Translate this page into:
Unraveling the mechanism of action of cepharanthine for the treatment of novel coronavirus pneumonia (COVID-19) from the perspectives of systematic pharmacology
⁎Corresponding authors at: Animal-Derived Food Safety Innovation Team, College of Animal Science and Technology, Anhui Agricultural University, Hefei 230036, China (F. Sun). 13141212481@163.com (Feifei Sun), wuyongning@cfsa.net.cn (Yongning Wu), lilin@ahau.edu.cn (Lin Li)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Abstract
Natural products play an irreplaceable role in the treatment of SARS-CoV-2 infection. Nevertheless, the underlying molecular mechanisms involved remain elusive. To better understand their potential therapeutic effects, more validation studies are needed to explore underlying mechanisms systematically. This study aims to explore the potential targets of action and signaling pathways of cepharanthine for the treatment of COVID-19. This study revealed that a total of 173 potential targets of action for Cepharanthine and 86 intersectional targets for Cepharanthine against COVID-19 were screened and collected. Gene Ontology enrichment analysis suggested that inflammatory, immune cell and enzyme activities were the critical terms for cepharanthine against COVID-19. Pathway enrichment analysis showed that five pathways associated with COVID-19 were the main signaling pathways for the treatment of COVID-19 via cepharanthine. Molecular docking and molecular dynamics simulations suggested that 6 core targets were regarded as potential targets for cepharanthine against COVID-19. In brief, the study demonstrates that cepharanthine may play an important role in the treatment of SARS-CoV-2 infection through its harmonious activity against SARS-CoV-2 pathways and multiple related targets. This article provides valuable insights required to respond effectively to concerns of western medical community.
Keywords
Natural products
COVID-19
Cepharanthine
Molecular mechanism
Network pharmacology
1 Introduction
The pandemic, Coronavirus Disease 2019 (COVID-19), has affected people around the world for over two years, causing millions of deaths around the world (Larkin, 2022, Jha et al., 2022). Novel coronavirus pneumonia is mainly transmitted directly or indirectly through the respiratory tract and is more likely to spread to the elderly, people with other diseases and those with immune-deficient systems. Different types of vaccines against novel coronavirus pneumonia have been widely promoted and administered to the population and their usefulness in preventing infection with novel coronavirus pneumonia is unquestionable (Chen et al., 2021a, Asselah et al., 2021). Many companies are developing specific drugs for novel coronavirus pneumonia, but given the complexity of novel coronaviruses and the efficacy of specific drugs, there is still a need for further development of more effective specific drugs.
Till now, the pharmaceutical community has made significant progress in mitigating the COVID-19 threat through the development of small-molecule drugs (Wang and Yang, 2022, Riva et al., 2020). Many compounds/antibodies have been authorized for COVID-19 treatment. These drugs clinically used against COVID-19 are being applied according to drug repositioning strategy. Gilead’s Veklury (Remdesivir) was conditionally approved to combat the outbreak (Beigel et al., 2020). Pfizer’s oral candidate Paxlovid (PF-07321332) (Mahase, 2021) and Merck’s Lagevrio (Molnupiravir) (Cully, 2021) raise hope for a COVID-19 cure. However, promising bullets still do not exist. As an indispensable resource for promising compounds, traditional medicine (Wang and Yang, 2021, Li et al., 2021a) and natural products (Wang et al., 2022, Yang and Wang, 2021) have attracted significant attention in countering COVID-19 infection. Besides, it has been reported that some traditional Chinese medicines including cepharanthine had shown good efficacy against novel coronavirus pneumonia (Fan et al., 2022, An et al., 2021, Fan et al., 2020). Cepharanthine is a leukoproliferative drug which works by promoting the proliferation of leukocytes in the body. However, like many traditional Chinese medicines, Cepharanthine can be used in new ways, it has been proved that have potential well efficacy against COVID-19 (Ohashi et al., 2021). However, the relevant mechanism of action needs to be further explored.
Network pharmacology is a new discipline based on systematic biology, which analyzes the interactions among the drug targets in biological organisms (Hopkins, 2008). Molecular docking and molecular dynamics simulations are increasingly employed in extensive disciplines and fields. Molecular docking can statically reveal drug-target and target-target interactions at the molecular level (Pinzi and Rastelli, 2019), while molecular dynamics simulations can dynamically simulate the specific binding processes and stability of drugs and targets in the organism. The combination of molecular docking and molecular dynamics can be used to obtain more accurate results on the mechanism of action and stability of drugs and targets.
Therefore, this study employs the methods of systematic pharmacology to supplement and verify the mechanism of the treatment of the novel coronavirus with cepharanthine in more detail.
2 Materials and methods
2.1 Database construction of cepharanthine targets
The SymMap database (https://www.symmap.org) (Wu et al., 2019), the pharmMapper database (https://www.lilab-ecust.cn/pharmmapper/) (Liu et al., 2010) and the SwissTarget Prediction database (https://www.swisstargetprediction.ch/) (Daina et al., 2019) were used to construct a database related to cepharanthine targets. In the SymMap database, cepharanthine was entered into the search field and its relevant targets were collected. The criteria used to screen the targets were as follows: Norm Fit score higher than 0.7 for pharmMapper database; probability higher than 0 for SwissTarget Prediction database. Finally, the ceparanthine targets were obtained by importing the targets from three databases into the Evenn website (https://www.ehbio.com/test/venn/) (Chen et al., 2021b) and removing the duplicates.
2.2 Database construction of COVID-19-related targets
Relevant targets for COVID-19 were analyzed and collected from the GEO Datasets of NCBI database (https://www.ncbi.nlm.nih.gov/) (Brown et al., 2015). GEO Datasets was used to screen and collect gene expression array datasets for severe COVID-19, and the GEO Series GSE164805 (PMID: 33679778) was finally selected as the microarray data for long non-coding RNAs (lncRNAs) differentially expressed in peripheral blood mononuclear cells of the severe COVID-19 group and the healthy group. Furthermore, the GEO2R web tool was employed to analyzed the genes that are differentially expressed between the two groups, and the analytical criteria for the genes are P value <0.05 and |log2(fold change)| >1. The genes screened by the analysis criteria were regarded as COVID-19-related targets. Finally, volcano maps of remarkably differentially expressed genes were visualized via the SRPLOT website (https://www.bioinformatics.com.cn/en) (Lu et al., 2022).
2.3 Overlapped targets between cepharanthine and COVID-19
The overlapped proteins encoded by disease genes are the potential drug targets for treatment. The overlapped targets of cepharanthine and COVID-19, acquired via the Evenn website, were used as the potential targets of cepharanthine against COVID-19.
2.4 Construction and analysis of protein–protein interaction (PPI) networks
PPI network was constructed by importing the overlapped targets into the STRING database (https://string-db.org) (Szklarczyk et al., 2021), where “Homo sapiens” was selected as the species, and the minimum required interaction score was set at the medium confidence level of 0.400. The PPI network is then imported into Cytoscape 3.9.1 software (Shannon et al., 2003) for optimization and refinement. Topological analysis of PPI networks was carried out according to three parameters including the Degree, maximal clique centrality (MCC) and maximum neighborhood component (MNC) in the CytoHubba plugin. Finally, the targets that scored above 1.5 times the mean value in Degree, MCC and MNC were considered as the core targets for the treatment of COVID-19 with cepharanthine.
