Translate this page into:
Virtual screening for the discovery of lawsone derivatives as PS Ⅱ inhibitors
⁎Corresponding authors. wcyang@gzu.edu.cn (Wenchao Yang), gxh200719@163.com (Xiuhai Gan)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
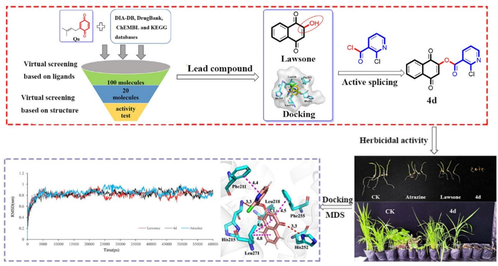
Abstract
Photosynthesis system Ⅱ (PS Ⅱ) inhibitors can block electron transport and disrupt plant photosynthesis and are used as herbicides. In order to develop new and effective PS II inhibitors, the lead structure lawsone was first found with remarkable herbicidal activity based on a virtual screening strategy, and its derivatives were designed and synthesized. The results of herbicidal activity evaluation showed that most of the compounds could achieve more than 80 % inhibitory effect on roots and stems of Portulaca oleracea at a dose of 100 μg/mL. Meanwhile, compound 4d showed more than 90 % inhibition on the stem of P. oleracea and on the root of Echinochloa crusgalli, which were superior to lawsone (64.9 and 67.6 %) and atrazine (63.6 and 65.9 %), respectively. Interestingly, 4d exhibited 100 % post-emergence herbicidal activity to Amaranthus retroflexus and Abutilon theophrasti at a dosage of 300 g a.i./ha, which was similar to atrazine but superior to lawsone. In addition, compound 4d can affect the efficiency of the photochemical process and strong bind PS II D1 protein. It revealed that compound 4d can be used as a promising PS II D1 inhibitor. This study provides a promising lead compound for developing new PS II inhibitors.
Keywords
Virtual screening
Photosystem II D1 protein
Lawsone
Herbicidal activity
Molecular docking
- PS II
-
Photosystem II
- PQ
-
plastoquinones
- MDS
-
Molecule dynamics simulation
- TLC
-
Thin-layer chromatography
- 1H NMR
-
1H nuclear magnetic resonance
- 13C NMR
-
13C nuclear magnetic resonance
- HRMS
-
High resolution mass spectrum
- RMSD
-
Root-mean-square deviation
Abbreviations
1 Introduction
Weeds not only compete with crops for sunlight, water, nutrients, and space but also hinder light and ventilation in the field, resulting in reduced grain production and economic losses (Lamberth et al., 2013). Weed control plays an important role in agricultural production. The use of synthetic herbicides is an economic and effective means to eliminate weeds in crop fields (Gou et al., 2021). However, long-term and repeated application of chemical herbicides leads to severe herbicide resistance and environmental contamination (Cummins et al., 2013; Alberto et al., 2016). It is urgent to develop innovative approaches for weed management to fulfill the current requirements of safe food production and environmental protection.
Plastoquinones (PQs), an important class of carriers for electron transfer in photosystem Ⅱ (PS Ⅱ), play a key role in maintaining normal photosynthesis (Borisova-Mubarakshina et al., 2018). D1 protein is an important protein subunit and combines D2 protein to form the basic framework of the PS II reaction center, which controls the electron transfer process in PS Ⅱ and affects the reduction of PQs directly (Singh, 2000). PS II-inhibiting herbicides block the electron transport from the primary quinone acceptor QA to the secondary quinone acceptor QB in PS II, disrupting photosynthesis and ultimately causing plant death (Kyle, 1985; Fuerst and Norman, 1991). As the most popular PS II-inhibiting herbicide, atrazine provides effective weed control at low-cost and about 33 million kilograms of it were used in agriculture in the US in 2019 (Yang et al., 2023). However, the long-term use of atrazine has brought severe herbicide resistance and environmental contamination (Svyantek et al., 2016; Marriage et al., 1975). It will be replaced by new, high-efficient, and low-toxic herbicides.
Natural products have attracted much attention due to their unique scaffolds, superior biological activity, lower toxicity and environmental-friendly, and easier degradation (Jin et al., 2023). Up to now, about 80 % of drugs on the market are natural products or their derivatives (Dayan et al., 2012). It is a crucial method for the development of novel herbicides based on the discovery of herbicidal active natural products and their derivatives (Lorsbach et al., 2019; Sparksa and Bryantb, 2022) and some of them have been used to manage weeds (Fig. 1). It was well-known that glyphosate and glufosinate are two important bioherbicides, which were aminophosphine analogues obtained by optimization on natural amino acid glycine (Zhou et al., 2020). Meanwhile, some natural products and their derivatives showed remarkable herbicidal activity and safety for crops, such as sorgoleone (Nimbal et al., 1996), and cinmethylin (Grossmann et al., 2012). In recent years, Patulin (Guo et al., 2021), antidesmone (Sampaio et al., 2016), citrinin (Yang et al., 2023), tenuazonic acid (Chen et al., 2017), and their derivatives (Yang et al., 2023; Wang et al., 2022) were found as PS II inhibitors via interacting with the binding pocket in the D1 protein. By comparing the structures of sorgoleone and antidesmone, it can see that they have similarly quinone structures, which can compete with plastoquinones at PS II and then disrupt photosynthesis. So, more excellent PS II inhibitors will be found based on quinones.
Some natural product herbicides.
The development of new synthetic pesticides was always difficult and expensive. However, the discovery of lead compounds by high-throughput virtual screening based on the structural characteristics and protein properties of ligands with cutting-edge computer-aided drug design methods can save cost and time and is considered an alternative method to traditional techniques for leading compound discovery. Among them, ligand-based virtual screening (LBVS) is a technique to identify compounds that may bind to one or several drug targets by similarity search, pharmacophore or quantitative structure–activity relationships model comparison (Singh et al., 2021). Meanwhile, structure-based virtual screening (SBVS) is used to identify compounds according to the structure of specific target proteins (De Azevedo, 2010). In recent years, there have been many successful cases of the development of new pesticides against different targets using LBVS/SBVS (Zhang et al., 2022; Fu et al., 2020; Lin et al., 2019). These successful examples showed that both LBVS and SBVS can promote the discovery of drug candidates by reducing the number of screened compounds, while improving the discovery efficiency of lead compounds.
