Translate this page into:
Synthesis, evaluation, and computational chemistry of novel selenenyl sulfides as 3C protease inhibitors with strong cell-based antiviral activity
⁎Corresponding authors at: State Key Laboratory of Mycology, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China (H.Q. Dai). daihq@im.ac.cn (Huan-Qin Dai), nkwjg@nankai.edu.cn (Jian-Guo Wang)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Abstract
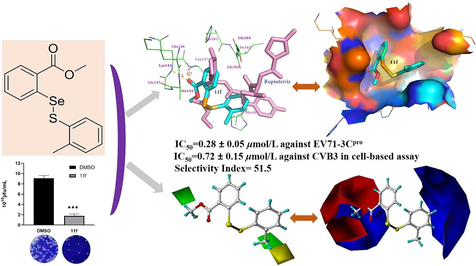
The 3C protease (3Cpro) is an important enzyme for developing therapeutic medicines against some severe viral infections. In order to discover new classes of antiviral agents, 42 novel selenenyl sulfides were prepared and characterized by 1H NMR, 13C NMR, 77Se NMR and HRMS. The target compounds demonstrated promising inhibitions against 3Cpro from either enterovirus 71 (EV71) or coxsackievirus B3 (CVB3), as well as desirable cell-based antiviral activity against these viruses. Among them, 11f displayed the most potent IC50 value of 0.28 ± 0.05 μM against EV71-3Cpro, and the strongest IC50 value of 0.72 ± 0.15 μM against CVB3, with a selectivity index of 51.5 against Hela cell, superior to the control drug rupintrivir. In addition, molecular docking was undertaken to predict a possible binding mode for compound 11f, and a comparative molecular field analysis (CoMFA) model was subsequently generated to understand the structure-activity relationships. Overall, the present research has provided some new insights to design a distinct family of antiviral compounds.
Keywords
3C protease
Selenenyl sulfides
Antiviral activity
Selectivity index
Computational chemistry
1 Introduction
Diseases caused by some viral infections have been threatening the health of human being seriously, such as severe acute respiratory syndrome (SARS), coronavirus disease 2019 (COVID-19), and enterovirus infections (Krilov, 2004; Li et al., 2021; Nayak et al., 2022). Enterovirus 71 (EV71) is the major pathogen responsible for hand, foot, and mouth disease (HFMD) (Salerno et al., 2023), while CVB3 is associated with aseptic meningitis, hepatitis, meningoencephalitis and chronic dilated cardiomyopathy (Zhu et al., 2023). Although the scales of EV71 and CVB3 outbreaks are not to the level of the COVID-19 pandemic, special attention should be also paid to them in that the existence of these viruses is long-standing and such infections are fatal to some patients. Even if different vaccines have been developed to prevent the prevalence of EV71 or CVB3 related infectious diseases, identification of novel chemical agents is still of vital importance since individuals are often unluckily infected by these picornaviruses.
Among the biological targets involved in the biological reactions of the picornavirus replication, a highly structurally conserved cysteine protease, namely 3C protease (3Cpro), does not exist in human bodies, and this feature has made it an popular target to design novel antiviral compounds (Ramajayam et al., 2011; Stubbing et al., 2023; Zeng et al., 2020). Some efforts were also made to develop covalent inhibitory antibody against human rhinovirus 14 (HRV14) 3C protease through unnatural amino acid mutagenesis in a recent study (Cheng et al., 2021). A similar enzyme from coronaviruses or caliciviruses, such as SARS-CoV or SARS-CoV-2, is called 3C-like protease (3CLpro). Continuous efforts have been devoted to identifying new agents that inhibit the proteases, especially in the battle against the COVID-19 pandemic (Han et al., 2022; Luz et al., 2023; Mohammad et al., 2020; Tan et al., 2023; Wang et al., 2022; Yoosefian et al., 2023; Zhang et al., 2020). Traditional inhibitors that target 3Cpro or 3CLpro include peptidomimetic compounds and natural products, such as rupintrivir (AG7088), nirmatrelvir (PF-07321332), baicalein, and DC07090 (Cannalire et al., 2022; Chen et al., 2023; Zang et al., 2022).
Some synthesized aromatic disulfides were identified as novel inhibitors (Example 1) of SARS-CoV-3CLpro but no covalent bond was observed to form with the enzyme (Wang et al., 2017). A clinical drug disulfiram was reported to exhibit some antiviral activity against COVID-19 (Jin et al., 2020). Selenium is an essential micronutrient for human health and insufficient intake of this element from daily diet will result in many disease (Sanmartin et al., 2011). For this reason, organoselenium compounds have attracted much attention and these compounds displayed versatile biological activities (Chipoline et al., 2022; Singh and Kunwar, 2021). As a representative selenium-containing organic compound, ebselen has been famous for its anti-inflammatory, antibacterial, and antioxidant activities (Maślanka and Mucha, 2023; Noguchi, 2016; Parnham and Sies, 2000). In the early stage of the COVID-19 outbreak, ebselen was identified as a potent inhibitor of SARS-CoV-2-3CLpro and it showed promising antiviral activity against SARS-CoV-2 in cell-based assay (Jin et al., 2020). Previously, some selenenyl sulfide compounds were synthesized and a few of them (Example 2) exhibited desirable activities against tobacco mosaic virus (TMV), showing a great potential for the development of new agrochemicals (Shang et al., 2023). Fig. 1 illustrates the chemical structures of all the examples mentioned above.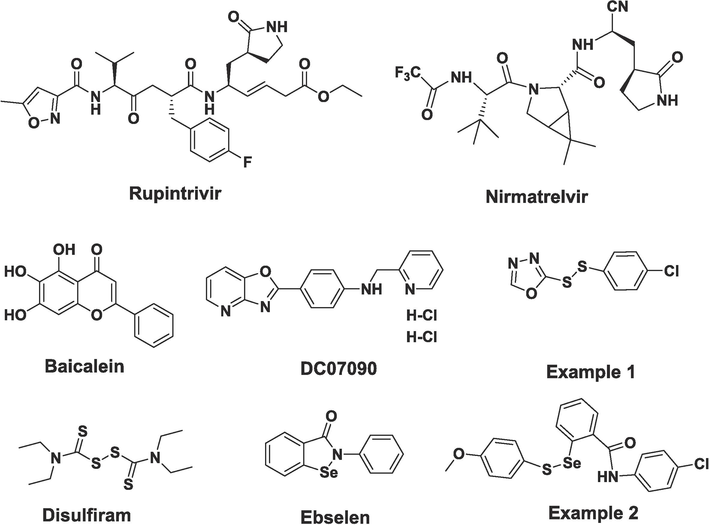
Molecular structures of Rupintrivir, Nirmatrelvir, Baicalein, DC07090, Examples 1&2, Disulfiram and Ebselen.
Given the advances in the biological activities of disulfides or selenenyl sulfide compounds that have been mentioned above and the fact that very limited drugs are clinically available for the treatment of EV71 or CVB3 infections and, it is worth exploring novel compounds containing selenenyl sulfide frameworks to identify different agents to combat these risky diseases. Bearing these in mind, synthesis of new selenenyl sulfides and evaluation of their biological activities against 3Cpro, together with the cell-based antiviral activity, is expected to be a preferable solution to offer some new chemical family of inhibitors that are effective upon EV71 or CVB3. This idea has been realized in this paper and 11f was identified as a stronger inhibitor than the control drug rupintrivir, as well as improved selectivity index. Since it is very challenging for the innovation of drugs that target 3Cpro, the current research will provide some insight for lead discovery against these viral infectious diseases for further consideration.
2 Results and discussion
2.1 Chemistry of the compounds
Aiming at investigating the influence of aliphatic groups (R1) and aromatic groups (R3) in the compounds, two different series, 7a-7u and 11a-11u were synthesized. The chemical routes are shown in Scheme 1 and Scheme 2, respectively. Although both compounds 7a-7u and 11a-11u were prepared via nucleophilic substitutions, corresponding reactions for these two series were different. Generally, 7a-7u were synthesized using a similar method to our previous work (shang et al., 2023), and a modification was that all intermediates 5 were synthesized from 4 in the presence of selenium powder, nano-Copper(I) iodide, and other related reagents. In comparison, compounds with close structure to 11a-11u have not been reported and the chemistry for the preparation of these compounds will be discussed. The starting material 8 (methyl 2-aminobenzoate or ethyl 2-aminobenzoate) was made into a diazonium salt 9 at iced temperature, after that potassium selenocyanate was added to the reactant in an acidic solution to give intermediate 10 (methyl 2-selenocyanatobenzoate or ethyl 2-selenocyanatobenzoate) in high yields, based on a known procedure (Diesendruck et al., 2009). Finally, 11a-11u were prepared by reacting 10 with different aromatic thiols 6 using aluminum oxide as a catalyst in dichloromethane at room temperature. The yields for 7a-7u were in the range of 7–85 % and those for 11a-11u were from 14 % to 67 %, respectively. Besides 1H NMR and 13C NMR, all the selenenyl sulfides were also characterized by 77Se NMR that is unique for the organoselenium compounds. The spectra of 1H NMR, 13C NMR, 77Se NMR, and HRMS for all the target compounds could be found in the supplementary data. Interestingly, for compounds from both 7 and 11 series, there existed a trend for the isomer compounds with only ortho or para position difference when R2 group was a substituted phenyl ring, that the ortho substituted compound had a smaller 77Se NMR chemical shift than the corresponding para substituted compound. Such phenomena could be observed from the following pairs of selenenyl sulfides: 7f (597.0) versus 7 g (566.7), 7i (597.8) versus 7 m (573.1), 7o (594.9) versus 7p (571.2), 11a (602.9) versus 11f (567.1), 11b (615.7) versus 11 g (594.1), 11c (597.0) versus 11 h (561.0), 11d (594.3) versus 11i (561.8), 11n (597.3) versus 11r (558.7), and 11o (594.2) versus 11 s (562.0), which was in agreement with the results in a previous study (Shang et al., 2023).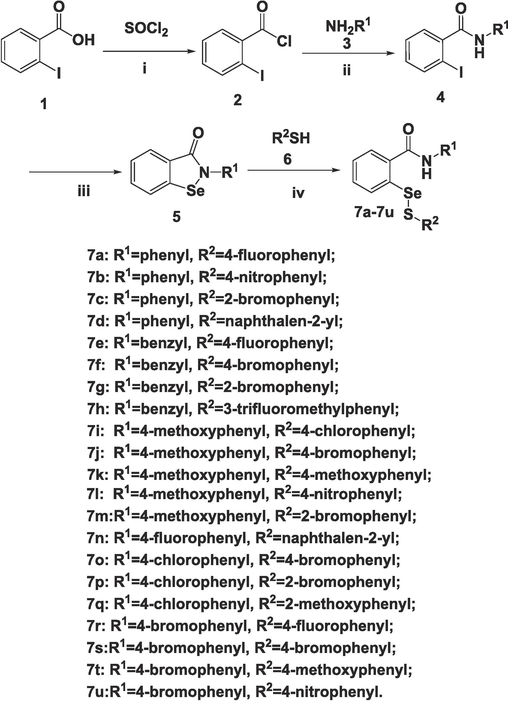
Reagents and conditions: (i) Reflux; (ii) Et3N, DCM, 0 °C to r.t.; (iii) Se, t-BuOK, nano-CuI, 1,10-phenanthroline, DMF, 110 °C; (iv) DCM, r.t.
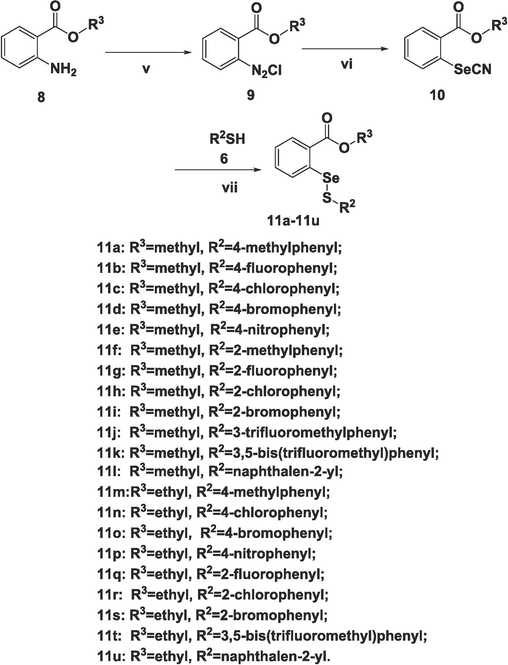
Reagents and conditions: (v) a. HCl, NaNO2, 0 °C; b. CH3COONa, 0 °C to r.t.; (vi) KSeCN, H2O, r.t.; (vii) Al2O3, DCM, r.t.
In order to confirm the chemical structures in the series of 11a-11u, single crystals of 11 s (CCDC number 2172212) suitable for X-ray diffraction were successfully obtained using a self-evaporation method in a mixed solution of hexane/dichloromethane(v/v = 100:1), after attempts on a few compounds. From Fig. 2, it can be seen that the molecule adopted an extended conformation in the crystal, from which a Se-S bond was successfully constructed between two phenyl rings and the ester part was linked with the left phenyl ring neighboring to the selenium atom (Se1 in the figure).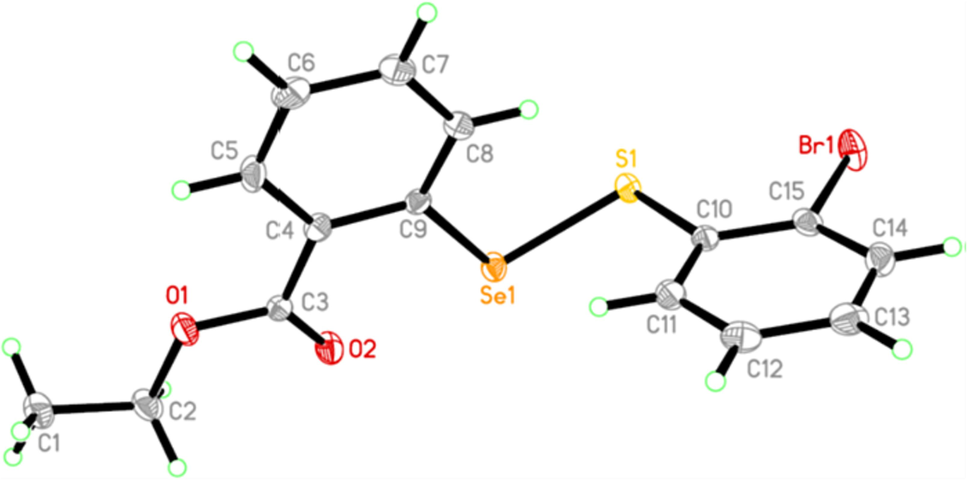
Crystal structure of compound 11 s.
