Translate this page into:
Synthesis and biological applications of some novel 8-Hydroxyquinoline urea and thiourea derivatives
⁎Corresponding authors. h.saadeh@uaeu.ac.ae (Haythem A. Saadeh)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
A number of novel urea and thiourea derivatives of 8-hydroxyquinoline have been designed, synthesized and evaluated for their anticancer activities. The structures of the new compounds were established by spectroscopic techniques, 1H NMR, 13C NMR, and mass spectrometry. The in vitro cytotoxicity against MCF7, and MDA-MB-231 cell lines were assessed by MTT assay. Six of the 11 compounds synthesized namely 5b, 5c, 5f, and 6b-d exhibited cytotoxicity with IC50 values ranged between 0.5 and 42.4 µM. Apoptotic features of cells treated with 5b compound were observed via florescent microscopy using DAPI and ethidium bromide/acridine orange staining against MCF-7 cells. Molecular docking of these molecules against 16 potential breast cancer protein revealed that these compounds could interact with the active site of poly (ADP-ribose) polymerase-1 (PARP1), B-cell lymphoma-extra large (Bcl-xL) and PARP5A (Tankyrase 1) by forming hydrogen bonds, π-π interactions and hydrophobic interactions. The docked poses of these molecules were observed to be similar in the active site of each of these targets.
Keywords
8-Hydroxyquinoline
Urea
Thiourea
Piperazine
Anticancer
Apoptosis
Molecular Docking
1 Introduction
8-Hydroxyquinoline is a privileged heterocycle that has drawn the attention of chemists, and medicinal chemists due to its unique physical and chemical properties (Saadeh et al., 2020). 8-Hydroxyquinoline and its derivatives are important types of compounds with diverse biological activities including antiviral agents (De la Guardia et al., 2018; Kos et al., 2019), antimicrobial agents (Capodagli et al., 2014; Eswaran et al., 2009; Vu et al., 2019), anticancer agents (Chan et al., 2013; Lescoat et al., 2012; Zhua et al., 2017), anti-Alzheimer agents (Crouch and Barnham, 2012; Wang et al., 2014; Yang et al., 2018) and anti-HIV agents (Cohen et al., 2012; Mekouar et al., 1998). Urea and Thiourea are organic compounds which possess a broad wide range of applications and their derivatives demonstrate a wide range of pharmacological potentials such as antidiabetic, antimicrobial, anticancer, and analgesic (Goffin et al., 2012; Naz et al., 2020; Vega-Pérez et al., 2012) which attracted the attention of many researchers in drug development. The bioactivity profile of 8-HQ derivatives and urea/thiourea derivatives prompted the design and synthesis of novel compounds containing these moieties as outlined in scheme 1 below.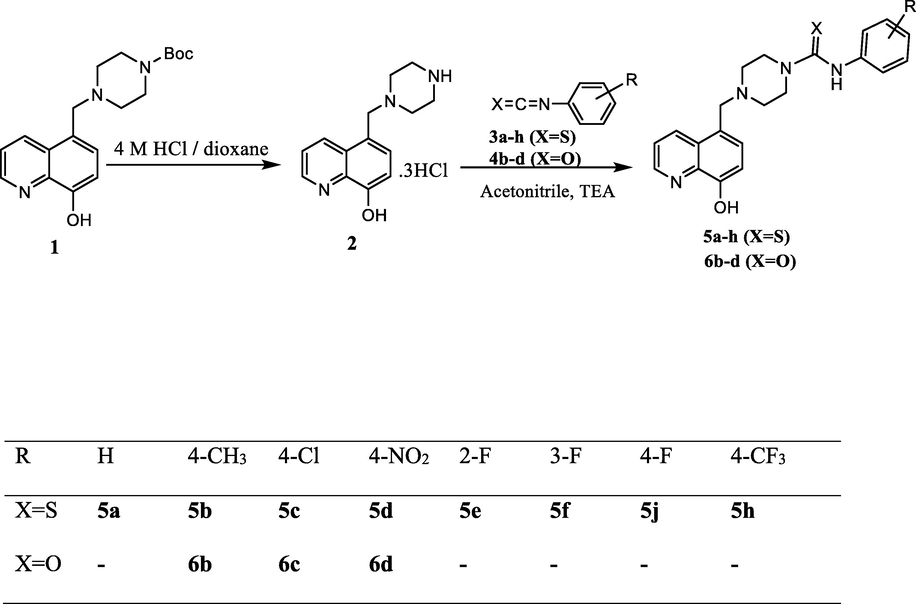
Synthesis of 8-hydroxyquinoline thiourea (5a-h) and hydroxyquinoline urea (6b-d) derivatives.
Around 19.3 million new cancer cases and 10.0 million cancer deaths in 2020 occurred globally, where female breast cancer is reported to be the most diagnosed cancer with around 2.3 million new cases (Sung et al., 2021). Chemotherapy, surgery, and other treatments have reduced cancer-associated deaths (Agrawal, 2014). Chemotherapy or other methods to counteract chemotherapy-induced harmful effects are often partly successful or may induce many side-effects that are added to patient discomfort (Abalo et al., 2017; Nurgali et al., 2018). Thus, research is being carried out to search for promising chemotherapy agents.
