Translate this page into:
Screening of small molecular compounds with carcinogenic inhibition function of HPV-16 E6
⁎Corresponding author at: Key Laboratory of Bio-Resources and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu 610065, Sichuan, PR China brainding@scu.edu.cn (Xianping Ding)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Cervical cancer is the second most malignant tumor among women of childbearing age, almost all cervical cancers are associated with high-risk (HR) human papillomavirus (HPV) persistent infection. HPV persistent infection and cell immortalization does not always cause cancer, but they increase risk and can lead to tumor formation and carcinogenesis in cooperation with other tumorigenic factors, and HR-HPV E6 oncoprotein is the mainly tumorigenic factors.
E6 binds to the highly conserved sequence “LXXLL” of E6AP to form a heterodimer, which recruits and induces the degradation of tumor suppressor protein p53, trigger the immortalized transformation of infected cells, and induce its carcinogenicity. Therefore, the small molecule compounds, who can compete the conservative and stable binding pocket of E6-E6AP, may inhibited the carcinogenicity risk of HR-HPV E6, and HPV-16 is one of the most important HR-HPV.
In this study, DOCK6, AutoDock Vina, Visual screen and GROMACS V4.5.7 were used to screen the candidate compounds from Specs library, and based on the pharmacokinetic properties and toxicity prediction by ADMSlab and ProtoX-II analytical tools, 20 E6-E6AP binding inhibition small molecule compounds were selected out. They were verified by molecular interactions, cell proliferation inhibition, apoptosis induction and p53/p21 protein content/expression on SiHa (HPV-16 infected line). Ultimately, compound 4 among them with good tumor suppressive potential was picked out, has the potential of medicine to HR-HPV related diseases.
Keywords
Cervical cancer
HR-HPV
Virtual screening
Inhibitors
Small molecule compounds
1 Introduction
Cervical cancer closely related to the persistent infection of HR-HPV, and is the major causes of death among women in developing countries (He and He, 2019). HPV prophylactic vaccine based on HPV VLP has been applied in clinic, with a certain effect on the prevention of HPV infection and related diseases, but, it has no therapeutic significance for patients who have already been infected with HPV or suffer from related diseases or even cervical cancer (Mark et al., 2009; Xu et al., 2019; Ennaifer et al., 2015). Cytotoxic drugs or surgical treatment, are mainly adopted for the cases of skin, genital warts, and advanced cervical precancerous lesion, to achieve rapid removal of lesions, but recurrent easy (Leaver, 2016; Ungár et al., 2011). Currently, there is no specific therapy methods, and effective antiviral drug is an urgent need to treat HPV infection and reduce the possibility that patients with HPV infection developing invasive cervical cancer.
p53 and Rb are famous tumor suppressor protein, HR-HPV E6 oncoprotein binds and ubiquitinates degrade p53 to inhibit its tumor suppressive function, and promote the immortality of keratinocytes (Yun et al., 2019; Sherr and Mccormick, 2002); E7 binds to pRb family proteins, activates and releases E2F transcription factor, initiates DNA synthesis and gene transcription, and finally induces the persistence of HR HPV infection (Yun et al., 2019; Liu et al., 2006); E6 and E7 oncoproteins disrupts the cell cycle, make a contribution to the non-morphogenetic transformation of HR-HPV infected cells, and paves the way for cell immortalization and carcinogenic ability (Ortiz-Ortiz et al., 2015; Roman and Munger, 2013; Moody and Laimins, 2010; Alvarez-salas, 2014), while the conversion induce efficiency of HPV E6 in infected cells into permanent is more obvious than that of E7, without E7, E6 can blind to the highly conserved sequence “LXXLL” of E6AP to form heterodimer, prompt ubiquitination degradation of p53 and induce the immortality of infected cells (Martinez-Zapien et al., 2016). α-9 HPV are almost all HR-HPV, HPV-16 is one of the most important HR-HPV, and its E6 is composed of 158 amino acids, that contains n-terminal domain, c-terminal domain and two Zinc finger domains three domains (Barbosa et al., 1989). Among them, the binding domains of two Zinc fingers form a deep pocket, which can bind to the “LXXLL” sequence of E6AP (Baleja et al., 2006). Therefore, the oncogenic induce function of HPV-16 can be inhibited by compete the binding pocket of E6 to E6AP.
Computer aided Drug Design (CADD) is widely used in inhibitors drug development (Tambunan and Parikesit, 2012). Ligand-based and structure-based virtual screening was used to simulate the binding process of drug targets and molecules through large-scale calculation by molecular docking and other technologies, evaluate the affinity of molecules to target, and finally screen out potential drug candidates with high affinity (Kumar et al., 2015; Najmanovich, 2017; Arnold et al., 2009). Virtual screening eliminates the steps of experimental screening, reduces the time and cost of drug development by conducting the main screening tasks and experimental verification through computer. The same approach has been used for small molecules screening for a variety of diseases, such as SARS-CoV-2 papain-like protease interaction small molecules, of which three compounds can be considered lead compounds that require further characterisation of potential antiviral effects in appropriate model systems (Epa et al., 2021).
