Translate this page into:
Heliotropium curassavicum extract: Potential therapeutic agent for liver cancer through cytotoxicity, apoptosis, and molecular docking analysis
⁎Corresponding author. nabutaha@ksu.edu.sa (Nael Abutaha)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Hepatocellular carcinoma is the fifth most common cancer worldwide, posing significant challenges due to drug resistance and adverse effects associated with current treatments. Plant extracts, known for their diverse bioactive compounds, offer promising alternatives for cancer treatment. The study aimed to investigate the potential of Heliotropium curassavicum by extracting its phytochemicals through Soxhlet extraction and maceration methods. The study also aimed to assess in-vitro cytotoxicity using the MTT assay, evaluate cell migration using scratch, and analyse apoptosis using fluorescent microscopy. Additionally, GC–MS analysis was performed to identify chemical compounds and in-silico analysis was conducted to predict the most potent anticancer compounds in the extracts. Only the maceration method using n-hexane (F4) and ethyl acetate extract (F5) showed cytotoxic activity against HuH7, HepG2, and MDA-MB-231. The F4 showed cytotoxic activity with IC50 values of 93.9, 121.7, and 142.2 µg/mL, respectively. Similarly, the F5 demonstrated cytotoxic effects with IC50 values of 144 µg/mL for HuH7, 74 µg/mL for HepG2, and 150 µg/mL for MDA-MB-231. The wound-healing assay demonstrated that the F5 extract significantly reduced the migration of HepG2 cells. Based on the acridine orange/ethidium bromide and DAPI staining, the F5 fraction exhibited apoptotic potential in HepG2 cells. In GC–MS analysis, 33 phytocompounds were identified in the F5 fraction, from which 9 compounds were chosen for drugability studies. Among them, phytol and oleic acid were the only ones that showed no hepatotoxicity, neurotoxicity, nephrotoxicity, cardiotoxicity, immunotoxicity, or carcinogenicity. Molecular docking studies revealed that phytol and oleic acid had the strongest binding affinities of −8.5 and −7.6 kcal/mol against 6OOY, respectively. This is followed by −7.2 kcal/mol (phytol) and −7.1 kcal/mol (oleic acid) against 1UOM. The phytochemicals identified in the F5 fraction demonstrate significant potential as therapeutic candidates for liver cancer, necessitating further investigation through additional studies.
Keywords
Apoptosis
Cytotoxicity
Drugability
Heliotropium curassavicum
Liver cancer
Molecular docking
1 Introduction
Cancer ranks as the second leading cause of mortality globally, with liver cancer emerging as the third most prevalent form of this disease. The highest incidence rates of liver cancer, surpassing 75 %, are documented in Asia and Africa. Every year, more than 750,000 new liver cancer cases are documented globally, with around 33,000 cases reported only in the United States. Common risk factors contributing to the development of liver cancer include excessive alcohol consumption, non-alcoholic fatty liver disease, and viral infections such as hepatitis C and B virus. These factors trigger chronic inflammation, leading to the progression of liver cirrhosis, ultimately culminating in hepatocellular carcinoma (HCC) (Marquardt et al., 2012, Liu et al., 2019, Alghamdi and Alghamdi, 2020). Radical treatments such as liver transplantation, ablation and surgery have improved the prognosis in recent years; however, current anticancer treatments for HCC face several significant challenges, including late diagnosis, drug resistance, limited efficacy of systemic therapies, liver function impairment, immune evasion, lack of reliable biomarkers, high propensity for metastasis and severe side effects (Kashif et al., 2018).
In 2022, breast cancer was the most prevalent cancer among women in 157 out of 185 countries, with higher incidence rates generally observed in developed nations compared to developing ones (WHO, 2024). The risk factors for breast cancer include genetic mutations, hormonal influences, age, family history, and lifestyle elements such as diet and physical activity (Obeagu and Obeagu, 2024). While advancements in screening, early detection, and treatment have led to improved survival rates, breast cancer continues to present several significant challenges, such as late-stage diagnosis, resistance to treatment, uneven access to care, adverse treatment effects, potential for recurrence, complex genetic profiles, and significant psychosocial effects (Waks and Winer, 2019, Łukasiewicz et al., 2021).
Natural products are increasingly being studied for their potential anticancer properties, with numerous compounds demonstrating promising results in preclinical studies. Researchers are particularly interested in the bioactive compounds found in plants, which offer diverse mechanisms of action, including apoptosis induction, cell cycle arrest, and inhibition of metastasis. These bioactive compounds also support DNA repair processes, and boost antioxidant defences (El Omari et al., 2021, Jang and Lee, 2023, Imtiaz et al., 2024). Integrating natural products into cancer treatment strategies is a potential avenue to treat cancer, overcome drug resistance and reduce side effects associated with conventional therapies (Kashif et al., 2018, Guo et al., 2024). Despite these findings, many plant sources remain largely unexplored for their potential therapeutic effects against malignancies (Barras et al., 2024, Zaghlol et al., 2024).
Heliotropium curassavicum L., a member of the Boraginaceae family, is native to salt marshes and sandy areas in Europe, Southeast Asia, and the Americas. Traditionally, it has been used to treat various health conditions, including diabetes, bacterial infections, constipation, erysipelas, gonorrhea, ulcers, wounds, and cancer (Hernandez et al., 2007, Akbar et al., 2023). The Heliotropium genus is known for its diverse biological activities, such as antispasmodic, insecticidal, antifungal, antibacterial, hepatotoxic, and anticancer effects (Okusa et al., 2007, Ghori et al., 2016, Akbar et al., 2023). However, despite the recognised biological activities of H. curassavicum, the full therapeutic potential of its biomolecules, particularly concerning anticancer and antiapoptotic mechanisms, has yet to be fully explored.
Computational methods have emerged as highly effective and cost-efficient tools to find promising candidates for drug discovery. These methods offer a crucial understanding of the therapeutic actions of bioactive compounds used in cancer treatment. In silico research aims to comprehend the characteristics of biologically active substances within organisms, mainly by evaluating their druggability and ADMET properties (absorption, distribution, metabolism, excretion, and toxicity). This approach aims to mitigate the risk of approximately 40 % rejection of selected pharmaceuticals during various trial stages.
Therefore, this study aimed to investigate the in-vitro cytotoxic and apoptotic effects of H. curassavicum collected from Saudi Arabia. Compounds from the F5 fraction were identified using GC–MS analysis. Following this, in silico approaches were employed to investigate the therapeutic potential of effective and safe biomolecules targeting hepatocellular carcinoma, aiming to identify promising new agents.
2 Materials and methods
2.1 Plant collection and extraction
H. curassavicum was harvested from Riyadh, Saudi Arabia, between September and December 2022. A voucher specimen (KSU no. 22-54R-10) was deposited in the Bioproduct Research Chair at King Saud University. The aerial parts, including leaves and stems, were cleaned with distilled water, dried at 25 °C, and powdered using a commercial grinder (LC, China). The plant powder was then subjected to extraction using Soxhlet and maceration methods. Methanol (99.9 % purity), ethyl acetate (99.5 % purity), n-butanol (99.5 % purity) and n-hexane (97 % purity) (VWR, BDH, Prolabo, EC), all of HPLC grade, were used for extraction, along with distilled water. All chemicals were utilized as received.
