Translate this page into:
Recent studies on advance spectroscopic techniques for the identification of microorganisms: A review
⁎Corresponding author. musharraf1977@yahoo.com (Syed Ghulam Musharraf)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
The continuous development of resistance to antibiotic drugs by microorganisms causes high mortality and morbidity. Pathogens with distinct features and biochemical abilities make them destructive to human health. Therefore, early identification of the pathogen is of substantial importance for quick ailments and healthcare outcomes. Several phenotype methods are used for the identification and resistance determination but most of the conventional procedures are time-consuming, costly, and give qualitative results. Recently, great focus has been made on the utilization of advanced techniques for microbial identification. This review is focused on the research studies performed in the last five years for the identification of microorganisms particularly, bacteria using advanced spectroscopic techniques including mass spectrometry (MS), infrared (IR) spectroscopy, Raman spectroscopy (RS), and nuclear magnetic resonance (NMR) spectroscopy. Among all the techniques, MS techniques, particularly MALDI-TOF/MS have been widely utilized for microbial identification. A total of 44 bacteria i.e., 6 Staphylococcus spp., 3 Enterococcus spp., 6 Bacillus spp., 4 Streptococcus spp., 6 Salmonella spp., and one from each genus including Escherichia, Acinetobacter, Pseudomonas, Proteus, Clostridioides, Candida, Brucella, Burkholderia, Francisella, Yersinia, Moraxella, Vibrio, Shigella, Serratia, Citrobacter, and Haemophilus (spp.) were discussed in the review for their identification using the above-mentioned techniques. Among all the identified microorganisms, 21% of studies have been conducted for the identification of E. coli, 14% for S. aureus followed by 37% for other microorganisms.
Keywords
MALDI-TOF/MS
FT-IR
SERS
NMR
- AMR
-
Antimicrobial Resistance
- AST
-
Antimicrobial Susceptibility Testing
- ANN
-
Artificial Neural Network
- AuNPs
-
Gold Nanoparticles
- AI
-
Artificial Intelligence
- BCs
-
Blood Cultures
- BCB
-
Blood Culture Bottle
- CFU
-
Colony Forming Unit
- CHO
-
Chinese Hamster Ovary
- CNN
-
Convolution Neural Network
- CNS
-
Central Nervous System
- DDA
-
Data-Dependent Analysis
- DCD-SERS
-
Drop-Coating Deposition Surface-Enhanced Raman Scattering
- DFA
-
Discriminant Function Analysis
- DIA
-
Data Independent Analysis
- ECDC
-
The European Centre for Disease Prevention and Control
- ESβL
-
Extended Spectrum β-lactamases
- ESI/MS
-
Electrospray Ionization Mass Spectrometry
- EU
-
Eurpean Union
- FT
-
Fourier Transform
- FAB
-
Fastidious Anaerobe Broth
- FcMBL@Fe3O4
-
Fragment Crystallizable Mannose Binding Lectin-modified
- Fe3O4 FT-IR
-
Fourier Transform Infrared spectroscopy
- GC-IMS
-
Gas Chromatography-Ion Mobility Spectrometer
- HCA
-
Hierarchical Cluster Analysis
- 1H NMR
-
Proton NMR
- IMS
-
Integrated Microfluidic System
- kNN
-
K-Nearest Neighbors
- KPCA-DT
-
Kernel Principal Component Analysis-Decision Tree
- LB
-
Luria Broth
- LC-MS
-
Liquid Chromatography Mass Spectrometry
- MALDI-TOF
-
Matrix Assisted Laser Desorption-Time-of-Flight
- MIC
-
Minimum Inhibitory Concentration
- ML
-
Machine Learning
- MLST
-
Multilocus Sequence Typing
- MRSA
-
Methicillin-Resistant S. aureus
- MSSA
-
Methicillin-Susceptible S. aureus
- MSP
-
Main Spectrum Profile
- MOBA
-
methylthio–2–oxobutyric acid
- NALDI-TOF MS
-
Nanotechnology-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry
- NIRS
-
Near-Infrared Spectroscopy
- NIST
-
The National Institute of Standards and Technology
- NMR
-
Nuclear Magnetic Resonance
- NRS
-
Normal Raman Scattering
- PCA
-
Principle Component Analysis
- PCR
-
Polymerase Chain Reaction
- PSA
-
Partial Least Square Analysis
- PMF
-
Peptide Mass Fingerprint
- PFGE
-
Pulsed-Field Gel Electrophoresis
- ppm
-
Parts Per Millions
- RTIs
-
Respiratory Tract Infections
- RS
-
Raman Spectroscopy
- SALDI-TOF
-
Surface-Assisted Laser Desorption/Ionization Time-of-Flight
- SERS
-
Surface Enhanced Raman spectroscopy
- SELDI-TOF
-
Surface-Enhanced Laser Desorption/Ionization Time-of-Flight
- SPME
-
Solid Phase Micro-Extraction
- ST
-
Sequence Type
- Strep. TSS
-
Streptococcal Toxic Shock Syndrome
- SVM
-
Multi-Support Vector Machine
- TOF
-
Time of Flight
- UHPLC-HRMS
-
Ultra-High Performance Liquid Chromatography-High Resolution Mass Spectrometry
- UTIs
-
Urinary Tract Infections
- VAP
-
Ventilator-Associated Pneumonia
- VOC
-
Volatile Organic Compounds
- OECD
-
Organization for Economic Co-operation and Development
- WGS
-
Whole Genome Sequencing
Abbreviations
1 Introduction
Infectious diseases are continuously growing and are the principal cause of mortality and morbidity throughout the globe by the resistance of pathogenic microorganisms to antibiotics. Antimicrobial resistance (AMR) is a common phenomenon that occurs when microbes are exposed to antimicrobials and the exchange of resistant characters happens (Sharma et al., 2018); (Rodgers et al., 2019). Antibiotic resistance is responsible for the worldwide deaths of more than 0.5 million people, out of which more than 40 % comprise newborn deaths every year (Foundation 2018). According to recent reports, in the European Union (EU), every year more than 33 K people lose life due to diseases stemming from antimicrobial-resistant bacteria (Anderson et al., 2019, Ben et al., 2019, Dadgostar 2019, Raoult et al., 2019) and costs an estimated annual economic burden of 1.5 billion euros including healthcare and production loss (Anderson et al., 2019). It is estimated by the latest Organization for Economic Co-operation and Development (OECD) report, that over the next 30 years, 2.4 million people will die due to antimicrobial-resistant pathogens in Europe, North America, and Australia and could cost up to US$ 3.5 billion annually (https://www.oecd.org/health/stemming-the-superbug-tide-9789264,307599-en.htm). This situation is already severe in low and middle-income regions, which are likely to rise significantly (Hofer 2019).
The identification of microorganisms based on traditional methods is estimated to require 2–5 days or more which includes morphological, physiological, chemical, and biochemical characterization. Furthermore, most of the phenotypic methods for microbial identification are time and material-consuming, and laborious (Bochner 2008). However, advanced spectroscopy techniques offer rapid and high-throughput analysis for microbial identification at the genus and species levels.
Since the last decade, matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) has changed the clinical microbiology field with a broader range, low cost, and early and rapid microbial identification from grown bacterial colonies. Microbial identification from BCs was further improved by applying it on the plate with a shorter incubation period allowing early and reliable identification (Idelevich et al., 2014, Kohlmann et al., 2015). The use of nanotechnology-assisted laser desorption/ionization time-of-flight mass spectrometry (NALDI-TOF MS) could be an innovative approach to enhance microbial analysis. This method uses a nanostructured silicon-based target plate instead of traditional organic matrices (Tatsuta et al., 2017). The primary function of nanomaterials in this technique is to enrich analyte particles and allow for effective desorption and ionization (Chu et al., 2018).
The surface-enhanced laser desorption/ionization time-of-flight (SELDI-TOF) technique is another method for identifying microorganisms. SELDI-TOF MS enables the direct study of bacterial lysates by allowing the selective absorption of proteins on the chromatographic array surface. Target protein homogenous binding enhances the repeatability of MS analyses. Consequently, both the mass-to-charge (m/z) ratio and the intensity values may be considered (Lundquist et al., 2005, Seibold et al., 2007). The biggest restriction of this approach is that most clinical microbiology laboratories do not have this expensive equipment. Similarly, Sunner and Chen proposed surface-assisted laser desorption/ionization mass spectrometry (SALDI-MS) (Law and Larkin 2011). SALDI is a matrix-free laser desorption/ionization method that replaces MALDI's organic matrix with various substrate surfaces such as graphite or nano silicon (Spraker et al., 2020). Because organic matrices are avoided, SALDI is an excellent candidate for use in low molecular-weight compounds (Song and Cheng 2020). Several applications of biological and microbial SALDI imaging on various silicon substrates have been developed during the last decade (Ronci et al., 2012, Chen et al., 2018). As an illustration, Wang et al 2022, created gold nanoparticles/thiol-cyclodextrin-functionalized TiO2 nanowires as the auxiliary surface for NP SALDI-MSI (Wang and Li 2021).
Various reviews have been published on physical, biochemical, and imaging techniques for microbial identification and susceptibility testing (Pulido et al., 2013, Van Belkum and Dunne Jr 2013, Idelevich and Becker 2019, Maugeri et al., 2019, Smith and Kirby 2019). A review for the designing and development of rapid detection of resistant microorganisms based on traditional agar methods, E-test, staining, kits, and MALDI methods has also been published (Leonard et al., 2018). Some reviews have focused on the use of individual techniques in microbiology such as MALDI-TOF techniques for detecting resistance biomarkers (Vrioni et al., 2018), identification of microorganisms from bloodstream infections by automated antimicrobial susceptibility testing (AST) (Wattal and Oberoi 2016, Nomura et al., 2020). Recently, comprehensive reviews on Omics approaches and novel techniques for microbial identification have been reported (Buszewski et al., 2021, Janiszewska et al., 2022). The use of electrospray ionization mass spectrometry (ESI-MS) for broad-range microbial identification (Kailasa et al., 2019), the progress of proteomics and MS application in clinical microbiology (van Belkum et al., 2015, Sanguinetti and Posteraro 2016). The use of infra-red spectroscopy (XU et al., 2007, Quintelas et al., 2018), Raman spectroscopy (Galvan and Yu 2018, Kaprou et al., 2021), and NMR (García-Álvarez et al., 2015), for bacterial typing and identification. However, all these articles are short of the applications of advanced spectroscopic techniques in terms of microbial identification.
This review aimed to comprehensively summarize and combine the applications of advanced spectroscopy techniques (MS, FT-IR, RS, and NMR) for microbial identification by utilizing recent examples of the past five years. Fig. 1 represents a schematic view of the review.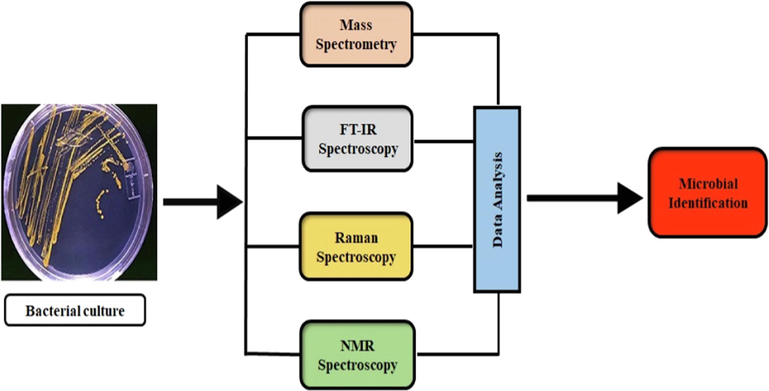
Scheme of the review.
2 Microbial pathogenicity and their associated effects
Pathogenic microorganisms cause severe diseases and harm to human beings. Worldwide, many diseases are reported due to the pathogenicity of microbes. Some of the famous are related to bloodstream infections, urinary tract infections (UTIs), respiratory tract infections (RTIs), ventilator-associated pneumonia (VAP), etc. Table 1 represents the comprehensive details related to the pathogenicity of microorganisms covered in the study that causes various infections and diseases.
No.
Microorganisms
Pathogenicity
1
Staphylococcus aureus
Causes skin infections, food poisoning, bone, and joint infections, etc.
2
Staphylococcus capitis
Bloodstream infections, nosocomial infections, etc.
3
Staphylococcus warneri
UTI’s
4
Staphylococcus haemolyticus
The nosocomial pathogen causes UTIs, sepsis,
5
Staphylococcus epidermidis
Inflammations, wound infections, sinus infections, endocarditis, etc.
6
Staphylococcus saprophyticus
UTIs, and cystitis in young women
7
Enterococcus faecalis
Bloodstream infections, periodontitis, UTIs, etc.
8
Enterococcus cloacae
UTIs, respiratory tract infections, endocarditis, septic arthritis, skin, and soft tissue, abdominal, etc.
9
Enterococcus faecium
UTIs, wound infections, prostatitis, bacteremia, endocarditis, and cellulitis.
10
Bacillus melitensis
Mastitis, abortion, stillbirth, and weak offspring in animals
11
Bacillus suis
swine brucellosis, and orchitis
12
Bacillus pseudomallei
Whitmore's disease in animals and humans.
13
Bacillus subtilis
Pneumonia, endocarditis, septicemia, and bacteremia
14
Bacillus anthracis
Anthrax
15
Bacillus cereus
The emetic (vomiting) and the diarrheal syndrome.
16
Streptococcus pyogenes
Streptococcal Toxic Shock Syndrome (Strep. TSS), myonecrosis, bacteremia, pneumonia, scarlet fever, and necrotizing fasciitis
17
Streptococcus agalactiae
Neonatal sepsis, and postpartum infection
18
Streptococcus milleri
Pyogenic infections, and bacteremia
19
Streptococcus pneumoniae
Pneumonia, middle ear infections, blood infection, and meningitis
20
Salmonella enterica
Nausea, vomiting, fever, abdominal pain, diarrhea, etc.
