Translate this page into:
New family of tetrazole-pyrazole compounds: Synthesis, characterization, antimicrobial activity and docking study
⁎Corresponding author. fouad_malek@yahoo.fr (Fouad Malek) f.malek@ump.ac.ma (Fouad Malek)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
New pyrazole-tetrazole hybrid molecules were obtained via two synthetic pathways. The first one is equimolar condensation of N-alkylated pyrazole with 1H-tetrazolic derivatives (yields in range 31–45%) while in the second route consists in a direct alkylation of the pyrazole-tetrazole derivatives at the N2 position of the pyrazolic ring (yields in range 63–75%). The structure of these compounds was characterized by spectroscopic and spectrometric methods. The evaluation of the antimicrobial activity of these compounds against four bacterial and six fungal strains was performed and the obtained findings show that they possess a medium activity (inhibition zone diameter in the range of 9–19 mm) compared to the used positive controls. Furthermore, the antibacterial and antifungal activities were affected by the substituents on the tetrazole rings and junction position between the two N-heterocycles. Some experimental results are also supported by a docking study.
Keywords
Antifungal activity
Anbacterial activity
Docking calculation
Hybrid
Pyrazole-tetrazole
1 Introduction
The chemistry of hybrid N-heterocyclic molecules has taken a great interest thanks to its implementation in different area including complexation (Yu et al., 2022), sensors (Da Lama et al., 2022) and energy (Xiong et al., 2019). In the medicinal chemistry field, the literature communicated several hybrid molecules as potent pharmacological agents. Indeed, they have showed a high antiviral (Jilloju et al., 2022), antibacterial (Chu et al., 2019), anticancer (Alzahrani et al., 2023) and enzyme inhibition (Akyüz et al., 2022) activities.
During the two last decades, our laboratory has a high interest towards the pyrazole and its derivatives. In fact, several architectures have been designed such as tripods (Malek et al., 2014; Elkodadi et al., 2007; Harit et al., 2012; Arrousse et al. 2020; Zerrouki et al. 2011), bipyrazoles (Bouabdallah et al., 2013; Bouabdallah et al., 2021; Harit et al., 2018a; Dahmani et al., 2021a; Bouabdallah et al., 2022) and macrocycles (Harit et al., 2020; Dahmani et al., 2021b; Harit et al., 2017a; Harit and Malek, 2017) and their biological potencies have been evaluated.
On the other hand, the tetrazolic derivatives are other class of nitrogen-containing heterocyclic compounds that have attracted a great attention by the specialist in the medicinal chemistry. The importance given to this N-heterocycle, come from its consideration as bioisoster of carboxylic acid due their similar pKa values and planar electronic delocalization (Zou et al., 2020). Therefore, the tetrazole and its derivatives showed a large spectrum of pharmacological properties including antimicrobial (Antypenko et al., 2016) antiviral (Mikolaichuk et al., 2021) and antidepressant (Wang et al., 2019) activities.
In this context, the assembly of these two N-heterocyclic rings in one skeleton appears to be a good approach to reach hybrid molecules with high pharmacological properties.
In addition, performing the docking calculations to support the biological activity evaluation is widely used. Indeed, this could give insight on the possible action mechanism of the compound under examination and thus establishing the structure activity relationships (SARs).
Therefore, the present paper is a continuous to our previous works (Cherfi et al., 2021; Cherfi et al., 2022; Oulous et al., 2022; Cherfi et al., 2023; Harit et al., 2022) and those that were communicated in the literature assigned to the synthesis of N-heterocycle hybrid products. In this study, we compare tow ways that allow the obtaining of a new series of hybrid tetrazole-pyrazole compounds. Their antibacterial and antifungal potencies were examined. Docking calculations were also conducted to verify the possible action mechanism of these molecules.
2 Materials and methods
2.1 Reagents and instruments
All chemicals were purchased from Aldrich Chemical Co. and used without further purification. NMR spectra were recorded on Bruker AC 500. Spin resonances are shown as chemical shifts (δ) in parts per million (ppm) and referenced to the residual peak of a CDCl3 solvent: 7.27 ppm (1H NMR), 77 ppm (13C NMR, central band). Spin multiplicity was shown by s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet. Mass spectrometry analysis was performed on a Platform II Micromass instrument (ESI+, CH3CN/H2O: 50/50) and Micromass Q-TOF micro MS spectrometer. Uncorrected melting Point was carried out in capillary on IA9100 (Electrothermal) apparatus. Elemental analysis was realised using EA 3000 Elemental Analyser. Compounds 1a-1d were synthesised as reported in our previous work (Oulous et al., 2022).
2.2 Synthesis
2.2.1 5-benzyl-2-((1-benzyl-5-methyl-1H-pyrazol-3-yl)methyl)-2H-tetrazole 2a
Route 1:
A mixture of 1a (19.6 mmol) and tBuOK (19.6 mmol) in DMF (60 mL) was heated under reflux for 1 h. To the hot mixture, a solution of benzyl bromide (19.6 mmol) in DMF (20 mL) was slowly added. The reaction mixture was then heated at 120 °C for 48 h. The resulting mixture was filtered and the solvent was evaporated to dryness. Components mixture were separated by column chromatography on silica gel (Diethyl ether, Rf = 0.51) to give 2a as yellow oil (75%).
Route 2:
A mixture of A (21.2 mmol) and tBuOK (21.2 mmol) in DMF (60 mL) was heated under reflux for 1 h. To the hot mixture, a solution of 1-benzyl-5-methyl-3-chloromethylpyrazole [ ] (21.2 mmol) in DMF (20 mL) was slowly added. The reaction mixture was heated at 120 °C for 48 h. The resulting mixture was filtered and the solvent was evaporated to dryness. The residue was purified by a silica column (diethylether, Rf = 0.51) to afford compound 2a as yellow oil (45%).