2.2 Soxhlet extraction
A total of 15 g of the powdered sample was extracted using n-hexane (F1), ethyl acetate (F2), and methanol (F3) via Soxhlet extraction for 24 h at 80 °C. Each solvent was individually introduced and subjected to reflux. Upon completion of the extraction period, the resulting extracts were collected and concentrated by evaporation at 45 °C under reduced pressure using a Heidolph rotary evaporator (Schwabach, Germany). The yields of the F1, F2, and F3 extracts were determined, and the extracts were subsequently stored in sealed containers at −4°C.
2.3 Maceration method
The powdered sample (50 g) was extracted using 70 % methanol (500 mL) in a media bottle, stirred, and sonicated for 10 min before being left to steep for 5 days at 25 °C. Subsequently, the solutions were filtered twice using muslin cloth, and the resulting filtrates were partially evaporated under reduced pressure at 45 °C until 100 mL was obtained. Following this, liquid–liquid extraction was performed to fractionate the extract into three fractions using solvents of varying polarity: n-hexane (F4), ethyl acetate (F5), and n-butanol (F6). The fractions were then filtered using a 0.25 filter, evaporated, and the resulting extracts were weighed to determine the extract yield as a percentage. Finally, the dried extracts were stored at 4 °C.
2.4 Cytotoxicity assay
HuH7 and HepG2 (human liver cancer cell lines), MDA-MB-231 (human breast cancer cells) and normal HUVEC (endothelial cell line) cells lines were obtained from the German Culture Collection (DSMZ), cultured in T-25 flasks (NEST, China) using Dulbecco's Modified Eagle Medium high (DMEM) glucose medium (Gibco, UK) supplemented with 10 % fetal bovine serum (Gibco, UK) and 1 % antibiotic (Penicillin-Streptomycin, Gibco, UK). The cells were maintained at 37 °C in a 5 % CO2 incubator (Sanyo, Japan) and subcultured as needed. Cell viability was assessed using the MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide) assay. Cells (5 × 104) were seeded in a 24-well plate and incubated with different concentrations (0.0–900 µg/mL (for 48 h. Post-treatment, the medium was replaced with a fresh medium. The MTT assay, as described by Abutaha (Abutaha et al., 2018), was conducted by adding MTT solution to each well and incubating it in the dark for 2 h. Following incubation, 0.01 % acidified isopropanol was added, and absorbance readings were taken at 570 nm with a microplate reader. (ChromMate, USA). Cell viability was expressed as a percentage compared to the vehicle control group (0.01 % methanol), and IC50 values were calculated using OriginPro 8.5 software (OriginLab, USA).
2.5 Calculation of selectivity indices
Selectivity indices were utilised to quantify the in vitro anticancer potential. The selectivity index (SI) was determined by calculating the ratio of the average IC50 value for the normal cell line (HUVEC) to the IC50 value for the cancer cell line (HuH7 and HepG2) from each experiment (Calderón-Montaño et al., 2021). An SI greater than 1 indicates higher selectivity for cancer cells, suggesting better therapeutic potential. An SI less than 1 means greater toxicity to normal cells. An SI around 1 suggests similar toxicity to both cell types, indicating poor selectivity(López-Lázaro, 2015; Calderón-Montaño et al., 2021).
2.6 In vitro wound scratch assay
We used the “in vitro scratch assay” method to examine cell migration. HepG2 cells were seeded in 24-well plates at a density of 5 × 104 cells\well and incubated for 24 h. After incubation, a scratch was made in each well (80 % confluent) using a 10 μL pipette tip. Images were then captured at the starting point (time 0) using a fluorescence microscope. (Leica, Germany). Afterwards, the cells were treated with an F5 fraction (70 µg/mL). Medium containing 0.01 % methanol served as the control. The plates were further incubated, and images were taken after 24 and 48 h. Image analysis was performed using Image J software. The percentage increase in wound closure compared to the initial value before treatment was calculated and reported as cell migration (Suarez-Arnedo et al., 2020).
2.7 Nuclear morphological assessment by DAPI staining
Nuclear morphology was evaluated using 4′,6-diamidino-2-phenylindole (DAPI) staining as reported by (Abutaha et al., 2022). Cancer cells were cultured at a density of 5 × 104 cells/mL in 24-well plates and exposed to 70 µg/mL of the treatment for 24 h. Medium containing 0.01 % methanol served as the control. After the treatment period, the cells were fixed with cold absolute ethanol at −20 °C for 10 min. Following fixation, the cells were washed twice with phosphate-buffered saline (PBS) to remove any residual fixative. The cells were then stained with 300 nM DAPI solution for 10 min at 25 °C. After staining, excess dye was removed by washing the cells twice with PBS.The stained cells were examined under a fluorescence microscope equipped with a DAPI filter.
2.8 Detection of apoptotic morphological changes using acridine orange–ethidium bromide (AO-EtBr) staining
Cancer cells were cultured at a density of 5 × 104 cells/mL in 24-well plates and exposed to 70 µg/mL of the treatment for 24 h. Medium containing 0.01 % methanol served as the control. Subsequently, the cells were stained with a mixture of AO-EtBr dye (1:1 v/v) at a concentration of 100 μg/mL in phosphate buffer saline and analysed using a fluorescent microscope (EVOS, USA) following the established protocol (Abutaha et al., 2022). Acridine orange stains both live and dead cells, while ethidium bromide marks cells with damaged membranes. Live cells appear uniformly green, early apoptotic cells show bright green nuclei with condensed or fragmented chromatin, late apoptotic cells have orange chromatin, and necrotic cells display normal nuclei stained orange/red by AO/EB (Behzad et al., 2016).
2.9 Gas chromatography-mass spectrometry (GC–MS) analysis
The F5 fraction was analysed using the Perkin-Elmer Clarus 680 system (Perkin-Elmer, Inc., USA). This system was equipped with an Elite-5MS capillary column made of fused silica. The column's dimensions were 30 m long, 250 µm in diameter, and 0.25 µm thick. Pure helium (99.99 %) served as the carrier gas, with a continuous flow rate of 1 mL per minute. The spectral detection in the GC–MS analysis used an electron ionisation method with a high ionisation energy of 70 electron volts (eV), a scan time of 0.2 s, and fragments in the range of 40 to 600 m/z. The injected volume was 1 µl, with a split ratio 10:1, and the injector temperature was consistently maintained at 250 degrees Celsius. The column oven temperature was initially set at 50 ◦C for 3 min, then gradually increased to 10 ◦C per minute until it reached 280 degrees Celsius. Finally, the temperature was ramped up to 300◦C and held for 10 min. The phytochemical constituents were identified by comparing them with the spectral database of verified compounds stored in the National Institute of Standards and Technology (NIST) library.
2.10 Molecular docking
The targets included MMP9 (PDB ID: 4XCT); PGR (PDB ID: 1A28); PIK3CA (PDB ID: 6PYS); PTGS2 (PDB ID: 5IKR); TNF (PDB ID: 6OOY); AR (PDB ID: 2AM9); SRC (PDB ID: 4 K11); EGFR (PDB ID: 5UGC); and ESR1 (PDB ID: 1UOM) were chosen as receptor proteins for molecular docking investigations. The three-dimensional structures of these receptors were obtained from the Protein Data Bank (https://www.rcsb.org/; accessed in March 2024). The ligands, oleic acid, and phytol were obtained from PubChem using a structure data file (SDF) format. The CB-DOCK 2 tool was used to dock ligands over receptors (https://cadd.labshare.cn/cb-dock2/php/index.php). Ligand and receptor molecules were uploaded as “pdb files” to CB-DOCK 2, and the docking was performed. PyMOL and PLIP tools were used to analyse the docked complexes, examining 2D and 3D interactions (Abutaha and Almutairi, 2023).
