Translate this page into:
Multi-wavelength HPLC fingerprint similarity metric for cold-hot nature identification of Chinese herbal medicines
⁎Corresponding authors. bmie530@163.com (Guohui Wei), zhenguow@126.com (Zhenguo Wang)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Cold-hot nature theory is the core basic theory of the nature of Chinese herbal medicines (CHMs). It is found that the material basis of cold-hot nature is CHM ingredients. In view of this, our group proposed a scientific hypothesis that “CHMs with similar nature should have similar material basis”. To demonstrate this hypothesis, we developed a novel multi-wavelength high performance liquid chromatography (HPLC) fingerprint similarity metric scheme for cold-hot nature identification. We explored a multi-wavelength distance metric learning model to compute the similarity of CHM ingredients, and developed an improved k-nearest neighbor algorithm based on multi-wavelength HPLC fusion (KMHF) to predict cold-hot nature of CHMs. Firstly, multi-wavelength HPLC fingerprints were used to extract the characteristic information of CHM ingredients. Secondly, we defined the similarity of CHM ingredients as semantic relevance and fingerprint similarity. We studied a multi-wavelength distance metric to measure the similarity of CHM ingredients. The learned distance metric could discover complementary characteristics of different wavelength HPLC through an optimization algorithm. Finally, an improved multi-wavelength k-nearest neighbor algorithm KMHF was proposed to analyze the relationship between cold-hot nature and CHM ingredients. Numerous experiments were designed to test the feasibility of the proposed KMHF algorithm. Experimental results indicate that the performance of our KMHF algorithm outperforms that of the compared algorithms. Experimental results demonstrate that the hypothesis that CHMs with similar cold-hot nature have similar material basis. The KMHF model is evaluated to be feasible for nature identification.
Keywords
Cold-hot nature
Nature identification
Chinese herbal medicines
Similarity metric
HPLC
1 Introduction
Cold-hot nature theory is the core theory of the nature of Chinese herbal medicines (CHMs), which has attracted the attention of investigators for many years (Gao et al., 2007). “Treating the cold syndrome with hot nature medicines and treating the hot syndrome with cold nature medicines” is the theoretical basis of clinical treatment of traditional Chinese medicine (TCM). This suggests that the concept of cold-hot nature patterns has been a guiding principle in TCM for administering CHMs (Ouyang et al., 2006; Wu et al., 2007). Therefore, correct discrimination of cold-hot nature of CHMs is the key to TCM research.
Numerous specialists attempted to explain the scientific connotation of cold-hot nature of CHMs from different perspectives. From one perspective, the material basis of cold-hot nature is confirmed as CHM ingredients (Fu et al., 2017; Wei et al., 2019b). Scientists developed numerous scientific hypotheses to demonstrate that the material basis of cold-hot nature was CHM ingredients, including Zhang’s hypothesis of “Three element” (Jin et al., 2014), Wang’s hypothesis of “Tri-element of property-effect-material”(Zhang, 2012), and Fu’s hypothesis of “Nature-Structure Relationship” (Fu et al., 2017). They constructed a lot of experiments to prove that the hypothesis was true. A typical method was to extract the ingredient information with chemical fingerprints, and establish the correlation between cold-hot nature and CHM ingredients with machine learning algorithms (Wei et al., 2019b). From another perspective, energy metabolism, such as oxygen consumption and ATPase activity, was introduced to study cold-hot nature of CHMs (Huang et al., 2014). CHMs with hot nature may increase the activity of SDH enzyme and promote the decomposition of muscle glycogen. These regulated the level of energy metabolism, so as to produce more ATP. CHMs with cold nature could significantly decrease the energy metabolism in normal rats (Wu et al., 2007). From the third perspective, some research analyzed cold-hot nature of CHMs with bioinformatics methods. Network pharmacology and in silico analysis were introduced to reveal the scientific connotation of cold-hot nature (Fu et al., 2017; Liang et al., 2013). Fu et al. proposed a hypothesis of “nature-structural relationship”, and integrated bioinformatics and network pharmacology methods to explore the scientific connotation of cold-hot nature of CHMs from molecular level (Shao et al., 2020). They found that CHMs with cold nature were related with mental and behavioural disorders diseases, and CHMs with hot nature were associated with endocrine, nutritional and metabolic diseases. As mentioned above, researches from different perspectives have made numerous achievements. However, the scientific connotation of cold-hot nature of CHMs needs further study. Our group attempts to utilize machine learning methods and chemical fingerprints to build a correlation between cold-hot nature and CHM ingredients to reveal the scientific connotation of CHM nature.
A number of studies have been performed to analyze the relationship between CHM ingredients and cold-hot nature (Fu et al., 2017; Wei et al., 2019b; Shao, 2020). It is found that the material basis of cold-hot nature of CHMs is chemical ingredients. Therefore, research on the relationship between cold-hot nature and CHM ingredients mainly includes ingredient information representation and nature classification. Ingredient information is the general characteristic of the ingredients contained in CHMs. Ingredient information representation has always been a research hotspot. Current research focused on chemical fingerprints and molecular descriptors of compounds. Chemical fingerprints, including infrared spectrum, ultraviolet spectrum, gas chromatography (GC), high performance liquid chromatography (HPLC), have been usually applied to study the ingredients of CHMs (Zhang, 2012). Wang et al. used HPLC fingerprint and gas chromatography fingerprint for systematic analysis of chemical compositions in Curcumae Rhizom and introduced chemometrics including unsupervised principal component analysis, supervised linear discriminant analysis, k-nearest neighbors (KNN) for the species authentication and quality control (Wang et al., 2021). CHMs are typically mixtures of compounds. Since chemical structure is the molecular basis of compound activity, characterization of molecular structure is essential to further understand CHM nature. Molecular descriptors were widely applied to extract the feature information of CHM compounds. Fu et al. computed compound-nature pairs of CHMs to study their physicochemical domain and introduced in silico target prediction to study differences related to their modes-of-action against proteins (Fu et al., 2017).