Simply from the view of chemistry, although limited compounds in the two series of selenenyl sulfides were synthesized in this study, more compounds with different structures could be considered for investigation. For example, the introduction of iodine atom to the structures might be an interesting topic, as previously shown as halide system (Lun et al., 2023).
2.2 Biological activity
The target selenenyl sulfides were all subjected to enzyme inhibitions against EV71-3Cpro and CVB3-3Cpro, and in vitro cell-based evaluations against these two viruses, the results of which have been summarized in Table 1. Generally, most compounds exhibited potent inhibitions against 3Cpro and ideal in vitro antiviral activities. In order to filter those with very weak inhibitions, the cutoff of IC50 value was set to 10 μM for 3Cpro and that was 30 μM for the cell-based activity. For comparison, rupintrivir was chosen as a control in this study. *Rup = rupintrivir.
Compound
IC50 (μM) for enzyme inhibition
IC50 (μM) for cell-based activity
EV71-3Cpro
CVB3-3Cpro
EV71
CVB3
7a
9.34 ± 0.56
>10
>30
21.2 ± 1.4
7b
2.16 ± 0.42
3.33 ± 0.24
13.4 ± 1.3
10.6 ± 1.1
7c
6.21 ± 0.21
9.64 ± 0.45
25.3 ± 1.1
14.6 ± 1.9
7d
8.79 ± 0.18
9.26 ± 0.53
12.6 ± 0.1
9.27 ± 1.97
7e
>10
>10
18.9 ± 2.1
26.4 ± 2.1
7f
8.72 ± 1.01
9.64 ± 0.27
22.4 ± 0.13
17.8 ± 1.5
7 g
9.85 ± 0.11
9.49 ± 0.31
>30
25.4 ± 0.41
7 h
>10
>10
>30
>30
7i
3.28 ± 0.47
9.64 ± 0.12
>30
>30
7j
>10
3.26 ± 0.09
16.7 ± 1.3
14.1 ± 1.7
7 k
>10
7.69 ± 0.26
24.5 ± 2.1
23.6 ± 1.01
7 l
1.78 ± 0.09
3.56 ± 0.21
9.24 ± 0.93
6.58 ± 0.02
7 m
>10
9.46 ± 0.12
20.6 ± 2.5
19.1 ± 2.06
7n
>10
>10
16.8 ± 1.6
15.6 ± 0.0
7o
4.31 ± 0.15
3.23 ± 0.03
25.8 ± 2.0
14.6 ± 0.1
7p
2.52 ± 0.20
8.46 ± 0.24
26.8 ± 0.9
14.6 ± 0.2
7q
3.21 ± 0.01
7.62 ± 0.21
9.82 ± 0.7
6.31 ± 0.11
7r
3.28 ± 0.73
2.56 ± 0.09
12.8 ± 0.4
5.42 ± 0.08
7 s
6.54 ± 0.37
>10
7.42 ± 1.51
6.21 ± 0.13
7 t
2.21 ± 0.06
>10
8.71 ± 0.18
9.62 ± 1.2
7u
>10
>10
20.8 ± 1.8
18.9 ± 2.1
11a
5.18 ± 0.01
2.54 ± 0.01
10.4 ± 0.4
12.4 ± 0.1
11b
5.32 ± 0.22
9.64 ± 0.29
25.7 ± 3.3
11.2 ± 0.2
11c
>10
5.65 ± 0.01
15.7 ± 2.4
24.4 ± 0.2
11d
>10
8.98 ± 0.99
23.9 ± 2.5
13.2 ± 0.2
11e
9.57 ± 0.12
3.01 ± 0.26
5.61 ± 0.21
1.15 ± 0.04
11f
0.28 ± 0.05
1.67 ± 0.31
2.48 ± 0.04
0.72 ± 0.15
11 g
2.31 ± 0.11
1.02 ± 0.12
7.28 ± 0.44
5.23 ± 0.11
11 h
>10
2.11 ± 0.05
19.4 ± 0.4
16.4 ± 0.6
11i
>10
9.52 ± 0.01
25.1 ± 0.11
13.2 ± 0.3
11j
1.24 ± 0.01
7.99 ± 0.22
>30
>30
11 k
7.59 ± 0.24
>10
>30
7.21 ± 1.26
11 l
>10
>10
>30
>30
11 m
8.21 ± 0.23
3.78 ± 0.05
11.1 ± 1.2
10.4 ± 1.2
11n
>10
9.64 ± 1.76
>30
>30
11o
8.91 ± 0.09
3.46 ± 0.57
>30
25.8 ± 3.01
11p
>10
6.63 ± 0.45
>30
17.9 ± 0.09
11q
3.89 ± 0.04
2.97 ± 0.07
9.59 ± 0.63
14.6 ± 0.8
11r
>10
2.64 ± 0.07
16.0 ± 0.1
22.1 ± 0.2
11 s
>10
7.98 ± 0.13
29.9 ± 9.5
>30
11 t
9.78 ± 0.11
9.65 ± 0.34
>30
18.2 ± 0.5
11u
2.46 ± 0.78
2.34 ± 0.45
2.35 ± 0.54
5.78 ± 0.11
Rup*
1.03 ± 0.53
2.01 ± 0.22
0.74 ± 0.22
6.52 ± 0.09
Amongst the compounds, 7b, 7c, 7d, 7f, 7 g, 7i, 7 l, 7o, 7p, 7q, 7r, 11a, 11b, 11e, 11f, 11 g, 11j, 11 m, 11o, 11q, 11 t, and 11u had IC50 values < 10 μM for both proteases; 7a, 7 s, 7 t, and 11 k had IC50 values < 10 μM for EV71-3Cpro; and 7j, 7k, 7m, 11c, 11d, 11h, 11i, 11n, 11p, 11r, and 11 s had IC50 values < 10 μM for CVB3-3Cpro, respectively. While for 7e, 7h, 7n, 7u and 11 l, both the IC50 values against EV71-3Cpro and CVB3-3Cpro were > 10 μM. For the EV71-3Cpro inhibitions, 7b, 7l, 7 t, 11g, 11j, and 11u and 11f had IC50 values < 2.5 μM, among which 11f had the best IC50 value of 0.28 ± 0.05 μM, much stronger than that of rupintrivir (1.03 ± 0.53 μM). For the CVB3-3Cpro inhibitions, 11f, 11g, 11h, and 11u had IC50 values < 2.50 μM, comparable to that of rupintrivir (2.01 ± 0.22 μM). Fig. 3a and 3b. display the inhibition curves of 11f against EV71-3Cpro and CVB3-3Cpro. Regarding a simple structure–activity relationship, when R2 was a 4-nitrophenyl group for compound 7, two of the compounds displayed the best inhibitions against EV71-3Cpro (7b and 7 l). However, this group did not give strong enzyme inhibitions for 7u, as well as the corresponding compounds for 11 (11e and 11p). For compound 11, when R2 was a 2-methylphenyl group, 2-fluorophenyl group or 3-trifluoromethylphenyl group, potent enzyme inhibitions were observed (11f, 11 g and 11j). However, 3-trifluoromethylphenyl group in 7 h showed very poor enzyme inhibition. When R2 was a naphthalene-2-yl, 11u gave strong inhibitions for both 3Cpro,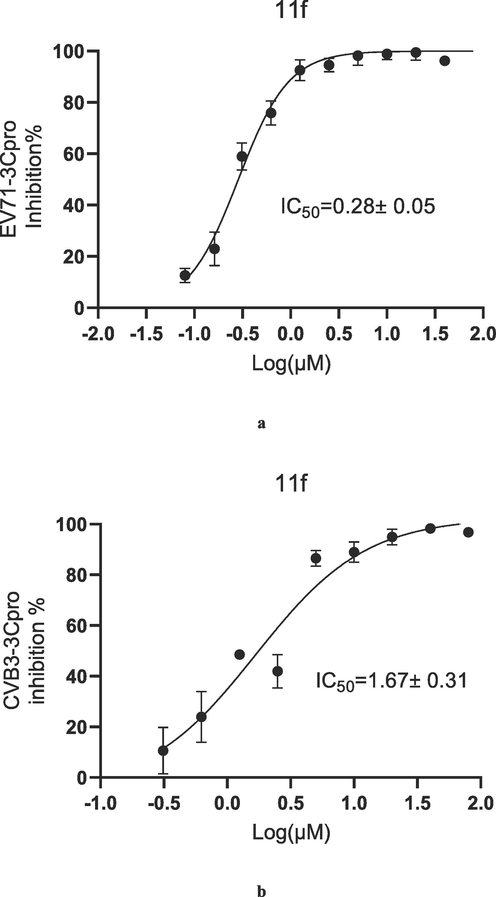
Inhibition curves of 11f against EV71-3Cpro (a) and CVB3-3Cpro (b).
nevertheless, 11 l exhibited no activity even at 10 μM.
It is a concern whether the target selenenyl sulfides inhibit the 3C protease through a covalent bonding manner. From the previous study, the aromatic disulfides did not form a covalent bond with the cysteine residue of SARC-CoV-3CLpro in the active site, which was evidenced by that fact that mass spectrometry of the protease had not been changed and the nonsymmetrical disulfides were reversible inhibitors (Wang et al., 2017). In another study, the ebselen-cysteine adduct was not found with Cys145 of SARS-CoV-2-3CLpro, using LC-MS/MS analysis of the tryptic digest of the enzyme, although such an adduct was observed in the ebselen-treated human glutathione S-transferase-pi (Amporndanai et al., 2021). In this study, inhibition type of the 11f was further determined by means of enzymatic kinetic behaviors, from which a competitive inhibition behavior was observed and it implied the inhibition was a reversible action (Fig S1). Due to all these reasons, it is quite likely that the selenenyl sulfides do not form covalent bond with the cysteine in the active site of the 3C proteases.
Based on the cell-based evaluation, none of 7 h, 7i, 11j, 11 l, and 11n had good activities against both EV71 and CVB3, 7a, 7 g, 11o, 11p, 11 k, 11 s and 11 t had one IC50 value > 30 μM against either EV71 or CVB3, and all the other selenenyl sulfides showed IC50 values of < 30 μM against the two viruses. For the cell-based activities against EV71, 11f and 11u displayed IC50 values of < 2.50 μM, which were the best ones among all the compounds but still weaker than that of rupintrivir (0.74 ± 0.22 μM). For the antiviral activities against CVB3, the IC50 values of 11e and 11f were 1.15 ± 0.04 μM and 0.72 ± 0.15 μM, respectively, which were several folds stronger the value of rupintrivir (6.52 ± 0.09 μM). Fig. 4a shows the dose–response curve of 11f against CVB3. Although the cell-based assay is a simple in vitro model, it is obvious that the properties of absorption, distribution, metabolism and excretion (ADME) should be taken for consideration for the cellular activity, since the cellular environment is much more complex than the pure enzyme inhibition. Therefore, it was not strange that there did not exist an absolute correction between the enzyme inhibition and the antiviral activity for the tested selenenyl sulfides. On the other hand, there exists such a possibility, that the antiviral activity of these selenenyl sulfides might involve targeting other viral proteins or host proteins in addition to 3Cpro. This is due to a fact that ebselen and some other compounds did not inhibit the viral replication of EV-A71, although those compounds inhibited cysteine proteases significantly (Ma et al., 2020).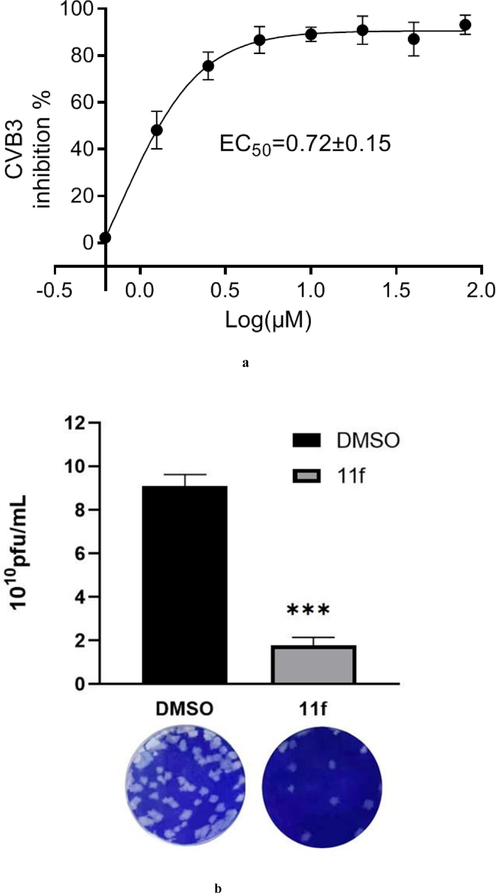
(a) Inhibition curve of 11f against CVB3. (b) Anti-CVB3 activity of 11f from viral plaque assay (the symbol *** means the significant difference p-value is no more than 0.001).
The antiviral activity was further supported by viral plaque assay, compound 11f showed very strong reduction of CVB3 particle formation activity at 5 µM, with 80 % inhibition as compared to DMSO controls (Fig. 4b). A subsequent cell cytotoxicity assay for 7q, 7r, 7 s, 11e, 11f, 11 g, 11u and rupintrivir against normal Hela cells revealed that selectivity indexes for these compounds were 6.6, 9.4, 8.2, 23.6, 51.5, 6.8, 7.0, and 7.6 for CVB3, respectively (Table 2). Taken together, 11f was identified as a potent antiviral agent targeting 3C/3CLpro with advantageous bio-safety. *N.D. = not determined (the IC50 for the anti-CVB3 data was > 30 μM).
Compound
Cell cytotoxicity data
(IC50, μM)Selectivity index
Compound
Cell cytotoxicity data
(IC50, μM)Selectivity index
7a
125.8 ± 10.4
5.9
11a
100.4 ± 13.8
8.1
7b
106.8 ± 12.5
10.0
11b
99.7 ± 11.0
8.9
7c
111.0 ± 8.8
7.6
11c
126.9 ± 18.7
5.2
7d
51.9 ± 7.3
5.6
11d
81.8 ± 9.9
6.2
7e
118.8 ± 11.4
4.5
11e
27.1 ± 3.2
23.6
7f
87.2 ± 7.8
4.9
11f
37.1 ± 2.5
51.5
7 g
81.3 ± 4.6
3.2
11 g
35.6 ± 6.2
6.8
7 h
N.D.*
N.D.
