTTGG
GOS-3
GGGTGGGTGGGTGGGT
GOS-4
G33
GOS-5
TTTTTTGGGTGGGTGGGTGGGTTTTTTT
GOS-6
GGTGGTGGTGGTTGTGGTGGTGGTGG
GOS-7
GGGGAGAAGGACGGGUCCAGUGCGAAACACGCACUGUUGAGUAGAGUGUGAGCUCCC
GOS-8
AGGGTTAGGGTTAGGGTTAGGG
GOS-9
GGGTAGGGCGGGTTGGG
GOS-10
CACTrAGGAAGAGATGTGGGTAGGGCGGGTTGGATCTCTGAAGTAGCGCCGCCGTAGTG
GOS-11
GGGTTGGGCGGGATGGGTATCATTACTATrAGGAAGAGAGATTACTAAACGATCGCCCA
GOS-12
GGGTTGGGCGGGATGGGTTTCACTrAGGCACTTTGGGTAGGG
GOS-13
GTTAGGGCATCACTATrAGGTTAGGGCATCACTAT
GOS-14
TCAGGGTTTTGGGTTTTTTGGGTTTTGGGAGAGTArATATCAGGGACACTT
GOS-15
TAGGGCCCCGGGCCCCCCGGGCCCCGGGAGAGCTrAGGCACTTTGGGTAGGG
GOS-16
BHQ-2-(CH2)6-GGCGCCGGAAGGTGTGGAAGGTCACCGGCGCC-(CH2)6-TAMRA
GOS-17
CGTCATGGGTGGGTGGGTGGGTGACGGGG
GOS-18
CCCTCCTTCCCTTCCTTTTCCAACCCAACCACCGGTTGGTGTGGTTGG
GOS-19
GGGCCGGCTTCGGCCGGCGGGG
GOS-20
GGAAGGTGTGGAAGG
GOS-21
NH2–(CH2)6-GGGTGGGTGGGTGGGT
GOS-22
HS-3(CH2CH2O)6-GGAAGGTGTGGAAGGTTCCAGACCT
GOS-23
NH2-GGGTGGGTGGGTGGGT
Label-free
Probe
Fluorophore
Detection linear range
LOD
Signal
Ref.
On-off
GOS-1
SYBR Green I
10–1000 nM
3 nM
Turn Off
(Zhang et al., 2013)
On-off
GOS-2
PG
4.83–4830 nM
4.83 nM
Turn Off
(Huang et al., 2016)
On-off
GOS-1
NMM
5–1000 nM
1 nM
Turn Off
(Guo et al., 2012)
On-off
GOS-1
ThT-E
0–1000 nM
0.76 nM
Turn Off
(Yang et al., 2019)
On-off
GOS-3
SYBR Green I
0–483 nM
18.31 nM
Turn Off
(Zhan et al., 2014)
On-off
GOS-4
Tb(III)
3–50 nM
1 nM
Turn Off
(Lin et al., 2011)
On-off
GOS-5
GeneFinderTM
0–386 nM
12.66 nM
Turn Off
(Zhan et al., 2015)
On-off
GOS-6
NMM
10–200 nM
5 nM
Turn Off
(Zhu et al., 2018)
Off-on
GOS-3
ZnPPIX
20–1000 nM
20 nM
Turn On
(Li et al., 2010)
Off-on
GOS-3
AMT
100–1000 nM
3.6 nM
Turn On
(Geng et al., 2020)
Off-on
GOS-6
TO
0–40 nM
8 nM
Turn On
(Jacobi et al., 2012)
Off-on
GOS-6
NMM
0–1000 nM
3 nM
Turn On
(Xu et al., 2015)
Off-on
GOS-7
DFHBI
5–500 nM
6 nM
Turn On
(DasGupta et al., 2015)
Off-on
GOS-8
MG
22.4–475 nM
6.7 nM
Turn On
(Li et al., 2018)
Off-on
GOS-9
Cationic perylene derivative
10–1000 nM
5 nM
Turn On
(Zhao et al., 2016)
Enzyme-assisted
GOS-10
AED
0.05–50 nM
0.0228 nM
Turn Off
(Zhao et al., 2013)
Enzyme-assisted
GOS-11
ZnPPIX
5–100 nM
3 nM
Turn On
(Fu et al., 2016)
Enzyme-assisted
GOS-12
ThT
10–1000 nM
0.06 nM
Turn On
(Wen et al., 2016)
Enzyme-assisted
GOS-13
CV
2.5–25000 nM
5 nM
Turn On
(Zhou et al., 2021)
Enzyme-assisted
GOS-14
ThT
0–0.05 nM
0.096 nM
Turn On
(Ravikumar et al., 2017)
Enzyme-assisted
GOS-15
ThT
3–9 nM
1.6 nM
Turn On
(Rajaji and Panneerselvam, 2020)
Enzyme-assisted
GOS-6
AUR
0–1000 nM
0.4 nM
Turn On
(Li et al., 2011)
Enzyme-assisted
GOS-2
TMB
0.5–460 nM
0.25 nM
Turn On
(Li et al., 2020)
In 2016, a novel approach utilizing GOS-2 (Table 1) was introduced to hinge on a comparable principle of interconversion between dsDNA and G-4 structures (Huang et al., 2016). This design enabled the detection of Pb(II) ions by extinguishing the fluorescence signal of PicoGreen (PG) (Fig. 1b, Table 2). In contrast to the sensor mentioned above, this method remarkably expedited Pb(II) detection. The detection process was achieved in 13 min, even at RT. The work addresses existing limitations and showcases the remarkable potential for refining and optimizing detection methodologies. The accelerated and simplified approach, exemplified by the GOS-2 design, is poised to leave a lasting impact on the Pb(II) ion detection landscape, enhancing efficiency and effectiveness in diverse settings.
To optimize the sensor for Pb(II) ion detection, the same sequence of GOS-1 (Table 1) was harnessed to design the sensor for Pb(II) ion detection (Guo et al., 2012). The introduction of K(I) ions led GOS-1 to assume a G-4 configuration, a state that exhibited heightened fluorescence upon interaction with NMM, which was renowned for its selectivity toward the G-4 structure. However, upon Pb(II) ions, these ions displaced K(I) ions, binding to the G-4 and generating a more stable G-4 structure. As a result, NMM’s binding affinity to this stable structure diminished, extinguishing the fluorescence signal and facilitating Pb(II) ion detection (Fig. 2a, Table 2). Similarly, Yang et al. capitalized on ethyl-substituted thioflavin T (ThT-E) (Yang et al., 2019). In their method, the presence of K(I) ions prompted GOS-1 to assemble into parallel G-4 with embedded ThT-E, showcasing robust fluorescence. ThT-E is a derivative of ThT, representing an advancement over ThT due to its heightened sensitivity and suitability for G-4 detection. In contrast, introducing Pb(II) ions induced the formation of antiparallel G-4 within the GOS-1, resulting in a reduced fluorescence signal. This approach boasted a swift detection response time of merely 5 min compared to 30 min incubation time of NMM, alongside an impressively low detection limit of 0.76 nM (Table 2), signifying remarkable sensitivity and considerable practical value (Fig. 2b). Without the assistance of K(I), Zhan et al. pioneered a novel sensor (Fig. 2c) that employed the fluorophore SYBR Green I, which was adept at binding to the GOS-3 probe through electrostatic interaction, yielding a stronger fluorescence signal (Zhan et al., 2014). However, the presence of Pb(II) ions instigated GOS-3 to adopt a G-4 conformation, causing the fluorescence excited by SYBR Green I to undergo a significant attenuation, facilitating the detection of Pb(II) ions. The sensor's fluorescence intensity exhibited a linear decrease proportionate to the rising Pb(II) ion concentration within the 0–100 ppb (0–483 nM). This sensor achieved a detection limit of 3.79 ppb (18.31 nM) (Table 2). By leveraging the interplay between Pb(II) ions and G-4 structures, researchers continue to push the boundaries of detection methodologies, advancing our ability to sensitively and efficiently monitor Pb(II) ions for various applications.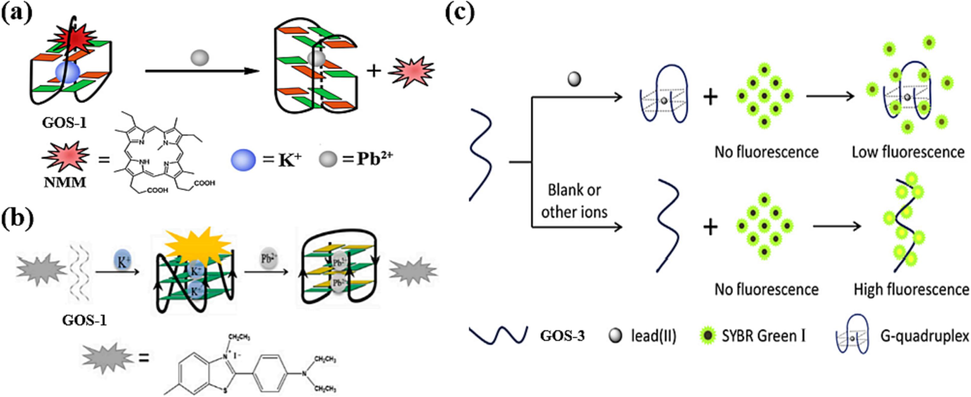
Formation of G-4-Pb(II) enables fluorescence release: (a) NMM as the fluorescence indicator (Guo et al., 2012); (b) ThT-E as the fluorescence indicator (Yang et al., 2019); (c) SYBR Green I as the fluorescence indicator (Zhan et al., 2014).
