Translate this page into:
Amiloride-modulated phosphorescence turn-off/on method for the detection of abasic site-containing dsRNA based on uridine triphosphate-capped Mn-doped ZnS quantum dots
⁎Corresponding author. crush_bc@163.com (Yan Gong)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
RNA plays a crucial role in biochemical processes. RNAs with abasic sites (AP-RNA), as one of RNA lesions, will cause serious negative consequences. Efficient and sensitive detection of AP-RNA deserves but does not receive enough attention. Water-soluble uridine triphosphate (UTP)-capped Mn-doped ZnS quantum dots (QDs), which have excellent room-temperature phosphorescence (RTP) properties, were prepared by wet-chemical method. The UTP on the surface of QDs could form a precipitation complex with amiloride (AMI) to quench the RTP. Meanwhile, abasic double-stranded RNA (AP-dsRNA) with a high affinity to AMI could dissociate AMI from the surface of UTP-QDs, resulting in RTP recovery. In this process, a highly sensitive and selective RTP-detection system for AP-dsRNA was constructed with AMI as the modulation factor and UTP-QDs as the luminescence matrix. RTP was turned off and on for the determination of AP-dsRNA, and the interference of background fluorescence and scattered light was effectively avoided during complex formation and aggregation. Steric hindrance of the conformation of AP-dsRNA provided efficient selectivity. The detection limit for AP-dsRNA was 0.86 nM, the relative standard deviation was 2.1%, and the recovery of biological samples with AP-dsRNA addition ranged from 95% to 103% at optimal conditions. The system is expected to provide new methods for environmental monitoring, early disease screening, and evaluation of gene editing efficiency.
Keywords
Uridine triphosphate
Quantum dot
Amiloride
Abasic double-stranded RNA
Phosphorescence
1 Introduction
RNA molecules play an important role in peptide bond formation (Nissen et al., 2000; Moore and Steitz, 2003), gene regulation (Bernstein et al., 2001; He and Hannon, 2004), and catalysis (Zhuang et al., 2000; Yamagami et al., 2019) in living organisms. However, the damage and repair of RNA molecular structures have not been studied as fully as those of DNA (Bellacosa and Moss, 2003). Abasic sites or apurinic/apyrimidinic (AP) sites, as a common form of RNA damage, can be formed under the effects of drug induction (Barbieri et al., 1993; Peumans et al., 2001), photochemical stimulation (Trzupek and Sheppard, 2005), viral reverse transcriptase (Küpfer and Leumann, 2005), and oxidation (Tanaka et al., 2011) due to the cleavage of N-glycosidic bond. AP sites can have a substantial negative impact on RNA structure and function, such as RNA-coding/folding properties (Trzupek and Sheppard, 2005) and protein recognition functions (Li et al., 2013; Hur, 2019), which are thought to be closely related to various neurodegenerative and other diseases (Calabretta et al., 2015). Most abasic RNAs (AP-RNAs) are single-stranded (AP-ssRNA) in their natural state, and the glycosidic bond of RNA is more stable than that of DNA (Garrett et al., 1966); hence, the detection of AP-RNA has not attracted much attention (Wang et al., 2018; Küpfer et al., 2007). AP-ssRNA analysis includes a few spectroscopic methods using fluorescent materials, such as aldehyde reactive probe (Tanaka et al., 2011) and nucleoside analog (Tanpure and Srivatsan, 2012).
AP double-stranded RNA (AP-dsRNA) are potent and allele-selective inhibitors of huntingtin (Huntington’s disease) and ataxin-3 (Machado–Joseph disease) expressions (Liu et al., 2013). A novel tool for gene silencing can be provided by introducing abasic substitutions as a tool for tailoring RNA duplexes (Hu et al., 2014). This abasic substitution of dsRNA prevents base pairing and steric hindrance and thus can greatly improve chemical stability (Küpfer and Leumann, 2006) and on-target activity (Lee et al., 2015). Effective quantitative studies on AP-dsRNA can be used to evaluate gene regulation and gene editing efficiency, assess the affinity of aptamers and molecular beacons (Xiang et al., 2009), and determine miRNA or siRNA (Bartel, 2004) to be applied in the early screening of diseases, efficacy evaluation, and toxicological (such as genotoxins and oxidative stress) detection. Unfortunately, only very few fluorescence (Sato et al., 2012; Sato et al., 2013; Sato et al., 2014) and noncovalent spin labeling (Kamble and Sigurdsson, 2018) have been reported for AP-dsRNA detection.