11 h
41.0 ± 5.1
2.5
7i
N.D.
N.D.
11i
99.4 ± 11.4
7.5
7j
173.4 ± 16.5
12.3
11j
N.D.
N.D.
7k
63.7 ± 4.8
2.7
11 k
35.3 ± 5.7
4.9
7l
15.1 ± 0.8
2.3
11 l
N.D.
N.D.
7m
108.9 ± 18.0
5.7
11 m
97.8 ± 13.3
9.4
7n
121.7 ± 17.3
7.8
11n
N.D.
N.D.
7o
110.1 ± 9.9
7.6
11o
38.7 ± 4.7
1.5
7p
156.2 ± 19.3
10.7
11p
35.8 ± 7.5
2.0
7q
41.6 ± 5.8
6.6
11q
99.3 ± 12.0
6.8
7r
50.8 ± 8.1
9.4
11r
99.5 ± 15.4
4.5
7 s
50.8 ± 6.1
8.2
11 s
N.D.
N.D.
7t
64.5 ± 7.5
6.7
11t
34.6 ± 4.0
1.9
7u
81.2 ± 6.7
4.3
11u
40.5 ± 6.3
7.0
Rup
49.6 ± 5.9
7.6
For a general comparison of the antiviral efficacies of between the two series 7a-7u and 11a-11u, compounds belonging to the second panel exhibited stronger cell-based antiviral activity. For the anti-CVB3 activity, the IC50 values of 11e and 11f were around 1 μM, however, none of the compounds from the first series was at the same level. This implied that more attention should be paid to compounds with aliphatic groups (R1) for the next round of molecular design to discover more potent compounds.
Regarding to whether the ester or the amide part of the selenenyl sulfides are stable in the cell-based assay, there is an assumption that the active component in the cellular environment is possible to be the hydrolyzed product. Nevertheless, this might not be true based on the following fact: 7b, 7 l, 7u, 11e and 11p had same R2 groups, however, the antiviral activities of them differed significantly. Otherwise, these compounds tended to demonstrate similar antiviral activity. Further experiments will be conducted to study the components after the cell-based assay for a detailed investigation.
In a previous result, we screened a big library of disulfide compounds against CVB3-3Cpro and evaluated their cell-based anti-CVB3 activities. Although some compounds showed submicromolar IC50 values against CVB3-3Cpro, the antiviral activities were very poor even at 10 μg/mL concentration (corresponding to approximately 30–40 μM based on different molecular weight of each compound), much weaker than that of rupintrivir at the same condition. That work was not published in a scientific paper, but shown in a patent application (Zhang et al., 2016). The present result suggested that although selenenyl sulfides are bioisosteres of the disulfide compounds, the former have more preferable biological behavior in the cell-based antiviral assay and have much greater potential for further development of novel antiviral agents.
Hundreds of papers have been published on the discovery of inhibitors of SARS-CoV-2 since the outbreak of COVID-19, however, there were very few reports on the identification of anti-CVB3 or anti-EV71 inhibitors. The preliminary result suggested that selenenyl sulfides in the present study were effective against CVB3 infection.
2.3 Computational chemistry
2.3.1 Proposed binding mode of active compound
Molecular docking has become a useful tool to predict the possible binding mode of inhibitor with biological target, when the co-crystal structure is unavailable (Bondock et al., 2023; Meng et al., 2022; Zhao et al., 2023;). To explore the plausible mode of action for this family of antiviral compounds, 11f was docked the active site of EV71-3Cpro (pdb code 3sjo) (Lu et al., 2011). This was because: (i) 11f was more potent than rupintrivir agaisnt EV71-3Cpro; (ii) Rupintrivir was in the binding pocket of this crystal structure; (iii) EV71-3Cpro and CVB3-3Cpro share fairly strong similarity. It should be noted that although this pdb file contained eight chains, these chains were identity in the sequence and the 3D structures superimposed one another perfectly. Therefore we choose chain A for the molecular simulation. Fig. 5a shows the interaction of 11f and the neighboring residues of EV71-3Cpro from the molecular simulation in comparison with rupintrivir in the crystal structure. As can be seen, 11f was much smaller than rupintrivir in size, and 11f overlaid with the left part of rupintrivin. The selenenyl sulfide inhibitor was involved in multiple hydrophobic contacts with His24, Phe25, His40, Lys143, Ala144, Gly145, His161, Ile162, Gly163, and Gly164, while Gln146 and Cys147 were found to form two hydrogen bonds with 11f. Fig. 5b depicts the three-dimensional molecular surface of the binding cavity generated by PyMOL and 11f adopted a relaxed conformation to inhabit in the cavity. Although fewer interactions were observed for 11f than for rupintrivir with the enzyme, much stronger inhibition of 11f against EV71 was determined experimentally. We will try the co-crystallization of this compound with EV71-3Cpro or CVB3-3Cpro to explain the exact mode of action in the future.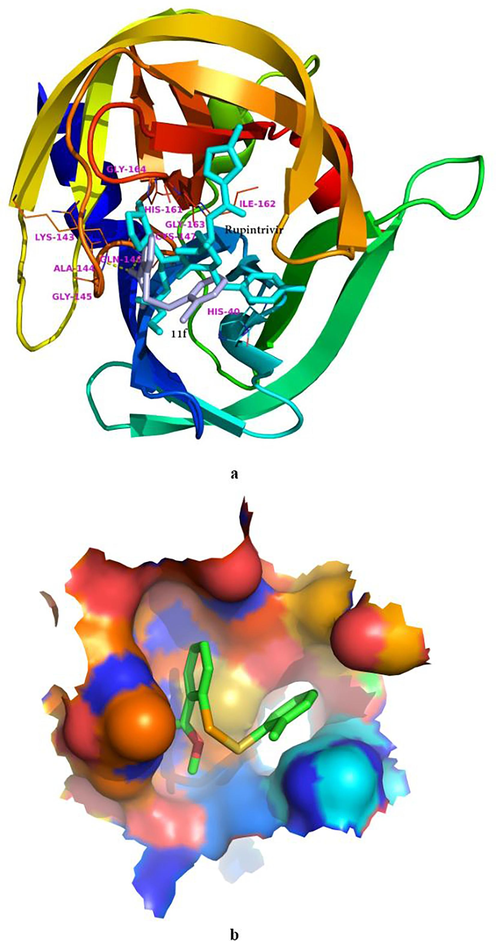
Predicted binding mode of 11f with EV71-3Cpro. (a) Comparison of 11f (lightblue) with rupintrivir (cyan), in which the full protein was shown in a cartoon model. (b) The molecular surface of the binding pocket (11f shown in stick model and colored by element).
2.3.2 Structure-activity relationship
Comparative field analysis (CoMFA) is powerful to analyze the quantitative structure–activity relationships (QSAR) of bioactive compounds by steric and electrostatic contributions using three-dimensional models (Cramer et al., 1988). The conformation of 11f from molecular docking was used to build all the molecular structures. For this study, the cell-based IC50 values against CVB3 were selected as the biological data to generate the CoMFA model because: (i) There was a bigger training set for building the model; (ii) CoMFA is not limited only to analyze the enzyme inhibitory data. Besides 7 h, 7i, 11j, 11 l, 11n and 11 s, the IC50 values of which were > 30 μM, compounds 7u, 11 t and 11u were also excluded from the training set because they were statistical outliers. The final training set gave a leave-one-out q2 of 0.519 when the number of optimum components was 6. The non-crossvalidated r2 was 0.931 with a standard error of estimate of 0.107 and F values of 18.118. This statistical result indicated that the CoMFA model was successful, even if the compounds belongs to two slightly different series. The steric contribution was 67.3 % and the electrostatic contribution was 32.7 %. The predicted biological data and experimental biological data from CoMFA were listed in Table 3. The most active compound 11f was used to explain the contour maps, as shown in Fig. 6. For the steric map (Fig. 6a), a bulky group in the green space is favorable for anti-CVB3 activity, and such a group in the yellow region is likely to decrease the antiviral activity. For the electrostatic map (Fig. 6b), an increase of the positive charge will give enhanced activity in the blue region, whereas in the red space, negatively charged group is likely to improve the cell-based inhibition. Inspired by the above computational chemistry, we will make further structural optimization to seek for compounds with improved activity. For example, it is practical in organic synthesis to introduce a halogen atom at 4-position of the phenyl ring that attached to the sulfur atom of 11f, and to change the ester moiety to ethyl or propyl group. The next round of molecular design is currently underway. *ED = experimental pIC50 data, PD = predicted pIC50 data, ER = error.
Compound
ED
PD
ER
Compound
ED
PD
ER
7a
4.77
4.58
0.19
7 t
5.02
4.97
0.05
7b
4.97
5.06
−0.09
11a
4.91
5.02
−0.11
7c
4.83
4.86
−0.03
11b
4.95
4.81
0.14
7d
5.03
5.00
0.03
11c
4.61
4.80
−0.19
7e
4.58
4.70
−0.12
11d
4.88
4.73
0.15
7f
4.75
4.65
0.10
11e
5.94
5.99
−0.05
7 g
4.60
4.58
0.02
11f
6.14
6.15
−0.01
7j
4.85
4.82
0.03
11 g
5.28
5.21
0.07
7 k
4.63
4.65
−0.02
11 h
4.78
4.83
−0.05
7 l
5.18
5.22
−0.04
11i
4.88
4.89
−0.01
7 m
4.72
4.75
−0.03
11 k
5.14
5.38
−0.24
7n
4.80
4.82
−0.02
11 m
4.98
4.90
0.08
7o
4.83
4.92
−0.09
11o
4.58
4.65
−0.07
7p
4.84
4.85
−0.01
11p
4.75
4.78
−0.03
7q
5.20
5.18
0.02
11q
4.84
4.93
−0.09
7r
5.27
5.19
0.08
11r
4.66
4.54
0.12
7 s
5.20
5.14
0.06
11 t
4.74
4.54
0.20
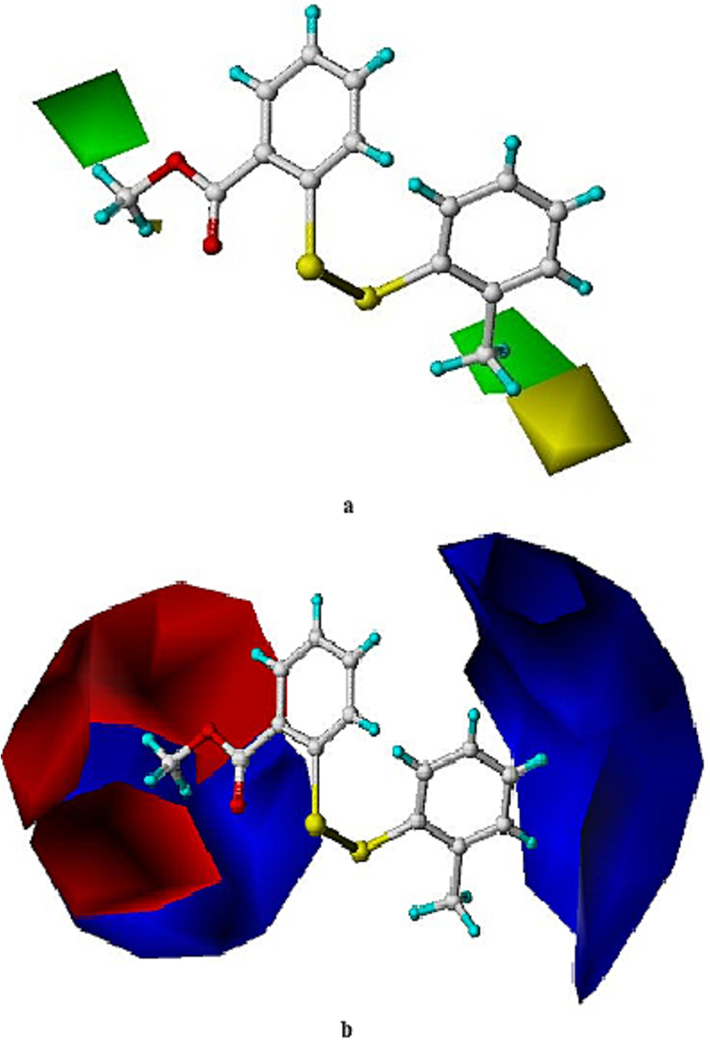
Three-dimensional contour maps of the anti-CVB3 activities from the CoMFA model for (a) steric field and (b) electrostatic field contributions.
3 Conclusion
In summary, two panels of 42 novel selenenyl sulfides were designed and synthesized via facile nucleophilic substitution reactions, and these compounds were fully characterized spectroscopically. These selenenyl sulfides were subjected to enzyme inhibitions EV71-3Cpro and CVB3-3Cpro, and 22 compounds among them displayed IC50 values below 10 μM against both proteases. The most potent one, being 11f, exhibited 0.28 ± 0.05 μM IC50 value against EV71-3Cpro, which was sevenfold more effective than that of rupintrivir. For the cell-based antiviral assay, 30 compounds showed IC50 values below 30 μM against both EV71 and CVB3. Compound 11f also offered the strongest antiviral activity against CVB3, and that was seven times stronger than rupintrivir. Further cytotoxicity data upon normal Hela cells indicated that 11f had a relatively high selectivity index of 51.5 against CVB3, and in contrast, rupintrivir only had a selectivity index of 7.6 at the same condition. These results indicated that 11f is a promising lead compound for further research on antiviral agents targeting 3Cpro. In addition, molecular docking was performed to predict a possible binding mode of 11f with EV71-3Cpro, and a CoMFA model was constructed to understand the structure–activity relationships of this class of antiviral compounds. The computational chemistry will provide meaningful guidance for further optimization of this family of inhibitors. For the next step, we will carry out gene knockout experiment to give a direct evidence that the antiviral activity was due to 3Cpro inhibition. Moreover, antiviral efficiency will also be evaluated based on an animal model to further investigate the behavior of this compound at a recognized laboratory. In conclusion, this study has brought some exciting results, which is meaningful for the discovery and innovation of novel antiviral inhibitors with different chemical skeletons to combat the risky diseases caused by picornavirus infections.
