Translate this page into:
Characterization of metabolites of five typical saponins from Caulophyllum robustum Maxim and their biotransformation in fibroblast-like synoviocytes by UHPLC-Q-Exactive-Plus-Orbitrap-MS
⁎Corresponding authors at: Heilongjiang University of Chinese Medicine, Harbin 150040, China(H. Kuang), School of Traditional Chinese Medicine, Guangdong Pharmaceutical University, Guangzhou 510024, China (Q. Wang). qhwang@hljucm.net (Qiuhong Wang), hxkuang15@hotmail.com (Haixue Kuang)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Abstract
Caulophyllum robustum Maxim (C. robustum) is a well-known traditional Chinese medicine (TCM) widely used for the treatment of rheumatoid arthritis (RA). Modern pharmacological studies have shown that saponins are the main material basis of C. robustum, of which Cauloside C, Cauloside D, Cauloside G, Cauloside H, and Leonticin D are typical representatives. Although they are known to enter the bloodstream as prototypes, metabolites, and sapogenins, their biotransformation in fibroblast-like synoviocytes (FLS) is not well understood. The UHPLC-Q-Exactive-Plus-Orbitrap-MS method was used to rapidly identify the prototypes and metabolites of C. robustum saponins in FLS and to explore the metabolic pathways of C. robustum saponins. As a result, deglycosylation, deacidification, deoxygenation, oxidation, sulfation, phosphorylation, hydrogenation, demethylation, and acetylation may be important pathways for saponins of C. robustum to function in FLS. Especially, we first discovered phosphorylated metabolites, oxidative metabolites, acetylated metabolites and deacidified metabolites. At the cellular level, saponins with different sugar chain structures of the same kind can be converted into each other. According to the structural analysis of the degradation products, the length of the sugar chain may affect the degradation of saponins in cells. This study aimed to analyze the degradation and transformation processes of C.robustum saponins in target cells and to provide scientific information for the discovery of new medicinal components.
Keywords
Caulophyllum robustum Maxim
Saponins
Cauloside G
Cauloside C
Metabolic profile
1 Introduction
Saponins are a class of glycosides attached to hydrophilic oligosaccharide groups by nonpolar triterpenes or steroids and are widely distributed in the plant kingdom as natural products (Dong et al.,2019). The content of saponins in traditional Chinese medicine(TCM) is relatively high, such as ginseng, Panax notoginseng, and Acanthopanax senticosus. In recent years, saponins have received extensive attention and have been applied in clinical practice because of their various pharmacological activities, such as anti-tumor, anti-oxidative, immunomodulatory, neuroprotective (Qu et al.,2022; Podolak et al.,2010). Moreover, saponins are expected to be one of the most promising drug candidates in the biomedical and pharmaceutical fields because of their obvious anti-inflammatory activities. Tian et al. combined LPS-induced RAW 264.7 cells and a mouse model of acute lung injury to systematically evaluate the anti-inflammatory effect of Tribulus terrestris L., and the results showed that saponins decreased the content of NO and TNF-α, reduced phagocytosis activity, and alleviated pulmonary edema of lung tissue(Tian et al.,2021). The anti-inflammatory properties of saponin-rich TTS extract also help alleviate insulin resistance and weight gain in obese female rats and improve gonadal hormone dysregulation and ovarian lesions (Abdel-Mottaleb et al.,2022). A novel triterpenoid saponin RT8 isolated from ginseng seeds can inhibit the production of ROS and NO and inhibit pro-inflammatory genes (IL-1β, IL-6, iNOS, COX2, and MMP-9) in a dose-dependent manner (Rho et al.,2020).
