Translate this page into:
Ligand-observed in-tube NMR in natural products research: A review on enzymatic biotransformations, protein–ligand interactions, and in-cell NMR spectroscopy
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Natural product-observed NMR methods have considerably expanded the potentialities for in-tube NMR monitoring of complex enzymatic biotransformations and investigation of protein-natural product interactions even in living cells. We review, herein, the significant advantages of ligand-observed in-situ NMR monitoring of enzymatic biotransformations without restoring to laborious and time-consuming chromatographic methods. Emphasis will be given to the potentialities of the use of the NMR bioreactor: (i) to investigate through saturation transfer difference (STD), the capacity of natural products to serve as enzyme substrates, (ii) to monitor multiple biotransformation products of natural products with the use of immobilized enzymes and (iii) to investigate interactions of biotransformed products with protein targets. The use of STD and its variants, transfer effect Noes for PHArmacophore Mapping (INPHARMA) NMR, in conjunction with computational methods, can provide excellent tools in investigating competitive binding modes even in proteins with multiple binding sites. The method has been successfully applied in the study of unsaturated free fatty acids (UFFAs)-serum albumin complexes in which the location and conformational states of UFFAs could not be determined accurately, despite numerous X-ray structural studies, due to conformational averaging. This combined method, thus, may find promising applications in the field of protein-natural product recognition research. The emerging concept of in-cell NMR and recent applications will be discussed since they can provide atomic level insights into natural product-protein interactions in living cells without the need of isotope labelled techniques.
Keywords
Biotransformations
STD NMR;
TR-NOESY NMR;
INPHARMA/ILOE NMR;
In-cell NMR
1 Introduction
Binding of low molecular weight (MW) natural products to proteins can play a major role in biocatalyzed reaction processes, regulation of biological processes cellular metabolism and designing novel bioactive molecules that can modulate protein–ligand and protein–protein interactions. Among the great variety of biophysical techniques, NMR spectroscopy is a primary method that can be used to investigate protein-natural product interactions and their reaction products due to the great variety of techniques that can be applied even at cellular level.
Monitoring of biocatalyzed reactions using NMR spectroscopy, has been of growing interest in research and development in the last few decades. Significant improvements in instrumentation and experimental techniques have made NMR spectroscopy a versatile technique in performing biotransformations not only outside the magnetic field but also by performing bioreactions for in situ monitoring in the NMR tube, without any prior fractionation or isolation of the individual analytes. Significant experimental and computational advances have also greatly facilitated investigation of protein–ligand interactions. There are two main approaches for protein-natural product interactions: the protein-observed and the natural product-observed NMR experiments. In protein-observed methods, the high-resolution 3D structures are resolved with the use of heteronuclear experiments on isotopically labeled (13C, 15N, 2D) proteins, especially for those with MW above 30 kDa. Natural products-observed NMR experiments require significantly less protein without isotope labelling and they are not limited to proteins of low MW. They are based on the modification of ligand NMR parameters, especially nuclear Overhauser effect, in the presence of protein receptors. The NMR parameters are population weighed average of the free and bound states, provided that dissociation constants are between 10-3-10-8 M. Ligand-observed NMR methods have also been utilized to provide atomic level insights into natural products-protein interactions in living cells without isotope labeled techniques.
This review was based on a plenary lecture which was delivered in the International Conference on Applied Chemistry and Biotechnology (ICACB-2022), Jordan, and focuses mainly on the advances in the period 2010–2022 for identification and quantification of enzymatic biotransformations of natural products, investigations of ligand-observed protein-natural product interactions in solution and in living cells.
2 In-tube NMR monitoring of enzymatic biotransformations
1H NMR spectroscopy has been extensively utilized as a versatile and non-invasive method for monitoring biocatalyzed transformations on purified enzymes, washed cell suspensions and bacterial cultures growing in NMR tubes (Brecker and Ribbons, 2000). There are four approaches for NMR reaction monitoring: (i) placing the reagents into a standard NMR tube; (ii) transfer of the mixture from the reaction vessel to the NMR tube (on-line monitoring); (iii) stopped-flow NMR for rapid kinetics and (iv) rapid injection NMR with the use of probe inserts for quick insertion of reagents into the NMR tubes. The first approach, which does not require specialized NMR equipment, can be conducted in deuterated solvents (due to a small NMR test tube volume) and non-deuterated solvents and has been shown to provide valuable structural and mechanistic information, although, kinetic data are strongly diffusion controlled (Foley et al., 2016).
In-situ-1H NMR was used to investigate the biosynthetic oxidation, with human UDP- glucose 6-dehydrogenase, of UDP-glucose into UDP-glucuronic acid (Eixelsberger et al., 2012). The combined use of wild-type enzyme and the Glu151 → Gln mutant, which is slow-reacting, showed that the UDP-glucuronic acid was the only product for both biotransformations. The absence of deuterium at C-5 was interpreated in terms of an intermediate UDP-gluco-hexodialdose which is trapped by thiohemiacetal adduct formation without incorporation of solvent deuterium (D2O).
Kyriakou et al. (2012) reported enzyme-catalyzed regioselective acylation of flavonoid aglycones and a novel rapid 1H NMR product screening of the reaction products. Although it has been reported in the literature that the Candida antarctica lipase B (CALB) enzyme has no detectable activity in flavonoid aglycones (Chebil et al., 2006; Chebil et al., 2007; De Oliveira et al, 2010; Bidouil et al, 2011), a significant regioselective acylation of the ring B of quercetin (Fig. 1) and narigenin was found by Kyriakou et al. (2012). The reaction was performed with quercetin (3 μmol), vinyl acetate (1 mmol) and 60 mg ml−1 of catalyst in solvent (200 μL) at 60° C. The reaction products were identified and quantified in the crude extract without any prior fractionation or isolation. The method was based on the resonances of the OH(5) protons, which are strongly deshielded in the region above 12 ppm, due to the formation of a strong intramolecular hydrogen bond interaction with the C(4) = O group of ring C (Fig. 1) (Siskos et al., 2013; 2015; 2017; Kontogianni et al., 2013). The resolution of the OH resonances was significantly improved with the addition of picric acid (Charisiadis et al., 2010; 2012; Nerantzaki et al., 2011) or trifluoroacetic acid (TFA) (Charisiadis et al., 2017; Mari et al., 2019) with line widths Δν1/2 ∼ 1.5 Hz (Fig. 2). The OH(5) resonances of unreacted quercetin (Q), 3΄-O acetylquercetin (Q1), 10 % yield, and 4΄-O acetylquercetin (Q2), 35 % yield, were clearly resolved, although the acylation sites are located eleven and twelve bonds apart. The excellent resolution of the OH(4΄) and OH(3΄) of the 3΄ and the 4΄–monoacylated derivatives allowed the application of 1H–13C HSQC and HMBC experiments for the complete assignment of the two derivatives (Fig. 3). For example, OH(4΄) shows 2J and 3J HMBC connectivities to C(3΄) and C(5΄), respectively, and the H(3΄΄) shows 2J and 4J HMBC connectivities to C(2΄΄) and C(3΄), respectively.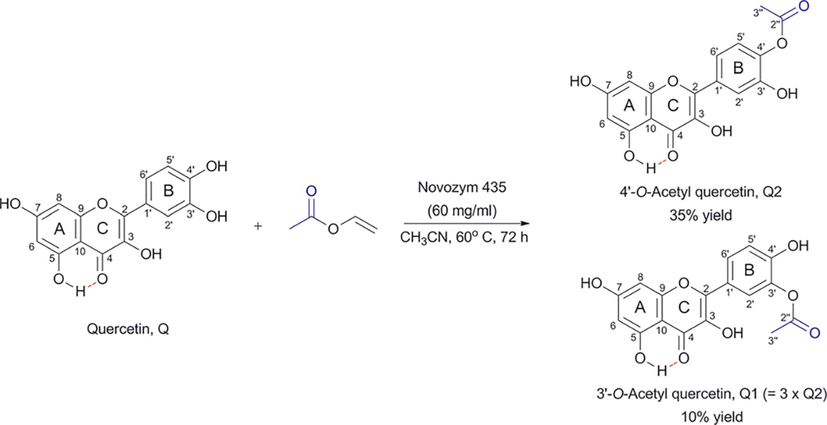
Regioselective acylation of quercetin with the use of CALB (Novozyme 435) leading to two products, Q1 (3΄-O-acetyl quercetin) and Q2 (4΄-O-acetyl quercetin). The OH(5) proton is implicated in the formation of an intramolecular hydrogen bond that is depicted with a red line (reprinted with permission of The Royal Society of Chemistry, from Kyriakou et al., 2012).
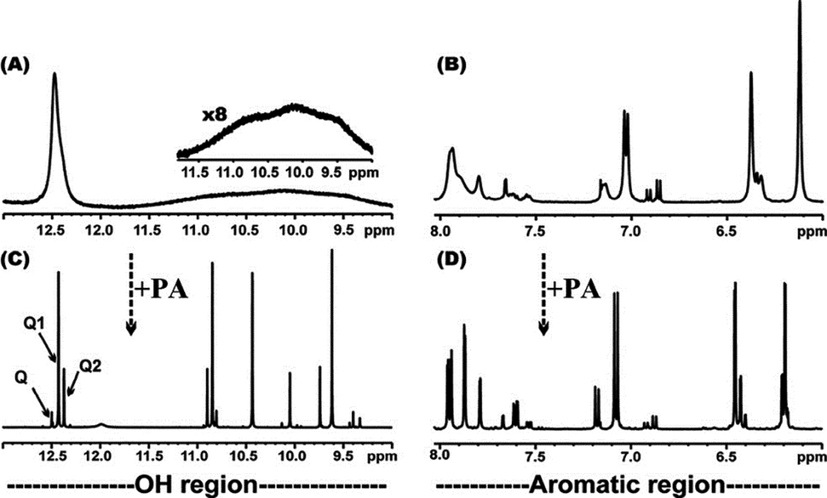
500 MHz 1D 1H NMR spectrum of the OH and aromatic regions of the enzymatically obtained monoacylated derivatives of quercetin in DMSO‑d6. The –OH and aromatic spectral regions are illustrated in (A), (B) before and in (C), (D) after the addition of picric acid, respectively. In (C) the absorptions of the OH(5) protons for Q (unreacted quercetin), Q1 and Q2 (Fig. 1) are indicated with arrows (reprinted with permission of The Royal Society of Chemistry, from Kyriakou et al., 2012).
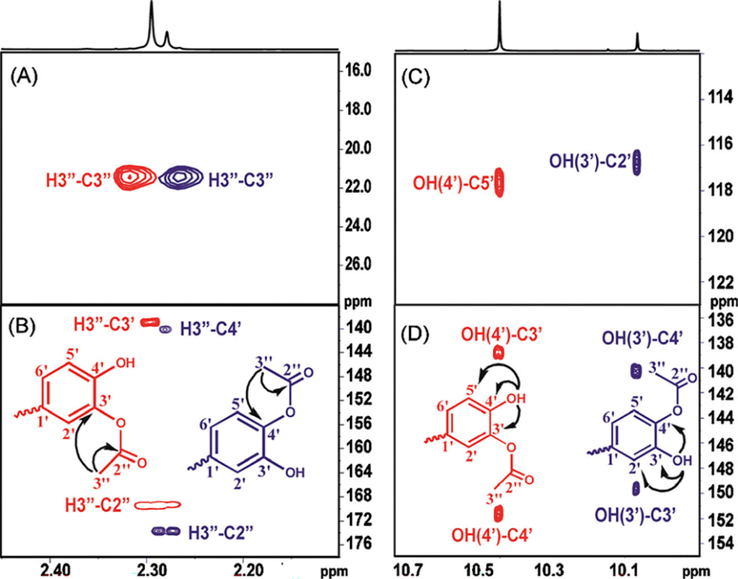
Selected regions of the 500 MHz 1H–13C HSQC (A) and 1H–13C HMBC (B)–(D) spectra of the solution used in Fig. 2(C). The arrows and cross peaks of importance for the unequivocal assignment of the 3′-and the 4′-monoacylated derivatives of quercetin are colored in red and blue, respectively (reprinted with permission of The Royal Society of Chemistry, from Kyriakou et al., 2012).