The goal of this study was to discover lead compounds for the development of new herbicides targeting PS II. As shown in Fig. 2, QB was used as a ligand of D1 protein, and compounds targeting PS II D1 were screened from different databases, and the top 20 compounds were obtained (Fig. 3) and the lead compound lawsone was confirmed with herbicidal activity evaluation. Meanwhile, lawsone derivatives were designed based on the analysis of the binding model of lawsone in PS Ⅱ D1. The target compounds were synthesized and their herbicidal activities were evaluated. In addition, the mechanisms of the lawsone derivative with the best herbicidal activity were investigated by chlorophyll fluorescence measurements, molecular docking, and molecular dynamic simulation methods.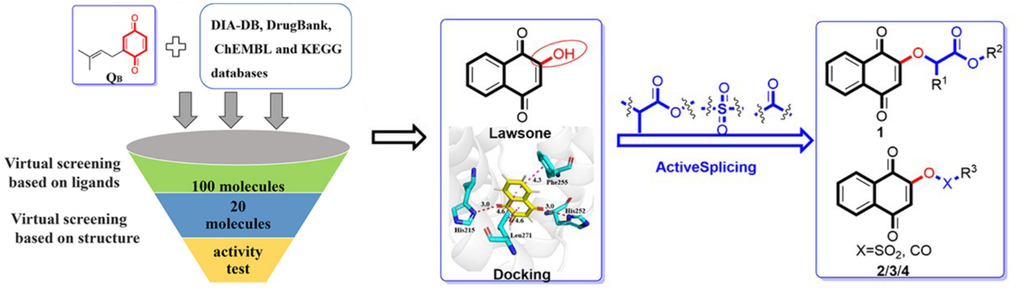
Design of the target compounds.
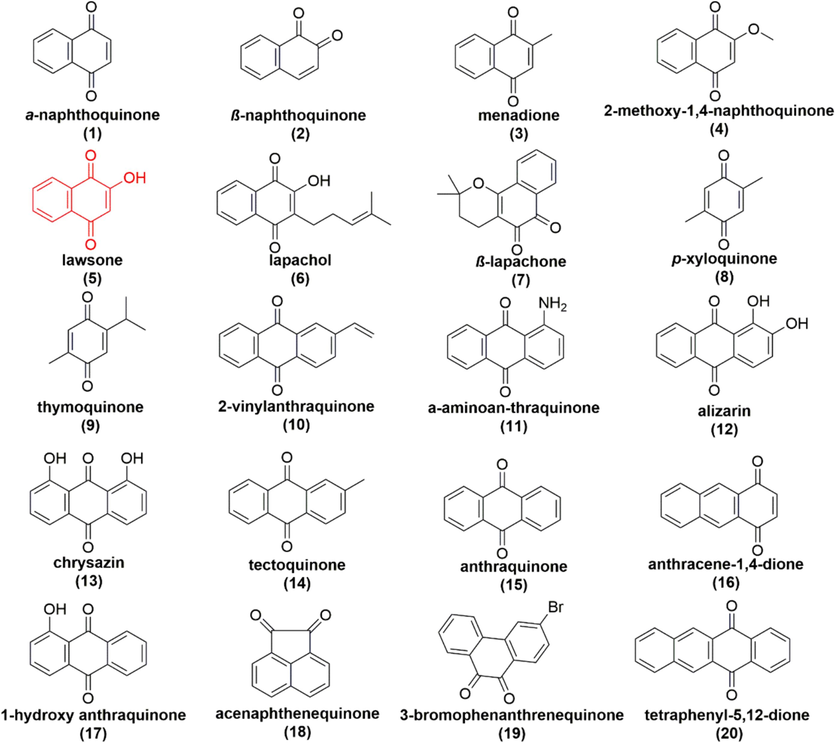
The structures of the top 20 molecules in score.
2 Materials and methods
2.1 Chemicals and instruments
All chemicals are purchased without any further purification from Aladdin (Aladdin Company, CHN). The raw materials needed for the reaction are directly used without further purification. Under the irradiation of ultraviolet lamp, the reaction was monitored on silica gel plate by thin layer chromatography. Melting point instrument (Shanghai INESA Optical Instrument Co., Ltd., China), without temperature correction. Using DMSO‑d6 or CDCl3 as solvent and TMS as internal standard, the 1H and 13C NMR spectra of the compounds were recorded on a Bruker DPX-400 or 500 spectrometers. High-resolution mass spectrometry (HRMS) was performed by Thermo Scientific Q-Exactive (Thermo, USA). The seeds of all weeds are purchased commercially and disinfected with 75 % alcohol.
2.2 Virtual screening based on ligands
The polar head quinone is the key structure for the interaction between QB and protein. Therefore, quinone was used as ligand and conducted virtual screening based on ligand on BRUSELAS (https://bio-hpc.ucam.edu/Bruselas/) (Banegas-Luna et al., 2019). The calculation mode is similarity search, and the algorithm selects pharmacophore and 3D scanning. The scanned databases include DIA-DB (186 compounds), Drug Bank (1,760 compounds), CHEMBL (1,578,131 compounds) and KEGG (23,667 compounds) (Wishart et al., 2006; Gaulton et al., 2012; Kanehisa et al., 2016; Pereira et al., 2019). The scoring function takes a weighted average, and the similarity threshold is 0.6 for screening.
2.3 Virtual screening based on structures
The program PyRx was used for virtual screening of the obtained small molecules based on D1 protein structure. The crystal structure of the D1 protein (PDB: 5XNL from Pisum sativum) was acquired from the RCSB Protein Data Bank (Battaglino et al., 2021), and further treated with the addition of hydrogen atoms and removal of water molecules with PyMOL. The binding behavior of the top 20 small molecules was analyzed according to the default score. Meanwhile, the physical and chemical properties including logP, pKa, hydrogen bond acceptor (HBAs), hydrogen bond donor (HBDs), and aromatic ring (ARs) of compounds were calculated with Discovery Studio v3.5, and the electronegativity was predicted by SYBYL-X 2.0 (Zhao et al., 2023).
2.4 Binding mode-based ligand design
In order to discover more active quinone derivatives, the binding mode of lead structure and D1 was determined by LeDock compared with QB (Zhang and Zhao, 2016). According to the molecular interaction modes of QB and lead structure binding to D1 protein (Fig. 4), some active substructures were introduced to lead structure, and the binding model and interaction energy of each ligand binding with QB site of D1 protein was performed by LeDock and Autodock Vina, respectively. The binding models of high active compounds to D1 were performed using the same method.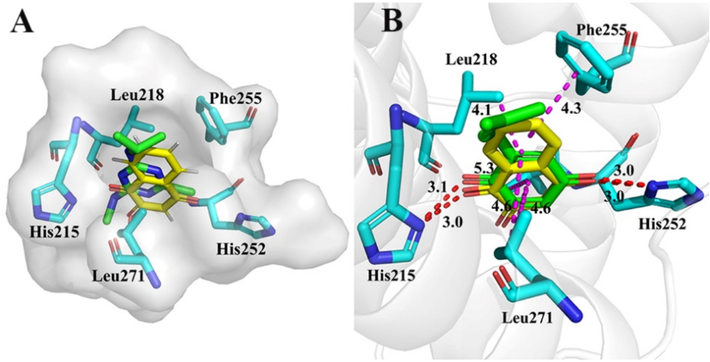
Simulated surface representation of Atrazine (green) and Lawsone (yellow) binding to D1 protein (A). In PsPS II D1 protein, QB (green) and Lawsone (yellow) combined with surrounding amino acids (B).