Nature classification introduces classical intelligent algorithms or builds machine learning algorithms to study the relationship between CHM ingredients and cold-hot nature. Classical intelligent algorithms, such as support vector machine, partial least square method and random forest, were usually introduced to predict cold-hot nature of CHMs. Xue’s group analyzed the CHMs with efficacy and indications, and constructed classical intelligent algorithms for cold-hot nature prediction (Zhang, 2012). Long et al. and Wang et al. calculated molecular descriptor of CHM compounds, and applied classical classifiers to discriminate cold-hot nature (Long et al., 2011; Wang et al., 2016). Nie et al. analyzed Metabonomics information of CHMs and used a random forest algorithm to identify the cold-hot nature of CHMs (Nie et al., 2015). Our group had made some explorations in the identification of cold-hot nature of CHMs (Wei et al., 2019b, 2021a). We introduced an extreme learning machine (ELM) algorithm to analyze CHM nature with molecular descriptors. Inspired by the similarity of CHM ingredients applied to evaluate the quality of CHMs (Wei et al., 2021a), our group explored the similarity of CHM ingredients to build machine learning algorithms for cold-hot nature prediction. For example, we proposed a novel multi-solvent UV spectrum similarity measure retrieval scheme for discriminating CHMs cold or hot (Wei et al., 2019b).
As mentioned above, numerous achievements have been made in the research of cold-hot nature. However, chemical fingerprint technology for nature prediction has not been comprehensively studied. Our group used UV spectrum and GC to analyze CHM ingredients for nature identification without considering HPLC (Wei et al., 2019b, 2021b). Compared with UV spectrum and GC, HPLC can better separate the components of CHMs and extract the information of CHM components (Qi et al., 2011). It is possible to obtain high prediction accuracy of cold-hot nature by studying the identification method of CHM nature based on HPLC. Furthermore, there is a hypothesis that CHMs with similar cold-hot nature have similar material basis. Designing a special nature identification algorithm according to this hypothesis and HPLC fingerprints may achieve higher prediction accuracy rates. In this work, HPLC fingerprints were applied to extract the information of CHM ingredients. With the obtained HPLC fingerprints, the similarity of CHM ingredients was defined as a Mahalanobis distance metric. This distance metric was learned by a constructed distance metric learning model. Finally, an improved multi-wavelength k-nearest neighbor algorithm was developed for predicting cold-hot nature of CHMs.
2 Materials and methods
2.1 CHM dataset
In this work, representative CHMs with clear nature were selected to study the correlation between CHM ingredients and cold-hot nature (Zhang, 2012). All selected CHMs were recorded in ‘Shen Nong’s Herbal Classic’ and the classical ‘Chinese Materia Medica’. The screening criteria of representative CHMs are as follows: (1) Traditional natural plant medicine only; (2) Clear CHM nature, high clinical recognition and no academic disputes. Finally, 61 CHMs were screened for nature identification, in which 30 CHMs are cold, and others are hot. The 61 CHMs are listed in Table 1.
Chinese Herbal Medicines
Nature
Source
Sampling area
Curculiginis Rhizoma
Hot
Hai Yao Ben Cao
Yibin, Sichuan
Pinelliae Rhizoma
Hot
Shen Nong’s Herbal Classic
Dazhou, Sichuan
Magnoliae Officinalis Cortex
Hot
Shen Nong’s Herbal Classic
Guangyuan, Sichuan
Euodiae Fructus
Hot
Shen Nong’s Herbal Classic
Tongren, Guizhou
Arisaematis Rhizoma
Hot
Shen Nong’s Herbal Classic
Heze, Shandong
Ephedrae Herba
Hot
Shen Nong’s Herbal Classic
Chifeng, Sichuan
Chuanxiong Rhizoma
Hot
Shen Nong’s Herbal Classic
Pengzhou, Sichuan
Zingiberis Rhizoma
Hot
Shen Nong’s Herbal Classic
Leshan, Sichuan
Corydalis Rhizoma
Hot
Paozhi Lun
Jinhua, Zhejiang
Chaenomelis Fructus
Hot
Shen Nong’s Herbal Classic
Xuancheng, Anhui
Aucklandiae Radix
Hot
Shen Nong’s Herbal Classic
Lijiang, Yunnan
Eucommiae Cortex
Hot
Shen Nong’s Herbal Classic
Mianyang, Sichuan
Santali Albi Lignum
Hot
Mingyi bielu
Guangdong
Epimedii Folium
Hot
Shen Nong’s Herbal Classic
Shanxi
Roasted Corydalis
Hot
Paozhi Lun
Jinhua, Zhejiang
Nardostachyos Radix et Rhizoma
Hot
Supplement to Materia Medica
Aba, Sichuan
Fructus Piperis Alba
Hot
Tang materia medica
Wenchang, Hainan
Mustard Seeds
Hot
Tang materia medica
Anhui
Carthami Flos
Hot
Tang materia medica
Xinxiang, Henan
Asari Radix et Rhizoma
Hot
Shen Nong’s Herbal Classic
Dandong, Liaoning
Notopterygii Rhizoma et Radix
Hot
Shen Nong’s Herbal Classic
Aba, Sichuan
Cinnamomi Cortex
Hot
Shen Nong’s Herbal Classic
Hechi, Guangxi
Atractylodis Rhizome
Hot
Shen Nong’s Herbal Classic
Jiangsu
Alpiniae Katsumadai Semen
Hot
Paozhi Lun
Hainan
Piperis Longi Fructus
Hot
Tang materia medica
Wenchang, Hainan
Ligustici Rhizoma et Radix
Hot
Shen Nong’s Herbal Classic
Aba, Sichuan
Psoraleae Fructus
Hot
Nature theory
Sichuan
Aconiti Lateralis Radix Praeparata
Hot
Shen Nong’s Herbal Classic
Jiangyou,Sichuan Province
Citri Reticulatae Pericarpium
Hot
Shen Nong’s Herbal Classic
Jiangmen,Guangdong
Alpiniae Officinarum Rhizoma
Hot
Mingyi bielu
Zhanjiang,Guangdong Province
Clematidis Radix et Rhizoma
Hot
Tang materia medica
Jiangsu
Platycladi Cacumen
Cold
Mingyi bielu
Linyi, Shandong
Kochiae Fructus
Cold
Shen Nong’s Herbal Classic
Feicheng, Shandong
Ecliptae Herba
Cold
Tang materia medica
Jinan, Shandong
Isatidis Folium
Cold
Mingyi bielu