4 Experimental section
4.1 Chemistry
The reagents and solvents for the chemical preparation were all of analytical grade quality, purchasing from the online mall for laboratory use in Nankai University (mall.nankai.edu.cn/labmai/cloudbean). All the liquid reagents and organic solvents were dried in advance and distilled using standard methods before use. All the reactions were conducted using oven dried glassware. Melting points were determined using an RT-2 melting apparatus (Shanghai PuZhe photoelectric Co., China) and were uncorrected. 1H NMR, 13C NMR, and 77Se NMR spectra were recorded using a Bruker Avance 400 MHz spectrometer (Bruker Corporation, Switzerland). The chemical shift data (δ) for the nuclear magnetic resonance spectra were expressed as parts per million (ppm), using deuterated chloroform (CDCl3) or dimethyl sulfoxide (DMSO‑d6) as solvent and tetramethylsilane (TMS) as an internal standard reference. High-resolution mass spectra were recorded on an FT-ICR mass spectrometer (Ionspec, 7.0 T) or a Varian QFT-ESI instrument (Varian Medical Systems, Crawley, UK). Single crystal X-ray diffraction was carried out on a Bruker Smart 1000 CCD diffractometer (Bruker Corporation, Switzerland). General silica gel (200–300 mesh) was used for the column chromatography purification.
4.2 General procedure for synthesis of intermediates and target compounds 7a-7u
4.2.1 Synthesis of intermediate 2
To a 25 mL flask, 2-iodobenzoic acid (1) (1.24 g, 5 mmol, 1.0 equiv.) was dissolved in freshly distilled thionyl chloride (5 mL) and the reaction mixture was stirred at reflux valufor 2 h. After that the thionyl chloride was removed via distillation at 100 °C over the course of 1 h and the resulting solid was dried in vacuum to give 2-iodobenzoyl chloride (2) in 95 % yield (1.26 g) as a yellow solid without further purification.
4.2.2 Synthesis of intermediate 4
Phenyl group as R1 was taken for an example here. To a solution of aniline (3) (1.24 g, 5 mmol, 1.0 equiv.) and triethylamine (2.1 mL, 15.0 mmol, 3.0 equiv.) was added 2-iodobenzoyl chloride (2) (1.06 g, 4 mmol, 0.8 equiv.) in 20 mL dichloromethane dropwise at iced temperature. After 2 h stirring, the mixture was changed to room temperature overnight. When the reaction completed, water (50 mL) was added and the organic layer was separated. The mixture was then washed with 2 mol/L hydrochloric acid (50 mL × 3), saturated sodium bicarbonate (50 mL × 3), and water (50 mL × 3), respectively. Subsequently the organic phase was collected, dried over anhydrous magnesium sulfate, and concentrated under reduced pressure. In the end, the residue was purified by column chromatography (petroleum ether/ethyl acetate = 5/1) to give 2-iodo-N-phenylbenzamide (4) (1.12 g, 86.7 % yield) as a white solid. The detailed experimental procedures were similar when R1 was changed to other groups in this study.
4.2.3 Synthesis of intermediate 5
Phenyl group as R1 was taken for an example here. 1,10-phenanthroline (2.97 g, 16.5 mmol, 3.3 equiv.) and nano copper(I) iodide (2.85 g, 15 mmol, 3 equiv.) were added to N, N-dimethylmethanamide (60 mL) and the mixture was stirred for 15 min at room temperature. Subsequently, 2-iodo-N-phenylbenzamide (4) (4.85 g, 15 mmol, 3 equiv.), selenium powder (1.78 g, 22.5 mmol, 4.5 equiv.), and potassium t-butoxide (2.86 g, 25.5 mmol, 5.1 equiv.) were added to the reactant mixture. The reaction was then heated to 110 °C for 12 h under nitrogen atmosphere and after that water (100 mL) was added to quench the reaction. In the next step, the organic phase was extracted with ethyl acetate (100 mL × 3) and the combined ethyl acetate extracts were dried over anhydrous sodium sulfate, filtered, and concentrated under reduced pressure. The intermediate 2-phenylbenzo[d][1,2]selenazol-3(2H)-one (5) was purified by column chromatography (petroleum ether/ethyl acetate = 4/1) as a pale-orange solid (2.01 g, 48.7 % yield). The experimental procedures were similar when R1 was changed to other groups in this study.
All the related intermediates are known compounds and the data for characterization were not provided in this paper (Bhabak and Mugesh, 2007; Dakova et al., 1991; Zhou and Chen, 1999).
4.2.4 Synthesis of target compound 7a-7u
All the substituted thiophenols (2-naphthalenethiol for 7d and 7n) were purchased from commercial suppliers. Generally, 1.0 equiv. of 5 was added to 5 mL dichloromethane, after that 1.0 equiv. of different substituted thiophenol was added to the reactant. The mixture was stirred for 1 h at room temperature and monitored by thin-layer chromatography. The target compound was finally purified by column chromatography (petroleum ether/ethyl acetate = 10/1) in different yields.
4.2.4.1 2-(((4-fluorophenyl)thio)selanyl)-N-phenylbenzamide (7a)
Pale-yellow solid; Yield 42 %; m.p.: 120–121 °C; 1H NMR (400 MHz, DMSO‑d6) δ 10.60 (s, 1H, NH), 8.24 (d, J = 7.8 Hz, 1H, Ar-H), 8.16 (d, J = 8.0 Hz, 1H, Ar-H), 7.74 (d, J = 8.4 Hz, 2H, Ar-H), 7.66 (t, J = 7.6 Hz, 1H, Ar-H), 7.52 (dq, J = 18.0, 7.4, 6.4 Hz, 3H, Ar-H), 7.40 (t, J = 7.8 Hz, 2H, Ar-H), 7.17 (q, J = 8.2, 7.4 Hz, 3H, Ar-H); 13C NMR (101 MHz, DMSO‑d6) δ 166.2, 162.5, 160.1, 138.1, 135.7, 132.6, 131.7, 131.3 (d, J = 8.3 Hz), 131.0, 128.8, 127.1, 126.4, 124.5, 121.1, 116.2 (d, J = 22.2 Hz); 77Se NMR (76 MHz, DMSO‑d6) δ 615.1; HRMS (ESI) m/z calcd for C19H14FNOSSe [M + H]+ 404.0018; found 404.0015.
4.2.4.2 2-(((4-nitrophenyl)thio)selanyl)-N-phenylbenzamide (7b)
Yellow solid; Yield 26 %; m.p. 165–166 °C; 1H NMR (400 MHz, CDCl3) δ 8.19 (d, J = 8.9 Hz, 1H, Ar-H), 8.07 (d, J = 9.0 Hz, 2H, Ar-H), 8.02 (d, J = 7.7 Hz, 1H, Ar-H), 7.78 (d, J = 7.3 Hz, 1H, Ar-H), 7.63 (m, 5H, Ar-H), 7.50 (t, J = 7.6 Hz, 1H, Ar-H), 7.41 (q, J = 8.0 Hz, 3H, Ar-H), 7.23 (t, J = 7.4 Hz, 1H, Ar-H); 13C NMR (101 MHz, CDCl3) δ 166.0, 146.1, 136.9, 132.8, 130.9, 129.3, 128.4, 128.1, 126.7 (d, J = 3.4 Hz), 126.4, 125.5, 124.5, 123.9, 120.8; 77Se NMR (76 MHz, CDCl3) δ 575.3; HRMS (ESI) m/z calcd for C19H14N2O3SSe [M−H]- 428.9818; found 428.9813.
4.2.4.3 2-(((2-bromophenyl)thio)selanyl)-N-phenylbenzamide (7c)
White solid; Yield 48 %; m.p. 123–124 °C; 1H NMR (400 MHz, CDCl3) δ 8.04 (d, J = 8.2 Hz, 1H, Ar-H), 7.98 (s, 1H, NH), 7.71 (d, J = 7.7 Hz, 1H, Ar-H), 7.61 (d, J = 7.8 Hz, 2H, Ar-H), 7.55 – 7.49 (m, 2H, Ar-H), 7.45 (t, J = 7.7 Hz, 1H, Ar-H), 7.41 – 7.28 (m, 3H, Ar-H), 7.16 (m, 2H, Ar-H), 6.98 (td, J = 7.8, 1.5 Hz, 1H, Ar-H); 13C NMR (101 MHz, CDCl3) δ 166.1, 139.8, 137.1, 132.7, 132.6, 131.2, 129.7, 129.6, 128.9, 128.1, 127.4, 126.6, 126.4, 125.3, 122.8, 120.7; 77Se NMR (76 MHz, CDCl3) δ 571.2; HRMS (ESI) m/z calcd for C19H14BrNOSSe [M + H]+ 463.9217; found 463.9213.
4.2.4.4 2-((naphthalen-2-ylthio)selanyl)-N-phenylbenzamide (7d)
White solid; Yield 76 %; m.p. 149–150 °C; 1H NMR (400 MHz, DMSO‑d6) δ 10.62 (s, 1H, NH), 8.24 (d, J = 7.6 Hz, 1H, Ar-H), 8.17 (d, J = 8.1 Hz, 1H, Ar-H), 8.06 (s, 1H, Ar-H), 7.85 (dd, J = 13.3, 8.5 Hz, 3H, Ar-H), 7.76 (d, J = 7.8 Hz, 2H, Ar-H), 7.62 (t, J = 8.1 Hz, 2H, Ar-H), 7.48 (t, J = 7.0 Hz, 3H, Ar-H), 7.41 (t, J = 7.3 Hz, 2H, Ar-H), 7.18 (t, J = 7.2 Hz, 1H, Ar-H); 13C NMR (101 MHz, DMSO‑d6) δ 166.3, 138.1, 135.9, 133.2, 133.0, 132.5, 131.6, 131.1, 128.9, 128.8, 128.7, 127.7, 127.3, 127.08, 127.03, 126.8, 126.4, 126.1, 124.5, 121.1; 77Se NMR (76 MHz, DMSO‑d6) δ 590.0; HRMS (ESI) m/z calcd for C23H17NOSSe [M + H]+ 436.0269; found 436.0268.
4.2.4.5 N-benzyl-2-(((4-fluorophenyl)thio)selanyl)benzamide (7e)
White solid; Yield 20 %; m.p. 111–112 °C; 1H NMR (400 MHz, CDCl3) δ 8.25 (d, J = 8.1 Hz, 1H, Ar-H), 7.57 (d, J = 7.7 Hz, 1H, Ar-H), 7.50 (q, J = 7.0, 6.1 Hz, 3H, Ar-H), 7.35 (d, J = 5.9 Hz, 5H, Ar-H), 7.26 (t, J = 7.5 Hz, 1H, Ar-H), 6.94 (t, J = 8.7 Hz, 2H, Ar-H), 6.71 (s, 1H, NH), 4.66 (d, J = 5.6 Hz, 2H, CH2); 13C NMR (101 MHz, CDCl3) δ 167.7, 163.2, 160.7, 137.5, 137.3, 132.2, 131.3 (d, J = 8.0 Hz), 130.5, 128.9, 128.3, 128.0, 127.9, 126.6, 126.1, 115.9 (d, J = 22.0 Hz), 44.3; 77Se NMR (76 MHz, CDCl3) δ 616.8; HRMS (ESI) m/z calcd for C20H16FNOSSe [M + Na]+ 439.9994; found 439.9998.
4.2.4.6 N-benzyl-2-(((4-bromophenyl)thio)selanyl)benzamide (7f)
White solid; Yield 19 %; m.p. 135–136 °C; 1H NMR (400 MHz, CDCl3) δ 8.13 (d, J = 8.1 Hz, 1H, Ar-H), 7.54 (d, J = 7.7 Hz, 1H, Ar-H), 7.46 (t, J = 7.0 Hz, 1H, Ar-H), 7.40 – 7.31 (m, 9H, Ar-H), 7.27 (t, J = 8.0 Hz, 1H, Ar-H), 6.53 (s, 1H, NH), 4.68 (d, J = 5.6 Hz, 2H, CH2); 13C NMR (101 MHz, CDCl3) δ 167.6, 137.4, 137.2, 136.1, 132.3, 131.8, 130.4, 128.9, 128.4, 128.1, 126.4, 126.1, 120.2, 44.4; 77Se NMR (76 MHz, CDCl3) δ 597.0; HRMS (ESI) m/z calcd for C20H16BrNOSSe [M + H]+ 477.9374; found 477.9368.
4.2.4.7 N-benzyl-2-(((2-bromophenyl)thio)selanyl)benzamide (7 g)
Pale-yellow solid; Yield 7 %; m.p. 135–136 °C; 1H NMR (400 MHz, CDCl3) δ 8.13 (d, J = 8.1 Hz, 1H, Ar-H), 7.54 (d, J = 7.7 Hz, 1H, Ar-H), 7.46 (t, J = 7.0 Hz, 1H, Ar-H), 7.40 – 7.31 (m, 9H, Ar-H), 7.27 (t, J = 8.0 Hz, 1H, Ar-H), 6.53 (s, 1H, NH), 4.68 (d, J = 5.6 Hz, 2H, CH2); 13C NMR (101 MHz, CDCl3) δ 167.7, 137.4, 137.2, 136.9, 132.7, 132.4, 130.5, 129.6, 128.9, 128.7, 128.1, 128.0, 127.9, 127.3, 126.4, 126.2, 122.7, 44.4, 29.7; 77Se NMR (76 MHz, CDCl3) δ 566.7; HRMS (ESI) m/z calcd for C20H16BrNOSSe [M + H]+ 477.9374; found 477.9370.
4.2.4.8 N-benzyl-2-(((3-(trifluoromethyl)phenyl)thio)selanyl)benzamide (7 h)
Pale-yellow solid; Yield 28 %; m.p. 100–101 °C; 1H NMR (400 MHz, CDCl3) δ 8.12 (d, J = 8.1 Hz, 1H, Ar-H), 7.76 (s, 1H, Ar-H), 7.68 (d, J = 7.7 Hz, 1H, Ar-H), 7.55 (d, J = 7.8 Hz, 1H, Ar-H), 7.49 – 7.43 (m, 1H, Ar-H), 7.41 – 7.27 (m, 8H, Ar-H), 6.56 (s, 1H, NH), 4.69 (d, J = 5.6 Hz, 2H, CH2); 13C NMR (101 MHz, CDCl3) δ 167.7, 138.4, 137.4, 136.9, 132.4, 131.9, 130.4, 129.3, 128.9, 128.2, 128.0, 127.9, 126.5, 126.2, 125.3 (q, J = 4.0 Hz), 125.3, 125.2, 123.0 (d, J = 3.7 Hz), 44.4; 77Se NMR (76 MHz, CDCl3) δ 590.4; HRMS (ESI) m/z calcd for C21H16F3NOSSe [M + H]+ 468.0143; found 468.0147.
