Translate this page into:
Identification and in silico molecular modelling study of newly isolated Bacillus subtilis SI-18 strain against S9 protein of Rhizoctonia solani
⁎Corresponding author. dwb@mail.hzau.edu.cn (Wubei Dong)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
The numerous bioactive components from Bacillus subtilis are commonly used as antimicrobial agents for reducing plant diseases caused by fungal pathovars. In this study, we isolated and identified B. subtilis SI-18 strain from twenty isolates of rhizosphere soil through morphological and molecular approaches, and explored its inhibitory activities against Rhizoctonia solani. According to morphological features and 16S rRNA and gyrB gene sequence analysis, B. subtilis SI-18 strain was identified. Additionally, the culture filtrate of B. subtilis SI-18 resulted in the suppression of R. solani mycelium growth and material leakage from the cells. Then, we have performed homology modelling and molecular docking study of S9 protein from R. solani where three potential compounds (D1, D2, and D3) were identified among 134 antimicrobial compounds derived from B. subtilis group based on higher binding energy and interaction at the active grove of the target protein. The D1 compound creates alkyl bond at Val48 whereas D2 also binds with Val48 by creating hydrogen bond. On the other hand, two hydrogen bonds were observed at Val48 and Ile52 by D3, which might be responsible for possible blocking of the target S9 protein of R. solani. To validate the docking study and understand the change in drug-ligand conformation, molecular dynamics simulation was assessed where rigid conformation was found for D1, D2 and D3 complexes. Moreover, ADMET study confirms that no toxicity and carcinogenicity were found for screened compounds. Based on our studies, we demonstrated that compounds D1, D2, and D3 derived from B. subtilis can be a potential inhibitor of S9 protein of R. solani that might be a possible strategy for fungal disease prevention.
Keywords
Bacillus subtilis
Antimicrobial potential
Rhizoctonia solani
S9 protein
Molecular docking
1 Introduction
The prevalence of systemic microbial infection has dramatically increased over the past decades, owing to the rising number of immunocompromised hosts. Meanwhile, nearly every microbial communities acquire resistance to eligible drugs over several years. So, the available anti-microbial drugs are either less effective or inadequate (Frieri et al., 2017). For these reasons, researchers are required for alternative antimicrobials or active compounds that offer feasible candidates to reduce multiple plants, animal and human-related diseases. Favourable bacteria are stated to be able to suppress phytopathogenic fungal growth by various approaches that induce plant cell immune response, including thickening of the cell wall (Benhamou et al., 1996), active oxygen bursts (Lu et al., 2017), deposition of callose, and enzyme accumulation dealing with the protection (Yang and Li, 2011). In order to thrive in unfavourable conditions, plants must exhibit a range of defensive mechanisms that allow them to ignore injuries to the tissue when microbes invade (Porter, 1985). In our lab, we screened many potential resistant genes from cDNA libraries using the B. subtilis expression system, as this system is a valuable method for acquiring resistant gene expression (Fu et al., 2020; Kong et al., 2018; Wu et al., 2020), but this technique is very time-consuming and confronts several hurdles throughout the testing. On account of this, in silico modelling approach was done to detect broad-scale antimicrobial peptides or compounds derived from B. subtilis species (Qutb et al., 2020). The primary goal of in silico molecular study is to identify bigger compounds databases into a smaller subset of expected bioactive compounds, allowing the optimization of key compounds by enhancing pharmacological activity (such as ADMET and affinity) and bind a nucleating site by integrating segments with optimized roles (Kapetanovic, 2008). However, many bioinformatics tools have been used to identify antimicrobial compounds from databases, including PubChem, ChEMBL-EMBL-EBI (Jinal and Amaresan, 2020).
The Bacillus genus comprises 541 species of rod-shaped Gram-positive bacteria ( https://www.bacterio.net/genus/Bacillus). Their tendency to develop endospores, their variety in morphological features and ability to generate various bioactive substances (AMCs) facilitate widespread distribution of arthropods and mammals in the soil, aquatic ecosystems, food and gut microbiome (Nicholson, 2002). The bacterial community of B. subtilis forms tiny vegetative cells (<1 to 0.5 μm wide) for which the strain B. subtilis 168 is considered as a model host (Belda et al., 2013). They are usually mesophilic and neutrophilic, although some can survive high pH conditions. For decades, the ability of B. subtilis community strains is to develop a broad range of antibiosis-mediating bioactive compounds. It is now estimated that almost 5% of its genome is dedicated to the formation of secondary metabolites for any specific strain of the B. subtilis group (Stein et al., 2002). Most of those substances are antimicrobial peptides (AMPs). Their structures are typically cyclic, hydrophobic and contain unusual moieties including D-amino acids (AA). In addition to AMPs, volatile substances also form a broad family of antimicrobial drugs which exhibit different metabolic and operational responsibilities. In recent time, Bacillus species derived bioactive substances which provides a much more efficient and eco-friendly supplement to soil-borne and other crop diseases (Sun et al., 2017). Many studies have also stated that rhizosphere species are ideal candidates for the control of certain adverse conditions responsible by R. solani on different crop such as rice, wheat, maize, tomato etc. (Ma et al., 2015; Muis and Quimio, 2016; Yu et al., 2017; Akinrinlola et al., 2018).
Plants must deal with a range of biotic and abiotic stress conditions when growing and evolving. For example, the ubiquitous soil-borne microbe R. solani along with plants is responsible for significant damage to many of the financially important agricultural and horticultural crop worldwide (Dean et al., 2012; Jia et al., 2007). R. solani strains grow anywhere and tend to be saprophytic and infectious to over 500 host plants. Sclerotia-dormant types of plant pathogens are immune to the severity of the climate and allow negative impacts to persist in the fungus (Kai et al., 2007). In China, the occurrence of the infection caused by R. solani is 10% to 30% overall in years and up to 50% in severe cases (Feng et al., 2017). Unfortunately, while the pathogen causes significant economic loss in many plants leading to decreased yields, there is presently no successful pathogen management approach feasible.
The S9 (40S ribosomal) protein acts a crucial role as initiation factors that resembles a primary rRNA-binding protein molecule which facilitates messenger RNAs scanning and protein synthesis initiation (Dharni et al., 2014). The ribosomal protein S9 is a vital protein located on the mRNA entry tunnel into the ribosome and performs an adequate function in the decoding process. Lindstrom and Zhang revealed that protein S9 is necessary for normal cell development and expansion, as degradation of S9 has led to reduced protein synthesis related with G1 cell cycle arrest (Lindström and Zhang, 2008). Recent evidence has shown that ribosomal protein S9 is found in the mRNA entry tunnel in the ribosomes and is engaged in mRNA translation regulation, probably termination of the translation (Pnueli and Arava, 2007). Further research on a particular antifungal agent targeting translation phase inhibition which has the effect of blocking protein synthesis and eventually its role will be of substantial concern for the future.
In this study, we screened out a total of twenty Bacillus strains and isolated the most prominent B. subtilis strain SI-18 that explored potential antimicrobial activity against R. solani and also demonstrated the mechanism of action of three B. subtilis derived compounds (D1, D2 and D3) inhibiting 40S ribosomal S9 protein of R. solani via docking strategies.
