Translate this page into:
Identification of the tannins in traditional Chinese medicine Paeoniae Radix Alba by UHPLC-Q-Exactive Orbitrap MS
⁎Corresponding author. 20120941161@bucm.edu.cn (Wei Cai)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Paeoniae Radix Alba (PRA, dried root of Paeonia lactiflora Pall.) is a widely used as traditional Chinese medicine and tannins are one of their main bioactive ingredients. However, there are rarely systematically investigated in this study. This study aimed to establish a rapid, high selective, high sensitive and effective method based on UHPLC-Q-Exactive Orbitrap MS for simultaneous identification the tannins in PRA. Separation was performed on Thermo Scientific Hypersil GOLD™ aQ (100 mm × 2.1 mm, 1.9 μm) using gradient elution consist of 0.1% formic acid acetonitrile and 0.1% formic acid water as the mobile phase at a flow rate of 0.3 mL/min. The mass spectrometer was operated with Q-Exactive Orbitrap spectrometer in negative ion mode. Finally, a total of 106 constituents were identified in PRA by UHPLC-Q-Exactive Orbitrap MS, 75 of those were reported from PRA for the first time. This result laid the foundation for in-depth research on the material basis efficacy and provided scientific basis for the selection of quality marker of PRA.
Keywords
Paeoniae Radix Alba
Tannins
UHPLC-Q-Exactive Orbitrap MS
1 Introduction
Paeoniae radix alba (PRA, Bai Shao in Chinese), the dried root of plant Paeonia lactiflora Pall. (Family Ranunculaceae), is a famous Traditional Chinese medicines (TCM) in China. RPA is produced by boiling the fresh root of the whole Paeonia lactiflora Pall. in water and peeling off the bark. PRA plays an important role in contribute to most biological activities that including subduing hyperactivity of the liver, relieving pain, reducing sweat, nourishing blood and regulating menstruation (State Pharmacopoeia Commission, 2020). So far, many compounds have been reported in PRA, including flavonoids, terpenes and volatile oils and so on. Its main active ingredients are paeoniflorin, albiflorin, and gallic acid, which have pharmacological effects such as liver protection, anti-inflammatory, anti-rheumatic, immune-regulating functions, etc (Ou et al., 2013., Wang et al., 2010). However, the research on chemical constituents of PRA is still insufficient.
Tannins, a class of polyphenolic compounds with complex structures, are widely exist in plants. According to the structure of tannins, tannins can be divided into three types: hydrolyzed tannins, condensed tannins and combined tannins of these two types (Xing et al., 2011). Glucogallins, a kind of hydrolyzed tanins, are a class of esters composed of gallic acids or their derivatives and polyols such as glucose, rhamnose and quinic acid, which generate gallic acids or polyols after hydrolysis (Chen, 2000). Most biological activities of glucogallins come from the hydrolysate. It is reported that gallic acids have multiple pharmacological activities including anti-inflammatory, antioxidant, anti-tumor, etc (Li et al., 2004., Ohno et al., 2001), and provide treatment for hypertension, myocardial infarction and diabetes among others (Dianat et al., 2014; Lee et al., 2015; Li et al., 2004; Punithavathi et al., 2011). Condensed tannins are mainly composed of catechin as the precursor through C—C bonds or C—O bonds condensation between each unit (Zhu and Jin, 2015), which have anti-oxidation, anti-tumor, anti-pathogen, anti-HIV and other pharmacological effects (Wang et al., 2013).
Although there are a lot of studies on the chemical components of PRA, they are mainly concentrated on monoterpene glycosides, and less on tannins. UHPLC-MS/MS is a powerful analytical technique used in detection and identification of chemical constituents in traditional Chinese medicine, drug, or biological samples (Cai et al., 2020; Hu et al., 2020; Ren et al., 2020). The characteristics such as the high separation efficiency of UHPLC and the high sensitivity and molecular structure elucidation ability by providing accurate mass measurement and abundant MSn fragment information of MS were integrated by UHPLC-MS/MS (Liao et al., 2020). Recently, UHPLC-Q-Exactive Orbitrap MS was widely used in the identification of chemical constitutions for its high selectivity, high sensitivity and effectivity. Therefore, it is necessary to systematically identify the type of tannins in PRA using UHPLC-Q-Exactive Orbitrap MS in the negative ion mode, Finally, a total of 106 compounds were found in PRA, 75 of which were reported for the first time. The result will benefit for the in-depth understanding of the pharmacological action of PRA and lay a foundation for the quality control of the drug in clinical use in the future.
2 Experimental
2.1 Chemicals and reagents
The chromatographic grade methanol (MeOH) and acetonitrile (ACN) were purchased from MACKIN company, the MS grade formic acid was purchased from Thermo Fisher Scientific Co., Ltd. (USA), purified water was obtained from Guangzhou Watsons Food & Beverage Co., Ltd. (China). Other solvents were of an analytical grade. Gallic acid (purity ≥ 98%), methyl gallate (purity ≥ 98%), (–)-epicatechin gallate (purity ≥ 98%), corilagin (galloy-HHDP-hexoside) (purity ≥ 98%) were purchased from Chengdu Purechem-Standard Co. Ltd. PRA samples were provide by Sun Ten Pharmaceutical Co., Ltd and turned into powder after being crushed and stored in vacuum packages.
2.2 Instruments and LC-MS/MS conditions
LC-MS/MS analyses, which performed on an Dionex Ultimate 3000 UHPLC (a quaternary pump, an LPG-3400SD vacuum degasser unit) and the UHPLC-Q-Exactive Orbitrap MS mass spectrometer equipped with an electrospray ionization (ESI) source, were used for simultaneous determination of tannins in PRA. The liquid chromatographic separations of all analyzed samples were achieved on a Thermo Scientific Hypersil GOLD™ aQ (100 mm × 2.1 mm, 1.9 μm) at 40 °C with a flow rate of 0.3 mL/min. The mobile phase consisted of (A) water containing 0.1% formic acid and (B) acetonitrile containing 0.1% formic acid. The gradient program was as follows: 0–2 min, 95% A; 2–5 min, 95–85% A; 5–20 min, 85–65% A; 20–22 min, 65–95% A; 22–25 min, 95% A. The sample injection volume was 1 μL.
The mass spectrometer analysis was operated in the negative electrospray ionization mode. High resolution mass spectrum was collected in the full scan mode in the mass range m/z 100–1500 at a resolution of 70, 000. The MS2 data at a resolution of 17, 500 was obtained by data-dependent MS2 scanning or parallel reaction monitoring (PRM) mode triggered by inclusion ions list, which was built by molecule predicted. The other conditions of MS analysis were as follows: The spray voltage was set to 3.2 kV; the sheath gas flow rate and the aux gas flow rate were set to 35 arb and 10 arb, respectively; the capillary temperature and the heater temperature were set to 320 °C and 350 °C, respectively; the S-lens RF level was 60.
2.3 Preparation of control and standard samples
Dried and pulverised PRA (1 g) was accurately weighed by electronic analytical balance. After 20 mL of 70% aqueous methanol was added, an extract was obtained by sonication for 1 h. The supernatants were removed using a syringe and filtered through a 0.22 μL nylon millipore filter and added to the liquid vial for further analysis.
The standard solutions including gallic acid, methyl gallate, (−)-epicatechin gallate, corilagin (galloy-HHDP-hexoside) were dissolved in methanol, respectively, to get reference standards solutions (0.1 mg/mL). All the standard solutions were stored at 4 °C before analysis.
2.4 Data processing and analysis
The Xcalibur software version 4.2 (Thermo Fisher Scientifific, California, USA) was used to obtain the raw data including the full-scan MS and MS2 data. The peaks detected with intensity over 10,000 were selected for identifications. The chemical formulas for all parent and fragment ions of the selected peaks were calculated from the accurate mass using a formula predictor by setting the parameters as follows: C [0–50], H [0–60], O [0–40], S [0–5], the mass tolerance of MS and MS2 was within 5 ppm, respectively.
3 Results and discussion
3.1 Establishment of analytical strategy
In order to screen and identify tannins systematically in PRA, an analytical strategy based on UHPLC-Q-Exactive Orbitrap MS was established in this study. Firstly, tannins in RPA were extracted and enriched by ultrasonic extraction with 70% methanol. Secondly, the sample contained tannins was injected into UHPLC-Q-Exactive Orbitrap MS to gain the high resolution mass data acquired by full MS scan with data dependence MS2 (Full Mass-ddMS2), which was processed by Compound Discover version 3.0 using high resolution extracted ion chromatography (HREIC) and expected compounds predicted. Thirdly, for the trace constituents precursor ions with relatively low content in the mass analyzer, especially when they co-eluted with higher content constituents, the subsequent fragments can be obtained by PRM mode triggered by inclusion ions list to make the tannins identification more sufficient in RPA. Fourth, diagnosis fragmentation ions (DFIs) data-mining techniques were adopted for the selective clarification of tannins that possessed similar mass fragmentation behaviors to those of reference standard. Finally, the compounds were identified based on the full scan MS, MS2 data, retention time and bibliography.
