Translate this page into:
Identification and characterization of antibacterial metal complexes in ZiHua DiDing (Viola philippica) aqueous extract
*Corresponding authors: E-mail addresses: pengliang8036@163.com (L. Peng); 15295786071@163.com (L. Zhang).
-
Received: ,
Accepted: ,
Abstract
The excessive use of antibiotics leading to bacterial resistance poses a significant threat to global public health, particularly the serious problem of resistance in Staphylococcus aureus, which requires the development of new antibiotics. Metal complexes can combat bacterial resistance due to their unique mode of action and minimal tendency to induce resistance. Exploring new antibacterial agents from traditional Chinese medicine mainly focuses on organic components. We aimed to discover metal complexes with strong antibacterial activity and low drug resistance from ZiHua DiDing (Viola philippica) aqueous extract (ZDAE). We identified metal complexes in ZDAE and demonstrated that the addition of metal ions, particularly manganese (Mn2+), significantly affects the antibacterial activity of the extract. We also detected the heptasaponin manganese complex (ESM) in ZDAE. Subsequent synthesis and characterization of ESM showed that it is a 2:1 complex of esculetin (ES) and Mn2+, with significantly higher antibacterial activity against various bacteria. Activity against S. aureus reduced four-fold from 4800 μmol/L to 1200 μmol/L. The antibacterial mechanism of ESM against S. aureus includes inhibition of biofilm formation and protein synthesis, reduction of energy production, and disruption of cell wall structure. In addition, the minimum inhibitory concentration of ESM in the drug-containing medium remained stable for 20 generations, indicating that the extract of V. philippica produced potent metal complexes with enhanced antibacterial properties and lower ability to induce bacterial resistance.
Keywords
Antibacterial
Esculetin
Managanese
Viola philippica
Complex
1. Introduction
The overuse of antibiotics has led to the emergence of highly resistant and infectious superbugs, such as methicillin-resistant S. aureus, drug-resistant Mycobacterium tuberculosis, and drug-resistant Streptococcus pneumoniae. These pathogens have acquired resistance to most clinically used antibiotics, posing a formidable threat to human health. The substantial delay in developing new antibacterial drugs further worsens bacterial resistance. Moreover, drug development over the past 20 years has been limited to the structural modifications of existing antibiotics, with minimal advancement in novel chemical structures and mechanisms [1]. Developing antibacterial drugs with novel structures, superior antibacterial activity, and low propensity for inducing bacterial resistance is therefore essential.
Metal complexes have shown considerable promise in antibacterial therapy, especially in combating drug resistance, due to their unique advantages. Their antibacterial mechanisms are multifaceted. One key pathway involves metal complexes binding to essential metal ions required by bacteria, forming complexes that enter the bacterial cells through absorption, transport, and storage systems, thus disrupting vital bacterial processes. For instance, Southwell et al [2]. demonstrated that the formation of metal complexes could lead to a deficiency of essential metals, effectively inhibiting bacterial growth. Furthermore, complexes containing non-essential metals can compete with essential metals within bacterial cells. Over time, this competition depletes the essential metals, ultimately causing bacterial death [3]. Another crucial mechanism involves metal complexes enhancing their affinity for bacterial membranes through the chelation of metal ions, thereby promoting their entry into bacterial cells [4,5]. Metal complexes can specifically target bacterial enzymes, interfering with their fundamental physiological functions. For example, synthesized manganese complexes, even at low concentrations, significantly enhance antibacterial activity by disrupting the bacterial respiratory chain by releasing carbon monoxide, thereby impacting bacterial energy metabolism [6]. Moreover, the low toxicity and high therapeutic index of manganese complexes suggest favorable safety profiles. Similarly, copper complexes exhibit potent antibacterial activity by disrupting bacterial cell membranes, affecting intracellular metabolic processes, and altering ion balance and energy metabolism. Notably, copper complexes did not induce resistance in selection experiments [7]. In conclusion, metal complexes interfere with bacterial growth through various mechanisms, offering significant potential in addressing drug resistance and showing promise for future clinical applications in antimicrobial therapy. Traditional Chinese medicine (TCM) and its active ingredients demonstrate substantial inhibitory effects on bacterial growth by interfering with bacterial metabolism and suppressing bacterial toxins [8-11]. Organic TCM components containing coordinating groups, such as amino, hydroxyl, and carboxyl, can form complexes with trace metals [12]. These metal complexes are stable and exhibit anti-inflammatory, antibacterial, antitumor, and antioxidant effects compared with the organic components [13-15]. Moreover, they prolong the retention time of TCM in the body, enhancing the overall efficacy [16].
ZiHua DiDing, the dried whole herb of the Violaceus plant Viola yedoensis Makino, possesses antibacterial, anti-inflammatory, and antiviral activities [17] and is commonly used to treat colds, dermatitis, eczema, and other diseases. V. philippica primarily contains various flavonoids, coumarins, organic acids [18], and trace metal elements, such as zinc (Zn), calcium (Ca), copper (Cu), iron (Fe), and manganese (Mn) [19]. Its primary coumarin compound, esculetin (ES), exhibits antibacterial, anti-inflammatory, antioxidant, and antitumor activities [20,21]. ES has a large, conjugated structure with a catechol group, giving it the ability to readily form complexes with metal ions.
Currently, the search for new antibacterial drugs from TCM focuses primarily on its organic components, with minimal investigation into the metal complexes formed by these components and trace metal elements during the decoction process. Therefore, we aim to identify metal complexes from ZiHua DiDing (Viola philippica) aqueous extract (ZDAE), which has a low propensity for inducing bacterial resistance. The selected metal complexes were further investigated for their structural characterization and antibacterial mechanisms.
2. Materials and Methods
2.1. Materials and instruments
2.1.1. Materials and reagents
For this study, ZiHua DiDing was purchased from Huaishuntang Pharmacy (Bozhou, China). ES with a purity ≥98% was obtained from Alfa Biotechnology Co., Ltd. (Chengdu, China). Metal chlorides, including MnCl2, ZnCl2, MgCl2, CaCl2, CuCl2, and crystal violet, were purchased from Xilong Chemical Co., Ltd. (Guangdong, China). Chromatography-grade formic acid and methanol were also sourced from Aladdin Co., Ltd. (Shanghai, China), and mass spectrometry-grade acetonitrile was acquired from Fisher, USA. D001 resin was supplied by Suqing Water Treatment Engineering Group Co., Ltd. (Jiangyin, China). The antibiotics used, namely ampicillin, ciprofloxacin, clindamycin hydrochloride, and gentamicin, were provided by Energy Chemical Co., Ltd. (Shanghai, China). The alkaline phosphatase (AKP) assay kit was procured from Nanjing Jiancheng Bioengineering Institute (Nanjing, China). Tryptic soy broth (TSB) and agar powder were supplied by Beijing Aobox Biotechnology Co., Ltd. (Beijing, China). Strains of S. aureus (S. aureus, ATCC 29213), Escherichia coli (E. coli, ATCC 25922), and Pseudomonas aeruginosa (P. aeruginosa, ATCC 27853) were obtained from Forhigh Biotechnology Co., Ltd. (Hangzhou, China).
2.1.2. Instruments and equipment
The analytical instruments utilized included a 1260 HPLC System (Agilent Technologies, Santa Clara, CA, USA) and a Xevo G2-XS UPLC-Q-TOF MS (Waters Corp., Milford, MA, USA). Samples were weighed using an XS203S precision electronic balance (Mettler Toledo GmbH, Columbus, OH, USA). A Hei-VAP rotary evaporator (Heidolph Instruments, Schwabach, Germany) was used for solvent evaporation. UV-visible spectroscopic analyses were performed using a UV-2550 spectrophotometer (UV-Vis) (Shimadzu Corp., Kyoto, Japan). Thermogravimetric analysis was performed using a TG 209F3 Tarsus thermos-microbalance (NETZSCH, Bavaria, Germany). Elemental analysis was carried out using the 7700 inductively coupled plasma mass spectrometer (ICP-MS) (Agilent Technologies, Santa Clara, CA, USA). Sterilization processes were managed using an LDZH-50KESS vertical pressure steam sterilizer (Shenan Medical Instrument Factory, Shanghai, China), and samples were incubated in an SPX-150 biochemical incubator (Ke Heng Industrial Co., Ltd. Shanghai, China).
2.2. Determination of minimum bactericidal concentration (MBC)
The experimental bacteria were inoculated onto solid media and incubated at 37°C for 24 h. Following incubation, a single colony was transferred to a Tryptic soy broth (TSB) liquid medium and incubated with continuous shaking at 120 rpm until the logarithmic growth phase was reached. The culture was diluted with fresh TSB liquid medium to a concentration of 105 CFU/mL.
The bacterial suspension (1.60 mL) and the test solution (0.40 mL) were added to each well of a sterilized 24-well plate. Following an 18-h incubation at 37°C, 50 μL of the bacterial suspension was removed from each plate and diluted a thousand-fold using sterile phosphate buffered solution (PBS), and 50 μL of the diluted solution was applied to solid media and incubated for another 18 h at 37°C. The plates were then examined and photographed. Minimum bactericidal concentration (MBC) is the lowest drug concentration with no observed bacterial colony growth.
2.3. Confirmation of metal complexes in ZDAE
2.3.1. Determination of organic components in ZDAE using different solvents
Dried V. philippica herb (10 g) was cut into small pieces and immersed in 200 mL purified water at 25°C for 30 mins, followed by reflux extraction at 98°C for 90 mins. After hot filtration, the ZDAE was obtained and dried under reduced pressure.
Two corresponding quantities of the dried ZDAE were dissolved in equal quantities of methanol and 15% formic acid-methanol (V:V). The resulting solutions were diluted tenfold with the respective solvents and filtered through a 0.22 μm membrane. The sample was then put through High-performance liquid chromatography (HPLC) detection. An injection volume of 10 μL, detection wavelength of 327 nm, column temperature of 30, and flow rate of 1 mL/min was used for the HPLC system with a YMC Triart C18 column (4.6 × 250 mm, 5 µm). Table S1 provides an overview of the elution parameters. The experiments were conducted in triplicate for each group.
2.3.2. Influence of metal ions on ZDAE antibacterial activity
2.3.2.1. Changes in the antibacterial activity of ZDAE after removal of metal ions
ZDAE was prepared as described in Section 2.3.1. Subsequently, 50 mL ZDAE was recovered and added to a glass chromatography column containing D001 cation-exchange resin and eluted sequentially with 300 mL purified water, 50% methanol, and 80% methanol. All eluates were collected, combined, and dried under reduced pressure to obtain ZDAE with metal trace elements removed (ZDAE-D001). An equal volume of ZDAE was also dried under reduced pressure to obtain the dried ZDAE.
