Translate this page into:
Prenylated sulfonyl amides from the leaves of Glycosmis pentaphylla and their potential anti-proliferative and anti-inflammatory activities
⁎Corresponding authors. meizhinan@mail.hzau.edu.cn (Zhinan Mei), yangguangzhong@mail.scuec.edu.cn (Guangzhong Yang)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Abstract
Six rare sulfur-containing amides were isolated from the leaves of Glycosmis pentaphylla. Compound 4 displayed remarkablely anti-inflammatory and anti-proliferative activities. These findings suggested that 4 could be promising candidate for the treatment of inflammation and cancer diseases.
Abstract
Six previously undescribed sulfur-containing amides were isolated from the leaves of Glycosmis pentaphylla. Their structures were delineated by HRESIMS, 1D and 2D NMR, electronic circular dichroism (ECD) calculations, and DP4+ analyses based on gauge-independent atomic orbital (GIAO) NMR calculations. Compounds 1–4 belong to the type of methylsulfonylpropenoic acid amides. Through different cyclization pathways of geranyloxy, compounds 1 and 2 carry uncommon cyclohexane-1, 3-diol and cyclohex-3-en-1-ol moiety, respectively. Compound 3 is the oxidation product of the double bond Δ6′′(7′′) of geranyloxy. Compound 5 is elucidated as the type of methylsulfonylpropanoic acid amide. Compound 6 represents a rare sulfur-containing amide possessing a morpholin-3-one moiety. All isolated compounds were evaluated for their anti-inflammatory and anti-proliferative activities. Compound 4 significantly inhibited lipopolysaccharide-induced nitric oxide (NO) production in mouse macrophage RAW 264.7 cells with the IC50 value of 0.55 µM. Moreover, compounds 3 and 4 exhibited different anti-proliferative activities against HepG-2 with IC50 values of 11.52 and 9.41 µM, respectively.
Keywords
Glycosmis pentaphylla
Rutaceae
Sulphur-containing amides
Anti-inflammatory activity
Anti-proliferative activity
1 Introduction
The genus Glycosmis belongs to the Rutaceae family, and many secondary metabolites with pharmacological and biological activities, such as alkaloids, flavonoids, and phenolic glycosides, are isolated from various parts of the Glycosmis plant (Yasir et al., 2019). Sulfur-containing amides are a special type of alkaloids, mainly derived from plants such as Glycosmis, and fungi, which are classified into four types (Hofer and Greger, 2000), including methylthiopropionic acid amides, methylsulfinylpropenoic acid amides, methylsulfonylpropenoic acid amides (MSPAAs), and methylthiocarbonic acid amides (Sutton et al., 1994). The structural characteristics of MSPAAs have a prenyloxy or geranyloxy in the para position of the benzene ring of the phenethylamide moiety. The structural diversity mainly comes from the oxidation of the geranyloxy side chain to form oxidation products such as hydroxyl, aldehyde, and ketone derivatives, or intramolecular cyclization to form dihydrofuranone rings containing ether and ketone functional groups (Yang et al., 2015; Greger et al., 1994; Hofer et al., 1995; Hofer et al., 2000; Hofer et al., 1998; Vajrodaya et al., 1998). Owing to their unique structure and biological activity, MSPAAs have attracted the attention of synthetic organic chemists, and the total synthesis of methylgerambullone has been completed (Moon et al., 2010).
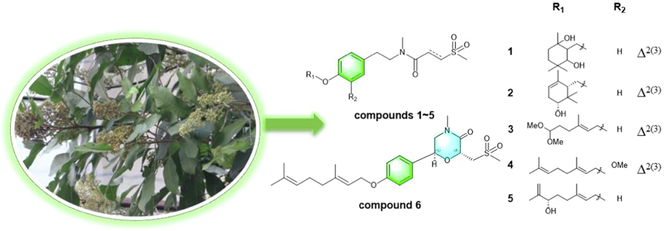
Glycosmis pentaphylla (Retz.) DC. is an evergreen shrub native to tropical and subtropical regions of China, India, and other countries, and is used as traditional medicine to treat a variety of chronic diseases including cough, fever, inflammation, bronchitis, rheumatism, diabetes, cancer, and more (Khandokar et al., 2021). Previously, our research group performed phytochemical studies of G. pentaphylla leaves and isolated 18 previously undescribed sulphur-containing amides. Methylgerambullin has the strongest inhibitory effect on the proliferation of HepG2 cells, and its inhibitory effect on NO production is about 37 times that of the positive drug dexamethasone. Moreover, the antitumor mechanism of methylgerambullin is based on the activation of mitochondrial and endoplasmic reticulum stress signaling pathways and inhibition of AKT and STAT3 pathways (Nian et al., 2020; Wu et al., 2021). Taken together, these findings prompted us to isolate more sulphur-containing amides from G. pentaphylla leaves and evaluate their biological activities. Thus, six previously undescribed sulphur-containing amides were separated from the leaves of G. pentaphylla (Fig. 1). In addition, all isolated compounds were evaluated for their anti-inflammatory and anti-proliferative properties.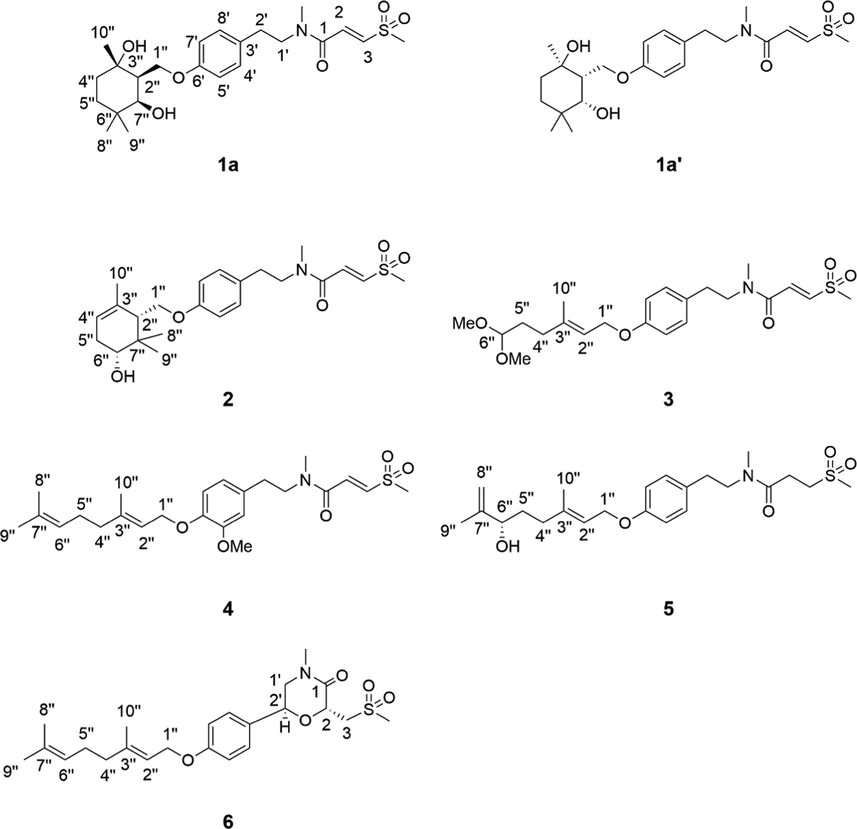
Structures of compounds 1–6.