Some researchers have attempted to design or screen E6-E6AP interactions inhibitors, but still in the research stage, such as Pitx2a, a dual-homologous domain transcription factor that can bind HPV-18 E6 protein to inhibit p53 degradation and induce cell cycle arrest, but with low activity. HR-HPV persistence infection induce the malignant tumors of reproductive system and other diseases, seriously threaten human health, and α-9 HPV persistence infection is associated with majority invasive cervical cancer, therefore it is of great practical significance to screen its E6-E6AP interaction inhibitors (Inhibitors et al., 2014; Inhibitors et al., 2014); can provide effective reference for the treatment of these diseases, and has positive significance for the treatment of HPV-related diseases.
2 Materials and methods
2.1 Molecular docking
X-ray crystal structure of HPV-16 E6 was obtained from RCSB protein databank (PDB code: 4XR8) in E6-E6AP-P53 complex crystal model, and the “LXXLL” binding pocket of E6 was selected as the virtual screening potential target (Waterhouse et al., 2018; Rodríguez-ruiz et al., 2019). E6 homologous comparision among HPV-16 was constrcuted by welogo 3.7.4, based on the all HPV-16 E6 protein from UniRef100.
The drug candidates were virtual screened from the Specs small molecule compound database, and the molecules with rotatable keys number is less than 10; relative molecular weight is less than 500; 2 < LogP < 5 is greater; hydrogen bond donors number is<5; hydrogen bond receptors number is less than 10 were qualified for virtual screening (Baleja et al., 2006).
DOCK6 was used for the primary rigid docking screen, Dock Prep tool of the Chimera program (https://www.cgl.ucsf.edu/chimera) was used to parameters manually set (Lu et al., 2018). The top 10% best docking results were selected for AutoDock Vina virtual screening, AutoDockTools software was used for parameter configuration of receptor protein and ligand, and the top 15% optimal molecules docking results were selected for subsequent study (Lu et al., 2018).
2.2 Visual screening, pharmacokinetic properties, toxicity prediction
In visual screening, E6-E6AP binding inhibitor candidate molecules were screened, based on the empirical rules (molecular surface geometric complementarity, physical and chemical complementarity, and other characteristics), the pharmacokinetic properties and toxicity of the selected candidate molecules were predicted by ADMSlab and ProtoX-II analytical tools, to evaluate the physicochemical characteristics, absorption, toxicity, and mutagenicity of these molecules.
2.3 Molecular dynamics simulation
The reliability and stability docking conformation of the candidate molecules with optimal binding conformation were evaluated by 30 ns molecular dynamics simulation in GROMACS V4.5.7 (Van Der Spoel et al., 2005). Root Mean Square Deviation (RMSD) is an important and intuitive index to evaluate the stability of protein ligand system in dynamic simulation. Lower the RMSD value is more stable the system, the target protein has a more stable conformation in the binding state with small molecules, indicate that the target protein and small molecules interacted well.
The topology file of the receptor protein and ligand molecule were generated by pdb2gmx program in GROMACS with AMBER99sb force field and Acpype program based on AMBERff99sb field (Van Der Spoel et al., 2005). Water molecular model was constructed by TIP3P water model. The coordinate files of these proteins and ligands were edited to construct the topology of the system;reconstruct the protein–ligand complex, and creat a rhombic dodecahedron periodic simulation box with a 1.0 nm minimum distance from the molecules to the box edge. A certain number of compensating ions was added to neutralize the net charge, and the whole system energy was minimized to 1000 kJ/mol/nm by steepest descent algorithm to reduce the unreasonable protein conformation. A two-step pre-balancing method was used to balance the system and 30 ns (nanosecond, i.e. 10–9 s) dynamics simulation was performed, as well as the system coordinates, energy information every 2 FS (femtoseconds, i.e., 10–15 s) were saved for subsequent analysis.
2.4 The validation of E6-E6AP binding inhibitors
Effective E6-E6AP binding inhibitors with biological activity against HPV-16 infected cells were validated by molecular interaction, cell proliferation inhibition, cell apoptosis, cell cycle and p53/p21 protein content and expression. SiHa (HPV-16 infected cells line) was used as experimental cells to study the effects of small molecule compounds, Luteolin was positive control, and DMSO was blank (Cherry et al., 2013).
2.4.1 Detection of cell proliferation inhibition
CCK8 kit was used to detect the inhibition of cell proliferation by E6-E6AP binding inhibitors. SiHa cells at logarithmic growth stage were inoculated on 96-well plates for 24 hours at 37 ℃. Culture medium was changed and E6-E6AP binding inhibitors were added to incubate for 48 h, then, 10% CCK8 cell culture medium was added and incubated for 45 min, to calculate the cell survival rate by detecting the absorbance at 450 nm with a microplate reader, according to the formula = [(experimental well - blank well)/(control well - blank well)].
2.4.2 Detection of molecular interaction
Molecular interactions of E6 and E6-E6AP binding inhibitors candidate were conducted by ForteBio OCTET K2 with NI-NTA sensor as following steps: firstly, loading HPV-16 E6 protein (HPV-16 E6 50ug/ml with 0.02% Tween-20 PBS) and inhibitors (inhibitors 20uM with 0.02% Tween-20 PBS) curing, then, baseline 120 s, association 60–90 s, disassociation 60–90 s, finally, analyse data by forteBIO Data analysis software (version: 10.0.0.5).