2.11 Drug Scanning through web-based tools
Assessing the druggability of compounds is essential to identify viable drug candidates. This study evaluated the main phytochemicals for their druggability using SwissADME, a web-based tool for assessing pharmacokinetic properties and drug-likeness. The ProTox-II online server was also employed to evaluate organ toxicity and predict potential adverse effects. Moreover, the adherence to Lipinski's Rule of Five (RO5) was assessed. This rule suggests that for a compound to be considered a good candidate for oral drugs, it should meet the following criteria: logP (partition coefficient) less than or equal to 5, molecular mass less than 500 g/mol, no more than 5 hydrogen bond donors, no more than 10 hydrogen bond acceptors, and a molecular refractivity within the range of 40–130. Compounds meeting all these criteria were deemed to have higher potential as drug candidates and could be further investigated for their therapeutic efficacy. The integration of SwissADME and ProTox-II evaluations, along with adherence to Ro5, provides a comprehensive assessment of the druggability and safety profile of these phytochemicals, facilitating the identification of promising drug candidates.
2.12 Statistical analysis
The results are expressed as mean ± standard deviation. Data analysis was carried out using Excel software (Microsoft, USA). Statistical evaluation employed a one-sample Student’s t-test, with significance determined at P<0.05.
3 Result
3.1 Yield of the extracts obtained
The yields obtained using Soxhlet extractor were 20 % for F1, 20 % for F2, and 46.66 % for F3. In contrast, the maceration method yielded 6.8 % for the n-hexane fraction (F4), 6 % for the ethyl acetate fraction (F5), and 17 % for the methanol fraction (F6). Among the solvents tested, polar solvents, namely n-butanol and methanol demonstrated the highest efficiency in extracting compounds.
3.2 Cytotoxicity Assay
Only the maceration method (F4 and F5 fractions) demonstrated cytotoxic activity against the tested cell lines. The F4 to F5 fractions of H. curassavicum exhibited varying levels of cytotoxic efficacy, showing a dose-dependent decrease in cell viability against HuH7 and HepG2 cell lines. The cytotoxicity of F4 fraction revealed cytotoxic activity against multiple cell lines, including HuH7, HepG2, and MDA-MB-231, with IC50 values of 93.9, 121.7, and 142.2 µg/mL, respectively. Similarly, the F5 demonstrated potent cytotoxic effects against these cell lines, with IC50 values of 144 µg/mL for HuH7, 74 µg/mL for HepG2, and 150 µg/mL for MDA-MB-231. The F5 fraction was particularly noteworthy, which exhibited the most potent cytotoxic effect compared to other tested extracts, with an IC50 value of 74 µg/mL (Fig. 1). The results also indicated that only the F5 fraction exhibited selective cytotoxicity towards the HepG2 cancer cell line. The SI for the F4 fraction was 0.77 for HepG2 and 0.66 for MDA-MB-231 cancer cell lines. Similarly, the SI for the F5 fraction was 1.26 for HepG2 and 0.62 for MDA-MB-231 cancer cell lines.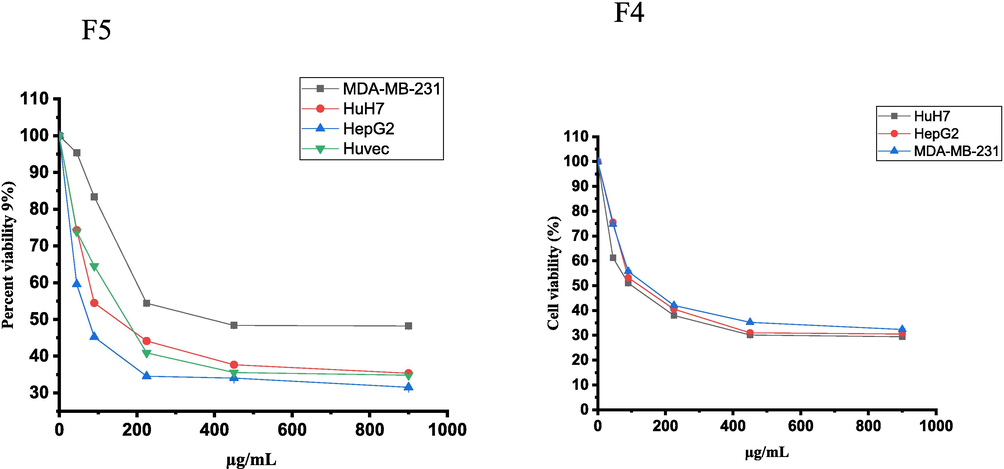
The cell viability inhibition of HepG2, HuH7 and MDA-MB-231 cancer cell lines and Huvec non-cancerous cell line treated with F4 and F5 fractions (0.0–900 µg/mL) of H. curassavicum aerial part for 24 h using the MTT assay. The data are expressed as the mean ± SD of three independent experiments.
3.3 Nuclear changes in HepG2 cells treated with F5
After 24 h of exposure to the F5 fraction (70 μg/mL), HepG2 cells exhibited an increased number of cells with condensed and fragmented nuclei compared to the control group, as shown in Fig. 2A. The treated cells displayed signs of apoptosis, including apoptotic body formation, nuclear fragmentation, membrane blebbing, and cell shrinkage. In contrast, untreated cells did not show these apoptotic features (Fig. 2B). Dual staining with AO-EtBr revealed more apoptotic cells in the F5-treated group than in the control. The F5 extract-treated cells showed early apoptotic characteristics, with yellow-green fluorescence and granular patterns in the nuclei, indicating apoptosis (Fig. 3).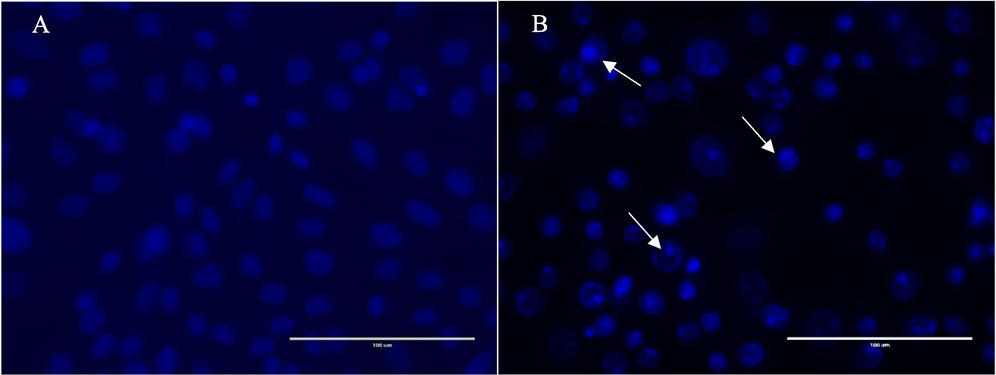
Nuclear staining using DAPI of HepG2 cells in the presence or absence of the of the F5 fraction of H. curassavicum. (a) Untreated HepG2 cells, (b) HepG2 cells treated with 70 µg/mL of the F5 fraction for 24 h. White arrows indicate nuclear fragmentation margination of the nucleus and cell shrinkage, all associated with the apoptotic mode of cell death.