4.2.4.9 2-(((4-chlorophenyl)thio)selanyl)-N-(4-methoxyphenyl)benzamide (7i)
White solid; Yield 75 %; m.p. 148–149 °C; 1H NMR (400 MHz, CDCl3) δ 8.17 (d, J = 8.1 Hz, 1H, Ar-H), 7.89 (s, 1H, NH), 7.70 (d, J = 7.6 Hz, 1H, Ar-H), 7.49 (t, J = 7.6 Hz, 3H, Ar-H), 7.43 (d, J = 8.2 Hz, 2H, Ar-H), 7.34 (t, J = 7.4 Hz, 1H, Ar-H), 7.18 (d, J = 8.4 Hz, 2H, Ar-H), 6.92 (d, J = 8.5 Hz, 2H, Ar-H), 3.82 (s, 3H, OCH3); 13C NMR (101 MHz, CDCl3) δ 165.9, 157.2, 137.3, 135.3, 132.4, 131.1, 130.2, 130.0, 128.9, 128.6, 126.5, 126.3, 122.7, 114.4, 55.6; 77SeNMR (76 MHz, CDCl3) δ 599.0; HRMS (ESI) m/z calcd for C20H16ClNO2SSe [M + H]+ 449.9828; found 449.9827.
4.2.4.10 2-(((4-bromophenyl)thio)selanyl)-N-(4-methoxyphenyl)benzamide (7j)
White solid; Yield 79 %; m.p. 149–150 °C; 1H NMR (400 MHz, CDCl3) δ 8.16 (d, J = 8.1 Hz, 1H, Ar-H), 7.87 (s, 1H, NH), 7.70 (d, J = 7.6 Hz, 1H, Ar-H), 7.50 (t, J = 8.8 Hz, 3H, Ar-H), 7.36 (q, J = 8.4 Hz, 5H, Ar-H), 6.93 (d, J = 8.4 Hz, 2H, Ar-H), 3.83 (s, 3H, OCH3); 13C NMR (101 MHz, CDCl3) δ 165.9, 157.2, 137.3, 135.9, 132.4, 131.9, 131.1, 130.4, 130.0, 128.6, 126.5, 126.2, 122.7, 120.2, 114.4, 55.5; 77Se NMR (76 MHz, CDCl3) δ 597.8; HRMS (ESI) m/z calcd for C20H16BrNO2SSe [M + H]+ 493.9323, found 493.9318.
4.2.4.11 N-(4-methoxyphenyl)-2-(((4-methoxyphenyl)thio)selanyl)benzamide (7 k)
White solid; Yield 67 %; m.p. 129–130 °C; 1H NMR (400 MHz, CDCl3) δ 8.33 (d, J = 8.1 Hz, 1H, Ar-H), 7.81 (s, 1H, NH), 7.64 (d, J = 7.7 Hz, 1H, Ar-H), 7.54 – 7.40 (m, 5H, Ar-H), 7.29 (t, J = 7.5 Hz, 1H, Ar-H), 6.88 (d, J = 9.0 Hz, 2H, Ar-H), 6.75 (d, J = 8.8 Hz, 2H, Ar-H), 3.79 (s, 3H, OCH3), 3.73 (s, 3H, OCH3); 13C NMR (101 MHz, CDCl3) δ 165.9, 159.1, 157.0, 137.6, 132.1, 132.0, 131.4, 130.1, 128.9, 127.7, 126.6, 126.1, 122.5, 114.6, 114.3, 55.5, 55.3; 77Se NMR (76 MHz, CDCl3) δ 633.7; HRMS (ESI) m/z calcd for C21H19NO3SSe [M + H]+ 446.0324; found 446.0326.
4.2.4.12 N-(4-methoxyphenyl)-2-(((4-nitrophenyl)thio)selanyl)benzamide (7 l)
Yellow solid; Yield 85 %; m.p. 189–190 °C. 1H NMR (400 MHz, DMSO‑d6) δ 8.05 (d, J = 9.0 Hz, 2H, Ar-H), 8.00 (d, J = 8.9 Hz, 1H, Ar-H), 7.92 (s, 1H, NH), 7.73 (d, J = 7.5 Hz, 1H, Ar-H), 7.60 (d, J = 9.0 Hz, 2H, Ar-H), 7.51 (d, J = 9.0 Hz, 2H, Ar-H), 7.49 – 7.44 (m, 1H, Ar-H), 7.37 (t, J = 7.9 Hz, 1H, Ar-H), 6.93 (d, J = 9.0 Hz, 2H, Ar-H), 3.81 (s, 3H, OCH3); 13C NMR (101 MHz, DMSO‑d6) δ 166.0, 156.3, 145.6, 135.2, 132.8, 130.9, 128.9, 128.2, 126.9, 126.7, 124.5, 124.1, 123.0, 122.2, 113.9, 55.2; 77Se NMR (76 MHz, DMSO‑d6) δ 577.3; HRMS (ESI) m/z calcd for C20H16N2O4SSe [M−H]- 458.9923; found 458.9920.
4.2.4.13 2-(((2-bromophenyl)thio)selanyl)-N-(4-methoxyphenyl)benzamide (7 m)
Pale-yellow solid; Yield 57 %; m.p. 154–155 °C; 1H NMR (400 MHz, CDCl3) δ 8.05 (d, J = 8.0 Hz, 1H, Ar-H), 7.92 (s, 1H, NH), 7.71 (d, J = 7.6 Hz, 1H, Ar-H), 7.52 (q, J = 10.2, 9.3 Hz, 4H, Ar-H), 7.46 (d, J = 7.6 Hz, 1H, Ar-H), 7.34 (t, J = 7.3 Hz, 1H, Ar-H), 7.16 (t, J = 7.5 Hz, 1H, Ar-H), 7.00 (t, J = 7.5 Hz, 1H, Ar-H), 6.93 (d, J = 8.5 Hz, 2H, Ar-H), 3.82 (s, 3H, OCH3); 13C NMR (101 MHz, CDCl3) δ 166.0, 157.2, 137.1, 132.7, 132.5, 131.2, 129.9, 129.6, 128.9, 128.1, 127.4, 126.4, 126.3, 122.7, 114.4, 55.6; 77Se NMR (76 MHz, CDCl3) δ 573.1; HRMS (ESI) m/z calcd for C20H16BrNO2SSe [M + H]+ 493.9323; found 493.9323.
4.2.4.14 N-(4-fluorophenyl)-2-((naphthalen-2-ylthio)selanyl)benzamide (7n)
White solid; Yield 27 %; m.p. 154–155 °C; 1H NMR (400 MHz, CDCl3) δ 8.30 (d, J = 8.2 Hz, 1H, Ar-H), 8.00 (s, 1H, Ar-H), 7.94 (s, 1H, NH), 7.79 (d, J = 7.8 Hz, 1H, Ar-H), 7.73 (t, J = 7.1 Hz, 3H, Ar-H), 7.62 (m, 3H, Ar-H), 7.47 (m, 3H, Ar-H), 7.36 (t, J = 7.5 Hz, 1H, Ar-H), 7.12 (t, J = 8.2 Hz, 2H, Ar-H); 13C NMR (101 MHz, CDCl3) δ 166.1, 161.1, 158.7, 137.8, 133.7 (d, J = 15.3 Hz), 133.1 (d, J = 2.7 Hz), 132.5, 132.2, 131.1, 129.0, 128.6, 127.7, 127.4, 127.4, 127.1, 126.6 (d, J = 3.8 Hz), 126.2, 125.9, 122.7 (d, J = 8.0 Hz), 116.0, 115.8; 77Se NMR (76 MHz, CDCl3) δ 590.4; HRMS (ESI) m/z calcd for C23H16FNOSSe [M−H]- 452.0029; found 452.0023.
4.2.4.15 2-(((4-bromophenyl)thio)selanyl)-N-(4-chlorophenyl)benzamide (7o)
White solid; Yield 57 %; m.p. 159–160 °C; 1H NMR (400 MHz, DMSO‑d6) δ 10.67 (s, 1H, NH), 8.25 (d, J = 7.7 Hz, 1H, Ar-H), 8.06 (d, J = 8.1 Hz, 1H, Ar-H), 7.80 – 7.71 (m, 3H, Ar-H), 7.64 (t, J = 7.6 Hz, 2H, Ar-H), 7.55 – 7.39 (m, 6H, Ar-H), 7.25 (t, J = 8.8 Hz, 3H, Ar-H); 13C NMR (101 MHz, DMSO‑d6) δ 166.7, 160.5, 158.1, 136.0, 134.9, 133.2, 132.5, 131.3, 131.1, 129.4, 127.6, 127.0, 123.6 (d, J = 8.0 Hz), 120.2, 115.9 (d, J = 22.3 Hz); 77Se NMR (76 MHz, DMSO‑d6) δ 594.9; HRMS (ESI) m/z calcd for C19H13BrClNOSSe [M−H]- 495.8682; found 495.8673.
4.2.4.16 2-(((2-bromophenyl)thio)selanyl)-N-(4-chlorophenyl)benzamide (7p)
White solid; Yield 22 %; m.p. 163–164 °C; 1H NMR (400 MHz, CDCl3) δ 8.07 (d, J = 8.1 Hz, 1H, Ar-H), 7.96 (s, 1H, NH), 7.71 (d, J = 7.6 Hz, 1H, Ar-H), 7.59 (d, J = 8.8 Hz, 2H, Ar-H), 7.53 (ddd, J = 7.9, 4.4, 1.4 Hz, 2H, Ar-H), 7.49 (t, J = 7.7 Hz, 1H, Ar-H), 7.36 (d, J = 8.7 Hz, 3H, Ar-H), 7.16 (t, J = 7.6 Hz, 1H, Ar-H), 7.01 (t, J = 7.6 Hz, 1H, Ar-H); 13C NMR (101 MHz, CDCl3) δ 166.0, 137.2, 136.9, 135.6, 132.8 (d, J = 6.4 Hz), 130.9, 130.4, 129.7, 129.3, 129.1, 128.1 127.5, 126.7, 126.4, 122.8, 121.9; 77Se NMR (76 MHz, CDCl3) δ 571.2; HRMS (ESI) m/z calcd for C19H13BrClNOSSe [M + H]+ 497.8828; found 497.8821.
4.2.4.17 N-(4-chlorophenyl)-2-(((2-methoxyphenyl)thio)selanyl)benzamide (7q)
White solid; Yield 55 %; m.p. 287–288 °C; 1H NMR (400 MHz, CDCl3) δ 8.20 (d, J = 8.1 Hz, 1H, Ar-H), 7.94 (s, 1H, NH), 7.66 (d, J = 7.6 Hz, 1H, Ar-H), 7.55 (d, J = 8.7 Hz, 2H, Ar-H), 7.48 – 7.42 (m, 2H, Ar-H), 7.30 (dd, J = 15.1, 8.0 Hz, 3H, Ar-H), 7.18 – 7.12 (m, 1H, Ar-H), 6.81 (m, 2H, Ar-H), 3.88 (s, 3H, OCH3); 13C NMR (101 MHz, CDCl3) δ 166.1, 157.4, 137.7, 135.8, 132.4, 131.3, 130.7, 130.2, 129.5, 128.1, 127.8, 126.5, 126.1, 121.8, 121.3, 110.6, 55.9; 77Se NMR (76 MHz, CDCl3) δ 561.6; HRMS (ESI) m/z calcd for C20H16ClNO2SSe [M + H]+ 449.9828; found 449.9835.
4.2.4.18 N-(4-bromophenyl)-2-(((4-fluorophenyl)thio)selanyl)benzamide (7r)
White solid; Yield 20 %; m.p. 161–162 °C; 1H NMR (400 MHz, CDCl3) δ 8.30 (d, J = 8.8 Hz, 1H, Ar-H), 7.92 (s, 1H, NH), 7.72 (d, J = 8.8 Hz, 1H, Ar-H), 7.59 – 7.48 (m, 7H, Ar-H), 7.39 (t, J = 7.4 Hz, 1H, Ar-H), 6.95 (t, J = 8.7 Hz, 2H, Ar-H); 13C NMR (101 MHz, CDCl3) δ 166.0, 163.3, 160.8, 137.6, 136.2, 132.6, 132.2, 131.5, 131.4, 130.9, 128.8, 126.8, 126.3, 122.2, 117.9, 116.1, 115.9; 77Se NMR (76 MHz, CDCl3) δ 615.0; HRMS (ESI) m/z calcd for C19H13BrFNOSSe [M + H]+ 481.9123; found 481.9115.
4.2.4.19 N-(4-bromophenyl)-2-(((4-bromophenyl)thio)selanyl)benzamide (7 s)
White solid; Yield 37 %; m.p. 284–285 °C; 1H NMR (400 MHz, CDCl3) δ 8.18 (d, J = 8.9 Hz, 1H, Ar-H), 7.91 (s, 1H, NH), 7.70 (d, J = 7.7 Hz, 1H, Ar-H), 7.56 – 7.48 (m, 5H, Ar-H), 7.35 (m, 5H, Ar-H); 13C NMR (101 MHz, CDCl3) δ 166.0, 137.5, 136.2, 135.8, 132.7, 132.2, 131.9, 130.8, 130.6, 129.4, 128.8, 126.6, 126.4, 122.1, 120.4, 118.0; 77Se NMR (76 MHz, CDCl3) δ 598.6; HRMS (ESI) m/z calcd for C19H13Br2NOSSe [M−H]- 539.8177; found 539.8170.
4.2.4.20 N-(4-bromophenyl)-2-(((4-methoxyphenyl)thio)selanyl)benzamide (7 t)
White solid; Yield 14 %; m.p. 112–113 °C; 1H NMR (400 MHz, CDCl3) δ 8.35 (d, J = 8.1 Hz, 1H, Ar-H), 7.94 (s, 1H, NH), 7.67 (d, J = 7.7 Hz, 1H, Ar-H), 7.55 – 7.43 (m, 7H, Ar-H), 7.31 (t, J = 7.4 Hz, 1H, Ar-H), 6.77 (dd, J = 8.7, 1.9 Hz, 2H, Ar-H), 3.74 (d, J = 2.0 Hz, 3H, OCH3); 13C NMR (101 MHz, CDCl3) δ 166.0, 159.3, 137.3, 136.4, 132.4, 132.1, 131.2, 129.2, 127.6, 126.7, 126.2, 122.1, 117.8, 114.6, 55.4; 77Se NMR (76 MHz, CDCl3) δ 633.2; HRMS (ESI) m/z calcd for C20H16BrNO2SSe [M−H]- 491.9178; found 491.9170.