Metal trace elements in 1 g of dry ZDAE and ZDAE-D001 were determined by ICP-MS under the detection conditions of 1.50 kV RF power, 1.80 V RF matching, 1.50 L/min auxiliary gas flow, and 1.10 L/min spray gas flow. Equal mass ZDAE and ZDAE-D001 were weighed, and MBC was determined according to Section 2.2. The final concentrations of the samples are 100, 50, 25, 12.50, and 6.25 mg/mL.
2.3.2.2. Effects of different metal ions on ZDAE antibacterial activity
Six 30 mL portions of ZDAE were prepared and 0.015 mmol each of MnCl2, ZnCl2, CaCl2, MgCl2, and CuCl2 were added to five of these portions. The samples were refluxed at 98°C with magnetic stirring for 1 h. After drying under reduced pressure, they were dissolved in 2.5 mL of purified water containing 20% dimethyl sulfoxide (DMSO). The solutions were then diluted five-fold with culture medium as the highest concentration, with subsequent concentrations diluted two-fold sequentially. An equal volume of ZDAE without additional metal ions underwent identical treatment. The minimum bactericidal concentration (MBC) of each test sample was determined as described in Section 2.2.
2.3.3. Detection of metal complexes in ZDAE
ZDAE (30 mL) was dried under reduced pressure, dissolved, and diluted with 14 mL of methanol. Further, 2 mL of the solution was retraced, centrifuged for 2 mins at 5000 × g, and analyzed using UPLC-Q-TOF MS. For Ultra-performance liquid chromatography (UPLC) analysis, a Waters CORTECS® UPLC® T3 (2.1 × 100 mm, 1.6 µm) column was used with flow rate at 0.3 mL/min, column temperature of 30°C, and injection volume of 2 μL (Table S2). Electrospray ionization (ESI) was used in MS analysis in negative ion mode, with data acquisition in the continuum. The mass range was set at 100–1200 Dalton, with collision energy from 10 to 45 V. Additional parameters included a capillary voltage of 2.0 V, a cone voltage of 40 V, an ion source temperature set at 100°C, a desolvation gas temperature maintained at 500°C, and a desolvation gas flow rate of 800 L/h.
2.4. Structural characterization of the Heptasaponin manganese complex (ESM)
2.4.1. Determination of the optimal molar ratio of ES to Mn2+
A total of 0.060 mmol of ES was weighed and mixed with varying amounts of MnCl2 (0.0075, 0.010, 0.015, 0.030, 0.060, 0.120, 0.240, and 0.480 mmol) in round bottom flasks. Each mixture was combined with 10 mL of distilled water, heated at 98°C for 2 h, and dried under reduced pressure. The reaction products were dissolved in 2.5 mL of purified water containing 20% dimethyl sulfoxide (DMSO) using ultrasonication. Subsequently, 0.80 mL of the solution was sequentially diluted two-fold. An equal amount of MnCl2 was similarly processed. The MBC of each specimen was calculated as described in Section 2.2.
2.4.2. Structural characterization of ESM
ES and MnCl2 were made to react in a 2:1 molar ratio. Subsequently, 1 mL of the reaction mixture was mixed with 9 mL of distilled water and centrifuged for 5 min at 5000 × g. Further, 1 mL of the supernatant was extracted to perform a UV-Vis spectrophotometric scan, measuring absorption spectra from 190 to 500 nm. The UV-Vis absorption spectra of ES were scanned using the same methodology. Next, 3 mL of the supernatant was analyzed using UPLC-Q-TOF MS. The sample was introduced via infusion and detected in the negative ion Electrospray ionization (ESI) mode, covering a mass detection range of 100–1500. Additional parameters included a drying gas flow rate of 800 L/h and a desolvation gas temperature of 500°C. The residual reaction mixture was dried under reduced pressure and dispatched to the BL14W1 beamline at the Shanghai Synchrotron Radiation Facility for X-ray absorption fine structure (XAFS) spectroscopy analysis. The thermogravimetric curves of ES, ESM, and MnCl2 were determined using thermogravimetric analysis (TGA). The TGA analysis was carried out under nitrogen protection at 30–800°C and a heating rate of 10°C/min.
The hypothetical structure of ESM was determined using quantum calculations in Gaussian 9.0 software. The B3LYP theoretical calculations were employed, using SMD pseudopotentials for the central metal atom and def2-SVP basis sets for the surrounding atoms. The geometric structure of ESM was optimized, and its vibrational frequencies were analyzed to obtain an energy-minimum structure with no imaginary frequencies. The structure was visualized using Gauss-View 6.0.
2.5. Investigation of the antibacterial activity of ESM
2.5.1. Preparation of test solutions
ESM was synthesized with an ES: Mn2+ molar ratio of 2:1. A stock solution was prepared by dissolving the product in 2.5 mL of purified water containing 20% DMSO, which was then diluted to the required concentrations with purified water for further use.
2.5.2. Determination of the MBC of ESM
The MBC of ESM was determined as described previously. The final concentrations of ESM (based on the molar mass of ES) were 4.8, 2.4, 1.2, 0.6, 0.3, and 0.15 mmol/L. ES and MnCl2 were used as controls, with final concentrations of 4.8 and 2.40 mmol/L, respectively.
2.5.3. Bactericidal kinetics of ESM
Test solutions were added to bacterial suspensions (105 CFU/mL) to achieve final concentrations of ESM at 1/2×, 1×, 2×, and 4× MBC. The mixtures were incubated at 37°C. Subsequently, 50 μL bacterial suspension was removed from each well at 0, 1, 2, 3, 4, 5, 6, 8, 10, 12, 14, and 16 h, and diluted 100–10,000-fold with sterile Phosphate buffered saline (PBS). A 50 μL volume of the diluted solution was then plated on solid media and incubated at 37°C for 18 h before colony counts were recorded. S. aureus was treated with water containing 20% DMSO and was established as the control group.
2.5.4. Effect of ESM on bacterial cell walls
Alkaline phosphatase (AKP) is typically situated between the bacterial cell wall and cell membrane, contained within the bacterial structure, and released into the culture medium only when the integrity of the bacterial cell wall is compromised. Monitoring AKP activity in the bacterial culture medium provides insights into the integrity of the bacterial cell wall. S. aureus in the logarithmic phase was centrifuged at 5000 × g for 5 mins. The resulting bacterial pellet was washed thrice with PBS and resuspended in PBS to an OD600 of 0.3. ESM, ES, and water containing 20% DMSO were individually added to the bacterial suspension and incubated at 37°C for 2 h. The mixture was centrifuged at 5000 × g for 10 mins, and AKP activity in the supernatant was measured following the AKP kit instructions. The blank control group consisted of untreated S. aureus. The final concentrations of ESM were 1/2×, 1×, 2×, 4×, and 8× MBC, whereas the corresponding final concentrations of ES were 9.6, 4.8, 2.4, 1.2, and 0.6 mmol/L.
2.5.5. Effect of ESM on bacterial biofilm formation
S. aureus suspension (105 CFU/mL) of 1.6 mL in the logarithmic phase was added to sterile 24-well plates, followed by the addition of 400 μL of either ESM, ES, or Mn2+. The control group received 400 μL of physiological saline with 20% DMSO. The final concentrations of ESM, ES, and Mn2+ were 0.60 mmol/L (1/2 × MBC), 0.60 mmol/L, and 0.30 mmol/L, respectively. Following 48 h of incubation at 37°C, the supernatant was discarded, and the residual solution was washed with PBS twice to eliminate any leftover planktonic bacteria. Biofilms underwent 15 mins of methanol fixation, 24 h of air drying, and 15 mins of 0.1% crystal violet staining. The excess stain was removed using deionized water, and 33% (W/V) acetic acid was used to dissolve the crystal violet. The experiments were conducted in triplicates for each group. The rate of inhibition of bacterial biofilm formation was determined by measuring the absorbance (OD595 nm) using a microplate reader:
2.5.6. Evaluation of the biosafety of ESM
Fresh sterile rabbit blood was washed thrice with sterile PBS and centrifuged at 2000 × g for 2 mins to obtain red blood cells. Subsequently, 50 μL of rabbit red blood cells and 150 μL of the test solution were combined with 1 mL of PBS, incubated at 37°C for 30 mins, and centrifuged at 2000 × g for 2 mins. The absorbance of the supernatant was measured at 543 nm. The hemolysis rate was then determined, with Triton X-100 (0.1%) serving as the positive control and PBS as the negative control:
Hemolysis rate (%) = (AExperiment - ANegative control) / (APositive control - ANegative control) × 100%. The 50% hemolytic complement (HC50) was calculated using SPSS.
Human normal liver cells (L-O2) in the logarithmic growth phase were seeded into sterile 96-well plates at 200 μL per well and incubated overnight at 37°C. After discarding the original medium, each well received 200 μL of medium containing the test compounds (ESM and ES at final concentrations of 5.617, 1.872, 0.624, 0.208, and 0.069 mmol/L, and DMSO final concentration ≤ 1%). DMSO was employed as a negative control at a 1% volume fraction. Following a 24 h incubation period at 37°C, the medium was swapped out with 20 µL PBS containing 5 mg/mL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), and the mixture was incubated further for 4 h at 37°C. The medium was discarded, and 150 μL of DMSO was added to each well to dissolve the formazan crystals. Absorbance (A) at 490 nm was subsequently measured to calculate the cell inhibition rates of ESM and ES:
2.5.7. Induction of S. aureus resistance by ESM and antibiotics
The test solution (40 μL) and bacterial solution (160 μL) were added to each well of a sterilized 96-well plate. The minimum inhibitory concentration (MIC) was determined as the lowest concentration of the test solution that prevented observable turbidity after 18 h of incubation at 37°C. The corresponding value was recorded as the MIC for the first passage. For subsequent passages, bacterial suspension from the well with ½ × MIC from the previous passage was cultured to the mid-logarithmic phase, and the suspension (105 CFU/mL) was added to the test solution and incubated at 37°C for 18 h. The MIC was recorded for each passage up to the 20th passage. In the first passage, the initial concentration of ESM (based on the molar mass of ES) was 4.80 mmol/L. The maximum final concentrations of the positive control antibiotics (ciprofloxacin, gentamicin, ampicillin, and clindamycin) were 40 μg/mL.
2.5.8. Multi-omics analysis of the antibacterial mechanism of ESM
S. aureus in the logarithmic phase was diluted with fresh Tryptic soy broth (TSB) liquid medium to a concentration of 5 × 105 CFU/mL. The diluted bacterial suspension was treated with ESM (final concentration 0.90 mmol/L) and ES (final concentration 0.90 mmol/L) using DMSO (final concentration 0.75%) as a blank control. The suspensions were incubated at 37°C for 5 h, followed by centrifugation at 5000 × g for 10 mins at 4°C. Subsequently, the bacterial pellets were collected, flash-frozen in liquid nitrogen, and stored at −80°C. Transcriptomic and proteomic analyses of the samples were performed by Beijing Novogene Co., Ltd.