Tangshan, Hebei
Rhei Radix et Rhizoma
Cold
Shen Nong’s Herbal Classic
Dingxi, Gansu
Asparagi Radix
Cold
Shen Nong’s Herbal Classic
Huairen, Guizhou
Fritillariae Cirrhosae Bulbus
Cold
Shen Nong’s Herbal Classic
Aba, Sichuan
Bupleuri Radix
Cold
Shen Nong’s Herbal Classic
Nanyang, Henan
Gardeniae Fructus
Cold
Shen Nong’s Herbal Classic
Zhangshu, Jiangxi
Rhizoma Anemarrhenae with Peet
Cold
Shen Nong’s Herbal Classic
Baoding, Hebei
Sargassum
Cold
Shen Nong’s Herbal Classic
Weihai, Shandong
Lophatheri Herba
Cold
Shen Nong’s Herbal Classic
Yuyao, Zhejiang
Trichosanthis Fructus
Cold
Shen Nong’s Herbal Classic
Feicheng, Shandong
Kansui Radix
Cold
Shen Nong’s Herbal Classic
Shanxi
Dried Rehmannia Root
Cold
Shen Nong’s Herbal Classic
Jiaozuo, Henan
Dianthi Herba
Cold
Shen Nong’s Herbal Classic
Laiwu, Shandong
Fraxini Cortex
Cold
Shen Nong’s Herbal Classic
Lingning
Arnebiae Radix
Cold
Shen Nong’s Herbal Classic
Urumqi, Xinjiang
Trachelospermi Caulis et Folium
Cold
Shen Nong’s Herbal Classic
Suzhou, Jiangsu
Aloe
Cold
Nature theory
Yunnan
Puerariae Lobatae Radix
Cold
Shen Nong’s Herbal Classic
Zibo, Shandong
Taraxaci_Herba
Cold
Tang materia medica
Linyi, Shandong
Menthae Haplocalycis Herba
Cold
Tang materia medica
Haimen, Jiangsu
Alizaris Radix
Cold
Tang materia medica
Zhenjiang, Jiangsu
Plantaginis Semen
Cold
Shen Nong’s Herbal Classic
Jiujiang, Jiangxi
Lonicerae Japonicae Flos
Cold
Tang materia medica
Linyi, Shandong
市
Stephaniae Tetrandrae Radix
Cold
Shen Nong’s Herbal Classic
Quzhou, Zhejiang
Phellodendri Chinensis Cortex
Cold
Shen Nong’s Herbal Classic
Bazhong, Sichuan
Coptidis Rhizome
Cold
Shen Nong’s Herbal Classic
Shizhu, Chongqing
Gentianae Radix et Rhizoma
Cold
Shen Nong’s Herbal Classic
Fushun, Liaoning
2.2 Hplc
In this work, we analyzed the ingredient information of CHMs with HPLC technology. The experimental methods of HPLC, including preparation of the test solution and chromatographic conditions, were configured in detail in Rf. Zhang, et al. 2012. We give a brief introduction as follows.
The preparation of the test solution is as follows: Firstly, we precisely measured about 0.5 g of the test medicinal powder, and put it in a tapered bottle with a stopper. Secondly, we precisely poured 50 ml of 50 % methanol into the bottle. We weighed and put it in 60℃ water bath for ultrasonic extraction for 30 min. After the extraction was completed, we cooled and weighed again, and supplemented the lost mass with 50 % methanol. Finally, we take the continuous filtrate to obtain a 50 % methanol extract. The chromatographic conditions are as follows: (1) Chromatographic column: Agilent XDB-C18 column (4.6 mm* 250 mm, 5 μm). (2) Mobile phase: acetonitrile–water (3:97) → acetonitrile–water (100:0), linear gradient elution for 90 min. (3) Flow rate: 1.0 ml/min. (4) Injection volume: 20 ml. (5) Column temperature: 35℃.
The test solution was obtained based on the given chromatographic conditions, and the DAD (diode array detector) was introduced for full wavelength scanning of 190–400 nm. Finally, each CHM was collected at 211 wavelengths of 190–400 nm, and the data were obtained for 6524 retention time points. Since the data are too large to allow further modeling and analysis, and the chromatographic data of the same CHM at adjacent wavelengths are highly correlated, the chromatographic data at representative wavelengths of each CHM were selected according to the characteristics of UV wavelength. In this study, the chromatographic data at three representative wavelengths of 210 nm, 227 nm, 236 nm were analyzed to build the nature classification model. We processed the representative fingerprints, and extracted the spectral interval with a prediction accuracy of more than 75 % based on a step length of 5 absorption values and an interval length of 95 absorption values. Finally, the absorption value of the fingerprint interval was adjusted in steps of 5.
2.3 HPLC fingerprint similarity
To analyze the relationship between CHM ingredients and cold-hot nature, our group developed a hypothesis that CHMs with similar cold-hot nature should have the similar material basis (Wei et al., 2021b). In our previous work, we had tested this hypothesis by characterizing the ingredient information with UV spectrum (Wei et al., 2019b, 2021a). In this work, our group attempted to reveal the relationship between CHM ingredients and cold-hot nature by testing this hypothesis with HPLC fingerprints. Therefore, CHMs with similar cold-hot nature should have similar HPLC fingerprints of CHMs. It means that if the HPLC fingerprints of two CHMs are similar, they are considered to be similar cold-hot nature.
The similarity of HPLC fingerprints had been widely investigated in studying CHM ingredients for quality evaluation of CHMs (Mao, 2020). In this work, the similarity of HPLC fingerprints was modeled for cold-hot nature prediction. Analyzing the definition of similarity, our group defined the similarity of HPLC fingerprints as semantic relevance and fingerprint similarity. Semantic relevance means the consistency of CHM cold-hot nature, which represents that if the cold-hot nature of two CHMs is similar, they are semantic similarity (Wei et al., 2018). Fingerprint similarity means the similarity of CHM HPLC fingerprints, which represents that two CHMs have similar ingredients related to cold-hot nature. We explored to learn a Mahalanobis distance to measure the similarity of HPLC fingerprints, which were both semantic relevance and fingerprint similarity. Smaller distance metric means more similar fingerprints.