Further improvements of the NMR tube bioreactor were also reported, by confining the immobilized enzyme (CALB) in the bottom of the NMR tube (Chatzikonstantinou et al. 2018; 2020). Although CALB was placed outside the active volume of the NMR coil, it was necessary to increase the resolution of the OH resonances with optimization of the amount of the immobilized enzyme, concentration of acetyl donor and use of low temperatures (Fig. 4). The optimized resolution of the OH resonances (Δν1/2 ≤ 4 Hz) allowed the real time monitoring of the biotransformation process (Fig. 5).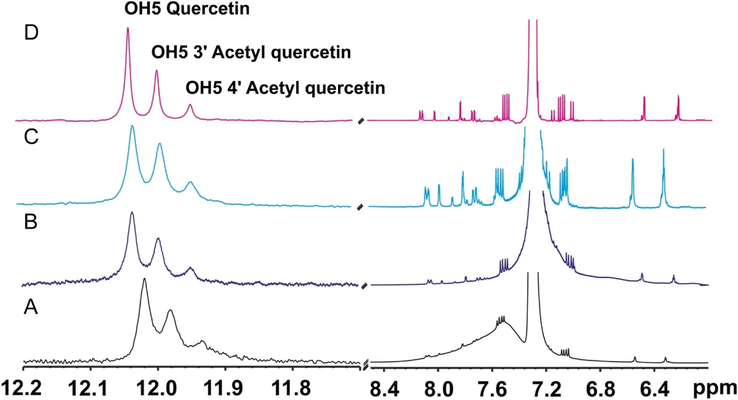
1H NMR spectra of in-situ esterification of quercetin catalyzed by CaLB. On a Bruker AV-400 spectrometer: (A) at 333 K, with 30 mg CaLB and 250 μL vinyl acetate; (B) at 298 K, with 30 mg CaLB and 250 μL vinyl acetate; (C) at 298 K, with 15 mg CaLB and 125 μL vinyl acetate. On a Bruker AV-500 spectrometer (D), at 282 K, with 15 mg CaLB and 125 μL vinyl acetate (reprinted with permission of Elsevier Inc., from Chatzikonstantinou et al., 2020).
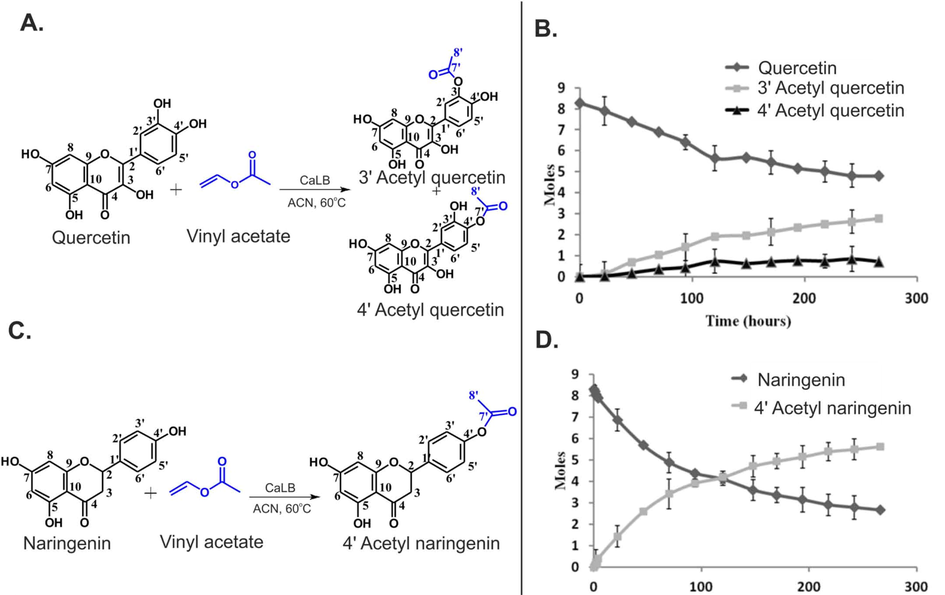
(A and C) The acetylation of quercetin and naringenin catalyzed by CaLB. (B, D) The real time monitoring of the biotransformation of quercetin and naringenin, respectively (reprinted with permission of Elsevier Inc., from Chatzikonstantinou et al., 2020).
Compared with the conventional method of monitoring biotransformation products, the NMR tube bioreactor has several advantages:
-
Eliminates fractionation, purification and separation steps of the reaction products.
-
Allows rapid optimization of enzymatic reaction conditions.
-
Facilitates kinetic and mechanistic studies.
-
Operates as a high throughput process in mixtures of putative enzymatic substrates.
-
The immobilized enzyme can be recycled for several reactions.
-
Can be easily complemented with ligand-observed NMR techniques (see Section 3.2).
Tsagogiannis et al. (2021) performed the in-situ monitoring of the reaction of protocatechuate 4,5-dioxygenase from pseudarthrobacter phenanthrenivorans sphe3 with protocatechuic acid (PCA) and gallate (Chatzikonstantinou et al., 2020). After 2 h, approximately 8 % of PCA was converted to the 4-carboxy-2-hydroxymuconate-6-semialdehyde (CHMS) product (Fig. 6ai); after 24 h, the PCA was totally consumed by 4,5-dioxygenase (Fig. 6aii). The reaction product CHMS exhibits characteristic resonances (Fig. 6b) due to keto-enol tautomerization process. The combination of 2D 1H–1H COSY, 1H–1H TOCSY, 2D 1H–13C HSQC and HMBC NMR spectra resulted in the complete resonance assignments for both the keto and enol forms. The equilibrium of the two tautomers was also investigated by the use of variable temperature (290–315 K) 1H NMR spectroscopy in the buffer solution at pH 8, which allowed the accurate determination of ΔHo = 25.58 kJ/mol and ΔSo = 0.10 kJ/mol K.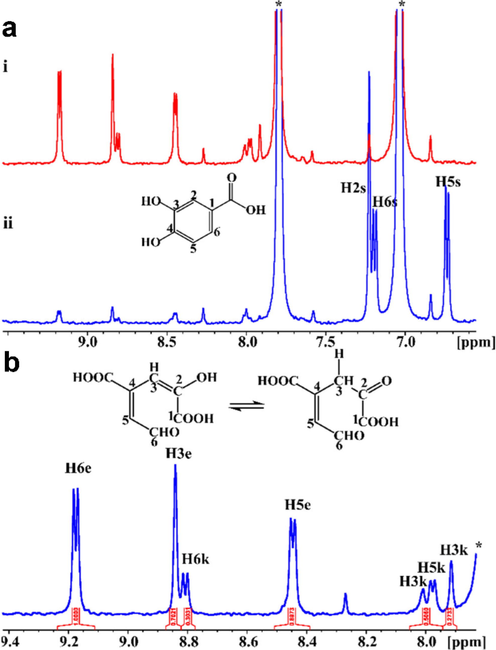
Biotransformation of PCA (1 mM) by PcaA (8 μΜ), inside the NMR tube. (a) 1H NMR spectra 2 h (blue spectrum, (i)) and 24 h (red spectrum, (ii)) after the addition of PCA to protocatechuate 4,5 dioxygenase. 1H NMR resonances of the substrate are shown in the blue spectrum. The asterisk (*) denotes protons of imidazole present in the buffer. (b) Expanded region (7.8–9.4 ppm) of the 1H NMR spectrum of Figure 6aii and the chemical structures of the enol (left) and keto (right) forms (Tsagogiannis et al., 2021).
Quantitative 1H NMR (q 1H NMR) was used to investigate steric and electronic effects of several monosubstituted benzoates as substrates of the Rieske dearomatizing dioxygenases (RDDs) in Ralstonia eutropha B9 (Bent et al., 2022). Higher activities were resulted with less bulky substituents, mainly in 3-position. The regioselectivity of the reaction was shown to have positive correlation with increasing size of substituents. These studies could be of importance in the synthesis of substrates for benzoate dioxygenases.
3 In-tube protein-natural products interactions with the combined use of STD, TR-NOESY and INPHARMA/ILOE NMR
3.1 Principles of NMR experiments
3.1.1 STD technique and its variants
The saturation transfer difference (STD) NMR technique (Meyer and Peters, 2003) is based on the nuclear Overhauser effect and has been widely used to characterize ligand-receptor interactions. It is very simple in the implementation and does not require proteins that are labelled with stable isotopes. Since the concentration of the protein is 50 to 1000 times lower than that of the ligands (μΜ–mM range) there is no need to use bacterial expression systems to produce high protein yields. In cases of problematic solubility of ligands in aqueous buffers, the use of DMSO‑d6 (concentration < 5 % ν/ν) is recommended to achieve concentrations of at least 100–300 μM, depending on the availability of high field instruments and high sensitivity cryoprobes. In the STD experiment the difference spectrum is obtained by subtracting a spectrum where the protein is selectively saturated (on-resonance) with a narrow selective pulse that does not excite the ligand, with signal intensity ISTD, from that without saturation (off-resonance) with intensities IO (Fig. 7). The difference in intensities ISTD = IO – ISAT of the ligand indicates an unambiguous epitope determination. The STD technique has shown to be efficient in detecting low-affinity ligands even in mixtures of homologues ligands and, thus, a powerful screening tool for characterizing ligand binding (Marcelo et al., 2012; Wagstaff et al., 2013). Thus, numerous STD studies have been published to characterize the binding of carbohydrates to lectins, enzymes and antibodies (Marcelo et al., 2012).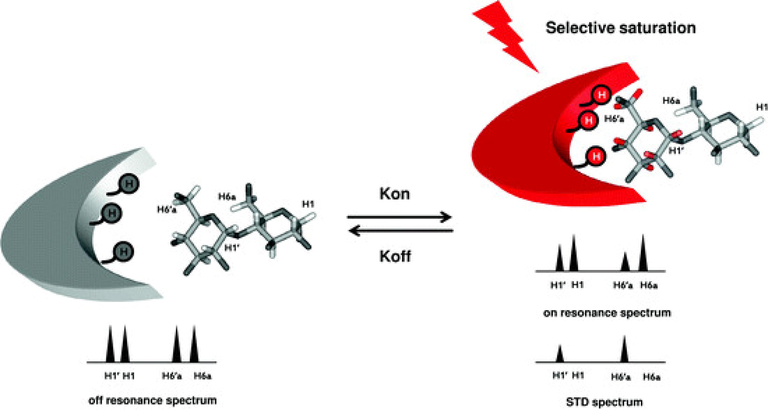
STD NMR experiment. Resonances of the protein are selectively saturated (red arrow) and rapidly transferred by spin diffusion throughout the protein. Intermolecular NOE transfer results in a decrease in the signal intensity of the protons of the ligand in close contact with the protein (protons in red). Graphically in the example the H1,1΄ and H6,6΄a resonances are shown. These changes are visual after subtraction of the on– to off-resonance spectrum in the STD spectrum (reprinted with permission of Springer-Verlag, from Marcelo et al., 2012).
Several variants of the standard STD experiment have been published based on the use of multiple on-resonance frequencies (Monaco et al., 2017; Watt et al., 2018; Monaco et al., 2020). The DiffErential EPitope mapping STD NMR (DEEP-STD NMR) can provide information on the nature of the aminoacids of the binding site surrounding the ligand. Provided that the high-resolution structure of the binding site is known, then, the orientation of the bound ligand can be inferred (Monaco et al., 2017). More recently an Inter-Ligand saturation transfer difference (IL-STD) NMR technique was used to investigate adjacent binding sites of the two ligands (Monaco et al., 2022). The technique is based on the selective proton saturation at a frequency δo (standard STD version) and a second frequency δ* to achieve selective saturation of the adjacent reporter ligand (Fig. 8). Significant STD differences of the ligand of the interest, demonstrate through space proximity of the two ligands. It has been claimed that the technique results in considerable saving of experimental time (one tenth) relative to the 2D inter-ligand NOE (see subsection 3.1.2 below).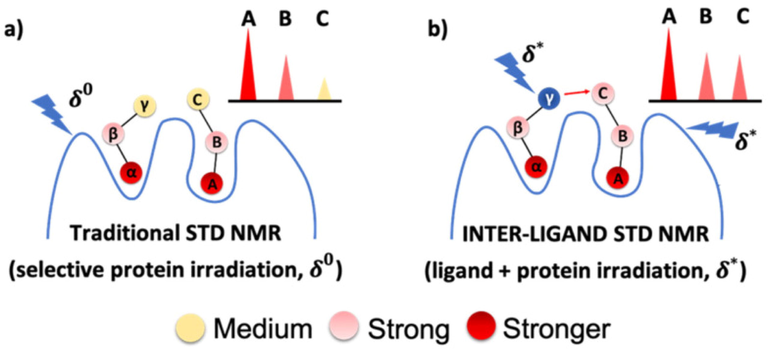
The IL-STD NMR approach: (a) STD NMR with selective irradiation (δ0) on protein protons; (b) STD NMR with selective irradiation (δ∗) on “reporter ligand” proton γ (supposed to be close to proton C of the adjacent “ligand of interest”) as well as on protein protons. The analysis of the IL-STD NMR experiment is focused exclusively on the protons (A, B, C) of the ligand of interest (Monaco et al., 2022).
