Translate this page into:
Network pharmacology and molecular docking approach to investigate the mechanism of a Chinese herbal formulation Yougui pills against steroid-related osteonecrosis of the femoral head
⁎Corresponding authors at: #208 Huanchengdong Road, Zhejiang Hospital of Integrated Traditional Chinese and Western Medicine, Hangzhou, Zhejiang, China. wangyifan8929@163.com (Yifan Wang), zhangpbs2017@163.com (Peng Zhang)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Yougui pills (YGPs) is a classical prescription formula for treating steroid-related osteonecrosis of the femoral head (SONFH) in traditional Chinese medicine (TCM). However, its key ingredients and mechanisms of action are still unknown. In the study, we used molecular docking and network pharmacology to comprehensively investigate the mechanism by YGPs impact on SONFH. TCMSP was used to identify the effective active components of YGPs. Targets associated with SONFH were extracted from GEO datasets in conjunction with GeneCards, OMIM, and DisGeNET databases. Protein-protein interaction (PPI) networks were built using the intersection targets by importing them into the STRING database. Furthermore, DAVID was used to conduct a pathway enrichment analysis for both GO and KEGG. Finally, molecular docking and molecular dynamics simulation was performed to verify the binding ability between key targets and active compound. Screening using relevant databases revealed 126 active compounds with 1078 targets in YGPs, mainly including helenalin, quercetin, stigmasterol, sesamin, etc. The topological analysis of the PPI network revealed that key targets included VEGFA, IL6, IL1B, TNF, ALB, and TP53. According to the findings of the GO enrichment analysis, genes were most significantly enriched in the inflammatory response, response to hypoxia, and positive regulation of angiogenesis. The effects of YGPs have been mostly linked to the HIF-1 signaling pathway and the PI3K-Akt signaling pathway, according to a KEGG pathway enrichment analysis. In addition, the results of molecular docking and molecular dynamics simulations demonstrated that the active chemicals in YGPs have high affinity with the relevant targets. This investigation integrates molecular docking and network pharmacology to identify the effective compounds, related targets, and potential mechanism of YGPs in the treatment of SONFH. This research may provide an integrative strategy to clarify the pharmacological mechanisms of traditional Chinese medicines.
Keywords
Steroid-related osteonecrosis of the femoral head
Yougui pills
Network pharmacology
Molecular docking
1 Introduction
Steroid-related osteonecrosis of the femoral head (SONFH) is a refractory disease caused by long-term or high-dose application of glucocorticoids, which leads to damage or interruption of the femoral head blood supply, resulting in local ischemic necrosis and collapse of the articular surface (Wang et al., 2018). Patients eventually require joint arthroplasty surgery among frequent femoral head collapse and hip joint dysfunction, which severely impacts the patient's quality of life (Chang et al., 2020). Nevertheless, exact pathophysiology and ideal treatment of SONFH remains unclear (Huang et al., 2021). To avoid the progression of osteonecrosis, it is vitally crucial to discover novel and effective treatments.
In recent years, traditional Chinese medicine (TCM) has progressively demonstrated its therapeutic effects and benefits in treating SONFH in recent years. TCM can be effective at improving patient symptoms, controlling disease progression, and improving joint function and life quality (Du et al., 2022). Yougui pills (YGPs), a classical TCM formula, show significant effects on treating SONFH. It is mainly comprised of Rehmannlae Radix Praeparata (Shudihuang, SDH), Dioscoreae Rhizoma (Shanyao, SY), Corni Fructus (Shanzhuyu, SZY), Lycii Fructus (Gouqi, GQ), Cuscutae Semen (Tusizi, TSZ), Eucommiae Cortex (Duzhong, DZ), Angelicae Sinensis (Danggui, DG), Aconiti Lateralis Radix Praeparata (Zhifuzi, ZFZ), Cinnamomi Cortex (Rougui, RG), Cervi Cornus Colla (Lujiaojiao, LJJ). Clinical investigations have demonstrated that YGPs can considerably enhance the treatment efficacy and clinical indicators of ONFH patients with Spleen and Kidney - Yang Deficiency Syndrome (Xie et al., 2021). Previous studies confirmed that YGPs could inhibit cartilage degradation, reduce osteoclastogenesis and promote bone formation (Zhang et al., 2017; Zhang et al., 2019).
Network pharmacology is a diverse discipline born under the background of biomedical big data and artificial intelligence (Hopkins, 2008). It uses database screening, computer simulation, and information mining to discern key targets, predict signal pathways and action mechanisms, better disclose the biological mechanisms underlying complicated diseases and the actions of drugs at the molecular level, quickly identify therapeutic targets and discover novel biomarkers (Li et al., 2022). Due to the complex composition of TCM, many targets and wide signal pathways of treatment, the in-depth study of TCM is time- and money-consuming (Li and Zhang, 2013). Fortunately, the properties of multi-channel, multi-component and multi collaborative research target of network pharmacology are well suited to studying TCM's underlying mechanisms (Wang et al., 2022c). Therefore, network pharmacology is increasingly used to investigate the potential TCM mechanisms for treating a variety of disorders (Fan et al., 2022; Wu et al., 2021).
Despite the researchers having carried out many basic studies previously, we are still unable to explain the YGPs specific target and exact mechanism in treating SONFH. In this study, we used bioinformatics approaches to investigate the major chemicals in YGPs and their possible targets and mechanism of action in treating SONFH. This study could serve as a theoretical base for future research on the YGPs pharmacodynamic material basis and mechanisms of action in treating SONFH. The flowchart of the research is shown in Fig. 1.
Workflow of the network pharmacological investigation strategy of YGPs in the treatment of SONFH.
2 Materials and methods
2.1 Data collection
2.1.1 Screening of active compounds and therapeutic targets of YGPs
The active compounds of SDH, SY, SZY, GQ, TSZ, DZ, DG, ZFZ, RG and LJJ were acquired from the TCMSP: https://old.tcmsp-e.com/tcmsp.php (Ru et al., 2014). Natural compound chemical, target, and pharmacokinetic data is made available through TCMSP platform. Oral bioavailability (OB) and drug-likeness (DL) were the primary pharmacokinetic parameters utilised to screen potential active ingredients (van der Graaf and Benson, 2011; Xu et al., 2012). The criteria for the bioactive ingredients screening is defined as: OB ≥ 30 % and DL ≥ 0.18 (He et al., 2022; Zhang et al., 2021). The YGPs active substances database was developed. We accessed PubChem (https://pubchem.ncbi.nlm.nih.gov/) to retrieve the compounds' canonical SMILES format. The Swiss Target Prediction Tools (STP, https://www.swissta rget.ch/) were utilised to search for the YGPs targets active compounds (Gfeller et al., 2014; Mu et al., 2021).
2.1.2 Establishment of a database relating to SONFH target
The differentially expressed genes (DEGs) related to SONFH were retrieved from GEO: https://www.ncbi.nlm.nih.gov/geo/), Series: GSE123568. The limma package was used to remove the batch effect and identify DEGs of SONFH. The selected criteria of DEGs were set as |logFC| > 1 and P < 0.05 (Gu et al., 2020). The R packages ggplot2 was utilised to visualise the volcanic charts. The heatmap was used to show the expression patterns of the top 30 DEGs sorted by their logFC values. In addition, disease targets related with SONFH were extracted using 3 databases, especially GeneCards (https://www.genecards.org) (Rebhan et al., 1997), the OMIM database (https://www.omim.org/) (Amberger et al., 2015), and the DisGeNET database (https://www.disgenet.org/) (Piñero et al., 2020). The keyword “osteonecrosis of the femoral head” was entered into the databases yielding the disease targets. Finally, the SONFH disease targets library was established by eliminating duplicate targets.
2.2 Establishment of PPI network
The overlapping between the SONFH-linked targets yielding the YGPs predicted targets by using Venny 2.1.0 (https://bioinfo.cnb.csic.es/tools/venny/index.html). To acquire PPI data, we imported the targets into the STRING database (https://string-db.org) (Szklarczyk et al., 2017). The organism's screening criterion was restricted to “Homo sapiens” and the confidence index was defined to ≥ 0.4 (Szklarczyk et al., 2019). Cytoscape3.7.2 (https://www.cytoscape.org/) was used to establish a PPI network and to perform a topological and cluster analysis (Shannon et al., 2003). Three topological indices, namely degree centrality (DC), betweenness centrality (BC), and closeness centrality (CC), were used to examine PPI network key targets (Song et al., 2018; Tang et al., 2015). The parameters of filter are configured to be more than twice median value (Jin et al., 2021). In order to specify the hub targets even further, the Cytohubba add-on for Cytoscape's network topology technique Maximum Clique Centrality (MCC) was employed (Chin et al., 2014; Fu et al., 2020). To further screen PPI network modules, Cluster analyses was done with the MCODE Cytoscape plugin (Jin et al., 2021; Zhang et al., 2020).
2.3 Selection and analysis of key targets
The key genes were obtained by the topological analysis of the PPI network and MCC algorithm of the Cytohubba plug-in. Protein expression profiles of key targets at the gene expression and tissue level were retrieved from database Human Protein Atlas (HPA, https://www.proteinatlas.org/) (Fan et al., 2020; Li et al., 2013).
2.4 GO and KEGG enrichment analyses
The Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/) is a tool for functional annotation that determines the biological significance of extensive gene lists (Huang da et al., 2009). The common targets of YGPs and SONFH were analyzed for GO and KEGG enrichment analyses through DAVID database. The GO and KEGG pathway enrichment analysis results were screened as per FDR < 0.05 and P < 0.05. Molecular function (MF), cellular component (CC), and biological process (BP) were the three sub-analyses that made up the GO enrichment analysis (Ye et al., 2021). The top 10 GO terms and top 20 KEGG pathway results with the lowest P-value were visualized using R software.
2.5 Network construction
Herb-compound-target (H-C-T) network and compound–target-pathway (C-T-P) network were performed to understand YGPs integral molecular mechanisms in treating SONFH. The networks were generated and shown through Cytoscape 3.7.2 software. The formation of the H-C-T network was based on the active compounds of YGPs and their corresponding common targets. The top 20 pathways and their corresponding targets and compounds were sorted out and constructed C-T-P network to better understand the relationship involving pathways, compounds, and targets.