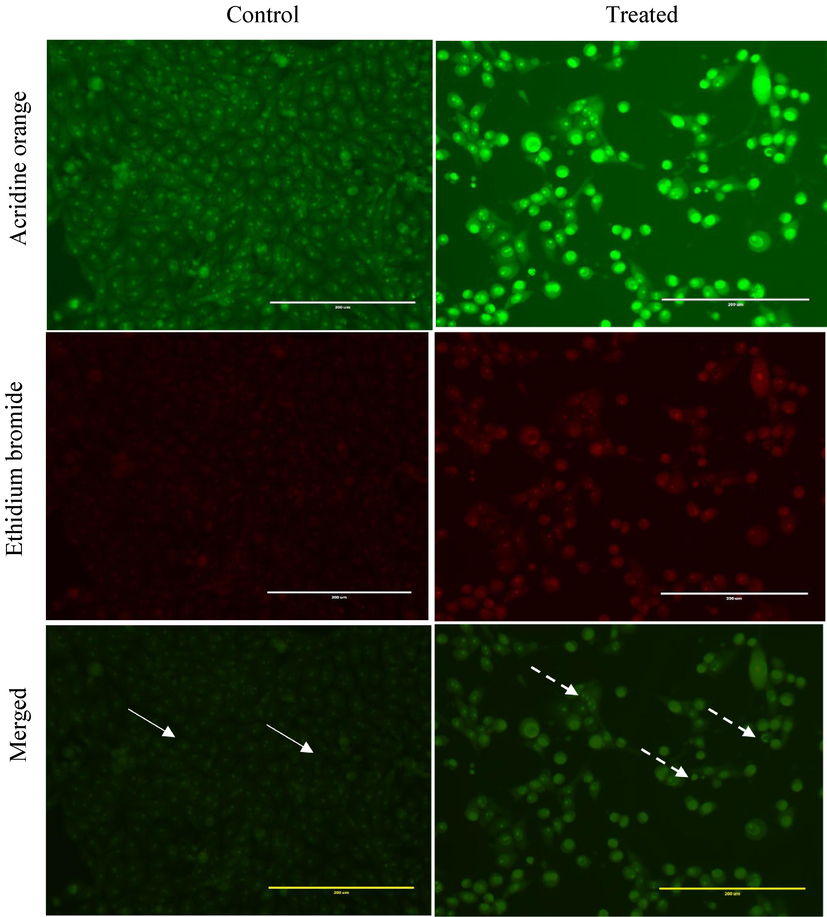
Dual acridine orange/ethidium bromide staining of HepG2 cells with characteristic symptoms of apoptosis: (i) 0.01 % methanol as negative control and cells treated with 70 µg/mL of the F5 fraction of H.curassavicum for 24 h. White arrow indicates live cells (uniformly green), and dashed arrow shows apoptotic cells (a distinctive yellow-green fluorescence in their nuclei with a concentrated crescent or granular pattern observed on one side of the cells). The images were taken with an EVOS fluorescence microscope at a magnification of 400x.
3.4 F5 fraction suppresses HepG2 cell migration
The wound healing assay was performed to evaluate the impact of the F5 fraction on the migration ability of HepG2 cells. Cells were cultured in a cell culture plate (6-well) and exposed to a 70 µg/mL concentration for 24 and 48 h. Cell migration was observed using a Leica phase contrast microscope (Leica, Germany). The results depicted in Fig. 4 demonstrate that cells exposed to F5 showed a significant decrease in migration (p < 0.05) following 48 h of incubation compared to the vehicle-treated group, where no inhibition of wound healing was observed, and the gap was nearly closed within 48 h.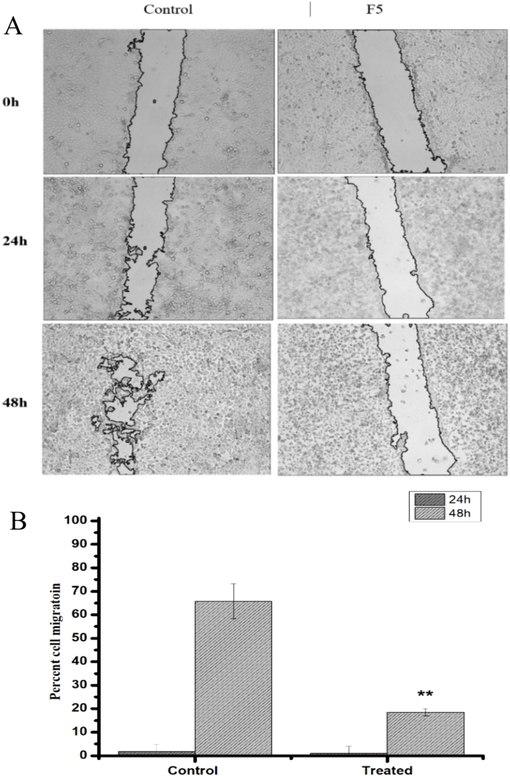
The Effect of the F5 fraction from Heliotropium curassavicum on HepG2 cell migration. (A) Images display the wounded HepG2 cell monolayer immediately post-wounding (t = 0h) and after 24 or 48 h of incubation. Cells were either mock-treated or exposed to the F5 fraction at 70 µg/mL. (B) The migration rate of the cells was determined as detailed in the materials and methods section. Statistical significance was assessed, with ∗ p < 0.05 indicating a significant difference compared to the mock-treated group. Data shown are representative of three independent experiments.
3.5 Gas chromatography-mass spectrometry (GC–MS) analysis
The GC–MS chromatogram of the F5 fraction of H. curassavicum revealed a total of 33 peaks. The compounds corresponding to these peaks were identified by matching their mass spectral fragmentation patterns with those of known substances in the NIST library. In total, the 33 phytocompounds were identified in the F5 fraction of H. curassavicum, as presented in Table 2 along with their retention times. The major compounds identified in the F5 fraction, along with their respective area percentages, are: 9-octadecenoic acid (Z)-, methyl ester (13.64 %), oleic acid (8.26 %), 8,11-octadecadienoic acid, methyl ester (7.54 %), 1-heptatriacotanol (6.34 %), ursodeoxycholic acid (5.96 %), hexadecenoic acid, 2,3-dihydroxypropyl ester (4.97 %), phytol (4.79 %), oxiraneoctanoic acid, 3-octyl-, methyl ester, trans (4.92 %), diisooctyl phthalate (4.12 %), 2-hydroxy-3-[(9e)-9-octadecenoyloxy]propyl (9e)-9-octadecenoate (3.51 %), and ursodeoxycholic acid (3.29 %) (Table 1).