Phosphorescence has better analytical performance than fluorescence in terms of photoluminescence detection. Phosphorescence has a longer life and a lower probability of spontaneous occurrence; hence, background interference, especially the spontaneous fluorescence of biomass materials, can be effectively avoided (Schulman, 2017). Mn-doped ZnS quantum dots (QDs) with room-temperature phosphorescence (RTP) have been widely used in analytical detection (Li et al., 2016; Zhang et al., 2018) and biological imaging (Zhou et al., 2014; Singhal et al., 2019). Although many studies have been conducted on the functionalization of QDs using inorganic or organic molecules (Yang and Bredol, 2008; Karakoti et al., 2015), the modification of and research on QDs using uridine triphosphate (UTP) have not been reported. Moreover, using a polyphosphate compound, such as adenosine triphosphate (Ren and Yan, 2012) or sodium polyphosphate (Wang et al., 2013), is practical in modifying QDs. In this study, UTP served as a surface passivator that provides the water stability of QDs and provides a possibility for the detection of AP-dsRNA.
As an excellent AP site-binding molecule, amiloride (AMI)’s chemical structure contains a guanidinium group containing pyrazine derivative (Scheme 1), which can bind strongly and selectively to AP-dsRNA, and its binding ability is two orders of magnitude stronger than that of AP-dsDNA (Sato et al., 2012). AMI can be combined with UTP to form a precipitated complex (Pettis et al., 2004). A sensitive AP-dsRNA detection system was constructed on the basis of the special properties of AMI by using AMI as a modulating substance and UTP-QDs as luminescence matrix. The supposed mechanism is that AMI could quench the RTP of UTP-QDs by electrostatic adsorption, and then AMI would dissociate from the surface of UTP-QDs with the introduction of AP-dsRNA under a stronger attraction of AP-dsRNA to AMI, resulting in RTP recovery. The system undergoes a process in which RTP is turned off and on (Scheme 1B).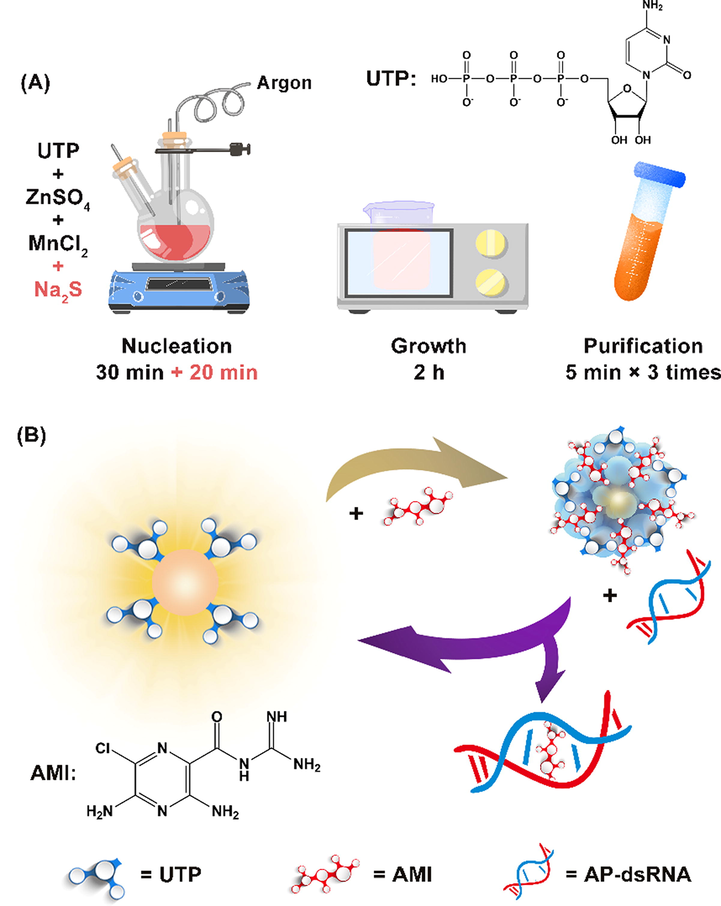
(A) Schematic diagram of UTP-QDs synthesis. (B) Mechanism of AP-dsRNA detection using UTP-QDs RTP modulated byAMI.
Mn-doped ZnS QDs with RTP properties were synthesized through wet-chemical method using UTP as the stabilizer and functional agent (Scheme 1A). AMI was used as a modulating material to establish a set of RTP off/on-sensitive detection methods for AP-dsRNA. This method exhibits strong specificity and high sensitivity and has been successfully applied to the analysis of AP-dsRNA in biological fluids to provide new methods for early disease screening and toxicity identification.