Rheumatoid arthritis (RA) is a disease characterized by the swelling of the synovial membrane of joints, and the target tissue is the synovium. Fibroblast-like synoviocytes (FLS) are major players in RA synovial inflammation(Mor et al.,2005), and the abnormal proliferation, migration, and invasion of FLS lead to the development of the disease(Wu et al.,2021; Németh et al.,2022). Caulophyllum robustum Maxim (C.robustum) is a folk medicine for RA in China (Lü et al.,2015). C.robustum extract can regulate the levels of inflammatory cytokines, regulate cellular immunity and humoral immune function, inhibit angiogenesis, slow down the formation and proliferation of synovial pannus, and reduce the erosion and damage of articular cartilage and bone(Wang et al.,2017). The detection results of the lncRNA-mRNA chip showed that there were 384 differentially expressed genes in C. robustum, of which 202 were upregulated and 182 were downregulated, affecting various inflammatory pathways, such as NF-κB, MAPK, and TNF-like receptor (Lü et al.,2020). The saponin content in the C. robustum extract was as high as 15.11%, of which Cauloside C (CLC), Cauloside D(CLD), Cauloside G (CLG), Cauloside H CLH), and Leonticin D (LD) are typical representatives of RA treatment (Fig. 1). Although many studies have been conducted, the biotransformation of C. robustum saponins remains unclear.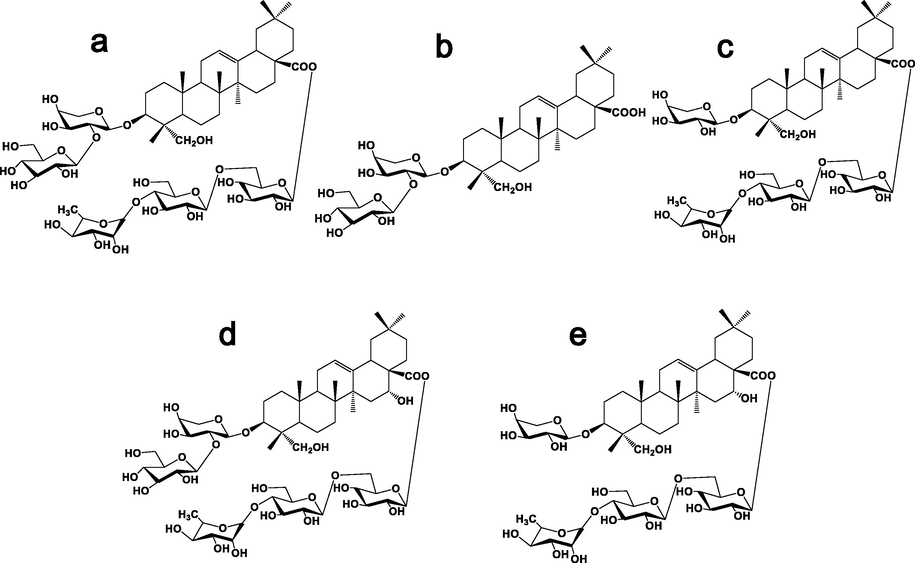
Chemical structure of five saponins from the active parts of Caulophyllum robustum Maxim(a: Cauloside G; b: Cauloside C;c: Cauloside D;d: Cauloside H; e:Leonticin D).
In recent years, research on the absorption and biotransformation of saponins in vivo has become a hot field. There are various biodegradation pathways for saponins that enter the body and are absorbed into the blood in various forms. Therefore, we studied the metabolism of total saponins from C.robustum in vivo. After the rats were given intragastric administration of Caulophyllum robustum Maxim effective part (CRME), 22 components were identified in rats, and metabolic transformations in bile, urine, and feces were characterized by deglycosylation of phase I metabolism, and metabolic transformations in bile, urine, and feces were characterized by glucuronidation and sulfation of phase II metabolism (Lü et al.,2019). Among them, the five saponins all showed a trend of fast absorption and slow elimination; the absorption mechanism of the small intestine was passive transport, and the duodenum had the highest permeability coefficient, which was mainly excreted in urine (Guo et al.,2020). However, what changes will occur in the monomer saponin components of C.robustum in the lesion site of RA is a question of concern.
Saponins produce new components and sapogenins through physiological and biochemical processes to exert their therapeutic effects. The basis and transformation mechanism should be revealed and elucidated. In summary, we aim to provide a rapid and sensitive UHPLC-Q-Exactive-Plus-Orbitrap-MS method to analyze metabolites of C. robustum saponins to explore the metabolic pathways in FLS.
