Translate this page into:
In silico study of novel niclosamide derivatives, SARS-CoV-2 nonstructural proteins catalytic residue-targeting small molecules drug candidates
⁎Corresponding authors at: TMU Research Center of Cancer Translational Medicine, Taipei Medical University, Taipei 11031, Taiwan (A.T.H. Wu). PhD Program for Cancer Molecular Biology and Drug Discovery, College of Medical Science and Technology, Taipei Medical University, Taipei 11031, Taiwan (H.-S. Huang). chaw1211@tmu.edu.tw (Alexander T.H. Wu), huanghs99@tmu.edu.tw (Hsu-Shan Huang)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2)-mediated coronavirus disease 2019 (COVID-19) infection remains a global pandemic and health emergency with overwhelming social and economic impacts throughout the world. Therapeutics for COVID-19 are limited to only remdesivir; therefore, there is a need for combined, multidisciplinary efforts to develop new therapeutic molecules and explore the effectiveness of existing drugs against SARS-CoV-2. In the present study, we reported eight (SCOV-L-02, SCOV-L-09, SCOV-L-10, SCOV-L-11, SCOV-L-15, SCOV-L-18, SCOV-L-22, and SCOV-L-23) novel structurally related small-molecule derivatives of niclosamide (SCOV-L series) for their targeting potential against angiotensin-converting enzyme-2 (ACE2), type II transmembrane serine protease (TMPRSS2), and SARS-COV-2 nonstructural proteins (NSPs) including NSP5 (3CLpro), NSP3 (PLpro), and RdRp. Our correlation analysis suggested that ACE2 and TMPRSS2 modulate host immune response via regulation of immune-infiltrating cells at the site of tissue/organs entries. In addition, we identified some TMPRSS2 and ACE2 microRNAs target regulatory networks in SARS-CoV-2 infection and thus open up a new window for microRNAs-based therapy for the treatment of SARS-CoV-2 infection. Our in vitro study revealed that with the exception of SCOV-L-11 and SCOV-L-23 which were non-active, the SCOV-L series exhibited strict antiproliferative activities and non-cytotoxic effects against ACE2- and TMPRSS2-expressing cells. Our molecular docking for the analysis of receptor-ligand interactions revealed that SCOV-L series demonstrated high ligand binding efficacies (at higher levels than clinical drugs) against the ACE2, TMPRSS2, and SARS-COV-2 NSPs. SCOV-L-18, SCOV-L-15, and SCOV-L-09 were particularly found to exhibit strong binding affinities with three key SARS-CoV-2’s proteins: 3CLpro, PLpro, and RdRp. These compounds bind to the several catalytic residues of the proteins, and satisfied the criteria of drug-like candidates, having good adsorption, distribution, metabolism, excretion, and toxicity (ADMET) pharmacokinetic profile. Altogether, the present study suggests the therapeutic potential of SCOV-L series for preventing and managing SARs-COV-2 infection and are currently under detailed investigation in our lab.
Keywords
SARS-CoV-2
ACE2
TMPRSS2
SCOV-L series, niclosamide derivatives
Nonstructural proteins
Antiproliferation
Data Availability Statement:
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
1 Introduction
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2)-induced coronavirus disease 2019 (COVID-19) is one of the most devastating global pandemic diseases in a century (Xu et al., 2020). Currently there have been more than 293 million reported cases and more than 5 million deaths globally, with these numbers continuing to dramatically increase daily. Furthermore, due to increased understanding of the virus cycle and global adoption of adequate management strategies, more than 255 million infected people have recovered from the virus (https://www.worldometers.info/coronavirus/).
Coronaviruses are non-segmented, positive-stranded RNA viruses of the Coronaviridae family (in the order Nidovirales). SARS-CoV-2 is one of the largest viral RNA genomes known to date (∼30 kb), encoding different structural (membrane, spike, envelope, and nucleocapsid) proteins, nonstructural proteins (NSPs of NSP1 ∼ NSP16), and five accessory proteins (open-reading frame 3a (ORF3a), ORF6, ORF7, ORF8, and ORF9) (Iftikhar et al., 2020). Angiotensin-converting enzyme-2 (ACE2) is a SARS-CoV-2 cellular receptor which facilitates viral entry into host cells via type II transmembrane serine protease (TMPRSS2) (Xie et al., 2005). Once the virus enters a host cell, the ORFs are translated into two polypeptides (pp1a and pp1b) (Sinha et al., 2020) which are further proteolytically divided into the 16 NSPs (Ziebuhr, 2005). The two viral proteases encoded by the SARS-CoV-2 (NSP3/papain-like protease and NSP5/3C-like protease) play vital roles in viral replication. The main protease (3C-like protease) mediates SARS-CoV replication and transcription via extensive proteolytic processing of the two replicase polyproteins (pp1a and pp1ab) to produce different functional proteins (Báez-Santos et al., 2015). The 3C-like protease, therefore, serves as an attractive target for anti-SARS drug discovery (Anand et al., 2003; Yang et al., 2003; Chou et al., 2003).
Lack of adequate understanding of the virus and inadequate effective treatments pose great challenges to the clinical management of SARS-COV-2 and thus highlight the critical need for discovering and developing new drugs. Although clinical drugs, including remdesivir, chloroquine, ivermectin, and favipiravir, have received appraisal for their promising effects for treating COVID-19 in experimental and clinical settings (Wang et al., 2020; Gao et al., 2020; Agrawal et al., 2020), remdesivir is the only drug that has so far been approved by the US Food and Drug Administration (FDA) for treating COVID-19 (Nih, 2021). It is therefore very important to search for other effective inhibitors as potential treatments of COVID-19. Niclosamide is a multipurpose clinical small molecule with several biological activities including antihelmintic, antioxidant, antimicrobial, anti-inflammatory, anticancer and immune-modulatory properties (Li et al., 2014; Kadri et al., 2018). Recent studies also identified the potential of niclosamide for treating viral infections (Xu et al., 2020) and reported its ability to inhibit replication of SARS-CoV by several investigators (Wu et al., 2004; Wen et al., 2007).
Our in-house synthesized small molecules have received great therapeutic success with translational relevance for the treatment of cancers, inflammations, oxidative stress, immune-related, and metabolic disorders (Lawal et al., 2021g; Huang et al., 2004b, 2004a, 2015; Khedkar et al., 2021; Lawal et al., 2021c, 2021d, 2021a, 2021e, 2021b; Lee et al., 2012, 2015a, 2015b, 2020; Liu et al., 2018; Madamsetty et al., 2019; Mokgautsi et al., 2021b, 2021a; Shen et al., 2019; Yadav et al., 2020; Wu et al., 2021; Lawal et al., 2022). In the present study, we report eight novel structurally related small-molecule derivatives of niclosamide (called the SCOV-L series) for their targeting potential against angiotensin-converting enzyme-2 (ACE2), type II transmembrane serine protease (TMPRSS2), and nonstructural protein (NSP) targets of SARS-CoV-2. Our in vitro study revealed that these compounds exhibited strict antiproliferative activities and non-cytotoxic effects against ACE2- and TMPRSS2-expressing cells. Exceptional results from SCOV-L-18, SCOV-L-15, and SCOV-L-09 demonstrated high ligand binding efficacies (compared to current clinical drugs) of these molecules against ACE2 and TMPRSS2 in receptor-ligand interaction studies. This SCOV-L series contains drug-like candidates with good absorption, distribution, metabolism, excretion, and toxicity (ADMET) pharmacokinetic properties and have potential for preventing and managing SARS-CoV-2 infection.
2 Methods
2.1 Analysis of ACE2 and TMPRSS expression across tissues and gene coexpression
We evaluated the tissue-specific expression and distribution patterns of ACE2 and TMPRSS2 across 63 tissues using the TissueDataExtractor modules of COXPRESdb (Obayashi et al., 2019). Entrez Gene IDs (ACE2: 59272 and TMPRSS2: 7113) were used to search and extract expression profiles of ACE2 and TMPRSS2 from the GTEx project dataset. All expression values were converted to base-2 logarithms with a pseudo count of 0.001, and visualized using line graphs. Term concept modules of the COREMINE medical server ( http://www.coremine.com/) were used to explore the gene coexpression and disease/drug/phenotype interactions and build biological mind maps of TMPRSS2 and ACE2.
2.2 Effects of ACE2 and TMPRSS2 messenger (m)RNA expression levels on tissue immune/immunosuppressive infiltrating cells
We used the immune module of GSCALite (https://bioinfo.life.hust.edu.cn/web/GSCALite/) server (Liu et al., 2018) and the Tumor Immune Estimation Resource (TIMER, vers. 2.0) (Li et al., 2017) to analyze the effects of ACE2 and TMPRSS2 mRNA expression levels on tissues infiltration levels of immune and immunosuppressive cells including cytotoxic lymphocytes (B cells, cluster of differentiation 4 (CD4) naïve, CD8 naïve, CD4 T, CD8 T, and γδ T cells), neutrophils, dendritic cells (DCs), macrophages, myeloid-derived suppressor cells (MDSCs), and regulatory T (Treg) cells. A correlation analysis was conducted using purity-corrected partial Spearman's rho values and statistical significance (p < 0.05).
2.3 MicroRNAs (miRNAs) regulatory network of SARS-CoV-2 by ACE2 and TMPRSS2
We queried the miRNA regulatory network of ACE2 and TMPRSS2 using a single package module containing six prediction tools (including DIANA-microT (2), MicroT4 (3), miRBridge (4), miRDB (5), miRMap (6), and PITA) (Liu et al., 2020). In addition, we created a self-mapping network of the identified miR regulatory network between ACE2 and TMPRSS2.
2.4 In silico analysis of the physicochemical and ADMET-pharmacokinetics properties of the SCOV-L series
Drug likeness, physicochemical properties, pharmacokinetics (ADMET), and medicinal chemistry of members of the SCOV-L series were evaluated using various in silico drug screening databases including the SwissADME, ADMETLab (https://admet.scbdd.com/calcpre/index_sys/), and ADMETSar (http://lmmd.ecust.edu.cn/admetsar2) databases (Liu et al., 2014). Drug likeness was determined using molecular descriptors including the molecular weight (MW), numbers of H-bond donors and acceptors, rotatable bond counts, topological polar surface area (TPSA), and partition coefficient (cLogP). ADMET pharmacokinetic properties analyzed included absorption (Caco-2 permeability, human intestinal absorption (HIA), P-glycoprotein (P-gp)-inhibitor/substrate, and bioavailability), distribution (volume distribution in a steady state (VDss), plasma protein binding, blood–brain barrier (BBB) and central nervous system (CNS) permeability), excretion (clearance rate and half-life time), and toxicity (AMES toxicity, human ether-a-go-go-related gene (hERG) inhibitor, and oral rat acute toxicity (expressed as the 50% lethal dose (LD50)), hepatotoxicity, and skin sensitization). Metabolism was assessed based on their tendency of cytochrome P450 inhibition and substrate. The drugs were also analyzed for pan-assay interference compounds (PAINS) alerts. Human intestinal absorption and BBB permeability were modeled using the BOILED EGG models, support vector machine_LiCABEDS algorithm model built based on the validity of four types of fingerprints of 1593 reported compounds (https://www.cbligand.org/BBB/)(Liu et al., 2014), and mathematical calculations based on the compounds' MW and number of H bonds (Pardridge, 1998).
2.5 Molecular docking analysis of the binding efficacies of members of the SCOV-L series with ACE2, TMPRSS2. and SARS-CoV-2 NSPs
2.5.1 Preparation of the ligand and receptor structures
Crystal structures of ACE2 (PDB: 1R42), TMPRSS2 (PDB: 7MEQ), chymotrypsin-like protease (3CLpro) (PDB: 6LU7), papain-like protease (NSP3: PLpro; PDB: 6WUU), and RNA-dependent RNA polymerase (RdRp_PDB: 7BV2) were downloaded from the RCSB PDB database All PDB were devoid of missising residues. The ligands (SCOV-L series) 2D structures of SCOV-L series were drawn in Chemdraw program. The ligands 2D structures were then converted to 3D structures with molecular mechanic optimizing by the Avogadro molecular builder and visualization tool vers. 1.XX (Marcus et al., 2012). The ligands (SCOV-L series) were generated in MOL 2 files which were subsequently converted to PDB format using the PyMOL Molecular Graphics System, vers. 1.2r3pre. All PDB files (SCOV-L compound series and targets) were converted to pdbqt format using AutoDock Vina (vers. 0.8, Scripps Research Institute, La Jolla, CA, USA) (Trott and Olson, 2010).