2.4.3 Detection of cell apoptosis
Apoptosis was analyzed by flow cytometry using PI and AnnexinV staining. 3 × 105 SiHa cells were inoculated in 6-well plates and treated with different drugs for 48 h. Cell culture medium was collected into the centrifuge tube respectively. After washing with PBS, the collected cells were digested with trypsin without EDTA and added into the centrifuge tube, and centrifuged at 1000g for 5 min.The supernatant was discarded to collect cells. 195 μl Annexin v-fitc binding solution, 5ul Annexin v-fitc staining solution and 10 μl PI staining solution were added in sequence, beaten gently, and incubate at room temperature in dark for 20 min, then placed in ice bath and detected by flow cytometry using Flowjo software to calculate the percentage of apoptosis.
2.4.4 Detection of cell cycle
3 × 105 SiHa cells were inoculated in 6-well plates with different drugs for 48 h. The cell were collected, washed and resuscitated with PBS, then, added anhydrous ethanol with 75% ethanol concentration to fix the cell at 4 ℃ overnight. Collect the fix cells and add Propidium iodide (PI) staining solution and RNase A. After the staining was completed, the sample was stored away from light, then detected by flow cytometry, and analyzed by Flowjo software.
2.4.5 Detection of protein content and expression of p53/p21
Protein content and expression of p53/p21 were measured by Western-bloting and qPCR. 6 × 105 SiHa cells were inoculated on a 6 cm cell culture dish for 24 h, changed the medium after cell adhesion, and the small molecule compounds were obtained at 37 ℃ with 5% CO2 for 48 h. Then, SiHa cell was collected and treated with 10 × lysis buffer (2% SDS, 100 mM Tris/HCl, pH 7.6) at 95 ℃ for 5 min and centrifuged at 16000 × g for 5 min. The supernatant was used as the total cell protein extract, protein concentration was determined by BCA method, and Western blotting was performed to analyse the p53/p21 protein (antibody incubation with rabbit anti-p53/p21 IgG and sleep anti-rabbit HRP IgG).
SiHa cells were inoculated in 6-well plates. After 24 h, the medium was replaced and added small-molecule drugs for co-incubation in cell culture box for 24 h. Total RNA extraction kit was used to extract RNA from each sample, and the RNA concentration was determined and adjusted. The transcription level of related genes was detected by q-PCR one-step kit, PCR products were analyzed by fluorescence quantitative PCR and agarose gel electrophoresis.
3 Results
3.1 HPV-16 E6 protein structure and E6-E6AP binding pattern
In homology comparison of HPV-16 E6 sequence from database of UniRef100, amino acid mutation of HPV16 E6 protein was mainly in 17th, 21th, 32th, 85th and 90th, however, amino acids in the key residues V38, Y39, L57, C58, V60, V69, L74, Y77, I80, L107 and surrounding E6 pockets of HPV16 E6 interaction with E6AP are relatively conserved and not easily mutated, few amino acid sites located at the binding slot edge have differences, indicating that the binding pocket of “LXXLL” is extremely conserved and stable. The “LXXLL” fragment in the binding groove is an α-helix, and three Leu side chains at the contact position between the helix and the bottom of binding groove, which can form a strong hydrophobic interaction with the binding groove (Fig. 1). Therefore, HPV-16 E6 protein crystal structure obtained from E6-E6AP-p53 binding model was regard as the receptor, to screen the small molecule compounds that could compete with E6-E6AP binding pocket of HPV-16 E6, and inhibit its carcinogenic function induction.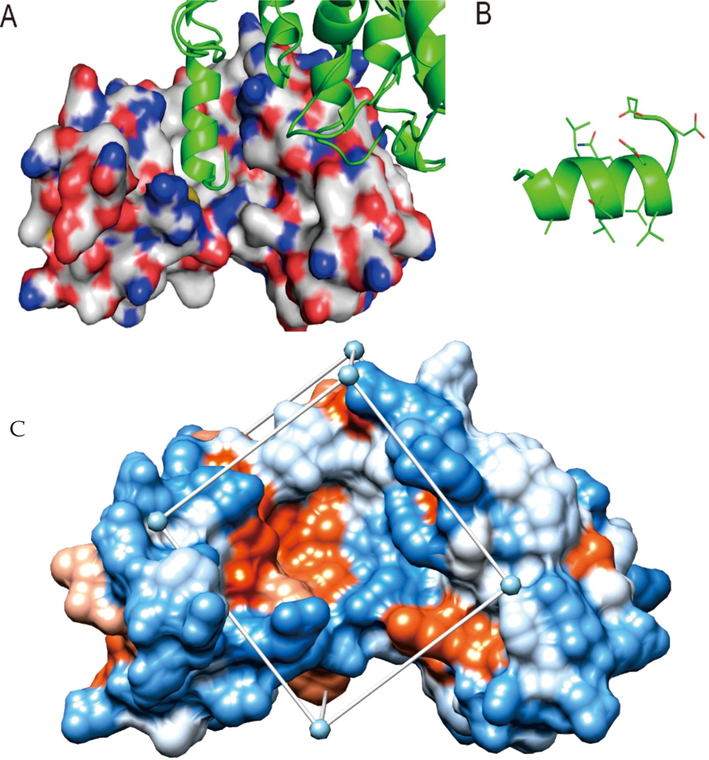
The binding mode of E6-E6AP, and crystal structure, docking space of E6 Note: A shows the binding mode of E6 and E6AP, in which the “LXXLL” segment in the binding groove is an α helix, B shows the main amino acid residues and their positions on the helix. C shows crystal structure and docking space of E6.; The box represents the docking space, which wraps around the target of E6 protein.