2 Material and methods
2.1 Collection of plant materials and culture of pathogens
The rhizosphere soil samples were collected from the wheat fields of Huazhong Agricultural University, Wuhan, Hubei Province, China. One gram of each soil samples were homogenized in 100 mL Erlenmeyer flasks containing 10 mL saline solution (0.006 mM FeSO4 7H2O; 0.01 mM CaCO3 7H2O; 0.08 mM MgSO4 7H2O; pH 7.0) and shaken at 200 rpm for 2 h to release the Bacillus cells from the colloidal fraction of the soil (Polanczyk, 2004). Then serially diluted (10−5 and 10−10, 100 μL) samples were taken and spread over the lysogenic broth (LB) agar plate (10 g of tryptone, 5 g of yeast extract, 10 g of NaCl, 20 g of agar 20 g prepared in 1000 mL of distilled 1000 mL where the pH was maintained at 7.3) and finally incubated the plates at 37 °C for overnight. On LB agar medium, the different morphological characteristics of the bacterial isolates were selected and purified (Verma et al., 2015). The optical density (OD) of the overnight liquid culture were measure of 1.7 at 600 nm. R. solani was cultivated at 28 °C on PDA (Potato dextrose agar) media (200 g of potato, 20 g of glucose, 20 g of agar, 1000 mL of distilled water, pH neutral) for three days in the dark.
2.2 Antagonistic assay of endophytic bacteria against R. Solani isolates
The isolated soil bacterial samples were re-streaked with similar media onto the new plates to ensure decontamination. By using the dual culture method, bacterial isolates were screened against R. solani (Zhang et al., 2017). To measure the bacterial effective inhibitory concentration and their viability, we inoculate the same number of bacterial cells into the PDA plates. In the meantime, the different strain cells (equal quantity) OD can also be calculated at 600 nm and roughly we obtained the similar number of cells. According to this findings, we assumed that the same number of cells of different strains should have the same viability. In our study, we placed 1 μL overnight bacterial isolates culture in the 2.5 cm edge of the PDA plates and incubated for 16 h at 25 °C. The next day 5 mm of R. solani fungal mycelium disks were placed in the center of each dish, where only R. solani fungal inoculated plate was considered as a ck (control). Each treatment was performed in triplicate and repeated the assay twice. The cultures were placed in an incubator at 25 °C under dark condition before the pathogen completely covered the control plates. The inhibitory ability was used to determine the antagonistic nature of bacterial isolates (Khedher et al., 2015). For further experiments, Bacillus isolates which exhibited the highest antimicrobial potential against R. solani were selected. The inhibition rate was computed using the following equation:
2.3 Molecular identification of SI-18 isolates
A universal primer pairs 27F/1492R was used to hybridize two conserved regions in 16S rRNA genes to amplify the 16S rRNA region, and multilocus sequence analysis (MLSA) was conducted with one housekeeping gene fragments such as DNA gyrase subunit B (gyrB) by using gyrB-F/gyrB-R primer pairs to investigate the relation between B. subtilis and reference strains (Tan et al., 2019; Wang et al., 2007). The volume of PCR reactions was as follows: DNA template 1 μL, PCR buffer 5 μL (10x), dNTPs 4 μL (4 mmol/L), MgCl2 3 μL (25 mmol/L), each primer 1 μL (1 mmol/L), Taq polymerase 0.5 μL (1.25 U) and double distilled water 34.5 μL. Each primer sequence and PCR conditions are listed in Table 1. In order to achieve the emergence of a single product of the predicted size, 1% agarose gel was used to run the PCR products. For PCR product purification, the QIA-quick PCR purification kit was used and sequenced done by Wuhan Tianyi Huiyuan Biological Technology Co., Ltd, Hubei, China. Nucleotide sequence homology enquiries were conducted via GenBank online search engine BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The nucleotide sequences of 16S rRNA and gyrB gene were deposited in the GenBank database, and the accession numbers were received (MT598066 and MW008865). For phylogenetic analysis, multiple sequence alignments and comparisons with reference strain for each of the genes were performed through the help of CLUSTALW and MEGA7 tools. All the sequences were aligned using MEGA7 (Kumar et al., 2016) multiple alignment output by the UPGMA method (Mohkam et al., 2016). The pairwise alignment parameters in MEGA7 software with following parameter gap opening penalty 16, extension penalty 6.67, delay cutoff 35%, and transition weight of DNA 35%; weight matrix IUB with 1000 bootstrap replicates. Nucleotide missing or fragmented data for every gene were erased (Kumar et al., 2016). A bootstrap analysis was also done for the determination of the statistical value of internal nodes in the phylogenetic tree. The corresponding sequence accession numbers are listed in Supplementary Table S1 for constructing phylogenetic tree analyses.
Target gene
Primer name
Primer sequence
PCR conditions
16S rRNA
(1490 bp)27-F
1492-R5′- AGAGTTTGATCCTGGCTCAG-3′
5′-GGTTACCTTGTTACGACTT-3′95 °C for 5 min; 94 °C for 30 s, 53 °C for 30 s and 72 °C for 30 s (30 cycles); and at 72 °C for 10 min in final extension
gyrB
(1200 bp)
gyrB-F
gyrB-R5′-GAAGGCGGNACNCAYGAAG-3′
5′-CTTCRTGNGTNCCGCCTTC-3′95 °C for 1 min; 94 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s (30 cycles); and at 72 °C for 5 min in final extension
2.4 Effects of B. Subtilis SI-18 cultures on R. Solani mycelial growth
The isolate was inoculated into 50 mL LB medium and inserted in a 170 rpm rotary shaker at 30 °C to identify degradation of the SI-18 culture filtrate against pathogenic R. solani fungus. The bacterial suspension was collected after overnight cultivation and centrifuged at 10,000 × g for 10 min at 4 °C. The precipitate was filtered with a 0.22 µm on filter sheet. Afterwards, the mycelium of R. solani was placed on PDA and filled with medium sheet agar, and then with the help of punch loops in the fungus-coated layer to obtain a 5 mm diameter R. solani mycelia disk. R. solani mycelium around 5 mm in diameter was diluted with SI-18 supernatant and incubated for 24 h at room temperature. As a control, the sterile water was added with about the same length of mycelium. Each treatment of the procedure was replicated 3 times. Finally, the sample was examined under a light microscope to detect changes in the mycelium (ZEISS, Germany).
2.5 Protein preparation and quality prediction
The crystal structure of the S9 (40S ribosomal) protein of R. solani was not found in the database ( https://www.rcsb.org/). Hence we have employed a homology modelling method to get the hypothetical protein structure from SWISS-MODEL (Guex and Peitsch, 1997; Schwede et al., 2003). The amino acid sequence of the protein was retrieved from the NCBI database ( http://ncbi.nlm.nih.gov/) (Geer et al., 2010) with an accession number ABE68880. The crystal structure of eukaryotic ribosome (4 V88) with A chain (2–186 sequence) with 3Ǻ resolution was used as a template. Furthermore, the model protein was subjected to energy minimization in the SWISS PDB Viewer software package to remove clashes among the atoms (Kaplan and Littlejohn, 2001). Finally, the quality of the model protein was assessed through Ramachandran plot analysis (Sheik et al., 2002), Verify3D (Eisenberg et al., 1997), and ERRAT (Colovos and Yeates, 1993) to check structural correctness. The active site of the predicted homology model protein was determined through COACH webserver (Yang et al., 2013).