4.2.4.21 N-(4-bromophenyl)-2-(((4-methoxyphenyl)thio)selanyl)benzamide (7u)
Yellow solid; Yield 69 %; m.p. 190–191 °C; 1H NMR (400 MHz, DMSO‑d6) δ 8.06 (d, J = 8.9 Hz, 2H, Ar-H), 8.01 (d, J = 8.2 Hz, 1H, Ar-H), 7.95 (s, 1H, NH), 7.74 (d, J = 7.8 Hz, 1H, Ar-H), 7.60 (d, J = 8.9 Hz, 2H, Ar-H), 7.51 (dd, J = 12.2, 2.9 Hz, 5H, Ar-H), 7.38 (t, J = 7.1 Hz, 1H, Ar-H); 13C NMR (101 MHz, DMSO‑d6) δ 167.0, 146.1, 145.9, 137.9, 135.8, 133.6, 132.1, 131.2, 129.6, 128.7, 127.5, 127.2, 124.6, 123.6, 122.9, 117.0; 77Se NMR (76 MHz, DMSO‑d6) δ 574.9; HRMS (ESI) m/z calcd for C19H13BrN2O3SSe [M + H]+ 508.9068; found 508.9059.
4.3 General procedure for synthesis of intermediates and target compounds 11a-11u
4.3.1 Synthesis of intermediate 9 and 10
Methyl group as R3 was taken for an example here. Methyl 2-aminobenzoate (3.02 g, 20 mmol, 4 equiv.) was added to a 250 mL round flask, hydrochloric acid (2 M, 12 mL) was then added slowly to the flask at ice bath condition. Under stirring, sodium nitrite solution (3 M, 8 mL) was added until the solution became transparent and clear. Subsequently, saturated sodium acetate was added to adjust the pH value to 4.8 and the temperature was changed to room temperature. The intermediate 9 was not separated since it was a diazo salt. Subsequently, potassium selenocyanate (2.88 g, 20 mmol, 4 equiv.) was added to the mixture when bubbles produced and orange solid precipitated. The reaction completed when no more bubbles came out. The product was filtered to give orange solid methyl 2-selenocyanatobenzoate (10) in 81.5 % yield.
The synthesis was in a similar manner when R3 was ethyl. These intermediates were known compounds and have not be characterized in this paper (Alberto et al., 2012; Jones et al., 2002; Dakova et al., 1991).
4.3.2 Synthesis of target compound 11a-11u
All the substituted thiophenols (2-naphthalenethiol for 11 l and 11u) were from the same commercial sources as those for the synthesis of 7a-7u. Generally, 1.0 equiv. of intermediate 10 was added to 5 mL dichloromethane, after that 1.0 equiv. of different substituted thiophenol and catalytic amout of aluminum oxide was added to the reactant. The mixture was stirred for 30 min at room temperature and monitored by thin-layer chromatography. The target compound was finally purified by column chromatography (n-hexane/ethyl acetate = 100/1) in different yields.
4.3.2.1 Methyl 2-((p-tolylthio)selanyl)benzoate (11a)
White solid; Yield 53 %; m.p. 51–52 °C; 1H NMR (400 MHz, CDCl3) δ 8.20 (d, J = 8.7 Hz, 1H, Ar-H), 8.04 (dd, J = 7.8, 1.4 Hz, 1H, Ar-H), 7.53 – 7.47 (m, 1H, Ar-H), 7.38 (d, J = 8.2 Hz, 2H, Ar-H), 7.27 (t, J = 7.0 Hz, 1H, Ar-H), 7.02 (d, J = 8.1 Hz, 2H, Ar-H), 3.95 (s, 3H, OCH3), 2.27 (s, 3H, CH3); 13C NMR (101 MHz, CDCl3) δ 168.0, 138.6, 136.8, 133.4, 132.9, 131.3, 129.7, 129.4, 127.5, 127.1, 125.9, 52.7, 20.9; 77Se NMR (76 MHz, CDCl3) δ 602.9; HRMS (ESI) m/z calcd for C15H14O2SSe [M + Na]+ 360.9772; found 360.9775.
4.3.2.2 Methyl 2-(((4-fluorophenyl)thio)selanyl)benzoate (11b)
White solid; Yield 14 %; m.p. 68–69 °C; 1H NMR (400 MHz, CDCl3) δ 8.18 (d, J = 8.3 Hz, 1H, Ar-H), 8.05 (d, J = 9.2 Hz, 1H, Ar-H), 7.52 (t, J = 8.4 Hz, 1H, Ar-H), 7.45 (dd, J = 8.8, 5.1 Hz, 2H, Ar-H), 7.29 (t, J = 7.5 Hz, 1H, Ar-H), 6.91 (t, J = 8.7 Hz, 2H, Ar-H), 3.96 (s, 3H, OCH3); 13C NMR (101 MHz, CDCl3) δ 168.1, 163.3, 160.8, 138.2, 133.5, 131.4, 131.3, 131.3, 127.3, 127.1, 126.1, 116.1, 115.9, 52.7; 77Se NMR (76 MHz, CDCl3) δ 615.7; HRMS (ESI) m/z calcd for C14H11FO2SSe [M + Na]+ 364.9521; found 364.9518.
4.3.2.3 Methyl 2-(((4-chlorophenyl)thio)selanyl)benzoate (11c)
Yellow solid; Yield 46 %; m.p. 72–73 °C; 1H NMR (400 MHz, CDCl3) δ 8.09 (d, J = 8.2 Hz, 1H, Ar-H), 8.06 (dd, J = 7.8, 1.3 Hz, 1H, Ar-H), 7.53 – 7.47 (m, 1H, Ar-H), 7.40 (d, J = 8.6 Hz, 2H, Ar-H), 7.29 (t, J = 7.5 Hz, 1H, Ar-H), 7.17 (d, J = 8.6 Hz, 2H, Ar-H), 3.97 (s, 3H, OCH3); 13C NMR (101 MHz, CDCl3) δ 168.1, 138.0, 134.9, 133.6, 132.6, 130.2, 129.0, 127.3, 127.2, 126.1, 52.9; 77Se NMR (76 MHz, CDCl3) δ 597.0; HRMS (ESI) m/z calcd for C14H11ClO2SSe [M + Na]+ 380.9226; found 380.9230.
4.3.2.4 Methyl 2-(((4-bromophenyl)thio)selanyl)benzoate (11d)
Yellow solid; Yield 42 %; m.p. 101–102 °C; 1H NMR (400 MHz, CDCl3) δ 8.12 – 8.06 (m, 2H, Ar-H), 7.51 (td, J = 8.2, 7.8, 1.5 Hz, 1H, Ar-H), 7.39 – 7.28 (m, 5H, Ar-H), 3.99 (s, 3H, OCH3); 13C NMR (101 MHz, CDCl3) δ 168.1, 137.9, 135.5, 133.6, 131.9, 131.4, 130.4, 127.3, 127.2, 126.2, 120.5, 52.8; 77Se NMR (76 MHz, CDCl3) δ 594.3; HRMS (ESI) m/z calcd for C14H11BrO2SSe [M + Na]+ 424.8721; found 424.8726.
4.3.2.5 Methyl 2-(((4-nitrophenyl)thio)selanyl)benzoate (11e)
Pale-red solid; Yield 43 %; m.p. 131–132 °C; 1H NMR (400 MHz, CDCl3) δ 8.13 (dd, J = 7.8, 1.3 Hz, 1H, Ar-H), 8.10 (d, J = 9.0 Hz, 2H, Ar-H), 7.95 (d, J = 8.1 Hz, 1H, Ar-H), 7.62 (d, J = 8.9 Hz, 2H, Ar-H), 7.55 – 7.49 (m, 1H, Ar-H), 7.36 (t, J = 7.1 Hz, 1H, Ar-H), 4.04 (s, 3H, OCH3); 13C NMR (101 MHz, CDCl3) δ 168.4, 145.5, 137.1, 133.8, 131.6, 128.0, 127.0, 126.6, 126.4, 124.4, 124.0, 53.0; 77Se NMR (76 MHz, CDCl3) δ 570.4; HRMS (ESI) m/z calcd for C14H11NO4SSe [M + Na]+ 391.9466; found 391.9470.
4.3.2.6 Methyl 2-((o-tolylthio)selanyl)benzoate (11f)
Yellow solid; Yield 56 %; m.p. 88–89 °C; 1H NMR (400 MHz, CDCl3) δ 8.14 (d, J = 8.1 Hz, 1H, Ar-H), 8.10 (dd, J = 7.8, 1.4 Hz, 1H, Ar-H), 7.56 – 7.49 (m, 1H, Ar-H), 7.46 (dd, J = 7.4, 1.6 Hz, 1H, Ar-H), 7.32 (t, J = 7.5 Hz, 1H, Ar-H), 7.18 (d, J = 6.8 Hz, 1H, Ar-H), 7.09 (pd, J = 7.2, 1.4 Hz, 2H, Ar-H), 4.01 (s, 3H, OCH3), 2.57 (s, 3H, CH3); 13C NMR (101 MHz, CDCl3) δ 168.1, 138.4, 137.2, 134.9, 133.5, 131.4, 130.1, 129.1, 127.7, 127.3, 126.7, 126.0, 52.7, 20.7; 77Se NMR (76 MHz, CDCl3) δ 567.1; HRMS (ESI) m/z calcd for C15H14O2SSe [M + Na]+ 360.9772; found 360.9775.
4.3.2.7 Methyl 2-(((2-fluorophenyl)thio)selanyl)benzoate (11 g)
Yellow solid; Yield 56 %; m.p. 78–79 °C. 1H NMR (400 MHz, CDCl3) δ 8.20 (d, J = 8.1 Hz, 1H, Ar-H), 8.05 (dd, J = 7.8, 1.3 Hz, 1H, Ar-H), 7.55 – 7.49 (m, 1H, Ar-H), 7.47 (td, J = 7.7, 1.4 Hz, 1H, Ar-H), 7.28 (t, J = 7.5 Hz, 1H, Ar-H), 7.15 (q, J = 7.8, 6.9 Hz, 1H, Ar-H), 7.00 (q, J = 8.1 Hz, 2H, Ar-H), 3.96 (s, 3H, OCH3); 13C NMR (101 MHz, CDCl3) δ 168.2, 162.1, 159.6, 138.2, 133.6, 132.4, 131.3, 128.9, 128.8, 127.6, 127.1, 126.1, 124.8, 124.7, 115.6, 115.4, 52.8; 77Se NMR (76 MHz, CDCl3) δ 594.1. HRMS (ESI) m/z calcd for C14H11FO2SSe [M + Na]+ 364.9521; found 364.9527.
4.3.2.8 Methyl 2-(((2-chlorophenyl)thio)selanyl)benzoate (11 h)
Yellow solid; Yield 41 %; m.p. 99–100 °C; 1H NMR (400 MHz, DMSO‑d6) δ 8.09 (d, J = 7.7 Hz, 1H, Ar-H), 7.94 (d, J = 8.1 Hz, 1H, Ar-H), 7.68 (t, J = 7.6 Hz, 1H, Ar-H), 7.48 (dq, J = 15.0, 7.3 Hz, 3H, Ar-H), 7.26 (p, J = 7.3 Hz, 2H, Ar-H), 3.96 (s, 3H, OCH3); 13C NMR (101 MHz, DMSO‑d6) δ 167.6, 136.1, 134.3, 133.5, 132.1, 131.4, 129.9, 129.7, 128.4, 128.3, 126.9, 126.7, 53.1; 77Se NMR (76 MHz, DMSO‑d6) δ 561.0. HRMS (ESI) m/z calcd for C14H11ClO2SSe [M + Na]+ 380.9226; found 380.9227.
4.3.2.9 Methyl 2-(((2-bromophenyl)thio)selanyl)benzoate (11i)
Pale-yellow solid; Yield 52 %; m.p. 115–116 °C; 1H NMR (400 MHz, CDCl3) δ 8.09 (d, J = 7.8 Hz, 1H, Ar-H), 7.98 (d, J = 8.1 Hz, 1H, Ar-H), 7.53 (d, J = 8.0 Hz, 1H, Ar-H), 7.48 (d, J = 8.0 Hz, 2H, Ar-H), 7.31 (t, J = 7.4 Hz, 1H, Ar-H), 7.16 (t, J = 8.3 Hz, 1H, Ar-H), 7.01 (t, J = 7.6 Hz, 1H, Ar-H), 4.01 (d, J = 1.5 Hz, 3H, OCH3); 13C NMR (101 MHz, CDCl3) δ 168.3, 137.6, 136.6, 133.7, 132.7, 131.4, 129.5, 128.1, 127.6, 127.5, 127.3, 126.2, 122.6, 52.9; 77Se NMR (76 MHz, CDCl3) δ 561.8; HRMS (ESI) m/z calcd for C14H11BrO2SSe [M + Na]+ 424.8721; found 424.8722.
4.3.2.10 Methyl 2-(((3-(trifluoromethyl)phenyl)thio)selanyl)benzoate (11j)
Yellow solid; Yield 27 %; m.p. 72–73 °C; 1H NMR (400 MHz, CDCl3) δ 8.06 (td, J = 7.0, 6.3, 1.0 Hz, 2H, Ar-H), 7.73 (s, 1H, Ar-H), 7.63 (d, J = 7.8 Hz, 1H, Ar-H), 7.53 – 7.47 (m, 1H, Ar-H), 7.38 (d, J = 7.8 Hz, 1H, Ar-H), 7.31 (q, J = 7.4, 6.9 Hz, 2H, Ar-H), 3.98 (s, 3H, OCH3); 13C NMR (101 MHz, CDCl3) δ 168.2, 137.8, 137.7, 133.7, 131.9, 131.5, 131.1, 129.4, 127.2, 126.3, 125.3 (q, J = 3.8 Hz), 125.1, 123.3 (q, J = 3.8 Hz), 122.4, 52.6; 77Se NMR (76 MHz, CDCl3) δ 586.1; HRMS (ESI) m/z calcd for C15H11F3O2SSe [M + Na]+ 414.9489; found 414.9495.
4.3.2.11 Methyl 2-(((3,5-bis(trifluoromethyl)phenyl)thio)selanyl)benzoate (11 k)
Yellow solid; Yield 22 %; m.p. 75–76 °C; 1H NMR (400 MHz, CDCl3) δ 8.14 (d, J = 7.8 Hz, 1H, Ar-H), 8.01 (d, J = 8.1 Hz, 1H, Ar-H), 7.93 (s, 2H, Ar-H), 7.66 (s, 1H, Ar-H), 7.56 (t, J = 8.4 Hz, 1H, Ar-H), 7.37 (t, J = 7.5 Hz, 1H, Ar-H), 4.05 (s, 3H, OCH3); 13C NMR (101 MHz, CDCl3) δ 168.4, 140.1, 137.0, 133.8, 132.2 (q, J = 33.4 Hz), 131.6, 128.3, 128.2, 127.2, 126.8, 126.5, 124.3, 121.6, 120.3, 120.24, 120.20, 120.17, 120.13, 53.0; 77Se NMR (76 MHz, CDCl3) δ 584.3; HRMS (ESI) m/z calcd for C16H10F6O2SSe [M + Na]+ 482.9363; found 482.9366.