To enhance the application of Pb(II) detection sensors, the multifunctional sensors were designed, which were capable of detecting two distinct types of metal ions, namely Pb(II) and Hg(II). In 2011, Lin et al. proposed the use of probes poly guanidine GOS-4 (Table 1) and terbium ion (Tb(III)) for the identification of Pb(II) ions, where the Tb(III) ion probe GOS-4 binds to the Tb(III) ion and changes its structure to a G-quarter structure, transferring energy from the nucleic acid sequence to Tb(III) and thus increasing the fluorescence signal; the Pb(II) ion competes with the Tb(III) ion to interact with GOS-4 forming a G-4 structure, which immediately decreases fluorescence and thus detects Pb(II) ions (Lin et al., 2011) (Fig. 3a). The team also found that the use of polythymidine (T33) and a benzothiazole-4-quinoline dimer derivative (TOTO-3) enabled the detection of mercury ions. This inexpensive method increases the detectable Pb(II) and Hg(II) concentrations to 3–50 nM and 25–500 nM. The detection limits were 1 nM for Pb(II) and 10 nM for Hg(II) (Table 2).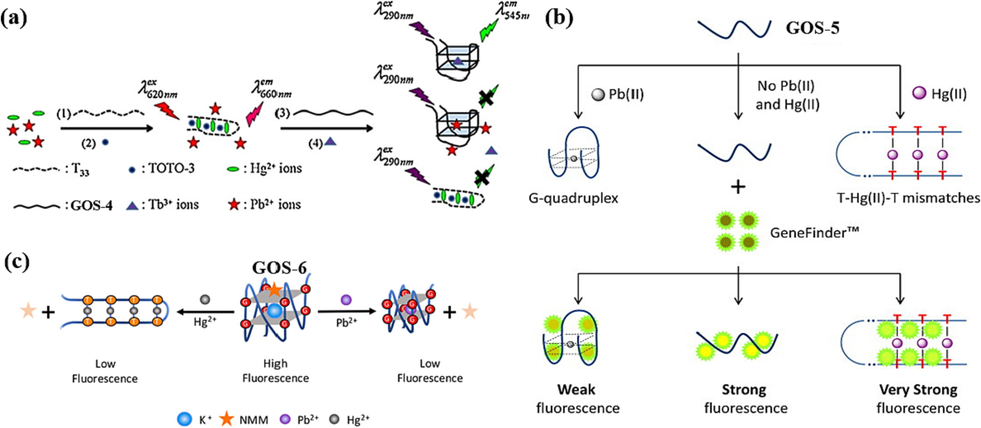
Dual function of DNA-based biosensors for Pb(II) and Hg(II) detection: (a) TOTO-3 and Tb(III) as the fluorescence indicator (Lin et al., 2011); (b) GeneFinder™ as the fluorescence indicator (Zhan et al., 2015); (c) NMM as the fluorescence indicator (Zhu et al., 2018).
Notably, in 2015, Zhan et al. engineered a novel structure for GOS-5 (Table 1), incorporating six T bases at both the 5′ and 3′-ends. This structural modification facilitated the concurrent detection of Pb(II) and Hg(II) ions (Zhan et al., 2015). The fluorescent indicator GeneFinderTM, distinguished by its heightened affinity for dsDNA compared to ssDNA, was instrumental in this configuration, eliciting a more robust fluorescence signal. It is effectively bound to GOS-5 by electrostatic interactions, producing moderate-intensity fluorescence. The presence of Pb(II) ions prompted the formation of a G-4 structure, leading to the stacking of the GeneFinderTM fluorophore and subsequent quenching of fluorescence. On the other hand, Hg(II) ions facilitated the creation of a T-Hg(II)-T hairpin structure with GOS-5, engendering more intense fluorescence when the fluorophore was excited (Fig. 3b). This approach, while effective, did involve a detection process lasting 45 min. Nevertheless, it's remarkable that the GOS-5 sensor demonstrated remarkable sensitivity, detecting Hg(II) and Pb(II) ions at concentrations as little as 2.33 ppb (11.62 nM) and 2.62 ppb (12.66 nM), separately (Table 2). This dual detection capability provided a pioneering avenue to detect multiple heavy metal ions simultaneously. Building on this progress, Zhu et al. further refined their efforts in 2018 by designing a bifunctional sensor (Fig. 3c, Table 2) capable of individually detecting Pb(II) and Hg(II) ions (Zhu et al., 2018). This novel approach relied on distinct fluorescence changes for each type of ion. This design significantly reduced detection time, achieving an impressive 10-minute reduction.
2.2 “Off-on” model
In the “off–on” model, the fluorescence of intercalated dyes is restored by binding to the target DNA structures due to the assistance of Pb(II). In 2010, Li et al. introduced a novel approach utilizing the GOS-3 probe in conjunction with the fluorophore ZnPPIX (Li et al., 2010). The method involved the formation of a dsDNA between GOS-3 and its complementary strand, with ZnPPIX exhibiting no recognition capability toward dsDNA. When Pb(II) existed, GOS-3 underwent a structural transformation, folding into a G-4 configuration. Intriguingly, ZnPPIX demonstrated an affinity for G-4, resulting in a significant enhancement in the fluorescence signal (Fig. 4a). Subsequently, after detecting Pb(II), the introduction of DOTA competitively bound to Pb(II), reinstating the sensor to its original state, thus allowing for multiple reuse cycles. The fluorescence intensity correlated linearly with Pb(II) concentrations from 20 nM to 1 µM (Table 2). Impressively, the detection limit for Pb(II) stood at 20 nM, underscoring the method's remarkable sensitivity. This innovative strategy facilitated the specific recognition of Pb(II) ions and demonstrated the potential for multiple reuse cycles through competitive binding.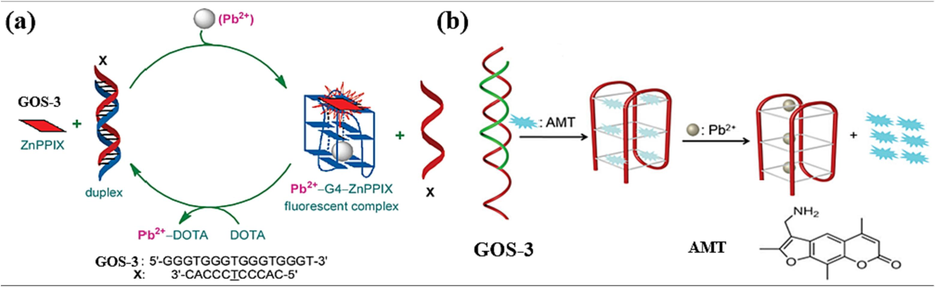
Based on the interplay between dsDNA and G-4 structural conversions: (a) ZnPPIX as the fluorescence indicator (Li et al., 2010); (b) AMT as the fluorescence indicator (Geng et al., 2020).
In 2020, a significant advancement was made by Geng et al. with the introduction of a sensor that harnessed the same sequence of the GOS-3 probe in combination with the fluorophore AMT (Geng et al., 2020). The methodology capitalized on AMT’s inherent propensity for strong fluorescence in solution. The interaction between AMT and the GOS-3 probe led to the formation of a G-4/AMT complex. The fluorescence was quenched because of electron transfer from the G bases in G-4 to AMT. However, when Pb(II) ions induced GOS-3 to fold into a G-4 structure, releasing AMT from G-4. This exchange triggered fluorescence excitation and subsequent Pb(II) ion recognition (Fig. 4b). The resulting fluorescence exhibited a linear relationship with increasing concentrations of Pb(II) ions within the range of 0.1 µM to 1 µM (Table 2). This property allowed for quantitative analysis of Pb(II) ions in clinical samples, presenting a powerful tool for clinical analysis.
Meanwhile, various other G-base-rich oligonucleotide sequences have been harnessed for Pb(II) detection, leading to diverse detection methodologies. Notably, in 2012, Jacobi et al. strategically employed GOS-6 for Pb(II) detection (Fig. 5a) (Jacobi et al., 2012). This method incorporated the fluorescent agent TO to stain GOS-6, manifesting as yellow and yielding a relatively weak fluorescent signal. However, upon the introduction of Pb(II), the two GOS-6 orchestrated the formation of a G-4 structure. This structural alteration induced a shift in staining color to green, accompanied by a significantly enhanced fluorescent signal. While effective, It is essential to note that this sensor demonstrated a relatively high detection limit of 8 nM, indicating potential for further sensitivity improvement (Table 2). Similarly, in 2015, Xu et al. pursued a parallel avenue (Fig. 5b), capitalizing on the same sequences of GOS-6 and embedded fluorescent agent NMM (Xu et al., 2015). Pb(II) ions were instrumental in guiding GOS-6 towards folding into a G-4 structure, thereby inducing fluorescence. Notably, this approach demonstrated an impressive reduction in the detection limit for Pb(II) ions, achieving sensitivity levels as low as 3 nM (Table 2). An additional advantage lay in the rapid binding kinetics of NMM to the G-4 structure catalyzed by Pb(II) ions, resulting in a fluorescence signal saturation time of less than 2 min. This efficiency enhancement effectively curtailed detection times and augmented the method's practical applicability.
Identify G-4-Pb(II) by various intercalators: (a) TO as the fluorescence indicator (Jacobi et al., 2012); (b) NMM as the fluorescence indicator (Xu et al., 2015); (c) DFHBI as the fluorescence indicator (DasGupta et al., 2015); (d) MG as the fluorescence indicator (Li et al., 2018); (e) compound 1 as the fluorescence indicator (Zhao et al., 2016).
An innovative breakthrough was achieved by DasGupta et al., who conceived the pioneering RNA probe GOS-7 (Table 1) (DasGupta et al., 2015). This development marked a significant departure, representing the first instance of an RNA aptamer being utilized for Pb(II) detection. The strategy hinged on the inherent structural dynamics of RNA sequences, leading to novel advantages and possibilities. Upon introducing a limited quantity of Pb(II) ions, GOS-7 exhibited a distinctive folding pattern, assembling into a two-layered G-4 structure. This structure, in turn, interacted with the fluorophore DFHBI, inducing a substantial fluorescence signal (Fig. 5c). The sensor’s fluorescence intensity demonstrated a linear correlation with Pb(II) ion concentrations within the range of 5 nM to 500 nM, with an impressive detection limit of 6 nM for Pb(II) ions (Table 2). It marked a pioneering foray into RNA-based Pb(II) detection, offering simplicity, ease of design, and operation. Furthermore, in 2018, Lu et al. revealed a novel application involving the probe GOS-8 (Table 1) and the fluorophore MG (Li et al., 2018). This strategy identified an antiparallel G-4 structure induced by Pb(II) ions. This complex interaction with Pb(II) ions yielded distinctive structural characteristics that could be harnessed for sensitive detection purposes (Fig. 5d). The outcomes of this study demonstrated a linear correlation between the fluorescence intensity difference and the concentration of Pb(II) ions. This linear relationship spanned the concentration range from 22.4 nmol/L to 475 nmol/L (Table 2). This achievement marked a significant stride toward enhanced detection methodologies for Pb(II) ions, offering insights into the dynamic interactions between specific nucleic acid structures and heavy metal ions.