2.5.2 Grid preparation and docking using AutoDock/Vina
Molecular docking was performed using AutoDock Vina (vers. 0.8) (Trott and Olson, 2010), and default settings of the software were adopted. Protocols described in previous studies (Lawal et al., 2021; Visualizer, 2020) were adopted for ligand and receptor preparations prior to docking. The protein structures were prepared by removing water and polar H-atoms, while the Kollman charges were added. Gasteiger charges were added to the ligands. Target receptors were set to be rigid, while all ligand bonds were allowed to freely rotate. Each of the grid boxes, X, Y, and Z, was set to 40 Å with spacing adjusted to 1.0 Å. The grid center was designated at dimensions (x, y, and z) for main protease (-10.85, 12.58 and 68.72), NSP3 (9.69, −8.42 and 17.13), and for the papain-like protease (21.83, 69.71 and 3.32) docking. All docking was performed with num_modes set to 10, an energy-range of 4, and docking exhaustiveness of 12. PDB files of the docked complexes were built using pyMOL software and analyzed for interaction affinities (expressed in kcal/mol). The pose with lowest energy of binding or binding affinity was extracted and aligned with receptor structure for further visualization analysis. Two-dimensional (2D) conformations of the receptor-ligand complexes and interaction distances between target amino acid residues and corresponding ligand atoms for conventional hydrogen bonds were visualized (expressed in Å) using Discovery studio visualizer vers. 19.1.0.18287 (BIOVIA, San Diego, CA, USA) (Shoemaker, 2006). A cut-off distance of 4 Å were used for the calculation of ligand–protein hydrogen bonds.
2.6 Cell lines and culture
ACE2- and TMPRSS2-expressing cell lines, including NCI-H322M, IGROV1, and A549, were obtained from the National Cancer Institute, National Institutes of Health (NIH-NCI, USA). Cells were cultured in 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin supplemented with RPMI-1640, and incubated in humidified 5% CO2 at 37 °C. Culture media were replaced after 73 h, and cells were subcultured when they reached 90% confluence.
2.7 In vitro antiproliferative analysis of members of the SCOV-L series against ACE2- and TMPRSS2-expressing cell lines
Members of the SCOV-L series (SCOV-L-02, SCOV-L-09, SCOV-L-10, SCOV-L-11, SCOV-L-15, SCOV-L-18, SCOV-L-22, and SCOV-L-23) were evaluated for in vitro activities against three ACE2- and TMPRSS2-expressing cell lines, including NCI-H322M, IGROV1, and A549. About 5000 ∼ 40,000 viable cells were seeded and incubated in 96-well plates at 37 °C with 95% air, 5% CO2, and 100% relative humidity for 24 h. Activities of the drugs were initially analyzed for their antiproliferative and cytotoxic natures by treatment with a therapeutic dose of 10 μM for 48 h (Holbeck et al., 2010; Vichai and Kirtikara, 2006). Cell viability and cell responses to drug treatment were determined using a sulforhodamine B (SRB) staining protocol (Thierry and Roch, 2020). The antiproliferative effect was assessed as the percentage growth inhibition compared to an untreated control after 48 h, while the cytotoxic effect was assessed based on the percentage cell killing compared to time zero. The compounds were further subjected to multiple-dose screening at five concentrations of 0, 0.1, 1.0, 10, and 100 μM. The activity of the drug on each cell line was calculated and expressed as the 50% lethal concentration (LC50) where Tz is the absorbance at time 0 and Ti is the absorbance of drug-treated cells after 48 h. When the maximum dose tested (100 μM) did not meet the required effect on the cell line, the LC50 was expressed as greater than the maximum drug concentration tested (>100 μM) (Lawal et al., 2021).
3 Results and discussion
3.1 Insight and biomedical mapping of ACE2 and TMPRSS2 regulatory process, gene-coexpression and therapy in SARS-CoV-2 infection
In order to have a general insight into the role of ACE2 and TMPRSS2 in SARs-COV2 infection, we mined coremine medical and construct the biomedical mind mapping of ACE2 and TMPRSS2 in SARS-CoV-2 infection with emphasis on biological process, molecular function, gene-coexpression, and drugs with proven efficacy for the treatment of the SARS-CoV-2 infection. Interestingly we found that ACE2 and TMPRSS2 concertedly regulated various biological processes including Tropism, membrane fusion, syncytium formation by plasma membrane fusion, extracellular structure organization, immune response, and inflammatory response in SARS-CoV-2 infection. The previous study has indicated that SARS-CoV-2 may evade the innate immune response, causing uncontrolled neutrophil extracellular traps formation and multi-organ failure (Doykov et al., 2020). The results of the present study therefore strongly suggested that the ACE2 and TMPRSS2 play a role in host immune response to SARS-CoV-2 infection thus contributing to multi-organ failure observed that are observed in severe COVID-19 patients (Sharma and Zhao, 2021). In addition, molecular functions regulated by these genes were receptor and protein binding, retinal dehydrogenase activity, peptidase activity, tRNA methyltransferase activity, and ion channel binding (Fig. 1). Through mining for potential target therapy, we identified several chemical drugs including; angiotensin I (1–7), Angiotensin II, camostat, Remdesivir, nafamostat, favipiravir, arbidol, Serine/Threonine Kinase Inhibitor XL418, Hydroxychloroquine, and 4-(4-guanidinobenzoyloxy) phenylacetic acid (Fig. 1), and several Chinese medicinal plants; Guang Guo Gan Cao, Xiang Hei Zhong Cao Zi, Qing Hao, Huang Si Yu Jin, Jiang Huang, Gan Cao, Huang Qin, Da Suan, Jiang Pi, and Sheng Jiang (Fig. 1) were found to exhibit strong potential for the treatment of SARS-CoV-2 infection via targeting ACE2 and TMPRSS2. Analysis of gene expression correlation revealed that the expression and activities of ACE2 and TMPRSS2 are associated with the activities of other proteins including CDSN, FURIN, ACE, AGT, AGTR1, TMPRSS4, MAS1, SLC6A19, AGTR2, and ADAM17, suggesting that these genes could play some role in the pathogenesis of SARS-CoV-2 infection and thus could be considered as potential targets in future drug design for SARS-CoV-2 infection.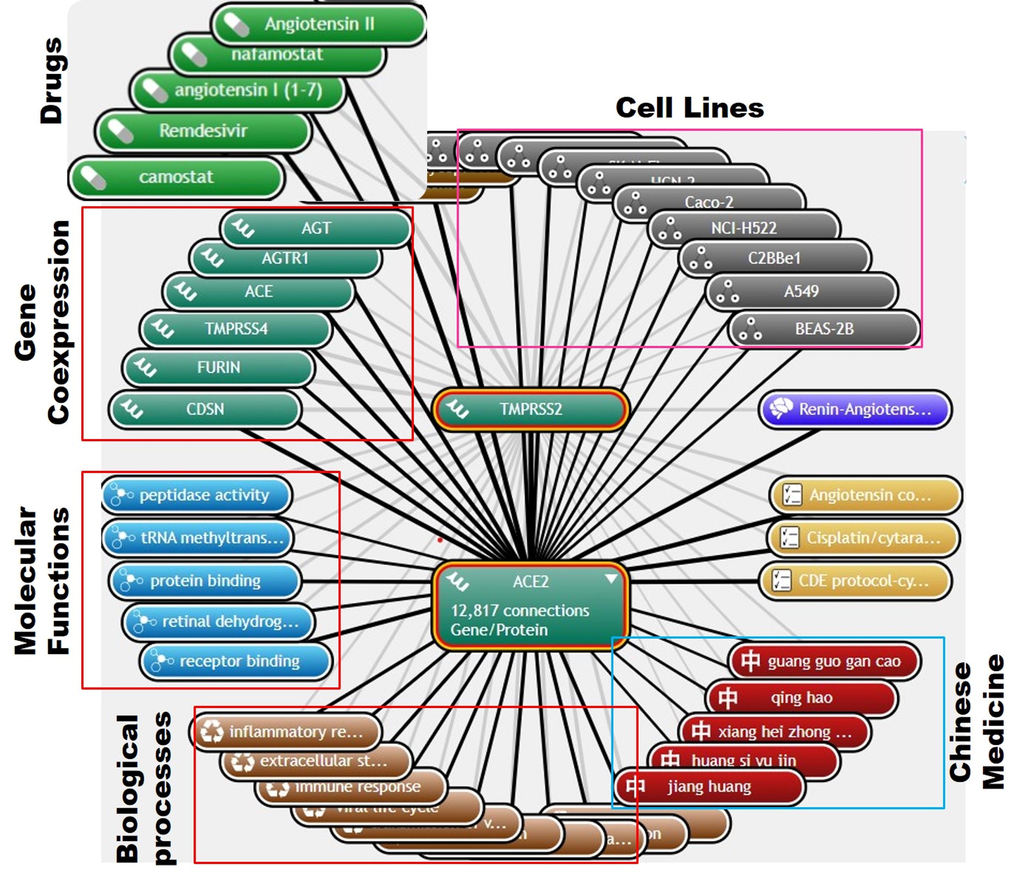
(A) Biomedical mindmap plot of the angiotensin-converting enzyme-2 (ACE2)/type II transmembrane serine protease (TMPRSS2 network in SARS-CoV-2 infection.
3.2 Differential ACE2 and TMPRSS2 expression across human tissues
SARS-CoV-2 may enter other tissues and organs through binding to ACE2, so we evaluated expression and distributions of ACE2, TMPRSS2, and CD147 across 62 tissues. Interestingly, we found that ACE2 was highly expressed in the testes, renal medulla, renal cortex, heart atrium, and heart ventricle (Fig. 2). TMPRSS2 was highly expressed in the prostate gland, salivary gland, renal medulla, renal cortex, stomach pyloric, and stomach cardiac. Expression levels of ACE2 and TMPRSS2 were >2 fold change (FC) higher in those tissue compared to the lungs and other organs. ACE2 and TMPRSS-2, are known to be involved in mediating the cellular entry of SARS-CoV-2 into tissues and organs. Binding of the S protein with the ACE2 receptor induces proteolytic cleavage by TMPRSS-2, a cellular transmembrane protein, that exposes the fusion peptide, which gets embedded in the target cell membrane and pulls viral membranes closer for fusion (Donoghue et al., 2000). Therefore, higher expression levels of ACE2 and TMPRSS2 in these tissues could be attributed to secondary complications of COVID-19, including cardiac injury, acute respiratory distress syndrome, arrhythmias, acute kidney injury, and multiple organ dysfunction syndromes (Donoghue et al., 2000; Petersen et al., 2010).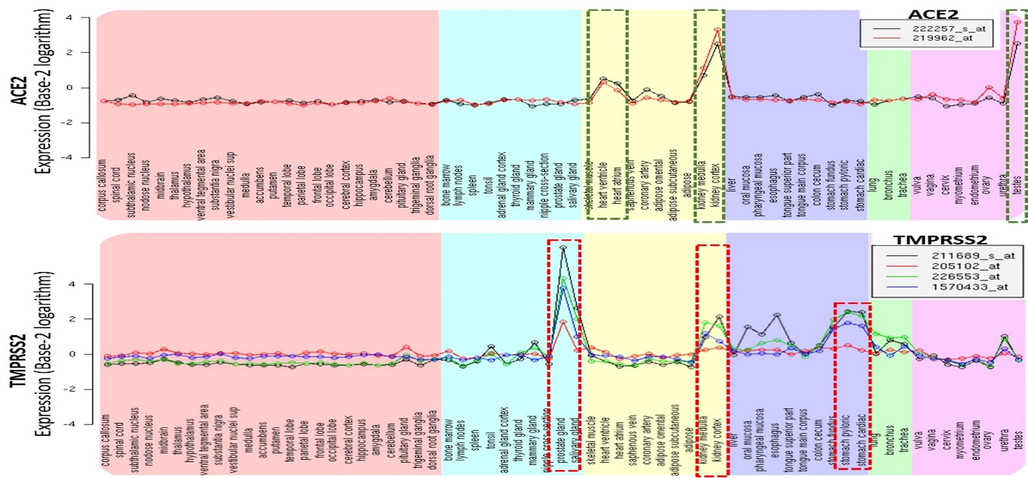
Tissue-specific expression plots of angiotensin-converting enzyme-2 (ACE2)/type II transmembrane serine protease (TMPRSS2) across 62 tissues. Expression values were converted to base-2 logarithms. The broken-line box within the plots indicate tissues with the highest expression levels.