3.2 Molecular docking, pharmacokinetic prediction
After DOCK, AutoDock Vina two-step virtual screening and visual screening, 20 candidate molecules were selected. The details of these molecules were shown in Table 1 and Supplementary Fig. 2, the bottom of binding site, constituted by 50th Leu, 62th Val and 67th Leu side chain groups is a hydrophobic surface, and all the selected compounds contact well with the target hydrophobic bottom of the binding site. In addition, some small molecules can also form hydrogen bond interactions with polar amino acid side chains (or main chains) around the binding pocket, and stabilized the binding conformation. Among the binding sites of target proteins, 32th Tyr, 71th Ser, 74th Ser, 107th Gln and 131th Arg in side chains can participate in hydrogen bonds formation, compounds 5, 7, 9, 15, 16, 17, 18 and 19 can form hydrogen bonds with 32th Tyr, compounds 6, 8, 10, 11, 18 and 19 can form hydrogen bond interactions with 71th or 74th Ser. All molecules have good intestinal tract absorption capacity, and have good water solubility except 4, 10, 11, 14 and 20, compounds 4, 7, 11, 15 and 18 had moderate toxicity, and the mutagenicity of small molecules was<0.5 except 7. Note: Compound ID mean the number of small compound, Affinity mean the best conformation affinity value that calculated by AutoDock Vina;SPECS ID indicates the number of the SPECS company database. LogPo/w mean the oil–water distribution coefficient predicted by XLOGP. −2.0 < LogP < 5 mean small molecules that have good druggability properties; LogS indicates water solubility of small molecule, insoluble <− 10 < poorly <− 6 < moderately <− 4 < soluble <− 2 < very < 0 < highly; Caco-2 mean the permeability coefficient log value of cacO-2 monolayer cells, representing the predicted value of permeability of human intestinal mucosa, which can reflect the absorption of small molecules in vivo, and the ideal value is greater than −5.15;LD50 mean the predicted value of half lethal dose, and toxicity grade was divided into: high toxicity: 1–50 mg/kg, moderate toxicity: 51–500 mg/kg, low toxicity: 501–5000 mg/kg, actual non-toxicity: 5000–15000 mg/kg;AMES Toxicity is mutagenicity prediction of pollutants; AMES toxicity mean the mutagenicity prediction of pollutants, and the corresponding relationship between probability value and grade is as follows: 0 ∼ 0.1 (--), 0.1 ∼ 0.3 (-), 0.3 ∼ 0.5 (-), 0.5 ∼ 0.7 (+), 0.7 ∼ 0.9 (++),
Compound
IDAffinity
(kcal/mol)SPECS
IDLogPo/w
LogS
Caco-2
(cm/s)LD50
(mg/kg)AMES toxicity
1
−9.6
AM-807/41929137
1.44
−3.03
−4.6
1190
---
2
−8.9
AO-022/43452926
4.24
−4.72
−4.832
1600
–
3
−9
AB-323/13887172
2.9
−4.48
−4.741
1900
–
4
−9.2
AO-022/42598600
4.43
−6.19
−5.024
680
---
5
−8.8
AE-641/30153058
3.83
−4.17
−4.298
754
–
6
−9.5
AG-690/11351745
3.46
−4.72
−4.689
600
–
7
−9.2
AK-968/37032076
4.46
−5.82
−4.813
500
+++
8
−9.3
AQ-390/42132708
3.62
−5.47
−5.135
1500
---
9
−8.9
AI-204/31696036
4.71
−5.65
−4.743
2000
---
10
−9.8
AP-906/41649303
4.46
−6.78
−4.62
1000
–
Compound
IDAffinity
(kcal/mol)SPECS
IDLog Po/w
Log S
Caco-2
(cm/s)LD50
(mg/kg)AMES
toxicity
11
−9.3
AM-807/42004712
4.57
−6.14
−4.842
500
---
12
−9.5
AK-968/15360515
3.14
−4.42
−4.999
1000
–
13
−9.0
AG-389/42091799
3.65
−5
−4.75
2500
–
14
−8.7
AN-329/42286180
4.55
−6.46
−4.701
1000
---
15
−9.0
AK-777/40838237
1.45
−3.53
−4.619
325
–
16
−9.6
AO-476/41339629
4.23
−5.47
−4.739
3160
–
17
−9.4
AG-205/37234018
4.24
−5.02
−4.913
678
–
18
−9.2
AP-906/42286090
3.5
−5.14
−4.641
200
---
19
−9.1
AG-690/09451038
4.31
−5.7
−4.965
2000
–
20
−9.1
AF-399/15539280
4.99
−6.69
−4.616
1000
---
21 (Luteolin)
−9.1
AM-721/20737006
2.53
−2.87
−4.722
522
+
3.3 Molecular dynamics simulation
The contribution of small molecule to target protein structural stability was analyzed (Supplementary Fig. 3). The 30 ns kinetic simulation results shown that 2, 4, 6, 8, 9, 10, 15, 17 and 20 were detected with a stronger binding stability and better occupy HPV-16 E6 “LXXLL” binding pocket. The average RMSD of unbound ligand target protein was about 0.4 nm after 10 ns, while the minimum RMSD of compounds 6 bound target protein was about 0.24 nm after 10 ns and the maximum was about 0.3 nm (the target protein bound by compounds 8 and 15). There was no big fluctuation during the whole simulation process, and shown good stability.