No
Retention Time
Name of the Compound
Area %
Molecular Formula
Molecular Weight
1
7.00
Tetradecane
0.80
C14H30
198
2
24.02
Ethanol, 2-(9-octadecenyloxy)-, (Z)-
0.81
C20H40O2
312
3
24.17
17-Octadecynoic acid
0.85
C18H32O2
280
4
25.47
2-Hexadecanol
1.03
C16H34O
242
5
25.64
Palmitic acid methyl ester
2.15
C17H34O2
270
6
26.41
Hexadecanoic acid, 2,3-dihydroxypropyl ester
4.97
C19H38O4
330
7
28.65
8,11-Octadecadienoic acid, methyl ester
7.54
C19H34O2
294
8
28.82
9-Octadecenoic acid (Z)-, methyl ester
13.64
C19H36O2
296
9
28.93
10-Octadecenoic acid, methyl ester
2.11
C19H36O2
296
10
29.15
Phytol
4.79
C20H40O
296
11
29.40
Oxiranoctanoic acid, 3-octyl-, methyl ester, trans-
4.92
C19H36O3
312
12
29.57
Oleic acid
8.26
C18H34O2
282
13
30.07
Oxiranoctanoic acid, 3-octyl-, cis-
1.66
C18H34O3
298
14
31.33
Tributyl acetylcitrate
1.10
C20H34O8
402
15
32.31
cis-13-Eicosenoic acid
0.79
C20H38O2
310
16
32.84
Oxiraneundecanoic acid, 3-pentyl-, methyl ester, cis-
1.11
C19H36O3
312
17
33.96
6,9,12,15-Docosatetraenoic acid, methyl ester
1.54
C23H38O2
346
18
34.09
2,3-Dihydroxypropyl elaidate
1.08
C21H40O4
356
19
35.82
Diisooctyl phthalate
4.12
C24H38O4
390
20
36.01
Docosanoic acid, methyl ester
0.90
C23H46O2
354
21
39.02
1,2-Benzenedicarboxylic acid
2.53
C24H38O4
390
22
41.58
Arabitol, pentaacetate
0.51
C15H22O10
362
23
41.93
Trilinolein
1.40
C57H98O6
878
24
42.19
2,4,6(1H,3H,5H)-Pyrimidinetrione, 5-(1-cyclohexen-1-yl)-5-ethyl-
2.38
C12H16N2O3
236
25
42.33
D-Galactitol, 1-deoxy-, pentaacetate
0.69
C16H24O10
376
26
42.55
Ursodeoxycholic acid
3.29
C24H40O4
392
27
42.97
Docosanoic acid, 8,9,13-trihydroxy-, methyl ester
1.28
C23H46O5
402
28
43.06
15,17,19,21-Hexatriacontatetrayne
0.90
C36H58
490
29
43.67
2-Hydroxy-3-[(9E)-9-octadecenoyloxy]propyl (9E)-9-octadecenoate
3.51
C39H72O5
620
30
44.23
Silane, trimethyl[[(3α)-stigmaster-5-en-3-yl]oxy]-
3.34
C32H58OSi
486
31
44.76
Ursodeoxycholic acid (repeated)
5.96
C24H40O4
392
32
44.85
Rhodopin
3.69
C40H58O
554
33
44.97
1-Heptatriacotanol
6.34
C37H76O
536
PubChem CID
5364509
14900
5319737
5280435
6454051
445639
33934
31401
537071
MW (g/mol) < 500
296.5
330.5
294.47
330.50
312.49
282.46
390.56
392.57
537
Heavy atoms
21
23
21
23
22
20
28
28
38
Arom. heavy atoms
0
0
0
0
0
0
6
0
0
Rotatable bonds
16
18
15
18
16
15
16
112.60
35
H-bond acceptors < 10
2
4
2
4
3
2
4
4
1
H-bond donors < 5
0
4
0
2
0
1
0
3
1
TPSA≤140 (A2)
26.30
66.76
26.30
66.76
38.83
37.30
52.60
77.76
20.23
Lipophilicity Log Po/w ≤ 5
4.75
4.64
5.76
4.64
5.42
5.65
6.5
3.6
13.02
Water solubility Log S (ESOL)
−5.32
−4.69
−4.97
−4.69
−5.21
−5.41
−6.66
−3.95
−12.51
GI absorption
High
High
High
High
High
High
High
High
Low
Drug-likeness(Lipinski)
Yes
Yes
Yes
Yes
Yes
Yes
Yes
Yes
No
Rat acute toxicity
3000
5000
1190
5000
16,000
48
1340
2000
1000
Hepatotoxicity
+
−
+
−
−
−
−
+
−
Neurotoxicity
−
−
+
−
−
−
−
−
−
Nephrotoxicity
−
+
−
−
+
−
+
+
−
Cardiotoxicity
−
−
−
−
−
−
+
−
Immunotoxicity
+
−
+
−
−
−
−
−
−
Carcinogenicity
−
−
−
−
−
+
−
−
3.6 Drugability Analyses
Among the top compounds analysed, 1-Heptatriacotanol was the only one that did not fully meet all the drug-like parameters specified by RO5 (Table 2). Furthermore, the admetSAR tool was employed to evaluate the pharmacological potential of the top-selected phytochemicals, focusing on ADMET properties relevant to medical applications. Only phytol and oleic acid demonstrated no toxicity among the compounds assessed across all tested parameters.
3.7 Docking studies
The binding affinities of oleic acid and phytol with MMP9, PGR, PIK3CA, PTGS2, TNF, AR, SRC, EGFR, ESR1, and 1YCR are shown in Fig. 5. In TNF (PDB ID: 6OOY), oleic acid exhibited a binding affinity of −7.6 kcal/mol. It formed in 11 hydrophobic interactions with the amino acid (aa) residues LEU-57 (2x), TYR-59 (4x), TYR-119 (3x), VAL-123, and LEU-157A. Additionally, it formed two hydrogen bonds with the aa residues ILE-58 and GLY-122 (Fig. 6B). Similarly, phytol displayed a binding affinity of −8.5 kcal/mol. It established 3 hydrogen bonds with the aa residues SER-60, LEU-120A (2x), and 12 hydrophobic interactions with the aa residues LEU-57 (2x), TYR-59 (5x), TYR-119 (3x), and TYR-151(Fig. 7A). For ESR1 (PDB ID: 1UOM), the binding affinity of oleic Acid to the protein was − 7.1 kcal/mol. It formed 12 hydrophobic interactions with the aa residues LEU-346 (2x), ALA-350, LEU-384, LEU-387, LEU-391 (2x), ILE-424 (2x), PHE-425, LEU-428, and HIS-524. Additionally, it exhibited two hydrogen bonds with the aa residues GLU-353 (2.82 Å) and ARG-394 (2.33 Å). Similarly, phytol displayed a binding affinity of −7.2 kcal/mol. It established 13 hydrophobic interactions with the aa residues LEU-346 (4x), TRP- 383, LEU- 387, PHE- 404 (2x), ILE- 424 (2x), PHE- 425, LEU- 428, and LEU- 525. In PIK3CA (PDB ID: 6PYS), the binding affinity of oleic acid to protein was − 6.7 kcal/ mol. It formed 6 hydrophobic interactions with the aa residues ILE-633 (2x), PHE- 666 (2x), MET- 811, GLN- 815 and a hydrogen bond with the aa residue of ARG- 818 (Figure h). In SRC (PDB ID: 4 K11), the binding affinity of oleic acid to protein was −6.1 kcal/mol. It formed 10 hydrophobic interactions with the aa residues LEU-273, VAL- 281 (3x), ALA- 293, LYS- 295, VAL- 323, LEU- 393 (2x), ALA- 403 and a hydrogen bond with the aa residue of LYS-339–295. In EGFR (PDB ID: 5UGC), the binding affinity of oxyresveratrol to protein was − 8.0 kcal/mol. It established 12 hydrophobic interactions with the aa residues LEU-718, PHE- 723 (2x), VAL=726 (2x), ALA- 743, MET-793A, LEU-844 (2x), and PHE-856(3x). it also formed a hydrogen bond with the aa residues of ATG- 841 and ASN- 842.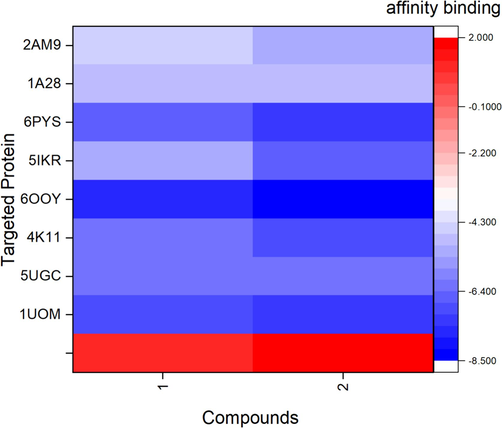
Heatmap illustrates the binding affinity of oleic acid (1) and phytol (2) (x-axis) against the 8 targeted proteins including 2AM9, 1A28, 6PYS, 5IKR, 6OOY, 4KI1, 5UGC, and 1UOM (y-axis).