2 Materials and methods
2.1 Chemicals and reagents
The roots and rhizomes of C. robustum were purchased from the Heilongjiang Province, China. The voucher samples (20120901, 20120902, and 20120903) were authenticated by Prof. Shaowa Lü and deposited at the College of Pharmacy, Heilongjiang University of Chinese Medicine, China. Cauloside C, Cauloside D, Cauloside G, Cauloside H, Leonticin D (self-made) were used as reference standards and showed purities of ≥ 98% by HPLC/ELSD analysis. β-Glucosidase was purchased from Shanghai Yuanye Biotechnology Co., Ltd. (Shanghai, China). LC-grade methanol and acetonitrile (ACN) were obtained from Fisher Scientific (USA). All other reagents used were of analytical grade. Basic Dulbecco’s modified Eagle’s medium (DMEM) basic (1X) from Gibco (Carlsbad, CA, USA). Fetal bovine serum (FBS) was obtained from Israel BI Bio Co. Ltd. (Israel). Trypsin was purchased from Biyuntian Biotechnology Co. Ltd. (Shanghai, China). Penicillin-streptomycin solution was purchased from HyClone (USA).
2.2 Preparation of reference solution
The test compounds Cauloside C,7.66 mg、Cauloside G,12.36 mg、Cauloside D,10.74 mg、Cauloside H,12.51 mg、Leonticin D,10.90 mg, respectively, were weighed precisely. The samples were dissolved in dimethyl sulfoxide (DMSO; 100 µL). All sample solutions were filtered through 0.22 m membrane filters, and the mother liquor (10 mM) was obtained by taking10 µL of the filter solution into volumetric flasks and dissolving it in DMEM supplemented with 1% penicillin/streptomycin (90 µL).
2.3 Cell lines and culture
Mouse L929 fibroblasts were used in this experiment, purchased from Shanghai Fuheng Biotechnology Co., Ltd. The cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin/ streptomycin and then incubated in a 5% CO2 atmosphere at 37 °C.
2.4 Drug administration and biosample collection
Well-grown cells were taken, digested, diluted with complete medium to obtain a cell suspension, counted, and inoculated into 15 T25 culture flasks(15 groups), 2 ml per well. Five groups were used as experimental groups, five groups were used as the corresponding blank groups, and the remaining five groups were used as the control group. All groups of cells were placed in a 5% CO2, 37 °C incubator for 24 h until the cells were completely adherent. After the cells adhered, the supernatant was discarded and washed 2–3 times with PBS. The experimental groups were administered 100 μM Cauloside C, Cauloside G, Cauloside D, Cauloside H and Leonticin D. The blank groups were cultured normally, and the control groups respectively were given 100 μM Cauloside C, Cauloside G, Cauloside D, Cauloside H, Leonticin D, and an appropriate amount of β-glucoside was incubated at 37 °C under 5% CO2.
The cells were counted after 2 h of culture. Then, 3 ml of pre-chilled PBS were added to wash off the medium thrice, and 1.5 ml of pre-chilled solvent (methanol:water = 80:20) were added for cell quenching. After repeated pipetting of the cells a few times, the adherent cells were scraped off. The T25 cell culture flask was placed in a − 20 °C refrigerator for 30 min. The suspension was transferred to a 2 ml centrifuge tube, vortexed for 2 min, and centrifuged at 12,000 r at 4 °C for 15 min. The sediment was discarded, and the supernatant was transferred into a centrifuge tube. Blank cell metabolite and negative control cell metabolite samples were collected in the same way, and all cell metabolite samples were stored at − 80 °C before analysis.
2.5 Preparation of cell samples
Cell metabolite samples were obtained via cryopreservation and lysed at room temperature. Cell metabolite solution (200 μL) was added four times the amount of methanol to precipitate the proteins. The supernatant was concentrated to dryness at 37 °C using a centrifugal concentrator. The samples were reconstituted in 100 μL methanol, vortexed, sonicated for 10 min, 4 °C (12000 r/min), centrifuged for 5 min, and the supernatant was collected for injection analysis.