2.5 GO enrichment analysis and KEGG pathway enrichment analysis
The overlapped targets were imported into the Metascape database (https://metascape.org/) (Zhou et al., 2019) for Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis (Kanehisa et al., 2017), respectively, where the P value was set at 0.01. GO enrichment analysis was performed in terms of cellular composition (CC), molecular function (MF) and biological processes (BP). The results were visualized on the SRPLOT website.
2.6 Molecular docking
The molecular docking were conducted between cepharanthine and the core targets of COVID-19. In addition, the molecular docking were performed between cepharanthine and the published targets such as ACE2, spike protein S1 (S), SARS-CoV-2 3CL hydrolase (3CL) and SARS-CoV-2 RNA-dependent RNA polymerase (RdRp), to verify the reliability. The 2D sdf structure of cepharanthine (Supplementary Fig. S1) was first obtained by entering the word cepharanthine into the PubChem website, and the obtained structure was imported into Chem3D software for minimization to obtain a molecularly dockable structure of cepharanthine. The targets were then imported into the RCSB PDB database (https://www.rcsb.org/) (Burley, 2019) and the UniProt database (https://www.uniprot.org) (UniProt, 2021) to obtain the crystal structures of the target proteins. Finally, AutoDockTOOLs 1.5.6 software was used to conduct molecular docking and the docking results were visualized in Pymol software (Yuan et al., 2017). AutoDockTOOLs 1.5.6 software was used as the molecular docking software for this study because of its advantages over other docking software, such as accurate docking (Gaillard, 2018).
2.7 Molecular dynamics simulations
MD simulations of the current study's molecular docking results were performed by Gromacs 2020.6 software (Pall et al., 2020). The above molecular docking results were used as the input conformation file for the MD simulations. Given that the molecular structure of cepharanthine is a multi-carbon ring skeleton structure, Charmm36 force field and TIP3P water model was chosen for MD simulations. And in order to simulate the environment of drug-target binding in the body, the systems of MD simulations were set to the sodium chloride solution at the temperature of 37 centigrade. During the MD simulations, processes involving hydrogen bonding are constrained using the Linear Constraint Solver (LINCS) algorithm with an integration step of 2 fs. Electrostatic interactions are calculated using the Particle-mesh Ewald (PME) method with the cut-off values set to 1.2 nm. In addition, the cut-off values for non-bonded interactions are 10 Å, updated every 10 steps. Before performing MD simulations, the molecular docking results were pre-processed. In order to obtain the initial conformation of the target in the solvent, the molecular docking results (hereafter referred to as the drug target complex) were first pre-equilibrated for 100 ps. Then, in order to warm up the drug target complex to 310 k with the solvent system, Isothermal-Isovolume (NVT) equilibrium of 100 ps was performed using a modified Berendsen temperature coupling algorithm with a coupling time constant of 0.1 ps. Subsequently, in order to equilibrate the drug target complex with the solvent system pressure, Isothermal-Isobaric (NPT) equilibration of 100 ps was performed using a Berendsen constant pressure to bring the system pressure up to 1 bar. Finally, a 50 ns MD simulation of the drug target complex was carried out. Furthermore, the data results from the MD simulations were visualized and analyzed through Origin 2021 sofware (Moberly et al., 2018).
3 Results
3.1 Cepharanthine targets and COVID-19-related targets
There are 10, 76 and 104 targets collected from SymMap database, pharmMapper database and SwissTargetPrediction database, respectively. These cepharanthine targets were combined and the duplicates were removed via the Evenn website. As a result, a total of 173 cepharanthine targets were obtained, shown in Fig. 1A. Additionally, volcano maps of remarkably differentially expressed genes (Fig. 1B) suggested that 18,306 genes (remove genes that blank gene name), down regulated or up regulated, were associated with severe COVID-19.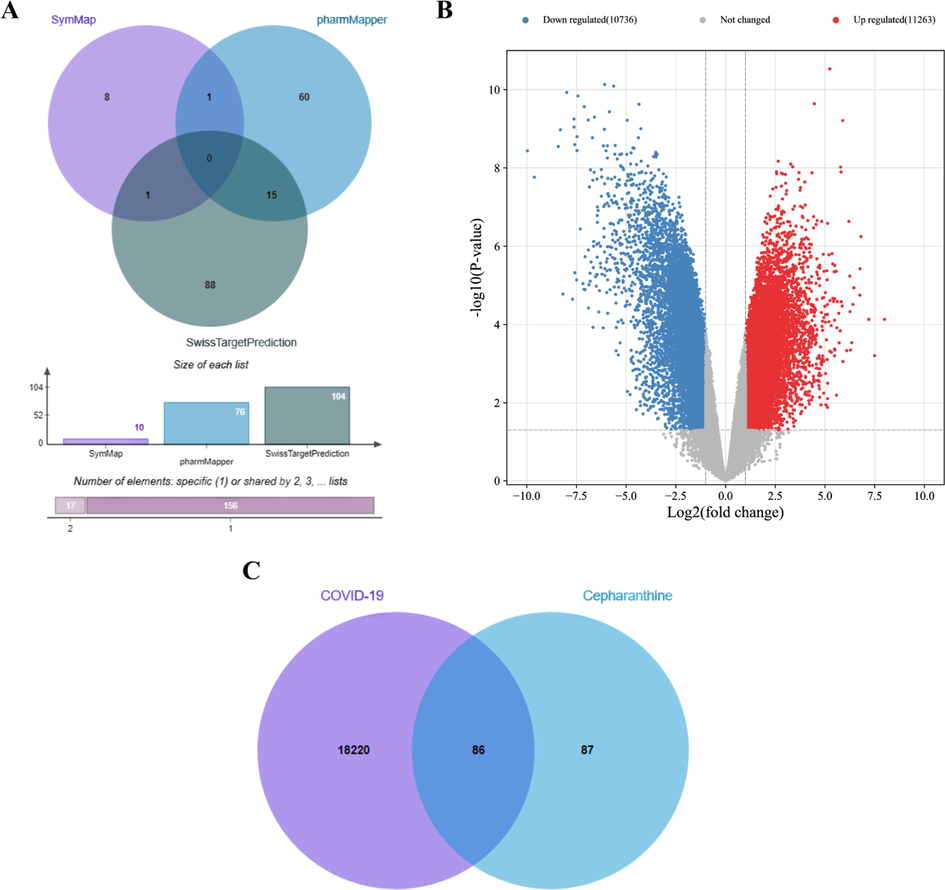
Database construction. A: Targets of cepharanthine. B: Volcano plot of COVID-19. C: Overlapped targets of cepharanthine for the treatment of COVID-19.
3.2 Overlapped targets between cepharanthine and COVID-19
In the current study, 86 overlapped targets between cepharanthine and COVID-19 were gathered via the Evenn website, which was described in Fig. 1C and Supplementary Table S1.