3.1.2 INPHARMA/ILOE technique
INPHARMA technique (Sanchez-Pedregal et al., 2005; Orts et al., 2008; Carlomagno, 2012) can be utilized to distinguish between competitive and allosteric binding modes of natural products bound to a macromolecular target if the binding mode of a second competition ligand is known from high resolution X-ray or NMR structural studies. The method is based on the 2D NOE magnetization transfer between two ligands that interact competitively to the same binding site with inter-ligand distances < 5 Å. The number of intermolecular NOEs can be utilized to obtain the relative positions of the two ligands within the binding pocket (Fig. 9). For the successful implementation of the INPHARMA technique, the two competing ligands should exhibit similar residence times with affinity ratios<10 (Otts et al., 2009). Despite the above limitations, the INPHARMA technique was found to be extremely useful even in the cases of proteins with multiple binding sites and ligands which exhibit multiple conformations (see discussion below).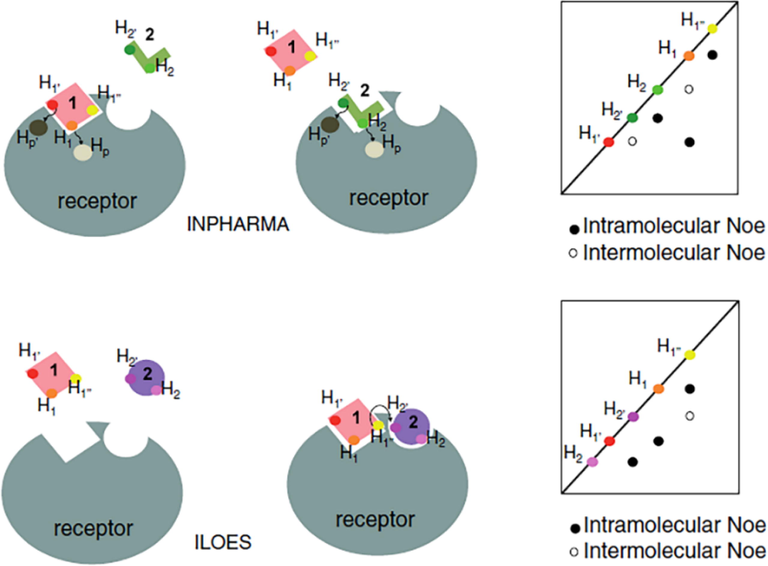
The inter-ligand nuclear Overhasuer effect (ILOE) versus the ILOE for pharmacophore mapping (INPHARMA) experiment. Hp, Hp΄, H1, H1′, H1″ and H2, H2′ represent protons of the protein (grey), the reference molecule (1, pink) and the fragment (2, green or violet). Arrows represent the magnetization transfer between two protons. Schematic representations of nuclear Overhasuer effect spectroscopy (2D NOESY) spectra are also shown. Black dots represent intramolecular nuclear Overhauser effects (NOEs) and white dots represent intermolecular NOEs (reprinted with permission of Springer, from Cala, 2014).
If two ligands bind simultaneously, with similar residence time in adjacent binding sites of a protein forming a ternary complex, then, inter-ligand NOEs (ILOEs) can be observed between the protons of ligands 1 and 2 that are in close contact (Fig. 9) (Li et al., 2001; Rademacher et al., 2011; Cala, 2014). For the observation of significant ILOE cross-peaks, mixing times of 600–800 ms are required, which are significantly longer than those used in INPHARMA experiments (70–300 ms) (Cala, 2014).
3.2 Selected applications
Chatzikonstantinou et al. (2018) reported further improvements of the NMR tube bioreactor by introducing three experimental steps (Fig. 10). The first one involves the application of 1D STD experiment to investigate the capacity of mixtures of natural products to serve as enzyme substrates. After identification of the most favorable substrates, the second step is applied for the 1D 1H NMR monitoring of multiple biotransformation products with the use of immobilized enzymes, as shown in detail in the previous section. The third step involves the evaluation of the interaction of the biotransformation products with specific protein targets.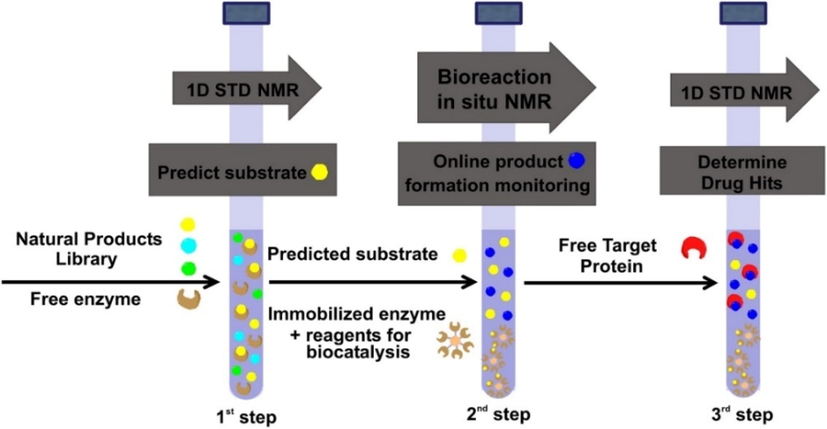
The three steps of the NMR tube bioreactor. 1st Step: Prediction of the capacity of natural products to serve as enzyme substrates with 1D STD NMR, 2nd Step: Multiple biotransformation products monitoring through 1D 1H NMR is exploited of the predicted substrates with immobilized enzymes, 3rd Step: Evaluation of the interaction of the enzymatic products with a pharmaceutical protein target with 1D STD NMR (reprinted with permission of Elsevier B.V., from Chatzikonstantinou et al., 2018).
Fig. 13 illustrates the advantages of the first step of the above method. An STD NMR experiment is applied in a mixture of natural products (quercetin and narigenin) which can serve as substrates for a given enzyme (CALB). Since the other components which are necessary for the biotransformation are not present in the mixture, the STD experiment can provide valuable information on the capacity of a natural product to interact with the enzyme. The STD experiment confirmed that the main interaction of quercetin with CALB is ring B (Fig. 11a). For narigenin, on the contrary, the main interactions are through the 6΄΄ methyl group and 4΄΄ protons of the sugar moiety (Fig. 11b).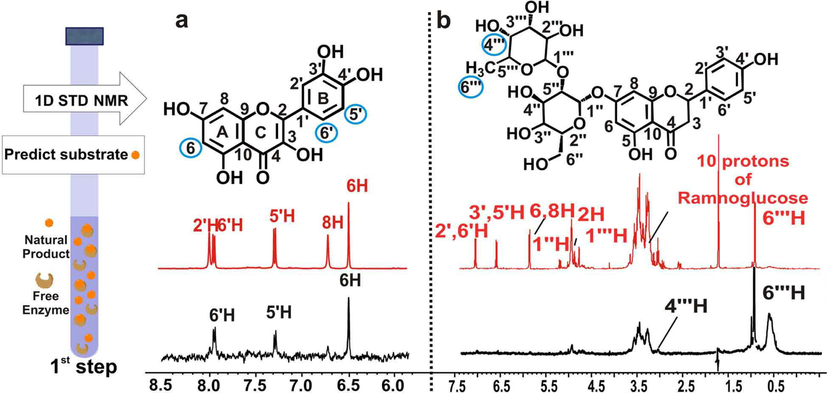
NMR based predictiοn of isolated compounds to act as enzymatic substrates. A schematic representation of the 1st Step applied in a single compound is shown on the left: Prediction of the capacity of natural products to serve as enzyme substrates through 1D STD NMR. Selected region of STD NMR spectra of a, quercetin, b, naringin, with free CALB in 65 % PBS in D2O, 35 % ACN d3 at 298 K. The reference 1H NMR spectra are colored in red and the difference 1H NMR spectra are colored in black. The protons of the putative enzymatic substrates identified to interact with the free enzyme are highlighted in filled blue circle (reprinted with permission of Elsevier B.V., from Chatzikonstantinou et al., 2018).
Having determined the favorable substrates that interact with CALB, the next step 2 involves the monitoring of multiple biotransformation products with the use of immobilized enzyme in the bottom of the NMR tube. Despite the use of 5 different substrates (luteolin-7-O-glucoside, rutin, narigenin, naringin and quercetin), the monitoring and quantification of 8 reaction products was achieved (Fig. 12). The 3rd step of the NMR tube bioreactor involves the rapid screening of the biotransformation products with the protein target. This step requires the following easily applicable protocol: (i) filtration of the immobilized enzyme; (ii) evaporation of the organic solvent, (iii) addition of the protein target which is dissolved in the appropriate buffer solution and (iv) evaluation with the use of 1D STD NMR of the interaction of the biotransformed products with the target protein.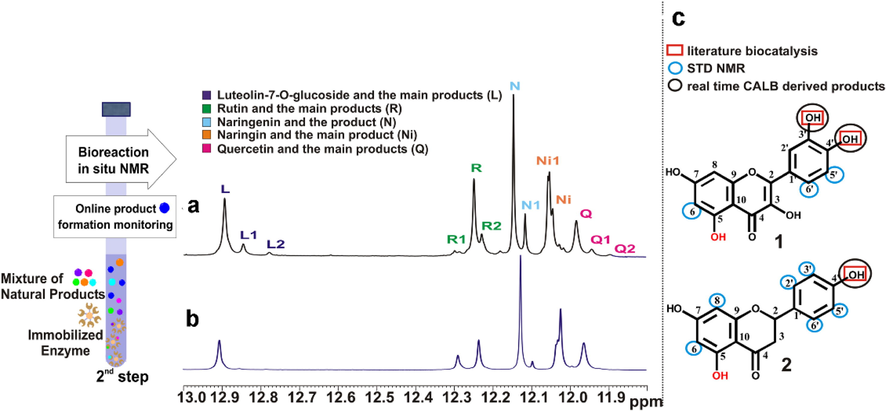
Real time multiple biotransformation products monitoring in 2nd Step of the NMR tube bioreactor. Selected region (OH5 protons) of the 1H NMR spectra of the crude CALB Novozyme 435 mediated biotransformation of a mixture of 5 flavonoids (quercetin (Q), naringenin (N), rutin (R) and luteolin-7-O-glucoside (L), naringin (Ni)) a, at 240 h and b, at 48 h, in ACN d3 at 290 K. c, The protons of quercetin (Q) and naringenin (N) identified to interact with the free enzyme are marked on the 2D structures by blue circles, the literature reported positions to be biotransformed are labeled in red squares, and the real time CALB Novozyme 435 derived products are labeled with black circles (reprinted with permission of Elsevier B.V., from Chatzikonstantinou et al., 2018).
Alexandri et al. (2022) applied ligand-observing STD, Tr-NOESY and INPHARMA techniques to probe the binding mode of free fatty acids (FFAs) with bovine and human serum albumin (BSA/HSA). Competition experiments were performed with warfarin and ibuprofen which are considered as stereotypical ligands for drug binding sites 1 and 2, respectively (Ghuman et al., 2005; Krenzel et al., 2013). The STD NMR spectra of caproleic, oleic (Fig. 13) and linolenic acids with BSA/HSA demonstrated extensive overlapping of all protons of the FFAs with protein aminoacid residues. Addition of warfarin at a molar ratio FFA/warfarin ≈ 1/1, resulted in a significant decrease in the STD signals (Fig. 13B). Similar results were obtained with the addition of ibuprofen (mole ratio FFA/ibuprofen ≈ 1/1) (Fig. 13C). The STD NMR spectra of α-linolenic acid in the presence of BSA and HSA and in competition experiments with warfarin and ibuprofen showed a minor reduction in the STD amplitude, contrary to the case of caproleic, oleic and linolenic acids. This minor effect, especially in the case of warfarin, can be interpreted in terms of weak binding affinity of α-linolenic acid to site FA7, which has been characterized as low affinity on the basis of numerous X-ray structural studies (Ghuman et al., 2005; Simard et al., 2005). An alternative interpretation of the minor STD competition effect can be attributed to higher affinity of α-linolenic acid relative to warfarin for the binding site FA7. This is in agreement with the significant reduction by 79 % of the STD NMR signal of warfarin in complexation with HSA upon the addition of warfarin (Fig. 14).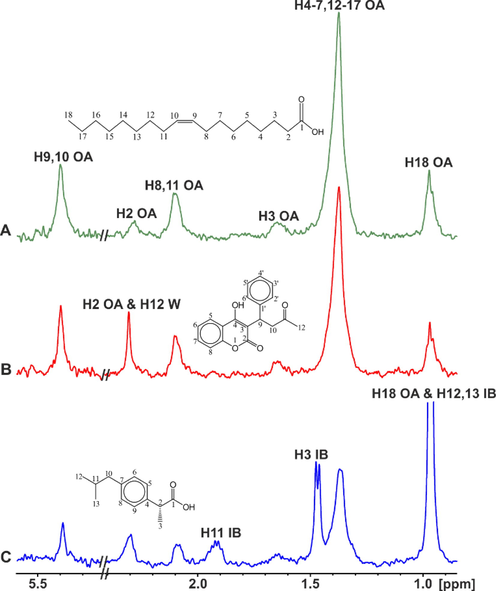
Selected regions of the STD NMR spectra of oleic acid (OA) (2.5 mM): (A) with BSA (25 μΜ) in 50 mM PBS buffer in D2O with 10 % DMSO‑d6; (B) after the addition of warfarin (W) (2.7 mM) in solution (A); (C) after the addition of ibuprofen (IB) (2.5 mM)) in solution (A) (T = 310 K) (reprinted with permission of Federation of European Biochemical Societies, from Alexandri et al., 2022).