3.2 Optimization of UHPLC-Q-Exactive Orbitrap MS condition
In order to obtain better chromatographic peak type and separation, variables factors were investigated in detection and identification process, including extraction solvent ranging from 60% to 100% methanol, the kind of mobile phase (acetonitrile/water, and acetonitrile containing 0.1% formic acid/water), the kind and content of acid (formic acid and acetic acid, 0.05, 0.1, and 0.2%), column (Thermo Scientific Hypersil GOLD™ aQ 100 mm × 2.1 mm, 1.9 μm and Waters ACQUITY BEH C18 column, 100 mm × 2.1 mm, 1.7 μm), flow rate of mobile phase (0.2, 0.3, and 0.4 mL/min), column temperature (30, 35, 40, 45 °C) and the mobile phase gradient. The MS parameters including the flow rate of sheath gas and auxiliary gas, the temperature of capillary and heater, spray voltage, et al. were examined. In the optimization condition of UHPLC-Q-Exactive Orbitrap MS, most of the tannins have shown efficient separation and parent/daughter ion pairs with high responses.
3.3 Establishment of DFIs
There are three types of tannins including hydrolyzed tannins, condensed tannins and compound tannins. It is easily understood that tannins with the same carbon skeletons will generate the similar fragmentations, which can be use as DFIs for the distinguish and characterization of tannins. The fragmentation patterns of 4 reference standards were investigated by UHPLC-Q-Exactive Orbitrap MS in negative mode to establish the DFIs, such as 169.0133 ([gallic acid-H]−), 125.0232 ([gallic acid-CO2-H]−) generated from gallic acid moiety, 300.9984 ([ellagic acid-H]−), 283.0457 ([ellagic acid-H- H2O]−) yielded by ellagic acid. Furthermore, there are fragment ions at m/z 321.0262 ([digallic acid–H]−), 331.0672 ([galloylglucose-H]−), 313.0580 (C13H14O9), 211.0426 (C13H7O3), 275.0192 (C13H7O7) and 183.0427 (C12H7O2). All the above can be used as DFIs of hydrolytic tannins. Fragment ions at m/z 289.0717 ([catechin acid-H]−) emerged from catechin moiety and can be used as DFIs of condensed tannins.
3.4 Characterization of the tannins in PRA
Under the LC-MS conditions of “2.3″, the extracted ion chromatogram (EIC) in negative ion mode was obtained as shown in Fig. 1 and the selected fragmentation pattern of components identified from PRA were shown in Fig. 2. As listed in Table 1, the chromatographic and mass data of those detected components are summarized though Xcalibur software version 4.2, which including retention times (tR), experimental Mass (negative-ion mode), molecular formula, error in ppm (between the theoretical mass and the experimental mass) of each tannins, as well as the MS/MS fragment ions. Eventually, a total of 106 tannins (75 first report) was accuracy or tentatively identified in PRA though UHPLC-Q-Exactive Orbitrap MS.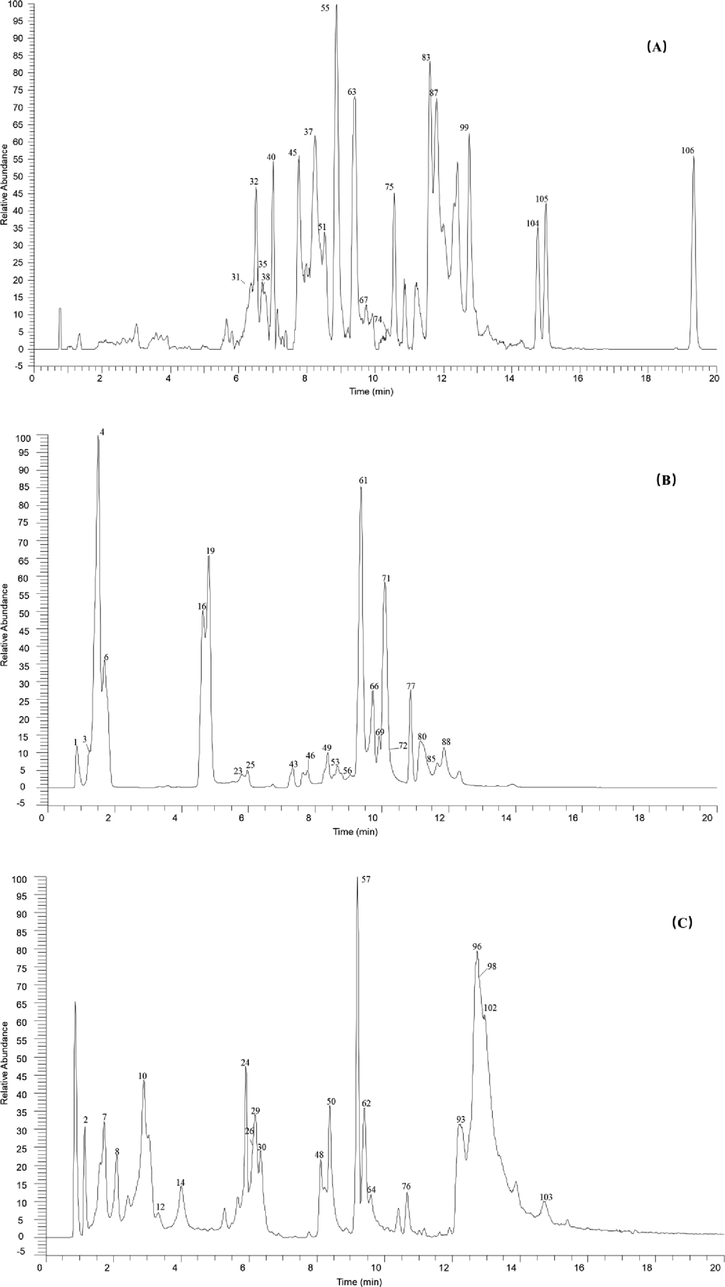
The high-resolution extracted ion chromatogram (HREIC) in 5 ppm for the multiple compounds in PRA. (A) m/z 481.09876, 315.01464, 783.10503, 873.15198, 343.04594, 935.07960, 703.16684, 599.10424, 329.03029, 694.12091, 801.0792, 461.07254, 473.03616; (B) m/z 463.05181, 321.02520, 787.09994, 545.05729, 335.04085, 493.11989, 939.11090, 183.02989, 631.16684, 169.01424; (C) m/z 477.06746, 621.05808, 483.07802, 937.09525, 721.14102, 315.07215, 343.06706, 801.11559, 461.10893, 633.07333; (D) 445.13514, 197.04554, 785.08429, 1243.13282, 345.08271, 487.05181, 495.07802, 715.13045, 491.08311; (E) 300.99899, 441.08271, 1091.12186, 783.17780, 635.08898, 451.12458, 527.14062.
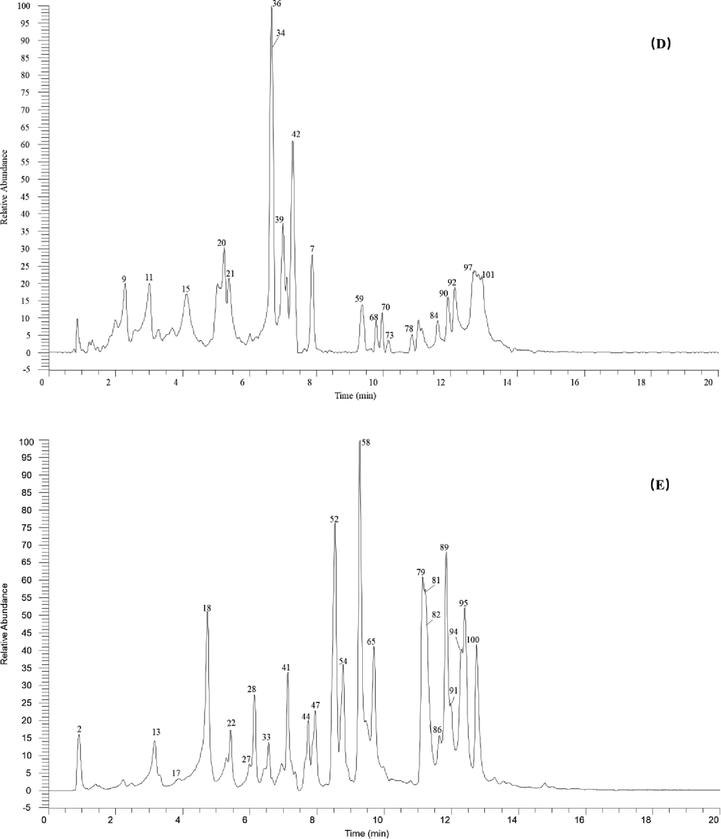
The high-resolution extracted ion chromatogram (HREIC) in 5 ppm for the multiple compounds in PRA. (A) m/z 481.09876, 315.01464, 783.10503, 873.15198, 343.04594, 935.07960, 703.16684, 599.10424, 329.03029, 694.12091, 801.0792, 461.07254, 473.03616; (B) m/z 463.05181, 321.02520, 787.09994, 545.05729, 335.04085, 493.11989, 939.11090, 183.02989, 631.16684, 169.01424; (C) m/z 477.06746, 621.05808, 483.07802, 937.09525, 721.14102, 315.07215, 343.06706, 801.11559, 461.10893, 633.07333; (D) 445.13514, 197.04554, 785.08429, 1243.13282, 345.08271, 487.05181, 495.07802, 715.13045, 491.08311; (E) 300.99899, 441.08271, 1091.12186, 783.17780, 635.08898, 451.12458, 527.14062.