3.4 The validation of HPV E6-E6AP binding inhibitors
3.4.1 Detection of cell proliferation inhibition, molecular interactions
Seventeen compounds (compounds 1–17) purchased from small molecule library were selected for experimental verification. In molecular interaction experiment, compounds 4, 7, and 12 have good molecular interaction ability to the target protein. The molecular interactions details of compounds to E6 protein were shown in Supplementary Fig. 4 and Table 2. Compound 4 showed the best inhibitory ability on the proliferation of SiHa cells, and the cell viability decreased with the increase of drug concentration, the IC50 of compounds was 23.87 uM (Fig. 2). Note: Compound ID is the number of small molecule compounds;IC50 was the semi-inhibitory concentration of candidate compounds and positive controls.
Compound ID
Structure
Molecular Weight (g/mol)
KD (μM)
1
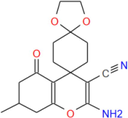
330.38
/
2
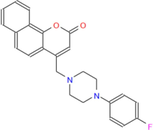
388.44
6.208E-03
3
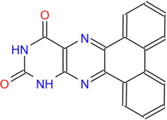
314.3
/
4
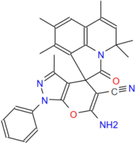
477.57
2.438E-05
5

311.33
/
6
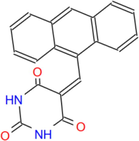
316.31
/
7
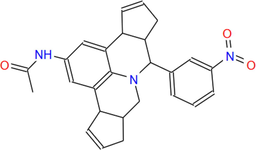
427.5
4.151E-06
Compound ID
Structure
Molecular Weight (g/mol)
KD (μM)
8
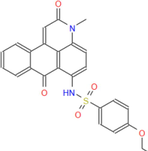
460.51
/
9

412.51
/
10
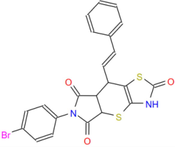
499.41
1.171E-03
11

400.33
7.721E-04
12
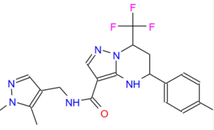
432.45
2.921E-06
13
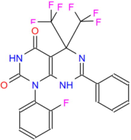
472.32
/
14
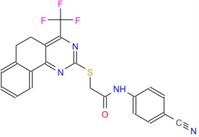
440.45
3.649E-03
Compound ID
Structure
Molecular Weight (g/mol)
KD (μM)
15
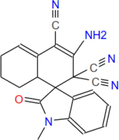
355.4
/
16
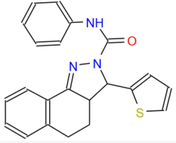
373.48
2.819E-04
17
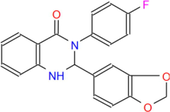
362.36
/
21 (Luteolin)
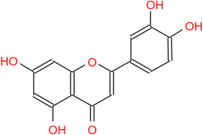
286.24
1.627E-03
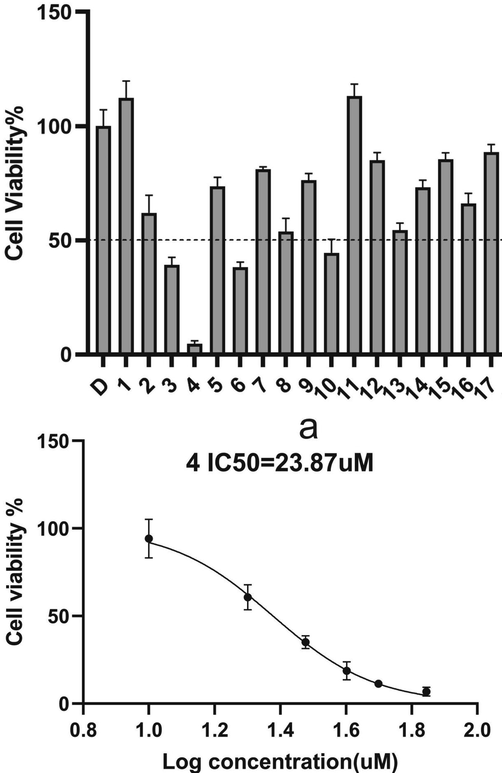
The cells proliferation inhibition after S4 added 48 h with different compounds concentration.