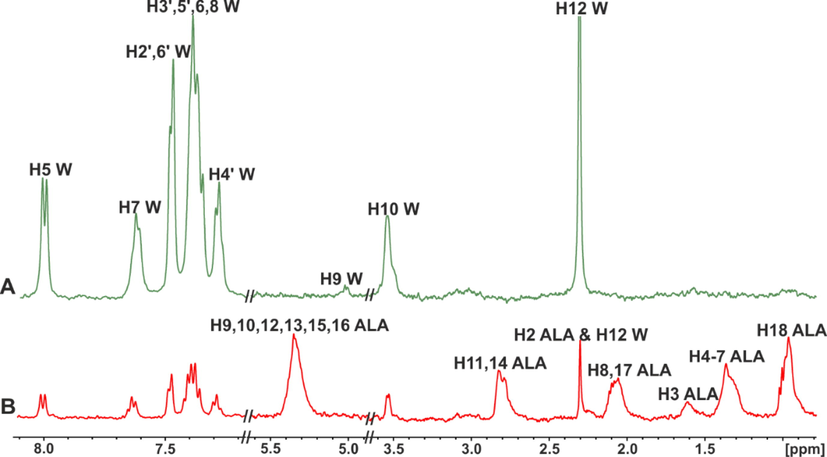
STD NMR spectra of: (A) warfarin (W) (2.5 mM) with HSA (25 μM) in 50 mM PBS buffer in D2O with 10 % DMSO‑d6; (B) warfarin (W) (2.5 mM) after its addition in the α-linolenic acid (ALA) (3.2 mM)/HSA (25 μΜ) complex in 50 mM PBS buffer in D2O with 10 % DMSO‑d6, T = 310 K (reprinted with permission of Federation of European Biochemical Societies, from Alexandri et al., 2022).
2D INPHARMA competition experiments with proteins with multiple binding sites, as in the case of HSA/BSA, were shown to be very informative. A significant number of negative inter-ligand NOEs were observed between FFAs and ibuprofen upon complexation with HSA/BSA (Fig. 15A). The common inter-NOEs between H2 and H3 of caproleic, oleic, linoleic and α-linolenic acids and H2 of ibuprofen demonstrate a common binding mode of the carboxylate groups of the ligands with HSA/BSA. Inter-NOE connectivities, however, were also observed between the terminal H11,12 of ibuprofen with H2 of α-linolenic acid, H11 of ibuprofen and H2 of oleic and H2,3 of linoleic acids, and H12,13 of ibuprofen with H2 of caproleic acid. The above inter-ligand distances (>5 Å) are beyond the limits of NOE experiments, under the hypothesis of a single conformational state of FFAs in the binding site FA4. Detailed docking calculations (Morris et al., 2009; Trott and Olson, 2010) confirmed the presence of two anchoring groups of amino acids in FA4 (Table 1). Fig. 16 shows that the two anchoring groups of amino acids orient the carboxylate groups of FFAs in antipodal positions (Alexandri et al., 2022).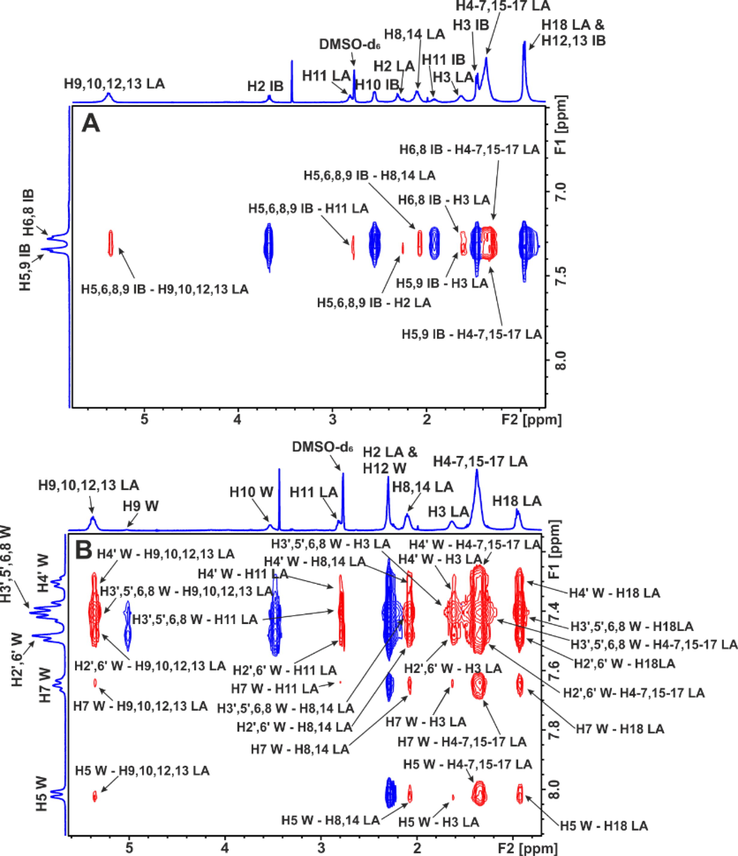
Selected regions of inter-ligand 2D Tr-NOESY NMR spectra of linoleic acid (LA) (2.5 mM) with BSA (25 μΜ) in 50 mM PBS buffer in D2O with 10 % DMSO‑d6: (A) after the addition of ibuprofen (IB) (3 mM). (B) After the addition of warfarin (W) (3 mM) (mixing time = 300 ms, T = 310 K). The red cross-peaks denote inter-ligand NOE connectivities (reprinted with permission of Federation of European Biochemical Societies, from Alexandri et al., 2022).
Ligand
R410
Y411
S419
T422
Group
Distance
(Å)
Group
Distance
(Å)
Pose/affinity (kcal/mol)
Group
Distance
(Å)
Group
Distance
(Å)
Pose/affinity (kcal/mol)
Anchor site 1
Anchor site 2
CA
NH2 η2
3.4
ΟΗ
2.2
1/-5.5
ΟΗ
5.2
ΟΗ
3.6
6/-5.3
OA
NH2 η2
2.4
ΟΗ
2.1
1/-6.8
ΟΗ
4.4
ΟΗ
3.2
5/-6.6
LA
NH2 η2
2.4
ΟΗ
1.8
2/-7.2
ΟΗ
4.3
ΟΗ
3.1
7/-7.1
ALA
NH2 η2
2.4
ΟΗ
2.0
8/-7.2
ΟΗ
3.8
ΟΗ
3.0
6/-7.3
IB
NH2 η2
2.9
ΟΗ
2.0
2/-7.3
–
–
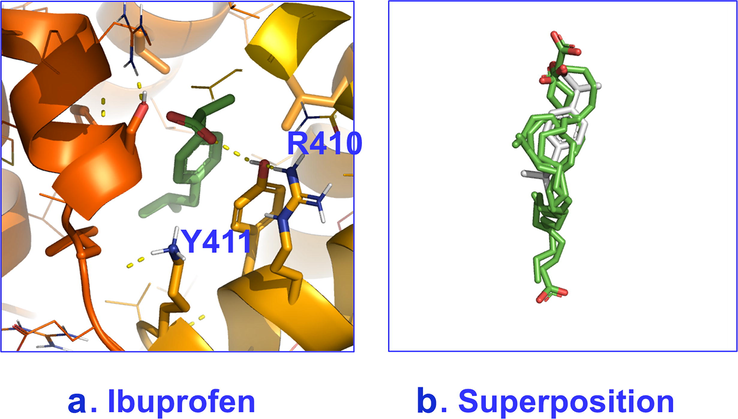
Poses with best scores for binding site 4 of HSA with: a. ibuprofen and b. superposition of caproleic, oleic, linoleic and α-linolenic acids and ibuprofen
(adopted from Alexandri et al., 2022).
A significant number of negative inter-ligand NOEs were also observed between caproleic, oleic, linoleic and α-linolenic acids and warfarin which demonstrate NOE transfer at the common binding site FA7 of HSA/BSA. Of particular interest are the 2D INPHARMA experiments when α-linolenic acid was added in the warfarin - HSA/BSA complex (Fig. 17B) instead of the reverse experiment in which warfarin was added to the α-linolenic acid HSA/BSA complex (Fig. 17A). In Fig. 17B a very significant increase in the magnitude of inter-ligand NOEs was observed which confirms the STD experiments that α-linolenic acid has higher affinity than warfarin at the binding site FA7. Strong NOEs were observed between the aromatic protons of the phenyl butyl and benzopyran ring and the H3 and terminal CH3 group of α-linolenic acid. Since the crystallographic distance of the centers of the two aromatic rings is > 6.9 Å, it was concluded, with the use of docking calculations, the presence of two docking sites (Table 2). This implies the presence of two conformational states of the FFAs at the FA7 binding site (Fig. 18). This conformational flexibility is, very probably, the reason that the location of FFAs could not be determined accurately, despite numerous available X-ray structural data (Ghuman et al., 2005; Simard et al., 2005).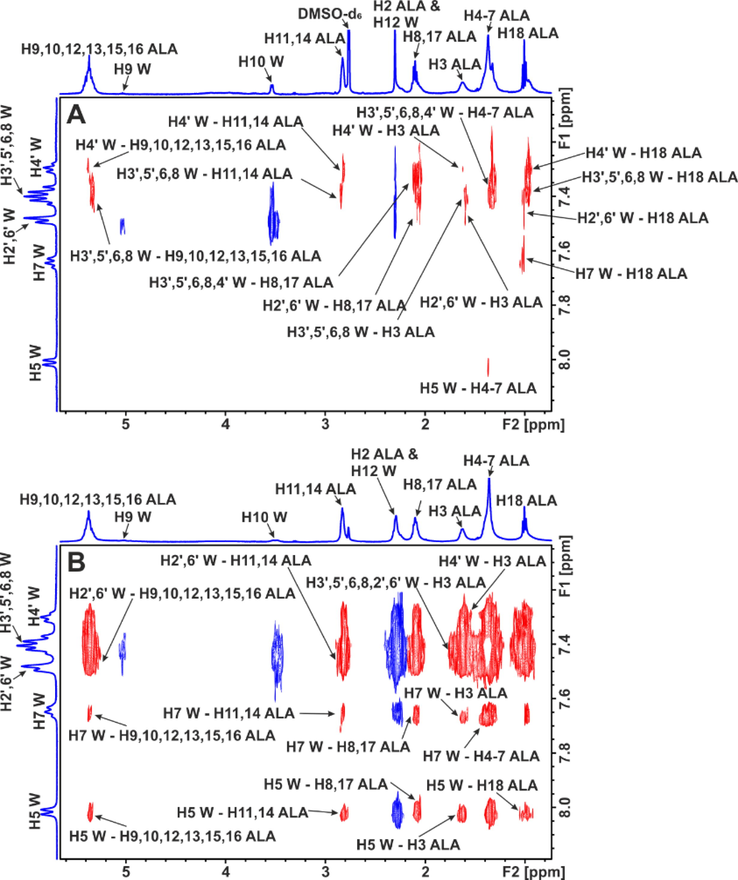
Selected regions of inter-ligand 2D Tr-NOESY NMR spectra of: (A) α-linolenic acid (ALA) (2.5 mM) with HSA (25 μΜ) in 50 mM PBS buffer in D2O with 10 % DMSO‑d6 after the addition of warfarin (W) (2.5 mM). (B) warfarin (W) (2.5 mM) with HSA (20 μΜ) in 50 mM PBS buffer in D2O with 10 % DMSO‑d6 after the addition of α-linolenic acid (ALA) (2.5 mM). The red cross-peaks denote inter-ligand NOE connectivities (reprinted with permission of Federation of European Biochemical Societies, from Alexandri et al., 2022).
Ligand
K199
R218
R222
H242
R257
Anchor site 1
Anchor site 2
Group
Dist.