2.5 General procedures for the preparation of compounds 1a–1e
As shown in Fig. 5, the lead compound lawsone (1.72 mmol) reacted with K2CO3 (2.58 mmol) in N, N-dimethylformamide (DMF) solution for 1 h. Subsequently, chlorine reagent was added and react at 50℃ for 1–2 h. After the reaction was completely monitored by TLC, the distilled water was added, and the mixture was extracted with ethyl acetate (3 × 50 mL) and the organic phase was dried over anhydrous Na2SO4, and then purified by column chromatography eluted with ethyl acetate/petroleum ether (1:5, V/V) to give target compounds 1a–1e.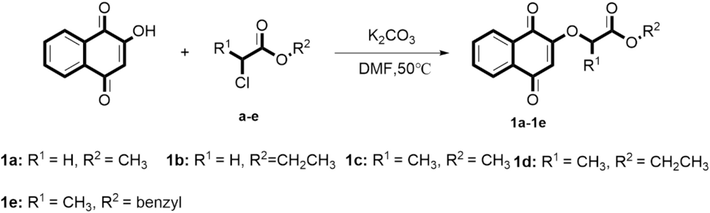
Synthesis route of the target compounds 1a − 1e.
2.6 General procedures for the preparation of compounds 2a–2e, 3a–3e and 4a–4 k
As shown in Fig. 6, lawsone (1.44 mmol) and dry Et3N (1.72 mmol) were added to the dry CH2Cl2, and then the aromatic sulfonyl chloride or acyl chloride (1.72 mmol) was added and stirred at 0 ℃ for 30 min, the mixture was allowed to reach room temperature and then stirred for 2 h. Then the solvent was removed under vacuum and the mixture was purified by column chromatography eluted with ethyl acetate/petroleum ether (1:10, V/V) to give target compounds 2a–2e, 3a–3e and 4a–4 k.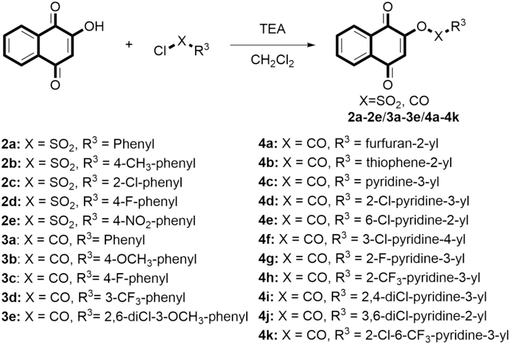
Synthesis route of the target compounds 2a − 2e, 3a − 3e and 4a − 4 k.
2.7 Petri dish assay
The herbicidal activities of target compounds were evaluated on P. oleracea and E. crusgalli using the reported methods (Sun et al., 2020; Cao et al., 2017). The seeds used in the experiment were disinfected and sterilized, then kept in a constant temperature incubator until they germinated. Dissolve the compound with 1 mL of DMF and prepared the required solution with 0.1 % tween-80. Subsequently, the germinated seeds are placed in 12-well plates, with 4–6 seeds in each hole (two filter papers are placed in each hole to keep moisture). Immediately after that, 1 mL of solution with a concentration of 100 μg/mL or 10 μg/mL was added to each well. The blank control group was a mixed solution of DMF and 0.1 % Tween-80 and the lead structure lawsone and commercial herbicides atrazine were used as the positive control group. After added drugs, the 12-well plates were placed in a manual climate incubator, and were automatically circulated in the light for 16 h (27 ℃) and in the dark for 8 h (25 ℃), and the relative humidity was 75 %. Each experiment had three parallel groups, and each experiment was repeated three times. After 5 days of observation, the length of stems and roots of weed seeds were measured and calculated the inhibition rate with follows formula.
Inhibition rate (%) = (Lcontrol–Ltreatment)/Lcontrol × 100 %.
2.8 Post-emergence herbicidal activity
The herbicidal activity of the target compounds against monocotyledonous grass (Digitaria sanguinalis, E. crusgalli, and Setaria viridis) and dicotyledonous grass (P. oleracea, A. retroflexus, and A. theophrasti) was performed with the previous method (Wang et al., 2005). The specific operation is to dissolve the compound in 1 mL DMF and dilute it to the required concentration with 0.1 % Tween −80. Lawsone and atrazine were used as the positive controls, and a mixture of DMF and 0.1 % Tween-80 was used as the blank control group. The sprouting weeds are planted in 8 cm plastic pots with two-thirds of which are organic soil. The compound is sprayed during the growth of monocotyledonous plants to the two-leaf stage and dicotyledonous plants to the three-leaf stage and cultivated in the greenhouse. After 15 days, the inhibition rate was visually measured compared with the blank control group. Each treatment was repeated three times.
2.9 Chlorophyll fluorescence measurements
The leaves of A. retroflexus were infiltrated with the 0.1 % Tween-80 solution of 3e and 4d with a concentration of 100 μM and kept in darkness for 30 min at room temperature. Then the initial fluorescence (FO) and the maximum fluorescence (FM) of the dark adaptation blade were measured with MINI-PAM-II fluorometer (Heinz Walz GmbH, Germany) under the setting of saturated pulse mode when the fluorescence value of the passing current reached a stable level. When the actinic light reached the steady-state fluorescence (FS), a saturation pulse was given to determine the maximum fluorescence (F'M) in the light-adapted leaves. Then the actinic light was removed, and the minimum fluorescence (F'O) in the leaves adapted to light was measured with far-red light for 3 sec. Finally, the maximum quantum efficiency (FV/FM) and photochemical quenching coefficient (qP) of PS II are calculated with follows formula (FM–FO)/FM and (F'M−FS)/(F'M−F'O), respectively (Chen et al., 2008; Thuillier et al., 2024).
2.10 Molecular dynamic simulation
The water molecules were removed from the PsPS II D1 protein (PDB: 5XNL), and then it was hydrogenated and its amino acid residues were corrected. The complexes of compound 4d, lawsone and atrazine with PsPS II D1 were performed with molecular dynamic simulation (MDS) by AMBER 18 (Adcock and McCammon, 2006; Yang et al., 2020). A water box was added around the ligand–protein complexes with a distance of 10 Å, and Cl- and Na+ were introduced to balance the system to electric neutrality. After energy minimization, the ligand–protein system was heated to 300 K and the pressure was raised to 0.1 MPa. Eventually, the MDS of each complex was executed lasting 60000 ps.
3 Results and discussion
3.1 Virtual screening
According to previous report (Battaglino et al., 2021), at the bottom of QB binding site, polar groups of His215, Ser264 and Phe265 form hydrogen bonds with carbonyl groups at the head of PQ, and Phe255, Ser264 and Leu271 in D1 participate in PQ binding and electron transfer in PS II. Therefore, on the basis of the structural characteristics of QB and the structure of quinones, we first conducted ligand-based virtual screening to screen thousands of molecules from the databases DIA-DB, DrugBank, ChEMBL, and KEGG, and selected the top 100 molecules. Secondly, the program PyRx was used to hydrogenate for the D1 (PDB: 5XNL) protein, remove water treatment, and carry out virtual screening based on protein structure to obtain the top 20 molecules in the score (Fig. 3). It can be seen that they are all of quinone derivatives and their affinity, physical and chemical properties were listed in Table 1. As is known that the physical and chemical properties of compounds affect their pharmacological efficacy and interaction with target enzymes (Zhao et al., 2023). From Table 1, we found that the compounds MW were within 136.15 to 287.11. Meanwhile, the logp, HBAs and ARs of these 20 compounds were similar. Compared with other compounds, lawsone, lapachol, α-aminoan-thraquinone, alizarin, chrysazin, and 1-hydroxy anthraquinone had HBDs, which indicated that they might form strong interactions with surrounding amino acids. In all the compounds, lawsone has the lowest logP and pKa values of 1.21 and 1.92, respectively, which are beneficial to the transmission and absorption of plants. Lawsone was identified as a promising lead structure for developing new PS Ⅱ inhibitors with the results of the pre-seedling herbicidal activity evaluation (Table 2).