2.6 Ligand preparation
For docking analysis, about 134 compounds were studied based on literature review, and all of the ligand molecules were antimicrobial compounds from B. subtilis (Supplementary Table S2) (Caulier et al., 2019). Among them, the compound CID-12754692 (3-Pentanone), CID-75581 (1-Methylideneindene), CID-31404 (2,6-Ditert-butyl-4-methylphenol) was denoted as D1, D2, and D3 were taken for docking analysis based on their binding affinity. The structure was extracted from the PubChem database as sdf format (Kim et al., 2016). The mmff94 force field exposed the ligand molecules to energy minimization (Halgren, 1996).
2.7 Molecular docking
Molecular docking study was carried out using AutoDock Vina (Forli et al., 2012; Goodsell et al., 1996; Trott and Olson, 2010) and Pyrx, to identify the potential small molecule inhibitors against S9 protein of R. solani. The energy minimization process was achieved via the field of universal force and the algorithm of the conjugating gradient. By default, the total number of steps and the existing steps for the update was set. The structure of proteins and ligands was translated to PDBQT format. The center of the grid box was set as (X: 265.411, Y:49.93, Z: 271.258) Ǻ while the dimension was (X: 53.68, 47.774, Z:70.108) Ǻ. The non-bonded interactions were explored via Discovery studio (Inc, 2012) and Pymol software package (DeLano, 2002).
2.8 Molecular dynamics simulation
The molecular dynamics (MD) simulation study was incorporated via YASARA dynamics software (Krieger et al., 2004). The docked complex was cleaned, and AMBER14 force field was applied (Dickson et al., 2014). A cubic simulation cell was created with a cell size of (92.80 × 92.80 × 92.80) Å. For short-range van der Waals and Coulomb interactions, a cut-off radius of 8Åwas considered. For long-range electrostatic interaction, the Particle Mesh Ewald method was applied (Krieger et al., 2006). The Berendsen thermostat was used for maintaining the temperature of the simulation trajectory. The total physiological condition of the simulation system was set as (temperature 310 K, 0.9% NaCl and pH 7.4). The simulation trajectory was saved after 100 ps interval to analyze the simulation data. Finally, the docked complex was simulated for 30 ns to analyze RMSD, RMSF, SASA and Rg (Islam et al., 2019; Mahmud et al., 2020).
2.9 ADMET properties prediction
AdmetSAR online database was utilized to predict the absorption, metabolic characteristics, and toxicity of two selected compounds (Cheng et al., 2012). Simplified molecular-input system (SMILES) and structure data file (SDF) used as an input format in the database to predict the properties of the selected compounds.
2.10 Statistical analysis
The data were supplied using mean ± standard deviation (SD) and calculated using SAS (version 8.1, 2017) via analysis of variance (ANOVA). In addition, t-tests were done to find out the mean between any two treatments at a significance level of p < 0.05.
3 Results and discussion
3.1 Isolation, screening and morphological features of B. subtilis SI-18 strains
A total of 20 rhizosphere soil Bacillus strains were isolated from the wheat field that exhibited potential inhibitory effects against R. solani (Table 2). To assess their antagonistic action against R. solani fungal isolates, the dual-culture approach was applied. Among those strains, the highest antimicrobial potential against R. solani was observed in SI-18. For further studies, strain SI-18 was selected. The strain SI-18 colony was yielded motile, purple, rod-shaped, and small to medium size clumps on the LB agar plates (Fig. 1a). The microscopic observation under 100X magnification showed that SI-18 strain was rod-shaped and purple colour which confirmed isolated strain was Gram-positive bacterium (Fig. 1b). Throughout this experiment, strain SI-18 had a noticeable antimicrobial effect on R. solani with an in vitro inhibition rate of 44.22%. Most notably, strain SI-18 inhibition levels against pathogens were far higher than others. These results indicated that strain SI-18 is a broad-spectrum antagonistic agent with possible implementations for fungal disease control. From the above findings, we speculated that rhizosphere-growing microorganisms can play a crucial function in plant protection and are potential sources of antimicrobial agents (Fan et al., 2016). Through the use of synthetic pesticide to mitigate the plant-fungal disease has been restricted due to drug resistance, contamination of the atmosphere and prohibited for use in organic farming (Wu et al., 2019). The several studies stated that B. subtilis has active bio-control agents that substantially prevent the growth of R. solani fungi (Huang et al., 2017; Islam et al., 2012; Mnif et al., 2016). (a-j) Small letters are for comparison of means in the same column. Data are means of three replicates (n = 3).
Isolates
Inhibition rate (%)
Isolates
Inhibition rate (%)
SI-1
30.85 ± 0.33 h
SI-11
32.25 ± 0.54 e
SI-2
33.28 ± 0.44 e
SI-12
32.33 ± 0.61 e
SI-3
36.52 ± 0.37 b
SI-13
36.45 ± 1.02 b
SI-4
31.11 ± 0.63 f
SI-14
27.26 ± 0.37 j
SI-5
31.10 ± 0.59 f
SI-15
31.50 ± 0.48 g
SI-6
29.25 ± 0.76 h
SI-16
29.88 ± 0.36 h
SI-7
35.50 ± 0.83 c
SI-17
33.08 ± 0.81 e
SI-8
34.25 ± 0.11 d
SI-18
44.22 ± 1.03 a
SI-9
36.17 ± 0.42 b
SI-19
34.73 ± 0.82 d
SI-10
32.76 ± 0.60 e
SI-20
35.66 ± 0.27 c
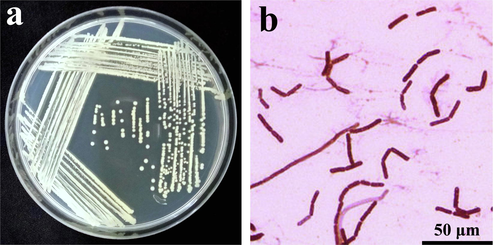
(a) B. subtilis SI-18 colony morphology on LB agar plates. (b) Gram staining microscopic observation of SI-18 strain (100x magnification), Bar = 50 μm.
