Translate this page into:
Synthesis and biological evaluation of coumarin derivatives containing oxime ester as α-glucosidase inhibitors
⁎Corresponding author at: School of Biotechnology and Health Sciences, Wuyi University, Jiangmen 529020, PR China. wyuchemxxt@126.com (Xue-Tao Xu)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
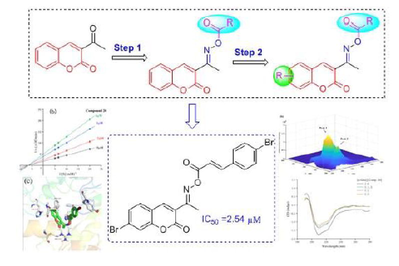
Abstract
In this study, potent coumarin derivatives containing oxime ester 1 ∼ 28 as α-glucosidase inhibitors were developed through a stepwise structure optimization, and the structure activity relationship was uncovered. Among them, compound 20 exhibited outstanding α-glucosidase inhibitory activity with IC50 of 2.54 ± 0.04 μM compared to 640.57 ± 1.13 μM of Acarbose. Kinetic study ascertained that compound 20 was a reversible and uncompetitive α-glucosidase inhibitor. 3D fluorescence results showed that the interaction of compound 20 with α-glucosidase caused the changes of microenvironments and polypeptide backbone structure of α-glucosidase. CD spectra results revealed that compound 20 decreased the α-helix content and increased the β-sheet content. Molecular docking analysis indicated that compound 20 well located into the active site and mainly bind with Phe157, His239, His279, Tyr71, and Arg312 to reduce the catalytic activity of α-glucosidase.
Keywords
Coumarin derivatives
a-Glucosidase
Inhibition mechanism
Molecular docking
1 Introduction
Type 2 diabetes mellitus (T2DM) is a metabolic disease characterized by hyperglycemia resulting from insulin resistance and insufficient insulin secretion by pancreatic β-cells. One of key reasons for the hyperglycemia is the enzymatic hydrolysis of carbohydrates. α-Glucosidase (EC 3.2.1.20), an exo-acting carbohydrase, locate in the human small intestine (Proença et al., 2017). It catalyzes hydrolysis of carbohydrate substrates to release α-D-glucopyranose (Ali et al., 2017). Excessive absorbed glucopyranose leads to the abnormal increase of blood glucose concentration, and long-term high blood glucose accelerates the non-enzymatic glycosylation to produce a large amount of advanced glycation end products (AGEs), thereby causing a series of complications of diabetes (Tsoutsouki et al., 2020). The inhibition of α-glucosidase activity leads to the retardation of carbohydrates digestion, and thereby resulting in the reduction of glucose absorption (Imran et al., 2015). The action mechanism of α-glucosidase inhibitors is attributed to their competitive binding to the carbohydrate-binding region of α-glucosidase (Sohretoglu et al., 2018). α-Glucosidase inhibitors have been reported to be used as effective agents in the treatment of carbohydrate metabolism related diseases, including diabetes, HIV, obesity, and dyslipidemia (Rasouli et al., 2017; Santos et al., 2018). Currently, some α-glucosidase inhibitor drugs have been used in the clinic, such as acarbose, voglibose, and miglitol. However, the clinical application found that those drugs still have some adverse side effects, which also limit their clinical use (Hedrington and Davis, 2019). Thence, it is urgent to develop more effective and lower toxic α-glucosidase inhibitors.
Coumarin, 2-γ-benzopyrones, is a vital core skeleton present in a large amount of natural and synthetic compounds (Stefanachi et al., 2018). Compounds containing coumarin moiety had been revealed to display various pharmacological activities, including anti-inflammatory, anti-bacterial, antiviral, anti-oxidation, anti-cancer (Wu et al., 2020; Matos et al., 2017; Sahni et al., 2021). In particularly, lots of natural and synthetic compounds containing coumarin moiety presented potential α-glucosidase inhibitory activity and hypoglycemic activity (Wang et al., 2016; Salar et al., 2016; Taha et al., 2018). Our previous study also demonstrated that coumarin moiety could act as the important core skeleton in designed α-glucosidase inhibitors (Xu et al., 2020). Some of the representative α-glucosidase inhibitors containing coumarin moiety were shown in Fig. 1. On the other hand, oxime ester serviced as an important active skeleton in many drugs, and showed many biological activities, such as bactericidal, anti-inflammatory and antiviral (Kassa et al., 2017; Schepetkin et al., 2021). Also, some oxime ester derivatives showed potent α-glucosidase inhibitory and hypoglycemic activity (Fig. 1) (Hosseini Ghazvini et al., 2018; Tran et al., 2022). Furthermore, benzoic acid and cinnamic acid also play important roles in many α-glucosidase inhibitors or hypoglycemic drugs (Fig. 1) (Hameed et al., 2019; Song et al., 2016).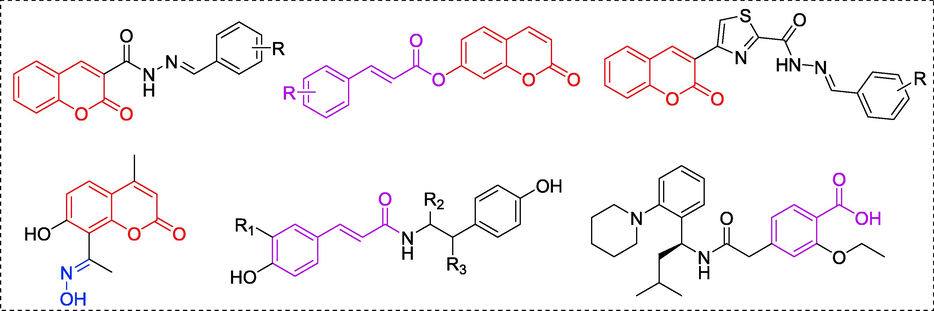
Chemical structures of some reported α-glucosidase inhibitors containing coumarin (Red color), oxime ester (Blue color), and benzoyl or cinnamic acyl moiety (Pink color).
According to the findings above, the hybridization of coumarin with cinnamic acid and benzoic acid through oxime linkage was developed aiming to obtain new α-glucosidase inhibitors (Fig. 2, design strategy). All synthesized compounds were assayed for their α-glucosidase.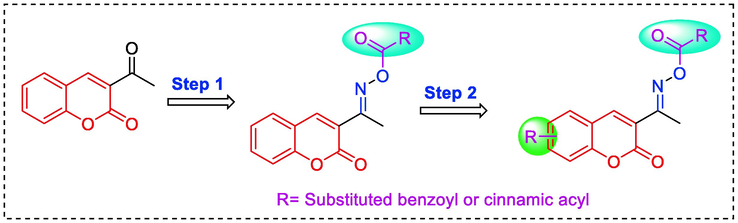
Design strategy of target coumarin compounds.
