Translate this page into:
Molecular mechanism of Yi-Qi-Yang-Yin-Ye against obesity in rats using network pharmacology, molecular docking, and molecular dynamics simulations
⁎Corresponding authors. wuyongning@cfsa.net.cn (Yongning Wu), lilin@ahau.edu.cn (Lin Li)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Abstract
The increasing prevalence of obesity globally, which as well as affecting people's daily lives and increasing the risk of obesity complications, also threatens the health of animal organisms simultaneously. It’s been reported that Yi-Qi-Yang-Yin-Ye had remarkable efficacy in the treatment of obesity. The specific underlying mechanism of action of Yi-Qi-Yang-Yin-Ye in treating obesity, however, remains ambiguous. Therefore, the innovative approach, which is network pharmacology combined with molecular docking and molecular dynamics simulations, was employed in the current research to explore the potential mechanism and promote further development in the treatment of obesity. The active ingredients and related targets of Yi-Qi-Yang-Yin-Ye and related targets of obesity were summarized from extensive public databases. Furthermore, network topology analysis and pathway enrichment analysis were performed to explore the complicated interactions between drug and targets. Finally, accurate validation methods composed of molecular docking and molecular dynamics simulations were conducted to elucidate the binding affinity of Yi-Qi-Yang-Yin-Ye with obesity-related targets. As a result, 13 main active ingredients and 5 core targets of Yi-Qi-Yang-Yin-Ye against obesity in rats were acquired through primary screening of network topology analysis. Pathway enrichment analysis demonstrated that intersectional targets were involved in multiple signaling pathways, where PI3K-Akt signaling pathway, MAPK signaling pathway, and Insulin resistance were the main pathways of Yi-Qi-Yang-Yin-Ye in treating obesity in rats. Finally, molecular docking indicated that the seven critical active ingredients displayed great binding affinity to the hub targets. Furthermore, molecular dynamics simulations further screened and obtained that five critical active ingredients acting on the Mapk1 target for Yi-Qi-Yang-Yin-Ye against obesity in rats. The innovative approach and the results achieved have further contributed to and revealed the molecular mechanisms for treating obesity, providing an alternative for treating obesity in animals and humans.
Keywords
Natural products
Yi-Qi-Yang-Yin-Ye
Obesity
Network pharmacology
Molecular docking
Molecular dynamics simulations
1 Introduction
Obesity has become one of the global concerns with the development of society and the improvement of living standards (Sljivic and Gusenoff, 2019). A significantly increased tendency toward obesity, which contributed to animal and human health in danger, was observed in both animals and humans. Moreover, obesity-induced diseases would have a significant influence on the economic loss of livestock and poultry industries (Klimentidis et al., 2011).
Natural products play an irreplaceable role in remedies due to their excellent therapeutic results and historical heritage. Yi-Qi-Yang-Yin-Ye (Y-Q-Y-Y-Y) is a traditional Chinese medicine prescription that is composed of four herbs, including Tribuli Herb, Astragali Radix, Rehmanniae Radix and Taxilli Herb. Tribuli Herb (He et al., 2008) is the main herb of Y-Q-Y-Y-Y, which is clinically used for the treatment of obesity-related diseases such as diabetes, systemic inflammation, and insulin resistance (Amin et al., 2006, Abdel-Mottaleb et al., 2022). Besides, the other components in this traditional Chinese medicine prescription also showed therapeutic effects on obesity-related diseases, where the extract of Astragali Radix has anti-diabetic and anti-inflammatory properties and promotes the metabolism of glucose and lipid (Hoo et al., 2010). Catalpol extracted from Rehmanniae Radix improves antidiabetic and antioxidant effects by balancing oxidative and antioxidant enzymes (Zhu et al., 2016). Extract of Taxilli Herb could increase glucose consumption and improve insulin sensitivity in vivo (Wang et al., 2006) as well as hypolipidemic and protect lipid from peroxidative effects (Luo et al., 2019). Furthermore, Y-Q-Y-Y-Y has been demonstrated to be effective for the treatment of metabolic syndrome and type II diabetes. However, the underlying mechanisms of action of Y-Q-Y-Y-Y for the treatment of obesity are unclear.
A Chinese medicine prescription is generally composed of several herbs with complex components and complicated mechanisms. The components with good pharmacokinetic properties in a prescription are the potential active compounds for the treatment of corresponding diseases. Considering the complex interaction between the prescription and the organism, network pharmacology and molecular docking were used to explore the active compound in the prescription, related targets, as well as the disease-related pathways, thereby revealing the mechanism of action of mode (Wang et al., 2021). Although network pharmacology and molecular docking have been adopted to explore the molecular mechanisms of molecular mechanisms in treating various diseases (Zhao et al., 2021, Liu et al., 2023). The innovative approach of network pharmacology combined with molecular docking and molecular dynamics (MD) simulations has not been fully exploited in investigating the molecular mechanisms of disease due to the rich analytical approach of these novelty methods as well as other advantages. MD simulation is a vigorous molecular simulation method that has emerged with the development of the times and has been widely employed in research in various scientific fields. MD simulation, with its remarkable dynamic simulation method and rich analytical measures, can be combined with many other analytical methods and experiments to further reveal the molecular mechanisms of scientific research accurately and efficiently. Moreover, the combined approach of molecular docking and MD simulations can further verify and screen the results of molecular docking (Pall et al., 2020). Therefore, this combined approach will further reveal the mechanism of herbal compounding in the treatment of diseases.
In the present study, with the purpose of further unraveling the potential molecular mechanisms of Y-Q-Y-Y-Y for the treatment of obesity and to facilitate the process of natural medicines in treating obesity, the current study adopted the above advanced approach to further investigate the molecular mechanisms of Y-Q-Y-Y-Y in treating obesity. In addition, with the advancement of science and technology, research methods and approaches are constantly being innovated and improved. Therefore, this study also aims to provide an alternative and inspiration for the subsequent natural drug treatment of obesity and other diseases.
2 Materials and methods
2.1 Database construction of active ingredients and corresponding targets of Y-Q-Y-Y-Y
The database of active ingredients of Y-Q-Y-Y-Y was constructed based on the TCMSP database (https://tcmspw.com/tcmsp.php) (Ru et al., 2014), ETCM database (https://www.tcmip.cn/ETCM/index.php) (Xu et al., 2019), SymMap database (https://www.symmap.org) (Wu et al., 2019) and TCMID database (https://www.megabionet.org/tcmid/) (Huang et al., 2018). In the TCMSP database, the screening criteria for the active ingredients of TCM were oral bioavailability (OB) ≥ 0.3 and drug-like (DL) ≥ 0.18. In the other three databases, OB ≥ 0.3 was used for preliminary screening, and then the screened active ingredients were imported into the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) (Kim et al., 2021), the TCMID database, and the TCMSP database for re-screening. Finally, the EVenn platform (https://www.ehbio.com/test/venn/) (Chen et al., 2021) was used to remove the duplicates of active ingredients obtained from the four databases. And the corresponding targets of the active ingredients were collected in the TCM database, the Uniprot database (UniProt Consortium, 2018) and the STRING database (https://string-db.org) (Szklarczyk et al., 2021).
2.2 Database construction of obesity-related targets in rats
To obtain a comprehensive list of obesity-related targets, five databases including DrugBank database (https://www.drugbank.ca) (Wishart et al., 2018), GeneCards database (https://www.genecards.org) (Safran et al., 2010), PHARMGKB database (https://www.pharmgkb.org/) (Barbarino et al., 2018), OMIM database (https://omim.org) (Amberger et al., 2015) and TTD database (https://db.idrblab.net/ttd/) (Wang et al., 2020) were selected. The collected targets from five databases were combined and the duplicates of obesity-related targets were removed via the EVenn web platform.
2.3 The intersectional targets between Y-Q-Y-Y-Y and obesity
The intersectional targets were obtained using the EVenn web platform, which would be the potential targets of Y-Q-Y-Y-Y for the treatment of obesity in rats.
2.4 Network establishment of active ingredients and the intersectional targets
Cytoscape is software that visualizes proteins, genes, and complex networks (Su et al., 2014). In the current study, the network between the active ingredients of Y-Q-Y-Y-Y and the intersectional targets was constructed using the open source platform, Cytoscape 3.9.1.
2.5 Protein-protein interaction (PPI) of intersectional targets and topological analysis
The PPI network of intersectional targets was constructed in the STRING database, while the minimum required interaction score was set to the highest confidence level (0.900). Then the PPI network was optimized and refined through Cytoscape 3.9.1 software. Additionally, the topological analysis was performed using the CytoHubba plugin.