2.6 ADME and toxicity predictions
It is very important to evaluate the drug's primary physiochemical and toxicological characteristics initially in the development phase. Hence, we evaluated the druggability of the selected compound. Absorption, distribution, metabolism, excretion, and toxicity (ADMET) characteristics of ten selected drugs were evaluated via the SwissADME (https://www.swissadme.ch/) and ADMETlab (https://admet.scbdd.com/) (Xiong et al., 2021).
2.7 Molecular docking verification
Molecular docking is a typical drug discovery technique, which is used to study the interaction between receptors and ligands and anticipate their manner of affinity and binding. Then, we utilised molecular docking to observe whether the selected compounds in YGPs identified from the H-C-T network bind to six key targets. AutoDockTools 1.5.6 and AutoDock Vina 4.2 were utilised for molecular docking (Trott and Olson, 2010). The docking procedure involved the following steps: firstly, the 3D structure format file of the active compounds(hydroxygenkwanin, helenalin, quercetin, isorhamnetin, beta-sitosterol, sitosterol, kaempferol, erythraline, stigmasterol, sesamin)was obtained from the PubChem databases (https://pubchem.ncbi.nlm.nih.gov/). The protein targets crystal structure of six key targets(VEGFA, IL6, TP53, TNF, ALB, ILB) were acquired from the RCSB Protein Data Bank (https://www.rcsb.org/) (Berman et al., 2000). The target proteins and the compounds were regarded as the receptor and the ligands, respectively. Secondly, AutoDockTools 1.5.6 software was used to perform the dehydration, hydrogenation, as well as calculating Gasteiger charges of the receptor protein. Both ligands and receptors were kept as PDBQT format. The evaluation of docking feasibility and stability was based on spatial location and binding capability. The docking grid box size and position of macromolecule were predicted using AutoDock Vina 4.2. All rotatable bonds of the ligands structure were permitted to rotate freely, whereas the receptor was treated as rigid. The smaller the docking score, the higher binding rate between the receptor and ligand. Finally, conformations exhibiting the most favorable binding energy were chosen to analyze the interaction between ligands and proteins. The results of molecular docking analyses were visualised using the software packages Discovery Studio 2020 and PyMOL through 2D and 3D images generation.
2.8 Molecular dynamics simulation
Molecular dynamics (MD) simulation is a widely applied computer simulation technique for investigating the atomic and molecular motions. Protein stability and interaction with the molecular docked active compound were assessed using MD modelling (Rakhshani et al., 2019; Van Der Spoel et al., 2005). The GROMACS software was used for MD simulation with CHARMM 36 force field for all atoms (Tripathi et al., 2016). The protein was solvated in a cubic box using a three-point transferable intermolecular potential (TIP3P) water model and periodic boundary conditions were set (Nayar et al., 2011). During the MD simulation process, the Particle-Mesh Ewald (PME) method was utilized for the assessment of the electrostatic interactions, and the non-bonded cut-off was kept at 10 Å (Agarwal et al., 2018). To maintain the constant temperature of the system at 300 K, the V-rescale temperature coupling method was used (Bussi et al., 2007). Similarly, the Berendsen method was applied to maintain the constant pressure at 1 bar (Eslami et al., 2010; Lin et al., 2017). MD simulation workflow involved the following: first, the energy of the system was reduced using the steepest descent method. Secondly, within 60 ps, the temperature system was gradually raised from 0 to 300 K. Then, the position restraint simulation of 5000 steps (2 fs each steep) was implemented under NVT (the constant Number of particles, Volume, and Temperature) and NPT (the constant Number of particles, Pressure, and Temperature) conditions. The TNF-stigmasterol complex system was finally subjected to a 100 ns MD simulation with a 2 fs time step, and the trajectory data was saved every 10 ps (Cruz et al., 2019; Zaman et al., 2021). The simulation results were visualised using the GROMACS embedded software and PyMOL (Makarewicz and Kaźmierkiewicz, 2013).
3 Results
3.1 Screening the active compounds and putative targets of YGPs and construction of the “H-C-T” network
126 chemical compounds from 8 herbs in YGPs were obtained from TCMSP databases, comprising 2, 28, 21, 45, 16, 20, 2, and 11 compounds from DG, DZ, FZ, GQ, SY, SZY, SDH, and TSZ, respectively. Basic information on 126 chemical compounds in YGPs were shown in Supplementary Table S1. 1078 targets were predicted among the 126 active compounds in YGPs.
We searched 722 DEGs in SONFH by analysis of the GSE123568 in GEO datasets, including 437 up-regulated genes and 285 down-regulated genes. The volcano map of the 722 DEGs was shown in Fig. 2A and the heatmap of the expression patterns of the top 30 DEGs is presented in Fig. 2B. The color depth changes according to their logic values. In addition, there were 525 SONFH-related targets retrieved from GeneCards, OMIM, and DisGeNET databases. Following eliminating the redundancy, 1237 disease targets were determined. Intersection analysis of 1078 YGPs compounds targets and 1237 disease targets identified 164 common targets (Fig. 2C and Supplementary Table S2).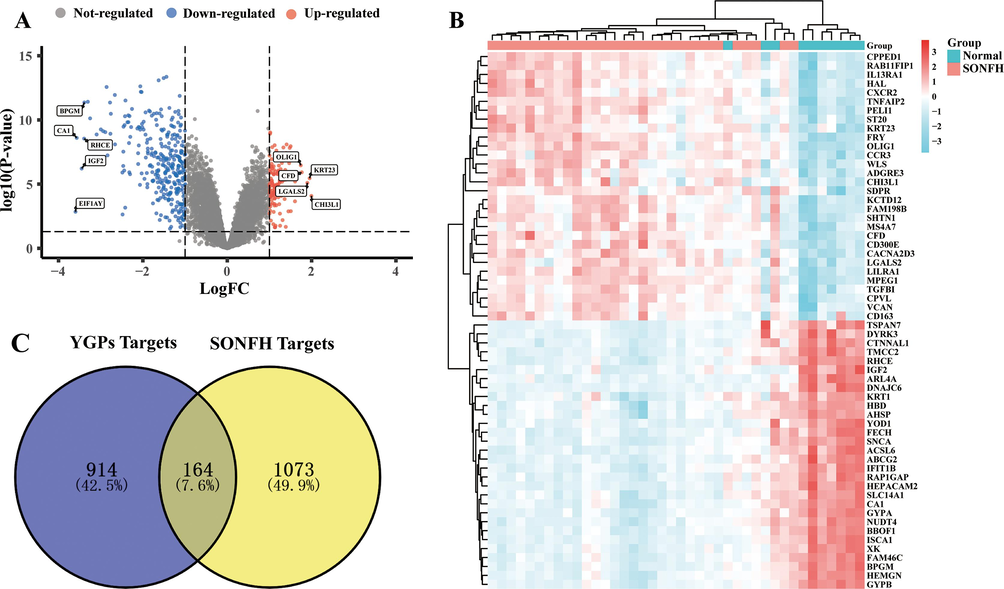
Screening of YGPs-SONFH common targets. (A) Differential genes volcano map shows the gene distribution in disease samples. Red and blue represent upregulated genes and downregulated genes, respectively, whereas grey indicates no significant difference. (B) Heatmaps exhibited the expression patterns of these 60 DEGs. Columns correspond to the samples and rows correspond to the genes. (C) Venn diagram of the 164 common targets between the active compounds targets of YGPs and the disease targets of SONFH.
Fig. 3 depicts the H-C-T network, which consisted of 283 nodes and 2195 edges. The network analysis showed that helenalin (degree: 29), hydroxygenkwanin (degree: 26), beta-sitosterol (degree: 25), kaempferol (degree: 24), isorhamnetin (degree: 24), stigmasterol (degree: 23), sesamin (degree: 23), sitosterol (degree: 23), quercetin (degree: 23), erythraline (degree: 23) have the highest number of connections to targets. The results indicate that each protein target is affected by multiple compounds and each compound could act on multiple protein targets. Thus, we speculate that YGPs may play a bone protective effect in SONFH through multi-component and multi-target.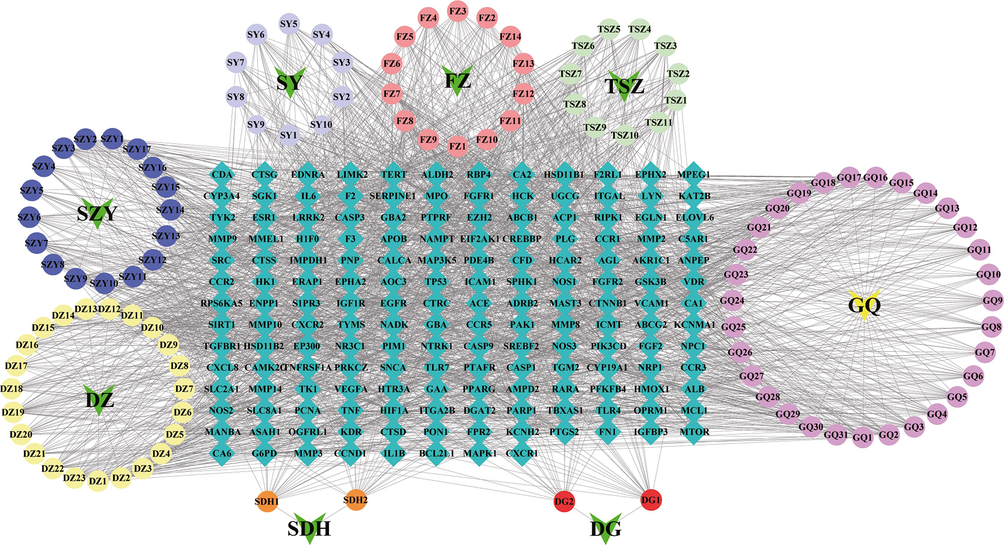
Herb–compound–target network. Green arrows represent the herbs in YGPs, circles node are compounds, green squares nodes represent the common targets. The edges represent the interaction between compounds and targets.