3.3 PPI network construction and analysis
The PPI network was obtained by importing 31 overlapped targets between cepharanthine and COVID-19 into the SRING database. The network was then introduced into Cytoscape 3.9.1 software for optimization and refinement, which was illustrated in Fig. 2A. Consequently, a PPI network with 86 nodes and 306 edges was constructed, where each node represented a target of cepharanthine for the treatment of COVID-19. The topological analysis of the PPI network according to the MCC, MNC and Degree parameters was performed via the CytoHubba plugin (Fig. 2B–D). In Fig. 2B–D, networks are ordered clockwise from the HSP90AA1 or ALB target, where the larger and darker the circle of the node, the closer link between the target and other targets. Finally, eight hub targets illustrated in Fig. 3A and Supplementary Table S2 were obtained, namely HSP90AA1, ALB, ESR1, ANXA5, XIAP, LCK, LYN and HCK.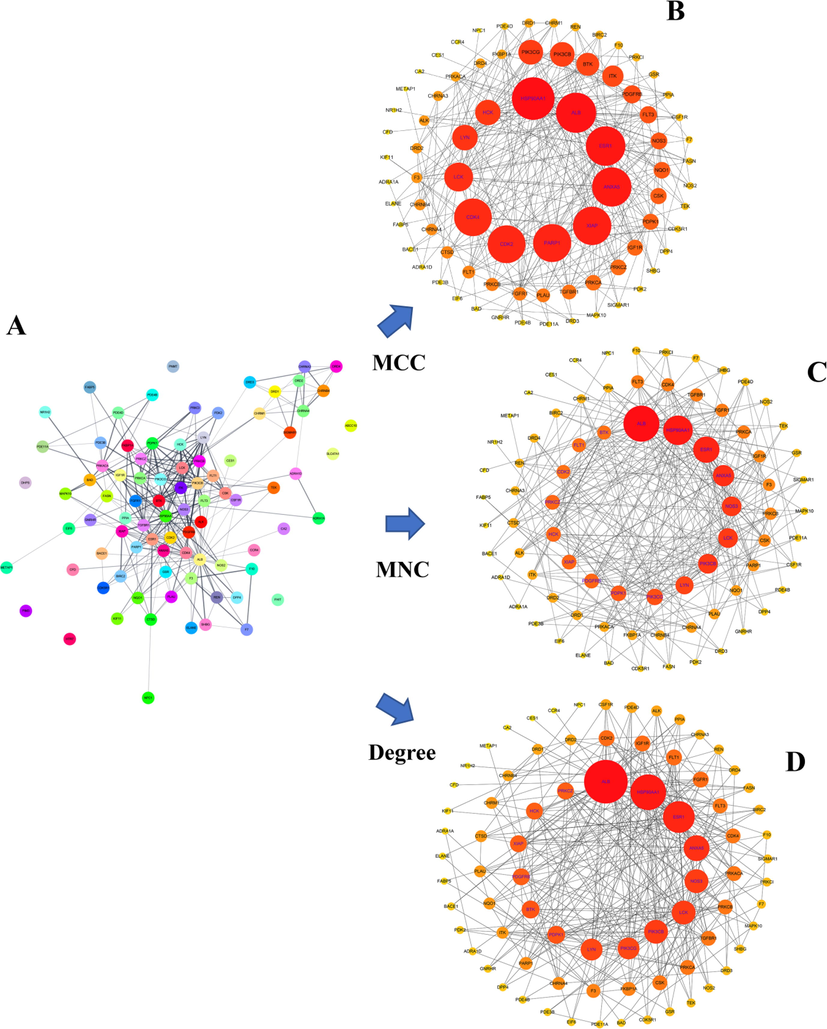
Information of PPI network. A: PPI network. B: PPI network under MCC parameter ranking. C: PPI network under MNC parameter ranking. D: PPI network under degree parameter ranking. In Figures B, C and D, networks are ordered clockwise from the HSP90AA1 or ALB target, where the larger and darker the circle of the node, the closer link between the target and other targets.
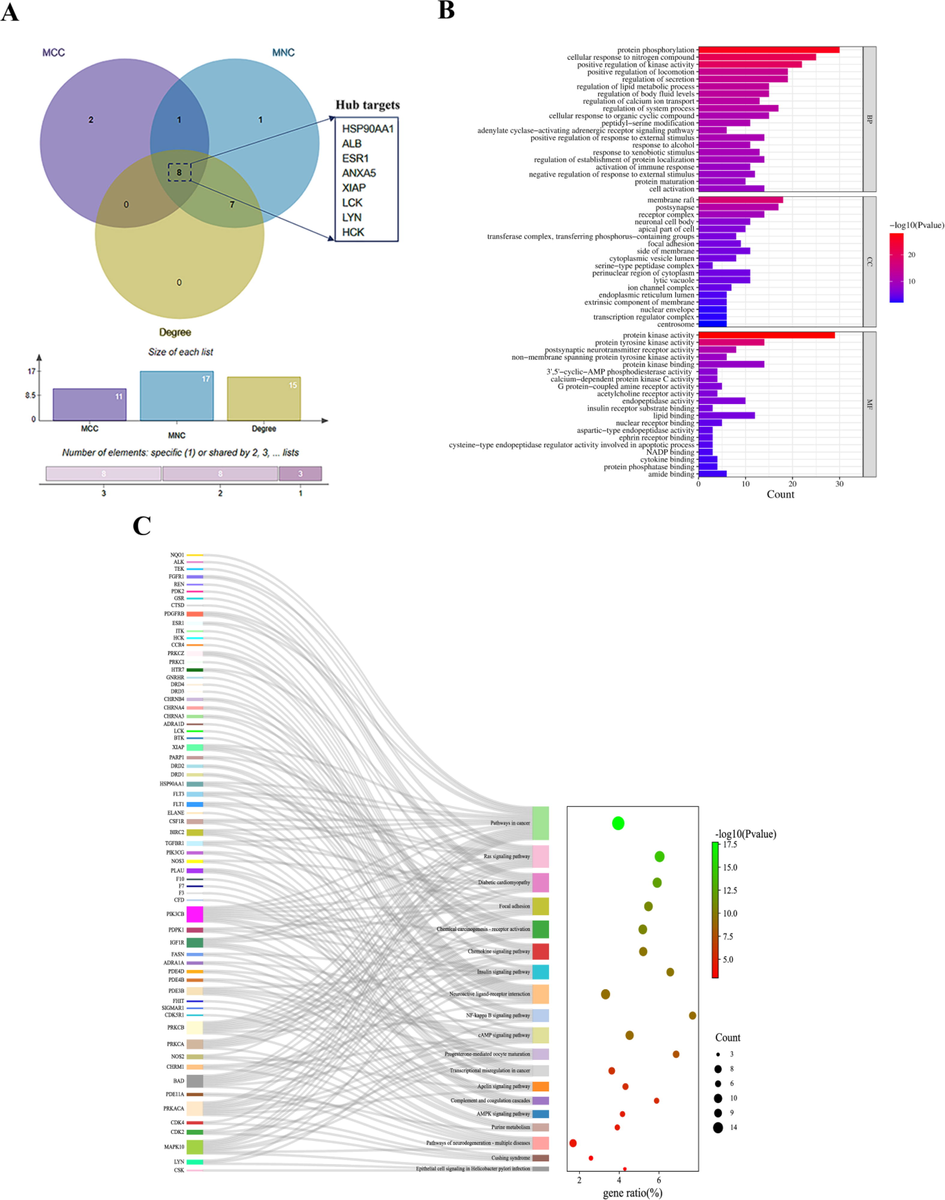
Information on the targets of cepharanthine for the treatment of COVID-19. A: Topological analysis of PPI networks by degree, MCC and MNC to filter hub targets. In the figure, the box on the right is indicate the hub targets of COVID-19 treatment by cepharanthine. B: GO enrichment analysis. C: KEGG pathway enrichment analysis. The left side of the figure reflects the pathway where each target is enriched.