2.6 GO and KEGG pathway enrichment analysis
The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed in Metascape database (https://metascape.org/) (Zhou et al., 2019) and the P Value Cutoff was set to 0.01. The obtained data were visualized and refined through the SRPLOT web platform.
2.7 Molecular docking
The active ingredients that are above the average degree values and one common active ingredient among Y-Q-Y-Y-Y as well as the only active ingredient of Rehmanniae Radix were selected for molecular docking with the hub obesity-related targets with high degree values from PPI network. The molecular docking was conducted in AutoDockTOOLs 1.5.6 software (Gaillard, 2018, Hill and Reilly, 2015), visualized in Pymol software and Discovery Studio software (Yuan et al., 2017), and refined through the SRPLOT web platform (https://www.bioinformatics.com.cn/en) (Lu et al., 2022).
2.8 Molecular dynamics simulations
To further validate and screen the results of molecular docking, MD simulations were performed by Gromacs 2020.6 software (Pall et al., 2020). Prior to conducting the MD simulations, the temperature and other conditions involved in the MD simulations’ processes were set to the conditions of drug-target binding in the organism. Considering that the hub ingredients in molecular docking were multi-carbon ring skeleton structures, all simulations were performed under the TIP3P water model and Charmm36 stance. During the simulations involving the action forces, the integration step involving hydrogen bonds was 2 fs and the LINCS algorithm was chosen for the constraints. The cut-off value of the electrostatic interaction was set to 1.2 nm and the interaction was calculated using the Particle-mesh Ewald (PME) method. In addition, the cut-off value for non-hydrogen bonding interactions was set to 10 Å. All simulations in this work were performed using the V-rescale temperature coupling method and the Berendsen method for temperature and pressure control, respectively. The specific simulations’ processes are as follows: First, to optimize the initial conformation of the target protein in the solvent, the molecular docking results of critical active ingredients and hub targets (subsequently called drug-target complexes) were pre-equilibrated at 100 ps. Then, the Isothermal-Isochoric (NVT) equilibrium of 100 ps and the Isothermal-Isobaric (NPT) equilibrium of 100 ps were performed to make the solvent system in which the drug target complex was located reach the proper temperature and pressure equilibrium, respectively. Finally, MD simulations were performed for 50 ns on the drug-targeted complexes obtained by molecular docking. Furthermore, all results of MD simulations obtained were visualized by Origin 2021 software.
3 Results
3.1 Collection of Y-Q-Y-Y-Y related and obesity-related targets
A total of 63 active ingredients of Y-Q-Y-Y-Y were collected from four Chinese medicine databases, of which 20 active ingredients were in Tribuli Herb, 37 in Astragali Radix, 1 in Rehmanniae Radix, and 5 in Taxilli Herb. The five intersectional active ingredients between the four herbs are illustrated in Fig. 1A. Of these, 3 active ingredients are intersectional active ingredients of Tribuli Herb and Astragali Radix; 1 active ingredient is an intersectional active ingredient of Tribuli Herb and Taxilli Herb; and 1 active ingredient is an intersectional active ingredient of Astragali Radix and Taxilli Herb (Fig. 1A and 2A, Supplementary Table S1). The detailed information for the active ingredients is available in Supplementary Table S1 and Fig. 1A. Similarly, the obesity-related targets collected from different public databases had intersectional targets as well. The obesity-related targets, including the intersectional targets, were illustrated in the Fig. 1B. Accordingly, 1629 Y-Q-Y-Y-Y-related targets and 9375 obesity-related targets were eventually collected.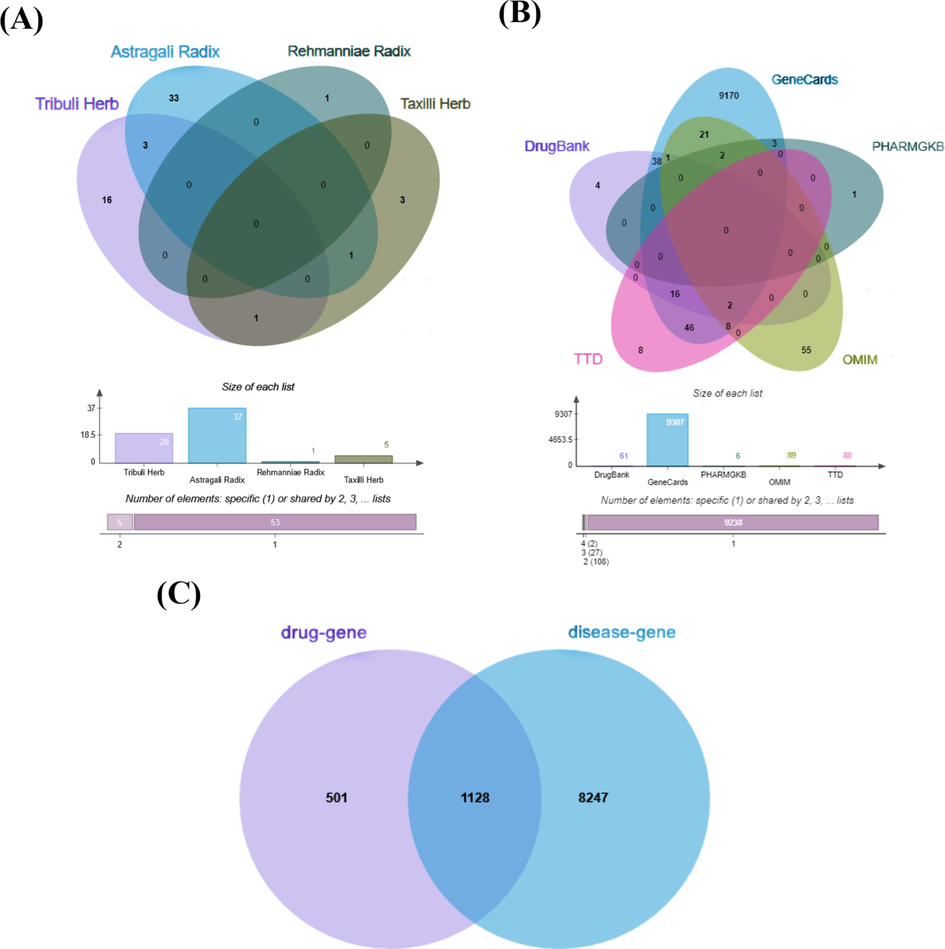
Databases construction. (A) Active ingredients of Y-Q-Y-Y-Y. (B) Rat obesity targets. (C) Intersectional targets of Y-Q-Y-Y-Y for the treatment of obesity in rats.