compound
affinity (kcal/mol)
MWa
LogPa
pKaa
HBAsa
HBDsa
ARsa
electronegativityb
1
−6.7
158.15
1.59
2.75
2
0
1
2
−6.6
158.15
1.48
2.74
2
0
1
3
−6.6
172.18
1.98
3.35
2
0
1
4
−6.5
188.18
1.43
2.56
3
0
1
5
−6.4
174.15
1.21
1.92
3
1
1
6
−6.4
256.30
2.93
4.13
3
1
1
7
−6.3
242.27
2.42
2.31
3
0
1
8
−6.3
136.15
1.22
3.97
2
0
0
9
−6.3
164.20
1.85
3.02
2
0
0
10
−6.3
234.25
3.27
4.68
2
0
1
11
−6.2
223.23
2.30
3.50
2
1
2
12
−6.2
240.21
2.02
3.10
4
2
2
13
−6.2
240.21
2.04
3.09
4
2
2

14
−6.2
222.24
3.01
4.67
2
0
2
15
−6.2
208.21
2.64
4.15
2
0
2
16
−6.1
208.21
2.55
4.26
2
0
2
17
−6.1
224.21
2.39
3.54
3
1
2
18
−6.1
182.17
2.05
3.72
2
0
2
19
−6.1
287.11
3.08
4.90
2
0
2

20
−6.0
258.27
3.57
5.60
2
0
3
compound
100 μg/mL
10 μg/mL
P.o.b
E.c.b
P.o.b
E.c.b
root
stem
root
stem
root
stem
root
stem
1
100
59.6 ± 0.2
53.6 ± 0.4
31.6 ± 0.5
76.2 ± 0.2
20.8 ± 0.2
24.8 ± 0.7
2.5 ± 0.2
2
100
41.6 ± 0.2
21.8 ± 0.4
26.4 ± 0.4
79.9 ± 0.4
37.7 ± 0.2
3.7 ± 0.6
10.4 ± 0.4
3
100
68.7 ± 0.1
59.5 ± 0.2
24.5 ± 0.3
83.1 ± 0.4
40.3 ± 0.3
21.6 ± 0.6
17.1 ± 0.2
4
100
54.9 ± 0.2
14.3 ± 0.3
31.9 ± 0.8
75.2 ± 0.4
29.9 ± 0.1
0
14.9 ± 0.5
5
100
64.9 ± 0.3
67.6 ± 0.2
45.4 ± 0.1
82.4 ± 0.2
50.6 ± 0.1
31.6 ± 0.3
26.9 ± 0.3
6
100
55.8 ± 0.1
29.5 ± 0.6
37.5 ± 0.1
74.9 ± 0.3
33.8 ± 0.1
0
17.9 ± 0.6
7
89.3 ± 0.4
49.7 ± 0.5
40.5 ± 0.4
40.9 ± 0.4
59.4 ± 0.1
13.3 ± 0.1
0.7 ± 0.4
13.9 ± 0.3
8
93.7 ± 0.3
63.7 ± 0.2
63.2 ± 0.2
36.8 ± 0.2
44.8 ± 0.1
41.8 ± 0.6
32.3 ± 0.4
14.9 ± 0.2
9
94.8 ± 0.1
53.5 ± 0.3
20.1 ± 0.2
37.5 ± 0.3
57.3 ± 0.4
29.4 ± 0.4
13.1 ± 0.6
21.1 ± 0.8
10
100
62.3 ± 0.2
21.8 ± 0.3
27.2 ± 0.6
69.8 ± 0.1
13.3 ± 0.1
0
13.5 ± 0.3
11
77.4 ± 0.5
73.1 ± 0.3
35.6 ± 0.2
36.3 ± 0.1
18.6 ± 0.1
46.5 ± 0.3
0
14.8 ± 0.6
12
84.7 ± 0.2
64.2 ± 0.3
68.1 ± 0.6
22.4 ± 0.1
77.9 ± 0.7
40.1 ± 0.7
26.6 ± 0.3
12.1 ± 0.3
13
75.5 ± 0.7
64.3 ± 0.2
24.4 ± 0.2
23.5 ± 0.4
65.8 ± 0.2
53.3 ± 0.6
0
10.1 ± 0.1
14
49.9 ± 0.4
53.4 ± 0.2
29.3 ± 0.4
24.5 ± 0.3
24.6 ± 0.2
45.7 ± 0.5
4.7 ± 0.7
14.5 ± 0.5
15
100
60.4 ± 0.1
69.8 ± 0.7
31.6 ± 0.6
32.3 ± 0.3
37.7 ± 0.4
27.3 ± 0.7
22.9 ± 0.7
16
100
64.1 ± 0.2
66.8 ± 0.2
26.9 ± 0.2
86.1 ± 0.5
35.1 ± 0.1
29.6 ± 0.3
10.2 ± 0.6
17
100
60.8 ± 0.2
28.3 ± 0.4
7.1 ± 0.3
85.9 ± 0.1
20.8 ± 0.2
23.2 ± 0.4
0
18
90.2 ± 0.1
50.7 ± 0.2
48.1 ± 0.1
11.8 ± 0.6
76.4 ± 0.2
0
20.6 ± 0.3
7.4 ± 0.5
19
40.5 ± 0.4
45.5 ± 0.1
22.1 ± 0.6
23.5 ± 0.4
10.7 ± 0.4
10.7 ± 0.2
0.2 ± 0.1
16.7 ± 0.2
20
34.4 ± 0.2
48.1 ± 0.1
33.1 ± 0.4
28.2 ± 0.2
0
0
0
20.5 ± 0.3
Atrazine
100
63.6 ± 0.2
65.9 ± 0.6
46.1 ± 0.3
91.9 ± 0.2
28.1 ± 0.5
4.2 ± 0.7
5.3 ± 0.3
3.2 Binding mode-based ligand design
PS II is a protein complex that includes many co-factors, of which, the hetero-dimer of D1 and D2 homologous protein is the core (Zabret et al., 2021). It is known that PS II herbicides interrupt the electron from QA to QB by binding the QB site between helix IV and V of D1 protein from Phe211 to Leu275 (Oettmeier, 1999). Based on the model of action between D1 protein and QB, the binding model of lawsone to D1 protein was performed by LeDock. The results indicated that lawsone can completely bind in the active pocket formed by QB (Fig. 4A). As well as QB, the two carbonyl and hydroxyl of lawsone form hydrogen-bond interaction to D1 protein with amino acids His 215 and His 252, respectively. Meanwhile, the quinone ring forms π-Alkyl action to amino acid Leu271. Surprisingly, the benzene ring of lawsone forms two π-Alkyl and π-π interactions with Leu218, Leu271, and Phe255 (Fig. 4B). It indicated that lawsone can inhibit electron transfer in PS II by occupying the binding site of QB. In order to discover the novel lawsone derivatives for the development of PS Ⅱ inhibitor herbicides, the aryl carboxylic acid substructures were introduced into the hydroxyl to design compounds 1a–1e, 2a–2e, 3a–3e, and 4a–4 k compounds on the basis of the model of action between D1 protein and lawsone. The results of the binding affinity of them showed that the order is 4a–4 k > 3a–3e > 2a–2e > 1a–1e, and stronger than the commercial PS Ⅱ inhibitor herbicide atrazine (Table 3). Among them, compound 4d has the strongest binding affinity with a value of 7.0 kcal/mol, which was obviously superior to lawsone (6.4 kcal/mol) and atrazine (5.4 kcal/mol). Theoretically, the better binding with D1 protein means more blocking electron transfer and the better herbicidal activity. However, the current conclusion is obtained by computer virtualization, and the actual results need to be verified by weeding activity tested.