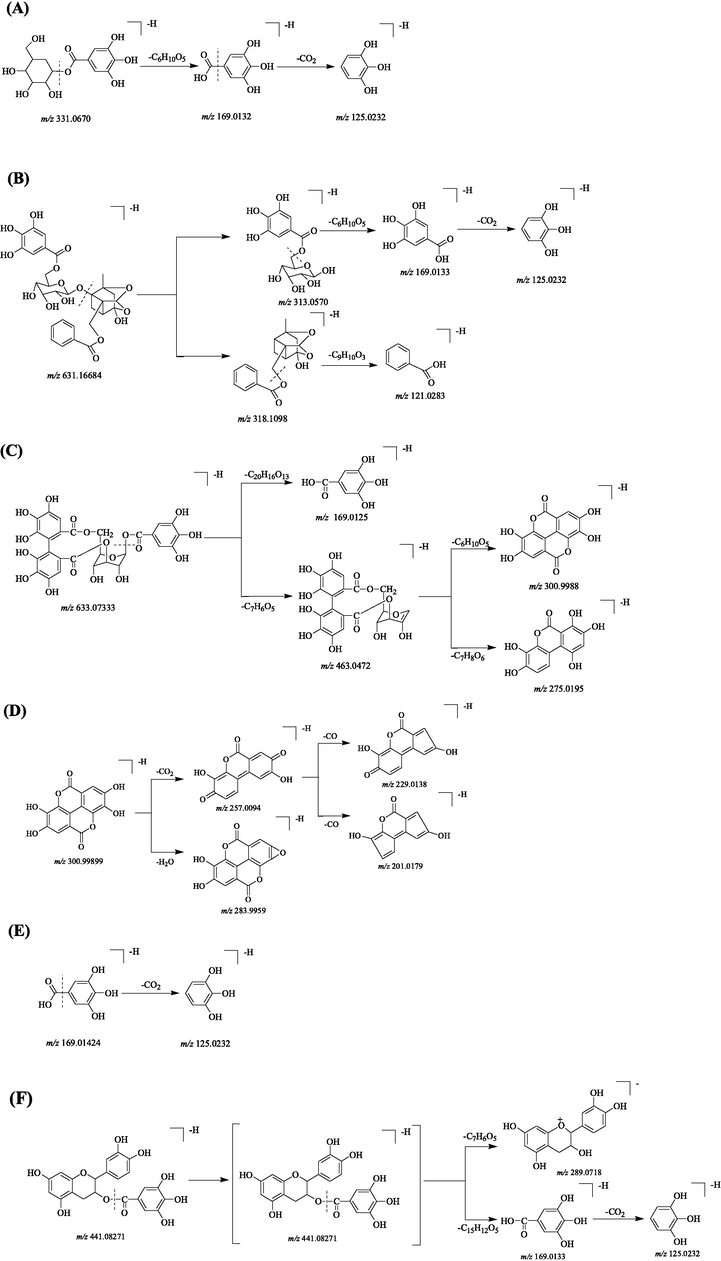
Proposed selected fragmentation pattern of components identified from PRA: Galloylglucose (A); Galloylpaeoniflorin(B); Corilagin (Galloy-HHDP-hexoside) (C); Ellagic acid (D); Gallic acid (E); (–)-Epicatechin gallate (F).
Peak
tR
Theoretical Mass m/z
Experimental Mass m/z
Error (ppm)
Formula
MS/MS fragment
Identification/Reactions
1
0.85
493.11989
493.11972
−0.351
C19H26O15
MS2[493]: 169.0133(100), 125.0233(82), 313.0657(31), 123.0076(19), 151.0027(14)
1′-O-galloylsucrose
2#
1.15
343.06706
343.06683
0.857
C14H16O10
MS2[343]: 169.0132(100), 191.0553(76), 125.0231(68), 107.0125(36), 109.0282(16), 85.0281(11)
galloylquinic acid isomer
3
1.24
493.11989
493.11963
−0.534
C19H26O15
MS2[493]: 169.0133(100), 125.0232(70), 331.0671(47), 123.075(10), 313.0564(10)
6′-O-galloylsucrose
4*
1.51
169.01424
169.01418
−0.394
C7H6O5
MS2[169]: 125.0232(100)
gallic acid
5#
1.53.
331.06706
331.06689
−0.543
C13H16O10
MS2[331]: 125.0232(100), 169.0132(86), 241.0349(19), 149.9947(17), 271.0457(16)
galloylglucose
6
1.70
493.11989
493.11954
−0.716
C19H26O15
MS2[493]: 169.0132(100), 129.0231(80), 313.0565(27), 283.0457(14)
6-O-galloylsucrose
7#
1.72
343.06706
343.06677
0.797
C14H16O10
MS2[343]: 169.0132(100), 125.0232(65), 173.0445(25), 191.0553(23), 93.0332(16)
galloylquinic acid isomer
8#
2.10
315.07215
315.07236
0.650
C13H16O9
MS2[315]: 108.0203(100), 152.0104(59), 109.0282(28), 153.0182(21)
galloylrhamnose isomer
9#
2.29
345.08271
345.08289
0.493
C14H18O10
MS2[345]: 183.0291(100), 139.0390(28), 107.0126(27), 225.0399(25)
methyl galloylglucose isomer
10#
2.89
315.07215
315.07227
0.364
C13H16O9
MS2[315]:109.0282(100), 153.0183(66), 169.0133(19), 123.0076(15), 125.0232(13)
galloylrhamnose isomer
11#
3.01
345.08271
345.08273
0.029
C14H18O10
MS2[345]: 183.0290(100), 124.0154(48), 139.0390(14)
methyl galloylglucose isomer
12#
3.10
315.07215
315.07233
0.555
C13H16O9
MS2[315]: 169.0133(100)., 123.0075(66), 125.0232(55), 151.0026(54), 107.0126(11), 139.0021(11)
galloylrhamnose isomer
13
3.17
527.14062
527.140691
−3.297
C23H28O14
MS2[527]: 169.0219(100), 165.0545(20), 313.0567(18), 61.09859(16), 125.0230(10)
6′-O-Galloyl-desbenzoylpaeoniforin isomer
14#
4.01
633.07333
633.07385
0.811
C27H22O18
MS2[633]: 300.9984(100), 275.0192(21), 169.0128(12), 125.0230(10)
gorilagin(Galloy-HHDP-hexoside) isomer
15#
4.13
495.07802
495.07822
0.387
C21H20O14
MS2[495]:109.0122(100), 137.0232(83), 169.0131(30), 125.0232(25), 313.0558(17)
di-O-galloylquinic acid
16*
4.64
183.02989
183.02980
−0.528
C8H8O5
MS2[183]: 168.0054(100), 124.0153(92), 140.0103(76), 111.0075(45), 139.0025(41)
methyl gallate
17#
4.74
451.12458
451.12445
0.962
C21H24O11
MS2[451]: 289.0717(100), 109.0282(27), 125.0232(19), 245.0817(19), 123.0439(18)
galloylsalidroside
18
4.76
527.14062
527.14032
−0.585
C23H28O14
MS2[527]: 479.1187(100), 271.0455(84), 497.1295(52), 313.0563(49), 169.0129(44)
6′-O-Galloyl-desbenzoylpaeoniforin
19
4.83
183.02989
183.02985
−0.255
C8H8O5
MS2[183]: 168.0055(100), 124.0153(94), 140.0104(79), 111.0074(50), 139.0025(38)
methyl gallate isomer
20
5.24
445.13514
445.13495
−0.448
C19H26O12
MS2[445]: 121.0283(100), 59.0125(12)
benzoylsucrose isomer
21#
5.40
785.08429
785.08490
0.770
C34H26O22
MS2[785]: 300.9988(100), 275.0201(44), 249.0401(37), 125.0233(27), 169.0137(23), 137.0233(18), 231.0286(15)
digalloyl-HHDP-glucose isomer
22
5.43
527.14062
527.14026
−0.699
C23H28O14
MS2[527]: 169.0130(100), 313.0559(60), 167.0336(30), 345.1181(26), 151.0022(26)
6′-O-Galloyl-desbenzoylpaeoniforin isomer
23#
5.82
321.02520
321.02509
−0.358
C14H10O9
MS2[321]: 169.0132(100), 125.0232(74)
digallic acid isomer
24#
5.91
483.07802
483.07797
−0.121
C20H20O14
MS2[483]: 169.0133(100), 125.0232(78), 211.0242(42), 271.0459(40), 313.0566(10)
Isomer of digalloylglucose
25#
5.98
321.02520
321.02512
−0.265
C14H10O9
MS2[321]: 169.0132(100), 125.0232(78)
digallic acid isomer
26*#
6.11
633.07333
633.07367
0.526
C27H22O18
MS2[633]: 300.9988(100), 275.0195(17), 125.0231(7), 169.0125(6)
Corilagin(Galloy-HHDP-hexoside)
27#
6.14
635.08898
635.08929
0.477
C27H24O18
MS2[635]: 169.0132(100), 125.0232(78), 465.0673(30), 313.0566(27), 123.0075(17)
Isomer of trigalloylglucose
28
6.15
527.14062
527.14056
−0.130
C23H28O14
MS2[527]: 169.0133(100), 125.0232(85), 123.0075(41), 151.0024(23), 107.0126(13)
6′-O-Galloyl-desbenzoylpaeoniforin isomer
29#
6.19
483.07802
483.07809
0.128
C20H20O14
MS2[483]: 125.0232(100), 169.0133(70), 151.0026(42), 439.0882(18), 107.0125(15)
Isomer of digalloylglucose
30#
6.34
483.07802
483.07813
0.210
C20H20O14
MS2[483]: 125.0232(100), 169.0133(43), 331.0672(33), 149.9948(31), 151.0027(16)
Isomer of digalloylglucose
31#
6.38
703.16684
703.16718
0.479
C36H32O15
MS2[703]: 61.9869(100), 289.0714(98), 251.0556(76), 125.0230(62), 287.0556(26),
theaflavin 3′-gallate isomer
32#
6.50
703.16684
703.16699
0.209
C36H32O15
MS2[703]: 61.9869(100), 289.0714(12), 251.0556(11), 125.0230(7)
theaflavin 3′-gallate isomer
33
6.57
527.14062
527.14056
−0.130
C23H28O14
MS2[527]: 347.0765(100), 345.1185(25), 169.0130(37), 125.0229(13)
6′-O-Galloyl-desbenzoylpaeoniforin isomer
34#
6.64
785.08429
785.08472
0.541
C34H26O22
MS2[785]: 300.9991(100), 275.0198(45), 249.0399(34), 125.0232(32), 169.0132(23)
digalloyl-HHDP-glucose isomer
35#
6.67
703.16684
703.16742
0.820
C36H32O15
MS2[703]: 61.9869(100), 289.0714(10), 125.0230(8)
theaflavin 3′-gallate isomer
36
6.68
445.13514
445.13501
−0.313
C19H26O12
MS2[445]: 121.0282(100), 135.0440(15)
benzoylsucrose isomer
37#
6.69
801.07920
801.07983
0.774
C34H26O23
MS2[801]: 125.0231(100), 289.0714(99), 121.0281(81), 96.9587(57), 151.0389(30), 169.0131(22)
punigluconin
38#
6.84
703.16684
703.16858
2.470
C36H32O15
MS2[703]: 61.9869(100), 289.0714(16), 251.0555(15), 125.0230(10)
theaflavin 3′-gallate isomer
39
7.00
445.13514
445.13489
−0.583
C19H26O12
MS2[445]: 121.02823(100)
benzoylsucrose isomer
40#
7.02
481.09876
481.09830
−0.964
C21H22O13
MS2[481]: 121.0280(100), 122.0314(39), 313.0559(27), 169.0129(24)
galloyvanilloy glucose isomer
41#
7.13
635.08898
635.08911
0.194
C27H24O18
MS2[635]: 169.0133(100), 125.0232(80), 465.0673(52),313.0565(20), 123.0075(13)
isomer of trigalloylglucose
42
7.29
197.04554
197.04495
−3.028
C9H10O5
MS2[197]: 169.0133(100), 125.0233(90), 140.0104(21), 111.0075(13)
ethyl gallate
43#
7.62
463.05181
463.05188
0.143
C20H16O13
MS2[463]: 300.9991(100), 89.0231(14), 101.0230(12), 59.0125(20)
