Translate this page into:
Saponins from Aesculus wilsonii seeds exert anti-inflammatory activity through the suppression of NF-κB and NLRP3 pathway
⁎Corresponding authors. zhwwxzh@tjutcm.edu.cn (Yi Zhang), wangtao@tjutcm.edu.cn (Tao Wang)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
The seeds of Aesculus wilsonii were reported to be rich in saponins and have anti-inflammatory activity. However, the phytochemistry investigation of its saponins is not clear yet, and the bioactivity is also rare. Therefore, the detailed constituents' study on saponins from A. wilsonii seeds was performed, and eleven new isolates, aeswilsaponins IA–IH (1–8), ⅡA (9), ⅡB (10) and ⅢC (11), along with twenty reported analogs (12–31) were yielded. All of them were examined for their inhibitory effects on NO release in LPS-induced RAW264.7 cells, indicating that 1, 4, 5, 12, 18, 22–24, 26, and 27 had potential anti-inflammatory activity. Moreover, NF-κB activity and the expression of NF-κB-linked genes were all examined on LPS-stimulated RAW 264.7 cells. The results suggested the anti-inflammation mechanism was at least partially related to the inhibition of NF-κB activity and consequent inhibition of p-IKK-α/β/IKK-α/β, iNOS, COX-2, IL-6, and TNF-α by compounds 4 and 5, as well as the inhibition of p-IKK-α/β/IKK-α/β, COX-2, and TNF-α by compound 1. Furtherly, in the LPS/ATP-induced peritoneal macrophages (PMs) cell model, the three tested ones had the inhibitory tendency on the NLRP3 inflammasome priming and assembling genes, NLRP3, pro-IL-1β, ASC, cleaved IL-1β, and cleaved caspase-1. Among them, compounds 4 and 5 inhibited the expression level of p-IKK-α/β/IKKα/β and p-p65/p65, and 1 reduced the expression level of p-p65/p65, clarifying that they may inhibit inflammatory response through NF-κB/NLRP3 signaling pathway. This work will provide a potential molecular mechanism of ''Suo Luo Zi'' and its saponins and supply novel candidates for treating inflammation-related diseases.
Keywords
Aesculus wilsonii
Anti-inflammation
NF-κB
NLRP3 pathway
1 Introduction
It is well known that inflammation is a major factor in the progression of various chronic diseases or disorders, including cancer, cardiovascular diseases, arthritis, autoimmune diseases, and inflammatory bowel disease (IBD) (Arulselvan et al., 2016). Therefore, reducing inflammatory reactions is essential for both disease prevention and treatment.
A key player in controlling the proper immune response during homeostatic function is nuclear factor-B (NF-κB). It is closely associated with the occurrence and development of IBD (Chen et al., 2019), rheumatoid arthritis (Jing, et al., 2021), chronic obstructive pulmonary disease (Galvão et al., 2021), and other inflammation-related diseases. IκB kinase (IKK) complex, which is an important element in NF-κB activation, allows the induction of NF-κB transcription and translocation (Afonina et al., 2017). External stimuli, such as lipopolysaccharide (LPS), can start the NF-κB signaling cascade. Once IKK is active, NF-κB dimers transfer into nuclear and promote the activation of inflammatory genes including interleukin (IL)-6, tumor necrosis factor (TNF)-α, cyclooxygenase (COX)-2, and inducible nitric oxide synthase (iNOS) (Karin and Greten, 2005; Kim, et al., 2015). NF-κB pathway, as a paradigm of inflammatory signaling, is also implicated in regulating the priming process of the Nod-like receptor pyrin domain containing 3 (NLRP3) inflammasome (Afonina et al., 2017). Inflammasomes are large intracellular multiprotein complexes that play a central role in innate immunity. NLRP3 inflammasome is essential in innate immune responses to pathogen-associated molecular patterns (PAMPs) or danger-associated molecular patterns (DAMPs) (Meylan et al., 2006; Yang et al., 2019). Two consecutive phases, priming and assembly, are required for the full activation of the NLRP3 inflammasome, and these steps are driven by the two signals PAMPs and DAMPs. During the priming step, NLRP3 inflammasome responds to PAMPs such as LPS, thus activating NF-κB signaling and promoting the synthesis of NLRP3 and pro-IL-1β. Then, during the assembly stage, DAMPs such as adenosine triphosphate (ATP) stimulate NLRP3 to recruit the adaptor molecule ASC and the dormant zymogen pro-caspase-1 to create the inflammasome, causing pro-caspase-1 to undergo autoproteolytic cleavage into active cleaved-caspase-1 and pro-IL-1 to undergo biologically active forms (Kelley et al., 2019; Wang et al., 2018). When NLRP3 is dysregulated, it has been connected to systemic inflammatory disorders such as cryopyrin-associated periodic syndromes, Alzheimer's disease, and auto-inflammatory diseases (Perri, 2022). Thus, inhibiting the inflammation through regulating NF-κB, NLRP3, and their relevant proteins serves as a potential strategy for the treatment of inflammation-related diseases.
Searching for anti-inflammatory drugs from natural products has become a hot topic in recent years. Pharmacological investigation suggests that escin, a natural mixture of triterpene saponins, extracted from the seeds of the traditional Chinese medicine ''Suo Luo Zi'' (Hippocastanaceae family) has a variety of activities such as anti-inflammatory (Cheng et al., 2015), anti-edema (Wang, et al., 2011), anti-viral (Kim, et al., 2017), and anti-tumor (Yang et al., 2019). Due to its protective effect on many edema disorders through anti-inflammatory and anti-edematous effects, escin monosodium salt (sodium aescinate) has achieved considerable success in the therapeutic treatment of limb or tissue swelling and chronic venous dysfunction (Gallelli et al., 2019). Though the seeds of Aesculus wilsonian Rehd, one of the major sources of “Suo Luo Zi” were reported to be rich in saponins, most of the studies on its anti-inflammatory activity were just limited to escin and/or escin IA, escin IB, isoescin IA, as well as isoescin IB. More extensive research on its other type pharmacological substances urgently needs to be carried out.
In the present study, the detailed phytochemistry investigation on triterpene saponins from A. wilsonii seeds was performed to yield eleven new isolates (1–11) along with twenty reported analogues (12–31). The NO production inhibitory activities of all the obtained saponins were evaluated on an LPS-stimulated RAW264.7 cells model, to screen potential anti-inflammatory constituents. And then, new compounds with anti-inflammatory activity were reported to inhibit inflammation by regulating NF-κB and NLRP3 signaling pathways in vitro. The details of the isolation, structure characterization, and bioactivity assay of saponins 1–31 were described as follows.
2 Materials and methods
2.1 Materials and methods for phytochemistry research
2.1.1 General experimental procedures
Analytical HPLC was performed on a Waters e2695 system equipped with a 2998 PDA detector (Waters). preparative HPLC (pHPLC) was carried out on a Shimadzu LC-8A system equipped with an SPD-20A detector (Shimadzu). Optical rotations were provided by a Rudolph Autopol V automatic polarimeter. UV spectra were acquired on a Varian Cary 50 UV–Vis (Varian, Inc.). IR spectra were recorded on a Varian 640-IR FT-IR spectrophotometer (Varian, Inc.). NMR spectra were collected on Bruker Ascend 600 MHz NMR spectrometers (Bruker BioSpin AG). Mass spectra were measured on the negative ion mode on a Thermo ESI-Q-Orbitrap MS spectrometer connected to an UltiMate 3000 UHPLC instrument via ESI interface (Thermo).
Column chromatography (CC) was carried out on Macroporous resin D101 (Haiguang Chemical Co., Ltd.), silica gel (48–75 μm, Qingdao Haiyang Chemical Co., Ltd.), YMC*Gel ODS-A-HG (50 μm, AAG12S50, YMC Co., Ltd.), and Sephadex LH-20 (Ge Healthcare Bio-Sciences). Cosmosil 5C18-MS-II (4.6 mm i.d. × 250 mm, 5 µm, Nakalai Tesque, Inc.) and Cosmosil 5C18-MS-II (20 mm i.d. × 250 mm, 5 µm, Nakalai Tesque, Inc.) columns were used for analytical and preparative isolation, respectively. All reagents used for phytochemical investigation were of analytical grade (Concord Technology Co. Ltd.).
2.1.2 Plant material
The seeds of Aesculus wilsonii Rehd were purchased from the Anguo Chinese medicine market (Hebei Province, China) on September 10, 2019, and were identified by Professor Lin Ma (Tianjin University of Traditional Chinese Medicine). A voucher specimen is kept at the Academy of Traditional Chinese Medicine of Tianjin University of TCM (No. 2019091014).
2.1.3 Extraction and isolation
The dried seeds of A. wilsonii (15.0 kg) were extracted under reflux for three times (3, 2, and 2 h) with 75, 60, and 60 L 70% EtOH, respectively. A residue (2.6 kg) was provided after the removal of the solvent under reduced pressure. A portion of it (2.2 kg) was loaded onto a D101 resin column and eluted with H2O and 95% EtOH sequentially to give the H2O (717.8 g) and 95% EtOH (SA, 847.5 g) eluates, respectively.
SA (400.0 g) was subjected to silica gel CC [CH2Cl2-MeOH (100:0 → 100:1 → 100:3 → 100:7 → 10:1 → 8:1 → 7:1 → 5:1 → 3:1 → 1:1 → 0:100, v/v) ] to obtain SA 1–SA 14. SA 11 (30.0 g) was fractionated by MCI gel CHP 20P CC [MeOH-H2O (20:80 → 30:70 → 40:60 → 50:50 → 60:40 → 70:30 → 80:20 → 100:0, v/v)] to yield SA 11–1–SA 11–19. SA11-14 (800.0 mg) was purified by pHPLC [MeOH-1% HAc (50:50, v/v)], and (3β,16α,21β,22α)-16,21,22,24,28-pentahydroxy-olean-12-en-3-O-β-d-glucopyranosiduronic acid (12, 59.8 mg, tR 71.6 min) was given. SA11-15 (1000.0 mg) was isolated by pHPLC [MeOH-1% HAc (65:35, v/v)] to give SA 11–15-1–SA 11–15-8. SA 11–15-5 (48.3 mg) was purified by pHPLC [CH3CN-1% HAc (28:72, v/v)] to produce aeswilsaponin IE (5, 17.3 mg, tR 30.1 min). Using the same pHPLC condition, aeswilsaponin IB (2, 17.5 mg, tR 44.4 min) was obtained from SA 11–15-6 (145.6 mg). SA 11–15-7 (121.7 mg) and SA 11–15-8 (161.3 mg) were separated by pHPLC [CH3CN-1% HAc (34:66, v/v)], as a result, aeswilsaponin ID (4, 13.7 mg, tR 27.0 min) and aeswilsaponin IA (1, 11.9 mg, tR 27.2 min) were gained from them, respectively. SA 11–16 (6.0 g) was subjected to Sephadex LH-20 CC [CH2Cl2-MeOH (1:1, v/v)] to obtain SA 11–16-1–SA 11–16-5. SA 11–16-2 (3000.0 mg) was purified by pHPLC [CH3CN-1% HAc (40:60, v/v)] to produce SA 11–16-2–1–SA 11–16-2–12. Among them, SA 11–16-2–5 was identified as 6′-methyleter-O-aesculiside A (30, 64.2 mg, tR 18.8 min). SA 11–16-2–2 (41.5 mg) was prepared by pHPLC [MeOH-1% HAc (68:32, v/v)], and aeswilsaponin IG (7, 13.7 mg, tR 18.5 min) was yielded. SA 11–16-2–10 (223.3 mg) was purified by pHPLC [CH3CN-1% HAc (41.5:58.5, v/v)] to obtain 21β-O-tigloyl-22α-O-acetylprotoaescigenin-3β-O-[β-d-glucopyranosyl (1 → 2)]-β-d-glucopyranosiduronic acid (13, 42.8 mg, tR 35.0 min). SA 11–16-2–11 (297.2 mg) was isolated by pHPLC [MeOH-1% HAc (75:25, v/v)] to gain 21β-O-angeloyl-22α-O-acetylprotoaescigenin-3β-O-[β-d-glucopyranosyl(1 → 2)]-β-d-glucopyranosiduronic acid (14, 49.1 mg, tR 18.2 min). SA 13 (100.0 g) was subjected to MCI gel CHP 20P CC [MeOH-H2O (20:80 → 30:70 → 40:60 → 50:50 → 60:40 → 70:30 → 80:20 → 100:0, v/v)] to give SA 13–1–SA 13–16. SA 13–4 (2.3 g) was separated by ODS CC [MeOH-H2O (30:70 → 40:60 → 50:50 → 70:30 → 80:20 → 100:0, v/v) ] to yield SA 13–4-1–SA 13–4-6. SA 13–4-5 (287.9 mg) was prepared by pHPLC [MeOH-1% HAc (62:38, v/v)] to produce desacylescin I (16, 40.8 mg, tR 15.3 min). SA 13–4-6 (127.0 mg) was isolated by pHPLC [MeOH-1% HAc (62:38, v/v)] to gain SA 13–4-6–1–SA 13–4-6–3. SA 13–4-6–2 was identified as (3β,16α,21β,22α)-16,21,22,24,28-pentahydroxyolean-12-en-3-yl-O-[β-d-glycopyranosyl(1 → 4)]-β-d-glucopyranosiduronic acid (15, 31.6 mg, tR 19.8 min). SA 13–4-6–1 (16.1 mg) was purified by pHPLC [CH3CN-1% HAc (26:74, v/v)], and aeswilsaponin IC (3, 10.0 mg, tR 14.4 min) was given. SA 13–5 (4.3 g) was separated by ODS CC [MeOH-H2O (35:65 → 45:55 → 55:45 → 70:30 → 80:20 → 100:0, v/v)] to gain SA 13–5-1–SA 13–5-8. SA 13–5-7 (574.9 mg) was purified by pHPLC [MeOH-1% HAc (62:38, v/v)] to obtain aesculusoside A (20, 103.5 mg, tR 59.9 min). Aeswilsaponin IF (6, 6.4 mg, tR 31.7 min) and aesculusoside C (17, 9.0 mg, tR 36.3 min) were produced from SA 13–5-8 (133.4 mg) by using the same isolation method. SA 13–7 (2.5 g) was separated by pHPLC [CH3CN-1% HAc (34:66, v/v)] to give SA 13–7-1–SA 13–7-6. SA 13–7-2 (486.2 g) was further fractionated by pHPLC [CH3CN-1% HAc (28:72, v/v)] to get SA 13–7-2–1–SA 13–7-2–5. SA 13–7-5 (418.5 mg) was purified by pHPLC [CH3CN-1% HAc (32:68, v/v)] to produce aesculusoside C (17, 7.9 mg, tR 20.0 min) and escin Ⅳ (21, 136.6 mg, tR 31.0 min). SA 13–8 (2.5 g) was separated by using CH3CN-1% HAc (32:68, v/v) and MeOH-1% HAc (62:38, v/v) as the HPLC mobile phase, and aeswilsaponin IIB (10, 12.4 mg, tR 23.0 min) was given. SA 13–9 (3.6 g) was isolated by pHPLC [CH3CN-1% HAc (36:64, v/v), Venusil PrepG C18] to gain SA 13–9-1–SA 13–9-7. SA 13–9-5 was identified as aesculiside A (25, 910.5 mg, tR 30.0 min). SA 13–9-2 (1601.4 mg) was separated by pHPLC [CH3CN-1% HAc (36:64, v/v)] to produce SA 13–9-2–1–SA 13–9-2–9. Among them, SA 13–9-2–2 was elucidated to be aesculusoside B (28, 44.2 mg, tR 12.5 min). SA 13–9-2–3 (61.8 mg) was further purified by pHPLC [CH3CN-1% HAc (30:70, v/v)] to get aeswilsaponin IIIA (11, 21.1 mg, tR 29.5 min). SA 13–11 (3.0 g) was fractionated by pHPLC [CH3CN-1% HAc (40:60, v/v)] to give escin Ia (23, 282.7 mg, tR 36.3 min), escin Ib (24, 134.6 mg, tR 42.5 min), isoescin Ib (27, 20.3 mg, tR 56.5 min), olean-12-ene-16α,21β,22α,24,28-pentol,3β-[(O-β-d-glucopyranosyl(1 → 2)-O-[β-d-glucopyranosyl-(1 → 4)]-β-d-glucopyranuronosyl)oxy]-,methyl ester,22-acetate 21-((Z)-2-methylcrotonate) (29, 39.1 mg, tR 69.5 min), and aeswilsaponin IH (8, 17.9 mg, tR 82.1 min). SA 13–12 (18.0 g) was subjected to pHPLC [MeOH-1% HAc (75:25, v/v), Venusil PrepG C18] to produce SA 13–12-1–SA 13–12-13. SA 13–12-5 (1596.4 mg) was isolated by pHPLC [CH3CN-1% HAc (36:64, v/v)] to give escin V (22, 42.5 mg, tR 32.5 min) and escin Ia (23, 831.2 mg, tR 39.9 min). SA 13–14 (2.0 g) was separated by pHPLC [CH3CN-1% HAc (41:59, v/v)] to obtain SA 13–14-1–SA 13–14-10. SA 13–14-2 and SA 13–14-5 were identified as aesculioside B (19, 102.2 mg, tR 15.2 min) and isoescin Ia (26, 440.3 mg, tR 35.1 min), respectively. SA 13–14-1 (113.9 mg) was purified by pHPLC [MeOH-1% HAc (79:21, v/v)] to gain aesculioside A (18, 67.4 mg, tR 13.7 min). SA 13–14-9 (127.9 mg) was purified by pHPLC [CH3CN-1% HAc (40:60, v/v)] to yield aeswilsaponin IIA (9, 90.0 mg, tR 46.0 min). SA 13–15 (3.0 g) was separated by pHPLC [CH3CN-1% HAc (43:57, v/v)] and [MeOH-1% HAc (78:22, v/v)] to give aesculioside G (31, 31.3 mg, tR 30.6 min).