1H NMR (CDCl3) δ: 2.17 (s, 3H, CH3Pz); 4.23 (s, 2H, Ph-CH2-Tz); 5.22 (s, 2H, Ph-CH2-Pz); 5.64 (s, 2H, Tz-CH2-Pz); 6.06 (s, 1H, PzH); 7.04–7.33 (m, 10H, PhH). 13C NMR (CDCl) δ: 11.33 (CH3Pz); 15.3 (CH3-CH2–); 31.89 (Ph-CH2-Tz); 51.04 (Tz-CH2-Pz); 52.92 (Ph-CH2-Pz); 105.38 (HCPz); 126.65, 126.78, 127.71, 128.42, 128.83 128.89, 136.84, 136.91 (CPh); 139.87 (N-C(CH3) = C); 143.52 (N = C(CH2)-Tz); 165.47 (CTz). MS m/z: calculed for C20H21N6 [M + H]+, 345.1828; found: 345.1828. Anal. Calcd for C20H20N6: C, 69.75; H, 5.85; N, 24.40. Found: C, 69.82 1; H, 5.91; N, 24.49.
2.2.2 5-benzyl-2-((1-benzyl-3-methyl-1H-pyrazol-5-yl)methyl)-2H-tetrazole 2b
Route 1:
A mixture of 1b (19.6 mmol) and tBuOK (19.6 mmol) in DMF (60 mL) was heated under reflux for 1 h. To the hot mixture, a solution of benzyl bromide (19.6 mmol) in DMF (20 mL) was slowly added. The reaction mixture was then heated under reflux for 48 h. The resulting mixture was filtered and the solvent was evaporated to dryness. Components mixture were separated by column chromatography on silica gel (Diethyl ether, Rf = 0.36) to afford compound 2b as yellow oil (67%).
Route 2:
Compound 2b was isolated during the purification of compound 2a, (Diethyl ether, Rf = 0.36) as yellow oil in 43 %. 1H NMR (CDCl3) δ: 2.14 (s, 3H, CH3Pz); 4.27 (s, 2H, Ph-CH2-Tz); 5.21 (s, 2H, Ph-CH2-Pz); 5.34 (s, 2H, Tz-CH2-Pz); 5.86 (s, 1H, PzH); 7.04–7.34 (m, 10H, PhH). 13C NMR (CDCl3) δ: 11.33 (CH3Pz); 29.34 (Ph-CH2-Tz); 45.33 (Tz-CH2-Pz); 53.19 (Ph-CH2-Pz); 105.28 (HCPz); 126.90, 127.57, 128.03, 128.91, 129.00, 129.01, 134.14, 136.51 (CPh); 140.70 (N-C(CH3) = C); 144.17 (N = C(CH2)-Tz); 154.00 (CTz). MS m/z: calculed for C20H21N6 [M + H]+, 345.1828; found: 345.1824. Anal. Calcd for C20H20N6: C, 69.75; H, 5.85; N, 24.40. Found: C, 69.91; H, 5.97; N, 24.63.
2.2.3 2-(2-((1-benzyl-5-methyl-1H-pyrazol-3-yl)methyl)-2H-tetrazol-5-yl)pyridine 2c
Route 1:
A mixture of 1c (19.6 mmol) and tBuOK (19.6 mmol) in DMF (60 mL) was heated under reflux for 1 h. To the hot mixture, a solution of benzyl bromide (19.6 mmol) in DMF (20 mL) was slowly added. The reaction mixture was then heated at 120 °C for 48 h. The resulting mixture was filtered and the solvent was evaporated to dryness. Components mixture were separated by column chromatography on silica gel (Diethyl ether, Rf = 0.46) to give 2c as yellow oil (63%).
Route 2:
A mixture of B (21.2 mmol) and tBuOK (21.2 mmol) in DMF (60 mL) was heated under reflux for 1 h. To the hot mixture, a solution of 1-benzyl-5-methyl-3-chloromethylpyrazole [ ] (21.2 mmol) in DMF (20 mL) was slowly added. The reaction mixture was heated at 120 °C for 48 h. The resulting mixture was filtered and the solvent was evaporated to dryness. The residue was purified by a silica column (diethylether, Rf = 0.46) to afford compound 2c as yellow oil (34%). 1H NMR (CDCl3) δ: 2.14 (s, 3H, CH3Pz); 5.26 (s, 2H, Ph-CH2-Pz); 6.07 (s, 2H, Tz-CH2-Pz); 6.13 (s, 1H, PzH); 7.02–7.38 (m, 5H, PhH); 7.43, 7.85, 8.24, 8.78 (m, 4H, Py). 13C NMR (CDCl3) δ: 11.32 (CH3Pz); 47.46 (Tz-CH2-Pz); 53.38 (Ph-CH2-Pz); 105.86 (HCPz); 125.81, 126.81, 127.79, 137.02 (CPh); 116.32, 124.46, 145.25, 145.59, 150.19 (CPy); 140.18 (CH3)CPz); 142.85 (CH2-CPz); 164.93 (CTz). HRMS m/z: calculed for C18H18N7 [M + H]+, 332.1624; found: 332.1624. Anal. Calcd for C18H17N7: C, 65.24; H, 5.17; N, 29.59. Found: C, 65.39; H, 5.35; N, 29.78.
2.2.4 2-(1-((1-benzyl-5-methyl-1H-pyrazol-3-yl)methyl)-1H-tetrazol-5-yl)pyridine 2d
Route 1:
A mixture of 1d (19.6 mmol) and tBuOK (19.6 mmol) in DMF (60 mL) was heated under reflux for 1 h. To the hot mixture, a solution of benzyl bromide (19.6 mmol) in DMF (20 mL) was slowly added. The reaction mixture was then heated at 120 °C for 48 h. The resulting mixture was filtered and the solvent was evaporated to dryness. Components mixture were separated by column chromatography on silica gel (Diethyl ether, Rf = 0.23) to give 2d as yellow oil (65%).