2 Materials and methods
2.1 General experimental procedures
Optical rotation in MeOH was determined using an Autopol IV polarimeter (Rudolph Research Analytical, Hackettstown, NJ, USA). UV spectra were obtained using a UH5300 UV–vis Double Beam spectrophotometer (Hitachi Co., Tokyo, Japan). 1D and 2D NMR spectra were recorded with CD3OD on a Bruker AVANCE IIITM 600 M Hz spectrometer (Bruker, Ettlingen, Germany) using tetramethylsilane (TMS) as the internal standard. Chemical shifts (δ) are expressed in ppm and coupling constants (J) are expressed in Hz. HR-ESI-MS data were acquired using a Thermo Scientific Q Exactive Orbitrap LC-MS/MS System (Thermo Scientific, Waltham, MA, USA). The preparation of compounds were performed on an Ultimate 3000 HPLC system (Thermo Fisher Scientific Dionex., Sunnyvale, CA, USA) with an Ultimate 3000 pump and an Ultimate 3000 Variable Wavelength detector. The HPLC conditions were described as follows: column: Nacalai Tesque 5C18-MS-II (250 × 10 mm, 5 μm), flow rate: 3 ml/min, detection wavelengths: UV, 220 and 254 nm. Silica gel for CC (200–300 mesh and 300–400 mesh) was obtained from the Qingdao HaiYang Chemical Group Co. ltd. (Qingdao, China). Chromatographic grade acetonitrile and methanol were purchased from Chang Tech Enterprise Co. ltd. (Taiwan, China). The human tumor cell line HepG2 and RAW264.7 murine macrophages were purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China). Cisplatin was purchased from Sigma-Aldrich (Saint Louis, MO, USA). Dexamethasone and lipopolysaccharide (LPS) were purchased from Sigma Chemical Co. ltd. (St. Louis, MO, USA). Cell Counting Kit (CCK-8) was purchased from Beyotime Biotechnology (Shanghai, China). The NO kit was purchased from Shanghai Biyuntian Biotechnology Co. ltd. (Shanghai, China). Dulbecco’s modified Eagle’s medium (DMEM) and penicillin–streptomycin solution were purchased from GE Healthcare Life Sciences (Logan, UT, USA). Fetal bovine serum (FBS) was purchased from Gibco, Life Technologies (Grand Island, NY, USA). Reagent grade DMSO was purchased from Vetec, Sigma Chemical Co. ltd. (St. Louis, MO, USA). Absorbance was recorded on a Multiskan GO microplate reader (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The organic solvent was acquired from Sinopharm Chemical Reagent Co., ltd. (Shanghai, China).
2.2 Plant material
The leaves of G. pentaphylla were collected from Mengyang Town, Jinghong City, Xishuangbanna Dai Autonomous Prefecture, Yunnan Province (north latitude 22°8′48″; east longitude 100°56′29″), and identified by Prof. Zhao Yinghong from Institute of Ethnic Medicine, Xishuangbanna Dai Autonomous Prefecture. The voucher specimen was deposited in the herbarium of the School of Pharmaceutical Sciences, South-Central Minzu University.
2.3 Extraction and isolation
12 kg of dried leaves of G. pentaphylla were powdered and extracted three times with 90 % EtOH at room temperature to obtain EtOH extract (860 g), which was then successively partitioned with petroleum ether (P.E.) and MeOH to acquire P.E. extract 100 g and MeOH extract 740 g. The MeOH extract (740 g) was subjected to polyamide column (80–100 mesh), and eluted with a gradient of EtOH-H2O (0:1, 3: 7, 1: 1, 7: 3, 95: 5) to give 5 subfractions (Fr.1-Fr.5). Fr.1 (100.9 g) was purified by a silica gel column chromatography (CC) eluted with CH2Cl2-MeOH (100:1, 50:1, 20:1, 9:1, 5:1) to provide 12 subfractions (Fr.1.1-Fr.1.12). Fr.1.4 (4.9 g) was subjected to a silica gel CC eluted with P.E.-EtOAc (8:2, 6:4, 4:6, 0:1), and then separated by semi-preparative HPLC (MeOH-H2O, 82:18, v/v) to yield compound 2 (1.1 mg, tR = 6.4 min). Fr. 1.6 (8.0 g) was chromatographed on silica gel column by eluting with P.E.-EtOAc (8:2, 6:4, 4:6, 2:8), and then further purified by semi-preparative HPLC (MeOH-H2O, 60:40, v/v) to yield compounds 1 (8.1 mg, tR = 12.2 min) which was further resolved by chiral column [column: CHIRALPAK® IC 4.6 × 250 mm 5 µm; mobile phase: MeCN: H2O (30:70); flow rate: 0.5 ml/min; detection: UV, 225 nm] to give a pair of enantiomer 1a and 1a′ at tR 54.3 and 46.9 min respectively, and 5 (8.2 mg, tR = 32.3 min). Fr. 2 (89.7 g) was subjected to a silica gel CC eluted with CH2Cl2-MeOH (100:1, 20:1, 9:1, 8:2, 0:1) to obtain 13 fractions (Fr. 2.1-Fr.2.13). Fr. 2.5 (9.3 g) was subjected to ODS CC eluted with MeOH-H2O (1:0, 7:3, 1:1, 3:7, 0:1) to obtain 7 fractions (Fr.2.5.1-Fr.2.5.7). Fr.2.5.4 (0.5 g) was further separated by a silica gel CC with a gradient of P.E.-EtOAc (9:1, 7:3, 4:6, 0:1), and then purified by semi-preparative HPLC (MeCN-H2O, 70:30, v/v) to give compound 4 (2.0 mg, tR = 14.5 min). Fr.2.6 (1.8 g) was subjected to silica gel CC by eluting with P.E.-EtOAc (8:2, 6:4, 2:8, 0:1), and then purified by semi-preparative HPLC (MeCN-H2O, 60:40, v/v) to afford compound 6 (3.8 mg, tR = 31.0 min). Fr.2.8 (31.6 g) was separated by a silica gel CC with a gradient of CH2Cl2-EtOAc (9:1, 7:3, 4:6, 0:1) to yield subfraction Fr.2.8.4 (6.9 g), which was further purified by a silica gel CC eluted with P.E.-EtOAc (8:2, 6:4, 4:6, 0:1) to obtain 10 subfractions (Fr.2.8.4.1-Fr.2.8.4.10). Fr.2.8.4.10 (2.95 g) was further purified by a silica gel CC eluted with P.E.-CH2Cl2 (1:0, 7:3, 3:7, 0:1) and CH2Cl2-MeOH (50:1, 9:1, 0:1), and then purified by semi-preparative HPLC (MeCN-H2O, 51:49, v/v) to give compound 3 (4.0 mg, tR = 15.1 min).