2.1.3.1 Aeswilsaponin IA (1)
White powder; [α]D25 –10.0 (c 0.10, MeOH); IR νmax (KBr) cm−1: 3371, 2943, 1721, 1616, 1454, 1421, 1374, 1257, 1162, 1106, 1072, 1027; 1H NMR (C5D5N, 600 MHz) δ: Table 1; 13C NMR (C5D5N, 150 MHz) δ: Table 2; ESI-Q-Orbitrap MS m/z 723.39764 [M − H]− (calcd for C38H59O13, 723.39502). o: overlapped.
No.
1
2
3
4
5
6
1
0.89 (m)
0.90 (m)
0.80 (m)
0.82 (m)
0.82 (m)
0.83 (m)
1.42 (m)
1.45 (m)
1.33 (m)
1.35 (m)
1.35 (m)
1.41 (m, o)
2
2.04 (m)
2.04 (m)
1.96 (m)
1.96 (m)
1.96 (m)
1.96 (m)
2.23 (m)
2.22 (m)
2.37 (m)
2.34 (m)
2.35 (m)
2.17 (m)
3
3.64 (dd, 5.4, 10.8)
3.62 (dd, 4.8, 10.8)
3.47 (dd, 3.0, 10.8)
3.48 (dd, 4.2, 11.4)
3.47 (dd, 4.0, 11.0)
3.54 (dd, 3.0, 10.2)
5
0.94 (br. d, ca. 12)
0.93 (m)
0.87 (m)
0.87 (br. d, ca. 11)
0.87 (br. d, ca. 12)
0.89 (m)
6
1.37 (m)
1.38 (m)
1.19 (m)
1.20 (m)
1.22 (m)
1.33 (m)
1.65 (m)
1.64 (m)
1.52 (m)
1.52 (m, o)
1.52 (m)
1.61 (m)
7
1.28 (m)
1.31 (m)
1.25 (m)
1.24 (m)
1.26 (m)
1.29 (m)
1.58 (m)
1.59 (m)
1.53 (m)
1.52 (m, o)
1.54 (m)
1.56 (m)
9
1.73 (m)
1.74 (m)
1.67 (m)
1.66 (m)
1.67 (m)
1.72 (m)
11
1.75 (m)
1.84 (m)
1.73 (m)
1.72 (t like, ca. 12)
1.79 (m)
1.82 (m)
1.89 (m, o)
1.92 (m)
1.86 (m)
1.85 (m)
1.85 (m)
1.91 (m)
12
5.38 (t like, ca. 3)
5.48 (t like, ca. 3)
5.38 (t like, ca. 3)
5.38 (t like, ca. 3)
5.45 (t like, ca. 3)
5.49 (m, o)
15
1.67 (br. d, ca. 11)
1.68 (br. d, ca. 15)
1.66 (m)
1.64 (br. d, ca. 15)
1.64 (br. d, ca. 15)
1.69 (br. d, ca. 15)
1.97 (dd, 3.0, 11.4)
1.94 (m)
2.06 (m)
1.95 (m)
1.92 (m)
1.93 (m)
16
4.89 (br. d, ca. 3)
4.83 (m)
5.05 (m)
4.87 (m, o)
4.81 (m)
4.82 (m)
18
2.95 (dd, 4.2, 13.2)
2.89 (dd, 3.6, 13.2)
2.82 (br. d, ca. 14)
2.91 (dd, 4.2, 13.8)
2.86 (dd, 3.0, 14.0)
2.89 (dd, 3.0, 13.2)
19
1.41 (dd, 4.2, 13.2)
1.43 (dd, 3.6, 13.2)
1.42 (m)
1.39 (dd, 4.2, 13.8)
1.42 (m)
1.42 (m)
3.11 (dd, 13.2, 13.2)
3.06 (dd, 13.2, 13.2)
3.05 (dd, 13.8, 13.8)
3.07 (dd, 13.8, 13.8)
3.03 (dd, 14.0, 14.0)
3.06 (dd, 13.2, 13.2)
21
6.43 (d, 10.2)
4.84 (d, 10.2)
4.84 (d, 9.6)
6.37 (d, 9.6)
4.79 (d, 9.0)
4.84 (d, 9.6)
22
4.84 (d, 10.2)
4.44 (d, 10.2)
4.68 (d, 9.6)
4.79 (d, 9.6)
4.40 (d, 9.0)
4.44 (d, 9.6)
23
1.57 (s)
1.55 (s)
1.36 (s)
1.37 (s)
1.36 (s)
1.50 (s)
24
3.67 (d, 11.4)
3.67 (d, 11.4)
3.36 (d, 10.2)
3.36 (d, 11.4)
3.36 (d, 11.5)
3.62 (d, 10.8)
4.41 (d, 11.4)
4.40 (d, 11.4)
4.34 (d, 10.2)
4.33 (d, 11.4)
4.33 (d, 11.5)
4.36 (d, 10.8)
25
0.79 (s)
0.81 (s)
0.67 (s)
0.68 (s)
0.69 (s)
0.77 (s)
26
0.84 (s)
0.90 (s)
0.83 (s)
0.80 (s)
0.95 (s)
0.97 (s)
27
1.89 (s)
1.90 (s)
1.87 (s)
1.82 (s)
1.84 (s)
1.89 (s)
28
3.69 (d, 10.2)
4.25 (d, 10.2)
3.72 (d, 10.2)
3.67 (d, 10.2)
4.24 (d, 10.5)
4.24 (d, 10.2)
3.98 (d, 10.2)
4.41 (d, 10.2)
4.03 (d, 10.2)
3.96 (d, 10.2)
4.38 (d, 10.5)
4.41 (d, 10.2)
29
1.13 (s)
1.36 (s)
1.35 (s)
1.10 (s)
1.34 (s)
1.36 (s)
30
1.32 (s)
1.41 (s)
1.41 (s)
1.29 (s)
1.39 (s)
1.41 (s)
1′
5.21 (d, 7.8)
5.19 (d, 7.8)
5.00 (d, 7.2)
4.99 (d, 7.8)
4.98 (d, 7.5)
5.08 (d, 7.2)
2′
4.18 (dd, 7.8, 8.4)
4.16 (dd, 7.8, 8.4)
4.28 (dd, 7.2, 8.4)
4.26 (dd, 7.8, 8.4)
4.24 (dd, 7.5, 8.5)
4.12 (dd, 7.2, 8.4)
3′
4.40 (dd, 8.4, 9.0)
4.39 (dd, 8.4, 9.0)
4.41 (dd, 8.4, 8.4)
4.41 (dd, 8.4, 9.6)
4.38 (dd, 8.5, 8.5)
4.33 (dd, 8.4, 9.0)
4′
4.68 (dd, 9.0, 9.6)
4.66 (dd, 9.0, 9.6)
4.60 (dd, 8.4, 9.6)
4.60 (dd, 9.6, 9.6)
4.59 (m)
4.55 (dd, 9.0, 10.2)
5′
4.80 (d, 9.6)
4.80 (d, 9.6)
4.64 (d, 9.6)
4.64 (d, 9.6)
4.61 (m)
4.68 (d, 10.2)
1′'
—
—
5.68 (d, 7.2)
5.64 (d, 7.8)
5.63 (d, 7.5)
5.24 (d, 7.2)
2′'
2.10 (s)
1.97 (s)
4.17 (dd, 7.2, 8.4)
4.14 (dd, 7.8, 8.4)
4.14 (dd, 7.5, 8.0)
4.09 (dd, 7.2, 8.4)
3′'
—
—
4.27 (dd, 8.4, 9.0)
4.25 (dd, 8.4, 9.0)
4.23 (dd, 8.0, 8.0)
4.31 (dd, 8.4, 9.0)
4′'
—
—
4.49 (dd, 9.0, 9.0)
4.45 (dd, 9.0, 9.6)
4.45 (m, o)
4.13 (dd, 8.4, 9.0)
5′'
—
—
3.78 (m)
3.77 (m)
3.77 (m)
4.00 (m)
6′'
—
—
4.40 (br. d, ca. 11)
4.38 (br. d, ca. 14)
4.39 (br. d, ca. 11)
4.23 (dd, 5.4, 12.0)
—
—
4.48 (dd, 3.0, 11.4)
4.44 (dd, 2.4, 13.8)
4.45 (m, o)
4.52 (br. d, ca. 12)
2′''
—
—
—
2.10 (s)
1.98 (s)
1.98 (s)
No.
1
2
3
4
5
6
7
8
9
10
11
1
38.6
38.6
38.5
38.5
38.6
38.5
38.4
38.5
38.8
38.8
38.7
2
26.9
26.9
26.7
26.7
26.7
26.7
26.5
26.6
26.6
26.5
26.5
3
88.8
88.8
90.7
90.7
90.7
88.9
91.2
91.2
89.3
89.3
89.4
4
44.4
44.4
43.7
43.7
43.7
44.3
43.7
43.7
39.5
39.5
39.5
5
56.0
56.0
56.1
56.1
56.1
56.0
56.1
56.1
55.6
55.7
55.6
6
18.7
18.7
18.5
18.5
18.5
18.7
18.5
18.5
18.4
18.4
18.4
7
33.4
33.4
33.2
33.2
33.3
33.4
33.2
33.2
33.1
33.1
32.8
8
40.0
40.0
39.9
39.9
39.9
39.9
39.9
39.9
40.0
40.0
40.1
9
46.8
46.8
46.8
46.8
46.8
46.8
46.7
46.7
46.9
46.9
47.8
10
36.5
36.4
36.4
36.4
36.4
36.5
36.3
36.4
36.7
36.8
36.7
11
24.1
24.1
24.0
24.0
24.1
24.1
24.0
24.0
23.9
23.9
23.9
12
123.3
123.8
122.9
123.3
123.6
123.6
123.7
123.6
124.0
124.2
123.8
13
143.5
143.3
144.0
143.5
143.3
143.3
142.9
142.9
142.8
142.5
143.1
14
41.9
41.9
42.0
41.8
41.9
41.9
41.6
41.7
41.8
41.8
42.1
15
34.4
34.6
34.3
34.4
34.6
34.5
34.6
34.6
34.7
34.6
25.7
16
67.9
68.0
67.8
67.9
68.1
68.0
67.9
68.0
67.6
67.8
18.9
17
48.1
46.5
47.3
48.1
46.5
46.6
48.0
48.0
47.1
46.0
42.8
18
40.4
40.8
41.1
40.4
40.8
40.8
40.0
40.1
40.6
41.5
41.4
19
47.7
47.7
48.2
47.8
47.7
47.7
47.2
47.2
47.3
47.4
46.3
20
36.1
36.5
36.5
36.1
36.4
36.5
36.2
36.3
36.4
36.8
36.4
21
82.0
78.6
78.6
82.0
78.6
78.6
79.5
78.9
81.5
76.2
76.8
22
72.6
73.6
77.0
72.8
73.7
73.7
74.3
74.4
71.0
78.1
71.9
23
23.3
23.3
22.5
22.5
22.5
23.2
22.4
22.4
28.0
28.1
28.0
24
63.3
63.2
63.4
63.4
63.4
63.2
63.2
63.3
16.7
16.9
16.7
25
15.4
15.5
15.6
15.6
15.6
15.4
15.5
15.5
15.7
15.7
15.6
26
16.8
16.9
16.7
16.6
16.9
16.9
16.6
16.7
17.0
16.7
17.0
27
27.4
27.4
27.4
27.4
27.4
27.4
27.4
27.4
27.4
27.4
26.3
28
65.8
66.9
68.1
66.0
67.0
66.9
63.7
63.8
66.4
68.3
66.8
29
29.9
30.6
30.6
29.8
30.5
30.5
29.4
29.5
29.8
30.2
30.3
30
20.2
19.4
19.5
20.2
19.3
19.4
20.1
20.3
20.2
19.4
19.1
1′
106.5
106.5
104.8
104.8
104.8
105.7
104.7
104.7
105.1
105.1
105.0
2′
75.4
75.4
81.6
81.6
81.8
74.8
79.2
79.4
81.0
81.1
80.9
3′
78.1
78.1
78.3
78.3
78.3
76.5
76.2
76.2
76.0
76.0
76.1
4′
73.6
73.5
73.1
73.0
73.0
83.5
82.0
81.8
82.3
82.1
82.2
5′
78.1
78.1
77.6
77.7
77.7
76.8
74.9
74.9
75.6
75.6
75.8
6′
172.8
172.9
173.0
172.5
172.6
ND
169.6
169.6
172.3
172.2
172.8
7′
52.7
52.7
1′'
171.5
170.9
105.0
105.0
105.2
104.9
104.2
104.2
105.4
105.4
105.3
2′'
21.4
20.8
75.8
75.8
75.8
75.0
75.7
75.7
77.1
77.1
77.0
3′'
78.2
78.2
78.3
77.8
78.1
78.1
77.9
77.9
77.9
4′'
69.8
69.9
69.9
71.6
69.7
69.8
71.6
71.7
71.8
5′'
78.4
78.4
78.5
78.5
78.4
78.4
78.4
78.3
78.3
6′'
61.6
61.6
61.6
62.4
61.6
61.6
62.3
62.4
62.4
1′''
171.6
170.8
171.0
105.2
105.1
104.8
104.7
104.6
2′''
21.4
20.8
20.8
74.5
74.5
74.9
74.9
74.9
3′''
78.2
78.2
78.1
78.1
78.0
4′''
71.5
71.5
71.5
71.5
71.5
5′''
78.6
78.5
78.5
78.4
78.4
6′''
62.3
62.3
62.7
62.8
62.8
1′'''
171.0
167.9
168.4
171.1
171.0
2′'''
21.1
129.0
129.8
21.3
20.8
3′'''
137.2
136.4
4′'''
15.9
14.2
5′'''
21.0
12.5
1′''''
171.0
171.0
170.8
170.8
2′''''
20.9
20.9
20.8
20.9
2.1.3.2 Aeswilsaponin IB (2)
White powder; [α]D25 –1.8 (c 0.11, MeOH); IR vmax (KBr) cm−1: 3392, 2936, 1721, 1599, 1446, 1415, 1379, 1245, 1161, 1110, 1076, 1027; 1H NMR (C5D5N, 600 MHz) δ: Table 1, 13C NMR (C5D5N, 150 MHz) δ: Table 2; ESI-Q-Orbitrap MS m/z 723.39795 [M -H]- (calcd for C38H59O13, 723.39502).