Route 2:
Compound 2d was isolated during the purification of compound 2c, (Diethyl ether, Rf = 0.23) as yellow oil in 31 % yield. 1H NMR (CDCl3) δ: 2.19 (s, 3H, CH3Pz); 5.19 (s, 2H, Ph-CH2-Pz); 5.90 (s, 1H, PzH); 6.24 (s, 2H, Tz-CH2-Pz); 6.92–7.35 (m, 5H, PhH); 7.43, 7.89, 8.34, 8.74 (m, 4H, Py). 13C NMR (CDCl3) δ: 11.32 (CH3Pz); 47.46 (Tz-CH2-Pz); 53.38 (Ph-CH2-Pz); 105.31 (HCPz); 125.41, 126.73, 128.85, 136.78 (CPh); 116.32, 124.69, 145.15, 145.44, 149.61 (CPy); 140.11 (CH3)CPz); 143.65 (CH2-CPz); 153.48 (CTz). HRMS m/z: calculed for C18H18N7 [M + H]+, 332.1624; found: 332.1623. Anal. Calcd for C18H17N7: C, 65.24; H, 5.17; N, 29.59. Found: C, 65.41; H, 5.28; N, 29.66.
2.3 Antimicrobial activity determination
The antifungal activity of the compounds 2a-2d was examined against four bacterial strains: Escherichia coli, Pseudomonas aeruginosa, Staphylocoque aureus and Listeria monocytogenes as well as six fungal strains: Geotrichum candidum, Aspergillus niger, Penicillium digitatum and Rhodotorula glutinis. Their potencies were determined through the measuring of their inhibition zone diameter obtained on agar culture media, as described in our recent works (Harit et al., 2017b). These experiments were performed in triplicate to check the results reproducibility. The used positive controls were the tetracyclin and the cycloheximide for the antibacterial and antifungal studies, respectively.
2.4 Docking calculations
Docking calculations were performed via the freely available software “iGemdock program” (Yang and Chen, 2004). The structures of fructofuranosidase from Rhodotorula dairenensis (8BEQ) and Biotin protein ligase in Staphylococcus aureus (3V7R) were obtained from Protein Data Bank (https://www.rcsb.org./pdb). ChemDraw 3D Ultra v.8.0 software was used to draw the 3D structure of molecules and the energy minimization was carried out by molecular mechanics (MM2), until the (RMS) gradient reaches a value under 0.1 kcal/mol. The 3D structure of (3aS,4S,6aR)-4-(5-{1-[4-(6-amino-9H-purin-9-yl)butyl]-1H-1,2,3-triazol-4-yl}pentyl)tetrahydro-1H-thieno[3,4-d]imidazol-2(3H)-one (Ref1) (70679452) from “National Library of Medicine; National Center for Biotechnology Information”, a free database of compounds (https://pubchem.ncbi.nlm.nih.gov/), while that of raffinose (Ref2) (ZINC6920405) was extracted from ZINC20 database, a free database of commercially-available compounds (https://zinc.docking.org/). iGemdock predicts the enzyme-molecule interaction profiles according to three kinds of interaction: electrostatic (Eelc), hydrogen-bonding (Hbond) and Van der Waal’s (VdW) interactions. The following equation: Binding energy = VdW + Hbond + Eelc, was used to calculate the empirical energy value. All calculations were performed under these parameters: population size: 200; generations: 70; solutions: 3 (Drug Screening); binding site radius: 6 Å. The 3D and 2D docked pose were visualized using RasMol 2.7.3 (https://www.rasmol.org) and BIOVIA Discovery Studio Visualizer 2021 v21.1.0.20298 (https://discover.3ds.com/discovery-studio-visualizer-download), respectively.
3 Results and discussion
3.1 Synthesis
The desired compounds were obtained following the pathway showed in the Scheme 1.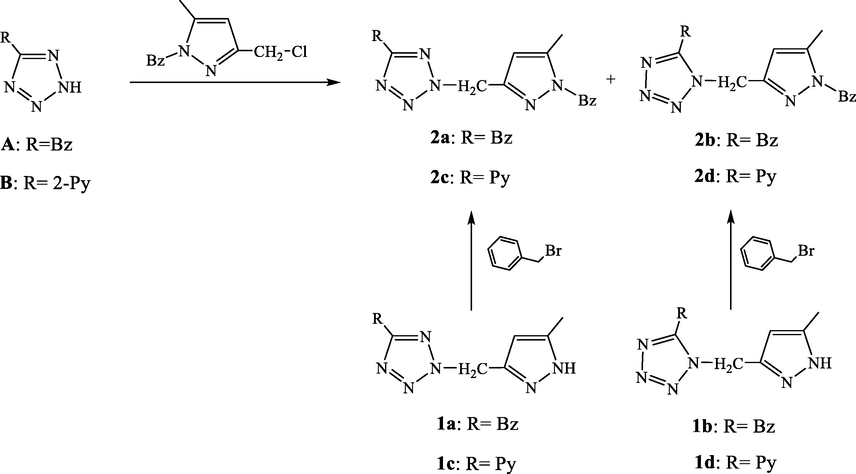
Pathway used for the synthesis of compounds 2a-2d.
