Metabolic Profiling of Mimusops elengi Linn. Leaves extract and in silico anti-inflammatory assessment targeting NLRP3 inflammasome
⁎Corresponding author. dinaphd888@gmail.com (D.F. Sayed)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Abstract
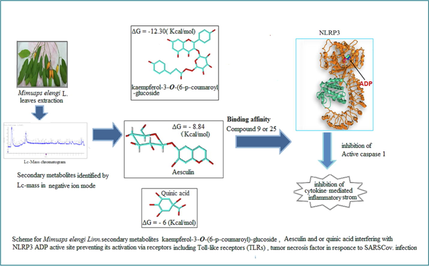
Mimusops elengi Linn. Secondary metabolites of flavonoids, phenolic acids, coumarin classes and stilbene were identified by UPLC/ESI-QTOF-HRMS/MS technique with negative ion detection. Major Mimusops elengi flavonoids included Myricitrin, Myricetin, and Kaempferol-3-O-alpha-L-rhamnoside. The most abundant Coumarin and phenolic acids detected in the chromatogram included aesculin and quinic acid respectively. Down regulation of NLRP3 inflammasome activation inhibits the severe inflammatory responses caused by virus infection. Studying in silicobinding affinity of flavonoids, coumarins and phenolic acid in M. elengi leaves extract against the ADP binding site of NLRP3 protein (PDB code: 6NPY) demonstrated that investigated compounds have docking scores ranged from −6.20 to −12.30 kcal/mol. The best score was achieved by kaempferol-3-O-(6-p-coumaroyl) -glucoside(Compound 9) followed by aesculin (Compound 25) while Quinic acid (Compound 20) showed the lowest affinity toward ADP-binding site of NLRP3.
Keywords
Mimusops elengi
Secondary metabolites
Phytochemistry
Corona virus
LC-Mass
- EtOAC
-
Ethyl acetate extract
- GMP-AMP
-
Cyclic guanosine monophosphate–adenosine monophosphate
- NOD2
-
Nucleotide-binding oligomerization domain-containing protein 2
- I IFNs
-
The type-I interferons
- IL-1β
-
Interleukin 1 beta (IL-1β) also known as leukocytic pyrogen or lymphocyte activating factor
- NLRP3
-
NLR family pyrin domain containing 3
- PAMPs
-
Pathogen-associated molecular patterns
- RT
-
Retention time
- UPLC
-
Ultra-performance liquid chromatography
Abbreviations
1 Introduction
The Sapotaceae are a suborder of the Ericales that includes flowering plants and splits into 53 genera and five tribes. Furthermore, 1250 species. Plants of this family have high economic value. They also give the locals a sizable source of support for their way of life. In addition to their economic worth, this family's extract and metabolites have medicinal activity like anti-inflammatory effects (Rodrigues et al., 2017) antiulcer (Baky et al., 2022), cytotoxic (Mogue et al., 2019), anticancer and scavenging free radicals (Ali et al., 2022). A medium-sized evergreen tree of the sapotaceae family called Mimusops elengi Linn, can be found in tropical forests, whose distribution extends to India (Warrier, 1993). Different parts of M. elengi have medicinal properties which possess antibacterial, (Moghadam et al., 2020) antifungal (Lee et al., 2018) and antioxidant properties (Boonyuen et al., 2009). Pulmonary inflammation is initiated through modulation of NLRP3 inflammasome by SARS-CoV related protein triggered in the macrophage via chemokines. As a result, using the NLRP3 inflammasome as a target for drugs that aim to reduce corona virus-related inflammation is beneficial. Plant-originated antioxidants such as phenolics (Lopez-Corona et al., 2022), flavonoids, vitamins, carotenes and tannins, play a vital role in protecting cells from inflammation with minimal side effects (Zhang et al., 2011). Gallic acid removes free radicals and protects cells from oxidative injury(Akhtar et al., 2010),(Gao et al., 2019).The initial line of defense for the host against microbial illnesses is the innate immune system (Stravalaci et al., 2022). The innate immune system plays a critical role in viral diseases by locating and eliminating infectious cells as well as creating an adaptive immune response (Diamond & Kanneganti, 2022). After a virus infection, host cells with pattern recognition receptors (PRRs) recognise PAMPs, or pathogen-associated molecular patterns and identify specific virus components like nucleic acids, protein, or lipids, and then immediately trigger immune cells (Li & Wu, 2021). The primary sensors of viral infection are retinoic acid-inducible gene-I (RIG-I) -like receptors, which are found on and inside immune cells (Rehwinkel & Gack, 2020). The activation of intracellular signalling cascades by Toll-like receptors, NOD-like receptors, and cyclic GMP-AMP synthase results in the release of type I IFNs as well as pro-inflammatory cytokines and chemokines (Mdkhana et al., 2021). These intracellular signalling cascades not only trigger an innate antiviral response but also cause the expression of co-stimulatory molecules crucial for the start of an adaptive immune response (Schultze & Aschenbrenner, 2021), (Primorac et al., 2022). Pyroptotic cell death controlled by the Nod-like receptor family, pyrin domain-containing 3 (NLRP3) inflammasome activation promotes the formation of intracellular multiprotein complexes known as inflammasomes, providing this essential extra endogenous adjuvant function (Jha & Ting, 2009).
2 Materials and methods
2.1 Chemicals
Organic solvents or Methanol (MeOH), dichloromethane, ethyl acetate, butanol, petroleum ether, Chloroform(CHCL3), and thin layer chromatography (TLC) Silica Gel 60F254 Plates were purchased from Sigma Aldrich. Sulphuric acid (H2SO4), and Ferric chloride from El-gomhouria Company. Silica gel G was obtained from Merck. While, sephadex LH-20 was obtained from Pharmacia, Uppsala, Sweden.
2.2 Plant material
Fresh Mimusops elengi L. leaves were collected from the raised plant in an Al Zohria garden, Cairo, Egypt. Dr. Abdelhaliem AbdelMogly Mohammed verified the authenticity of plant leaves at the department of fluorescence research and plant taxonomy and confirmed by professor Ahmed Mohammed Fawzy, head of the department of Fluorescence, at the agricultural research center. Certificate specimens are stored in the Department of Pharmacognosy's herbarium in the Faculty of Pharmacy under Registration Number ME-2016., the plant material was maintained in a container that was firmly sealed and air dried.
2.3 Preparation of M. elengi extracts
The dried powder of Mimusops elengi Linn. Leaves (1.5 kg) were subjected to exhaustive extraction with 70% (MeOH) solution (3 × 7L). The methanolic extracts were concentrated to dryness using a rotary evaporator with a vacuum pump and water bath temperature at 40OC. The dried methanolic extract (228 g) was suspended in distilled water and was defatted with petroleum-ether successfully. The defatted crude extract (192 g) was partitioned successively using a separating funnel with ethyl acetate (EtOAC) and n-butanol, to yield 25 g and 17.9 g of dry residue, respectively. The EtOAC extract was subjected to fractionation on silica gel column (110 × 3 cm, 25 g) using a stepwise gradient elution from Chloroform/ Methanol (CHCL3/MeOH) mixtures up to pure MeOH. Based on thin layer chromatography (TLC) using methanol/H2SO4 (10%) as a spray reagent, the individual 200 fractions (each 50 ml) were pooled into six collective fractions.
2.4 Liquid chromatography–electrospray ionization–tandem mass spectrometry (LC-ESI-MS/MS) metabolic Profiling of M.elengi extract.
The Metabolomics & Proteomics Unit at Cancer Hospital of Children conducted the LC-ESI-MS/MS study. The test procedure was performed according to the method described by Alqahtani et al., (2022) and Mohammed et al., (2021).The sample was prepared by macerating M. Elengi powder leaves at room temperature in petroleum ether. Then filtered and dried. The dried defatted powder was extracted with MeOH till exhaustion, then the extract was evaporated under vacuum at 40 ◦C. A weighed amount of the dry residue (50 mg) was then added to a 1 ml solution of deionized water: -methanol:acetonitrile (50:25:25). The test sample was subjected to two minutes of vortexing, ten minutes of ultrasonication, and a further ten minutes of centrifugation at 1000 rpm in this solvent mixture. The reconstitution solvent was used to dilute the sample solution, which was then injected in 10 L at a concentration of 1 g/L. To enable the filtration process, separation was done using an Axion AC system (Kyoto, Japan) attached to an autosampler system and an In-Line filter discs precolumn (0.5 m 3.0 mm, Phenomenex, USA). 3 min at 1 g/mL of the starting combination. Peaks extraction with a signal-to-noise ratio of more than 5 (non-targeted analysis) and more than 3 feature intensities of the sample-to-blank ratio were viewed using the master view for total ion chromatogram (TIC). Applying MS-DIAL 4.6 (RIKEN® Tokyo, Japan) and Reifycs Abf (Analysis Base File) Converter (Reifycs®, Tokyo, Japan) for Wiff file conversion enabled data interpretation. Electrospray ionisation (ESI) mode and a Triple TOFTM 5600 + system with a Duo-SprayTM system were used for the MS (AB SCIEX, Concord, Canada). PeakViewTM software version 2 1 was used to compare retention duration and m/z values obtained by MS and MS2 in order to identify compounds. The Manager of XIC in PeakViewTM software was used to calculate peak area values. Resulted ion chromatograms (XICs) for each targeted analyte were automatically exhibited and compared to threshold defined by user.