2 Experimental section
2.1 Materials and chemicals
All kinds of reagents related to RNA and DNA (see SI for sequence) were synthesized and purified by Sangon Biotech Co., Ltd. (Shanghai, China). Uridine 5′-triphosphate trisodium salt dihydrate, zinc sulfate heptahydrate, manganese chloride, and sodium sulfide nonahydrate were purchased from Aladdin Biotechnology Co., Ltd. (Shanghai, China). Phosphoric acid, boric acid, acetic acid, and sodium hydroxide were used to prepare Britton–Robinson (BR) buffer solution (J&K scientific Ltd., Beijing, China). All chemicals were analytical grade and were used as received without further purification. Ultrapure water (18.2 MΩ cm) was prepared via a Milli-Q water integration system (Merck KGaA, Darmstadt, Germany).
2.2 Apparatus
The microstructure was characterized using a laser particle size analyzer (Zetasizer Nano ZS, Malvern, UK), a transmission electron microscope (TEM, Tecnai G2 F20, FEI, US), an atomic force microscope (AFM, SPA400, Seiko, Japan), and an X-ray diffractometer (XRD, D8 ADVANCE, Bruker, Germany). Material composition data were obtained via X-ray photoelectron spectroscopy (XPS, ESCALAB 250Xi, Thermo, US). Surface group properties were determined using an infrared spectrophotometer (IR, Varian 640, US) with KBr pellets as the sample matrix in the frequency range of 500–4000 cm−1. Establishment of the detection system was mainly achieved by setting the fluorescence spectrophotometer (Eclipse, Varian, US) to phosphorescence mode at the excitation wavelength of 300 nm, and the slit widths of excitation and emission were 10 and 20 nm, respectively. An ultraviolet spectrophotometer (UV-3600, Shimadzu, Japan) was used for auxiliary characterization.
2.3 Synthesis of UTP-capped Mn-ZnS QDs
UTP-capped Mn-doped ZnS QDs were synthesized through a one-step wet-chemical method. Added 2.5 mL ZnSO4 (0.1 M) and 1 mL MnCl2 (0.01 M) to 50 mL UTP solution (0.01 M) in sequence. The reaction was stirred for 30 min under an argon atmosphere. 2.5 mL Na2S (0.1 M, the solution was prepared upon use) was quickly added after the UTP stably combined with Zn2+ and Mn2+, and then stirring was continued for 20 min. The reaction solution was then transferred to a beaker and bathed at 50 °C for 2 h under air atmosphere. Centrifuged (3000 rpm, 5 min) and washed (ethanol, v/v = 1:1) three times, then vacuum dry to obtain the desired UTP-QD product.
2.4 Measurement procedure
BR buffer solution (0.5 mL, 0.1 M, Ph 5.8), UTP-QDs (0.5 mL, 0.12 mg mL−1), NaCl (0.5 mL, 0.1 M), and equal volumes of AMI standard solutions of different concentrations were sequentially added to a 10 mL calibrated test tube (final concentration range of 10–625 nM) to examine the quenching behavior of AMI on the RTP of UTP-QDs.
BR buffer solution (0.5 mL, 0.1 M, pH 5.8), UTP-QDs (0.5 mL, 0.12 mg mL−1), NaCl (0.5 mL, 0.1 M), and AMI (500 μL, 6 mM) were sequentially added to a 10 mL calibrated test tube to detect AP-dsRNA based on its RTP recovery, which was quenched by AMI-induced. After the mixture was left for 5 min, AP-dsRNA standard solution (or real sample) was added. Then, left for another 10 min to measure RTP.
2.5 Biological sample treatment
Serum samples were collected at a local hospital (The Second People’s Hospital, Linfen, China), and plant lysate samples, which came from tobacco (Nicotiana tabacum, K326) leaves infected with cucumber mosaic virus, were provided by Biofavor Biotech Services Ltd. (Wuhan, China). An appropriate (150-fold) dilution of serum and plant lysate was made before detection.