3.4 GO enrichment analysis
GO enrichment analysis was performed by Metascape software on 86 overlapped targets between cepharanthine and COVID-19 in terms of cellular composition (CC), molecular function (MF) and biological process (BP). In the three types of enrichment analysis, all categories enrichment analysis was performed based on the top 20 terms in terms of -log10 (P value). As seen in Fig. 3B, for cellular composition, the main enrichments were in various membrane components, receptor complex, protein kinase complex, neuronal cell body, ion channel complex, apical part of cell, cytoplasmic vesicle lumen and transcription regulator complex. For molecular function, the main enrichments were in various protease activities, neurotransmitter receptor activity, G protein-coupled amine receptor activity, cysteine-type endopeptidase regulator activity involved in apoptotic process and binding to another important receptor. For biological processes, the main enrichments were in activation of immune response, positive regulation of programmed cell death, positive regulation of cytokine production, protein phosphorylation and response to xenobiotic stimulus.
3.5 KEGG pathway enrichment analysis
The overlapped 86 targets were imported into Metascape software for KEGG pathway enrichment analysis. As shown in Fig. 3C, it clearly showed which pathway each target belongs to, and the targets were mainly enriched in Pathways in cancer, Ras signaling pathway, Chemokine signaling pathway, NF-kappa B signaling pathway, cAMP signaling pathway, Progesterone-mediated oocyte maturation, complement and coagulation cascades, AMPK signaling pathway and Epithelial cell signaling in Helicobacter pylori infection.
3.6 Molecular docking of core targets
The 2D sdf structure of cepharanthine was imported into AutoDockTOOLs 1.5.6 software individually for molecular docking. The PDB IDs of the core protein crystal structures obtained through the RCSB PDB database and the UniProt database were shown in Table 1. Autodock related literature suggested that good docking results are indicated when the docking bond energy is less than −5 kca.mol−1. Therefore, the binding energies were in detail available in Table 1, where the binding energy less than −6.4 kca.mol−1 indicated that cepharanthine exerted good binding capacity. In addition, the molecular docking was visualized by Pymol software in Fig. 4.
Target (PDB ID)
Binding site
Binding free energy
HSP90AA1 (3T0H)
ALA-21
−6.85 kcal.mol−1
ALB (7DJN)
LYS-262
−6.4 kcal.mol−1
ESR1 (7UJO)
GLU-380, ARG-515
−7.2 kcal.mol−1
ANXA5 (6K25)
ASP-265
−8.25 kcal.mol−1
XIAP (5OQW)
ASN-249, TRP-323
−7.96 kcal.mol−1
LCK (3MPM)
HIS-297, GLU-495
−7.33 kcal.mol−1
LYN (5XY1)
GLU-471
−7.4 kcal.mol−1
HCK (5H0B)
HIS-319, ASP-517
−7.96 kcal.mol−1
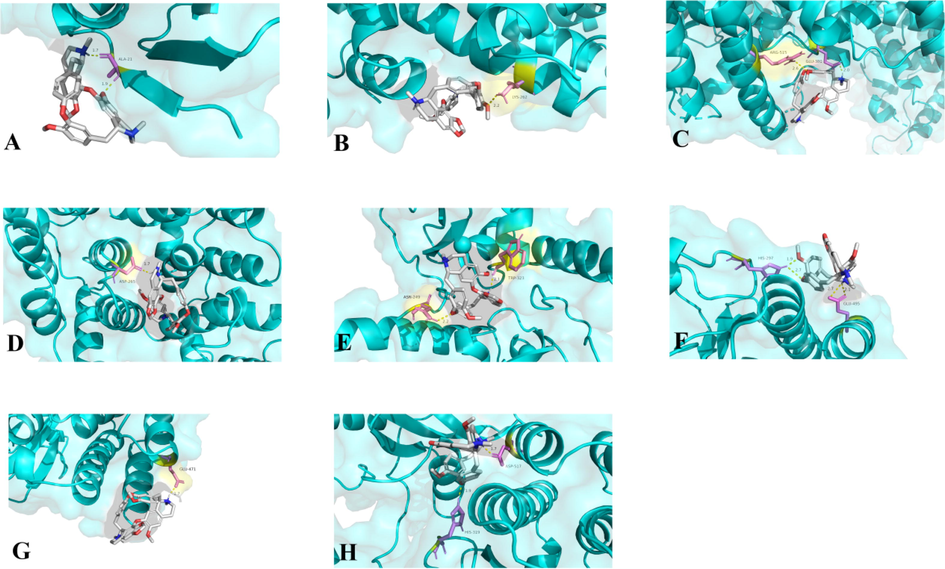
Molecular models of cepharanthine binding to core targets. Proteins A: HSP90AA1 (3T0H), B: ALB (7DJN), C: ESR1 (7UJO), D: ANXA5 (6K25), E: XIAP (5OQW), F: LCK (3MPM), G: LYN (5XY1), H: HCK (5H0B) are shown interacting with cepharanthine molecule. The yellow dashed line showed the hydrogen bond, the molecular in purple were the amino acids as acceptors while the molecular in white were cepharanthine.
3.7 Molecular docking of cepharanthine with ACE2, spike protein S1, SARS-CoV-2 3CL hydrolase, SARS-CoV-2 RNA-dependent RNA polymerase
To further accurately elucidate the mechanism of action of cepharanthine for the treatment of COVID-19, cepharanthine was molecularly docked to the hub targets of COVID-19 by AutoDockTOOLs 1.5.6 software. The results were available in Table 2, where the binding energy of cepharanthine to ACE2 was −8.33 kca.mol−1, suggesting a stable binding capacity. Additionally, the results demonstrated that cepharanthine had good binding affinity to SARS-CoV-2 3CL hydrolase (3CL) and spike protein S1 (S) and SARS-CoV-2 RNA-dependent RNA polymerase (RdRp). The molecular docking was visualized in Fig. 5.