21
Salmonella typhi
Malaise, anorexia, typhoid fever, non-productive cough, headache, relative bradycardia, and constipation or diarrhea
22
Salmonella paratyphi A
23
Salmonella paratyphi B
24
Salmonella paratyphi C
25
Salmonella typhimurium
Gastroenteritis, bloodstream, infections in mice
26
Escherichia coli
UTIs, pneumonia, diarrhea, neonatal meningitis, bloodstream infections, cholecystitis, cholangitis, etc.
27
Klebsiella pneumoniae
Pneumonia, meningitis, bloodstream, wound infections, etc.
28
Acinetobacter baumannii
Pneumonia, wound, UTIs, bloodstream infections
29
Pseudomonas aeruginosa
Pneumonia, septicemia, endophthalmitis, endocarditis, meningitis, and malignant external otitis
30
Proteus mirabilis
UTIs, bloodstream infections
31
Clostridioides difficile
Intestinal diseases, severe diarrhea, food poisoning
32
Listeria monocytogenes
Fever, miscarriage, muscle aches, CNS diseases
33
Candida albicans
Candidiasis
34
Morganella morganii
Skin and soft tissue infections, UTIs, septic arthritis, gastroenteritis, etc.
35
Brucella abortus
Brucellosis
36
Burkholderia mallei
Glanders in animals.
37
Francisella tularensis
Tularemia in animals
38
Yersinia pestis
Bubonic plague in humans and animals.
39
Moraxella catarrhalis
Acute bacterial rhino sinusitis, and chronic obstructive pulmonary disease
40
Vibrio parahaemolyticus
gastroenteritis, sepsis, and wound infections
41
Shigella sonnei
Shigellosis
42
Serratia marcescens
UTIs, pneumonia, bloodstream infection, lower respiratory tract infection, meningitis, and wound infection.
43
Citrobacter freundii
wound infections, UTIs, meningitis, and sepsis
44
Haemophilus influenzae
Ear infections, bloodstream infections
3 Use of advance spectroscopic techniques in the identification of microorganisms
Different spectroscopic techniques have been used for microbial identification. The use of mass spectrometry was first reported in 1975 for the identification of bacteria (Anhalt and Fenselau 1975). The analysis of proteins by MS had to look out for the arrival of soft ionization techniques (MALDI and ESI) because of their larger size and magnitudes (Sauer and Kliem 2010). By the mid of 1990 s, it was being used for bacterial identification due to its suitability in microbiological research laboratories (Claydon et al., 1996, Holland et al., 1996). Moreover, MALDI-TOF/MS and other hyphenated techniques (, LC-MS, GC–MS, etc.) have been widely used for the identification of various microorganisms.
IR spectroscopy was utilized for the identification and discrimination of bacteria in the 1950 s and 1960 s (Whetsel 1991). Early methods of bacterial analysis by FT-IR were impractical and laborious however, in the 1980 s significance of FT-IR spectroscopy for biological application was resumed with the advancement of modern interferometer and multivariate statistical analysis tools (Burgula et al., 2007); (Preisner et al., 2007). The analysis of biological samples by Raman spectroscopy (RS) was reported for the first time in the late1980s by focusing on the resonance Raman Effect (Howard et al., 1980). Later on, different Raman techniques were applied for the identification (Chauvet et al., 2017), and discrimination of bacterial isolates (Jarvis and Goodacre 2004). Proton NMR (1H NMR) was used for the first time to differentiate bacteria at the genera level based on their cellular metabolite composition (Delpassand et al., 1995).
A comprehensive Table 2 is presented for comparing the possible advantages and disadvantages of the techniques related to microbial identification. However, the choice for choosing any of the techniques for studying microorganisms depends upon the mindset and skills of the researcher in his field.
Mass Spectrometry Techniques
IR
Raman
NMR
MALDI-TOF MS
LC-MS
GC–MS
Advantages
-Fast analysis
-High-throughput-High sensitivity
(can detect as low as 103 CFU mL−1)
Automated e.g., MALDI Biotyper®, VITEK®MS SARAMIS™ AndromasDirect identification
(Sepsityper and SELTERS kits)
(Lévesque et al., 2015, Jang and Kim 2018)
(Freiwald and Sauer 2009, Marko et al., 2012)
- Rapid analysis
- High sensitivity
-Good separation of polar compounds
- Determination of IC50 values- Direct identification
(shotgun proteomics method)
-Targeted analysis
(Tracz et al., 2013, Berendsen et al., 2017, Roux-Dalvai et al., 2019, Lasch et al., 2020)
-Fast analysis and sensitivity
-Good separation efficiency-Spectral database available (Wiley and National Institute Standard and Technology (NIST)
libraries mass spectral database)
(Garcia et al., 2008)
-Fast and sensitive screening
- High-throughput- Automated microbial typing methods are available e.g., S
(IR-Biotyper®)
-The whole organism fingerprinting
-Cell lysis is not necessary for analysis
–non-invasive
-little sample needed
(Kosa et al., 2017, Hu et al., 2021)
- High specificity
- Little sample preparation
- Allows AST
- SERS identification of specific biomarkers
helps to determine MICs and AST
-Culture-free identification
-The whole organism fingerprinting
(Stöckel et al., 2016, Weiss et al., 2019, Kumar et al., 2020)
-Intrinsically quantitative
-Little sample preparation
-The same sample can be used repeatedly
-High reproducibility
-Reliable assignment of structure
- Determination of MIC
-Monitoring of living systems
(García-Álvarez et al., 2015, García-Álvarez et al., 2019)
Disadvantages
-High initial costs of equipment-Database development (spectra from resistant and susceptible strains)
should be developed
- Biomarker discovery for AMR needed
- Not applicable for MIC determination
- Lower discrimination power between closely related species
(Rodrigues et al., 2017, Grenga et al., 2019)
-High initial costs of equipment- Database requirement (spectra from resistant and susceptible strains)
not available
- Biomarker discovery for AMR needed
(Gowda and Djukovic 2014, Aszyk et al., 2018)
-Time-consuming sample preparation steps
- Complex sample preparation
-sample volatility requirements
- Applicable for only non-polar compound
-High Temperatures usage
- Derivatization steps are required in the case of non-volatile compounds
(Wittmann 2007, Lu et al., 2008)
-Testing of a purified single strain,-Databases (spectra from resistant and susceptible strains)
should be developed
- Biomarker discovery for AMR needed
- IR data vary by culture conditions
-Not applicable for MIC determination
-Multivariate statistical analysis is a must
(Ami et al., 2012)
-Poor sensitivity of NRS
-Databases not available
(Eberhardt et al., 2015)
-Large equipment costs
-Lower sensitivity- Low limit of detection
(can’t detect below 103 CFU ml−1)
- Databases not available
(Pan and Raftery 2007, Gupta et al., 2009)
Applications of the advanced spectroscopic techniques related to microbial studies are focused on below in detail.
3.1 MALDI-TOF mass spectrometry
MALDI-TOF is a mass spectrometry technique, that was introduced with great success in clinical diagnostics for the identification of pathogens, a decade ago. Franz Hillenkamp and Michael Karas developed matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) in 1985 (Karas et al., 1985). In 1988, the first time their group reported the detection of labile biopolymers - proteins with this technology (Fuh et al., 2017). John Fenn and Koichi Tanaka work on MALDI and its application to protein analysis and earned the 2002 Nobel Prize in Chemistry (Tanaka et al.,). MALDI coupled with a time-of-flight (TOF) analyzer was used in microbiology to differentiate intact bacterial cells in the late 1990 s (Buszewski et al., 2021). MALDI became one of the primary methods for the identification of proteins and, consequently, for proteomics almost immediately after its discovery.
3.1.1 Mechanism of laser desorption ionization (LDI)
MALDI-TOF MS is a soft ionization technique that enables the ionization of samples to charge molecules to measure their m/z. Before analysis, samples are mixed with a small organic compound known as a matrix, which facilitates energy transfer to test samples, ionization, and analysis of non-volatile, high molecular weight, and polar substances. The matrix absorbs UV radiation well, quickly sublimates, and after the desorption process, provides large amounts of ions in both positive and negative ionization modes required for the ionization of the test substance (Park et al., 2014). A test substance is deposited on a steel plate (target plate) having spots for several different samples to be applied, followed by matrix deposition and left for dryness. The sample spots are irradiated with a short burst of a laser beam, and energetically remove matrix particles from the sample surface absorb the laser energy and transfer the analyte particles to the gas phase. Analyte molecules are often ionized into singly charged positive ions [M + H]+ or negative ions [M-ַַH]- with neighboring matrix molecules during the ablation process (Gao and Cassady 2008), followed by the detection of molecular weight and time-of-flight (TOF). TOF works on the basis that ions of varying m/z are time-scattered as they travel down a field-less drift route of known length. Assuming that all of the ions begin their travel at the same moment, the lighter ions will reach the detector before the heavier ones. The analysis results in a spectrum, which indicates the masses of the produced ions, and the signals are ordered in an increase in mass.
3.1.2 Microbial identification based on MALDI-TOF MS
Identification of microorganisms by MALDI-TOF/MS is based on four commercially available systems and with their databases (a) the MALDI Biotyper (Bruker Daltonics, Bremen, Germany); (b) the Spectral Archive and Microbial Identification System (SARAMIS™) (AnagnosTec, Potsdam, Germany); (c) the Andromas (Andromas, Paris, France) and (d) the Vitek MS (bioMérieux, Marcy l’Etoile, France) (online:, online:). Most of the installed systems in routine diagnostics are the MALDI Biotyper and the Vitek MS which are approved by FDA for microbial identification (Posteraro et al., 2013). Both systems are different in instrumentation, identification algorithms, and databases (Carroll and Patel 2015). Data reveals that both systems perform similarly and the identification rate at the genus level is very high 97–99 % while varies from 85 to 97 % at the species level (Cherkaoui et al., 2010, Marko et al., 2012, Alby et al., 2013, Mancini et al., 2013, Kärpänoja et al., 2014, Mather et al., 2014).
The MALDI Biotyper employs a similarity pattern measure (Cassini et al., 2019), with a database of references known as the Main Spectrum Profile (MSP). When comparing the obtained and reference spectra, the similarity is expressed as “log (scoring),” where a score of 2.3 indicates a “high confidence identification,” a score between 2.0 and 2.3 indicates a “secure genus identification,” a score between 1.7 and 2 indicates a “low confidence identification,” and a score of 1.7 is considered to indicate “no reliable identification.” A further measure to assess the identification is the consistency of the top 10 findings. The following criteria are used in the case of mycobacteria: Confidence levels of 1.6 for low and 1.8 for high-level identification (Rodríguez-Sánchez et al., 2016). The VITEK®-MS uses a machine learning-based algorithm “Advanced Spectra Classifier”. Spectra ranging from 3000 to 17,000 Da are separated into 13,000 fragments and weighted based on their relevance in identifying a certain bacterial species. Unknown spectra are subjected to the same procedure and are compared sequentially with the Vitek MS database. The acquired findings are expressed as percentages: 99.9 %—a perfect match, 60 % to 99.8 %—a good match, and 60 %—no identification. Matching is determined in the SARAMIS system based on common strains that incorporate intraspecific species diversity. Unknown strains are identified by comparing their spectra to those in the “SuperSpectra” database, and confidence levels range from high (>98 %) to medium (85 % to 98 %) to low (75 % to 85 %) (Leyer et al., 2017). According to the research, both methods have equal rates of identification (Lévesque et al., 2015, Lee et al., 2017).
Its precision though is heavily reliant on the database coverage of commercially available MALDI-TOF MS equipment. Identification of Brucella was not achievable since this genus was not listed in the databases of the two major MALDI-TOF MS system manufacturers (Rudrik et al., 2017, Tracz et al., 2017). Bacteria that are not included in the database, likewise cannot be identified. As a result, if a species cannot be recognized using one approach, it must be verified using another. If the species to be identified is not found in the database, the score value of a similar species is high, and if the species is displayed at the species level, the identification is incorrect. Furthermore, enough biomass is necessary for good identification outcomes. Although some researchers propose a detection limit of 6 × 103 CFU/spot a limit of 1 × 105 CFU/spot is frequently necessary (Hsieh et al., 2008, Kirkpatrick and Viollier 2012). In the future, these limitations are needed to be addressed. It can be helpful to build in-house libraries of various microbial strains by research laboratories and its integration with standard databases will further improve the identification of microorganisms. Moreover, sample preparation procedures can also be modified for better results. More importantly, the maintenance and calibration of the instrument by skillful personnel are vital.
3.1.3 Sample preparation approaches
The most important component of each “Omics” approach is the step of sample preparation. The synthesis of ethanol-formic acid protein extracts, direct transfer, and direct transfer with formic acid are among the most common sample preparation techniques used to identify bacteria using MALDI-TOF MS. The gold standard for creating a reference database is ethanol-formic acid extraction (Drevinek et al., 2012). The three procedures were compared in a study for Gram-positive rod identification, which revealed that the results obtained from direct formic acid transfer were comparable with ethanol-formic acid (Schulthess et al., 2014). Therefore, in the routine clinical analysis, the direct transfer approach is more successful. There are no appreciable changes in the identification rates between the direct sample transfer and the extraction process for numerous environmental bacteria, including Legionella spp. (Pascale et al., 2020). It gives promising identification results for rod-shaped Gram-negative bacteria (Tsuchida et al., 2020a). However, anaerobic bacteria, Gram-positive bacteria, and certain Mycobacteria had worse results. It has been reported that B. Subtilis was misidentified as Bacillus mojavensis and vice versa, this misguided identification may have resulted from the two bacteria's highly similar mass spectra (Huang et al., 2016, Wang et al., 2021). Gram-positive bacteria with a thick cell wall can be recognized in a wider variety of methods, albeit not necessarily down to the species level. With these strains, it is challenging to produce a homogenous, sufficient number of bacterial cells in a smear (Veloo et al., 2014). Thus, the extraction techniques are preferred for MALDI detection of Gram-positive bacteria due to better protein recovery, especially for spore-forming bacteria.