compound
affinity
(kcal/mol)100 μg/mL
10 μg/mL
P.o.b
E.c.b
P.o.b
E.c.b
root
stem
root
stem
root
stem
root
stem
1a
6.4
100
93.5 ± 0.3
60.7 ± 0.5
30.9 ± 0.7
74.2 ± 0.2
72.4 ± 0.2
4.5 ± 0.8
18.3 ± 0.2
1b
−6.0
100
75.3 ± 0.2
41.7 ± 0.7
36.2 ± 0.2
72.8 ± 0.1
61.8 ± 0.2
23.4 ± 0.4
17.7 ± 0.3
1c
−6.0
100
69.4 ± 0.1
35.6 ± 0.2
34.1 ± 0.2
69.1 ± 0.1
57.3 ± 0.1
15.7 ± 0.7
15.7 ± 0.5
1d
−6.0
100
77.9 ± 0.1
41.9 ± 0.8
35.9 ± 0.1
37.1 ± 0.1
46.8 ± 0.1
22.5 ± 0.3
11.1 ± 0.2
1e
−6.1
90.5 ± 0.1
81.6 ± 0.3
32.1 ± 0.7
37.8 ± 0.2
50.9 ± 0.2
59.2 ± 0.4
5.4 ± 0.3
30.3 ± 0.4
2a
−6.3
100
66.5 ± 0.1
46.1 ± 0.9
67.5 ± 0.4
80.7 ± 0.1
60.3 ± 0.9
33.8 ± 0.7
39.6 ± 0.2
2b
−6.0
100
89.1 ± 0.6
51.3 ± 0.5
34.7 ± 0.2
81.8 ± 0.2
66.1 ± 0.4
13.1 ± 0.2
22.9 ± 0.4
2c
−6.1
100
69.9 ± 0.3
43.1 ± 0.6
35.1 ± 0.4
35.3 ± 0.1
57.9 ± 0.2
0
0
2d
−6.1
94.9 ± 0.2
56.1 ± 0.1
41.7 ± 0.7
41.5 ± 0.6
86.4 ± 0.1
35.1 ± 0.1
21.1 ± 0.4
11.2 ± 0.1
2e
−6.3
100
87.1 ± 0.3
30.9 ± 0.4
25.1 ± 0.3
86.5 ± 0.5
70.4 ± 0.1
13.4 ± 0.7
20.3 ± 0.2
3a
−6.3
100
90.5 ± 0.3
48.7 ± 0.2
18.5 ± 0.4
83.7 ± 0.3
63.9 ± 0.2
9.4 ± 0.2
8.6 ± 0.3
3b
−6.0
95.6 ± 0.2
87.1 ± 0.1
60.7 ± 0.7
42.4 ± 0.9
82.8 ± 0.2
47.5 ± 0.2
3.3 ± 0.5
0
3c
−6.1
83.3 ± 0.3
29.6 ± 0.3
41.5 ± 0.3
41.8 ± 0.3
37.3 ± 0.4
10.6 ± 0.3
8.4 ± 0.2
23.1 ± 0.3
3d
−6.2
92.9 ± 0.1
68.8 ± 0.5
67.5 ± 0.2
62.5 ± 0.4
21.8 ± 0.6
12.9 ± 0.1
33.9 ± 0.7
28.8 ± 0.2
3e
−6.5
100
89.6 ± 0.2
82.4 ± 0.2
67.8 ± 0.7
89.5 ± 0.1
70.4 ± 0.2
53.1 ± 0.4
28.5 ± 0.4
4a
−6.0
100
81.1 ± 0.1
51.8 ± 0.9
25.2 ± 0.3
100
58.7 ± 0.2
23.4 ± 0.4
20.2 ± 0.7
4b
−6.1
100
71.4 ± 0.1
40.6 ± 0.7
20.3 ± 0.4
100
56.1 ± 0.3
5.8 ± 0.9
5.9 ± 0.5
4c
−6.4
100
58.4 ± 0.3
59.7 ± 0.7
27.4 ± 0.2
100
40.5 ± 0.3
35.1 ± 0.6
25.4 ± 0.7
4d
−7.0
100
93.7 ± 0.4
91.3 ± 0.2
82.1 ± 0.3
100
80.3 ± 0.4
54.6 ± 0.5
49.2 ± 0.3
4e
−6.5
97.4 ± 0.4
78.2 ± 0.2
60.7 ± 0.7
49.2 ± 0.3
73.5 ± 0.1
60.4 ± 0.3
28.8 ± 0.7
31.6 ± 0.6
4f
−6.6
100
76.9 ± 0.1
80.6 ± 0.2
24.5 ± 0.3
85.1 ± 0.3
61.7 ± 0.1
51.9 ± 0.6
16.1 ± 0.2
4 g
−6.3
93.1 ± 0.6
79.4 ± 0.1
53.6 ± 0.5
33.8 ± 0.1
76.2 ± 0.1
59.4 ± 0.5
34.7 ± 0.7
23.8 ± 0.4
4 h
−6.4
94.5 ± 0.2
77.4 ± 0.6
55.3 ± 0.4
37.5 ± 0.3
82.1 ± 0.2
41.3 ± 0.2
38.9 ± 0.2
26.3 ± 0.4
4i
−6.4
100
58.2 ± 0.3
61.8 ± 0.6
48.9 ± 0.6
87.1 ± 0.3
11.4 ± 0.3
23.4 ± 0.7
29.1 ± 0.2
4j
−6.3
94.5 ± 0.2
86.5 ± 0.5
67.5 ± 0.6
27.2 ± 0.1
80.5 ± 0.1
76.8 ± 0.3
15.5 ± 0.7
15.8 ± 0.1
4 k
−6.3
100
80.5 ± 0.1
52.9 ± 0.4
34.7 ± 0.2
90.7 ± 0.2
70.9 ± 0.2
37.5 ± 0.6
15.6 ± 0.7
Lawsone
−6.4
100
64.9 ± 0.3
67.6 ± 0.2
45.4 ± 0.1
82.4 ± 0.2
50.6 ± 0.1
31.6 ± 0.3
26.9 ± 0.3
Atrazine
−5.4
100
63.6 ± 0.2
65.9 ± 0.6
46.1 ± 0.3
91.9 ± 0.2
28.1 ± 0.5
4.2 ± 0.7
5.3 ± 0.3
3.3 Chemistry
The compounds 1a–1e, 2a–2e, 3a–3e and 4a–4k via binding mode-based ligand design were synthesized. As shown in Fig. 5, the target compounds 1a–1e was obtained by etherification of starting materials lawsone and chlorine reagents (directly purchased) with the assistance of K2CO3 in DMF, respectively. In addition, the lead structure lawsone was esterified with substituted sulfonyl chloride/acyl chloride to obtain the target compounds 2a–2e, 3a–3e and 4a–4 k, respectively (Fig. 6). The structures of the target compounds were confirmed by 1H NMR, 13C NMR, and HRMS data, as listed in the Supporting Information.