ellagic acid hexose
44#
7.75
635.08898
635.08917
0.288
C27H24O18
MS2[635]: 169.0133(100), 125.0232(92), 313.0565(13),
Isomer of trigalloylglucose
45#
7.76
935.07960
935.08142
1.942
C41H28O26
MS2 [935]: 300.9985(100), 633.0729(78), 302.0016(14), 275.0195(11), 125.0230(10), 169.0129(8)
galloyl-bis-HHDP-D-glucopyranose
46#
7.78
335.04085
335.04089
0.104
C15H12O9
MS2[335]: 183.0291(100), 168.0056(8)
methyl digallate isomer
47#
7.95
441.08271
441.08270
−0.045
C22H18O10
MS2[441]: 289.0718(100), 109.0282(35), 125.0232(27), 123.0440(23), 245.0817(23), 203.0707(22), 137.0233(20)
(–)-Epicatechin gallate isomer
48#
8.11
477.06746
477.06760
0.286
C21H18O13
MS2[477]: 298.9834(100), 314.0070(93), 270.9885(67), 312.9991(24), 285.0047(18)
methylellagic acid glucopyranoside isomer
49#
8.38
787.09994
787.10040
0.578
C34H28O22
MS2[787]: 169.0133(100), 125.0232(76)
tetragalloylglucose isomer
50#
8.39
937.09525
937.09589
0.679
C41H30O26
MS2[937]:300.9989(100), 275.0196(27), 169.0134(47), 125.0232(24), 249.0403(14)
trigalloyl-HHDP-glucose isomer
51#
8.52
473.03616
473.03589
−0.578
C21H14O13
MS2[473]: 59.0125(100), 71.0125(53), 125.0233(50), 169.0133(39), 101.0231(27)
trigallic acid
52#
8.54
441.08271
441.08273
0.023
C22H18O10
MS2[441]: 289.0716(100), 125.0232(41), 137.0232(39), 109.0282(37), 245.0817(24)
(–)-Epicatechin gallate isomer
53#
8.66
787.09994
787.10059
0.819
C34H28O22
MS2[787]: 169.0133(100), 125.0232(76), 123.0076(17)
tetragalloylglucose isomer
54#
8.78
441.08271
441.08289
0.386
C22H18O10
MS2[441]: 169.0133(100), 125.0232(87), 289.0716(35), 109.0283(13), 245.0815(7), 203.0706(7)
(–)-Epicatechin gallate isomer
55#
8.87
481.09876
481.09906
0.616
C21H22O13
MS2[481]: 121.0283(100), 168.0550(53), 125.0232(43), 122.0316(39), 149.9948(30), 59.0125(15)
Galloyvanilloy glucose isomer
56#
9.04
787.09994
787.10065
0.895
C34H28O22
MS2[787]: 169.0133(100), 125.0232(86), 123.076(16), 617.0789(13)
tetragalloylglucose isomer
57#
9.20
477.06746
477.06747
0.013
C21H18O13
MS2[477]: 229.9911(100), 315.0147(76), 298.9832(28), 270.9883(20),
methylellagic acid glucopyranoside isomer
58
9.28
300.99899
300.99884
−0.500
C14H6O8
MS2[300]: 229.0138(100), 247.0085(84), 185.0235(39), 283.9959(37), 201.0179(30)
Ellagic acid
59#
9.36
715.13045
715.12921
−1.745
C36H28O16
MS2[715]: 82.9528(100), 169.0132(22), 121.0282(14), 125.0233(13)
theaflavin-3-Gallate isomer
60
9.37
631.16684
631.16681
−0.053
C30H32O15
MS2[631]: 169.01329(100), 125.0232(78), 121.0283(45), 123.0075(23), 211.0240(21), 313.0563(17), 271.0462(17)
galloylpaeoniflorin isomers
61#
9.38
335.04085
335.04068
−0.523
C15H12O9
MS2[335]: 183.0290(100), 168.0056(7)
methyl digallate isomer
62#
9.40
721.14102
721.13950
−2.111
C35H30O17
MS2[721]:169.0.0133(100), 125.0233(70), 461.1089(36), 211.0241(27), 313.0565(23), 123.0075(16)
thonningianin B
63#
9.42
783.10503
783.10773
3.447
C35H28O21
MS2[783]: 169.0133(100), 125.0232(92), 121.0283(72), 631.1673(34), 151.0027(22)
myricetin 3-O-(2′',3′'-digalloyl)-β-D-galactopyranoside
64#
9.60
937.09525
937.09595
0.743
C41H30O26
MS2[937]: 300.9990(100), 275.0199(22), 169.0133(19), 125.0231(18), 229.0129(11)
trigalloyl-HHDP-glucose isomer
65*#
9.69
441.08271
441.08255
−0.385
C22H18O10
MS2[441]: 289.0717(100), 125.0232(62), 169.0132(45), 137.0232(25), 203.0706(22), 245.0818(22)
(–)-Epicatechin gallate
66#
9.72
335.04085
335.04080
−0.165
C15H12O9
MS2[335]: 183.0290(100), 168.0055(7)
methyl digallate isomer
67#
9.72
461.07254
461.07260
0.110
C21H18O12
MS2[461]: 271.9177(100), 182.0210(93), 285.0400(82), 183.0287(50), 273.9148(33), 89.0229(31)
3,3′-Di-O-methyl-4-O-((β-D-xylopyranosyl)ellagic acid)
68#
9.79
491.08311
491.08307
−0.089
C22H20O13
MS2[491]: 328.0220(100), 312.9987(49), 169.0130(3), 125.0231(1)
dimethylellagic acid glucoside
69
9.91
631.16684
631.16693
0.137
C30H32O15
MS2[631]: 169.0133(100), 125.0232(89), 121.0283(55), 123.0075(38), 107.0126(21), 151.0026(18), 313.0569(13)
galloylpaeoniflorin isomers
70#
9.96
491.08311
491.08316
0.094
C22H20O13
MS2[491]: 169.0133(100), 125.0232(73)
dimethylellagic acid glucoside isomer
71
10.08
939.11090
939.11145
0.581
C41H32O26
MS2[939]: 169.0133(100), 125.0232(800), 123.0075(38), 95.0125(11)
pentagalloylglucose
72
10.15
631.16684
631.16705
0.327
C30H32O15
MS2[631]: 169.0133(100), 125.0232(93), 121.0283(56), 123.0075(37), 107.0125(23), 151.0025(15), 313.0570(12)
galloylpaeoniflorin isomers
73#
10.16
715.13045
715.12903
−1.997
C36H28O16
MS2[715]: 82.9528(100), 631.1674(67), 169.0134(7)
Theaflavin-3-Gallate isomer
74#
10.35
694.12091
694.12047
−0.646
C30H31O17S
MS2[694]: 169.0133(100), 125.0232(73), 121.0283(45), 123.0075(23), 631.1670(22)
Galloypaeoniforin sulfonate
75#
10.56
599.10424
599.10474
0.829
C28H24O15
MS2[599]: 169.0130(100), 313.0562(74), 285.0400(46), 284.0324(43), 241.0358(12)
kaempferol-3-O-(2′'-O-galloyl)-β-D-glucopyranoside
76#
10.66
801.11559
801.11627
0.842
C35H30O22
MS2[801]: 125.0232(100), 169.0133(98), 183.0290(62), 123.0076(30), 139.0390(20), 107.0125(15)
methyl tetragalloylglucose
77
10.85
631.16684
631.16711
0.423
C30H32O15
MS2[631]: 125.0232(100), 121.0283(97), 169.0133(79), 123.0076(59), 631.1675(39)
galloylpaeoniflorin isomers
78#
10.85
715.13045
715.12988
−0.808
C36H28O16
MS2[715]: 82.9528(100), 631.1674(46), 169.0133(8)
Theaflavin-3-Gallate isomer
79
11.13
1091.12186
1091.12244
0.529
C48H36O30
MS2[1091]: 939.1113(100), 169.0133(32), 769.0895(24)
hexagalloyl glucose isomer
80
11.15
545.05729
545.05743
0.137
C24H18O15
MS2[545]: 169.0133(100), 125.0232(76), 469.0517(65), 123.0075(11), 393.0457(8)
Dihydroxybenzoic acetate-digallate derivative
81#
11.20
783.17780
783.17847
0.853
C37H36O19
MS2[783]: 169.0134(100), 125.0232(84), 121.0283(48), 631.1669(29), 123.0076(26), 211.0243(22)313.0565(18)
digalloylpaeoniflorin isomer
82
11.25
1091.12186
1091.12244
0.529
C48H36O30
MS2[1091]: 939.1112(100), 169.0133(34), 769.0895(23)
hexagalloyl glucose isomer
83#
11.60
315.01464
315.01480
0.507
C15H8O8
MS2[315]: 299.9912(100)
methylellagic acid isomer
84#
11.61
487.05181
487.05090
−0.914
C22H16O13
MS2[487]: 169.0129(100), 125.0229(21), 183.0285(5)
methyl trigallate isomer
85
11.65
545.05729
545.05762
0.327
C24H18O15
MS2[545]: 169.0133(100), 125.0232(77), 469.0517(63), 123.0075(11), 393.0462(9)
Dihydroxybenzoic acetate-digallate derivative
86
11.65
1091.12186
1091.12268
0.749
C48H36O30
MS2[1091]: 169.0133(100), 939.1123(27), 769.0891(17)
hexagalloyl glucose isomer