3 Results and discussion
3.1 Characterization of UTP-capped Mn-doped ZnS QDs
Morphological characterization results showed that the QDs have an average particle size of 3.6 nm (Fig. 1A) and a thickness of 3.9 nm (Fig. 1B). The XRD pattern of QDs (Fig. 1C) showed obviously (1 1 1), (2 2 0), and (3 1 1) planes of typical zinc blend structure. The XPS pattern of UTP-QDs exhibited the peaks of Zn (2p) at 1022 and 1045 eV (Fig. 1D), which are attributed to the presence of Zn2+ that matches the Zn–S bonding energy (Shin et al., 2011). Weak Mn 2p spectra in the range of 630–660 eV shown in Fig. 1D (inset) prove the existence of Mn2+ in the form of zinc atom substitutes or interstitial impurities (Yang et al., 2001).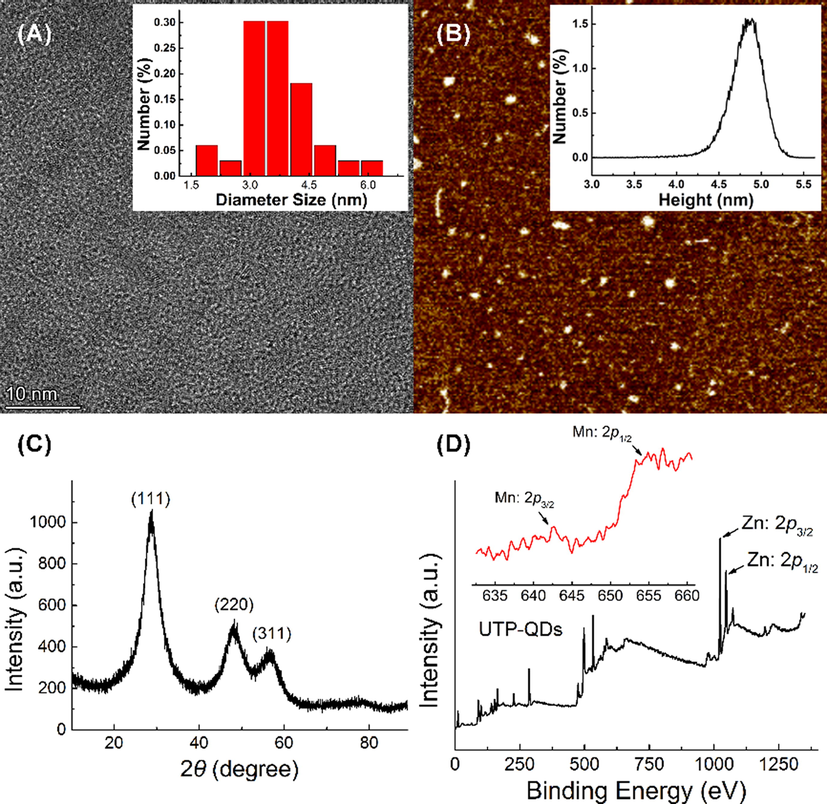
Characterization of UTP-capped Mn-doped ZnS QDs: (A) TEM image and particle size distribution (inset); (B) AFM image and height distribution (inset); (C) XRD patterns; (D) XPS images and Mn 2p spectra (inset).
As an inorganic material, Mn-doped ZnS QDs has relatively few infrared characteristic peaks; hence, the infrared characteristic peak at 1000–4000 cm−1 is mainly contributed by UTP (Fig. 2). Two distinct characteristic peak shifts, including the carbonyl group on amide shifting from 1702 cm−1 to 1637 cm−1 and the PO2− peak shifting from 1255 cm−1 to 1236 cm−1, were observed by comparing UTP with UTP-QDs. This shifting indicates that amide and phosphate bonds are involved in the modification of UTP-capped Mn-doped ZnS QDs (Ren and Yan, 2012). The attribution of detailed infrared characteristic peaks is shown in Table S1.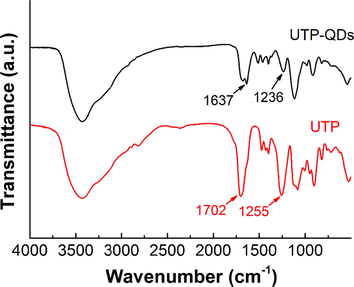
Infrared spectra of UTP-QDs and UTP.
RTP characterization results showed that the largest excitation peak and the largest emission peak of the UTP-QDs were 300 and 590 nm, respectively, with an RTP lifetime of 2.54 ms (Fig. S1) and an absolute quantum yield of 14.78% (Fig. S2). The mechanism of RTP generation is attributed to the energy transfer from the band gap of ZnS to the Mn2+ dopant and the subsequent transition from the triplet state (4T1) to the ground state (6A1) of the Mn2+ incorporated into the ZnS host lattice (Chung et al., 2001) (Fig. S3). The maximum phosphorescent excitation peak of UTP-QDs slightly blue-shifted from 290 nm to 300 nm compared with other materials that modified Mn-doped ZnS QDs, suggesting that UTP binds to the lower energy sites of the QDs (Sakthivel et al., 2015).