3.2 Network construction of intersectional targets
In the present study, 1128 intersectional targets of Y-Q-Y-Y-Y- for the treatment of obesity in rats were collected, which were shown in Fig. 1C. In terms of percentage, the intersectional targets of the drug and disease occupy a larger proportion of the total targets of the drug and a smaller proportion of the total targets of the disease. It is evident that the collection of targets is sufficient and reliable. The active ingredients-targets network is one of the vital analytical methods utilized in network pharmacology to elucidate the degree of interaction between active ingredients and targets. In the current study, the active ingredients-targets network with 2040 edges and 1186 nodes is available in Fig. 2A. The degree in the network indicated the number of adjacent genes in the network that directly interact with the gene. The degree value is positively correlated to the core connectivity. Thus, the higher the degree value of the active ingredient, the more intersectional targets it interacts with. The major active ingredients above the average degree values were Hexose, quercetin, calycosin, kaempferol, Oleanolic Acid, Jaranol, Beta-Sitosterol, 7-O-methylisomucronulatol, 4′-Methyl-N-Methylcoclaurine, isorhamnetin and formononetin, which were in detail described in Table 1. In addition, to comprehensively explore the mechanism, sitosterol, an intersectional active ingredient that is not included in the core active ingredients, and docosahexaenoic acid (the active ingredient of Rehmanniae Radix) are also considered the core active ingredients of Y-Q-Y-Y-Y. The specific information and chemical structures of the 13 main active ingredients are illustrated in Supplementary Table S2 and Supplementary Figure S3.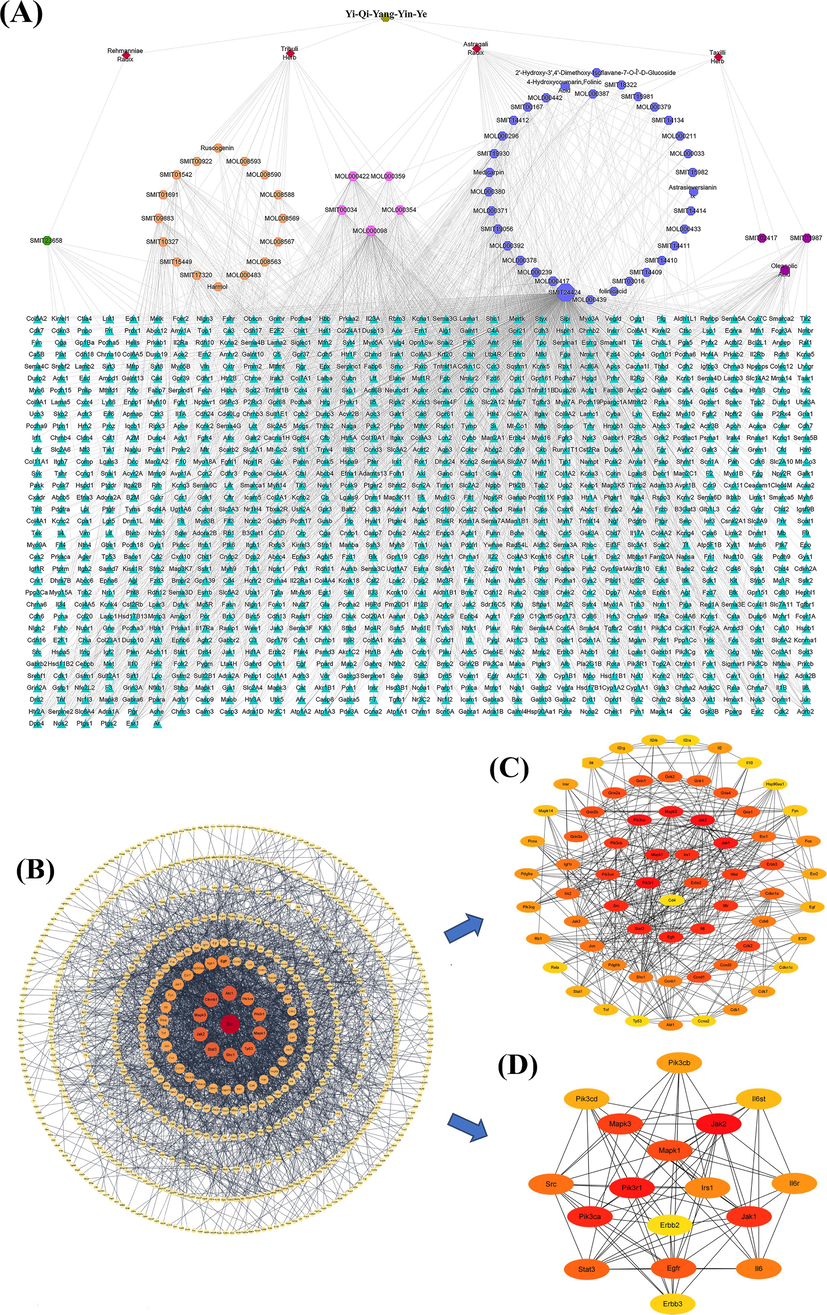
Topology analysis. (A) Active ingredients-targets network. Red diamonds represent the traditional Chinese medicine ingredients in Y-Q-Y-Y; Green circles represent the active ingredient of Rehmanniae Radix; Orange circles represent the active ingredient of Tribuli Herb; Dark blue circles represent the active ingredient of Astragali Radix; purple circles represent the active ingredient of Taxilli Herb; pink circles represent the active ingredients shared by the four traditional Chinese medicines, respectively; Cyan triangles represent intersectional targets. The active ingredients in the figure were indicated by the ID of each herbal database. (B) Construction and analysis of PPI network. (C) CytoHubba top100 network. (D) CytoHubba top18 network. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
ID
Active ingredient Name
InChI Key
Degree
SMIT24424
Hexose
WQZGKKKJIJFFOK-UHFFFAOYSA-N
901
MOL000098
quercetin
REFJWTPEDVJJIY-UHFFFAOYSA-N
161
MOL000417
Calycosin
ZZAJQOPSWWVMBI-UHFFFAOYSA-N
70
MOL000422
kaempferol
IYRMWMYZSQPJKC-UHFFFAOYSA-N
68
Oleanolic Acid
MIJYXULNPSFWEK-GTOFXWBISA-N
61
MOL000239
Jaranol
BJBUTJQYZDYRMJ-UHFFFAOYSA-N
55
SMIT00034
Beta-Sitosterol
KZJWDPNRJALLNS-VJSFXXLFSA-N
52
MOL000378
7-O-methylisomucronulatol
BLHQCBJSTMDZQA-LBPRGKRZSA-N
44
SMIT09883
4′-Methyl-N-Methylcoclaurine
HHYXHNSYWAYIPS-UHFFFAOYSA-N
43
MOL000354
isorhamnetin
IZQSVPBOUDKVDZ-UHFFFAOYSA-N
41
MOL000392
formononetin
HKQYGTCOTHHOMP-UHFFFAOYSA-N
38
MOL000359
sitosterol
KZJWDPNRJALLNS-ZFVHJZABSA-N
24
SMIT23658
Docosahexaenoic Acid
MBMBGCFOFBJSGT-KUBAVDMBSA-N
12
3.3 Construction and analysis of PPI network
The PPI network of the intersectional targets yielding 2317 edges was constructed, as was shown in Fig. 2B. Each node in the network represented a target for Y-Q-Y-Y-Y treatment of obesity in rats. In the UniProt database, targets with above-average extent values and crystal structures are Mapk1, Pik3r1, Irs1, Cd4, and Erbb2. Furthermore, the topological analysis, including the degree, was analyzed using the CytoHubba plugin, while the top 18 and top 66 nodes were selected to construct the network (available in Fig. 2C and Fig. 2D). As demonstrated in Fig. 2B, the larger the circle and the darker the color of the target, the closer the interaction between the target and other targets, and the figure clearly demonstrates the interaction size between each cross-target and other targets by the above principle. Similarly, in Fig. 2C and 2D, the interaction values of the targets are also shown according to the principle of color depth, and the darker the color, the more interaction with other targets. Therefore, the above five targets (Mapk1, Pik3r1, Irs1, Cd4, and Erbb2) were selected as the core targets of Y-Q-Y-Y-Y for the treatment of obesity in rats. And the five core targets’ specific information and chemical structures were demonstrated in Supplementary Table S3 and Supplementary Figure S4.
3.4 GO enrichment analysis
The GO enrichment analysis on the intersectional targets was in terms of biological processes (BP), cellular components (CC), and molecular functions (MF), as shown in Fig. 3A. For BP, the intersectional targets were mainly enriched in the regulation of MAPK cascade, the circulatory system process, the regulation of secretion, and the inflammatory response, as well as other processes. For CC, the intersectional targets were enriched in receptor complexes, membrane rafts, neuronal cell bodies, and protein kinase complexes, as well as other components. For MF, the intersectional targets were enriched in neurotransmitter receptor activity, G protein-coupled peptide receptor activity, hormone binding, and any other functions such as protein kinase binding and carbohydrate binding.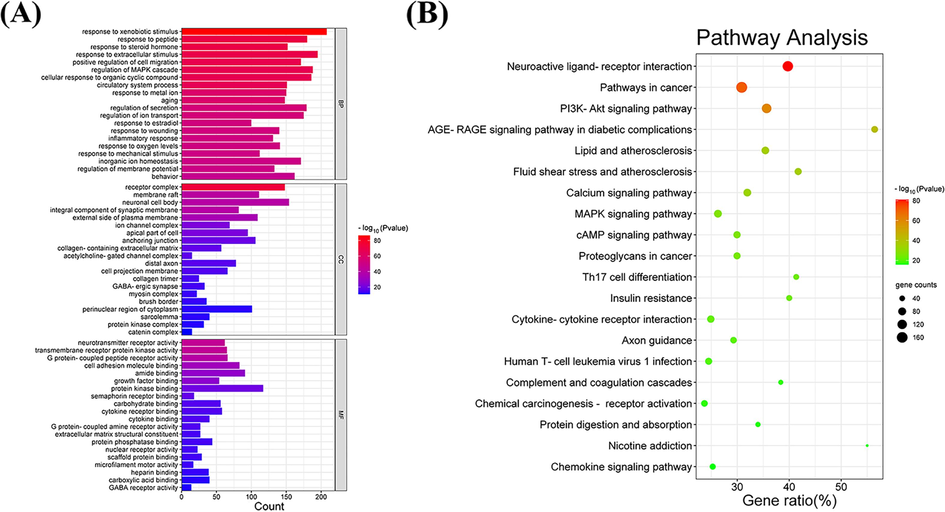
Enrichment analysis. (A) GO enrichment analysis. (B) KEGG enrichment analysis.