Two synthetic pathways were evaluated to reach the desired compounds 2a-2d. The first one consists in an alkyation of the compound A (Oulous et al., 2022) by the chlorinated derivative B (Harit et al., 2016; Harit et al., 2018b) in DMF and in the presence of sodium hydroxide that was used as base. This reaction leads to two position isomers in (1:1) ratio which evidences that the reaction was not regioselective. However, the separation of the two isomers were found to be not complicated due the obvious difference in their rapport frontal Rf. On the other hand, the differencing between the two isomers can easily performed via the carbon NMR spectroscopy. Indeed tetrazolic carbon is shifted around 164 ppm, when the alkylation was occurred at the N2 position and around 153 ppm in the case of N1 alkylation (Butler and Fleming, 1997). Thus, compounds 2a (N1) and 2b (N2) have the rapport frontals values 0.23 and 0.57, respectively, in contrast to the isomers compound 2c and 2d where the (N1) position structure is attributed to the compound with the high rapport frontal value. The structures of different compounds were also ensured by High resolution mass spectrometry and elemental analysis.
For the second route, the benzylation of compounds 1a-1d through the benzyl bromide gives in the four case one major product as displayed in the corresponding thin layer chromatograms (TLC). As described above, the benzylation position could also be determined using 13C NMR spectroscopy (Harit et al., 2016). In the case of pyrazole, the signal assigned to the carbon atom linked to the methyl group appears at ≈148 and 138 ppm for a benzylation at the 1,3 and 1,5 pattern, respectively (Fig. 1). Therefore, the apparition of signals around 140 ppm for the major products evidences that the alkylation was performed at the 1,5 pattern. Finally, the same compounds were obtained through the two routes but in different yields. In fact, the second route allows the obtaining of compounds in satisfactory yields better than the first one.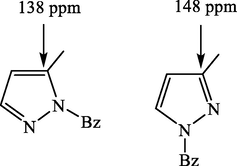
Possible isomers resulting from the N-alkylation of the pyrazolic ring.
Structures of the obtained compounds were also ensured by 1H NMR spectroscopy and HRMS analysis. We give in Fig. 2. The 1H and 13C NMR spectra of compound 2b.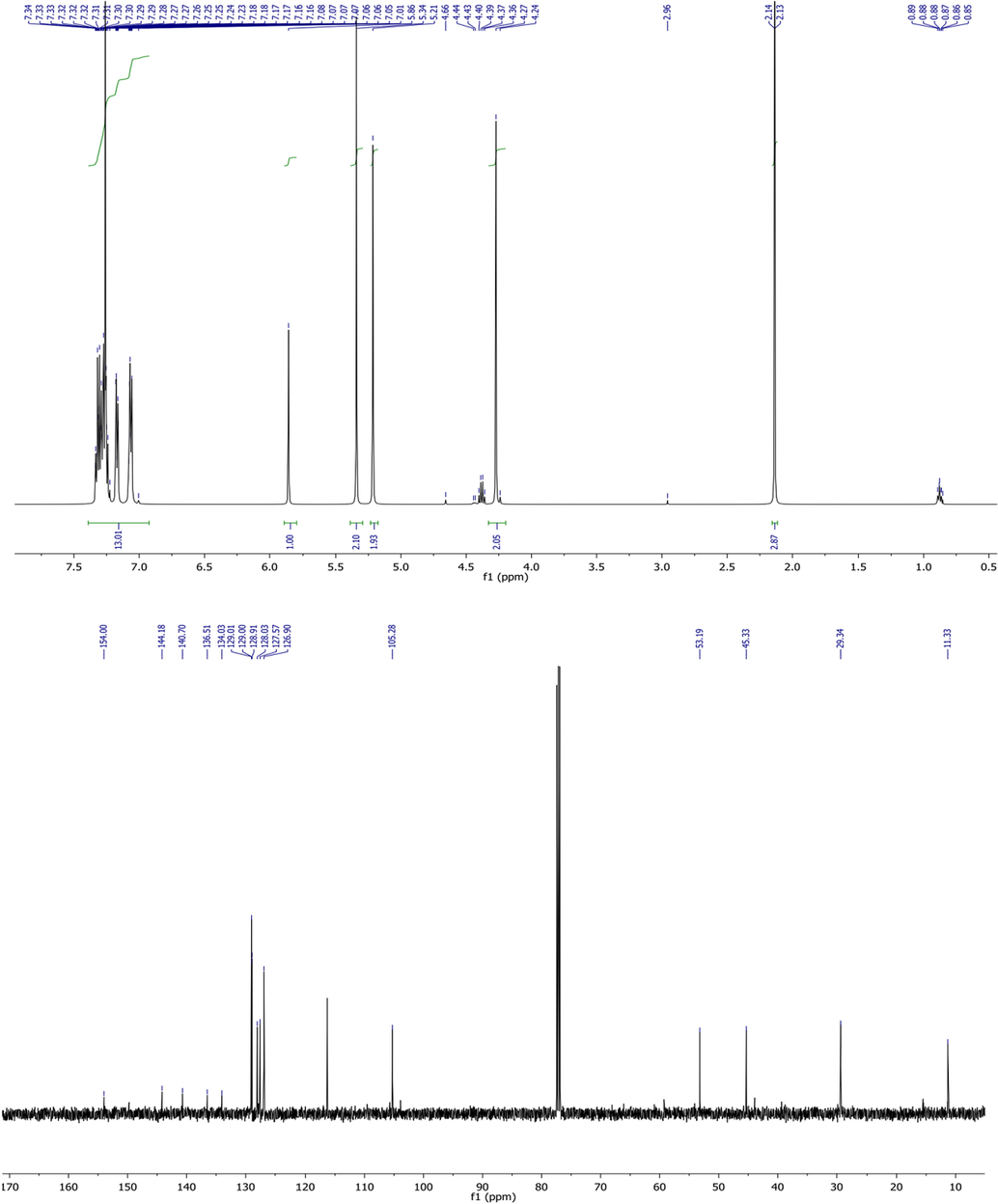
1H and 13C NMR spectra of compound 2b.