3.2 Molecular identification of B. subtilis SI-18 strain
Using 16S rRNA gene sequencing and the MLSA method, molecular identification of the SI-18 strain was carried out. Sequencing reactions performed with two sets of primer pairs (27F/1492R and gyrB-F/gyrB-R) and amplified −1490 bp and −1200 bp fragments , respectively (Supplementary Fig. S1). To find out the best sequence homology, and the evolutionary relationship of the B. subtilis strains, the BlastN search in the NCBI database was applied. It showed 99% homology from the NCBI database with B. subtilis strain BCRC 14,716 (EF423595.1). The phylogenetic trees of the 13 Bacillus strains built from the 16S rRNA and gyrB gene sequences are shown in Fig. 2. Using the UPGMA method, the gyrB gene sequence generated a data set and showed 100% sequence similarity with corresponding sequences of reported B. subtilis strains (GenBank accession numbers: MG025594.1, JX513923.1, KF224942.1, and MN662266.1). All the B. subtilis strains formed a monophyle clade with 100% bootstrap support and 99.6–100% of the gyrB gene sequence similarities were found among them. From the phylogenetic analysis based on these two gene cluster sequences, more than 99% of the 16S rRNA gene sequence similarity was speculated where the gyrB gene exhibits 100% sequence similarity which indicates that the gyrB gene is a better genetic marker for the analysis of phylogenetic and taxonomic correlations at the species level in B. subtilis group.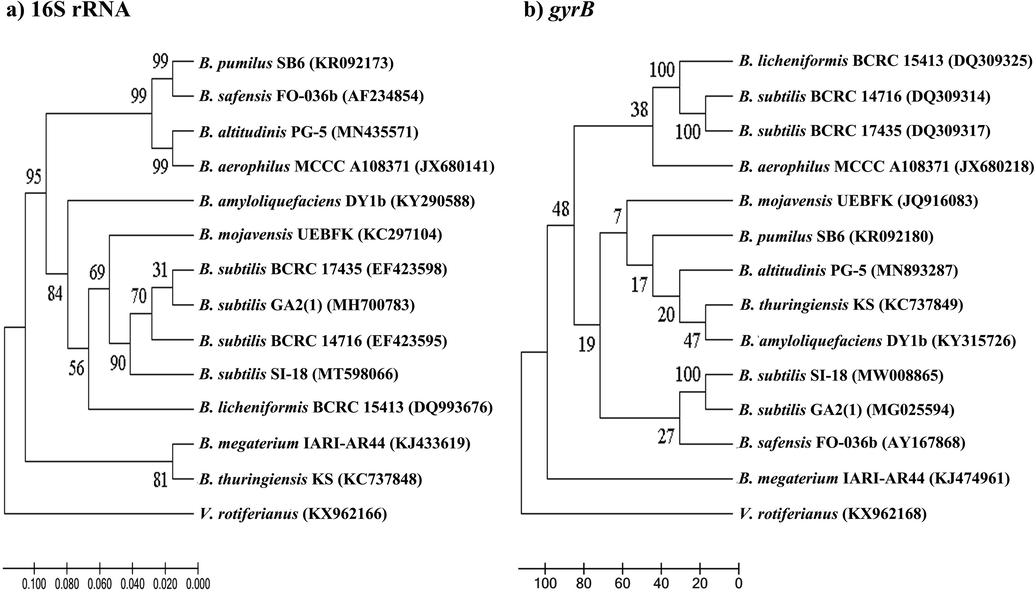
Phylogenetic tree showing the evolutionary relationship based on 16S rRNA (a) and gyrB (b) gene sequences among 13 Bacillus and our isolated strains. The trees were constructed using the UPGMA method. The bootstrap values (n = 1000) higher than 50% are visible at the internodes in the tree, and evolutionary distances were calculated using the p-distance method. As the outer group, Vibrio rotiferianus was used.
3.3 Suppression of R. solani mycelium growth using B. subtilis SI-18 culture filtrates
The degradation of R. solani mycelium growth by the SI-18 filtrate was visualized using a light microscope. The findings exhibited high vascularization, drainage on protoplasm, injury to the cell wall and pathogen deformation as compared with the controlled growth (Fig. 3a) treated with SI-18 filtrate after 16 h on pathogenic hyphae (Fig. 3b). A significant number of abnormal expansion, misalignment, and rupture of R. solani mycelium were likely to be attributed when treated for 24 h (Fig. 3c). Various Bacillus spp. have been reported as possible antimicrobials for plant disease (Ali et al., 2016; Gu et al., 2017). They have antagonistic features because such microorganisms generate bioactive components of broad-spectrum level (Huang et al., 2014). Secondary metabolites from Bacillus defend plants by either directly suppressing the pathogens or by enhancing systemic resistance (ISR) in the host organism (Asaka and Shoda, 1996). SI-18 demonstrated antagonistic to R. solani in this analysis. This study revealed that the SI-18 strain could secrete a large number of antimicrobial compounds. In terms of understanding the mechanisms of biological regulation via the PubChem database, we tried to predict the antifungal substances obtained by B. subtilis and incorporate them into the docking studies.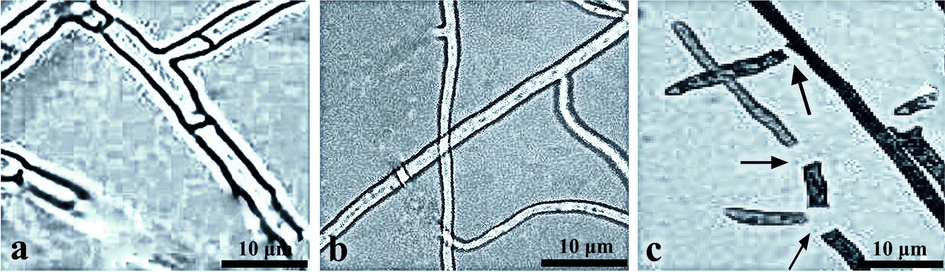
The suppression of mycelial growth of R. solani by SI-18 strain filtrates in different treatments. (a) control, (b) 16 h and (c) 24 h time intervals.