In this study, the in vitro efficacy of the novel 8-Hydroxyquinoline urea and thiourea derivatives against MCF-7 and MDA-MB-231 breast cancer cells and its potential to induce apoptosis were investigated. In silico molecular docking of these 8-HQ derivatives was performed against 16 potential breast cancer targets identified through an extensive literature analysis with the aim of getting a better understanding of the target and binding behavior of these novel derivatives.
2 Materials and methods
2.1 Chemistry
Melting points were assessed in an open capillary tube using Melting Point apparatus (Sanyo Gallenkamp MPD 350-BM 3.5, UK) and are uncorrected. FT-IR spectra were recorded using FT-IR spectrophotometer (Thermo Nicolet Nexus 470, USA). 1H NMR and 13C NMR spectra were recorded, using Varian-400 MHz (USA), at 25 °C in CDCl3 or DMSO‑d6 at 400 MHz using solvent peaks [CDCl3: 7.26 (D), 77.2 © ppm and DMSO‑d6 2.50 (D) and 39.7 (C) ppm] as internal references. The assignment of chemical shifts is based on standard NMR experiments (1H, 13C, 1H–1H COSY, 1H–13C HSQC, HMBC). TLC analyses were performed on silica F254 and detection at 254 nm by UV light. High resolution mass spectra (HRMS) were obtained as [M−H]- using electrospray ion trap (ESI) negative ion mode technique by collision-induced dissociation on a Bruker APEX-4 (7-Tesla) instrument. Column chromatographies were carried out on silica Gel 60 (230 mesh). Chemicals and reagents were obtained from ACROS ORGANICS, USA and Sigma Aldrich Chemical Co.. All chemicals and reagents were used as received without further purification. Compound 1 was prepared according to the procedures described in the literature (Agrawal, 2014; Sung et al., 2021). Synthesis of 8-hydroxyquinoline-carbothioamide (5a-h) and 8-hydroxyquinoline carboxamide derivatives (6b-d) have been provided in the supporting information file.
2.2 Biology
2.2.1 Cell culture
The human breast cancer cell lines MDA-MB-231 and MCF7 (DSMZ, Germany) were used. The cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) in 25 cm2 flasks (NIST, China). The DMEM were supplemented with fetal bovine serum (FBS) (Gibco, USA). The plates were incubated at 37 °C and 5% CO2 in a humidified incubator.
2.2.2 Cytotoxicity studies
MCF7 and MDA-MB-231 cells were grown in 96-well plates (NIST, China) at a density of 50,000 cells/well and incubated for 24 h at 37 °C. The test was carried out following a 48 h incubation with different concentrations of the compounds. Then, 20 µL of MTT (Thermo, USA) solution was added to each well and incubated for 2 h. The optical densities were analyzed at 520 nm using a multi-plate reader (ChroMate, USA). The results are expressed as the percentage of the control and were calculated as the concentration that reduce the proliferation of cells to 50% (IC50) using Origin 8. Each compound was tested in triplicate (Elnakady et al., 2017).
2.2.3 DAPI staining
The DAPI staining method was used for the morphological analysis of DNA. The cells were seeded and treated at the IC50 concentration of the compounds as in previous section. Then, the medium was aspirated, and fixed with ice cold ethanol for 15 min. After 24 h, the wells were washed with PBS, and then the stained with DAPI (1 µg/mL) (Themo, USA). After staining, wells were washed with PBS and observed under a fluorescent microscope (Evos, USA).
2.2.4 Acridine Orange/Ethidium bromide (AO/EB) staining
AO/EB staining was used to detect apoptosis. MCF7 and MDA-MB-231 cells were exposed to IC50 concentration of the compounds for 24 h. The wells were washed with PBS and 1 μL of AO/EB (10% of acridine orange and 10% of ethidium bromide in PBS) and incubated for 5 min. The cells were then imaged under fluorescence microscope (Evos, USA). Images were merged using Image j software.
2.3 In silico molecular docking
8-hydroxyquinoline urea and thiourea derivatives that showed cytotoxic potential against breast cancer cell lines (5b, 5c, 5f, and 6b-d), were subjected to molecular docking analysis. Since the molecular target(s) of these compounds are currently not known, potential breast cancer targets were identified through an extensive literature analysis. A total of 16 proteins were identified that were shown to have a potential role in the progression of breast cancer (Table 5). Three-dimensional X-ray crystal structure of these proteins was downloaded from the Protein Data Bank (PDB; https://www.rcsb.org). The retrieved protein structures were pre-processed using the Protein Preparation Wizard of Schrödinger Suite 2021–1. The preparation stage covered several steps, including assignment of bond orders, proper orientation of disorientated groups, setting of ionization states, creating disulfide bonds, deletion of water molecules, co-factors, and metal ions, and capping termini assignment of partial charges. Missing atoms and side chains of residues were also added. Finally, geometrically stable protein structures were obtained by optimizing and minimizing the protein structure. A receptor grid was generated with default parameters around the active site of each of the structures. Structures of the ligands to be docked were built using the 2D sketcher module of Schrödinger Maestro. The ligand molecules were pre-processed and conformers were generated using Schrödinger Ligprep. Processing of ligands included converting 2D structures to 3D, generation of tautomer and ionization states, the addition of hydrogen atoms, neutralization of charged groups, and finally optimization of the geometry of the molecules. All prepared protein structures and ligand molecules were subjected to the virtual screening workflow of Schrödinger Maestro.