3.2 Antimicrobial screening
3.2.1 Antibacterial activity
The antibacterial activity of compounds 2a-2d was examined against four bacterial strains: Escherichia coli and Pseudomonas aeruginosa as representative of gram negative bacteria, while Staphylocoque aureus and Listeria monocytogenes as representative of gram negative bacteria. The obtained results are regrouped in Table 1. *NA: No activity.
Compound
Escherichia
coli
Pseudomonas aeruginosa
Staphylocoque aureus
Listeria monocytogenes
2a
10
10
12
12
2b
9
09
13
13
2c
10
08
12
11
2d
10
09
13
10
Tetracyclin
20
21
20
22
DMSO
NA*
NA
NA
NA
By comparing results of couples (2a,2b) and (2c,2d) which differ by the position of the junction between the pyrazole and tetrazole rings, we observe that antibacterial activity was not affected by this parameter against both gram negative and positive bacterial strains. Furthermore, compounds 2a and 2b with benzyl group on the tetrazol fragment were partially less efficiencies against the gram positive bacteria than the gram negative ones. Replacing the benzyl group by the pyridine one (2c and 2d) do not influence practically the activity of such compounds. Nevertheless, the activity of these compounds was found to be less than of that corresponding to the used positive control.
3.2.2 Antifungal activity
The antifungal activity of these compounds was also examined against six fungal strains: Geotrichum candidum, Aspergillus niger, Penicillium digitatum, Rhodotorula glutinis, Saccharomyces, Candida Albicans. In this case, cycloheximide was used as positive control.
Results showed in Table 2, evidenced that all compounds possess some antifungal activity against the four strains in different potency. However, their activities were found to be less than cycloheximide. In contrast of the antibacterial activity results, we observed that the antifungal potency of the tested compounds was considerably affected by group nature on the tetrazolic moiety as well as the position of the junction. Furthermore, these compounds showed no activity against Penicillium digitatum. In the case of Aspergillus niger strain, both compounds 2a and 2b displayed some activity in contrast to compounds 2c and 2d where no activity was observed. This result suggests that the replacing of the benzyl group by the pyridine on the tetrazolic fragment decreases the activity in this kind of structures. Moreover, all molecules displayed a good activity against Rhodotorula glutinis, with an inhibition zone diameter in the range of 14–19 mm. *NA: No activity.
Compound
Geotrichum candidum
Aspergillus niger
Penicillium digitatum
Rhodotorula glutinis
Saccharomyces
Candida Albicans
2a
15
14
NA*
14
11
16
2b
14
12
NA
18
11
13
2c
12
NA
NA
16
12
10
2d
13
NA
NA
19
13
13
Cycloheximide
30
31
30
30
27
35
DMSO
NA
NA
NA
NA
NA
NA
3.3 Docking study
We reported above, that all examined compounds showed some antimicrobial activity against Staphylocoque aureus and Rhodotorula glutinis strains. In this context, docking calculations were performed to take insight on the possible mechanism of action of such molecules.
In the case of Staphylocoque aureus, the Biotin protein ligase in Staphylococcus aureus (SaBPL) was chosen because it is considered as potential antibacterial target according to the literature (Jitrapakdee et al., 2008; Duckworth et al. 2011). Before the docking of compounds 2a- 2d on active site of this enzyme, the protocol was first validated by redoking the reference ligand. The results figured out in Fig. 3 evidenced that the docked reference ligand interacts with the active site via almost the same interactions compared to those experimentally reported in the literature.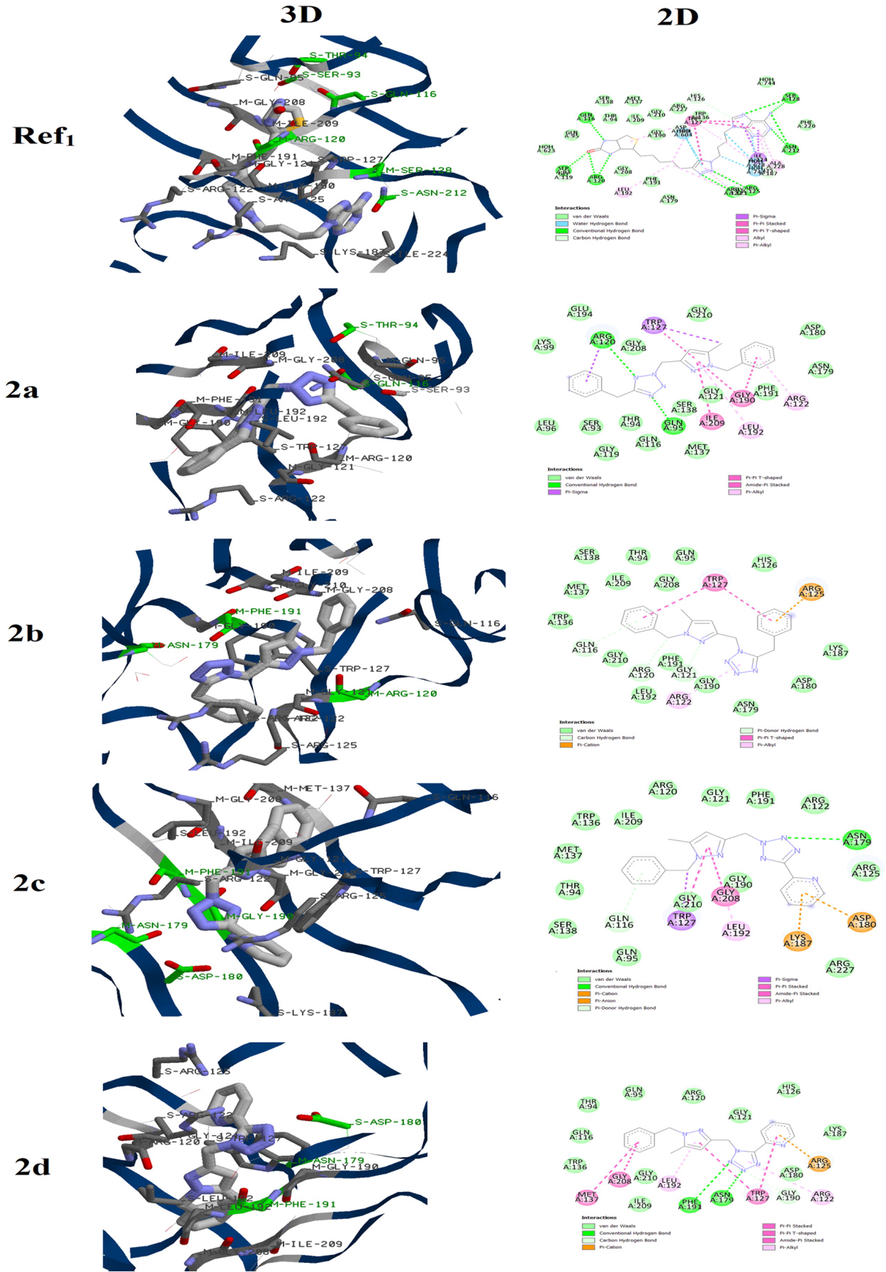
3D and 2D binding mode of the Biotin protein ligase in Staphylococcus aureus active site with Ref1 and compounds 2a-2d.