Calculation of equilibrium dissociation constant KD value (KD = Koff/Kon), reflecting the strength of the binding ability of interaction, larger the Kon is, smaller the dissociation Koof is, and the smaller the KD value is, the stronger the binding ability is. Blue line is the initial experimental data, and the blue line is the initial experimental data, and red analysis software fits. The details of molecular interactions of HPV16 E6 and compounds shown that compound 4, 10, 20, luteolin with positive control compound, and positive control compound 21 had significant binding and dissociation effect.Therefore, they are thought to interact with each other. For example, compound 4, which has the best inhibitory effect at the cellular level, has the lowest equilibrium dissociation constant with E6 protein. At the same time, we also measured the binding between protein E6 and compound 4 of different concentrations, as shown in Fig. 3. As the concentration of small molecule 4 increased from 10 μM to 30 μM, the binding between E6 protein and small molecule 4 became stronger. So we're going to go with compound 4.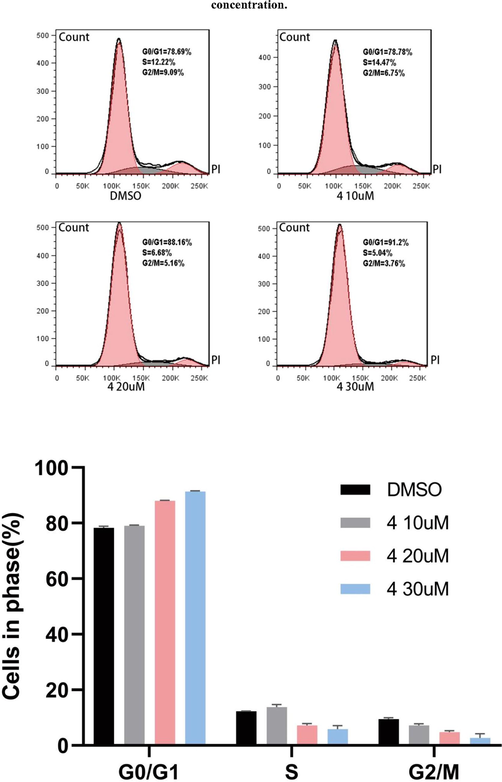
The cells apoptosis after S4 added 48 h with different compounds concentration.
Combined with previous experiment results, compound 4 showed potential druggability and was selected for subsequent experimental verification.
3.4.2 Cell cycle and apoptosis assay
In SiHa cells, the percentage of G0/G1, S, G2/M were 78.69%, 12.22%, 9.09%, when DMSO were added; 10 uM compound 4 were added, they were 78.78%, 14.47%, 6.75%; 20 uM compound 4 were added, they were 88.16%, 6.68%, 5.16%; and 30 uM compound 4 were added, they were 91.2%, 5.94%, 3.76%. G1 was arrested by compound 5 addition concentration increase (Fig. 3).
Apoptosis rates of SiHa cells were 2.53%, when DMSO added; they were 4.51%, 11.0% and 20.9%, when compound 4 added at 10 μM, 20 μM and 40 μM; and they were 3.13%, 4.51% and 6.14%, when Luteolin added at 10 μM, 20 μM and 40 μM (Fig. 4). The above data showed that compound 4 induced apoptosis of SiHa cells significantly stronger than Luteolin, and increased with drug concentration, shown a statistical significance (P < 0.05).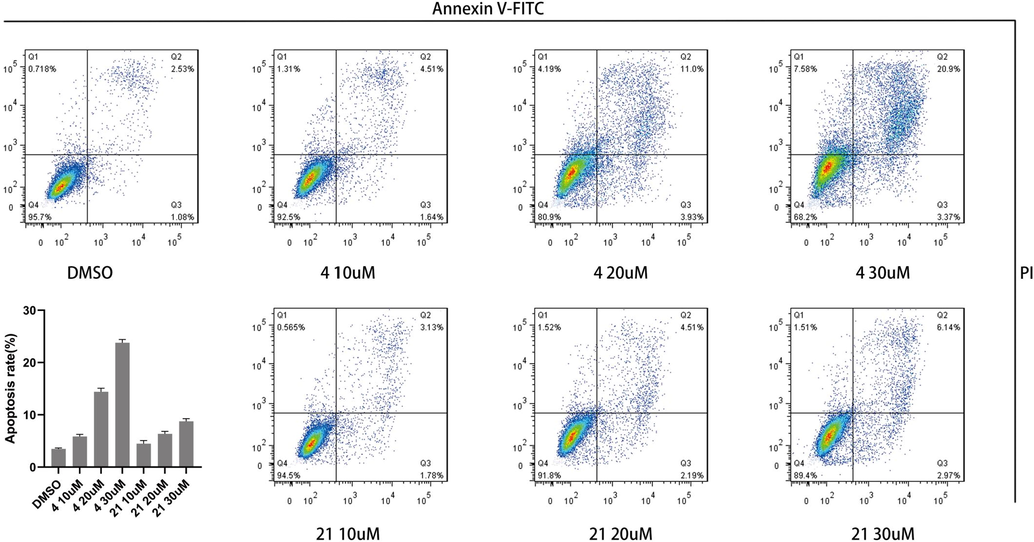
The cells apoptosis after S4 added 48 h with different compounds concentration.
3.4.3 p53/p21 expression in HPV-related cells
SiHa cells were treated with compound 4 and Luteolin at 10 μM, 20 μM and 40 μM to detect p53/p21 by Western-blotting, and β-actin was internal reference. As shown in Fig. 5, the p53/ p21 protein content in SiHa cells increased with compound 4 and Luteolin concentration.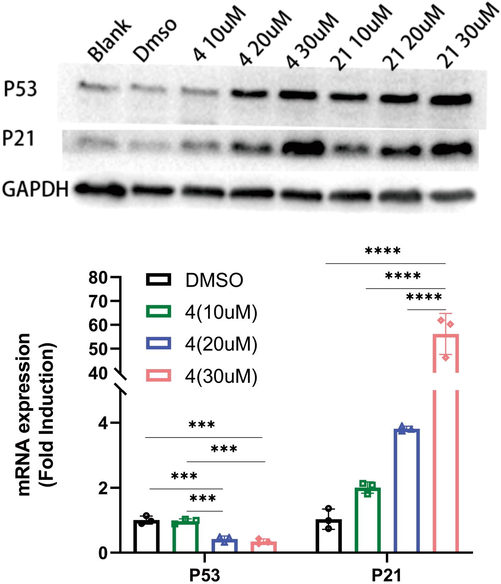
Effect of compounds on p53/p21 concentration and relative expression levels on SiHa cells with different compounds concentration. Note: The expression data in the figure are the percentage of expression relative to DMSO group, * represents P value, *** represents p < 0.001, ** represents p < 0.01, *represents p < 0.05.