2.1.3.3 Aeswilsaponin IC (3)
White powder; [α]D25 –14.6 (c 0.04, MeOH); IR νmax (KBr) cm−1: 3400, 2948, 1631, 1454, 1414, 1378, 1308, 1249, 1163, 1073, 1030; 1H NMR (C5D5N, 600 MHz) δ: Table 1; 13C NMR (C5D5N, 150 MHz) δ: Table 2; ESI-Q-Orbitrap MS m/z 843.43860 [M − H]− (calcd for C42H67O17, 843.43728).
2.1.3.4 Aeswilsaponin ID (4)
White powder; [α]D25 –7.4 (c 0.11, MeOH); IR νmax (KBr) cm−1: 3377, 2945, 1718, 1614, 1415, 1373, 1260, 1164, 1073, 1035; 1H NMR (C5D5N, 600 MHz) δ: Table 1; 13C NMR (C5D5N, 150 MHz) δ: Table 2; ESI-Q-Orbitrap MS m/z 885.45148 [M − H]− (calcd for C44H69O18, 885.44784).
2.1.3.5 Aeswilsaponin IE (5)
White powder; [α]D25 –10.0 (c 0.04, MeOH); IR νmax (KBr) cm−1: 3390, 2945, 1724, 1645, 1453, 1419, 1381, 1245, 1168, 1074, 1031; 1H NMR (C5D5N, 500 MHz) δ: Table 1; 13C NMR (C5D5N, 125 MHz) δ: Table 2; ESI-Q-Orbitrap MS m/z 885.45160 [M − H]− (calcd for C44H69O18, 885.44784).
2.1.3.6 Aeswilsaponin IF (6)
White powder; [α]D25 –4.1 (c 0.24, MeOH); IR νmax (KBr) cm−1: 3374, 2944, 1727, 1611, 1410, 1384, 1244, 1159, 1074, 1032; 1H NMR (C5D5N, 600 MHz) δ: Table 1; 13C NMR (C5D5N, 150 MHz) δ: Table 2; ESI-Q-Orbitrap MS m/z 885.45142 [M − H]− (calcd for C44H69O18, 885.44784).
2.1.3.7 Aeswilsaponin IG (7)
White powder; [α]D25 –14.4 (c 0.21, MeOH); IR νmax (KBr) cm−1: 3377, 2947, 1733, 1624, 1435, 1374, 1261, 1159, 1071, 1033; 1H NMR (C5D5N, 600 MHz) δ: Table 3; 13C NMR (C5D5N, 150 MHz) δ: Table 2; ESI-Q-Orbitrap MS m/z 1103.53040 [M − H]− (calcd for C53H83O24, 1103.52688). o: overlapped.
No.
7
8
9
10
11
1
0.76 (m)
0.78 (m)
0.83 (m)
0.83 (m)
0.77 (m)
1.35 (m)
1.35 (m)
1.39 (m)
1.39 (m)
1.36 (m, o)
2
1.90 (m)
1.91 (m)
1.85 (m)
1.85 (m)
1.83 (m, o)
2.13 (m)
2.16 (m)
2.18 (m)
2.16 (m)
2.15 (m)
3
3.38 (dd, 4.8, 11.4)
3.39 (dd, 4.2, 9.6)
3.24 (dd, 3.6, 11.4)
3.24 (dd, 4.2, 11.4)
3.22 (dd, 3.0, 10.2)
5
0.82 (br. d, ca. 13)
0.82 (m)
0.71 (m)
0.71 (br. d, ca. 12)
0.68 (br. d, ca. 11)
6
1.19 (m)
1.22 (m)
1.29 (m, o)
1.29 (m, o)
1.29 (m)
1.53 (m)
1.52 (m)
1.48 (m)
1.47 (m)
1.47 (m)
7
1.24 (m)
1.26 (m)
1.29 (m, o)
1.29 (m, o)
1.26 (m)
1.49 (m)
1.50 (m)
1.55 (m)
1.55 (m)
1.44 (m)
9
1.63 (m)
1.64 (m)
1.70 (m)
1.70 (m)
1.57 (dd, 8.4, 8.4)
11
1.71 (m)
1.74 (m)
1.88 (m)
1.85 (m, o)
1.83 (m, o)
1.84 (m, o)
1.85 (m, o)
12
5.39 (t like, ca. 3)
5.40 (t like, ca. 3)
5.48 (t like, ca. 3)
5.45 (t like, ca. 3)
5.35 (t like, ca. 3)
15
1.61 (m)
1.61 (m)
1.68 (br. d, ca. 14)
1.64 (br. d, ca. 14)
1.04 (m)
1.84 (m, o)
1.85 (m, o)
1.93 (m)
1.88 (m, o)
1.69 (m)
16
4.46 (m)
4.47 (m, o)
4.80 (m)
4.60 (m)
2.02 (m)
—
—
—
—
2.13 (m)
18
3.08 (dd, 3.0, 12.0)
3.07 (dd, 3.0, 11.4)
2.90(dd, 4.2, 13.2)
2.82 (dd, 4.2, 13.8)
2.71 (dd, 3.6, 14.4)
19
1.38 (dd, 3.0, 12.0)
1.41 (dd, 3.0, 11.4)
1.42 (dd, 4.2, 13.2)
1.41 (dd, 4.2, 13.8)
1.36 (m, o)
3.07 (dd, 12.0, 12.0)
3.08 (dd, 11.4, 11.4)
3.16 (dd, 13.2, 13.2)
3.05 (dd, 13.8, 13.8)
2.16 (dd, 14.4, 14.4)
21
6.54 (d, 9.6)
6.63 (d, 10.2)
6.55 (d, 10.2)
4.95 (d, 9.6)
3.82 (d, 9.6)
22
6.25 (d, 9.6)
6.23 (d, 10.2)
4.57 (d, 10.2)
5.91 (d, 9.6)
4.22 (d, 9.6)
23
1.33 (s)
1.33 (s)
1.24 (s)
1.23 (s)
1.23 (s)
24
3.34 (d, 9.6)
3.34 (d, 11.4)
1.08 (s)
0.94 (s)
1.07 (s)
4.28 (d, 9.6)
4.28 (d, 11.4)
25
0.65 (s)
0.67 (s)
0.84 (s)
0.85 (s)
0.82 (s)
26
0.78 (s)
0.81 (s)
1.00 (s)
0.79 (s)
0.99 (s)
27
1.82 (s)
1.82 (s)
1.82 (s)
1.85 (s)
1.31 (s)
28
3.40 (d, 10.2)
3.40 (d, 9.6)
4.30 (d, 10.8)
4.09 (d, 11.4)
4.48 (d, 10.8)
3.62 (d, 10.2)
3.62 (d, 9.6)
4.36 (d, 10.8)
4.12 (d, 11.4)
4.53 (d, 10.8)
29
1.09 (s)
1.10 (s)
1.14 (s)
1.31 (s)
1.29 (s)
30
1.31 (s)
1.33 (s)
1.35 (s)
1.38 (s)
1.29 (s)
1′
4.92 (d, 7.8)
4.91 (d, 7.8)
4.99 (d, 7.2)
4.95 (d, 7.8)
4.93 (d, 6.0)
2′
4.31 (m, o)
4.27 (dd, 7.8, 9.0)
4.43 (dd, 7.2, 7.8)
4.39 (dd, 7.8, 8.4)
4.38 (dd, 6.0, 8.4)
3′
4.35 (dd, 8.4, 9.0)
4.34 (dd, 8.4, 9.0)
4.42 (dd, 7.8, 9.6)
4.38 (dd, 8.4, 9.0)
4.36 (dd, 7.2, 9.0)
4′
4.49 (dd, 9.0, 9.0)
4.47 (m, o)
4.59 (dd, 9.6, 9.6)
4.54 (dd, 9.0, 9.6)
4.52 (dd, 7.8, 9.6)
5′
4.57 (d, 9.0)
4.55 (d, 9.6)
4.66 (d, 9.6)
4.62 (d, 9.6)
4.59 (d, 9.6)
7′
3.93 (s)
3.92 (s)
—
—
—
1′'
5.69 (d, 7.8)
5.64 (d, 7.2)
5.46 (d, 7.2)
5.42 (d, 7.8)
5.42 (d, 7.2)
2′'
4.14 (dd, 7.8, 9.0)
4.10 (dd, 7.2, 8.4)
4.13 (dd, 8.4, 8.4)
4.08 (dd, 7.8, 8.4)
4.08 (dd, 7.2, 7.8)
3′'
4.25 (dd, 9.0, 9.0)
4.23 (dd, 8.4, 9.0)
4.29 (m, o)
4.26 (dd, 8.4, 8.4)
4.25 (dd, 7.8, 9.0)
4′'
4.55 (dd, 9.0, 9.6)
4.50 (dd, 9.0, 9.6)
4.39 (dd, 9.6, 9.6)
4.33 (dd, 8.4, 9.0)
4.30 (dd, 9.0, 9.0)
5′'
3.72 (m)
3.71 (m)
3.95 (m)
3.92 (m)
3.93 (m)
6′'
4.37 (br. d, ca. 11)
4.35 (dd, 6.0, 11.4)
4.29 (m, o)
4.27 (m)
4.24 (m, o)
4.48 (br. d, ca. 11)
4.45 (br. d, ca. 11)
4.51 (dd, 4.2, 11.4)
4.49 (dd, 3.0, 12.0)
4.48 (br. d, ca. 11)
1′''
5.05 (d, 7.8)
5.03 (d, 7.8)
5.23 (d, 7.2)
5.19 (d, 7.2)
5.17 (d, 7.2)
2′''
4.00 (dd, 7.8, 8.4)
3.98 (dd, 7.8, 8.4)
4.08 (dd, 7.2, 7.8)
4.05 (dd, 7.2, 8.4)
4.04 (dd, 7.2, 7.8)
3′''
4.22 (m, o)
4.19 (m, o)
4.24 (m, o)
4.20 (dd, 8.4, 9.0)
4.23 (dd, 7.8, 9.0)
4′''
4.22 (m, o)
4.19 (m, o)
4.24 (m, o)
4.19 (dd, 9.0, 9.0)
4.17 (dd, 9.0, 9.0)
5′''
4.03 (m)
4.00 (m)
4.02 (m)
3.99 (m)
3.97 (m)
6′''
4.31 (m, o)
4.29 (dd, 3.0, 10.8)
4.54 (m)
4.46 (dd, 4.2, 11.4)
4.46 (dd, 4.2, 11.4)
4.54 (dd, 2.4, 11.4)
4.53 (br. d, ca. 11)
—
4.51 (dd, 3.0, 11.4)
4.53 (m, o)
2′'''
2.16 (s)
—
—
2.00 (s)
1.98 (s)
3′'''
—
6.00 (q, 7.2)
7.04 (q, 7.2)
—
—
4′'''
—
2.12 (d, 7.2)
1.60 (d, 7.2)
—
—
5′'''
—
2.04 (s)
1.87 (s)
—
—
2′''''
2.00 (s)
1.94 (s)
2.04 (s)
2.14 (s)
—
2.1.3.8 Aeswilsaponin IH (8)
White powder; [α]D25 –25.0 (c 0.03, MeOH); UV λmax (MeOH) nm (log ε): 250 (4.34), 256 (4.37), 262 (4.22); IR νmax (KBr) cm−1: 3364, 2952, 1716, 1651, 1456, 1370, 1246, 1161, 1072, 1045, 1027; 1H NMR (C5D5N, 600 MHz) δ: Table 3; 13C NMR (C5D5N, 150 MHz) δ: Table 2; ESI-Q-Orbitrap MS m/z 1143.56177 [M − H]− (calcd for C56H87O24, 1143.55818).
2.1.3.9 Aeswilsaponin IIA (9)
White powder; [α]D25 –1.9 (c 0.74, MeOH); UV λmax (MeOH) nm (log ε): 250 (4.24), 256 (4.27), 262 (4.11); IR νmax (KBr) cm−1: 3371, 2929, 1718, 1609, 1411, 1383, 1267, 1158, 1072, 1041; 1H NMR (C5D5N, 600 MHz) δ: Table 3; 13C NMR (C5D5N, 150 MHz) δ: Table 2; ESI-Q-Orbitrap MS m/z 1113.55151 [M − H]− (calcd for C55H85O23, 1113.54761).
2.1.3.10 Aeswilsaponin IIB (10)
White powder; [α]D25 –5.6 (c 0.14, MeOH); IR νmax (KBr) cm−1: 3375, 2942, 1725, 1613, 1412, 1379, 1255, 1160, 1073, 1040; 1H NMR (C5D5N, 600 MHz) δ: Table 3; 13C NMR (C5D5N, 150 MHz) δ: Table 2; ESI-Q-Orbitrap MS m/z 1073.52014 [M − H]− (calcd for C52H81O23, 1073.51631).
2.1.3.11 Aeswilsaponin IIIA (11)
White powder; [α]D25 –1.6 (c 0.13, MeOH); IR νmax (KBr) cm−1: 3367, 2943, 1733, 1612, 1416, 1381, 1241, 1161, 1072, 1042; 1H NMR (C5D5N, 600 MHz) δ: Table 3; 13C NMR (C5D5N, 150 MHz) δ: Table 2; ESI-Q-Orbitrap MS m/z 1015.51404 [M − H]− (calcd for C50H79O21, 1015.51084).