2 Results and discussion
2.1 Chemistry
The target coumarin derivatives 1 ∼ 28 were synthesized according to the synthetic route illustrated in Schemes 1. Substituted salicylaldehyde underwent perkins reaction with ethyl acetaacetate to obtain substituted acetyl coumarin, which reacted with hydroxylamine hydrochloride to produce coumarin containing oxime. Finally, target coumarin derivatives 1 ∼ 28 were obtained from the esterification of coumarin containing oxime with substituted benzoic acid or substituted cinnamic acid. All coumarin derivatives 1 ∼ 28 were identified by 1H NMR, 13C NMR and HRMS.
Synthesis of coumarin derivatives 1 ∼ 28. Reagents and condition: (a) Ethyl Acetaacetate, Pi3peridine, EtOH; (b) Hydroxylamine hydrochloride, Pyridine, EtOH; (c) Substituted benzoic acid chloride or substituted cinnamic acid chloride, triethylamine, DCM.
2.2 α-Glucosidase inhibition assay and structure–activity relationships (SAR) analysis
At the beginning, to develop more potent α-glucosidase inhibitors and ascertain the SAR, compounds 1 ∼ 9 (containing benzoyl) and compounds 10 ∼ 18 (containing cinnamic acyl) were preferable synthesized through the scaffold hybridization strategy. Then the synthesized coumarin derivatives were assayed for their in vitro α-glucosidase inhibitory activity with commercially α-glucosidase inhibitor acarbose as a positive control. The results showed that coumarin derivatives 1 ∼ 18 demonstrated excellent and potent inhibitory activity against α-glucosidase with the IC50 range of 5.06 ± 0.08 ∼ 93.23 ± 0.25 μM, as compared to the standard acarbose (IC50 = 640.57 ± 1.13 μM) (Table 1). These results showed that the hybridization of coumarin with substituted cinnamic acid or benzoic acid through oxime linkage could obtain potential α-glucosidase inhibitors. Values are shown in mean ± SD. #Indicating comparisons between the compound and acarbose (#P < 0.05).
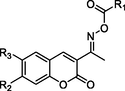
Compound
R1
R2
R3
α-glucosidase inhibition
(IC50 μM)
1
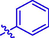
H
H
93.23 ± 0.25#
2
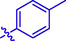
H
H
15.91 ± 0.25#
3
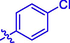
H
H
10.95 ± 0.11#
4
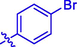
H
H
6.46 ± 0.05#
5

H
H
29.21 ± 0.32#
6
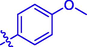
H
H
19.34 ± 0.22#
7
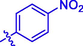
H
H
26.12 ± 0.26#
8
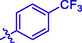
H
H
26.16 ± 0.26#
9
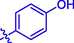
H
H
26.14 ± 0.26#
10
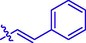
H
H
34.35 ± 0.31#
11
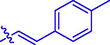
H
H
13.39 ± 0.09#
12
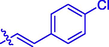
H
H
6.34 ± 0.05#
13
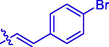
H
H
5.06 ± 0.08#
14
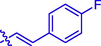
H
H
11.55 ± 0.13#
15
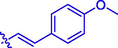
H
H
15.09 ± 0.10#
16
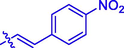
H
H
8.71 ± 0.21#
17
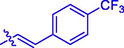
H
H
19.66 ± 0.04#
18
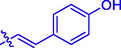
H
H
21.54 ± 0.02#
19
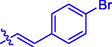
Cl
H
3.42 ± 0.07#
20
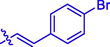
Br
H
2.54 ± 0.04#
21
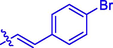
F
H
3.92 ± 0.02#
22
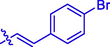
CH3
H
5.24 ± 0.16#
23
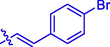
OCH3
H
6.43 ± 0.06#
24
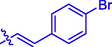
H
Cl
4.25 ± 0.02#
25
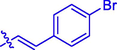
H
Br
3.81 ± 0.14#
26
H
F
3.73 ± 0.10#
27
H
CH3
6.17 ± 0.04#
28
H
OCH3
7.84 ± 0.14#
Acarbose
640.57 ± 1.13
Then the SAR was analyzed based on activity data. For compounds 1 ∼ 9 (containing benzoyl), compound 1 without substituent group presented an IC50 of 93.23 ± 0.25 μM. Compounds 2 ∼ 9 with different substituent group showed stronger inhibitory activity than compound 1, suggesting the introduction of substituent group (as shown by CH3, Cl, Br, F, OCH3, NO2, CF3, or OH) could effectively enhance the inhibitory activity. Among them, Bromine substituent (Br) showed the strongest enhancement on the inhibitory activity. Similar for compounds 10 ∼ 18 (containing cinnamic acyl), it was also observed that compounds 11 ∼ 18 with substituent group (CH3, Cl, Br, F, OCH3, NO2, CF3, or OH) at benzene ring of cinnamic acid displayed stronger inhibitory activity than compound 10 (without substituent group), revealing that the introduction of substituent group was vital for the α-glucosidase inhibition. Furthermore, compounds 10 ∼ 18 (containing cinnamic acyl) showed higher α-glucosidase inhibitory activity than compounds 1 ∼ 9 (containing benzoyl) with the same substituent group, respectively, suggesting that cinnamic acyl was more beneficial for α-glucosidase inhibition than benzoyl. Among them, compound 13 (containing bromcinnamic acyl) showed the highest inhibitory activity (IC50 = 5.06 ± 0.08 μM).
Then, coumarin fragment of the compound was further optimized based on compound 13, subsequently producing compounds 19 ∼ 28. Their α-glucosidase inhibition results showed that the introduction of substituent group (Cl, Br, or F) at C-6 or C-7 position of coumarin could efficiently increase the inhibitory activity. However, substituent group (CH3, or OCH3) was introduced at C-6 or C-7 position of coumarin, leading to a negative effect. In particularly, compound 20 containing Br at C-7 position of coumarin fragment showed the best α-glucosidase inhibitory activity with an IC50 of 2.54 ± 0.04 μM. The physicochemical properties of 3f were predicted and shown in Table 2.
Molecular formula
Molecular weight
Rotatable bonds
H-bond acceptoer atoms
H-bond donnor atoms
Polar surface area (Å2)
LogPo/w
Water solubility
compound 20
C20H13Br2NO4
491.13
5
5
0
68.87
5.01
Poorly soluble
From aforementioned results, it could easily be extracted that the α-glucosidase inhibitory activity of coumarin derivatives 1 ∼ 28 was depending upon the characteristic of oxime ester (benzoyl or cinnamic acyl) and substituent group on benzoyl, cinnamic acyl or coumarin.