In a departure from commercially available dyes, Zhao et al. introduced a homemade fluorescent indicator called “compound 1”, a water-soluble cationic perylene derivative (Zhao et al., 2016). This innovative strategy (Fig. 5e) showcased a distinct pathway to achieve sensitive Pb(II) detection, capitalizing on the interaction between compound 1 and the GOS-9 probe, as outlined in Table 1. In this methodology, the strong affinity between compound 1 and GOS-9 led to the accumulation of compound 1 and consequent fluorescence quenching. However, upon adding Pb(II) ions, GOS-9 transformed from an irregular coil structure to a G-4 configuration. This structural shift significantly weakened the affinity between GOS-9 and compound 1, resulting in the release of compound 1 and the subsequent restoration of fluorescence. One noteworthy achievement of this method was extending the range of detectable Pb(II) ion concentration from 10 nM to 1 µM. The detection limit was also impressively reduced to 5 nM (Table 2). The practicality of this method was further emphasized by its potential for yielding a unique response in the presence of Pb(II) ions, demonstrating the versatility and creativity of contemporary sensor design.
2.3 Enzyme-assisted methods
The emergence of DNAzymes as functional nucleic acids with both catalytic activity and specificity for divalent metal ions has garnered substantial interest in recent years, particularly for their application in label-free sensor design. In the presence of Pb(II), the substrate chain undergoes an inducement to cleave at the designated cleavage site rA. This cleavage liberates G-rich sequences, subsequently facilitating the assembly of a G-4 structure. This structural alteration triggers the recognition of an intercalator dye, thereby eliciting a fluorescence signal. In 2013, Zhao et al. contributed to this evolving field by designing a novel sensor with distinct capabilities (Zhao et al., 2013). The essence of this sensor lay in its response to the presence of Pb(II) and K(I) ions. No significant fluorescence alteration was observed when each ion was present individually. However, a unique response emerged when Pb(II) and K(I) were simultaneously introduced. In this scenario, Pb(II) initiated the fracture of the GR-5 DNAzyme strand, while K(I) prompted the folding of the poly-G loop of the GOS-10 probe (Table 1) into G-4. As a result, the fluorescence of the aloe-emodin derivative (AED) was quenched (Fig. 6a). Remarkably, this method exhibited a shallow detection limit for Pb(II), reaching an impressive 22.8 pM. A robust linear relationship between ion concentration and fluorescence burst efficiency was observed within 50 pM to 50 nM (Table 2).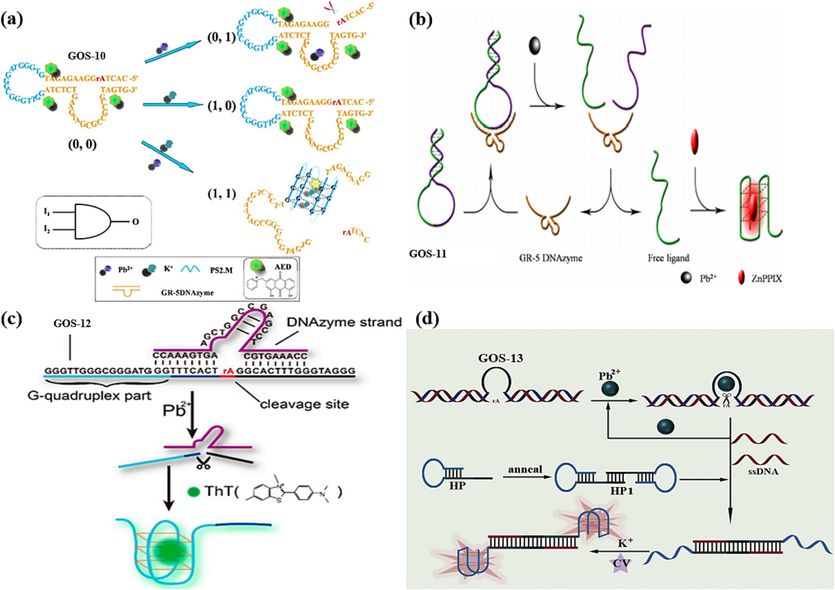
DNA-based Biosensors for Pb(II) detection using DNAzyme: (a) AED as the fluorescence indicator (Zhao et al., 2013); (b) ZnPPIX as the fluorescence indicator (Fu et al., 2016); (c) ThT as the fluorescence indicator (Wen et al., 2016); (d) CV as the fluorescence indicator (Zhou et al., 2021).
In 2016, a parallel innovation was introduced by Fu et al., who devised a sensor also grounded in the GR-5 DNAzyme, as depicted in Fig. 6b (Fu et al., 2016). This ingenious design harnessed the DNAzyme's catalytic abilities to affect a responsive detection mechanism. The sensor’s operation was elegantly orchestrated: the presence of Pb(II) ions led to the cleavage of the GOS-11 probe (Table 1) at the GR-5 DNAzyme stem. Consequently, the G-rich sequences of GOS-11 underwent a pivotal transformation, folding into a G-4 structure. ZnPPIX is incorporated within this structure, resulting in a heightened and more pronounced fluorescent signal. The critical attribute of this method lay in its impressive detection limit for Pb(II) ions, which stood at a mere 3 nM. Additionally, the range of detectable concentrations was notably expanded, encompassing the spectrum from 5 nM to 100 nM (Table 2).
Similar work was carried out by Wen et al., leveraging the synergy between DNAzymes and ThT, and a dynamic and responsive sensor was created, as outlined in Fig. 6c (Wen et al., 2016). The sensor's architecture revolved around the GOS-12 probe (Table 1), which formed a hybrid structure with the DNAzyme chain, effectively inhibiting the form of the G-4 structure. However, the introduction of Pb(II) ions had a transformative effect. Pb(II) cleaved the GOS-12 and liberated G-rich sequences. Subsequently, these liberated sequences were skillfully induced to fold into a G-4 configuration through the interaction with ThT. This event culminated in the manifestation of a strong fluorescent signal. A linear correlation between the fluorescence signal and the Pb(II) ion concentration logarithm was established within the concentration range of 10 nM to 1000 nM. Significantly, the detection limit achieved was a mere 0.06 nM (Table 2).
Recently, Zhou et al. employed a Pb(II) ion-dependent 8–17 DNA enzyme to activate the cleavage site rA, resulting in the liberation of an ss-DNA fragment (Fig. 6d) (Zhou et al., 2021). This ss-DNA strand served as a catalyst, effectively opening the dumbbell-shaped hairpin DNA by orchestrating them folding into two G-4 structures. Upon the subsequent introduction of K(I) ions and crystalline violet (CV), a synergistic effect occurred, leading to the encapsulation of CV as a fluorescent agent within a CV/G-4 conjugate complex, releasing the fluorescent signal. Remarkably, the developed sensor exhibited a robust linear correlation between the fluorescence signal and Pb(II) concentration within 5 nM to 2.5 µM and 2.5 µM to 25 µM. The sensor’s detection capabilities showcased an impressive breadth, as summarized in Table 2.
However, the GR-5 DNAzyme exhibits superior selectivity compared to the 8–17 DNAzyme (Liang et al., 2017). This heightened selectivity is attributed to the GR-5 DNAzyme’s interaction with the graphene oxide (GO) surface, which is mediated by hydrogen bonding and π-stacking interactions. In 2017, Ravikumar et al. orchestrated a dual-metal ion detection system targeting Pb(II) and Hg(II) (Ravikumar et al., 2017). The design hinged on the strategic interplay between the GOS-14 probe (Table 1), GR-5 DNAzyme strand, and GO, leading to a responsive sensing mechanism. Initially, GOS-14 formed a complex with the DNAzyme, followed by binding to the GO surface, culminating in creating the GO-DNAzyme complex. When Pb(II) was introduced, a transformative cascade was set in motion. The presence of Pb(II) prompted the cleavage of the GOS-14 complex, leading to the release of the DNA chains containing GT. Subsequently, this liberated sequence folded into a G-4 structure, inducing a strong fluorescent signal through ThT. On this basis, the introduction of Hg(II) ushered in a discernible response. Hg(II) was pivotal in creating a distinctive T-Hg(II)-T structure. This structural shift disrupted the pre-existing G-4 configuration, yielding a pronounced reduction in the emitted fluorescent signal. Consequently, this unique alteration in fluorescence behavior provided a means for accurately detecting Hg(II), as demonstrated in Fig. 7a. Notably, this biosensor exhibited an impressive detection prowess, boasting remarkably low detection limits for both Pb(II) and Hg(II), quantified at 96 pM and 356 pM, respectively (Table 2). The fluorescence intensity gradually enhanced, increasing Pb(II) ion concentration within the range of 0 nM to 0.05 nM. Conversely, it gradually declined with the elevation in Hg(II) ions concentration. While the quantitative range was relatively limited, the sensitivity of the method held significant promise for sensitive dual-metal ion detection.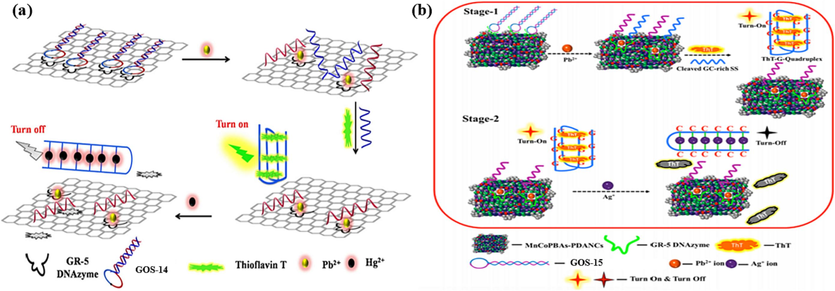
DNAzyme-based biosensors for two heavy metal ions detection: (a) ThT as the fluorescence indicator and GO as the quencher for the Pb(II) and Hg(II) detection (Ravikumar et al., 2017); (b) ThT as the fluorescence indicator and MnCoPBAs-PDANCs as the quencher for the Pb(II) and Ag(I) detection (Rajaji and Panneerselvam, 2020).