3 Results and discussion
3.1 Chemistry
A series of hydroxyquinoline-carbothioamide (thiourea) (5a-h) and hydroxyquinoline –carboxamide (urea) derivatives (6b-d) were synthesized as highlighted in scheme 1. The synthesis of target thiourea 5a-j and urea 6b-d was accomplished in two steps: deprotection of N-Boc protected piperazine-8-hydroxyquinoline 1 (Yang et al., 2018) by HCl/dioxan to give piperazine-8-hydroxyquinoline salt 2 (Youdim et al., 2005) which then reacted with substituted isothiocyanates 3a-h and isocyanates 4b-d in the presence of excess triethylamine to give thiourea (5a-h) and urea (6a-h) respectively in good yields. All intermediates and final derivatives were purified by column chromatography, recrystallized, before identification by spectroscopic techniques.
The newly synthesized compounds (5a-h) and (6b-d) were characterized by melting point, 1H and C13 NMR, IR and MS spectral data. The detailed data found in the experimental part are in agreement with the suggested structures. The 1H NMR showed the protons of substituted benzene ring, piperazine and the 8-hydroxyquinoline moiety with correct integration. The thioamide and amide N-H appeared around 9.80 ppm. The 13C NMR data revealed the presence of a singlet around 180 ppm for compounds (5a-h) and around 156 ppm for (6b-d) which correspond to C = S and C = O, respectively. In the IR spectra, Peaks correlated with C = S and C = O stretching vibrations appear between 1520 and 1590 cm−1 for compounds (5a-h) and between 1630 and 1650 cm−1 for compounds (6b-d) respectively. The mass spectra display the correct molecular ion peaks as [M−H]- for which the measured high-resolution (HRMS) data are in good agreement with the calculated values.
3.2 Biology
3.2.1 Cytotoxicity assay
The cytotoxicity of compounds was assessed against two tumor cell lines (MCF-7 and MDA-MB-231) via MTT assay. As revealed in Fig. 1, only compounds 5b, 5c, 5f, and 6b-d exhibited inhibitory activities against the two cell lines tested. The IC50 values of tested compounds were summarized in Table 1. All the six compounds showed potent cytotoxicity against the two cancer cell lines tested compared with the control. The results indicated that compounds 5b exhibited the most potent activity against MCF-7 cells lines; therefore, it was selected for further studies.
Effect of 5b, 5c, 5f and 6b-d on growth inhibition of MCF7, and MDA-MB-231 cell lines. The cells were treated with increasing concentrations of compounds, the cytotoxicity was measured using the MTT test after 48 h. The data were analyzed using Origin 8 Pro.
MDA-MB-231
IC50 in µMMCF-7
IC50 in µM
5b
9.7
0.5
5c
22.8
5.8
5f
5.5
2.5
6b
1.5
2.9
6c
42.4
4.8
6d
6.6
9.1
3.2.2 DAPI staining
DAPI staining was carried out to investigate the nuclear morphology of MCF-7 cells caused by 5b compound. DAPI is a fluorescent dye that labels DNA. The nuclei of live cells were evenly stained with light blue, whereas apoptotic nuclei were stained with bright blue due to chromatin condensation. As shown in Fig. 2, a higher level of apoptosis with nuclear fragmentation, condensation, and higher brightness were detected post-treatment with IC50 (0.5 µM).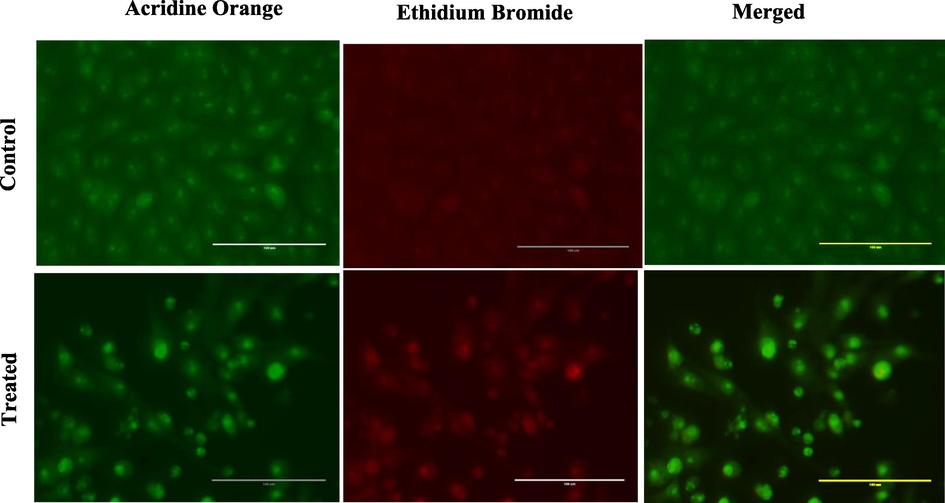
Dual staining of MCF-7 by Acridine orange and Ethidium bromide shows the apoptotic activity of compound 5b on MCF-7 cell (control and treated) after 24 h of treatment.