87#
11.80
315.01464
315.01471
0.221
C15H8O8
MS2[315]: 299.9912(100)
methylellagic acid isomer
88
11.85
545.05729
545.05756
0.267
C24H18O15
MS2[545]: 169.0133(100), 125.0232(79), 469.0517(69), 123.0075(11), 393.0460(10)
Dihydroxybenzoic acetate-digallate derivative
89
11.85
1091.12186
1091.12256
0.639
C48H36O30
MS2[1091]: 169.0133(100), 939.1115(31), 769.0903(19)
hexagalloyl glucose isomer
90#
11.93
487.05181
487.05106
−0.754
C22H16O13
MS2[487]: 183.0290(100), 169.0133(81), 125.0232(66), 395.0323(21), 123.0075(20)
methyl trigallate isomer
91#
12.01
783.17780
783.17865
1.083
C37H36O19
MS2[783]: 169.0133(100), 125.0232(87), 121.0283(25), 123.0076(17), 151.0024(11), 107.0126(14)
digalloylpaeoniflorin isomer
92#
12.13
487.05181
487.05127
−0.544
C22H16O13
MS2[487]: 169.0133(100), 125.0232(76), 183.0290(72), 125.0232(76)
methyl trigallate isomer
93
12.23
621.05808
621.05758
−0.806
C22H22O21
MS2[621]: 469.0516(100), 169.0133(98), 125.0232(69)
galloy-valoneic acid bilactone isomer
94#
12.30
783.17780
783.17834
0.687
C37H36O19
MS2[783]: 169.0134(100), 125.0233(98), 121.0283(45), 123.0075(24), 107.0126(16),
digalloylpaeoniflorin isomer
95#
12.41
783.17780
783.17853
0.930
C37H36O19
MS2[783]: 169.0133(100), 125.0232(90), 121.0283(23), 123.0076(19), 107.0126(13)
digalloylpaeoniflorin isomer
96
12.71
621.05808
621.05763
−0.726
C22H22O21
MS2[621]: 469.0517(100), 169.0133(93), 125.0232(67)
galloy-valoneic acid bilactone isomer
97
12.71
1243.13282
1243.13391
0.876
C55H40O34
MS2[1243]: 939.1108(100), 769.0879(11), 1091.1224(8)
heptagalloy glucose
98#
12.74
461.10893
461.10910
1.262
C22H22O11
MS2[461]: 169.0133(100), 125.0232(98)
cinnamoylgalloy glucose isomers
99#
12.75
873.15198
873.15129
−0.791
C42H34O21
MS2[873]: 125.0232(100), 169.0133(83), 123.0074(24), 121.0284(16), 211.0240(12)
thonningianin A
100#
12.76
783.17780
783.17859
1.006
C37H36O19
MS2[783]: 125.0232(100), 169.0133(94), 121.0283(37), 123.0075(36),151.0026(14), 631.1684(11)
digalloylpaeoniflorin isomer
101#
12.96
487.05181
487.05194
0.126
C22H16O13
MS2[487]: 183.0290(100), 335.0411(13)
methyl trigallate isomer
102#
12.97
461.10893
461.10922
1.382
C22H22O11
MS2[461]: 151.0026(100), 125.0232(62), 169.0134(29), 83.0125(23), 107.0125(23)
cinnamoylgalloy glucose isomers
103#
14.70
461.10893
461.10913
1.292
C22H22O11
MS2[461]: 125.0232(100), 169.0055(94), 149.9949(64), 89.0231(14)
cinnamoylgalloy glucose isomers
104#
14.76
329.03029
329.03058
0.880
C16H10O8
MS2[329]: 314.0065(100), 298.9833(20)
dimethylellagic acid isomer
105#
15.00
329.03029
329.03046
0.515
C16H10O8
MS2[329]: 314.0068(100), 298.9834(80), 270.9883(76)
dimethylellagic acid isomer
106#
19.33
343.04594
343.04608
0.407
C17H12O8
MS2[343]: 312.9995(100), 328.0221(94), 297.9757(55)
trimethyl ellagic acid
3.4.1 Identification of hydrolyzed tannins
3.4.1.1 Identification of gallotannins
Gallic acid derivatives and gallotannins, composed of monomer galloyl moiety and multiple galloyl moieties linked to polyols were identified based on the DFIs at m/z 169.0133 ([gallic acid-H]−) and 125.0232 ([gallic acid-CO2-H]−) as well as the neutral loss of a dehydrated galloyl moiety (152 Da) (Erşan et al., 2016).
Compound 1, 3 and 6 were found at 0.85, 1.24, 1.70 min, which show common precursor ion at [M−H]− m/z 493.119. The major fragment at m/z 331.0672 due to loss of a glucose reside and m/z 313.0566 attributed to neutral loss of a glucose moiety, which further gave rise to the product ions at m/z 169.0133 and 125.0232. Then they were tentatively characterized as 1′-O-galloylsucrose, 6′-O-galloylsucrose and 6-O-galloylsucrose, respectively (Li et al., 2009). Compounds 2 and 7 appeared at a retention time (tR) of 1.15 min and 1.72 min respectively, which tentatively identified as galloylquinic acid isomers. The parent ions at m/z 343.066 due to the loss of galloyl moieties (152 Da) and further generated the characteristic fragments (m/z 169.0133 and 125.0232) of gallic acid moiety and characteristic fragments (m/z 191.0553) of quinic acid moiety (Erşan et al., 2016). Similarly, compound 15 was observed at 4.13 min, appeared similar losses to galloylquinic acid isomers with an extra loss of galloyl moiety (152 Da) and deduced as di-O-galloyl quinic acid.
Compound 5 was found at 1.53 min, possessing the quasi-molecular ion [M−H]− at m/z 331.0668 and tentatively identified as galloylglucose. The daughter ion at m/z 169.0132 attributed to the loss of a hexose moiety and further generated the ion at m/z 125.0232 by loss of CO2 (Erşan et al., 2016). The proposed fragmentation pathway of galloylglucose was shown in Fig. 2(A). The compound 24, 29, 30 with the molecular formula C20H20O14 were found at 5.91, 6.19, 6.34 min and having the quasi-molecular ions at m/z 483.078 in the ESI-mode, which were been tentatively proposed as isomers of digalloylglucose. (Erşan et al., 2016). Compounds 27, 41, and 44 yielded a quasi-molecular ion [M−H]− at m/z 635.089 and were eluted at 6.14, 7.13 and 7.75 min, respectively. All of those compounds showed the fragment ions at m/z 169.0133 (C7H5O5), 125.0232 (C6H5O3), 313.0565 (C13H14O9), 123.0075 (C5H3O3) and might be considered as isomer of trigalloylglucose. Compounds 49, 53, and 56 were eluted at 8.38, 8.66 and 9.04 min and shared the same empirical molecular formula C34H28O22 at m/z 787.100, matched to that of tetragalloylglucose isomers. The fragments of [M−gallic acid]−, [M−3gallic acids-108]−, [M−3gallic acids-152]− were found at m/z 617.0789, 169.0133, 125.0232 in tetragalloylglucose isomers (Erşan et al., 2016). Similarly, compound 71 was eluted at 10.08 min, yielded a deprotonated ion [M−H]− m/z 939.1114 and deduced as pentagalloylglucose. Compounds 79, 82, 86 and 89 at m/z 1091.122 having the same molecular formula C48H36O30 and appeared at a retention time (tR) of 11.13, 11.25, 11.65 and 11.85 min, which tentatively identified as hexagalloyl glucose isomers. Compound 97 was observed at 12.71 min, yielded a precursor ion at m/z 1243.133 have been characterized as heptagalloy glucose. The typical fragment ions of [M−H−galloyl]−, [M−H−2galloyl]−, [M−H−2galloyl−Glu]− were found at m/z 1091.1224, 939.1108 and 769.0879. In short, The mono-, di-, tri-, tetra-, penta-, hexa- and hepta- exhibited sequential losses of galloyl moieties (152 Da) from their parent ions at m/z 331.0668, 483.078, 635.089, 787.100, 939.1114, 1091.122 and 1243.133, respectively.