Spectroscopic Data
Glycopentamide S (1): colorless oil; UV (MeOH) λmax (log ε) 225 (4.05), 270 (3.60); 1H NMR data see Table 1 and 13C NMR data see Table 3; HRESIMS m/z 476.20764 [M + Na]+ (calcd for C23H35NO6SNa, 476.20773).
Glycopentamide S (1a): +0.049 (MeOH, c = 0.05); ECD (1.05 × 10-4 M, MeOH) λ(θ) 206 (-0.25), 229 (+2.29), 242 (+0.09), 275 (+1.13) nm;
Glycopentamide S (1a′): −0.051 (MeOH, c = 0.05); ECD (1.05 × 10-4 M, MeOH) λ(θ) 212 (+0.06), 228 (-2.09), 245 (-0.11), 277 (-1.01) nm;
Glycopentamide T (2): colorless oil; +49.26 (MeOH, c = 0.03); UV (MeOH) λmax (log ε) 225(3.97), 270 (3.44); ECD (6.55 × 10-5 M, MeOH) λ(θ) 200 (+16.20), 209 (-0.96), 223 (+3.79) nm; 1H NMR data see Table 1 and 13C NMR data see Table 3; HRESIMS m/z 458.19687 [M + Na]+ (calcd for C23H33NO5SNa, 458.19716).
Glycopentamide U (3): colorless oil; UV (MeOH) λmax (log ε) 230 (4.21), 270 (4.02); 1H NMR data see Table 1 and 13C NMR data see Table 3; HRESIMS m/z 462.19199 [M + Na]+ (calcd for C22H33NO6SNa, 462.19208).
Glycopentamide V (4): colorless oil; UV (MeOH) λmax (log ε) 235 (3.89), 275 (3.83); 1H NMR data see Table 2 and 13C NMR data see Table 3; HRESIMS m/z 450.23120 [M + H]+ (calcd for C24H36NO5S, 450.23087).
Glycopentamide W (5): colorless oil; +7.78 (MeOH, c = 0.05); UV (MeOH) λmax (log ε) 225(4.06), 275 (3.24); ECD (1.09 × 10-4 M, MeOH) λ(θ) 231 (+0.71), 260 (-0.07), 275 (+0.18) nm;1H NMR data see Table 2 and 13C NMR data see Table 3; HRESIMS m/z 460.21243 [M + Na]+ (calcd for C23H35NO5SNa, 460.21282).
Glycopentamide X (6): colorless oil; +566.7 (MeOH, c = 0.01); UV (MeOH) λmax (log ε) 205 (4.35), 225 (4.19), 275 (3.30); ECD (2.18 × 10-5 M, MeOH) λ(θ) 200 (+2.43), 208 (-0.76), 217 (+0.90), 231 (-2.11) nm; 1H NMR data see Table 2 and 13C NMR data see Table 3; HRESIMS m/z 458.19693 [M + Na]+ (calcd for C23H33NO5SNa, 458.19716).
2.4 NMR calculations
Computational NMR data were obtained from the IEFPCM model at the mPW1PW91/6–311 + G(2d,p) level in methanol using the GIAO (gauge-independent atomic orbital) method. (Detailed NMR calculations are provided in Supplementary Information).
2.5 ECD calculations
The ECD in methanol was calculated using the IEFPCM model with time-dependent density functional theory (TD-DFT). (See Supplementary Information for detailed ECD calculations).
2.6 Anti-proliferative activity bioassay
The anti-proliferative activity of the isolated compounds on HepG-2 cell line was measured by the CCK-8 method according to the previously described protocol (Chen et al., 2019). In short, 5 × 103 HepG2 cell lines per well (in 100 μL of culture medium) were seeded in 96-well plates. After being treated with gradient concentrations (20 μM, 10 μM, 5 μM, 2.5 μM and 1.25 μM) of each compound at 37 °C for 24 h, the supernatant was discarded. Then, 100 μL cell culture medium containing 10 % CCK-8 solution was added to each well at 37 °C for 1 h. The absorbance values of each well at 450 nm were measured on a microplate spectrophotometer. Cisplatin was used as the positive control.
2.7 Anti-inflammatory activity bioassay
The cell viability was firstly determined by the CCK-8 method. RAW264.7 cells were seeded into 96-well plates (2 × 105 cells per well) and cultured overnight. After treated with LPS (1.0 μg/mL) for 24 h, the culture medium was replaced by new medium containing gradient concentrations of each compound (20 μM, 10 μM, 5 μM, 2.5 μM and 1.25 μM). Dexamethasone was used as the positive control. After 24 h incubation, the supernatant was collected to determine NO content using Griess reagent. (Teng et al., 2019).
3 Results and discussion
All isolated compounds are obtained as colorless oils and belong to the sulphur-containing amides through HR-ESI-MS data, which are the characteristic components of this plant. Moreover, two sets of resonance signals are observed in the 1H and 13C spectrum of compounds 1–5 (Tables 1-3), indicating that these compounds are composed of two inseparable conformers. A comparison of NMR data of 1–4 with those of methyldambullin indicates that compounds 1–4 contain (E)-3-(methylsulfonyl)propenoic acid moiety and N-methyltyramine part. The rotation of the C—N amide bond is restricted due to the presence of the N-methyl group, resulting in two conformers, (a) s-cis and (b) s-trans, in which the ratio of the two conformers a/b ranges from 50 % to 75 % by 1H NMR spectrum analysis. To facilitate structural analysis, we describe the structural identification of compounds 1–5 based on NMR data of conformer (a) (Greger et al., 1994).
No.