3.2.3 Acridine orange/ethidium bromide doubles (AO/EB) staining
AO/EB staining was carried out to investigate the morphological alternation in cell death. As shown in Fig. 3, the apoptotic cells had green or orange particles in their nuclei due to condensed or fragmented chromatin, and the non-apoptotic control cells had green chromatin with organized structures. According to the results, the 5b compounds induced apoptosis in MCF-7 cancer cell lines. Chemotherapeutic agents have been reported to induce apoptosis in different cancer types.
Apoptotic activity of compound 5b on MCF-7 cell (A: control and B: treated) after 24 h of treatment. White arrows represent fragmented and apoptotic nucle.
Apoptosis is a process in which abnormal cells are discarded. Consequently, apoptosis induction is an appealing target to kill cancer cells; therefore, it is a helpful strategy in cancer treatment (Brown and Attardi, 2005; Kerr et al., 1972). In the current investigation, the morphological features of apoptotic cells were assessed using fluorescence microscopy. After DAPI and AO/ EB staining, the control cells exhibited normal morphology while the treated cells showed the characteristic feature of apoptosis, indicating a promising anticancer activity of the 5b compound.
3.3 In silico molecular docking
The virtual screening workflow revealed that, out of 16 proteins evaluated here, poly (ADP-ribose) polymerase-1(PARP1), B-cell lymphoma-extra large (Bcl-xL) and PARP5A (Tankyrase 1) exhibited good docking scores, binding energies, and interactions. PARP family of enzymes regulate a wide range of cellular functions and PARP 1 is a key member of DNA repair mechanisms and high expression of PARP1 is implicated in several malignancies, including breast cancer (Cortesi et al., 2021). Similarly, PARP5A which is also known as Tankyrase 1 is mainly involved in telomere maintenance and Wnt/β-catenin pathway (Bao et al., 2012; Okada-Iwasaki et al., 2016). The hyperactivation of the Wnt/β-catenin pathway is observed in many forms of cancer including breast cancer. Cellular homeostasis and development are highly regulated by the interplay of pro-apoptotic and anti-apoptotic proteins. Overexpression of Bcl-xL is highly correlated with the progression of multiple solid cancer types.