2.5.9. qPCR validation
In this experiment, three groups were established: blank, ES, and ESM, each consisting of three parallel replicates. Total RNA was extracted from S. aureus samples using the Transzol Up Plus RNA Kit, following the manufacturer’s protocol. All samples were cultured under conditions identical to those used for the transcriptomic samples. Next, a reverse transcription reaction was performed in a 20 µL reaction mixture, which included 3 µL of total RNA/mRNA, 4 µL of 5× EasyScript® All-in-One SuperMix for quantitative polymerase chain reaction (qPCR), and 1 µL of gDNA Remover. The reverse transcription procedure followed a two-step process: incubation at 42°C for 15 mins, followed by inactivation at 85°C for 5 seconds to generate cDNA templates of the target RNA. Subsequently, real-time quantitative PCR was conducted using the synthesized cDNA templates. The amplification reaction mixture consisted of 10 µL of 2× Real PCR-EasyTM-MIX-SYBR, 2 µL of RNase-free ddH2O, 1 µL of forward and reverse primers, and 6 µL of cDNA template. The PCR cycling conditions were as follows: an initial denaturation at 95°C for 2 mins, followed by 40 amplification cycles, each consisting of denaturation at 95°C for 5 seconds, annealing at 60°C for 34 seconds, and extension at 95°C for 15 seconds. After the experiment, the amplification and melting curves were analyzed to evaluate the efficiency and specificity of the amplification reaction. The relative mRNA expression levels were calculated using the 2–△△Ct method. The specific primer sequences have been shown in Table 1.
| Gene name | Primer name | Primer sequence (5’-3’) | Product length (bp) |
|---|---|---|---|
| 16S | 16S F | AGACCAAAACGCTGATGTAAGTCA | 230 |
| 16S R | TGCAAATCGTAACATCAACTGGCA | ||
| thrs | thrs F | TGCGCGCCAATTACAAGATGAAT | 195 |
| thrs R | ACTGTTTCTTGGTCTTGCGATCCA | ||
| norB | norB F | TGGGAAAGATAGACAACGCGCTT | 73 |
| norB R | AAATGAACAAACACCAGAGCCGC | ||
| lytM | lytM F | AACGACAAACACCCAACAAGCAC | 248 |
| lytM R | TCACATATGTTTGGCCGCTTCCA | ||
| adhP | adhP F | TGGTAAAGTCATCGAAGTTGCGGA | 268 |
| adhP R | AGTTGTCACACCTGCGCATGTAA |
2.6. Statistical analysis
SPSS Statistics 27 (IBM Corporation, Armonk, USA) was used for statistical analysis. A t-test was performed on the antibacterial activity-related data of ESM and ES. After running each experiment three times, parallel experimental data is obtained. The significant differences between groups are represented by assigning letters to the data of different groups. The same letter indicates no significant differences between groups, while different letters indicate significant differences between groups. The reference group for both transcriptomics and proteomics included S. aureus subsp. aureus NCTC8325.
3. Results and Discussion
3.1. Identification of antibacterial metal complexes in ZDAE
3.1.1. Comparative analysis of ZDAE composition in different solvents
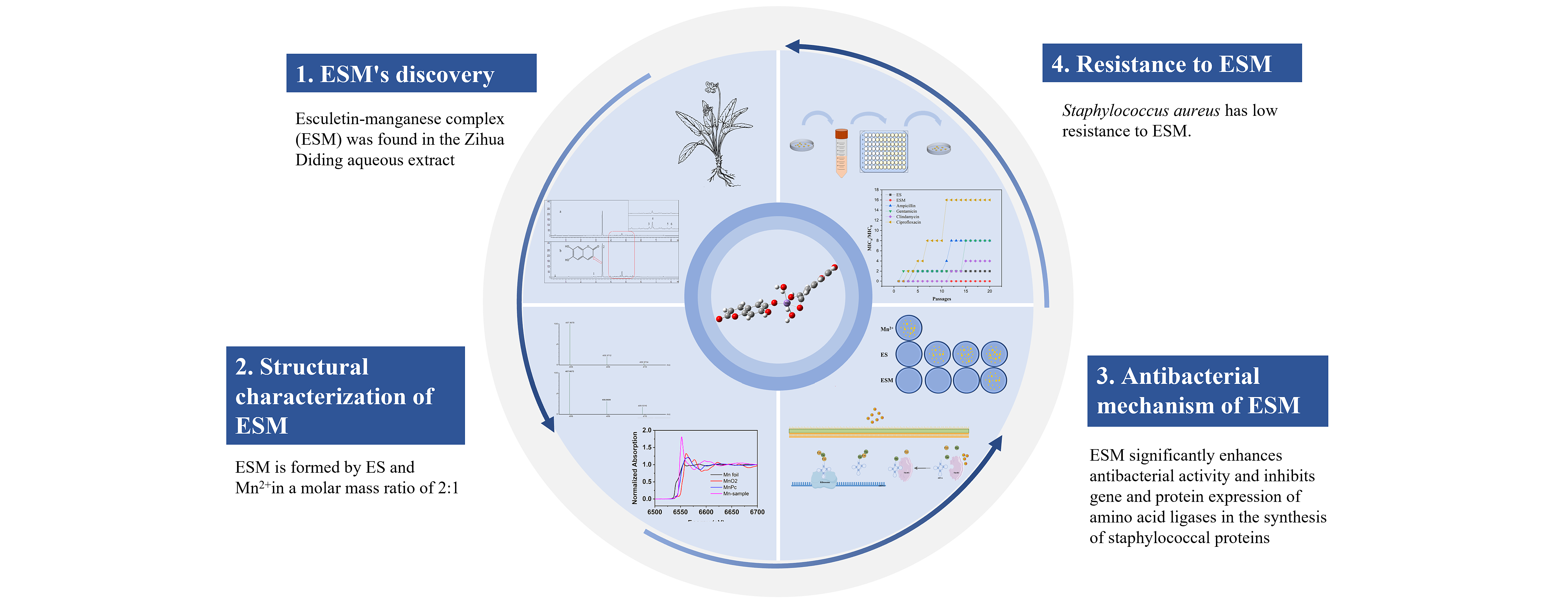
In Figure 1, subpart Figure 1a shows the HPLC chromatograms of dried ZDAE dissolved in 15% formic acid-methanol and methanol. The compounds in dried ZDAE samples dissolved in 15% formic acid-methanol and methanol were nearly identical with similar retention times, suggesting that adding the right amount of formic acid to the solvent did not change the composition of compounds in the samples. The peak areas of all compounds in samples dissolved in 15% formic acid-methanol were notably higher than those in methanol (Figure 1b). Specifically, the peak areas of compounds labeled peaks 2 and 4 were amplified by 49.84% and 130.54%, respectively. Comparison with reference standards revealed that the compound with the highest content (represented by peak 2) was ES.
- Antibacterial metal complexes in ZDAE. (a) HPLC chromatograms of organic content of dried ZDAE samples dissolved in different solvents (327 nm) - (i) organic content of samples dissolved in methanol and (ii) organic content of samples dissolved in 15% methanol acetic acid. (b) Changes in organic content of ZDAE dissolved in methanol and 15% formic acid methanol. (c) Decline in metal element content in ZDAE after treatment with D001 resin. (d) During the D001 cation exchange resin treatment, the antibacterial activity of ZDAE decreased (from left to right, final concentrations of ZDAE are 100, 50, 25, 12.5, and 6.25 mg/mL). (e) Ultra-performance liquid chromatography quadrupole time-of-flight mass spectrometer (UPLC-Q-TOF MS) results of the C18H10O8Mn complex in ZDAE.
3.1.2. Effects of metal ions on the antibacterial activity of ZDAE
3.1.2.1. Changes in the metal ion content and antibacterial activity of ZDAE post-metal ion removal
The inductively coupled plasma mass spectrometer (ICP-MS) analysis results (Figure 1c) showed that after treatment with D001 resin, the content of all 14 metal ions in ZDAE decreased. Especially essential metal elements such as Ca, K, Mg, Mn, and Zn, were lowered by 90.39%, 99.58%, 98.49%, 96.38%, and 58.04%, respectively. Further experimental results (Figure 1d) showed that the minimum inhibitory concentration of ZDAE dry extract was 25 mg/mL, while the MBC of ZDAE-D001 after treatment increased to 100 mg/mL. This indicates that the D001 resin treatment leads to a decrease in the antibacterial activity of ZDAE, which may be related to a significant reduction in the aforementioned metals.