4.3.2.12 Methyl 2-((naphthalen-1-ylthio)selanyl)benzoate (11 l)
Yellow solid; Yield 23 %; m.p. 68–69 °C; 1H NMR (400 MHz, CDCl3) δ 8.21 (d, J = 7.6 Hz, 1H, Ar-H), 8.08 (dd, J = 7.8, 1.4 Hz, 1H, Ar-H), 7.96 (s, 1H, Ar-H), 7.77 (d, J = 7.6 Hz, 1H, Ar-H), 7.71 (m, 2H, Ar-H), 7.59 (dd, J = 8.7, 1.9 Hz, 1H, Ar-H), 7.54 – 7.47 (m, 1H, Ar-H), 7.43 (pd, J = 6.9, 1.4 Hz, 2H, Ar-H), 7.33 – 7.26 (m, 1H, Ar-H), 4.00 (s, 3H, OCH3); 13C NMR (101 MHz, CDCl3) δ 168.2, 138.4, 133.6, 133.5, 133.5, 132.2, 131.4, 128.7, 127.7, 127.6, 127.3, 127.2, 126.9, 126.6, 126.0, 125.9, 52.7; 77Se NMR (76 MHz, CDCl3) δ 586.5; HRMS (ESI) m/z calcd for C18H14O2SSe [M + Na]+ 396.9772; found 396.9775.
4.3.2.13 Ethyl 2-((p-tolylthio)selanyl)benzoate (11 m)
Yellow solid; Yield 67 %; m.p. 54–55 °C; 1H NMR (400 MHz, CDCl3) δ 8.07 (dd, J = 8.1, 0.8 Hz, 1H, Ar-H), 7.93 (dd, J = 7.8, 1.4 Hz, 1H, Ar-H), 7.36 (ddd, J = 8.6, 7.3, 1.5 Hz, 1H, Ar-H), 7.25 (d, J = 8.2 Hz, 2H, Ar-H), 7.14 (td, J = 7.8, 1.1 Hz, 1H, Ar-H), 6.90 (d, J = 8.0 Hz, 2H, Ar-H), 4.29 (q, J = 7.1 Hz, 2H,CH2), 2.14 (s, 3H,Ph-CH3), 1.28 (t, J = 7.1 Hz, 3H, CH3); 13C NMR (101 MHz, CDCl3) δ 167.6, 138.5, 136.8, 133.3, 132.9, 131.3, 129.7, 129.4, 127.5, 125.8, 61.8, 20.9, 14.5;77Se NMR (76 MHz, CDCl3) δ 602.3; HRMS (ESI) m/z calcd for C16H16O2SSe [M + Na]+ 374.9928; found 374.9932.
4.3.2.14 Ethyl 2-(((4-chlorophenyl)thio)selanyl)benzoate (11n)
White solid; Yield 19 %; m.p. 55–56 °C; 1H NMR (400 MHz, CDCl3) δ 8.12 (dd, J = 5.3, 1.1 Hz, 1H, Ar-H), 8.11 – 8.08 (m, 1H, Ar-H), 7.55 – 7.48 (m, 1H, Ar-H), 7.46 – 7.40 (m, 2H, Ar-H), 7.31 (t, J = 7.5 Hz, 1H, Ar-H), 7.23 – 7.16 (m, 2H, Ar-H), 4.45 (q, J = 7.1 Hz, 2H, CH2), 1.44 (t, J = 7.1 Hz, 3H, CH3); 13C NMR (101 MHz, CDCl3) δ 167.7, 137.9, 134.9, 133.5, 132.6, 131.4, 130.2, 129.0, 127.4, 127.2, 126.1, 62.0, 14.3; 77Se NMR (76 MHz, CDCl3) δ 597.3; HRMS (ESI) m/z calcd for C15H13ClO2SSe [M + Na]+ 394.9382; found 394.9382.
4.3.2.15 Ethyl 2-(((4-bromophenyl)thio)selanyl)benzoate (11o)
Yellow solid; Yield 29 %; m.p. 57–58 °C; 1H NMR (400 MHz, CDCl3) δ 8.09 (d, J = 9.2 Hz, 2H, Ar-H), 7.50 (t, J = 8.4 Hz, 1H, Ar-H), 7.38 – 7.32 (m, 4H, Ar-H), 7.30 (d, J = 7.0 Hz, 1H, Ar-H), 4.45 (q, J = 7.1 Hz, 2H, CH2), 1.44 (t, J = 7.1 Hz, 3H, CH3); 13C NMR (101 MHz, CDCl3) δ 167.7, 137.8, 135.6, 133.5, 131.9, 131.4, 130.4, 127.5, 127.2, 126.1, 120.5, 61.9, 14.4; 77Se NMR (76 MHz, CDCl3) δ 594.2; HRMS (ESI) m/z calcd for C15H13BrO2SSe [M + Na]+ 438.8877; found 438.8877.
4.3.2.16 Ethyl 2-(((4-nitrophenyl)thio)selanyl)benzoate (11p)
White solid; Yield 26 %; m.p. 99–100 °C. 1H NMR (400 MHz, CDCl3) δ 8.13 (d, J = 9.1 Hz, 1H, Ar-H), 8.08 (d, J = 8.9 Hz, 2H, Ar-H), 7.92 (d, J = 8.1 Hz, 1H, Ar-H), 7.60 (d, J = 8.9 Hz, 2H, Ar-H), 7.49 (t, J = 7.7 Hz, 1H, Ar-H), 7.34 (t, J = 8.0 Hz, 1H, Ar-H), 4.48 (q, J = 7.1 Hz, 2H, CH2), 1.46 (t, J = 7.1 Hz, 3H, CH3); 13C NMR (101 MHz, CDCl3) δ 168.0, 145.6, 137.0, 133.7, 131.5, 128.0, 127.6, 126.9, 126.5, 124.0, 62.2, 14.3; 77Se NMR (76 MHz, CDCl3) δ 570.6; HRMS (ESI) m/z calcd for C15H13NO4SSe [M + Na]+ 405.9623; found 405.9628.
4.3.2.17 Ethyl 2-(((2-fluorophenyl)thio)selanyl)benzoate (11q)
White solid; Yield 39 %; m.p. 86–87 °C; 1H NMR (400 MHz, CDCl3) δ 8.21 (d, J = 7.9 Hz, 1H, Ar-H), 8.08 (dd, J = 7.8, 1.5 Hz, 1H, Ar-H), 7.56 – 7.45 (m, 2H, Ar-H), 7.33 – 7.27 (m, 1H, Ar-H), 7.16 (dtd, J = 9.4, 5.1, 1.7 Hz, 1H, Ar-H), 7.05 – 6.97 (m, 2H, Ar-H), 4.43 (q, J = 7.1 Hz, 2H, CH2), 1.42 (t, J = 7.1 Hz, 3H, CH3); 13C NMR (101 MHz, CDCl3) δ 167.8, 162.1, 159.6, 138.1, 133.5, 132.4, 131.3, 128.8, 127.5, 126.0, 124.7, 123.7, 115.4, 62.0, 14.3; 77Se NMR (76 MHz, CDCl3) δ 593.7; HRMS (ESI) m/z calcd for C15H13FO2SSe [M + Na]+ 378.9678; found 378.9680.
4.3.2.18 Ethyl 2-(((2-chlorophenyl)thio)selanyl)benzoate (11r)
White solid; Yield 23 %; m.p. 62–63 °C; 1H NMR (400 MHz, CDCl3) δ 8.10 (dd, J = 7.8, 1.4 Hz, 1H, Ar-H), 8.04 – 7.98 (m, 1H, Ar-H), 7.48 (td, J = 7.1, 2.0 Hz, 2H, Ar-H), 7.37 – 7.33 (m, 1H, Ar-H), 7.30 (td, J = 7.8, 1.1 Hz, 1H, Ar-H), 7.14 – 7.06 (m, 2H, Ar-H), 4.46 (q, J = 7.1 Hz, 2H, CH2), 1.45 (t, J = 7.1 Hz, 3H, CH3); 13C NMR (101 MHz, CDCl3) δ 167.8, 137.6, 134.8, 133.6, 132.9, 131.4, 129.8, 129.5, 127.6, 127.5, 127.5, 127.4, 126.1, 62.0, 14.3; 77Se NMR (76 MHz, CDCl3) δ 558.7; HRMS (ESI) m/z calcd for C15H13ClO2SSe [M + Na]+ 394.9382; found 394.9385.
4.3.2.19 Ethyl 2-(((2-bromophenyl)thio)selanyl)benzoate (11 s)
White solid; Yield 31 %; m.p. 74–75 °C; 1H NMR (400 MHz, CDCl3) δ 8.10 (dd, J = 7.8, 1.4 Hz, 1H, Ar-H), 8.00 – 7.96 (m, 1H, Ar-H), 7.53 (dd, J = 7.9, 1.2 Hz, 1H, Ar-H), 7.51 – 7.45 (m, 2H, Ar-H), 7.31 (td, J = 7.8, 1.0 Hz, 1H, Ar-H), 7.16 (td, J = 8.0, 1.3 Hz, 1H, Ar-H), 7.01 (td, J = 7.7, 1.5 Hz, 1H, Ar-H), 4.47 (q, J = 7.1 Hz, 2H, CH2), 1.45 (t, J = 7.1 Hz, 3H, CH3); 13C NMR (101 MHz, CDCl3) δ 167.8, 137.5, 136.7, 133.6, 132.7, 131.4, 129.5, 128.1, 127.6, 127.5, 127.5, 126.2, 122.7, 62.0, 14.4; 77Se NMR (76 MHz, CDCl3) δ 562.0; HRMS (ESI) m/z calcd for C15H13BrO2SSe [M + Na]+ 438.8877; found 438.8875.
4.3.2.20 Ethyl 2-(((3,5-bis(trifluoromethyl)phenyl)thio)selanyl)benzoate (11 t)
Yellow solid; Yield 21 %; m.p. 93–94 °C; 1H NMR (400 MHz, CDCl3) δ 8.13 (dd, J = 7.8, 1.4 Hz, 1H, Ar-H), 7.99 (d, J = 7.7 Hz, 1H, Ar-H), 7.92 (s, 2H, Ar-H), 7.64 (s, 1H, Ar-H), 7.57 – 7.49 (m, 1H, Ar-H), 7.38 – 7.32 (m, 1H, Ar-H), 4.48 (q, J = 7.1 Hz, 2H, CH2), 1.46 (t, J = 7.1 Hz, 3H, CH3); 13C NMR (101 MHz, CDCl3) δ 167.95, 140.15, 136.94, 133.71, 132.19 (q, J = 33.5 Hz), 131.6, 128.3, 128.3, 127.6, 126.8, 126.5, 124.3, 121.6, 120.2 (p, J = 3.7 Hz), 62.2, 14.3; 77Se NMR (76 MHz, CDCl3) δ 584.6; HRMS (ESI) m/z calcd for C17H12F6O2SSe [M + Na]+ 496.9520; found 496.9529.
4.3.2.21 Ethyl 2-((naphthalen-2-ylthio)selanyl)benzoate (11u)
Yellow solid; Yield 22 %; m.p. 75–76 °C; 1H NMR (400 MHz, CDCl3). δ 8.23 (d, J = 8.1 Hz, 1H, Ar-H), 8.11 (dd, J = 7.8, 1.3 Hz, 1H, Ar-H), 7.98 (s, 1H, Ar-H), 7.77 (d, J = 7.6 Hz, 1H, Ar-H), 7.72 (dd, J = 8.0, 4.3 Hz, 2H, Ar-H), 7.61 (dd, J = 8.7, 1.8 Hz, 1H, Ar-H), 7.52 – 7.48 (m, 1H, Ar-H), 7.47 – 7.39 (m, 2H, Ar-H), 7.30 (t, J = 7.5 Hz, 1H, Ar-H), 4.47 (q, J = 7.1 Hz, 2H, CH2), 1.46 (t, J = 7.1 Hz, 3H, CH3); 13C NMR (101 MHz, CDCl3) δ 167.7, 138.3, 133.6, 133.5, 133.5, 132.1, 131.5, 128.7, 127.6, 127.5, 127.3, 126.9, 126.6, 126.0, 125.9, 61.9, 14.4; 77Se NMR (76 MHz, CDCl3) δ 586.6; HRMS (ESI) m/z calcd for C19H16O2SSe [M + Na]+ 410.9928; found 410.9935.
4.4 Biological reagents and conditions
Sodium chloride, potassium chloride, disodium hydrogen phosphate dodecahydrate, potassium dihydrogen phosphate and dimethyl sulfoxide were purchased from National Pharmaceutical Group Chemical Reagent Co., Ltd. Tris-(hydroxymethyl)-aminomethane was purchased from Novon Scientific. Imidazole was purchased from MP Biomedicals. Dithiothreitol was purchased from Beijing Xinjingke Biotechnology Co.,Ltd. Bovine serum albumin and glycerol were purchased from Sigma. Rupintrivir was purchased from Toronto Research Chemicals. Cell counting Kit-8 was purchased from Dojindo Chemicals. Substrate for the 3Cpro/3CLpro, H-Asp-{Glu(EDANS)}-Met-Ser-Ala-Ile-Phe-Gln-Gly-Pro-Ile-Ser-{Lys(DABCYL)}-Asp-OH was purchased from GL Biochem in Shanghai. 96-well microtiter plates were purchased from Corning Co. Ltd. Enzyme inhibition and cell based activity was recorded on a Spectra max 190 microplate reader (Molecular Devices Co. Ltd.). All the experiments were conducted in a standard biosafety shelter laboratory 2 (BSL-2).