2.5 MTT cell viability assessment on U937 cell line
2.5.1 MTT assay according to Van de Loosdreecht et al. (1994) cell culture
1- A full monolayer sheet formed after 24 h of incubation at 37 °C with 1 X 105 cells/ml (100 UL) in the 96-well tissue culture plate. After a confluent sheet of cells had grown, the 96 well micro titer plates' growth material was decanted, and the cell monolayer had been twice washed with wash media. The test sample was diluted twice in RPMI medium with 2% serum (maintenance medium). In various wells, 0.1 ml of each dilution was examined, leaving 3 wells as controls that merely received maintenance media. The plate was tested after 37 °C of incubation. The physical characteristics of toxicity, such as partial or total loss of the monolayer, rounding, shrinkage, or cell granulation, were examined in the cells. The MTT solution (5 mg/ml in PBS) (BIO). To each well, 20ul of the MTT solution was added, and were shaken at 150 rpm for five minutes to properly blend the MTT into the media. 1 to 5 h were spent in incubation (37 °C, 5% CO2) to allow the MTT to break down. The media were disgruntled. (If required, dry plates on paper towels to remove residue). In 200Ul of DMSO, formazan (a metabolic product of the MTT) was reconstituted 5 min on a shaking table at 150 rpm to completely combine the formazan and solvent. At 620 nm, the background was subtracted while the optical density was read at 560 nm. Then, optical density and cell number had a direct correlation (Slater et al., 1963).
2.6 Docking study
The X-ray co-crystal structure of NLRP3 bound to ADP (PDB code: 6NPY) was downloaded from Protein Data Bank. The receptor was prepared by deletion of water molecules and other hetatoms, addition of polar hydrogen and Kollman charges and energy minimization and then saved in pdbqt format. All ligands were sketched using ChemBioDraw Ultra 14.0 software (CambridgeSoft Corporation), then energy minimized by MMFF94x Force Field in gas and saved in PDBQT format. Docking simulation was performed using PyRx AutoDock VINA. A grid box with dimensions of (25 × 25 × 25) Å and center coordinates X = 81.6, Y = 100 and Z = 91.5 was created. The Lamarckian Genetic Algorithm (LGA) method has been employed for docking calculations (Trott & Olson, 2010). The generated docking poses were ranked according to their docking scores, and the best energy pose was selected. The 3D and 2D interactions between the ligands and the receptor have been visualized using Discovery Studio Visualizer in order to identify the most significant interactions.
3 Results
3.1 Isolated compounds from M.elengi ethyl acetate extract
Compound (a): colourless crystal (28 mg), Molecular weight m/z 420, molecular formula C30H50O, pink-violet colour with Liebermann Burchard test (Liebermann, 1985) m.p. 215 correspondingly 1H NMR (CDCl3,400 MHz): 4.70, 4.55 (2H, s, H-29a, 29b), 3.2 (1H, m, H-3), 0.77, 0.79, 0.85, 0.94, 0.97, 1.05, and 1.65 (3H, s), respectively, 13C NMR (CDCl3, 100 MHz): δ151.0 (C-20), 109.33(C-29), 79.0(C-3), 55.30(C-5), 50.44(C-9), 48.71(C18), 48.31(C-19), 48.0(C-17), 42.19(C-14), 40.01(C-8), 39.22 (C-22), 38.76(C-4), 38.71(C-1), 38.05(C-13), 36.71(C10), 35.59(C-16), 34.28(C-7), 29.85(C-21), 27.99(C-23), 27.4(C-2), 27.04(C-15), 25.56(C-12), 21.64(C-11), 20.93(C30), 19.31(C-6), 18.32(C-28), 18.01(C-25), 17.71(C-26), 16.31(C-24), 16.05(C-27). These assignments are in agreement with the structure of lupeol (Abdullahi et al., 2013).
Compound (b1): was obtained as an amorphous white solid compound with a melting point: 190 EI-MS m/z, 426. 1H NMR diagnostic peaks occurred at 5.10 (IH, J = 3.6 Hz, H-12), 3.16 (1H, m, H-3). The 1-H NMR(400 MHz,DMSO, δ ppm) analysis 1.5601–1.4895(H1), 1.610 (H-2), 4.614 dd (J = 4.4, 11.5) (H3), 0.8013(H-5), 1.505, 1.319 (H-6), 1.5439(H-8), 1.89(H-11), 5.10 t (3.6) (H-12), 1.2893(H-18), 1.319 m(H-19), 1.9502(H-20), 0.8686 s(H-23), 0.8429 s(H-24), 0.9399 s(H-25), 0.9600 s(H-26), 1.016 s(H-27) 0.78 s(H-28), 0.7621 d(H-29), 0.842 d(H-30)13C NMR (100 MHz, CDCl3, δ ppm): 38.59 (C1), 27.2 (C2), 79.04 (C3), 38.79 (C4), 55.17 (C5), 18.38 (C6), 32.65 (C7), 40.01 (C8), 47.23 (C9), 36.95 (C10), 23.55 (C11), 121.73 (C12), 145.01 (C13), 41.72 (C14), 26.16 (C15), 26.94 (C16), 32.50 (C17), 47.64 (C18), 46.93 (C19), 31.10 (C20), 37.15 (C21), 34.74 (C22), 15.5 (C23), 28. 10 (C24), 15.60 (C25), 16.91 (C26), 26.01 (C27), 28.41 (C28), 33.36 (C29), 23.71(C30) the information above fits the structure of α - amyrin (3β - hydroxy - urs – 12 – en – 3 - ol). Obtained in mixture form with β. Amyrin (Alam & Haque, 2020).
Compound (b2): whitish, amorphous substance with a melting point between 190 and 192 °C. Amyrin triterpenoid mass is compatible with EI-MS m/z 426.3 (M + ).1H- NMR diagnostic peaks occurred in 5.04 (IH, J = 3.56 Hz, H-12), 3.13 (1H, m, H-3). The 1-H NMR (400 MHz,DMSO, δ ppm) analysis 1.438 (Hb-1), 1.378 (Ha-1), 1.408 (Hb-2), 1.438 (Ha-2) (H2), 3.1374 dd (J = 4.4, 11.5)(H-3), 0.598 (H-5), 1.478 (Hb-6), 1.8935 (Ha-6)(H-6), 1.838(H-9) 1.7950(H-11), 5.048 t (3.56) (H-12), 1.803 (H-18) 1.478 (H-19) 1.5439 (H-21) 0.658 s (H-23), 0.8563 s (H-24), 0.8095 s (H-25), 0.828 s (H-26) 1.001 s (H-27), 0.6908 s (H-28), 0.7304 s (H-29), 0.7304 s (H-30).13C NMR (CDCl3,100 MHz) δ ppm: 38.4(C1), 23.6 (C2) 80.98, (C3)37.7 (C4)55.26 (C5)18.25 (C6), 32.9 (C7), 40.0 (C8), 47.64 (C9), 36.79 (C10), 23.40 (C11), 124.43 (C12), 139.4 (C13), 42.2 (C14), 28.09 (C15),26.7 (C16) 33.75 (C17), 59.06 (C18),39.61 (C19), 39.64(C20), 31.27 (C21), 41.5 (C22), 28.07 (C23), 16.95 (C24)15.76 (C25), 17.51 (C26), 23.23 (C27), 28.96(C28), 16.92 (C29), 21.44 (C30).The presence of. β. Amyrin peaks in the 13C- NMR spectrum allow for the identification of component 2(b) as β. Amyrin in a mixture. (Okoye et al., 2014).
Compound (c):
White needles, m.p. 336 °C; MS: HR-EIMS m/z 457.441calculated. for C30H48O3); 1H NMR (DMSO, 400 MHz): 5.13 (1H, br s, H-12), 4.29 (1H, br s, OH), 3.00 (1H, dd, J = 12, 6.4 Hz, H-3), 2.099 (1H, d, J = 11.28 Hz, H-18), 1.04 (3H, s, Me-23), 0.92 (3H, s, Me-27), 0.87 (3H, s, Me-26), 0.85 (3H, s, Me-24), 0.90 (3H, d, J = 7.5 Hz, Me-30), 0.81 (3H, d, J = 6.36 Hz, Me-29), 0.68(3H, s, Me-25). 13C NMR (DMSO, 100 MHz): 39.9 (C-1), 28.7 (C-2),76.9 (C-3), 38.72 (C-4), 55.2 (C-5), 18.4 (C-6), 33.3 (C-7), 40.6 (C-8), 47.5 (C-9), 36.99 (C-10), 23.7 (C-11), 125.0 (C-12), 138.6 (C-13), 42. 1 (C-14), 28.01 (C-15), 24.2 (C-16), 47.2 (C-17), 52.8(C-18), 39.3 (C-19), 39.3 (C-20), 30.6(C-21), 36. 9 (C-22), 28.7 (C23), 15.6 (C-24), 16.4 (C-25), 17.4 (C-26), 23.7 (C-27), 178.8 (C-28), 17.4 (C-29), 21.5 (C30), Using the information provided, component (c) was determined to be 3-hydroxyolean-12-en-1–8-28-oic acid. (18α-oleanolic acid) ursolic acid (Chen et al., 2019).