3.2 PPI network of common targets
In order to investigate the mechanism that is responsible for the therapeutic effects of YGPs against SONFH, we constructed a PPI network by importing the 164 common targets into the STRING database. There were a total of 160 nodes and 2023 edges in the network. We identified 14 core targets according to the degree values, BC, and CC (Fig. 4A and Fig. 4C). As shown in Fig. 4B, a cluster analysis was done utilizing MCODE to construct a highly connected sub-network, and the targets were divided into 5 groups. The CytoHubba plug-in was utilised to choose the top 10 genesranked by the MCC method as hub genes (Fig. 4D).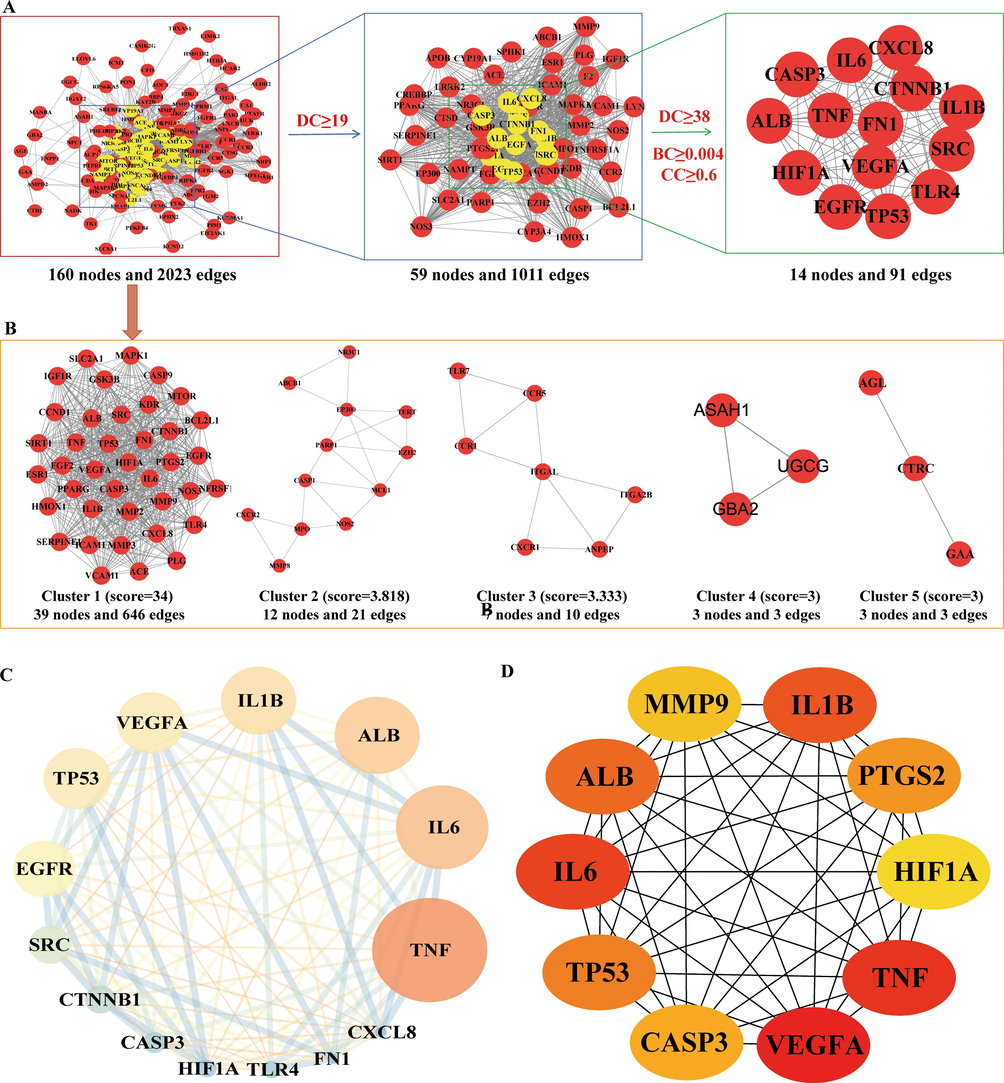
Identification of candidate targets via protein–protein interaction (PPI) analysis. (A) The process of topological screening for the PPI network. (B) PPI network based on clustery analysis using the MCODE plug-in. (C) The 14 core targets obtained by screening 164 common targets through DC, BC, CC. The node size is proportional to the target degree in the network. (D) The hub genes selected from the PPI network using the CytoHubba plugin. The node color was from pale yellow to red, and the corresponding degree gradually larger.
3.3 Selection and analysis of key targets
The top six targets with the highest MCC scores and degree values were TNF, IL6, ALB, IL1B, VEGFA, and TP53, which were considered the key targets. To determine whether these targets were differentially expressed during osteogenesis, we utilized the HPA database to compare the protein and gene expression levels of key targets between human bone marrow and adipose tissue. The results showed that TNF, IL6, ALB and VEGFA, were highly expressed in bone marrow, while the expression of IL1B and TP53 were not detected in the bone marrow and adipose tissue (Fig. 5A). The levels of gene expression for all six of the most important targets were found to be much higher in the bone marrow than in the adipose tissue (Fig. 5B). Therefore, we believed that the expression of these six critical targets may differ between bone tissue and adipose tissue and that they may be essential genes in the osteogenic differentiation process.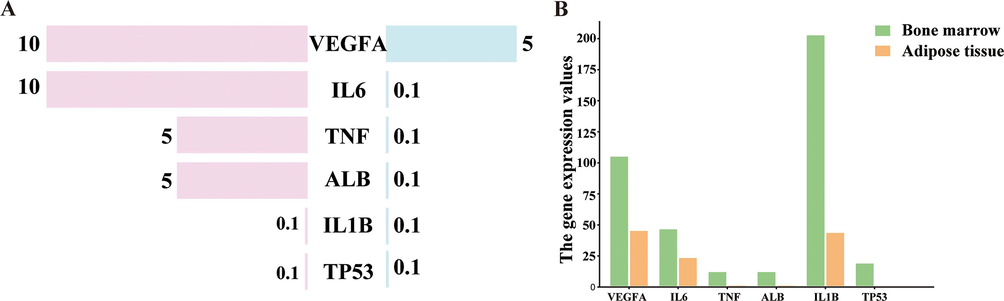
The expression analysis of key targets in human tissues. The analysis and summary of protein expression (A) and gene expression (B) of key targets in human tissues from HPA database. (A) Bubble chart of the top 10 biological process (BP) terms, cellular component (CC) terms, and molecular function (MF) terms of GO enrichment analysis. (B) Column chart of the top 10 biological process (BP) terms, cellular component (CC) terms, and molecular function (MF) terms of GO enrichment analysis are shown as green, orange, and purple bars, respectively.
3.4 GO enrichment analysis
On the basis of (BP), (CC), and (MF), GO enrichment of 164 potential targets was done to further investigate the diverse mechanisms of YGPs in the treatment of SONFH. There were a total of 425 GO terms that were enriched: 276 terms for BP, 110 terms for MF, and 39 terms for CC. These findings are shown in Supplementary Tables S3, S4, and S5. The top 10 enriched BP terms, MF terms, and CC terms are shown in Fig. 6A. In Fig. 6B, a column chart was used to display the terms that had the highest gene counts in each of the categories. The results demonstrated that BP terms were mainly associated with a cytokine-mediated signaling pathway, inflammatory response, response to hypoxia, positive regulation of angiogenesis. The MF results demonstrated that several targets involved in protein binding, protein activity, and protein homodimerization activity. According to the results of the CC, the majority of targets appear to be located on the cell surface, extracellular space, and plasma membrane.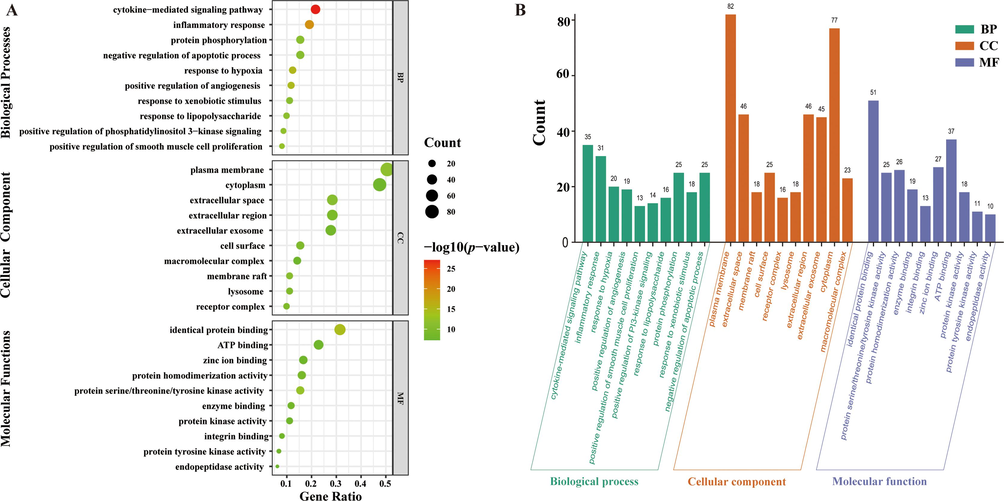
Results of GO enrichment analysis. (A) Bubble chart of the top 10 biological process (BP) terms, cellular component (CC) terms, and molecular function (MF) terms of GO enrichment analysis. (B) Column chart of the top 10 biological process (BP) terms, cellular component (CC) terms, and molecular function (MF) terms of GO enrichment analysis are shown as green, orange, and purple bars, respectively.
3.5 KEGG analysis
KEGG pathway analysis was used to determine the pathways that were significantly altered by YGPs in SONFH treatment; the significance level used was P < 0.05, and the results showed that 146 pathways were significantly enriched (Supplementary Table S6). Depending on gene ratios and P-values, the top 20 highly enriched pathways were filtered (Table 1 and Fig. 7A-B). Using Cytoscape 3.7.2, we chose the top 20 pathways to build a C-T-P network with 183 nodes and 1024 edges (Fig. 7C). A Sankey diagram was created depending on the relationships between enriched pathways and targets utilizing a free online platform (https://www.bioinformatics.com.cn) (Fig. 7D). KEGG enrichment analysis suggested that YGPs pharmacological mechanisms in SONFH therapy may be mostly associated with the HIF-1 signaling pathway, TNF signaling pathway, EGFR tyrosine kinase inhibitor resistance, thyroid hormone signaling pathway, PI3K-Akt signaling pathway, apoptosis. We visualized the HIF-1 signaling pathway and PI3K-Akt signaling pathway utilising “pathview” in R (Fig. 8A-B).