3.1.2.2. Identification of metal ions enhancing antibacterial activity in ZDAE
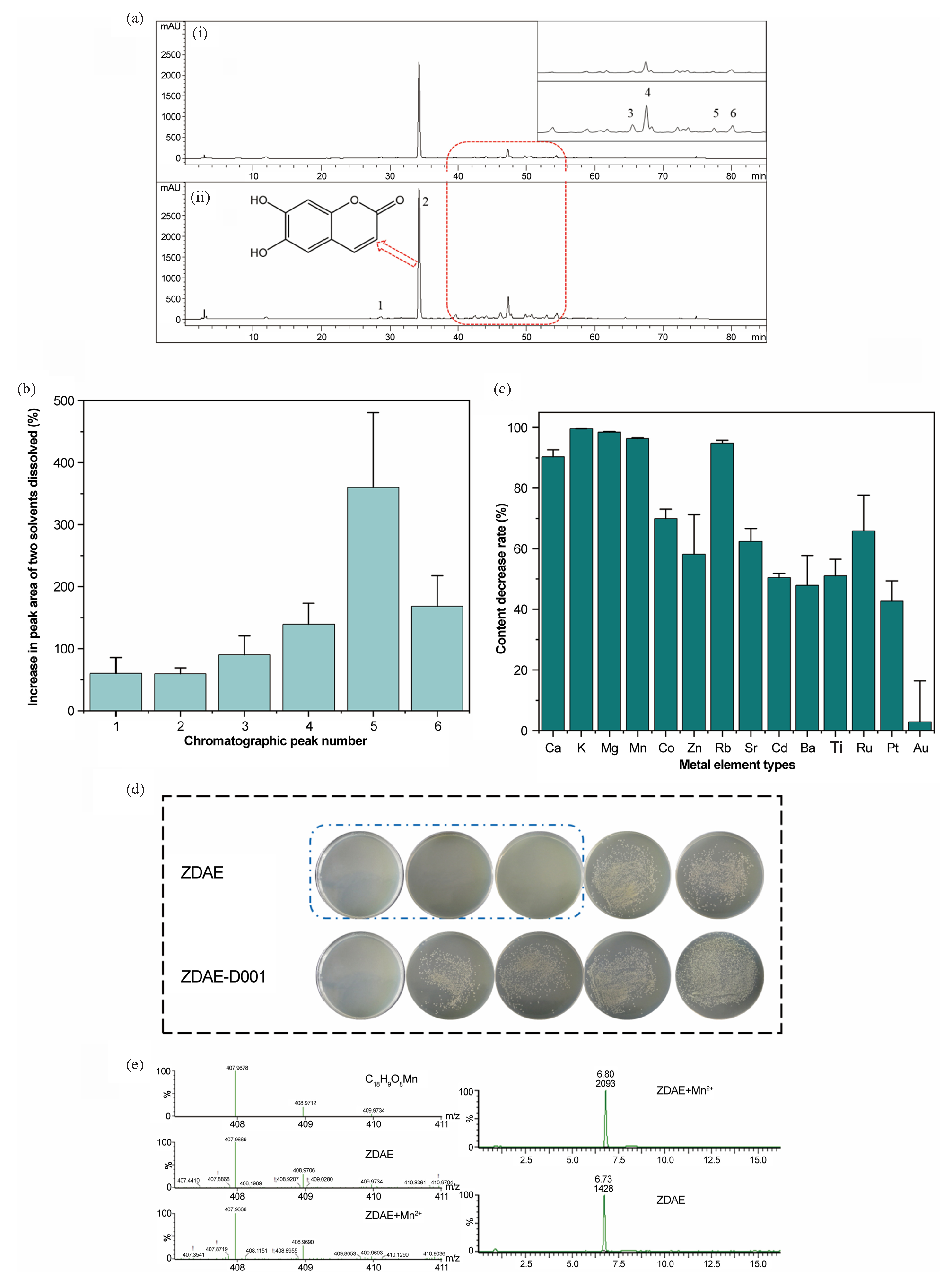
The metal ion addition method was used to identify metal ions that enhance the antibacterial potency of ZDAE. Five essential metal elements, Ca2+, Mg2+, Zn2+, Cu2+, and Mn2+, were individually introduced into ZDAE, and the corresponding changes in antibacterial activity were monitored (Figure 2). Initially, metal ions at a final concentration of 1.2 mmol/L lacked antibacterial efficacy. However, Mn2+ notably augmented the antibacterial activity of ZDAE. Zn2+ also enhanced the activity to some extent (at 12.5 mg/mL, bacterial count was significantly lower than in ZDAE). In contrast, Ca2+ and Mg2+ did not significantly alter inhibitory activity, whereas Cu2+ addition notably reduced inhibitory potential.
- The antibacterial activity of ZDAE to different metal ions. Metal ions (Mn2+, Zn2+, Ca2+, Mg2+, Cu2+) were individually added to ZDAE to assess their impact on antibacterial activity. From left to right, the final concentrations of five metal ions are 1.20, 0.60, 0.30, and 0.15 mmol/L, respectively, and the final concentrations of ZDAE are 50, 25, 12.50, and 6.25 mg/mL.
3.1.2.3 Detection of metal complexes in ZDAE
Using UPLC-Q-TOF MS in negative ion mode, a compound with a molecular weight of 407.9668 was detected in ZDAE. Theoretical calculations and comparison with isotope distributions (Figure 1e) revealed this compound to be the quasi-molecular ion of C18H9O8Mn, that is, a metal complex formed by ES and Mn2+ in a molar ratio of 2:1. To confirm this, Mn2+ was added to ZDAE, and UPLC-Q-TOF MS was repeated under the same conditions, revealing a considerable increase in the peak area of the compound (Figure 1e).
3.2. Synthesis and structural characterization of ESM
3.2.1. Determination of the optimal molar ratio of ES to Mn2+
The inhibitory effects of ESM at different ratios on S. aureus (Figure S1). ES alone, with an MBC of 4.80 mmol/L, exhibited some antibacterial activity. Following the reaction with Mn2+, the MBC decreased to between 2.4 and 1.2 mmol/L, indicating enhanced antibacterial activity. These results suggest that Mn2+ augments the antibacterial effect of ES, possibly due to alterations in their chemical properties resulting from the interaction between Mn2+ and ES, thereby amplifying the inhibitory effect on S. aureus. The highest antibacterial activity occurred when the ratio of ES to Mn2+ was 2:1. This ratio likely results in the formation of the most stable complex between ES and Mn2+, maximizing the antibacterial activity. Lower Mn2+ concentrations may reduce ESM complex formation, whereas higher concentrations may result in less stable alternative coordination forms, thereby diminishing antibacterial activity.
3.2.2. UV-visible absorption spectra of ESM
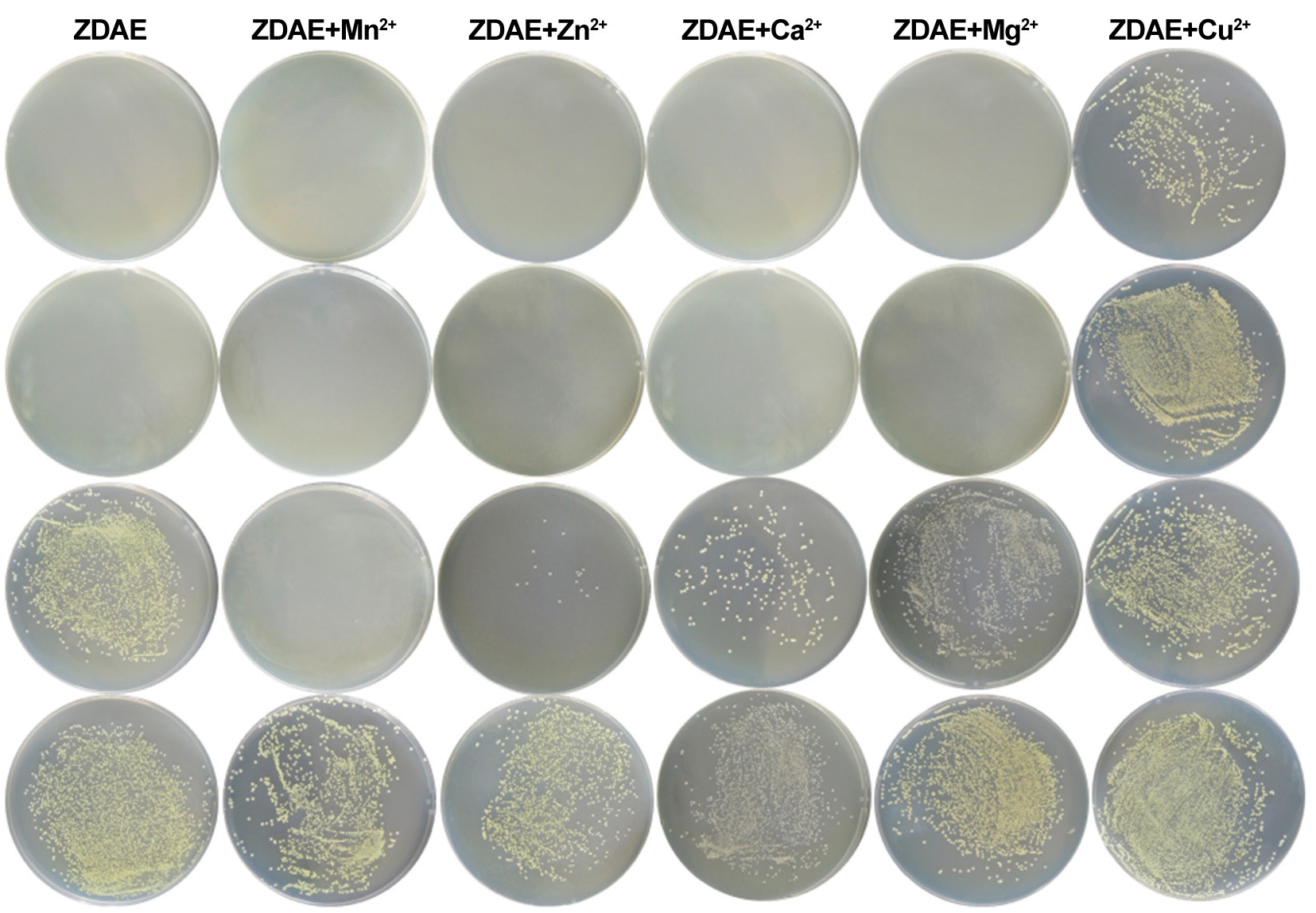
ES displayed two prominent absorption peaks at 205 and 345 nm, corresponding to the lactone group and 2-hydroxycinnamoyl group, respectively (Figure 3a). The UV-visible absorption spectra of ESM and ES in water have equivalent peak forms and a significant overlap. However, compared to that of ES, the absorbance of ESM was lower at 203 and 344 nm.