In summary, these results confirm that this method successfully prepares UTP-capped Mn-doped ZnS QDs with RTP and uniform morphology.
3.2 AMI turns-off the RTP of UTP-QDs
The RTP quenching of UTP-QDs occurred rapidly within 5 min after adding AMI and reached a stable value, which was dependent on pH (Fig. S4). Meanwhile, the RTP lifetime of QDs slowly decreased with the increase in AMI concentration (Fig. S1). Two classical quenching models, namely, dynamic and static models (Valeur, 2003), were analyzed to explain the quenching process of photoluminescence. The inverse of the RTP variable (P0 − P)−1 (P0 and P represent the RTP intensity of UTP-QDs in the absence and presence of AMI, respectively) to the inverse of the concentration of AMI was plotted (Fig. 3A) and found to be consistent with Lineweaver–Burk equation (Eq. (1)) (Lakowicz and Weber, 1973).
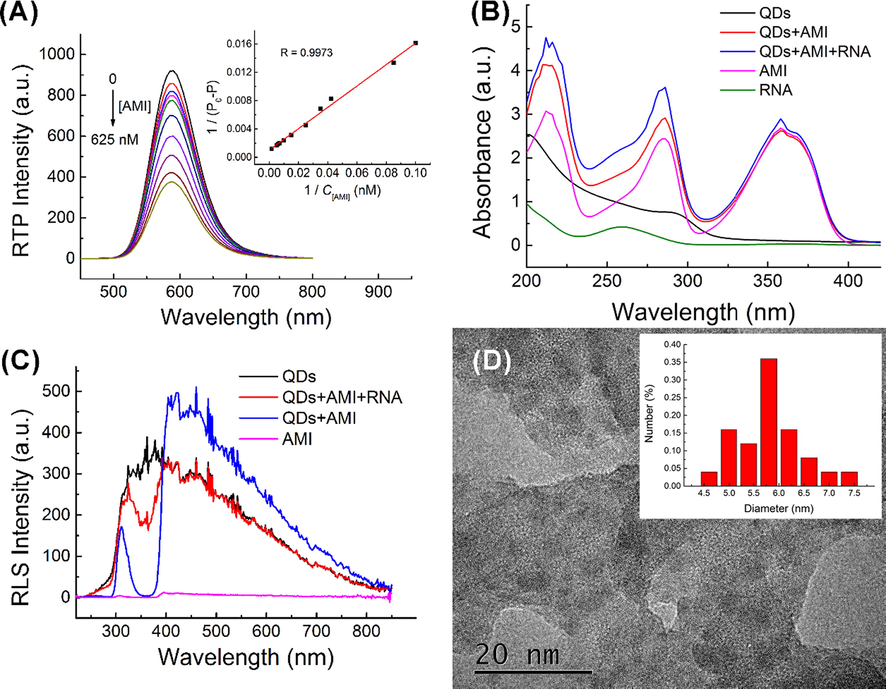
(A) Lineweaver–Burk plot of interaction between UTP-QDs (60 mg L−1) and AMI (0–625 nM) in BR buffer (50 mM, pH 5.8) containing 100 mM NaCl; (B) UV absorption of the correlation between UTP-QDs, AMI and AP-dsRNA; (C) RLS spectra of the correlation between UTP-QDs, AMI and AP-dsRNA; (D) TEM image of the precipitate formed by UTP-QDs and AMI.
This result implies that the RTP quenching process followed the static quenching (Lakowicz, 2013).
Two main causes contributed to this static quenching model. (1) The UV–vis spectrum characterization results showed that the absorption peak of AMI extensively overlapped with the RTP excitation peak of UTP-QDs, indicating that the internal filtering effect easily occurred (Fig. 3B). (2) Resonance light scattering (RLS) spectra (Fig. 3C) and TEM images (Fig. 3D) showed that obvious RLS signals appeared in the system with the addition of AMI, indicating the generation of aggregates. This finding is in line with theoretical expectations that UTP on the surface of QDs can coordinate with AMI from a non-luminous complex precipitate in the presence of NaCl (Pettis et al., 2004).