2.3.1 Distance metric learning
Define
to represent the CHM fingerprint dataset in the input space, in which
being the total number of CHMs and
being the ith sample. Denote superscript T as the transpose of a vector or a matrix, the Mahalanobis distance between xi and xj can be defined as (Weinberger et al., 2009):
Eq. (2) illustrates that calculating Mahalanobis distance between and is equivalent to learning a transformation of Euclidean distance in the input space. In this work, the transformation matrix is learned based on the HPLC similarity of CHMs. After learning matrix , Mahalanobis distance between and in Eq. (2) can be obtained.
2.3.2 Similarity metric
In this work, a Mahalanobis distance was learned to quantify the similarity of HPLC fingerprints (Liu et al., 2010). However, previous distance metric learning studies mainly focused on the analysis of semantic relevance, ignoring the study of fingerprint similarity. We defined the similarity of CHM HPLC fingerprints as semantic relevance and fingerprint similarity. Therefore, the transformation matrix in Eq. (2) contained two sources: the transformation matrix of semantic relevance and the transformation matrix of fingerprint similarity.
The conception of semantic relevance describes the separability of cold and hot categories. This requires that the class separability increases when the inter class divergence matrix increases or the intra class divergence matrix decreases. We modeled the semantic relevance with differential scatter discriminant criterion (DSDC) algorithm (Wei et al., 2016), the formula is as follows:
Our model uses the variation of DSDC:
In Eq. (4), is the inter class divergence matrix, is the intra class divergence matrix. is a nonnegative balance parameter, which tunes the relative merits of maximizing the inter class divergence to the minimization of the intra class divergence. The obtained matrix. is the transformation matrix from semantic relevance.
According to the definition of fingerprint similarity, it describes the similarity of HPLC fingerprints. This represents the similarity of CHM ingredients. In previous studies, feature similarity of pulmonary nodule images had been modeled as patch alignment frameworks. Inspired by the definition of feature similarity, we explored the patch alignment framework to study the similarity of CHM HPLC fingerprints.
Define a HPLC fingerprint dataset in input space
, for each
, its local patch is
.
is the
nearest neighbor of
based on Euclidean distance. For each patch
, there is a transformation model
,
is the set of sample maps in feature space
. To solve such transformation model
, one solution is to minimize the error between the linear mapping of patch
and the feature representation
, and then align all the patches (Zhang et al., 2009). We minimized the local patch errors as:
In Eq. (3), is the local mapping matrix, denotes the bias, is the vector of all ones, is a regularization parameter.
There is an assumption that the fingerprint samples are centered, i.e.,
.To calculate the optimal solution of Eq. (5), we set the derivatives of the objective function in regard to
and
to zeros. The solution is:
In (7),
, and then, the global alignment is (Zhang et al., 2009):
In (9), is a selection matrix such that .
Define
is a representation of dataset
in feature space. On the basis of the assumption of linearization that
, the global patches errors is as follows:
Therefore, semantic relevance produced the transformation matrix
in Eq. (4), and fingerprint similarity learned the transformation matrix
in Eq. (10). By integrating Eq. (4) and Eq. (10), we obtained a similarity metric model. The model is as follows:
2.3.3 Projection learning
To calculate the optimal transformation matrix
in Eq.(11) for similarity metric, the low dimensional representation of HPLC fingerprints should avoid redundancy as much as possible. To solve this problem, an orthogonal projection learning method was studied. We learned the transformation matrix
as follows:
To solve Eq. (12), eigenvalue decomposition on matrix was used to obtain the optimal projections. We built the optimal solution matrix with eigenvectors of corresponding to the smallest eigenvalues.
2.4 Multi-wavelength HPLC fingerprint fusion
In this study, three wavelength HPLC fingerprints were used to analyze the ingredient information of CHMs. Different wavelength HPLC fingerprints mine different characteristics of CHM ingredients, which usually have different physical properties. Therefore, it is perhaps not optimal to concatenate three wavelength HPLC fingerprints straightforwardly into a long fingerprint vector (Yu et al., 2012). This would cause curse-of-dimensionality and over-fitting problems. In particular, it is difficult to learn a robust distance measure in a high-dimensional feature space if the number of CHM fingerprints is not large enough. To solve this problem, multi-wavelength HPLC fingerprint fusion scheme was explored for nature identification.
In this section, we extended single wavelength HPLC fingerprint similarity metric to multi-wavelength feature spaces. We utilized multi-wavelength HPLC fingerprints to learn multiple transformation matrices to build multi-wavelength distance metric. We linearly integrated the similarity metrics learned from multi-wavelength HPLC fingerprints with the weights
and added a regularizer to the weights. Thus, the objective function is as follows:
Therefore, the objective function (12) was constructed to learn a distance metric for each wavelength fingerprint data, while the objective function (13) was built to integrate the information of the multi-wavelength HPLC fingerprints with the combination weights. This scheme mitigates the over-fitting problem and decreases the complexity of the model.