Target (PDB ID)
Binding site
Binding free energy
ACE2 (1R42)
GLN-102, GLU-398, ASP-509, SER-511
−8.33 kcal.mol−1
Spike protein S1 (3BGF)
TRP-103
−6.41 kcal.mol−1
SARS-CoV-2 3CL hydrolase (6LU7)
GLN-110, ASP-153, THR-292
−7.33 kcal.mol−1
RNA-dependent RNA polymerase (6M71)
TRP-290, TYR-294
−6.43 kcal.mol−1
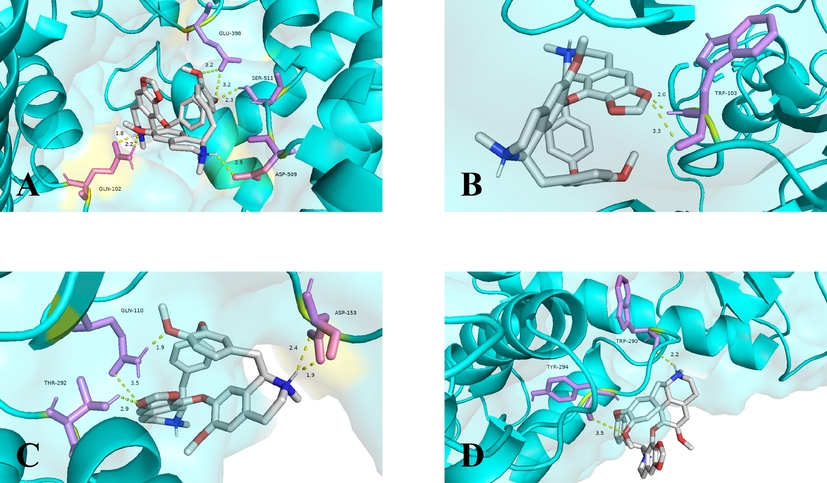
Molecular models of cepharanthine binding to the hub targets of COVID-19. Proteins (A) ACE2 (1R42), (B) Spike protein S1 (3BGF), (C) SARS-CoV-2 3CL hydrolase (6LU7), RNA-dependent RNA polymerase (6M71) are shown interacting with cepharanthine molecule. The yellow dashed line showed the hydrogen bond, the molecular in purple were the amino acids as acceptors while the molecular in white were cepharanthine.
3.8 Molecular dynamics simulations
To further investigated and verified the binding of cepharanthine to core targets, analysis of Root Mean Square Deviation (RMSD), Root Mean Square Fluctuation (RMSF) and Free Energy Landscape (FEL) were obtained by Gromacs 2020.6 software and Origin 2021 sofware. The RMSD analysis plots reflect the dynamic binding stability of cepharanthine to the corresponding targets. In 50 ns MD simulations, the binding trajectory deviated by less than 0.5 nm, indicating stable drug-target binding and a stable trajectory. Fig. 6A shows that among the 8 key targets, the binding of cepharanthine to ALB, HSP90AA1 and XIAP was unstable. Cepharanthine binds stably to the other 5 core targets. Fig. 6B shows that among the COVID-19 key related targets, cepharanthine binds stably to ACE2 and 3CL. The RMSF analysis plots reflect the flexibility and the degree of drastic action of protein residues throughout MD simulations during the binding of core targets to cepharanthine. Supplementary Fig. S2 shows that most of the target protein residues have greater flexibility and specific action in the simulated binding process when cepharanthine dynamically binds to key targets. FEL analysis plots demonstrate the conformational free energy changes of cepharanthine and the core targets during the simulations and characterize the adequacy of sampling by analyzing the energy trap. The energy trap is interpreted as a graph of Gibbs free energy, RMSD and Gyration, similar to energy release, and is primarily used to reflect drug-target binding energy changes and sampling adequacy. In addition, the radius of gyration (Rg) in the FEL diagram characterize the tightness of the protein structure, and similarly the radius of gyration can be relied upon to characterize the change in peptide chain relaxation of the protein during the simulation. As illustrated in Supplementary Fig. S3, each of the core targets have energy trap when bound to cepharanthine, indicating that the free energy of binding changes and these simulations were well sampled for characterization and the Rg of each core target is appropriate for drug-target binding.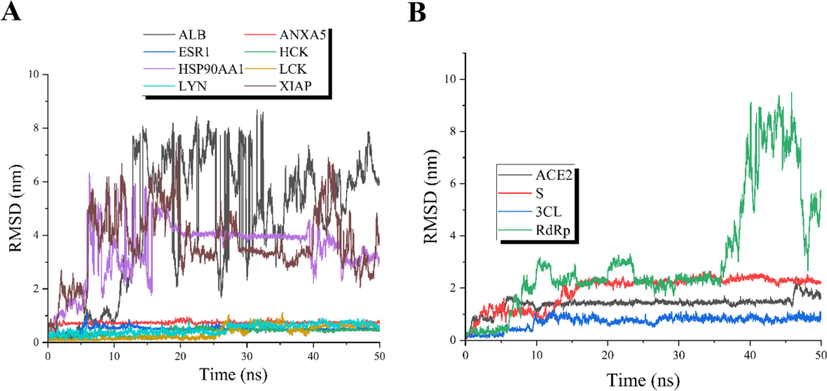
RMSD plots. A: RMSD plots of eight core targets for cepharanthine against COVID-19. B: RMSD plots of the hub targets of COVID-19. The plots show that the trajectory profile of the dynamic binding of the drug target at a time duration of 50 ns.
4 Discussion
Cepharanthine is the active ingredient in many herbal medicines, like Stephania epigaea Lo. As a leukoproliferative drug, cepharanthine could increase white blood cells and promote the proliferation of the bone marrow tissue. Cepharanthine was reported to have potential anti-COVID-19 effects when used alone or in combination with nelfinavir (Ohashi et al., 2021). And Li et al demonstrated that Part of the mechanism of the treatment of COVID-19 with cepharanthine (Li et al., 2021b). Moreover, cepharanthine was granted as a national patent due to its efficacy against COVID-19 virus (Zhang et al., 2022, Biswas et al., 2021). However, the detailed mechanism remains unclear and needs in-depth study.