(Å)
Pose / affinity (kcal/mol)
Group
Dist.
(Å)
Pose / affinity (kcal/mol)
Group
Dist.
(Å)
Pose / affinity (kcal/mol)
Group
Dist.
(Å)
Pose / affinity (kcal/mol)
Group
Dist.
(Å)
Pose / affinity (kcal/mol)
CA
NHε-
2.5
2/-5.2
NHε-
3.0
1/-5.3
OA
NH2 η1
3.2
9/-5.6
NHε
2.5
1/-6.0
NHτ
2.6
3/-5.8
NH2 η1
2.8
7/-5.7
LA
NH2 η1
3.0
7/-5.9
NHε
2.4
2/-6.2
NHτ
2.4
6/-6.0
NHε
2.8
1/-6.2
ALA
NH2 η2
2.3
1/-7.1
NHτ
3.3
4/-7.0
NH2 η1
3.5
4/-7.0
W
O(4)--–NH3+
2.6
7/-7.5
NH2 η1,2- C(11) = O
2.2
7/-7.5
NH2 η1- C(11) = O
2.5
4/-7.7
C⚌O ring- Hε2
2.3
4/-7.7
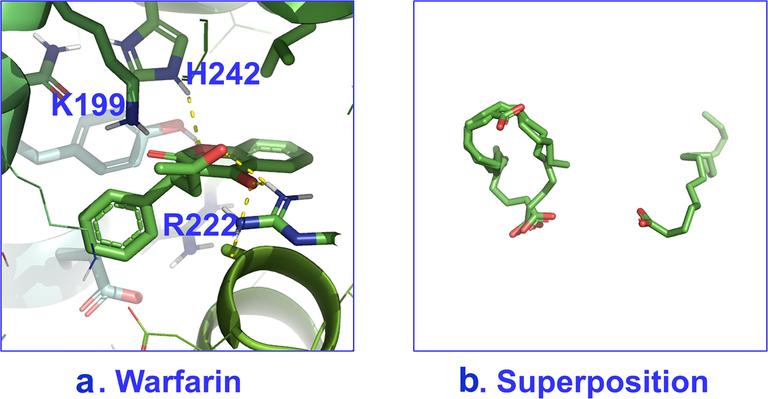
Poses with best scores for binding site 7 of HSA with: a. warfarin and b. superposition of caproleic, linoleic and α-linolenic acids; for α-linolenic acid, spatial deviation is observed with respect to the rest of the FFAs.
(adopted from Alexandri et al., 2022).
Hernychova et al. (2022) investigated the non-covalent vs covalent modes of interaction of 9/10-nitro-oleic acid (NO2-OA) with HSA. The STD spectrum of oleic acid in the presence of HSA shows a strong signal of the olefinic H9,10 protons (Fig. 19A(b)). In contrast, a very significant reduction of the STD signal of oleic acid was observed which became indistinguishable from the noise (Fig. 19A(c)). This demonstrates that NO2-OA has a significantly higher affinity to HSA than that of oleic acid. The STD spectrum of warfarin in the presence of HSA shows strong signals of the aromatic protons (Fig. 19B(b)). The STD signal of warfarin bound to HSA upon the addition to the NO2-OA/HSA complex was significantly reduced (>80 %) (Fig. 19B(c)) which shows the primary role of non-covalent competitive binding mode. The STD spectrum of ibuprofen in the presence of HSA shows strong signals of the aromatic protons (Fig. 19B(b)). The STD amplitude of ibuprofen upon the addition to the NO2-OA/HSA shows a minor reduction (Fig. 19C(c)), which demonstrates that NO2-OA does not have a higher affinity for HSA than that of ibuprofen. 2D-INPHARMA experiment demonstrated strong inter-ligand NOE connectivities between the aromatic protons of ibuprofen and the olefinic H2,8,11 and H3 of NO2-OA. Both ligands, therefore, share a common binding site (<4.5 Å) and, thus, are competitive rather than allosteric, towards binding site FA4.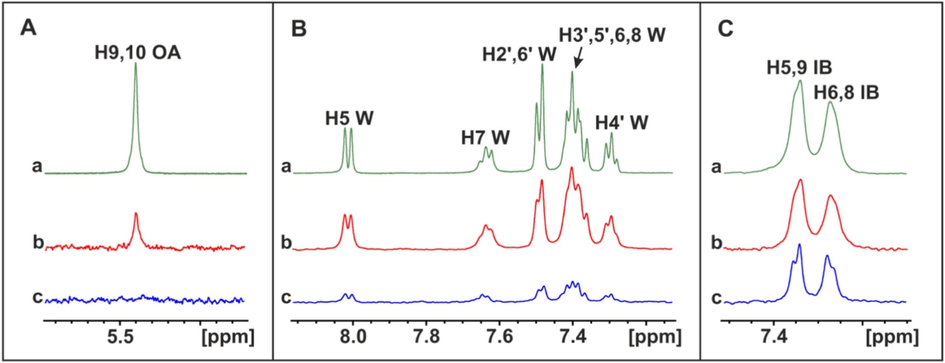
Selective regions of 1H NMR spectra (500 MHz) of: A(a) oleic acid (OA) (1.25 mM) with native HSA (25 μM). (b) STD NMR spectrum of (a). (c) STD NMR spectrum of oleic acid (1.25 mM) after addition to the NO2-OA (1.25 mM)/HSA (25 μM) complex. B(a). warfarin (W) (1.25 mM) with native HSA (25 μM). (b) STD NMR spectrum of (a). (c) STD NMR spectrum of warfarin (1.25 mM) after addition to the NO2-OA (1.25 mM)/HSA (25 μM) complex. C(a) ibuprofen (IB) (1.25 mM) with native HSA (20 μM). (b) STD NMR spectrum of (a). (c) STD NMR spectrum of ibuprofen (1.25 mM) after addition to the NO2-OA (1.25 mM)/HSA (25 μM) complex (reprinted with permission of Elsevier B.V., from Hernychova et al., 2022).
The structural basis of artemisinin binding sites in serum albumin was investigated with the combined use of NMR and docking calculations (Primikyri et al. 2022). Fig. 20 shows efficient binding of artemisinin to BSA since STD amplification factors (ASTD) of all protons are above 60 %. Competition experiments were also performed with warfarin and ibuprofen. After the addition of equimolar concentration of warfarin in the complex of artemisinin with BSA/HSA, the resulting STD signal intensities (warfarin/artemisinin 3/1) demonstrated a stronger STD effect of warfarin which implies a higher affinity of warfarin relative to artemisinin. Similar results were also obtained with competition experiments with ibuprofen. 2D INPHARMA experiments revealed significant inter-ligand NOE connectivities between the aromatic protons of warfarin and several protons of artemisinin (Fig. 21). Thus, 14-, 13- and 15-CH3 groups of artemisinin show strong and strong-to-medium NOEs with the benzoyl group of warfarin. Site specific docking simulations (pose number 2 of Fig. 22) are in excellent agreement with the experimental inter-ligand NOEs. Similarly, strong inter-ligand NOEs were observed between the aromatic protons of ibuprofen with 14-, 13- and 15-CH3 groups of artemisinin, in excellent agreement with site-specific docking calculations. The significant superposition of warfarin, which is in the anionic form at pH ≈ 7.4, with artemisinin, which is highly hydrophobic, emphasizes the extraordinary ligand binding properties of HSA (Fasano et al. 2005).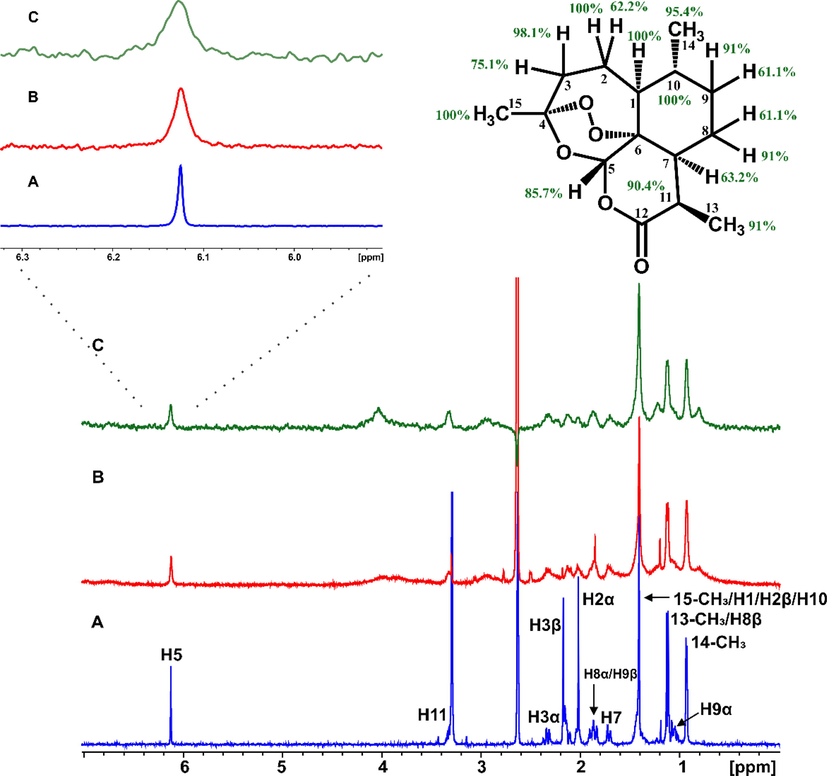
(A) 1H NMR of 2 mM artemisinin in PBS, pH 7.4, D2O with 10 % DMSO‑d6. (B) 1H NMR of 2 mM artemisinin with 20 μM BSA in PBS, pH 7.4, D2O with 10 % DMSO‑d6. (C) STD NMR of 2 mM solution (B) (Primikyri et al., 2022).
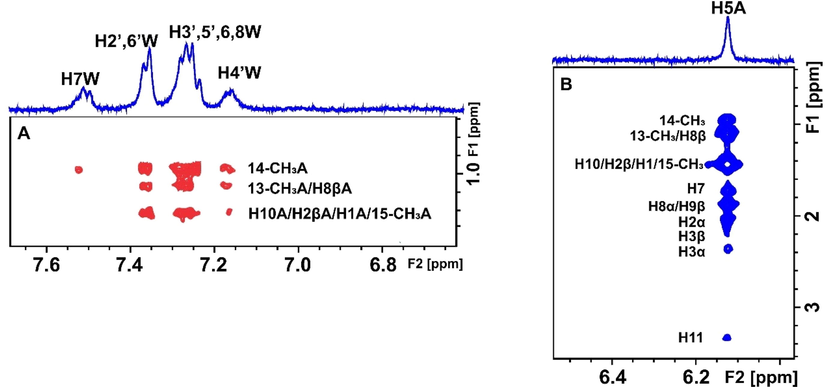
Selective region of 2D Tr-NOESY NMR spectrum of 2 mM artemisinin and 20 μΜ BSA after the addition of 2 mM warfarin in PBS buffer solution in D2O, pD 7.4 with 10 % DMSO‑d6. (A) Red cross-peaks correspond to inter-NOEs between warfarin and artemisinin and (B) blue cross-peaks correspond to intra-NOEs of artemisinin, mixing time = 300 ms (Primikyri et al., 2022).
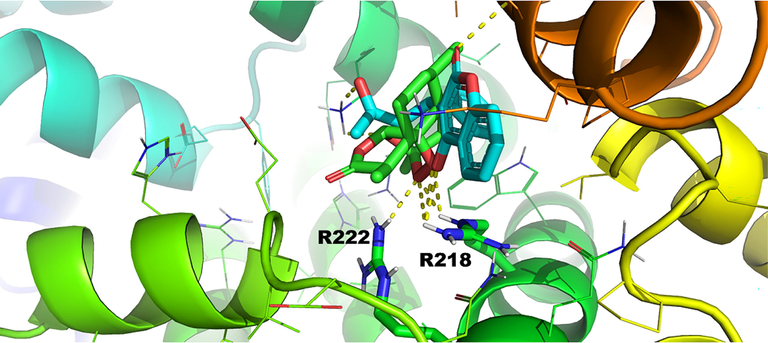
Pose number 2 of warfarin and artemisinin for binding site FA7 of HSA (1BM0.pdb) (Primikyri et al., 2022).
In the case of ligands that bind simultaneously to adjacent protein pockets (Fig. 9), the inter-ligand NOEs (ILOEs) may require significantly longer mixing times (600–800 ms) for a sufficient S/N ratio. This often leads to the dilemma of whether or not the intensities of the NOEs reflect the true inter-NOEs or spin-diffusion process or both. Sledz et al. (2010) performed a careful optimization of inter-ligand NOEs to inhibitory discovery against Mycobacterium tuberculosis pantothenate synthetase (PtS). For highly hydrophobic inhibitors negative inter-NOEs were observed, even in the case of perdeuterated protein (d PtS). This was attributed to high concentrations of hydrophobic and isoluble inhibitors that give rise to non-specific interactions.