3.1.4 Identification based on commercial kits
To further enhance direct microbial identification from positive BCs using MALDI-TOF MS, several protocols have been established. These protocols aim to remove blood cells and host proteins from BCs before MALDI-TOF MS analysis. Currently, there are various in-house methods as well as some commercial kits available for BCs sample preparation making it suitable for MALDI-TOF-MS analysis. The developed in-house protocols are based on obtaining pure microbial cells through the use of diverse lysis substances like saponin, sodium dodecyl sulfate (SDS), and ammonium chloride, or through stepwise centrifugation to separate blood cells from bacteria (Maelegheer and Nulens 2017, Kayin et al., 2019, Tsuchida et al., 2020b, Zengin Canalp and Bayraktar 2021). Additionally, three kits commercially available are in use right now: the rapid BACpro® II (Nittobo Medical Co., Tokyo, Japan), the Vitek MS blood culture kit (bioMérieux, Marcy-l'Étoile, France), and the Sepsityper® Kit (Bruker Daltonics GmbH, Bremen, Germany) [(Martiny et al., 2012, Kayin et al., 2019). However, the Sepsityper®Kit is the most popular and FDA-approved kit among the three commercially available kits (Morgenthaler and Kostrzewa 2015). Marina., et al. studied the application of rapidBACpro® II kit for bacterial identification from positive blood cultures (BCs) using MALDI-TOF/MS. A total of 801 microbial isolates were screened by rapidBACpro® II kit and 80.0 % of isolates were identified correctly at the species level (92.3 % of the Gram-negative and 72.4 % of the Gram-positive bacteria (Oviaño et al., 2021). The developed method was further evaluated for the identification of gram-negative microbes in positive BCs with the detection of extended-spectrum β-lactamases (ESβL) and carbapenemases production using MALDI-TOF/MS analysis (Roncarati et al., 2021). A comparison of the performances of two commercial kits, named SepsiTyper™ Kit and the SELTERS (Treibmann., et al. 2015) were also assessed and proved to be comparable for the identification of BCs microbes (Di Gaudio et al., 2018).
3.1.5 Identification based on various extraction protocols
Various extraction protocols have been established and employed for microbial identification from blood cultures. The direct identification of microbes from BCs was achieved for the management of pediatric patients through a cost-effective rapid method using MALDI-TOF/MS (Vitek MS bioMérieux). Each positive BC was centrifuged to remove erythrocytes from bacterial cells followed by the addition of triton (10-X) to the supernatant. From a total of 360 BCE samples, 85 % were identified at the species level among mono-microbial cultures. The method correctly identified 99 % of gram-negative isolates at both genus and species levels while for gram-positive 84 % and 81 %, respectively (Samaranayake et al., 2020). Similarly, direct microbial identification from BCs was achieved by adding 30 µL of 5 % saponin and 1 mL distilled water followed by centrifugation, transfer, and re-centrifugation of the supernatant. Resulted pellets were constituted in formic acid and screened by Clin-TOF/MS with 100 % accuracy. The estimated method cost was $0.5 per sample in just 20 min turnaround time (Huang et al., 2019).
The VACUETTE® Z Serum Sep Clot Activator tube comprising a sterile gel was also used to identify microbes from BCs. After centrifugation, the supernatant was discarded and pellets were collected from the surface of the inert gel followed by MALDI-TOF/MS analysis. In comparison with the routine methods, the developed method accurately identified 90 % while the SepsiTyper kit identified 99 % of the isolates (Azrad et al., 2019). Direct identification from positive BCs was achieved by optimizing the sample processing methodology. 3 mL of blood was transferred to a tube containing separating gel followed by centrifugation. Addition of deionized water in the supernatant followed by re-centrifugation. The resulting bacterial cell membrane was subjected to MALDI-MS analysis. A total of 829 samples were collected from which 7 false-positive samples were excluded. The rate of accuracy of the optimized method for gram-negative bacteria was 91.5 %, gram-positive 88.3 %, fungi 84.8 %, and other anaerobic and rare bacteria 80 and 66.6 %, respectively (Yuan et al., 2020).
In one of the studies, direct identification of 80 % bacteria in positive BCs was achieved by developing a 10 min extraction protocol by adding 200 µL of blood in 1 mL solution of Triton X-100 followed by centrifugation, deposition of target plate, and MS analysis. In 632 blood culture bottles (BCBs), 80 % of direct identification of bacteria (96 % of Enterobacteriaceae spp., 95 % of S. aureus, 92 % of enterococci spp., and 62 % of streptococci spp.) was achieved with a log (score) threshold ≥ of 1.5 (Simon et al., 2019). A protocol for direct microbial identification using MALDI-TOF/MS from positive BCs after a short-term incubation on a solid medium has also been studied. The protocol was evaluated to directly recognize microbes from 162 positive BCs at different incubation periods i.e., 3, 5, and 24 h. The identification of bacteria at the species level was 64.1, 85.0, and 94.1 % at 3, 5, and 24 h, respectively (Curtoni et al., 2017).
3.1.6 Identification based on bacterial cell enrichment
Different methodologies were applied to enrich bacterial cells before MALDI-MS analysis. A polyallylamine–polystyrene copolymer was used for the enrichment of bacterial cells from positive BCs followed by identification with MALDI-TOF/MS analysis. By using representative species Escherichia (E.) coli and Staphylococcus (S.) capitis, it was found that polyallylamine–polystyrene copolymer can form aggregates with protocol processing time as shorter as 15 min. The identification from BCs by analyzing 17 strains of 5 species of E. coli, Klebsiella (K.) pneumoniae, Enterococcus (E.) faecalis, Staphylococcus (S.) aureus, and S. capitis was satisfactory (Ashizawa et al., 2017). The use of fragment crystallizable mannose-binding lectin-modified Fe3O4 (FcMBL@Fe3O4) for capturing bacteria from aqueous solution and bovine blood followed by MALDI-TOF/MS analysis has been reported. It suggests that the release of bacteria from functional material can increase the accuracy of identification (Sun et al., 2021). Similarly, an uncommon pathogen, Vibrio alginolyticus has been identified by using Fc-MBL@Fe3O4 enrichment with MALDI-TOF MS profiling in liquid-cultured samples (Ying et al., 2021).
3.1.7 Protein chip techniques
ProteinChip Arrays with surfaces that nourish certain proteins were developed by BioRad. Therefore, MALDI-TOF MS was renamed to SELDI-TOF-MS by the company. The ProteinChip method is a de novo method for discovering proteins that do not require an early understanding of specific proteins. ProteinChip arrays, ProteinChip reader, and specialized software make up the core components of the technology mentioned. According to Shah et al., three types of matrices, hydrophobic (H50), strong anion exchange (SAX/Q10), or mild cationic (CM10), can give wide proteome coverage in all microorganisms. ProteinChip arrays are created employing various chemical characteristics of the surface (Shah et al., 2010). Biological materials, like cell lysates, extracts, or bodily fluids, are applied to the ProteinChip Array, allowing proteins to attach to the surface depending on chromatographic characteristics or specifically tailored biological affinity. The ProteinChip Reader and SELDI-TOF MS are used to analyze and identify proteins that remained on the template surface, unbound molecules have been flushed away. The resulting MS spectra are analyzed employing alternative protein mapping techniques, which compare the relative expression levels of distinct molecular weights using statistical and bioinformatics methodologies (Reddy and Dalmasso 2003).
Rajakarun's research employing the CM10 ProteinChip Array achieved a broader spectrum of S. aureus isolates (Rajakaruna 2010). This was further supported by the research of Shah et al., who distinguished accurately between S. aureus strains with varying levels of methicillin resistance using the SELDI-TOF MS method and CM10 (Shah et al., 2011). A hydrophobic reversed-phase H50 surface was employed by Schmid et al. to recognize the gonorrhea-causing Neisseria gonorrhoeae. SELDI-TOF MS may be able to identify minor changes in the protein level between strains, as per initial research on N. gonorrhoeae strains that indicated tiny differences in mass spectral patterns (Schmid et al., 2005).
Similarly, a microchannel silicon nanowire microfluidic chip was used to capture bacteria in urine samples followed by MALDI-TOF/MS screening. Bacteria can be identified by their method without a culture with a concentration of 106 CFU mL−1 under optimum conditions, they identified with low as 103 CFU mL−1 concentration of bacteria incubated for 4 h (Li et al., 2021). Direct microbial identification was achieved by sample harvesting in 1 mL of fastidious anaerobe broth (FAB) followed by centrifugation and removal of the supernatant. The pellet was smeared on the target plate followed by 1 µL of 70 % formic acid, organic matrix, and MALDI analysis. An overall sensitivity of 70.4 % was achieved when comparing MALDI-TOF/MS and routine procedures (Jaworski et al.,).
3.1.8 Identification of UTIs microbes
The high-throughput screening was carried out for uropathogen in innate urine samples using MALDI-TOF/TOF tandem mass spectrometry. The results obtained from the direct approach were reliable at the genus level for single microbial samples and also suitable for clinical settings with single-organism infectious etiologies (Oros et al., 2020). Direct identification of carbapenemase-producing Enterobacteriaceae spp. in urine samples was achieved by MALDI-TOF/MS in 90 min. The assay reliably identified 91 % of the samples with 100 % sensitivity (Oviaño et al., 2017). Identification of UTI-causing microbes by the direct screening of urine specimens was also reported by using MALDI-TOF/MS. A total of 307 out of 1638 bacterial species were identified and the most dominating pathogens were E. coli (43.23 %), K. pneumoniae (15.28 %), and Enterococcus spp. (13.97 %) (Lee et al., 2019). Identification of UTI microbes directly in urine samples collected in 2015–2017 was also successfully achieved by using MALDI-MS (Kitagawa et al., 2018).
Apart from biological sample analysis, a variety of Staphylococcus species including S. aureus the dominant species (79.1 %) followed by Staphylococcus (S.) warneri (12.5 %) and Staphylococcus (S.) haemolyticus (8.3 %), respectively, isolated from mobile phones were identified using MALDI-TOF/MS (Noumi et al., 2020). Similarly, discrimination of Clostridium spp. by analyzing 123 strains using MALDI-TOF/MS with a multivariate statistical analysis method has been conducted (Schaumann et al., 2018). The discovery of specific biomarker peaks for discriminating and identification of Clostridioides (C.) difficile genotype ST37 based on MALDI-TOF/MS has been described. A set of specific peaks at m/z 3,242 and 3286 appeared to be specific for C. difficile genotype ST37 and can be distinguished from non-ST37 genotypes (Li et al., 2018).
3.1.9 Artificial intelligence with MALDI
MALDI-TOF MS has changed the face of microbiology by making it possible to identify species with incredible accuracy and speed. The use of machine learning has been increasing to improve species identification and fast antimicrobial resistance determination. Machine learning techniques have recently been applied to extract as much useful information as possible from MALDI-TOF MS (De Bruyne et al., 2011, Fangous et al., 2014). Machine learning techniques are capable of identifying statistical relationships in the data while also taking non-linear interactions and feature interactions into account. Thus, using machine learning approaches, new or hidden information that is present in MALDI-TOF mass spectra can be uncovered. This knowledge has been valuable for identifying and distinguishing species, especially those that are phylogenetically close to one another and sub-lineages of those species (Florio et al., 2018, Weis et al., 2020).
Recently developed ML models (e.g., SVM, RF, and ANN) enable fast classification of group B. streptococcus serotypes (Wang et al., 2019), distinguish between E. coli and Shigella species (Ling et al., 2019), types of Staphylococcus haemolyticus strains (Chung et al., 2019), and distinguish between Clostridium (Schaumann et al., 2018), and Klebsiella species (Rodrigues et al., 2018). Moreover, Desaire and Hua, used a machine learning method designed for glycomics and glycoproteomics data classification to accurately identify between closely related bacteria using MALDI-TOF MS. The authors claimed that, on average, the model performed better than previous standards (Desaire and Hua 2019).
In addition, (Tomachewski et al., 2018), introduced a protein-based bacterial classifying method. To do this, it uses a library of more than 28,500 bacterial taxonomic records to compare m/z data from MALDI-TOF MS analysis to ML models. The m/z values of 13 r-proteins from 116 bacterial strains were analyzed, and the results showed an accuracy of 95.7 %. In addition, successful systems utilizing a combination of MALDI-TOF MS data and ML algorithms have been reported for the detection of extended-spectrum beta-lactamase-producing E. coli strains (Sousa et al., 2020), rapid detection of cfiA metallo-b-lactamase-producing B. fragilis strains (Ho et al., 2017), and identification of fluconazole resistance in C. albicans strains (Delavy et al., 2020).