3.4 Homology modelling
The three-dimensional structure of 40S ribosomal protein S9 of R. solani was predicted from the SWISS-MODEL server. The sequence identity was found as 77.14% between template and model structure, and sequence coverage was 0.99. However, the protein structure derived from SWISS-MODEL was not ready to use for docking study; hence the model structure was subjected to energy minimization. After that, MolProbity Ramachandran plot analysis revealed the hypothetical protein had 155 (90.1%) amino acid in the favoured region, 12 (7%) in the allowed region and 5 (2.9%) in the outlier zone which once again verified the consistency of the predicted S9 protein model (Fig. 4). Furthermore, the protein quality was found as 82.5806% in ERRAT, and maximum amino acid residue of the model protein was below 99% error and 95% warning threshold. These results suggest a high resolution of the protein structure and better protein quality (Fig. 5). Three-dimensional structure of protein complexes was illustrated in Supplementary Fig. S2 (obtained from SWISS-Model). The protein synthesis is primarily based on the ribosome, where genetic codes are converted into proteins while their roles vary in the process of translation of prokaryotes and eukaryotes (Gkarmiri et al., 2015). The ribosomal protein S9 has become well known for its diverse functions, such as an indicator of nucleolar localization and its involvement in many tumour forms, and can play a vital role in ribosome assembly by connecting and strengthening r-proteins and encouraging their interaction with rRNA influences rDNA transcription (Lindström and Zhang, 2008). This S9 protein is also known to correspondence with ribosomal subunits (both small and large) (Yu et al., 2006). However, during the phase of disease progression, 40S ribosomal protein S9 from R. solani links to the fully mature rRNA where it attached spontaneously to the 5′ ends of the 16S rRNA resulting in complications in plant growth (Gerstner et al., 2001). To reduce this drawback, we carried out molecular docking of 134 compounds and based on binding affinities best three ligand molecules, D1, D2, and D3 compounds from the bacteria B. subtilis was screened.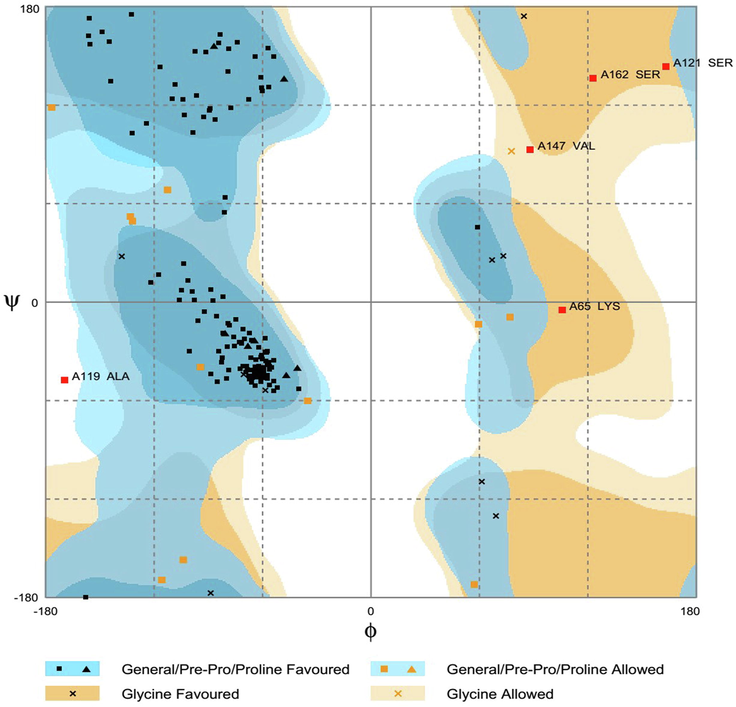
Ramachandran plot analysis of the predicted S9 protein of R. solani where 90.1% residues are found in the favoured region.
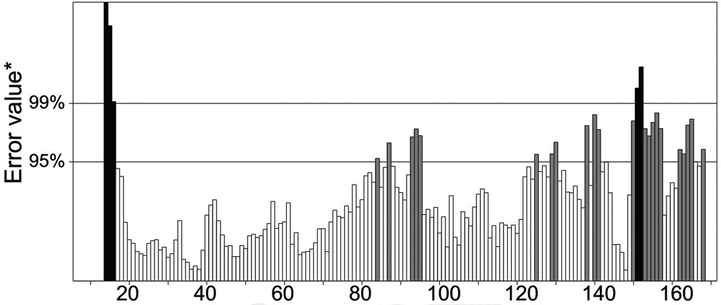
ERRAT analysis of the hypothetical protein.
3.5 Molecular docking
Molecular docking calculation was performed in AutoDock Vina, where the best three ligand candidates were selected based on binding affinity. The D1 compound had −8.2 Kcal/mol energy in docking study and showed notable non-bonded interaction with 40S ribosomal protein S9 of R. solani. Besides, one Pi-Pi-T-shaped interaction at Phe104 and five alkyl bonds at Ile83, Val147, Arg79, Leu80, and Val48 was seen in D1 and protein complex (Fig. 6a and Table 3). On the other hand, D2 ligand molecules had four hydrogen bonds at Arg44, Leu42, Val48, and Phe104. Moreover, Ile45, Val48, Ile83, Arg107, and Val85 created an alkyl bond for D2, whereas a single pi-alkyl bond was formed in Val147 (Fig. 6b and Table 3). However, a similar binding pattern was observed for D3 complex where five hydrogen bond was found at Val48, Ile45, Ile52 and Ile83 and Leu80. This D3 and 40S ribosomal protein S9 of R. solani also stabilized by one alkyl bond at Val147 and one pi-alkyl bond at Phe104 (Fig. 6c and Table 3). Identification of the hydrogen and hydrophobic interaction of the biological complex widely applied due to fast and accurate prediction and reduced lab cost (Bappy et al., 2020; Islam et al., 2019; Mahmud et al., 2019). The COACH server predicts the binding sites of the protein in Val48, Leu49, Ser50, Ile52, Arg53, Arg54, Ala55, Ala56, Leu59. The literature suggests that 1-hydroxyphenazine and 40S ribosomal protein