2.3 Inhibitory mechanism and type assay
To analyze the action mechanisms of α-glucosidase inhibition of all coumarin derivatives, enzyme kinetic analysis was performed on the most potent compound 20. As illustrated in Fig. 3a, the plots of residual enzyme activity versus enzyme concentration in the presence of compound 20 (0 ∼ 4 μM) gave a family of straight lines with common intersection at the origin, indicating that compound 20 is a reversible inhibitor (Ali et al., 2017).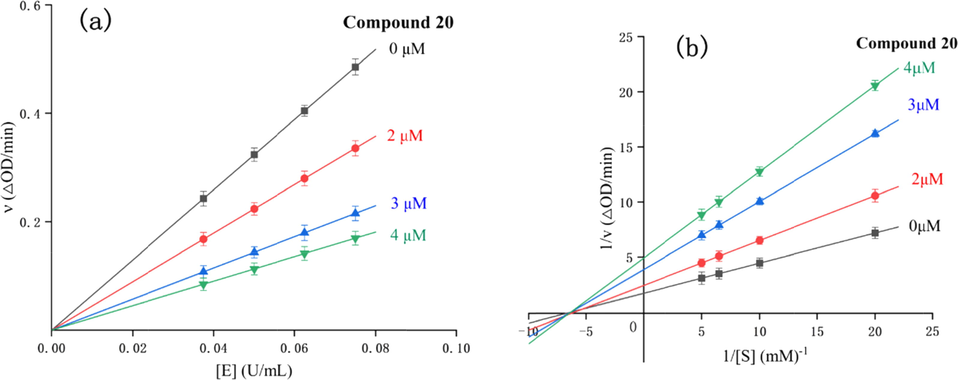
Inhibitory mechanism (a) and type assay (b) of compound 20 against α-glucosidase.
The kinetic type of compound 20 on α-glucosidase inhibition was also performed using Lineweaver-Burk plots of residual enzyme activity versus substrate concentration (Fig. 3b). As revealed from the Lineweaver-Burk plots, the data lines of compound 20 intersected in the x axis thereby indicating a noncompetitive inhibition (Tsoutsouki et al., 2020). Moreover, thire slop versus inhibitor concentrations of compound 20 to produce the inhibition constant (KI) was calculated as 1.5 μM.
2.4 Three-dimensional (3D) fluorescence spectra assay
The 3D fluorescence spectra was used to analyze the conformational changes of α-glucosidase interacting with compound 20. In the three-dimensional fluorescence spectrum of α-glucosidase alone (Fig. 4a), Peak 1 (λex = 276 nm, λem = 330 nm) reflected the fluorescence spectra of Tyr with Trp residues, and Peak 2 (λex = 232 nm, λem = 332 nm) denoted the fluorescence feature of polypeptide backbone structure. After treated with compound 20 (concentration 2 μM), the fluorescence intensity of Peak 1 reduced by 13.45%, and Peak 2 decreased by 37.04% (Fig. 4b), suggesting that the interaction of compound 20 with α-glucosidase caused the changes of microenvironments of the tyrosine and tryptophan residues and the polypeptide backbone structure of α-glucosidase.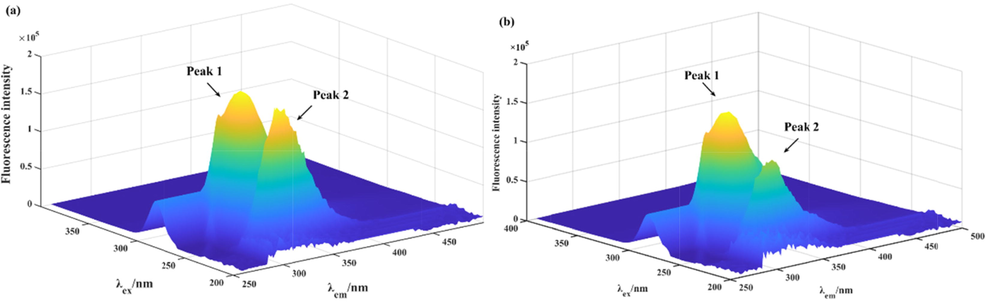
The 3D fluorescence spectra of α-glucosidase (a) and α-glucosidase with compound 20 system (b).
2.5 CD spectra assay
The conformational changes, especially the secondary structures changes, of α-glucosidase was monitored by the CD spectra. As shown in Fig. 5, two negative bands appeared at 210 and 222 nm, which were defined as the typical features of α-helixes. While treatment of compound 20 lead to the obviously reduce of negative bands intensity (Table 3). Treatment of compound 20 (molar ratios: 3:1) caused a decrease in the content of the α-helix (from 10.40 to 9.40%), β-turn (from 20.10 to 19.40%), and random coil (from 33.20 to 32.00%), while an increase in antiparallel (from 35.10 to 39.30%), respectively. These results revealed that compound 20 bound to α-glucosidase and rearranged its secondary structure.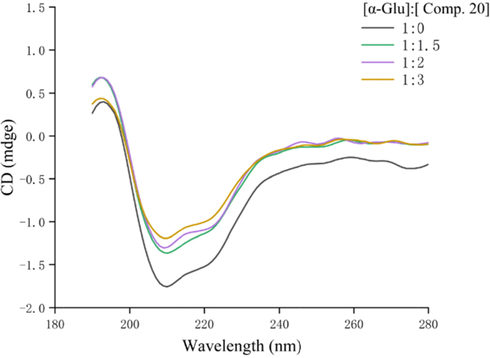
CD spectra of α-glucosidase in the presence of compound 20.
Molar ratio
[α-Glu]:[Comp.20]α-Helix
(%)β-Sheet
(%)β-Turn
(%)Rndm Coil
(%)
1:0
10.40
35.10
20.10
33.20
1:1.5
10.00
39.00
19.50
31.80
1:2
9.90
39.20
19.50
31.80
1:3
9.40
39.30
19.40
32.00
2.6 Molecular docking
Molecular docking was performed to analyze the binding mode of compound 20 to α-glucosidase. The docking results between compound 20 and α-glucosidase was shown in Fig. 6. Compound 20 well located into the active site with a “U shaped” conformation, and cinnamic acid part nested inside of the active site (Fig. 6a∼c). The detailed interaction results were shown out using 3D view (Fig. 6c) and 2D view (Fig. 6d), respectively. It was observed that the carbonyl of coumarin and nitrogen of oxime formed hydrogen bonds with Arg312, respectively (bond length: 1.9 Å and 2.4 Å), which was recognized as the key interaction between compound 20 and α-glucosidase. The bromcoumarin fragment formed Pi-Pi stacking with Phe157 (4.4 Å) and Pi-Alkyl interactions His239 and His279. Moreover, the bromcinnamic acyl formed Pi-Alkyl interactions with Tyr71. All above interactions helped compound 20 to anchor in the active site of the α-glucosidase.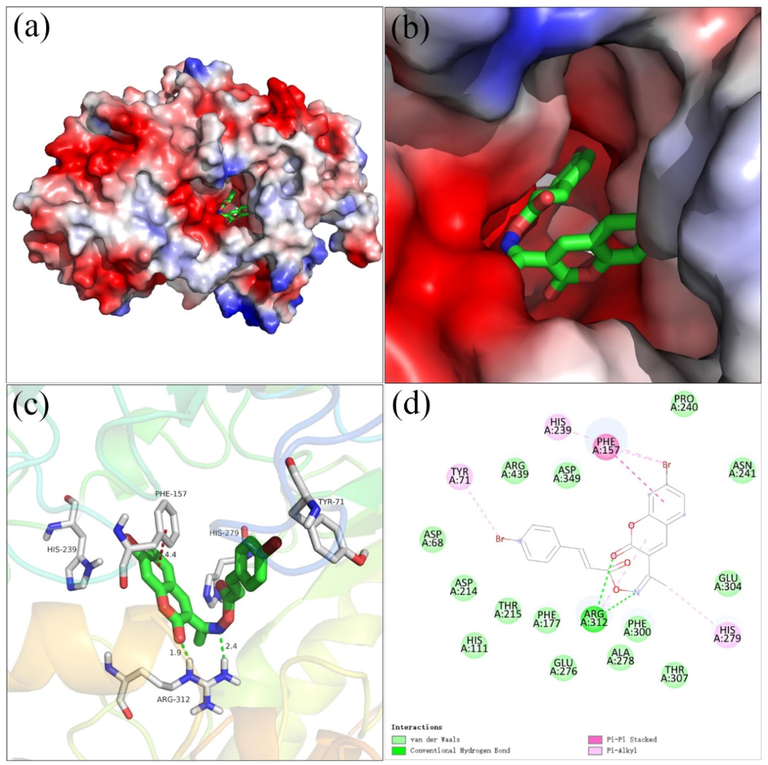
The molecular docking of compound 20 and α-glucosidase, (a, b) compound 20 in the electrostatics active pocket, (c) 3D view of compound 20 with α-glucosidase, (d) 2D view of compound 20 with α-glucosidase.