2.1.4 Acid hydrolysis and determination of the absolute configuration of sugars of 1–11
The solution of compounds 1–11 (3.0 mg each) in 2 M HCl (6.0 mL) was heated under reflux for 3 h, respectively. The reaction mixture was partitioned with EtOAc (3 × 6.0 mL), and the obtained aqueous phase was evaporated to dryness using N2. The residues and authentic sugar samples (d-glucose and d-glucuronic acid) were dissolved in anhydrous pyridine (1.0 mL) containing l-cysteine methyl ester hydrochloride (1.0 mg) and heated at 60 °C for 1.5 h, respectively, then O-tolyisothiocyanate (1.0 mL) was added to the mixture and heated further for 1.5 h. After that, the reaction mixture was analyzed by HPLC [column, Cosmosil 5C18-MS-II column, 4.6 mm × 250 mm; mobile phase, CH3CN-H2O (25:75, v/v); flow rate, 0.8 mL/min)]. As results, the presence of d-glucuronic acid in compounds 1 and 2, as well as d-glucuronic acid and d-glucose in 3–11 was clarified by comparing the retention times (tR) of their derivatization reaction products with those of authentic sugar samples (d-glucuronic acid, tR 20.3 min; d-glucose, tR 18.7 min).
2.1.5 Absolute configuration determination of aglycon of 1
2.1.5.1 Alkaline hydrolysis of compound 1
A solution of compound 1 (10.0 mg) in 1% NaOH aqueous (3.0 mL) was stirred at 30 °C for 1 h. Then their reaction mixture was partitioned with n-butanol-H2O (1:1, v/v) to obatin n-butanol extraction, which was purified by pHPLC [CH3CN-1% HAc (26:74, v/v)] to provide (3β,16α,21β,22α)-16,21,22,24,28-pentahydroxyolean-12-en-3-O-β-d-glucopyranosiduronic acid (12). A solution of 12 in 1 M HCl (5.0 mL) was stirred at 90 °C for 5 h. The reaction mixture was extracted with EtOAc. Then, the EtOAc extraction was separated by pHPLC [CH3CN-1% HAc (45:55, v/v)] to obtain aglycon, 3β,16α,21β,22α,24,28-hexahydroxyolean-12-en (1a, tR 14.6 min, see supplymentary materials).
2.1.5.2 Snatzke's reaction of 1a
Compound 1a (2.0 mg) dissolved in dimethyl sulfoxide (DMSO) (spectrally pure, 0.5 mL) was subjected to ECD measurement as blank firstly. Then 1a (2.0 mg) and Mo2(AcO)4 (1.7 mg) were added to DMSO (1.0 mL), and the ECD of their reaction product was scanned every 10 min after the reaction for 10 min at room temperature until the ICD spectrum was stationary. After subtracting blank control, the absolute configuration of 1a was elucidated by the diagnostic band at 310–340 nm in the ICD spectrum.
2.1.6 Alkaline hydrolysis of 2, 4, and 5–10
The solution of compounds 2, 4, and 5–10 (5.0 mg each) in 1% NaOH aqueous (1.0 mL) was stirred at 30 °C for 1 h. Then their reaction mixture was partitioned with n-butanol-H2O (1:1, v/v) to obtain n-butanol extraction, respectively. The n-butanol extraction of 2 was purified by pHPLC [CH3CN-1% HAc (26:74, v/v)] to gain (3β,16α,21β,22α)-16,21,22,24,28-pentahydroxyolean-12-en-3-O-β-d-glucopyranosiduronic acid (12, tR 23.3 min). By using the same separation method, 3-O-β-d-glucopyranosyl(1 → 2)-β-d-glucuronopyranosyl-3β,16α,21β,22α,24,28-hexahydroxyolean-12-en (3, tR 14.5 min) was produced from compounds 4 and 5. The n-butanol extractions of both compounds 7 and 8 were separated by pHPLC [MeOH-1% HAc (62:38, v/v)] to provide desacylescin I (16, tR 15.1 min). And 3-O-[β-d-glucopyranosyl(1 → 2)][β-d-glucopyranosyl(1 → 4)]-β-d-glucuronopyranosyl-3β,16α,21β,22α,28-pentahydroxyolean-12-en (9a, tR 22.5 min, see supplementary materials) was yielded from n-butanol extractions of 9 and 10 by using pHPLC separation [CH3CN-1% HAc (26:74, v/v)].
2.2 Materials and methods for anti-inflammatory assay
2.2.1 Reagents
3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT), LPS, adenosine triphosphate (ATP), and dexamethasone (DEX) were ordered from Sigma-Aldrich (St. Louis, MO). The NO kit was purchased from Shanghai Biyuntian Biotechnology Co. ltd. (Shanghai, China). Mouse IL-1β ELISA kits were supplied by Elabscience Biotechnology Co., Ltd. (Wuhan, China). RIPA lysis buffer and sample buffer were ordered from Solarbio Science and Technology (Beijing, China). Protease (1% phenylmethanesulfonyl fluoride in isopropanol) and phosphatase inhibitor were supplied by Sangon Biotech. (Shanghai, China). Dulbecco's modified Eagle's medium (DMEM), Roswell Park Memorial Institute medium (RPMI), and fetal bovine serum (FBS) were purchased from Biological Industries (Beit HaEmek, Israel). Bicinchoninic acid assay (BCA) protein quantification kit, penicillin, and streptomycin solution were purchased from Thermo Fisher Scientific (Waltham, MA). Immunoblot polyvinylidene fluoride (PVDF) membranes were purchased from Merck Millipore Ltd. (Darmstadt, Germany). Primary antibodies for NF-κB (ab16502), phosphorylated (p)-NF-κB (ab16502), iNOS (ab3523), COX-2 (ab52237), TNF-α (ab6671), IL-6 (ab9324), NLRP3 (ab270449), pro-caspase1 + p10 + p12 (ab179515), β-actin (ab8227) were purchased from Abcam (Cambridge, MA, USA). Primary antibodies against IKK (2682S) and p-IKK (2697), IL-1β (#12242) and HRP-linked goat anti-rabbit IgG (#7074) were purchased from Cell Signaling Technology (Danvers, USA). Primary antibodies for ASC (YT0365) were purchased from ImmunoWay Biotechnology Company (TX, USA).
2.2.2 General experimental procedures of compounds on RAW264.7 cells
The cytotoxicity analysis and NO production inhibitory assay of isolated saponins on RAW264.7 cells as well as the statistical analysis of them, were conducted by using the method have been reported by us previously (Li et al., 2022).
The procedure for western blot assay is as following: The experiment was divided into four groups including normal group, LPS group, the positive drug DEX group and the tested groups. Firstly, RAW 264.7 cells were cultured in the 6-cell plates (4 × 106 cells/mL) for 24 h to allow cells adherent to the wall. Then the supernatants of normal group was replaced with serum-containing medium, LPS (0.5 μg/mL) was administered to LPS group, LPS (0.5 μg/mL) combined with positive drug DEX (1.5 μg/mL) was given to DEX group and the tested compounds 1, 4, 5 (each 50 μM) together with LPS (0.5 μg/mL) were applied to the tested groups, respectively, further culturing for 18 h. Finally, all groups of RAW 264.7 cells were separated and collected to 1.5 mL centrifuge tubes for the preparation of extracting protein. RIPA cell lysate and protease at the ratio of 100:1 were added to the cells for incubating for 30 min on ice. After incubation, the cells were centrifuged at 4 °C and 12,000 g for 5 min to collect proteins. Then BCA protein quantification kit was used to determine the concentration of protein and PBS was added to adjust the same final protein concentration of each sample. Next, 4 × loading buffer was mixed with protein sample at the ratio of 1:3, incubating for 5 min at 100 °C to denaturate the protein thoroughly. When the protein sample was cooled to room temperature, they were loaded to 10% or 15% sodium dodecyl sulfate and then transferred to polyvinylidene fluoride (PVDF) membranes. The PVDF membranes were blocked with 5% skim milk for 1 h at room temperature. After being washed three times with TBST, PVDF membranes were incubated with primary antibodies against iNOS, IL-6, TNF-α and COX-2 at 4 °C overnight, followed by being washed three times with TBST. Then PVDF membranes were incubated with secondary antibody and washed. Finally, the protein membranes were visualized using a ChemiDoc MP Imaging System.
Experimental procedures for the determination of proteins involved in the NF-κB signaling pathway were similar to the method above. The only difference was that the LPS stimulation time was 8 h.
2.2.3 General experimental procedures of compounds 1, 4, and 5 on PMs cells
2.2.3.1 Cell preparation
C57BL/6J mice were used for peritoneal macrophages (PMs) cells extraction according to the related literature (Li et al., 2022; Zhang et al., 2008). Mice were intraperitoneally injected with 2 mL 4% thioglycolate broth per mouse to allow the inflammatory response to proceed for 5 days. Then the mice were cervical dislocated rapidly and infiltrated with 70% alcohol for 5 min. Next, by cutting a small incision with sterile scissors, the cells were collected by peritoneal lavage with 10 mL of cold PBS and repeated. The collected cells were centrifuged at 4 °C and 1000 rpm for 5 min. Then, PMs cells were cultured in RPMI 1640 medium containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin in each well of a 96-well plate (5 × 104 cells/mL) or 6-well plate (2 × 106 cells/mL) and changed fresh medium after 4 h, incubating overnight in a 5% CO2 humidified incubator at 37 °C for subsequent experiments.
2.2.3.2 Cytotoxic analysis
Using the similar method reported by Zhang et al (Zhang et al., 2008), PMs cells were seeded in 96-well plate (5 × 104 cells/mL) and treated with 50 μM 1, 4, and 5 for 18 h, respectively. Then, the supernatant of each culture well was removed and 100 μL MTT solution (500 μg/mL) was added for 4 h at 37 °C. At the end of the treatment period, the formazan crystals were dissolved with DMSO and the absorbance was measured at 490 nm by using a BioTek Cytation five-cell imaging multi-mode reader (Winooski, VT, USA).
2.2.3.3 ELISA assay
The concentrations of IL-1β in PMs cells culture supernatants were quantified using ELISA kits according to the manufacturer’s instructions.
2.2.3.4 Western blot assay
PMs cells were seeded in the 6-cell plates (2 × 106 cells/mL) overnight. The inflammatory activation assay was conducted reported with decorations (Pan, et al., 2021; Shen et al., 2021). The experimental cells were divided into six groups, normal group (N), LPS group, LPS plus ATP (LPS/ATP) group, compound 1 (50 μM) + LPS/ATP group, compound 4 (50 μM) + LPS/ATP group and compound 5 (50 μM) + LPS/ATP group. First, the supernatant of the normal group was replaced with serum-containing RPMI 1640 medium, and the other five groups were replaced with LPS (1 μg/mL) for 4 h. Then the supernatants of the tested compounds groups were discarded and 1, 4, and 5 were added and incubated for 4 h, respectively. In the end, the supernatants of all six groups were discarded and incubated with 5 mM ATP for 30 min. The protein in supernatants and the protein in cells were collected separately. Cells were lysized with lysis buffer, then centrifuged at 4 °C and 12,000 g for 5 min. The cells were lysed by using RIPA lysis buffer supplemented with protease (1% phenylmethanesulfonyl fluoride in isopropanol) and phosphatase inhibitor with the ratio of 100:1:1 for 30 min on ice followed by centrifugation for 5 min at 12,000 g, 4 °C. Meanwhile, the protein in supernatants was mixed with trichloroacetic acid at the ratio of 4:1. After incubating for 1 h, the mixture was centrifuged at 4 °C and 4000 g for 15 min. Next, the supernatant was discarded, and 500 μL of pre-chilled acetone was added to the precipitate, then centrifuged at 4 °C and 4000 g for 15 min. Then the protein concentrations in the lysis solution and the protein concentrations in the supernatant were determined by using a BCA protein assay kit, respectively. After that, the protein sample was combined with 4 × sample buffer, heating at 100 °C for 5 min. Equal amounts of protein (10–40 μg) were loaded onto 12% sodium dodecyl sulfate (SDS) polyacrylamide gel. The fraction above 60 kDa was stained with Komas Brilliant Blue for 20–30 min, then the decolorizing solution was used to wash it off to a clear blue background. On the other hand, the rest fraction was transferred onto PVDF membranes. They were incubated with primary antibodies against NF-κB, p-NF-κB, IKK, p-IKK, NLRP3, pro-caspase1 + p10 + p12, IL-1β, ASC, and β-actin at 4 °C overnight, followed by treatment with horseradish peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin G (IgG) at room temperature for 1 h. Finally, the immunoreactive protein bands were developed by Immobilon Western Chemilumescent HRP Substrate (Millipore, Massachusetts, USA), and detected using a ChemiDoc MP Imaging System (Bio-Rad Laboratories, Hercules, USA). The band intensities were analyzed using Image J software (Version 1.0, National Institutes of Health, Bethesda, MD, USA).
2.2.4 Statistical analysis
Data were expressed as the mean ± SD. Significant differences among groups were determined using one-way ANOVA with Dunnett's multiple comparisons test. Data were considered significant when *P < 0.05, **P < 0.01, and ***P < 0.001. Data analyses were performed using GraphPad Prism 8.0 (GraphPad Software, Inc., La Jolla, CA, USA).
3 Results
3.1 Structural characterization
The 70% ethanol extract of air-dried A. wilsonii seeds was fractionated by D101 resin CC eluted with a gradient of EtOH in water. The 95% EtOH part was separated consequently by silica gel, Sephadex LH-20, MCI gel CHP 20P, and ODS CC, as well as pHPLC under the guidance of thin layer chromatography (TLC) and HPLC analysis. As a result, aeswilsaponins IA–IH (1–8), ⅡA (9), ⅡB (10), and ⅢC (11) together with twenty known ones (12–31) (Fig. 1) were afforded and identified.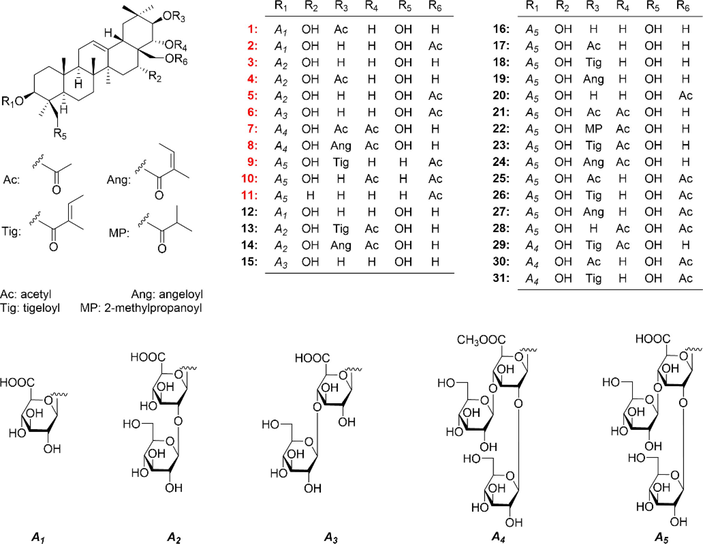
The saponins (1–31) obtained from A. willion seeds.