Pathway ID
Pathway name
Count
Genes
P-Value
hsa05200
Pathways in cancer
41
GSK3B, CXCL8, ITGA2B, SLC2A1, PIK3CD, PTGS2, FGF2, HIF1A, EGFR, IGF1R, CASP9, EDNRA, RPS6KA5, CCND1, TERT, CASP3, PIM1, HMOX1, MAPK1, EP300, CAMK2G, NTRK1, EGLN1, CREBBP, NOS2, MMP2, FN1, F2, MMP9, ESR1, TGFBR1, MTOR, VEGFA, IL6, RARA, CTNNB1, PPARG, TP53, FGFR2, FGFR1, BCL2L1
5.48E-15
hsa05167
Kaposi sarcoma-associated herpesvirus infection
26
GSK3B, CXCL8, SRC, PIK3CD, PTGS2, FGF2, HIF1A, ICAM1, CASP9, CCND1, CASP3, MAPK1, EP300, CCR5, CCR3, CCR1, LYN, CREBBP, TYK2, MTOR, TNFRSF1A, VEGFA, HCK, IL6, CTNNB1, TP53
9.26E-15
hsa04933
AGE-RAGE signaling pathway in diabetic complications
19
VCAM1, CXCL8, NOS3, MMP2, SERPINE1, FN1, PIK3CD, F3, TNF, PRKCZ, TGFBR1, ICAM1, VEGFA, IL6, CCND1, IL1B, CASP3, PIM1, MAPK1
1.92E-13
hsa05417
Lipid and atherosclerosis
25
GSK3B, CXCL8, SRC, PIK3CD, TNF, ICAM1, CASP9, CASP3, CASP1, MAPK1, APOB, CAMK2G, MAP3K5, LYN, VCAM1, NOS3, MMP3, MMP9, TNFRSF1A, IL6, IL1B, PPARG, TP53, TLR4, BCL2L1
8.38E-13
hsa04066
HIF-1 signaling pathway
19
EGLN1, CREBBP, NOS2, NOS3, SERPINE1, SLC2A1, PIK3CD, HIF1A, EGFR, MTOR, IGF1R, HK1, VEGFA, IL6, EP300, HMOX1, MAPK1, CAMK2G, TLR4
9.06E-13
hsa05205
Proteoglycans in cancer
23
SRC, MMP2, FN1, PIK3CD, FGF2, HIF1A, ESR1, TNF, MMP9, EGFR, MTOR, IGF1R, VEGFA, PAK1, CCND1, CASP3, KDR, MAPK1, CTNNB1, CAMK2G, TP53, TLR4, FGFR1
1.82E-11
hsa05163
Human cytomegalovirus infection
23
CCR1, GSK3B, CXCL8, SRC, PIK3CD, PTGS2, TNF, EGFR, MTOR, TNFRSF1A, VEGFA, CASP9, IL6, CCND1, IL1B, CASP3, CXCR2, MAPK1, CTNNB1, RIPK1, CCR5, TP53, CCR3
1.15E-10
hsa05215
Prostate cancer
16
GSK3B, CREBBP, MMP3, PIK3CD, MMP9, EGFR, MTOR, IGF1R, CASP9, CCND1, EP300, MAPK1, CTNNB1, TP53, FGFR2, FGFR1
2.10E-10
hsa05418
Fluid shear stress and atherosclerosis
18
VCAM1, NOS3, SRC, MMP2, ITGA2B, PIK3CD, TNF, PRKCZ, MMP9, ICAM1, TNFRSF1A, VEGFA, IL1B, KDR, HMOX1, CTNNB1, TP53, MAP3K5
5.52E-10
hsa04668
TNF signaling pathway
16
VCAM1, MMP3, PIK3CD, PTGS2, TNF, MMP9, ICAM1, TNFRSF1A, MMP14, IL6, RPS6KA5, IL1B, CASP3, MAPK1, RIPK1, MAP3K5
1.68E-09
hsa01521
EGFR tyrosine kinase inhibitor resistance
13
GSK3B, SRC, PIK3CD, FGF2, EGFR, MTOR, IGF1R, VEGFA, IL6, KDR, MAPK1, FGFR2, BCL2L1
1.81E-08
hsa04919
Thyroid hormone signaling pathway
15
GSK3B, CREBBP, SRC, SLC2A1, PIK3CD, HIF1A, ESR1, MTOR, CASP9, KAT2B, CCND1, EP300, MAPK1, CTNNB1, TP53
4.11E-08
hsa05219
Bladder cancer
10
RPS6KA5, CXCL8, CCND1, SRC, MMP2, MAPK1, TP53, MMP9, EGFR, VEGFA
4.20E-08
hsa05230
Central carbon metabolism in cancer
12
NTRK1, G6PD, SLC2A1, MAPK1, PIK3CD, HIF1A, TP53, EGFR, FGFR2, MTOR, FGFR1, HK1
5.08E-08
hsa05164
Influenza A
17
CREBBP, CXCL8, PIK3CD, PLG, TYK2, TNF, ICAM1, TNFRSF1A, CASP9, IL6, IL1B, CASP3, CASP1, EP300, MAPK1, TLR7, TLR4
8.90E-08
hsa04151
PI3K-Akt signaling pathway
24
NTRK1, GSK3B, NOS3, ITGA2B, FN1, PIK3CD, FGF2, EGFR, MTOR, IGF1R, VEGFA, CASP9, IL6, CCND1, KDR, MAPK1, SGK1, TP53, TLR4, FGFR2, MCL1, EPHA2, FGFR1, BCL2L1
1.17E-07
hsa05152
Tuberculosis
17
CREBBP, NOS2, SRC, VDR, SPHK1, TNF, CTSS, TNFRSF1A, CASP9, IL6, IL1B, CASP3, EP300, MAPK1, CTSD, CAMK2G, TLR4
1.82E-07
hsa04210
Apoptosis
15
NTRK1, PARP1, PIK3CD, TNF, CTSS, TNFRSF1A, CASP9, CASP3, MAPK1, RIPK1, CTSD, TP53, MCL1, BCL2L1, MAP3K5
1.82E-07
hsa05161
Hepatitis B
16
CREBBP, PCNA, CXCL8, SRC, PIK3CD, TYK2, TNF, MMP9, TGFBR1, CASP9, IL6, CASP3, EP300, MAPK1, TP53, TLR4
2.65E-07
hsa04064
NF-kappa B signaling pathway
12
LYN, VCAM1, CXCL8, PARP1, IL1B, RIPK1, PTGS2, TNF, TLR4, BCL2L1, ICAM1, TNFRSF1A
3.05E-07
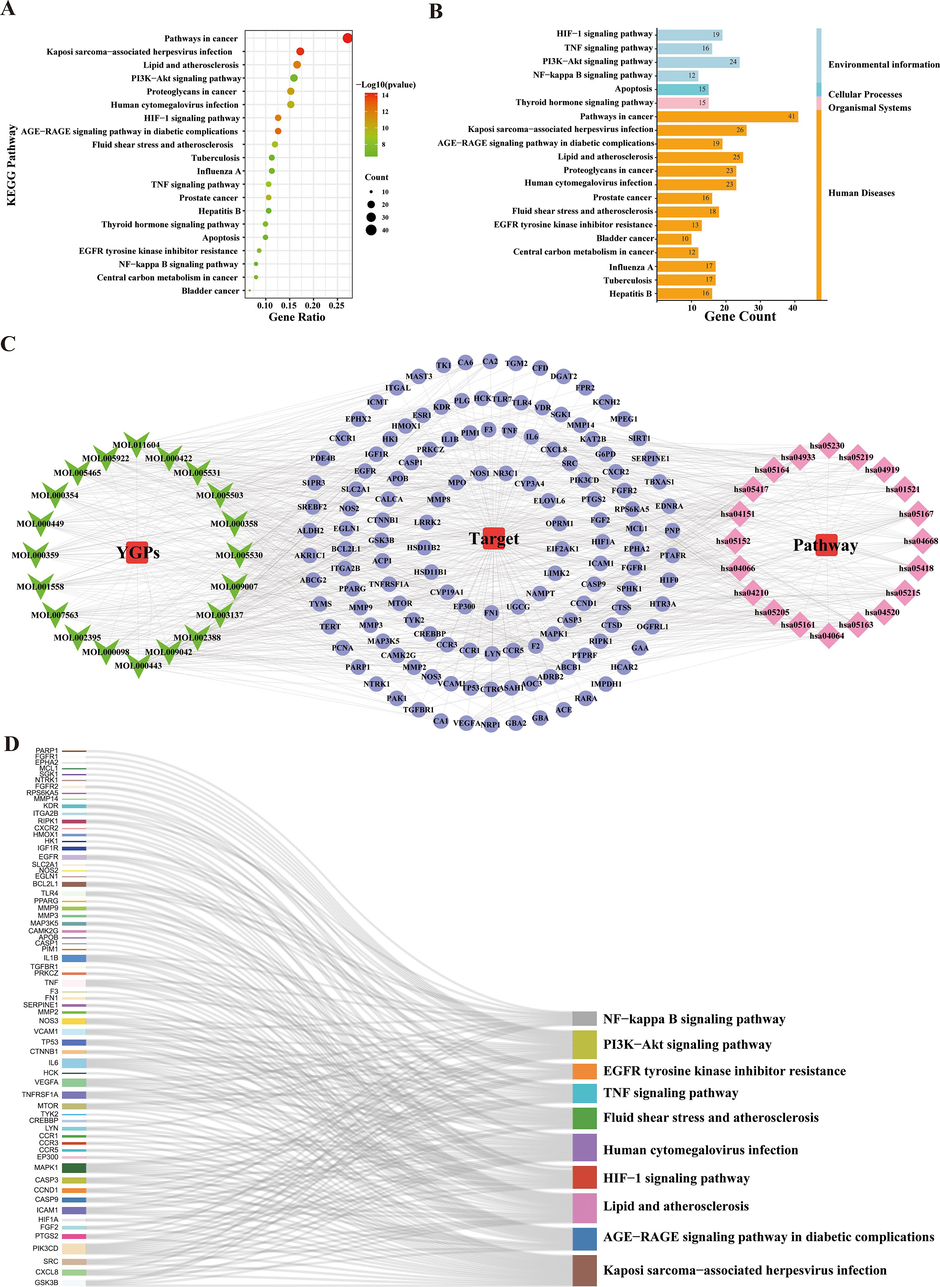
Results of KEGG enrichment analysis and key pathway network construction. (A) The bubble chart of the top 20 pathways based on KEGG enrichment analysis. (B) The KEGG type of the top 20 pathways based on KEGG enrichment analysis. (C) The compound–target–pathway(C-T-P) network implicated in the mechanism of YGPs in SONFH treatment. The purple nodes represent the targets; the pink nodes represent the pathways, whereas the green nodes represent the compounds. (D) The sankey diagram of the KEGG pathway analysis of the therapeutic targets of YGPs in SONFH treatment. The left rectangle nodes of the sankey diagram represent the therapeutic targets, the right rectangle nodes of the sankey diagram represent the KEGG pathways, and the lines represent the ownership of targets and pathways.