1 a
1 a
2 a
2 a
3 a
3 a
2
6.83 (d, 15.0)
7.34 (d, 15.0)
6.85 (d, 15.0)
7.34 (d, 15.0)
6.82 (d, 15.0)
7.34 (d, 15.0)
3
6.96 (d, 15.0)
7.38 (d, 15.0)
6.95 (d, 15.0)
7.38 (d, 15.0)
6.96 (d, 15.0)
7.39 (d, 15.0)
-SCH3
2.94 (s)
3.09 (s)
2.94 (s)
3.09 (s)
2.94 (s)
3.09 (s)
–NCH3
3.06 (s)
3.05 (s)
3.06 (s)
3.07 (s)
3.06 (s)
3.06 (s)
1′
3.68 (t, 6.3)
3.63 (t, 7.5)
3.69 (t, 6.3)
3.64 (t, 7.5)
3.68 (t, 6.3)
3.63 (t, 7.2)
2′
2.84 (t, 6.3)
2.82 (t, 7.5)
2.85 (t, 6.3)
2.83 (t, 7.5)
2.84 (t, 6.3)
2.82 (t, 7.2)
4′, 8′
7.04 (d, 8.4)
7.15 (d, 8.4)
7.06 (d, 8.4)
7.17 (d, 8.4)
7.04 (d, 8.4)
7.15 (d, 8.4)
5′, 7′
6.87 (d, 8.4)
6.90 (d, 8.4)
6.84 (d, 8.4)
6.86 (d, 8.4)
6.83 (d, 8.4)
6.85 (d, 8.4)
1′′
4.21 (m)
4.21 (m)
4.26 (dd, 3.6, 9.6); 4.02 (dd, 4.8, 9.6)
4.26 (dd, 3.6, 9.6);4.02 (dd, 4.8, 9.6)
4.53 (d, 6.6)
4.53 (d, 6.6)
2′′
2.20 (m)
2.20 (m)
2.15 (br s)
2.15 (br s)
5.46 (t, 6.6)
5.46 (t, 6.6)
4′′
1.78 (m); 1.51 (m)
1.78 (m); 1.51 (m)
5.39 (br s)
5.39 (br s)
2.10 (t, 7.8)
2.10 (t, 7.8)
5′′
1.69 (m); 1.16 (m)
1.69 (m); 1.16 (m)
2.26 (m) /2.05 (m)
2.26 (m) /2.05 (m)
1.72 (m)
1.72 (m)
6′′
3.43 (dd, 5.4, 7.2)
3.43 (dd, 5.4, 7.2)
4.34 (m)
4.34 (m)
7′′
3.60 (m)
3.60 (m)
8′′
0.97 (s)
0.97 (s)
1.06 (s)
1.07(s)
9′′
1.00 (s)
1.00 (s)
0.94 (s)
0.95(s)
10′′
1.24 (s)
1.24 (s)
1.73 (s)
1.73 (s)
1.75 (s)
1.75 (s)
6′′-OMe
3.30 (s)
3.30 (s)
6′′-OMe
3.30 (s)
3.30 (s)
No.
4 a
4 a
5 a
5 a
6
2
6.78 (d, 15.0)
7.34 (d, 15.0)
2.47 (t, 7.2)
2.86 (t, 7.2)
4.83 (dd, 2.7, 10.5)
3
6.96 (d, 15.0)
7.39 (d, 15.0)
3.14 (t, 7.2)
3.39 (t, 7.2)
4.04 (dd, 10.2, 15.0) / 3.41 (d, 15.6)
-SCH3
2.92 (s)
3.09 (s)
2.95 (s)
2.96 (s)
2.92 (s)
–NCH3
3.08 (s)
3.07 (s)
2.86 (s)
2.98 (s)
3.03 (s)
1′
3.71(t, 6.0)
3.66 (t, 7.5)
3.59 (t, 6.6)
3.56 (t, 7.5)
3.70 (dd, 9.0, 12.6); 3.51 (dd, 3.0, 12.6)
2′
2.84 (t, 6.0)
2.82 (t, 7.5)
2.84 (t, 6.6)
2.76 (t, 7.5)
5.12 (dd, 3.3, 9.3)
4′
6.76(d, 1.8)
6.88(d,1.8)
7.12 (d, 8.4)
7.13 (d, 7.8)
7.37 (d, 8.4)
5′
6.84 (d, 8.4)
6.86 (d, 8.4)
6.93 (d, 8.4)
7′
6.83(d, 7.8)
6.86 (d, 7.8)
6.84 (d, 8.4)
6.86 (d, 8.4)
6.93 (d, 8.4)
8′
6.59 (dd, 8.4,1.8)
6.76 (dd, 8.4,1.8)
7.12 (d, 8.4)
7.17 (d, 7.8)
7.37 (d, 8.4)
1′′
4.55 (d, 6.0)
4.55 (d, 6.0)
4.54 (d, 6.0)
4.52 (d, 6.0)
4.56 (d, 6.6)
2′′
5.45(t, 6.6)
5.45(t, 6.6)
5.47 (t, 6.3)
5.47 (t, 6.3)
5.44 (t, 6.0)
4′′
2.06 (t, 7.5)
2.06 (t, 7.5)
2.12 (m); 2.05 (m)
2.12 (m); 2.05 (m)
2.07 (t, 7.2)
5′′
2.13(q, 6.6)
2.13(q, 6.6)
1.65 (m)
1.65 (m)
2.13 (q, 7.2)
6′′
5.10(t, 6.6)
5.10(t, 6.6)
3.98 (t, 6.6)
3.98 (t, 6.6)
5.09 (m)
8′′
1.61(s)
1.61(s)
4.81 (br s); 4.91 (br s)
4.81 (br s); 4.91 (br s)
1.66 (s)
9′′
1.66(s)
1.66(s)
1.71 (s)
1.71 (s)
1.60 (s)
10′′
1.72 (s)
1.72 (s)
1.75 (s)
1.75 (s)
1.74 (s)
5′-OMe
3.83(s)
3.83(s)
No.
1 a
1 a
2 a
2 a
3 a
3 a
4 a
4 a
5 a
5 a
6
1
165.6
165.1
165.6
165.2
165.6
165.1
165.6
165.1
171.7
171.5
168.2
2
133.3
134.4
133.7
134.5
133.3
134.4
133.0
134.4
27.0
27.8
73.3
3
139.8
140.9
139.8
140.9
139.9
140.9
139.9
140.9
51.4
51.3
56.3
-SCH3
42.6
42.4
42.6
42.4
42.5
42.4
42.5
42.4
41.0
41.3
43.1
–NCH3
34.6
36.9
34.5
36.9
34.6
36.9
34.6
36.7
34.3
36.5
35.1
1′
53.4
51.7
53.4
51.7
53.4
51.7
53.3
51.6
53.0
51.6
55.0
2′
34.5
33.4
34.5
33.4
34.5
33.4
34.8
33.9
34.4
33.7
71.8
3′
131.1
131.9
131.5
132.3
131.3
132.1
132.3
132.3
131.2
132.4
130.4
4′
131.6
131.0
131.6
131.1
131.5
131.0
114.2
114.1
131.3
131.0
129.2
5′
116.2
115.9
116.2
115.9
116.3
116.1
151.3
151.3
116.3
116.0
116.1
6′
160.0
159.7
159.3
159.0
159.5
159.1
148.3
148.3
159.4
159.0
160.7
7′
116.2
115.9
116.2
115.9
116.3
116.1
115.5
115.8
116.3
116.0
116.1
8′
131.6
131.0
131.6
131.1
131.5
131.0
122.9
122.3
131.3
131.0
129.2
1′′
66.2
66.2
68.4
68.3
65.9
65.9