In the present study, 86 overlapped targets which were the potential drug targets for treatment were collected. According to the PPI network and three different topological analysis parameters, eight core targets including HSP90AA1, ALB, ESR1, ANXA5, XIAP, LCK, LYN, HCK were obtained. HSP90AA1 is heat shock protein HSP 90-alpha, which is involved in signal transduction and the cell cycle. It has been reported that HSP90AA1 can also mediate the binding of TOMM70 to the receptor, thereby promoting an antiviral response (Wei et al., 2015). ALB is an albumin that could regulate plasma osmolality and bind inorganic ions and other substances in the organism. Hypoalbuminemia is one of the risk factors that makes COVID-19 devastating, due to the critical role in maintaining the osmolality and in reducing the thromboembolism (Ramadori, 2021). ESR1 is estrogen receptor 1, which plays an important role in the growth, sexual development and other reproductive functions of the body. It has been documented that ESR1 has the ability to disrupt NF-kappa-B DNA binding activity and inhibit IL6 promoter expression, thereby regulating inflammatory responses (Liu et al., 2005). Furthermore, recent studies suggest that ESR1 may be an important participant in the suppression of COVID-19 inflammatory and immune responses (Li et al., 2022). ANXA5 is an anticoagulant protein that acts as an inhibitor of the prothrombin complex and is involved in the coagulation cascade. Studies have shown significant changes in Annexin A5 in COVID-19 patients compared to healthy subjects, suggesting that ANXA5 is an important target for the treatment of COVID-19 and deserves further investigation (Ercan et al., 2021). XIAP is an X-linked inhibitor of apoptosis that regulates not only apoptosis and cystein, but also immunity, inflammatory signaling and cell proliferation, as well as cell invasion and metastasis. XIAP can act as a receptor that regulates NF-kappa-B signaling, thereby promoting the inflammatory and immune responses of the body activation (Grau-Expósito et al., 2022, Damgaard and Gyrd-Hansen, 2011, Lu et al., 2007). In addition, studies have shown that XIAP-related factors are increased in patients with COVID-19 (Zhu et al., 2020). Interesting, in the hub targets, LCK, LYN and HCK all belong to the Src family of tyrosine kinases and are associated with the body's immune cells and inflammation. The targets associated with the body’s immune cells and inflammation would be required to recovery of the patient and a blockage of them may be have no benefit. But the following references demonstrate that these targets may be very important for treat COVID-19 and can be regarded as potential therapeutic targets. LCK, Tyrosine-protein kinase Lck, plays an important role not only in T cell maturation and selection and in the function of mature T cells, but also in T cell antigen receptor (TCR)-related signaling pathways (Wang et al., 2011, Gelkop et al., 2005). Studies that have analyzed the relationship between COVID-19 severity and immune cells have shown that T cells play an extremely important antiviral role in the organism of COVID-19 patients (Lee et al., 2022). LYN is Tyrosine-protein kinase Lyn, which plays an important role in the developmental maturation of B cells, the positive and negative regulation of inflammatory responses and the co-catalysis of T cells (Brian and Freedman, 2021). LYN has been less studied in relation to COVID-19, but recent related studies have shown that LYN has an important relationship with severe COVID-19 (Chen et al., 2022). HCK is the tyrosine protein kinase HCK, which plays an important role in hemopoiesis, regulation of the innate immune response and release of inflammatory molecules (Podar et al., 2004). HCK has been identified as an important pharmacological target for improving lung function in patients with COVID-19 and has been significantly associated with COVID-19 in studies comparing healthy individuals with patients with severe COVID-19 (Chen et al., 2022, Chakraborty et al., 2021). In short, the selected hub targets elucidate the pharmacological mechanisms of action of cepharanthine for the treatment of COVID-19 at the level of antiviral, organismal metabolism, anticoagulation, and regulation of inflammatory and immune responses.
To investigate the specific mechanism of action, the GO enrichment analysis and KEGG pathway enrichment analysis of the overlapped targets were carried out. In the GO enrichment analysis, in terms of CC, MF, and BP, the top 20 terms of −log10(p value) were selected, respectively. GO enrichment analysis indicated that cepharanthine might act through protein kinase complex, receptor complex, serine-type peptidase complex, various membrane components, ion channel complex, and transcription regulator complex and other cellular components, and it showed protein kinase activity, protein tyrosine kinase activity, cysteine-type endopeptidase regulator activity involved in apoptotic process and other molecular functions. Additionally, cepharanthine is involved in activation of immune response, positive regulation of programmed cell death, positive regulation of cytokine production, protein phosphorylation and response to xenobiotic stimulus and other biological processes. The terms that related to regulate inflammatory response, immune cell and various enzyme activities ranked top, suggesting that cepharanthine may treat COVID-19 through modulating the inflammatory response, formation and function of immune cells and enzyme activity. Moreover, in KEGG pathway enrichment analysis, five pathways associated with COVID-19 were enriched including NF-kappa B signaling pathway, Chemokine signaling pathway, cAMP signaling pathway, Complement and coagulation cascades and AMPK signaling pathway. COVID-19 causes severe damage to the body, which is caused by the activation of the COVID-19 virus leading to a “cytokine storm” in the body. The NF-kappa B signaling pathway plays an important role in the mechanism by which the virus causes the body to produce a “cytokine storm”, which causes the body to produce large amounts of inflammatory and chemokines, leading to severe damage. Therefore, inhibition of the NF-kappa B signaling pathway and the Chemokine signaling pathway is important in the mitigation and treatment of COVID-19 (Gudowska-Sawczuk and Mroczko, 2022, Hariharan et al., 2021, Attiq et al., 2021). The cAMP signaling pathway and the AMPK signaling pathway regulate COVID-19 have been relatively poorly reported, but recent literature has uncovered a role for the cAMP signaling pathway in the treatment and prognosis of COVID-19 (Bergantin, 2022, Zhuang et al., 2021). Similarly, AMPK plays an important role in viral infections. AMPK is a regulator of cell proliferation and growth, and of the body's immune function, and plays an important role in promoting and suppressing viruses (Cai et al., 2021, Bhutta et al., 2021). In addition, complement and coagulation cascades and platelet activation were verified to be associated with COVID-19 (Bumiller-Bini et al., 2021). Notably, other signaling pathways such as diabetes, Focal adhesion and various other diseases also ranked relatively high in the enrichment analysis, and the reason for the high ranking of these signaling pathways may be that the severe COVID-19 patients collected in this study were already in their old age. However, the potential exists for these pathways to regulate COVID-19 as well, which is also of great importance.
To further validate the reliability of the core targets, molecular docking was used to show the binding affinity. The results implied that the docking binding energies of cepharanthine to targets enriched in the coronavirus disease-COVID-19 pathway were all less than −6.4 kca.mol−1, suggesting the potential targets of cepharanthine for the treatment of COVID-19.
ACE2 is the initiation binding protein for COVID-19 virus invasion in humans (Gross et al., 2020). Spike protein S1 is a subunit of the most important surface membrane protein of COVID-19 virus, which is responsible for recognition and binding to ACE2 (Huang et al., 2020). SARS-CoV-2 3CL hydrolase and SARS-CoV-2 RNA-dependent RNA polymerase is hydrolases and RNA replicase of COVID-19 virus (Mouffouk et al., 2021). The molecular docking results showed that cepharanthine had a stable binding affinity to ACE2 and SARS-CoV-2 3CL hydrolase, implying that cepharanthine may take effects through binding to ACE2 and SARS-CoV-2 3CL hydrolase.
In addition, the study has employed molecular docking and infection assay to demonstrate molecular docking mimics cepharanthine's preference for binding to spike S1 and NPC1 targets, and that infection assays demonstrated cepharanthine's therapeutic effects (Hijikata et al., 2022). And according to the study, NPC1 target is very important in the treatment of viral infections (Sturley et al., 2020). The above study adopted molecular docking to show more stable binding of cepharanthine to the relevant target. However, the molecular docking is only a static simulation and it needs further validation and screening. Besides, Rogosnitzky et al reviewed the antiviral effects of cepharanthine (Rogosnitzky et al., 2020). The review showed that in vitro effects of cepharanthine against COVID-19 and indicated that further studies are needed to investigate the mechanism of cepharanthine in the treatment of COVID-19 in vivo. The present study did not research from the NPC1 target, but the study did focus on the pharmacological targeting of cepharanthine in the treatment of COVID-19 in vivo and also investigated the interaction of cepharanthine with notable COVID-19 targets and proteases by static and dynamic methods. Therefore, in our work, the combination approach of molecular docking and molecular dynamics simulation was employed to screen and validate the hub targets of cepharanthine for the treatment of COVID-19. It is evident from the above literature that NPC1 has been shown to be a potential target for the treatment of viral infections and therefore, the results of this study need to be combined with the NPC1 target for further in-depth study.