Aeswilsaponin IA (1) was obtained as a white powder. Its molecular formula, C38H60O13 (m/z 723.39764 [M – H]−) was determined by HR-ESI-MS. Its IR spectrum showed characteristic absorptions assignable to hydroxyl (3371 cm−1), olefinic bond (1616 cm−1), and ether function (1072 cm−1). Acid hydrolysis of 1 yielded d-glucuronic acid, which was established with HPLC analysis by comparing the retention time with that of its authentic sugar sample after derivatization (Tanaka et al., 2007; Zhang et al., 2020). Comparing its 1H (C5D5N, Table 1) and 13C NMR (C5D5N, Table 2) spectra with those of (3β,16α,21β,22α)-16,21,22,24,28-pentahydroxyolean-12-en-3-O-β-d-glucopyranosiduronic acid (12) (Kim et al., 2017) furtherly consolidated the presence of one β-d-glucopyranuronosyl [δH 5.21 (1H, d, J = 7.8 Hz, H-1′); δC 73.6 (C-4′), 75.4 (C-2′), 78.1 (C-3′ and 5′), 106.5 (C-1′), 172.8 (C-6′)]. Meanwhile, the signals at δH 2.10 (3H, s, H3-2′') and δC 171.5 (C-1′') indicated the existence of acetyl, which was confirmed by the HMBC correlation from δH 2.10 (H3-2′') to δC 171.5 (C-1′'). Thirty-eight carbon signals displayed in its 13C NMR spectrum, except for the above-mentioned ones, the remaining thirty carbon signals suggested that aeswilsaponin IA (1) was a triterpene saponin. The 1H NMR spectrum exhibited signals assignable to six angular methyls at δ 0.79, 0.84, 1.13, 1.32, 1.57, 1.89 (3H each, all s, H3-25, 26, 29, 30, 23, and 27), two oxygenated methylene at δ 3.67, 4.41 (1H each, both d, J = 11.4 Hz, H2-24), 3.69, 3.98 (1H each, both d, J = 10.2 Hz, H2-28), one olefinic proton at δ 5.38 (1H, t like, ca. J = 3 Hz, H-12), and four oxygenated methines at δ 3.64 (1H, dd, J = 5.4, 10.8 Hz, H-3), 4.84 (1H, d, J = 10.2 Hz, H-22), 4.89 (1H, br. d, ca. J = 3 Hz, H-16), 6.43 (1H, d, J = 10.2 Hz, H-21), indicating it was one of oleanane-type triterpene saponin derivatives. The planar structure of its aglycon was consolidated by the cross-peaks found in the 1H 1H COSY spectrum and the correlations from H-12 to C-13; H2-15 to C-13, C-17; H-18 to C-13; H2-19 to C-13, C-17; H-22 to C-17; H3-23 to C-3–5, C-24; H2-24 to C-3–5, C-23; H3-25 to C-1, C-5, C-9, C-10; H3-26 to C-7–9, C-14; H3-27 to C-8, C-13–15; H2-28 to C-16–18, C-22; H3-29 to C-19–21, C-30; H3-30 to C-19–21, C-29 showed in its HMBC spectrum (Fig. 2). Meanwhile, the HMBC correlations displayed from δH 6.43 (H-21) to δC 171.5 (C-1′'); δH 5.21 (H-1′) to δC 88.8 (C-3) suggested that the linkage positions of acetyl and β-d-glucopyranuronosyl were at C-21 and C-3, respectively. The coupling constant between H-21 and H-22 was 10.2 Hz, indicating that they were in the trans conformation. Then, aeswilsaponin IA (1) was subjected to alkaline hydrolysis, acid hydrolysis, Snatzke's reaction, and NOESY spectrum determination successively to elucidate the absolute configuration. It was deacetylated by 1% NaOH to afford (3β,16α,21β,22α)-16,21,22,24,28-pentahydroxyolean-12-en-3-O-β-d-glucopyranosiduronic acid (12), which was hydrolyzed by 1 M HCl to obtain aglycon 1a. The absolute configuration of C-21 and C-22 in 1a was determined by Mo2(AcO)4-induced circular dichroism (ICD). The ICD showed a negative cotton effect at 315 nm (Fig. 3), indicating the 21R,22R configuration according to the Snatzke's rule (Sun et al., 2015; Zhang et al., 2020). Finally, H-3, H-5, H-9, 23-CH3, and 27-CH3 were all consolidated to be α oriented by NOE correlations between δH 6.43 (H-21) and δH 3.11 (H-19α); H-19α and δH 1.89 (H3-27); H3-27 and δH 1.73 (H-9); H-9 and δH 0.94 (H-5); H-5 and δH 1.57 (H3-23), 3.64 (H-3). Moreover, the NOE cross-peaks between δH 4.84 (H-22) and δH 2.95 (H-18), 3.69, 3.98 (H2-28); H2-28 and δH 0.84 (H3-26), 4.89 (H-16); δH 0.79 (H3-25) and δH 3.61, 4.41 (H2-24) suggested that H-16, H-18, 25-CH3, 26-CH3, 24-CH2OH, and 28-CH2OH were all β oriented (Fig. 4). Based on the above-mentioned evidence, the structure of aeswilsaponin IA (1) was elucidated to be 3-O-β-d-glucuronopyranosyl-21-acetyl-3β,16α,21R,22R,24,28-hexahydroxyolean-12-en.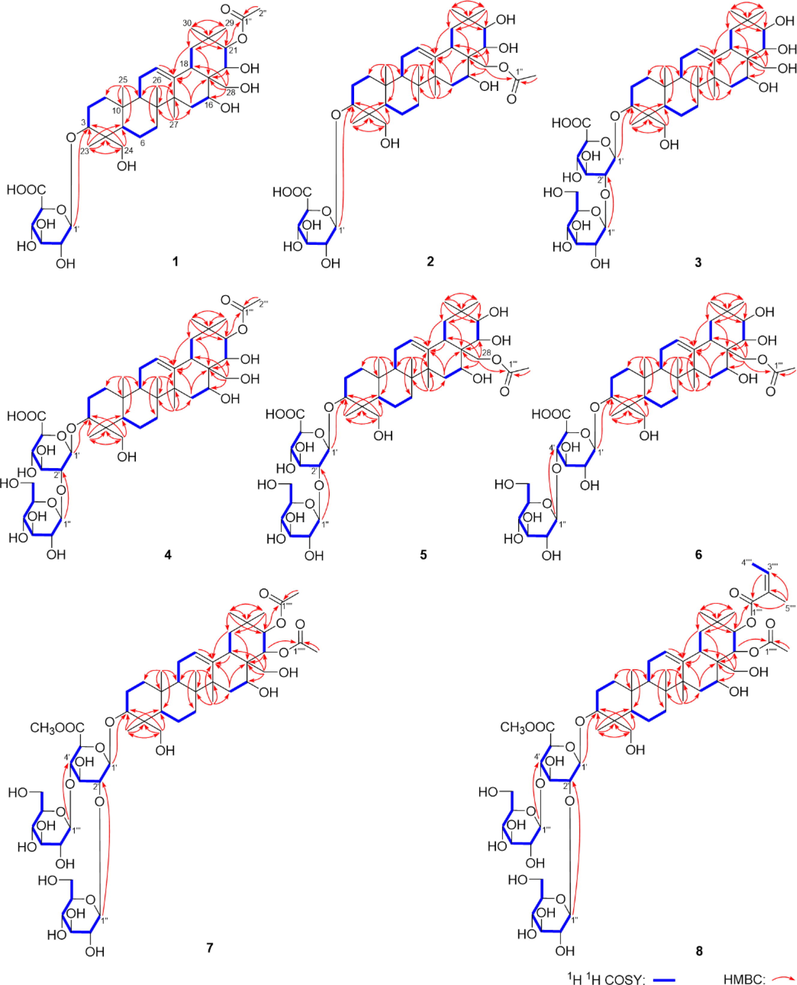
The main 1H 1H COSY and HMBC correlations of compounds 1–8.
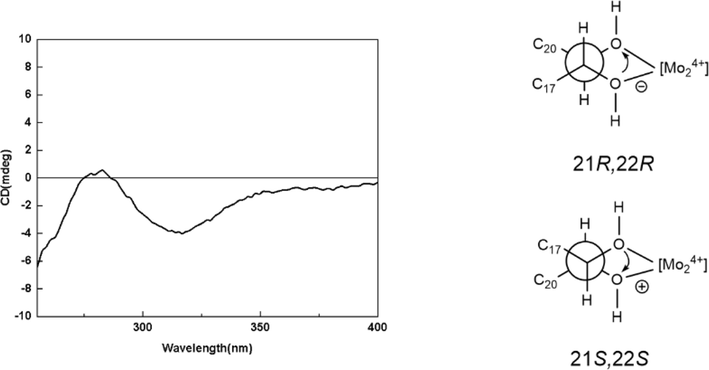
The ICD spectrum of 1a.
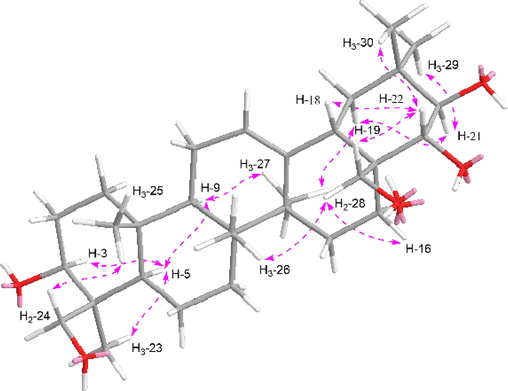
The main NOE correlations of aglycon part of compounds 1–8.
Aeswilsaponin IB (2) was revealed to have the same molecular formula, C38H60O13 (m/z 723.39795 [M – H]−) as that of compound 1 by HR-ESI-MS analysis. The 1D and 2D NMR spectra indicated it also had (3β,16α,21β,22α)-16,21,22,24,28-pentahydroxyolean-12-en, β-d-glucopyranuronosyl, and acetyl groups. However, the NMR resonance signals in C-17, 21, 22, 28–30 were significantly different between them (Table 2). Compared with 1, the signal of H-21 was upfield shifted by 1.59 ppm, while the signals of H2-28 were downfield shifted by 0.43 and 0.56 ppm, indicating that the acetyl was attached to C-28, which was confirmed by the HMBC correlation from H2-28 to C-1′'. Similar to compound 1, (3β,16α,21β,22α)-16,21,22,24,28-pentahydroxyolean-12-en-3-O-β-d-glucopyranosiduronic acid (12) was provided by the alkaline hydrolysis of 2. Then, the structure of aeswilsaponin IB (2) was clarified to be 3-O-β-d-glucuronopyranosyl-28-acetyl-3β,16α,21β,22α,24,28-hexahydroxyolean-12-en.
The HR-ESI-MS data of aeswilsaponin IC (3) showed a [M – H]− ion at m/z 843.43860 corresponding to a molecular formula of C42H68O17. Acid hydrolysis suggested that d-glucose and d-glucuronic acid existed in 3. It was elucidated to have the same aglycon, 3β,16α,21β,22α,24,28-hexahydroxyolean-12-en as 1 by comparing its 1H (C5D5N, Table 1) and 13C NMR (C5D5N, Table 2) data with compound 12 and the information provided by 2D NMR spectra (Fig. 2). In addition, β-d-glucopyranuronosyl was also presented in aeswilsaponin IC (3). Unlike compound 1, the signals related to acetyl disappeared and those belonging to β-d-glucopyanosyl [δH 5.68 (1H, d, J = 7.8 Hz, H-1′'); δC 61.6 (C-6′'), 69.8 (C-4′'), 75.8 (C-2′'), 78.2 (C-3′'), 78.4 (C-5′'), 105.0 (C-1′')] appeared in 3. Beyond this, the 13C NMR signals in C-1 and C-2 of the β-d-glucopyranuronosyl were upfield shifted and downfield shifted [δC 75.4 (C-2′), 106.5 (C-1′) for 1; δC 81.6 (C-2′), 104.8 (C-1′) for 3], respectively, suggesting that the β-d-glucopyanosyl linked with C-2 of β-d-glucopyranuronosyl. The HMBC cross-peak from δH 5.68 (H-1′') to δC 81.6 (C-2′) confirmed that speculation. Moreover, the substitution position of β-d-glucopyranuronosyl was revealed by the correlation from δH 5.00 (H-1′) to δC 90.7 (C-3). Thus, the structure of aeswilsaponin IC (3) was determined as 3-O-β-d-glucopyranosyl(1 → 2)-β-d-glucuronopyranosyl-3β,16α,21β,22α,24,28-hexahydroxyolean-12-en.
Both aeswilsaponins ID (4) and IE (5) were obtained as white powder. The same molecular formula, C44H70O18 (m/z 885.45148 [M – H]− for 4, 885.45160 [M – H]− for 5) was established on the HR-ESI-MS analysis. The 1H (C5D5N, Table 1), 13C NMR (C5D5N, Table 2), and 1H 1H COSY, HSQC, as well as HMBC (Fig. 2) spectra suggested that the aglycon of them was 3,16,21,22,24,28-hexahydroxyolean-12-en. Meanwhile, C-3 of them was substituted by β-d-glucopyranosyl(1 → 2)-β-d-glucopyranuronosyl. Compared with 3, compounds 4 and 5 had one additional acetyl. The HMBC correlations from δH 6.37 (H-21) to δC 171.6 (C-1′'') and δH 4.24, 4.38 (H2-28) to δC 170.8 (C-1′'') for 4 and 5 (Fig. 2) indicated that the acetyl linked with C-21 and C-28, respectively. The NOE cross-peaks between H-19α and H-21, H3-27; H-9 and H-5, H3-27; H-5 and H-3, H3-23; H-22 and H-18, H2-28; H2-28 and H-16, H3-26; H3-25 and H2-24 (Fig. 4) suggested the aglycon's relative configurations of 4 and 5 were consistent with those of 3. Moreover, after alkaline hydrolysis, compound 3 was obtained from 4 and 5. Consequently, the structures of aeswilsaponins ID (4) and IE (5) were identified as 3-O-β-d-glucopyranosyl(1 → 2)-β-d-glucuronopyranosyl-21β-acetyl-3β,16α,21β,22α,24,28-hexahydroxyolean-12-en and 3-O-β-d-glucopyranosyl(1 → 2)-β-d-glucuronopyranosyl-28-acetyl-3β,16α,21β,22α,24,28-hexahydroxyolean-12-en, respectively.
HR-ESI-MS analysis showed the molecular formula of aeswilsaponin IF (6) was also C44H70O18 (m/z 885.45142 [M – H]−). In addition, 1D and 2D NMR spectra suggested that it had the same units including 28-acetyl-3β,16α,21β,22α,24,28-hexahydroxyolean-12-en, β-d-glucopyranosyl, and β-d-glucuronopyranosyl as 5. However, the correlation from δH 5.24 (H-1′') to δC 83.5 (C-4′) clarified that β-d-glucopyranosyl linked with C-4 of β-d-glucuronopyranosyl. Therefore, aeswilsaponin IF (6) was identified as 3-O-β-d-glucopyranosyl(1 → 4)-β-d-glucuronopyranosyl-28-acetyl-3β,16α,21β,22α,24,28-hexahydroxyolean-12-en.