67.1
67.2
65.9
66.0
66.0
2′′
47.9
47.8
50.8
50.8
121.6
121.7
121.3
121.4
121.5
121.6
121.3
3′′
72.9
72.9
135.2
135.2
141.4
141.4
142.1
142.1
141.9
141.7
142.1
4′′
39.4
39.4
121.4
121.4
35.5
35.5
40.8
40.8
36.7
36.7
40.8
5′′
32.7
32.7
32.7
32.7
32.0
32.0
27.5
27.5
34.2
34.2
27.5
6′′
35.6
35.6
74.9
74.9
105.9
105.9
125.2
125.2
76.2
76.2
125.1
7′′
75.2
75.2
38.4
38.4
132.7
132.7
148.9
148.9
132.7
8′′
28.6
28.6
26.9
26.9
17.9
17.9
111.7
111.7
26.1
9′′
25.2
25.2
18.6
18.7
26.1
26.1
17.8
17.8
17.9
10′′
24.4
24.4
22.8
22.7
16.8
16.8
16.8
16.8
16.8
16.8
16.8
5′-OMe
56.4
56.6
6′′-OMe
53.6
53.6
6′′-OMe
53.6
53.6
The molecular formula of 1 was deduced to be C23H35NO6S through HR-ESI-MS (Fig. S9, Supplementary Information abbreviated SI) at m/z 476.20764 [M + Na]+ (calcd for C23H35NO6SNa, 476.20773), which was composed of two conformers a and b in 2.5:1. In addition to the characteristic signals of (E)-3-(methylsulfonyl)propenoic acid moiety and N-methyltyramine part, compound 1 also contained 10 carbon signals, including three methyls [δC 28.6 (q), 25.2 (q), 24.4 (q)], two methylenes [δC 39.4 (t), 32.7 (t)], one oxygenated methylene [δC 66.2 (t)], one methine [δC 47.9 (d)], one oxygenated methine [δC 75.2 (d)], one sp3 quaternary carbon [δC 35.6 (s)], and one oxygenated sp3 tertiary carbon [δC 72.9 (s)]. Careful analysis of the 2D NMR (Fig. S4-S6, SI) yielded two structural fragments (A) –CH2(4′′)CH2(5′′)- and (B) –CH2(1′′)(O)CH(2′′)CH(7′′)(O)-. HMBC correlations (Fig. 2) from CH3-10′′ [δH 1.24 (s)] to δC 72.9 (s, C-3′′), 39.4 (t, C-4′′), and 47.9 (d, C-2′′), and from CH3-8′′ [δH 0.97 (s)] and CH3-9′′ [δH 1.00 (s)] to δC 75.2 (d, C-7′′), 35.6 (s, C-6′′), and 32.7 (t, C-5′′) suggested that two structural fragments A and B were connected to C-3′′ through C-4′′ and C-2′′, and C-6′′ through C-5′′ and C-7′′ respectively. Therefore, the remaining carbon signals could establish 2-oxymethylene-1, 4, 4-trimethylcyclohexane-1, 3-diol moiety. Moreover, HMBC correlations from CH2-1′′ [δH 4.21] to δC 160.0 (s, C-6′) implied that this moiety was connected to C-6′. In ROESY spectrum (Fig. S7, SI), the correlation of H3-10′′ with H2-1′′ implied that 3′′–OH and H-2′′ were co-facial. The relative configuration of C-7′′ could not be determined due to the lack of useful ROESY correlations. Therefore, to further verify the relative configuration of C-7′′, two isomers (2′′S*, 3′′S*, 7′′R*)-1a and (2′′S*, 3′′S*, 7′′S*)-1b were subjected to NMR calculations with the DP4 + analysis. These results showed that (2′′S*, 3′′S*, 7′′R*)-1a (Figs. 4-5, Tables S3, S5, and S7-S8, Fig. 65, SI) was the most likely structure for 1 with a 100.00 % DP4 + probability based on all NMR data (Marcarino et al., 2020). This determined the relative configuration of 1. Compound 1 has a small optical rotation value and no cotton effect in the CD spectrum, indicating that they may be a racemic mixture, which was further resolved by chiral column [column: CHIRALPAK® IC 4.6 × 250 mm 5 µm; mobile phase: MeCN: H2O (30:70); flow rate: 0.5 ml/min; detection: UV, 225 nm] to give a pair of enantiomer 1a and 1a′ at tR 54.3 and 46.9 min respectively. By comparing the experimental ECD data with the calculated results, the absolute configurations of 1a/1a′ were assigned as (2′′S, 3′′S, 7′′R)-1a and (2′′R, 3′′R, 7′′S)- 1a′ (Fig. 6). Therefore, the structure of 1 was determined as shown in Fig. 1, and denoted as glycopentamide S.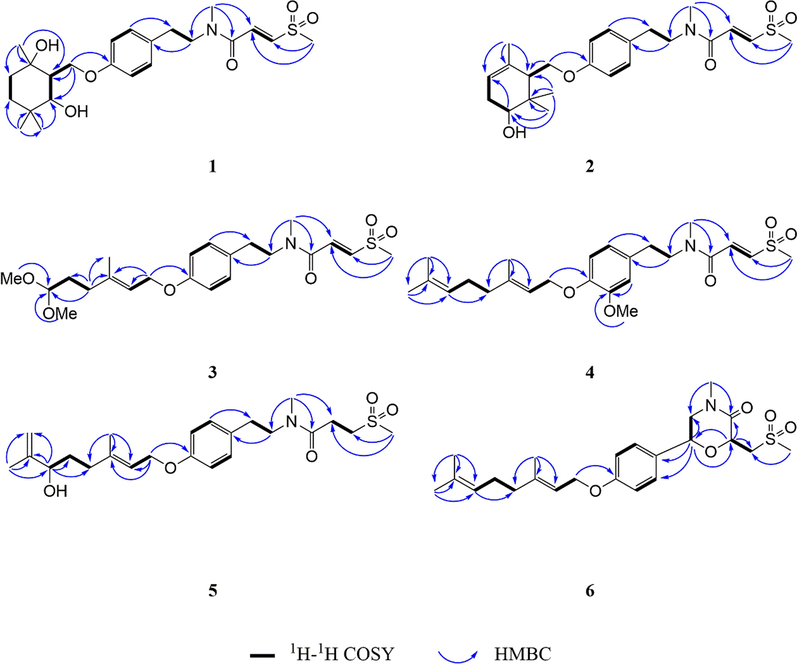
Key HMBC and 1H–1H COSY correlations of compounds 1–6.
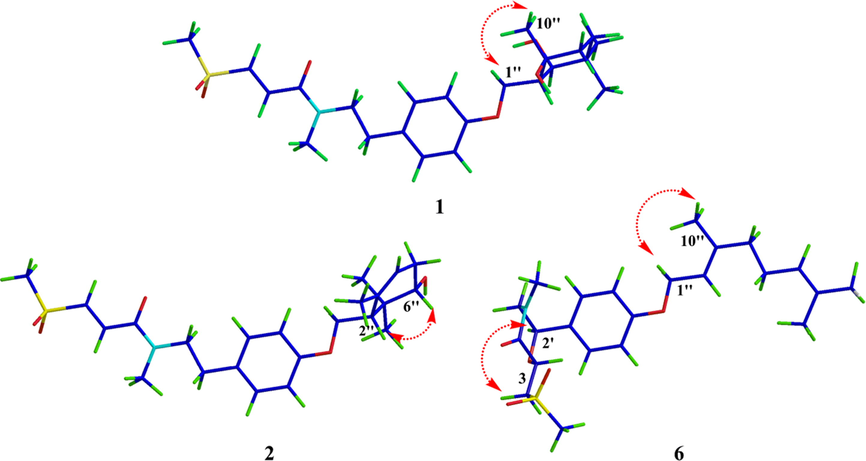
ROESY correlations for compounds 1, 2 and 6.