Structural data have previously revealed the importance of P2 and P4 pockets in Bcl-xL for the tight binding of BH3 peptides and inhibitors. These pockets are lined by residues Glu96, Tyr101, Ser106, Asp107, Leu108, Arg139, and Tyr195 Analysis of docking results revealed that all molecules interacted with the active site of Bcl-xL protein by forming hydrogen bonds, π-π interactions and hydrophobic interactions (Table 2). The docked molecules exhibited a similar pose in the active site (Fig. 4). The 8-hydroxyquinoline moiety of these compounds consistently interacted with Tyr101 and Tyr195 of the P4 pocket by forming π-π interactions.
Compound
GlideScore (kcal/mol)
MM-GBSA binding energy (kcal/mol)
Residues that formed polar interactions
5b
−7.30
−76.75
Ala 93, Tyr101, Arg139, Tyr195
5c
−7.30
−81.06
Tyr101, Phe105, Arg139
5f
−7.11
−67.73
Ala93, Tyr101, Arg139, Tyr195
6b
−7.61
−88.44
Ala93, Tyr101, Arg139, Tyr195
6c
−7.58
−81.06
Ala93, Tyr101, Arg139, Tyr195
6d
−6.99
−77.88
Phe105, Arg132, Arg139
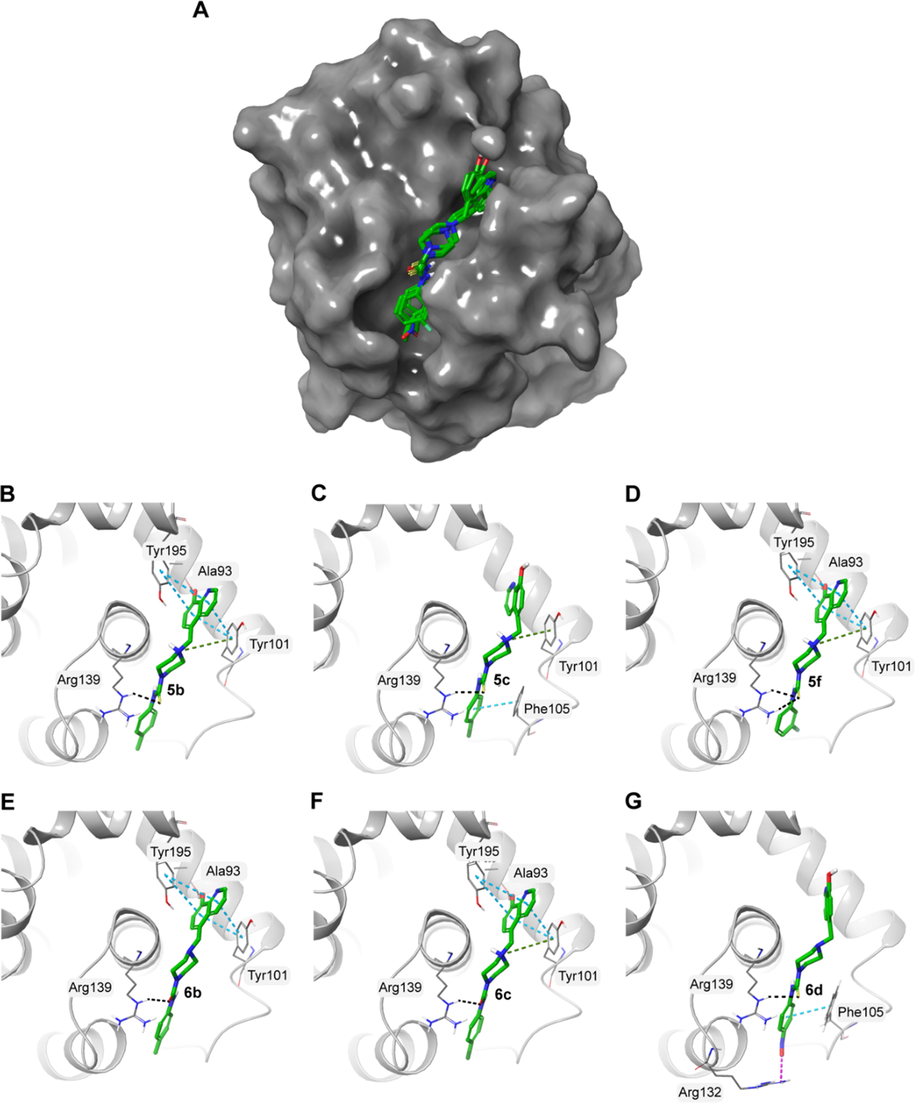
A) Surface representation of Bcl-xL (PDB ID: 4QVX) with the docked molecules shown in green stick representation. Docked molecules and the polar residues they interact with: B) 5b; C) 5c; D) 5f; E) 6b; F) 6c; G) 6d. Black dashed lines represent hydrogen bonds and green dashed lines represent π-π interactions.
The PARP family of proteins are a large group of proteins involved in various cellular processes such as DNA repair, genome maintenance and cell death. PARP1 is a multidomain protein consisting of three N-terminal zinc finger domains and a catalytic (CAT) domain at the C-terminal. The CAT domain is highly conserved across PARP family and it comprises of a helical subdomain (HD) and a ADP-ribosyl transferase (ART) subdomain (Steffen et al., 2013). Studies have shown that non-selective inhibitors of PARP proteins bind to the nicotinamide (NI) pocket of the highly conserved region of the catalytic domain of PARP1. This region is made up of Gly863, Tyr896, Ser904, Tyr907 and Glu988. Here, the docking data indicated that all molecules interacted with the catalytic domain in a similar orientation by interacting with these residues (Table 3 and Fig. 5). Similar to nicotinamide interaction, the hydroxyl group of 8-hydroxyquinoline molecule made hydrogen bond interactions with serine and glycine backbone atoms of the NI pocket. The benzene rings also made π–π stacking interactions with a tyrosine residue (Tyr 907).