For compound 2a, we noted that this molecule was attached to the active site via two conventional hydrogen bounds between the tetrazolic ring and residues ARG120 and GLN95. The pyrazolic moiety also participate in the fixation of the molecules by forming π- π T-shaped and π -alkyl interactions with GLY190, ILE209 and LEU192 residues. Changing the position of the methylene chain that links the two azoles, results in obvious alteration of the interactions between 2b and the active site. In fact this compound was mainly attached to the pocket through Vander walls interactions with several residues such as GLN116, LYS187 and MET137 while no conventional hydrogen bound was observed. Furthermore, we observed π -cation interaction between the ARG125 and the benzyl attached to the tetrazolic ring which is absent in the case of compound 2a. Replacing the benzyl group tethered to the tetrazolic ring in compound 2a by the pyridine one (compound 2c) also influences the binding mode to the active site. In addition to the interactions between the pyrazole and residues TRP127 and GLY208, the complex enzyme-2c is mainly stabilized via conventional hydrogen bound between the tetrazole and residue ASN179 as well as several Vander walls interactions.
In the molecule 2d, the benzyl of compound 2b is also substituted by the pyridine group. This structural modification led to two conventional hydrogen bounds between the tetrazolic ring and residues ASN179 and PHE191. Nevertheless, the π -cation and π-π T-shaped interactions with the ARG125 and TRP127, respectively, were kept. Finally, the binding energies of these molecules with the active site of the Biotin protein ligase in Staphylococcus aureus are regrouped in Table 3. The comparison between results of these molecules showed that the binding energy was weakly affected. This suggests that the antibacterial activity against Staphylocoque aureus may be similar which is in good agreement with the obtained experimental data.
Compound
Binding energy
Kcal/mol
2a
−151.531
2b
−154.584
2c
−148.661
24
−151.734
For the Rhodotorula glutinis, the β–Fructofuranosidases from Rhodotorula glutinis var. dairenensis was examined as possible target. The first step was redoking raffinose at the active site. As displayed in Fig. 4, the docked raffinose is attached to the active via the same interactions reported for the complex β–Fructofuranosidases-raffinose.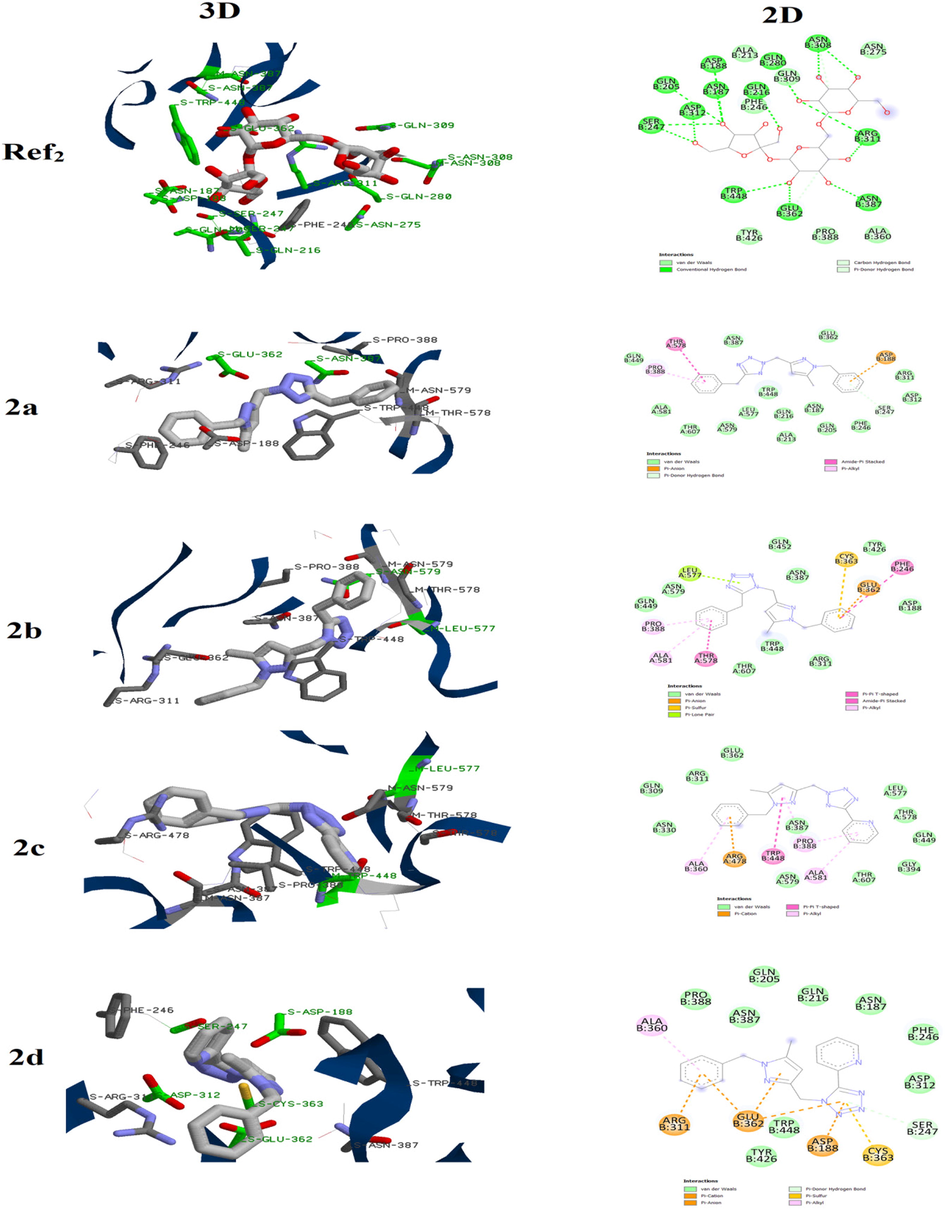
3D and 2D binding mode of the β–Fructofuranosidases active site with Ref2 and compounds 2a-2d.