3 Conclusion
In summary, coumarin derivatives containing oxime ester 1 ∼ 28 were s ynthesized and evaluated for inhibitory activities against α-glucosidase. All the synthesized coumarin derivatives showed potent inhibitory activities against α-glucosidase and stronger than acarbose. The inhibition kinetic studies, CD spectra, 3D fluorescence, and docking simulation indicated the binding between compound 20 with α-glucosidase. Considering all these experimental data, coumarin derivatives containing oxime ester might be used as leading compound to find effective drugs for the management of T2D.
4 Experimental
4.1 Materials and methods
α-Glucosidase from Saccharomyces cerevisiae (EC 3.2.1.20) was purchased from Sigma-Aldrich. p-Nitrophenyl-α-D-galactopyranoside (PNPG) was obtained from Abcam. All other reagents and solvents were commercial available. 1H NMR and 13C NMR spectra were recorded in CDCl3 on 500 MHz instruments. High-resolution mass spectral analysis (HRMS) data were measured on the Apex II by means of the ESI technique.
4.2 General procedure for the synthesis of coumarin derivatives 1 ∼ 28
A mixture of substituted salicylaldehyde (1.0 mmol), ethyl acetoacetate (1.1 mmol), and piperidine (0.02 mmol) in ethanol (10 mL) was stirred at 70 °C for 2 h. After the reaction was completed, the mixture was poured into ice-cold water, followed by the filtration to obtain the crude product. Then the substituted acetyl coumarin was yield by recrystallization from ethanol.
The mixture of substituted acetyl coumarin (1.0 mmol), hydroxylamine hydrochloride (3.0 mmol), and pyridine (0.04 mmol) in ethanol (10 mL) was stirred at room temperature for 20 h. The produced solid was filtered and washed with ethanol to produce substituted acetyl coumarin containing oxime.
To the ice bath solution of substituted acetyl coumarin containing oxime (1.0 mmol) and triethylamine (1.1 mmol) in DCM (3 mL), the substituted benzoic acid chloride or substituted cinnamic acid chloride in DCM (3 mL) was added, and then reacted for 12 h after warming to room temperature. After quenched by water, the mixture was extracted with dichloride for three times, and washed with brine, dried by MgSO4. The solvent was removed under vacuum to obtain the crude product, followed by purification through column chromatography to yield the corresponding coumarin derivatives 1 ∼ 28.
(1, C18H13NO4). White sold; Yield 72%; m.p. 157–159 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.20 (s, 1H), 8.13 (dd, J = 8.1, 1.5 Hz, 2H), 7.66–7.57 (m, 3H), 7.51 (t, J = 7.7 Hz, 2H), 7.39–7.30 (m, 2H), 2.55 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 163.60, 162.71, 159.25, 154.37, 143.51, 133.67, 133.02, 129.76, 129.09, 128.71, 128.66, 124.98, 123.48, 118.53, 116.70, 77.33, 77.07, 76.82, 16.03; HRMS (ESI) [M + H]+ calcd. for C18H13NO4:308.0914; found: 308.0926.
(2, C19H15NO4). White sold; Yield 70%; m.p. 160–161 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.20 (s, 1H), 8.02 (d, J = 8.0 Hz, 2H), 7.59 (t, J = 7.6 Hz, 2H), 7.37 (d, J = 8.3 Hz, 1H), 7.35–7.28 (m, 3H), 2.55 (s, 3H), 2.44 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 163.76, 162.54, 159.21, 154.35, 144.86, 143.46, 133.06, 129.07, 128.47, 126.00, 125.97, 124.99, 123.42, 118.49, 117.96, 116.70, 77.32, 77.07, 76.81, 15.97; HRMS (ESI) [M + H]+ calcd. for C19H15NO4: 322.1071; found:322.1084.
(3, C18H12ClNO4). White sold; Yield 70%; m.p. 185–186 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.19 (s, 1H), 8.06 (dd, J = 8.5, 1.6 Hz, 2H), 7.61–7.57 (m, 2H), 7.49 (dd, J = 8.4, 1.6 Hz, 2H), 7.39–7.31 (m, 2H), 2.55 (d, J = 1.5 Hz, 3H); 13C NMR (126 MHz, CDCl3) δ 162.98, 162.81, 159.23, 154.38, 143.59, 140.23, 133.12, 131.13, 129.12, 127.07, 125.02, 123.33, 118.48, 116.74, 77.31, 77.26, 77.05, 76.80, 16.09; HRMS (ESI) [M + H]+ calcd. for C18H12ClNO4: 342.0525; found: 342.0536.
(4, C18H12BrNO4). White sold; Yield 69%; m.p. 180–182 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.18 (s, 1H), 8.00–7.97 (m, 2H), 7.66–7.64 (m, 2H), 7.62–7.57 (m, 2H), 7.38–7.31 (m, 2H), 2.54 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 163.01, 162.94, 159.20, 154.38, 143.58, 133.12, 132.11, 131.22, 129.11, 128.91, 127.53, 125.02, 123.31, 118.48, 116.73, 77.32, 77.07, 76.81, 16.08; HRMS (ESI) [M + H]+ calcd. for C18H12BrNO4: 387.9997; found: 388.0004.
(5, C18H12FNO4). White sold; Yield 68%; m.p. 208–209 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.19 (s, 1H), 8.17–8.14 (m, 2H), 7.61–7.58 (m, 2H), 7.39–7.31 (m, 2H), 7.19 (t, J = 8.6 Hz, 2H), 2.55 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 167.17, 165.13, 162.82, 162.67, 159.23, 154.39, 143.54, 133.08, 132.41, 132.33, 129.10, 125.00, 124.89, 124.86, 123.38, 118.51, 116.73, 116.08, 115.90, 77.30, 77.25, 77.04, 76.79, 16.04; HRMS (ESI) [M + H]+ calcd. for C18H12FNO4: 326.0820; found: 326.0832.