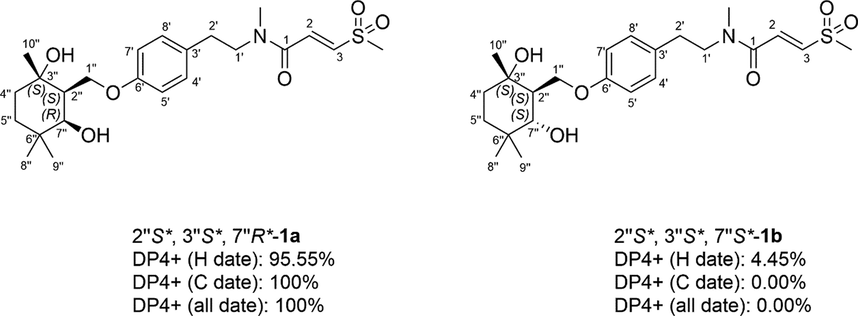
DP4 + analysis of two possible diastereomers (1a and 1b) for 1.
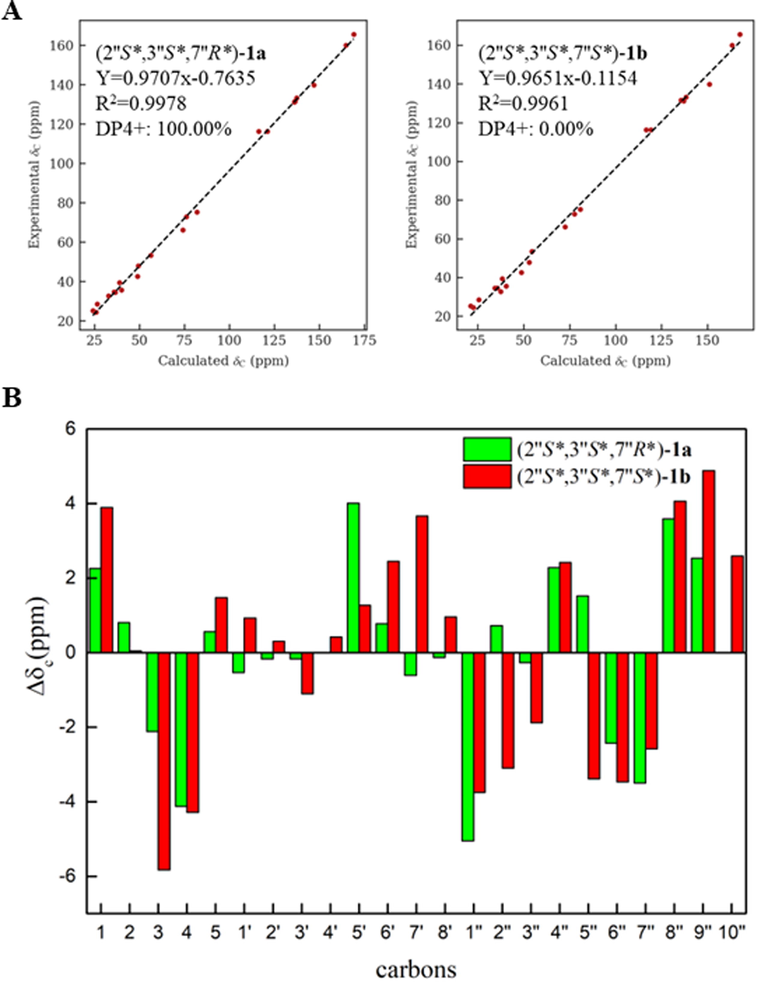
(A) Linear regression fitting of calculated 13C chemical shifts of two possible isomers of 1a and 1b with the experimental values (B) Deviation between calculated 13C chemical shifts of two possible isomers of 1a and 1b with the experimental values.
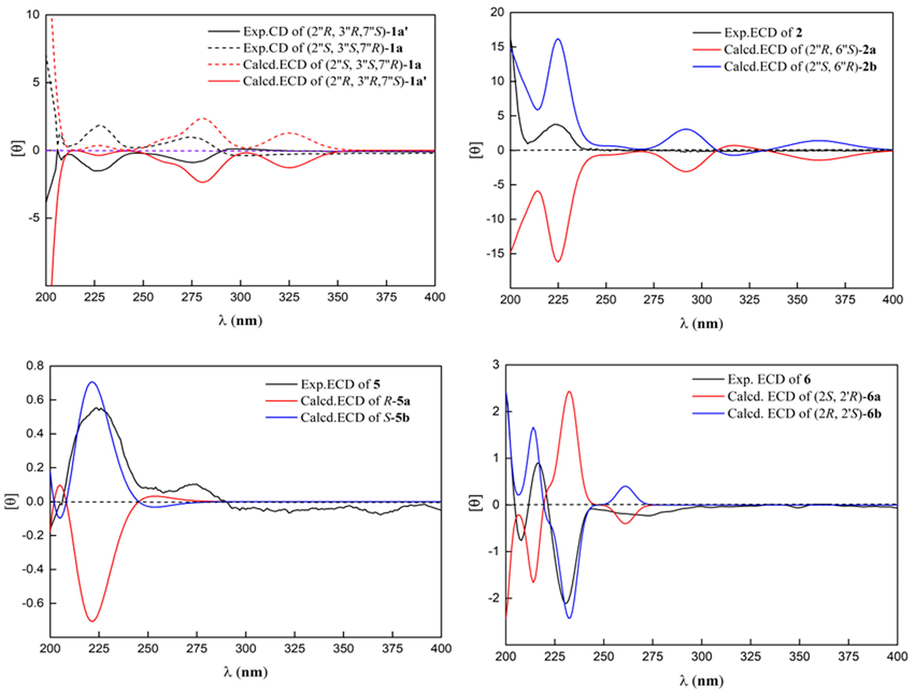
Calculated and experimental ECD spectra of 1, 2, 5 and 6.