Then, compounds 2a-2d were docked using the same parameters and results are showed in Fig. 2. It is clear that the nature of group as well as the junction position between the two azoles affect the binding of such family of molecules to the active site.
Compound 2a is mainly attached to the active site via Vander walls interactions in addition to π -anion interaction with ASP188. For its position isomer 2b, it showed additional types of interactions such π -lone pair and π -sulfur with LEU577 and CYS363, respectively. In the case of 2c, a π -anion interaction between the benzyl group and ARG478 was observed. Furthermore, several Vander walls interactions were established to give more stability to the enzyme-2c complex. They pyridine group also participates in the binding of 2c to the active site via π -alkyl interactions with PRO388 and ALA581. These two interactions were not observed in its isomer 2d. However we note the presence of π -anion, π -cation and π -sulfur interactions leading to more stable complex.
In the other hand the binding energies compounds 2a-2d with the active site of the the β–Fructofuranosidases from Rhodotorula glutinis var. dairenensis (Table 4) increases in this order 2a ≈ 2c > 2b ≈ 2d. This allows us to suggest that the antifugal activity against Rhodotorula glutinis augments in this order 2d ≈ 2b > 2c ≈ 2a which is also in good agreement with results experimentally obtained.
Compound
Binding energy
Kcal/mol
2a
−115.346
2b
−124.456
2c
−117.953
2d
−126.743
4 Conclusion
During this work, four pyrazole-tetrazole hybrid compounds were obtained via two different pathways and in different yields. The structure of these molecules was ensured by NMR, and elemental analysis as well mass spectrometry. All compounds possess both antifungal and antibacterial activities. The structure reactivity relationship (SAR) study evidenced the effect of the substituent nature on the tetrazolic ring and the junction position on the antimicrobial activity. A docking study on the Biotin protein ligase in Staphylococcus aureus and β–Fructofuranosidases from Rhodotorula glutinis var. dairenensis supports findings obtained experimentally. Further studies will be conducted to evaluate the antitumoral activity of this family of compounds.
CRediT authorship contribution statement
Ahlam Oulous: Investigation, Methodology. Tarik Harit: Investigation, Writing – original draft, Methodology, Writing – review & editing. Meryem Idrissi Yahyaoui: Investigation, Methodology. Abdeslam Asehraou: Formal analysis, Writing – review & editing. Fouad Malek: Writing – review & editing, Supervision, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Synthesis and docking studies of novel benzimidazole derivatives containing thiophene and triazole rings as potential urease inhibitors. Russ. J. Bioorg. Chem.. 2022;48(Suppl 1):S87-S95.
- [CrossRef] [Google Scholar]
- Synthesis, biological evaluation, and molecular docking of new benzimidazole-1, 2, 3-triazole hybrids as antibacterial and antitumor agents. Polycycl. Aromat. Compd.. 2023;43(4):3380-3391.
- [CrossRef] [Google Scholar]
- The inhibition behavior of two pyrimidine-pyrazole derivatives against corrosion in hydrochloric solution: experimental, surface analysis and in silico approach studies. Arab. J. Chem.. 2020;13(7):5949-5965.
- [CrossRef] [Google Scholar]
- Hartree-Fock and density functional theory studies on tautomerism of 5, 5′-diisopropyl-3, 3′-bipyrazole in gas phase and solution. Chem. Phys. Lett.. 2013;588:208-214.
- [CrossRef] [Google Scholar]
- Substituent effects in 3, 3’bipyrazole derivatives. x-ray crystal structures, molecular properties and DFT analysis. Acta Chim. Slov.. 2021;68(3):718-727.
- [Google Scholar]
- A new C, C-linked functionalized bipyrazole: synthesis, crystal structure, spectroscopies and DFT studies. Evaluation of the antibacterial activity and catalytic properties. Heterocycles. 2022;104(3):14592.
- [CrossRef] [Google Scholar]
- meta-Benzenotetratetrazolophanes. J. Heterocycl. Chem.. 1997;34(2):691-693.
- [CrossRef] [Google Scholar]
- Synthesis and characterization of new pyrazole–tetrazole derivatives as new vasorelaxant agents. Drug Dev. Res.. 2021;82(7):1055-1062.