Compound (d): Yellow amorphous powder (100 mg). When exposed to extended UV radiation, a yellow fluorescence patch changed to yellowish orange with Naturstoff and faint blue with FeCl3. 1H NMR (DMSO‑d6, 400 MHz): δ ppm 6.89 (2H, s, H-2‘/6‘), 6.376 (1H, brs, H-8), 6.209 (1H, brs, H-6), 3.984 (1H, brs, H3‘‘),5.2(1H, brs, H-1‘‘) 0.835 (3H, brd, H-6′' d, J = 6.4 Hz,), 3.17–4.1 (remaining sugar protons). 13C NMR (DMSO‑d6, 100 MHz): δ ppm 178.07 (C-4), 164.63 (C-7), 161.78 (C-5), 157.96 (C-2),156.87(C-9), 146.24 (C-3‘/5‘), 136.91 (C-3), 134.75 (C-4′), 120.07 (C-1‘), 108.36 (C2‘/6‘), 102.41 (C-10, C-1‘‘), 99.11 (C-6), 93.98 (C-8), 71.74 (C-4‘‘), 71.02 (C-2‘‘), 70.85 (C-3‘‘), 70.48 (C-5‘‘), 17.99 (C-6‘‘). Negative ESI-MS/MS (EI-, relative intensity %), m/z 463.2 (M - H+) (cal: 464.126) for a MF: C21H19O12, Accordingly, compound (d) was identified as myricitrin (Motlhatlego et al., 2020).
Compound (e): An amorphous yellow powder with an Rf value of 0.41 was coloured using diluted sodium hydroxide; the result was a strongly yellow solution that went colourless when diluted acid was applied, indicating the presence of flavonoids. Negative HRESI-MS/MS, (EI-, relative intensity%), m/z 479.03 (M - H + ), (cal: for molecular formula C21H20O13.1H NMR (DMSO‑d6, 400 MHz): δ 3.27∼3.63(m, sugar protons), 5.30 (1H, d, J = 7.2 Hz, H-1″), 6.202 (1H, d, J = 2.0 Hz, H-6), 6.38 (1H, d, J = 2.08 Hz, H-8),7.19 (2H, s, H-2′/H-6′); 13C NMR(DMSO‑d6, 100 MHz): δ 60.5 (C-6″),68.45 (C-4″), 71.74 (C-2″), 73.75 (C-3″),76.64 (C-5″), 93.88 (C-8), 99.15 (C-6),102.4 (C-1″),104.46, 104.30 (C-10), 108.88 (C-2′/6′), 120.28 (C-1′), 134.21 (C-3),137.30 (C-4′), 145.95 (C-3′/5′), 156.70(C-2), 156.5 (C-9), 161.68 (C-5), 164.78(C-7), 177.74 (C-4) Accordingly, compound (e) was identified as Myricetin 3-O-β-galactopyranoside that was confirmed by comparison with reported literature (Gürbüz et al., 2015).
Compound (f): was isolated as a yellow, amorphous powder that, when exposed to ammonia vapour or sprayed with AlCl3, transformed from purple to yellow under UV light. Complete acid hydrolysis resulted in the production of two sugar moieties (glucose and rhamnose) in the aqueous phase and myricetin in the organic phase. 1H-NMR (DMSOd6): (ppm) = 6.20 (1H, d, J = 1.8 Hz) H-6, 6.389 (1H, d, J = 1.8 Hz) H-8, 7.2044 (2H, s) H-2′, 7.2044(2H, s) C-6′, 5.3730 (1H, d, J = 6 Hz)H-1″, 3.24 ∼ 3.42 (m, sugar protons) H-2″, 3.4160 1H, d, J = 8.8 Hz (H-5″), 5.208 (1H) (H-1″′).13C-NMR (DMSO‑d6):157.93 (C-2), 134.21 (C-3), 178.24(C-4), 162.43(C-5), 99.15(C-6), 164.78(C-7), 94.00(C-8), 158.01(C-9), 104.46(C-10), 120.42(C-1′),109.00(C-2′),145.8 (C-3′),137.17(C-4′),145.8(C-5′), 109.00(C-6′),104.35(C-1″),73.90(C-2″),76.40 (C-3″), 71.66 (C-4″), 76.64 (C-5″), 66.57 (C-6″),102.49 (C-1″),70.85(C-2″′),70.48(C-3″′),73.5 (C-4″′),69.81 (C-5″′),17.99(C-6″′). The presence of glucose and rhamnose was deduced from the 1 H-NMR and 13C-NMR spectra. Comparing the reported data with those reported in the literature (Fossen et al., 1998) revealed that compound (f) was Myricetin-3-O-α-L-rhamnopyranosyl(1→6) glucopyranoside.
Compound (g): It is a dark yellow amorphous powder that fails the Molisch test and fluoresces brightly yellow when exposed to ammonia vapour, orange when exposed to the Naturstoff reagent, and green when exposed to the FeCl3 spray reagent. 1H NMR spectral data (400 MHz, DMSO‑d6) δ ppm 7.7 (1H, d, J = 2.2 Hz, H-2′), 7.54 (1H, dd, J = 2.0, 8.42 Hz, H-6′), 6.91 (1H, d, J = 8.48 Hz, H-5′), 6.42 (1H, d, J = 1.76 Hz, H-8), 6.20 (1H, d, J = 1.8 Hz, H-6). 13C NMR (DMSO‑d6, 100 Hz) δ C: 176.28 (C, C-4), 164.35 (C, C-7), 161.17(C, C-9), 156.6 (C, C-5), 148.14 (C, C-2), 146.5 (C, C-4′), 145.50 (C, C-3′), 136.19 (C, C-3), 122.46 (C, C-1′), 120.47 (CH, C-6′), 116.08 (CH, C-2′), 115.54 (CH, C-5′), 103.48 (C, C-10), 98.66 (CH, C-6), 93.83. As a result, component (g) was recognised as quercetin, which was verified by comparison with published data. (Hu et al., 2013).
Compound (h) was discovered as an off-white amorphous powder with a melting point of 257 °C and was seen to fluoresce faintly violet under short-wave UV light before turning deep blue when FeCl3 spray reagent was applied. A molecular ion peak at m/z 169 [MH] was visible in negative ESI-MS and corresponded to a molecular formula. Two symmetrical protons of C7H6O5 were found in the aromatic area at H 6.93 ppm in the 1H NMR spectrum data (400 MHz, DMSO‑d6) (2H, s, H-2 and H-6). Aromatic carbons were detected by 13C NMR (100 MHz, DMSO‑d6) at δC 109.1 ppm (C2, C6), 120.9 ppm (C1), 138.4 ppm (C4), 145.8 ppm (C3, C5) and carbonyl carbon at δC 167.9 ppm (–CO) (Lim et al., 2004). Compound (h) was identified as 3,4, 5-trihydroxy-benzoic acid based on the mentioned NMR data and literature comparison (Gallic acid).
Compound (I) Melting point: 227 °C; obtained as white ovoid crystals; soluble in alcohol and water but insoluble in organic solvents; negative ESI-MS analysis revealed molecular ion peak 163.24 with chemical formula C6H12O5. While Compound (I) does not diminish Fehling's solution and appears to be isomeric with rhamnose, this suggests that it is a cyclitol. The signal in its 1H NMR spectrum reveals that the structure of (I) is monocyclic. Compound (I) acetylation with acetic anhydride in the presence of triethylamine produce an oily pentacetate proved that it had five hydroxyl groups.; 1H NMR (400 MHz, D2O): δ1.78 (H6ax., ddd, J = 14, 11.5,3Hz),1.8(H6 eq.,dddd,J = 14,4.8,3.5,0.9 Hz), 3.5(H-2,t,J = 9.2 Hz), 3.64 (H-3, dd, J = 9.4, 3.1 Hz), 3.8 (H-1, ddd, J = 11.2,9,4.9 Hz),3.95(H-4, td,J = 3.2,0.9 Hz),4.04(H-5,4,5 = 3. 3 Hz) 13C NMR (100 MHz,D2O) δ33.2 (C-6), 68.5 (C-5), 68.8 (C-l), 70.9 (C-3), 72.2 (C-4), 75. 74 (C-2). The data suggest (i) to be pentahydroxy cyclohexane [(+) – quercitol](Lavaud et al., 1994).