Compound
GlideScore (kcal/mol)
MM-GBSA binding energy (kcal/mol)
Residues that formed polar interactions
5b
−9.38
−59.82
Gly863, Ser904, Tyr907, Tyr896
5c
−8.39
−66.04
Thr887, Tyr896, Ser904, Tyr907
5f
−9.37
−59.16
Gly863, Tyr896, Ser904, Tyr907
6b
−8.05
−75.18
Gly863, Tyr896, Ser904, Tyr907
6c
−8.84
−65.87
Thr887, Tyr896, Ser904, Tyr907
6d
−9.50
−58.78
Gly863, Tyr896, Ser904, Tyr907
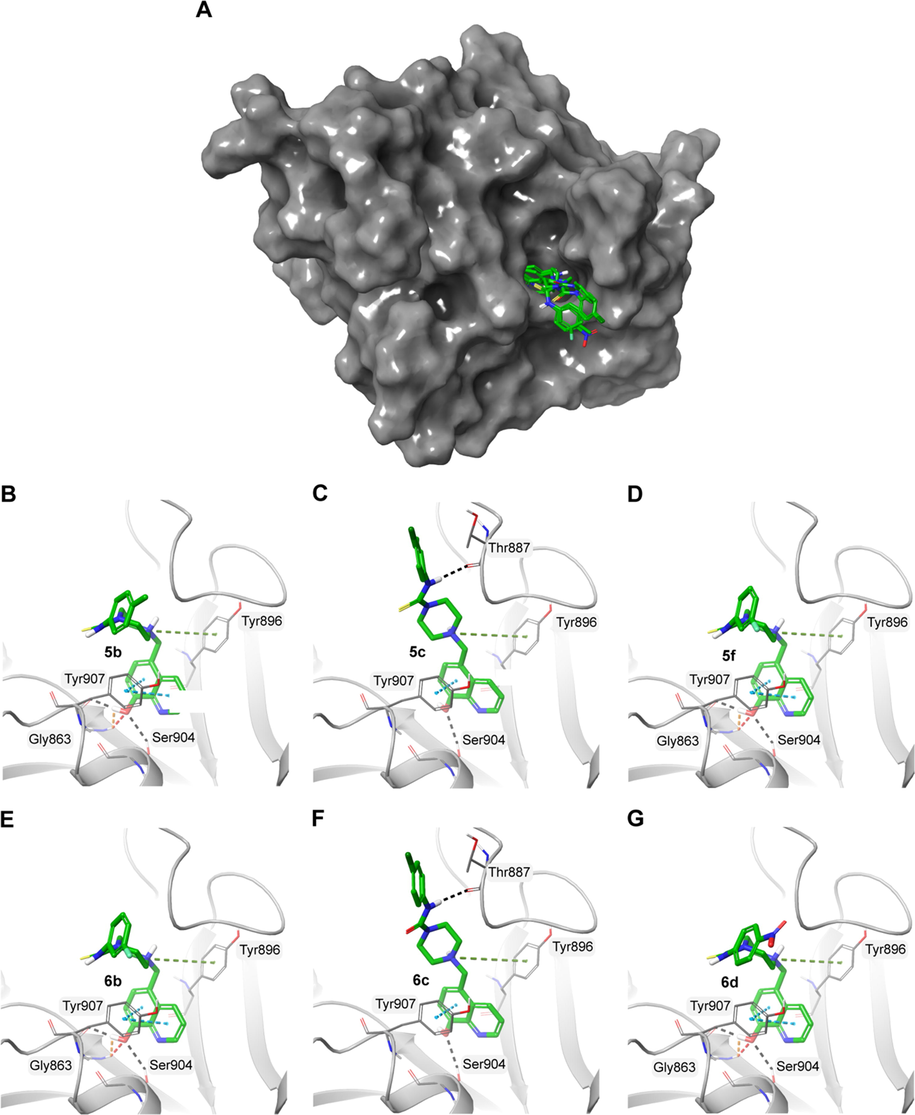
A) Surface representation of PARP1 (PDB ID: 6NRF) with the docked molecules shown in green stick representation. Docked molecules and the polar residues they interact with: B) 5b; C) 5c; D) 5f; E) 6b; F) 6c; G) 6d. Black dashed lines represent hydrogen bonds and green dashed lines represent π-π interactions.
Tankyrases (TNKS1 and TNKS2) are two highly homologous human PARP superfamily members. Similar to other PARP members, they contain a catalytic ART domain at the C-terminal but HD domain is absent in tankyrases. It was also observed that the NI site of PARP 5A is very similar to other PARP proteins. Structural data reveals that PARP 5A (TNKS1) catalytic domain binding pockets can be categorised into nicotinamide pocket and adenosine pocket (Gunaydin et al., 2012). Here, the docking data demonstrated that the molecules could bind to the nicotinamide-binding pocket by forming interactions with Asp1198, Tyr1231, Tyr1224, and Ser1221 of PARP5A (Table 4 and Fig. 6). As nicotinamide pocket is highly conserved among the PARP family, molecules that bind to this region could not only inhibit these specific proteins but could also potentially inhibit other members of the PARP family.
Compound
GlideScore (kcal/mol)
MM-GBSA binding energy (kcal/mol)
Residues that formed polar interactions
5b
−9.22
−93.50
Met 1207, Tyr1213
5c
−7.85
−88.98
Tyr1213, Tyr1224
5f
−8.10
−82.56
Asp1198, His1201, Met1207, Tyr1213
6b
−8.39
−79.74
Asp1198, His1201
6c
−7.82
−88.27
Asp1198, Met1207
6d
−7.50
−77.99
Phe1188, Asp1198, Lys1220, Ser1221, Tyr1224
No.