2.6 Instruments and conditions
2.6.1 Analysis of triterpene saponins by UHPLC
Chromatographic analysis was performed using a Vanquish UHPLC Ultra High-Performance Liquid Chromatograph (Thermo Scientific Corporation, USA). Sample separation was achieved on a Waters XBridge BEH C18 column (2.5 μm, 2.1 × 100 mm) with a constant flow rate of 0.30 ml/min at 35 °C. The mobile phases were 0.1% (v/v) formic acid in water (A) and Acetonitrile (B) using a gradient elution of 2 %–2% (B) at 0–1 min, 2%–20% (B) at 1.0–5.0 min, 20%–50% (B) at 5.0–10.0 min, 50%–80% (B) at 10.0–15.0 min, 80%–95% (B) at 15.0–20.0 min, 95%–95% (B) at 20.0–25.0 min, 95%–2% (B) at 25.0–26.0 min, 2%–2% (B) at 26.0–30.0 min. The injected sample volume was set at 2.0 μL.
2.6.2 Q-Exactive-Plus-Orbitrap-MS analysis
Detection was performed using a Q Exactive-Plus-Orbitrap mass spectrometer with an electrospray ionization (ESI) probe in both positive and negative modes. The acquisition parameters were as follows: nitrogen was used as the sheath gas and auxiliary gas (purity > 99.99%). The sheath gas flow rate was set at 30 arb. The auxiliary gas flow rate was set to 15 arb and the sweep gas flow rate at 3 arb. The capillary temperature was 263 °C, the RF level was 55%, and the collision energy was 30 V. The ion spray voltage was set to 2.5 kV for the negative mode and 3.5 KV for the positive mode. The mass range was recorded from m/z 150 to 2000, according to saponins reported in C.robustum.
2.7 Data processing and analysis