Ligand-observed NMR techniques have also been applied in extracts of natural products. STD NMR, Tr-NOESY and STD-TOCSY experiments were utilized to investigate the binding potential of the ethyl acetate extract of Stryphnodendron polyphyllum with human serum albumin (HSA) (Tanoli et al., 2015). However, a hyphenated system comprising LC-SPE NMR was utilized to facilitate the assignment of the complex 1H NMR spectra. STD NMR was utilized to investigate the interaction of a diethyl ether extract of Artemisia annua with bovine serum albumin (Primikyri et al., 2022). Artemisinin was shown to be the principal analyte involved in the interaction. Further STD experiments with selective irradiation of the characteristic singlet H5 resonance (δ = 6.26 ppm) of artemisinin facilitated the identification of discrete STD signals of artemisinin even in overcrowded regions of the spectrum (2.5–0.9 ppm).
From the examples shown above it can be concluded that: (i) the STD NMR and its variants, due to high sensitivity, can be very useful as a preliminary step for ligand screening prior to the application of the more demanding INPHARMA NMR. Further improvement of the STD method includes the investigation of ligand structures, within the protein binding site, with the use of complete relaxation and conformation exchange matrix (CORCEMA) calculations which were applied for the screening of carbohydrates and peptides (Jayalakshmi and Rama Krishna, 2004; Maity et al., 2019). (ii) In the case of extracts of natural products, the use of multiple on-resonance saturation frequencies in less crowded spectral regions (Monaco et al., 2017; Watt et al., 2018; Monaco et al., 2020) could facilitate the interpretation of the results, without the need of hyphenated LC-SPE NMR techniques. (iii) The 2D INPHARMA experiments, in combination with docking and/or molecular dynamics simulations, can be successfully applied in investigating competitive binding in natural product-protein interactions, even in cases of proteins with multiple binding sites.
4 In-cell NMR of protein-natural products interactions
4.1 In-cell enzymatic biotransformations
Enzymes are the targets of many natural products and drugs, therefore, NMR can be used to investigate their simulation or inhibition in cellular environments (Siegel and Selenko, 2019) by monitoring biotransformed products. In-cell enzymatic activity was reported for the antibiotics for meropenem and imipenem which can be hydrolyzed by the new Delhi metallo-β-lactamase subclass 1 (NDM-1) produced by antibiotic resistant bacterial strain (Ma et al., 2014). Time resolved 1H NMR spectra of cell suspension of E.coli expressing the β-lactamase NDM-1, showed clearly distinguished resonances of the hydrolysis products. The same method was used for the screening of several drug candidates in a 96-well plate containing E.coli cells (Ma et al., 2015).
19F has found several applications in fragment-based drug discovery since ∼ 25 % of the approved drugs contain fluorine atoms, 19F is a spin ½ nucleus with 100 % natural abundance, sensitivity of 83 % relative to that of 1H and a chemical shift range of several hundred ppm (Norton et al., 2016). In-cell enzymatic hydrolysis of a fluorinated anandamide analog catalyzed by human fatty acid amide hydrolase, overexpressed in HEK 293 cells, was followed by real-time 19F NMR (Veronesi et al., 2016). The fluorinated substrate and the fluorinated product showed significantly different chemical shifts. Real-time 19F NMR was used to investigate the phosphorylation of kemptide containing (2S, 4R)-perfluoro-tert-butyl-Hyp by the protein kinases pkA activity in HeLA cell extracts (Tressler et al., 2020). Drug related cellular 19F NMR studies, therefore, may proved particularly promising (Buchholz et al., 2021).
4.2 Protein-natural products interactions
NMR spectroscopy is a versatile tool for investigating biomolecular structures in a breadth of cellular systems from prokaryotic to eukaryotic cells. Readers can consult recent publications for comprehensive and critical discussion on sample preparation, isotopic labeling, NMR techniques and a wide range of applications (Banci et al., 2013; Hamantsu et al., 2013; Freedberg and Selenko, 2014; Siegal and Selenko, 2019; Nishida et al., 2020; Theillet and Luchinat, 2022; Luchinat and Banchi, 2022; Luchinat et al., 2022; Theillet, 2022). In this Review we focus only to ligand-observed cellular NMR since it can provide fast analysis using mainly 1H NMR, with significantly lower preparation costs of the label-free samples.
Protein-ligand interactions can be investigated in living cells with the saturation transfer double difference (STDD) method, which involves a second STD experiment which is performed with the same sample, but in the absence of the ligand to remove unwanted STD signals due to the cellular matrix (Fig. 23) (Claasen et al., 2005). Due to high sensitivity of the STD technique, very good quality spectra were obtained in<1 h. The technique was used to investigate the binding of the highly potent (KD = 5 μΜ) cyclic pentapeptide cyclo (RGDfV) to integrin a11bβ3 which is the most abundant glycoprotein of intact blood platelets. The significant 5-fold increase of the STD amplitude with integrin a11bβ3 in native platelets than in liposomes emphasizes the importance of investigating interactions of membrane proteins in their natural environment. Although both systems show similar STD effects, Arg4δ,δ΄ and GlyHα showed significantly different STD amplitudes, presumably due to slightly different binding modes. The STDD method was also used to investigate the interaction of the dendritic-cell specific ICAM-3 grabbing non-integrin (DC-SIGN) receptor with Saccharomyces cerevisiae mannan (Marie et al., 2005) and to screen marine natural products against cannabinoid G-protein coupled receptors (Pereira et al., 2009).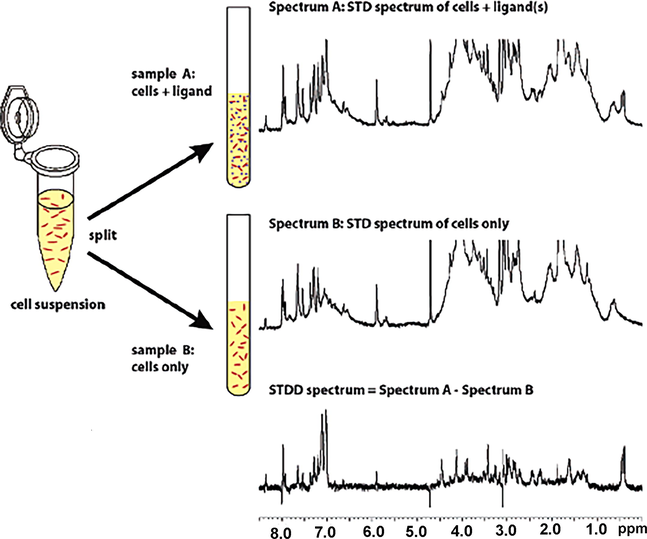
Schematic overview of the Saturation Transfer Double Difference (STDD) NMR method to detect binding of ligands (denoted by purple dots) to membrane-bound proteins in living cells (denoted by red dots). A cell suspension is split up into two NMR tubes. Sample A contains also the added ligands. Sample B contains only the cell suspension. Top: STD NMR spectrum of ligand and cells containing the transmembrane receptor. The spectrum contains the STD signals from the added ligand and the other binding processes in the cell. Middle: STD NMR spectrum of cells containing the transmembrane receptor. The STD NMR spectrum contains all signals from binding processes within the cell. No ligand receptor interactions can occur. The STDD spectrum (bottom) is obtained by subtracting the cell STD spectrum (middle) from the cell/ligand STD spectrum (top) (reprinted with permission of the American Chemical Society, from Claasen et al., 2005).
Potenza et al. (2011) investigated the interaction of two cyclic mimetics of the Arg-Gly-Asp motif with ECV304 bladder cancer cells in which integrin avβ3 is highly expressed. STD experiments were used to identify the protons of the ligands in closest contact with the protein, and Tr-NOESY to define the preferred conformation of the bound ligands. Complementary use of docking calculations showed that the two peptides, which differ in the configuration of two carbons, exhibit different modes of interaction with the protein.
Mari et al. (2010), investigated the binding of various cyclopeptides containing the RGD and NGR motifs onto two human cancer cell lines, including a non-small cell lung carcinoma (MR300) and a melanoma (MSR3) cell line, which display different phenotypes for CD13 and avβ3. Both cell lines express avβ3 but differ in the expression of CD13; the MR300 cell express CD13(avβ3+ CD13+ cells) whereas MSR3 cells are negative (avβ3+ CD13- cells). Literature structural and biochemical data showed that the RGD and iso DGR motifs can effectively bind (low μΜ affinities) to avβ3, unlike the NGR peptide which interacts with CD13 on tumor vessels, thus, indicating that NGR and RGD/isoDGR are separate moieties binding to different receptors. In the presence of avβ3sh CD13- cells in which efficient silencing (>70 %) of avβ3 integrin was achieved, the CRGDC and CisoDGRC resulted in positive NOE effects (Fig. 24). It was concluded that in experiments using living cells, the change in sign of the NOE cross-peaks is determined primarily by the ligand–protein interactions rather than due to changes in medium viscosity.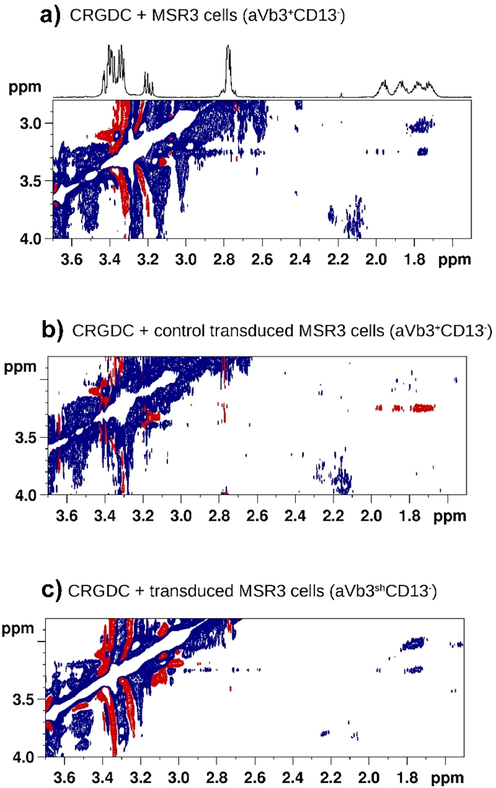
A) Tr-NOESY spectrum (mixing time = 100 ms) of CRGDC in the presence of αvβ3+ CD13+ cells: the crosspeaks deriving from correlations of the arginine γ/δ protons of the ligand are negative (blue), with the same sign as the diagonal. The flipped sign in NOE cross-peaks indicates that CRGDC interacts with a cell surface receptor. b) Tr-NOESY spectrum (mixing time = 600 ms) of CRGDC in the presence of β3-silenced (αvβ3shCD13-) cells: the cross-peaks (red) deriving from correlations of the arginine γ/β and γ/δ protons of the ligand have opposite phase with respect to the diagonal, indicating that the molecule is free in solution. c) As expected, CRGDC binds to its receptor also in mock transduced cells (Tr-NOESY control experiment) (reprinted with permission of Wiley-VCH, from Mari et al., 2010).
The binding of cyclic RGD peptidomimetic to a5B1 integrin, overexpressed in breast cancer cells, was investigated with the combined use of NMR (STD and 2D Tr-NOESY) and docking calculations (Guzzetti et al., 2017). The ligand with the larger number of STD signals showed the highest potency with nMI50 value. The binding of an antituberculosisa non-pathogenic bacterial system was investigated by NMR (Bouvier et al., 2019). The variable intensities of the experimental STD signals of the ligand were compared with the computational values for a set of conformations for a model building of the hit-target interaction at atomic resolution.