Protein
PDB ID
1
Phosphoinositide3-kinases (PI3K)
1E8Z
2
Bromodomain-containing protein 4 (BRD 4)
4NUD
3
B-cell lymphoma-extra large (Bcl-xL)
3ZLR
4
Epidermal growth factor receptor (EGFR)
1 M17
5
Pim 1Kinase
5VUA
6
B-cell lymphoma 2 (Bcl-2)
4MAN
7
Cathepsin B
3AI8
8
Poly(ADP-Ribose) Polymerase 1)PARP1
6NRF
9
Histone deacetylases (HDAC)
3MAX
10
Cyclin Dependent Kinase 4 (CDK4)
6OQO
11
Janus kinase 2 (JAK2) (triple mutant)
4E6Q
12
Janus kinase 2 (JAK2)
3KRR
13
Janus kinase 1 (JAK1)
4E4N
14
Maternal Embryonic Leucine Zipper Kinase (MELK)
4UMT
15
Tankyrase (PARP5A)
5ETY
16
Platelet-derived growth factor receptor alpha (PDGFRA)
6JOL
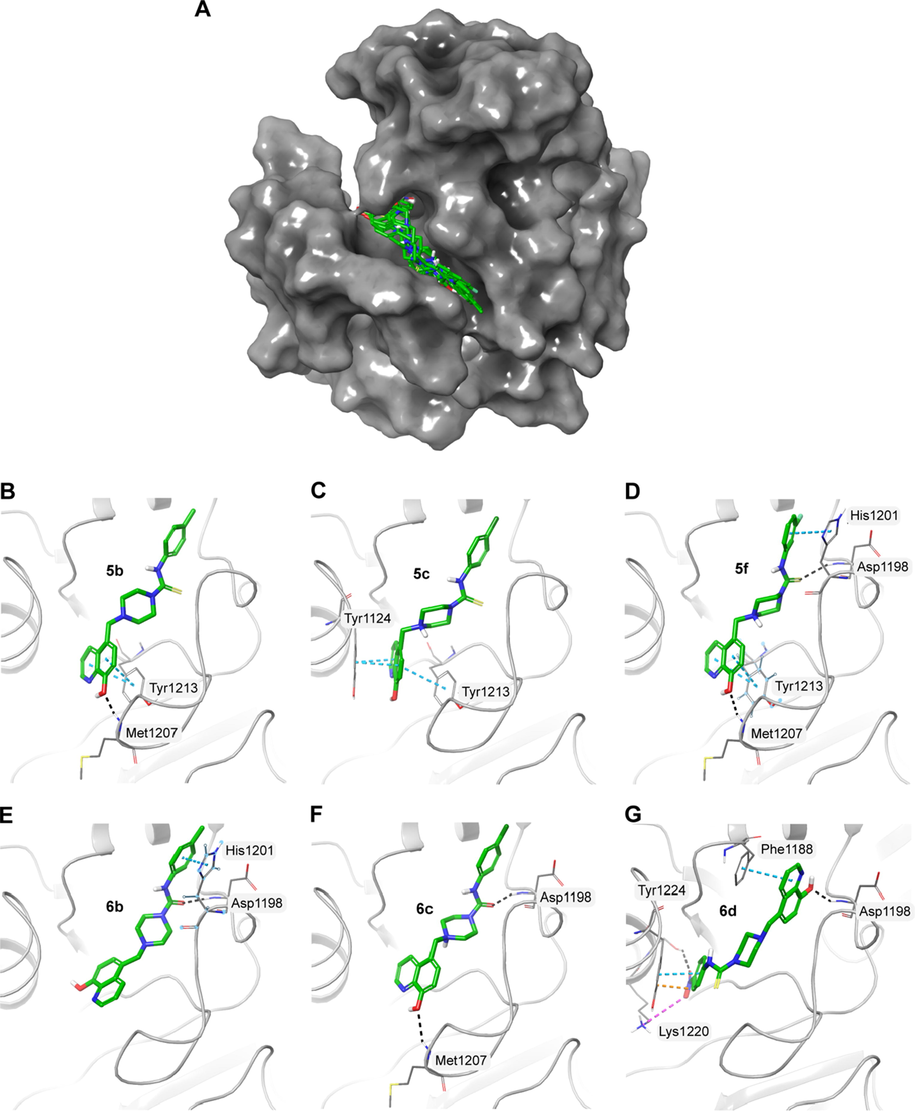
A) Surface representation of PARP5A (PDB ID: 5ETY) with the docked molecules shown in green stick representation. Docked molecules and the polar residues they interact with: B) 5b; C) 5c; D) 5f; E) 6b; F) 6c; G) 6d. Black dashed lines represent hydrogen bonds and green dashed lines represent π-π interactions.
4 Conclusion
This study includes the design and synthesis of novel compounds (5a-i) and (6b-d) containing 8-HQ, urea, thiourea and piperazine moieties to investigate their biological potential and uses. The structures of these compounds were confirmed by different spectroscopic tools including 1H NMR, 13C NMR, IR and high-resolution mass spectrometry. The compounds were assessed for their anti-proliferative potentials against the human breast cancer cells. Many of those compounds (5b, 5c, 5f, and 6b-d) revealed a promising potential against MCF-7, and MDA-MB-231cells and apoptosis seems to be involved in the mechanism of action. The promising compounds were docked against a collection of potential breast cancer targets. Bcl-xL, PARP1 and PARP5A were identified as potential targets based on docked pose and binding energy.
Acknowledgments
Princess Nourah bint Abdulrahman University Researchers, Supporting Program number (PNURSP 2022R82), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- May cannabinoids prevent the development of chemotherapy-induced diarrhea and intestinal mucositis? Experimental study in the rat. Neurogastroenterol. Motil.. 2017;29:e12952
- [Google Scholar]
- Late effects of cancer treatment in breast cancer survivors. South Asian J. Cancer. 2014;3:112-115.