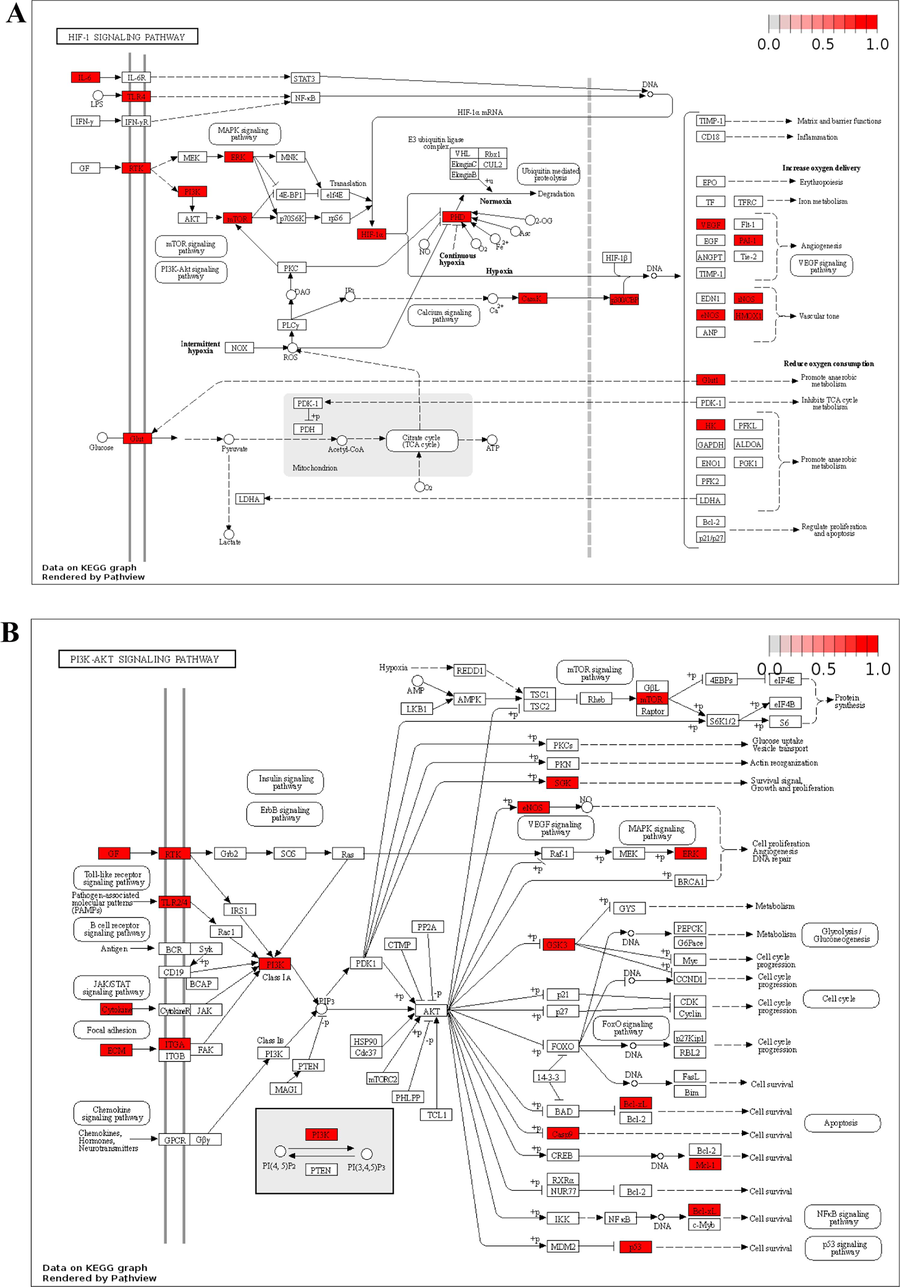
Distribution of key targets in the most related pathways. (A) Distribution of key targets in the HIF-1 signaling pathway. (B) Distribution of key targets in the PI3K/AKT signaling pathway. The red rectangle stands for the key targets. The putative targets and the genes implicated in the pathway are shown in red.
3.6 ADME and toxicity analysis
A number of potential medications failed preclinical and clinical testing due to insufficient ADME and high toxicity characteristics. Consequently, assessing the key physiochemical and toxicity properties during the initial phase of drug development was a crucial process. It is well acknowledged that ADMET prediction plays a crucial role in the simplicity and acceleration of the drug design process. So, ADMET values of ten selected compounds were calculated and shown in Supplementary Table S7.
3.7 Molecular docking
Molecular docking was used to verify the intermolecular interactions mode of the six key targets with the top ten active compounds. The details of targets and compounds, as well as the spatial coordinates of docking, are provided in Supplementary Table S8. The binding energies of key targets and active compounds are shown in Fig. 9. Usually, it is believed that the smaller docking binding energy leads to a more stable conformation.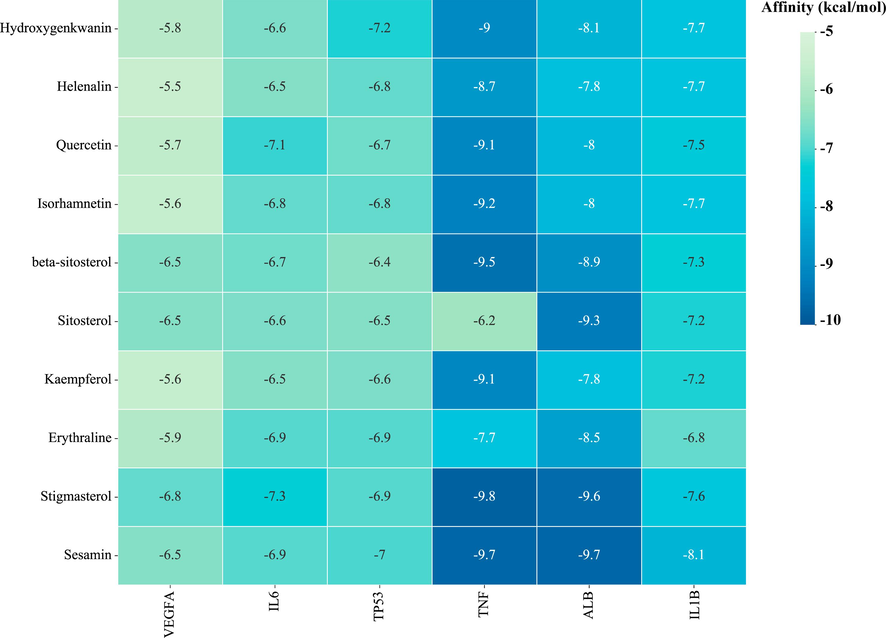
Heatmap of molecular docking score. Affinity energy(kcal/mol) of key targets and active compounds of herbs. TNF- Stigmasterol(A1), ALB- Sesamin (B1), IL1B- Sesamin (C1), TP53- Hydroxygenkwanin (D1), IL6- Stigmasterol (E1), VEGFA- Stigmasterol (F1). (A2), (B2), (C2), (D2), (E2) and (F2): Two dimensional patterns of bond, respectively.
It was observed that the binding energy of docking was in range from −5.8 to −9.8 kcal/mol. These results indicated that ten compounds have good binding activities with the target proteins. Among six key targets, TNF exhibited the lowest binding energy with each ingredient. Among the ten ligands docking with TNF, stigmasterol showed the lowest energies (-9.8 kcal/mol). Stigmasterol can form a hydrogen bond with the active pocket amino acid residue Glu-116 (Fig. 10A1 and 10A2). Sesamin showed the lowest binding energy (-9.7 kcal/mol) with ALB, the interplay among them was depicted in Fig. 10B1 and 10B2, two hydrogen bonds were observed with Phe-134, Tyr-161 residue. Stigmasterol performed the lowest energy (-7.3 kcal/mol) with IL6. As shown in Fig. 10E1 and 10E2, in the binding pocket of IL6, Met-117 residue forms a hydrogen bond with stigmasterol. The free binding energy of hydroxygenkwanin with TP53 was −7.2 kcal/mol. The binding affinity was contributed by the hydrogen bonding with the His-115, Pen-113, Asp-268, and Ser-269 residue (Fig. 10D1 and 10D2). Stigmasterol exhibited the best affinity (-6.8 kcal/mol) with VEGFA. The binding affinity was contributed by the Pi-Sigma interactions with the Tyr-21 residue (Fig. 10F1 and 10F2). Sesamin formed three hydrogen bonding with Ser-5, Ser-43, Glu-64 residue and demonstrated -8.1 kcal/mol binding affinity by molecular docking (Fig. 10C1 and 10C2). The docking results for each ligand–protein interaction were summarized in Supplementary Table S9.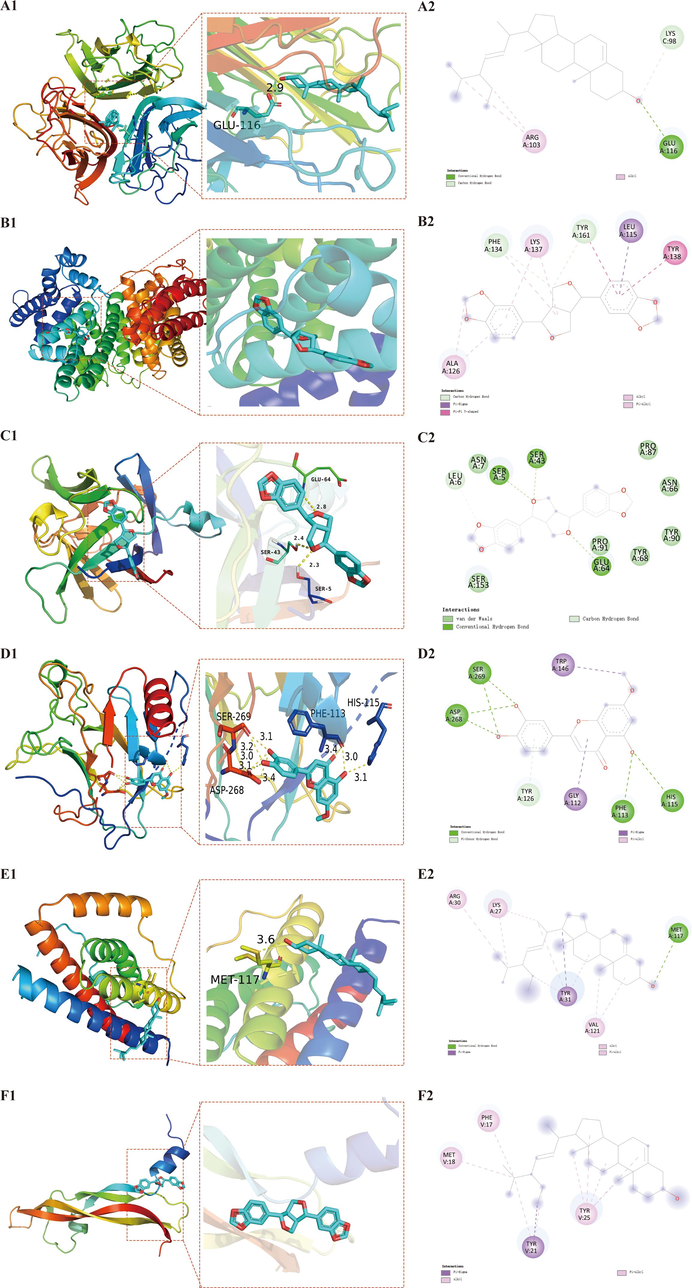
Docking patterns of key targets and specific active compounds. TNF- Stigmasterol(A1), ALB- Sesamin (B1), IL1B- Sesamin (C1), TP53- Hydroxygenkwanin (D1), IL6- Stigmasterol (E1), VEGFA- Stigmasterol (F1). (A2), (B2), (C2), (D2), (E2) and (F2): Two dimensional patterns of bond, respectively.