The molecular formula of 2 was deduced to be C23H33NO5S through HR-ESI-MS (Fig.S21, SI) at m/z 458.19687 [M + Na]+ (calcd for C23H33NO5SNa, 458.19716), which was composed of two conformers a and b in 2.3:1. Careful comparison of the NMR data of compounds 1 and 2 revealed that compound 2 was the absence of one oxygenated tertiary carbon and methylene, but had one more tri-substituted double bond. It was speculated that compound 2 might be the dehydration product of 1. However, through careful analysis of the 2D NMR (Fig. S17-S18, SI), two structural fragments (A) –CH(4′′)CH2(5′′)CH(6′′)(O)- and (B) –CH2(1′′)(O)CH(2′′)- were obtained which were completely different from the dehydration product of 1. In the same way, HMBC correlations from CH3-10′′ [δH 1.73 (s)] to δC 135.2 (s, C-3′′), 121.4 (d, C-4′′), and 50.8 (d, C-2′′), and from CH3-8′′ [δH 1.06 (s)] and CH3-9′′ [δH 0.94 (s)] to δC 74.9 (d, C-6′′), 38.4 (s, C-7′′), and 50.8 (d, C-2′′) constructed 5- oxymethylene-4,6,6-trimethylcyclohex-3-en-1-ol moiety, which was connected to C-6′ by HMBC correlation from CH2-1′′ [δH 4.26, 4.02] to δC 159.3 (s, C-6′). The relative configuration of 2 was determined by interpreting the ROESY spectrum (Fig. 3, Fig. S19, SI), where the correlation of H-2′′ with H-6′′ indicated that these protons were co-facial. NMR calculations further confirmed the relative configuration of 2 (Tables S4 and S6, SI). At the B3LYP/6–31 + G(d) level, the absolute configuration of 2 was determined by the ECD calculations using the TDDFT/ECD method. The calculated ECD curve of (2′′S, 6′′R)-2 was in good agreement with the experimental ECD spectrum (Fig. 6). Therefore, the absolute configuration of 2 was shown in Fig. 1, and denoted as glycopentamide T.
The molecular formula of 3 was deduced to be C22H33NO6S through HR-ESI-MS (Fig. S31, SI) at m/z 462.19199 [M + Na]+ (calcd for C22H33NO6SNa, 462.19208), which was composed of two conformers a and b in 2.6:1. Its 1H and 13C NMR data (Fig. 23–25, SI) were similar to those of methyldambullin (Greger et al., 1994), except for the presence of a 1,1-dimethoxyethyl group at C-4′′ [δH 1.72 (2H, m), 4.34 (1H, m), 3.30 (6H, s); δC 32.0 (t), 105.9 (d), 2 × 53.6 (q)] in 3 instead of a prenyl group at C-4′′ in methyldambullin. Thus, compound 3 was the oxidation product of the double bond Δ6′′(7′′) of methyldambullin, resulting in the loss of three carbon atoms. These result was further supported by HMBC correlations (Fig. 27, SI) from CH3O– [δH 3.30(s)] to δC 105.9 (d, C-6′′) and from H2-4′′ [δH 2.10] to δC 105.9 (d, C-6′′), and 1H–1H COSY of –CH(6′′)CH2(5′′)CH2(4′′)-. Moreover, the configuration of double bond Δ2′′(3′′) was assigned as E based on ROESY correlation of Me-10′′/H2-1′′. Thus, the structure of 3 was determined as shown in Fig. 1, and denoted as glycopentamide U.
The molecular formula of 4 was deduced to be C24H35NO5S through HR-ESI-MS (Fig. S40, SI) at m/z 450.23120 [M + H]+ (calcd for C24H36NO5S, 450.23087), which was composed of two conformers a and b in 2.6:1. By comparing with the NMR data (Fig. S32-34, SI) of methyldambuline and methylgerambullin, it was found that the substituent pattern of 4 on the benzene ring was different from that of methyldambuline and methylgerambullin (Greger et al., 1994). The para-substitution pattern of the benzene ring of methyldambullin and methylgerambullin was replaced by the 1,2,4-trisubstitution benzene ring of 4. HMBC correlations (Fig. S36, SI) from H-4′ [δH 6.76] and MeO [δH 3.83] to δC 151.3 (s, C-5′), from H-1′′ [δH 4.55] to δC 148.3 (s, C-6′), and ROESY correlations (Fig. S38, SI) from MeO to H-4′ confirmed the methoxyl and geranyloxy were attached to C-5′ and C-6′, respectively. Moreover, the configuration of double bond Δ2′′(3′′) was established as E through ROESY correlation of Me-10′′ with H2-1′′. Thus, the structure of 4 was determined as shown in Fig. 1, and denoted as glycopentamide V.
The molecular formula of 5 was deduced to be C23H35NO5S through HR-ESI-MS (Fig. S48, SI) at m/z 460.21243 [M + Na]+ (calcd for C23H35NO5SNa, 460.21282), which was composed of two conformers a and b in 1:1. The NMR data of compound 5 (Fig. S41-43, SI) were similar to those of glycopentamide B (Nian et al., 2020), except that glycopentamide B had a trans double bond Δ1(2) [δH 6.80 (d, J = 15.0 Hz), δC 133.3; δH 6.95 (d, J = 15.0 Hz), δC 139.8], while 5 had two methylene signals [δH 3.14 (t, J = 7.2 Hz), δC 51.4; δH 2.47 (t, J = 7.2 Hz), δC 27.0], indicating that 5 was the saturated product of the double bond Δ1(2) of glycopentamide B. This deduction was further confirmed by HMBC correlations (Fig. S45, SI) from H2-2 [δH 2.47] and H2-3 [δH 3.14] to δC 171.7 (s, C-1), and 1H–1H COSY of –CH2(2)CH2(3)-. The absolute configuration of 5 was assigned as 6′S based on the ECD calculations. Thus, the structure of 5 was determined as shown in Fig. 1, and denoted as glycopentamide W.
The molecular formula of 6 was deduced to be C23H33NO5S through HR-ESI-MS (Fig. S58, SI) at m/z 458.19693 [M + Na]+ (calcd for C23H33NO5SNa, 458.19716). Interestingly, a comparison of the NMR data of 1–5 with that of 6 (Fig. S50-52, SI) showed that 6 displayed only one set of NMR signals, indicating that 6 lacked the s-cis and s-trans conformers. A comparison of its NMR data with those of methyldambullin and methylgerambullin implied that they possessed para-geranyloxy phenyl group (Greger et al., 1994). Apart from the abovementioned signals, the remaining signals included two methyls [δC 43.1 (q), 35.1(q)], two oxygenated methines [δC 73.3 (d); 71.8 (d)], two methylenes [δC 55.0 (t); 56.3 (t)], and one carbonyl [δC 168.2 (s)]. Detailed analysis of 13C NMR data of compounds 1–6 suggested that the two methyls were connected to N atom and sulfonyl respectively. Two structural fragments (A) –CH2(1′)CH(2′)(O)- and (B) –CH2(3)CH(2)(O)- were obtained by careful analysis of the 2D NMR (Fig. S53-55, SI). HMBC correlations from –NCH3 [δH 3.03 (s)] to δC 55.0 (t, C-1′) and 168.2 (s, C-1), and from H-2 [δH 4.83] to δC 71.8 (d, C-2′), 168.2 (s, C-1) and 56.3 (t, C-3) suggested that two structural fragments A and B were connected to N through C-1′ and C-1, and connected by ether bond through C-2 to C-2′. Therefore, 4-methyl-2-((methylsulfonyl)methyl) morpholin-3-one moiety could be established. HMBC correlations from H-2′ [δH 5.12] to δC 129.2 (d, C-4′, 8′) confirmed the morpholin-3-one moiety was connected to C-3′. The relative configuration of 6 was determined by interpreting the ROESY spectrum (Fig. S56, SI), where the correlation of H-2′ with H2-3 indicated that these protons were co-facial and H-2′ and H-2 were assigned trans. The absolute configuration of 6 was determined by comparing the experimental and calculated ECD spectra. Consequently, the absolute configuration of 6 was defined as (2R, 2′S)-6. Therefore, the absolute configuration of 6 was shown in Fig. 1, and denoted as glycopentamide X.