- [Google Scholar]
- Inhibition of tankyrases induces Axin stabilization and blocks Wnt signalling in breast cancer cells. PLoS ONE. 2012;7:e48670
- [Google Scholar]
- The role of apoptosis in cancer development and treatment response. Nat. Rev. Cancer. 2005;5:231-237.
- [Google Scholar]
- A noncompetitive inhibitor for Mycobacterium tuberculosis’s class IIa fructose 1, 6-bisphosphate aldolase. Biochemistry. 2014;53:202-213.
- [Google Scholar]
- Synthesis of 8-hydroxyquinoline derivatives as novel antitumor agents. ACS Med. Chem. Lett.. 2013;4:170-174.
- [Google Scholar]
- Preparation of raltegravir-chelator derivatives as HIV integrase inhibitors. WO: PCT Int Appl.; 2012. p. :A2.
- An Overview of PARP Inhibitors for the Treatment of Breast Cancer. Targeted Oncol. 2021:1-28.
- [Google Scholar]
- Therapeutic redistribution of metal ions to treat Alzheimer’s disease. Acc. Chem. Res.. 2012;45:1604-1611.
- [Google Scholar]
- Antiviral activity of novel quinoline derivatives against dengue virus serotype 2. Molecules. 2018;23:672.
- [Google Scholar]
- Characteristics, chemical compositions and biological activities of propolis from Al-Bahah, Saudi Arabia. Sci. Rep.. 2017;7:1-13.
- [Google Scholar]
- Synthesis and antimicrobial activities of novel quinoline derivatives carrying 1, 2, 4-triazole moiety. Eur. J. Med. Chem.. 2009;44:4637-4647.
- [Google Scholar]
- Benzopyrans display in vitro anticancer activity and selectivity on apoptosis-resistant glioblastoma cells: screening, synthesis of simplified derivatives, and structure-activity relationship analysis. Eur. J. Med. Chem.. 2012;54:834-844.
- [Google Scholar]
- Novel binding mode of a potent and selective tankyrase inhibitor. PLoS ONE. 2012;7:e33740
- [Google Scholar]
- Apoptosis: a basic biological phenomenon with wideranging implications in tissue kinetics. Br. J. Cancer. 1972;26:239-257.
- [Google Scholar]
- 8-Hydroxyquinoline-2-Carboxanilides as Antiviral Agents Against Avian Influenza Virus. ChemistrySelect. 2019;4:4582-4587.
- [Google Scholar]
- Antiproliferative and iron chelating efficiency of the new bis-8-hydroxyquinoline benzylamine chelator S1 in hepatocyte cultures. Chem. Biol. Interact.. 2012;195:165-172.
- [Google Scholar]
- Styrylquinoline derivatives: a new class of potent HIV-1 integrase inhibitors that block HIV-1 replication in CEM cells. J. Med. Chem.. 1998;41:2846-2857.
- [Google Scholar]
- Synthesis, characterization, and pharmacological evaluation of thiourea derivatives. Open Chem.. 2020;18:764-777.
- [Google Scholar]
- Adverse effects of cancer chemotherapy: Anything new to improve tolerance and reduce sequelae? Front. Pharmacol.. 2018;9:245.
- [Google Scholar]
- The discovery and characterization of K-756, a novel Wnt/β-catenin pathway inhibitor targeting tankyrase. Mol. Cancer Ther.. 2016;15:1525-1534.
- [Google Scholar]
- Recent advances in the synthesis and biological activity of 8-hydroxyquinolines. Molecules. 2020;25:4321.
- [Google Scholar]
- Structural implications for selective targeting of PARPs. Front. Oncol.. 2013;3:301.
- [Google Scholar]
- Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.. 2021;71:209-249.
- [Google Scholar]
- Isoprenyl-thiourea and urea derivatives as new farnesyl diphosphate analogues: Synthesis and in vitro antimicrobial and cytotoxic activities. Eur. J. Med. Chem.. 2012;58:591-612.
- [Google Scholar]
- In vitro activities of a new fluoroquinolone derivative highly active against Chlamydia trachomatis. Bioorg. Chem.. 2019;83:180-185.
- [Google Scholar]
- Donepezil+ propargylamine+ 8-hydroxyquinoline hybrids as new multifunctional metal-chelators, ChE and MAO inhibitors for the potential treatment of Alzheimer's disease. Eur. J. Med. Chem.. 2014;80:543-561.
- [Google Scholar]
- Novel 8-hydroxyquinoline derivatives targeting β-amyloid aggregation, metal chelation and oxidative stress against Alzheimer’s disease. Bioorg. Med. Chem.. 2018;26:3191-3201.
- [Google Scholar]
- Pharmaceutical compositions comprising iron chelators for the treatment of neurodegenerative disorders and some novel iron chelators. Google Patents. 2005
- [Google Scholar]
- Synthesis and structure-activity relationships study of a-aminophosphonate derivatives containing a quinoline moiety. Chin. Chem. Lett.. 2017;28:1514-1518.
- [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2022.103905.
Appendix A
Supplementary data
The following are the Supplementary data to this article:Supplementary Data 1
Supplementary Data 1