(6, C19H15NO5). White sold; Yield 65%; m.p. 165–167 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.20 (s, 1H), 8.10 – 8.08 (m, 2H), 7.59 (d, J = 7.7 Hz, 2H), 7.39–7.31 (m, 2H), 7.00–6.97 (m, 2H), 3.90 (s, 3H), 2.54 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 163.93, 163.38, 162.21, 159.33, 154.36, 143.45, 132.97, 131.90, 129.08, 124.97, 123.60, 120.79, 118.57, 116.70, 114.00, 77.30, 77.25, 77.05, 76.79, 55.56, 16.00; HRMS (ESI) [M + H]+ calcd. for C19H15NO5: 338.1019; found: 338.1033.
(7, C18H12N2O6). Yellow sold; Yield 65%; m.p. 244–245 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.38 – 8.36 (m, 2H), 8.32 – 8.29 (m, 2H), 8.19 (s, 1H), 7.64–7.59 (m, 2H), 7.40–7.33 (m, 2H), 2.58 (s, 3H);13C NMR (126 MHz, CDCl3) δ 163.82, 161.81, 159.13, 154.43, 150.89, 143.74, 134.15, 133.30, 130.89, 129.16, 127.73, 125.09, 123.89, 123.80, 123.04, 118.40, 116.78, 77.30, 77.25, 77.05, 76.79, 16.20; HRMS (ESI) [M + H]+ calcd. for C19H15NO5: 338.1019; found: 338.1033.
(8, C19H12F3NO4). White sold; Yield 69%; m.p. 168–170 °C; 1H NMR (500 MHz, DMSO‑d6) δ 8.37 (s, 1H), 8.03 – 8.01 (m, 2H), 7.91 (dd, J = 7.8, 1.7 Hz, 1H), 7.83–7.81 (m, 2H), 7.71 (ddd, J = 8.8, 7.4, 1.7 Hz, 1H), 7.51–7.47 (m, 1H), 7.43 (td, J = 7.5, 1.1 Hz, 1H), 2.45 (s, 3H); 13C NMR (126 MHz, DMSO) δ 163.93, 162.59, 159.07, 154.24, 143.92, 133.74, 132.69, 131.84, 130.08, 128.58, 127.85, 125.48, 123.43, 118.83, 116.71, 40.45, 40.38, 40.29, 40.21, 40.12, 40.04, 39.95, 39.87, 39.79, 39.62, 39.45, 16.35; HRMS (ESI) [M + H]+ calcd. for C19H15NO4: 402.0944; found: 402.0951.
(9, C18H13NO5). White sold; Yield 68%; m.p. 159–160 °C; 1H NMR (500 MHz, DMSO‑d6) δ 10.53 (s, 1H), 8.36 (s, 1H), 7.98–7.94 (m, 2H), 7.91 (dd, J = 7.8, 1.6 Hz, 1H), 7.74–7.69 (m, 1H), 7.50 (d, J = 8.3 Hz, 1H), 7.43 (t, J = 7.5 Hz, 1H), 6.96–6.90 (m, 2H), 2.43 (s, 3H); 13C NMR (126 MHz, DMSO) δ 163.08, 162.98, 162.77, 159.17, 154.20, 143.71, 133.63, 132.28, 130.02, 125.46, 123.71, 118.94, 118.88, 116.69, 116.18, 40.45, 40.37, 40.28, 40.21, 40.12, 40.04, 39.95, 39.87, 39.78, 39.62, 39.45, 16.22; HRMS (ESI) [M + H]+ calcd. for C18H13NO5: 324.0863; found: 324.08.
(10, C20H15NO4). White sold; Yield 67%; m.p. 161–162 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.16 (s, 1H), 7.88 (d, J = 16.0 Hz, 1H), 7.62–7.55 (m, 4H), 7.45–7.38 (m, 3H), 7.38–7.29 (m, 2H), 6.59 (d, J = 16.0 Hz, 1H), 2.49 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 164.32, 162.14, 159.26, 154.34, 146.84, 143.39, 134.14, 132.96, 130.85, 129.06, 129.02, 128.35, 124.95, 123.58, 118.55, 116.68, 115.29, 77.32, 77.06, 76.81, 15.94; HRMS (ESI) [M + H]+ calcd. for C20H15NO4: 334.1070; found: 334.1074.
(11, C21H17NO4). White sold; Yield 62%; m.p. 154–155 °C; 1H NMR (500 MHz, DMSO‑d6) δ 8.34 (s, 1H), 7.90 (dd, J = 7.8, 1.6 Hz, 1H), 7.81 (d, J = 16.1 Hz, 1H), 7.69 (dd, J = 7.5, 4.0 Hz, 3H), 7.48 (d, J = 8.3 Hz, 1H), 7.42 (td, J = 7.5, 1.1 Hz, 1H), 7.27 (d, J = 7.8 Hz, 2H), 6.79 (d, J = 16.0 Hz, 1H), 2.38 (s, 3H), 2.34 (s, 3H). 13C NMR (126 MHz, DMSO) δ 164.18, 162.56, 159.13, 154.19, 146.68, 143.72, 141.46, 133.63, 131.70, 130.07, 130.02, 129.14, 125.45, 123.64, 118.87, 116.68, 114.93, 40.48, 40.39, 40.31, 40.23, 40.14, 40.05, 39.98, 39.88, 39.81, 39.64, 39.47, 21.56, 16.15. HRMS (ESI) [M + H]+ calcd. for C21H17NO4: 348.1229; found: 348.1233.
(12, C20H14ClNO4). White sold; Yield 65%; m.p. 248–249 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.15 (s, 1H), 7.83 (d, J = 16.0 Hz, 1H), 7.63–7.55 (m, 2H), 7.55–7.49 (m, 2H), 7.41–7.29 (m, 4H), 6.56 (d, J = 16.0 Hz, 1H), 2.48 (s, 3H). 13C NMR (126 MHz, CDCl3) δ 164.09, 162.28, 159.24, 154.32, 145.34, 143.43, 136.78, 133.02, 132.60, 129.52, 129.32, 129.07, 124.98, 123.49, 118.50, 116.69, 115.86, 77.35, 77.09, 76.84, 15.97. HRMS (ESI) [M + H]+ calcd. for C20H14ClNO4: 368.0682, found: 368.0687.
(13, C20H14BrNO4). White sold; Yield 69%; m.p. 182–183 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.15 (s, 1H), 7.81 (d, J = 16.0 Hz, 1H), 7.62–7.53 (m, 4H), 7.52 – 7.43 (m, 2H), 7.38–7.30 (m, 2H), 6.58 (d, J = 16.0 Hz, 1H), 2.48 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 164.10, 162.31, 159.26, 154.34, 145.44, 143.45, 133.03, 132.29, 129.71, 129.09, 125.20, 124.99, 123.50, 118.52, 116.71, 115.97, 77.31, 77.26, 77.06, 76.80, 15.98. HRMS (ESI) [M + H]+ calcd. For C20H14BrNO4: 412.0187; found: 412.0181.