3.1.10 Alternate approaches
An alternate procedure for identifying pathogens is to use signals conserved from certain proteins present in microbial cells. Due to their abundance, high degree of conservation, and chromosomal gene encoding, ribosomal proteins are one of the finest biomarkers (Ziegler et al., 2015). Although extremely stable, interspecies and inter-strain variances in microbes can be exploited for typing and subtyping. In MALDI-TOF MS, reference databases including predicted masses of microbial ribosomal subunits determined directly from genome sequences became an alternative to pattern-based identification of bacteria. PAPMIDTM (Mabritec AG, Riehen, Switzerland) was constructed as a database of probable protein weights for identification, which was proved to support reference databases like SARAMISTM (Mabritec, Riehen, Switzerland) (Ziegler et al., 2015). Suarez et al. classified Neisseria meningitidis strain based on ribosomal signals into six groupings related to sequence types (Suarez et al., 2013), S. agalactiae (Rothen et al., 2019), and E. coli (Matsumura et al., 2014), complexes were effectively separated into subspecies using this method in the MALDI-TOF MS study. Toh et al. successfully employed this procedure to distinguish between Acinetobacter haemolyticus and Acinetobacter genomic species, including 13BJ/14 T strains (Toh et al., 2015). Additionally, the MALDI technology also allows microbial identification based on the profiling of other biological molecules like lipids or nucleic acids. There is now a significant surge in interest in the use of microbial lipid profiling for taxonomic classification (Gidden et al., 2009). It allowed the discrimination of Bacillus and Brevibacillus, with an average correct identification rate of 62.23 % (AlMasoud et al., 2016). Similarly, the Bacillus spp., discrimination based on conventional procedures is very difficult in comparison with lipid fingerprinting (Shu et al., 2012).
3.1.11 Yeast identification
Over the past decade, substantial technological advancements have been made in the field of clinical mycology. With MALDI-TOF MS, identifying yeasts and molds down to the species level is not only possible but also extremely feasible, even within species complexes and cryptic organisms. When these systems are widely used in diagnostic settings, they will likely improve patient outcomes by allowing for earlier diagnosis and treatment. MALDI-TOF MS is ideally suited for low-resource situations due to its ease of use and cheap consumables (Sow et al., 2015). Pathogenic Candida species can be difficult to identify based on growth and biochemical reactions alone, hence MALDI is employed commonly to distinguish between them (Santos et al., 2011). MALDI-TOF MS can be used to differentiate between species within the Candida complexes such as the C. parapsilosis complex, the C. glabrata complex, and the C. haemulonis complex, depending on the available database. New taxonomy allows us to differentiate between species within the C. neoformans and C. gattii complexes (Walsh and McCarthy 2019). It is possible to identify species from a single CFU using MALDI-TOF MS.
3.2 Other mass spectrometry techniques
In the last few years’ progress has been made in testing other proteome and MS-based techniques such as liquid chromatography-mass spectrometry (LC-MS), tandem mass spectrometry (MS/MS), Gas chromatography-mass spectrometry (GC–MS), etc. for the identification of pathogenic microorganisms.
3.2.1 Identification based on LC-MS analysis
LC-MS with electrospray ionization is one of the important techniques for the analysis of microbial proteome and metabolome. Direct identification of pathogenic microorganisms based on multiple discriminatory peptides was performed by developing an LC-MS/MS-based scheme. The method was found feasible for the identification of Bacillus (B.) anthracis, Brucella (B.) abortus, Bacillus (B.) melitensis, Bacillus (B.) suis, Bacillus (B.) pseudomallei, Burkholderia (B.) mallei, Francisella (F.) tularensis, and Yersinia (Y.) pestis directly from positive blood culture flasks (Berendsen et al., 2020). An LC-MS/MS shotgun proteomics method was developed for 33 aerobic cultures, with 100 % microorganisms while in 28 anaerobic cultures 96 % of microorganisms were accurately identified (Berendsen et al., 2017). Direct detection of extended-spectrum beta-lactamases (CTX-M) in positive BCs using a saponin extraction workflow followed by an LC-MS/MS bottom-up proteomics approach was also established. It was applied to BBs containing E. coli and K. pneumoniae. The proteome analysis identified 95 % ESβLs CTX-M of the isolates directly from BCs (Fleurbaaij et al., 2017).
The use and discovery of biomarkers are of great importance in the diagnosis of infectious illnesses and clinical applications. Therefore, an LC-MS/MS in combination with a machine learning approach has been applied to 15 UTIs pathogens. Peptide libraries were obtained from pure bacterial colonies in data-dependent analysis (DDA) mode followed by verification by data-independent analysis (DIA) mode in urine samples. Machine learning classifiers (NaiveBayes, BayesNet, and Hoeffding tree) were used to express a peptide marker to discriminate from each other in less than 4 h (Roux-Dalvai et al., 2019). The use of LC-MS/MS-based proteotyping for S. aureus, Moraxella (M.) catarrhalis, Haemophilus (H.) influenza, and Streptococcus (S.) pneumoniae which are commonly responsible for RTIs was also reported. Species-unique peptides were initially found on pure cultures of reference strains in the discovery phase, followed by spiking negative samples in the qualification phase while positive samples were analyzed to find species-unique peptides in the verification phase. Positive samples were analyzed by using the targeted-MS method for the selected peptide (Karlsson et al., 2020).
The early identification of VAP-causing bacteria i.e., Acinetobacter (A.) baumannii, E. coli, S. aureus, S. pneumoniae, Pseudomonas (P.) aeruginosa, and H. influenza by targeted bottom-up proteomics approach was also established. Strain-specific peptide identification was performed in DDA mode using LC-ESI-Q-TOF-MS (Bardet et al., 2021). An LC-MS-based bottom-up approach and in silico peptide mass data have been used for microbial identification. The MS data were tested against the in-house build library which was calculated from the UniProt Knowledgebase, Swiss-Prot, and TrEMBL databases. Identification was carried out from the calculation of spectral distances between instrumental and in silico peptide mass data (Lasch et al., 2020). The identification of pathogenic microorganisms from public libraries was achieved by using a proteomic approach. A total of 42 collected samples were grown in Luria Broth (LB) medium in the presence of ampicillin or kanamycin followed by trypsin digestion and analysis by LC-ESI-MS/MS. Identification at the species level can be done by species-unique peptides with a Python-based script which allows the detection of such unique peptides (Jung et al., 2019).
3.2.2 Identification based on GC–MS analysis
Microorganisms in a variety of conditions whether it competes with other microbes or lack sufficient nutrients, produce compounds or metabolites like toxins or antibiotics generally known as secondary metabolites. However, it also produces some low molecular weight compounds known as volatile organic compounds. Therefore, the detection of these metabolites is important in the identification or characterization of the microbes.
Drees., et al. reported early and fast pathogen differentiation from BCs using gas chromatography-ion mobility spectrometry (GC-IMS) analysis. Samples of E. coli (DSM 25944), S. aureus (DSM 13661), and P. aeruginosa (DSM 1117) cultures were incubated for 8 h and the point of differentiation was determined. The differentiation was based on the intensities of the detected signals in the investigated species were found to be possible by performing the principal component analysis (PCA) (Drees et al., 2019).
Similarly, S. aureus, E. coli, and Candida (C.) albicans, responsible for respiratory tract infections (RTIs) were identified from their VOCs profile by using GC–MS coupled to solid-phase micro-extraction (SPME) fiber. The utmost common volatile compound formed by E. coli was indole, S. aureus produced 2,3-pentandione by, cis-dihydro-α-terpinyl acetate, 1-decyne, 1,3-heptadiene, 2,5-dimethyl pyrazine, ethyl butanoate, and cyclohexene,4-ethenyl while, C. albicans major compounds were alcohols (Karami et al., 2017). Similarly, S. aureus, Vibrio (V.) parahaemolyticus, and Shigella (S.) sonnei were also identified based on their metabolite profiling by SPME coupled with GC–MS analysis. A total of 32 VOCs including 17 for S. aureus, 13 for V. parahaemolyticus, and 14 for S. sonnei were identified (Wang et al., 2018). The studies suggest that the screening of microbial VOC can be helpful in the diagnosis and identification of microorganisms by GC–MS analysis.
3.3 FT-IR spectroscopy
The infra-red (IR) region of the electromagnetic spectrum is consisting of three regions near-, mid, and far-IR. The mid-IR region (400–4000 cm-1) is the most frequently utilized region for the acquisition of bacterial analysis. The principle of this technique for analyses of different samples is that when radiations of IR are conceded through a sample (e.g., bacteria cell) it causes excitation and vibration of different functional groups thus; characteristic spectral peaks originate on IR spectra. All bacterial species have a complex cell arrangement specific to a particular strain and present a specific pattern of fingerprint on the FT-IR spectrum (Davis and Mauer 2010). Five spectral windows have been reported to correspond to absorption expressed in wavenumbers, including, the spectral region of 3000–2800 cm−1 is commonly dominated by fatty acids-related compounds, the spectral region of 1700–1500 cm−1 by carbonyl residual proteins, the carbonyl group of the peptide at about 1650 and 1500 cm−1 for C⚌O and —C—O— respectively, and polysaccharides and free amino acids in the region of (1450–1400 cm_1). The window 900–700 cm−1 is referred to as the fingerprint region and contains information significant to strain-specific discrimination (Maity et al., 2013). Today time-domain spectroscopy is used and radiant power data is recorded as a function of time which is achieved by Fourier Transform (FT) (Baravkar et al., 2011).
FT-IR spectroscopy has already been successfully applied for rapid microbial identification. Many studies have reported its application in combination with artificial intelligence (AI) systems, such as artificial neural networks (ANN), which is a powerful tool in microbial diagnostics.
3.3.1 Identification based on various algorithms
Many studies have been conducted for microbial identification based on various algorithms. In one of the studies, four gram-negative bacilli, P. aeruginosa, K. pneumoniae, Enterococcus (E.) cloacae, and A. baumannii, were discriminated by IR-biotyper within 3hr. The congruence of IR spectral clusters was compared with two reference methods, multilocus sequence Typing (MLST) and (Pulsed-field gel electrophoresis) PFGE. It was found that FT-IR spectroscopy correctly clustered P. aeruginosa, K. pneumoniae, and E. cloacae isolates, belonging to the same Sequence Type (ST) (Martak et al., 2019).
The discriminatory power of FT-IR spectroscopy was evaluated as a fast technique for typing K. pneumoniae clinical isolates, and compared to whole-genome-sequencing (WGS). An average linkage algorithm was used to generate clusters for FT-IR spectral data showing that the similarity of Klebsiella strains can be quickly calculated by FT-IR spectroscopy with high resolution that displays high congruence with WGS typing (Dinkelacker et al., 2018). Automated analysis of microbial FT-IR spectra which identify the spectral components that were determined by the strain genotype and not by culture conditions has been reported. The algorithm has also been tried out on the clinical isolates of S. aureus against several bacterial isolates causing infection, including, E. faecalis, Enterococcus (E.) faecium, K. pneumoniae, E. coli, Serratia (S.) marcescens, E. cloacae, A. baumannii, P. aeruginosa, Staphylococcus (S.) epidermidis, and C. albicans cultured in different media for diverse times, and found reliably discriminated from rest of the bacterial isolates (Suntsova et al., 2018). FT-IR spectroscopy with multivariate analysis was used for the discrimination of clinically relevant serogroups, sub-serogroups, and serotypes of non-typhoid Salmonella. The serogroups determination is based on the polysaccharide’s composition of O-antigen. Sharp differences were reported in the polysaccharide region in the spectra which were used in subsequent salmonella typing. Salmonella enterica isolates belonging to Sero-groups (B, C, D, and E) were discriminated against with high accuracy (Campos et al., 2018).
3.3.2 Identification based on artificial neural Network (ANN)
FT-IR hyperspectral imaging combined with ANN-based image segmentation was used for the identification of Gram-positive and Gram-negative bacteria through FT-IR micro spectroscopic imaging. Spectral data were resolved with the help of supervised modular ANN classifiers for hyperspectral image segmentation. The resultant segmentation maps suggest a taxonomic determination below the species level (Lasch et al., 2018).
Cordovana., et al. reported the identification of biochemically verified Salmonella isolates associated with typhoid and paratyphoid fever based on FT-IR Biotyper. Isolates of Salmonella (S.) Typhi, Salmonella (S.) paratyphi A, Salmonella (S.) paratyphi B, Salmonella (S.) paratyphi C, and other phylogenetically closely related Salmonella serovars from serogroup O:2, O:4, O:7, and O:9 was analyzed based on each O-serogroups. ANN was used to build the classifiers to differentiate between typhoidal and non-typhoidal serovars within each of the four serogroups. The correctness of the classifiers was 99.9 %, 87.0 %, 99.5 %, and 99.0 % for S. Typhi, S. Paratyphi A, B, and C, respectively (Cordovana et al., 2021), ANN-assisted FT-IR spectroscopy-based rapid identification of the Bacillus (B.) cereus group by performing multivariate data analysis using the deep learning toolbox of MATLAB to construct an ANN allowing the differentiation of B. cereus group members. The model resultedin being 100 % correct for the identification of the training set and 95.5 % for overall identification (Bagcioglu et al., 2019). FT-IR in combination with ANN was performed on clinical isolates of the E. cloacae complex. This study reports the development of ANN that was trained to recognize if the two isolates belong to the same serotype (ST) assuming the differences between two IR spectra (Vogt et al., 2019).
3.4 Raman spectroscopy
Raman spectroscopy (RS) is a non-invasive optical technique that was first discovered in 1928. It works on the principle of the excitation of electron clouds for scattering using a non-ionizing laser. When the vibrational state of the molecule changes, energy transfers either from the molecule to the photon or from the photon to the molecule, and is called Raman scattering. (Tu and Chang 2012, Bumbrah and Sharma 2016).
Studies revealed that Surface Enhanced Raman spectroscopy (SERS) emerged as a potentially powerful technique for pathogenic bacterial detection (Hong et al., 2018, Kirchhoff et al., 2018, Novelli-Rousseau et al., 2018). It is also advantageous because it consents to the identification of pathogens as well as AST in a similar assay in just less than 3 h. SERS identification of specific biomarkers helps to determine MICs and AST of bacterial species (Athamneh et al., 2014, Liu et al., 2017, Puttaswamy et al., 2018).