It is noteworthy that stigmasterol and sesamin showed a higher binding affinity with target proteins, which implied that these compounds may play critical roles in the therapeutic effects of YGPs. Considering the ADMET properties of the ten active compounds, stigmasterol and sesamin can be used as candidate drug molecules.
3.8 Molecular dynamics simulation
To determine the stability of protein–ligand complexes, a simulation of molecular dynamics was conducted. TNF(1tnf)-stigmasterol was exposed to 100 ns of molecular dynamics simulation to analyse the mobility, trajectory, and conformational changes of molecules based on the docking data.
Root mean square deviation (RMSD) can be used to evaluate the conformational stability of protein and ligands, as well as the degree of atomic position deviation from the starting position lower deviation in RMSD values, shows better conformational stability. Fig. 11A depicted that TNF protein showed good stability in the whole simulation process, and ligands tended to be stable from 50 ns and persisted till the end of the simulation. TNF-stigmasterol complex underwent a conformational change that lasted for a few simulation frames, returning to its stable conformation after 30 nm. The mean deviation for the TNF-stigmasterol complex was observed to be 0.552 Å. Accordingly, the conformation of the protein–ligand systems remained balanced without significant changes and the combination is relatively stable. The Root Mean Square Fluctuation (RMSF) is utilized to analyse local variations along protein chain residues and changes in ligand atom positions at specified temperatures and pressures. The RMSF plot represents that the fluctuation of certain residues is similar (Fig. 11B). The peak in the plot of the RMSF value shows that residues in these regions fluctuated the most during the whole simulation time. This value ranged between 0.042 Å and 0.633 Å. TNF-stigmasterol complex has lower fluctuations which revealed that the interaction between TNF and stigmasterol was strong and stable. Together, the result strongly supports the validity of the docking results.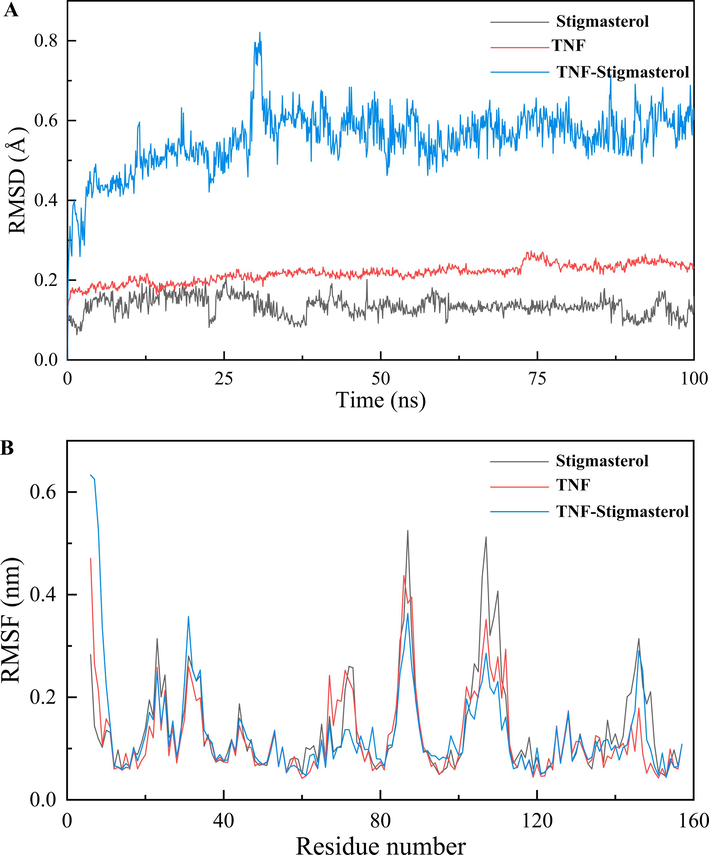
RMSD and RMSF of MD simulation. (A)The RMSD of TNF-Stigmasterol. (B) The RMSF of TNF-Stigmasterol.
4 Discussion
SONFH is a severe orthopaedic disorder, caused by long-term glucocorticoid therapy (Wu et al., 2019). It is a degenerative bone condition that, in the end, causes the femoral head to collapse, which in turn destroys the hip joint (Chen et al., 2020). Current treatments for SONFH are aimed at preventing femoral head collapse. Nevertheless, the optimal treatment is still unknown (Fu et al., 2019; Kubo et al., 2016).
TCM has been used for thousands of years to treat bone-related diseases. YGPs is a classic formula and has been used as a treatment option for SONFH in the clinic (Zhang et al., 2017; Zhang et al., 2019). However, the bioactive compounds and the underlying mechanisms are still unclear. Accordingly, we aimed to detect the possible compounds and mechanisms of YGPs in SONFH using the network pharmacology and molecular docking strategy.
126 active compounds of YGPs were screened using TCMSP database and predicted 1078 targets through SwissTargetPrediction webserver. Based on the analysis of the constructed herb-compound-target network, hydroxygenkwanin, helenalin, quercetin, isorhamnetin, beta-sitosterol, sitosterol, kaempferol, erythraline, stigmasterol, sesamin were the active ingredients associated with most of the targets. Several previous research have demonstrated the effectiveness of these compounds in treating SONFH. Helenalin, a lactone compound, has been found to be a chemical inhibitor of NF-κB. Notably, helenalin can clearly suppress the expression of P2X7R, MMP-13, SP, and PGE2 in osteoarthritic rats (Hu et al., 2016). To exert chondroprotective effects, quercetin can inhibit chondrocyte inflammation and apoptosis and modulate synovial macrophage polarisation to M2 macrophages (Hu et al., 2019). Isorhamnetin, a flavonoids compound, can remedy cartilage damages by suppressing excessive osteoclast activity and chondrocyte programmed cell death (Zhou et al., 2019). β-sitosterol, a phytosterols compound, demonstrated promise efficacy in preventing glucocorticoid-induced osteoporosis by protecting osteoblasts and inhibiting osteoclastogenesis (Wang et al., 2022). By modulating the XIST/miR-130a/STAT3 axis in chondrocytes, Kaempferol, a tetrahydroxyflavone compound, could suppress inflammation and extracellular matrix degradation (Xiao et al., 2021). Stigmasterol, is a plant sterol that can attach to the membranes of chondrocytes and has potential anti-inflammatory and anti-catabolic effects. Stigmasterol counteracts the expression of cartilage-degrading MMPs and has an anti-inflammatory effect on the pro-inflammatory mediator PGE2 (Gabay et al., 2010). Deng S et al. have reported that sesamin prevents osteonecrosis of the femoral head, as described by blocking ROS-induced osteoblast apoptosis (Deng et al., 2018). Therefore, multiple active compounds of YGPs can effectively treat SONFH through multiple channels.
Six key targets (TNF, IL6, ALB, IL1B, VEGFA, TP53) were methodically obtained through the target interaction network. Several of these target proteins had highly degrees in cluster network. The TNF is at the core of the whole network. It is wildly known that TNF-α is an important inflammatory factor. According to previous studies, TNF-α is linked to the proliferation and maturation of osteoclasts. The polymorphisms of TNF-α were presented as risk factors for SONFH (Liu et al., 2015). A recent study demonstrated that the pathophysiology of SONFH was influenced by a TNF-a-mediated alteration in M1/M2 macrophage polarisation (Wu et al., 2015). IL6 is a major pro-inflammatory cytokine that played a crucial role in bone disease. It is reported that IL6 rs2069837 is significantly linked to ONFH occurrence (Wang et al., 2022). IL1B is the most important inducer of pro-inflammatory immune responses, which could cause inflammation and bone diseases. According to a study, IL1B rs2853550 raised the risk of SONFH, whereas IL1B rs1143630 decreased the risk (Yu et al., 2019). VEGFA is reportedly an essential factor for chondrocyte death, angiogenesis, and bone formation (Gerber et al., 1999). Studies have demonstrated that significant expression differences of VEGFA were detected in several human tissues for various genotypes of rs2010963. Significant association between SNP rs2010963 and ONFH (Ma et al., 2018). TP53 protein is a tumor suppressor which plays significant roles in the suppression of bone progression. A study proved that downregulating TP53 could significantly improve the reparative effect of BMSCs on early SONFH (Zhang et al., 2020).