The seamless combination of molecular docking results and results of MD simulations shows that the MD simulation further screened and confirmed the key targets for the treatment of COVID-19 with cepharanthine. In the RMSD plots, most of the core targets with stable dynamic trajectories have RMSD deviations of less than 0.5 nm and below 1 nm. And in the RMSF plots, most of the core targets have an RMSF of less than 0.3 nm. The FEL analysis plots indicate that the binding energy changes corresponding to the molecular docking results and the MD simulations characterization in this study are well sampled. And the peptide chain is loosened to the right degree when the drug target binds. Therefore, from the combined analysis of RMSD, RMSF and FEL, six core targets, namely ESR1, ANXA5, LCK, LYN, HCK and 3CL, were finally identified through further screening and validation of MD simulations as potential core targets of action of cepharanthine for the treatment of COVID-19.
This study provides a theoretical basis for further exploration of the mechanism of cepharanthine for the treatment of COVID-19. Though relevant experiments in humans are needed for further study, this study provides novel insights for the development of natural-product-inspired chemicals as potential counters to SARS-CoV-2 infection.
5 Conclusions
This study is the first time to unravel the mechanism of cepharanthine against COVID-19 by network pharmacology, molecular docking along with molecular dynamics simulations. The critical targets and pathways of cepharanthine for the treatment of COVID-19 were obtained, which facilitate and provide direction for further research of cepharanthine against COVID-19. The results suggest that cepharanthine exerts efficacy through a multi-target and multi-pathway manner. And these results provide novel insights for the development of natural-product-inspired chemicals as potential counters to SARS-CoV-2 infection.
CRediT authorship contribution statement
Feifei Sun: Conceptualization, Project administration, Funding acquisition. Jinde Liu: Investigation, Software. Ali Tariq: Validation. Zhonglei Wang: Visualization. Yongning Wu: Supervision. Lin Li: Supervision.
Acknowledgements
This research was funded by Foundation of the higher education institutions of Anhui Province (No. KJ2021A0147), Natural Science Foundation of Anhui Agricultural University (No. 2019zd17), and the Animal-Derived Food Safety Innovation Team (ANRC2021040).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- The direct evidence and mechanism of traditional Chinese medicine treatment of COVID-19. Biomed. Pharmacother.. 2021;137:111267
- [CrossRef] [Google Scholar]
- COVID-19: Discovery, diagnostics and drug development. J. Hepatol.. 2021;741:168-184.
- [CrossRef] [Google Scholar]
- The triumvirate of NF-κB, inflammation and cytokine storm in COVID-19. Int. Immunopharmacol.. 2021;101(Pt B):108255.
- [CrossRef] [Google Scholar]
- Remdesivir for the Treatment of Covid-19 - Final Report. N. Engl. J. Med.. 2020;38319:1813-1826.
- [CrossRef] [Google Scholar]
- Mental Disorders and Poor COVID-19 Prognosis: Reevaluating the Relationship through Ca2+/cAMP Signalling. Curr. Top. Med. Chem.. 2022;2215:1215-1218.
- [CrossRef] [Google Scholar]
- Candidate antiviral drugs for COVID-19 and their environmental implications: a comprehensive analysis. Environ. Sci. Pollut. Res. Int.. 2021;2842:59570-59593.
- [CrossRef] [Google Scholar]
- The Src-family Kinase Lyn in Immunoreceptor Signaling. Endocrinology 2021:16210.
- [CrossRef] [Google Scholar]
- Gene: a gene-centered information resource at NCBI. Nucleic Acids Res.. 2015;43D1:D36-D42.
- [CrossRef] [Google Scholar]
- MASPs at the crossroad between the complement and the coagulation cascades - the case for COVID-19. Genet. Mol. Biol.. 2021;441(Suppl 1):e20200199.
- [Google Scholar]
- Rcsb Protein Data Bank: Sustaining a Living Digital Data Resource that Enables Breakthroughs in Scientific Research and Biomedical Education. Biophys. J.. 2019;1163:329a-a.
- [CrossRef] [Google Scholar]
- The pharmacological mechanism of Huashi Baidu Formula for the treatment of COVID-19 by combined network pharmacology and molecular docking. Ann. Palliat. Med.. 2021;104:3864.
- [CrossRef] [Google Scholar]
- Selective targeting of the inactive state of hematopoietic cell kinase (Hck) with a stable curcumin derivative. J. Biol. Chem.. 2021;296:100449
- [CrossRef] [Google Scholar]
- Bioinformatics analysis of potential pathogenesis and risk genes of immunoinflammation-promoted renal injury in severe COVID-19. Front. Immunol.. 2022;13:950076
- [CrossRef] [Google Scholar]
- Aging in COVID-19: Vulnerability, immunity and intervention. Ageing Res. Rev.. 2021;65:101205
- [CrossRef] [Google Scholar]
- EVenn: Easy to create repeatable and editable Venn diagrams and Venn networks online. J. Genet.. 2021;489:863-866.
- [CrossRef] [Google Scholar]
- A tale of two antiviral targets-and the COVID-19 drugs that bind them. Nat. Rev. Drug Discov.. 2021;21:3-5.
- [CrossRef] [Google Scholar]
- SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res.. 2019;47W1:W357-W364.
- [CrossRef] [Google Scholar]
- Inhibitor of apoptosis (IAP) proteins in regulation of inflammation and innate immunity. Discov. Med.. 2011;1158:221-231.
- [Google Scholar]
- Platelet Phenotype Analysis of COVID-19 Patients Reveals Progressive Changes in the Activation of Integrin αIIbβ3, F13A1, the SARS-CoV-2 Target EIF4A1 and Annexin A5. Front. Cardiovasc. Med.. 2021;8:779073
- [CrossRef] [Google Scholar]
- Cepharanthine: A Promising Old Drug against SARS-CoV-2. Adv. Biol.. 2022;612:e2200148.
- [Google Scholar]
- Repurposing of clinically approved drugs for treatment of coronavirus disease 2019 in a 2019-novel coronavirus-related coronavirus model. Chin. Med. J. (Engl.). 2020;1339:1051-1056.
- [CrossRef] [Google Scholar]
- Evaluation of AutoDock and AutoDock Vina on the CASF-2013 Benchmark. J. Chem. Inf. Model.. 2018;588:1697-1706.
- [CrossRef] [Google Scholar]
- T cell activation-induced CrkII binding to the Zap70 protein tyrosine kinase is mediated by Lck-dependent phosphorylation of Zap70 tyrosine 315. J. Immunol. (Baltimore, Md.: 1950). 2005;17512:8123-8132.
- [CrossRef] [Google Scholar]
- Evaluation of SARS-CoV-2 entry, inflammation and new therapeutics in human lung tissue cells. PLoS Pathog.. 2022;181:e1010171.
- [Google Scholar]
- ACE2, the receptor that enables infection by SARS-CoV-2: biochemistry, structure, allostery and evaluation of the potential development of ACE2 modulators. ChemMedChem. 2020;1518:1682-1690.