The Xcalibur 4.2 software was used for mass spectrometry data processing, and the following results were obtained: the extracted ion chromatogram of the compound, the retention time of the compound, the response value, the measured value of the exact molecular weight of the compound, and the secondary mass spectrometry information of the compound. For compounds with controls, confirmation was performed by comparing relative retention times, quasimolecular ions, and secondary fragment ions of each control. For compounds without controls, speculation was performed by combining the literature reports; if the mass spectrometry information of the compound could not be obtained in the literature, the molecular composition of the compound could be reversely inferred by combining the measured secondary fragment ions with the fragmentation law of the compound, and new components could be found.
3 Results
3.1 Fragmentation behaviors of five reference substances
Cauloside G exhibited a quasi-molecular ion peak at m/z 1235.6068 [M − H]−. The main fragment ions observed in the mass spectra were m/z 765.4472 [M − Rha − Glc − Glc]−, 603.3911[M-Rha-Glc-Glc-Glc-Glc-H]−,471.3482[M-Rha-Glc-Glc-Glc-Ara-H]−, 585.3778 [M − Rha − Glc − Glc − Glc − H2O]−and 423.3326 [M − Rha − Glc − Glc − Glc − Ara − H2O − HCOOH]−. The ions at m/z 471.3482 corresponded to hederagenin, which was generated by the loss of a-A.ra from m/z 603.3911 (Fig. 2a). Possible cleavage pathways are shown in Fig. 3a.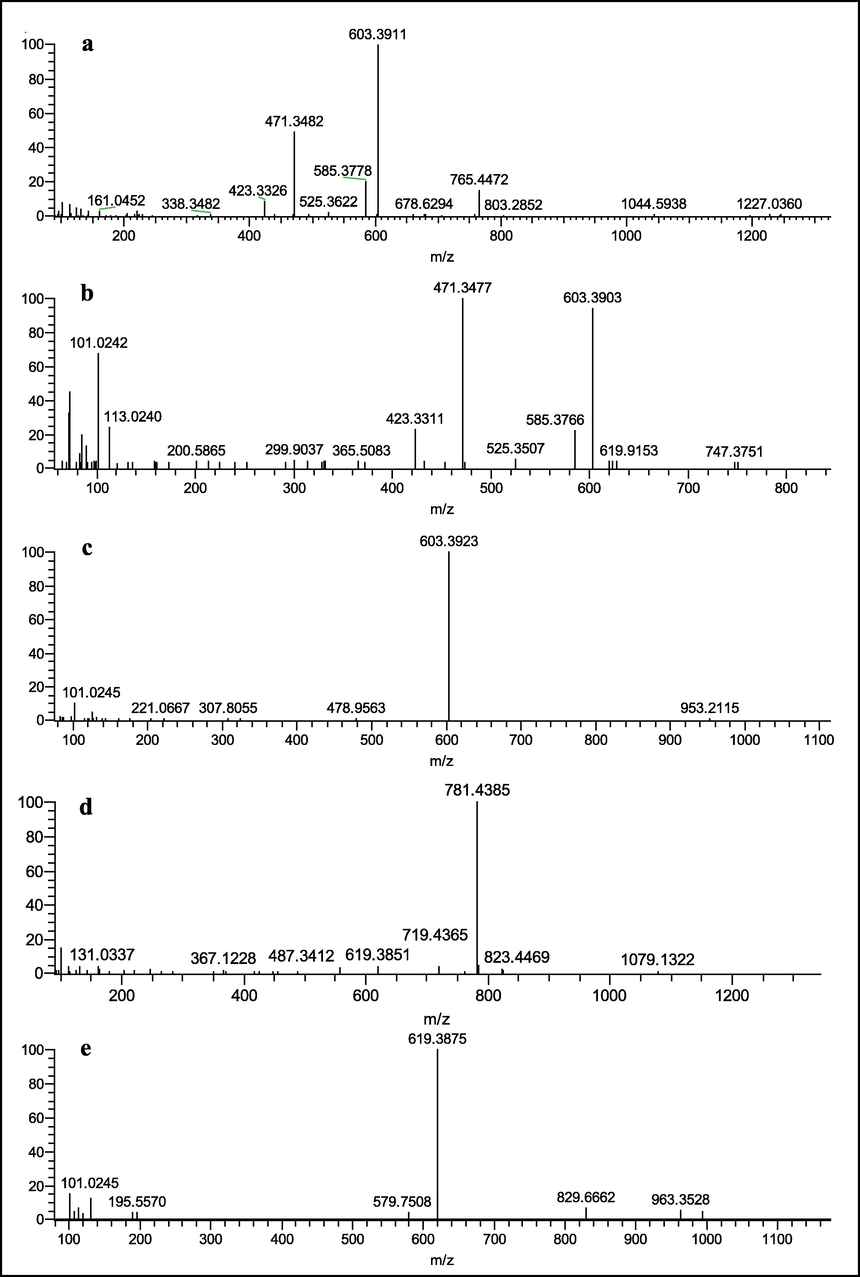
ESI-MS 2 spectrum of Cauloside G (a), Cauloside C (b), Cauloside D (c), Cauloside H(d), Leonticin D (e) (Negative ion mode).