Analysis of p53/p21 expression in SiHa cells after administration 48 h, β-actin is the housekeeping gene. The expression details of p53/p21 were shown in Fig. 5. The expression level of p53 showed down-regulated with compound 4 addition, down-regulation was enhanced with drug concentration increasing, and the difference between 10 µM and 20 µM dose groups shown a statistically significant (P < 0.01), indicated that compound 4 did not up-regulate the expression of p53 in SiHa cells, and up-regulate the expression of p21.
4 Discussion
HR-HPV persistent infection is closely related to the occurrence of cervical cancer and other malignant tumors, and the carcinogenicity induction of HR-HPV may realize by its E6 protein, through E6-E6AP-p53 binding mediate p53 ubiquitination degradation, and p53 is one of the most important tumor suppressor protein. α-9 HPV is almost all high-risk HPV, and has a high prevalence in the population, therefore related drug development is extremely important. The α-9 HPV E6 structure and binding pattern of E6-E6AP indicate that the “LXXLL” binding pocket of HPV E6 between α-9 genus was extremely stable and conservative, and the crystal model of HPV-16 E6 was selected as the target to screen competitive inhibitor of E6-E6AP binding pocket, that may inhibit oncogenic risk of α-9 genus HPV.
After two-step virtual screening by DOCK, AutoDock Vina and visual screening based on the empirical rules, 20 candidate molecules of E6-E6AP binding inhibitor were screened from 277,405 molecules in the Specs database. According to the molecular docking results of computer simulation, these molecules binding to E6 can fully occupy the binding sites of E6 and E6AP, and their binding stability were evaluated by molecular dynamics simulation. Virtual screening is a rapid and efficient method for drug screening, but currently, there is a few crystal structure data of complete E6 protein, and no co-crystallization of small molecule compounds with E6 was reported, therefore, virtual screening of drugs remains many risks. And in this study, the candidate compounds were screened based on the structure characteristics of “LXXLL” of E6AP to HPV-16 E6. In addition, the accuracy of the DOCK and AutoDock Vina have some limitations. Although the two software have their own scoring functions, some unsatisfactory molecular binding modes with E6 may be very good, and the range of visual screening also affects the hit rate of binding inhibitors. Virtual screening includes screening methods based on ligand and pharmacophore models, and this screening is a structure-based virtual screening. If there are known small molecule inhibitors of E6, we can also conduct virtual screening through molecular structure similarity. During visual screening, the success rate of screening active E6-E6AP-binding inhibitors can be greatly increased by analyzing whether the screened inhibitor candidate molecules contain pharmacophore favorable for binding to the E6 binding pocket, and the use of a variety of molecular docking programs or software. And pharmacokinetic properties and toxicity prediction, molecular dynamics simulation and biochemical experiments were performed to verify the reliability and available of E6-E6AP binding inhibitor.
p53 is a famous tumor suppressor protein, which can regulate cell cycle, promote apoptosis of infected cells and clear infected cells. In HPV infected cells, E6-E6AP mediates ubiquitination degradation of p53, inhibits infected cells apoptosis, and induce the immortalization of infected cells. To verify the effectiveness and activity of E6-E6AP binding inhibitor candidate molecules obtained by virtual screening, biochemical and cell experiment were conducted, and compound 4 were selected out. The apoptosis and cell cycle assay shown that G1 phase of the SiHa cells arrest increased with compound 4 addition, as well as the apoptosis of SiHa cells, all indicated that the compound 4 has a strong effect on the HPV-16 infected cell. The content of p53 in SiHa cells increased significantly with compound 4 concentration increase, and p53 mRNA in SiHa cells showed that compound 4 addition did not up-regulate the expression of p53, and p21 increased. The above experiments showed that the content of p53 increased by reducing the degradation of p53 rather than increasing its expression. This conclusion is consistent with our design and expectation for screening of HR-HPV E6 carcinogenic inhibitory small molecule compounds. The increase of p53, one of the most common tumor suppressor proteins, can increase the cell tumor suppressor function, reduce the risk of disease, and due to the similarities and differences of drug targets structure among α-9 genus HPV, small molecular compounds design and selection, according to HPV-16 E6, have a certain curative effect on the other α-9 genus HPV infection.
The expression level change of p53/p21 may be related to the damage of small molecular compounds to cells. Therefore, it is necessary to carry out necessary chemical modification or group modification on the compound 4 to reduce its toxicity and increase its molecular activity, stability, water solubility. Finally, it is necessary to determine whether it is suitable for the treatment HPV-16 and other α-9 genus HPV related diseases according to the performance of animal and clinical trials, and turn it into a real anticancer drug, to provide powerful help for the prevention and treatment of related clinical diseases.