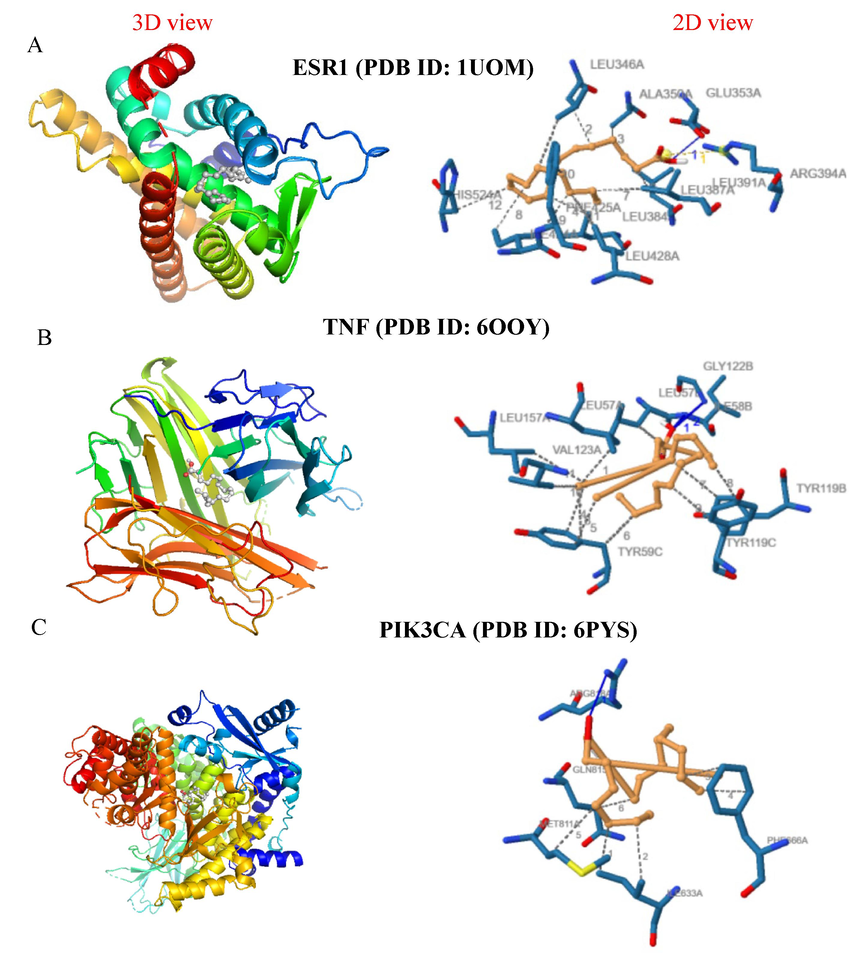
Molecular docking analysis showing the 3D and 2D interaction diagrams of the oleic acid with target proteins: (a) ESR1 (PDB ID: 1UOM), (b) TNF (PDB ID: 6OOY), and (c) PIK3CA (PDB ID: 6PYS).
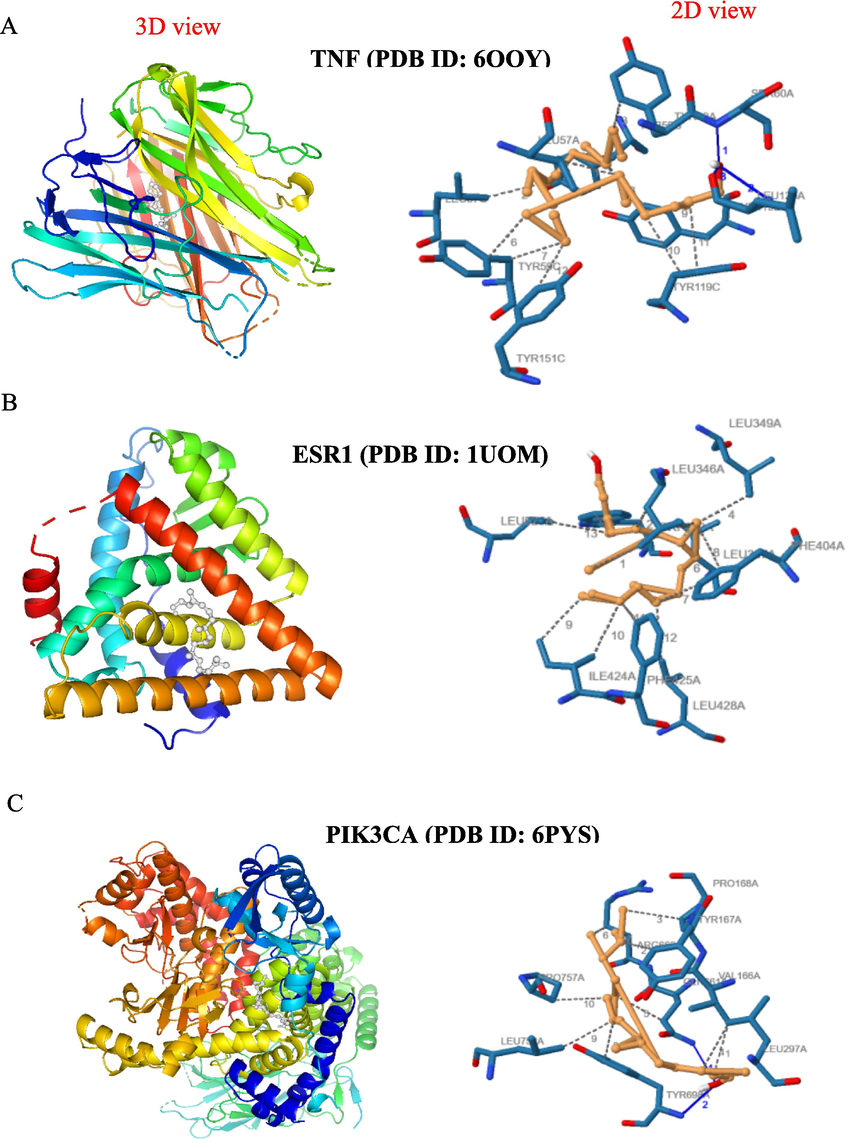
Molecular docking analysis showing the 3D and 2D interaction diagrams of the phytol with target proteins: (a) TNF (PDB ID: 6OOY); (b) ESR1 (PDB ID: 1UOM); (c) PIK3CA (PDB ID: 6PYS).
4 Discussion
Plants have been used in cancer treatment for centuries, with around 3000 species known to have anticancer effects. Many anticancer drugs today are derived from plant-based compounds (Kluwe et al., 1982, Hirschfeld et al., 1983, Asma et al., 2022). Plant-based anticancer drugs include compounds like taxanes, steroidal saponins, vinca alkaloids, diterpenoids, and epipodophyllotoxin lignans and their derivatives. These substances are commonly used in cancer treatment due to their effectiveness against cancer cells (Puri et al., 2023).