3.3 ACE2 and TMPRSS2 are associated with the modulation of host immune response via regulation of immune-infiltrating cells at the site of tissue/organs entries
Expression levels of ACE2 are associated with immune-infiltrating cells in various tissues. However, in line with high expression levels of ACE2 in the testes, kidneys, and heart, a unique pattern of immune infiltration was observed in kidney and testicular tissues; ACE2 was associated with negative infiltration of lymphocytes including B and T cells in testicular, kidney, and liver tissues. Similarly, high expression levels of TMPRSS2 negatively regulated the infiltration of T and B cells in lung, testicular, and kidney tissues (Fig. 3). In addition, expression levels of ACE2 and TMPRSS2 were negatively associated with infiltration levels of various immunosuppressive cells including the MDSCs and Treg cells in various tissues (Fig. 4). However, positive correlations of gene expression and infiltration of DCs and neutrophils in kidney and testicular tissues were observed (Fig. 4). In summary, our analysis revealed that ACE2 and TMPRSS2 expression were negatively correlated with immune cell infiltration, especially B cells, T cells, and DCs, in various tissues. Since DCs are important antigen-presenting cells (APCs) that trigger T-cell-mediated immune responses (Sullivan and Ganem, 2005), impaired function of tissue-infiltrating DCs, and T cells may seriously affect the body's immune response and hence increase the severity of COVID-19 infection. Therefore, targeting ACE2 and TMPRSS2 could modulate the immune response towards SARS-CoV-2 infection.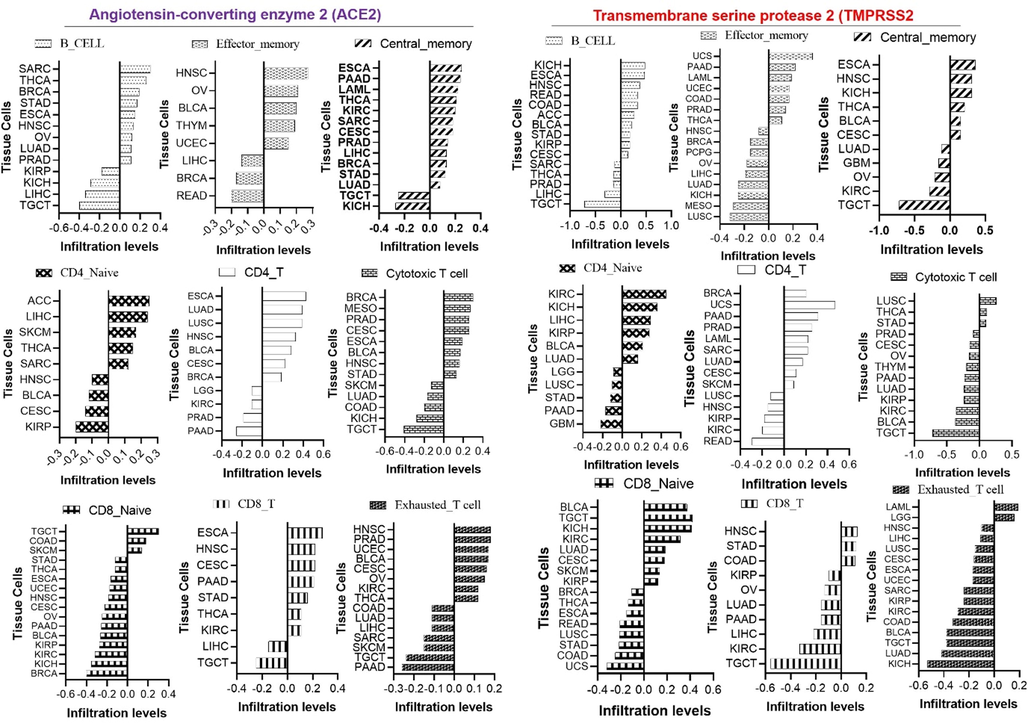
Effects of angiotensin-coverting enzyme-2 (ACE2) and type II transmembrane serine protease (TMPRSS2) mRNA expression levels on tissue infiltration levels of immune and immunosuppressive cells.
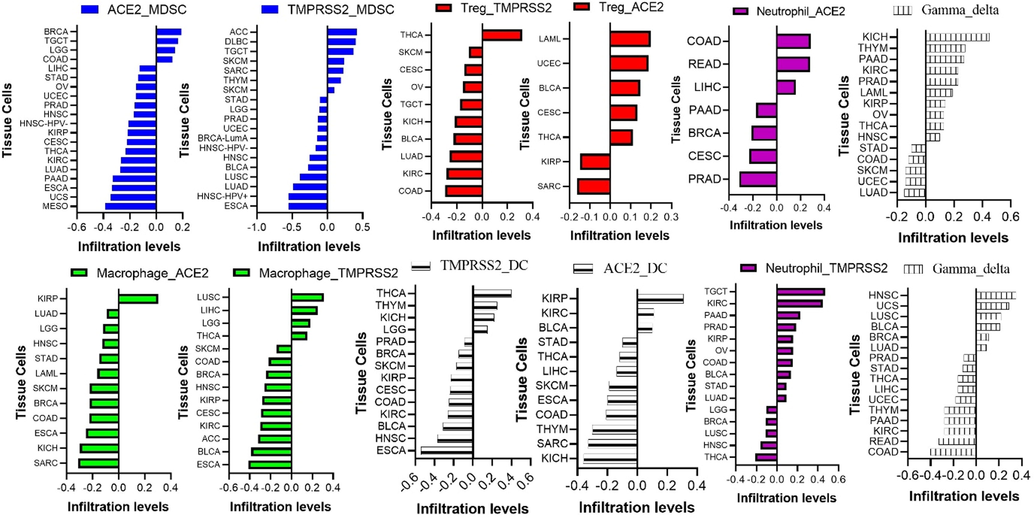
Effects of angiotensin-converting enzyme-2 (ACE2) and type II transmembrane serine protease (TMPRSS2) mRNA expression levels on tissue infiltration levels of immune and immunosuppressive cells.
3.4 Deciphering TMPRSS2 an ACE2 microRNAs target regulatory network in SARS-CoV-2 infection
MicroRNAs (miRNAs) are small, noncoding RNAs that regulate gene expression and translation in various organisms including viruses (Demirci and Adan, 2020). Studies have reported that RNA viruses could generate miRNAs with regulatory functions on the viral replication, translation and even modulating the host expression to the infections (Hu et al., 2017). Therefore, deciphering the miRNAs network associated with SARS-CoV-2 infection would provide an additional avenue for therapeutic targets. Interestingly, our miR regulatory network of SARS-CoV-2 infection identified miR-214-3p, miR-335-3p, miR-181c-5p, miR-452-5p, miR-3065-3p, miR-204-5p, and miR-185-5p as TMPRSS2 target network, while miR-149-5p, miR-1301-3p, miR-225-5p, miR-218-5p, and miR-520a-5p are ACE2 target regulatory network in SARS-CoV-2 infection (Fig. 5). The present study, therefore, unravels the TMPRSS2 and ACE2 microRNAs target regulatory network in SARS-CoV-2 infection and thus open up a new window for microRNAs based therapy for the treatment of SARS-CoV-2 infection.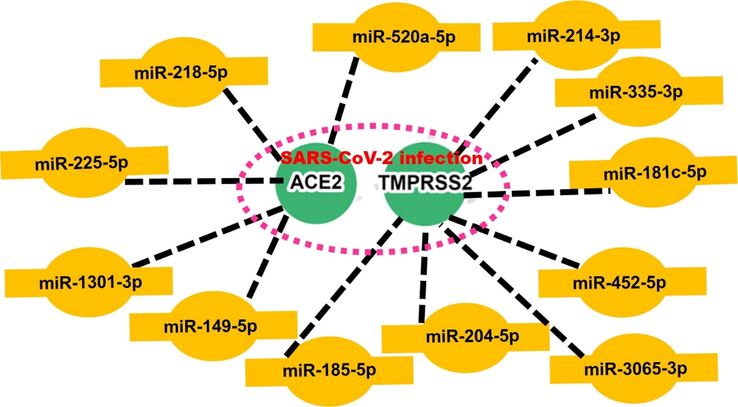
MicroRNA (miR) regulatory network of ACE2 and TMPRSS2 in SARS-CoV-2 infection.
3.5 Rationale for the Scaffold-hopping based design and synthesis of niclosamide derivatives, the SCOV-L series of compounds
Scaffold-hopping of bioactive compounds containing different core structures is an important approach in medicinal chemistry for the design and development of new drugs (Lawal et al., 2017). Medicinal plants have been used for the effective treatment of microbial infections and several kinds of human diseases (Lawal et al., 2015; Iwu, 1993; Wu et al., 2022; Onikanni et al., 2021; Yeh et al., 2021; Wu et al., 2021; Salleh et al., 2021), and are considered to produce least or no adverse effects in clinical trials when compared to conventional drugs (Spiller and Sawyer, 2006; Samples, 2004). Biphenyl, flavones, and isoflavones are important natural product backbones, and several bioactive compounds containing these backbones were reported to have a vast range of biological activities, including antioxidative, anti-atherosclerosis, muscle relaxant, antimicrobial, anti-inflammatory, anticancer, and antiviral effects (Erlund, 2004; Saakre et al., 2021). Quercetin, a naturally occurring dietary flavonoid, is well known for its antioxidants and potential for treatment of several inflammations, metabolic, chronic and infectious diseases (Wang et al., 2016; Onikanni et al., 2022; Azmi et al., 2021). Accumulating evidence has indicated the potential of quercetin for the treatment of COVID 19 infection. It inhibits the ACE2, PLpro, 3CLpro, and NTPase/helicase, thus interfering with the various stages of the coronavirus entry and replication (Pan et al., 2020; Muchtaridi et al., 2020; Snetkov et al., 2021).
A number of clinical drugs, e.g., diflunisal a salicylic acid derivative with several pharmacological activities (anticancer, anti-arthritis, analgesic, and anti-inflammatory properties) (Fung and Babik, 2021) and entrectinib (an anticancer drug), contain difluorophenyl as an important component responsible for their bioactivity. Niclosamide is a multipurpose compound with proven efficacy in treating several diseases, including oxidative stress, infections, metabolic disorders, inflammation, and cancers (Li et al., 2014; Kadri et al., 2018). Niclosamide is a multipurpose clinical small molecule with several biological activities including antihelmintic, antioxidant, antimicrobial, anti-inflammatory, anticancer and immune-modulatory properties (Li et al., 2014; Kadri et al., 2018). Recent studies also identified the potential of niclosamide for treating viral infections (Xu et al., 2020) and reported its ability to inhibit replication of SARS-CoV by several investigators (Wu et al., 2004; Wen et al., 2007). Therefore, due to the various activities mentioned above, these compounds and their derivatives may represent target compounds worthy of future clinical trials against coronavirus infections. Hence, we conducted a scaffold-hopping of these bioactive compounds (flavones and isoflavones), and marketed drugs (biphenyl, difluorophenyl, and niclosamide), leading to the discovery and synthesis of 8 structurally related novel small molecules (Fig. 5): N-(2,4-difluorophenyl)-2′,4′-difluoro-4-hydroxybiphenyl-3-carboxamide (SCOV-L02), N-(3-cyanophenyl)-2′,4′-difluoro-4-hydroxy-[1,1′-biphenyl]-3-carboxamide (SCOV-L09), N-(4-cyanophenyl)-2′,4′-difluoro-4-hydroxy-[1,1′-biphenyl]-3-carboxamide (SCOV-L10), N-(3,4-dimethoxyphenyl)-2′,4′-difluoro-4-hydroxy-[1,1′-biphenyl]-3-carboxamide (SCOV-L10), 6-(2,4-difluorophenyl)-3-(3,4-difluorophenyl)-2H-benzo[e] (Xu et al., 2020; Xie et al., 2005) oxazine-2,4(3H)-dione (SCOV-L15), 6-(2,4-Difluorophenyl)-3-(3-(trifluoromethyl)phenyl)-2H-benzo[e](Xu et al., 2020; Xie et al., 2005)oxazine-2,4(3H)-dione (SCOV-L18), 4-(6-(2,4-Difluorophenyl)-2,4-dioxo-2H-benzo[e](Xu et al., 2020; Xie et al., 2005)oxazin-3(4H)-yl)benzonitrile (SCOV-L22), and 6-(2,4-Difluorophenyl)-3-(3,4-dimethoxyphenyl)-2H-benzo[e](Xu et al., 2020; Xie et al., 2005)oxazine-2,4(3H)-dione (SCOV-L23) (Fig. 6).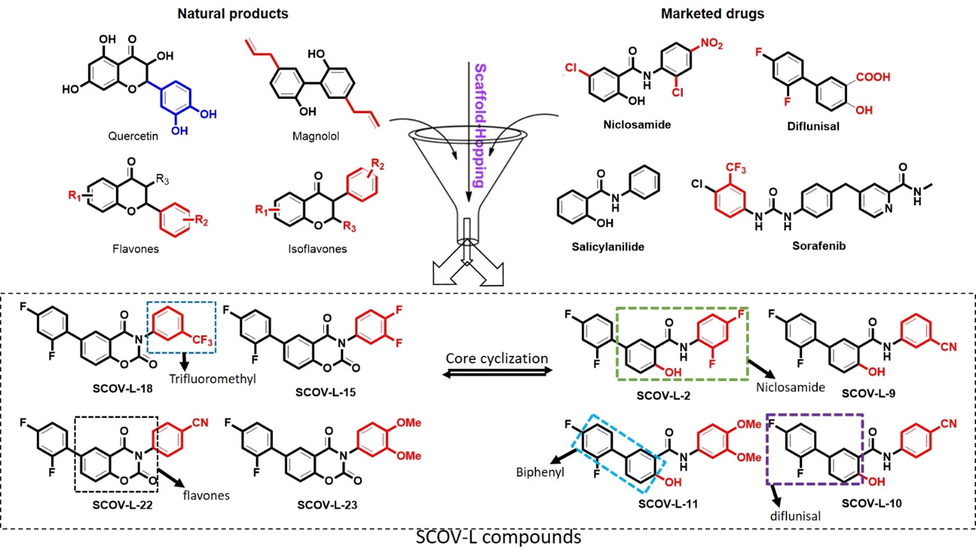
Scaffold-hopping approach for the design of niclosamide derivatives (SCOV-L compounds).