3.2 UPLC-QTOF-ESI-MS/MS metabolic profiling
In the current study, the polyphenolic 2ry metabolites in the M. elengi leaf extract were identified using UPLC-QTOF-ESI -MS/MS''. The RT, % abundance, MS fragmentation, and MS fragment ion were represented in Table 1, Fig. 2. Identification of each peak of compounds was carried out by comparing authentic reference compound spectra and literature, fragmentation patterns in negative mode. Compounds were identified in M. elengi extract and characterized as flavonoids; Myricitrin (1), Myricetin (2), Kaempferol-3-O-alpha-L-rhamnoside (3), Quercitrin (4), Kaempferol-3-Glucuronide(5), Daidzein-8-C-glucoside (6) Phlorizin (7) Isorhamnetin-3-O-glucoside (8), Kaempferol-3-O-(6-p-coumaroyl)-glucoside (9),Okanin-4′-O-glucoside (10) Quercetin-3-Glucuronide (11), Baicalein-7-O-glucuronide (12), phenolic acid: D-(-)-Quinic acid (20) and major identified Coumarins Aesculin (25) Fig. 1.
| No. | RT (min.) | * % of abundance | [M−H]- | Molecular formula | Fragmentation | Tentatively identified compounds | References |
|---|---|---|---|---|---|---|---|
|
|||||||
| 1 | 6.42 | 40.82 | 463.0888 | C21H20O12 | 317.0309, 316.0238, 271.0260 | Myricitrin | (Saldanha et al., 2013) |
| 2 | 8.78 | 8.55 | 317.0549 | C15H10O8 | 317.043, 300.04, 271.026, 211.071, 179.0102, 112.985 | Myricetin | (Gates & Lopes, 2012) |
| 3 | 7.40 | 4.03 | 431.0982 | C21H20O10 | 285.04, 284.033, 255.03 | Kaempferol-3-O-α-L-rhamnoside | (Ibrahim et al., 2016),(Farid et al., 2022) |
| 4 | 6.74 | 2.61 | 447.0963 | C21H20O11 | 301.036, 300.0295 | Quercitrin | (Llorent-Martínez et al., 2015) |
| 5 | 5.51 | 0.75 | 461.1302 | C21H18O12 | 415.12, 392.902, 324.915, 256.92, 167.036 | Kaempferol-3-Glucuronide | (Kajdžanoska et al., 2010) |
| 6 | 8.15 | 0.46 | 415.1956 | C21H20O9 | 295, 267 | Daidzein-8-C-glucoside | (Zhang et al., 2005; Prasain et al., 2004) |
| 7 | 7.38 | 0.22 | 435.1275 | C21H24O10 | 273.08, 167.03, 123.04 | Phlorizin | (Buzgaia et al., 2021) |
| 8 | 6.56 | 0.16 | 477.1062 | C22H22O12 | 315, 179, 151 | Isorhamnetin-3-O-glucoside | (Brito et al., 2014) |
| 9 | 5.24 | 0.15 | 593.0786 | C30H26O13 | 447, 307, 285, 271 | Kaempferol-3-O-(6-p-coumaroyl)-glucoside | (Felipe et al., 2014) |
| 10 | 7.25 | 0.15 | 449.1478 | C21H22O11 | 316.02, 281.06, 256.92 | Okanin-4′-O-glucoside | (Alotaibi et al., 2022) |
| 11 | 5.78 | 0.13 | 477.0671 | C21H18O13 | 301, 112.98 | Quercetin-3-glucuronide | (Song et al., 2021) |
| 12 | 5.62 | 0.09 | 445.1425 | C21H18O11 | 269 | Baicalein-7-O-glucuronide | (Li et al., 2014) |
| 13 | 9.22 | 0.08 | 271.0618 | C15H12O5 | 185, 151, 125, 119 | Naringenin | (Chen et al., 2016) |
| 14 | 6.38 | 0.06 | 433.0002 | C20H18O11 | 301, 300, 271, 151 | Quercetin-3-D-xyloside | (Eltamany et al., 2020) |
| 15 | 9.59 | 0.04 | 298.9434 | C16H12O6 | 298.9418 | 3 5 7-trihydroxy-4′-methoxyflavone | (Brito et al., 2014) |
| 16 | 8.39 | 0.04 | 315.0686 | C16H12O7 | 315.0400 | 3′-Methoxy-4′,5,7-trihydroxyflavonol | (Elmongy et al., 2022) |
| 17 | 8.03 | 0.03 | 283.1053 | C16H12O5 | 283.0922 | Acacetin | (El-sayed et al., 2021) |
| 18 | 5.72 | 0.02 | 507.093 | C23H24O13 | 135.03372 | Syringetin-3-O-galactoside | (Flamini et al., 2015) |
| 19 | 6.16 | 0.02 | 609.1451 | C27H30O16 | 300.02704, 180.96916 | Luteolin-3′, 7-di-O-glucoside | (van Der Klift et al., 2021) |
|
|||||||
| 20 | 1.18 | 31.56 | 191.0564 | C7H12O6 | 173.044, 127.039, 111.044, 93.0346, 85.029 | D- (-)-Quinic acid | (Zhu et al., 2022; Gouveia & Castilho, 2011) |
| 21 | 1.27 | 2.79 | 179.0556 | C9H8O4 | 161.0439, 143.036, 131.036, 89.024, 81.0385, 78.9569 | Caffeic acid | (Abdelaziz et al., 2020) |
| 22 | 1.43 | 1.79 | 353.1978 | C16H18O9 | 191, 85 | Chlorogenic acid | (Managa et al., 2020) |
| 23 | 1.66 | 0.23 | 163.0391 | C9H8O3 | 143.044, 119.04, 71.022 | 3-(4- hydroxyphenyl) prop-2-enoic acid | (Liu et al., 2007) |
|
|||||||
| 24 | 8.57 | 4.28 | 405.1019 | C20H22O9 | 243.0646, 242.0590, 151.0043, 112.9839 | E-3,4,5′-Trihydroxy-3′-glucopyranosylstilbene (Astringin) | (Flamini & De Rosso, 2019) |
|
|||||||
| 25 | 6.70 | 0.18 | 339.2029 | C15H16O9 | 339, 183 | Esculin | (Elhady et al., 2022) |
| 26 | 10.13 | 0.08 | 177.0219 | C9H6O4 | 133.03,177.02 | Daphnetin | (Jiménez-López et al., 2017) |
|
|||||||
| 27 | 5.15 | 0.29 | 183.0298 | C8H8O5 | 124.01,140.01, 168.0, 183.031 | 3,4-Dihydroxymandelate | (Sule et al., 2017) |
| 28 | 1.10 | 0.35 | 245.0291 | C5H12O7P2 | 153.99, 165.06,169.01, 188.942, 245.03 | Gamma-Dimethyallyl pyrophosphate ammonium salt | (Lee et al., 2011) |
| 29 | 6.99 | 0.04 | 167.0398 | C5H4N4O3 | 167.03 | Uric acid | (Zhu et al., 2022) |
*percentage of abundance calculated from peak area (peak area of the compound / total peaks area of the identified secondary metabolites X 100.
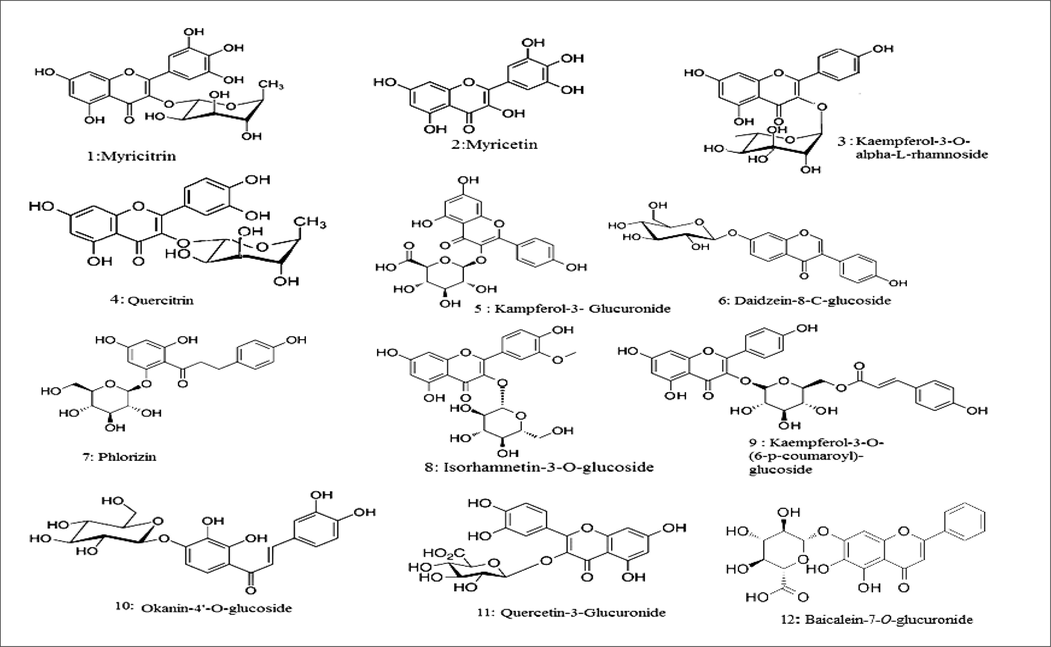
- Main compounds identified by(LC-ESI-MS/MS) Metabolite Profiling of M.elengi extract.
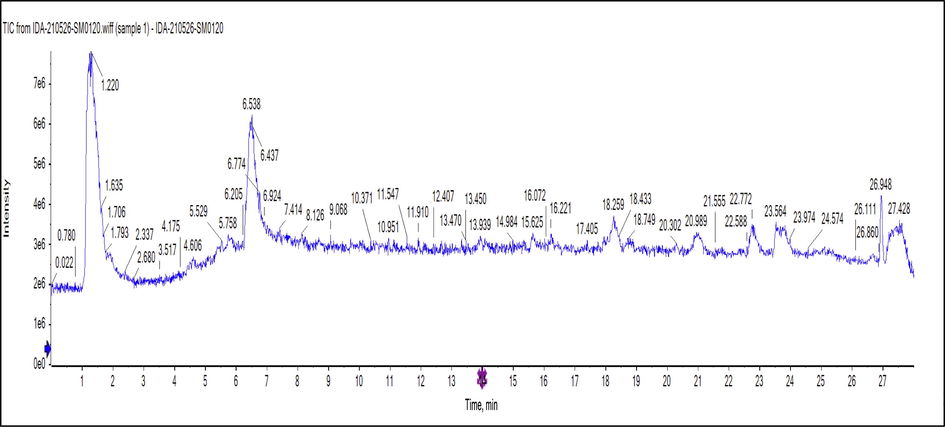
- Negative ion mode TIC mass chromatogram of M. elengi methanol extract.