Similarly, in 2020, Panneerselvam et al. contributed to the advancement of dual-metal ion detection, specifically targeting Pb(II) and Ag(I) (Rajaji and Panneerselvam, 2020). The sensor's architecture rested on DNAzyme/MnCoPBAs-PDANCs complexes and the GOS-15 probe, as outlined in Table 1. The binding between GOS-15 and the DNAzyme strand facilitated the attachment to MnCoPBAs-PDANCs surfaces, inhibiting the fluorescence signal. The introduction of Pb(II) initiated a complex sequence of events: the cleavage of the complex, the release of a GC-containing sequence, and its subsequent folding into a G-4 structure, leading to a pronounced fluorescence enhancement through ThT. Remarkably, the presence of Ag(I) disrupted this sequence, forming a C-Ag(I)-C dsDNA and inducing a drastic reduction in fluorescence (Fig. 7b). The sensing element showcased linearity within the Pb(II) concentration range of 3 nM to 9 nM and the Ag(I) concentration range of 6 nM to 20 nM, with a detection limit of 1.6 nM and 4.2 nM (Table 2).
The formation of a steady complex between the G-4 structure and Hemin yields the G-4/hemin DNAzyme, renowned for its robust catalytic activity, often harnessed for identifying heavy metals like Pb(II). In 2011, a notable study by Li et al., a Pb(II) detection strategy employing the GOS-6 designed sensor was introduced (Fig. 8a) (Li et al., 2011). In this approach, Pb(II) induces the GOS-6 probe to adopt a G-4 structure. Subsequently, this G-4 configuration interacts with hemoglobin, catalyzing H2O2-mediated oxidation of Amplex UltraRed (AUR), resulting in a striking augmentation of fluorescence intensity. The concentration of Pb(II) ions exhibited a linear correlation with fluorescence intensity within a range spanning 0 to 1000 nM, achieving a remarkably low detection limit of a mere 0.4 nM, as detailed in Table 2. This technique showcases exceptional sensitivity and selectivity in detecting Pb(II).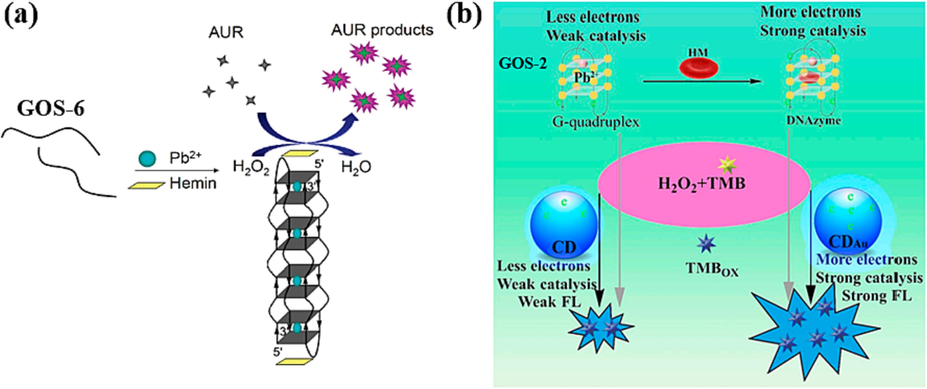
Based on G-4/hemin DNAzyme: (a) AUR products as the fluorescence indicator (Li et al., 2011); (b) CDAu as the fluorescence indicator (Li et al., 2020).
In 2020, Li et al. achieved a significant breakthrough in the realm of Pb(II) detection by harnessing Gold-doped carbon dots (CDAu) (Li et al., 2020). The pivotal concept revolved around the coupling of the GOS-2 sequence, which was adsorbed onto the surface of CDAu. This coupling resulted in a non-fluorescent state of the system. The introduction of Pb(II) induces the folding of the Pb(II)-aptamer into a G-4 structure, leading to the detachment of the G-4 from CDAu. The liberated G-4 molecule, in conjunction with heme, served as the foundation for a G-4/heme DNAzyme entity. This DNAzyme configuration catalyzed the oxidation of H2O2-TMB, ultimately yielding the production of the oxidized TMB compound (TMBOX) with a fluorescence signal, as depicted in Fig. 8b. The method exhibited a commendable capability in detecting Pb(II) concentrations spanning from 0.0005 to 0.46 µM, and showcased an impressive limit of detection, reaching 0.25 nM. The sensitivity of this sensor marks a significant advancement compared to previously reported sensing approaches, as outlined in Table 2. This innovation holds promise for enhanced Pb(II) detection, signifying a step forward in heavy metal ion analysis.
3 Labeled-methods
Labeling involves attaching a fluorescent dye and a quenching group to a probe. The configuration of the DNA probes, induced by Pb(II), undergoes a transition into a G-4 structure, thereby triggering a change in fluorescence that serves as the basis for Pb(II) detection. Commonly employed quenchers encompass 4-([4-(dimethylamino)phenyl]azo)benzoic acid (DABCYL) (Liu et al., 2009) and members of the black hole quencher (BHQ) series (Lu et al., 2019). In subsequent investigations, it has been determined that consecutive G bases, along with G-based G-4 structures (Zhan et al., 2013, Zhang et al., 2018), can also serve as effective quenchers. Moreover, nanomaterials have gained prominence in constructing DNA biosensors due to their robust quenching capabilities. Notably, materials such as GO (Li et al., 2013) and gold nanoparticles (Chung et al., 2013) have been harnessed in the context of Pb(II) detection.
3.1 Common quenchers
In 2009, Liu et al. introduced a GOS-2 probe, as outlined in Table 1, which was labeled with fluorescein (FAM) at one end and the quencher DABCYL at the other (Liu et al., 2009). Upon interaction with Pb(II), GOS-2 underwent a conformational shift from an unbound state to a G-4 structure. The resulting fluorescence resonance energy transfer (FRET) between FAM and DABCYL led to fluorescence quenching. Similarly, when exposed to Hg(II), GOS-2 adopted a double helix configuration, resulting in analogous FRET-induced fluorescence attenuation, depicted in Fig. 9a. Pb(II) and Hg(II) were detected as low as 300 pM and 5 nM, respectively. The fluorescence response exhibited a linear correlation with ion concentration within the ranges of 0.5–30 nM for Pb(II) and 10–200 nM for Hg(II), as indicated in Table 3. Notably, this marks the advent of the first sensor with the remarkable capability to concurrently detect both Hg(II) and Pb(II), thereby introducing a novel avenue for designing sensing elements.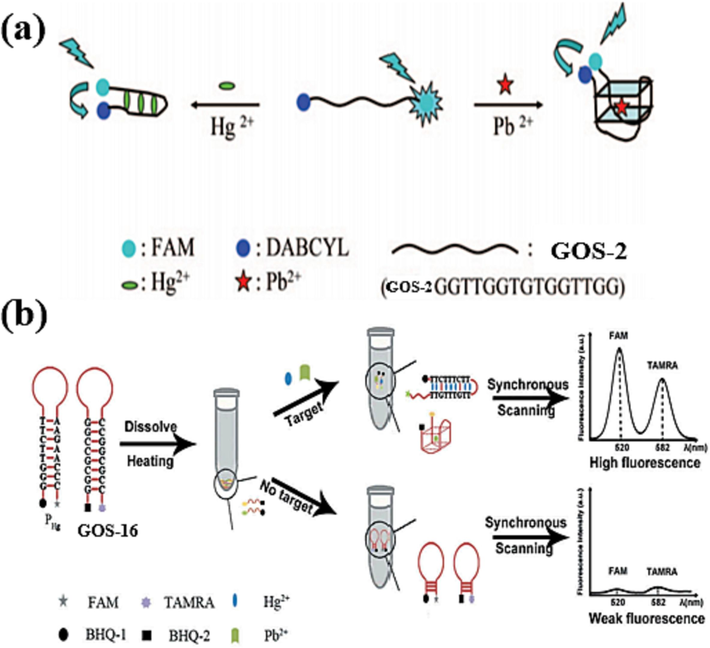
Fluorescence dyes quenched by common quenchers: (a) DABCYL as the fluorescence quencher (Liu et al., 2009); (b) BHQ-1 and BHQ-2 as the fluorescence quenchers (Lu et al., 2019).
Probe
Fluorophore
Quencher
Detection linear range
LOD
Signal
Ref.