4.5 Enzyme inhibition assay
EV71-3Cpro and CVB3-3Cpro were expressed and purified similar to the previous method (Lu et al., 2011; Wan et al., 2023). The DNA encoding EV71-3C or CVB3-3C protease was amplified and inserted into pET28a (Novagen). The resulting plasmid encodes an N-terminal His-tagged EV71-3C or CVB3-3C protease. For the expression and purification, the plasmid was transformed into Rosetta™ (DE3) Competent Cells (Novagen). Bacteria culture was grown in LB culture at 37 °C until the density OD600 reached 1.0, after that IPTG was added to the culture (final concentration of 0.5 mM) to induce expression, which was shaken for another 20 h at 18 °C. Bacterial cells were collected by centrifugation at 4000 g, and resuspended in the lysis buffer that contained 50 mM Tris-HCl (pH 8.0), 150 mM NaCl and 20 mM imidazole. After that the bacterial cells were disrupted by ultra-sonication and cell debris was removed by centrifugation at 20,000 g for 1 h before the supernatant was loaded to Ni-nitrilotriacetic acid resin (GE Healthcare, USA) pre-equilibrated with the lysis buffer. Nonspecific contaminants were further removed by washing the resin with 10x bed volume of the wash buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 20 mM imidazole. For the next step the recombinant 3C proteins were eluted with the elution buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 300 mM imidazole. 6xHis-tag was cleaved by Thrombin (Sigma). In the end 3C protease was purified by gel-filtration (Superdex 200 column, GE Healthcare, USA) pre-equilibrated with the gel-filtration buffer that contained 25 mM HEPES (pH 8.0), 150 mM NaCl. Fractions containing 3C protease were pooled and concentrated to 5 mg/mL.
CVB3-3Cpro inhibition assay was taken as an example here and the assay for EV71-3Cpro was highly similar. The tested compounds were dissolved in DMSO stock solutions of 50 mM concentration. The total volume of the reaction buffer was 50 μL, which included 2.3 μM of CVB3-3Cpro, 25 μM of substrate and PBS-BSA buffer (PBS buffer × 2 with 2 mg/mL BSA and 5.85 mg/mL NaCl), and 0–200 μM of the tested compounds of double concentration gradient. Rupintrivir was selected as a control for comparison. The reaction was incubated at 37 °C for 60 min, followed by the fluorescence absorbance recorded by the microplate reader. The excitation/emission light was 340/490 nm. The IC50 values were calculated by Eq. (1). v = v0 / (1 + [I] /IC50)(1)
where v is the inhibited rate, v0 is the uninhibited rate, and [I] indicates the concentration of the inhibitor. All the assays were finished in triplicate.
4.6 Cell-based antiviral evaluations
The recombinant CVB3 and EV71 viruses were constructed using reverse genetic approaches from 12 plasmid system and restored in the Key Laboratory of Pathogenic Microorganisms and Immunology, Institute of Microbiology, Beijing. Good conditioned Hela cells were cultured in 12-well plates and incubated at 37 °C and 5 % CO2 environmental condition for 24 h, and then the culture medium was changed to DMEM medium for 1 h. After that the inhibitors were added to the medium in different concentrations and incubated for 40 min. Subsequently, the Hela cells were infected by CVB3 or EV71 with a multiplicity of infection (MOI) of 0.1. The positive control only contained the Hela cells and did not contain the inhibitor and the virus, and the negative control contained the Hela cells and the virus. A further 16 h incubation was performed until the negative control exhibited obvious cytopathic effect (CPE) and then the cell viability was measured using the MTS reagent. The IC50 values were calculated by equation (2). IC50 = [CL(IH −50) + CH(50 - IL)] / (IH - IL)(2)
where CL means low concentration and IL means inhibitory data at this concentration, while CH means high concentration and IH means inhibitory data at this condition. All the assays were evaluated in triplicate.
Viral titer was determined by the viral plaque assay. In detail, when Hela cells reached 95 % density, the cells were infected by different concentrations of CVB3 for 1 h. After washed by PBS, DMEM containing 1.5 % low melting point agarose and 2 mg/L pancreatic enzyme was added. After 3 d incubation, the viral plaque was counted.
The cytotoxicity assay was similar to the above cell-based antiviral experiment, and the only difference was that the Hela cells were not infected by the virus.
4.7 Molecular docking
The molecular structure of the 11f was constructed within Sybyl 7.3 (Tripos Inc., St Louis, MO), based on the crystal structure of 11 s. The structure model was assigned Gasteiger-Hückel charges and minimized by the Tripos force field. FlexX module was used to carry out the molecular docking task. The crystal structure of EV71-3Cpro in complex with rupintrivir (pdb code 3sjo) was retrieved from the pdb databank. All water molecules were removed, and hydrogen atoms were added in the standard geometry. Any amino acid residue within 5.0 Å of the location of rupintrivir was considered to be in the binding pocket. Cscore calculation was enabled and set to serial mode. The scoring procedures were performed using the default parameters in the program.
4.8 Comparative field analysis
All the 3D structures of the compounds were built based on the structure of 11f from molecular docking result and treated using same manner within Sybyl 7.3. All the parameters were used the default value within CoMFA module and the column filtering was set to 2.0 kcal/mol. The “leave-one-out” (LOO) cross validation method was tried to determine the optimum number of partial least squares (PLS) components. Finally, the non-cross validated method was used to derive the model to explain the quantitative structure-activity relationship.
CRediT authorship contribution statement
Jin-Yin Tang: Methodology, Investigation. Shengwang Dai: Methodology, Investigation. Xiaofang Wang: Visualization, Investigation. Mengting Zhang: Data curation, Resources. Jin-Rui Shi: Validation, Formal analysis. Yong-Xuan Hong: Formal analysis. Zhi-Juan Sun: Data curation. Huan-Qin Dai: Funding acquisition, Writing – review & editing, Project administration, Software, Supervision. Jian-Guo Wang: Conceptualization, Funding acquisition, Writing – original draft, Writing – review & editing, Project administration, Software, Supervision.
Acknowledgment
This research was financially supported by the National Key Research and Development Program of China (No. 2017YFE0108200), National Natural Science Foundation of China (No. 22277060 and No. 22277135), and Department of New Drug Registration, Hebei Immune Cell Application Engineering Research Center/Baoding Newish Technology Co., Ltd.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Imidazolium-containing diselenides for catalytic oxidations with hydrogen peroxide and sodium bromide in aqueous solutions. Tetrahedron. 2012;68(51):10476-10481.
- [CrossRef] [Google Scholar]
- Inhibition mechanism of SARS-CoV-2 main protease by ebselen and its derivatives. Nat. Commun.. 2021;12(1):3061.
- [CrossRef] [Google Scholar]
- Synthesis, characterization, and antioxidant activity of some ebselen analogues. Chem. A Eur. J.. 2007;13(16):4594-4601. https://
- [Google Scholar]
- Design, synthesis, cytotoxic evaluation and molecular docking of novel 1, 3, 4-thiadiazole sulfonamides with azene and coumarin moieties as carbonic anhydrase inhibitors. Arab. J. Chem.. 2023;16(8):104956.
- [CrossRef] [Google Scholar]
- Targeting SARS-CoV-2 proteases and Polymerase for COVID-19 treatment: state of the art and future opportunities. J. Med. Chem.. 2022;65(4):2716-2746.
- [CrossRef] [Google Scholar]
- Advances in research on 3C-like protease (3CLpro) inhibitors against SARS-CoV-2 since 2020. RSC Medicinal Chemistry. 2023;14(1):9-21.
- [CrossRef] [Google Scholar]
- A CDR-based approach to generate covalent inhibitory antibody for human rhinovirus protease. Bioorg. Med. Chem.. 2021;42:116219
- [CrossRef] [Google Scholar]
- Synthesis and investigation of the trypanocidal potential of novel 1,2,3-triazole-selenide hybrids. Eur. J. Med. Chem.. 2022;243:114687
- [CrossRef] [Google Scholar]
- Comparative molecular field analysis (CoMFA). 1. effect of shape on binding of steroids to carrier proteins. J. Am. Chem. Soc.. 1988;110(18):5959-5967.
- [CrossRef] [Google Scholar]
- Electrochemical behavior of pharmacologically interesting seleno-organic compounds - 2. 7-substituted-N-aryl-1,2-benzisoselenazol-3(2H)-one. Electrochim. Acta. 1991;36(3–4):631-637.
- [CrossRef] [Google Scholar]
- Predicting the cis-trans dichloro configuration of group 15–16 chelated ruthenium olefin metathesis complexes: a DFT and experimental study. Inorg. Chem.. 2009;48(22):10819-10825.
- [CrossRef] [Google Scholar]
- Structure-based optimization of ML300-derived, noncovalent inhibitors Targeting the severe acute respiratory syndrome coronavirus 3CL protease (SARS-CoV-2 3CLpro) J. Med. Chem.. 2022;65(4):2880-2904.
- [CrossRef] [Google Scholar]
- Structure of mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289-293.
- [CrossRef] [Google Scholar]
- Methyl 2-selenocyanatobenzoate. Acta Crystallogr. Sect. E: Struct. Rep. Online. 2002;58(11):o1298-o1300.
- [CrossRef] [Google Scholar]
- Emerging infectious disease issues in international adoptions: severe acute respiratory syndrome (SARS), avian influenza and measles. Curr. Opin. Infect. Dis.. 2004;17(5):391-395.
- [CrossRef] [Google Scholar]
- The emergence, genomic diversity and global spread of SARS-CoV-2. Nature. 2021;600(7889):408-418.
- [CrossRef] [Google Scholar]
- Enterovirus 71 and coxsackievirus A16 3C proteases: binding to rupintrivir and their substrates and anti-hand, foot, and mouth disease virus drug design. J. Virol.. 2011;85(19):10319-10331.
- [CrossRef] [Google Scholar]
- Introducing ferroelasticity into 1D hybrid Lead HalideSemiconductor by halogen substitution strategy. Small. 2023;19:2303127.
- [CrossRef] [Google Scholar]
- Luz, A.B.S., de Medeiros. A.F., Bezerra. L.L., Lima. M.S.R., Pereira. A.S., E. Silva, E.G.O., Passos, T.S., Monteiro, N.K.V., Morais. A,H,A., 2023. Prospecting native and analogous peptides with anti-SARS-CoV-2 potential derived from the trypsin inhibitor purified from tamarind seeds. Arab. J. Chem. 16 (8), 104886. 10.1016/j.arabjc.2023.104886.
- Ebselen, disulfiram, Carmofur, PX-12, tideglusib, and shikonin are nonspecific promiscuous SARS-CoV-2 Main protease inhibitors. ACS Pharmacology & Translational Science. 2020;3(6):1265-1277.
- [CrossRef] [Google Scholar]
- Chemical preparation, degradation analysis, computational docking and biological activities of novel sulfonylureas with 2,5-disubstituted groups. Pestic. Biochem. Physiol.. 2022;188:105261
- [CrossRef] [Google Scholar]
- Identification of high-affinity inhibitors of SARS-CoV-2 main protease: Towards the development of effective COVID-19 therapy. Virus Res.. 2020;288:198102.
- [CrossRef] [Google Scholar]
- Global emergence of enterovirus 71: a systematic review. BeniSuef University Journal of Basic and Applied Sciences. 2022;11(1):78.
- [CrossRef] [Google Scholar]
- Ebselen, a useful tool for understanding cellular redox biology and a promising drug candidate for use in human diseases. Arch. Biochem. Biophys.. 2016;595:109-112.
- [CrossRef] [Google Scholar]
- Ebselen: prospective therapy for cerebral ischaemia. Expert Opin. Invest. Drugs. 2000;9(3):607-619.
- [CrossRef] [Google Scholar]
- Recent development of 3C and 3CL protease inhibitors for anti-coronavirus and anti-picornavirus drug discovery. Biochem. Soc. Trans.. 2011;39(5):1371-1375.
- [CrossRef] [Google Scholar]
- Rational design of novel nucleoside analogues reveals potent antiviral agents for EV71. Eur. J. Med. Chem.. 2023;246:114942
- [CrossRef] [Google Scholar]
- Selenium and clinical trials: new therapeutic evidence for multiple diseases. Curr. Med. Chem.. 2011;18(30):4635-4650.
- [CrossRef] [Google Scholar]
- Facile synthesis, crystal structure, quantum calculation, and biological evaluations of novel selenenyl sulfide compounds as potential agrochemicals. Pest Manag. Sci.. 2023;79(5):1885-1896.
- [CrossRef] [Google Scholar]
- Redox reactions of organoselenium compounds: implication in their biological activity. Free Radic. Res.. 2021;55(6):641-654.
- [CrossRef] [Google Scholar]
- P1 glutamine isosteres in the design of inhibitors of 3C/3CL protease of human viruses of the Pisoniviricetes class. RSC Chemical Biology. 2023;4(8):533-547.
- [CrossRef] [Google Scholar]
- SARS-CoV-2 Main protease drug design, assay development, and drug resistance studies. Acc. Chem. Res.. 2023;56(2):157-168.
- [CrossRef] [Google Scholar]
- Comparison of Target pocket Similarity and Progress into Research on inhibitors of picornavirus 3C proteases. Chem. Biodivers.. 2023;20(3):e202201100.
- [Google Scholar]
- Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: chemical synthesis, biological evaluation, molecular docking and 3D-QSAR study. Eur. J. Med. Chem.. 2017;137:450-461.
- [CrossRef] [Google Scholar]
- Enterovirus A71 antivirals: past, present, and future. Acta Pharm. Sin. B. 2022;12(4):1542-1556.
- [CrossRef] [Google Scholar]
- A suitable drug structure for interaction with SARS-CoV-2 main protease between boceprevir, masitinib and rupintrivir; a molecular dynamics study. Arab. J. Chem.. 2023;16(9):105051
- [CrossRef] [Google Scholar]
- High-throughput screening of SARS-CoV-2 main and papain-like protease inhibitors. Protein Cell. 2022;14(1):17-27.
- [CrossRef] [Google Scholar]
- Synthesis and biological evaluation of berberine derivatives as a new class of broad-spectrum antiviral agents against coxsackievirus B. Bioorg. Chem.. 2020;95:103490
- [CrossRef] [Google Scholar]
- Zhang, L.X., Wang, J.G., Dai, H.Q., Zhang, X.M., Wang, Q.Q., Song, F.H., Shang, J., Wang, L.Q., Ma, R., 2016. Application of asymmetric aromatic disulfide compound as virus 3C protease inhibitor in preparing antiviral agent, CN 105640951 A.
- Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409-412.
- [CrossRef] [Google Scholar]
- Structure-directed expansion of biphenyl-pyridone derivatives as potent non-nucleoside reverse transcriptase inhibitors with significantly improved potency and safety. Chin. Chem. Lett.. 2023;34(12):108261
- [CrossRef] [Google Scholar]
- Synthesis and crystal structure of 2-phosphonoalkyl-1,2-benzisoselenazol-3(2H)-ones and their antitumor activities. Heteroat. Chem. 1999;10(3):247-254.
- [CrossRef] [Google Scholar]
- Antiviral activity of the HSP90 inhibitor VER-50589 against enterovirus 71. Antiviral Res.. 2023;211:105553
- [CrossRef] [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2024.105713.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary data 1
Supplementary data 1