3.4.1 Identification based on algorithms
Identification of microorganisms has been achieved using various Raman spectroscopy-based approaches including some deep learning procedures and mathematical algorithms.
A Raman dataset for common pharmaceutical microorganisms was used as a deep learning strategy known as convolution neural network (CNN) to classify bacterial contaminations. The successful classification of different samples containing individual bacteria and bacteria mixed with Chinese Hamster Ovary (CHO) cells with 95 %–100 % accuracy (Maruthamuthu et al., 2020), a database of Raman bacterial spectra applying deep learning analysis for the identification of commonly occurring pathogenic bacteria. The proposed method is culture-free, and label-free, based on single-cell analysis for the phenotypic identification of bacterial strains (Ho et al., 2019). A study included 115 different bacterial strains and the obtained spectra were evaluated by one-way analysis of variance and differentiated peaks representing the different biochemical compositions. Spectral differences were identified with 89.5 % accuracy by studying Raman spectra of nine bacterial strains of P. aeruginosa, Enterococcus spp., S. aureus, E. cloacae, Morganella (M.) morganii, K. pneumoniae, E. coli, Listeria (L.) monocytogenes, Proteus (P.) mirabilis species (Oliveira et al.,).
3.4.2 Identification based on Surface-Enhanced Raman spectroscopy (SERS)
SERS with some modifications in nano-composites along with mathematical algorithms has been used for the identification of various microbial strains.
A SERS method was reported for the identification and discrimination of bacterial strains using SERS substrate of uncoated spherical gold nanoparticles (AuNPs) with PCA and partial least square analysis (PSA) (Akanny et al., 2020). The screening of Salmonella (S.) typhimurium based on a novel three-dimensional DNA walker method on gold-surfaced magnetic nanoparticles has also been reported (Yang et al., 2021). A spectroscopic database based on spectral signatures by typing several Mycobacterium species, E. coli, Bacillis (S.) subtilis, K. pneumoniae, and many other bacterial species was developed. The spectral signatures of live and dead bacteria were differentiated from spectra of treated and untreated mycobacteria (Kumar et al., 2020).
3.4.3 Identification based on isotope labeling
Few tag-free and isotope labeling methods along with algorithms have also been reported for the identification of microorganisms using Raman spectroscopy.
The identification of pathogenic E. coli in less than 1 h by using gold-enabled substrate in SERS scattering to evaluate the spectra of quinolone-resistant E. coli isolated strains by processing spectral data with the help of PCA along with the selected multi-support vector machine (SVM) algorithm (Kim et al., 2019). The quantitative differentiation of bacteria labeled with varying concentrations of 12C/13C-glucose and 14N/15N-ammonium chloride has been achieved using SERS involving in situ synthesis of silver nanoparticles, along with multivariate chemometrics of the resultant SERS spectra (Chisanga et al., 2017). A drop-coating deposition surface-enhanced Raman scattering (DCD-SERS) procedure coupled with a multivariate statistical method for the identification of quinolone-resistant K. pneumoniae has also been reported (Cheong et al., 2017). A SERS approach for the label-free detection of pathogenic bacteria based on specific DNA aptamer binding with customized silver nanoparticles has also been reported (Chen et al., 2017, Gao et al., 2017).
3.5 Nuclear magnetic resonance (NMR) spectroscopy
Nuclear magnetic resonance (NMR) spectroscopy is a dominant technique widely used for the structural elucidation of unknown molecules or compounds that are isotopically enriched.
The current status of NMR for studying pathogenic microorganisms is not so impressive. However, it has been used in the last decades for the identification of microbes and AST. Therefore, some examples from the past are given below to make a clear idea about the potential of NMR for microbial identification.
The first use of 1H NMR for microbial identification was reported in 1995. The identification of S. aureus, Staphylococcus (S.) epidermidis, E. faecalis, Streptococcus (S.) pneumoniae, Streptococcus (S.) pyogenes, Streptococcus (S.) agalactiae, and the Streptococcus (S.) milleri was achieved by Bourne et al., 2001, (Bourne et al., 2001) by combining proton magnetic resonance spectroscopy (1H MRS) with multivariate statistical analysis. Isolates were identified based on consistent high-probability classification of spectra from duplicate cultures with 92 % of agreement with conventional identification methods.
In the same year, Ohara. et al. evaluated 50 strains of methicillin-resistant S. aureus from 42 patients using 1H NMR, and DNA fingerprints were acquired by pulsed-field gel electrophoresis (PFGE). Each strain was found to have 8 to 9 specific resonance features in the spectral region of 0.5–4.5 ppm but with different intensities among the strains (Ohara et al., 2001).
The applications of 1H NMR in the diagnosis of UTIs have been reported by Gupta et al. In the first study, the potential of 1H NMR for the detection of P. aeruginosa in UTIs. The diagnosis of P. aeruginosa was achieved from its particular property of metabolizing nicotinic acid to 6-hydroxynicotinic acid (6-OHNA). The produced quantity of 6-OHNA is associated with the viable bacterial count while other UTIs-causing bacteria do not possess this property (Gupta et al., 2005). Similarly, 1H NMR was utilized for diagnosing K. pneumoniae in UTIs from its specific property to metabolize glycerol to 1,3-propanediol (1,3-PD), acetate, ethanol, and succinate. The amount of 1,3-PD correlates with K. pneumoniae counts while the rest of the UTI cause bacteria to lack this property (Gupta et al., 2006). Furthermore, 1H NMR was utilized for the qualitative and quantitative diagnosis of E. coli, P. aeruginosa, K. pneumoniae, and Proteus (P.) mirabilis in urine samples suspected of UTIs. For the quantification of P. aeruginosa, NA was added to the suspension while glycerol, lactose, and methionine were added to K. pneumoniae, E. coli, and P. mirabilis suspensions, respectively. From 1H NMR spectra, it was found that P. aeruginosa only metabolizes NA while the rest of the others do not metabolize. Similarly, K. pneumoniae particularly metabolize glycerol into 1,3-propanediol. E. coli produce lactate through lactose metabolism while P. mirabilis metabolizes methionine to 4–methylthio–2–oxobutyric acid (MOBA) while the rest of the other UTIs bacteria do not metabolize (Gupta et al., 2009).
Moreover, 1H NMR was utilized to quantify acetate, lactate, ethanol, succinate, citrate, creatinine, glycine, trimethylamine, trimethylamin-N-oxide, urea, and hippurate as the metabolic products for the identification of E. coli, P. aeruginosa, K. pneumoniae, Enterobacter ssp., Acinetobacter spp., P. mirabilis, Citrobacter (S.) frundii and gram-positive E. faecalis, Bacillus spp., and Staphylococcus (S.) saprophyticus in urine samples infected with UTI. Univariate and Multivariate discriminant function analysis (DFA) was also performed to describe the key biomarkers that separate the infected group from the control group. DFA revealed that acetate, lactate, succinate, and formate were able to discriminate healthy controls from UTI patients with 99.5 % accuracy (Gupta et al., 2012b).
For the last few years, there is a brake to the application of NMR in microbiology from the research community related to microbial identification. It might be due to the high per-sample cost of the technique or some other limitations of it. Anyhow, NMR is a powerful technique having the capabilities for broad applications in microbiological fields, novel, and advanced studies might be expected in near future.
4 General perspectives
The review has been conducted to explore the applications of advanced spectroscopic techniques i.e., MS, FT-IR, RS, and NMR in the field of clinical microbiology. All these techniques (excluding NMR because of no recent studies in the last 5 years) have been extensively used for the development of new methodologies related to bacterial identification. Fig. 2 shows the number of publications in the last five years i.e., 2017–2021. The figure clearly shows that MS tools and RS are widely used for microbial identification followed by FT-IR and NMR spectroscopy. Similarly, Fig. 3 represents the number of studies conducted in the last five years against bacterial identification through advanced spectroscopy techniques. It reveals that about 21 % of studies have been conducted only for E. coli, 14 % for S. aureus, 11 % for K. pneumoniae, and 37 % for other microorganisms. The conducted data provides insight into the scope and importance of advanced spectroscopic techniques for microbial identification for better health and research outcomes.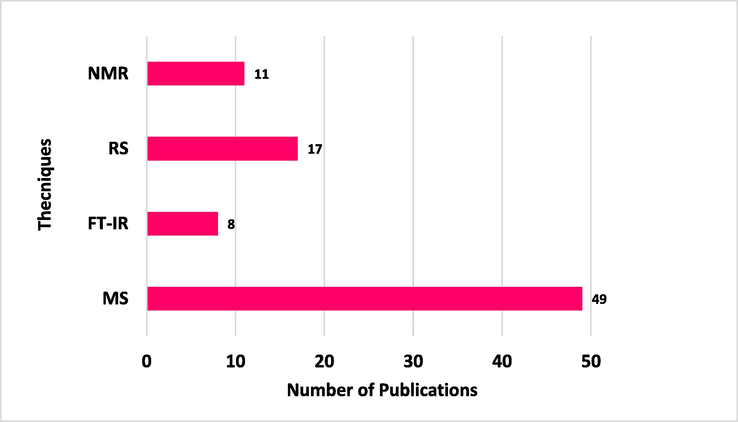
Number of publications based on advanced spectroscopic techniques related to microbial identification in the last five years.
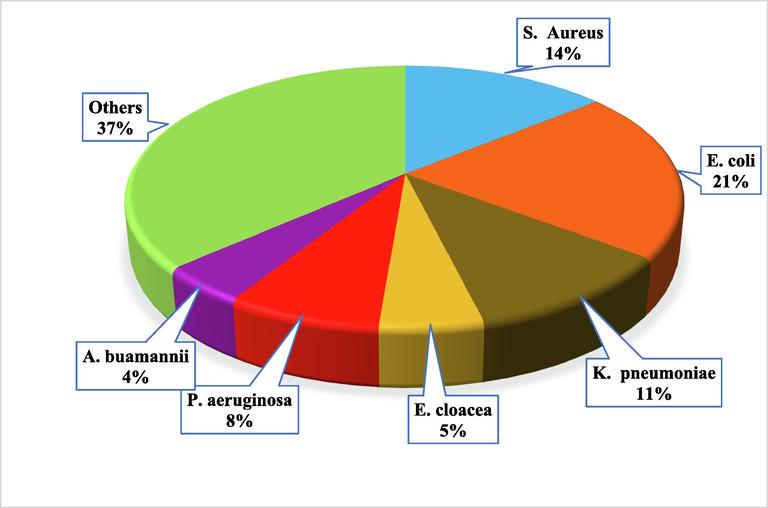
Number of studies in the last five years related to individual microorganisms based on advanced spectroscopic techniques.
It is well understood that MALDI-TOF/MS has a lot of contributions in the field but, it has also some limitations in comparison to its counterparts. Sometimes issues occur due to the use of a laser and a chemical matrix (organic acid) associated with it. In comparison, ESI-MS is a technique that ionizes samples at high atmospheric pressure and analyzes it in a liquid state irrespective of the laser as same as in MALDI-TOF/MS. Due to this feature of ESI-MS, it has a broad range of applications concerning microbial identification (Smith et al., 1995, Vaidyanathan and Goodacre 2006, Sampath et al., 2007). LC-MS usually has lower temperatures required regardless of the sample volatility as in the case of GC–MS. It is used for microbial identification in clinical diagnosis due to its simple sample preparations, and lower cost, and has also been applied for the determination of whole metabolome coverage of Saccharomyces (S.) cerevisiae, and the detection of commercially available metabolites of the in silico metabolome of Bacillus (B.) subtilis, and E. coli (Bakhtiar et al., 2002). GC–MS offers better separation, sensitivity, robustness, simple handling with lower cost, and numerous linearity range with access to commercial and, public libraries and generally has been used for the analysis of non-polar compounds. (Christie 1998, Dettmer et al., 2007, Garcia et al., 2008, Franco-Duarte et al., 2019). GC–MS demands sample volatility as most of the compounds are not volatiles and thus need complex and time-consuming derivatization steps.
FT-IR allows a cost-effective depiction of the complex biological system that comprises undefiled cells, tissues, and even whole-model organisms (Ami et al., 2012). The basic applications of the FT-IR are associated with the possibility to screen many samples at the same time, do not require cell lysis, being environment-friendly, and executing high-throughput screening and monitoring real-time processes (Kosa et al., 2017). RS is differentiable from other techniques due to its low cost, fast screening, and extensive report about the chemical composition, structures, and interactions of metabolites in microbes (Walsh et al., 2011, Stöckel et al., 2016). Vibrational spectroscopy differentiates microbes based on their biochemical composition; it is very advantageous for discriminating between slight differences among the same species. Both IR and Raman techniques are forms of vibrational spectroscopy and can offer “whole organism fingerprinting (Lu et al., 2011). In comparison to other methods, NMR can be used in a congenital mode. It has low sensitivity and a lower limit of detection (if the concentration is lower than 103 CFU/mL) some metabolites could not be detected and thus can give false-negative results (Gupta et al., 2012a). However, these limitations are adjusted by their quantitative nature (Pan and Raftery 2007). The application of NMR is its non-destructive nature which means that the same sample can be used several times if needed. Furthermore, the NMR tube can be used as an incubator for the development of AST and the monitoring of living system processes (García-Álvarez et al., 2015).
5 Future challenges and perspective
In the future, there will be not only a need for rapid diagnoses and standardization of testing but also for the detection of new pathogens and the development of diagnostic tests for new diseases that have a high social impact, as was the case with the outbreak of severe acute respiratory syndrome (SARS) and more recently COVID-19. Scientific responses to new emerging threats are more rapid than administrative responses, and there are often prolonged delays in the approval of new diagnostic tests for use outside research laboratories.