3.4 Herbicidal activity
In order to further confirm the lead compound, the pre-seedling herbicidal activities of 20 molecules obtained by virtual screening were tested using Petri dish and the results are shown in Table 2. As can be seen from Table 2, when the concentration is 100 μg/mL, most of them inhibit the growth of P. oleracea roots by more than 80 %. Especially, compounds 1–6, 10, 15–17 exhibited 100 % inhibitory rate on P. oleracea, which was similar to atrazine. Meanwhile, compounds 3, 5, 16, 17 showed more than 80 % than inhibitory effect at the concentration of 10 μg/mL. It indicated that the inhibitory effect of roots were obviously better than stems. In addition, compounds 5, 8, 12, 15, 16 showed remarkable inhibitory (more than 60 %) effect on the root growth of E. crusgalli, which was as well as atrazine. Overall, lawsone (compound 5) showed excellent inhibitory activity on the root and stem growth of P. oleracea and E. crusgalli at 100 and 10 μg/mL. It proved that lawsone is a promising lead structure for the development of herbicides.
As shown in Table 3, compounds 1a–1d, 2a–2c, 2e, 3a, 3e, 4a–4d, 4f, 4i and 4 k showed 100 % inhibition on the root growth of P. oleracea at 100 μg/mL, which were similar to lawsone and atrazine. Meanwhile, compounds 1a, 3a, and 4d exhibited more than 90 % inhibitory effect on stem growth of P. oleracea, which were better than lawsone (64.9 %) and atrazine (63.6 %). Even at the concentration of 10 μg/mL, compounds 4a–4d had 100 % inhibitory activity against the growth roots of P. oleracea, better than lawsone (82.4 %) and atrazine (91.9 %). Only compound 4d showed more than 80 % inhibitory activity on the growth stem of P. oleracea. In addition, compounds 3e (82.4 %), 4d (91.3 %), and 4f (80.6 %) showed better inhibitory effects on root of E. crusgalli than lawsone (67.6 %) and atrazine (65.9 %) at 100 μg/mL (Fig. 7). It is worth noting that compound 4d (82.1 %) had better inhibitory effects on stem of E. crusgalli than lawsone (45.4 %) and atrazine (46.1 %). Furthermore, compound 4d showed 54.6 and 49.2 % inhibition on the roots and stems of E. crusgalli at 10 μg/mL, superior to lawsone (31.6 and 26.9 %) and atrazine (4.2 and 5.3 %). To sum up, compound 4d showed excellent inhibitory effects on P. oleracea and E. crusgalli. As shown in Table 4, compounds 3e and 4d showed 100 % post-emergence herbicidal activity against P. oleracea, A. retroflexus, and A. theophrasti at 450 g a.i./ha, which were similar to atrazine but significantly superior to lawsone (lower than 10 %). Compound 4d exhibited over 100 %, 100 % and 90 %, 70 % inhibitory effect on A. retroflexus and A. theophrasti at a dose of 300 and 150 g a.i./ha, respectively. It indicated that compound 4d can be used as pre- and post-emergence candidate herbicide.
Symptoms of Echinochloa crusgalli treated by compound 4d at 100 μg/mL, compared with the positive control (Atrazine), lead compound (Lawsone) and control check (CK).
compound
dosage
(g a.i./ha)inhibition rateb(%)
E.c.
D.s.
S.v.
P.o.
A.r.
A.t.
1a
450
G
G
G
E
D
F
1b
450
G
G
G
E
E
F
1c
450
G
G
G
F
F
E
1d
450
G
G
G
F
F
F
1e
450
G
G
G
F
G
F
2a
450
G
G
G
G
F
G
2b
450
G
G
G
F
G
F
2c
450
G
G
G
E
F
F
2d
450
G
G
G
G
G
F
2e
450
G
G
G
F
F
F
3a
450
G
G
G
G
G
G
3b
450
G
G
G
F
G
F
3c
450
G
G
G
G
G
F
3d
450
G
G
G
F
G
F
3e
450
F
F
F
A
A
A
300
F
F
G
B
B
A
4a
450
G
G
G
F
F
F
4b
450
G
G
G
G
G
G
4c
450
G
G
G
G
G
G
4d
450
F
F
F
A
A
A
300
F
F
F
B
A
A
150
G
G
G
D
B
C
4e
450
G
G
G
G
G
G
4f
450
G
G
G
G
G
G
4 g
450
G
G
G
E
G
F
4 h
450
G
G
G
G
G
G
4i
450
G
G
G
D
F
G
4j
450
G
G
G
G
G
F
4 k
450
G
G
G
G
G
F
Lawsone
450
G
G
G
G
G
G
Atrazine
450
F
F
F
A
A
A
300
G
G
G
A
A
A
150
G
G
G
D
D
C
3.5 Structure-activity relationship
In terms of herbicidal activity data, compounds 3e and 4d have good herbicidal activity against P. oleracea, A. retroflexus and A. theophrasti. The structure–activity relationships were analyzed according to the inhibitory activities of compounds on the growth stem of E. crusgalli at 100 μg/mL. When X is sulfonyl or acyl, electron-donating aryl favor for inhibitory effect, such as 2b (R=4-CH3-Phenyl, 51.3 %) > 2a (R=Phenyl, 46.1 %) > 2d (R=4-F-Phenyl, 41.7 %) > 2e (R=4-NO2-Phenyl, 30.9 %) and 3b (R=4-OCH3-Phenyl, 60.7 %) > 3a (R=Phenyl, 48.7 %) > 3c (R=4-F-Phenyl, 41.5 %). At the same time, the introduction of pyridyl and furyl is beneficial to the activity, and the introduction of thienyl is unfavorable to the activity, which is consistent with the orde 4c (R=pyridine-3-yl, 59.7 %) > 4a (R=furfuran-2-yl, 51.8 %) > 3a (R=Phenyl, 48.7 %) > 4b (R=thiophene-2-yl, 40.6 %). Among the compounds 4a–4d containing pyridinyl, the compound with 2-Cl-pyridine showed the better activity than others, such as 4d (R=2-Cl-pyridine-3-yl, 91.3 %) > 4 h (R=2-CF3-pyridine-3-yl, 55.3 %) ≈ 4 g (R=2-F-pyridine-3-yl, 53.6 %). It indicated that introducing the chlorine-containing pyridine groups is an effective strategy to discover compounds with high herbicidal activity.