The molecular formula of aeswilsaponin IG (7) was revealed to be C53H84O24 (m/z 1103.53040 [M – H]−) by HR-ESI-MS analysis. The occurence of d-glucuronic acid and d-glucose in 7 was comfirmed by acid hydrolysis method similar to that of compounds 1–6. The existence of one β-d-glucuronopyranosyl and two β-d-glucopyranosyl was consolidated by combining the three anomeric proton signals at δ 4.92 (1H, d, J = 7.8 Hz, H-1′), 5.05 (1H, d, J = 7.8 Hz, H-1′''), 5.69 (1H, d, J = 7.8 Hz, H-1′') (Table 3), sugar related 13C NMR signals at δC 62.3–105.2, 169.6 (Table 2), as well as the cross-peaks displayed in its 1H 1H COSY and HSQC spectra. On the other hand, the signals δH 3.93 and δC 52.7 suggested the presence of methoxyl. The correlation from δH 3.93 (H3-7′) to δC 169.6 (C-6′) indicated that C-6 of β-d-glucuronopyranosyl had been methylated. Its planar structure of aglycon was consistent with that of 1–6. The presence of fifty-five carbons were confirmed by its 13C NMR and MS determination. In addition to the above aglycon and glycosyl carbon signals, the remaining four ones at δC 20.9, 21.1, 171.0, 171.0 (Table 2) suggested that it owned two acetyl groups. According to the HMBC correlations from δH 6.25 (H-22) and 6.54 (H-21) to δC 171.0 (C-1′''' and C-1′'''') (Fig. 2), the substitution positions of two acetyls were deduced to be C-21 and C-22. Moreover, based on the HMBC cross-peaks from δH 4.92 (H-1′) to δC 91.2 (C-3); δH 5.69 (H-1′') to δC 79.2 (C-2′); δH 5.05 (H-1′'') to δC 82.0 (C-4′) (Fig. 2), two β-d-glucopyranosyl were identified as lateral sugars attached to C-2 and C-4 of β-d-glucuronopyranoide methyl ester, which was linked to C-3 of the aglycon.
The molecular formula of aeswilsaponin IH (8), C56H88O24 (m/z 1143.56177 [M – H]−) was established on the HR-ESI-MS. The existence of angeloyl was consolidated according to the proton and proton cross-peak between δH 6.00 (H-3′''') and δH 2.12 (H-4′'''), as well as the HMBC correlations from δH 6.00 (H-3′''') to δC 21.0 (C-5′'''), 167.9 (C-1′'''); δH 2.12 (H-4′''') to δC 129.0 (C-2′'''); δH 2.04 (H-5′''') to δC 129.0 (C-2′'''), 137.2 (C-3′'''), 167.9 (C-1′''') (Fig. 2). The NMR data of 8 were very similar to those of 7 except for the disappearance of one acetyl, and the appearance of one angeloyl moiety. Whose linkage position was revealed to be C-21 by the HMBC correlation from δH 6.63 (H-21) to δC 167.9 (C-1′''') (Fig. 2).
The NOE correlations of 7 and 8 (Fig. 3) suggested that the relative configuration of their aglycon was the same as that of 1–6. Desacylescin I (16) (Yoshikawa et al., 1996) was obtained by alkaline hydrolysis of them. The resonance signals of 7 and 8 on A–D rings were correlated with those of compound 3, indicating their aglycons were identical. Then, the structures of aeswilsaponins IG (7) and IH (8) were elucidated.
The aglycons of 1–8 were the same as each other. Its absolute configuration was deduced to be 3S,4S,5R,8R,9R,10R,14S,16R,17R,18S,21R,22R.
Aeswilsaponin IIA (9) was obtained as white powder with molecular formular, C55H86O23 (m/z 1113.55151 [M – H]−). d-glucuronic acid and d-glucose were found in it by the acid hydrolysis. The anomeric proton signals at δ 4.99 (1H, d, J = 7.2 Hz, H-1′), 5.23 (1H, d, J = 7.2 Hz, H-1′''), 5.46 (1H, d, J = 7.2 Hz, H-1′') (Table 3) and carbon signals displayed at δC 62.3–105.4, 172.3 (Table 2) suggested the existence of one β-d-glucuronopyranosyl and two β-d-glucopyranosyl. Meanwhile, the signals assignable to one tigeloyl [δH 1.60 (3H, s, H3-4′'''), 1.87 (3H, s, H3-5′'''), 7.04 (1H, q, J = 7.2 Hz, H-3′'''); δC 168.4 (C-1′''')] and one acetyl [δH 2.04 (3H, s, H3-2′''''); δC 170.8 (C-1′'''')] were also found in its 1D NMR spectra. The NMR resonance signals of its aglycon were one more methyl and one less oxymethylene than those of compounds 1–––8. The aglycon of 9 was suggested to be distinguished from those of 1–––8 by the fact that C-24 was methyl rather than hydroxymethyl according to the HMBC correlations from δH 1.24 (H3-23) to δC 16.7 (C-24), 39.5 (C-4), 55.6 (C-5), 89.3 (C-3); δH 1.08 (H3-24) to δC 28.0 (C-23), 39.5 (C-4), 55.6 (C-5), 89.3 (C-3) (Fig. 5). Furtherly, the linkage relationships between tigeloyl, acetyl, β-d-glucuronopyranosyl, β-d-glucopyranosyl, and aglycon were elucidated through the cross-peaks from δH 6.55 (H-21) to δC 168.4 (C-1′'''); δH 4.30, 4.36 (H2-28) to δC 170.8 (C-1′''''); δH 4.99 (H-1′) to δC 89.3 (C-3); δH 5.46 (H-1′') to δC 81.0 (C-2′); δH 5.23 (H-1′'') to δC 82.3 (C-4′) (Fig. 5). Compound 9 was hydrolyzed by 1% NaOH to yield 3-O-[β-d-glucopyranosyl(1 → 2)][β-d-glucopyranosyl(1 → 4)]-β-d-glucuronopyranosyl-3β,16α,21β,22α,28-pentahydroxyolean-12-en (9a). Its chemical shifts of proton and carbon in D and E rings were consistent with those of 2. Combined with the biosynthesis rule, they were presumed to have the same absolute configurations at C-16–––18, 21, 22. The configurations of the other chiral centers were further clarified by the NOE correlations between H-19α and H-21, H3-27; H-9 and H-5, H3-27; H-5 and H-3, H3-23; H2-28 and H3-26; H3-25 and H3-24 (Fig. 6). Then, the structure of aeswilsaponin IIA (9) was determined as 3-O-[β-d-glucopyranosyl(1 → 2)][β-d-glucopyranosyl(1 → 4)]-β-d-glucuronopyranosyl-21β-tigeloyl-28-acetyl-3β,16α,21β,22α,28-pentahydroxyolean-12-en.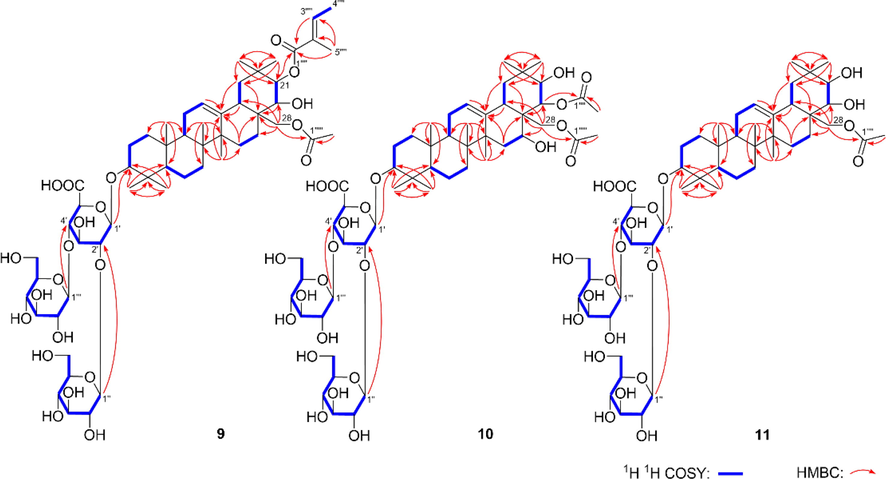
The main 1H 1H COSY and HMBC correlations of compounds 9–11.
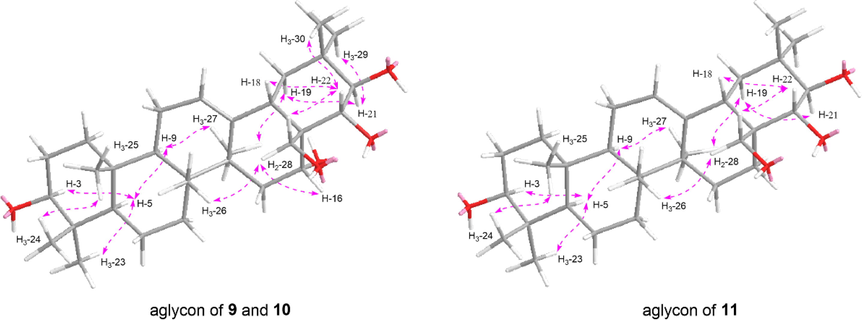
The main NOE correlations of aglycon parts of 9–11.
The HR-ESI-MS of aeswilsaponin IIB (10) displayed a [M – H]− ion at m/z 1073.52014 (calcd for C52H81O23, 1073.51631) corresponding to the molecular formula, C52H82O23. The NMR data of 10 was similar to those of 9 except for the disappearance of a tigeloyl, and the appearance of an acetyl. The linkage positions of the two acetyl groups were clarified according to the HMBC correlations from H-22 to C-1′''' and H2-28 to C-1′'''' (Fig. 5). Then, the structure of aeswilsaponin IIB (10) was identified as 3-O-[β-d-glucopyranosyl(1 → 2)][β-d-glucopyranosyl(1 → 4)]-β-d-glucuronopyranosyl-22α,28-diacetyl-3β,16α,21β,22α,28-pentahydroxyolean-12-en.
Aeswilsaponin IIIA (11) was assigned the molecular formula C50H80O21 based on an ion peak at m/z 1015.51404 [M – H]− in the HR-ESI-MS. Similar to 9 and 10, the 1H (C5D5N, Table 3) and 13C NMR (C5D5N, Table 2) spectra of 10 implied the occurrence of one [β-d-glucopyranosyl(1 → 2)][β-d-glucopyranosyl(1 → 4)]-β-d-glucuronopyranosyl [δ 4.93 (1H, d, J = 6.0 Hz, H-1′), 5.17 (1H, d, J = 7.2 Hz, H-1′''), 5.42 (1H, d, J = 7.2 Hz, H-1′')] and one acetyl [δH 1.98 (3H, s, H3-2′'''); δC 171.0 (C-1′''')]. Fifty carbon signals displayed in its 13C NMR spectrum. Except for the signals related to the above-mentioned moieties, the remaining thirty ones suggested compound 11 was one of triterpene saponins. Moreover, its 1H NMR spectrum displayed resonances attributable to seven methyl [δ 0.82, 0.99, 1.07, 1.23, 1.29, 1.29, 1.31 (3H each, all s, H3-25, 26, 24, 23, 29, 30 and 27)], one singlet olefinic methine [δ 5.35 (1H, t like, ca. J = 3 Hz, H-12)], one oxymethylene [δ 4.48, 4.53 (1H each, both d, J = 10.8 Hz, H2-28)], and three oxymethine [δ 3.22 (1H, dd, J = 3.0, 10.2 Hz, H-3), 3.82 (1H, d, J = 9.6 Hz, H-22), 4.22 (1H, d, J = 9.6 Hz, H-21)] (Table 3), indicating 11 was also one of oleanane-type triterpene saponins. The planar structure of its aglycon was elucidated according to the proton and proton cross-peaks and the long-range correlations shown in Fig. 5.
In addition, the substitution positions of acetyl and [β-d-glucopyranosyl(1 → 2)][β-d-glucopyranosyl(1 → 4)]-β-d-glucuronopyranosyl were determined to be C-28 and C-3 through the HMBC cross-peaks from δH 4.48, 4.53 (H2-28) to δC 171.0 (C-1′''') and δH 4.93 (H-1′) to δC 89.4 (C-3) (Fig. 5), respectively. Based on the consistency of NMR signals at positions 1–12 and 23–26, the substituents and configurations of compounds 10 and 11 on the A–C ring were inferred to be identical. Moreover, the configurations of C-14, C-17, C-18, C-21, and C-22 were consolidated according to the NOE correlations between H3-27 and H-19α; H-19α and H-21; H2-28 and H-22, H3-26; H-22 and H-18 (Fig. 6). Therefore, the structure of aeswilsaponin IIIA (11) was clarified.
Comparied the spectroscopic data with those reported in literatures, the structures of known compounds 12–28, 30, and 31 were identified as (3β,16α,21β,22α)-16,21,22,24,28-pentahydroxy-olean-12-en-3-O-β-d-glucopyranosiduronic acid (12) (Kim et al., 2017), 21β-O-tigloyl-22α-O-acetylprotoaescigenin-3β-O-[β-d-glucopyranosyl (1 → 2)]-β-d-glucopyranosiduronic acid (13) (Zhao et al., 2012), 21β-O-angeloyl-22α-O-acetylprotoaescigenin-3β-O-[β-d-glucopyranosyl(1 → 2)]-β-d-glucopyranosiduronic acid (14) (Zhao et al., 2012), (3β,16α,21β,22α)-16,21,22,24,28-pentahydroxyolean-12-en-3-yl-O-[β-d-glycopyranosyl(1 → 4)]-β-d-glucopyranosiduronic acid (15) (Kim et al., 2017), desacylescin I (16) (Yoshikawa et al., 1996), aesculusoside C (17) (Cheng et al., 2018), aesculioside A (18) (Zhang et al., 1999), aesculioside B (19) (Zhang et al., 1999), aesculusoside A (20) (Cheng et al., 2018), escin IV (21) (Yoshikawa et al., 1998), escin V (22) (Yoshikawa et al., 1998), escin Ia (23) (Yoshikawa et al., 1996), escin Ib (24) (Yoshikawa et al., 1996), aesculiside A (25) (Wei et al., 2004), isoescin Ia (26) (Zhang et al., 1999), isoescin Ib (27) (Zhang et al., 1999), aesculusoside B (28)(Cheng et al., 2018), 6′-methyleter-O-aesculiside A (30) (Yang et al., 2019), and aesculioside G (31) (Zhang et al., 1999), respectively. The NMR data (see supplymentary materials) of olean-12-ene-16α,21β,22α,24,28-pentol,3β-[(O-β-d-glucopyranosyl(1 → 2)-O-[β-d-glucopyranosyl(1 → 4)]-β-d-glucopyranuronosyl)oxy]-,methyl ester,22-acetate 21-((Z)-2-methylcrotonate) (29) was reported here firstly.