S9 of R. solani had non-covalent interaction in Leu76 and Arg69 (Dharni et al., 2014) residues whereas our study suggests more non-bonded interactions against 40S ribosomal protein. More importantly, previous studies had no interactions in the active groove of the target protein, which is the key to possible inhibition of the target protein (Dharni et al., 2014). In our study, ligand molecules D1 and D2 had alkyl and hydrogen bond with Val48, which belongs to the active pockets. Moreover, two more non-bonded interactions (Val48 and Ile52) at the active sites were found for D3 which suggests better binding compared to the previous study. However, the non-bonded interaction between ligand–protein complex gives insights about possible binding interaction and helps to work on the further molecular modelling process.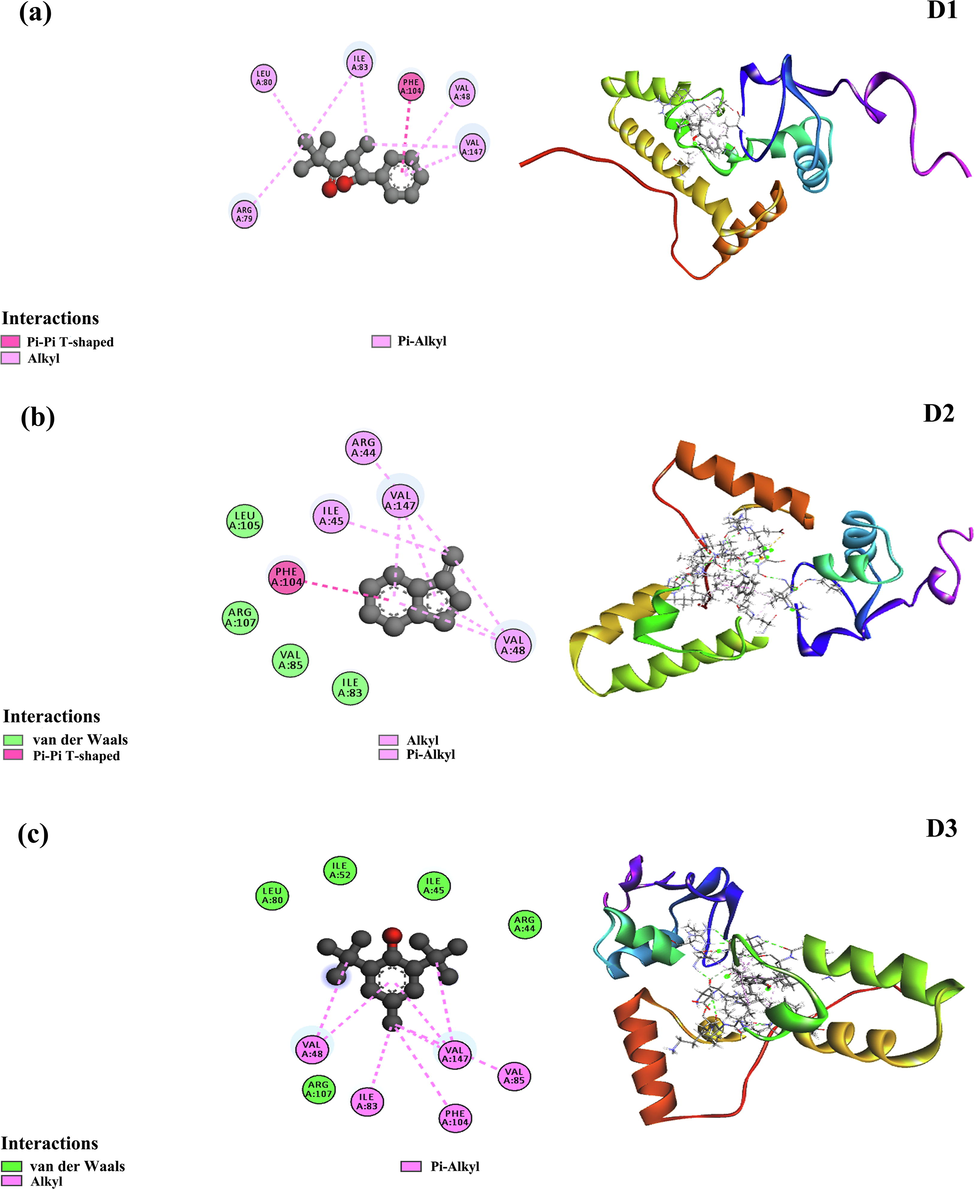
The top binding pose obtained by molecular docking simulation both in 2D binding pose and 3D docked pose of the D1, D2 and D3 complex (a,b,c). Residues involved in interactions with van der Waals, alkyl, Pi-alkyl, and Pi-Pi T-shaped are illustrated in various colors shown in the inset.
Compound
Binding energy(Kcal/mol)
Amino acid residue
Distance
Bond type
D1
−8.2
Phe104
Ile83
Val147
Arg79
Leu80
Val485.14
4.23
4.70
4.26
4.29
5.36PPT
A
A
A
A
A
D2
−8.4
Arg44
Leu42
Val48
Phe104
Ile45
Val48
Val147
Ile83
Arg107
Val851.88
2.2
1.86
2.98
4.61
4.30
4.81
5.12
4.86
4.77H
H
H
H
A
A
PA
A
A
A
D3
−8.2
Val48
Ile45
Ile52
Ile83
Leu80
Val147
Phe1041.80
2.69
2.24
2.76
2.14
3.87
4.62H
H
H
H
H
A
PA
3.6 Binding stability and rigidity of the docking complex
The root mean square deviation of the docked complex analyzed to understand the stability and rigidity of the complex. The RMSD descriptors of D1 and D2 complex were initially followed the upper trend from 0 to 8 ns and stabilized subsequently. Although D3 complex had lower RMSD at the initial phase, it also followed the same direction from 0 to 10 ns. However, D3 complex had slightly higher RMSD value than D1 and D2, but they did not over exceed and below than 2.5 Å. On the other hand, the root mean square fluctuations (RMSF) was determined to understand the flexibility over amino acid residue. Overall, maximum amino acid residue exhibit lower RMSF peak except, Pro2, Arg3, Al4, Pro5, Lys65, Lys120, Lys138, Gly166, Val172, Lys169, Arg175, Ala174 residues. From Fig. 7 it was illustrated that the all three docked complex D1, D2 and D3 complexes follow the same aptitude. Moreover, the radius of gyration from the simulation trajectory was evaluated to determine the degree of protein compactness. The higher rg value tells the folding probabilities of the protein whereas the lower radius of gyration value exhibit compactness of the protein. The D1 and D2 complex has more Rg value which indicates the more mobile nature of the protein. However, the D3 complex had lower Rg than D1 and D2 which indicates the tight packaging of the system. On the other hand, the solvent-accessible surface area was determined to understand the change in the surface area. The D1, D2 and D3 complexes had initial fluctuation from the beginning which indicates the increase of the docked complex surface area. However, D1, D2 complex followed the same trend from 10 to 25 ns and thereafter increase the surface area. The D3 complex has shrunken the surface area which illustrated in Fig. 7. Overall, the stable nature of the protein–ligand complex was observed in the MD trajectory which describes the complex stability.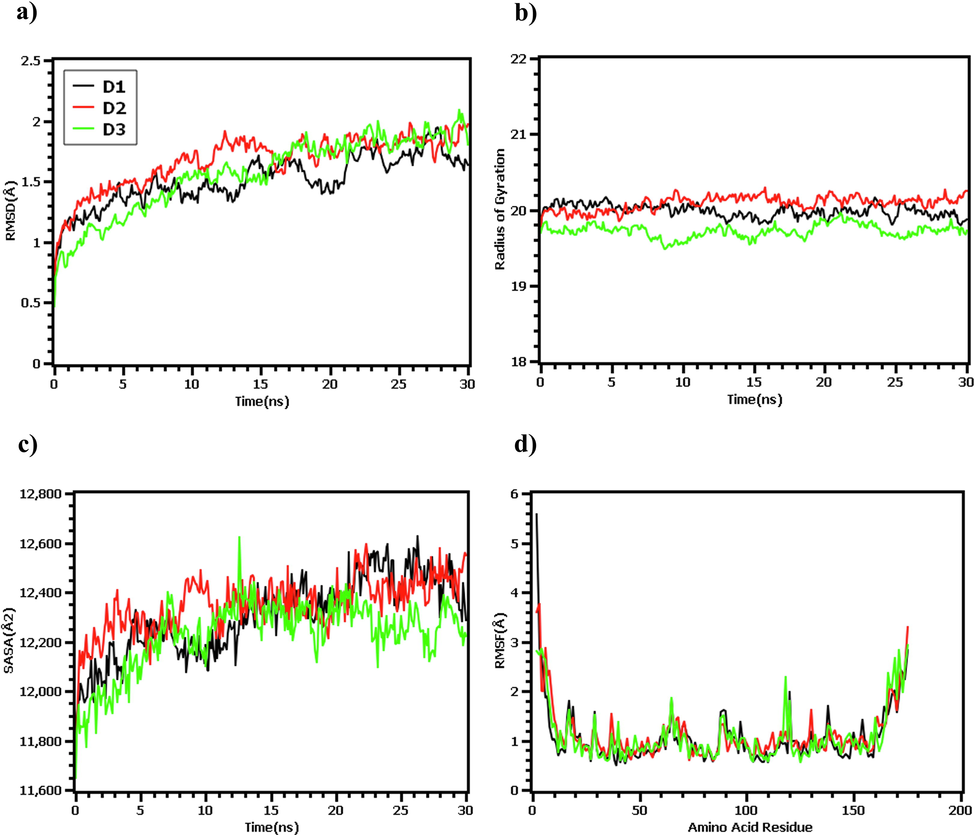
The molecular dynamics simulation of the docked complex. Here (a) root mean square fluctuations of the c-alpha atom of the complex, (b) radius of gyration to understand the mobile nature of the complex, (c) solvent accessible surface area of the complexes, (d) root mean square fluctuation to demonstrate the flexibility in the amino acid residue.