3.6 Exploring the therapeutic potential of the SCOV-L series for the prevention/treatment of SARS-CoV-2 by targeting ACE2 and TMPRSS2 expression
3.6.1 Delineating cell line models for ACE2 and TMPRSS2 expression
It is known that SARS-CoV-2 is internalized to tissues via binding to ACE2 and cleavage by TMPRSS2. Cancer patients exhibited worse prognoses and severe complications of SARS-CoV-2 infections (Montopoli et al., 2020), and several clinical trials with anticancer drugs proved their efficacy in decreasing the risk of SARS-CoV-2 infection and ameliorating its complications (Patel et al., 2020; Cao et al., 2019; Bakouny et al., 2020). In order to obtain a reliable cell line model with high expression levels of ACE2 and TMPRSS2 for experimental studies, we mined the DTP database of the National Cancer Institute (NCI) for expression levels of ACE2 and TMPRSS2 across 60 cell lines of the NCI-60 panel of human cell lines. Interestingly, we found that ACE2 was more than 5-fold overexpressed in NCI-H322M a NSCLC cell line and over 10-fold overexpressed in IGROV1, an ovarian cancer cell line compared to the majority of the NCI-60 cell lines. However, the majority of non-small cell lung cancer (NSCLC) cell lines (NCI-H226, NCI-H23, NCI-H322M, NCI-H460, and NCI-H522), melanoma cell lines (SK-MEL-2, SK-MEL-28, SK-MEL-5, UACC-257, and UACC-62), ovarian cancer cell lines (IGROV1, OVCAR-3, and OVCAR-4), renal cancer cell lines (A498, ACHN, TK-10, and UO-31) and a prostate cancer cell line (Du-145) showed stable expression of TMPRSS2 (Fig. 7).
Angiotensin-converting enzyme-2 (ACE2) and type II transmembrane serine protease (TMPRSS2) panels across 60 cell lines of the National Cancer Institute (NCI)-60 panel of human cell lines, to establish cell line models for ACE2 and TMPRSS2 expression.
3.7 Members of the SCOV-L series have potential for preventing and treating COVID-19 infection via antiproliferative activities against ACE2- and TMPRSS2-expressing cell lines
The severity of COVID-19 complications can be attributed to increased expression levels of TMPRSS2 and ACE2 in over-actively dividing cells, leading to compromised immune responses and a worse prognosis of COVID-19 infection. Therefore, chemotherapeutic agents with antiproliferative effects could be useful in inhibiting the activities of ACE2 and TMPRSS2 and alleviating COVID-19 complications. With that in mind, we evaluated the antiproliferative activities of members of the SCOV-L series against the hyper-proliferation of ACE2 and TMPRSS2 in certain cell lines. Interestingly, we found that at a therapeutic dose of 10 µM, all SCOV-L series members (excepting SCOV-L-11 and SCOV-L-23) were strictly antiproliferative and non-cytotoxic against ACE2- and TMPRSS2-expressing cell lines, including A549, NCI-H322M, and ICROV1 cells. Among the eight compounds, SCOV-L-18, SCOV-L-15, and SCOV-L-09 demonstrated higher antiproliferative effects than the other compounds (Fig. 8A). In addition, we evaluated dose-dependent effects of the compounds, and found that at concentrations of <10 µM, the compounds demonstrated no activities against the cell lines. Despite cytotoxic effects of the compounds being observed at a very high dose of 100 µM, LC50 values of the compounds were greater than 100 µM against all the ACE2- and TMPRSS2-expressing cell lines (Fig. 8B). Therefore, by translation, the antiproliferative effects of ACE2- and TMPRSS2-expressing cell lines by the SCOV-L series at a therapeutic dose of 10 µM strongly suggested these compounds could limit the tissue internalization of SARS-CoV-2 via ACE2 binding, decreasing the risk of SARS-CoV-2 infection and its severe complications. Collectively these findings strongly suggested that the SCOV-L series at a therapeutic dose could be safely explored for preventing and treating COVID-19 infection.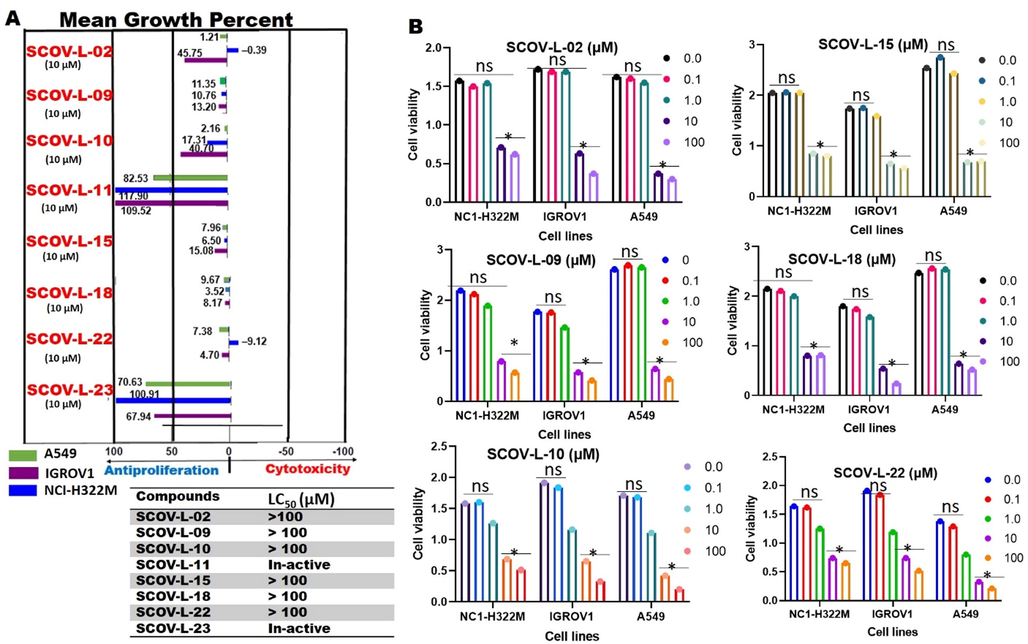
Members of the SCOV-L series have antiproliferative properties against angiotensin-converting enzyme-2- (ACE2) and type II transmembrane serine protease (TMPRSS2)-expressing cell lines.
3.8 Molecular docking analysis correlated with the antiproliferative activities and revealed the potential of SCOV-L series for targeting ACE2, TMPRSS2, and NSPs
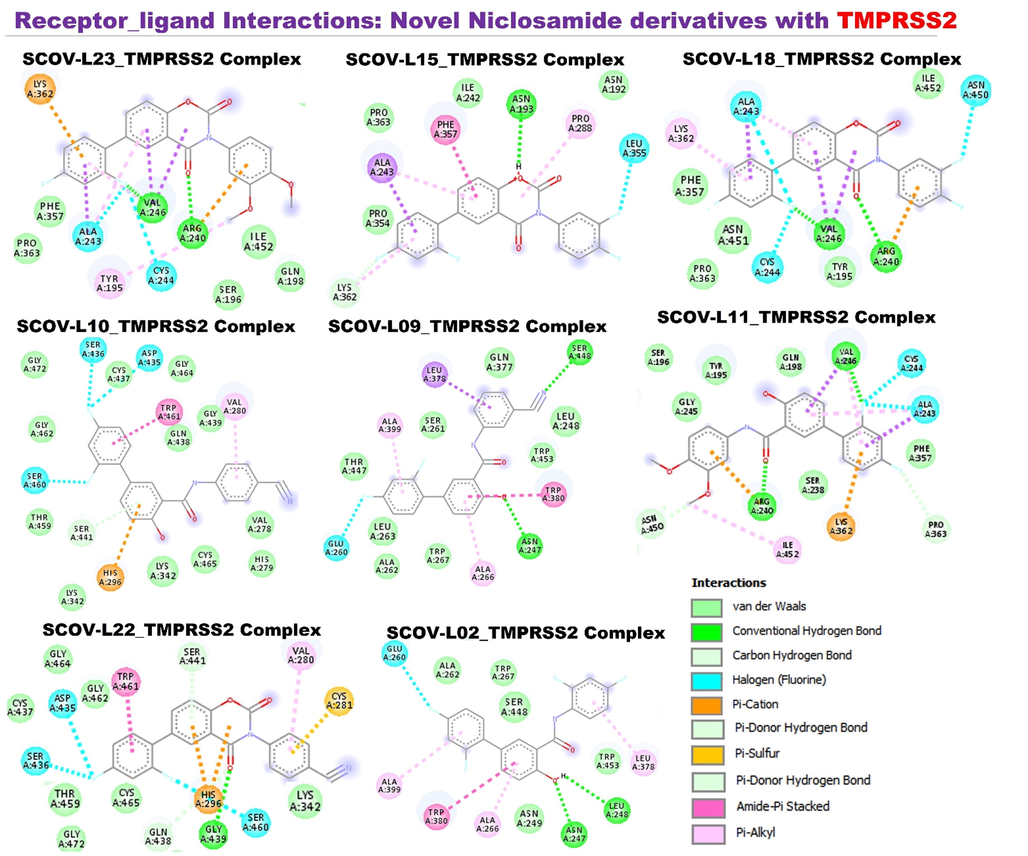
Molecular docking is a structural simulation of binding affinities and interactions between a target protein molecule and a small-molecule drug candidate for target inhibition of the protein (Onikanni et al., 2021; Shifeng et al., 2022). Hence, we explored the molecular docking to analyze the binding affinities and potential of members of the SCOV-L series to target ACE2, TMPRSS2, and SARS-CoV-2 NSPs including NSP5 (3CLpro), NSP3 (PLpro), and RdRp. A total of 12 compound including 4 standard drugs (Favipiravir, Chloroquine, Remdesivir, and Niclosamide) and the 8 in-house synthesize niclosamide derivatives (SCOV-L series) were subjected the docking analysis. Interestingly, our findings collaborated the in vitro antiproliferative trend of the SCOV-L series, revealing high binding efficacies of the SCOV-L series, particularly SCOV-L-18, SCOV-L-15, and SCOV-L-09 against ACE2, TMPRSS2, and SARS-CoV-2 NSPs (Table 1, Figs. 9–13). In line with our observation that SCOV-L11 and SCOV-L23 demonstrated no antiproliferative activities against ACE2- and TMPRSS2-expressing cell lines, these two compounds demonstrated the least interactions and binding efficacies towards ACE2, TMPRSS2, and SARS-CoV-2 NSPs. Collectively, our molecular docking analysis hinted that member of the SCOV-L series, particularly SCOV-L-18, SCOV-L-15, and SCOV-L-09, had strong potential for limiting the tissue internalization of SARS-CoV-2 via ACE2 binding, decreasing the risk of SARS-CoV-2 infection and severe complications (Table 1). RdRp, RNA-dependent RNA polymerase; ACE2, angiotensin-converting enzyme-2; TMPRSS2, type 2 transmembrane serine protease; ΔG, binding free energy.
Compounds
NSP5: 3CLpro
ΔG (kcal/mol)NSP3: PLpro
ΔG (kcal/mol)RdRp
ΔG (kcal/mol)ACE2
ΔG(kcal/mol)TMPRSS2
ΔG (kcal/mol)
SCOV-L-02
−8.10
−9.50
−8.20
−9.50
−7.5
SCOV-L-09
−8.40
−9.60
−8.50
−9.10
−7.8
SCOV-L-10
−8.00
−9.70
−8.20
−8.50
−7.3
SCOV-L-11
−7.80
−9.60
−7.80
−8.20
−6.9
SCOV-L-15
−8.60
−10.50
−8.20
−9.30
−7.4
SCOV-L-18
−8.50
−10.30
−8.20
−9.40
−7.6
SCOV-L-22
−8.10
−10.90
−8.10
−9.10
−7.7
SCOV-L-23
−8.20
−10.23
−7.60
−8.70
−7.2
Niclosamide
−7.00
−8.00
7.40
−7.40
−6.5
Remdesivir
−8.10
−8.70
7.60
−8.60
−6.4
Chloroquine
−5.90
−6.60
5.90
−6.20
−6.2
Favipiravir
−4.80
−5.60
5.60
−5.50
−5.3
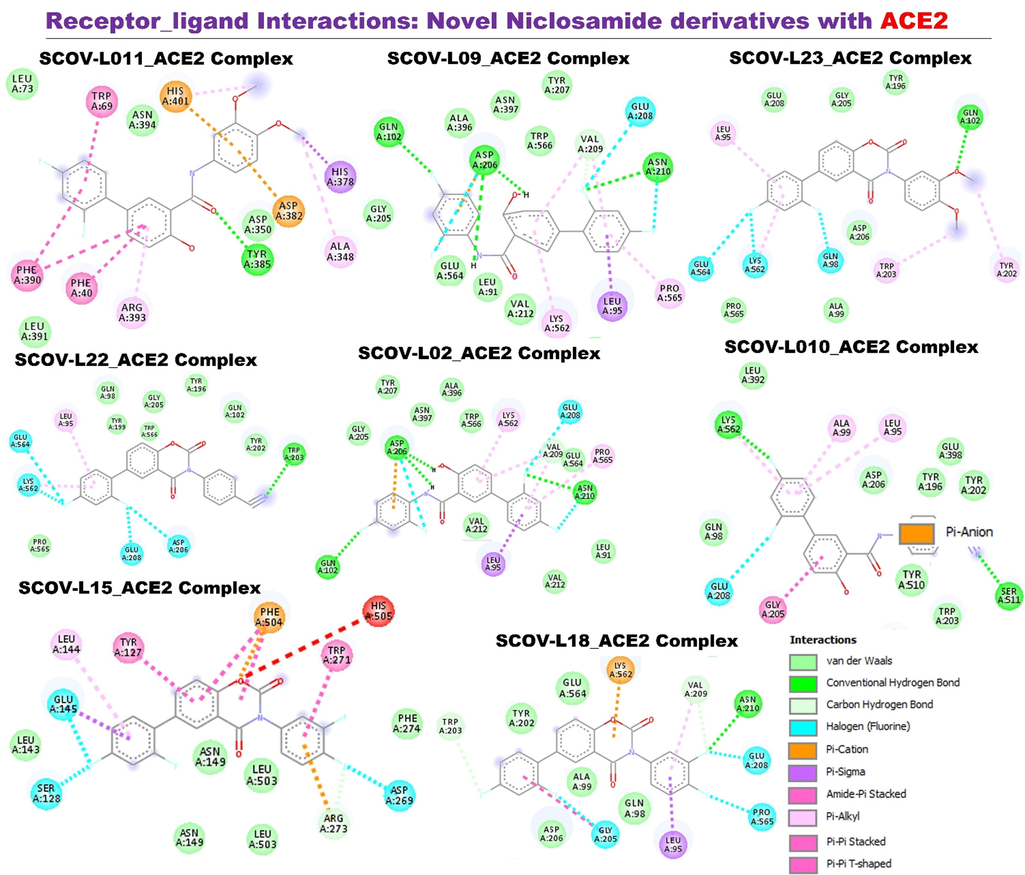
Receptor-ligand interaction of members of the SCOV-L series with angiotensin-converting enzyme-2 (ACE2). Image represents a two-dimensional (2D) view of the optimal docked complex.