- [CrossRef] [Google Scholar]
- The Role of Nuclear Factor Kappa B (NF-κB) in Development and Treatment of COVID-19: Review. Int. J. Mol. Sci.. 2022;239
- [CrossRef] [Google Scholar]
- The Role and Therapeutic Potential of NF-kappa-B Pathway in Severe COVID-19 Patients. Inflammopharmacology. 2021;291:91-100.
- [CrossRef] [Google Scholar]
- Evaluating cepharanthine analogues as natural drugs against SARS-CoV-2. FEBS Open Bio. 2022;121:285-294.
- [CrossRef] [Google Scholar]
- Network pharmacology: the next paradigm in drug discovery. Nat. Chem. Biol.. 2008;411:682-690.
- [CrossRef] [Google Scholar]
- Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin.. 2020;419:1141-1149.
- [CrossRef] [Google Scholar]
- KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res.. 2017;45D1:D353-D361.
- [CrossRef] [Google Scholar]
- Global COVID-19 Death Toll May Be Triple the Reported Deaths. J. Am. Med. Assoc.. 2022;32715:1438.
- [CrossRef] [Google Scholar]
- Integrated analysis of plasma and single immune cells uncovers metabolic changes in individuals with COVID-19. Nat. Biotechnol.. 2022;401:110-120.
- [CrossRef] [Google Scholar]
- Estrogen hormone is an essential sex factor inhibiting inflammation and immune response in COVID-19. Sci. Rep.-Uk. 2022;121
- [CrossRef] [Google Scholar]
- Transcriptome analysis of cepharanthine against a SARS-CoV-2-related coronavirus. Brief. Bioinform.. 2021;222:1378-1386.
- [CrossRef] [Google Scholar]
- Potential treatment of COVID-19 with traditional Chinese medicine: What herbs can help win the battle with SARS-CoV-2? Engineering 2021
- [CrossRef] [Google Scholar]
- Estrogen receptor inhibits interleukin-6 gene expression by disruption of nuclear factor kappaB transactivation. Cytokine. 2005;314:251-257.
- [CrossRef] [Google Scholar]
- PharmMapper server: a web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res.. 2010;38(suppl_2):W609-W614.
- [CrossRef] [Google Scholar]
- Study on the mechanism of action of Ephedra Herba Decoction against influenza A virus based on network pharmacology. TMR Modern Herbal Med.. 2022;52:10.
- [CrossRef] [Google Scholar]
- XIAP induces NF-kappaB activation via the BIR1/TAB1 interaction and BIR1 dimerization. Mol. Cell. 2007;265:689-702.
- [CrossRef] [Google Scholar]
- Covid-19: Pfizer's paxlovid is 89% effective in patients at risk of serious illness, company reports. BMJ (Clin. Res. Ed.). 2021;375:n2713.
- [CrossRef] [Google Scholar]
- Flavonols as potential antiviral drugs targeting SARS-CoV-2 proteases (3CL(pro) and PL(pro)), spike protein, RNA-dependent RNA polymerase (RdRp) and angiotensin-converting enzyme II receptor (ACE2) Eur. J. Pharmacol.. 2021;891:173759
- [CrossRef] [Google Scholar]
- Potential anti-COVID-19 agents, cepharanthine and nelfinavir, and their usage for combination treatment. iScience. 2021;244:102367
- [CrossRef] [Google Scholar]
- Heterogeneous parallelization and acceleration of molecular dynamics simulations in GROMACS. J. Chem. Phys.. 2020;15313:134110
- [CrossRef] [Google Scholar]
- Molecular Docking: Shifting Paradigms in Drug Discovery. Int. J. Mol. Sci.. 2019;2018
- [CrossRef] [Google Scholar]
- Critical role for hematopoietic cell kinase (Hck)-mediated phosphorylation of Gab1 and Gab2 docking proteins in interleukin 6-induced proliferation and survival of multiple myeloma cells. J. Biol. Chem.. 2004;27920:21658-21665.
- [CrossRef] [Google Scholar]
- Albumin Infusion in Critically Ill COVID-19 Patients: Hemodilution and Anticoagulation. Int. J. Mol. Sci.. 2021;2213
- [CrossRef] [Google Scholar]
- Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;5867827:113-119.
- [CrossRef] [Google Scholar]
- Cepharanthine: a review of the antiviral potential of a Japanese-approved alopecia drug in COVID-19. Pharmacol. Rep.: PR. 2020;726:1509-1516.
- [CrossRef] [Google Scholar]
- Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res.. 2003;1311:2498-2504.
- [CrossRef] [Google Scholar]
- Potential COVID-19 therapeutics from a rare disease: weaponizing lipid dysregulation to combat viral infectivity. J. Lipid Res.. 2020;617:972-982.
- [CrossRef] [Google Scholar]
- The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res.. 2021;49D1:D605-D612.
- [CrossRef] [Google Scholar]
- UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res.. 2021;49D1:D480-D489.
- [CrossRef] [Google Scholar]
- Bioactive Natural Products in COVID-19 Therapy. Front. Pharmacol.. 2022;2982
- [CrossRef] [Google Scholar]
- Chinese herbal medicine: Fighting SARS-CoV-2 infection on all fronts. J. Ethnopharmacol.. 2021;270:113869
- [CrossRef] [Google Scholar]
- Broad-spectrum prodrugs with anti-SARS-CoV-2 activities: Strategies, benefits, and challenges. J. Med. Virol.. 2022;944:1373-1390.
- [CrossRef] [Google Scholar]
- RhoH modulates pre-TCR and TCR signalling by regulating LCK. Cell. Signal.. 2011;231:249-258.
- [CrossRef] [Google Scholar]
- Tom70 mediates Sendai virus-induced apoptosis on mitochondria. J. Virol.. 2015;897:3804-3818.
- [CrossRef] [Google Scholar]
- SymMap: an integrative database of traditional Chinese medicine enhanced by symptom mapping. Nucleic Acids Res.. 2019;47D1:D1110-D1117.
- [CrossRef] [Google Scholar]
- Natural products, alone or in combination with FDA-approved drugs, to treat COVID-19 and lung cancer. Biomedicines. 2021;96:689.
- [CrossRef] [Google Scholar]
- Using PyMOL as a platform for computational drug design. Wiley Interdiscip. Rev.: Comput. Mol. Sci.. 2017;72:e1298.
- [Google Scholar]
- Comparison of viral RNA-host protein interactomes across pathogenic RNA viruses informs rapid antiviral drug discovery for SARS-CoV-2. Cell Res.. 2022;321:9-23.
- [CrossRef] [Google Scholar]
- Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun.. 2019;101:1523.
- [CrossRef] [Google Scholar]
- Single-Cell Sequencing of Peripheral Mononuclear Cells Reveals Distinct Immune Response Landscapes of COVID-19 and Influenza Patients. Immunity. 2020;533(685–696):e683.
- [Google Scholar]
- Exploring the Potential Mechanism of Shufeng Jiedu Capsule for Treating COVID-19 by Comprehensive Network Pharmacological Approaches and Molecular Docking Validation. Comb. Chem. High T Scr.. 2021;249:1377-1394.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary material
Supplementary material to this article can be found online at https://doi.org/10.1016/j.arabjc.2023.104722.
Appendix A
Supplementary material
The following are the Supplementary material to this article:Supplementary data 1
Supplementary data 1