3.5 KEGG pathway enrichment analysis
KEGG pathway enrichment analysis was conducted to investigate which pathways the intersectional targets were specifically enriched in, thus further exploring and validating the molecular mechanism of Y-Q-Y-Y-Y for the treatment of obesity in rats. As illustrated in Fig. 3B, six of the top 20 pathways were obesity-associated pathways, which were the PI3K-Akt signaling pathway, the AGE-RAGE signaling pathway in diabetic complications, Lipid and atherosclerosis, the MAPK signaling pathway, Th17 cell differentiation, and Insulin resistance. Besides, these six pathways associated with obesity in rats were also ranked highly in terms of the number of genes involved in the pathway.
3.6 Molecular docking results
Molecular docking facilitates the understanding of the binding affinity of core active ingredients and hub targets. The PDB IDs for the obesity-related targets were 4S32 (Mapk1), 1FU5 (Pik3r1), 5WRM (Irs1), 1CID (Cd4), and 1N8Y (Erbb2), respectively. The InChI Key for the structure of the active ingredients from the PubChem database is shown in Table 1. The results’ heat map of all molecular docking is illustrated in Fig. 4. The binding free energy in molecular docking is a vital parameter for evaluating the stability of drug-target binding, and the smaller the value of the binding free energy, the more stable the binding of the drug to the target. After further analysis, the molecular docking results indicated that the seven critical active ingredients exhibited excellent binding affinity for the three central targets compared to other active ingredients and targets (Table 2 and Fig. 5A). In the results of molecular docking, Sitosterol, the active ingredient both in Tribuli Herb and Taxilli Herb, showed the best binding performance to the hub target. In addition, Oleanolic Acid, the active ingredient of Taxilli Herb, binds very closely to the hub target; the three active ingredients (kaempferol, Beta-Sitosterol, isorhamnetin) in Tribuli Herb and Astragali Radix and quercetin, the active ingredient in Astragali Radix and Taxilli Herb, also bind closely to the hub target. Consequently, the 7 active ingredients (quercetin, kaempferol, oleanolic acid, beta-sitosterol, isorhamnetin, sitosterol and docosahexaenoic acid) and three targets (Mapk1, Pik3r1 and Irs1) were regarded as critical active ingredients and hub targets for Y-Q-Y-Y-Y against obesity in rats.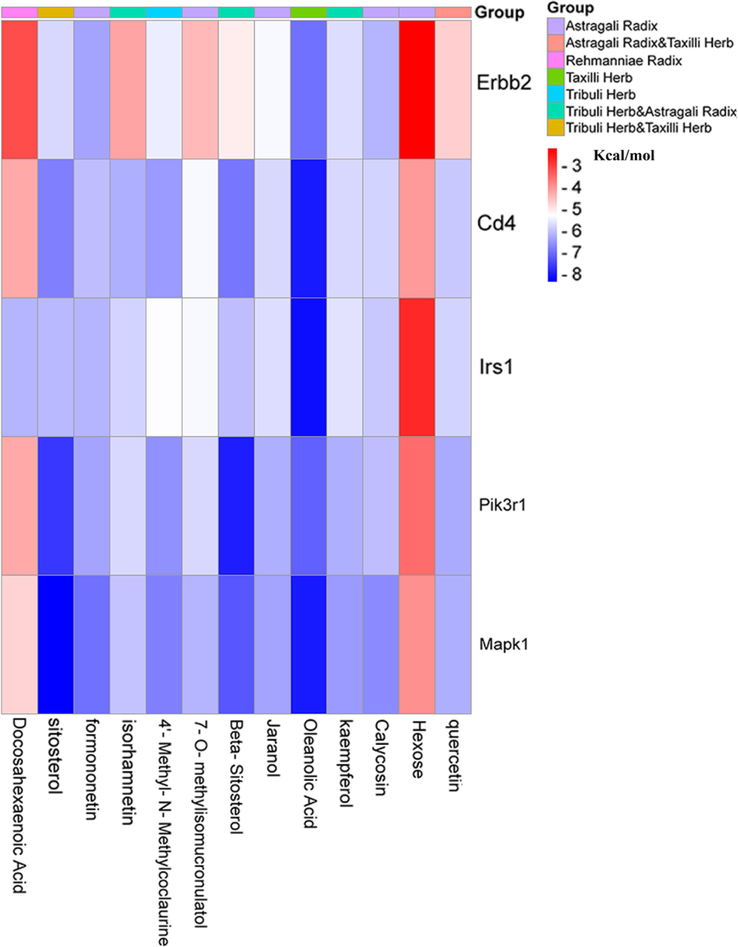
Molecular docking clustering heat map. Horizontal letters and vertical letters indicate key active ingredients and hub targets, respectively. The number on the right in the figure indicates the affinity energy (Unit: kcal.mol−1). The smaller the binding energy, the better the binding. The group on the right in the figure indicates which herbal medicine the active ingredient belongs to.
Target (PDB ID)
Active ingredient
Binding site
Affinity energy
Mapk1 (4S32)
quercetin
ARG-68, PHE-329, LEU-333, ASP-334
−6.23 kcal·mol−1
kaempferol
ARG-68, ALA-172, PHE-329, ASP-334
−6.47 kcal·mol−1
Oleanolic Acid
PHE-352
−7.98 kcal·mol−1
Beta-Sitosterol
ARG-89, GLN-353
−7.28 kcal·mol−1
isorhamnetin
ARG-68, VAL-171, PHE-329
−5.96 kcal·mol−1
sitosterol
GLN-95, LYS-97, ASP-98
−8.33 kcal·mol−1
Pik3r1 (1FU5)
Beta-Sitosterol
ILE-85
−7.96 kcal·mol−1
Irs1 (5WRM)
Oleanolic Acid
LYS-238, LYS-400, ARG-402
−8.1 kcal·mol−1
Docosahexaenoic Acid
LYS-238, LYS-400, ARG-402
−6.16 kcal·mol−1
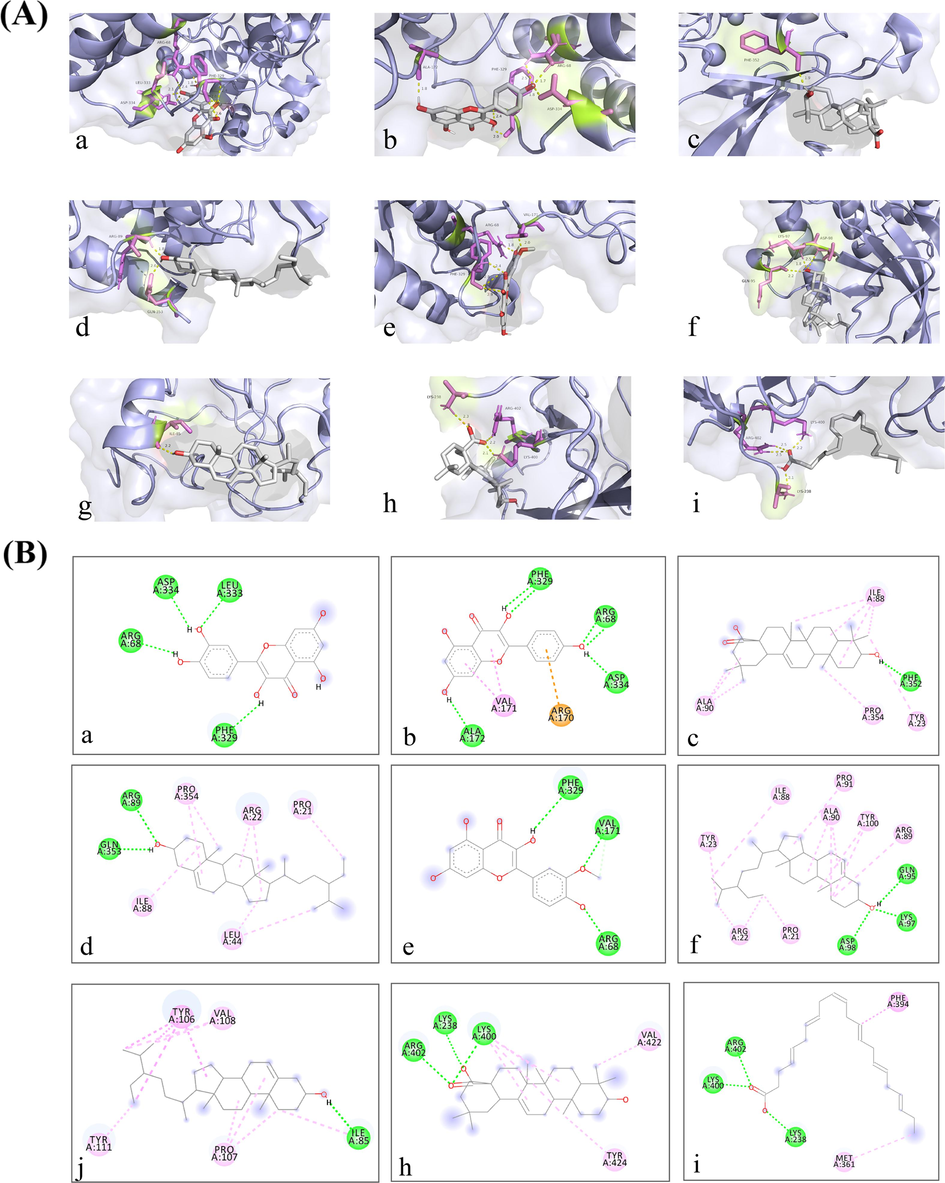
Molecular models of critical active ingredients of Y-Q-Y-Y-Y binding to core targets. (A) Molecular docking results (detailed hydrogen bonds illustrated in 3D diagram). The yellow dashed line showed the hydrogen bond, the molecular in purple were the amino acids as acceptors while the molecular in white were active ingredient. (B) specific information of binding site (detailed hydrogen bonds, electrostatic forces and hydrophobic forces illustrated in 2D diagram). The green lines stand for hydrogen bonds. The orange line stands for electrostatic force. The pink lines stand for hydrophobic forces. The different colored spheres indicate the amino acids specifically bound by the active ingredient at that target. The gray molecules are the critical active ingredients. In the Figure A and B, the diagrams (a-i) have the same sub-signs indicating the same active ingredient and target binding molecule docking results. a) quercetin binding to Mapk1. b) kaempferol binding to Mapk1. c) Oleanolic Acid binding to Mapk1. d) Beta-Sitosterol binding to Mapk1. e) isorhamnetin binding to Mapk1. f) sitosterol binding to Mapk1. g) Beta-Sitosterol binding to Pik3r1. h) Oleanolic Acid binding to Irs1. i) Docosahex-aenoic Acid binding to Irs1. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In the exploration of the binding ability of the active ingredient to the target, the research additionally visualized and analyzed the hydrogen bond formation in the molecular docking results. The research further analyzed the other forces involved in the binding between the drug and the target in the molecular docking results mentioned above, namely, electrostatic forces and hydrophobic forces, by using the visual analysis tool (Fig. 5B).
3.7 Molecular dynamics simulations
Root mean squared deviation (RMSD), root mean squared fluctuations (RMSF), radius of gyration (Rg), solvent accessible surface area (SASA), and H-bond analysis were obtained by analyzing and integrating the results of MD simulations. RMSD demonstrates the stability of the dynamic binding of active ingredients to the targets in the organism. The RMSD plot in this work (Fig. 6A) reflected that the RMSD curves of most of the critical active ingredients and hub targets fluctuate below 1 nm, which suggested that their binding is stable in the body environment. However, the RMSD curves of Docosahexaenoic Acid-Irs1, Beta-Sitosterol-Pik3r1, Oleanolic Acid-Irs1, and Quercetin-Mapk1 exhibited very drastic fluctuations in their trajectories in the 50 ns simulation, indicating that their dynamic binding in the organism is unstable. RMSF is typically used to reveal the flexible size of amino acid residues in target proteins when the active component binds to the target protein. As shown in Supplementary Fig. S1, the RMSF curves showed that most of the protein amino acid residues in the drug-target complexes had a slight jitter and were less than 0.3 nm. It is noticeable that most of the active ingredients bound Mapk1 protein with relatively mild jittering of protein amino acid residues, but the complexes formed by Oleanolic Acid and Isorhamnetin with Mapk1 were more violently jittered (Supplementary Fig. S1A). And all RMSF curves of the Irs1 target were slightly jittered and approximate (Supplementary Fig. S1B). Besides, the RMSF curve’s jitters for the Pik3r1 target (Supplementary Fig. S1C) were significantly larger than those of the above two targets, and the jitters were concentrated in the middle part of the curve. To exhibit the changes in the overall structure of the drug-target complex upon binding and the tightness of the target protein structure, the measurement of Rg was performed. The greater fluctuation of the Rg curve demonstrates that the binding of the active ingredient has a strong effect on the target. As illustrated in Fig. 6B, the Rg curves of Mapk1 and Irs1 targets were slightly fluctuating and distributed at 2.15 nm and 2.4 nm on average. However, the Rg curve of the Pik3r1 target fluctuated sharply. SASA was performed to calculate the area of the target exposed to the solvent system during the simulations. It is generally considered that the less violent the jittering of the SASA curve, the more tightly and stably the active ingredient binds to the target. The SASA plot (Fig. 6C) also showed that the curves of both Mapk1 and Irs1 targets are relatively smooth and approximate, but the curve of the pik3r1 target is jittering dramatically, which is consistent with the results of RMSF and Rg. H-bond analysis is a statistical analysis of the number of hydrogen bonds between the active ingredient and the target complex during the simulation. The plot of H-bond analysis (Supplementary Fig. S2) showed that most of the active ingredients formed hydrogen bonds with the targets during the dynamic simulations, and the hydrogen bonds were stable during the simulations. However, the H-bond curves of Oleanolic Acid-Mapk1 and Docosahexaenoic Acid-Irs1 had more vacancies that represented no hydrogen bond formation. Moreover, the H-bond analysis plot of Sistosterol-Mapk1 indicated that it had a slight hydrogen bond vacancy during the simulation.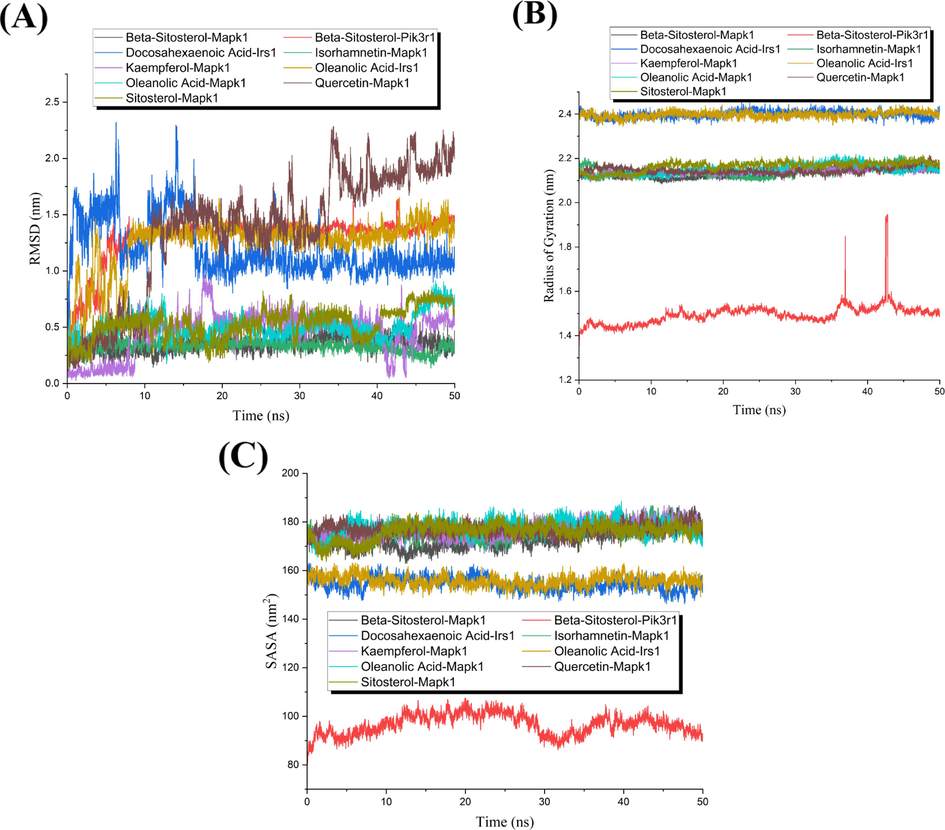
The analysis plots of MD simulations. (A) RMSD plot. (B) Rg Plot. (C) SASA plot. As illustrated in the figure, different color curves distinguished the different drug-target complexes.