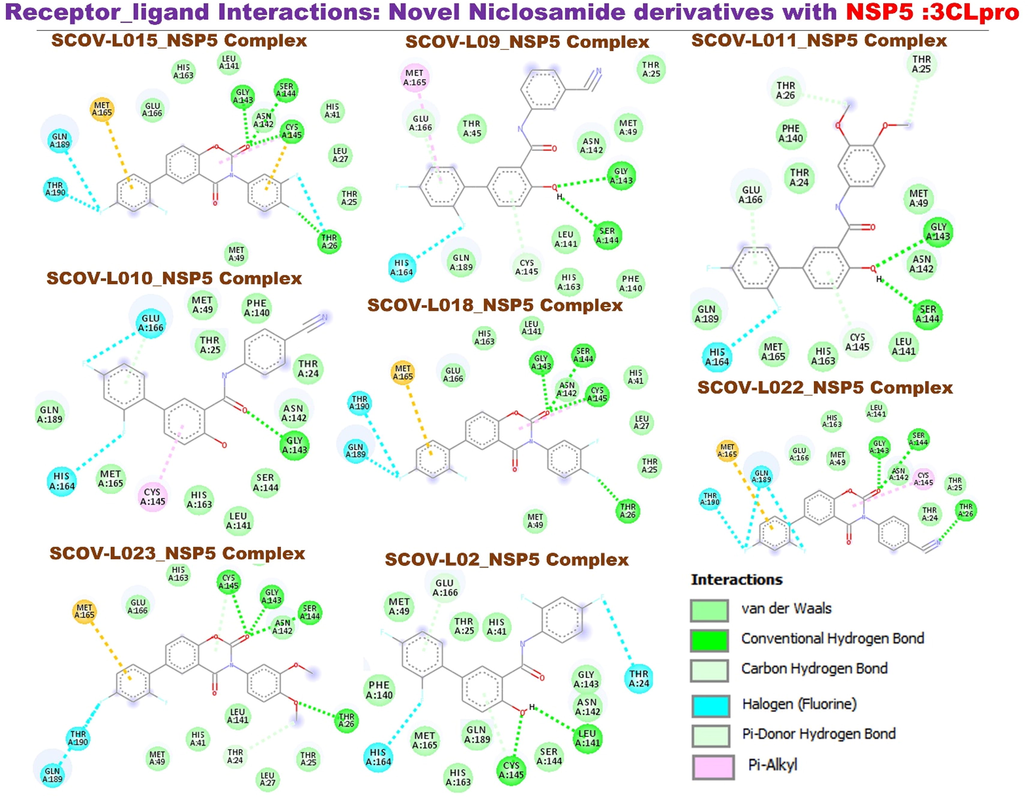
Receptor-ligand interactions of members of the SCOV-L series with nonstructural protein 5 (NSP5). Image represents a two-dimensional (2D) view of the optimal docked complex.
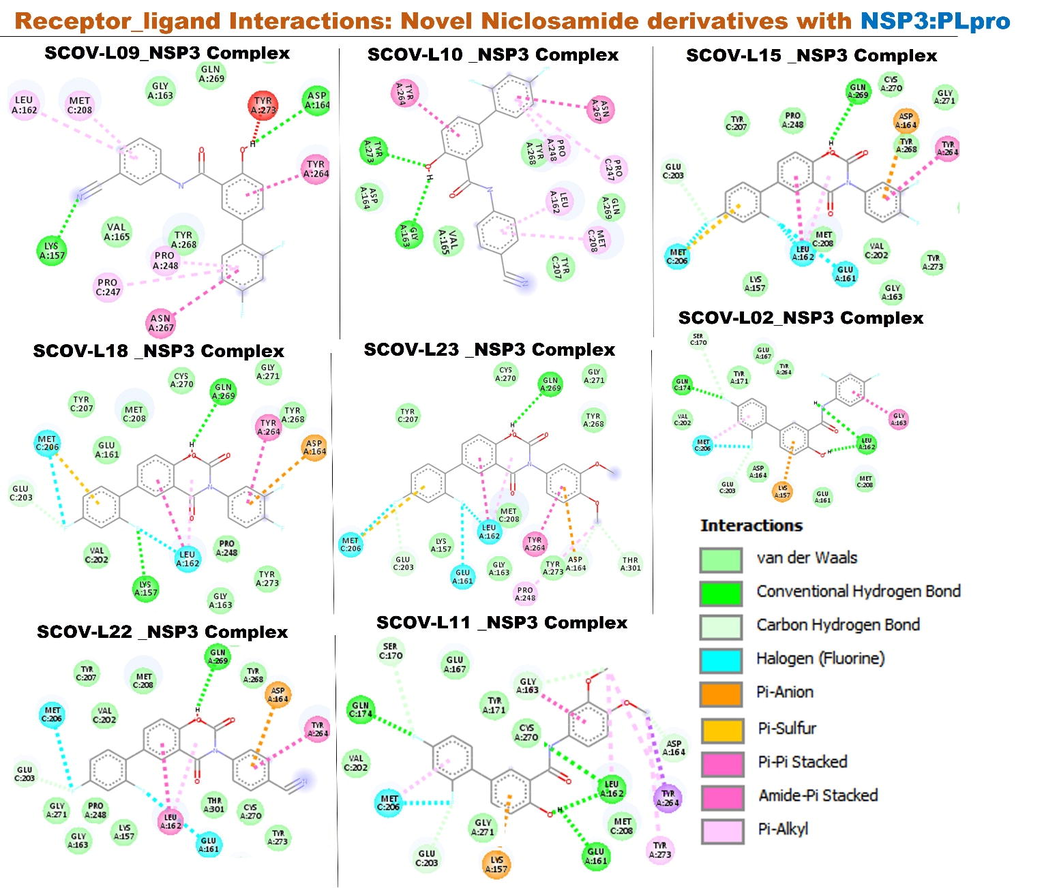
Receptor-ligand interactions of members of the SCOV-L series with nonstructural protein 3 (NSP3) papain-like protease. Image represents the two-dimensional (2D) view of the optimal docked complex.
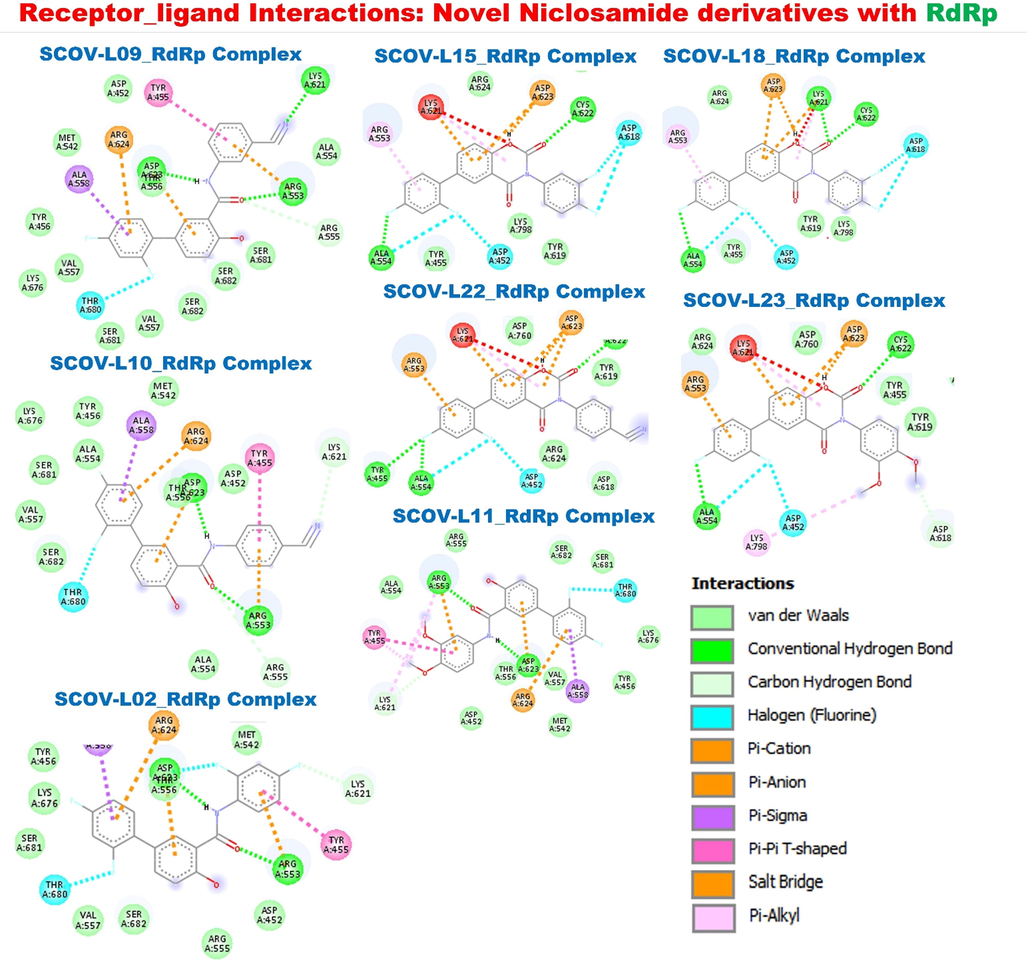
Receptor-ligand interactions of members of the SCOV-L series with RNA-dependent RNA polymerase (RdRp). The image represents a two-dimensional (2D) view of the optimal docked complex.
Receptor-ligand interaction of members of the SCOV-L series with nonstructural protein 5 (NSP5). The image represents a two-dimensional (2D) view of the optimal docked complex.
Molecular docking of members of the SCOV-L series with ACE2: ACE2 plays a vital role in regulating viral attachment and entry. Upon fusion of ACE2 with RBD, the s1 unit of the spike protein is dissociated, and the s2 unit will further undergo structural rearrangement. therefore, inhibition of the post-fused s2 subunit is very important to avoid fusion and prevent further viral infection and replication (Yan et al., 2020). Furthermore, mechanism of SARS-CoV-2 fusion relies on the interaction of heptad repeat 1 and 2 (HR1 and HR2) regions of the spike protein. Therefore, the HR2 domains were considered a significant target. Interestingly, our results revealed that the SCOV-L -ACE2 docked complex have the interaction fingerprint with those of critical residues as part of ACE2 binding interface (Veerasamy and Karunakaran, 2022).
All members of the SCOV-L series docked well to the binding cavity of ACE2 with binding affinities ranging –8.2 ∼ –9.5 kcal/mol. SCOV-L02 and SCOV-L18 demonstrated the highest binding affinities, while SCOV-L11, SCOV-L23, and SCOV-L10 demonstrated the least affinities for ACE2 binding (Table 1). ACE2_SCOV-L series complexes were bonded by several H-bonds, halogen and alkyl interactions, and several π-interactions. It was observed that SCOV-L18 exhibited higher numbers of hydrogen and halogen bonds with amino acid residues (ASN210, VAL209, TRP203, GLU208, PRO565, and GLY205) of the ACE2 binding pocket; while SCOV-L02 H-bonded to GLN102, ASN210, VAL209, and ASP206 with much-closer proximities than exhibited by other members of the series. These higher interactions thus could be responsible to the higher affinities that SCOV-L02 and SCOV-L18 had for ACE2 than other members of the series. In addition, the interaction complex formed between ACE2, and each member of the SCOV-L series was also supported by various van der Waal forces between the ligand and amino acid residues of the ACE2 binding pocket (Fig. 9). However, the parental compound (niclosamide) and the SARS-CoV-2 clinical drugs tested, including remdesivir, chloroquine, and favipiravir, exhibited lower affinities for ACE2 (−5.5 ∼ 8.60 kcal/mol) compared to members of the SCOV-L series. This study shows that SCOV-L compounds particularly, SCOV-L18 might be promising ACE2 inhibitors.