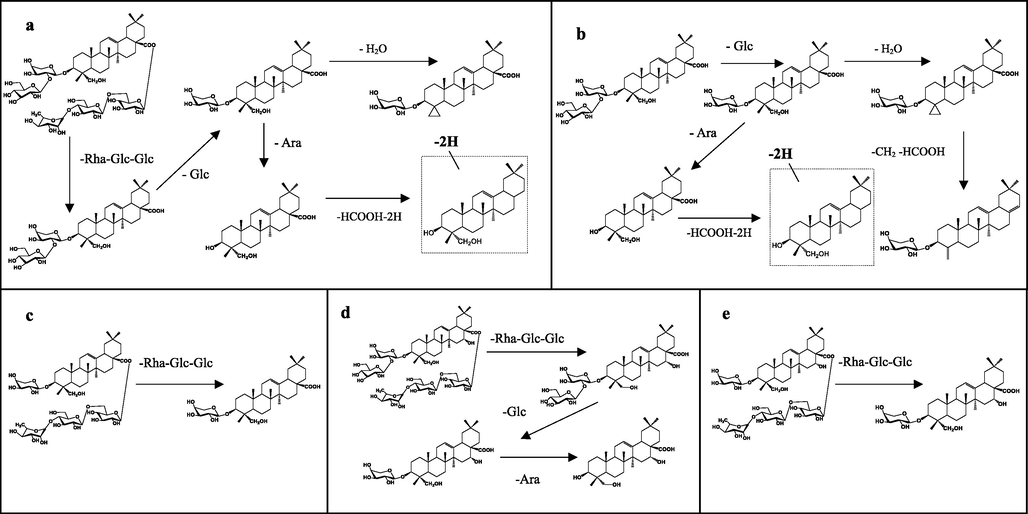
Proposed fragmentation pathways of Cauloside G (a), Cauloside C (b), Cauloside D (c), Cauloside H(d), Leonticin D (e) in the negative mode.
Cauloside C produced an [M − H]− ion at m/z 765.4417, which produced fragment ions at m/z 603.3903, 585.3766,525.3507,471.3477 and 423.3311 (Fig. 2b). The ion at m/z 603.3903 corresponds to the loss of Glc from the ion at m/z 765.4417. The ions at m/z 585.3766 resulted from 603.3903 by loss of H2O, and the fragment ion at m/z 525.3507 resulted from the cleavage of CH2 and HCOOH. The ions at m/z 471.3477 corresponded to hederagenin, which was generated by the loss of a-Ara from m/z 603.3903. The ions at m/z 423.3311 resulted from 471.3477 owing to the loss of 2H and HCOOH. The proposed fragmentation pathway is illustrated in Fig. 3b.
Cauloside D yielded a deprotonated [M − H]− ion at m/z 1073.5487. In the negative ion mode, the product ions at m/z 603.3923 [M − Rha − Glc − Glc − H] − were produced by the loss of Rha(1 → 4)Glc(1 → 6)Glc − from C-28 (Fig. 2c). The proposed fragmentation pathway is illustrated in Fig. 3c.
Cauloside H exhibited a quasi-molecular ion [M − H]− at m/z 1251.6039. The main fragment ions of the mass spectra were observed at m/z 781.4385[M-Rha-Glc-Glc-H]−, 619.3851[M-Rha-Glc-Glc-Glc-H]−, and 487.3412 [M − Rha − Glc − Glc − Glc − Ara − H]−(Fig. 2d), The possible fragmentation pathways are shown in Fig. 3d.
Leonticin D produced an [M − H]− ion at m/z 1089.5466, which produced fragment ions at m/z 619.3875 and 579.7508(Fig. 2e). The ions at m/z 619.3875 resulted from 1089.5466 by the loss of Rha(1 → 4)Glc(1 → 6)Glc-. The proposed fragmentation pathway is illustrated in Fig. 3e.
In conclusion, the C-3 and C-28 oligosaccharide chains of the saponin molecule are capable of deglycosylation, with differences of 132 Da for deglycosylation of Ara, 162 Da for deglycosylation of Glc, and 146 Da for deglycosylation of Rha. The differential value of the dehydration reaction susceptible to saponin elements is 18 Da. In the negative ion mode, Cauloside G produced characteristic fragment ions such as m/z 1235.6068, 765.4472, 603.3911, 585.3778, 471.3482, and 423.3326; Cauloside C produced characteristic fragment ions such as m/z 765.4417, 603.3903, 585.3766, 471.3477, and 423.3311; Cauloside D produced characteristic fragment ions such as m/z 1073.5487 and 603.3923; Leonticin D produced characteristic fragment ions such as m/z 1089.5466 and 619.3875 because it contains a lateral hydroxyl group; and Cauloside H produced characteristic fragment ions such as m/z 1251.6039, 781.4385, 619.3851, and 487.3412.