3.7 Pharmacological properties study
The potential ligand molecules need to pass several pharmacological and pharmacodynamics properties to ensure safety and efficacy. Herein, in silico ADMET prediction can act as a valid alternative by offering fast and low cost. The docked ligand was assessed through ADMETSAR tools to calculate absorption, toxicity, donor and acceptor of the hydrogen bond, and molecular weight. All the ligand molecules showed a positive response towards the blood–brain barrier and human intestinal absorption probability. Similarly, D1, D2, and D3 molecules have no toxic profile on the hepatotoxicity scale, although the D3 compound had a positive response on the carcinogenicity scale (Table 4). However, many drugs have been considered till now despite having carcinogenicity profile or any violation in pharmacological properties assessment. ‘+’ sign indicates the positive results; ‘–’ signs indicates the negative results; ‘NC’ indicates no-carcinogen whereas ‘C’ indicates carcinogen.
Properties
D1
D2
D3
Human Intestinal Absorption
0.9944(+)
0.9746(+)
0.9928(+)
Blood Brain Barrier
0.9973(+)
0.9919(+)
0.9946(+)
Carcinogenicity
0.7451(NC)
0.6380(NC)
0.6286(C)
Hepatotoxcity
0.7500(NC)
0.6250(NC)
0.6790(NC)
Molecular Weight
220.31 g/mol
128.17 g/mol
220.36 g/mol
Number of H bond Acceptor
2
0
1
Number of H bond donor
1
0
1
4 Conclusions
In conclusion, soil-borne pathogenic fungi R. solani strains are responsible for the severe loss to economic crops across the globe. Bacillus species play a significant role in minimizing these drawbacks, as it has many bioactive components. In this study, we identified SI-18 strain from twenty different soil samples and predicted the Bacillus D1, D2, and D3 compounds as potential inhibitors for S9 protein of R. solani. Finally, a comprehensive molecular docking approach used to recognize key residues responsible for the possible dynamics of R. solani S9 protein inhibition and interaction.
Acknowledgements
We express our deepest gratitude to Molecular-Plant Microbe Interactions Laboratory, Huazhong Agricultural University, Wuhan, China, for all facilities and support.
Funding
This work is supported by the National Major Project for Transgenic Organism Breeding (2016ZX08003-001).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Akinrinlola, R.J., Yuen, G.Y., Drijber, R.A., Adesemoye, A.O., 2018. Evaluation of Bacillus strains for plant growth promotion and predictability of efficacy by in vitro physiological traits. Int. J. Microbiol. 2018. ID: 5686874.
- Ex Vivo Application of Secreted Metabolites Produced by Soil-Inhabiting Bacillus spp. Efficiently Controls Foliar Diseases Caused by Alternaria spp. Appl. Environ. Microbiol.. 2016;82:478-490.
- [Google Scholar]
- Biocontrol of Rhizoctonia solani damping-off of tomato with Bacillus subtilis RB14. Appl. Environ. Microbiol.. 1996;62:4081-4085.
- [Google Scholar]
- Extensive immunoinformatics study for the prediction of novel peptide-based epitope vaccine with docking confirmation against envelope protein of Chikungunya virus: a computational biology approach. J. Biomol. Struct. Dyn. 2020:1-16.
- [Google Scholar]
- An updated metabolic view of the Bacillus subtilis 168 genome. Microbiology. 2013;159:757-770.
- [Google Scholar]
- Induction of defense-related ultrastructural modifications in pea root tissues inoculated with endophytic bacteria. Plant Physiol.. 1996;112:919-929.
- [Google Scholar]
- Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Front. Microbiol.. 2019;10:302.
- [Google Scholar]
- admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J. Chem. Inf. Model.. 2012;52:3099-3105.
- [Google Scholar]
- Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci.. 1993;2:1511-1519.
- [Google Scholar]
- The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol.. 2012;13:414-430.
- [Google Scholar]
- Pymol: An open-source molecular graphics tool. CCP4 Newsl. protein Crystallogr.. 2002;40:82-92.
- [Google Scholar]
- The interaction pattern between a homology model of 40S ribosomal S9 protein of Rhizoctonia solani and 1-Hydroxyphenaize by docking study. Int: Biomed Res; 2014. p. :2014.
- [20] VERIFY3D: assessment of protein models with three-dimensional profiles. Methods in Enzymology. Elsevier 1997:396-404.
- [Google Scholar]
- Diversity, distribution, and antagonistic activities of rhizobacteria of Panax notoginseng. J. Ginseng Res.. 2016;40:97-104.
- [Google Scholar]
- Survival of Rhizoctonia solani AG-1 IA, the Causal Agent of Rice Sheath Blight, under Different Environmental Conditions. J. Phytopathol.. 2017;165:44-52.
- [Google Scholar]
- Forli, W., Halliday, S., Belew, R., Olson, A.J., 2012. AutoDock Version 4.2.
- Two antimicrobial genes from Aegilops tauschii Cosson identified by the Bacillus subtilis expression system. Sci. Rep.. 2020;10:1-11.
- [Google Scholar]
- Recognition of 16S rRNA by ribosomal protein S4 from Bacillus stearothermophilus. Biochemistry. 2001;40:7165-7173.
- [Google Scholar]
- Transcriptomic changes in the plant pathogenic fungus Rhizoctonia solani AG-3 in response to the antagonistic bacteria Serratia proteamaculans and Serratia plymuthica. BMC Genomics. 2015;16:630.
- [Google Scholar]
- Automated docking of flexible ligands: applications of AutoDock. J. Mol. Recognit.. 1996;9:1-5.
- [Google Scholar]
- Bacillomycin D produced by Bacillus amyloliquefaciens is involved in the antagonistic interaction with the plant-pathogenic fungus Fusarium graminearum. Microbiol: Appl. Environ; 2017. p. :83.
- SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714-2723.
- [Google Scholar]
- Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem.. 1996;17:490-519.
- [Google Scholar]
- Suppression of bacterial wilt of tomato by bioorganic fertilizer made from the antibacterial compound producing strain Bacillus amyloliquefaciens HR62. J. Agric. Food Chem.. 2014;62:10708-10716.
- [Google Scholar]
- Rhizospheric Bacillus subtilis exhibits biocontrol effect against Rhizoctonia solani in pepper (Capsicum annuum). Int: Biomed Res; 2017. p. :2017.
- Inc, A.S., 2012. Discovery studio modeling environment, release 3.5. Accelrys Discov. Stud. Accelrys Softw. Inc, San Diego.