For rapid microbial identification, culture time is a rate-limiting step, reducing this time for microbial identification will be a major challenge in the future. Other challenges for the future are the desire to have even more rapid analysis by removing the need to enrich and isolate the bacteria before analysis. Advances in instrumentation and methods will support the finding of accurate means of microbial identification.
Mass spectrometry technology has improved to a point where it can make improvements in bacterial identification. In the future, the identification of microbial agents for the diagnosis of the disease can be achieved by searching the recorded MS data for the particular peptide that is a marker of a specific disease strain from the positive culture, as an alternative to today’s laboratory diagnostic tests. Fully automated sample preparation and dedicated software for microbial data analysis have to be developed before spectroscopy-based diagnostics can be applied as routine analysis in clinical microbiology. IR spectroscopy and Raman spectroscopy in parallel with multivariate chemometrics may play a crucial role in rapid and sensitive screening. NMR might also play a vital role if sensitivity issues are resolved without exceeding the overall instrumental costs.
The global exchange of laboratory data should be promoted which could significantly assist infection control, guide therapy, characterize resistance epidemiology, identify errors in laboratory testing, and promote collaboration in surveillance activities through the data exchange.
6 Conclusion
Advanced spectroscopic techniques have emerged as rapid tools for microbial identification in the last decades. Despite the limitations of some techniques, great attention has been made to exploring the application of advanced spectroscopy techniques in microbiological fields. MS techniques particularly, MALDI-TOF/MS and LC-MS have shown to be convenient, rapid, and simple techniques for microbial identification. However, it is used for pure samples, since the complex samples may promote background interference. Vibrational spectroscopy techniques i.e., FT-IR and RS with fast analysis, and simple sample preparation procedures are used. Although, NMR with a lower limit of detection has been used for bacterial identification and the low sensitivity issues might be resolved in the future. In today’s growing world of microbial infections and resistance to antibiotics, high-throughput advanced spectroscopy tools will be the right options for developing new methodologies for accurate and rapid microbial identification.
CRediT authorship contribution statement
Muhammad Ramzan: Conceptualization, Writing – review & editing. Ali Raza: Writing – review & editing. Zaib un Nisa: Writing – review & editing. Syed Ghulam Musharraf: Conceptualization.
Acknowledgment
The authors would like to acknowledge Higher Education Commission (HEC) Pakistan, for their financial support to Z.N under the HEC Indigenous Fellowship program (520-164404-2PS6-82), and PSF project No. PSF/Res/S-ICCBS/Chem (601).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Surface-enhanced Raman spectroscopy using uncoated gold nanoparticles for bacteria discrimination. J. Raman Spectrosc.. 2020;51:619-629.
- [Google Scholar]
- Comparison of matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry platforms for the identification of Gram-negative rods from patients with cystic fibrosis. J. Clin. Microbiol.. 2013;51:3852-3854.
- [Google Scholar]
- Classification of Bacillus and Brevibacillus species using rapid analysis of lipids by mass spectrometry. Anal. Bioanal. Chem.. 2016;408:7865-7878.
- [Google Scholar]
- Ami, D., Natalello, A., Doglia, S.M., 2012. Fourier transform infrared microspectroscopy of complex biological systems: from intact cells to whole organisms. Intrinsically disordered protein analysis, Springer: 85-100.
- Anderson, M., C. Clift, K. Schulze, et al., 2019. Averting the AMR crisis: What are the avenues for policy action for countries in Europe?
- Applications of copolymer for rapid identification of bacteria in blood culture broths using matrix-assisted laser desorption ionization time-of-flight mass spectrometry. J. Microbiol. Methods. 2017;139:54-60.
- [Google Scholar]
- Main strategies, analytical trends and challenges in LC-MS and ambient mass spectrometry–based metabolomics. TrAC Trends Anal. Chem.. 2018;108:278-295.
- [Google Scholar]
- Phenotypic profiling of antibiotic response signatures in Escherichia coli using Raman spectroscopy. Antimicrob. Agents Chemother.. 2014;58:1302-1314.
- [Google Scholar]
- Cheap and rapid in-house method for direct identification of positive blood cultures by MALDI-TOF MS technology. BMC Infect. Dis.. 2019;19:1-7.
- [Google Scholar]
- Detection and Identification of Bacillus cereus, Bacillus cytotoxicus, Bacillus thuringiensis, Bacillus mycoides and Bacillus weihenstephanensis via Machine Learning Based FTIR Spectroscopy. Front Microbiol.. 2019;10:902.
- [CrossRef] [Google Scholar]
- High-throughput mass spectrometric analysis of xenobiotics in biological fluids. J. Liq. Chromatogr. Relat. Technol.. 2002;25:507-540.
- [Google Scholar]
- FT-IR spectroscopy: principle, technique and mathematics. Int J Pharm. Bio. Sci. 2011;2:513-519.
- [Google Scholar]
- Early and specific targeted mass spectrometry-based identification of bacteria in endotracheal aspirates of patients suspected with ventilator-associated pneumonia. Eur. J. Clin. Microbiol. Infect. Dis.. 2021;40:1291-1301.
- [Google Scholar]
- Human health risk assessment of antibiotic resistance associated with antibiotic residues in the environment: a review. Environ. Res.. 2019;169:483-493.
- [Google Scholar]
- Identification of microorganisms grown in blood culture flasks using liquid chromatography–tandem mass spectrometry. Future Microbiol.. 2017;12:1135-1145.
- [Google Scholar]
- Untargeted accurate identification of highly pathogenic bacteria directly from blood culture flasks. Int. J. Med. Microbiol.. 2020;310:151376
- [Google Scholar]
- Global phenotypic characterization of bacteria. FEMS Microbiol. Rev.. 2008;33:191-205.
- [Google Scholar]
- Identification of Enterococcus, Streptococcus, and Staphylococcus by multivariate analysis of proton magnetic resonance spectroscopic data from plate cultures. J. Clin. Microbiol.. 2001;39:2916-2923.
- [Google Scholar]
- Raman spectroscopy–Basic principle, instrumentation and selected applications for the characterization of drugs of abuse. Egypt. J. Forensic Sci.. 2016;6:209-215.
- [Google Scholar]
- Review of mid-infrared fourier transform-infrared spectroscopy applications for bacterial detection. J. Rapid Methods Autom. Microbiol.. 2007;15:146-175.
- [CrossRef] [Google Scholar]
- A new approach to identifying pathogens, with particular regard to viruses, based on capillary electrophoresis and other analytical techniques. TrAC Trends Anal. Chem.. 2021;139:116250
- [Google Scholar]
- Discrimination of non-typhoid Salmonella serogroups and serotypes by Fourier Transform Infrared Spectroscopy: a comprehensive analysis. Int J Food Microbiol.. 2018;285:34-41.
- [CrossRef] [Google Scholar]
- Systems for identification of bacteria and fungi. Manual of Clinical Microbiology. 2015:29-43.
- [Google Scholar]
- Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect. Dis.. 2019;19:56-66.
- [Google Scholar]
- Microbiological identification by surface-enhanced Raman spectroscopy. Appl. Spectrosc. Rev.. 2017;52:123-144.
- [Google Scholar]
- Exploration of fungal metabolic interactions using imaging mass spectrometry on nanostructured silicon. J. Nat. Prod.. 2018;81:1527-1533.
- [Google Scholar]
- Label-free SERS detection of salmonella typhimurium on DNA aptamer modified AgNR substrates. J. Food Meas. Charact.. 2017;11:1773-1779.
- [Google Scholar]
- Rapid label-free identification of Klebsiella pneumoniae antibiotic resistant strains by the drop-coating deposition surface-enhanced Raman scattering method. Spectrochim. Acta A Mol. Biomol. Spectrosc.. 2017;183:53-59.
- [Google Scholar]
- Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J. Clin. Microbiol.. 2010;48:1169-1175.
- [Google Scholar]
- Quantitative detection of isotopically enriched E. coli cells by SERS. Faraday Discuss.. 2017;205:331-343.
- [Google Scholar]
- Gas chromatography-mass spectrometry methods for structural analysis of fatty acids. Lipids. 1998;33:343-353.
- [Google Scholar]
- Nanoparticle-based laser desorption/ionization mass spectrometric analysis of drugs and metabolites. J. Food Drug Anal.. 2018;26:1215-1228.
- [Google Scholar]
- Incorporating statistical test and machine intelligence into strain typing of Staphylococcus haemolyticus based on matrix-assisted laser desorption ionization-time of flight mass spectrometry. Front. Microbiol.. 2019;10:2120.
- [Google Scholar]
- The rapid identification of intact microorganisms using mass spectrometry. Nat. Biotechnol.. 1996;14:1584-1586.
- [Google Scholar]
- Classification of Salmonella enterica of the (Para-)Typhoid Fever Group by Fourier-Transform Infrared (FTIR) Spectroscopy. Microorganisms.. 2021;9:853.
- [CrossRef] [Google Scholar]
- Rapid identification of microorganisms from positive blood culture by MALDI-TOF MS after short-term incubation on solid medium. Curr. Microbiol.. 2017;74:97-102.
- [Google Scholar]
- Antimicrobial resistance: implications and costs. Infection Drug Resistance. 2019;12:3903.
- [Google Scholar]
- Fourier transform infrared (FT-IR) spectroscopy: a rapid tool for detection and analysis of foodborne pathogenic bacteria. Curr. Res. Technol. Education Topics Appl. Microbiol. Microbial Biotechnol.. 2010;2:1582-1594.
- [Google Scholar]
- Bacterial species identification from MALDI-TOF mass spectra through data analysis and machine learning. Syst. Appl. Microbiol.. 2011;34:20-29.
- [Google Scholar]
- Machine learning approach for Candida albicans fluconazole resistance detection using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Front. Microbiol.. 2020;10:3000.
- [Google Scholar]
- Rapid identification of common human pathogens by high-resolution proton magnetic resonance spectroscopy. J. Clin. Microbiol.. 1995;33:1258-1262.
- [Google Scholar]
- Adaption of the Aristotle classifier for accurately identifying highly similar bacteria analyzed by MALDI-TOF MS. Anal. Chem.. 2019;92:1050-1057.
- [Google Scholar]
- Improvement of a rapid direct blood culture microbial identification protocol using MALDI-TOF MS and performance comparison with SepsiTyper kit. J. Microbiol. Methods. 2018;155:1-7.
- [Google Scholar]
- Typing and species identification of clinical klebsiella isolates by fourier transform infrared spectroscopy and matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol.. 2018;56
- [CrossRef] [Google Scholar]
- GC-IMS headspace analyses allow early recognition of bacterial growth and rapid pathogen differentiation in standard blood cultures. Appl. Microbiol. Biotechnol.. 2019;103:9091-9101.
- [Google Scholar]
- Evaluation of sample preparation methods for MALDI-TOF MS identification of highly dangerous bacteria. Lett. Appl. Microbiol.. 2012;55:40-46.
- [Google Scholar]
- Advantages and limitations of Raman spectroscopy for molecular diagnostics: an update. Expert Rev. Mol. Diagn.. 2015;15:773-787.
- [Google Scholar]
- Classification algorithm for subspecies identification within the Mycobacterium abscessus species, based on matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol.. 2014;52:3362-3369.
- [Google Scholar]
- Direct detection of extended-spectrum beta-lactamases (CTX-M) from blood cultures by LC-MS/MS bottom-up proteomics. Eur. J. Clin. Microbiol. Infect. Dis.. 2017;36:1621-1628.
- [Google Scholar]
- Recent advances and ongoing challenges in the diagnosis of microbial infections by MALDI-TOF mass spectrometry. Front. Microbiol.. 2018;9:1097.
- [Google Scholar]
- Foundation, A. t. M., 2018. Antimicrobial resistance benchmark 2018.
- Advances in chemical and biological methods to identify microorganisms—from past to present. Microorganisms.. 2019;7:130.
- [Google Scholar]
- Phylogenetic classification and identification of bacteria by mass spectrometry. Nat. Protoc.. 2009;4:732-742.
- [Google Scholar]
- MALDI mass spectrometry in medical research and diagnostic routine laboratories. Int. J. Mass Spectrom.. 2017;416:96-109.
- [Google Scholar]
- Surface-Enhanced Raman Scattering for Rapid Detection and Characterization of Antibiotic-Resistant Bacteria. Adv. Healthc. Mater.. 2018;7:1701335.
- [Google Scholar]
- Negative ion production from peptides and proteins by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Communications in Mass Spectrometry: Int. J. Devoted Rapid Dissemination Up-to-the-Minute Res. Mass Spectrometry.. 2008;22:4066-4072.
- [Google Scholar]
- Intuitive label-free SERS detection of bacteria using aptamer-based in situ silver nanoparticles synthesis. Anal. Chem.. 2017;89:9836-9842.
- [Google Scholar]
- Separation and mass spectrometry in microbial metabolomics. Curr. Opin. Microbiol.. 2008;11:233-239.
- [Google Scholar]
- Proton nuclear magnetic resonance spectroscopy as a technique for gentamicin drug susceptibility studies with Escherichia coli ATCC 25922. J. Clin. Microbiol.. 2015;53:2433-2438.
- [Google Scholar]
- Proton nuclear magnetic resonance for antimicrobial drug susceptibility studies: why has progress been slow? Future Medicine.. 2019;14:1175-1177.
- [Google Scholar]
- Lipid compositions in Escherichia coli and Bacillus subtilis during growth as determined by MALDI-TOF and TOF/TOF mass spectrometry. Int. J. Mass Spectrom.. 2009;283:178-184.
- [Google Scholar]
- Overview of mass spectrometry-based metabolomics: opportunities and challenges. Mass Spectrometry in Metabolomics. 2014:3-12.
- [Google Scholar]
- Pathogen proteotyping: a rapidly developing application of mass spectrometry to address clinical concerns. Clin. Mass Spectrometry. 2019;14:9-17.