Molecular docking of members of the SCOV-L series with TMPRSS2: Members of the SCOV-L series also bonded to TMPRSS2 by several H-bonds and halogen interactions, a few alkyl interactions, and multiple van der Waal forces. These compounds demonstrated the least target binding affinities against TMPRSS2 compared to their affinities for other targets tested. Except for SCOV-L11 and SCOV-L-23 which demonstrated the lowest binding efficacies to the target, all other members of the SCOV-L series exhibited similar binding affinities to TMPRSS2 (−7.4 ∼ − 7.7 kcal/mol). However, the parental compound (niclosamide) and SARS-CoV-2 clinical drugs tested, including remdesivir, chloroquine, and favipiravir, exhibited lower affinities for TMPRSS2 (−5.3 ∼ 6.5 kcal/mol) compared to the SCOV-L series (Fig. 10).
Molecular docking of members of the SCOV-L series with papain-like protease (NSP3: PLpro): PLpro is another enzyme that is critical for SARS-CoV-2 replication by the action of its catalytic triad and responsible for the inflammation of host cells (Shin et al., 2020). The viral replication and inflammation of the host cell was shown to be hindered by compounds that bind with the catalytic triad (Rut et al., 2020). Our docking analysis revealed that the SARS-CoV-2 PLpro exhibited the top rank of favored target proteins for the ability to bind to SCOV-L series ligands. All members of the SCOV-L series docked well to the binding cavity of NSP3 with very high affinities ranging –9.5 ∼ –10.9 kcal/mol. Interestingly, the affinities of the compounds against SARS-CoV-2 PLpro followed a unique pattern: SCOV-L02 to SCOV-L-11 exhibited lower binding affinities than SCOV-L15 to SCOV-L-23 which demonstrated the highest affinities (–10.3 ∼ –10.9 kcal/mol) against the target. Furthermore, a similar mode of ligand-receptor interactions involving specific hydrogen and/or halogen bonds with GLN269, LYS157, LEU162, ASP164, MET206, GLU161, and GLN174 residues of the NSP3 binding cavity were observed in the complex formed between SARS-CoV-2 PLpro and each member of the SCOV-L series. Furthermore, several other interactions involving Met208, Pro247, Pro248, Thr301, Asn267, Tyr268, Gln269 residues were found in the docked complexes. Nevertheless, all members of the SCOV-L series exhibited higher target binding affinities to SARS-CoV-2 PLpro compared to target affinities demonstrated by the parental compound (niclosamide) and SARS-CoV-2 clinical drugs tested, including remdesivir, chloroquine, and favipiravir (Fig. 11, Table 1).
In compliance with our findings, previous study by a group of Poland and USA researcher (te Velthuis et al., 2011); reported that the side chain interaction of VIR250 and VIR251 (inhibitor of CoV-2 PLpro) with the PLpro backbone carbonyl oxygen of Tyr268 projects the ligand toward Met208, Pro247, Pro248, and Thr301 where it engages in an interaction network of van der Waals forces. This interaction network is facilitated by a 1.5-Å shift of the β14-β15 loop (Asn267, Tyr268, and Gln269) toward the ligand, thus facilitating several new contacts that would be unable to occur in the absence of this shift (te Velthuis et al., 2011). The similar residual interaction observed in the complex formed by SCOV-L compounds and PLpro strongly suggested that SCOV-L compounds could be a novel inhibitor of PLpro and thus serve as potential drug candidate for SARS-CoV-2 infection.
Molecular docking of members of the SCOV-L series with RdRp: SARS-CoV-2 RdRp catalyzes the synthesis of new viral RNA and plays a crucial role in the SARS-CoV-2 replication cycle (Grein et al., 2020). Our molecular docking analysis revealed that members of the SCOV-L series also bonded by H-bonds to RdRp at its catalytic residue ARG553, in addition to several other H-bonds (TYR455, ALA554, CYS621, CYS622, ASP623,) halogen interactions, a few alkyl interactions, pi-pi T shapes, and pi-sigma, pi-cation, pi-anion, and multiple van der Waal forces including contact with the RdRp catalytic residue ASP750. Similar to the affinities of members of the SCOV-L series with TMPRSS2, SCOV-L11 and SCOV-L23 exhibited the least affinities for RdRp. The remaining compounds demonstrated similar binding affinities to RdRp (−8.1 ∼ − 8.5 kcal/mol). However, the parental compound (niclosamide) and SARS-CoV-2 clinical drugs tested, including remdesivir, chloroquine, and favipiravir, exhibited lower affinities for RdRp (−5.6 ∼ 7.6 kcal/mol) compared to members of the SCOV-L series (Fig. 12). When compared with remdesivir a clinical anti-COVID-19 that inhibits viral RNA polymerases (Zhang et al., 2020), a same interaction involving the hydrogen bonding, van der walls and pi- interaction were found in complex between SCOV-L series and SARS-CoV-2 RdRp. This strongly suggest that SCOV-L series could be a novel candidate for targeting remdesivir in the fight against COVID-19 infection.
Molecular docking of members of the SCOV-L series with the main protease (NSP5: 3CLpro): The SARS-CoV-2 main protease (3C-like protease) mediates viral replication and transcription via extensive proteolytic processing of the two replicase polyproteins (pp1a and pp1ab) to produce different functional proteins (Báez-Santos et al., 2015). The 3C-like protease, therefore, serves as an attractive target for anti-SARS drug discovery (Anand et al., 2003; Yang et al., 2003; Chou et al., 2003). All SCOV-L series interactions with the SARS-CoV-2 main protease (NSP5: 3CLpro) had similar patterns, trends, and binding affinities as the compounds exhibited against RdRp. In the same trend, SCOV-L11 exhibited the least affinity against the main protease (NSP5: 3CLpro), while SCOV-L15 and SCOV-L18 demonstrated higher activities. Remdesivir demonstrated higher affinity than SCOV-L11 However, the parental compound (niclosamide) and SARS-CoV-2 clinical drugs tested, including chloroquine and favipiravir, exhibited lower affinities for NSP3 (−4.8 and −7.0 kcal/mol) compared to members of the SCOV-L series (Fig. 13).
The active site of the main protease contains catalytic dyad (Cys145 and His41), located between the two domains of the protease (Jin et al., 2020). This catalytic site is highly conserved among all types of coronaviruses, and thus inhibitors that target this site exhibit a broad spectrum of anti-SARS-CoV-2 activity (Semenov and Krivdin, 2022). Our analysis of interactions with 3CLpro residues showed that the SCOV-L compounds are located deeply within the binding pocket of the 3CLpro active site, being in the cleft between the two protease domains and constituting the catalytic dyad Cys145–His41 (Pillaiyar et al., 2016). This finding indicates the fact that SCOV-L compounds have the potential to covalently bind to amino acid residues in this region of the main protease. This ability to interact with a main protease provides additional benefits in suppressing viral activity.
Specifically, the compounds formed hydrogen bond interaction with the Cys145 and van der Waals force with His41 of the binding site. Other hydrogen bonding that stabilizes the complex include Ser144, Gly143, Thr26, and Leu141. Several other interactions including π-alkyl, π–π stacking interactions, and several van der Waals were formed between the compound and amino acid residues. Our results agree with the study of Semenov and Krivdin (Pillaiyar et al., 2016) who reported that natural product inhibitors of SARS-CoV-2 main proteas formed interaction with Cys145 and His41 residues of the binding sites. It has been reported that the inhibitors exhibit their action via two steps- (i) they first bind non-covalently to the catalytic residue of the enzyme. (ii) this is followed by a nucleophilic attack by Cys145 that results in the covalent bond formation, thereby inhibiting the enzyme reversibly or irreversibly (Amin, 2013). Collectively, our in silico study strongly suggested the potential of SCOV-L series particularly, the SCOV-L15 and SCOV-L18 to impede the activity of SARS-CoV-2 main protease. However, the limitation of our study must be acknowledged. The absence of invitro data to corroborate our findings marked a critical area that required further studies. Analysis of therapeutic potential of the active compounds in Realtime assay or ELISA based assay is necessary for the clinical applicability of our findings.
3.9 Members of the SCOV-L series satisfied the criteria of a drug-like candidate and demonstrated good ADMET pharmacokinetic properties suitable for therapeutic application
Drug likeness: Drug pharmacokinetic and ADMET properties are important features that define the clinical success of a therapeutic agent. Analyses of drug pharmacokinetic and ADMET properties are therefore an integral part of drug design and development. The drug likeness and pharmacokinetics properties of members of the SCOV-L series were evaluated using in silico studies. Interestingly all eight compounds satisfied the criteria of drug-like candidates based on the Lipinski rule. Although all of the compounds (except SCOV-L23; 10.431 μg/mL) had poor solubilities (0.354 ∼ 8.93 μg/mL), the calculated logP values (3.23 ∼ 4.86) indicated a lipophilic character, while high TPSA values (49.33 ∼ 75.17) suggested that all of the compounds we evaluated were good permeates of the various biological barriers, with plasma protein affinities of >80%.
Absorption: All compounds had high values of HIA (>40%) and bioavailability of >50% indicating good permeation across membranes. In addition, our ADMET lab calculations indicated that the compounds were good permeates of colorectal adenocarcinoma cells, with permeability coefficients ranging − 5.126 ∼ − 4.501 cm/s, justifying the good intestinal absorption evaluated in the consensus. None of the tested compounds was a substrate for P-gp, a multidrug-resistance protein 1 (MDR1), which pumps drugs and various compounds out of cells (Robert and Jarry, 2003). These factors suggest the good absorption, permeability, retention, and optimal drug delivery by members of the SCOV-L series. Furthermore, as demonstrated in Fig. 14, SCOV-L15, SCOV-L18, SCOV-L22, and SCOV-L23 exhibited potential inhibitory roles against P-gp. This selective blockade of the action of P-gp by these members of the SCOV-L series suggests improved drug uptake and bioavailability (Xu et al., 2012).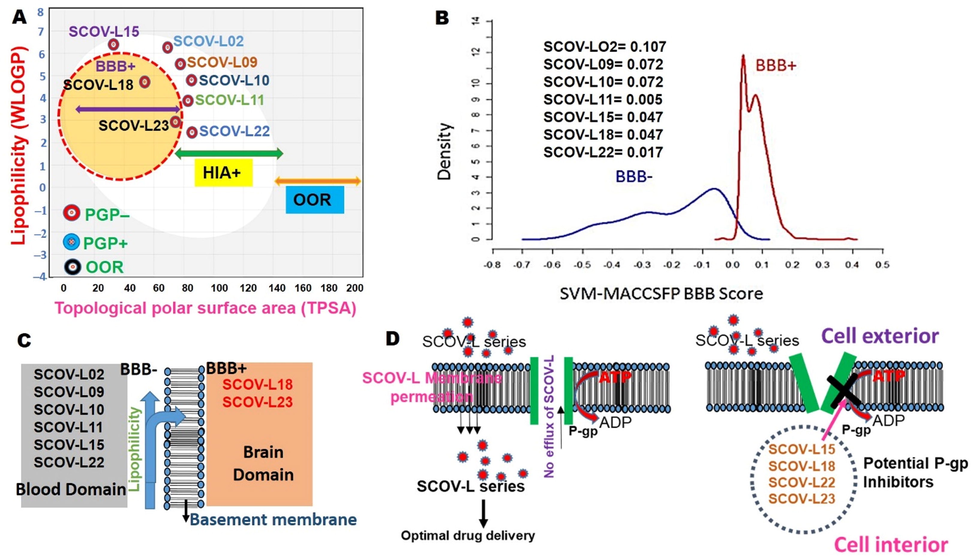
Modeling of membrane permeation and the ability to serve as a P-glycoprotein (P-gp) substrate by members of the SCOV-L series.
Distribution: We used three mathematical modeling approaches to identify the BBB permeation ability of members of the SCOV-L series. The first approach was a model based on the numbers of H-bonds (<8 ∼ 10) and MW (<400 ∼ 500) (Pardridge, 1998), while the second approach was a model based on the principle of the support vector machine-based LiCABEDS algorithm (Liu et al., 2014). Using these approaches, our results revealed good BBB permeation abilities of SCOV-L series compounds (Table 2, Fig. 14). However, using a stricter approach of BBB modeling based on lipophilicity and TPSA values, we found that all members of the SCOV-L series had high HIA potential; however, only SCOV-L18 and SCOV-L23 were able to permeate the BBB. In line with these observations, only SCOV-L18, SCOV-L22, and SCOV-L23 demonstrated < 90% plasma protein binding, suggesting the good bioavailability and high therapeutic index of SCOV-L18, SCOV-L22, and SCOV-L23 compared to other members of the SCOV-L series. MW, molecular weight; HB, hydrogen bond; TPSA, Topological polar surface area; Papp, Predicting apparent passive permeability; Caco-2, colorectal adenocarcinoma cells; P-gp, P-glycoprotein; HIA, human intestinal absorption); PPB, plasma protein binding; VD, volume distribution; BBB, blood–brain barrier; CL, clearance rate; hERG, human ether-a-go-go-related gene; SkinSen, skin sensitization; LD50, acute toxicity as the 50% lethal dose; PAINS, Pan Assay Interference Compounds; +, positive result; –, negative result.