3.3 AP-dsRNA-induced RTP restoration of AMI-modulated UTP-QDs
The RTP intensity of UTP-QDs was almost unchanged in the presence of AP-dsRNA (Fig. S5), meaning that the interaction between AP-dsRNA and UTP-QDs is weak due to steric hindrance in AP-dsRNA (Varshavsky, 2006), which makes it difficult for UTP-QDs to bind to it. However, AP-dsRNA was able to recover the AMI-quenched RTP (Fig. 4). Both the considerable variation in UV (Fig. 3B) and reduction in RLS (Fig. 3C) after adding AP-dsRNA to the solution of UTP-QDs indicated the de-aggregation of QDs in the presence of AMI, which was also verified by TEM image (Fig. S6).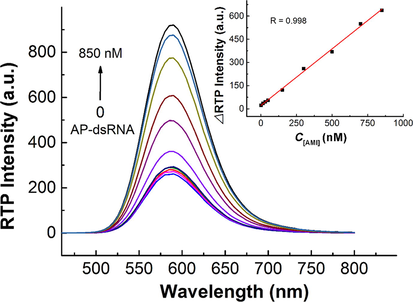
Concentration-dependent RTP intensity of AMI (300 nM)-modulated UTP-QDs (60 mg L−1) in the presence of AP-dsRNA at various concentrations and the corresponding calibration curve (inset). All solutions were prepared in 50 mM BR buffer at pH 5.8 containing 100 mM NaCl.
The phosphorescence recovery intensity (ΔP) showed a good linearity against the AP-dsRNA concentration (C, in the range of 1.4–850 nM) with a linear regression equation of ΔP = 0.7325C + 25.8815 (R = 0.9988). The relative standard deviation for the 11 replicate detections of 300 nM AP-dsRNA was 2.1%, whereas the detection limit (3 s) for AP-dsRNA was 0.86 nM (Fig. 4). RTP suffers from less background interference by biological fluids; hence, this system can detect AP-dsRNA in real samples without complex pretreatments. A comparison of its analysis performance with other methods is shown in Table 1.
Method
Linear range (nM)
LOD (nM)
RSD (%)
Ref.
Fluorescence
5000
–
–
Li et al., 2016
Fluorescence
4.7–18
–
–
Saenger, 2013
Fluorescence
nonlinear
1.2
0.25
Sakthivel et al., 2015
RTP
1.4–850
0.86
2.1
This work
3.4 Factors affecting the detection system
The pH of the system affected the AMI-induced RTP quenching (Fig. S7A) and the subsequent RTP recovery by AP-dsRNA (Fig. S7B). The recovered RTP intensity remarkably increased with the increase in pH from 3.0 to 5.8, remained unchanged in the pH range of 5.8–6.8, and slowly decreased and fluctuated in the range of 6.8–9.0. The optimal pH range of 5.8–6.8 favored the AMI-induced RTP quenching of the UTP-QDs, and mono-cation formed by amidino (pKa 8.7) protonation easily form ion pairs with abasic sites in dsRNA, thereby increasing RTP recovery. Moreover, the ionization of UTP (quaternary anion) is also minimal under this condition.
The concentration of NaCl can substantially enhance the binding affinity of AMI to UTP and AP-dsRNA; this binding affinity also plays an important role in quenching and recovering RTP in the system. When the concentration of NaCl is 100 mM, the quenching efficiency and recovery efficiency can be properly balanced in the detection system (Fig. S8). Accordingly, subsequent experiments were performed in 50 mM BR buffer containing 100 mM NaCl at pH 5.8. The optimal RTP-restoration by AP-dsRNA is reached in the concentration range of 250–350 nM of AMI (Fig. S9). The middle concentration (300 nM) was selected for subsequent experiments.
3.5 Selectivity of the RTP detection for AP-dsRNA
The potential interference of structural analogs and co-existing substances in the determination of AP-dsRNA was investigated to evaluate the practicability of the RTP method for RNA detection (Fig. S10). Results showed that AP-dsRNA has obvious recovery ability to the RTP of UTP-QD-AMI compared with other co-existing materials; therefore, this system has excellent selectivity. AP-dsDNA showed certain RTP recovery ability, but this recovery was limited by the different spatial conformation of RTP with AP-dsRNA, which determines the specificity of this detection system to AP-dsRNA. This can be attributed to differences in the double helix structure of DNA and RNA, that is, RNA is A-form and DNA is B-form, and the addition of UTP-QD-AMI can change the conformation of AP-dsRNA but not of AP-dsDNA. The larger inclination of the A-form than the B-form (1°) in nucleic acids (19°) allows more extensive base-to-base overlap and provides more efficient stacking interactions of nucleotides (Saenger, 2013), which translates into an efficient binding affinity for AP-dsDNA (Sato et al., 2012). This process explains why double-stranded nucleic acids (dsDNA and dsRNA) without AP-sites and single-stranded nucleic acids (ssDNA and ssRNA) with AP-sites do not effectively restore the quenched RTP.