Based on GO functional analysis, biological processes involving the cytokine-mediated signaling pathway, inflammatory response, response to hypoxia, and positive regulation of angiogenesis, were associated with the effects of YGPs in treating SONFH. Previous study has demonstrated that increased inflammatory response is one of the major characteristics of SONFH (Wang et al., 2021). Another study showed that human umbilical cord mesenchymal stem cell therapy could enhance trabecular bone integrity of the femoral head and prevent necrosis of the femoral head and glucocorticoid-induced injury by promoting angiogenesis (Tian et al., 2021). The KEGG enrichment analysis indicated that YGPs pharmacological mechanisms in SONFH therapy might be mostly associated with the HIF-1 signaling pathway, TNF signaling pathway, EGFR tyrosine kinase inhibitor resistance, PI3K-Akt signaling pathway, and apoptosis. HIF1- is a fundamental regulator of collagen synthesis that enables chondrocytes in the hypoxic growth plate to preserve their role as professional secretory cells (Bentovim et al., 2012). A previous study has demonstrated that the HIF-1 signaling pathway can regulate the expression of osteogenic and angiogenic factors under normoxic conditions in vitro (Zou et al., 2011). PI3K/AKT pathway plays a crucial role in a variety of fundamental cellular processes, such as survival, proliferation, growth, and differentiation (Guntur and Rosen, 2011). This study indicated that salidroside inhibited dex-induced osteoblast apoptosis by stimulating the PI3K/Akt signaling pathway and decreasing osteoblast caspase-3 expression (Xue et al., 2018). Another study has shown that total flavonoids of Rhizoma Drynariae stimulated osteoblast proliferation, suppressed apoptosis, and decreased ROS levels through activating the PI3K/AKT signaling pathway (Lv et al., 2021). These results suggested that the pathways of immune responses, inflammation, apoptosis, as well as metabolism may be associated with ONFH.
Finally, molecular docking results verified that the key targets have good binding properties with active compounds. The MD simulation results further suggested that TNF structures (1tnf1)-stigmasterol complexes were very stable. The findings of this study will demonstrate that stigmasterol could perhaps contribute to the therapeutic effects of YGPs in SONFH.
Although some important preliminary findings were obtained, this study still has some limitations. To investigate the involvement of YGPs in the treatment of SONFH, we used only advanced bioinformatics and computational techniques. Therefore, the reliability and accuracy of the predictions need to be further in vivo animal experiments verified.
5 Conclusion
The study combined systematic pharmacology and molecular docking to explore the active compounds, candidate targets, and pharmaceutical mechanisms of YGPs against SONFH. Our results indicated that YGPs might exert their therapeutic effects on SONFH by an inflammatory response and positive regulation of angiogenesis. Moreover, our results may help to illuminate YGPs pharmacodynamic material and mechanism in SONFH therapy and may be used as a basis for future in vivo animal experiments studies.
Funding
This work was supported by the National Natural Science Foundation of China (No. 82205136), Zhejiang Traditional Chinese Medicine Administration (No. 2017ZKL017, No. 2022ZA129), Hangzhou Biomedical and Health Industry Development Support Technology Special Project (No.2022WJCY175, No.2022WJC176) and Clinical Research Foundation of Zhejiang Provincial Medical Association(2020ZYC-A113).
Summary
Steroid-related osteonecrosis of the femoral head (SONFH) is a refractory disease caused by long-term or high-dose application of glucocorticoids, which leads to damage or interruption of the femoral head blood supply, resulting in local ischemic necrosis and collapse of the articular surface. Yougui pills (YGPs), a classical TCM formula, show significant effects on treating SONFH. Clinical investigations have demonstrated that YGPs can considerably enhance the treatment efficacy and clinical indicators of ONFH patients with Yang deficiency in the spleen and kidney. However, the targets and related biological processes that mediate its effects against SONFH have not been systematically investigated. A network pharmacology approach was applied to explore the underlying mechanisms of YGPs against SONFH. Transcriptome data, systematic pharmacology analysis, GO and KEGG enrichment analyses, molecular docking were used in this study. The present study predicted the active components, candidate targets and pharmaceutical mechanisms of YGPs against SONFH. We identified 126 active compounds with 1078 targets in YGPs. 1237 SONFH-related were determined from GEO datasets in conjunction with GeneCards, OMIM, and DisGeNET databases. Among the 164 common targets, 6 key targets were identified by a topological analysis. According to the findings of the GO enrichment analysis, genes were most significantly enriched in the inflammatory response, response to hypoxia, and positive regulation of angiogenesis. The effects of YGPs have been mostly linked to the HIF-1 signaling pathway and the PI3K-Akt signaling pathway, according to a KEGG pathway enrichment analysis. In addition, the results of molecular docking and molecular dynamics simulations demonstrated that the active chemicals in YGPs have high affinity with the relevant targets. Our results indicated that YGPs might exert its therapeutic effects on SONFH by inflammatory response and positive regulation of angiogenesis. Moreover, our study may help to illuminate pharmacodynamic material and mechanism of YGPs in SONFH treatment and provide an integrative strategy to clarify the pharmacological mechanisms of traditional Chinese medicines.
CRediT authorship contribution statement
Ying Wang: Conceptualization, Funding acquisition, Data curation, Writing – original draft, Visualization, Investigation, Validation, Formal analysis, Project administration, Software. Tengfei Xu: Conceptualization, Writing – original draft, Validation, Formal analysis. Xueying Chen: Funding acquisition, Data curation, Visualization, Methodology, Resources. Yang Ye: Visualization, Project administration, Software. Liqin Liu: Funding acquisition, Data curation, Resources. Yifan Wang: Writing – review & editing, Methodology, Supervision, Resources, Project administration, Software. Peng Zhang: Funding acquisition, Writing – review & editing, Investigation, Formal analysis, Methodology, Supervision, Resources, Project administration, Software.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- An integrated computational approach of molecular dynamics simulations, receptor binding studies and pharmacophore mapping analysis in search of potent inhibitors against tuberculosis. J. Mol. Graph. Model.. 2018;83:17-32.
- [Google Scholar]
- Amberger, J.S., Bocchini, C.A., Schiettecatte, F., Scott, A.F., and Hamosh, A., 2015. OMIM.org: Online Mendelian Inheritance in Man (OMIM(R), an online catalog of human genes and genetic disorders. Nucleic Acids Res 43, D789-798.
- HIF1α is a central regulator of collagen hydroxylation and secretion under hypoxia during bone development. Development. 2012;139:4473-4483.
- [Google Scholar]
- The pathogenesis, diagnosis and clinical manifestations of steroid-induced osteonecrosis. J. Autoimmun.. 2020;110:102460
- [Google Scholar]
- Steroid-induced osteonecrosis of the femoral head reveals enhanced reactive oxygen species and hyperactive osteoclasts. Int. J. Biol. Sci.. 2020;16:1888-1900.
- [Google Scholar]
- cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol.. 2014;8(Suppl 4):S11.
- [Google Scholar]
- Molecular dynamics simulation and binding free energy studies of novel leads belonging to the benzofuran class inhibitors of Mycobacterium tuberculosis Polyketide Synthase 13. J. Biomol. Struct. Dyn.. 2019;37:1616-1627.
- [Google Scholar]
- Sesamin protects the femoral head from osteonecrosis by inhibiting ROS-induced osteoblast apoptosis in rat model. Front. Physiol.. 2018;9:1787.
- [Google Scholar]
- The system research of the molecular mechanism of quyushengxin capsule in the treatment of osteonecrosis of the femoral head. Evid. Based Complement. Alternat. Med.. 2022;2022:2968075.
- [Google Scholar]
- Molecular dynamics simulation with weak coupling to heat and material baths. J. Chem. Phys.. 2010;133:084105
- [Google Scholar]
- Network pharmacology and experimental validation to reveal the pharmacological mechanisms of chongcaoyishen decoction against chronic kidney disease. Front. Mol. Biosci.. 2022;9:847812
- [Google Scholar]
- Bioinformatics analysis of the biological changes involved in the osteogenic differentiation of human mesenchymal stem cells. J. Cell Mol. Med.. 2020;24:7968-7978.
- [Google Scholar]
- Mechanisms and molecular targets of the Tao-Hong-Si-Wu-Tang formula for treatment of osteonecrosis of femoral head: A network pharmacology study. Evid. Based Complement. Alternat. Med.. 2020;2020:7130105.
- [Google Scholar]
- Early diagnosis and treatment of steroid-induced osteonecrosis of the femoral head. Int. Orthop.. 2019;43:1083-1087.
- [Google Scholar]
- Stigmasterol: a phytosterol with potential anti-osteoarthritic properties. Osteoarthr. Cartil.. 2010;18:106-116.
- [Google Scholar]
- VEGF couples hypertrophic cartilage remodeling, ossification and angiogenesis during endochondral bone formation. Nat. Med.. 1999;5:623-628.
- [Google Scholar]
- Gfeller, D., Grosdidier, A., Wirth, M., Daina, A., Michielin, O., and Zoete, V., 2014. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic acids research 42, W32-38.
- Mechanisms of indigo naturalis on treating ulcerative colitis explored by GEO gene chips combined with network pharmacology and molecular docking. Sci. Rep.. 2020;10:15204.
- [Google Scholar]
- The skeleton: a multi-functional complex organ: new insights into osteoblasts and their role in bone formation: the central role of PI3Kinase. J. Endocrinol.. 2011;211:123-130.
- [Google Scholar]
- Network pharmacology-based approach to understand the effect and mechanism of Danshen against anemia. J. Ethnopharmacol.. 2022;282:114615
- [Google Scholar]
- Network pharmacology: the next paradigm in drug discovery. Nat. Chem. Biol.. 2008;4:682-690.
- [Google Scholar]
- Quercetin alleviates rat osteoarthritis by inhibiting inflammation and apoptosis of chondrocytes, modulating synovial macrophages polarization to M2 macrophages. Free Radic. Biol. Med.. 2019;145:146-160.
- [Google Scholar]
- Blocking of the P2X7 receptor inhibits the activation of the MMP-13 and NF-κB pathways in the cartilage tissue of rats with osteoarthritis. Int. J. Mol. Med.. 2016;38:1922-1932.