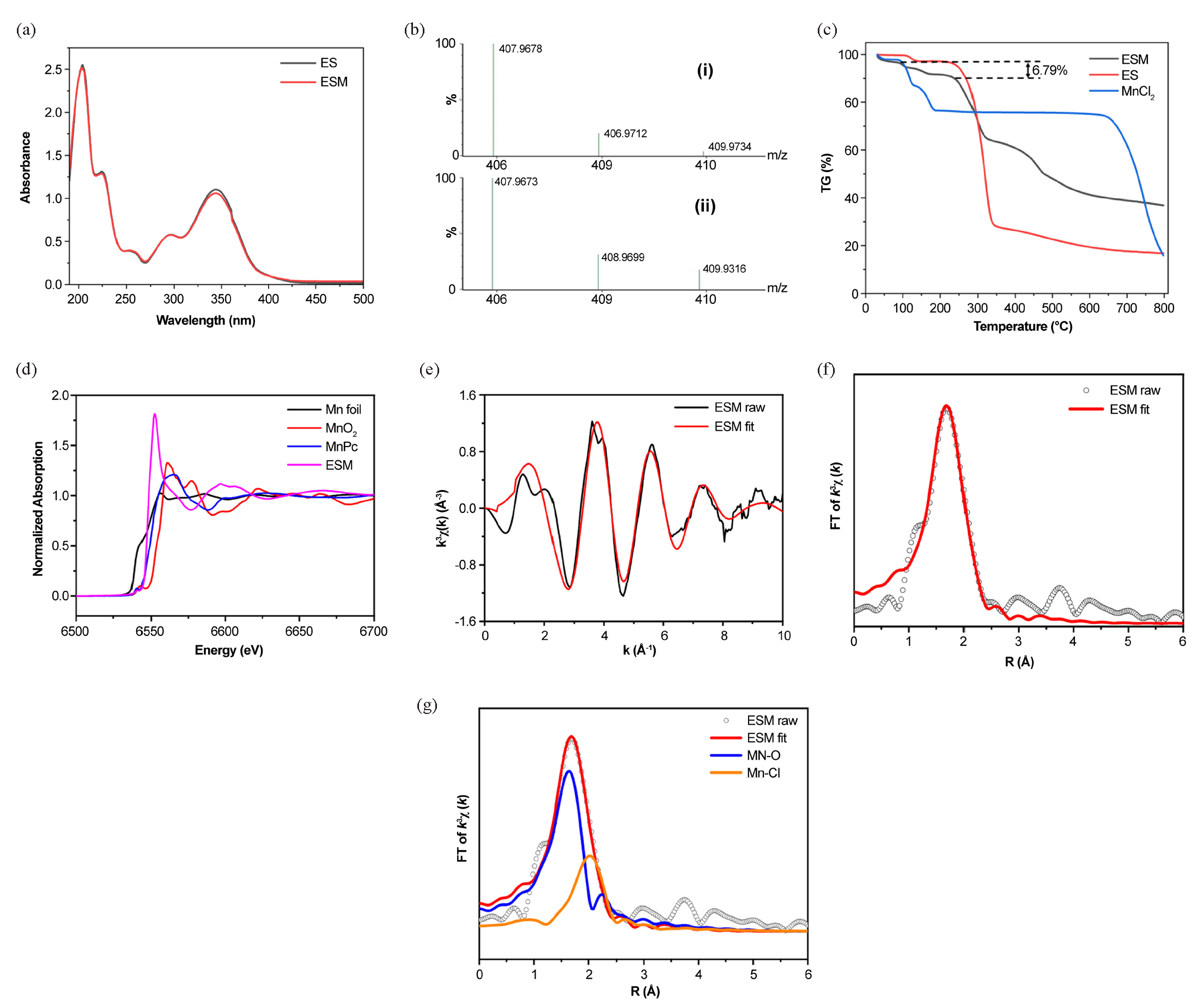
- Structural characterization of ESM. (a) UV-visible absorption spectra of ES and ESM in water. (b) Theoretical (i) and experimental (ii) isotope distributions of C18H9O8Mn. (c) Thermogravimetric curves of ES, ESM, and MnCl2. (d) EXAFS spectra of ESM, MnO2, MnPc, and Mn foil. (e) K-space fitting of ESM. (f) R-space fitting of ESM. (g) Coordination bonds of Mn in ESM.
3.2.3. TOF MS results of ESM
The supernatant from the reaction of ES with Mn2+ in hot water was analyzed using TOF MS (Table 2). Under the ESI negative ion mode, the partially reacted ES (m/z=177.0188) and a compound with an m/z of 407.9678 was detected. Precise molecular weight determination and isotopic distribution comparison (Figure 3b) identified the compound as the [M-H]- quasi-molecular ion of the ES and Mn2+ complex in a 2:1 molar ratio.
| Sample name | m/z | Molecular formula | Possible structure | Comparison between theoretical and experimental isotopic distributions | |
|---|---|---|---|---|---|
| Theoretical value | Measured value | ||||
| ES | 177.0188 | 177.1920 | C9H5O4 | [ES-H]- | |
| ES-Mn2+ | 407.9678 | 407.9673 | C18H9O8Mn | [2(ES-H)+Mn2+-H]- | Figure 3b (i,ii) |
3.2.4. TGA results of ESM
Based on the speculated structure of ESM from its mass spectrometry, it is inferred that ESM may contain bound water molecules due to the synthesis method involving aqueous reaction [22]. Therefore, TGA was conducted to determine the bound water content in ESM.
The thermogravimetric curves of ES and ESM were measured from 30 to 800°C under a flowing N2 atmosphere at a rate of 20 mL/min (Figure 3c). ES exhibited two distinct weight loss steps: one with a 2.8% weight loss rate between 100 and 150°C, and another with a 67% weight loss rate between 210 to 330°C. Overall, ES exhibited a total weight loss rate of 83.19%, leaving a residue of 16.81%. In contrast, ESM showed three weight loss steps. The first step, with a weight loss rate of 6.79%, occurred at 50–230°C, likely due to the loss of two crystal water molecules (theoretical weight loss rate of 7.20%). The second step, with a weight loss rate of 27.3%, occurred between 210 and 330°C. The third step, with a weight loss rate of 24.8%, was observed between 360 and 700°C. As the temperature increased, the complex underwent continuous degradation, with the ligands gradually decomposing and the framework collapsing, forming chlorinated and manganese-containing oxides.
3.2.5. X-ray absorption fine structure (XAFS) of ESM
The synchrotron radiation absorption spectroscopy (XAFS) test data can be divided into three parts: pre-edge, near edge (XANES), and extended edge (EXAFS). Therefore, normalized Mn K-edge XANES spectra of standard Mn foil, MnO2, manganese phthalocyanine (MnPc), and ESM were collected. As shown in the XANES spectrum (Figure 3d), the ESM edge spectrum exhibits a pre-edge transition at 6542 eV, similar to MnPc (6542 eV), but distinct from Mn foil and MnO2. These observations suggest that the Mn in ESM is in the +2 oxidation state. The local atomic structure and relative bond lengths with regard to the absorbing atom were determined by fitting the EXAFS data in K and R space (Figure 3e-g). Table 3 presents the structural characteristics and shell parameters. The results suggest that in ESM, Mn coordinates with O atoms in the ES structure to form Mn-O bonds. Within the complex, Mn is in the +2 oxidation state and exhibits an Mn-O4 coordination form.
| Samples | Path | CN[a] | R(Å)[b] | σ2(´10-3Å [2])[c] | ΔE(eV)[d] | R factor[e] |
|---|---|---|---|---|---|---|
| Mn sample | Mn-O | 4.1±0.30 | 2.19±0.02 | 7.4±0.80 | 0.7±8.20 | 0.02 |
| Mn-Cl | 1.6±0.40 | 2.51±0.02 | 9.8±2.30 |
Wavelet transform analysis of the sample’s R and K space contour maps (Figure S2) revealed maximum intensities of ESM near 4.56 Å-1 and 1.67 Å, corresponding to smaller atomic serial numbers. This finding is consistent with the bond lengths determined in R space, confirming the presence of Mn-O bonds in ESM.
3.2.6. Quantitative calculation of ESM
As direct determination of the exact structure of ESM using instrumentation was not feasible, quantitative calculations were utilized to identify the most likely structure of ESM. First, the geometric structure of ES was optimized, and its electron cloud density was analyzed. The results (Figure S3) indicated that electron density was primarily concentrated on the phenolic hydroxyl groups at positions 6 and 7, suggesting that Mn2+ in ESM likely coordinates with the oxygen atoms of these hydroxyl groups.
When ES and Mn form a complex at a 2:1 ratio, there are three possible coordination modes (Table 4). Based on this, the structure of ESM was quantitatively calculated. All atoms except the central metal Mn atom were set with the def2-SVP basis, whereas the Mn used the Stuttgart/Dresden effective core potential basis set. Solvation effects were accounted for during energy calculations, with parameters set according to reaction temperature and time to optimize the structure of ESM at a 2:1 molar ratio. The coordination of Mn2+ with the oxygen atoms on the 6- and 6ʹ-hydroxyl groups of ES (Compound A) resulted in the lowest binding energy and bond lengths that closely matched the XANES results (Table 4; Figure S4). Therefore, it is speculated that the optimal structure of C18H10O8Mn is where Mn2+ is coordinated with the oxygen atoms on the 6- and 6ʹ-hydroxyl groups of ES, along with the oxygen atoms of two water molecules (Figure 4).
| Compound | Binding energy/Hartree | Bond | Bond length (Å) |
|---|---|---|---|
|
A |
-1.071 | Mn-O | 1.981 |
| 2.007 | |||
| Mn-O-H2 | 2.101 | ||
| 2.138 | |||
|
B |
-1.065 | Mn-O | 1.968 |
| 2.015 | |||
| Mn-O-H2 | 1.664 | ||
| 1.664 | |||
|
C |
-1.041 | Mn-O | 1.904 |
| 2.040 | |||
| Mn-O-H2 | 1.937 | ||
| 2.664 |
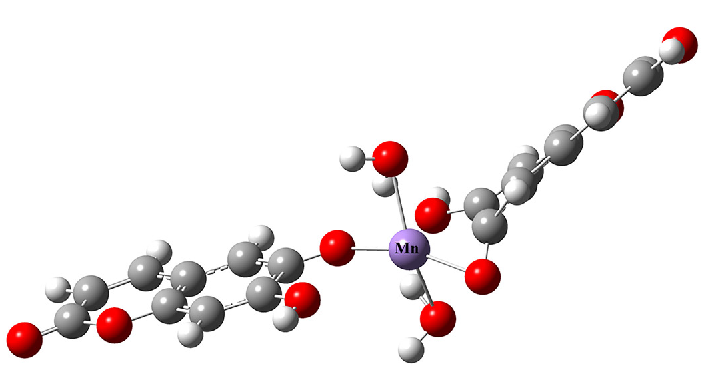
- Optimal structure of ESM (C18H10O8Mn). (White dots denote H atoms, gray dots denote C atoms, red dots denote O atoms, and purple dot denotes Mn atom.)
3.3. Antibacterial activity of ESM
3.3.1. MBC of ESM
Mn2+ alone did not exhibit significant antibacterial activity against S. aureus, Escherichia coli, and P. aeruginosa, even at its maximum concentration (Figure S5). In contrast, ESM demonstrated significantly enhanced antibacterial activity compared with ES against all three bacterial strains. Specifically, ESM exhibited four times the antibacterial activity of ES against S. aureus and twice the effectiveness against E. coli and P. aeruginosa.
3.3.2. Bactericidal kinetics of ESM
The bactericidal rate of ESM depends on both concentration and time (Figure 5a). ESM at a concentration of ½ × MBC demonstrated no inhibitory effect on S. aureus. Conversely, ESM with a concentration equal to MBC showed no significant difference in bacterial count in the culture medium compared with that of the control group from 0 to 7 h of exposure. However, the bacterial count began to rapidly decrease after 7 h, and by 14 h, all S. aureus cells were eradicated. At 2× and 4× the MBC concentrations of ESM, maximum bacterial counts were observed at 4 and 2 h of exposure, respectively. By 8 and 5 h, respectively, all bacteria were eliminated.