(14, C20H14FNO4). White sold; Yield 67%; m.p. 166–170 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.14 (s, 1H), 7.84 (d, J = 16.0 Hz, 1H), 7.58 (ddd, J = 9.4, 4.8, 2.3 Hz, 4H), 7.38–7.28 (m, 2H), 7.10 (t, J = 8.6 Hz, 2H), 6.51 (d, J = 16.0 Hz, 1H), 2.48 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 165.21, 164.20, 163.20, 162.18, 159.24, 154.33, 145.47, 143.38, 132.98, 130.43, 130.40, 130.33, 130.26, 129.06, 124.96, 123.55, 118.52, 116.68, 116.30, 116.12, 115.04, 115.02, 77.33, 77.27, 77.07, 76.82, 15.93; HRMS (ESI) [M + H]+ calcd. for C20H14FNO4: 352.0975; found: 352.0982.
(15, C21H17NO5). White sold; Yield 68%; m.p. 150–152 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.15 (s, 1H), 7.83 (d, J = 16.0 Hz, 1H), 7.59–7.53 (m, 4H), 7.37–7.30 (m, 2H), 6.94–6.92 (m, 2H), 6.45 (d, J = 16.0 Hz, 1H), 3.85 (s, 3H), 2.48 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 164.70, 161.83, 159.30, 154.32, 146.52, 143.34, 132.92, 130.12, 129.05, 126.91, 124.94, 123.67, 118.57, 116.67, 114.44, 112.54, 77.32, 77.27, 77.07, 76.81, 55.46, 15.91; HRMS (ESI) [M + H]+ calcd. for C21H17NO5: 364.1178; found: 364.1182.
(16, C20H14N2O6). Yellow sold; Yield 70%; m.p. 221–222 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.30 – 8.27 (m, 2H), 8.16 (s, 1H), 7.91 (d, J = 16.0 Hz, 1H), 7.76–7.74 (m, 2H), 7.60 (ddd, J = 14.2, 8.0, 1.6 Hz, 2H), 7.39–7.31 (m, 2H), 6.73 (d, J = 16.1 Hz, 1H), 2.50 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 163.40, 162.80, 159.19, 154.37, 148.78, 143.74, 143.51, 140.12, 133.13, 129.09, 128.95, 125.02, 124.29, 123.35, 119.71, 118.46, 116.75, 77.30, 77.25, 77.05, 76.79, 16.02; HRMS (ESI) [M + H]+ calcd. for C20H14N2O6: 379.0923; found: 379.0927.
(17, C21H14F3NO4). White sold; Yield 75%; m.p. 169–171 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.15 (s, 1H), 7.88 (d, J = 16.1 Hz, 1H), 7.71–7.65 (m, 4H), 7.61–7.56 (m, 2H), 7.37–7.30 (m, 2H), 6.66 (d, J = 16.1 Hz, 1H), 2.49 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 163.76, 162.54, 159.21, 154.35, 144.86, 143.46, 133.06, 129.07, 128.47, 126.00, 125.97, 124.99, 123.42, 118.49, 117.96, 116.70, 77.32, 77.07, 76.81, 15.97; HRMS (ESI) [M + H]+ calcd. for C21H14F3NO4: 402.0944; found: 402.0951.
(18, C20H15NO5). White sold; Yield 75%; m.p. 156–158 °C; 1H NMR (500 MHz, DMSO‑d6) δ 10.12 (d, J = 23.3 Hz, 1H), 8.33 (d, J = 23.1 Hz, 1H), 7.93 (d, J = 8.4 Hz, 1H), 7.78–7.48 (m, 5H), 6.83 (d, J = 8.5 Hz, 2H), 6.61 (d, J = 16.0 Hz, 1H), 2.37 (s, 4H); 13C NMR (126 MHz, DMSO) δ 164.49, 162.21, 160.73, 159.15, 154.18, 146.98, 143.66, 133.60, 131.22, 130.00, 125.51, 125.44, 123.73, 118.89, 116.68, 116.30, 116.26, 111.91, 40.56, 40.47, 40.39, 40.30, 40.22, 40.13, 40.06, 39.97, 39.89, 39.80, 39.63, 39.46, 16.12; HRMS (ESI) [M + H]+ calcd. for C20H15NO5: 350.1018; found: 350.1025.
(19, C20H13ClBrNO4). White sold; Yield 70%; m.p. 201–203 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.12 (s, 1H), 7.81 (d, J = 16.0 Hz, 1H), 7.57–7.53 (m, 2H), 7.51 (d, J = 8.3 Hz, 1H), 7.47–7.43 (m, 2H), 7.37 (d, J = 1.9 Hz, 1H), 7.30 (dd, J = 8.4, 2.0 Hz, 1H), 6.57 (d, J = 16.0 Hz, 1H), 2.47 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 164.00, 161.98, 158.54, 154.54, 145.53, 142.56, 139.10, 133.00, 132.30, 129.81, 129.70, 125.68, 125.24, 123.42, 117.09, 117.08, 115.88, 77.31, 77.26, 77.05, 76.80, 15.87; HRMS (ESI) [M + H]+ calcd. for C20H13ClBrNO4: 483.9343; found: 483.9348.
(20, C20H13Br2NO4). White sold; Yield 70%; m.p. 190–191 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.11 (s, 1H), 7.81 (d, J = 16.0 Hz, 1H), 7.58–7.53 (m, 3H), 7.48–7.42 (m, 4H), 6.57 (d, J = 16.0 Hz, 1H), 2.47 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 164.00, 162.00, 158.48, 154.44, 145.56, 142.65, 132.99, 132.31, 129.89, 129.71, 128.52, 127.21, 125.25, 123.68, 120.05, 117.43, 115.86, 77.30, 77.25, 77.05, 76.80, 15.88; HRMS (ESI) [M + H]+ calcd. for C20H13Br2NO4: 489.9282; found: 489.9286.
(21, C20H13BrFNO4). White sold; Yield 70%; m.p. 186–188 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.13 (s, 1H), 7.81 (d, J = 16.0 Hz, 1H), 7.56 (dd, J = 7.0, 5.0 Hz, 3H), 7.46 (d, J = 8.2 Hz, 2H), 7.08 (dt, J = 8.3, 1.7 Hz, 2H), 6.57 (d, J = 16.0 Hz, 1H), 2.47 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 166.22, 164.18, 164.03, 162.06, 158.79, 155.63, 155.53, 145.49, 142.81, 133.01, 132.30, 132.20, 130.81, 130.73, 129.70, 125.23, 122.31, 122.28, 115.91, 115.27, 115.24, 113.45, 113.27, 104.53, 104.32, 77.30, 77.05, 76.79, 15.89; HRMS (ESI) [M + H]+ calcd. for C20H13BrFNO4: 430.0085; found: 430.0085.