To calculate the solution of objective function (13), firstly, the optimal transformation matrices
were learned on the basis of Eq. (12) corresponding to different wavelengths. Secondly, the optimum solution was obtained by introducing the lagrangian multiplier method. Based on a Lagrange multiplier
, the objective function is transforms to:
To solve this problem, we took the partial derivatives of
with regard to
and
to be zeros. We obtained:
Integrating the equations in (15), we obtained:
Since
, we get the solution of
as follows:
Putting the solution of
into (15), we can obtain:
With multi-wavelength HPLC fingerprint fusion, we could get the Mahalanobis distance
between
and
and weight value
corresponding to the
th wavelength HPLC fingerprints. Finally, we obtained a multi-wavelength Mahalanobis distance
between fingerprints
and
as follows:
2.5 Cold-hot nature identification scheme
As mentioned above, a multi-wavelength Mahalanobis distance
was learned to measure the similarity of CHM HPLC fingerprints. With this similarity metric, an improved k-nearest neighbor algorithm based on multi-wavelength HPLC fusion (KMHF) was proposed for cold-hot nature identification. Fig. 1 displays the cold-hot nature identification based on similarity metric of multi-wavelength HPLC. For a query CHM with unclear nature, we firstly studied the ingredient information of this CHM with multi-wavelength HPLC fingerprints. The multi-wavelength distance metric was learned to measure the similarity of HPLC fingerprints. With the learned distance metric, we then computed the Mahalanobis distances between the query CHM and CHM dataset. The calculated Mahalanobis distances were sorted from small to large in monotonically increasing order. The most similar
CHMs corresponding to the smallest
Mahalanobis distances were chosen to analyze cold-hot nature of the query CHM. Finally, we introduced a cold nature probability (
) to evaluate the cold-hot nature of this query CHM with the most similar
CHMs. The cold nature probabilities of the query CHMs represented their cold degree, which could be calculated by the ratio of the weights of cold CHMs to the total weights of CHMs retrieved. The cold nature probability was calculated as: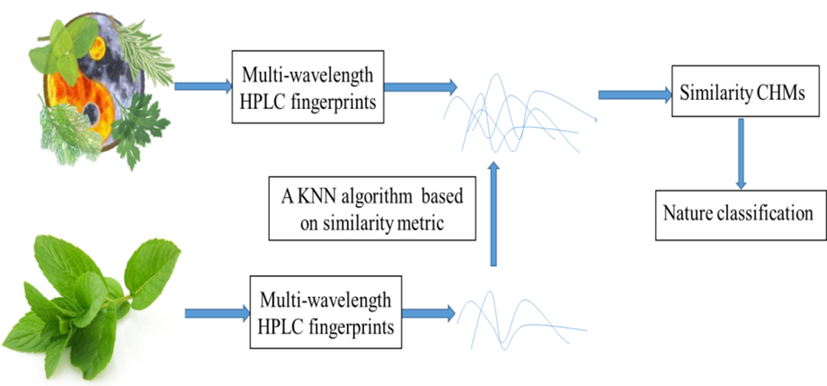
Cold-hot nature identification based on similarity metric of multi-wavelength HPLC.
2.6 The proposed KMHF scheme for nature identification
An improved k-nearest neighbor algorithm based on multi-wavelength HPLC fusion (KMHF).
Given a CHM HPLC dataset , is the wavelength HPLC fingerprints, , and the number of nature classes .
Transformation matrics construction. Calculate the matrix , which is corresponding to the th wavelength HPLC fingerprints. Eigenvalue decomposition on matrix to get the m eigenvalues of corresponding to the smallest m eigenvectors. Construct the transformation matrix with the smallest m eigenvectors.
Mahalanobis distance learning. Calculate between HPLC fingerprints and with the transformation matrices and the weight value based on Eq.(2) and (19).
Similarity metric. Retrieve the k most similar CHMs, which have the smallest k Mahalanobis distances between the query CHM and the CHM dataset.
Cold-hot nature identification. Compute the ratio of the weights of cold CHMs to the total weights of CHMs retrieved.
2.7 Performance evaluation
In this subsection, the feasibility and effectiveness of the KMHF scheme for cold-hot nature identification were evaluated with numerous built experiments. We compared the prediction identification performance of KMHF scheme with that of some classical algorithms, such as the classical distance metric learning algorithms (large margin nearest neighbor (LMNN) (Weinberger et al., 2009), information-theoretic metric learning (ITML)) (Davis et al., 2007), the cold-hot nature classification schemes (Pearson correlation coefficient (PCC) (Wei et al., 2021c), retrieval system (RS) (Wei et al., 2019a), and extreme learning machine (ELM)). All evaluation experiments were performed in the environment of multi-wavelength HPLC fingerprint dataset. The similar CHMs with clear nature were calculated to discriminate the cold-hot nature of CHMs with unclear nature. We firstly used multi-wavelength HPLC fingerprints to analyze the ingredient information of CHMs. Secondly, we developed a KMHF scheme to discriminate the cold-hot nature of CHMs. Finally, numerous experiments were built to evaluate the feasibility and effectiveness of the KMHF scheme.
In our experiments, we introduced stability evaluation to estimate the identification performance of our KMHF scheme. Stability evaluation describes the proportion of calculated similar CHMs that are semantic relevance with the query CHMs. Leave-one-CHM-out approach was introduced to assess the stability evaluation in the whole CHM multi-wavelength HPLC fingerprints. In each case, one CHM was left as the test query CHM, and remaining 60 CHMs were selected as the reference training CHM dataset. Because every CHM should be selected as a query-one, this process was performed 61 times. Cold probability of each test CHM was calculated to represent the extent that the nature of this CHM belongs to cold. In our scheme, we found
reference CHMs which were most similar to the query CHM, and calculated the cold probability of the query CHM. As a result, 61 cold probabilistic values were obtained. By varying the threshold of the cold probability, a Receiver Operating Characteristic (ROC) curve was drawn. The area under the ROC curve (AUC) and identification accuracy (ACC) were used to test the performance of our KMHF scheme. The formula of identification accuracy is defined as:
In (20), represents a ratio of the number of accurately identified CHMs from the th query CHM to the number of the most similar CHMs . Therefore, is a function of number , and ACC value changes with the change of number . Finally, the AUC and ACC values were calculated to analyze the performance of our KMHF scheme.
3 Results
3.1 Parameter configurations
In this study, several parameters in KMHF scheme should be optimized for cold-hot nature identification. The balance parameter in (4), parameter in (5), parameter in (13), parameter k in (20) need to be studied for the optimal identification model. All parameters are configured in the environment of multi-wavelength HPLC fingerprints.
Our experiments introduced stability evaluation to study the parameters for the optimal KMHF scheme. AUC and ACC were used as the evaluating indicators to evaluate the performance of our KMHF scheme with varying the values of parameters (
,
,
and k). As a result, AUC and ACC values were studied as functions of the tuning parameters (
,
,
and k) to describe more comprehensive curves for the optimal KMHF scheme. We tuned the parameter
in (4) in the range of [10-8, 10-6, 10-4, 10-2, 1, 102, 104, 106, 108]. Fig. 2 displays the AUC and ACC curves for nature identification of CHM HPLC fingerprints when the tuning parameter
varies from 10-8 to 108. From Fig. 2, our KMHF scheme is not suitable for a larger parameter
. If
, the identification performance of KMHF scheme is poor. If
, the identification performance is relatively stable. By studying AUC and ACC curves, our KMHF scheme reaches optimal when defining the parameter
. In the experiments, parameter
in (5) is set as 103, parameter
in (13) is set as 10, the number of retrieved CHMs k in (20) is set as 7.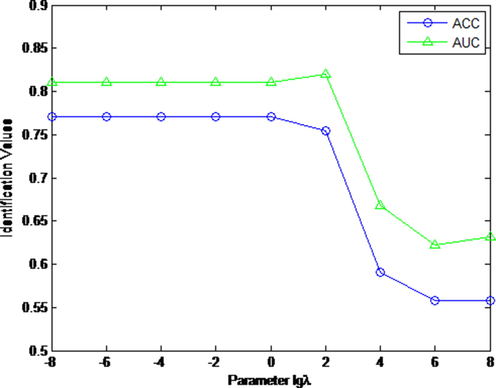
The curves of AUC and ACC values with different
.