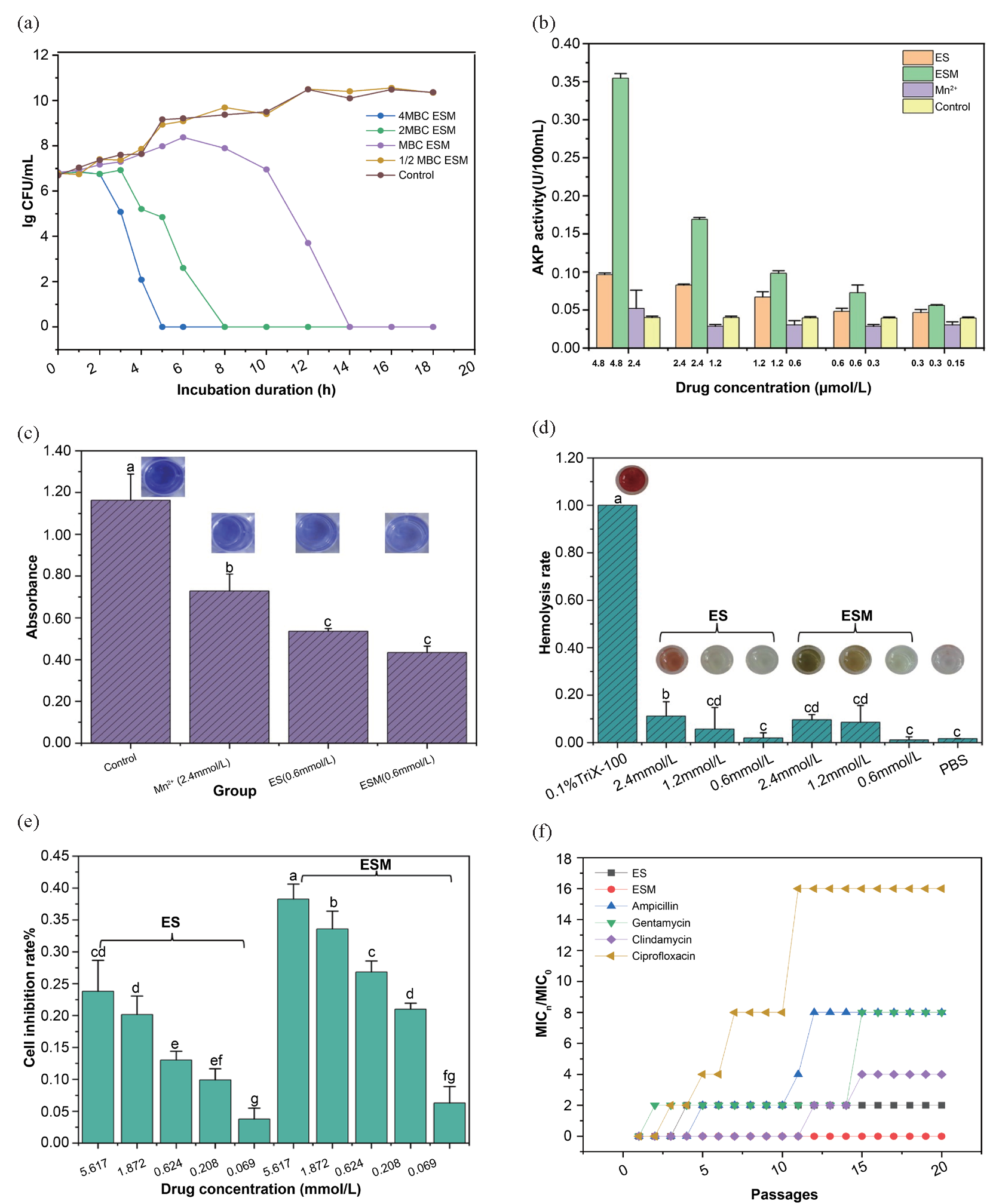
- Results of antibacterial activity test for ESM. (a) Bactericidal kinetics of ESM against S. aureus (with an MBC at 1.20 mmol/L); (b) AKP activity in the culture medium after treatment with ES and ESM; (c) Inhibitory effects of ES and ESM on the formation of S. aureus biofilm. Note: The concentration of ESM, ES, and Mn2+ is 0.60 mmol/L, 0.60 mmol/L, and 2.40 mmol/L, respectively; (d). Hemolytic effects of ES and ESM on rabbit erythrocytes; (e) Inhibitory effects of ES and ESM on the proliferation of L-O2 cells; (f) S. aureus resistance to ES, ESM, and four antibiotics (In the data analysis of this article, the abc non-parametric annotation method is used to analyze the differences between multiple sample groups through multiple comparisons using ANOVA (analysis of variance). Each group is assigned a letter marker (e.g., a, b, c, d). If the mean difference between the two groups is significant, their labeling will be different. If there is no significant difference, their labels are the same.) (AKP: Alkaline phosphatase; ES: Esculetin; ESM: Esculetin-manganese complex.)
3.3.3. Effect of ESM on the cell wall
Mn2+ at all tested concentrations did not augment AKP activity in the culture medium (Figure 5b), suggesting that Mn2+ alone did not disrupt the bacterial cell wall. In contrast, ES and ESM treatments increased AKP activity in the culture medium, with higher concentrations showing more pronounced effects. At equivalent concentrations, the ESM group exhibited substantially greater extracellular AKP activity than the ES group.
3.3.4. Inhibitory effect of ESM on biofilm formation
As ½ × MBC of ESM did not inhibit the growth of S. aureus, its inhibitory effect on the formation of S. aureus biofilm was assessed (Figure 5c). Compared with the control group, ES and Mn2+ showed a certain degree of inhibitory effect on biofilm formation, with an inhibition rate of 53.5% ± 6.4% for ES and 62.5% ± 2.4% for ESM. ESM exhibits a significantly stronger inhibitory effect on biofilm formation.
3.3.5. Biosafety of ESM
To evaluate the hemolytic activity of ESM, sterile rabbit blood was used, and the HC50 value of ESM against rabbit erythrocytes was calculated to assess the biosafety of ESM. ES and ESM exhibited significantly lower hemolytic activity compared to that of the positive control, 0.1% Triton X-100 (Figure 5d). Moreover, ESM generally demonstrates lower hemolytic activity than ES at equivalent concentrations, with a hemolysis rate of 10.62% at a concentration of 2× MBC compared to 16.97% for ES at the same concentration. The calculated HC50 value for ESM exceeded 200 μg/mL. Therefore, ESM demonstrated superior biocompatibility and exerted minimal toxicity on normal red blood cells.
To assess the toxicity of the compound in normal cells, a thiazolyl blue (MTT) assay was conducted on normal liver cells (L-O2). At a concentration of 1.20 mmol/L, ESM exhibited a cell inhibition rate of 23.17%, whereas ES showed 12.65% inhibition at the same concentration (Figure 5e). The IC50 value of ESM for L-O2 cells exceeded 100 μM, suggesting minimal inhibitory effects on the proliferation of normal liver cells and low toxicity.
3.3.6. Induction of S. aureus resistance by ESM and antibiotics
We found that S. aureus developed resistance to gentamicin, ciprofloxacin, ampicillin, and clindamycin after 2, 3, 5, and 12 passages, respectively (Figure 5f). Following 20 passages, the MIC of the four antibiotics increased 16-fold (ciprofloxacin), 8-fold (ampicillin and gentamicin), 4-fold (clindamycin), and 2-fold (ES), whereas the MIC of ESM remained unchanged. These results imply that the development of ESM resistance in S. aureus is less likely.
3.4. Potential antibacterial mechanisms of ESM
3.4.1. Transcriptomic analysis
A total of 694 differentially expressed genes in the ESM treatment group were compared with the ES treatment group (Figure S6a and S6b), of which 341 were significantly upregulated, whereas the remaining 353 were significantly downregulated.
Gene Ontology analysis revealed that all three categories of biological functions (namely, biological process [BP], cellular component, and molecular function [MF]) were enriched in the differentially expressed genes between ESM and ES treatments. Following ESM treatment, metabolic activities, cationic antimicrobial peptide resistance, and aminoacyl-tRNA production were among the pathways KEGG analysis identified (Figure S6c and S6d).
3.4.2. Proteomics analysis
In the ESM treatment group, 158 proteins showed significant changes in expression levels compared to the ES treatment group, with 33 proteins significantly upregulated and 125 proteins substantially downregulated (Figure S7a). Comparative analysis indicated distinct differences in protein expression levels among the ESM, ES, and control groups (Figure S7b).
Significant enrichments in cellular amino acid metabolism, siderophore biosynthesis, and carboxylic acid activities in the BP category were observed in the ESM treatment group compared to those of the ES group. The MF category showed enrichments in the hydrolase and iron-sulfur binding. Differentially expressed proteins in ESM were associated with pathways, such as glycerol metabolism and S. aureus infection, whereas downregulated proteins were associated with different metabolic pathways and secondary metabolite biosynthesis (Figure S7c and d).
3.4.3. Integrative analysis of transcriptomics and proteomics
The integrative analysis identified 37 differentially expressed proteins (7 upregulated, 30 downregulated), the speculated protein expression trends of which were consistent with their corresponding genes. Functional analysis of genes and proteins showing similar expression patterns and related functions revealed key mechanisms underlying the antibacterial activity of ESM (Table 5).