3.6 Chlorophyll fluorescence measurements
Environmental stress including thermal damage, photo-inhibition and others factors can lead to the structural change of PS II pigment level and then affect the FO level (Krause and Weis, 1984). The FO and FM values were determined when all reaction centers of PS II are opened after 30 min of dark reaction and turned off by intense pulsed light illumination, respectively. As shown in Table 5, the ratio of FV/FM has no significant difference of compounds 3e, 4d, lawsone, and atrazine, which indicates that these compounds have no influence on the efficiency of light energy transfer between antenna pigment molecules or from these molecules to PS II reaction center. In addition, the qP value indicates the proportion of excitons captured by open traps and converted into chemical energy at the PS II reaction center, which depends on the presence of the oxidation state QA Chen et al., 2008; Krause and Weis, 1984) The qP values of compounds 3e (0.71), 4d (0.66) and atrazine (0.43) were significantly lower than CK (0.95), which indicated that they affected the efficiency of the entire photochemical process and the functional state of PS II. The present works demonstrated that the lawsone derivative 4d can be considered as a promising PS II inhibitor. aFluorescence parameters in table were measured using MINI-PAM-Ⅱ after the leaves coated with drugs in the presence of 3e, 4d, Lawsone and Atrazine. Each data is the average of 5 ∼ 8 repetitions ± standard error (SE). Different small letters (a–c) indicate values significantly different within treatments (p < 0.05).
parameter
control
3e
4d
Lawsone
Atrazine
FO
0.04 ± 0.03a
0.03 ± 0.01a
0.03 ± 0.05a
0.03 ± 0.03 a
0.04 ± 0.01 a
FM
0.22 ± 0.01 a
0.18 ± 0.02 a
0.14 ± 0.02 a
0.15 ± 0.03 a
0.21 ± 0.04 a
FV/FM
0.82 ± 0.06b
0.80 ± 0.01b
0.79 ± 0.03b
0.80 ± 0.05b
0.81 ± 0.03b
qP
0.95 ± 0.03c
0.71 ± 0.03c
0.66 ± 0.01c
0.81 ± 0.01c
0.43 ± 0.01c
3.7 Molecular docking analysis
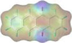
The molecular docking was performed and shown in Fig. 8. It can be seen from Fig. 8A, atrazine binds to QB binding site in D1 protein, and N of aromatic ring forms hydrogen bond with His215 (3.4 Å). Besides, aromatic rings form π-Alkyl hydrophobic interactions with Leu218 (4.5 Å) and Leu271 (4.5 Å). Lawsone can bind to the D1 protein (Fig. 8B), the two carbonyl groups form hydrogen bonds with His215 (3.0 Å) and His252 (3.0 Å) and the aromatic rings form one π-π and two π-Alkyl hydrophobic interactions with Phe255 (4.3 Å) and Leu271 (4.6 Å of both), respectively. As well as lawsone, the D1 protein was intensively bound with compound 4d (Fig. 8C), which forms two hydrogen bonds His215 (3.3 Å) and His252 (3.3 Å) with two carbonyl groups, respectively. Meanwhile, the aromatic rings of 4d form one π-π and three π-Alkyl hydrophobic interactions with Phe255 (4.5 Å), Leu 218 (5.1 Å), and Leu271 (4.6 and 4.8 Å), respectively. In addition, the pyridine ring of 4d forms a new π-π interaction on Phe211 (4.4 Å) with D1 protein.
Binding models of the commercial herbicide Atrazine (A), Lawsone (B) and compound 4d (C) with PsD1 (PDB: 5XNL). The purple line indicates hydrophobic interactions between compounds and surrounding residues. The red line indicates hydrophilic hydrogen-bond interactions.
3.8 Molecular dynamic simulation
During the simulation of 60,000 picoseconds, the RMSD mode of protein and ligand was shown in Fig. 9. Obviously, at the beginning, D1 protein has some fluctuations of 0.8 Å displacement. The RMSDs fluctuates about 0.2 Å–1 Å around a certain thermal average during the whole simulation time, which accurately explains the equilibrium simulation. Lawsone maintains roughly the same RMSD levels as compound 4d, while atrazine had slightly higher mean levels than compound 4d. In general, the fluctuation of protein and ligand remains unchanged after 5000 picoseconds.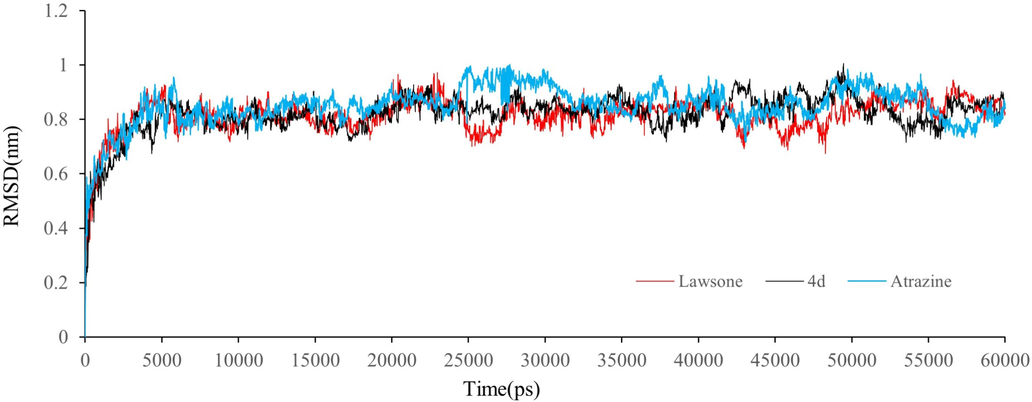
RMSD trajectories of D1-ligand complex during 60000 ps simulations.
4 Conclusion
In this paper, lawsone as lead compound was found and confirmed with virtual screening and herbicidal activity evaluation. Then, some hydrophobic substructures were introduced into lawsone to obtain a series of natural-based naphthoquinone derivatives. Compound 4d showed the best pre-emergence herbicidal activities at 100 and 10 μg/mL against P. oleracea. Meanwhile, it has 100 % post-emergence herbicidal activities to A. retroflexus and A. theophrasti at the dosage of 300 g a.i./ha, which was equivalent to the commercial herbicide atrazine but significantly better than lawsone. The preliminary mechanisms indicated lawsone and 4d can strongly bind the D1 protein and be considered as PS Ⅱ D1 inhibitors. This work provides useful guidance for rapid and rational design of PS Ⅱ inhibitors, and natural-based naphthoquinones can be used as a potential lead structure for the development of new PS Ⅱ inhibitors.
CRediT authorship contribution statement
Qingqing Wang: Writing – original draft, Investigation, Data curation. Wei Zhang: Investigation, Data curation. Fangping Zhong: Investigation, Data curation. Wenchao Yang: Writing – review & editing, Conceptualization. Xiuhai Gan: Writing – review & editing, Resources, Funding acquisition, Conceptualization.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2021YFD1700102), the Guizhou Provincial Fundation for Excellent Scholars Program (GCC [2023]069), and the Central Government Guides Local Science and Technology Development Fund Projects (Qiankehezhongyindi [2023] 001).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Molecular dynamics: survey of methods for simulating the activity of proteins. Chem. Rev.. 2006;106:1589-1615.