The combination of AMI and UTP-QDs cannot be broken nor can the recovery of the quenched RTP of UTP-QDs by AP-dsRNA be affected; therefore, the detection performance of the system is less affected by other nucleotides and co-existing substances. These substances are found in low concentrations in biological fluids or can be appropriately diluted to eliminate interference.
3.6 Application of the RTP-recovery system in AP-dsRNA detection
The AMI-modulated UTP-capped Mn-doped ZnS QD-based turn-off–on RTP-system was used for AP-dsRNA detection in serum and plant lysate samples. The phosphorescence background of biological fluids samples was negligible after proper dilution (150 times), and obtained spiked recovery rates of 95%–103% (Table S2). The developed turn-off–on RTP systems can be employed in the detection of AP-dsRNA in biological fluids.
4 Conclusion
UTP-functionalized Mn-doped ZnS QDs having excellent phosphorescent properties were synthesized by a wet-chemical method. AMI can form a precipitated complex with UTP on the surface of the QDs and quench RTP. Then AP-dsRNA can dissociate AMI from the surface of UTP-QDs through its strong affinity with AMI, resulting in RTP recovery. Based on this mechanism, a method for detecting AP-dsRNA based on UTP-QD-AMI system was established. This method exhibits strong specificity and high sensitivity and has been successfully applied to the analysis of AP-dsRNA in biological fluids to provide new methods for early disease screening and toxicity identification.
Acknowledgments
This work was supported by Natural Science Foundation of Shanxi Province (20210302124477).
Author contributions
All authors contributed to literature research and manuscript writing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363.
- [Google Scholar]
- The effect of RNA base lesions on mRNA translation. Nucleic Acids Res.. 2015;43:4713-4720.
- [Google Scholar]
- Formation and distinctive decay times of surface-and lattice-bound Mn2+ impurity luminescence in ZnS nanoparticles. J. Phys. Chem. B.. 2001;105:4128-4132.
- [Google Scholar]
- The Acid-Catalyzed Solvolysis of Pyrimidine Nucleosides1. J. Org. Chem.. 1966;31:2219-2227.
- [Google Scholar]
- MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet.. 2004;5:522.
- [Google Scholar]
- Allele-selective inhibition of mutant atrophin-1 expression by duplex and single-stranded RNAs. Biochemistry. 2014;53:4510-4518.
- [Google Scholar]
- Double-stranded RNA sensors and modulators in innate immunity. Annu. Rev. Immunol.. 2019;37:349-375.
- [Google Scholar]
- Purine-derived nitroxides for noncovalent spin-labeling of abasic sites in duplex nucleic acids. Chem. Eur. J.. 2018;24:4157-4164.
- [Google Scholar]
- Surface functionalization of quantum dots for biological applications. Adv. Colloid Interface Sci.. 2015;215:28-45.
- [Google Scholar]
- Trans-lesion synthesis and RNaseH activity by reverse transcriptases on a true abasic RNA template. Nucleic Acids Res.. 2007;35:6846-6853.
- [Google Scholar]
- RNA Abasic Sites: Preparation and Trans-Lesion Synthesis by HIV-1 Reverse Transcriptase. ChemBioChem. 2005;6:1970-1973.
- [Google Scholar]
- The chemical stability of abasic RNA compared to abasic DNA. Nucleic Acids Res.. 2006;35:58-68.
- [Google Scholar]
- Principles of fluorescence spectroscopy. Springer Science & Business Media; 2013.
- Quenching of fluorescence by oxygen. Probe for structural fluctuations in macromolecules. Biochemistry. 1973;12:4161-4170.
- [Google Scholar]
- Abasic pivot substitution harnesses target specificity of RNA interference. Nat. Comm.. 2015;6:10154.
- [Google Scholar]
- Plasma-assisted quadruple-channel optosensing of proteins and cells with Mn-doped ZnS quantum dots. Nanoscale.. 2016;8:4291-4298.
- [Google Scholar]
- Combination of pentafluorophenylhydrazine derivatization and isotope dilution LC-MS/MS techniques for the quantification of apurinic/apyrimidinic sites in cellular DNA. Anal. Bioanal. Chem.. 2013;405:4059-4066.
- [Google Scholar]
- RNA duplexes with abasic substitutions are potent and allele-selective inhibitors of huntingtin and ataxin-3 expression. Nucleic Acids Res.. 2013;41:8788-8801.