| Mode of action | DEP | Protein function | Transcriptome expression | Protein expression |
|---|---|---|---|---|
| Cell wall | atl | Peptidoglycan amidase activity, autolysis-related gene. The expression of autolysin protein Atl does not directly mediate the initial adhesion of bacteria; instead, their primary function lies in degrading the peptidoglycan layer of the cell wall [ 23, 24] | Up | Up |
| lytM | lytM is an autolysin produced by S. aureus, belonging to the peptide chain endonuclease. It is secreted outside the cell during bacterial growth and specifically hydrolyzes the peptide bond between glycine and peptidoglycan in the cell wall of S. aureus, leading to bacterial lysis. [ 25- 27] | Up | Up | |
| clfA | Cell wall protein, virulence factor. Can mediate the adhesion of S. aureus to matrix proteins, thereby infecting the host [ 28] | Down | Down | |
| aaa | Peptidoglycan hydrolase is involved in membrane division during cell division [ 29] | Down | Up | |
| Drug resistance-related | manA | Phosphomannose isomerase is involved in carbohydrate and biofilm synthesis [ 30] | Down | Down |
| norB | Norfloxacin-resistant S. aureus efflux pump gene. Mediating S A develops resistance to hydrophilic quinolone drugs (such as ciprofloxacin and norfloxacin) and hydrophobic quinolone drugs (such as moxifloxacin). [ 31] | Down | Down | |
| Protein synthesis | thrs | Threonyl-tRNA synthetase (thrRS) [ 32] | Down | Down |
| cysS, ybaK | Cysteine-tRNA synthesis [ 30] | Up | Down | |
| thrC | Seryl-tRNA synthetase [ 33] | Down | Down | |
| metE | Cystathionine gamma-synthase II (cobalamin-independent), encoding homocysteine S-methyltransferase [ 34] | Down | ||
| Ribosomal-related genes rplN, rimP | Inhibit the activity of ribosome and protein synthesis by affecting the structure and function of the ribosome, further impacting intracellular protein synthesis and cellular functions [ 35] | Up | Down | |
| clpP | Plays an essential role in cellular protein metabolism by facilitating the refolding or degradation of damaged proteins [ 36] | Down | Down | |
| Energy production and metabolism | pflB, pflA | Formate acetyltransferase, pflB, and pflA, collectively form the operon pfl, which catalyzes the reversible conversion of pyruvate to formate during bacterial sugar metabolism under anaerobic conditions [ 37, 38] | Down | Down |
| sdhB | Succinate dehydrogenase/fumarate reductase, composed of the three coding genes sdhA, sdhB, and sdhC, plays a pivotal role in bacterial energy metabolism by participating in the electron transport chain and the tricarboxylic acid cycle [ 39] | Down |
The transcriptomic and proteomic analysis indicated that ESM affected bacterial life processes through multiple mechanisms, ultimately leading to bacterial death (Figure 6). ESM can activate autolytic enzymes lytM and atl in S. aureus, upregulating their gene and protein expressions, enhancing autolysis of the bacterial cell wall, causing cell wall damage and leakage of cellular contents, thereby triggering bacterial death. ESM can inhibit the gene and protein expressions of various aminoacyl-tRNA synthetases involved in bacterial protein syntheses, such as thrs, cysS, thrC, metE, and dapD, affecting their protein translation and significantly reducing protein levels. Clp, together with ATPases, such as clpX and clpA, forms the ClpP protease complex, which can recognize and degrade misfolded or aged proteins, thereby maintaining intracellular protein homeostasis, crucial for the survival and environmental adaptation of bacterial cells [40]. Downregulation of clpP expression disrupts protein metabolism, further weakening bacterial survival and ultimately causing death.
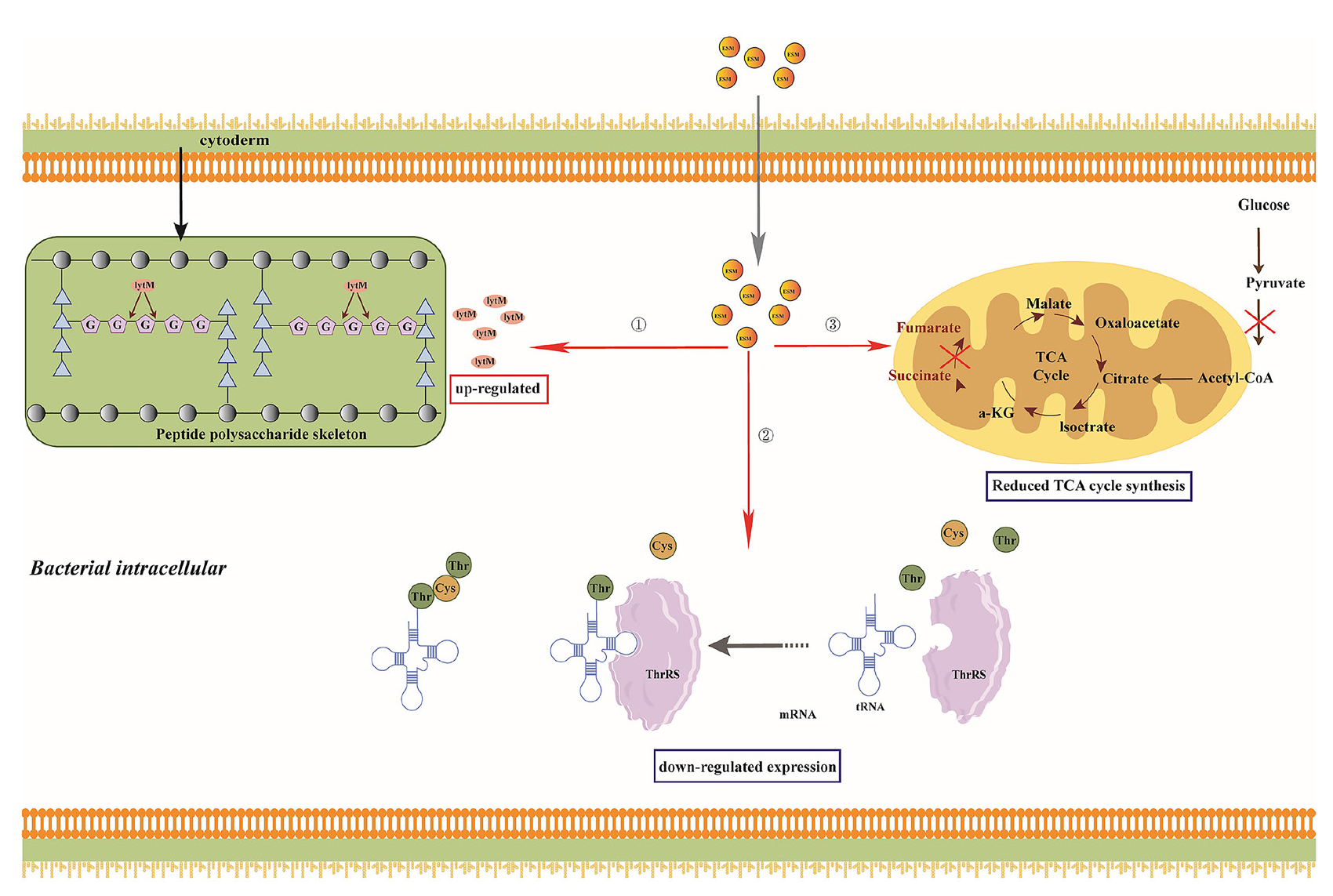
- Partial antibacterial mechanisms of ESM. The figure depicts partial mechanisms that explain how ESM partially inhibits S. aureus: ① ESM breaks down peptidoglycan linkages in the bacterial cell wall by upregulating lytM expression; ②. ESM diminishes the expression of Thrs, which reduces protein expression; ③ ESM mitigates the expression of important TCA cycle enzymes, which reduces energy production. ESM, esculetin-manganese complex.
Compared to ES, ESM exhibits a significant inhibitory effect by downregulating the expression of the norB gene, which encodes an efflux pump member of the MFS family. This leads to increased resistance to hydrophilic quinolone and tetracycline antibiotics [29]. Additionally, ESM has been shown to inhibit biofilm formation in bacteria, particularly by suppressing the expression of manA gene mRNA and proteins in S. aureus, thereby disrupting its biofilm structure. Further studies have demonstrated that Mn(II) complexes, at concentrations ranging from 0.25 to 0.5 mM, significantly inhibit biofilm formation in Pseudomonas aeruginosa. This inhibition may be related to specific interactions between the Mn(II) complex and bacterial surfaces or biofilm structures. The Mn(II) complex also inhibits the secretion of pyocyanin, an important toxin produced by Pseudomonas aeruginosa, which is often linked to its virulence and biofilm formation. Thus, Mn(II) complexes not only physically impede biofilm formation but also indirectly affect biofilm stability by reducing toxin secretion [41]. Moreover, studies have found that manganese-containing nanoparticle complexes, when exposed to green light, significantly enhance the production of reactive oxygen species (ROS), such as hydrogen peroxide and superoxide anion, which inhibit the regeneration of bacteria within biofilms. By promoting ROS generation, these nanoparticles effectively reduce the regenerative capacity of bacteria in biofilms [42]. Additionally, metal-tdda-phen complexes disrupt biofilm structure and inhibit bacterial aggregation and colonization by interacting with extracellular DNA and extracellular polysaccharides. This interaction enhances their antibacterial activity [43]. Manganese salicylate complexes exhibit significant antimicrobial activity against Gram-positive bacteria, particularly S. aureus and its methicillin-resistant strain (MRSA). At low concentrations, these complexes effectively inhibit bacterial growth and, through molecular docking studies, have been shown to inhibit the activity of DNA gyrase (GyrB) in S. aureus, leading to DNA structural damage and preventing bacterial proliferation and biofilm formation [44]. Therefore, the multifaceted antimicrobial mechanisms of manganese complexes position them as promising new agents with low resistance potential, warranting further investigation and development. To validate the transcriptomic results, real-time quantitative PCR (qPCR) was performed on genes showing significant differential expression. Four genes were selected for qPCR validation, and the results were compared with the transcriptomic sequencing data. The comparison revealed that the resultant trends were consistent in both the transcriptomic sequencing and qPCR (Figure S8, further confirming the reliability of the sequencing data.
4. Conclusions
The findings of this study highlight the need to consider the role of metal complexes formed between organic components during the extraction process and the trace metal elements contained in them when studying the active ingredients of TCM. These metal complexes, as potential candidate drugs, may play an important role in the development and application of TCM, providing new perspectives and experimental evidence for the modernization of TCM. However, the antibacterial activity of ESM in vivo has not been systematically evaluated, and its mechanism has not been further validated through protein expression-level experiments. Future research will focus on these two aspects for an in-depth exploration of the antibacterial mechanism and drug resistance development process of ESM.
Acknowledgment
We would like to thank Editage (www.editage.cn) for English language editing. This work was supported by the National Natural Science Foundation of China (Grant No. 82360763) and the Jiangxi Provincial Natural Science Foundation (Grant No. 20212BAB206005).
CRediT authorship contribution statement
Liang Peng: Writing – review & editing, Project administration; Lin Zhang: Writing – review & editing; Chenxi Jiang and Zhimin Li: Writing – original draft, Methodology; Jintao Wang, Xiangwen Liao and Yanshi Xiong: Data curation; Wenying Guo and Jiayi Liu: Validation.
Declaration of competing interest
There are no conflicts of interest to declare.
Data availability
The data used to support the findings of this study are available from the corresponding author upon request.
Declaration of Generative AI and AI-assisted technologies in the writing process
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript and no images were manipulated using AI.
Supplementary data
Supplementary material to this article can be found online at https://dx.doi.org/10.25259/AJC_99_2024.
References
- Bacteria-responsive nanoliposomes as smart sonotheranostics for multidrug resistant bacterial infections. ACS Nano. 2019;13:2427-2438. https://doi.org/10.1021/acsnano.8b09336.
- [Google Scholar]
- Experimental methods for evaluating the bacterial uptake of trojan horse antibacterials. Chemical Medicinal Chemist. 2021;16:1063-1076. https://doi.org/10.1002/cmdc.202000806.
- [Google Scholar]
- Coordination complexes to combat bacterial infections: Recent developments, current directions and future opportunities. Chemistry (Weinheim an der Bergstrasse, Germany). 2021;27:7340-7350. https://doi.org/10.1002/chem.202004822.