(22, C21H16BrNO4). White sold; Yield 70%; m.p. 198–201 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.14 (s, 1H), 7.83 (d, J = 16.0 Hz, 1H), 7.59–7.55 (m, 2H), 7.48–7.45 (m, 3H), 7.20–7.13 (m, 2H), 6.60 (d, J = 16.0 Hz, 1H), 2.50 (s, 6H); 13C NMR (126 MHz, CDCl3) δ 164.11, 162.51, 159.51, 154.52, 145.32, 144.68, 143.46, 133.06, 132.28, 129.69, 128.74, 126.25, 125.15, 122.21, 116.84, 116.16, 116.06, 77.30, 77.25, 77.05, 76.80, 22.03, 15.97; HRMS (ESI) [M + H]+ calcd. for C21H16BrNO4: 426.0336; found: 426.0335.
(23, C21H16BrNO5). White sold; Yield 70%; m.p. 205–210 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.11 (s, 1H), 7.80 (d, J = 16.0 Hz, 1H), 7.57–7.53 (m, 2H), 7.49–7.43 (m, 3H), 6.88 (dd, J = 8.6, 2.4 Hz, 1H), 6.83 (d, J = 2.4 Hz, 1H), 6.57 (d, J = 16.0 Hz, 1H), 3.90 (s, 3H), 2.47 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 164.16, 163.91, 162.61, 159.61, 156.44, 145.25, 143.58, 133.08, 132.27, 130.14, 129.68, 125.13, 119.67, 116.12, 113.44, 112.20, 100.53, 77.30, 77.25, 77.05, 76.79, 55.93, 15.93; HRMS (ESI) [M + H]+ calcd. for C21H16BrNO5: 442.0251; found: 442.0285.
(24, C20H13ClBrNO4). White sold; Yield 70%; m.p. 201–203 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.06 (s, 1H), 7.82 (d, J = 16.0 Hz, 1H), 7.58–7.53 (m, 5H), 7.46 (d, J = 8.2 Hz, 2H), 7.33–7.30 (m, 1H), 6.58 (d, J = 16.0 Hz, 1H), 2.47 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 163.98, 161.87, 158.62, 152.67, 145.61, 142.03, 132.99, 132.94, 132.31, 130.28, 129.72, 128.06, 125.26, 124.73, 119.52, 118.18, 115.82, 77.30, 77.25, 77.05, 76.79, 15.92; HRMS (ESI) [M + H]+ calcd. for C20H13ClBrNO4: 447.9771; found: 447.9769.
(25, C20H13Br2NO4). White sold; Yield 65%; m.p. 171–172 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.08 (s, 1H), 7.61–7.57 (m, 3H), 7.55–7.51 (m, 3H), 7.49–7.47 (m, 2H), 6.60 (d, J = 16.0 Hz, 1H), 2.50 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 164.00, 161.85, 158.57, 153.14, 145.62, 141.94, 135.74, 132.99, 132.31, 131.13, 129.72, 125.26, 124.69, 120.01, 118.45, 117.55, 115.82, 77.30, 77.25, 77.04, 76.86, 76.79, 15.92; HRMS (ESI) [M + H]+ calcd. for C20H13Br2NO4: 489.9282; found: 489.9284.
(26, C20H13BrFNO4). White sold; Yield 65%; m.p. 192–193 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.08 (s, 1H), 7.81 (d, J = 15.9 Hz, 1H), 7.55 (d, J = 8.3 Hz, 2H), 7.45 (d, J = 8.3 Hz, 2H), 7.37–7.29 (m, 2H), 7.24 (d, J = 2.9 Hz, 1H), 6.57 (d, J = 16.0 Hz, 1H), 2.47 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 163.98, 161.95, 159.87, 158.83, 157.92, 150.50, 150.48, 145.57, 142.32, 142.30, 132.99, 132.30, 129.71, 125.25, 124.70, 120.64, 120.44, 119.23, 119.16, 118.38, 118.32, 115.84, 114.19, 114.00, 77.31, 77.26, 77.05, 76.80, 15.91; HRMS (ESI) [M + H]+ calcd. for C20H13BrFNO4: 430.0089; found: 430.0085.
(27, C21H16BrNO4). White sold; Yield 70%; m.p. 182–183 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.09 (s, 1H), 7.85–7.78 (m, 1H), 7.58–7.53 (m, 2H), 7.48–7.43 (m, 2H), 7.39 (dd, J = 8.4, 2.1 Hz, 1H), 7.34 (d, J = 2.1 Hz, 1H), 7.25–7.21 (m, 1H), 6.58 (d, J = 16.0 Hz, 1H), 2.47 (s, 3H), 2.42 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 164.12, 162.47, 159.48, 152.52, 145.39, 143.43, 134.76, 134.14, 133.05, 132.29, 129.70, 128.70, 125.17, 123.33, 118.27, 116.41, 116.01, 77.30, 77.25, 77.05, 76.79, 20.82, 15.99; HRMS (ESI) [M + H]+ calcd. for C21H16BrNO4: 426.0342; found: 426.0335.
(28, C21H16BrNO5). White sold; Yield 70%; m.p. 251–253 °C; 1H NMR (500 MHz, Chloroform-d) δ 8.11 (s, 1H), 7.81 (d, J = 16.0 Hz, 1H), 7.56 (d, J = 8.2 Hz, 2H), 7.46 (d, J = 8.2 Hz, 2H), 7.29 (d, J = 9.1 Hz, 1H), 7.17 (dd, J = 9.1, 2.9 Hz, 1H), 6.98 (d, J = 2.9 Hz, 1H), 6.58 (d, J = 16.0 Hz, 1H), 3.85 (s, 3H), 2.48 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 164.10, 162.40, 159.41, 156.36, 148.87, 145.42, 143.26, 133.03, 132.29, 132.27, 129.70, 125.19, 123.72, 121.08, 118.86, 117.75, 115.99, 110.54, 77.30, 77.25, 77.05, 76.80, 55.89, 15.95; HRMS (ESI) [M + H]+ calcd. for C21H16BrNO5: 442.0251; found: 442.0285.
4.3 a-Glucosidase inhibition and kinetics assay
The α-glucosidase inhibitory activity of coumarin derivatives 1–28 was assayed using PNPG as substrate (Taha et al., 2018; Carreiro et al., 2014). The α-glucosidase (0.1 U/mL) and test compound was added into phosphate buffer saline (0.1 M, pH 6.8), followed by an incubation for 10 min at 37 °C. After PNPG (0.1 U/mL) was added, the absorbance change was measured at 405 nm. The test compound was dissolved in DMSO, and acarbose was used as a positive control. All experiments were performed 4 times.
The enzyme inhibitory kinetics (Deng et al., 2022) of compound 20 were obtained by the plots of enzymatic reaction rate vs enzyme concentration with or without compound 20, and the substrate inhibitory kinetics (Hu et al., 2021) were measured using the Lineweaver-Burk plot of enzymatic reaction rate vs substrate concentration with or compound 20.
4.4 3D fluorescence spectra assay
Compound 20 (2 μM) was added into α-glucosidase (5 μM) and incubated for 5 min. Then the 3D fluorescence spectra of the mixture were recorded (Zeng et al., 2016). The excitation and emission wavelengths were 200–600 nm. The slit width is 2.5 nm. The data was imported into Matlab for processing.