3.2 Anti-inflammatory activity screening and structure–activity relationships analysis of saponins from A. wilsonian seeds
The nitric oxide (NO) production inhibitory activities of obtained triterpene saponins (1–31) were evaluated on an LPS-stimulated RAW264.7 cells model. Based on the results of MTT assay (Fig. S86, S87), the NO release inhibitory effects screening of compounds 1–31 implied that new isolates, 1, 4, and 5, as well as known ones, 12, 18, 22–24, 26, and 27 could inhibit NO production at the measured concentration (The bioassay concentrations were 10 μM for 8, 13, 14, 29; 30 μM for 9, 23, 24, 31, and 50 μM for 1–7, 10, 11, 12, 15–28, 30, Table S1). Moreover, compounds 23 and 24 displayed a concentration-dependent manner at 3, 10, 30 μM, 1, 4, 5, 12, 18, 22, 26, and 27 showed the similar phenomenon at concentration of 10, 25, 50 μM (Fig. 7). The structure–activity relationships (SARs) suggested that tigloyl and angeloyl substitution could significantly increase the inhibition of NO release in RAW264.7 cells (23, 24 > 22; 17, 18, 19 > 16). Meanwhile, both C-22 acetylation and more glycosylation enhanced the activity (23 > 18, 26; 24 > 19, 27; 23 > 13; 24 > 14); However, the activity was decreased by methyl glucuronate (23 > 29; 24 > 8).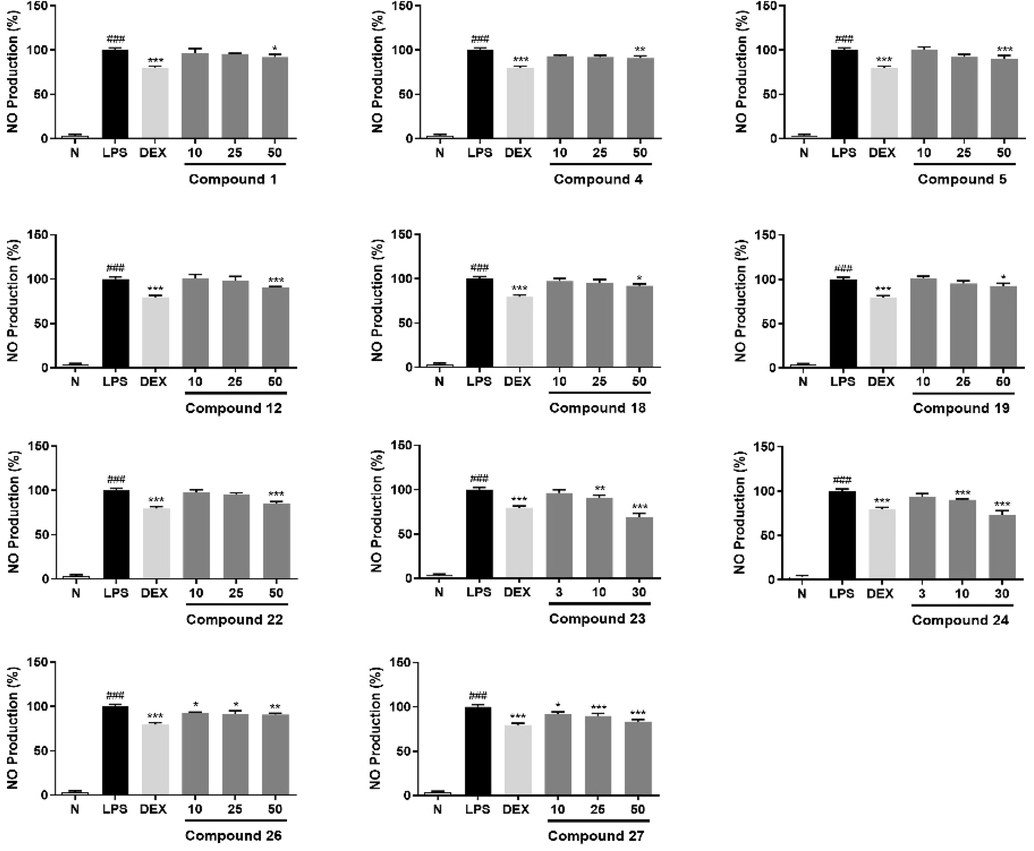
Inhibitory effects of compounds 1, 4, 5, 12, 18, 19, 22, 26, and 27 at concentration of 10, 25, 50 μM; 23 and 24 at concentration of 3, 10, 30 μM on NO production in RAW264.7 cells, respectively. Nitrite relative concentration (NRC): percentage of control group (set as 100%). Values represent the mean ± SD of six determinations. *P < 0.05; **P < 0.01; ***P < 0.001 (Differences between compound-treated group and control group). ###p < 0.001 (Differences between control group and normal group).
3.3 Saponins from A. wilsonian seeds suppressed LPS-induced inflammation on RAW 264.7 cells via NF-κB pathway
Additionally, the anti-inflammatory signaling pathway of new compounds 1, 4, 5 were further researched by western blot assay in RAW 264.7 macrophage cell model. As shown in Fig. 8A, the protein expression of inflammation-related factors, iNOS, COX-2, IL-6, and TNF-α increased significantly in the model group. Compared with model, compounds 4 and 5 could down-regulated the level of iNOS, COX-2, IL-6, and TNF-α at 50 μM. While compound 1 could reduce the expression of COX-2 and TNF-α, but had no effect on the production of IL-6 and iNOS, furtherly suggesting that more glycosylation enhanced the anti-inflammatory activity. The level changes of IL-6, TNF-α, COX-2, and iNOS were closely related to the regulation of NF-κB signaling pathway (Karin and Greten, 2005; Kim, et al., 2015). To clarify it, western blot assay was conducted on the protiens of p65, p-p65, IKKα/β, and p-IKK-α/β. Our study demonstrated that LPS significantly elevated the ratios of p-p65 to p65 (p-p65/p65) and p-IKK-α/β to IKKα/β (p-IKK-α/β/IKKα/β) on RAW 264.7 cells. However, the values of p-IKK-α/β/IKKα/β and p-p65/p65 were decreased after treating with compounds 1, 4, and 5 (Fig. 8A), indicating that the NF-κB signaling pathway was involved in the process of regulating inflammatory factor release by the above-tested compounds.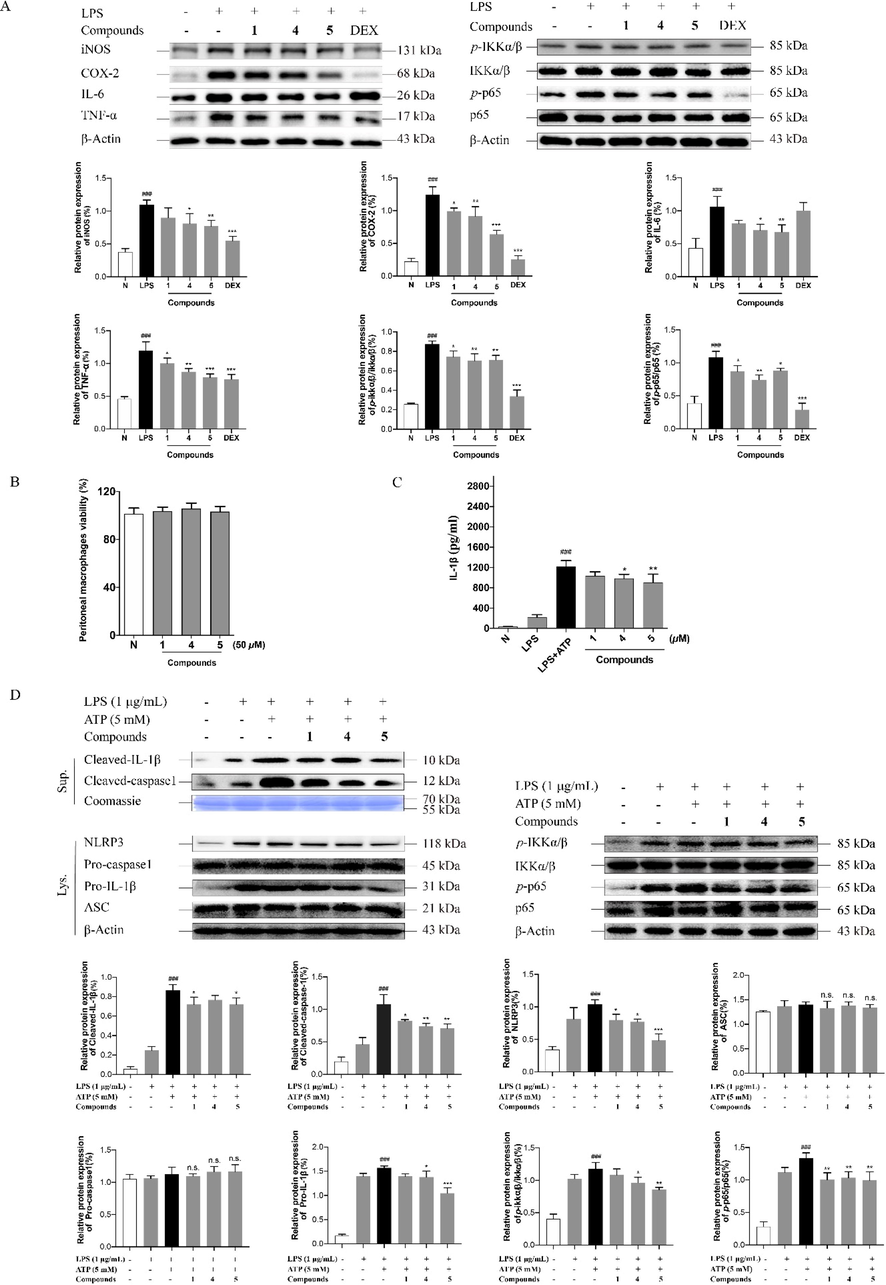
Compounds 1, 4, and 5 exert anti-inflammatory activity through NF-κB and NLRP3 signaling pathway. TNF-α, IL-6, iNOS, COX-2 levels in RAW 264.7 cells treated with the tested compounds or LPS for 18 h and IKK-α/β, p-IKK-α/β, NF-κB/p65 and p-p65 levels in RAW 264.7 cells treated with the tested compounds or LPS for 8 h were analyzed by Weatern blot analysis (A); Effects of 1, 4, and 5 in PMs cells viability (B); The levels of IL-1β in supernatants in PMs cells (C); The protein expression of IKK-α/β, p-IKK-α/β, NF-κB/p65, p-p65, cleaved caspase-1, cleaved IL-1β, NLRP3, Pro-caspase-1, Pro-IL-1β and ASC in LPS/ATP-induced PMs cells (D). Fig. 8A: N: normal group without LPS, DEX and tested samples; LPS: model group with 0.5 μg/mL LPS; DEX: positive drug group with 0.5 μg/mL LPS + 1.5 μg/mL DEX; tested compound groups were treated with 0.5 μg/mL LPS + 50 μM compounds 1, 4, and 5, respectively. Fig. 8D: N: normal group without LPS, ATP and tested compounds, LPS: model group with 1 μg/mL LPS; LPS/ATP: model group with 1 μg/mL LPS plus 5 mM ATP; tested compound groups were treated with 1 μg/mL LPS plus 5 mM ATP + 50 μM compounds 1, 4, and 5, respectively. Values represent the mean ± SD of three determinations. *P < 0.05; **P < 0.01; ***P < 0.001 (Differences between compound-treated group and control group). ##P < 0.01; ###P < 0.001 (Differences between LPS-treated group and control group).
3.4 Saponins from A. wilsonian seeds suppressed LPS/ATP-induced inflammation on PMs cells via NF-κB/NLRP3 pathway
NLRP3 is a large multiprotein complex composed of sensor protein NLRP3, adaptor protein ASC, and caspase-1 (Lu et al., 2014). ASC is responsible for the assembly of inflammasomes. However, RAW 264.7 macrophage cells lack ASC gene expression (He et al., 2015). Then, LPS/ATP-induced PMs cell model (Cai et al., 2022) was chosen to verify how compounds 1, 4, and 5 regulate NF-κB/NLRP3 pathway.
The MTT assay results implied that compounds 1, 4, and 5 showed no effect on cellular viability at the concentration of 50 μM on PMs cells (Fig. 8B). ELISA results demonstrated that the concentration of IL-1β in supernatant were significantly increased after LPS/ATP treatment. Nevertheless, 50 μM of compounds 4 and 5 treatment reduced the production of inflammatory cytokines IL-1β. Compound 1 showed a tendency to inhibit the secretion of IL-1β, while there was no significant difference compared with the model group (Fig. 8C). In addition, the NLRP3 signaling relevant proteins were assayed. As shown in Fig. 8D, all of the three tested compounds had the inhibitory tendency on the NLRP3 inflammasome priming and assembling genes, NLRP3, pro-IL-1β, and ASC, as well as on its activation-related proteins, cleaved IL-1β and cleaved caspase-1. However, compound 1 significantly down-regulated the protein expression of NLRP3, cleaved IL-1β, and cleaved caspase-1; the level of NLRP3, pro-IL-1β, and cleaved caspase-1 was reduced by compound 4. Compound 5 could remarkably inhibit the expression of NLRP3, pro-IL-1β, and cleaved caspase-1 in comparison with compounds 1 and 4. NF-κB signaling activation status was tested to further validate whether the compounds 1, 4, and 5 inhibited NF-κB-mediated regulation on the NLRP3 inflammasome by western blot assay. After LPS/ATP stimulation in PMs cells, the levels of IKKα/β, p-IKK-α/β, p65, and p-p65 were profoundly elevated. Nevertheless, compounds 4 and 5 reduced the values of p-IKK-α/β/IKKα/β and p-p65/p65, 1 decreased the value of p-p65/p65, remarkably (Fig. 8D), revealing compounds 1, 4, and 5 suppressed NLRP3 expression partly through NF-κB signaling pathway.
4 Discussion
A. wilsonii, A. chinensis Bge, and A. chinensis Bge. var. chekiangensis (Hu et Fang) are the major sources of the traditional Chinese medicine ''Suo Luo Zi'' included in the Pharmacopoeia of the People's Republic of China (Cao et al., 2023). Though ''Suo Luo Zi'' have been proved to be rich in polyhydroxyoleanene triterpene saponins, little is known about the chemical constituents and the biological activity of A. wilsonii compared with the other two species. In the process of investigating the triterpene saponins, eleven unreported isolates, aeswilsaponins IA–IH (1–8), ⅡA (9), ⅡB (10) and ⅢC (11), together with twenty known ones (12–31) were afforded. The current work has given researchers a deeper understanding of the materials of ''Suo Luo Zi''. For the first time, we have completed the systematic phytochemical study of saponin components in A. wilsonii.
When the obtained saponins from A. wilsonii seeds were compared to those from A. chinensis and A. chinensis var. chekiangensis, it was discovered that while the major saponins in A. wilsonii seeds and the other two are comparable, A. wilsonii seeds have a different composition. What’s more, only glucuronic acid and glucose substitutions have been found in A. wilsonii saponins so far, while the saponins from the seeds of the latter two have a wide variety of substituted glycosyls, including glucuronic acid, glucose, xylose, and galactose (Lu et al., 2016; Zhang et al., 2020). Moreover, known compounds, 12 and 15 have been only isolated from the seeds of A. wilsonii. All these characteristics can be used to distinguish the seeds of A. wilsonii, A. chinensis, and A. chinensis var. chekiangensis.