The cytotoxic effects observed in the F4 and F5 fractions against HuH7, HepG2, and MDA-MB-231 cell lines are consistent with findings from other studies on plants within the same family. For instance (Erdoğan et al., 2020) reported that extracts from Paracaryum bingoelianum exhibited cytotoxicity against MCF-7 and HT-29 cancer cell lines, with IC50 values of 581.4 μg/mL and 473.2 μg/mL, respectively. Similarly, Labbozzetta et al. found that extracts from Glandora rosmarinifolia demonstrated cytotoxic effects on HL-60 (IC50: 36.75 μg/mL), HL-60R (IC50: 37.0 μg/mL), and hTERT RPE-1 (IC50 > 50.0 μg/mL) cancer cells (Labbozzetta et al., 2022).
The apoptosis-inducing effects observed in HepG2 cells treated with the F5 fraction, such as nuclear fragmentation and cell shrinkage, align with findings reported in the literature on natural compounds. Rajabi et al (Rajabi et al., 2021) documented similar apoptotic features in cancer cells treated with extracts from various plants. Additionally, studies on plants from the same family support these findings; for example, Erdoğan et al. (2020) reported that Paracaryum bingoelianum extracts induced apoptosis in MCF-7 and HT-29 cancer cell lines. The desired outcome of an anticancer agent is to induce cell death through apoptosis. This process can be observed using DAPI, a widely used fluorescent dye that binds to adenine–thymine-rich regions in DNA. Fragmented and condensed nuclei visible using DAPI staining indicate apoptosis (Kapuscinski, 1995). Alternatively, the AO/EB double staining method differentiates between stages of apoptosis and necrosis. Viable cells display a uniform green nucleus; early apoptosis is indicated by fragmented and condensed nuclei, and late apoptosis is marked by orange nuclear fragments (Fernando et al., 2018). In Fig. 3, an increased number of apoptotic bodies were viewed in cell lines.
The selectivity index (SI) values for the F5 fraction, especially its higher SI for HepG2 cells, align with the findings of Calderón-Montaño et al. (2022), that reported selectivity towards various cancer cell lines in a study involving different plant extracts. The authors reported Various phytochemicals that may contribute to the selective anticancer activity, including cardiac glycosides, taxane-type diterpenes like paclitaxel, ferruginol, and sandaracopimaric acid, as well as monoterpenes such as limonene and carveol, and sesquiterpenes like β-caryophyllene and humulene (Calderón-Montaño et al., 2021).
GC–MS analysis identified several compounds in the extract, including oleic acid and phytol, which were reported in various studies to induce apoptosis in multiple cancer cell lines. Oleic acid has shown it can trigger cell death in cancer cells, possibly by increasing the production of reactive oxygen species (ROS) inside the cells or by increasing the activity of caspase 3 (Carrillo Pérez, 2012). Similarly, phytol has shown promising anticancer effects by inducing apoptosis and necrosis in sarcoma 180 (S-180) and leukaemia (HL-60) cell lines (de Alencar et al., 2023). It also exhibited anti-angiogenic potential by causing apoptosis by depolarising mitochondrial membrane potential in A549 cells (Rajab et al., 1998).
Metastasis poses a significant challenge to effective cancer treatment, making compounds that inhibit cancer cell migration crucial for disease management and prognosis. The anticancer potential of the F5 fraction is demonstrated by its wound-healing effect on HepG2 cells, suggesting its ability to hinder cancer cell migration. In a similar study, extracts of Andrographis paniculata significantly impaired the wound-healing ability of HeLa cells, reducing their migration by 40 % and preventing wound closure. The extracts also showed anti-angiogenic effects, as they reduced blood vessel formation in hen eggs (Anoor et al., 2022).
Toxicity and adverse effects of compounds are responsible for approximately 20 % of failures in drug development. Toxicity testing typically involves animal trials, which are complex, costly, and time-consuming. However, in silico toxicity analysis offers a quick and inexpensive alternative that does not require animal trials, making it a valuable tool for supporting preclinical drug development. (Zhou et al., 2016). Consequently, the toxicity profiles of the top nine selected compounds were analysed using in silico methods. To be considered potential drug candidates, compounds must adhere to at least four criteria outlined in RO5, which predicts a high likelihood of oral bioavailability and supports further experimental investigations (Veber et al., 2002). Among the top compounds analysed, 1-Heptatriacotanol was the only one that did not fully meet all the drug-like parameters specified by RO5 (Table 2). Compounds that break none or only one of these rules can still be regarded as potential drug candidates (Mustafa and Winum, 2022).
Only phytol and oleic acid demonstrated no toxicity among the compounds assessed across all tested parameters. These two compounds met the criteria for bioavailability, showing no signs of hepatotoxicity, neurotoxicity, nephrotoxicity, cardiotoxicity, immunotoxicity, or carcinogenicity. In contrast, other compounds exhibited at least one type of toxicity. Based on comprehensive drug profiling, the findings suggest that phytol and oleic acid, as highlighted in this study, fulfil the criteria for potential drug candidacy due to their safe profiles and compliance with RO5. Therefore, these two compounds were chosen for molecular docking.
Molecular docking analysis revealed that phytol and oleic acid exhibit strong binding affinity and high docking scores with targeted proteins ESR1 (1UOM) and TNF (6OOY). ESR1, a steroid receptor, is crucial in regulating tumorigenesis, particularly in breast cancer (Carausu et al., 2019). If mutation occurs, ESR1 pre-exists in primary malignancy and can be triggered during metastasis. Thus, suppression of ESR1 gene activation may be exploited as an anticancer strategy. Similarly, TNF-α is a cytokine with significant roles in promoting and suppressing liver cancer. As a pro-inflammatory molecule, TNF-α contributes to liver cancer development through various mechanisms. It enhances cancer-related inflammation, leading to the recruitment and differentiation of immune suppressor cells. These cells help tumours evade immune surveillance, promoting cancer progression and metastasis (MDPI)). TNF-α is also associated with the regulation of cell death and survival pathways. It can induce apoptosis in cancer cells under certain conditions, yet paradoxically, it also promotes cell proliferation and survival in the tumour microenvironment by activating pathways such as NF-κB, which are associated with inflammation and cell proliferation (Laha et al., 2021).
5 Conclusion
In conclusion, the phytochemicals extracted from Heliotropium curassavicum, particularly those in the F5 fraction, exhibit strong potential as therapeutic candidates for hepatocellular carcinoma. The study's findings highlight the selective cytotoxicity, inhibition of cell migration, and induction of apoptosis in HepG2 cells by the F5 fraction. Additionally, the identification of non-toxic compounds like phytol and oleic acid, with strong binding affinities through molecular docking, highlights their promise as anticancer agents. These results warrant further investigation on systemic toxicity, pharmacokinetics, and therapeutic efficacy to determine its safety and effectiveness as a potential anticancer agent. Additionally, understanding its long-term effects, possible side effects, and optimal dosage will be essential steps in translating these findings into clinical applications.
6 Research funding
Researchers Supporting Project number (RSPD2024R757), King Saud University, Riyadh, Saudi Arabia.
CRediT authorship contribution statement
Nael Abutaha: Writing – review & editing, Methodology, Investigation, Funding acquisition, Formal analysis, Conceptualization. Raed Alghamdi: Software, Methodology, Data curation. Omair Alshahrani: Methodology. Muhammad Al- Wadaan: Writing – review & editing, Supervision, Data curation.