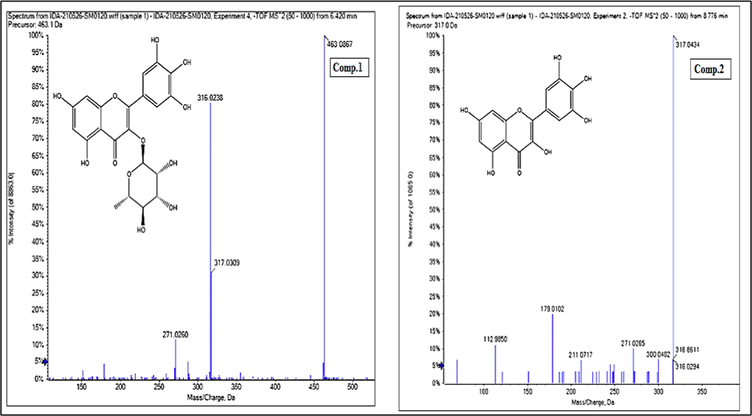
- Mass spectrum of Myricitrin (Comp.1)and Myricetin aglycone(Comp.2).
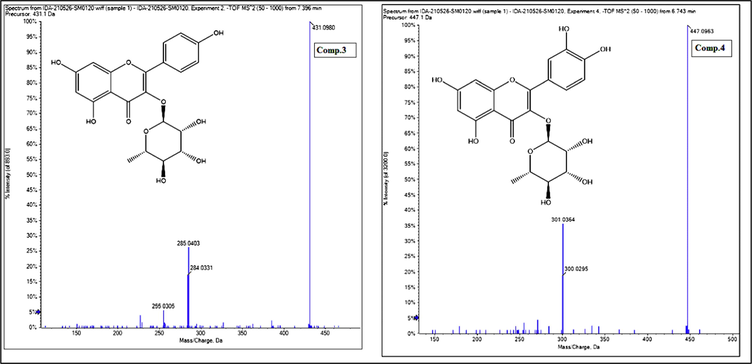
- Mass spectrum of Kaempferol-3-O-αL-rhamnoside (Comp.3) and Quercitrin(Comp.4).

- Mass spectrum of Quinic acid (Comp.20)and Caffeic acid (Comp.21).
3.3 Cytotoxicity evaluation of extract using (MTT) with 6 concentrations 1000, 500, 250, 125, 62.5, 31.25 μg/ml
To determine the ethyl acetate (EtOAC) Extract IC50, the dose–response curve was plotted. The number of viable U937 (Histocytic lymphoma) cells grown in repeated extract dilutions in comparison to control was statistically analysed. It was found as seen in Fig. 6 that the Extract, with an IC50 value of 55.8 μg/mL, had the highest cytotoxicity on the U937 cell line.
3.4 Molecular docking
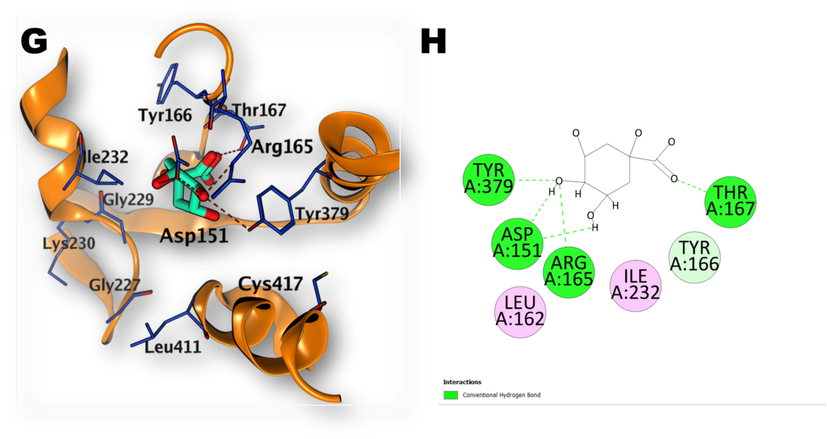
Docking simulation was performed in order to study the binding affinity and binding poses of the investigated compounds with the target protein. An induced fit method has been employed to dock each compound in the ADP binding site of the NLRP3 protein (PDB code: 6NPY). The docking scores and interactions with the active site residues are summarized in Table 2, Figs. 8–10. Compound 9 has the highest binding affinity record −12.30 (ΔG in Kcal/mol). Compound 25 Aesculin recorded −8.84 (ΔG in Kcal/mol). While compound 20 D-(-)-Quinic acid reported: −6.2 (ΔG in Kcal/mol) Table 2: Docking results of compounds (9, 25, 20) with the ADP binding site of NLRP3 (PDB code: 6NPY).
| Ligand |
Binding affinity (ΔG in Kcal/mol) |
Interaction parameters | |||
|---|---|---|---|---|---|
| Interaction | Residue | δ(Å) | E (Kcal/mol) | ||
|
Compond 9 (Kaempferol-3-O-(6-p-coumaroyl)-glucoside |
−12.30 | H-donor | Asp 151 | 2.87 | −4.5 |
| H-donor | Asp 303 | 2.70 | −2.9 | ||
| H-acceptor | His 520 | 2.85 | −1.5 | ||
| Pi-Pi | Trp 414 | 3.52 | |||
|
Compond 25 (Aesculin) |
−8.84 | H-donor | Asp 151 | 2.72 | −5.1 |
| H-acceptor | Thr 167 | 3.11 | −2.1 | ||
| H-acceptor | Thr 167 | 2.84 | −1.5 | ||
| H-acceptor | Arg 165 | 3.09 | −1.7 | ||
| Pi-Pi | Trp 414 | 3.81 | |||
| Compound20(quinic acid) | −6.20 | H-donor | Asp 151 | 2.93 | −2.1 |
| H-donor | Asp 151 | 3.21 | −1.5 | ||
| H-acceptor | Thr 167 | 3.11 | −1.2 | ||
| H-acceptor | Arg 165 | 3.04 | −2.1 | ||
| H-acceptor | Tyr 379 | 3.22 | −0.8 | ||
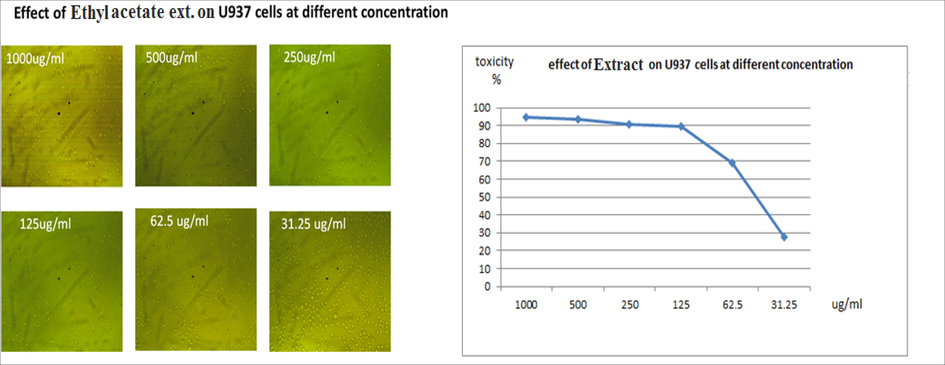
- Effect of ethyl acetate extract at different concentration on u937cell line with ic50 graph showing that concentration 55.8 had highest cytotoxic activity.
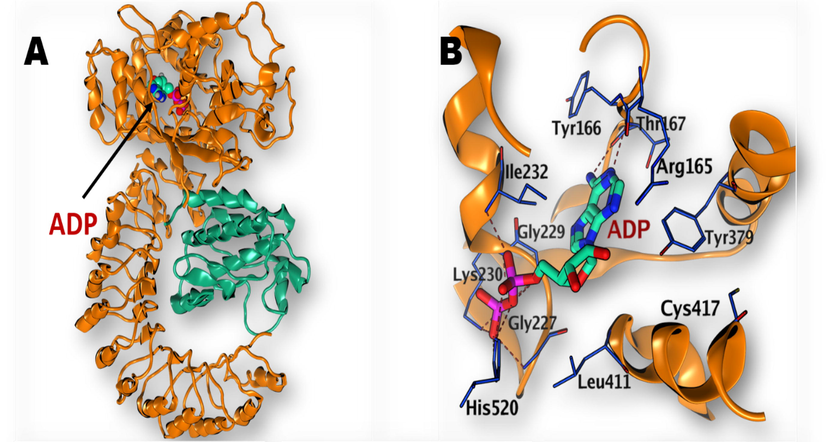
-
(A) Structure of NLRP3 with bound ADP (PDB: 6NPY) (B) Magnification of the active site of NLRP3 with bound ADP.