The discovery of anti-inflammatory drugs and the treatment of inflammation are crucial since inflammation is a major factor for the progression of various chronic diseases or disorders (Arulselvan et al., 2016). NO is an important inflammatory mediator secreted by activated macrophages, and its secretion level is a vital indicator for various inflammatory models (Oishi et al., 2020). There are many macrophage or macrophage-like cell lines for screening anti-inflammatory constituents in vitro, which including RAW 264.7 cells, THP-1 cells (Chanput et al., 2010), and PMs cells (Wang et al., 2009), etc.. Among them, RAW 264.7 cell is a common cell line for studying microbiological immunology and other related research fields because of its strong ability to adhere to phagocytosis antigens (Ruan et al., 2019). Additionally, it possesses the qualities of stability and simplicity of use. Therefore, it is suitable for rapid activity screening of large numbers of natural products. In this study, LPS-stimulated RAW264.7 cell model was used to evaluate the potential anti-inflammatory activity of the obtained thirty-one triterpene saponins. As a result, new compounds 1, 4, and 5, as well as known ones, 12, 18, 22–24, 26, and 27 were found to inhibit NO release. Among them, escin Ⅰa (23), escin Ⅰb (24), isoescin Ⅰa (25), and isoescin Ⅰb (26), the main saponins in A. wilsonian seeds, showed strong inhibitory activities, which was consistent with literature reports (Cheng et al., 2015; Gallelli et al., 2019). However, the selective pressure exerted by the continuous passage of these cell lines usually leads to the loss of some genes, which are not important for cell proliferation, but are crucial for the immune function of macrophages, especially making it difficult for such passage of cells to show the true cell physiological activity of primary macrophages. Therefore, in order to better study the immune response of macrophages after being infected by pathogens, primary cell lines are often used. At present, the three main sources of primary mouse macrophages are PMs, alveolar macrophages (AMs), and bone marrow-derived macrophages (BMDMs) (Ma et al., 2022). Among them, PMs are the major cell type of peritoneal cells that participate in multiple aspects of innate and acquired immunity in the peritoneal cavity. Owing to their strong ability to release a large number of inflammatory cytokines, they play a critical role in various mechanism studies for multiple diseases closely related to inflammation response (Liu et al., 2018). What’s more, considering that the acquisition of PMs is relatively easy, PMs were used for mechanism research in this study. Herein, the potential anti-inflammatory pharmaceutical substances of A. wilsonii have been clarified to be its saponins for the first time.
Escin is a triterpene saponins mixture, which mainly consists of escin A, B, C, and D, extracted from ''Suo Luo Zi''. Escin can suppress the activation of NF-κB in the skin of rats with paw edema and capillary permeability by increasing the expression of the glucocorticoid receptor, which further inhibits the expression of TNF-α and IL-1β (Zhao et al., 2018). On the other hand, it could also lessen concanavalin A-induced autoimmune hepatitis in mice by lowering neutrophil infiltration and blocking TNF-α/NF-κB signaling pathways (Elshal and Hazem, 2022). What's more, combined pretreatment with coenzyme Q10 and escin enhanced anti-inflammatory effect via suppressing the production of TNF-α and IL-1β as well as the expression of NF-κB and NLRP3 in LPS-induced lung injury experiments (Ali et al., 2021). According to literature review, A. wilsonii seeds was reported to be rich in saponins, but most of the studies on the anti-inflammatory activity of it were limited to escin. Though there are many researches on the saponins of ''Suo Luo Zi'', the further bioactivity of the monomer components, especially the in-depth study of their action mechanism is very rare. On the basis of comprehensive phytochemistry investigation, we investigated the anti-inflammatory effects of mono-saponins from ''Suo Luo Zi'' based on the NF-κB and NLRP3 pathway for the first time. In LPS-stimulated RAW 264.7 cells, compounds 4 and 5 intervention could down-regulated the expression level of inflammation-related factors such as iNOS, COX-2, IL-6, and TNF-α, and 1 treatment could reduce the expression of COX-2 and TNF-α, which may be realized by inhibiting the phosphorylation of Ikk-α/β and p-p65 to p65, in NF-κB signaling pathway. Furtherly, in the LPS/ATP-induced PMs cell model, the three tested ones had the inhibitory tendency on the NLRP3 inflammasome priming and assembling genes, NLRP3, pro-IL-1β, ASC, cleaved IL-1β, and cleaved caspase-1. Among them, compounds 4, 5 inhibited the values of p-IKK-α/β/IKKα/β and p-p65/p65, and 1 reduced the value of p-p65/p65, clarifying they inhibit inflammatory response through NF-κB/NLRP3 signaling pathway (Fig. 9). The NLRP3 pathway is an emerging inflammation-related pathway, the extraordinary regulatory effects of new saponins in A. wilsonii may provide new ideas for its application in the research and development of anti-inflammatory Drug development.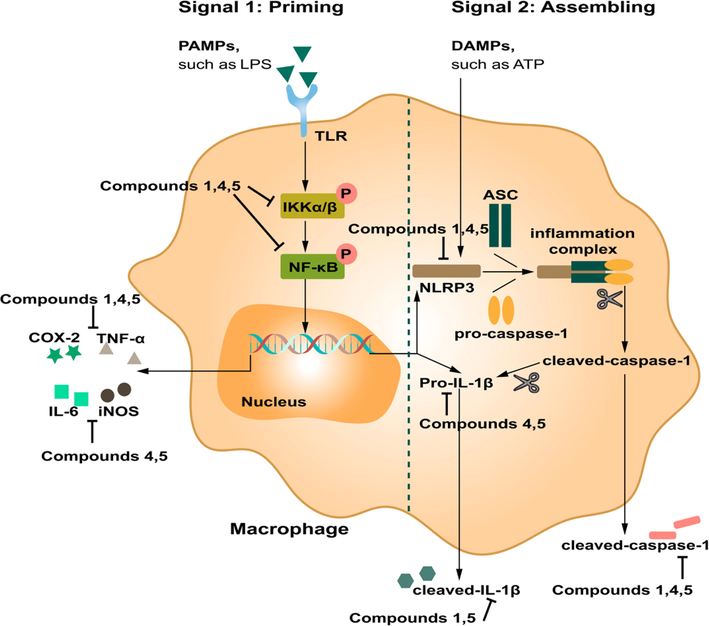
The mechanism diagram of compounds 1, 4, and 5 exerting anti-inflammatory activity.
5 Conclusion
In the process of investigating the triterpene saponins, eleven unreported isolates, aeswilsaponins IA–IH (1–8), ⅡA (9), ⅡB (10), and ⅢC (11) together with twenty known ones (12–31) were afforded. Among the known isolates, 29 was gained from Aesculu genus firstly, 12–15, 17, 18, 20–22, 28, and 31 were obtained from this species for the first time. Additionaly, the characteristics of saponins in the seeds of A. wilsonii, A. chinensis, and A. chinensis var. chekiangensis were summarized to distinguish them. Compounds 1, 4, 5, 12, 18, 22–24, 26, and 27 showed NO release inhibitory activity on LPS-stimulated RAW 264.7 cells. SARs study suggested that C-21 tigloyl or angeloyl substitution, C-22 acetylation, and the increase of glycosyl groups might play the important role for the activity. The further western blot assay demonstrated that new saponins 1, 4, and 5 suppressed inflammatory response partly via NF-κB/NLRP3 signaling pathway. This work will provide potential molecular mechanism of ''Suo Luo Zi'' and its saponins, as well as supply novel candidates for treating inflammation-related diseases. In the future studies, in addition to exploring the in-depth molecular mechanism of ''Suo Luo Zi'' saponin in vitro, its in vivo study on the inflammation-related disease models should be carried out as well.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant no. 82074118).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Limiting inflammation-the negative regulation of NF-κB and the NLRP3 inflammasome. Nat. Immunol.. 2017;18(8):861-869.
- [Google Scholar]
- Pretreatment with coenzyme Q10 combined with aescin protects against sepsis-induced acute lung injury. Cells Tissues Organs.. 2021;210(3):195-217.
- [Google Scholar]
- Role of antioxidants and natural products in inflammation. Oxid. Med. Cell Longev.. 2016;2016:5276130.
- [Google Scholar]
- USP5 attenuates NLRP3 inflammasome activation by promoting autophagic degradation of NLRP3. Autophagy. 2022;18(5):990-1004.
- [Google Scholar]
- NO release inhibitory activity of flavonoids from Aesculus wilsonii seeds through MAPK (p38), NF-κB and STAT3 cross-talk signaling pathways. Planta Med.. 2023;89(1):46-61.
- [Google Scholar]
- Transcription profiles of LPS-stimulated THP-1 monocytes and macrophages: a tool to study inflammation modulating effects of food-derived compounds. Food Funct.. 2010;1(3):254-261.
- [Google Scholar]
- NEK7 interacts with NLRP3 to modulate the pyroptosis in inflammatory bowel disease via NF-κB signaling. Cell Death Dis.. 2019;10(12):906.
- [Google Scholar]
- Triterpenoid saponins from the seeds of Aesculus chinensis and their cytotoxicities. Nat. Prod. Bioprospect.. 2018;8(1):47-56.
- [Google Scholar]
- Escin increases the survival rate of LPS-induced septic mice through inhibition of HMGB1 release from macrophages. Cell Physiol. Biochem.. 2015;36(4):1577-1586.
- [Google Scholar]
- Escin suppresses immune cell infiltration and selectively modulates Nrf2/HO-1, TNF-α/JNK, and IL-22/STAT3 signaling pathways in concanavalin A-induced autoimmune hepatitis in mice. Inflammopharmacology. 2022;30(6):2317-2329.
- [Google Scholar]
- Escin: a review of its anti-edematous, anti-inflammatory, and venotonic properties. Drug Des. Devel. Ther.. 2019;13:3425-3437.
- [Google Scholar]
- A microRNA-21-mediated SATB1/S100A9/NF-κB axis promotes chronic obstructive pulmonary disease pathogenesis. Sci. Transl. Med.. 2021;13(621):eaav7223.
- [Google Scholar]
- Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res.. 2015;25(12):1285-1298.
- [Google Scholar]
- Celastrol inhibits rheumatoid arthritis through the ROS-NF-κB-NLRP3 inflammasome axis. Int. Immunopharmacol.. 2021;98:107879
- [Google Scholar]
- NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat. Rev. Immunol.. 2005;5(10):749-759.
- [Google Scholar]
- The NLRP3 inflammasome: an overview of mechanisms of activation and regulation. Int. J. Mol. Sci.. 2019;20(13):3328.
- [Google Scholar]
- Antiviral escin derivatives from the seeds of Aesculus turbinata Blume (Japanese horse chestnut) Bioorg. Med. Chem. Lett.. 2017;27(13):3019-3025.
- [Google Scholar]
- Intestinal anti-inflammatory activity of Sasa quelpaertensis leaf extract by suppressing lipopolysaccharide-stimulated inflammatory mediators in intestinal epithelial Caco-2 cells co-cultured with RAW 264.7 macrophage cells. Nutr. Res. Pract.. 2015;9(1):3-10.
- [Google Scholar]
- Polyphenols from the peels of Punica granatum L. and their bioactivity of suppressing lipopolysaccharide-stimulated inflammatory cytokines and mediators in RAW 264.7 cells via activating p38 MAPK and NF-κB signaling pathways. Molecules. 2022;27(14):4622.
- [Google Scholar]
- The Chinese medicine babaodan suppresses LPS-induced sepsis by inhibiting NLRP3-mediated inflammasome activation. J. Ethnopharmacol.. 2022;292:115205
- [Google Scholar]
- The peritoneal macrophages in inflammatory diseases and abdominal cancers. Oncol. Res.. 2018;26(5):817-826.
- [Google Scholar]
- Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156(6):1193-1206.
- [Google Scholar]
- Research progress on the chemical constituents and bioactivity of Aesculus chinensis Bunnge var. chinensis. Xibei Yaoxue Zazhi. 2016;31(6):651-654.
- [Google Scholar]
- Establishment and identification of bone marrow-derived macrophage culture model. Chin. J. Biol.. 2022;35(6):706-717.
- [Google Scholar]
- Intracellular pattern recognition receptors in the host response. Nature. 2006;442(7098):39-44.
- [Google Scholar]
- Role of type2 inflammatory biomarkers in chronic obstructive pulmonary disease. J. Clin. Med.. 2020;9(8):2670.
- [Google Scholar]
- SARS-CoV-2 N protein promotes NLRP3 inflammasome activation to induce hyperinflammation. Nat. Commun.. 2021;12(1):5306.
- [Google Scholar]
- The NLRP3-inflammasome in health and disease. Int. J. Mol. Sci.. 2022;23(21):13103.
- [Google Scholar]
- Bioactive constituents from the roots of Eurycoma longifolia. Molecules. 2019;24(17):3157.
- [Google Scholar]
- Punicalin ameliorates cell pyroptosis induced by LPS/ATP through suppression of ROS/NLRP3 pathway. J. Inflamm. Res.. 2021;14:711-718.
- [Google Scholar]
- Subergorgiaols A-L, 9,10-secosteroids from the south China sea gorgonian Subergorgia rubra. Steroids. 2015;94:7-14.
- [Google Scholar]
- Facile discrimination of aldose enantiomers by reversed-phase HPLC. Chem. Pharm. Bull.. 2007;55(6):899-901.
- [Google Scholar]
- Screening anti-inflammatory components from Chinese traditional medicines using a peritoneal macrophage/cell membrane chromatography-offline-GC/MS method. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci.. 2009;877(27):3019-3024.
- [Google Scholar]
- Escin attenuates cerebral edema induced by acute omethoate poisoning. Toxicol. Mech. Methods. 2011;21(5):400-405.
- [Google Scholar]
- REV-ERBα integrates colon clock with experimental colitis through regulation of NF-κB/NLRP3 axis. Nat. Commun.. 2018;9(1):4246.
- [Google Scholar]
- Antiinflammatory triterpenoid saponins from the seeds of Aesculus chinensis. Chem. Pharm. Bull.. 2004;52(10):1246-1248.
- [Google Scholar]
- 16-Tigloyl linked barrigenol-like triterpenoid from Semen aesculi and its anti-tumor activity in vivo and in vitro. RSC Adv.. 2019;9(54):31758-31772.
- [Google Scholar]
- Recent advances in the mechanisms of NLRP3 inflammasome activation and its inhibitors. Cell Death Dis.. 2019;10(2):128.
- [Google Scholar]
- Yoshikawa, M., Murakami, T., Matsuda, H., Yamahara, J., Murakami, N., Kitagawa, I., 1996. Bioactive saponins and glycosides. III. Horse chestnut. (1): The structures, inhibitory effects on ethanol absorption, and hypoglycemic activity of escins Ia, Ib, IIa, IIb, and IIIa from the seeds of Aesculus hippocastanum L. Chem. Pharm. Bull. 44 (8), 1454–1464.
- Yoshikawa, M., Murakami, T., Yamahara, J., Matsuda, H., 1998. Bioactive saponins and glycosides. XII. Horse chestnut.1) (2): Structures of escins IIIb, IV, V, and VI and isoescins Ia, Ib, and V, acylated polyhydroxyoleanene triterpene oligoglycosides, from the seeds of horse chestnut tree (Aesculus hippocastanum L., Hippocastanaceae). Chem. Pharm. Bull. 46 (11), 1764–1769.
- Zhang, X., Goncalves, R., Mosser, D. M., 2008. The isolation and characterization of murine macrophages. Curr. Protoc. Immunol. Chapter 14, 14.1.1–14.1.14.
- New saponins from the seeds of Aesculus chinensis. Chem. Pharm. Bull.. 1999;47(11):1515-1520.
- [Google Scholar]
- Bioactive triterpenoid saponins from the seeds of Aesculus chinensis Bge. var. Chekiangensis. Front. Chem.. 2020;7:908.
- [Google Scholar]
- Chemical constituents in zymolyte of Aesculi Semen extract. Xiandai Yaowu Yu Linchuang. 2012;27(5):442-445.
- [Google Scholar]
- Anti-inflammatory effect of external use of escin on cutaneous inflammation: possible involvement of glucocorticoids receptor. Chin J Nat Med.. 2018;16(2):105-112.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary materials associated with this article include the NMR and HRESIMS spectra of compounds 1–11, cell viability assay on RAW 264.7 cell model of compounds 1–31, the raw data for western blot assays, and the physical and chemical data of compounds 29, 1a, and 9a. Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2023.105077.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary data 1
Supplementary data 1