Acknowledgements
The authors express their sincere appreciation to the Researchers Supporting Project number (RSPD2024R757), King Saud University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- S-phase cell cycle arrest, and apoptotic potential of Echium arabicum phenolic fraction in hepatocellular carcinoma HepG2 cells. J. King Saud Univ.-Sci.. 2022;34:101735
- [Google Scholar]
- Exploring the therapeutic potential of GC–MS separated compounds from Dracaena cinnabari against dengue virus and Aedes aegypti using in silico tools. J. King Saud Univ. Sci.. 2023;35:102478
- [Google Scholar]
- Anti-proliferative and anti-inflammatory activities of entophytic Penicillium crustosum from Phoenix dactylifer. Pak. J. Pharm. Sci.. 2018;31
- [Google Scholar]
- Evaluation of bioactive compounds as antimicrobial and antidiabetic agent from the crude extract of Heliotropium curassavicum L. Biocatal. Agric. Biotechnol.. 2023;50:102745
- [Google Scholar]
- The incidence rate of liver cancer in saudi arabia: an observational descriptive epidemiological analysis of data from the Saudi Cancer Registry (2004–2014) Cancer Manag. Res. 2020:1101-1111.
- [Google Scholar]
- Methanol extraction revealed anticancer compounds Quinic Acid, 2 (5H)-Furanone and Phytol in Andrographis paniculata. Mol. Clin. Oncol.. 2022;17:1-13.
- [Google Scholar]
- Natural products/bioactive compounds as a source of anticancer drugs. Cancers. 2022;14:6203.
- [Google Scholar]
- Recent Advances in Chemistry and Antioxidant/Anticancer Biology of Monoterpene and Meroterpenoid Natural Product. Molecules. 2024;29:279.
- [Google Scholar]
- Primula auriculata extracts exert cytotoxic and apoptotic effects against HT-29 human colon adenocarcinoma cells. Iranian J. Pharmaceut. Res.: IJPR. 2016;15:311.
- [Google Scholar]
- Screening for selective anticancer activity of 65 extracts of plants collected in Western Andalusia. Spain Plants. 2021;10:2193.
- [Google Scholar]
- Manipulation of Amino Acid Levels with Artificial Diets Induces a Marked Anticancer Activity in Mice with Renal Cell Carcinoma. Int. J. Mol. Sci.. 2022;23:16132.
- [Google Scholar]
- ESR1 mutations: a new biomarker in breast cancer. Expert Rev. Mol. Diagn.. 2019;19:599-611.
- [Google Scholar]
- Carrillo Pérez, C., M. d. M. Cavia Camarero and S. Alonso de la Torre, 2012. Antitumor effect of oleic acid; mechanisms of action. A review. Nutrición Hospitalaria, 2012, v. 27, n. 6 (Noviembre-Diciembre), p. 1860-1865.
- Anticancer effects of phytol against Sarcoma (S-180) and Human Leukemic (HL-60) cancer cells. Environ. Sci. Pollut. Res.. 2023;30:80996-81007.
- [Google Scholar]
- Anticancer mechanisms of phytochemical compounds: focusing on epigenetic targets. Environ. Sci. Pollut. Res.. 2021;28:47869-47903.
- [Google Scholar]
- The antiproliferative potential of isolated emodin and aloe-emodin from Rheum ribes on different cancer cell lines. Biol. Div. Conserv.. 2020;13:160-168.
- [Google Scholar]
- Apoptotic and antiproliferative effects of Stigmast-5-en-3-ol from Dendronephthya gigantea on human leukemia HL-60 and human breast cancer MCF-7 cells. Toxicol. In Vitro. 2018;52:297-305.
- [Google Scholar]
- Ethnopharmacological, phytochemical and pharmacognostic potential of genus Heliotropium L. Turk. J. Pharm. Sci.. 2016;13:143-168.
- [Google Scholar]
- Therapeutic Effects of Natural Products on Liver Cancer and Their Potential Mechanisms. Nutrients. 2024;16:1642.
- [Google Scholar]
- Antimicrobial activity of the essential oil and extracts of Cordia curassavica (Boraginaceae) J. Ethnopharmacol.. 2007;111:137-141.
- [Google Scholar]
- Assessing personality: effects of the depressive state on trait measurement. Am. J. Psychiatry. 1983;140:695-699.
- [Google Scholar]
- Traditional and contemporary herbal medicines in management of cancer: A scoping review. J. Ayurveda Integr. Med... 2024;15:100904
- [Google Scholar]
- Mechanisms of phytochemicals in anti-inflammatory and anti-cancer. MDPI. 2023;24:7863.
- [Google Scholar]
- In vitro antiproliferative and apoptosis inducing effect of a methanolic extract of Azadirachta indica oil on selected cancerous and noncancerous cell lines. Asian Pac. J. Trop. Med.. 2018;11:555-561.
- [Google Scholar]
- Carcinogenicity testing of phthalate esters and related compounds by the National Toxicology Program and the National Cancer Institute. Environ. Health Perspect.. 1982;45:129-133.
- [Google Scholar]
- Antitumor effect of Glandora rosmarinifolia (Boraginaceae) essential oil through inhibition of the activity of the Topo II enzyme in acute myeloid leukemia. Molecules. 2022;27:4203.
- [Google Scholar]
- The role of tumor necrosis factor in manipulating the immunological response of tumor microenvironment. Front. Immunol.. 2021;12:656908
- [Google Scholar]
- The trends in incidence of primary liver cancer caused by specific etiologies: results from the Global Burden of Disease Study 2016 and implications for liver cancer prevention. J. Hepatol.. 2019;70:674-683.
- [Google Scholar]
- Two preclinical tests to evaluate anticancer activity and to help validate drug candidates for clinical trials. Oncoscience. 2015;2:91.
- [Google Scholar]
- Breast cancer—epidemiology, risk factors, classification, prognostic markers, and current treatment strategies—an updated review. Cancers. 2021;13:4287.
- [Google Scholar]
- Molecular diagnosis and therapy of hepatocellular carcinoma (HCC): an emerging field for advanced technologies. J. Hepatol.. 2012;56:267-275.
- [Google Scholar]
- The importance of sulfur-containing motifs in drug design and discovery. Expert Opin. Drug Discov.. 2022;17:501-512.
- [Google Scholar]
- Direct and indirect antimicrobial effects and antioxidant activity of Cordia gilletii De Wild (Boraginaceae) J. Ethnopharmacol.. 2007;112:476-481.
- [Google Scholar]
- Organization, W. H., 2024. Breast cancer. https://www.who.int/news-room/fact-sheets/detail/breast-cancer.
- Puri, S., N. Hegde, S. Sawant, et al., 2023. Applications of phytochemicals in cancer therapy and anticancer drug development. Recent Frontiers of Phytochemicals, Elsevier: 335-351.
- Antimycobacterial activity of (E)-phytol and derivatives: a preliminary structure-activity study. Planta Med.. 1998;64:2-4.
- [Google Scholar]
- The most competent plant-derived natural products for targeting apoptosis in cancer therapy. Biomolecules. 2021;11:534.
- [Google Scholar]
- An image J plugin for the high throughput image analysis of in vitro scratch wound healing assays. PLoS One. 2020;15:e0232565.
- [Google Scholar]
- Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem.. 2002;45:2615-2623.
- [Google Scholar]
- Unveiling the anti-cancer potential of Euphorbia greenwayi: cytotoxicity, cell migration, and identification of its chemical constituents. Fut. J. Pharmaceut. Sci.. 2024;10:24.
- [Google Scholar]
- Systems pharmacology in small molecular drug discovery. Int. J. Mol. Sci.. 2016;17:246.
- [Google Scholar]