- [Google Scholar]
- Synthesis, spectroscopic characterisation, thermal analysis, DNA interaction and antibacterial activity of copper(I) complexes with n, n′- disubstituted thiourea. Journal of Molecular Structure. 2016;1106:352-365. https://doi.org/10.1016/j.molstruc.2015.10.010.
- [Google Scholar]
- Antibacterial, antifungal and in vitro antileukaemia activity of metal complexes with thiosemicarbazones. Journal of Cellular and Molecular Medicine. 2015;19:865-878. https://doi.org/10.1111/jcmm.12508.
- [Google Scholar]
- Discovery of antibacterial manganese(i) tricarbonyl complexes through combinatorial chemistry. Chemical Science. 2024;15:3907-3919. https://doi.org/10.1039/d3sc05326a.
- [Google Scholar]
- A novel pyrazolone complex p-FAH-cu-bpy induces death of escherichia coli and staphylococcus aureus by disrupting cell structure and blocking energy. Archives of Microbiology. 2023;205:376. https://doi.org/10.1007/s00203-023-03714-6.
- [Google Scholar]
- Discovery of metal complexes with antibacterial properties in aqueous extracts of Radix scutellariae and a study of the antibacterial properties of the baicalin–manganese complex. Inorganic Chemistry Frontiers. 2023;10:6506-6518. https://doi.org/10.1039/d3qi01034a.
- [Google Scholar]
- The antimicrobial property of JY-1, a complex mixture of traditional chinese medicine, is linked to it abilities to suppress biofilm formation and disrupt membrane permeability. Microbial Pathogenesis. 2024;189:106573. https://doi.org/10.1016/j.micpath.2024.106573.
- [Google Scholar]
- The pharmaceutical applications of total flavonoids extract from isatis tinctoria l. leaves. Pharmacological Research - Modern Chinese Medicine. 2022;3:100122. https://doi.org/10.1016/j.prmcm.2022.100122.
- [Google Scholar]
- Antibacterial activity mechanism of coptisine against pasteurella multocida. Frontiers in Cellular and Infection Microbiology. 2023;13:1207855. https://doi.org/10.3389/fcimb.2023.1207855.
- [Google Scholar]
- Biological properties of metal complexes of curcumin. BioFactors (Oxford, England). 2019;45:304-317. https://doi.org/10.1002/biof.1504.
- [Google Scholar]
- Synthesis, characterization and biochemical analysis of azo-metal complex of embelin with second group transition metals. Current Organocatalysis. 2022;9:155-162. https://doi.org/10.2174/2213337208666211102104240.
- [Google Scholar]
- Design, synthesis, and biological activity of 18β-glycyrrhetinic acid derivatives and their metal complexes. Medical Sciences Forum. 2022;14:129. http://dx.doi.org/10.3390/ECMC2022-13193.
- [Google Scholar]
- Homo- and heterometallic complexes of zn(II), {Zn(II)Au(I)}, and {Zn(II)Ag(I)} with pentadentate schiff base ligands as promising anticancer agents. Dalton Transactions. 2023;52:12282-12295. https://doi.org/10.1039/d3dt01749d.
- [Google Scholar]
- A critical review on quercetin bioflavonoid and its derivatives: Scope, synthesis, and biological applications with future prospects. Arabian Journal of Chemistry. 2023;16:104881. https://doi.org/10.1016/j.arabjc.2023.104881.
- [Google Scholar]
- In vitro antibacterial and antioxidant activities and molecular docking analysis of phytochemicals from cadia purpurea roots. Journal of Tropical Medicine. 2022;2022:4190166. https://doi.org/10.1155/2022/4190166.
- [Google Scholar]
- Protective effect of n-butanol extract from viola yedoensis on immunological liver injury. Chemistry & Biodiversity. 2021;18:e2001043. https://doi.org/10.1002/cbdv.202001043.
- [Google Scholar]
- UPLC-q-TOF-MS profiling of viola stocksii boiss. and evaluation of aphrodisiac potential and risk factors associated with erectile dysfunction. Journal of Ethnopharmacology. 2024;321:117477. https://doi.org/10.1016/j.jep.2023.117477.
- [Google Scholar]
- Pharmacological activities of esculin and esculetin: A review. Medicine (Baltimore). 2023;102:e35306. https://doi.org/10.1097%2FMD.0000000000035306.
- [Google Scholar]
- Lignans, flavonoids and coumarins from viola philippica and their α-glucosidase and HCV protease inhibitory activities. Natural Product Research. 2019;33:1550-1555. https://doi.org/10.1080/14786419.2017.1423305.
- [Google Scholar]
- Design and synthesis of pyridine bis-hydrazone metal complexes of co(II), cu(II), and hg(II): Spectral, gaussian, electrochemical, biological, drug-likeness and molecular docking investigations. Inorganic Chemistry Communications. 2024;162:112188. https://doi.org/10.1016/j.inoche.2024.112188.
- [Google Scholar]
- Autolysin-mediated peptidoglycan hydrolysis is required for the surface display of staphylococcus aureus cell wall-anchored proteins. Proceedings of the National Academy of Sciences of the United States of America. 2023;120:e2301414120. https://doi.org/10.1073/pnas.2301414120.
- [Google Scholar]
- A defect in cell wall recycling confers antibiotic resistance and sensitivity in staphylococcus aureus. The Journal of Biological Chemistry. 2022;298:102473. https://doi.org/10.1016/j.jbc.2022.102473.
- [Google Scholar]
- Staphylococcus aureus sacculus mediates activities of M23 hydrolases. Nature Communications. 2023;14:6706. https://doi.org/10.1038/s41467-023-42506-w.
- [Google Scholar]
- Chemical shift assignments of the catalytic domain of staphylococcus aureus lytM. Biomolecular NMR Assignments. 2024;18:1-5. https://doi.org/10.1007/s12104-023-10161-3.
- [Google Scholar]
- Differential binding of human and murine igGs to catalytic and cell wall binding domains of staphylococcus aureus peptidoglycan hydrolases. Scientific Reports. 2021;11:13865. https://doi.org/10.1038/s41598-021-93359-6.
- [Google Scholar]
- Protein-conjugated microbubbles for the selective targeting of S. aureus biofilms. Biofilm. 2022;4:100074. https://doi.org/10.1016/j.bioflm.2022.100074.
- [Google Scholar]
- Characterization of the modular design of the autolysin/adhesin aaa from staphylococcus aureus. PloS one. 2012;7:e40353. https://doi.org/10.1371/journal.pone.0040353.
- [Google Scholar]
- In vitro selection identifies staphylococcus aureus genes influencing biofilm formation. Infection and Immunity. 2023;91:e0053822. https://doi.org/10.1128/iai.00538-22.
- [Google Scholar]
- Biosynthesis of Fe3O4@Ag nanocomposite and evaluation of its performance on expression of norA and norB efflux pump genes in ciprofloxacin-resistant staphylococcus aureus. Biological Trace Element Research. 2019;191:522-530. https://doi.org/10.1007/s12011-019-1632-y.
- [Google Scholar]
- ThrRS-mediated translation regulation of the RNA polymerase III subunit RPC10 occurs through an element with similarity to cognate tRNA ASL and affects tRNA levels. Genes. 2023;14:462. https://doi.org/10.3390/genes14020462.
- [Google Scholar]
- Phenotypic and genomic characteristics of oxacillin-susceptible mecA-positive staphylococcus aureus, rapid selection of high-level resistance to beta-lactams. European Journal of Clinical Microbiology & Infectious Diseases: Official Publication of the European Society of Clinical Microbiology. 2023;42:1125-1133. https://doi.org/10.1007/s10096-023-04646-1.
- [Google Scholar]
- Lysine and threonine biosynthesis from aspartate contributes to staphylococcus aureus growth in calf serum. Applied and Environmental Microbiology. 2016;82:6150-6157. https://doi.org/10.1128/AEM.01399-16.
- [Google Scholar]
- Carrier-free binary self-assembled nanomedicines originated from traditional herb medicine with multifunction to accelerate MRSA-infected wound healing by antibacterial, anti-inflammation and promoting angiogenesis. International Journal of Nanomedicine. 2023;18:4885-4906. https://doi.org/10.2147/IJN.S422944.
- [Google Scholar]
- Antimicrobial potential of chlorella sorokiniana on MRSA – an in vitro study and an in silico analysis on clpP protease. Journal of King Saud University - Science. 2023;35:102668. https://doi.org/10.1016/j.jksus.2023.102668.
- [Google Scholar]
- The staphylococcus aureus arlRS two-component system regulates virulence factor expression through mgrA. Molecular Microbiology. 2020;113:103-122. https://doi.org/10.1111/mmi.14404.
- [Google Scholar]
- Carbon source-dependent reprogramming of anaerobic metabolism in staphylococcus aureus. Journal of Bacteriology. 2021;203:e00639-e00620. https://doi.org/10.1128/JB.00639-20.
- [Google Scholar]
- Transposon mutagenesis identifies novel genes associated with staphylococcus aureus persister formation. Frontiers in Microbiology. 2015;6:1437. https://doi.org/10.3389/fmicb.2015.01437.
- [Google Scholar]
- Structure of Staphylococcus aureus ClpP bound to the covalent active‐site inhibitor cystargolide A. Angewandte Chemie. 2023;136:e202314028. https://doi.org/10.1002/anie.202314028.
- [Google Scholar]
- Tuning anti-biofilm activity of manganese(II) complexes: Linking biological effectiveness of heteroaromatic complexes of alcohol, aldehyde, ketone, and carboxylic acid with structural effects and redox activity. International Journal of Molecular Sciences. 2021;22:4847. https://doi.org/10.3390/ijms22094847.
- [Google Scholar]
- Manganese complex‐Gold nanoparticle hybrid for biofilm inhibition and eradication. Chemical Biology Chemist. 2024;25 https://doi.org/10.1002/cbic.202400500.
- [Google Scholar]
- The antibacterial and anti-biofilm activity of metal complexes incorporating 3,6,9-trioxaundecanedioate and 1,10-phenanthroline ligands in clinical isolates of pseudomonas aeruginosa from irish cystic fibrosis patients. Antibiotics (Basel, Switzerland). 2020;9:674. https://doi.org/10.3390/antibiotics9100674.
- [Google Scholar]
- Promising antifungal and antibacterial agents based on 5‐Aryl‐2,2′‐bipyridines and their heteroligand salicylate metal complexes: Synthesis, bioevaluation, molecular docking. Chemical Medicinal Chemist. 2022;17 https://doi.org/10.1002/cmdc.202100577.
- [Google Scholar]