Through combining and analyzing the above five results of MD simulations, it can be summarized that the Pik3r1 target jittered more violently in the plots of RMSD, RMSF, Rg, and SASA, indicating that the dynamic binding of the Pik3r1 target and the active ingredient was unstable. And the Irs1 target complexes and some of the Mapk1 target complexes were also unstable in dynamic simulations. Therefore, five critical active ingredients (kaempferol, Oleanolic Acid, Beta-Sitosterol, isorhamnetin and sitosterol) act on hub target Mapk1 were further verified and obtained for the treatment of obesity in rats based on the results of MD simulations.
4 Discussion
In the present study, a total of 58 active ingredients in Y-Q-Y-Y-Y were screened, which may serve as the material base for the treatment of obesity in rats. The core active ingredients were obtained through Cytoscape analysis, and the hub targets were screened according to PPI network analysis. The molecular docking results of the core active ingredients and the hub targets revealed that quercetin, kaempferol, oleanolic acid, beta-sitosterol, isorhamnetin, sitosterol and docosahexaenoic acid were the critical components in Y-Q-Y-Y-Y for the treatment of obesity in rats. These critical active ingredients belong to four groups, including flavonoids, pentacyclic triterpenoids, sterols, and fatty acids. Noticeably, most of these selected as main active ingredients are intersectional active ingredients of individual herbs, which suggests that each herb has an integral role in the treatment of obesity by Y-Q-Y-Y-Y. It also indicates the accuracy of this method to further investigate the core active ingredients acting as significant components in the treatment of obesity by Y-Q-Y-Y-Y. The active ingredients of flavonoid are quercetin, kaempferol, and isorhamnetin, which exhibit the efficacy of anti-inflammatory, antioxidant and facilitate the improvement of obesity-related diseases (Panche et al., 2016, Maleki et al., 2019). Quercetin can improve insulin resistance through anti-inflammatory effects and treat obesity by adjusting the intestinal flora (Chen et al., 2016, Sato and Mukai, 2020). Besides, quercetin could improve obesity by activating the non-shivering thermogenesis of brown adipose tissue and the browning of white adipose tissue. It can also activate brown adipose tissue non-shivering thermogenesis and white adipose tissue browning to improve obesity (Arias et al., 2017, Perdicaro et al., 2020). Kaempferol could exert anti-inflammatory effects by inhibiting gene activity and protein expression, improving hyperlipidemia and reducing visceral fat in rats by increasing hepatic PPARα levels (Chang et al., 2011, Alam et al., 2020). Isorhamnetin inhibits adipogenesis by down-regulating PPAR-γ gene expression and attenuates pro-inflammatory cytokine gene expression, thereby resulting in hypolipidemic and anti-inflammatory effects (Lee et al., 2009, Gong et al., 2020). The active ingredient of pentacyclic triterpenoids is Oleanolic Acid, which has anti-inflammatory, anti-diabetic, hepatoprotective, and hypolipidemic effects (Tian et al., 2002, Dzubak et al., 2006, Castellano et al., 2022). The active ingredients of sterols group are beta-sitosterol and sitosterol. Natural phytosterols could lower blood lipids and improve obesity-related diseases (Vezza et al., 2020). Beta-sitosterol may improve obesity in rats through anti-inflammation, up-regulation of insulin sensitivity, down-regulation of IKKβ/NF-κB and JNK signaling pathways (Gumede et al., 2020, Jayaraman et al., 2021). The active ingredient in the fatty acid group is docosahexaenoic acid. The fatty acids, especially unsaturated fatty acids, are known to regulate blood sugar, participate in lipid metabolism and inflammation, and improve obesity-related diseases (Wrzosek et al., 2022). Additionally, docosahexaenoic acid is negatively associated with fat accumulation (Zhuang et al., 2019). Overall, the above-related literature also further suggested that the screened main active ingredients have significant roles in the treatment of obesity in rats by Y-Q-Y-Y-Y, which deserves further in-depth research and exploration of its molecular mechanism. In addition, other active ingredients explored by network topology analysis should not be neglected and may also have therapeutic roles.
Through further exploration, 1128 targets were selected as intersectional targets for Y-Q-Y-Y-Y for the treatment of obesity in rats. From Fig. 1C, it can be clearly seen that the intersectional targets account for a large percentage of all drug targets but a smaller percentage of all disease targets. This percentage analysis also side by side verifies the accuracy of the targets collected through the extensive database, and that Y-Q-Y-Y-Y may be able to exert excellent therapeutic effects on obesity in rats through these intersectional targets. Among these intersectional targets, five core targets of Y-Q-Y-Y-Y for the treatment of obesity in rats were obtained through PPI network analysis, which were Mapk1, Pik3r1, Irs1, Cd4, and Erbb2. Mapk1 is mitogen-activated protein kinase 1, which is involved in cell growth, survival, activation, and apoptosis, as well as mitosis, and plays an important role in response to external stimulation by insulin (Pearson et al., 2001, Kyriakis and Avruch, 2001). Pik3r1 is phosphatidylinositol 3-kinase regulatory subunit alpha. As a component required for insulin-stimulated increases in glucose uptake and glycogen synthesis. Pik3r1 also plays a role in improving glucose tolerance by modulating the cellular response to ER stress (Draznin, 2006). Irs1 is the insulin receptor substrate 1 that mediates the control of various cellular processes by insulin. The expression of Irs1 is dramatically downregulated in obesity-related conditions such as diabetes (Biddinger and Kahn, 2006). Cd4 is the T-cell surface glycoprotein CD4. Cd4+ T cells in adipose tissue are associated with inflammation and insulin resistance in the body (McLaughlin et al., 2014, Zhao et al., 2018) and Cd4+ T cells contribute to obesity memory (Zou et al., 2018). Erbb2, tyrosine-protein kinase erbB-2, is a gene associated with cancer. Furthermore, Erbb2 is involved in the upstream of the PI3K-Akt signaling pathway and MAPK signaling pathway, which may play a role in the treatment of obesity.
In GO enrichment analysis, the intersectional targets are enriched in terms of BP, CC, and MF, respectively. The results indicated that the active ingredients may be enriched in the organism through receptor complexes, glycolipid-rich structural domains, neuronal cell bodies, and other cellular components such as protein kinase complexes, exerting molecular functions such as neurotransmitter receptor activity, G protein-coupled peptide receptor activity, hormone binding, protein kinase binding, carbohydrate binding, and participating in MAPK cascade regulation, negative regulation of the inflammatory response, circulatory system processes, and secretion regulation of the body, and a series of other biological processes. Equally, the KEGG enrichment analysis revealed that six of the top 20 pathways were associated with obesity in rats, which were the PI3K-Akt signaling pathway, the AGE-RAGE signaling pathway in diabetic complications, Lipid and atherosclerosis, MAPK signaling pathway and Th17 cell differentiation, and Insulin resistance (Endo et al., 2015, Suriagandhi and Nachiappan, 2022, Dong et al., 2022). Proteins interact with each other, and a drug acting on a single target may influence a large number of targets through the target it is acting on. Similarly, other obesity-related pathways and other top-ranked enriched pathways also have a great potential to be acted upon by a single target to cause a great effect, thus achieving the effect of treating obesity in rats. Therefore, while exploring the main enriched pathways and the basic enriched pathways, other enriched pathways also exist as a pathway to explore the molecular mechanism of disease treatment. Interestingly, the results suggested that the PI3K-Akt signaling pathway and MAPK signaling pathway may be the basic pathways of action of the active ingredients in Y-Q-Y-Y-Y for the treatment of obesity in rats.