In this work, we evaluated the performance of cold-hot nature identification with different parameter
in Eq.(5). We tuned the parameter
in (5) in the range of [10-3, 10-2, 10-1, 1, 101, 102, 103, 104, 105]. Fig. 3 displays the AUC and ACC value curves for nature identification with different
. From Fig. 3, our KMHF scheme is sensitive to parameter
. By studying AUC and ACC curves, our KMHF scheme reaches optimal when defining the parameter
. In this experiment, the parameter
is set as 1, parameter
in (13) is set as 10, the number of retrieved CHMs k in (20) is set as 7.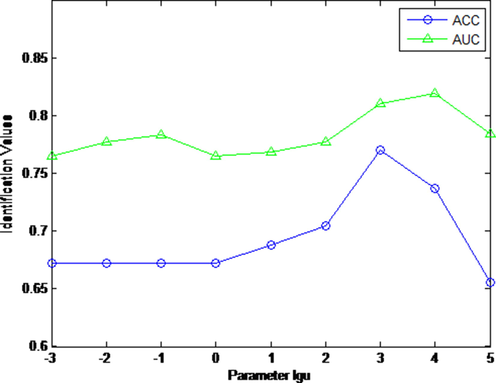
The AUC and ACC curves with different
.
In this study, we analyzed the impact of different parameter
on our scheme for cold-hot nature identification. We tuned the parameter
in (13) in the range of [10-3, 10-2, 10-1, 1, 101, 102, 103]. Fig. 4 displays the AUC and ACC value curves for nature identification with different
. From Fig. 4, our KMHF scheme is more suitable for a large parameter
. The AUC and ACC curves both have a peak when
. Comprehensively analyzing the AUC and ACC curves, we define
as 10. when
, our scheme is optimal. In this experiment, the parameter
is set as 1, parameter
in (5) is set as 103, the number of retrieved CHMs k in (20) is set as 7.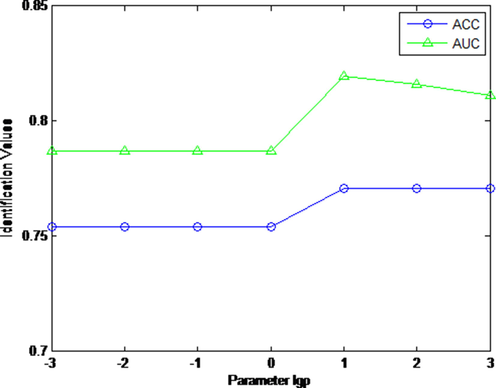
The AUC and ACC curves with different
.
Furthermore, we configured the number of retrieved CHMs k in (20) for evaluating the identification performance of our scheme. We tuned the parameter k in (20) in the range of [1, 3, 5, 7, 10, 12, 15, 20]. Fig. 5 shows the AUC and ACC curves with different parameter k. According to this figure, AUC and ACC curves have fluctuations when parameter k takes different values, which indicates that the performance of our KMHF scheme fluctuates slightly with the increase of k. Comprehensively analyzing the AUC and ACC curves, our KMHF scheme reaches optimal performance when k = 7. In this experiment, the tradeoff parameter
is set as 1, the parameter
is set as
, parameter
in (13) is set as 10.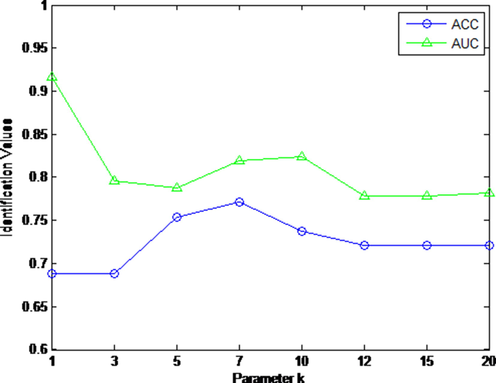
The AUC and ACC curves with different k.
3.2 Performance evaluation
Performance evaluation was performed to verify the feasibility of our KMHF scheme with the stability evaluation. Leave-one-CHM-our method was constructed to perform the stability evaluation. Several classical identification schemes were introduced to compare the cold-hot nature classification performance with our KMHF scheme, including our nature identification schemes (RS, PCC and ELM) and distance metric learning models (i.e., LMNN, ITML). RS and PCC schemes have been used for nature classification of CHMs with UV fingerprints. ELM has been utilized to analyze the nature of CHM compounds. A long fingerprint vector from straightforwardly concatenating three wavelength HPLC fingerprints was included as a comparative reference. The similarity metric from optimal solution matrix
in Eq. (12) was used to tackle this long fingerprint vector. We marked this scheme as LFV. Table 2 displays the performance comparison of our KMHF scheme and several classical identification schemes. From the comparison results in Table 2, we obtained the following conclusions. Firstly, our KMHF scheme outperforms the comparative LFV in nature identification. This demonstrates our inference that it is not optimal for straightforwardly concatenating three wavelength HPLC fingerprints as a long vector. Secondly, the classification performance of our KMHF scheme is better than that of the comparison classification schemes. This indicates that our scheme comprehensively analyzes the semantic relevance and fingerprint similarity of CHMs, and has better identification performance of cold-hot nature. Especially, different wavelength HPLC fingerprints can better mine the ingredient information of CHMs for cold-hot nature identification. Thirdly, the identification performance of our KMHF scheme outperforms that of RS. Compared with our scheme, RS does not consider the fingerprint similarity. Therefore, fingerprint similarity is an effective complement to semantic relevance. Fourth, the similarity metric methods (including ITML, LMNN and RS) have better classification performance than PCC and ELM. This shows that similarity metric methods are more effective for nature classification of CHMs. Furthermore, the similarity metric experiments confirm the proposed theoretical hypothesis that CHMs with similar cold-hot nature have similar material basis. Fifth, the performance of ELM algorithm is poor for cold-hot nature prediction. Finally, in summary, the constructed experiments verify the feasibility of our KMHF scheme.