- [Google Scholar]
- The structural basis of large ribosomal subunit function. Annu. Rev. Biochem.. 2003;72:813-850.
- [Google Scholar]
- The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920-930.
- [Google Scholar]
- Ionic interaction of amiloride and uridine 5′-triphosphate in nebulizer solutions. J. Pharm. Sci.. 2004;93:2399-2406.
- [Google Scholar]
- Ribosome-inactivating proteins from plants: more than RNA N-glycosidases? FASEB J.. 2001;15:1493-1506.
- [Google Scholar]
- Ultrasonic assisted synthesis of adenosine triphosphate capped manganese-doped ZnS quantum dots for selective room temperature phosphorescence detection of arginine and methylated arginine in urine based on supramolecular Mg2+–adenosine triphosphate–arginine ternary system. Talanta. 2012;97:16-22.
- [Google Scholar]
- Principles of nucleic acid structure. Springer Science & Business Media; 2013.
- Structural, band gap and photoluminescence behaviour of Mn-doped ZnS quantum dots annealed under Ar atmosphere. J. Mater. Sci. M.. 2015;26:1533-1542.
- [Google Scholar]
- Strong and selective binding of amiloride to an abasic site in RNA duplexes: thermodynamic characterization and microRNA detection. Angew. Chem. Int. Ed.. 2012;51:6369-6372.
- [Google Scholar]
- Abasic site-binding ligands conjugated with cyanine dyes for “off–on” fluorescence sensing of orphan nucleobases in DNA duplexes and DNA–RNA hybrids. Chem. Commun.. 2014;50:515-517.
- [Google Scholar]
- 2, 4-Diamino-6, 7-dimethylpteridine as a fluorescent ligand for binding and sensing an orphan cytosine in RNA duplexes. Chem. Commun.. 2013;49:9983-9985.
- [Google Scholar]
- Fluorescence and phosphorescence spectroscopy: physicochemical principles and practice. Elsevier; 2017.
- Effect of different annealing conditions on the properties of chemically deposited ZnS thin films on ITO coated glass substrates. Sol. Energy Mater. Sol. Cells.. 2011;95:856-863.
- [Google Scholar]
- Synthesis and characterisation of functional manganese doped ZnS quantum dots for bio-imaging application. Adv. Appl. Ceram.. 2019;118:321-328.
- [Google Scholar]
- An assay for RNA oxidation induced abasic sites using the Aldehyde Reactive Probe. Free Radical Res.. 2011;45:237-247.
- [Google Scholar]
- Synthesis and photophysical characterisation of a fluorescent nucleoside analogue that signals the presence of an abasic site in RNA. ChemBioChem. 2012;13:2392-2399.
- [Google Scholar]
- Photochemical generation of ribose abasic sites in RNA oligonucleotides. Org. Lett.. 2005;7:1493-1496.
- [Google Scholar]
- Room-temperature phosphorescent discrimination of catechol from resorcinol and hydroquinone based on sodium tripolyphosphate capped Mn-doped ZnS quantum dots. Anal. Chem.. 2013;85:1920-1925.
- [Google Scholar]
- Wang, I., R. Loechli, Y. Liu, et al., 2018. Abasic Sites in RNA of Yeast and Human. SSRN 3227917.
- Abasic site-containing DNAzyme and aptamer for label-free fluorescent detection of Pb2+ and adenosine with high sensitivity, selectivity, and tunable dynamic range. J. Am. Chem. Soc.. 2009;131:15352-15357.
- [Google Scholar]
- Cellular concentrations of nucleotide diphosphate-chelated magnesium ions accelerate catalysis by RNA and DNA enzymes. Biochemistry. 2019;58:3971-3979.
- [Google Scholar]
- Surface passivation and photoluminescence of Mn-doped ZnS nanocrystals. Adv. Mater. Sci. Eng. 2008
- [Google Scholar]
- Photoluminescence characteristics of ZnS nanocrystallites co-doped with Co2+ and Cu2+. Inorg. Chem. Commun.. 2001;4:734-737.
- [Google Scholar]
- Aggregation-induced phosphorescence enhancement of Mn-doped ZnS quantum dots: the role of dot-to-dot distance. Nanoscale.. 2018;10:9236-9244.
- [Google Scholar]
- Low-toxic Mn-doped ZnSe@ZnS quantum dots conjugated with nano-hydroxyapatite for cell imaging. Nanoscale.. 2014;6:14319-14325.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2022.104050.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary data 1
Supplementary data 1