- [CrossRef] [Google Scholar]
- Synthesis, antimicrobial activity and in-silico docking of two macrocycles based on pyrazole-tetrazole subunit. J. Mol. Struct.. 2022;1261:132947
- [CrossRef] [Google Scholar]
- New tetrapodal pyrazole-tetrazole ligands: synthesis, characterization, and evaluation of the antibacterial activity. Polycycl. Aromat. Compd.. 2023;43(6):5735-5746.
- [CrossRef] [Google Scholar]
- Discovery of novel triazole-containing pyrazole ester derivatives as potential antibacterial agents. Molecules. 2019;24(7):1311.
- [CrossRef] [Google Scholar]
- Synthesis and structural analysis of push-pull imidazole-triazole based fluorescent bifunctional chemosensor for Cu2+ and Fe2+ detection. Dyes Pigm.. 2022;205:110539
- [CrossRef] [Google Scholar]
- Synthesis, characterization, X-ray structure and in vitro antifungal activity of triphenyltin complexes based on pyrazole dicarboxylic acid derivatives. J. Mol. Struct.. 2021;1225:129137
- [CrossRef] [Google Scholar]
- Two novel macrocyclic organotin (IV) carboxylates based on bipyrazoledicarboxylic acid derivatives: syntheses, crystal structures and antifungal activities. J. Organomet. Chem.. 2021;948:121913
- [CrossRef] [Google Scholar]
- Bisubstrate adenylation inhibitors of biotin protein ligase from Mycobacterium tuberculosis. Chem. Biol.. 2011;18(11):1432-1441.
- [CrossRef] [Google Scholar]
- New synthesis of two tridentate bipyrazolic compounds and their cytotoxic activity tumor cell lines. Nat. Prod. Res.. 2007;21(11):947-952.
- [CrossRef] [Google Scholar]
- Synthesis and enzyme inhibitory activities of some new pyrazole-based heterocyclic compounds. Med. Chem. Res.. 2012;21(10):2772-2778.
- [CrossRef] [Google Scholar]
- Fluorinated polymers based on pyrazole groups for fuel cell membranes. Eur. Polym. J.. 2016;79:72-81.
- [CrossRef] [Google Scholar]
- New generation of tetrapyrazolic macrocycles: synthesis and examination of their complexation properties and antibacterial activity. Tetrahedron. 2017;73(34):5138-5143.
- [CrossRef] [Google Scholar]
- Synthesis, characterization, antimicrobial activity, and docking studies of new triazolic tripodal ligands. Chem. Biodivers.. 2017;14(12):e1700351.
- [Google Scholar]
- New copper complexes with bipyrazolic ligands: synthesis, characterization and evaluation of the antibacterial and catalytic properties. J. Mol. Struct.. 2018;1163:300-307.
- [CrossRef] [Google Scholar]
- Synthesis and characterization of new fluorinated copolymers based on azole groups for fuel cell membranes. Solid State Ion.. 2018;317:108-114.
- [CrossRef] [Google Scholar]
- Synthesis, characterization, antibacterial properties and DFT studies of two new polypyrazolic macrocycles. Polycycl. Aromat. Compd.. 2020;40(5):1459-1469.
- [CrossRef] [Google Scholar]
- Hybrid pyrazole-tetrazole derivatives with high α-amylase inhibition activity: synthesis, biological evaluation and docking study. ChemistrySelect. 2022;7(48):e202203757.
- [Google Scholar]
- Elaboration of new thin solid membrane bearing a tetrapyrazolic macrocycle for the selective transport of lithium cation. Sep. Purif. Technol.. 2017;188:394-398.
- [CrossRef] [Google Scholar]
- Discovery of (±)-3-(1 H-pyrazol-1-yl)-6, 7-dihydro-5 H-[1, 2, 4] triazolo [3, 4-b][1, 3, 4] thiadiazine derivatives with promising in vitro anticoronavirus and antitumoral activity. Mol. Divers.. 2022;26(3):1357-1371.
- [CrossRef] [Google Scholar]
- Structure, mechanism and regulation of pyruvate carboxylase. Biochem. J. 2008;413(3):369-387.
- [CrossRef] [Google Scholar]
- Tridentate bipyrazole compounds with a side-arm as a new class of antitumor agents. Res. Chem. Intermed.. 2014;40(2):681-687.
- [CrossRef] [Google Scholar]
- Synthesis, structure, and antiviral properties of novel 2-adamantyl-5-aryl-2H-tetrazoles. Chem. Heterocycl. Compd.. 2021;57(4):442-447.
- [CrossRef] [Google Scholar]
- New pyrazole-tetrazole hybrid compounds as potent α-amylase and non-enzymatic glycation inhibitors. Bioorganic Med. Chem. Lett.. 2022;69:128785
- [CrossRef] [Google Scholar]
- Synthesis and evaluation of antidepressant activities of 5-Aryl-4, 5-dihydrotetrazolo [1, 5-a] thieno [2, 3-e] pyridine derivatives. Molecules. 2019;24(10):1857.
- [CrossRef] [Google Scholar]
- Energetic furazan and triazole moieties: a promising heterocyclic cation. ChemistrySelect. 2019;4(30):8876-8881.
- [CrossRef] [Google Scholar]
- GEMDOCK: a generic evolutionary method for molecular docking. Proteins: Struct. Funct. Bioinf.. 2004;55(2):288-304.
- [CrossRef] [Google Scholar]
- Zinc (II) and cobalt (II) complexes with unusual coordination of mixed imidazole-1, 2, 4-triazole ligand in a protonated cationic form. Polyhedron. 2022;217:115741
- [CrossRef] [Google Scholar]
- Synthesis of new derivatized pyrazole based ligands and their catecholase activity studies. Arab. J. Chem.. 2011;4(4):459-464.
- [CrossRef] [Google Scholar]
- Bioisosteres in drug discovery: focus on tetrazole. Future Med. Chem.. 2020;12(2):91-93.
- [CrossRef] [Google Scholar]