Compound 8, 10 and 12 were found at 2.10, 2.89, 3.10 min, possessing the same quasi-molecular ions [M−H]− at m/z 315.072 and tentatively identified as galloylrhamnose isomers. The main product ions at m/z 169.0133 were obtained by the loss of a rhamnose residue (146 Da) at [M−H]− and then obtain product ions at m/z 125.0232 by the loss of two CO2 (Sobeh et al., 2019).
Compound 9 and 11 exhibited the quasi-molecular ions at m/z 345.082 and appeared similar losses to galloylglucose with an extra loss of methyl moiety (14 Da). Based on major fragment ions at m/z 183.0290 ([M−H−hexose]−), 139.0390 ([M−H−hexose−CO2]−), these compounds were tentatively assigned to methyl galloylglucose isomers. Similarly, The compound 76 with the molecular formula C35H30O22 and having the deprotonated ions at m/z 801.1162 in the ESI-mode, which were been tentatively proposed as methyl tetragalloylglucose. It showed the presence of tetragalloylglucose fragment ions like 125.0232 and 169.0133. The fragment ion at m/z 183.0290 indicated the presence of a methyl gallate.
Compound 17 was found at 4.74 min, yielded parent ion [M−H]− m/z 451.1244, consisted of a galloyl moiety (152 Da) and a salidroside moiety (299 Da) and deduced as galloylsalidroside according to the MS and MS/MS spectra (Liu et al., 2017).
Compounds 18 was observed at 4.76 min, yielded a deprotonated ion [M−H]− m/z 527.1403 and tentatively identified as 6′-O-Galloyl-desbenzoylpaeoniforin (Li et al., 2016b). The MS/MS spectrum presented [M−H−HCHO]−, [M−H−HCHO−H2O]−, [M−H−HCHO−H2O−gallic−2HCHO−CO2]− ion at m/z 497.1295, 479.1187 and 271.0455. Compounds 13, 22, 28 and 33 were observed at 3.17, 5.43, 6.15 and 6.57 min, which appeared same precursor ions at m/z 527.140 with compounds 18 and yielded fragment ion at m/z 169.0130, 313.0559, 125.0232. The fragment ion at m/z 313.0563 and 169.0129 indicated the presence of a galloyl glucose. These compounds were tentatively proposed as isomers of 6′-O-Galloyl-desbenzoylpaeoniforin.
Compound 20, 36, and 39, were detected at 5.24, 6.68 and 7.00 min, with the same empirical molecular formula C19H25O15, matched to that of benzoylsucrose isomers and consistent with reference (Li et al., 2009).
Compound 40 and 55 were observed at 7.02 and 8.87 min, possessing the same quasi-molecular ions [M−H]− at m/z 481.099, matched to that of galloyvanilloy glucose isomers. The fragment at m/z 313.0559 generated by the loss of a vanilloy group, which further gave rise to the product ions at m/z 169.0129 and 125.0232. Similarly, compounds 98, 102 and 103 were observed at 12.74, 12.97 and 14.70 min, possessing the same quasi-molecular ions [M−H]− at m/z 461.109, matched to that of cinnamoylgalloy glucose isomers (Wang et al., 2020). The main daughter ion at m/z 151.0026 may be due to loss of a galloyl glucose (313 Da). The DFIs 169.0133 ([gallic acid-H]−) and 125.0232 ([gallic acid-CO2-H]−) were found in the UHPLC-Q-Exactive Orbitrap MS analysis. Compound 63 was eluted at 9.42 min, yielded parent ion [M−H]− m/z 783.1077 and deduced as myricetin 3-O-(2′',3′'-digalloyl)-β-D-galactopyranoside and had an MS/MS fragment ion at m/z 631.1673 caused by loss of a galloyl moiety (152 Da), showed characteristic fragments (m/z 169.0133 and 125.0232) of gallic acid moiety and characteristic fragments (m/z 151.0027) of myricetin moiety (Abu-Reidah et al., 2015). Compound 75 was detected at 10.56 min, yielded a parent ion [M−H]− m/z 599.1047 in the ESI-mode, which produced typical daughter ions at m/z 313.0562 and 285.0400 indicated the presence of a galloyl glucose and kaempferol, respectively. So it was tentatively identified as kaempferol-3-O-(2′'-O-galloyl)-β-D-glucopyranoside (Zehl et al., 2011).
Compound 60, 69, 72 and 77 were detected at 9.37, 9.91, 10.15 and 10.85 min, possessing the same quasi-molecular ions [M−H]− at m/z 631.166, matched to that of galloylpaeoniflorin isomers. MS/MS fragment ion at m/z 313.0570 and 121.0283 indicated the presence of a galloyl glucose and a benzoyl group, respectively (Xu et al., 2006) . Then the fragment at m/z 313.0570 gave rise to 169.0133, 125.0232 by successive loss of a glucose reside and a CO2. The proposed fragmentation pathway of galloylpaeoniflorin was shown in Fig. 2 (B). Similarly, compounds 81, 91, 94, 95 and 100 were detected at 11.20, 12.01, 12.30, 12.41 and 12.76 min, possessing the same precursor ions [M−H]− at m/z 783.178, matched to that of digalloylpaeoniflorin isomers and had one more dehydrated galloyl moiety (152 Da) than galloylpaeoniflorin. Therefore, their MS/MS fragmentation pattern were very similar to galloylpaeoniflorin. Besides, compounds 74 was eluted at 10.35 min, with the molecular formula C30H31O17S at m/z 694.1204. Among them, its fragment ion m/z 631.1670 proved the existence of a galloylpaeoniflorin. Therefore, this compound was determined as galloypaeoniforin sulfonate.
3.4.1.2 Identification of ellagitannins
Compound 26 with quasi-molecular ions [M−H]− at m/z 633.073 was eluted at 6.11 min and unambiguously identified as corilagin (galloy-HHDP-hexoside), which was affirmed by comparison to commercial reference standard. MS/MS fragment ion at m/z 300.9988 due to sequential loss of a gallic acid (170 Da) and a glucose residue (162 Da). The proposed fragmentation pathway of corilagin was shown in Fig. 2(C). Compound 14 also had the same [M−H]− m/z 633.073 and characteristic fragment m/z 300.9984, 275.0193, 169.0128 with compound 26, which was tentatively inferred as corilagin (galloy-HHDP-hexoside) isomer. Similarly, compound 21, 34 had quasi-molecular ions at m/z 785.084 and compound 50, 64 yielded same deprotonated molecule [M−H]− m/z 937.095. These compounds appeared similar losses to galloy-HHDP-hexoside isomers with an extra loss of galloyl moiety (152 Da) or two extra loss of galloyl moieties (304 Da), and all generated the characteristic fragment ions at m/z 300.9989 (C14H5O8), 275.0198 (C13H7O7), 125.0230 (C6H5O3), 169.0132 (C7H5O5), which been tentatively characterized as digalloyl-HHDP-glucose isomers and trigalloyl-HHDP-glucose isomers, respectively (Khaled et al., 2019). Compound 45 was found at 7.76 min and possessing the deprotonated ion [M−H]− at m/z 935.0798. The one of main fragments at 633.0724 m/z shows the loss of one HHDP group and fragment ions at m/z 300.9985 and 275.0195 indicated the presence of a HHDP group. The characteristic fragments m/z 169.0129 ([gallic acid-H]−), 125.0230 ([gallic acid-CO2-H]−) also can be observed. Based on these MS data, compound 45 was deduced as galloyl-bis-HHDP-D-glucopyranose and consistent with reference (Glasenapp et al., 2019).
Compound 37 was eluted at 6.69 min, yielded a deprotonated ion [M−H]− m/z 801.0798 and fragment ions at m/z 125.0231, 289.0714, 121.0281, 96.9587, 151.0389, 169.0131. Based on these MS data, compound 37 was suggested as punigluconin and consistent with reference (Chan et al., 2018).
Compounds 48 and 57 were eluted at 8.11 and 9.20 min, possessing the same quasi-molecular ions [M−H]− at m/z 477.067 and deduced as methylellagic acid glucopyranoside isomers (Chen et al., 2017). The main daughter ion at m/z 315.0147 was attributed to the loss of a glucose residue (162 Da), which further gave rise to the product ions at m/z 298.9832 due to loss of a methyl radical (15 Da).
Compound 58 was eluted at 9.28 min, yielded parent ion [M−H]− m/z 300.9988 and deduced as ellagic acid according to the MS and MS/MS spectra. The typical daughter ions at m/z 229.0138 [M−2H]− and 283 [M−H- H2O]− were attributed to loss of a H− or a H2O and then gave rise to the ions at m/z 169.0134 and 125.0232 (Abu-Reidah et al., 2015). The proposed fragmentation pathway of ellagic acid was shown in Fig. 2(D). Similarly, compound 43, which was eluted at 7.62 min, exhibited quasi-molecular ions [M−H]− at m/z 463.0518, generated main fragments at m/z 300.9991 by loss of a hexose (162 Da) and identified as ellagic acid hexose (Yang et al., 2012).