The inhibitory effect of these compounds on HepG2 cell proliferation was determined by CCK-8 method, and their anti-inflammatory effects were assessed by inhibiting the activity of NO production stimulated by lipopolysaccharide in mouse macrophage RAW 264.7 cells (Table S9). As a result, only compound 4 significantly inhibited lipopolysaccharide-induced NO production in mouse macrophage RAW 264.7 cells with the IC50 value of 0.55 µM (dexamethasone as a positive control, IC50: 9.24 µM). The remaining compounds were inactive showing IC50 values in excess of 20 μM. Moreover, compounds 3 and 4 exhibited different anti-proliferative activities against HepG-2 with IC50 values of 11.52 and 9.41 µM, respectively (cisplatin as a positive control, IC50: 5.96 µM). The other compounds showed no obviously anti-proliferative activities with IC50 values in excess of 20 μM. Based on previous research and our results (Nian et al., 2020), the geranyloxy group at C-6′ of these methylsulfonylpropenoic acid amides might be crucial for the anti-proliferative and anti-inflammatory activities.
4 Conclusions
Sulfur-containing amides are a special type of alkaloids, and mainly isolated from the genus Glycosmis. This compounds showed novel structure and exhibited diversity bioactivity, like antimicrobial, cytotoxic, antitrypanosomal, antiviral activities. In this paper, six previously undescribed sulfur-containing amides with structural diversities were isolated from the leaves of G. pentaphylla. It is noteworthy that 6 represents the first example of sulfur-containing amide possessing a morpholin-3-one moiety. Moreover, Compound 4 had the strongest proliferative activity on HepG2 cells and had a significant inhibitory effect on NO production. The relationship between structure and activity indicates that the geranyloxy group at at C-6′ plays an important role in the anti-inflammatory and anti-proliferative activities of these alkaloids. These findings indicate that among the active ingredients of G. pentaphylla that exert anti-inflammatory and anticancer activities, in addition to the flavonoids reported in the previous literature (Shoja et al., 2015; Shoja et al., 2016), sulfur-containing amides also play an important role. These findings could explain the traditional use of G. pentaphylla to treat liver cancer and inflammatory diseases and provide a scientific basis for traditional medicine knowledge.
CRediT authorship contribution statement
Wenli Xie: Investigation, Formal analysis, Writing – original draft. Hefeng Nian: Investigation, Formal analysis. Xueni Li: Formal analysis. Jing Xu: Formal analysis. Yu Chen: Formal analysis. Zhinan Mei: Supervision, Writing – review & editing. Guangzhong Yang: Supervision, Writing – review & editing.
Acknowledgements
This work was supported by the National Key Research and Development Program (2022YFC3502200), the Key Research and Development Program of Hubei Province (2021ACB003), and the Major Scientific and Technological Project of Hubei Province (2020ACA007).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Adamantyl and homoadamantyl derivatives from Garcinia multiflora fruits. RSC Adv.. 2019;9:12291-12299.
- [Google Scholar]
- Sulphones derived from methylthiopropenoic acid amides from Glycosmis angustifolia. Phytochemistry. 1994;37:1305-1310.
- [Google Scholar]
- Hofer, O., & Greger, H. 2000. Sulfur-containing amides from Glycosmis species (Rutaceae). in: Herz W., Falk H., Kirby G.W., Moore R. E. (Eds.), Prog. Chem. Org. Nat. Prod, 187-223.
- New anthranilic and methylsulfonylpropenoic acid amides from Thai Glycosmis species. Liebigs Ann. 1995;1995:1789-1794.
- [Google Scholar]
- Phenethylamides with an unusual 4-oxo-2-oxolenyl terpenoid side chain from Glycosmis species. Monatsh. Chem.. 1998;129:213-219.
- [Google Scholar]
- Prenylated sulfonyl amides from Glycosmis species. Phytochemistry. 2000;54:207-213.
- [Google Scholar]
- Ethnomedicinal uses, phytochemistry, pharmacological activities and toxicological profile of Glycosmis pentaphylla (Retz.) DC.: A review. J. Ethnopharmacol.. 2021;278:114313
- [Google Scholar]
- NMR calculations with quantum methods: development of new tools for structural elucidation and beyond. Acc. Chem. Res. 2020;53:1922-1932.
- [Google Scholar]
- Total synthesis and biological evaluation of methylgerambullone. Bioorg. Med. Chem. Lett.. 2010;20:52-55.
- [Google Scholar]
- Anti-inflammatory and antiproliferative prenylated sulphur-containing amides from the leaves of Glycosmis pentaphylla. Fitoterapia. 2020;146:104693
- [Google Scholar]
- Glycosmis pentaphylla (Retz.) DC arrests cell cycle and induces apoptosis via caspase-3/7 activation in breast cancer cells. J. Ethnopharmacol. 2015;168:50-60.
- [Google Scholar]
- In vitro mechanistic and in vivo anti-tumor studies of Glycosmis pentaphylla (Retz.) DC against breast cancer. J. Ethnopharmacol. 2016;186:159-168.
- [Google Scholar]
- In vivo immunosuppressive activity of gliotoxin, a metabolite produced by human pathogenic fungi. Infect. Immun. 1994;62:1192-1198.
- [Google Scholar]
- Homoadamantane polycyclic polyprenylated acylphloroglucinols from the fruits of Garcinia multiflora. Fitoterapia. 2019;137:104245
- [Google Scholar]
- Organ-specific chemical differences in Glycosmis trichanthera. Phytochemistry. 1998;48:897-902.
- [Google Scholar]
- Methylgerambullin derived from plant Glycosmis pentaphylla (Retz) correa. Mediates anti-hepatocellular carcinoma cancer effect by activating mitochondrial and endoplasmic reticulum stress signaling and inhibiting AKT and STAT3 pathways. Food Chem. Toxicol.. 2021;149:112031
- [Google Scholar]
- Five new sulphur-containing amides from Glycosmis lucida with antifeedant activity against Tribolium castaneum. Ind. Crop. Prod.. 2015;74:628-634.
- [Google Scholar]
- The genus Glycosmis [Rutaceae]: a comprehensive review on its phytochemical and pharmacological perspectives. The Nat. Prod. J.. 2019;9(2):98-124.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2022.104528.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary data 1
Supplementary data 1