- [Google Scholar]
- Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc.. 2009;4:44-57.
- [Google Scholar]
- Steroid-induced osteonecrosis of the femoral head: Novel insight into the roles of bone endothelial cells in pathogenesis and treatment. Front. Cell Dev. Biol.. 2021;9:777697
- [Google Scholar]
- Network pharmacology-based and molecular docking prediction of the active ingredients and mechanism of ZaoRenDiHuang capsules for application in insomnia treatment. Comput. Biol. Med.. 2021;135:104562
- [Google Scholar]
- Clinical and basic research on steroid-induced osteonecrosis of the femoral head in Japan. J. Orthop. Sci. : Off. J. Japanese Orthop. Assoc.. 2016;21:407-413.
- [Google Scholar]
- Heart research advances using database search engines, Human Protein Atlas and the Sydney Heart Bank. Heart Lung Circ.. 2013;22:819-826.
- [Google Scholar]
- Network pharmacology prediction and molecular docking-based strategy to explore the potential mechanism of Huanglian Jiedu Decoction against sepsis. Comput. Biol. Med.. 2022;144:105389
- [Google Scholar]
- Traditional Chinese medicine network pharmacology: theory, methodology and application. Chin. J. Nat. Med.. 2013;11:110-120.
- [Google Scholar]
- Application of Berendsen barostat in dissipative particle dynamics for nonequilibrium dynamic simulation. J. Chem. Phys.. 2017;146:124108
- [Google Scholar]
- Combined effect of tnf-α polymorphisms and hypoxia on steroid-induced osteonecrosis of femoral head. Int. J. Clin. Exp. Path.. 2015;8:3215-3219.
- [Google Scholar]
- Lv, W., Yu, M., Yang, Q., Kong, P., and Yan, B., 2021. Total flavonoids of Rhizoma drynariae ameliorate steroid‑induced avascular necrosis of the femoral head via the PI3K/AKT pathway. Molecular medicine reports 23.
- Relationship of common variants in VEGFA gene with osteonecrosis of the femoral head: A Han Chinese population based association study. Sci. Rep.. 2018;8:16221.
- [Google Scholar]
- Molecular dynamics simulation by GROMACS using GUI plugin for PyMOL. J. Chem. Inf. Model.. 2013;53:1229-1234.
- [Google Scholar]
- Potential compound from herbal food of Rhizoma Polygonati for treatment of COVID-19 analyzed by network pharmacology: Viral and cancer signaling mechanisms. J. Funct. Foods. 2021;77:104149
- [Google Scholar]
- Comparison of tetrahedral order, liquid state anomalies, and hydration behavior of mTIP3P and TIP4P water models. J. Chem. Theory Comput.. 2011;7:3354-3367.
- [Google Scholar]
- Piñero, J., Ramírez-Anguita, J.M., Saüch-Pitarch, J., Ronzano, F., Centeno, E., Sanz, F., and Furlong, L.I., 2020. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic acids research 48, D845-d855.
- Enhanced GROMACS: toward a better numerical simulation framework. J. Mol. Model.. 2019;25:355.
- [Google Scholar]
- GeneCards: integrating information about genes, proteins and diseases. Trends Genet. : TIG. 1997;13:163.
- [Google Scholar]
- TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminf.. 2014;6:13.
- [Google Scholar]
- Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res.. 2003;13:2498-2504.
- [Google Scholar]
- Uncovering the mechanism of Maxing Ganshi Decoction on asthma from a systematic perspective: A network pharmacology study. Sci. Rep.. 2018;8:17362.
- [Google Scholar]
- Szklarczyk, D., Morris, J.H., Cook, H., Kuhn, M., Wyder, S., Simonovic, M., Santos, A., Doncheva, N.T., Roth, A., Bork, P., et al., 2017. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic acids research 45, D362-d368.
- Szklarczyk, D., Gable, A.L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., Simonovic, M., Doncheva, N.T., Morris, J.H., Bork, P., et al., 2019. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic acids research 47, D607-d613.
- CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems. 2015;127:67-72.
- [Google Scholar]
- Human umbilical cord mesenchymal stem cells prevent glucocorticoid-induced osteonecrosis of the femoral head by promoting angiogenesis. J. Plast. Surg. Hand Surg. 2021:1-7.
- [Google Scholar]
- Structural investigations into the binding mode of novel neolignans Cmp10 and Cmp19 microtubule stabilizers by in silico molecular docking, molecular dynamics, and binding free energy calculations. J. Biomol. Struct. Dyn.. 2016;34:1232-1240.
- [Google Scholar]
- AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem.. 2010;31:455-461.
- [Google Scholar]
- Systems pharmacology: bridging systems biology and pharmacokinetics-pharmacodynamics (PKPD) in drug discovery and development. Pharm. Res.. 2011;28:1460-1464.
- [Google Scholar]
- Wang, T., Li, S., Yi, C., Wang, X., and Han, X., 2022. Protective Role of β-Sitosterol in Glucocorticoid-Induced Osteoporosis in Rats Via the RANKL/OPG Pathway. Alternative therapies in health and medicine.Oct;28(7):18-25.
- An intron SNP rs2069837 in IL-6 is associated with osteonecrosis of the femoral head development. BMC Med. Genomics. 2022;15:5.
- [Google Scholar]
- Bone morphogenetic protein 2 controls steroid-induced osteonecrosis of the femoral head via directly inhibiting interleukin-34 expression. J. Mol. Endocrinol.. 2021;68:1-9.
- [Google Scholar]
- The pathogenesis of steroid-induced osteonecrosis of the femoral head: A systematic review of the literature. Gene. 2018;671:103-109.
- [Google Scholar]
- Mechanisms underlying the therapeutic effects of Qingfeiyin in treating acute lung injury based on GEO datasets, network pharmacology and molecular docking. Comput. Biol. Med.. 2022;145:105454
- [Google Scholar]
- TNF-a mediated inflammatory macrophage polarization contributes to the pathogenesis of steroid-induced osteonecrosis in mice. Int. J. Immunopathol. Pharmacol.. 2015;28:351-361.
- [Google Scholar]
- Noncoding RNAs in steroid-induced osteonecrosis of the femoral head. Biomed Res. Int.. 2019;2019:8140595.
- [Google Scholar]
- Exploring the pharmacological mechanisms of Xihuang Pills against prostate cancer via integrating network pharmacology and experimental validation in vitro and in vivo. Front. Pharmacol.. 2021;12:791269
- [Google Scholar]
- Kaempferol attenuates the effects of XIST/miR-130a/STAT3 on inflammation and extracellular matrix degradation in osteoarthritis. Future Med. Chem.. 2021;13:1451-1464.
- [Google Scholar]
- Xie, Y.M., Liu, H., Jiang, J.J., Wei, X., Shen, H., Zhi, Y.J., Sun, J., Li, J.Y., Bao, X.X., Shi, W., et al., 2021. [Clinical practice guideline for postmenopausal osteoporosis with traditional Chinese medicine]. Zhongguo Zhong yao za zhi .Nov;46(22):5992-5998.
- ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res.. 2021;49:W5-w14.
- [Google Scholar]
- A novel chemometric method for the prediction of human oral bioavailability. Int. J. Mol. Sci.. 2012;13:6964-6982.
- [Google Scholar]
- Salidroside inhibits steroid-induced avascular necrosis of the femoral head via the PI3K/Akt signaling pathway: In vitro and in vivo studies. Mol. Med. Rep.. 2018;17:3751-3757.
- [Google Scholar]
- Exploring the molecular mechanism of action of Yinchen Wuling powder for the treatment of hyperlipidemia, using network pharmacology, molecular docking, and molecular dynamics simulation. Biomed. Res. Int.. 2021;2021:9965906.
- [Google Scholar]
- Yu, Y., Zhang, Y., Wu, J., Sun, Y., Xiong, Z., Niu, F., Lei, L., Du, S., Chen, P., and Yang, Z., 2019. Genetic polymorphisms in IL1B predict susceptibility to steroid-induced osteonecrosis of the femoral head in Chinese Han population. Osteoporosis international : a journal established as result of cooperation between the European Foundation for Osteoporosis and the National Osteoporosis Foundation of the USA 30, 871-877.
- Zaman, Z., Khan, S., Nouroz, F., Farooq, U., and Urooj, A., 2021. Targeting protein tyrosine phosphatase to unravel possible inhibitors for Streptococcus pneumoniae using molecular docking, molecular dynamics simulations coupled with free energy calculations. Life Sci. Jan 1;264:118621.
- Zhang, F., Peng, W., Zhang, J., Dong, W., Wu, J., Wang, T., and Xie, Z., 2020. P53 and Parkin co-regulate mitophagy in bone marrow mesenchymal stem cells to promote the repair of early steroid-induced osteonecrosis of the femoral head. Cell death & disease. Jan 20;11(1):42.
- Zhang, L., Han, L., Wang, X., Wei, Y., Zheng, J., Zhao, L., and Tong, X., 2021. Exploring the mechanisms underlying the therapeutic effect of Salvia miltiorrhiza in diabetic nephropathy using network pharmacology and molecular docking. Bioscience reports 41.
- A bioinformatics investigation into molecular mechanism of Yinzhihuang granules for treating hepatitis B by network pharmacology and molecular docking verification. Sci. Rep.. 2020;10:11448.
- [Google Scholar]
- Yougui Pills attenuate cartilage degeneration via activation of TGF-β/Smad signaling in chondrocyte of osteoarthritic mouse model. Front. Pharmacol.. 2017;8:611.
- [Google Scholar]
- Yougui pills exert osteoprotective effects on rabbit steroid-related osteonecrosis of the femoral head by activating β-catenin. Biomed. Pharmacother. = Biomed. Pharmacother.. 2019;120:109520
- [Google Scholar]
- Isorhamnetin attenuates osteoarthritis by inhibiting osteoclastogenesis and protecting chondrocytes through modulating reactive oxygen species homeostasis. J. Cell Mol. Med.. 2019;23:4395-4407.
- [Google Scholar]
- In vitro study of enhanced osteogenesis induced by HIF-1α-transduced bone marrow stem cells. Cell Prolif.. 2011;44:234-243.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2024.105609.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary data 1
Supplementary data 1