GOS-2
FAM
DABCYL
0.5–30 nM
0.3 nM
Turn Off
(Liu et al., 2009)
GOS-16
TAMRA
BHQ-2
0.2–24 nM
0.16 nM
Turn On
(Lu et al., 2019)
GOS-3
FAM
G-4
4.83–96.62 nM
3.72 nM
Turn Off
(Zhan et al., 2013)
GOS-3
NMM, FAM
G-4
60–300 nM
28 nM
Turn Off
(Liu et al., 2021)
GOS-17
FAM
G bases
0.5–500 nM
0.4 nM
Turn Off
(Wang et al., 2013)
GOS-18
HEX
G-4
1–100 nM
0.096 nM
Turn On
(Zhang et al., 2018)
GOS-19
HEX
G bases
1–20 nM
0.092 nM
Turn Off
(Zhang et al., 2021)
GOS-20
FAM
GO
2–50 nM
0.400 nM
Turn On
(Li et al., 2013)
GOS-3
FAM
GO
0.005–0.07 nM; 0.07–20 nM
0.0005 nM
Turn On
(Khoshbin et al., 2019)
GOS-21
CdSe/ZnS QD
GO
0.1–10 nM
0.09 nM
Turn On
(Li et al., 2013)
GOS-21
rGQDs
GO
9.9–435 nM
0.6 nM
Turn On
(Qian et al., 2015)
GOS-22
Cy 5.5TM
AuNPs
0.01–1000 nM
0.027 nM
Turn On
(Chung et al., 2013)
GOS-23
UCNPs
AuNPs
0.1–100 nM
0.05 nM
Turn On
(Wu et al., 2014)
In 2019, Lu et al. introduced an innovative approach by incorporating BHQ-2 as the quencher to enhance assay sensitivity (Lu et al., 2019). The fluorophore tetramethyl-6-carboxy rhodamine (TAMRA) was conjugated to the probe GOS-16 (Table 1). As shown in Fig. 9b, the probe showcased a hairpin configuration wherein TAMRA and BHQ-2 existed close, resulting in quenched fluorescence. Upon the presence of Pb(II), the GOS-17 probe underwent a structural shift from a hairpin to a G-4 arrangement. This transition facilitated the separation of TAMRA from BHQ-2, subsequently reinstating the fluorescence signal. Additionally, a similar probe was designed to enable Hg(II) detection, wherein the fluorophore FAM and the quencher BHQ-1 were incorporated. Upon the introduction of Hg(II), a T-Hg(II)-T structure formed, akin to the previously discussed mechanism, leading to the separation of FAM from BHQ-1 and the consequent restoration of the fluorescence signal. The fluorescence response of Pb(II) exhibited a linear correlation with TAMRA fluorescence between 0.2 and 24 nmol/L concentration. Similarly, the fluorescence of Hg(II) displayed a linear relationship within the concentration range of 0.7 to 84 nmol/L. The calculated detection limits were 0.16 nmol/L for Pb(II) and 0.36 nmol/L for Hg(II), respectively, as illustrated in Table 3. This research demonstrated a creative and sensitive approach to simultaneous Pb(II) and Hg(II) detection.
3.2 G bases as quenchers
Although conventional quenchers have good quenching ability, they have some disadvantages, such as poor stability and short life. One of the primary concerns is the high cost and complexity associated with the labeling process, which involves attaching the quenchers to DNA sequences. This procedure can be time-consuming and technically demanding, potentially leading to an increase in price, and the purity of the probes was also affected. Therefore, methods that utilize DNA structural changes to quench the fluorescence have attracted much attention from researchers.
In 2013, Zhan et al. achieved a substantial advancement in Pb(II) ion detection through the utilization of a FAM-labeled GOS-3 probe (Table 1) (Zhan et al., 2013). This innovative sensor design exhibited a broad Pb(II) ion detection range: the labeled FAM exhibited a strong fluorescence signal in its free state. Upon introducing Pb(II) ions, the probe underwent a conformational change to form a G-4 structure. This structural alteration brought the FAM-Pb(II) distance in proximity to the Pb-O distance, facilitating reasonable FRET and resulting in fluorescence quenching, as illustrated in Fig. 10a. Within the concentration range of 1–20 ppb (4.83–96.62 nM), the fluorescence signal demonstrated a gradual decline with increasing Pb(II) ion concentration, thereby establishing a robust linear relationship, the detection limit for Pb(II) was only 0.77 ppb (3.72 nM) (Table 3). Furthermore, while K(I) does possess the capability to induce G-rich oligonucleotides to form G-4 structures, the distance of K-O, O-O and the vertical separation of G-4 is typically longer than those of the stable G-4 formed by Pb(II). Consequently, the FRET rate with K(I) is significantly lower than that observed with Pb(II), rendering the assay highly specific for Pb(II) ion detection. A notable advantage of this method is its rapid reaction time of only 5 min, contributing to a considerable reduction in detection time. The method achieves detection by generating a G-4 structure and changing the conformation of GOS-3 to quench the fluorescent dye.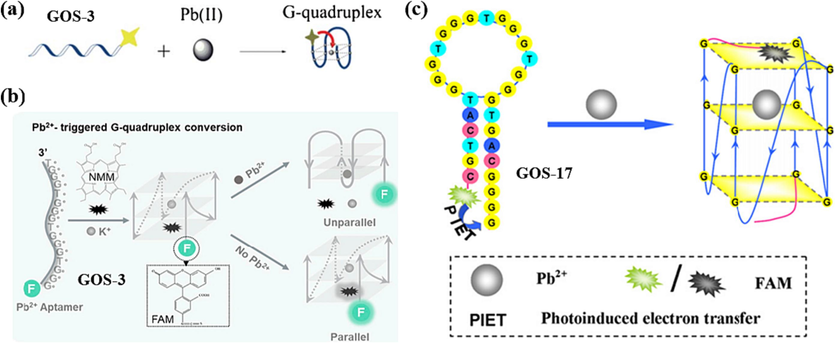
G bases as the fluorescence quenchers: (a) Pb(II)-stabilized G-4 formation (Zhan et al., 2013); (b) a combination of labeled and unlabeled methods (Liu et al., 2021); (c) based on different PIET interactions between G bases and G-quartet (Wang et al., 2013).
In 2021, Liu et al. devised an innovative sensor design with more accurate detection capabilities. The sensor's functioning is structured as follows: K(I) triggers the GOS-3 probe (Table 1) to assume a parallel G-4 structure that can bind to NMM, generating a strong fluorescent signal. However, when Pb(II) ions are introduced, the G-4 transforms into an antiparallel G-4 structure. In this conformation, it cannot effectively bind to NMM, leading to the extinguishing of the fluorescent signal, as depicted in Fig. 10b (Liu et al., 2021). Furthermore, FAM, which is labeled at the end of the GOS-3 probe, serves as a fluorescent reference for the proportional determination of Pb(II). The proportional assay remains unaffected by changes in probe concentration and instrumental variations. The concentration of Pb(II) ions exhibited a robust linear relationship with fluorescence intensity in the range of 0.06–0.3 µM, with a remarkable detection limit of just 0.028 µM, as detailed in Table 3. While NMM is commonly utilized as a fluorescent agent for recognizing G-4, its utilization as a single signal can render assay outcomes susceptible to fluctuations in probe concentration and instrumental factors. This method ingeniously combines the stability-enhancing qualities of NMM with the FAM fluorophore positioned at the probe's end to significantly improve the accuracy and reliability of Pb(II) detection. This method demonstrates that G bases have a specific quenching ability, but their quenching effect is affected by the distance between the G base and the fluorophore.
Some researchers have already found that G bases have good bursting ability, which can be detected for metal ions by photo-induced electron transfer (PIET) between the fluorescent dye and the G bases, leading to fluorescence quenching (Knemeyer et al., 2000). In 2013, Wang et al. devised an innovative approach utilizing the GOS-17 probe (Wang et al., 2013), outlined in Table 1. Their strategy capitalized on the distinct quenching abilities between G bases and G-quartet structures. They incorporated three G bases at the 3′ end of the GOS-17, and FAM was introduced at the 5′ end. The hairpin DNA structure brought the three G bases and FAM in close proximity, promoting electron transfer and leading to decreased fluorescence intensity. However, in the presence of Pb(II) ions, the hairpin DNA structure transformed into a G-4 arrangement. In this configuration, FAM was able to stack onto the G-4 structure, enhancing the electron transfer capability and consequently leading to a further reduction of the fluorescence intensity (Fig. 10c). This method exhibited a remarkable capacity to detect Pb(II) ions at a sensitivity of 0.4 nM, demonstrating both cost-effectiveness and high sensitivity. The fluorescence quenching efficiency exhibited a linear relationship with ion concentration within the range of 0.5–500 nM for Pb(II) (Table 3). This approach presented a unique Pb(II) detection strategy, showcasing the potential for sensitive and selective analysis.
In 2018, Zhang et al. developed a sensor that capitalizes on the G-4′s ability to quench fluorophores (Zhang et al., 2018). This sensor, based on the GOS-18 probe (Table 1), exhibited a unique dual-detection capability for Pb(II) and Ag(I). When solely Pb(II) is present, the G-rich segment of GOS-18 forms a G-4 structure. At this point, the fluorophore HEX is distanced from the G-4 structure, resulting in a strong fluorescent signal from HEX. However, when Ag(I) is also introduced, the formation of a C-Ag(I)-C complex with the C base facilitates photoinduced electron transfer (PET) in the proximity of HEX and the G-4, effectively quenching the fluorescence signal, illustrated in Fig. 11a. Similarly, in cases where only Ag(I) is present, a hairpin structure is formed, leading to a strong fluorescence signal from HEX. Upon introducing Pb(II), induced folding of GOS-18 into a G-4 structure, resulting in the quenching of the fluorescence signal. The sensor demonstrated a linear correlation between fluorescence intensity and the concentrations of both ions in the range of 1–100 nmol/L. Notably, the detection limits were reported as 96 pmol/L for Pb(II) and 21 pmol/L for Ag(I), as summarized in Table 3. This innovative approach provides a promising avenue for simultaneously detecting these two metal ions with high sensitivity.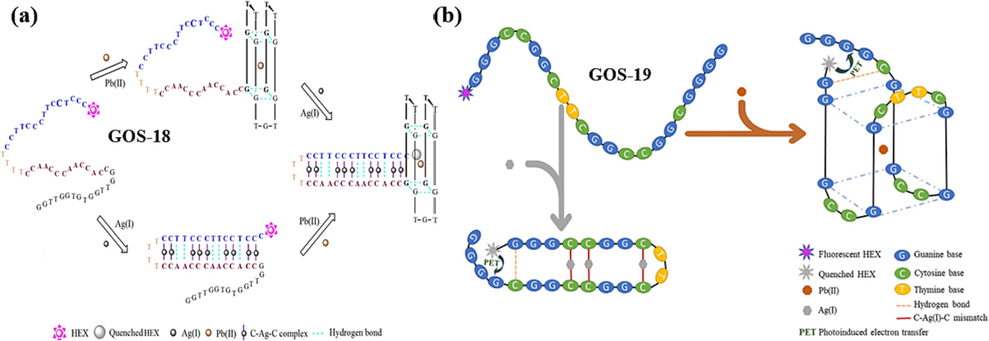
Fluorophore quenched by the formation of G-4 for two heavy metal ions detection: (a) the simultaneous detection of trace Pb(II) and Ag(I) ions using a HEX-labeled GOS-18 (Zhang et al., 2018); (b) the simultaneous detection of Pb(II) and Ag(I) ions using a HEX-labeled GOS-19 probe (Zhang et al., 2021).