4.5 CD spectroscopy
Compound 20 (35 μM) was added into α-glucosidase (31 μM) and incubated for 5 min. Then the CD spectrum of the mixture was recorded (Peng et al., 2015). The CDNN was used to analyze the proportion of secondary conformation of protein.
4.6 Molecular docking
SYBYL software was used to simulate the interaction between compound 20 and α-glucosidase (Yu et al., 2018). The homology model of α-glucosidase was constructed according to previous works (Wang et al., 2017; Wang et al., 2018) and optimized by procedure of removing water molecules, adding hydrogen atoms, adding charge, and repairing end residues, followed by the generation of active pocket. Compound 20 was charged with Gasteiger-Hückle and prepared by energy minimization program. Thus, the docking between compounds and target protein was operated in the default format, and the results were visualized by Pymol and Discover studio software.
Acknowledgements
This work was financially supported by the Fundamental and Applied Basic Research Fund of Guangdong Province (No. 2022A1515011657), Department of Education of Guangdong Province (Nos. 2019KZDXM035, 2021KTSCX135, 2021KCXTD044), and Jiangmen Science and Technology Plan Project (2021030103150006664).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Hydrazinyl arylthiazole based pyridine scaffolds: Synthesis, structural characterization, in vitro α-glucosidase inhibitory activity, and in silico studies. Eur. J. Med. Chem.. 2017;138:255-272.
- [Google Scholar]
- 3-Hydroxypyrrolidine and (3, 4)-dihydroxypyrrolidine derivatives: Inhibition of rat intestinal α-glucosidase. Bioorg. Chem.. 2014;54:81-88.
- [Google Scholar]
- Synthesis and bioactivities evaluation of oleanolic acid oxime ester derivatives as alpha-glucosidase and alpha-amylase inhibitors. J. Enzyme. Inhib. Med. Chem.. 2022;37:451-461.
- [Google Scholar]
- Synthesis of benzotriazoles derivatives and their dual potential as alpha-amylase and alpha-glucosidase inhibitors in vitro: Structure-activity relationship, molecular docking, and kinetic studies. Eur. J. Med. Chem.. 2019;183:111677
- [Google Scholar]
- Considerations when using alpha-glucosidase inhibitors in the treatment of type 2 diabetes. Expert. Opin. Pharmaco.. 2019;20:2229-2235.
- [Google Scholar]
- Synthesis, characterization, anti-diabetic potential and DFT studies of 7-hydroxy-4-methyl-2-oxo-2H-chromene-8-carbaldehyde oxime. Spectrochim. Acta A.. 2018;205:111-131.
- [Google Scholar]
- Novel cinnamic acid magnolol derivatives as potent alpha-glucosidase and alpha-amylase inhibitors: Synthesis, in vitro and in silico studies. Bioorg. Chem.. 2021;116:105291
- [Google Scholar]
- Synthesis of novel flavone hydrazones: In-vitro evaluation of α-glucosidase inhibition, QSAR analysis and docking studies. Eur. J. Med. Chem.. 2015;105:156-170.
- [Google Scholar]
- The evaluation of the reactivating and neuroprotective efficacy of two newly prepared bispyridinium oximes (K305, K307) in Tabun-poisoned rats-a comparison with trimedoxime and the oxime K203. Molecules. 2017;22:1152.
- [Google Scholar]
- Inhibitory kinetics and mechanism of kaempferol on alpha-glucosidase. Food Chem.. 2015;190:207-215.
- [Google Scholar]
- α-Glucosidase inhibition by flavonoids: An in vitro and in silico structure-activity relationship study. J. Enzyme. Inhib. Med. Ch.. 2017;32:1216-1228.
- [Google Scholar]
- Differential alpha-amylase/alpha-glucosidase inhibitory activities of plant-derived phenolic compounds: A virtual screening perspective for the treatment of obesity and diabetes. Food Funct.. 2017;8:1942-1954.
- [Google Scholar]
- Overview of coumarins and its derivatives: Synthesis and biological activity. Lett. Org. Chem.. 2021;18:880-902.
- [Google Scholar]
- Syntheses of new 3-thiazolyl coumarin derivatives, in vitro alpha-glucosidase inhibitory activity, and molecular modeling studies. Eur. J. Med. Chem.. 2016;122:196-204.
- [Google Scholar]
- A comprehensive review on xanthone derivatives as alpha-glucosidase inhibitors. Eur. J. Med. Chem.. 2018;157:1460-1479.
- [Google Scholar]
- Oximes: Novel therapeutics with anticancer and anti-inflammatory potential. Biomolecules. 2021;11:777.
- [Google Scholar]
- Discovery of potent alpha-glucosidase inhibitor flavonols: Insights into mechanism of action through inhibition kinetics and docking simulations. Bioorg. Chem.. 2018;79:257-264.
- [Google Scholar]
- Cinnamic acid amides from Tribulus terrestris displaying uncompetitive alpha-glucosidase inhibition. Eur. J. Med. Chem.. 2016;114:201-208.
- [Google Scholar]
- Coumarin: A natural, privileged and versatile scaffold for bioactive compounds. Molecules. 2018;23:250.
- [Google Scholar]
- Synthesis, α-glucosidase inhibition and molecular docking study of coumarin based derivatives. Bioorg. Chem.. 2018;77:586-592.
- [Google Scholar]
- Synthesis, α-glucosidase inhibition and molecular docking of coumarin based derivatives. Bioorg. Chem.. 2018;77:586-592.
- [Google Scholar]
- Maydisone, a novel oxime polyketide from the cultures of Bipolaris maydis. Nat. Prod. Res.. 2022;36:102-107.
- [Google Scholar]
- Advances in the management of diabetes: therapies for type 2 diabetes. Postgrad. Med. J.. 2020;96:610-618.
- [Google Scholar]
- Design, synthesis and biological evaluation of novel coumarin thiazole derivatives as α-glucosidase inhibitors. Bioorg. Chem.. 2016;65:167-174.
- [Google Scholar]
- Discovery of 3,3-di(indolyl)indolin-2-one as a novel scaffold for α-glucosidase inhibitors: In silico studies and SAR predictions. Bioorg. Chem.. 2017;72:228-233.
- [Google Scholar]
- Synthesis, biological evaluation, and docking studies of novel 5,6-diaryl-1,2,4-triazine thiazole derivatives as a new class of α-glucosidase inhibitors. Bioorg. Chem.. 2018;78:195-200.
- [Google Scholar]
- Synthesis and biological evaluation of coumarin derivatives as alpha-glucosidase inhibitors. Eur. J. Med. Chem.. 2020;189:112013
- [Google Scholar]
- Comparison of two docking methods for peptide-protein interactions. J. Sci. Food. Agric.. 2018;98:3722-3727.
- [Google Scholar]
- Inhibitory Mechanism of apigenin on alpha-glucosidase and synergy analysis of flavonoids. J. Arg Food Chem.. 2016;64:6939-6949.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2022.104072.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary data 1
Supplementary data 1