Primikyri et al. (2018) reported a combined STD and 2D-NOESY experiment to monitor the binding of natural products and synthetic analogues to the anti-apoptotic Bcl-2 protein (Primikyri et al., 2014) inside living human T-leukemic cells. Experiments were performed with quercetin and a quercetin-alanine bioconjugate. Quercetin uptake in jurkat cells was shown to be very rapid and effective (Fiorani et al., 2010). The quercetin-alanine bioconjugate showed 2-bold increased permeability compared to quercetin in model cell line (Kim et al., 2009; Kellici et al., 2017). The STD spectrum shows that all protons of 3΄-quercetin-alanine were involved in interactions with intracellular receptors (Fig. 25A). Addition of the inhibitor HA14-1, which binds selectively to the BH3 pocket of Bcl-2 with IC50 = 9 μM (Wang et al., 2000; Manero et al., 2006) involved in interactions with intracellular receptors (Fig. 25A) shows a significant reduction of the STD signal intensities of the quercetin-amino acid hybrid by a factor of ∼ 60 %. This result shows that the selective inhibitor HA14-1 competes favorably the hybrid molecule to the BH3 domain of Bcl-2. Further confirmation of this interaction was obtained when a similar STD experiment was performed in Jurkat Puro cells which do not overexpress the Bcl-2 protein. In this case the STD signal intensities of the hybrid molecule were reduced by 35 % compared to the STD intensities in jurkat cells. Primikyri et al. (2018), furthermore, investigated conformational changes of 3΄ quercetin-alanine upon binding to Bcl-2 at a cellular level. Strong negative Tr-NOEs, between the aromatic protons H5΄– Η6΄, H2΄– Η5΄ and H2΄– Η8 and between the methyl and Hα protons of the alanine residue were observed. It is worth noting that H2΄– Η5΄ and H2΄– Η8 cross-peaks were not present in the spectrum of the conjugated molecule in buffer solution. 2D Tr-NOESY experiments, therefore, are very effective in investigating the bound conformation of natural products in their native cellular environment. Spectral acquisition parameters, however, should ensure cell viability (∼2h) throughout the Tr-NOESY experiment. Of particular interest are also similar experiments with taxifolin. The STD spectrum shows that all protons of taxifolin are involved in intracellular interactions. Addition of the selective inhibitor HA4-1 did not affect the STD amplitude of taxifolin which demonstrated that this natural product does not interact with Bcl-2 at the cellular level.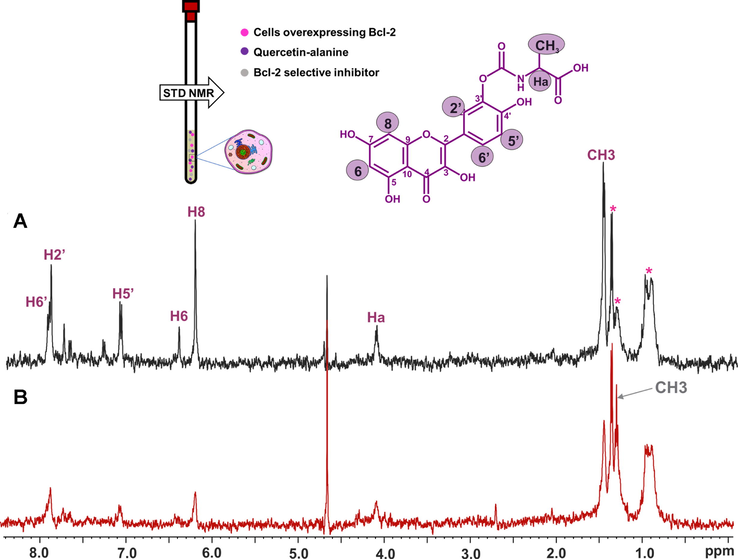
STD 500 MHz in-cell NMR spectra of (A) 3′ quercetin-alanine (purple color annotation) (1.4 mm) within Jurkat Bcl-2 cells and (B) 3′ quercetin-alanine (1.4 mm) within Jurkat Bcl-2 cells in the presence of the selective inhibitor HA14-1 (gray color annotation) (0.2 mm). Residual signals from the cells appear at 0.97 and 1.35 ppm annotated with asterisks. The multiplet at 1.4 ppm annotated with an asterisk is attributed to alanine and N-carboxy alanine due to hydrolysis of the quercetin conjugate (reprinted with permission of Federation of European Biochemical Societies, from Primikyri et al., 2018).
4.3 Bioreactors for real-time in-cell NMR
The sensitivity problems associated with in-cell NMR necessitate the need of high cell densities in the NMR tube which result in fast depletion of oxygen, of cellular nutrients, intracellular pools of ATP, glucose and NADPH. This may affect structure–function of proteins and increase of cell apoptosis within 1 to 2 h (Siegal and Selenko, 2019; Luchinat et al., 2022). To overcome the problem of low cell viability and maintain a stable metabolic state of cells for several hours or even days, flow-probe bioreactors were developed for investigating cellular metabolism (Freyer et al., 1990; Gillies et al., 1993). Emphasis has been given to the design of bioreactors for narrow-bore (5 mm) magnets (Kubo et al., 2013; Breindel et al., 2018; Cerofolini et al., 2019; Luchinat et al., 2020). Using a commercially available sealed flow unit, a 90 % cell viability and metabolic activity up to 72 h was achieved for HEK 293 T cells encapsulated in agarose hydrogels (∼3 x107 cells) while a nutrient flow was maintained with the use of an FPLC pumb (Luchinat et al., 2020). This bioreactor set up allowed the investigation of the binding of two inhibitors (acetazolamide and methazolamide) to human carbonic anhydrase (CA2) using real-time 1D 1H NMR spectra. Time-dependent binding curves were interpreted in terms of different cell membrane permeability of the two inhibitors. Although further research is needed to understand the effect of different hydrogels on cell phenotype (Luchinat et al., 2020; Mateos et al., 2020), the use of bioreactors for real-time in-cell NMR has a promising future.
5 Summary and outlook
In-tube NMR monitoring is an established method for obtaining information about catalyzed reactions of immobilized enzymes, investigation of the capacity of natural products to serve as enzyme substrates and an excellent tool for studies of interactions of biotransformed products with protein targets. Investigation of biocatalyzed reactions with the use of immobilized microbes or cell suspensions is also a fruitful future research area. The NMR methods of investigating enzymatic biotransformations, of extracellular (Tomaselli et al., 2023) and intracellular metabolites can be complemented with comprehensive computational tools for small molecule metabolism prediction and identification (Djoumbou-Feunang et al., 2019).
The ligand-observed STD and INPHARMA/ΙLOE NMR techniques, when used in combination, were shown to be of broad applicability since they are: (i) not limited by the protein molecular weight, (ii) can be utilized for multiple natural product binding screening and (iii) are capable for investigating atomic level bound conformation (in conjugation with computations), provided that a reference bound molecule of known structure is available.
Ligand-observed STD and 2D NOESY in cell NMR experiments were shown to be a fruitful approach for protein–ligand interactions in the field of natural products since they utilize label-free samples, they can provide fast analysis, and are less constrained in terms of types of cells and protein-target expression compared to protein-observed in-cell NMR methods with selectively labeled proteins. The problem of cell viability can be alleviated with the use of bioreactors for real-time in-cell NMR which is a promising field for further investigation. Contrary to protein-observed approaches, however, information of the protein-target is not specifically assessed, unless competition experiments with protein-specific inhibitors at a cellular level and protein overexpression can be achieved.
The above ligand-observed methods are not limited to protein–ligand interactions, since they have been successfully applied to DNA-ligand interaction even at cellular level (Krafcikova et al., 2019). In the long term it would be expected that the ligand-observed potentialities will be further expanded with ultra-high field instrumentation equipped with cryoprobes.
Acknowledgements
I would like to thank Prof. A.G. Tzakos, Prof. H. Stamatis, Prof. A.I. Koukou, Dr. A. Primikyri, Dr. G. Papamokos and Dr. E Alexandri (University of Ioannina) that have contributed with their research work to the elaboration of this review. The research has been co-financed by the Operational Program “Human Resources Development, Education and Lifelong Learning” by the European Union (European Social Fund) and Greek National Funds (EDBM34 MIS 82309) and from the Hellenic Foundation for Research and Innovation (H.F.R.I.) under the “First Call for H.F.R.I. Research Projects to support Faculty members and Researchers and the procurement of high-cost research equipment grant” (Project Number: 2050).
References
- NMR and computational studies reveal novel aspects in molecular recognition of unsaturated fatty acids with non-labeled serum albumin. FEBS Journal. 2022;289:5617-5636.
- [CrossRef] [Google Scholar]
- Atomic-resolution monitoring of protein maturation in live human cells by NMR. Trends Nat. Chem. Biol.. 2013;9:297-299.
- [CrossRef] [Google Scholar]
- Quantitative 1H-NMR analysis reveals steric and electronic effects on the substrate specificity of benzoate dioxygenase in Ralstonia eutropha B9. J. Industr Microb. Biotechnol.. 2022;49:kuac006.
- [CrossRef] [Google Scholar]
- Combined docking and molecular dynamics simulations to enlighten the capacity of Pseudomonas cepacia and Candida antarctica lipases to catalyze quercetin acetylation. J. Biotechnol.. 2011;156:203-210.
- [CrossRef] [Google Scholar]
- Target engagement and binding mode of an antituberculosis drug to its bacterial target deciphered in whole living cells by NMR. Biochemistry. 2019;58(6):526-533.
- [CrossRef] [Google Scholar]
- Biotransformations monitored in situ by proton nuclear magnetic resonance spectroscopy. Trends Biotechnol.. 2000;18:197-202.
- [CrossRef] [Google Scholar]
- Real-time in-cell Nuclear Magnetic Resonance: Ribosome-targeted antibiotics modulate quinary protein interactions. Biochemistry. 2018;57:540-546.
- [CrossRef] [Google Scholar]
- 19F NMR viewed through two different lenses: Ligand-observed and protein-observed 19F NMR applications for fragment-based drug discovery. RSC Chem. Biol.. 2021;2(5):1312-1330.
- [CrossRef] [Google Scholar]
- NMR-based analysis of protein–ligand interactions. Anal. Bioanal. Chem.. 2014;406:943-956.
- [CrossRef] [Google Scholar]
- NMR in natural products: understanding conformation, configuration and receptor interactions. Nat. Prod. Rep.. 2012;29:536-554.
- [CrossRef] [Google Scholar]
- Real-time insights into biological events: In-cell processes and protein-ligand interactions. Biophys. J.. 2019;116:239-247.
- [CrossRef] [Google Scholar]
- Exploring the “forgotten”- OH NMR spectral region in natural products. Chem. Commun.. 2010;46:3589-3591.
- [CrossRef] [Google Scholar]
- Rapid and direct low micromolar nmr method for the simultaneous detection of hydrogen peroxide and phenolics in plant extracts. J. Agric. Food Chem.. 2012;60:4508-4513.
- [CrossRef] [Google Scholar]
- Determination of polyphenolic phytochemicals using highly deshielded –OH 1H-NMR signals. Phytochem. Anal.. 2017;28(3):159-170.
- [CrossRef] [Google Scholar]
- Enriching the biological space of natural products, through real time biotransformation monitoring: The NMR tube bioreactor. BBA - General Subjects. 2018;1862:1-8.
- [CrossRef] [Google Scholar]
- The NMR tube bioreactor. Method. Enzymol.. 2020;633:71-101.
- [CrossRef]
- Enzymatic acylation of flavonoids: Effect of the nature of the substrate, origin of lipase, and operating conditions on conversion yield and regioselectivity. J. Agric. Food Chem.. 2007;55:9496-9502.
- [CrossRef] [Google Scholar]
- Direct observation of ligand binding to membrane proteins in living cells by a Saturation Transfer Double Difference (STDD) NMR spectroscopy method shows a significantly higher affinity of integrin αIIbβ3 in native platelets than in liposomes. J. Am. Chem. Soc.. 2005;127:916-919.
- [CrossRef] [Google Scholar]
- An approach based on Density Functional Theory (DFT) calculations to assess the Candida antarctica lipase B selectivity in rutin, isoquercitrin and quercetin acetylation. J. Mol. Cat. B: Enzym.. 2010;66:325-331.
- [CrossRef] [Google Scholar]
- Biotransformer: a comprehensive computational; tool for small molecule metabolism prediction and metabolite identification. J. Cheminform.. 2019;11:1-25.
- [CrossRef] [Google Scholar]
- Catalytic mechanism of human UDP-glucose 6-dehydrogenase: in situ proton NMR studies reveal that the C-5 hydrogen of UDP-glucose is not exchanged with bulk water during the enzymatic reaction. Carbohydr. Res.. 2012;356:209-214.
- [CrossRef] [Google Scholar]
- The extraordinary ligand properties of human serum albumin. IUBMB Life. 2005;57(12):787-796.
- [CrossRef] [Google Scholar]
- Mitochondria accumulate large amounts of quercetin: prevention of mitochondrial damage and release upon oxidation of the extramitochondrial fraction of the flavonoid. J. Nutr. Biochem.. 2010;21:397-404.
- [CrossRef] [Google Scholar]
- Reaction monitoring using online vs tube NMR spectroscopy: seriously different results. Magn. Reason. Chem.. 2016;54:451-456.
- [CrossRef] [Google Scholar]
- Live cell NMR. Ann. Rev. Biophys.. 2014;43:171-192.
- [CrossRef]
- A system for viably maintaining a stirred suspension of multicellular spheroids during NMR spectroscopy. NMR Biomed.. 1990;3:195-205.