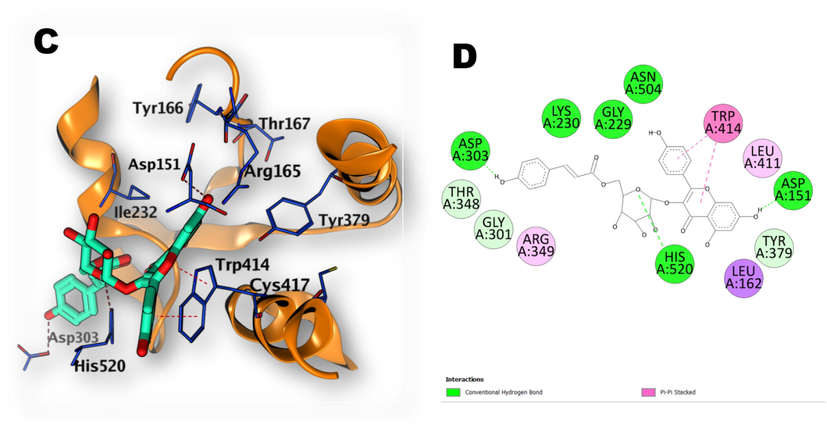
-
(C, D), 3D illustration of the docking poses of compound (9): kaempferol-3-O-(6-p-coumaroyl)-glucoside, (in ADP binding site of NLRP3 with 2D depiction of the binding interaction between compound 9 and the active site).

-
(E, F): 3D illustration of the docking poses of compound (25): Aesculin (in ADP binding site of NLRP3 with 2D depiction of the binding interaction between compound 25 and the active site).
-
(G, H): 3D illustration of the docking poses of compound (20): Quinic acid (in ADP binding site of NLRP3 with 2D depiction of the binding interaction between compound 20 and the active site).
4 Discussion
The UPLC/ESI-QTOF-HRMS/MS technique in its negative ionization mode permitted the tentative identification of 29 metabolites in M. elengi methanol extract (Fig. 2). The majority of these metabolites were flavonoids and their O- and C- glycosides, phenolic and organic acids, stilbenes, and coumarins. The identified metabolites were displayed (Table 1) based on their relative abundance in the extract, which was calculated using the total peak area of the identified metabolites. Myricitrin was the most abundant metabolite (40.82%) among all identified compounds, followed by quinic acid (31.56 %), myricetin (8.55 %), astringin (4.28 %), kaempferol-3-O-α-L-rhamnoside (4.03 %), caffeic acid (2.79%), and quercitrin (2.61 %).
4.1 Identification of flavonoids
In the negative ionisation mode of UPLC/ESI-QTOF-HRMS/MS, diagnostic mass fragments at positions 285, 301, 315, and 317 identified the aglycones as kaempferol, quercetin, isorhamnetin, and myricetin, respectively. The identification of pentosides (xylose or arabinose), hexosides (glucose or galactose), deoxyhexosides (rhamnose), and glucuronic acid, respectively, was made possible by the neutral losses of 132, 162, 146, and 176 mass units. The retro Dies-Alder (RDA) reactions of flavon-3-ols with a dihydroxylated A ring are characteristic of values of m/z lower than the aglycone (i.e., m/z 317), and m/z 137 is a typical fragment of the trihydroxylated B ring (Saldanha et al., 2013).
Generally, characteristic fragmentations of precursor ions of C-glycosides are shown to be cross-ring cleavages of the glycoside moiety [M−H– (60/90/120)] - whereas O-glycosides are shown to eliminate the sugar moiety from the aglycone unit. To differentiate between 6- and 8-C-glycosides, 6-C-glycosides exhibit [M−H−18] - as a characteristic ion due to the removal of a water molecule, as well as a higher abundance of [M−H−90] - ions than 8-C glycosides, while the relative intensity of the [M−H−120] - ions is higher in flavonoid 8-C-glycosides than flavonoid 6-C-glycosides. Flavonoid 6,8-di-C-glycosides exhibit cross-ring cleavages of the sugar attached to the C-6 position preferential (Singh et al., 2015). Of the flavonoids identified from M. elengi methanol extract, compound 1 (retention time 6.42) exhibited a [M – H] − at m/z 463 corresponding to the molecular formula C21H20O12 and a fragment ion at m/z 317 indicating the loss of rhamnosyl moiety (146 Da) yielding the aglycone part. The fragment ion at m/z 316 indicates [M−2H−rhamnose]-. Based on published data and library databases, this compound was identified as myricitrin, (Fig. 3). In addition, compound 2 (retention time 8.78) exhibited a [M – H] − at m/z 317 corresponding to the molecular formula C15H10O8 and fragment ions at m/z 316 [M−2H] - and [1,2A−] at m/z 179. Based on published data and library databases, this compound was identified as myricetin aglycone (Fig. 3).
Compound 3 revealed a molecular ion peak at m/z 431 [M – H]− at a retention time of 7.40 min, of 7.40 min, corresponding to the molecular formula C21H20O10. The MS/MS fragmentation produced the product ions at m/z 285 [M – 146 – H] -, indicating the loss of rhamnosyl moiety, and at m/z 284 [M – 146 – 2H] - suggesting the presence of kaempferol derivative. This compound was identified as kaempferol-3-O-α-L-rhamnoside based on a comparison with data from library databases and previously published literature, (Fig. 4). Compound 4 revealed a precursor ion at m/z 447 at a retention time of 6.74 min. The second-generation of the precursor ion at m/z 447 [M – H] − produced the product ions at m/z 301 [M – 146 – H] -, indicating the loss of rhamnosyl moiety, and at m/z 300 [M – 146 – 2H] -. Such a fragmentation pattern confirmed the presence of a quercetin rhamnosyl derivative. Based on published data and library databases, this compound was identified as quercitrin, (Fig. 4). These four compounds represent the major identified flavonoids of M. elengi methanol extract (Table 1).
4.2 Identification of phenolic and organic acids
Results of UPLC/ESI-QTOF-HRMS/MS analysis of M. elengi methanol extract in negative ionization mode revealed the presence of both phenolic and organic acids. Commonly, in the spectra of all phenolic acids and organic acids, the molecular ion peak, [M−H] -, has been seen. MS2 spectra also show fragment ions characteristic for decarboxylation (–CO2, 44 Da) and dehydration (–H2O, 18 Da). Caffeic acid, chlorogenic acid, and 3-(4-hydroxyphenyl) prop-2-enoic acid were detected to be the major phenolic acids in the extract, whereas quinic acid was the most abundant organic acid, representing 31.56% of the identified compounds. Compound 21 exhibited a molecular ion peak [M−H] − at m/z 179, corresponding to the molecular formula C9H8O4 with a characteristic daughter ion at m/z 161 [M−H−18]-, corresponding to the neutral loss of a water moiety. This compound was tentatively identified as caffeic acid based on published data and library database, (Fig. 5). Compound 20 exhibited a molecular ion peak [M−H]− at m/z 191, corresponding to molecular formula C7H12O6 with characteristic daughter ions at m/z 173 [M−H−18] -, corresponding to the neutral loss of a water moiety, and 127 [M−H−64] -, corresponding to neutral losses of 2H2O and CO. This compound was identified as quinic acid based on published data and library database, (Fig. 5).
4.3 Identification of stilbenes
Only one stilbene was emerged in the output LC/MS data and was detected at a retention time of 8.57 min. This compound revealed a molecular ion peak [M−H] − at m/z 405, corresponding to the molecular formula C20H22O9 with characteristic daughter ions at m/z 243 [M−H−162] -, corresponding to the loss of glucose moiety, and 242 [M−2H−162] -. Based on published data and library databases, this compound was identified as astringin.
4.4 Identification of coumarins
Coumarins, like all other identified phenolic types, showed experimental molecular ion peaks that were in complete accord with their own predicted accurate masses and molecular formulas. As presented (Table 1), two coumarins were detected in the methanol extract of M. elengi. Compound 25 was expected to be a di-hydroxycoumarin hexoside, where it showed a molecular ion peak at m/z 339 [M−H] –, corresponding to molecular formula C15H16O9, and its MS2 spectrum gave the characteristic fragment ion at m/z 177 [M−H−162] –, corresponding to the loss of hexoside moiety. This compound was tentatively identified as esculin consistence with the conventional library database and the literature, (Table 1). Finally, the recorded data was in complete accordance with the conventional library database and reported literature for daphnetin.