Classifiers
AUC
ACC
ITML
0.739
0.672
LMNN
0.766
0.681
ELM
0.625
0.592
RS
0.789
0.650
PCC
0.604
0.581
LFV
0.808
0.734
KMHF
0.819
0.771
3.3 Nature identification examples
Leave-one-CHM-out method was introduced to give the examples of nature identification. Two representative CHMs, including Lophatheri Herba (cold) and Rhizoma Arisaematis (hot), were selected as query instances to illustrate nature identification. Table 3 reports two query CHM examples obtained from our KMHF scheme, in which query CHMs are listed in the second row and top k = 7 similar CHMs are showed in other rows. The top k = 7 similar CHMs are arranged in monotonically increasing order of Mahalanobis distance. Lophatheri Herba is chosen as a typical cold medicine to explain nature identification. The calculated similar CHMs include six cold reference CHMs and one hot reference CHM. Its cold nature probability we obtained is 92.5 %, which indicates that Lophatheri Herba is probably cold. Rhizoma Arisaematis is chosen as a typical hot medicine to analyze the nature identification. The calculated similar CHMs include five hot reference CHMs and two cold reference CHMs. Its cold nature probability we obtained is 3.1 %, which indicates that Rhizoma Arisaematis is probably hot. The nature identification instances demonstrate the relationship between CHM ingredients and cold-hot nature.
Identification Instances
CHMs with cold nature
CHMs with hot nature
Query CHMs
Lophatheri Herba (cold)
Rhizoma Arisaematis (hot)
The similar reference CHMs
Anemarrhena Asphodeloides Bunge (cold)
Clematidis Radix et Rhizoma (hot)
Rhei Radix et Rhizoma (cold)
Mustard Seeds (hot)
Dianthi Herba (cold)
Corydalis Rhizoma (hot)
Notopterygii rhizoma et radix (hot)
Ligustici Rhizoma et Radix (hot)
Gardeniae Fructus (cold)
Aconiti Lateralis Radix Praeparata (hot)
Stephaniae Tetrandrae Radix (cold)
Ecliptae Herba (cold)
Puerariae Lobatae Radix (cold)
Trachelospermi Caulis et Folium (cold)
3.4 Overall identification performance
In this work, the overall identification performance of our KMHF scheme was analyzed with four evaluation indices, including confusion matrix, F-score, precision, and recall. All evaluation indices were obtained from leave-one-CHM-out method. Table 4 displays the confusion matrix from nature identification of 61 CHMs. The identification accuracy rate of hot CHMs is 64.5 % (20/31), while the prediction accuracy rate of cold CHMs is 90.0 % (27/30). Therefore, the total prediction accuracy rate of 61 CHMs is 77.0 % (49/61). According to Table 4, our scheme has higher identification accuracy for cold CHMs, but poor identification accuracy for hot CHMs. Table 5 shows the precision, recall, and F-score of nature identification of 61 CHMs. By summarizing Table 4 and Table 5, we conclude that our scheme is effective in nature identification of 61 CHMs with HPLC fingerprints. The ingredient information can be used to analyze cold-hot nature of CHMs.
Ground Truth
Identification
Cold
Hot
Cold
27
3
Hot
11
20
Cold
Hot
Recall
90.0 %
64.5 %
Precision
71.1 %
87.0 %
F-score
79.4 %
74.1 %
4 Discussion
HPLC has been widely used in the analysis of CHM ingredients, including nature identification and quality evaluation. Its advantage is that it can quantitatively and qualitatively test CHM ingredients. Our group has done a lot of work in the nature identification, but mainly focused on the UV spectrum. Therefore, this study explores to introduce HPLC to analyze the CHM ingredients, and build a classification model for nature evaluation. Multi-wavelength HPLC fingerprints are used to extract different characteristics of CHM ingredients. The experimental results find that HPLC can extract more feature information of CHM ingredients related to CHM nature.
Multi-wavelength HPLC fingerprints challenge the classical classifiers. These algorithms may not be able to mine the feature information of CHM components, resulting in low accuracy of cold-hot nature identification. Especially, different wavelength HPLC fingerprints represent different physical properties of CHM ingredients. The classical classifiers cannot adapt to this data feature. It perhaps leads to low identification accuracy by straightforwardly concatenating three wavelength HPLC fingerprints into a long vector. In this study, our scheme introduced a multi-wavelength distance metric learning algorithm for cold-hot nature identification. The experimental results demonstrate that our scheme can better mine characteristic information of CHM ingredients related to CHM nature.
The theoretical basis of this study comes from a hypothesis that CHMs with similar cold-hot nature have the same or similar material basis. This study introduced a multi-wavelength distance metric learning algorithm to measure the similarity of CHM ingredients and proposed an improved KNN scheme for cold-hot nature evaluation. Experimental results indicate that there is a close relationship between CHM nature and its ingredients. From the perspective of HPLC, our experiments find that CHMs with similar ingredients related to cold-hot nature have similar cold-hot nature. Our experimental results support the hypothesis.
Distance metric learning algorithms, such as LMNN, ITML and RS, mainly focus on semantic relevance of CHMs without considering fingerprint similarity of CHM HPLC. However, semantic relevance of CHMs cannot reflect the whole concept of similarity measure. Our group defines the similarity metric as semantic relevance and fingerprint similarity. Experiments indicate that fingerprint similarity is an important complement to similarity measurement, which can improve the identification accuracy of cold-hot nature.
However, there are some problems to be solved in the future. Firstly, multi-wavelength HPLC fingerprints are used to analyze CHM ingredients. Gas chromatography and spectroscopy are not taken into account. Multi-fingerprints technology can extract different characteristic information of CHM ingredients, and perhaps improve the accuracy of nature identification. As a result, nature identification scheme based on multi-fingerprints fusion is the focus of future research. Secondly, the dataset in this study is a small sample and high-dimensional dataset. This poses a challenge to most classifiers. It is the direction of future research by designing special classifiers according to the characteristics of data. Thirdly, in this study, a HPLC data of 61 CHMs was used to test the proposed KMHF scheme. However, this is a primary assessment, which needs more CHM HPLC fingerprints to verify. Consequently, an extended dataset and an independent testing dataset are needed in the future studies.