- [CrossRef] [Google Scholar]
- Structural basis of the drug-binding specificity of human serum albumin. J. Mol. Biol.. 2005;353:38-52.
- [CrossRef] [Google Scholar]
- Design and application of NMR-compatible bioreactor circuits for extended perfusion of high-density mammalian cell cultures. NMR Biomed.. 1993;6:95-104.
- [CrossRef] [Google Scholar]
- Insights into the binding of cyclic RGD peptidomimetics to α5β1 integrin by using live-cell NMR and computational studies. ChemOpen. 2017;6:128-136.
- [CrossRef] [Google Scholar]
- Discovery of small molecule WWP2 ubiquitin ligase inhibitors. Chem. Eur. J.. 2018;24:17677-17680.
- [CrossRef] [Google Scholar]
- Hamatsu, J., O’Donovan, D., Tanaka, T., Shirai, T., Hourai, Y., Mikawa, T., Ikeya, T., Mishima, M., Boucher, W., O. Smith, B. Laue, E.D., Shirakawa, M., Ito, Y., 2013. High-resolution heteronuclear multidimensional NMR of proteins in living insect cells using a baculovirus protein expression system. J. Am. Chem. Soc. 135, 1688-1691. https://doi.org/10.1021/ja310928u.
- Serum albumin as a primary non-covalent binding protein for nitro-oleic acid. Int. J. Biomol. Micromol.. 2022;203:116-129.
- [CrossRef] [Google Scholar]
- CORCEMA refinement of the bound ligand conformation within the protein binding pocket in reversibly forming weak complexes using STD-NMR intensities. J. Magn. Reason.. 2004;168(1):36-45.
- [CrossRef] [Google Scholar]
- Rational design and structure-activity relationship studies of quercetin-amino acid hybrids targeting the anti-apoptotic protein Bcl-xL. Org. Biomol. Chem.. 2017;15:7956-7976.
- [CrossRef] [Google Scholar]
- In vitro solubility, stability and permeability of novel quercetin-amino acid conjugates. Bioorg. Med. Chem.. 2009;17:1164-1171.
- [CrossRef] [Google Scholar]
- Kontogianni, V., Charisiadis, P., Primikyri, A., Pappas, C.G., Exarchou, V., Tzakos, A.G., Gerothanassis, I.P., 2013. Ηydrogen bonding probes of phenol -OH groups. Org. Biomol. Chem. 11, 1013-1025. https://doi.org/10.1039/C2OB27117F.
- Monitoring DNA−ligand interactions in living human cells using NMR spectroscopy. J. Am. Chem. Soc.. 2019;141:13281-13285.
- [CrossRef] [Google Scholar]
- Correspondence of fatty acid and drug binding sites on human serum albumin: A two-dimensional nuclear magnetic resonance study. Biochemistry. 2013;52:1559-1567.
- [CrossRef] [Google Scholar]
- A Gel-encapsulated bioreactor system for NMR Studies of protein–protein interactions in living mammalian cells. Angew. Chem. Int. Ed.. 2013;52:1208-1211.
- [CrossRef] [Google Scholar]
- Unexpected enzyme-catalyzed regioselective acylation of flavonoid aglycones and rapid product screening. Org. Biomol. Chem.. 2012;10:1739-1742.
- [CrossRef] [Google Scholar]
- Inter ligand Overhauser effects in type II dihydrofolate reductase. Biochemistry. 2001;40:4242-4252.
- [CrossRef] [Google Scholar]
- In-cell NMR: From target structure and dynamics to drug screening. Curr. Op. Struct. Biol.. 2022;74:102374
- [CrossRef] [Google Scholar]
- Real-time quantitative in-cell NMR: Ligand binding and protein oxidation monitored in human cells using multivariate curve resolution. Anal. Chem.. 2020;92:9997-11006.
- [CrossRef] [Google Scholar]
- Radio signals from live cells: The coming of age of in-cell solution NMR. Chem. Rev.. 2022;122:9267-9306.
- [CrossRef] [Google Scholar]
- Ma, J., Cao, Q., McLeod, S.M., Ferguson, K., Gao, N., Breeze, A.L., Hu, J., 2015. Target-based whole-cell screening by 1H NMR spectroscopy. Angew. Chem. Int. Ed. 54, 4764-4767. Angew. Chem. Int. Ed. 54, 4764-4767. https://doi.org/10.1002/anie.201410701.
- Real-Time monitoring of new delhi metallo-b-lactamase activity in living bacterial cells by 1H NMR spectroscopy. Angew. Chem. Int. Ed.. 2014;53:2130-2133.
- [CrossRef] [Google Scholar]
- NMR methods to characterize protein-ligand interactions. Angew. Nat. Prod. Commun.. 2019;14(5):1-17.
- [CrossRef] [Google Scholar]
- The small organic compound HA4-1 prevents Bcl-2 interaction with Bax to sensitize malignant glioma cells to induction of cell death. Cancer Res.. 2006;66:2757-2764.
- [CrossRef] [Google Scholar]
- The interaction of saccharides with antibodies. A 3D view by using NMR. In: Kosma P., Müller-Loennies S., eds. Anticarbohydrate Antibodies. Vienna: Springer; 2012.
- [CrossRef] [Google Scholar]
- 1D Saturation transfer difference NMR experiments on living cells: the DC-SIGN/oligomannose interaction. Angew. Chem. Int. Ed.. 2005;27:296-298.
- [CrossRef] [Google Scholar]
- 2D TR-NOESY experiments interrogate and rank ligand–receptor interactions in living human cancer cells. Angew. Chem. Int. Ed.. 2010;49:1071-1074.
- [CrossRef] [Google Scholar]
- Solvent-dependent structures of natural products based on the combined use of DFT calculations and 1H-NMR chemical shifts. Molecules. 2019;2:2290.
- [CrossRef] [Google Scholar]
- NMR characterization of surface receptor protein interactions in live cells using methylcellulose hydrogels. Angew. Chem. Int. Ed.. 2020;59:3886-3890.
- [CrossRef] [Google Scholar]
- NMR spectroscopy techniques for screening and identifying ligand binding to protein receptors. Angew. Chem. Int. Ed.. 2003;42:864-890.
- [CrossRef] [Google Scholar]
- Differential epitope mapping by STD NMR spectroscopy to reveal the nature of protein–ligand contacts. Angew. Chem. Int. Ed.. 2017;56:15289-15293.
- [CrossRef] [Google Scholar]
- Exploring multi-subsite binding pockets in proteins: DEEP-STD NMR fingerprinting and molecular dynamics unveil a cryptic subsite at the GM1 binding pocket of cholera toxin B. Chem. Eur. J.. 2020;26:10024-10034.
- [CrossRef] [Google Scholar]
- Inter-ligand STD NMR: An efficient 1D NMR approach to probe relative orientation of ligands in a multi-subsite protein binding pocket. Pharmaceuticals. 2022;15:1030.
- [CrossRef] [Google Scholar]
- AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem.. 2009;30:2785-2791.
- [CrossRef] [Google Scholar]
- Novel determination of the total phenolic contents in crude plant extracts by the use of 1H NMR of the -OH spectral region. Anal. Chim. Acta. 2011;688:54-60.
- [CrossRef] [Google Scholar]
- In situ structural biology using in-cell NMR. Biochim. Biophys. Acta Gen Subj.. 2020;1864:129364
- [CrossRef] [Google Scholar]
- Crystallography-independent determination of ligand binding modes. Angew. Chem. Int. Ed.. 2008;47:7736-7740.
- [CrossRef] [Google Scholar]
- The INPHARMA technique for pharmacophore mapping: a theoretical guide to the method. J. Magn. Reson.. 2009;200:64-73.
- [CrossRef] [Google Scholar]
- Functional cell-based screening and saturation transfer double-difference NMR have identified haplosamate A as a cannabinoid receptor agonist. ACS Chem. Biol.. 2009;4:139-144.
- [CrossRef] [Google Scholar]
- STD and Tr-NOESY NMR study of receptor-ligand interactions in living cancer cells. Chem. Bio. Chem.. 2011;12:695-699.
- [CrossRef] [Google Scholar]
- Direct binding of Bcl-2 family proteins by quercetin triggers its pro-apoptotic activity. ACS Chem. Biol.. 2014;9:2737-2741.
- [CrossRef] [Google Scholar]
- Probing the Interaction of a quercetin bioconjugate with bcl-2 in living human cancer cells with in-cell NMR spectroscopy. FEBS Letters.. 2018;592:3367-3379.
- [CrossRef] [Google Scholar]
- Structural basis of artemisinin binding sites in serum albumin with the combined use of NMR and docking calculations. Molecules. 2022;27:5912.
- [CrossRef] [Google Scholar]
- Targeting norovirus infection-multivalent entry inhibitor design based on NMR experiments. Chem Eur. J.. 2011;17:7442-7453.
- [CrossRef] [Google Scholar]
- The INPHARMA method: Protein-mediated interligand NOEs for pharmacophore mapping. Angew. Chem. Int. Ed.. 2005;44:4172-4175.
- [CrossRef] [Google Scholar]
- Locating high-affinity fatty acid-binding sites of albumin by X-ray crystallography and NMR spectroscopy. Proc. Nat. Acad. Sci. USA. 2005;102:17958-17963.
- [CrossRef] [Google Scholar]
- Siskos, M.G., C.G., Tzakos, A.G., Gerothanassis, I.P., 2015. Accurate ab initio calculations of O–H⋯O and O–H⋯-O proton chemical shifts: Towards elucidation of the nature of the hydrogen bond and prediction of hydrogen bond distances. Org. Biomol. Chem. 13, 8852-8868. https://doi.org/10.1039/C5OB00920K.
- Investigation of solute-solvent interactions in phenol compounds: Accurate ab initio calculations of solvent effects on 1H NMR shieldings. Org. Biomol. Chem.. 2013;11:7400-7411.
- [CrossRef] [Google Scholar]
- Hydrogen atomic positions of O-H···O hydrogen bonds in solution and in the solid state: The synergy of quantum chemical calculations with 1H-NMR chemical shifts and X-ray diffraction methods. Molecules. 2017;22(3):415.
- [CrossRef] [Google Scholar]
- Optimization of the interligand Overhauser effect for fragment linking: Application to inhibitor discovery against Mycobacterium tuberculosis pantothenate synthetase. J. Am. Chem. Soc.. 2010;132:4544-4545.
- [CrossRef] [Google Scholar]
- Human serum albumin-specific recognition of the natural herbal extract of Stryphnodendron polyphyllum through STD NMR, hyphenations and docking simulation studies. RSC Adv.. 2015;5:23431-23442.
- [Google Scholar]
- Theillet, F.-X. and Luchinat E., 2022. In-cell NMR: Why and how? Progr. NMR Spectrosc. 132-133, 1-112. https://doi.org/10.1016/j.pnmrs.2022.04.002.
- Theillet, F.-X., 2022. In-Cell structural biology by NMR: The benefits of the atomic scale. Chem. Rev.122, 1-9497-9570. https://doi.org/10.1021/acs.chemrev.1c00937.
- Bacteria as sensors: Real-time NMR analysis of extracellular metabolites detects sub-lethal amounts of bactericidal molecules released from functionalized materials. Biochim. Biophys. - Gen. Subj.. 2023;1867(1):130253
- [CrossRef] [Google Scholar]
- Perfluoro- tert-Butyl hydroxyprolines as sensitive, conformationally responsive molecular probes: detection of protein kinase activity by 19F NMR. ACS Chem. Biol.. 2020;15(4):1096-1103.
- [CrossRef] [Google Scholar]
- Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem.. 2010;31:455-461.
- [CrossRef] [Google Scholar]
- Tsagogiannis, E., Vandera, E., Primikyri, A., Asimakoula, S., Tzakos, A.G., Gerothanassis, I.P., Koukkou, A.-I., 2021. Characterization of protocatechuate 4,5-dioxygenase from pseudarthrobacter phenanthrenivorans sphe3 and in situ reaction monitoring in the NMR tube. Int. J. Mol. Sci. 22, 9647. DOI: 10.3390/ijms22179647.
- Fluorine nuclear magnetic resonance-based assay in living mammalian cells. Anal. Biochem.. 2016;495:52-59.
- [CrossRef] [Google Scholar]
- Recent developments and applications of saturation transfer difference nuclear magnetic resonance (STD NMR) spectroscopy. Mol. BioSyst.. 2013;9:571-577.
- [CrossRef] [Google Scholar]
- Structure-based discovery of an organic compound that binds Bcl-2 protein and induces apoptosis of tumor cells. Proc. Natl. Acad. Sci.. 2000;97:7124-7129.
- [CrossRef] [Google Scholar]