- [Google Scholar]
- 1H NMR spectroscopy in the diagnosis of Klebsiella pneumoniae‐induced urinary tract infection. NMR in Biomedicine: An International Journal Devoted to the Development and Application of Magnetic Resonance In vivo. 2006;19(8):1055-1061.
- [Google Scholar]
- 1H-nuclear magnetic resonance spectroscopy for identifying and quantifying common uropathogens: a metabolic approach to the urinary tract infection. BJU Int.. 2009;104:236-244.
- [Google Scholar]
- Metabolomics of urinary tract infection: a new uroscope in town. Expert Rev. Mol. Diagn.. 2012;12:361-369.
- [Google Scholar]
- Broad identification of bacterial type in urinary tract infection using 1 H NMR spectroscopy. J. Proteome Res.. 2012;11:1844-1854.
- [Google Scholar]
- 1H NMR spectroscopy in the diagnosis of Pseudomonas aeruginosa‐induced urinary tract infection. NMR in Biomedicine. 2005;18(5):293-299.
- [Google Scholar]
- Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning. Nat. Commun.. 2019;10:1-8.
- [Google Scholar]
- Rapid detection of cfiA metallo-β-lactamase-producing Bacteroides fragilis by the combination of MALDI-TOF MS and CarbaNP. J. Clin. Pathol.. 2017;70:868-873.
- [Google Scholar]
- Hofer, U., 2019. The cost of antimicrobial resistance. Nature Reviews Microbiology. 17, 3-3.
- Rapid identification of intact whole bacteria based on spectral patterns using matrix-assisted laser desorption/ionization with time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom.. 1996;10:1227-1232.
- [Google Scholar]
- Antibiotic susceptibility determination within one cell cycle at single-bacterium level by stimulated Raman metabolic imaging. Anal. Chem.. 2018;90:3737-3743.
- [Google Scholar]
- A resonance Raman method for the rapid detection and identification of bacteria in water. Appl. Spectrosc.. 1980;34:72-75.
- [Google Scholar]
- Highly efficient classification and identification of human pathogenic bacteria by MALDI-TOF MS. Mol. Cell. Proteomics. 2008;7:448-456.
- [Google Scholar]
- Evaluation of the IR Biotyper for Klebsiella pneumoniae typing and its potentials in hospital hygiene management. J. Microbial. Biotechnol.. 2021;14:1343-1352.
- [Google Scholar]
- Establishment and application of an analytical in-house database (IHDB) for rapid discrimination of Bacillus subtilis group (BSG) using whole-cell MALDI-TOF MS technology. Mol. Cell. Probes. 2016;30:312-319.
- [Google Scholar]
- Evaluation of an in-house MALDI-TOF MS rapid diagnostic method for direct identification of micro-organisms from blood cultures. J. Med. Microbiol.. 2019;68:41-47.
- [Google Scholar]
- How to accelerate antimicrobial susceptibility testing. Clin. Microbiol. Infect.. 2019;25:1347-1355.
- [Google Scholar]
- Rapid identification of microorganisms from positive blood cultures by MALDI-TOF mass spectrometry subsequent to very short-term incubation on solid medium. Clin. Microbiol. Infect.. 2014;20:1001-1006.
- [Google Scholar]
- Rapid and robust MALDI-TOF MS techniques for microbial identification: a brief overview of their diverse applications. J. Microbiol.. 2018;56:209-216.
- [Google Scholar]
- “Omic” Approaches to Bacteria and Antibiotic Resistance Identification. Int. J. Mol. Sci.. 2022;23:9601.
- [Google Scholar]
- Discrimination of bacteria using surface-enhanced Raman spectroscopy. Anal. Chem.. 2004;76:40-47.
- [Google Scholar]
- Jaworski, M., J. Haiko and B. Saeedi, Rapid identification of bacterial species directly from enrichment broth by MALDI-TOF mass spectrometry. Biomed. J. 1, 4.
- Identification of pathogenic bacteria from public libraries via proteomics analysis. Int. J. Environ. Res. Public Health. 2019;16:912.
- [Google Scholar]
- Progress of electrospray ionization and rapid evaporative ionization mass spectrometric techniques for the broad-range identification of microorganisms. Analyst. 2019;144:1073-1103.
- [Google Scholar]
- Initial study of three different pathogenic microorganisms by gas chromatography-mass spectrometry. F1000Research. 2017;6
- [Google Scholar]
- Influence of the wavelength in high-irradiance ultraviolet laser desorption mass spectrometry of organic molecules. Anal. Chem.. 1985;57:2935-2939.
- [Google Scholar]
- Discovery of species-unique peptide biomarkers of bacterial pathogens by tandem mass spectrometry-based proteotyping. Mol. Cell. Proteomics. 2020;19:518-528.
- [Google Scholar]
- Evaluation of two matrix-assisted laser desorption ionization–time of flight mass spectrometry systems for identification of viridans group streptococci. Eur. J. Clin. Microbiol. Infect. Dis.. 2014;33:779-788.
- [Google Scholar]
- Comparison of rapid BACpro® II, Sepsityper® kit and in-house preparation methods for direct identification of bacteria from blood cultures by MALDI-TOF MS with and without Sepsityper® module analysis. Eur. J. Clin. Microbiol. Infect. Dis.. 2019;38:2133-2143.
- [Google Scholar]
- A rapid tag-free identification of Escherichia coli antibiotic-resistant isolates using Raman scattering. Anal. Methods. 2019;11:5381-5387.
- [Google Scholar]
- Simple ciprofloxacin resistance test and determination of minimal inhibitory concentration within 2 h using Raman spectroscopy. Analytical Chem.. 2018;90:1811-1818.
- [Google Scholar]
- Improved bacterial identification directly from urine samples with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Clin. Lab. Anal.. 2018;32:e22301.
- [Google Scholar]
- MALDI-TOF mass spectrometry following short incubation on a solid medium is a valuable tool for rapid pathogen identification from positive blood cultures. Int. J. Med. Microbiol.. 2015;305:469-479.
- [Google Scholar]
- FTIR spectroscopy as a unified method for simultaneous analysis of intra-and extracellular metabolites in high-throughput screening of microbial bioprocesses. Microb. Cell Fact.. 2017;16:1-11.
- [Google Scholar]
- Rapid detection of bacterial infection and viability assessment with high specificity and sensitivity using Raman microspectroscopy. Anal. Bioanal. Chem.. 2020;412:2505-2516.
- [Google Scholar]
- FT-IR hyperspectral imaging and artificial neural network analysis for identification of pathogenic bacteria. Anal Chem.. 2018;90:8896-8904.
- [CrossRef] [Google Scholar]
- Identification of microorganisms by liquid chromatography-mass spectrometry (LC-MS1) and in silico peptide mass libraries. Mol. Cell. Proteomics. 2020;19:2125-2139.
- [Google Scholar]
- Recent advances in SALDI-MS techniques and their chemical and bioanalytical applications. Anal. Bioanal. Chem.. 2011;399:2597-2622.
- [Google Scholar]
- An integrated microfluidic system for antimicrobial susceptibility testing with antibiotic combination. Lab Chip. 2019;19:2699-2708.
- [Google Scholar]
- Comparison of the Bruker Biotyper and VITEK MS matrix-assisted laser desorption/ionization time-of-flight mass spectrometry systems using a formic acid extraction method to identify common and uncommon yeast isolates. Ann. Lab. Med.. 2017;37:223.
- [Google Scholar]
- Recent advances in the race to design a rapid diagnostic test for antimicrobial resistance. ACS Sensors. 2018;3:2202-2217.
- [Google Scholar]
- A side by side comparison of Bruker Biotyper and VITEK MS: utility of MALDI-TOF MS technology for microorganism identification in a public health reference laboratory. PLoS One. 2015;10:e0144878.
- [Google Scholar]
- Comparison of Saramis 4.12 and IVD 3.0 Vitek MS matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of mycobacteria from solid and liquid culture media. J. Clin. Microbiol.. 2017;55:2045-2054.
- [Google Scholar]
- Capture and detection of urine bacteria using a microchannel silicon nanowire microfluidic chip coupled with MALDI-TOF MS. Analyst. 2021;146:1151-1156.
- [Google Scholar]
- Identification and characterization of Clostridium difficile sequence type 37 genotype by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol.. 2018;56:e01990-e11917.
- [Google Scholar]
- A novel short-term high-lactose culture approach combined with a matrix-assisted laser desorption ionization-time of flight mass spectrometry assay for differentiating Escherichia coli and Shigella species using artificial neural networks. PLoS One. 2019;14:e0222636.
- [Google Scholar]
- Label and label-free based surface-enhanced Raman scattering for pathogen bacteria detection: A review. Biosens. Bioelectron.. 2017;94:131-140.
- [Google Scholar]
- Application of mid-infrared and Raman spectroscopy to the study of bacteria. Food Bioproc. Tech.. 2011;4:919-935.
- [Google Scholar]
- Comparative evaluation of software for deconvolution of metabolomics data based on GC-TOF-MS. TrAC Trends Anal. Chem.. 2008;27:215-227.
- [Google Scholar]
- Discrimination of Francisella tularensis subspecies using surface enhanced laser desorption ionization mass spectrometry and multivariate data analysis. FEMS Microbiol. Lett.. 2005;243:303-310.
- [Google Scholar]
- Same-day identification and antibiotic susceptibility testing on positive blood cultures: a simple and inexpensive procedure. Eur. J. Clin. Microbiol. Infect. Dis.. 2017;36:681-687.
- [Google Scholar]
- Identification and discrimination of bacteria using Fourier transform infrared spectroscopy. Spectrochim Acta A Mol Biomol Spectrosc.. 2013;116:478-484.
- [CrossRef] [Google Scholar]
- Comparative evaluation of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry systems for identification of yeasts of medical importance. J. Clin. Microbiol.. 2013;51:2453-2457.
- [Google Scholar]
- Evaluation of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization–time of flight mass spectrometry systems for identification of nonfermenting Gram-negative bacilli isolated from cultures from cystic fibrosis patients. J. Clin. Microbiol.. 2012;50:2034-2039.
- [Google Scholar]
- Fourier-Transform infrared spectroscopy can quickly type gram-negative bacilli responsible for hospital outbreaks. Front Microbiol.. 2019;10:1440.
- [CrossRef] [Google Scholar]
- Comparison of an in-house method and the commercial Sepsityper™ kit for bacterial identification directly from positive blood culture broths by matrix-assisted laser desorption-ionisation time-of-flight mass spectrometry. Eur. J. Clin. Microbiol. Infect. Dis.. 2012;31:2269-2281.
- [Google Scholar]
- Raman spectra-based deep learning: a tool to identify microbial contamination. MicrobiologyOpen.. 2020;9:e1122.
- [Google Scholar]
- Comparison of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization–time of flight mass spectrometry systems for identification of mycobacteria using simplified protein extraction protocols. J. Clin. Microbiol.. 2014;52:130-138.
- [Google Scholar]
- Detection of extended-spectrum-β-lactamase-producing Escherichia coli ST131 and ST405 clonal groups by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol.. 2014;52:1034-1040.
- [Google Scholar]
- Identification and antibiotic-susceptibility profiling of infectious bacterial agents: a review of current and future trends. Biotechnol. J.. 2019;14:1700750.
- [Google Scholar]
- Rapid identification of pathogens in positive blood culture of patients with sepsis: review and meta-analysis of the performance of the sepsityper kit. Int. J. Microbiol. 2015
- [Google Scholar]
- Mass spectrometry-based microbiological testing for blood stream infection. Clin. Proteomics. 2020;17:1-11.
- [Google Scholar]
- Phenotypic and genotypic characterization with MALDI-TOF-MS based identification of Staphylococcus spp. isolated from Mobile phones with their antibiotic susceptibility, biofilm formation, and adhesion properties. Int. J. Environ. Res. Public Health. 2020;17:3761.
- [Google Scholar]
- Culture-free antibiotic-susceptibility determination from single-bacterium Raman spectra. Sci. Rep.. 2018;8:1-12.
- [Google Scholar]
- Analysis of methicillin-resistant Staphylococcus aureus isolates by proton magnetic resonance spectroscopy. J. Infect.. 2001;43:116-121.
- [Google Scholar]
- Oliveira, F.S.d.S.E., da Silva, A.M., Pacheco, M.T.T., et al., Biochemical characterization of pathogenic bacterial species using Raman spectroscopy and discrimination model based on selected spectral features. Lasers in medical science.
- online:, V. M. b. A., http://www.biomerieux-diagnostics.com/sites/clinic/files/9300819-002-gb-a_vitek-ms.pdf.
- Identification of pathogens from native urine samples by MALDI-TOF/TOF tandem mass spectrometry. Clin. Proteomics. 2020;17:1-9.
- [Google Scholar]
- Rapid direct detection of carbapenemase-producing Enterobacteriaceae in clinical urine samples by MALDI-TOF MS analysis. J. Antimicrob. Chemother.. 2017;72:1350-1354.
- [Google Scholar]
- Oviaño, M., A. Ingebretsen, A. K. Steffensen, et al., 2021. Evaluation of the rapidBACpro® II kit for the rapid identification of microorganisms directly from blood cultures using MALDI-TOF MS. bioRxiv.
- Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Anal. Bioanal. Chem.. 2007;387:525-527.
- [Google Scholar]
- Current and emerging technologies for rapid detection and characterization of Salmonella in poultry and poultry products. Food Microbiol.. 2014;38:250-262.
- [Google Scholar]
- Evaluation of MALDI–TOF mass spectrometry in diagnostic and environmental surveillance of Legionella species: a comparison with culture and mip-gene sequencing technique. Front. Microbiol.. 2020;11:589369
- [Google Scholar]
- MALDI-TOF mass spectrometry in the clinical mycology laboratory: identification of fungi and beyond. Expert Rev. Proteomics. 2013;10:151-164.