SCOV-L02
SCOV-L09
SCOV-L10
SCOV-L11
SCOV-L15
SCOV-L22
SCOV-L23
SCOV-L18
C19H11F4NO2
C20H12F2N2O2
C20H12F2N2O2
C21H17F2NO4
C20H9F4NO3
C21H12F2N2O3
C22H17F2NO5
C20H11F4NO3
MW
361.294
350.324
350.334
385.366
387.288
378.334
413.376
389.304
Lipophilicity (LogP)
4.868
4.461
4.461
4.607
4.167
3.235
3.381
3.642
HB acceptor
2
3
3
4
4
3
4
3
HB donor
2
2
2
2
0
1
1
1
TPSA
49.33
73.12
73.12
67.79
52.21
75.17
69.84
51.38
Drug likeness
0.594
0.356
0.428
0.418
0.398
0.40
0.368
0.37
Physicochemical properties
LogS (Solubility)
0.354 μg/mL
0.61 μg/mL
0.539 μg/mL
0.997 μg/mL
2.275 μg/mL
8.93 μg/mL
10.431 μg/mL
2.525 μg/mL
LogD7.4 (Distribution coefficient D)
1.09
0.89
0.879
1.105
2.661
0.907
1.057
1.073
LogP (Distribution coefficient P)
4.868
4.461
4.607
4.167
3.235
3.381
3.642
Absorption
Papp (Caco-2 permeability)
−4.92 cm/s
−4.995 cm/s
−5.033 cm/s
−5.103 cm/s
−4.501 cm/s
−5.088 cm/s
−5.126 cm/s
−5.003 cm/s
P-gp inhibitor
---(0.24)
-(0.484)
-(0.349)
-(0.459)
++(0.743)
+(0.538)
+(0.634)
+(0.544)
P-gp substrate
---(0.061)
---(0.022)
---(0.021)
---(0.152)
---(0.056)
---(0.033)
---(0.158)
---(0.081)
HIA
+(0.64)
+(0.619)
+
+(0.541
++(0.785)
+(0.526)
-(0.435)
+(0.532)
F (20% bioavailability)
++(0.87)
++(0.87)
++(0.87)
++(0.767)
++(0.791)
++(0.738)
+(0.582)
++(0.737)
Distribution
PPB (%)
94.759 %
94.499 %
94.296 %
92.457 %
92.344 %
88.615 %
88.382 %
88.534 %
VD (L/kg)
−0.861
−0.713
−0.71
−0.587
−0.61
−0.626
−0.772
−0.849
BBB
++(0.831)
++
++(0.722)
++(0.886)
+++(0.973)
++(0.815)
+++(0.909)
++(0.941)
Metabolism
P450 CYP1A2 substrate
+(0.504)
-(0.498)
+(0.536)
+(0.654)
+(0.528)
+(0.596)
+(0.608)
+(0.624)
P450 CYP3A4 inhibitor
---(0.223)
+(0.638)
---(0.247)
-(0.49)
---(0.216)
---(0.253)
-(0.488)
---(0.229)
P450 CYP3A4 substrate
-(0.334)
-(0.458)
+(0.512)
-(0.474)
+(0.634)
+(0.582)
+(0.538)
+(0.544)
P450 CYP2C9 substrate
+(0.514)
-(0.391)
-(0.48)
+(0.576)
+(0.501)
-(0.436)
+(0.511)
-(0.433)
P450 CYP2C19 substrate
---(0.228)
---(0.25)
---(0.256)
-(0.34)
(0.35)
-(0.36)
-(0.446)
-(0.336)
P450 CYP2D6 inhibitor
-(0.351)
-(0.368)
-(0.36)
-(0.365)
---(0.293)
-(0.386)
-(0.389)
-(0.405)
P450 CYP2D6 substrate
-(0.419)
-(0.387)
-(0.458)
-(0.493)
-(0.388)
-(0.471)
+(0.547)
-(0.48)
Excretion
T 1/2 (half-life time)
1.865 h
1.906 h
1.915 h
1.939 h
1.951 h
1.811 h
1.799 h
1.949 h
CL (mL/min/kg)
1.49
1.618
1.597
1.669
1.266
1.234
1.471
1.084
Toxicity/Alert
hERG (hERG blocker)
++(0.867)
++(0.804)
++(0.832)
+(0.656)
++(0.734)
++(0.713)
++(0.726)
++(0.768)
H-HT (human hepatotoxicity)
++(0.854)
+++(0.936)
+++(0.932)
+++(0.942)
++(0.742)
-(0.396)
-(0.726)
-(0.39)
AMES (AMES mutagenicity)
---(0.234)
---(0.29)
---(0.29)
-(0.306)
-(0.344)
---(0.244)
---(0.266)
---(0.282)
SkinSen
-(0.436)
-(0.423)
-(0.423)
---(0.223)
---0.236
---(0.26)
---(0.202)
---(0.265)
LD50 (mg/kg)
880.766
962.653
994.191
999.709
376.733
475.199
302.235
342.996
PAINS
0 alert
0 alert
0 alert
0 alert
0 alert
0 alert
0 alert
0 alert
Metabolism: Cytochrome P450 consists of a group of isozymes responsible for the metabolism and biotransformation of drugs and chemicals. Inhibition of CYP450 isoforms may inhibit drug metabolism and induce drug-drug interactions in which co-administered drugs accumulate to toxic levels. Fortunately, our results showed that members of the SCOV-L series exhibited high negative tendencies as inhibitors of CYP3A4 and CYP2D6. In addition, results also showed tendencies of members of the SCOV-L series to be substrates of CYP1A2 and CYP450 3A4. The presence of these isoform in the liver and intestines indicates that these organs are sites of clearance of the SCOV-L series. However, the compounds were predicted to be neither a substrate nor an inhibitor of CYP2D6 and CYP2C19, indicating good metabolic stability against these isoforms.
Toxicity: Our ADMET lab calculations of potential hERG blockers based on Combined Multiple Pharmacophores and Machine Learning Approaches (Alves et al., 2015) showed that the compounds had medium tendencies to inhibit hERG ion channels and to present hepatotoxic risks. The AMES test database for mutagenicity revealed negative toxicity tendencies of all members of the SCOV-L series in this toxicity model (Alves et al., 2015). Furthermore, our exploration of the quantitative structure–activity relationship (QSAR) model of skin sensitization (SkinSen) and its application to identify potentially hazardous compounds (Alves et al., 2015) indicated negative SkinSen tendencies of all members of the SCOV-L series. In addition, it is worth noting that SCOV-L02, SCOV-L09, SCOV-L10, and SCOV-L11 demonstrated high levels of acute safety profiles with LD50 values of >880 mg/kg. However, the later series of SCOV-L15, SCOV-L18, SCOV-L22 and SCOV-L23 demonstrated less-acute safety profiles with LD50 values of 376, 342, 475, and 302 mg/kg, suggesting that these four members of the series could be toxic, but only in high doses. In addition, these compounds demonstrated no alerts for PAINS criteria.
Clearance: All members of the SCOV-L series exhibited low half-life times (1.799 ∼ 1.949 h) and clearance rates (1.084 ∼ 1.669 mL/min/kg). Conclusively, despite other members of the SCOV-L series exhibiting good to moderate profiles, it can be concluded that compounds SCOV-L15, SCOV-L18, SCOV-L22, and SCOV-L23 presented more-satisfactory drug-like and ADMET properties.
4 Conclusions
In conclusion, results of the present study increase our understanding of the immunosuppressive roles of ACE2 and TMPRSS2. To our delight, a series of small-molecule niclosamide derivatives of the SCOV-L series demonstrated high binding efficacies against ACE2, TMPRSS2, and NSPs. The SCOV-L series, particularly SCOV-L-18, SCOV-L-15, and SCOV-L-09 were found to exhibit strong binding affinities with three key SARS-CoV-2 proteins: 3CLpro, PLpro, and RdRp. These compounds bind to the several catalytic residues of the proteins hence, suggesting the ability to impede their catalytic activities and preventing or alleviating SARS-CoV-2 infection. Interestingly, among the eight compounds, SCOV-L-18, SCOV-L-15, and SCOV-L-09 also mostly satisfied the criteria of drug-like candidates, having good adsorption, distribution, metabolism, excretion, and toxicity (ADMET) pharmacokinetic profile. Further studies are ongoing to fully ascertain the potential of these compound for the treatment of SARS-CoV-2 infection.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Funding
H.-S. Huang is funded by the National Science and Technology Council, Taiwan, grant number NSTC111-2314-B-038-017 and the Ministry of Science and Technology, Taiwan, grant number MOST111-2314-B-038-122. Alexander TH Wu is funded by the National Science and Technology Council, Taiwan (111-2314-B-038 -142 and 111-2314-B-038 -098).
CrediT Author Contribution Statement
Bashir Lawal: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. Sheng-Kuang Tsai: Conceptualization, Data curation, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. Alexander T.H Wu: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Writing – review & editing.Hsu-Shan Huang: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Writing – review & editing.
Acknowledgements
Bashir Lawal acknowledged the Hillman Family Foundation for the award of the Hillman Postdoctoral Fellowship for Innovative Cancer Research
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Favipiravir: a new and emerging antiviral option in COVID-19. Med. J. Armed Forces India. 2020;76:370-376.
- [Google Scholar]
- Predicting chemically-induced skin reactions. Part I: QSAR models of skin sensitization and their application to identify potentially hazardous compounds. Toxicol. Appl. Pharmacol.. 2015;284:262-272.
- [Google Scholar]
- P-glycoprotein inhibition for optimal drug delivery. Drug Target Insights. 2013;7:27-34.
- [Google Scholar]
- Coronavirus main proteinase (3CL<sup>pro</sup>) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763-1767.
- [Google Scholar]
- Utilization of quercetin flavonoid compounds in red onion (Allium cepa L.) as inhibitor of spike sars-CoV-2 protein against ACE2 receptors. Biosaintifika: J. Biol. Biol. Edu.. 2021;13:356-362.
- [Google Scholar]
- The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res.. 2015;115:21-38.
- [Google Scholar]
- COVID-19 and cancer: current challenges and perspectives. Cancer Cell. 2020;38:629-646.
- [Google Scholar]
- Ruxolitinib in treatment of severe coronavirus disease 2019 (COVID-19): A multicenter, single-blind, randomized controlled trial. J. Allergy Clin. Immunol.. 2019;146(2020):137-146.e133.
- [Google Scholar]
- Binding mechanism of coronavirus main proteinase with ligands and its implication to drug design against SARS. Biochem. Biophys. Res. Commun.. 2003;308:148-151.
- [Google Scholar]
- Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform.. 2012;4:17.
- [Google Scholar]
- Computational analysis of microRNA-mediated interactions in SARS-CoV-2 infection. PeerJ. 2020;8:e9369.
- [Google Scholar]
- A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ. Res.. 2000;87:e1-e9.
- [Google Scholar]
- 'The long tail of Covid-19' – the detection of a prolonged inflammatory response after a SARS-CoV-2 infection in asymptomatic and mildly affected patients. F1000Research. 2020;9:1349.
- [Google Scholar]
- Review of the flavonoids quercetin, hesperetin, and naringenin, Dietary sources, bioactivities, bioavailability, and epidemiology. Nutr. Res.. 2004;24:851-874.
- [Google Scholar]
- COVID-19 in immunocompromised hosts: what we know so far. Clin. Infect. Dis.: Off. Publ. Infect. Dis. Soc. Am.. 2021;72:340-350.
- [Google Scholar]
- Gao, J., Tian, Z., Yang, X., 2020. Breakthrough: CHLOROQUINE phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends.
- Compassionate use of remdesivir for patients with severe Covid-19. N. Engl. J. Med.. 2020;382:2327-2336.
- [Google Scholar]
- Analysis of Food and Drug Administration-approved anticancer agents in the NCI60 panel of human tumor cell lines. Mol. Cancer Ther.. 2010;9:1451-1460.
- [Google Scholar]
- Structure-based design and synthesis of regioisomeric disubstituted aminoanthraquinone derivatives as potential anticancer agents. Helv. Chim. Acta. 2004;87:999-1006.
- [Google Scholar]
- Synthesis and structure–activity correlations of the cytotoxic bifunctional 1,4-diamidoanthraquinone derivatives. Bioorg. Med. Chem.. 2004;12:6163-6170.
- [Google Scholar]
- Huang, H.-s., Yu, D.-s., Chen, T.-C., 2015. Thiochromeno [2, 3-c] quinolin-12-one derivatives, preparation method and application thereof. Google Patents.
- Identification of potential inhibitors of three key enzymes of SARS-CoV2 using computational approach. Comput. Biol. Med.. 2020;122:103848
- [Google Scholar]
- African medicinal plants. Maryland: CRC Press; 1993.
- Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289-293.
- [Google Scholar]
- In-silico evaluation of genetic alterations in ovarian carcinoma and therapeutic efficacy of NSC777201, as a novel multi-target agent for TTK, NEK2, and CDK1. Int. J. Mol. Sci.. 2021;22:5895.
- [Google Scholar]
- Potential antimalarials from African natural products: a reviw. J. Intercul. Ethnopharmacol.. 2015;4:318.
- [Google Scholar]
- African natural products with potential antioxidants and hepatoprotectives properties: a review. Clin. Phytosci.. 2017;2:1-66.
- [Google Scholar]
- mTOR/EGFR/iNOS/MAP2K1/FGFR/TGFB1 are druggable candidates for N-(2,4-difluorophenyl)-2′,4′-difluoro-4-hydroxybiphenyl-3-carboxamide (NSC765598), with consequent anticancer implications. Front. Oncol.. 2021;11
- [Google Scholar]
- Pro-oncogenic c-Met/EGFR, biomarker signatures of the tumor microenvironment are clinical and therapy response prognosticators in colorectal cancer, and therapeutic targets of 3-phenyl-2H-benzo[e][1,3]-oxazine-2,4(3H)-dione derivatives. Front. Pharmacol.. 2021;12
- [Google Scholar]
- In vivo pharmacokinetic and anticancer studies of HH-N25, a selective inhibitor of topoisomerase I, and hormonal signaling for treating breast cancer. J. Inflamm. Res.. 2021;14:4901-4913.
- [Google Scholar]
- BC-N102 suppress breast cancer tumorigenesis by interfering with cell cycle regulatory proteins and hormonal signaling, and induction of time-course arrest of cell cycle at G1/G0 phase. Int. J. Biol. Sci.. 2021;17:3224-3238.
- [Google Scholar]
- Pharmacoinformatics and preclinical studies of NSC765690 and NSC765599, potential STAT3/CDK2/4/6 inhibitors with antitumor activities against NCI60 human tumor cell lines. Biomedicines. 2021;9:92.
- [Google Scholar]
- mTOR/EGFR/iNOS/MAP2K1/FGFR/TGFB1 Are druggable Candidates for N-(2, 4-difluorophenyl)-2′, 4′-difluoro-4-hydroxybiphenyl-3-carboxamide (NSC765598), with consequent anticancer implications. Front. Oncol.. 2021;11
- [Google Scholar]
- Transcriptomic-based identification of the immuno-oncogenic signature of cholangiocarcinoma for HLC-018 multi-target therapy exploration. Cells. 2021;10:2873.
- [Google Scholar]
- Identification of a novel immune-inflammatory signature of COVID-19 infections, and evaluation of pharmacokinetics and therapeutic potential of RXn-02, a novel small-molecule derivative of quinolone. Comput. Biol. Med.. 2022;148:105814
- [Google Scholar]
- Design, synthesis and evaluation of telomerase inhibitory, hTERT repressing, and anti-proliferation activities of symmetrical 1,8-disubstituted amidoanthraquinones. Eur. J. Med. Chem.. 2012;50:102-112.
- [Google Scholar]
- Novel inhibitors of RANKL-induced osteoclastogenesis: design, synthesis, and biological evaluation of 6-(2,4-difluorophenyl)-3-phenyl-2H-benzo[e][1,3]oxazine-2,4(3H)-diones. Bioorg. Med. Chem.. 2015;23:4522-4532.
- [Google Scholar]
- Discovery of 5-(2′,4′-difluorophenyl)-salicylanilides as new inhibitors of receptor activator of NF-κB ligand (RANKL)-induced osteoclastogenesis. Eur. J. Med. Chem.. 2015;98:115-126.
- [Google Scholar]
- HNC0014, a multi-targeted small-molecule, inhibits head and neck squamous cell carcinoma by suppressing c-Met/STAT3/CD44/PD-L1 oncoimmune signature and eliciting antitumor immune responses. Cancers (Basel). 2020;12
- [Google Scholar]
- TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res.. 2017;77:e108-e110.
- [Google Scholar]
- Multi-targeted therapy of cancer by niclosamide: a new application for an old drug. Cancer Lett.. 2014;349:8-14.
- [Google Scholar]
- GSCALite: a web server for gene set cancer analysis. Bioinformatics. 2018;34:3771-3772.
- [Google Scholar]
- Arthroprotective effects of Cf-02 sharing structural similarity with quercetin. Int. J. Mol. Sci.. 2018;19
- [Google Scholar]
- DriverDBv3: a multi-omics database for cancer driver gene research. Nucleic Acids Res. 2020;48:D863-D870.
- [Google Scholar]
- AlzPlatform: an Alzheimer’s disease domain-specific chemogenomics knowledgebase for polypharmacology and target identification research. J. Chem. Inf. Model.. 2014;54:1050-1060.
- [Google Scholar]
- Design and evaluation of PEGylated liposomal formulation of a novel multikinase inhibitor for enhanced chemosensitivity and inhibition of metastatic pancreatic ductal adenocarcinoma. Bioconjug. Chem.. 2019;30:2703-2713.
- [Google Scholar]
- An integrated bioinformatics study of a novel niclosamide derivative, nsc765689, a potential gsk3β/β-catenin/stat3/cd44 suppressor with anti-glioblastoma properties. Int. J. Mol. Sci.. 2021;22:2464.
- [Google Scholar]
- Network pharmacological analysis through a bioinformatics approach of novel NSC765600 and NSC765691 compounds as potential inhibitors of CCND1/CDK4/PLK1/CD44 in cancer types. Cancers. 2021;13:2523.
- [Google Scholar]
- Molecular docking of bioactive compounds against BRCA and COX Proteins, Prog. Drug Res. 71 (2016) 181-183.
- Androgen-deprivation therapies for prostate cancer and risk of infection by SARS-CoV-2: a population-based study N = 4532. Ann. Oncol.: Off. J. Eur. Soc. Med. Oncol.. 2020;31:1040-1045.
- [Google Scholar]
- Natural flavonoids as potential angiotensin-converting enzyme 2 inhibitors for anti-SARS-CoV-2. Molecules. 2020;25:3980.
- [Google Scholar]
- NIH, COVID-19 Treatment Guidelines, Antiviral Drugs That Are Approved or Under Evaluation for the Treatment of COVID-19 National Institute of Health, 2021, pp. 1–61.
- COXPRESdb v7: a gene coexpression database for 11 animal species supported by 23 coexpression platforms for technical evaluation and evolutionary inference. Nucleic Acids Res.. 2019;47:D55-D62.
- [Google Scholar]
- Sterculia tragacantha Lindl leaf extract ameliorates STZ-induced diabetes, oxidative stress, inflammation and neuronal impairment. J. Inflamm. Res.. 2021;14:6749-6764.
- [Google Scholar]
- Therapeutic efficacy of Clompanus pubescens leaves fractions via downregulation of neuronal cholinesterases/Na+-K+ATPase/IL-1 β, and improving the neurocognitive and antioxidants status of streptozotocin-induced diabetic rats. Biomed. Pharmacother.. 2022;148:112730
- [Google Scholar]
- Chinese herbal compounds against SARS-CoV-2: puerarin and quercetin impair the binding of viral S-protein to ACE2 receptor. Comput. Struct. Biotechnol. J.. 2020;18:3518-3527.
- [Google Scholar]
- CNS drug design based on principles of blood-brain barrier transport. J. Neurochem.. 1998;70:1781-1792.
- [Google Scholar]
- Does androgen deprivation therapy protect against severe complications from COVID-19? Anna. Oncol.: Off. J. Eur. Soc. Med. Oncol.. 2020;31:1419-1420.
- [Google Scholar]
- Tumor antigen presentation by dendritic cells. Crit. Rev. Immunol.. 2010;30:345-386.
- [Google Scholar]
- An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem.. 2016;59:6595-6628.
- [Google Scholar]
- Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: a framework for anti-COVID-19 drug design. Sci. Adv.. 2020;6
- [Google Scholar]
- Perspectives on plant flavonoid quercetin-based drugs for novel SARS-CoV-2. Beni-Suef University J. Basic Appl. Sci.. 2021;10:1-13.
- [Google Scholar]
- Systematic review of medicinal plants used for treatment of diabetes in human clinical trials: an ASEAN perspective. Evid. Based Complement Alternat. Med.. 2021;2021:5570939.
- [Google Scholar]
- Serial review: flavonoids and isoflavones (photoestrogens): absorption, metabolism, and bioactivity. Free Radic. Biol. Med.. 2004;37:1324-1350.
- [Google Scholar]
- Combined computational NMR and molecular docking scrutiny of potential natural SARS-CoV-2 Mpro inhibitors. J. Phys. Chem. B. 2022;126:2173-2187.
- [Google Scholar]
- Updates on clinical trials evaluating the regenerative potential of allogenic mesenchymal stem cells in COVID-19, npj. Regen. Med.. 2021;6:37.
- [Google Scholar]
- RV-59 suppresses cytoplasmic Nrf2-mediated 5-fluorouracil resistance and tumor growth in colorectal cancer. Am. J. Cancer Res.. 2019;9:2789-2796.
- [Google Scholar]
- Molecular docking and dynamics simulation studies of ginsenosides with SARS-CoV-2 host and viral entry protein targets. Nat. Prod. Commun.. 2022;17 1934578X221134331
- [Google Scholar]
- Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657-662.
- [Google Scholar]
- The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer. 2006;6:813-823.
- [Google Scholar]
- Identification of bioactive compounds from Glycyrrhiza glabra as possible inhibitor of SARS-CoV-2 spike glycoprotein and non-structural protein-15: a pharmacoinformatics study. J. Biomol. Struct. Dyn. 2020:1-15.
- [Google Scholar]
- Toxicology of oral antidiabetic medication. Am. J. Health Syst. Pharm.. 2006;63:929-938.
- [Google Scholar]
- The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res.. 2011;40:1737-1747.
- [Google Scholar]
- SARS-CoV2 may evade innate immune response, causing uncontrolled neutrophil extracellular traps formation and multi-organ failure. Clin. Sci.. 2020;134:1295-1300.
- [Google Scholar]
- AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem.. 2010;31:455-461.
- [Google Scholar]
- Molecular docking unveils the potential of andrographolide derivatives against COVID-19: an in silico approach. J. Genet. Eng. Biotechnol.. 2022;20:58.
- [Google Scholar]
- Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc. 2006;1:1112-1116.
- [Google Scholar]
- BIOVIA, Dassault Systèmes, BIOVIA Workbook, Release 2020; BIOVIA Pipeline Pilot, Release 2020. San Diego: Dassault Systèmes; 2020.
- Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res.. 2020;30:269-271.
- [Google Scholar]
- The biological activities, chemical stability, metabolism and delivery systems of quercetin: a review. Trends Food Sci. Technol.. 2016;56:21-38.
- [Google Scholar]
- Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus. J. Med. Chem.. 2007;50:4087-4095.
- [Google Scholar]
- A preclinical investigation of GBM-N019 as a potential inhibitor of glioblastoma via exosomal mTOR/CDK6/STAT3 signaling. Cells. 2021;10:2391.
- [Google Scholar]
- Inhibition of severe acute respiratory syndrome coronavirus replication by niclosamide. Antimicrob. Agents Chemother.. 2004;48:2693-2696.
- [Google Scholar]
- Multiomics identification of potential targets for alzheimer disease and antrocin as a therapeutic candidate. Pharmaceutics. 2021;13:1555.
- [Google Scholar]
- Identification of a novel theranostic signature of metabolic and immune-inflammatory dysregulation in myocardial infarction, and the potential therapeutic properties of ovatodiolide, a diterpenoid derivative. Int. J. Mol. Sci.. 2022;23:1281.
- [Google Scholar]
- Follow-up study on pulmonary function and lung radiographic changes in rehabilitating severe acute respiratory syndrome patients after discharge. Chest. 2005;127:2119-2124.
- [Google Scholar]
- In silico prediction of chemical Ames mutagenicity. J. Chem. Inf. Model. 2012;52:2840-2847.
- [Google Scholar]
- Broad spectrum antiviral agent niclosamide and its therapeutic potential. ACS Infect. Dis.. 2020;6:909-915.
- [Google Scholar]
- SARS-CoV-2 induces transcriptional signatures in human lung epithelial cells that promote lung fibrosis. Respir. Res.. 2020;21:182.
- [Google Scholar]
- Preclinical evaluation of the novel small-molecule MSI-N1014 for treating drug-resistant colon cancer via the LGR5/β-catenin/miR-142-3p network and reducing cancer-associated fibroblast transformation. Cancers (Basel). 2020;12
- [Google Scholar]
- Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444-1448.
- [Google Scholar]
- The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci.. 2003;100:13190-13195.
- [Google Scholar]
- Identification of NSP3 (SH2D3C) as a prognostic biomarker of tumor progression and immune evasion for lung cancer and evaluation of organosulfur compounds from Allium sativum L. as therapeutic candidates. Biomedicines. 2021;9:1582.
- [Google Scholar]
- Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved & #x3b1;-ketoamide inhibitors. Science. 2020;368:409-412.
- [Google Scholar]