In 2021, Zhang et al. made further refinements and introduced the GOS-19 probe (Table 1) (Zhang et al., 2021). Again, the fluorophore HEX was labeled at the 5′ end of the probe based on the formation of a G-4 structure by Pb(II) and a C-Ag(I)-C mismatch structure by Ag(I). The difference, however, is that the identification of Pb(II) and Ag(I) is independent, and the two ions do not need to rely on the other to achieve detection. The probe has four consecutive G bases near the fluorophore HEX and undergoes PET-quenched fluorescence when a hairpin structure is formed in the presence of Ag(I). This innovative strategy enabled a single probe to simultaneously detect two distinct metal ions, as depicted in Fig. 11b. The fluorescence intensity displayed a linear relationship with ion concentration for Pb(II) and Ag(I) within the 1–20 × 10-9 mol/L concentration range. The detection limits achieved were 92 × 10-12 mol/L for Pb(II) and 82 × 10-12 mol/L for Ag(I), as summarized in Table 3. This advancement marked a significant stride in multiplex metal ion detection, underlining the potential for dual-metal ion sensing using a single probe.
3.3 Nanomaterials as quenching groups
Although G base as a quencher has the advantages of inexpensive and straightforward operation, it has certain limitations in probe design, which not only has more stringent requirements on the spatial structure of the probe but also has lower quenching efficiency, leading to curtailing the sensitivity of ion detection. To overcome these limitations, researchers have utilized various nanomaterials as alternatives (Dil et al., 2021, Mehrabi and Ghaedi, 2023). Nanomaterials such as GO and nanoparticles offer distinct advantages over traditional labeling methods. Many nanomaterials have been developed in recent years, such as GO (Li et al., 2013, Khoshbin et al., 2019), quantum dots (Li et al., 2013, Qian et al., 2015), and nanoparticles (Chung et al., 2013, Wu et al., 2014), due to their better quenching ability, high stability, long lifetime, and biocompatibility.
GO has become an essential nanomaterial in biosensing due to its distinctive properties, including its capability to quench fluorescence through π-π interactions with adsorbed ss-DNA. This quenching can occur through mechanisms like FRET (Li et al., 2013, Khoshbin et al., 2019) or phototransfer electron (Qian et al., 2015) transfer. One of the applications of GO in biosensing is its role in quenching the fluorescence of labeled probes, which can then be modulated based on the presence of specific target molecules. In 2013, Li et al. utilized a modified G-rich oligonucleotide sequence called GOS-20 (Table 1) (Li et al., 2013). The FAM fluorophore, labeled on the GOS-20, is brought into proximity to the GO surface due to the high affinity between ss-GOS-20 and GO, resulting in a significant reduction in fluorescence signal. As shown in Fig. 12a, in the presence of Pb(II) ions, the GOS-20 assumes a specific G-4 complex structure that weakens its affinity for GO, leading to a reduced quenching effect. The signal reduction correlates with Pb(II) ions, enabling the sensor to detect the ions. This method takes 10 min for Pb(II) detection, demonstrating high specificity and efficiency. It indicates that the sensor's fluorescence signal exhibited a linear relationship with the concentration of Pb(II) ions when present in the range of 2–50 nM, and the detection limit was 400 pM (Table 3).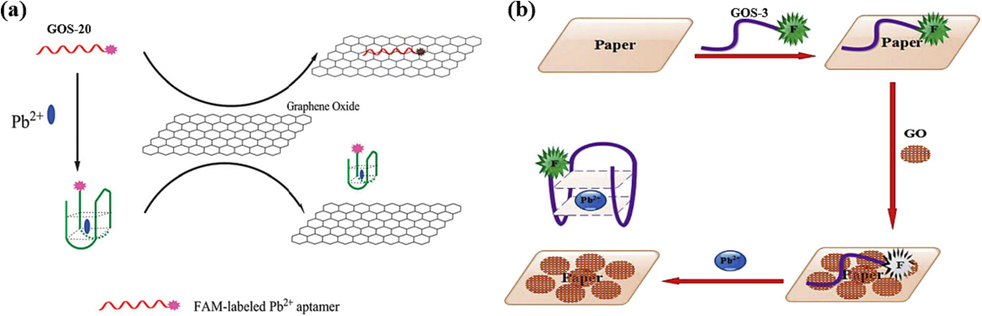
GO as the fluorescence quencher: (a) a FAM-labeled GOS-20 probe for Pb(II) detection (Li et al., 2013); (b) paper-based sensors for Pb(II) detection (Khoshbin et al., 2019).
In 2019, Khoshbin et al. developed a paper-based sensing sensor, as illustrated in Fig. 12b (Khoshbin et al., 2019). The FAM-labeled nucleic acid chain binds to GO by non-covalent bonding, at which time the FAM fluorophore is transferred to the surface of GO by FRET, and fluorescence quenching occurs. The presence of Pb(II) induced the folding of GOS-3 from freestyle to a G-4 structure, at which point the charge density was enhanced, thereby reducing the binding of the nucleic acid chain to the GO, and the fluorescence signal was restored. Using fluorophore FAM-labeled nucleic acid chains GOS-3 (Table 1) reduced the detection limit to 0.5 pM. The Pb(II) concentration was linearly correlated with the fluorescence intensity at 5–70 pM and 0.07–20 nM ion concentrations (Table 3). Paper sensors have the advantage of low cost and real-time detection (Jiang et al., 2017). Nowadays, it has been applied in many fields, such as environmental monitoring and medical diagnosis. The method combines nanomaterials with paper-based components, resulting in a notable increase in detection sensitivity.
Conventional organic dyes have high fluorescence intensity but have the disadvantage of being susceptible to photobleaching. However, this problem is avoided by using inorganic quantum dots (QDs), which offer advantages such as good resistance to photobleaching and high fluorescence quantum yields (Zhang et al., 2022). In a study conducted by Li et al. in 2013, they developed a biosensor that harnessed the properties of CdSe/ZnS QDs and GO, as shown in Fig. 13a (Li et al., 2013). They designed nucleotide chains labeled as GOS-21 (Table 1). They incorporated them into an aqueous solution containing GO, forming a stable GO/aptamer-QD complex that exhibited relatively weak fluorescence. However, introducing Pb(II) ions into this system induced the formation of G-4/Pb(II) complexes within the nucleic acid chains, causing a significant increase in the fluorescence signal. This phenomenon effectively “turned on” the fluorescence signal, enabling the detection of Pb(II) ions. This rapid detection method requires only 25 min for the entire process, and it can be applied for the trace detection of Pb(II) ions in a river water environment, with an impressive detection limit of 90 pM. The fluorescence signal displayed a linear correlation with the increase in Pb(II) ion concentration within the range of 0.1–10 nM, as detailed in Table 3.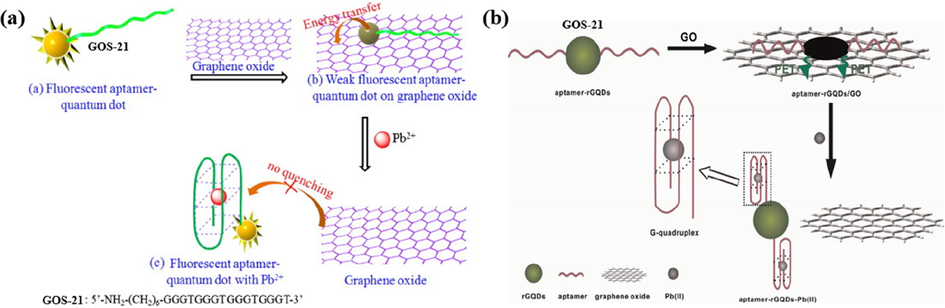
GO as the fluorescence quencher: (a) CdSe/ZnS QDs were labeled to GOS-21 for Pb(II) detection (Li et al., 2013); (b) rGQDs were labeled to GOS-21 for Pb(II) detection (Qian et al., 2015).
Graphene quantum dots (GQDs), known for their biocompatibility, hold great potential in biosensor construction (Qian et al., 2014). In 2015, Qian et al. developed a novel sensing method utilizing GOS-21 (Table 1) for the detection of Pb(II) ions (Qian et al., 2015), illustrated in Fig. 13b. This method employed a unique combination of GOS-21, reduced graphene quantum dots (rGQDs), and GO to achieve fluorescence-based Pb(II) detection. When Pb(II) ions were introduced, it caused the conformational change of GOS-21 and formed a stable G-4/Pb(II) complex. This structural alteration of the aptamer allowed it to detach from GO, resulting in fluorescence recovery. As a result, this method exhibited a wide detectable concentration range for Pb(II) ions, spanning from 9.9 nM to 435 nM. It achieved an impressive low detection limit of only 0.6 nM (Table 3).