- [Google Scholar]
- Herbicide-related signaling in plants reveals novel insights for herbicide use strategies, environmental risk assessment and global change assessment challenges. Sci. Total. Environ.. 2016;1:1618-1628.
- [Google Scholar]
- BRUSELAS: HPC generic and customizable software architecture for 3D ligand-based virtual screening of large molecular databases. J. Chem. Inf. Model.. 2019;59:2805-2817.
- [Google Scholar]
- Binding properties of photosynthetic herbicides with the QB site of the D1 protein in plant photosystem II: a combined functional and molecular docking study. Plants. 2021;10:1008-1501.
- [Google Scholar]
- Oxidation of the plastoquinone pool in chloroplast thylakoid membranes by superoxide anion radicals. FEBS Lett.. 2018;592:3221-3228.
- [Google Scholar]
- Kresoxim-methyl derivatives: synthesis and herbicidal activities of (pyridinylphenoxymethylene) phenyl methoxyiminoacetates. J. Agric. Food Chem.. 2017;65:6114-6121.
- [Google Scholar]
- Recent advances in tenuazonic acid as a potential herbicide. Pest. Biochem. Physiol.. 2017;143:252-257.
- [Google Scholar]
- Action of tenuazonic acid, a natural phytotoxin, on photosystem II of spinach. Environ. Exp. Bot.. 2008;62:279-289.
- [Google Scholar]
- Key role for a glutathione transferase in multiple-herbicide resistance in grass weeds. Proc. Natl. Acad. Sci. USA. 2013;110:5812-5817.
- [Google Scholar]
- Rationale for a natural products approach to herbicide discovery. Pest Manag. Sci.. 2012;68:519-528.
- [Google Scholar]
- Based on the virtual screening of multiple pharmacophores, docking and molecular dynamics simulation approaches toward the discovery of novel HPPD inhibitors. Int. J. Mol. Sci.. 2020;21:5546.
- [Google Scholar]
- Interactions of herbicides with photosynthetic electron transport. Weed Sci.. 1991;39:458-464.
- [Google Scholar]
- Show moreChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res.. 2012;40:1100-1107.
- [Google Scholar]
- On the mode of action of the herbicides cinmethylin and 5-benzyloxymethyl-1,2-isoxazolines: putative inhibitors of plant tyrosine aminotransferase. Pest Manag. Sci.. 2012;68:482-492.
- [Google Scholar]
- Action mode of the mycotoxin patulin as a novel natural photosystem II inhibitor. J. Agric. Food Chem.. 2021;69:7313-7323.
- [Google Scholar]
- Structure-based virtual screening of natural products and optimization for the designand synthesisof novel CeCht1 inhibitorsas nematicide candidates. J. Agric. Food Chem.. 2023;71:244-254.
- [Google Scholar]
- KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res.. 2016;44:457-462.
- [Google Scholar]
- Chlorophyll fluorescence as a tool in plant physiology II interpretation of fluorescence signals. Photosynth. Res.. 1984;5:139-157.
- [Google Scholar]
- The 32000 dalton QB protein of photosystem II. Photochem. Photobiol.. 1985;41:107-116.
- [Google Scholar]
- Current challenges and trends in the discovery of agrochemicals. Science. 2013;341:742-746.
- [Google Scholar]
- Crystal structure of 4-hydroxyphenylpyruvate dioxygenase in complex with substrate reveals a new starting point for herbicide discovery. Research. 2019;2602414
- [Google Scholar]
- Natural products: a strategic lead generation approach in crop protection discovery. Pest Manag. Sci.. 2019;75:2301-2309.
- [Google Scholar]
- Residues of atrazine, simazine, linuron and diuron after repeated annual applications in a peach orchard. Weed Res.. 1975;15:373-379.
- [Google Scholar]
- Herbicidal activity and site of action of the natural product sorgoleone. Pestic. Biochem. Phys.. 1996;54:73-83.
- [Google Scholar]
- Herbicide resistance and supersensitivity in photosystem II. Cell. Mol. Life Sci.. 1999;55:1255-1277.
- [Google Scholar]
- Exploring african medicinal plants for potential anti-diabetic compounds with the DIA-DB inverse virtual screening web server. Molecules. 2019;24:2002.
- [Google Scholar]
- Evaluation of antidesmone alkaloid as a photosynthesis inhibitor. Pest. Biochem. Physiol.. 2016;134:55-62.
- [Google Scholar]
- Turnover of D1 protein encoded by psbA gene in higher plants and cyanobacteria photosynthetic efficiency to maintain plant productivity under photoinhibitory irradiance. Photosynthetica. 2000;38:161-169.
- [Google Scholar]
- Virtual screening web servers: designing chemical probes and drug candidates in the cyberspace. Brief. Bioinforma.. 2021;22:1790-1818.
- [Google Scholar]
- Impact of natural products on discovery of, and innovation in, crop protection compounds. Pest Manag. Sci.. 2022;78:399-408.
- [Google Scholar]
- Design, synthesis, and herbicidal activity of N-benzyl-5-cyclopropyl-isoxazole-4-carboxamides. J. Agric. Food Chem.. 2020;68:15107-15114.
- [Google Scholar]
- Target and nontarget resistance mechanisms induce annual bluegrass (Poa annua) resistance to atrazine, amicarbazone, and diuron. Weed Technol.. 2016;30:773-782.
- [Google Scholar]
- Mode-of-action of the natural herbicide radulanin A as an inhibitor of photosystem II. Pest Manag. Sci.. 2024;80:156-165.
- [Google Scholar]
- Synthesis, crystal structure and herbicidal activity of mimics of intermediates of the KARI reaction. Pest Manag. Sci.. 2005;61:407-412.
- [Google Scholar]
- Structure-based ligand design and discovery of novel tenuazonic acid derivatives with high herbicidal activity. J. Adv. Res.. 2022;40:29-44.
- [Google Scholar]
- Drugbank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res.. 2006;34:668-672.
- [Google Scholar]
- Action of the fungal compound citrinin, a bioherbicide candidate, on photosystem II. Pest Manag. Sci.. 2023;79:2301-2309.
- [Google Scholar]
- LARMD: integration of bioinformatic resources to profile ligand-driven protein dynamics with a case on the activation of estrogen receptor. Brief. Bioinform.. 2020;21:2206-2218.
- [Google Scholar]
- Enriching screening libraries with bioactive fragment space. Bioorg. Med. Chem. Lett.. 2016;26:3594-3597.
- [Google Scholar]
- Discovery of (5-(benzylthio)-4-(3-(trifluoromethyl) phenyl) -4H-1,2,4-triazol-3-yl) methanols as potent phytoene desaturase inhibitors through virtual screening and structure optimization. J. Agric. Food Chem.. 2022;70:10144-10157.
- [Google Scholar]
- Design, synthesis and bioactivity of novel 2–(arylformyl)cyclohexane-1,3-dione derivatives as HPPD inhibitors. J. Agric. Food Chem.. 2023;71:17678-17688.
- [Google Scholar]
- C-P Natural products as next-generation herbicides: chemistry and biology of glufosinate. J. Agric. Food Chem.. 2020;68:3344-3353.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2024.105985.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary Data 1
Supplementary Data 1