5 Conclusions
In this study, a KMHF scheme was developed to fuse multi-wavelength HPLC fingerprints for cold-hot nature identification. Multi-wavelength HPLC fingerprints of CHMs were used to analyze the characteristic information of CHM ingredients. An improved KNN scheme was proposed for nature identification. Numerous experiments demonstrate that cold-hot nature of CHMs is closely related to CHM ingredients. Comparative experiments indicate that the nature identification performance of our scheme is the best. Therefore, our scheme can better mine the ingredient information related to cold-hot nature. Furthermore, our experiments support the scientific hypothesis that CHMs with the same cold-hot nature have similar material basis.
Author contributions
G.W. conceived and designed the project, M. Q., analyzed the Chinese medicine data sets. Z.W. collected data and provided expert knowledge. All authors read and approved the final manuscript.
Acknowledgments
The research is supported by the national key basic research development program (973Program) (No. 2007CB512600); National Natural Science Foundation of China (No. 81473369); Qi Huang Scholars Support Projecting; Shandong Province medical and health science and technology development plan (No. 202109040649); Shandong Provincial Natural Science Foundation (No. ZR2022MH203).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Davis, J.V., Kulis, B., Jain, P., Sra, S., Dhillon, I. S., 2007. Information-theoretic Metric Learning. In: Proc. of the International Conference on Machine Learning, Corvallis, Oregon, USA, pp. 209–216.
- Toward understanding the cold, hot, and neutral nature of Chinese medicines using in silico mode-of-action analysis. J. Chem. Inf. Model.. 2017;57:468-483.
- [Google Scholar]
- Discussion on scientific connotation of four natures of Chinese Materia Medica. Acta Univ Tradit. Med. Sin. Pharmacol. Shanghai. 2007;21:16-18.
- [Google Scholar]
- Study on discrimination mode of cold and hot properties of traditional chinese medicines based on biological effects. China J. Chin. Meter. Med.. 2014;39:3353-3358.
- [Google Scholar]
- Mathematical exploration of essence of herbal properties based on “Three-Elements” theory. China J. Chin. Mater. Med.. 2014;39:4060-4064.
- [Google Scholar]
- Molecular network and chemical fragment-based characteristics of medicinal herbs with cold and hot properties from chinese medicine. J. Ethnopharmacol.. 2013;148:770-779.
- [Google Scholar]
- A boosting framework for visuality-preserving distance metric learning and its application to medical image retrieval. IEEE Trans. Pattern Anal. Mach. Intell.. 2010;32:30-44.
- [Google Scholar]
- A Combination system for prediction of Chinese materia medica properties. Comput. Methods Programs Biomed.. 2011;101:253-264.
- [Google Scholar]
- Study on Quality Evaluation Standard of the Flower of Chrysanthemum Morifolium Ramat based on the Correlation of Ingredients and Efficacy. Beijing: China academy of Chinese medical sciences; 2020.
- The research for metabolomics discriminant method for cold and hot property of traditional Chinese medicine based on random forest. J. Jiangxi Univ. Tradit. Chin. Med.. 2015;27:82-86.
- [Google Scholar]
- Researech thinking and method of modern study on four properties theory of Chinese materia medica. J. Beijing Univ. Tradit. Chin. Med.. 2006;29:592-594.
- [Google Scholar]
- Application of the Bayesian network in Chinese herbal medicine property recognition. J. Shandong Univ. (Health Sci.). 2011;49:147-152.
- [Google Scholar]
- Research on the Relationship of “Nature-Structure” based on Information of Literature and Chemical Biology. Jinan: Shandong Univ; 2020.
- High performance liquid chromatography fingerprint and headspace gas chromatography-mass spectrometry combined with chemometrics for the species authentication of Curcumae Rhizoma. J. Pharmaceut. Biomed.. 2021;202:114144
- [Google Scholar]
- Classification of mixtures of Chinese herbal medicines based on a Self-organizing Map (SOM) Mol. Inform.. 2016;35:109-115.
- [Google Scholar]
- Similarity measurement of Lung Masses for medical image retrieval using kernel based semisupervised distance metric. Med. Phys.. 2016;43:6259-6269.
- [Google Scholar]
- Content-based image retrieval for lung nodule classification using texture features and learned distance metric. J. Med. Syst.. 2018;42:13.
- [Google Scholar]
- Similarity measurement of chinese medicine ingredients for cold-hot nature identification. TMR Mod. Herb. Med.. 2019;002:183-191.
- [Google Scholar]
- Multisolvent similarity measure of Chinese herbal medicine ingredients for cold-hot nature identification. J. Chem. Inf. Model.. 2019;59:5065-5073.
- [Google Scholar]
- Nature identification of Chinese herbal medicine compounds based on molecular descriptors. J. AOAC INT.. 2021;104:1754-1759.
- [Google Scholar]
- Cold–hot nature identification based on GC similarity analysis of Chinese herbal medicine ingredients. RSC Adv.. 2021;11:26008-26015.
- [Google Scholar]
- Cold-hot nature identification of Chinese medicine based on an ultraviolet chemical fingerprint. Spectroscopy. 2021;36:23-29.
- [Google Scholar]
- Distance metric learning for large margin nearest neighbor classification. J. Mach. Learn. Res.. 2009;10:207-244.
- [Google Scholar]
- Understanding ZHENG in traditional Chinese medicine in the context of neuro-endocrine-immune network. IET Syst. Biol.. 2007;1:51-60.
- [Google Scholar]
- Semisupervised multiview distance metric learning for cartoon synthesis. IEEE T. Image Process.. 2012;21:4636-4648.
- [Google Scholar]
- Research on Pattern Recognition for Chmp-markers based on Multi-dimensional and Multi-data Characteristic Fingerprint. Jinan: Shandong Univ; 2012.
- Patch alignment for dimensionality reduction. IEEE Trans. Knowl. Data Eng.. 2009;21:1299-1313.
- [Google Scholar]