In 2013, Chung et al. developed a versatile sensor that employs gold nanoparticles (AuNPs) as quenchers, allowing for the dual detection of Hg(II) and Pb(II) ions (Chung et al., 2013), as shown in Fig. 14a. A probe called GOS-22 (Table 1), which was coupled to AuNPs and hybridized to a Cy 5.5TM-labeled partially complementary DNA chain. In the absence of Pb(II), this setup caused the Cy 5.5TM to shift toward the AuNPs, effectively quenching the fluorescence signal. However, in the presence of Pb(II) ions, GOS-22 formed a G-4 structure, increasing the distance between Cy 5.5TM and AuNPs, resulting in the recovery of the fluorescence signal. And the same story to the Hg(II) detection. This sensor achieved simultaneous detection of two different metal ions and successfully demonstrated its applicability in serum containing DNA enzymes. The fluorescence intensity of both Pb(II) and Hg(II) increased linearly with their respective ion concentrations in the range of 10 pM to 1 µM, with detection limits of 27 pM for Pb(II) and 51 pM for Hg(II) (Table 3). High sensitivity and dual-ion capability make this sensor highly suitable for various applications, particularly environmental monitoring and clinical diagnostics.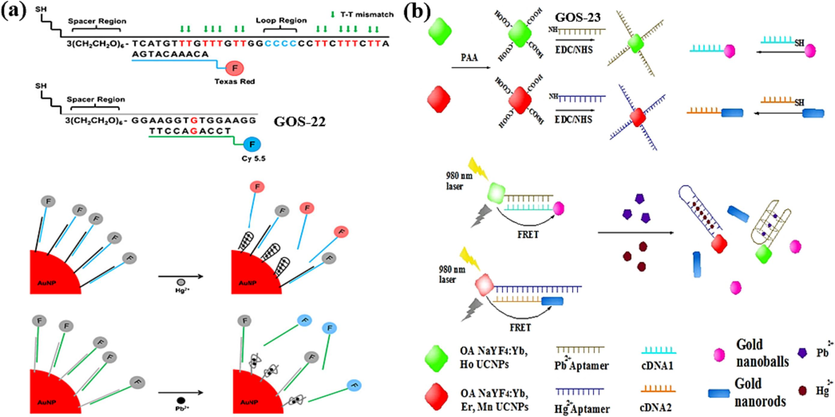
AuNPs as the fluorescence quencher for Hg(II) and Pb(II) detection: (a) Texas Red and Cy 5.5 as the fluorescence indicators (Chung et al., 2013); (b) UCNPs as the fluorescence indicators (Wu et al., 2014).
In 2014, Wu et al. introduced a dual FRET sensor that enabled the detection of both Pb(II) and Hg(II) ions (Wu et al., 2014). The sensor design involved an amino-modified GOS-23 (Table 1), which was initially linked to upconversion nanoparticles (UCNPs) and later bound to complementary DNA with gold nanoparticles (AuNPs). In this configuration, FRET occurred between UCNPs and AuNPs, leading to fluorescence quenching. However, when Pb(II) ions were introduced, they altered the conformation of GOS-23, causing it to fold into a G-4 structure. This structural transition led to the de-hybridization of the gold nanospheres and the subsequent fluorescence restoration. A similar principle was applied to detect Hg(II) ions. The sensor demonstrated a linear relationship between the Pb(II) concentration and the corresponding fluorescence intensity from 0.1 nM to 100 nM. Similarly, it exhibited a linear relationship between the Hg(II) concentration and the resultant fluorescence intensity, gracefully extending its sensitivity across the vast span of 0.5 nM to a commanding 500 nM. The method exhibited remarkable sensitivity, with detection limits of 50 pM for Pb(II) and 150 pM for Hg(II) (Table 3). This dual FRET sensor offered excellent sensitivity and selectivity, making it suitable for various environmental monitoring and analytical chemistry applications.
4 Conclusions
Fluorescent DNA biosensors possess unique properties that make them suitable for simple operation, high accuracy, and cost-effectiveness. These advantages, alongside their high application value and development potential, position them as a valuable tool for environmental detection and analysis, such as Pb(II) detection. This review explores various methods for detecting Pb(II) ions by forming Pb/G-4 structures using Pb(II)-specific oligonucleotides. These methods can be broadly categorized into non-fluorescently labeled probes and fluorescently labeled probes, and their construction principles are further categorized based on these distinctions. Among the non-labeling methods, there are subcategories such as signal on–off/off–on models. Pb(II) detection is accomplished using fluorophores embedded in DNA that bind differently to various structures, such as G-4 and DNA duplex, resulting in distinct fluorescent signals. The enzyme-assisted method employs DNA enzymes to cleave the substrate strand, facilitating the formation of the G-quadruplex structure, and has demonstrated significant advancements in lowering the limit of detection for Pb(II). In the labeling method, quenching groups play a crucial role, and these groups are further categorized into conventional quenchers, continuous G bases, and nanomaterials. The probes incorporate a conventional quencher and fluorescent dye, resulting in a robust quenching response by altering the DNA structure. However, the DNA probes have lower yields and higher costs. Using G bases as quenchers can simplify the DNA probe synthesis, but it may limit the diversification of DNA probe design, and fluorescence quenching ability may be compromised. Nanomaterials, including GO and AuNPs, are known for their high stability, excellent quenching ability, and strong biocompatibility. They are extensively used as fluorescent quenchers in Pb(II) detection, and this area of research is likely to be a prominent focus for future investigations. This review serves as a valuable reference platform for researchers interested in designing convenient and ultra-sensitive biosensors. These biosensors hold the potential to yield results of unparalleled precision and sensitivity in the realm of Pb(II) ion detection, thereby illuminating the path forward in this critical domain. The methods discussed, particularly those utilizing fluorescence, offer practical approaches for achieving real-time, sensitive, and specific Pb(II) ion detection. Such detection methods are crucial for various applications, including environmental monitoring, food analysis, and disease detection.
Funding
This work was supported by “the Foundational Research Funds of Zhejiang Sci-Tech University under Grant (No. 23042137-Y).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Recyclable Target Metal-Enhanced Fluorometric Naked Eye Aptasensor for the Detection of Pb2+ and Ag+ Ions Based on the Structural Change of CaSnO3@PDANS-Constrained GC-Rich ssDNA. Acs Omega.. 2021;6:30580-30597.
- [CrossRef] [Google Scholar]
- Dual fluorometric biosensor based on a nanoceria encapsulated metal organic framework and a signal amplification strategy of a hybridization chain reaction for the detection of melamine and Pb2+ ions in food samples. New J Chem.. 2022;46:12952-12967.
- [CrossRef] [Google Scholar]
- A Turn-ON fluorometric biosensor based on ssDNA immobilized with a metal phenolic nanomaterial for the sequential detection of Pb(II) and epirubicin cancer drug. RSC Adv.. 2021;11:12361-12373.
- [CrossRef] [Google Scholar]
- Bismuth as a general internal standard for lead in atomic absorption spectrometry. Anal Chim Acta.. 2014;831:24-30.
- [CrossRef] [Google Scholar]
- Contents of cadmium, mercury and lead in fish from the Atlantic sea (Morocco) determined by atomic absorption spectrometry. Food Chem.. 2014;147:357-360.
- [CrossRef] [Google Scholar]
- Fluorescence and electrochemical assay for bimodal detection of lead ions based on Metal-Organic framework nanosheets. Talanta.. 2021;232:122405
- [CrossRef] [Google Scholar]
- Fluorometric determination of lead(II) by using aptamer-functionalized upconversion nanoparticles and magnetite-modified gold nanoparticles. Microchim Acta.. 2020;187:85.
- [CrossRef] [Google Scholar]
- Small molecular fluorescent probes for the detection of lead, cadmium and mercury ions. Coord Chem Rev.. 2021;429:213691
- [CrossRef] [Google Scholar]
- Nuclease-resistant DNA aptamer on gold nanoparticles for the simultaneous detection of Pb2+ and Hg2+ in human serum. Biosens Bioelectron.. 2013;41:827-832.
- [CrossRef] [Google Scholar]
- Spinach RNA aptamer detects lead(II) with high selectivity. Chem Commun (camb).. 2015;51:9034-9037.
- [CrossRef] [Google Scholar]
- Magnetic dual-template molecularly imprinted polymer based on syringe-to-syringe magnetic solid-phase microextraction for selective enrichment of p-Coumaric acid and ferulic acid from pomegranate, grape, and orange samples. Food Chem.. 2020;325:126902
- [CrossRef] [Google Scholar]
- Simultaneous selective enrichment of methylparaben, propylparaben, and butylparaben from cosmetics samples based on syringe-to-syringe magnetic fluid phase microextraction. Talanta.. 2021;221:121547
- [CrossRef] [Google Scholar]
- Highly selective magnetic dual template molecularly imprinted polymer for simultaneous enrichment of sulfadiazine and sulfathiazole from milk samples based on syringe-to-syringe magnetic solid-phase microextraction. Talanta.. 2021;232:122449
- [CrossRef] [Google Scholar]
- Genome-scale genetic screen of lead ion-sensitive gene deletion mutations in Saccharomyces cerevisiae. Gene.. 2015;563:155-159.
- [CrossRef] [Google Scholar]
- Combining whispering gallery mode optofluidic microbubble resonator sensor with GR-5 DNAzyme for ultra-sensitive lead ion detection. Talanta.. 2020;213:120815
- [CrossRef] [Google Scholar]
- A label-free DNAzyme fluorescence biosensor for amplified detection of Pb2+-based on cleavage-induced G-quadruplex formation. Talanta.. 2016;147:302-306.
- [CrossRef] [Google Scholar]
- An improved structure-switch aptamer-based fluorescent Pb2+ biosensor utilizing the binding induced quenching of AMT to G-quadruplex. Chem Commun (camb).. 2020;56:10517-10520.
- [CrossRef] [Google Scholar]
- A G-quadruplex based label-free fluorescent biosensor for lead ion. Biosens Bioelectron.. 2012;35:123-127.
- [CrossRef] [Google Scholar]
- Highly sensitive and selective optical detection of lead(ii) using a label-free fluorescent aptasensor. RSC Adv.. 2016;6:90300-90304.
- [CrossRef] [Google Scholar]
- Visual detection of lead(II) using a label-free DNA-based sensor and its immobilization within a monolithic hydrogel. Analyst.. 2012;137:704-709.
- [CrossRef] [Google Scholar]
- A mercury free electrode based on poly O-cresophthalein complexone film matrixed MWCNTs modified electrode for simultaneous detection of Pb(II) and Cd(II) Microchem J.. 2019;148:92-101.
- [CrossRef] [Google Scholar]
- Zwitterion-functionalized polymer microspheres as a sorbent for solid phase extraction of trace levels of V(V), Cr(III), As(III), Sn(IV), Sb(III) and Hg(II) prior to their determination by ICP-MS. Microchim Acta.. 2018;185:228.