Additionally, the molecular docking of the core active ingredients with the hub targets was performed by Autodock software. The results showed that the seven critical active ingredients present in the four herbs of Y-Q-Y-Y-Y had excellent binding capability with hub targets, especially with Pik3r1, Mapk1, and Irs1. Therefore, the 7 active ingredients (quercetin, kaempferol, oleanolic acid, beta-sitosterol, isorhamnetin, sitosterol, and docosahexaenoic acid) and three targets (Mapk1, Pik3r1, and Irs1) were considered as critical active ingredients and hub targets of Y-Q-Y-Y-Y for the treatment of obesity in rats. Besides, these core active ingredients bind to the amino acids on the core target site through multiple hydrogen bonds, indicating their stable binding affinity. To further plainly analyze the binding ability of the molecular docking results, the core active ingredients with the hub targets of the specific molecular docking sites about bond formation and forces are further shown in Fig. 5B. Hydrogen bond formation is one of the most crucial parameters for investigating the binding ability of an active ingredient to a target. The forces formed at the molecular docking site should not be ignored, however. The diagram clearly shows the formation of numerous hydrogen bonds, electrostatic forces, and hydrophobic forces between the critical active ingredients and the hub targets. Among them, the green ones are hydrogen bonds, the orange ones are electrostatic forces, and the pink ones are hydrophobic forces. The number of hydrophobic groups generating hydrophobic forces varies among the different docking results, whereas the more hydrophobic groups are available from the binding free energy of the combined molecular docking results, the more stable the binding between the active ingredient and the target site is made to be. It may be attributed to the fact that the hydrophobic groups provide the critical active ingredient with a hydrophobic environment suitable for its stable binding. In addition, electrostatic forces may also be of importance in stabilizing the binding of drug targets. Therefore, by analyzing the binding free energy, hydrogen bonding, and various forces mentioned above, the 7 active ingredients and 3 targets mentioned above were selected as the critical active ingredients and core targets for the treatment of obesity in rats with Y-Q-Y-Y-Y.
MD simulations were performed based on the results of molecular docking, which were available for further screening and validating hub targets and critical active ingredients for Y-Q-Y-Y-Y against obesity in rats. The seamless combination of molecular docking and MD simulations can further explore and validate the research results. In the RMSD plot, the stable and unstable drug-target complexes were clearly distinguished and exhibited. And the plots of RMSF, Rg, and SASA demonstrated that most of the Mapk1 target complexes and Irs1 target complexes curves displayed consistent and slight jitter. However, the above three plots all showed that the curves of the Pik3r1 target complex fluctuated greatly, which indicated that the active ingredient binding to the Pik3r1 target was not steady. Through the overall analysis, the possible reason for Pik3r1 target complexes’ violent jitter is not only the irregular curl of the Pik3r1 target itself but also the unstable binding of Pik3r1 target complexes. Moreover, in the RMSF plots of Mapk1 target, Oleanolic Acid and Isorhamnetin showed the most violent jitter during dynamic binding to Mapk1. Nevertheless, when analyzed together with the RMSD curves, the reason for their violent jitter may be due to the tighter binding of these two active ingredients to the Mapk1 target, resulting in conformational or flexible residue changes in the Mapk1 target. And the fluctuations of the RMSF curves formed by the binding of Oleanolic Acid to Mapk1 were mostly at the maximum of the fluctuations of the binding curves of other active ingredients to Mapk1, which also indicated that Oleanolic Acid was more tightly bound to Mapk1. Interestingly, quercetin showed specific peaks in the RMSF curves with Mapk1 compared to other active ingredients. The reason for the peak specificity of quercetin in combination with the RMSD curves may be that during the simulation it was always moving between the cavities of the Mapk1 target, constantly searching for new binding sites, resulting in a certain effect on many flexible peptides of the target. Despite the plots of RMSF, Rg, and SASA showing that the curves of the Irs1 target complexes were consistent and slightly jittering, the RMSD plot and H-bond analysis of the Irs1 target complexes showed that the dynamic binding of the Irs1 target to the active ingredients is unstable. From the H-bond analysis plots, most of the target complexes showed stable hydrogen bond formation throughout the dynamic binding processes. However, oleanolic acid exhibited a prolonged hydrogen bond-free combination to Mapk1 and Irs1 during the dynamic binding, and their RMSD curves also showed that dynamic binding of these drug-target complexes was unstable. As a consequence, 5 critical active ingredients (kaempferol, Oleanolic Acid, Beta-Sitosterol, isorhamnetin and sitosterol) acting on Mapk1 target were obtained for Y-Q-Y-Y against obesity in rats by comprehensive analysis of MD simulations.
The underlaying mechanism of action was explored based on a network pharmacology approach combined with molecular docking and MD simulations. The critical active ingredients of Y-Q-Y-Y-Y, the core obesity-related targets, and the acting mechanisms of the drug-target complexes, as well as the potential enriched pathways, were revealed, which could lay a theoretical basis for an in-depth exploration of the mechanism of action of Y-Q-Y-Y-Y in the treatment of obesity and provide new insights for the treatment of obesity-related diseases.
5 Conclusions
The research unraveled the underlying mechanism of Y-Q-Y-Y-Y for the treatment of obesity in rats through the innovative approach of network pharmacology combined with molecular docking and MD simulations. The results of the present study further revealed the potential critical active ingredients, targets, and pathways of Y-Q-Y-Y-Y for the treatment of obesity in rats, as well as the potential mechanism of action between the critical active ingredients and the hub targets. In summary, the results indicated that Y-Q-Y-Y-Y may exert pharmacological effects in a multi-component, multi-target, multi-pathway way. The main pathways of action of Y-Q-Y-Y-Y in treating obesity in rats may be the MAPK signaling pathway, the PI3K-Akt signaling pathway, and the insulin resistance signaling pathway. The advanced research approach employed by the current study can provide an alternative and inspiration for other research about natural products treating diseases. Furthermore, the obtained results would facilitate further research on the mechanism of action of Y-Q-Y-Y-Y in treating obesity in animals and humans, and provide new insights for the treatment of obesity-related diseases.
CRediT authorship contribution statement
Feifei Sun: Conceptualization. Jinde Liu: Conceptualization. Jingfei Xu: Software, Data curation. Ali Tariq: Validation. Yongning Wu: Supervision, Project administration, Funding acquisition. Lin Li: Supervision, Project administration, Funding acquisition.
Acknowledgments
We would like to thank the financial support from the major projects of the National Natural Science Foundation of China (No. 22193064), Foundation of the higher education institutions of Anhui Province (No. KJ2021A0147), National undergraduate training program for innovation and entrepreneurship (No.202210364024), and the Animal-Derived Food Safety Innovation Team (ANRC2021040).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Saponin-rich extract of Tribulus terrestris alleviates systemic inflammation and insulin resistance in dietary obese female rats: Impact on adipokine/hormonal disturbances. Biomed. Pharmacother.. 2022;147:112639
- [CrossRef] [Google Scholar]
- Kaempferol as a dietary anti-inflammatory agent: current therapeutic standing. Molecules. 2020;2518
- [CrossRef] [Google Scholar]
- Amberger, J. S., Bocchini, C. A., Schiettecatte, F., Scott, A. F. and Hamosh, A., 2015. OMIM.org: Online Mendelian Inheritance in Man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 43Database issue, D789-798. 10.1093/nar/gku1205.
- The protective effect of Tribulus terrestris in diabetes. Ann. N. Y. Acad. Sci.. 2006;1084:391-401.
- [CrossRef] [Google Scholar]
- A combination of resveratrol and quercetin induces browning in white adipose tissue of rats fed an obesogenic diet. Obesity (Silver Spring). 2017;251:111-121.
- [CrossRef] [Google Scholar]
- PharmGKB: A worldwide resource for pharmacogenomic information. Wiley Interdiscip. Rev. Syst. Biol. Med.. 2018;104:e1417.
- [Google Scholar]
- From mice to men: insights into the insulin resistance syndromes. Annu. Rev. Physiol.. 2006;68:123-158.
- [CrossRef] [Google Scholar]
- Oleanolic acid: extraction, characterization and biological activity. Nutrients. 2022;143
- [CrossRef] [Google Scholar]
- Chang, Chia Ju, Tzeng, Thing-Fong, Liou, Shorong-Shii, Chang, Yuan-Shiun and Liu, I-Min, 2011. Kaempferol regulates the lipid-profile in high-fat diet-fed rats through an increase in hepatic PPARα levels. Planta medica. 7717, 1876-1882. 10.1055/s-0031-1279992.
- Therapeutic effects of quercetin on inflammation, obesity, and type 2 diabetes. Mediators Inflamm.. 2016;2016:9340637.
- [CrossRef] [Google Scholar]
- EVenn: Easy to create repeatable and editable Venn diagrams and Venn networks online. J. Genet. Genomics. 2021;489:863-866.
- [CrossRef] [Google Scholar]
- Hua-Tan-Sheng-Jing decoction treats obesity with oligoasthenozoospermia by up-regulating the pi3k-akt and down-regulating the jnk mapk signaling pathways: at the crossroad of obesity and oligoasthenozoospermia. Front. Pharmacol.. 2022;13:896434
- [CrossRef] [Google Scholar]
- Molecular mechanisms of insulin resistance: serine phosphorylation of insulin receptor substrate-1 and increased expression of p85alpha: the two sides of a coin. Diabetes. 2006;558:2392-2397.