5 Conclusion
Compound 4 was screened from 277,405 molecules in the Specs database by virtual screening and experimental verification, that specificity target HPV-16 infected cells, inhibit E6-E6AP blinding and p53 ubiquitin degradation, increase the contents of tumor suppressor protein p53, eventually inbibit the oncogenic risk of HR-HPV E6. By chemically modifying its water solubility and toxicity, it is hoped that new anticancer drugs targeting HPV E6 protein can be developed.
Author contributions
J.H., Y.L., Q.L. and X.D. conceived and designed the study; J.H., Y.L., Q.L., C.C., N.L., Y.C., T.L., X.W. and S.F. collected the samples; J.H., Y.L. and Q.L. performed the experiments. J.H., Q.L. and X.D. wrote the manuscript. All authors read, edited, and approved the final manuscript for submission.
Acknowledgments
The authors thank the Affiliate Reproductive Hospital Genitalia Hygiene Research Center (Chengdu, Sichuan, China) and all patients and control subjects for their participation in this study.
References
- Alvarez-salas, L.M., 2014. Inhibition of HPV-16 E6 / E7 immortalization of normal keratinocytes by hairpin ribozymes Inhibition of HPV-16 E6 - E7 immortalization of normal keratinocytes by hairpin ribozymes.
- Identification of inhibitors to papillomavirus type 16 E6 protein based on three-dimensional structures of interacting proteins. Antiviral Res.. 2006;72:49-59.
- [Google Scholar]
- Papillomavirus polypeptides E6 and E7 are zinc-binding proteins. J. Virol.. 1989;63:1404-1407.
- [Google Scholar]
- Structure based identification and characterization of flavonoids that disrupt human papillomavirus-16 E6 function. PLoS One. 2013;8:1-21.
- [Google Scholar]
- Type-specific human papillomavirus distribution in invasive squamous cervical carcinomas in Tunisia and vaccine impact. Asian Pacific. J. Cancer Prev.. 2015;16
- [Google Scholar]
- Interaction of small molecules with the SARS-CoV-2 papain-like protease: In silico studies and invitro validation of protease activity inhibition using an enzymatic inhibition assay. J. Mol. Graph. Model.. 2021;104
- [Google Scholar]
- Distribution of high-risk HPV types among women in Sichuan province, China: a cross-sectional study. BMC Infect Dis. BMC Infect. Dis.. 2019;19:1-8.
- [Google Scholar]
- Inhibitors, P.E., Malecka, K.A., Fera, D., Schultz, D.C., Hodawadekar, S., Reichman, M., et al., 2014. Identification and characterization of small molecule human papillomavirus E6 inhibitors.
- Identification and characterization of small molecule human papillomavirus E6 inhibitors. Chem. Biol. 2014
- [Google Scholar]
- Virtual screening for potential inhibitors of high-risk human papillomavirus 16 E6 protein. Interdisc. Sci. Comput. Life Sci.. 2015;7:136-142.
- [Google Scholar]
- Structure of the human Papillomavirus E7 oncoprotein and its mechanism for inactivation of the retinoblastoma tumor suppressor. J. Biol. Chem.. 2006;281:578-586.
- [Google Scholar]
- Identification of two potential glycogen synthase kinase 3β inhibitors for the treatment of osteosarcoma. Acta Biochimica et Biophysica Sinica. 2018;50(5):456-464.
- [Google Scholar]
- Comparison of the immunogenicity and safety of cervical cancer vaccines in healthy women aged 18–45 years. Hum. Vaccine 2009:705-719.
- [Google Scholar]
- Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53. Nature. 2016;529:541-545.
- [CrossRef] [Google Scholar]
- Moody, C.A., Laimins, L.A., 2010. Human papillomavirus oncoproteins: pathways to transformation 10.
- Evolutionary studies of ligand binding sites in proteins. Curr. Opin. Struct. Biol.. 2017;45:85-90.
- [CrossRef] [Google Scholar]
- Association of human papillomavirus 16 E6 variants with cervical carcinoma and precursor lesions in women from Southern Mexico. Virol. J. 2015:12.
- [Google Scholar]
- In silico prediction of structural changes in human papillomavirus type 16 (HPV16) E6 oncoprotein and its variants. BMC Mol. Cell Biol. 2019:1-12.
- [Google Scholar]
- Sherr, C.J., Mccormick, F., 2002. The RB and p53 pathways in cancer 2 103–12.
- Tambunan, U.S.F., Parikesit, A.A., 2012. HPV Bioinformatics: In Silico Detection, Drug Design and Prevention Agent Development. Top Cerv Cancer with an Advocacy Prev 237–252. Available from: http://www.intechopen.com/articles/show/title/hpv-bioinformatics-in-silico-detection-and-prevention-agent-development.
- Gynecologic Oncology Surgical treatment of lymph node metastases in stage IB cervical cancer. The laterally extended parametrectomy (LEP) procedure: Experience with a 5 year follow-up. Gynecol. Oncol.. 2011;123:337-341.
- [CrossRef] [Google Scholar]
- SWISS-MODEL: homology modelling of protein structures and complexes. Oxford University Press; 2018. p. :296-303.
- Prophylactic vaccination against human papillomaviruses to prevent vulval and vaginal cancer and their precursors. Expert Rev. Vaccines. 2019;18:1157-1166.
- [Google Scholar]
- Structural basis for recognition of the tumor suppressor protein PTPN14 by the oncoprotein E7 of human papillomavirus. PLoS Biol.. 2019;17:1-28.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2023.104759.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary data 1
Supplementary data 1