Compounds 67 appeared at a retention time (tR) of 9.72 min, possessing the quasi-molecular ions [M−H]− at m/z 461.0726 and tentatively identified as 3,3′-Di-O-methyl-4-O-(β-D-xylopyranosyl) ellagic acid. The MS/MS spectrum presented [M−H−189 Da]−, [M−H−187 Da]−, [methyl gallate–H]−, [methyl gallate–2H]− ion at m/z 273.9148, 271.9177, 183.0287 and 182.0210 (Singh et al., 2016).
Compounds 68 was eluted at 9.79 min, with the molecular formula C22H20O13 at m/z 491.083. The fragment ions m/z 328.0220 and 312.9987 indicated the presence of a dimethyl ellagic acid and methyl ellagic acid, respectively. Based on these MS data, compound 70 are suggested as dimethylellagic acid glucoside. Compounds 70 had same MS/MS spectra date with compounds 68 and characterized as the isomer of dimethylellagic acid glucoside.
Compounds 83 and 87 were eluted at 11.60 and 11.80 min, possessing the same quasi-molecular ions [M−H]− at m/z 315.014 and deduced as methylellagic acid isomers. The main daughter ion at m/z 299.9912 was attributed to the loss of a methyl radical (15 Da). Similarly, compound 104, 105 with the same precursor ions at m/z 329.030 have been characterized as dimethylellagic acid isomers and had an MS/MS fragment ion at m/z 314.0065, 298.9833 due to sequential loss of a methyl radical (15 Da) in MS2 (Zehl et al., 2011). Compounds 106 was eluted at 19.33 min, and possessing the deprotonated ion [M−H]− at m/z 343.046 and deduced as trimethyl ellagic acid. The product ions at m/z 328.0221, 312.9995 and 297.9757 were attributed to the loss of successive loss of a methyl radical (15 Da).
3.4.1.3 Identification of gallic acid derivatives
Gallic acid (4, tR 1.51 min) was the major tannins of PRA and exhibited quasi-molecular ion [M−H]− at m/z 169.0141, generated main characteristic fragments at m/z 125.0232 ([gallic acid-CO2-H]−), which was identified by comparison with reference substances. The proposed fragmentation pathway of gallic acid was shown in Fig. 2(E). Therefore, multiple galloyl moieties were found at m/z 321.025 (23, tR 5.82 min and 25, tR 5.98 min) and m/z 473.035 (51, tR 8.52 min), which were deduced as digallic acid isomers and trigallic acid, respectively. The product ions at m/z 321.0262 and 169.0147 were obtained by successive loss of gallic acid moieties from a main ion at m/z 473.0358 in the UHPLC-Q-Exactive Orbitrap MS analysis.
Compounds 16 was eluted at 4.64 min, possessing quasi-molecular ion [M−H]− at m/z 183.0298, showed characteristic fragments at (m/z 168.0056) of a demethylated product ion, and was accurately characterized as methyl gallate, which was identified by comparison with reference substances. Compounds 19 had the same mass spectrum information and identified as methyl gallate isomer. Similarly, compounds (46, tR 7.78 min, 61, tR 9.38 min and 66, tR 9.72 min) and compounds (84, tR 11.61 min, 90, tR 11.93 min, 92, tR 12.13 min, and 101, tR 12.96 min) were tentatively characterized as methyl digallate isomers and methyl trigallate isomers due to sequential loss of galloyl moieties (152 Da) from their parent ions at [M−H]− m/z 335.0408 and [M−H]− m/z 487.051, respectively, showing that the characteristic fragments m/z 169.0133 ([gallic acid-H]−), 125.0232 ([gallic acid-CO2-H]−). One of major fragment at m/z 183.0291 could be yielded by the loss of a galloyl moiety (152 Da). Compound 42 was eluted at 7.29 min, yielded a deprotonated ion [M−H]− m/z 197.044 and deduced as ethyl gallate.
Compounds 80, 85 and 88 were observed at 11.15, 11.65, and 11.85 min, possessing the some quasi-molecular ions [M−H]− at m/z 545.057, matched to that of Dihydroxybenzoic acetate-digallate derivative. MS/MS fragment ion at m/z 469.0517 ([M−H−72]−) and 393.0462 ([M−H−152]−) were obtained owing to neutral losses of acetyl + H2O and galloyl moieties from the precursor ion (m/z 545), respectively. In addition, The product ions at m/z 169.0133, 125.0232 and 123.0075 indicated the presence of a gallic acid (Dienaitė et al., 2019).
Compound 93 and 96 were detected at 12.23 and 12.71 min, possessing the same quasi-molecular ions [M−H]− at m/z 621.057, matched to that of galloy-valoneic acid bilactone isomers. The dominant product ions at m/z 469.0516 attributed to neutral loss of galloyl moiety (152 Da), indicating valoneic acid bilactone in structure (Dienaitė et al., 2019). The characteristic fragments m/z 169.0133 ([gallic acid-H]−), 125.0232 ([gallic acid-CO2-H]−) also can be observed.
3.4.2 Identification of condensed tannins
Compound 65 was eluted at 9.69 min, yielded deprotonated ion [M−H]− m/z 441.082 and unambiguously identifed as (–)-Epicatechin gallate, which was affirmed by comparison to commercial reference standard. There compounds (47, tR 7.95 min, 52, tR 8.54 min and 54, tR 8.78 min) had the same [M−H]− m/z 441.082 and characteristic fragment m/z 289.0716 (C15H13O6), 169.0133 (C7H5O5), 125.0232 (C6H5O3), 245.0817 (C14H13O4) with compounds 65. So, they were tentatively assigned as (–)-Epicatechin gallate isomers. The proposed fragmentation pathway of (–)-Epicatechin gallate was shown in Fig. 2(F).
Compound 31, 32, 35 and 38 were found at 6.38, 6.50, 6.67 and 6.83 min, possessing the same parent ions [M−H]− at m/z 703.167 and tentatively deduced as theaflavin 3′-gallate isomers. One of main fragment at m/z 125.0230 probably produced by sequential two loss of catechin moiety (289 Da). The product ions at m/z 61.9869, 289.0714 emerged from catechin moiety (Poon, 1998).
Compounds 59, 73 and 78 at m/z 715.129 having the same molecular formula C36H28O16 and appeared at a retention time (tR) of 9.36, 10.16, and 10.85 min. Based on these MS2 data, compound 61, 75 and 79 are suggested as theaflavin-3-gallate isomers and consistent with reference (Kuhnert et al., 2010).
Compound 62 (tR 9.40 min) had quasi-molecular ion at m/z 721.1395 and compound 99 (tR 12.75 min) yielded deprotonated ion [M−H]− at m/z 873.1512. The molecular weight of Compound 62 is 152 Da less than that of compound 99, whereby the difference lies in the different substituents present at the C-3 position of glucose, whereby one is the H and the other is the galloyl, and all generated the characteristic fragment ions at m/z 169.0133 (C7H5O5), 125.0233 (C7H5O5), 211.0241 (C13H7O3), which been tentatively characterized as thonningianin B and thonningianin A, respectively (Wong et al., 2020).
3.5 Pharmacological activity of tannins in PRA
According to reports, tannins have antiviral, antitumour, antihypertensive, uraemic toxin decreasing and renal failure-improving actions, and have great potential as a new pharmaceutical resource (Adderson et al., 1961; Nishioka, 1983).
Gallotannins represent the simplest class of hydrolyzable tannins, containing gallic acid substituents esterified with a polyol residue. As highmolecular-weight tannins, penta-, hexa- and heptagalloylglucose, have greatest potential to reduce blood glucose in an insulin resistant state (Juan et al., 2011). Moreover, there are many studies on the biological activity of pentagalloylglucose , which have seen traditional use in the treatment of diseases related to skin barrier disruption (Kim et al., 2020) Furthermore, pentagalloylglucose has other main physiological activities including anti-inflammatory, anti-allergic, antitumor, antiviral, and antibacterial effects (Parker et al.,2016), and recently been highlighted as the most important and is currently being developed into a therapeutic for cancer and diabetes (He et al., 2010).
Ellagitannins have antibacterial and antioxidant properties, and can be used as chemical defense barriers due to the unique particularities of their structure and the remarkable selectivity of their embedded chemical reactivity, sush as galloy-HHDP-hexoside (Quideau et al., 2010., Li et al., 2016a).
Gallic acid, an abundant hydrolyzable tannin found in RPA, has been extensively studied for its antioxidant and antiviral activity (Parker et al.,2016).
Condensed tannins, also referred to as proanthocyanidins, have been shown to have the potential beneficial effects on human health, including immunomodulatory, anti-inflammatory, anticancer, antioxidant, cardioprotective and antithrombotic properties (Sieniawska, 2015., Smeriglio et al., 2014). In particular, theaflavin 3′-gallate and theaflavin-3-gallate would be potential compounds for treating platinum-resistant ovarian cancer (Pan et al., 2017).
4 Conclusion
In this research, an efficient strategy based on UHPLC Q-Exactive Orbitrap MS in the negative ion mode was established to detect tannins components in PRA. Finally, a total of 106 constituents, among them, 75 compounds were first reported in PRA, including gallotannins, ellagitannins, gallic acid derivatives and condensed tannins were detected and identified based on their chromatographic retention, MS and MS2, and bibliography data. According to previous studies, gallic acids are the main component of RPA, which have multiple pharmacological activities including anti-inflammatory, antioxidant, antibacterial, antitumor, and provide treatment for hypertension, myocardial infarction and diabetes. Overall, the result laid the foundation for in-depth research on the Pharmacodynamic material basis of PRA.