- [CrossRef] [Google Scholar]
- Pharmacological activities of natural triterpenoids and their therapeutic implications. Nat. Prod. Rep.. 2006;233:394-411.
- [CrossRef] [Google Scholar]
- Obesity drives th17 cell differentiation by inducing the lipid metabolic kinase, acc1. Cell Rep.. 2015;126:1042-1055.
- [CrossRef] [Google Scholar]
- Evaluation of AutoDock and AutoDock Vina on the CASF-2013 Benchmark. J. Chem. Inf. Model.. 2018;588:1697-1706.
- [CrossRef] [Google Scholar]
- Isorhamnetin: a review of pharmacological effects. Biomed. Pharmacother.. 2020;128:110301
- [CrossRef] [Google Scholar]
- Beta-sitosterol shows potential to protect against the development of high-fructose diet-induced metabolic dysfunction in female rats. J. Med. Food. 2020;234:367-374.
- [CrossRef] [Google Scholar]
- The protective effect of Yi-Qi-Yang-Yin-Ye, a compound of traditional Chinese herbal medicine in diet-induced obese rats. Am. J. Chin. Med.. 2008;364:705-717.
- [CrossRef] [Google Scholar]
- The effective fraction isolated from Radix Astragali alleviates glucose intolerance, insulin resistance and hypertriglyceridemia in db/db diabetic mice through its anti-inflammatory activity. Nutr. Metab. (Lond.). 2010;7:67.
- [CrossRef] [Google Scholar]
- TCMID 2.0: a comprehensive resource for TCM. Nucleic Acids Res.. 2018;46D1:D1117-D1120.
- [CrossRef] [Google Scholar]
- β-sitosterol circumvents obesity induced inflammation and insulin resistance by down-regulating IKKβ/NF-κB and JNK signaling pathway in adipocytes of type 2 diabetic rats. Molecules. 2021;267:2101.
- [CrossRef] [Google Scholar]
- PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res.. 2021;49D1:D1388-D1395.
- [CrossRef] [Google Scholar]
- Canaries in the coal mine: a cross-species analysis of the plurality of obesity epidemics. Proc. Biol. Sci.. 2011;2781712:1626-1632.
- [CrossRef] [Google Scholar]
- Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev.. 2001;812:807-869.
- [CrossRef] [Google Scholar]
- Isorhamnetin represses adipogenesis in 3T3-L1 cells. Obesity (Silver Spring). 2009;172:226-232.
- [CrossRef] [Google Scholar]
- Exploring the mechanism of ellagic acid against gastric cancer based on bioinformatics analysis and network pharmacology. J. Cell Mol. Med. 2023
- [CrossRef] [Google Scholar]
- Lu, YQ, Ding, H, Shi, ZL, Lin, HB and Zhang, GC, 2022. Study on the mechanism of action of Ephedra Herba Decoction against influenza A virus based on network pharmacology. TMR Modern Herbal Medicine. 52, 10. 10.53388/MHM2022A0502001.
- Improvement effects of ethanol extract from Taxillus sutchuenensis on blood glucose level, liver and renal complications in type 2 diabetes mellitus model mice and its mechanism. China Pharm. 2019:796-801.
- [Google Scholar]
- T-cell profile in adipose tissue is associated with insulin resistance and systemic inflammation in humans. Arterioscl. Throm. Vas.. 2014;3412:2637-2643.
- [CrossRef] [Google Scholar]
- Heterogeneous parallelization and acceleration of molecular dynamics simulations in GROMACS. J. Chem. Phys.. 2020;15313:134110
- [CrossRef] [Google Scholar]
- Pearson, Gray, Robinson, Fred, Beers Gibson, Tara, Xu, Bing-e, Karandikar, Mahesh, Berman, Kevin and Cobb, Melanie H, 2001. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev. 222, 153-183. 10.1210/edrv.22.2.0428.
- Quercetin attenuates adipose hypertrophy, in part through activation of adipogenesis in rats fed a high-fat diet. J. Nutr. Biochem.. 2020;79:108352
- [CrossRef] [Google Scholar]
- TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform.. 2014;6:13.
- [CrossRef] [Google Scholar]
- Safran, M., Dalah, I., Alexander, J., Rosen, N., Iny Stein, T., Shmoish, M., Nativ, N., Bahir, I., Doniger, T., Krug, H., Sirota-Madi, A., Olender, T., Golan, Y., Stelzer, G., Harel, A. and Lancet, D., 2010. GeneCards Version 3: the human gene integrator. Database (Oxford). 2010, baq020. 10.1093/database/baq020.
- Modulation of chronic inflammation by quercetin: the beneficial effects on obesity. J. Inflamm. Res.. 2020;13:421-431.
- [CrossRef] [Google Scholar]
- The obesity epidemic and bariatric trends. Clin. Plast. Surg.. 2019;461:1-7.
- [CrossRef] [Google Scholar]
- Su, G., Morris, J. H., Demchak, B. and Bader, G. D., 2014. Biological network exploration with Cytoscape 3. Curr Protoc Bioinformatics. 47, 8 13 11-24. 10.1002/0471250953.bi0813s47.
- Therapeutic target analysis and molecular mechanism of melatonin - treated leptin resistance induced obesity: a systematic study of network pharmacology. Front. Endocrinol.. 2022;13:927576
- [CrossRef] [Google Scholar]
- The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res.. 2021;49D1:D605-D612.
- [CrossRef] [Google Scholar]
- Survey of pharmacology of oleanolic acid. Zhongguo Zhong Yao Za Zhi. 2002;2712(884–886):901.
- [Google Scholar]
- UniProt Consortium, T., 2018. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 465, 2699. 10.1093/nar/gky092.
- Phytosterols: nutritional health players in the management of obesity and its related disorders. Antioxidants (Basel). 2020;912
- [CrossRef] [Google Scholar]
- TCM network pharmacology: A new trend towards combining computational, experimental and clinical approaches. Chin. J. Nat. Med.. 2021;191:1-11.
- [CrossRef] [Google Scholar]
- Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res.. 2020;48D1:D1031-D1041.
- [CrossRef] [Google Scholar]
- Effects of extract of herba Taxilli on glucose consumption in cultured HePg2 cells of human. Chin. Arch. Tradit. Chin. Med. 2006:442-443.
- [Google Scholar]
- DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res.. 2018;46D1:D1074-D1082.
- [CrossRef] [Google Scholar]
- Impact of fatty acids on obesity-associated diseases and radical weight reduction. Obes. Surg.. 2022;322:428-440.
- [CrossRef] [Google Scholar]
- SymMap: an integrative database of traditional Chinese medicine enhanced by symptom mapping. Nucleic Acids Res.. 2019;47D1:D1110-D1117.
- [CrossRef] [Google Scholar]
- ETCM: an encyclopaedia of traditional Chinese medicine. Nucleic Acids Res.. 2019;47D1:D976-D982.
- [CrossRef] [Google Scholar]
- Using PyMOL as a platform for computational drug design. Wiley Interdiscip. Rev.: Comput. Mol. Sci.. 2017;72:e1298.
- [Google Scholar]
- CD4(+) T cells in obesity and obesity-associated diseases. Cell. Immunol.. 2018;332:1-6.
- [CrossRef] [Google Scholar]
- Network pharmacology combined with bioinformatics to investigate the mechanisms and molecular targets of astragalus radix-panax notoginseng herb pair on treating diabetic nephropathy. Evid. Based Complement. Alternat. Med.. 2021;2021:9980981.
- [CrossRef] [Google Scholar]
- Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun.. 2019;101:1523.
- [CrossRef] [Google Scholar]
- Antidiabetic and antioxidant effects of catalpol extracted from Rehmannia glutinosa (Di Huang) on rat diabetes induced by streptozotocin and high-fat, high-sugar feed. Chin. Med.. 2016;11:25.
- [CrossRef] [Google Scholar]
- Differential anti-adipogenic effects of eicosapentaenoic and docosahexaenoic acids in obesity. Mol. Nutr. Food Res.. 2019;6314:e1801135.
- [Google Scholar]
- CD4+T cells memorize obesity and promote weight regain. Cell. Mol. Immunol.. 2018;156:630-639.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2023.105390.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary data 1
Supplementary data 1