4.5 NLRP3 inflammasome assembly complex in response to viral infection
Given the importance of the NLRP3 inflammasome in many domains of human health and disease, it is crucial to comprehend the processes involved in activation (Nakagawa et al., 2015). Developing targeted therapeutics for the treatment of NLRP3-driven disorders will be made easier with a better knowledge of the activation and assembly of the NLRP3 inflammasome complex (Fig. 7). It takes two steps for macrophages to activate the NLRP3 inflammasome, which must be “primed” either in advance of or concurrently with a secondary NLRP3-specific activation of receptors that signal via MyD88/TRIF, such as Toll-like receptors (TLRs), IL-1R, tumour necrosis factor receptor, and NOD2 (TIR-domain-containing adapter-inducing interferon) (Alishahi et al., 2019) (Wang et al., 2022). The activation of caspase-1, however, does not require additional protein translation as priming occurs quickly (Xie et al., 2019). According to a study by Surabhi et al.,(2022) activating the NLRP3 inflammasome results in an increase in the release of IL-1β (Surabhi et al., 2022), (Miller et al., 2022). Moreover, a previous investigation showed that IL-1β mediated by NLRP3 may cause asthma exacerbations (Im & Ammit, 2014). Inhibitors of NLRP3 inflammasome activation result in anti-inflammatory effects (Hung et al., 2019). Data findings showed that an Ethyl acetate extract causes the induction of apoptosis in U937 cell lines with IC50 55.8 μg/mL, resulting in growth inhibition in a concentration-dependent manner via apoptotic cell death induction. Compound 9 kaempferol-3-O-(6-p-coumaroyl) -glucoside possesses antioxidant properties (Dall’Acqua et al., 2008), suppresses cytokines (Allam, 2021). Resulting data showed that kaempferol-3-O-(6-p-coumaroyl) -glucoside has the strongest affinity fitting NLRP3 ADP-binding site. Moreover, Aesculin showed a docking configuration with high affinity −8.84 (ΔG in Kcal/mol) interacting with the NLRP3 inflammasome ADP active site.
4.6 Docking experiment
The investigated compounds have demonstrated docking scores ranging from -6.20 to −12.30 kcal/mol. The best affinity towards ADP-binding site of NLRP3 was achieved by kaempferol-3-O-(6-p-coumaroyl) -glucoside (Compound 9). The phenolic hydroxyl group of the p-coumaryl moiety of the compound (9) has formed a hydrogen bond with Asp303 whereas 7-OH of the flavonoid part, has established another hydrogen bond with Asp151. The oxygen atom of the sugar ring also formed a hydrogen bond with His520. The flavonoid part ring B and C have shown Pi-Pi interaction with Trp 414 (Fig. 8). Regarding the interaction of compound (25) (Aesculin) with the binding site, the OH groups of the sugar ring have established three hydrogen bonds interactions with Arg165 and Thr167 while the coumarin OH group formed a hydrogen bond with Asp151. The coumarin ring also formed Pi-Pi interaction with Trp414 (Fig. 9). Compound (20) quinic acid has interacted with the binding site through formation of one hydrogen bond by 3-OH groups with Asp151 and three hydrogen bonds by 4-OH with Asp151, Arg165 and Tyr379. Also, the carbonyl oxygen atom formed one hydrogen bond withThr167 (Fig. 10).
4.7 Conclusion
Both phenolic compounds and flavonoids are generally recognized as secondary metabolites of plants, and they have been investigated for their potential to improve human health and treat or prevent a variety of diseases. Flavonoids can reduce inflammatory mediators by interfering with the macrophage NLRP3 inflammasome. The current study revealed that kaempferol-3-O-(6-p-coumaroyl)-glucoside displayed significant binding affinity and high energy activities interacting with NLRP3 active site.
Acknowledgments
The Pharmacognosy Department at Al-Azhar University (Girls), Cairo, Egypt, provided support for this work, with the facilities provided by National Center for Radiation Research and Technology at the Atomic Energy Authority's National Center for Drug Research.
Funding
No state industry provided funding for this study in the form of a grant.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Ultra performance liquid chromatography-tandem mass spectrometeric analysis of ethyl acetate fraction from saudi Lavandula coronopifolia Poir and evaluation of its cytotoxic and antioxidant activities. Journal of Herbmed Pharmacology. 2020;9(3):268-276.
- [Google Scholar]
- Isolation of Lupeol from the Stem-bark of Lonchocarpus sericeus (Papilionaceae) Scholars Academic Journal of Biosciences. 2013;1(1):18-19.
- [Google Scholar]
- Gallic acid esters from the stem bark of Mimusops elengi L. Nat. Prod. Res.. 2010;24(10):962-972.
- [Google Scholar]
- Alam, S., & Haque, M. R. 2020. Phytochemical screening of Colocasia gigantea and Colocasia affinis (Family: Araceae) using 1H-NMR and 13C-NMR techniques. BioRxiv.
- Anticancer and photocatalytic activities of zinc oxide nanorods synthesized from Manilkara littoralis leaf extract. Mater. Chem. Phys.. 2022;277:125541
- [Google Scholar]
- NLRP3 inflammasome in ischemic stroke: as possible therapeutic target. Int. J. Stroke. 2019;14(6):574-591.
- [Google Scholar]
- Suppression of cytokine production by newly isolated flavonoids from pepper. Fitoterapia. 2021;151:104903
- [Google Scholar]
- Alotaibi, B., Negm, W. A., Elekhnawy, E., El-Masry, T. A., Elharty, M. E., Saleh, A., Abdelkader, D. H., & Mokhtar, F. A. 2022. Antibacterial activity of nano zinc oxide green-synthesised from Gardenia thailandica triveng. Leaves against Pseudomonas aeruginosa clinical isolates: in vitro and in vivo study. Artificial Cells, Nanomedicine, and Biotechnology, 50(1), 96-106.
- Encephalartos villosus Lem. Displays a Strong In Vivo and In Vitro Antifungal Potential against Candida glabrata Clinical Isolates. Journal of Fungi. 2022;8(5):521.
- [Google Scholar]
- Baky, M. H., Elsaid, M. B., & Farag, M. A. 2022. Phytochemical and biological diversity of triterpenoid saponins from family Sapotaceae: A comprehensive review. Phytochemistry, 113345.
- Antioxidant capacity and phenolic content of Mimusops elengi fruit extract. Agriculture and Natural Resources. 2009;43(1):21-27.
- [Google Scholar]
- HPLC-UV-MS profiles of phenolic compounds and antioxidant activity of fruits from three citrus species consumed in Northern Chile. Molecules. 2014;19(11):17400-17421.
- [Google Scholar]
- Antioxidant Activity, α-Glucosidase Inhibition and UHPLC–ESI–MS/MS Profile of Shmar (Arbutus pavarii Pamp) Plants. 2021;10(8):1659.
- [Google Scholar]
- Analysis of flavonoids in Rhamnus davurica and its antiproliferative activities. Molecules. 2016;21(10):1275.
- [Google Scholar]
- Ursolic acid isolated from Isodon excisoides induces apoptosis and inhibits invasion of GBC-SD gallbladder carcinoma cells. Oncol. Lett.. 2019;18(2):1467-1474.
- [Google Scholar]
- Evaluation of in vitro antioxidant properties of some traditional Sardinian medicinal plants: Investigation of the high antioxidant capacity of Rubus ulmifolius. Food Chem.. 2008;106(2):745-749.
- [Google Scholar]
- Innate immunity: the first line of defense against SARS-CoV-2. Nat. Immunol.. 2022;23(2):165-176.
- [Google Scholar]
- Metabolic Profiling, Chemical Composition, Antioxidant Capacity, and In Vivo Hepato-and Nephroprotective Effects of Sonchus cornutus in Mice Exposed to Cisplatin. Antioxidants. 2022;11(5):819.
- [Google Scholar]
- Antidiarrheal and Antibacterial Activities of Monterey Cypress Phytochemicals. In Vivo and In Vitro Approach. Molecules. 2022;27(2):346.
- [Google Scholar]
- UPLC-ESI-MS/MS Profile of The Ethyl Acetate Fraction of Aerial Parts of Bougainvillea'Scarlett O'Hara'Cultivated in Egypt. Egypt. J. Chem.. 2021;64(2):793-806.
- [Google Scholar]
- Eltamany, E. E., Elhady, S. S., Ahmed, H. A., Badr, J. M., Noor, A. O., Ahmed, S. A., & Nafie, M. S. 2020. Chemical profiling, antioxidant, cytotoxic activities and molecular docking simulation of Carrichtera annua DC.(Cruciferae). Antioxidants, 9(12), 1286.
- Farid, M. M., Ibrahim, F. M., Ragheb, A. Y., Mohammed, R. S., Hegazi, N. M., Shabrawy, M. O. E., Kawashty, S. A., & Marzouk, M. M. 2022. Comprehensive phytochemical characterization of Raphanus raphanistrum L.: In vitro antioxidant and antihyperglycemic evaluation. Scientific African, 16, e01154.
- Phytochemical analysis of Pfaffia glomerata inflorescences by LC-ESI-MS/MS. Molecules. 2014;19(10):15720-15734.
- [Google Scholar]
- High-resolution mass spectrometry and biological properties of grapevine and wine stilbenoids. Stud. Nat. Prod. Chem.. 2019;61:175-210.
- [Google Scholar]
- Study of grape polyphenols by liquid chromatography-high-resolution mass spectrometry (UHPLC/QTOF) and suspect screening analysis. J. Anal. Methods Chem.. 2015;2015
- [Google Scholar]
- Myricetin 3-rhamnosyl (1→ 6) galactoside from Nymphaéa x marliacea. Phytochemistry. 1998;49(7):1997-2000.
- [Google Scholar]
- A role of gallic acid in oxidative damage diseases: A comprehensive review. Nat. Prod. Commun.. 2019;14(8) 1934578X19874174
- [Google Scholar]
- Characterisation of flavonoid aglycones by negative ion chip-based nanospray tandem mass spectrometry. International journal of analytical chemistry. 2012;2012
- [Google Scholar]
- Characterisation of phenolic acid derivatives and flavonoids from different morphological parts of Helichrysum obconicum by a RP-HPLC–DAD-(−)–ESI-MSn method. Food Chem.. 2011;129(2):333-344.