Acknowledgements
This work was financially supported by the the Scientific Research Fund of Hunan Provincial Education Department (no. 19A353), the Scientific research cultivation project of Hunan University of Medicine (20KJPY01) and the Hunan University of Medicine High-Level Talent Introduction Startup Funds (no. 15001).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- HPLC–DAD–ESI-MS/MS screening of bioactive components from Rhus coriaria L. (Sumac) fruits. Food Chem.. 2015;166:179-191.
- [Google Scholar]
- The pharmacognosy of the root of Rauwolfia ligustrina Roem. and Schult. J. Pharm. Pharmacol.. 1961;13:224-239.
- [Google Scholar]
- A systematic strategy for rapid identification of chlorogenic acids derivatives in Duhaldea nervosa using UHPLC-Q-Exactive Orbitrap mass spectrometry. Arab. J. Chem.. 2020;13(2):3751-3761.
- [Google Scholar]
- Enhancing antioxidant capacity of Lactobacillus acidophilus-fermented milk fortified with pomegranate peel extracts. Food Biosci.. 2018;26:185-192.
- [Google Scholar]
- Current situation and prospect of chemical utilization of gallic tannin in China. Chem. Ind. Forest Prod. 2000:71-82.
- [Google Scholar]
- In vitro gastrointestinal digestion promotes the protective effect of blackberry extract against acrylamide-induced oxidative stress. Sci. Rep.. 2017;13:7-40514.
- [Google Scholar]
- Protective effects of co-administration of gallic Acid and cyclosporine on rat myocardial morphology against ischemia/reperfusion. Jundishapur J. Nat. Pharm. Prod.. 2014;9:17186-17795.
- [Google Scholar]
- Isolation of strong antioxidants from paeonia officinalis roots and leaves and evaluation of their bioactivities. Antioxidants (Basel). 2019;8:249.
- [Google Scholar]
- Identification of phenolic compounds in red and green pistachio (Pistacia vera L.) hulls (exo-and mesocarp) by HPLC-DAD-ESI-(HR)-MS. J. Agric. Food Chem.. 2016;64:5334-5344.
- [Google Scholar]
- Anti-adhesion activity of tannins isolated from the mangrove laguncularia racemosa. Chem. Biodivers.. 2019;16(5):e1800632.
- [CrossRef] [Google Scholar]
- Phytochemical and biological studies of Paeoniaceae. Chem. Biodivers. 2010;7(4):805-838.
- [Google Scholar]
- Challenges in analysis of hydrophilic metabolites using chromatography coupled with mass spectrometry. J. Anal. Test.. 2020;4:140-162.
- [Google Scholar]
- Pharmacological evaluation of insulin mimetic novel suppressors of PEPCK gene transcription from Paeoniae Rubra Radix. J. Ethnopharmacol.. 2011;1371:592-600.
- [Google Scholar]
- A biochemometric approach for the assessment of Phyllanthus emblica female fertility effects as determined via UPLC-ESI-qTOF-MS and GC-MS. Food & function. 2019;10(8):4620-4635.
- [Google Scholar]
- Penta-O-galloyl-β-D-glucose from Paeonia lactiflora Pall. root extract enhances the expression of skin barrier genes via EGR3. J. Ethnopharmacol. 2020;248:112337.
- [CrossRef] [Google Scholar]
- Oxidative cascade reactions yielding polyhydroxy-theaflavins and theacitrins in the formation of black tea thearubigins: evidence by tandem LC-MS. Food Funct.. 2010;1(2):180.
- [CrossRef] [Google Scholar]
- Gallic acid isolated from Spirogyra sp. improves cardiovascular disease through a vasorelaxant and antihypertensive effect. Environ. Toxicol. Pharmacol.. 2015;39(2):764-772.
- [Google Scholar]
- Identification and quantification of the bioactive components in Osmanthus fragrans roots by HPLC-MS/MS. J. Agric. Food Chem.. 2020;66:359-367.
- [Google Scholar]
- High-resolution MALDI mass spectrometry imaging of gallotannins and monoterpene glucosides in the root of Paeonia lactiflora. Sci. Rep.. 2016;6:36074.
- [Google Scholar]
- HPLC-ESI-MS∼ N method was used to identify the chemical constituents in serum and brain of rats after oral administration of extract of Paeonia lactiflora. Chin. Tradit. Herbal Drugs. 2016;47:2475-2481.
- [Google Scholar]
- Chemical profiling of Radix Paeoniae evaluated by ultra-performance liquid chromatography/photo-diode-array/quadrupole time-of-flight mass spectrometry. J. Pharm. Biomed. Anal.. 2009;49(2):253-266.
- [Google Scholar]
- Qualitative and quantitative analysis of major constituents from Dazhu Hongjingtian capsule by UPLC/Q-TOF-MS/MS combined with UPLC/QQQ-MS/MS. Biomed. Chromatogr.. 2017;31(6):e3887.
- [CrossRef] [Google Scholar]
- Research progress on the biological effects of gallic acid. China Pharm.. 2004;7:767-769.
- [Google Scholar]
- Cytotoxic activity of gallic acid against liver metastasis of mastocytoma cells P-815. Anticancer Res.. 2001;21:3785-3880.
- [Google Scholar]
- Chemistry and biological activities of tannins. Yakugaku zasshi: J. Pharmac. Soc. Jpn.. 1983;103:125-142.
- [Google Scholar]
- Simultaneous determination of four monoterpene glycosides in total glycosides of Paeonia paeoniae by HPLC. Chin. Med. Mat.. 2013;36:423-425.
- [Google Scholar]
- Inhibitory effect of black tea pigments, theaflavin–3/3'-gallate against cisplatin-resistant ovarian cancer cells by inducing apoptosis and G1 cell cycle arrest. Int. J. Oncol.. 2017;51:1508-1520.
- [Google Scholar]
- A pharmacological review of bioactive constituents of paeonia lactiflora pallas and paeonia veitchii lynch. Phytother. Res. : PTR. 2016;30:1445-1473.
- [Google Scholar]
- Analysis of catechins in tea extracts by liquid chromatography– electrospray ionization mass spectrometry. J. Chromatogr. A. 1998;794(1-2):63-74.
- [Google Scholar]
- Antihyperglycaemic, antilipid peroxidative and antioxidant effects of gallic acid on streptozotocin induced diabetic Wistar rats. Eur. J. Pharmacol.. 2011;650(1):465-471.
- [Google Scholar]
- Ellagitannins-an underestimated class of plant polyphenols: chemical reactivity of C-glucosidic ellagitannins in relation to wine chemistry and biological activity. Recent Adv. Polyphenol. Res.. 2010;2:81-137.
- [Google Scholar]
- Characterization of acidic glycosphingolipid changes in C6 glioma rats treated with temozolomide using ultra-high-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry. J. Anal. Test.. 2020;4:217-225.
- [Google Scholar]
- Health effects of Vaccinium myrtillus L.: evaluation of efficacy and technological strategies for preservation of active ingredients. Mini Rev. Med. Chem.. 2014;14:567-584.
- [Google Scholar]
- Albizia anthelmintica: HPLC-MS/MS profiling and in vivo anti-inflammatory, pain killing and antipyretic activities of its leaf extract. Biomed. Pharmacother.. 2019;115:108882.
- [CrossRef] [Google Scholar]
- Activities of tannins-from in vitro studies to clinical trials. Nat. Prod. Commun.. 2015;10:1877-1884.
- [Google Scholar]
- Profiling of gallic and ellagic acid derivatives in different plant parts of Terminalia arjuna by HPLC-ESI-QTOF-MS/MS. Nat. Prod. Commun.. 2016;11:239-244.
- [Google Scholar]
- State Pharmacopoeia Commission, 2020. Pharmacopoeia of the People's Republic of China: A edition.
- Chemical characterisation and quantification of the major constituents in the Chinese herbal formula Jian-Pi-Yi-Shen pill by UPLC-Q-TOF-MS/MS and HPLC-QQQ-MS/MS. Phytochem. Anal.. 2020;31(6):915-929.
- [Google Scholar]
- Comparison of pharmacological effects between Radix Paeonia lactiflora and Radix Paeonia lactiflora. Chin. J. Exp. Tradit. Med. Form.. 2010;16:112-114.
- [Google Scholar]
- Biological activities of selected plants and detection of bioactive compounds from ardisia elliptica using UHPLC-Q-exactive orbitrap mass spectrometry. Molecules. 2020;25:3067.
- [Google Scholar]
- Analysis of hydrolyzable tannins and other phenolic compounds in emblic leafflower (Phyllanthus emblica L.) fruits by high performance liquid chromatography–electrospray ionization mass spectrometry. J. Agric. Food Chem.. 2012;60(35):8672-8683.
- [Google Scholar]
- Research progress of condensed tannins. China Tradit. Herb. Drugs. 2013;44:1687-1699.
- [Google Scholar]
- Characterization of compounds in the Chinese herbal drug Mu-Dan-Pi by liquid chromatography coupled to electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom.. 2006;20(22):3275-3288.
- [Google Scholar]
- Identification and quantification of flavonoids and ellagic acid derivatives in therapeutically important Drosera species by LC–DAD, LC–NMR, NMR, and LC–MS. Anal. Bioanal. Chem.. 2011;400(8):2565-2576.
- [Google Scholar]