- [Google Scholar]
- Fourier transform infrared (FT-IR) spectroscopy in bacteriology: towards a reference method for bacteria discrimination. Anal Bioanal Chem.. 2007;387:1739-1748.
- [CrossRef] [Google Scholar]
- Progress on the development of rapid methods for antimicrobial susceptibility testing. J. Antimicrob. Chemother.. 2013;68:2710-2717.
- [Google Scholar]
- A comprehensive review of the present and future antibiotic susceptibility testing (AST) systems. Arch Clin Microbiol.. 2018;9
- [Google Scholar]
- An overview of the evolution of infrared spectroscopy applied to bacterial typing. Biotechnol. J.. 2018;13:1700449.
- [Google Scholar]
- Rajakaruna, L.K., 2010. Proteomics as a Tool for the Characterisation of Nosocomial Pathogens, Nottingham Trent University (United Kingdom).
- Attributable deaths caused by infections with antibiotic-resistant bacteria in France. Lancet Infect. Dis.. 2019;19:128-129.
- [Google Scholar]
- SELDI ProteinChip (R) array technology: protein-based predictive medicine and drug discovery applications. J. Biomed. Biotechnol.. 2003;2003:237-241.
- [Google Scholar]
- Can the legacy of industrial pollution influence antimicrobial resistance in estuarine sediments? Environ. Chem. Lett.. 2019;17:595-607.
- [Google Scholar]
- Elucidating constraints for differentiation of major human Klebsiella pneumoniae clones using MALDI-TOF MS. Eur. J. Clin. Microbiol. Infect. Dis.. 2017;36:379-386.
- [Google Scholar]
- Identification of Klebsiella pneumoniae, Klebsiella quasipneumoniae, Klebsiella variicola and related phylogroups by MALDI-TOF mass spectrometry. Front. Microbiol.. 2018;9:3000.
- [Google Scholar]
- Evaluation of MALDI Biotyper Mycobacteria Library v3. 0 for identification of nontuberculous mycobacteria. J. Clin. Microbiol.. 2016;54:1144-1147.
- [Google Scholar]
- Rapid identification and detection of β-lactamase-producing Enterobacteriaceae from positive blood cultures by MALDI-TOF/MS. J. Global Antimicrobial Resistance. 2021;24:270-274.
- [Google Scholar]
- Mass spectrometry imaging on porous silicon: investigating the distribution of bioactives in marine mollusc tissues. Anal. Chem.. 2012;84:8996-9001.
- [Google Scholar]
- Subspecies typing of Streptococcus agalactiae based on ribosomal subunit protein mass variation by MALDI-TOF MS. Front. Microbiol.. 2019;10:471.
- [Google Scholar]
- Fast and accurate bacterial species identification in urine specimens using LC-MS/MS Mass Spectrometry and Machine Learning*[S] Mol. Cell. Proteomics. 2019;18:2492-2505.
- [Google Scholar]
- Safety and accuracy of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of highly pathogenic organisms. J. Clin. Microbiol.. 2017;55:3513-3529.
- [Google Scholar]
- Rapid direct identification of positive paediatric blood cultures by MALDI-TOF MS technology and its clinical impact in the paediatric hospital setting. BMC. Res. Notes. 2020;13:1-8.
- [Google Scholar]
- Rapid identification of emerging infectious agents using PCR and electrospray ionization mass spectrometry. Ann. N. Y. Acad. Sci.. 2007;1102:109.
- [Google Scholar]
- Mass spectrometry applications in microbiology beyond microbe identification: progress and potential. Expert Rev. Proteomics. 2016;13:965-977.
- [Google Scholar]
- Matrix-assisted laser desorption/ionization time-of-flight intact cell mass spectrometry to detect emerging pathogenic Candida species. Diagn. Microbiol. Infect. Dis.. 2011;71:304-308.
- [Google Scholar]
- Mass spectrometry tools for the classification and identification of bacteria. Nat. Rev. Microbiol.. 2010;8:74-82.
- [Google Scholar]
- Discrimination of human pathogen Clostridium species especially of the heterogeneous C. sporogenes and C. botulinum by MALDI-TOF mass spectrometry. Curr. Microbiol.. 2018;75:1506-1515.
- [Google Scholar]
- New approaches to identification of bacterial pathogens by surface enhanced laser desorption/ionization time of flight mass spectrometry in concert with artificial neural networks, with special reference to Neisseria gonorrhoeae. J. Med. Microbiol.. 2005;54:1205-1211.
- [Google Scholar]
- Evaluation of the Bruker MALDI Biotyper for identification of Gram-positive rods: development of a diagnostic algorithm for the clinical laboratory. J. Clin. Microbiol.. 2014;52:1089-1097.
- [Google Scholar]
- Optimized application of surface-enhanced laser desorption/ionization time-of-flight MS to differentiate Francisella tularensis at the level of subspecies and individual strains. FEMS Immunol. Med. Microbiol.. 2007;49:364-373.
- [Google Scholar]
- Changing concepts in the characterisation of microbes and the influence of mass spectrometry. Mass Spectrometry for Microbial Proteomics. 2010:1-34.
- [Google Scholar]
- Tracing the transition of methicillin resistance in sub-populations of Staphylococcus aureus, using SELDI-TOF mass spectrometry and artificial neural network analysis. Syst. Appl. Microbiol.. 2011;34:81-86.
- [Google Scholar]
- Antimicrobial resistance: its surveillance, impact, and alternative management strategies in dairy animals. Front. Veterinary Sci.. 2018;4:237.
- [Google Scholar]
- Lipid fingerprinting of Bacillus spp. using online MALDI-TOF mass spectrometry. Anal. Methods. 2012;4:3111-3117.
- [Google Scholar]
- Direct identification of 80 percent of bacteria from blood culture bottles by matrix-assisted laser desorption ionization–time of flight mass spectrometry using a 10-minute extraction protocol. J. Clin. Microbiol.. 2019;57:e01278-e11218.
- [Google Scholar]
- Characterization of bacterial phospholipids by electrospray ionization tandem mass spectrometry. Anal. Chem.. 1995;67:1824-1830.
- [Google Scholar]
- Desorption and ionization mechanisms and signal enhancement in surface assisted laser desorption ionization mass spectrometry (SALDI-MS) Appl. Spectrosc. Rev.. 2020;55:220-242.
- [Google Scholar]
- Putative protein biomarkers of Escherichia coli antibiotic multiresistance identified by MALDI mass spectrometry. Biology. 2020;9:56.
- [Google Scholar]
- Usefulness of MALDI-TOF mass spectrometry for routine identification of Candida species in a resource-poor setting. Mycopathologia. 2015;180:173-179.
- [Google Scholar]
- Imaging mass spectrometry for natural products discovery: a review of ionization methods. Nat. Prod. Rep.. 2020;37:150-162.
- [Google Scholar]
- The application of Raman spectroscopy for the detection and identification of microorganisms. J. Raman Spectrosc.. 2016;47:89-109.
- [Google Scholar]
- Ribosomal proteins as biomarkers for bacterial identification by mass spectrometry in the clinical microbiology laboratory. J. Microbiol. Methods. 2013;94:390-396.
- [Google Scholar]
- Releasing bacteria from functional magnetic beads is beneficial to MALDI-TOF MS based identification. Talanta. 2021;225:121968
- [Google Scholar]
- Identification of microorganisms by Fourier-transform infrared spectroscopy. Bull. Russian State Medical Univ. 2018:50-57.
- [CrossRef] [Google Scholar]
- Tanaka, K., H. Waki, Y. Ido, et al., Protein and Polymer Analyses up to m/z 100000 by Laser Ionization Time-of-flight Mass.
- Nanoparticle-assisted laser desorption/ionization mass spectrometry (Nano-PALDI MS) with Py-Tag for the analysis of small molecules. Mass Spectrometry.. 2017;6:S0069-S.
- [Google Scholar]
- Differentiation of Acinetobacter genomic species 13BJ/14TU from Acinetobacter haemolyticus by use of matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) J. Clin. Microbiol.. 2015;53:3384-3386.
- [Google Scholar]
- Ribopeaks: a web tool for bacterial classification through m/z data from ribosomal proteins. Bioinformatics. 2018;34:3058-3060.
- [Google Scholar]
- A simple shotgun proteomics method for rapid bacterial identification. J. Microbiol. Methods. 2013;94:54-57.
- [Google Scholar]
- Custom database development and biomarker discovery methods for MALDI-TOF mass spectrometry-based identification of high-consequence bacterial pathogens. J. Microbiol. Methods. 2017;134:54-57.
- [Google Scholar]
- An in-house centrifugation and membrane filtration technique for identifying microorganisms from positive blood culture bottles with high identification rates using matrix-assisted laser desorption ionization–Time-of-flight mass spectrometry: a preliminary report. J. Infect. Chemother.. 2020;26:266-271.
- [Google Scholar]
- Current status of matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (MALDI-TOF MS) in clinical diagnostic microbiology. Molecules. 2020;25:4775.
- [Google Scholar]
- Diagnostic applications of Raman spectroscopy. Nanomed.: Nanotechnol. Biol. Med.. 2012;8:545-558.
- [Google Scholar]
- High-throughput microbial characterizations using electrospray ionization mass spectrometry and its role in functional genomics. CHEMICAL ANALYSIS-NEW YORK-INTERSCIENCE THEN JOHN WILEY-.. 2006;169:229.
- [Google Scholar]
- Progress in proteomics for clinical microbiology: MALDI-TOF MS for microbial species identification and more. Expert Rev. Proteomics. 2015;12:595-605.
- [Google Scholar]
- Next-generation antimicrobial susceptibility testing. J. Clin. Microbiol.. 2013;51:2018-2024.
- [Google Scholar]
- Veloo, A., Elgersma, P., Friedrich, A., et al., 2014. The influence of incubation time, sample preparation and exposure to oxygen on the quality of the MALDI-TOF MS spectrum of anaerobic bacteria. Clinical Microbiology and Infection. 20, O1091-O1097.
- Fourier-Transform Infrared (FTIR) spectroscopy for typing of clinical enterobacter cloacae complex isolates. Front. Microbiol.. 2019;10:2582.
- [CrossRef] [Google Scholar]
- MALDI-TOF mass spectrometry technology for detecting biomarkers of antimicrobial resistance: current achievements and future perspectives. Annals Translational Medicine. 2018;6
- [Google Scholar]
- The expanding use of matrix-assisted laser desorption/ionization-time of flight mass spectroscopy in the diagnosis of patients with mycotic diseases. Expert Rev. Mol. Diagn.. 2019;19:241-248.
- [Google Scholar]
- Molecular detection and species-specific identification of medically important Aspergillus species by real-time PCR in experimental invasive pulmonary aspergillosis. J. Clin. Microbiol.. 2011;49:4150-4157.
- [Google Scholar]
- Rapid classification of group B Streptococcus serotypes based on matrix-assisted laser desorption ionization-time of flight mass spectrometry and machine learning techniques. BMC Bioinf.. 2019;20:1-17.
- [Google Scholar]
- Monolithic Gold Nanoparticles/Thiol-β-cyclodextrin-Functionalized TiO2 Nanowires for Enhanced SALDI MS detection and imaging of natural products. Anal. Chem.. 2021;94:952-959.
- [Google Scholar]
- Rapid identification of Staphylococcus aureus, Vibrio parahaemolyticus and Shigella sonnei in foods by solid phase microextraction coupled with gas chromatography–mass spectrometry. Food Chem.. 2018;262:7-13.
- [Google Scholar]
- Evaluation of three sample preparation methods for the identification of clinical strains by using two MALDI-TOF MS systems. J. Mass Spectrom.. 2021;56:e4696.
- [Google Scholar]
- Microbial identification and automated antibiotic susceptibility testing directly from positive blood cultures using MALDI-TOF MS and VITEK 2. Eur. J. Clin. Microbiol. Infect. Dis.. 2016;35:75-82.
- [Google Scholar]
- Machine learning for microbial identification and antimicrobial susceptibility testing on MALDI-TOF mass spectra: a systematic review. Clin. Microbiol. Infect.. 2020;26:1310-1317.
- [Google Scholar]
- Surface-enhanced Raman spectroscopy of microorganisms: limitations and applicability on the single-cell level. Analyst. 2019;144:943-953.
- [Google Scholar]
- The first fifty years of near-infrared spectroscopy in America. NIR news. 1991;2:4-5.
- [Google Scholar]
- Progress on the application of infrared spectroscopy in microbiology research. China Brewing. 2007;3
- [Google Scholar]
- A novel surface-enhanced Raman scattering (SERS) strategy for ultrasensitive detection of bacteria based on three-dimensional (3D) DNA walker. Biosens. Bioelectron.. 2021;172:112758
- [Google Scholar]
- Application of MALDI-TOF MS profiling coupled with functionalized magnetic enrichment for rapid identification of pathogens in a patient with open fracture. Front. Chem.. 2021;9:672744
- [Google Scholar]
- Evaluation of an optimized method to directly identify bacteria from positive blood cultures using MALDI-TOF mass spectrometry. J. Clin. Lab. Anal.. 2020;34:e23119.
- [Google Scholar]
- Zengin Canalp, H. and B. Bayraktar, 2021. Direct Rapid Identification from Positive Blood Cultures by MALDI-TOF MS: Specific Focus on Turnaround Times. Microbiology Spectrum. 9, e01103-01121.
- Ribosomal protein biomarkers provide root nodule bacterial identification by MALDI-TOF MS. Appl. Microbiol. Biotechnol.. 2015;99:5547-5562.
- [Google Scholar]