- [Google Scholar]
- Isolation and structure elucidation of uncommon secondary metabolites from Cistus salviifolius L. Rec. Nat. Prod.. 2015;9(2):175.
- [Google Scholar]
- Analysis of flavonoids from leaves of Acanthopanax brachypus harms. J. Chil. Chem. Soc.. 2013;58(1):1549-1552.
- [Google Scholar]
- Bavachin attenuates LPS-induced inflammatory response and inhibits the activation of NLRP3 inflammasome in macrophages. Phytomedicine. 2019;59:152785
- [Google Scholar]
- The NLRP 3 inflammasome: role in airway inflammation. Clin Exp Allergy. 2014;44(2):160-172.
- [Google Scholar]
- Inflammasome-associated nucleotide-binding domain, leucine-rich repeat proteins and inflammatory diseases. J. Immunol.. 2009;183(12):7623-7629.
- [Google Scholar]
- Rosa rubiginosa and Fraxinus oxycarpa herbal teas: characterization of phytochemical profiles by liquid chromatography-mass spectrometry, and evaluation of the antioxidant activity. New J. Chem.. 2017;41(15):7681-7688.
- [Google Scholar]
- HPLC-DAD-ESI-MSn identification of phenolic compounds in cultivated strawberries from Macedonia. Maced. J. Chem. Chem. Eng.. 2010;29(2):181-194.
- [Google Scholar]
- Electrospray-mass spectrometric analysis of plasma pyrophosphates separated on a multi-modal liquid chromatographic column. Mass Spectrometry Letters. 2011;2(4):92-95.
- [Google Scholar]
- Antifungal properties of Mimusops elengi seeds against paddy seed-borne fungi and selected pathogenic fungi. Malaysian Journal of Microbiology. 2018;14(2):152-158.
- [Google Scholar]
- Influence of mefenamic acid on the intestinal absorption and metabolism of three bioactive flavones in Radix Scutellariae and potential pharmacological impact. Pharm. Biol.. 2014;52(3):291-297.
- [Google Scholar]
- Pattern recognition receptors in health and diseases. Signal Transduct. Target. Ther.. 2021;6(1):1-24.
- [Google Scholar]
- Antiplatelet activity of gallic acid and methyl gallate. Food Sci. Biotechnol. 2004
- [Google Scholar]
- Biotransformation of p-Coumaric Acid (=(2E)-3-(4-Hydroxyphenyl) prop-2-enoic Acid) by Momordica charantia Peroxidase. Helv. Chim. Acta. 2007;90(6):1117-1132.
- [Google Scholar]
- Analysis of phenolic compounds in leaves from endemic trees from Madeira Island. A contribution to the chemotaxonomy of Laurisilva forest species. Ind. Crop. Prod.. 2015;64:135-151.
- [Google Scholar]
- Antioxidant, Anti-Inflammatory and Cytotoxic Activity of Phenolic Compound Family Extracted from Raspberries (Rubus idaeus): A General Review. Antioxidants. 2022;11(6):1192.
- [Google Scholar]
- Effects of different drying methods on untargeted phenolic metabolites, and antioxidant activity in Chinese cabbage (Brassica rapa L. subsp. chinensis) and nightshade (Solanum retroflexum Dun.) Molecules. 2020;25(6):1326.
- [Google Scholar]
- Nucleic acid-sensing pathways during SARS-CoV-2 infection: expectations versus reality. J. Inflamm. Res.. 2021;14:199.
- [Google Scholar]
- Human fetal membrane IL-1β production in response to bacterial components is mediated by uric-acid induced NLRP3 inflammasome activation. J. Reprod. Immunol.. 2022;149:103457
- [Google Scholar]
- Current herbal medicine as an alternative treatment in dentistry: In vitro, in vivo and clinical studies. Eur. J. Pharmacol.. 2020;889:173665
- [Google Scholar]
- Two new polyhydroxylated pentacyclic triterpenes with cytotoxic activities from Manilkara pellegriniana (Sapotaceae) Phytochem. Lett.. 2019;31:161-165.
- [Google Scholar]
- Phytochemical profiling, in vitro and in silico anti-microbial and anti-cancer activity evaluations and Staph GyraseB and h-TOP-IIβ receptor-docking studies of major constituents of Zygophyllum coccineum L. Aqueous-ethanolic extract and its subsequent fractions: An approach to validate traditional phytomedicinal knowledge. Molecules. 2021;26(3):577.
- [Google Scholar]
- Inhibitory effect of Newtonia extracts and myricetin-3-o-rhamnoside (myricitrin) on bacterial biofilm formation. BMC Complementary Medicine and Therapies. 2020;20(1):1-10.
- [Google Scholar]
- Somatic NLRP3 mosaicism in Muckle-Wells syndrome. A genetic mechanism shared by different phenotypes of cryopyrin-associated periodic syndromes. Ann. Rheum. Dis.. 2015;74(3):603-610.
- [Google Scholar]
- beta-Amyrin and alpha-amyrin acetate isolated from the stem bark of Alstonia boonei display profound anti-inflammatory activity. Pharm. Biol.. 2014;52(11):1478-1486.
- [Google Scholar]
- Identification of Puerarin and its metabolites in rats by liquid chromatography− tandem mass spectrometry. J. Agric. Food Chem.. 2004;52(12):3708-3712.
- [Google Scholar]
- RIG-I-like receptors: their regulation and roles in RNA sensing. Nat. Rev. Immunol.. 2020;20(9):537-551.
- [Google Scholar]
- Triterpenes from Pouteria ramiflora (Mart.) Radlk. Leaves (Sapotaceae) Food Chem. Toxicol.. 2017;109:1063-1068.
- [Google Scholar]
- Characterization of flavonoids and phenolic acids in Myrcia bella cambess. Using FIA-ESI-IT-MSn and HPLC-PAD-ESI-IT-MS combined with NMR. Molecules. 2013;18(7):8402-8416.
- [Google Scholar]
- Structural characterization of flavonoid C-and O-glycosides in an extract of Adhatoda vasica leaves by liquid chromatography with quadrupole time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom.. 2015;29(12):1095-1106.
- [Google Scholar]
- Studies on succinate-tetrazolium reductase systems: III. Points of coupling of four different tetrazolium salts III. Points of coupling of four different tetrazolium salts. BBA. 1963;77:383-393.
- [Google Scholar]
- Discovery and Characterization of Phenolic Compounds in Bearberry (Arctostaphylos uva-ursi) Leaves Using Liquid Chromatography-Ion Mobility–High-Resolution Mass Spectrometry. J. Agric. Food Chem.. 2021;69(37):10856-10868.
- [Google Scholar]
- Recognition and inhibition of SARS-CoV-2 by humoral innate immunity pattern recognition molecules. Nat. Immunol.. 2022;23(2):275-286.
- [Google Scholar]
- The norepinephrine metabolite 3, 4-dihydroxymandelic acid is produced by the commensal microbiota and promotes chemotaxis and virulence gene expression in enterohemorrhagic Escherichia coli. Infect. Immun.. 2017;85(10):e00431-e1417.
- [Google Scholar]
- Hydrogen peroxide is crucial for NLRP3 inflammasome-mediated IL-1β production and cell death in pneumococcal infections of bronchial epithelial cells. J. Innate Immun.. 2022;14(3):192-206.
- [Google Scholar]
- AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem.. 2010;31(2):455-461.
- [Google Scholar]
- A tetrazolium-based colorimetric MTT assay to quantitate human monocyte mediated cytotoxicity against leukemic cells from cell lines and patients with acute myeloid leukemia. J. Immunol. Methods. 1994;174(1–2):311-320.
- [Google Scholar]
- Microextraction of reseda luteola-dyed wool and qualitative analysis of its flavones by uhplc-uv, nmr and ms. Molecules. 2021;26(13):3787.
- [Google Scholar]
- Pinocembrin suppresses oxidized low-density lipoprotein-triggered NLRP3 inflammasome/GSDMD-mediated endothelial cell pyroptosis through an Nrf2-dependent signaling pathway. Sci. Rep.. 2022;12(1):1-16.
- [Google Scholar]
- Indian medicinal plants: a compendium of 500 species. Vol Vol. 5. Orient Blackswan; 1993.
- Complement membrane attack complexes assemble NLRP3 inflammasomes triggering IL-1 activation of IFN-γ–primed human endothelium. Circ. Res.. 2019;124(12):1747-1759.
- [Google Scholar]
- Antioxidant and anti-inflammatory activities of selected medicinal plants containing phenolic and flavonoid compounds. J. Agric. Food Chem.. 2011;59(23):12361-12367.
- [Google Scholar]
- High-performance liquid chromatography–tandem mass spectrometry for identification of isoflavones and description of the biotransformation of kudzu root. Anal. Bioanal. Chem.. 2005;383(5):787-796.
- [Google Scholar]
- Systematic Screening of Chemical Constituents in the Traditional Chinese Medicine Arnebiae Radix by UHPLC-Q-Exactive Orbitrap Mass Spectrometry. Molecules. 2022;27(9):2631.
- [Google Scholar]
Further reading
- Chemical characterisation of non-defective and defective green arabica and robusta coffees by electrospray ionization-mass spectrometry (ESI-MS) Food Chem.. 2008;111(2):490-497.
- [Google Scholar]