- [CrossRef] [Google Scholar]
- A paper-supported aptasensor for total IgE based on luminescence resonance energy transfer from upconversion nanoparticles to carbon nanoparticles. Sensors Actuat B-Chem.. 2017;239:319-324.
- [CrossRef] [Google Scholar]
- A simple paper-based aptasensor for ultrasensitive detection of lead (II) ion. Anal Chim Acta.. 2019;1071:70-77.
- [CrossRef] [Google Scholar]
- Probes for detection of specific DNA sequences at the single-molecule level. Anal Chem.. 2000;72:3717-3724.
- [CrossRef] [Google Scholar]
- Fluorescence spectroscopy for revealing mechanisms in biology: Strengths and pitfalls. J Biosciences.. 2018;43:555-567.
- [CrossRef] [Google Scholar]
- A Lead(II)-Driven DNA Molecular Device for Turn-On Fluorescence Detection of Lead(II) Ion with High Selectivity and Sensitivity. J Am Chem Soc.. 2010;132:13156-13157.
- [CrossRef] [Google Scholar]
- Fluorescence detection of lead(II) ions through their induced catalytic activity of DNAzymes. Anal Chem.. 2011;83:225-230.
- [CrossRef] [Google Scholar]
- Detection of radon with biosensors based on the lead(II)-induced conformational change of aptamer HTG and malachite green fluorescence probe. J Environ Radioactiv.. 2018;195:60-66.
- [CrossRef] [Google Scholar]
- A “turn-on” fluorescent sensor for detection of Pb2+ based on graphene oxide and G-quadruplex DNA. Phys Chem Chem Phys.. 2013;15:12800-12804.
- [CrossRef] [Google Scholar]
- A novel fluorescence aptamer biosensor for trace Pb(II) based on gold-doped carbon dots and DNAzyme synergetic catalytic amplification. J Lumin.. 2020;221:117056
- [CrossRef] [Google Scholar]
- Detection of lead (II) with a “turn-on” fluorescent biosensor based on energy transfer from CdSe/ZnS quantum dots to graphene oxide. Biosens Bioelectron.. 2013;43:69-74.
- [CrossRef] [Google Scholar]
- DNAzyme-based biosensor for detection of lead ion: A review. Microchem J.. 2017;131:145-153.
- [CrossRef] [Google Scholar]
- Fluorescence detection of mercury(II) and lead(II) ions using aptamer/reporter conjugates. Talanta.. 2011;84:324-329.
- [CrossRef] [Google Scholar]
- Highly selective DNA-based sensor for lead(II) and mercury(II) ions. Anal Chem.. 2009;81:2383-2387.
- [CrossRef] [Google Scholar]
- Ratiometric G-Quadruplex Assay for Robust Lead Detection in Food Samples. Biosensors.. 2021;11:274.
- [CrossRef] [Google Scholar]
- A label-free DNAzyme-based nanopore biosensor for highly sensitive and selective lead ion detection. Anal Methods.. 2016;8:7040-7046.
- [CrossRef] [Google Scholar]
- Simultaneous detection of lead (II) and mercury (II) ions using nucleic acid aptamer molecular beacons. Int J Environ an Ch.. 2019;101:1922-1934.
- [CrossRef] [Google Scholar]
- Magnetic nanofluid based on green deep eutectic solvent for enrichment and determination of chloramphenicol in milk and chicken samples by high-performance liquid chromatography-ultraviolet: Optimization of microextraction. J Chromatogra a.. 2023;1689:463705
- [CrossRef] [Google Scholar]
- Magnetic nanofluid based on hydrophobic deep eutectic solvent for efficient and rapid enrichment and subsequent determination of cinnamic acid in juice samples: Vortex-assisted liquid-phase microextraction. Talanta.. 2023;260:124581
- [CrossRef] [Google Scholar]
- Ratiometric fluorescent detection of lead ions in aquatic environment and living cells using a fluorescent peptide-based probe. J Hazard Mater.. 2022;427:128161
- [CrossRef] [Google Scholar]
- Simultaneous preconcentrations of Co2+, Cr6+, Hg2+ and Pb2+ ions by Bacillus altitudinis immobilized nanodiamond prior to their determinations in food samples by ICP-OES. Food Chem.. 2017;215:447-453.
- [CrossRef] [Google Scholar]
- DNA nanosensor based on biocompatible graphene quantum dots and carbon nanotubes. Biosens Bioelectron.. 2014;60:64-70.
- [CrossRef] [Google Scholar]
- A fluorescent nanosensor based on graphene quantum dots-aptamer probe and graphene oxide platform for detection of lead (II) ion. Biosens Bioelectron.. 2015;68:225-231.
- [CrossRef] [Google Scholar]
- A Novel Polydopamine Grafted 3D MOF Nanocubes Mediated GR-5/GC DNAzyme Complex with Enhanced Fluorescence Emission Response toward Spontaneous Detection of Pb(II) and Ag(I) Ions. ACS Omega.. 2020;5:25188-25198.
- [CrossRef] [Google Scholar]
- Fluorometric determination of lead(II) and mercury(II) based on their interaction with a complex formed between graphene oxide and a DNAzyme. Microchim Acta.. 2017;185:2.
- [CrossRef] [Google Scholar]
- DNAzyme Based Amplified Biosensor on Ultrasensitive Fluorescence Detection of Pb (II) Ions from Aqueous System. J Fluoresc.. 2017;27:2101-2109.
- [CrossRef] [Google Scholar]
- Paper-Based Fluorescence Chemosensors for Metal Ion Detection in Biological and Environmental Samples. BioChip J.. 2021;15:216-232.
- [CrossRef] [Google Scholar]
- A novel colorimetric potassium sensor based on the substitution of lead from G-quadruplex. Analyst.. 2013;138:856-862.
- [CrossRef] [Google Scholar]
- Developing Novel G-Quadruplex Ligands: from Interaction with Nucleic Acids to Interfering with Nucleic Acid-Protein Interaction. Molecules.. 2019;24:396.
- [CrossRef] [Google Scholar]
- A colorimetric lead (II) ions sensor based on selective recognition of G-quadruplexes by a clip-like cyanine dye. Talanta.. 2015;136:210-214.
- [CrossRef] [Google Scholar]
- Single-labeled hairpin probe for highly specific and sensitive detection of lead(II) based on the fluorescence quenching of deoxyguanosine and G-quartet. Biosens Bioelectron.. 2013;41:137-142.
- [CrossRef] [Google Scholar]
- A Sensitive and Label-Free Pb(II) Fluorescence Sensor Based on a DNAzyme Controlled G-Quadruplex/Thioflavin T Conformation. Sensors.. 2016;16:2155.
- [CrossRef] [Google Scholar]
- Dual fluorescence resonance energy transfer assay between tunable upconversion nanoparticles and controlled gold nanoparticles for the simultaneous detection of Pb2+ and Hg2+. Talanta.. 2014;128:327-336.
- [CrossRef] [Google Scholar]
- Turn-on and label-free fluorescence detection of lead ions based on target-induced G-quadruplex formation. Chem Commun (camb).. 2015;51:8165-8168.
- [CrossRef] [Google Scholar]
- A Lead (II) Ion Sensor Based on Selective Recognition of G-quadruplex for Ethyl-substitutive Thioflavin T. ChemistrySelect.. 2019;4:10787-10791.
- [CrossRef] [Google Scholar]
- Evaluation of liquid cathode glow discharge-atomic emission spectrometry for determination of copper and lead in ores samples. Talanta.. 2017;164:216-221.
- [CrossRef] [Google Scholar]
- A simple fluorescent assay for lead(II) detection based on lead(II)-stabilized G-quadruplex formation. RSC Adv.. 2013;3:16962-16966.
- [CrossRef] [Google Scholar]
- Label-free fluorescent sensor for lead ion detection based on lead(II)-stabilized G-quadruplex formation. Anal Biochem.. 2014;462:19-25.
- [CrossRef] [Google Scholar]
- Fluorescent detection of Hg2+ and Pb2+ using GeneFinder and an integrated functional nucleic acid. Biosens Bioelectron.. 2015;72:95-99.
- [CrossRef] [Google Scholar]
- Simultaneous detection of trace toxic metal ions, Pb2+ and Ag+, in water and food using a novel single-labeled fluorescent oligonucleotide probe. Sensor Actuat B-Chem.. 2018;261:58-65.
- [CrossRef] [Google Scholar]
- Dual-band fluorescence detection of double-stranded DNA with QDs-Mn2+-pefloxacin. Colloid Surface b.. 2022;217:112649
- [CrossRef] [Google Scholar]
- Bifunctional single-labelled oligonucleotide probe for detection of trace Ag(I) and Pb(II) based on cytosine-Ag(I)-cytosine mismatches and G-quadruplex. Anal Chim Acta.. 2021;1151:338258
- [CrossRef] [Google Scholar]
- Label-free DNA sensor for Pb2+ based on a duplex–quadruplex exchange. Anal Methods.. 2013;5:6100-6105.
- [CrossRef] [Google Scholar]
- The fluorescence regulation of a tri-functional oligonucleotide probe HEX-OND in detecting Pb(II), cysteine, and K(I) based on two G-quadruplex forms. Anal Bioanal Chem.. 2023;415:2763-2774.
- [CrossRef] [Google Scholar]
- Cationic-perylene-G-quadruplex complex based fluorescent biosensor for label-free detection of Pb2+. Talanta.. 2016;149:98-102.
- [CrossRef] [Google Scholar]
- Input-dependent induction of G-quadruplex formation for detection of lead (II) by fluorescent ion logic gate. Biosens Bioelectron.. 2013;43:231-236.
- [CrossRef] [Google Scholar]
- Double-stranded DNA nanobridge enhanced fluorescence of crystal violet/G-quadruplex complex for detection of lead ions and crystal violet. Sensor Actuat B-Chem.. 2021;340:129968
- [CrossRef] [Google Scholar]
- Duplex functional G-quadruplex/NMM fluorescent probe for label-free detection of lead(II) and mercury(II) ions. J Hazard Mater.. 2018;355:50-55.
- [CrossRef] [Google Scholar]