- Assessment of structurally and functionally high-risk nsSNPs impacts on human bone morphogenetic protein receptor type IA (BMPR1A) by computational approach. Comput. Biol. Chem.. 2019;80:31-45.
- [Google Scholar]
- Isolation and identification of antifungal compounds from Bacillus subtilis C9 inhibiting the growth of plant pathogenic fungi. Mycobiology. 2012;40:59-65.
- [Google Scholar]
- Rapid determination of rice cultivar responses to the sheath blight pathogen Rhizoctonia solani using a micro-chamber screening method. Plant Dis.. 2007;91:485-489.
- [Google Scholar]
- Characterization of medicinal plant-associated biocontrol Bacillus subtilis (SSL2) by liquid chromatography-mass spectrometry and evaluation of compounds by in silico and in vitro methods. J. Biomol. Struct. Dyn.. 2020;38:500-510.
- [Google Scholar]
- Volatiles of bacterial antagonists inhibit mycelial growth of the plant pathogen Rhizoctonia solani. Arch. Microbiol.. 2007;187:351-360.
- [Google Scholar]
- Computer-aided drug discovery and development (CADDD): in silico-chemico-biological approach. Chem. Biol. Interact.. 2008;171:165-176.
- [Google Scholar]
- Efficacy of Bacillus subtilis V26 as a biological control agent against Rhizoctonia solani on potato. C. R. Biol.. 2015;338:784-792.
- [Google Scholar]
- Antimicrobial genes from Allium sativum and Pinellia ternata revealed by a Bacillus subtilis expression system. Sci. Rep.. 2018;8:1-12.
- [Google Scholar]
- Making optimal use of empirical energy functions: force-field parameterization in crystal space. Proteins Struct. Funct. Bioinforma.. 2004;57:678-683.
- [Google Scholar]
- Fast empirical pKa prediction by Ewald summation. J. Mol. Graph. Model.. 2006;25:481-486.
- [Google Scholar]
- MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol.. 2016;33:1870-1874.
- [Google Scholar]
- Ribosomal protein S9 is a novel B23/NPM-binding protein required for normal cell proliferation. J. Biol. Chem.. 2008;283:15568-15576.
- [Google Scholar]
- Evaluation of the Potential of five Housekeeping Genes for Identification of Quarantine Pseudomonas syringae. J. Phytopathol.. 2017;165:73-81.
- [Google Scholar]
- Microencapsulation of Bacillus subtilis B99–2 and its biocontrol efficiency against Rhizoctonia solani in tomato. Biol. Control. 2015;90:34-41.
- [Google Scholar]
- Exploring the potent inhibitors and binding modes of phospholipase A2 through in silico investigation. J. Biomol. Struct. Dyn.. 2020;38:4221-4231.
- [Google Scholar]
- Exploring the potent inhibitors and binding modes of phospholipase A2 through in silico investigation. Dyn: J. Biomol. Struct; 2019.
- Purification and identification of Bacillus subtilis SPB1 lipopeptide biosurfactant exhibiting antifungal activity against Rhizoctonia bataticola and Rhizoctonia solani. Environ. Sci. Pollut. Res.. 2016;23:6690-6699.
- [Google Scholar]
- Identification of Bacillus probiotics isolated from soil rhizosphere using 16S rRNA, recA, rpoB gene sequencing and RAPD-PCR. Probiotics Antimicrob. Proteins. 2016;8:8-18.
- [Google Scholar]
- Biological control of banded leaf and sheath blight disease (Rhizoctonia solani Kuhn) in corn with formulated Bacillus subtilis BR23. Indones. J. Agric. Sci.. 2016;7:1-7.
- [Google Scholar]
- Roles of Bacillus endospores in the environment. Cell. Mol. Life Sci. C.. 2002;59:410-416.
- [Google Scholar]
- Genome-wide polysomal analysis of a yeast strain with mutated ribosomal protein S9. BMC Genomics. 2007;8:285.
- [Google Scholar]
- Polanczyk, R.A., 2004. Estudos de Bacillus thuringiensis Berliner visando ao controle de Spodoptera frugiperda (JE Smith).
- Physicochemical and biophysical panel symposium biologically active secondary metabolites. Pestic. Sci. 1985;16:422-427.
- [Google Scholar]
- Prediction and Characterization of Cationic Arginine-Rich Plant Antimicrobial Peptide SM-985 From Teosinte (Zea mays ssp. mexicana) Front. Microbiol.. 2020;11:1353.
- [Google Scholar]
- SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res.. 2003;31:3381-3385.
- [Google Scholar]
- Two different lantibiotic-like peptides originate from the ericin gene cluster of Bacillus subtilis A1/3. J. Bacteriol.. 2002;184:1703-1711.
- [Google Scholar]
- Identification and biocontrol potential of antagonistic bacteria strains against Sclerotinia sclerotiorum and their growth-promoting effects on Brassica napus. Biol. Control. 2017;104:35-43.
- [Google Scholar]
- Isolation and identification of a Bacillus subtilis HZ-72 exhibiting biocontrol activity against flax seedling blight. Eur. J. Plant Pathol.. 2019;153:825-836.
- [Google Scholar]
- AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem.. 2010;31:455-461.
- [Google Scholar]
- Assessment of genetic diversity and plant growth promoting attributes of psychrotolerant bacteria allied with wheat (Triticum aestivum) from the northern hills zone of India. Ann. Microbiol.. 2015;65:1885-1899.
- [Google Scholar]
- Comparison of gyrB gene sequences, 16S rRNA gene sequences and DNA–DNA hybridization in the Bacillus subtilis group. Int. J. Syst. Evol. Microbiol.. 2007;57:1846-1850.
- [Google Scholar]
- Cell Membrane-Interrupting Antimicrobial Peptides from Isatis indigotica Fortune Isolated by a Bacillus subtilis Expression System. Biomolecules. 2020;10:30.
- [Google Scholar]
- Bio-control of Rhizoctonia solani via induction of the defense mechanism and antimicrobial compounds produced by Bacillus subtilis SL-44 on pepper (Capsicum annuum L.) Front. Microbiol.. 2019;10:2676.
- [Google Scholar]
- Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics.. 2013;29:2588-2595.
- [Google Scholar]
- miRDeep-P: a computational tool for analyzing the microRNA transcriptome in plants. Bioinformatics. 2011;27:2614-2615.
- [Google Scholar]
- An improved strategy for stable biocontrol agents selecting to control rice sheath blight caused by Rhizoctonia solani. Microbiol. Res.. 2017;203:1-9.
- [Google Scholar]
- Nucleophosmin is essential for ribosomal protein L5 nuclear export. Mol. Cell. Biol.. 2006;26:3798-3809.
- [Google Scholar]
- Screening and characterization of endophytic Bacillus for biocontrol of grapevine downy mildew. Crop Prot.. 2017;96:173-179.
- [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2020.09.044.
Appendix A
Supplementary data
The following are the Supplementary data to this article:Supplementary data 1
Supplementary data 1