3.2 Characterization of five representative saponin-related metabolites in FLS
The in vitro cell model can easily simulate the physiological environment of the human body by controlling the culture conditions and has good stability. At the same time, combined with the high separation ability of chromatography and the ability of mass spectrometry to accurately measure the quality, it can quickly screen active ingredients suitable for the study of the pharmacodynamic material basis of the complex system of TCM. Positive and negative ion modes were used for scanning. Owing to the presence of acidic groups in the saponin structure, it is easy to lose H+ and form the quasi-molecular ion peak of [M − H]−. Therefore, to compare the (TIC) total ion chromatograms of each group of samples, negative ion mode data were selected for analysis (Fig.S1). Scan cell samples for potential metabolic components to identify target metabolites and metabolic pathways. After comparison with the blank samples, the components detected in the cell samples are listed in Table 1, and the chemical structures of each component are shown in Fig. S2 (Supplementary Data).
NO
Observed
Identification
Formula
Experimental Molecular Weight
Theoretical Molecular Weight
Diff (ppm)
Adducts
MS/MS Fragment ions (m/z)
Status
RT (min)
Cauloside G
M0
9.04
Cauloside G
C59H96O27
1235.6068
1235.6065
0.24
-H
1281.6131、1235.6041、765.4472、603.3910
Prototype
M1
11.97
CLG-Rha-Glc-Glc
C41H66O13
765.4422
765.4419
0.39
-H
811.4516、765.4422、603.3920、471.3509
Metabolites
M2
12.71
CLG-Rha-Glc-Glc-Glc
C35H56O8
603.3908
603.3902
0.99
-H
603.3908、471.3481、585.3815、423.3273
Metabolites
M3
14.66
CLG-Rha-Glc-Glc-Glc-Ara
C30H48O4
471.3482
471.3479
0.64
-H
471.3482、423.3273、439.3226
Metabolites
M4
20.73
CLG-Rha-Glc-Glc-Glc-Ara-HCOOH-2H
C29H44O2
423.3277
423.3268
2.13
-H
423.3277
Metabolites
M5
9.04
CLG + SO3-O + 2H2
C60H102O31S
1349.599
1349.6042
−3.71
+HCOO
1349.5990、765.4425、603.3918、471.3477
Metabolites
M6
9.04
CLG-CH2O + HPO3-O + H2
C58H97O28P
1271.5844
1271.5831
1.02
-H
1271.5844、765.4419、603.3920、471.3487
Metabolites
M7
20.6
CLG-Rha-Glc-Glc-Glc-Ara-CH2OH
C29H44O3
439.3226
439.3218
1.82
-H
439.3226
Metabolites
M8
9.74
CLG + C2H2O
C61H98O28
1277.6167
1277.6171
−0.31
-H
1277.6167
Metabolites
Cauloside C
M0
11.97
Cauloside C
C41H66O13
765.4417
765.4419
−0.26
-H
811.4513、603.3907、471.3491
Prototype
M1
12.74
CLC-Glc
C35H56O8
603.3912
603.3902
1.66
-H
603.3912、471.3494
Metabolites
M2
14.67
CLC-Glc-Ara
C30H48O4
471.3494
471.3479
3.18
-H
471.3494、
Metabolites
M3
20.72
CLC-Glc-Ara- HCOOH-2H
C29H44O2
423.3277
423.3268
2.13
-H
423.3277
Metabolites
M4
11.97
CLC + SO3-O + 2H2
C41H70O15S
833.4311
833.4351
−5.52
-H
879.4376、833.4311、603.3883、471.3467
Metabolites
M5
11.97
CLC-CH2O + HPO3-O + H2
C40H67O14P
801.4219
801.4915
2.99
-H
801.4219、603.3930、585.3815、471.3492
Metabolites
M6
11.63
CLC-O + H2
C42H66O16
825.4306
825.4267
4.72
+HCOO
825.4306
Metabolites
M7
10.99
CLC + 4O
C42H68O19
875.4306
875.4271
3.99
+HCOO
875.4306
Metabolites
Cauloside D
M0
9.31
Cauloside D
C53H86O22
1073.5487
1073.5527
−3.72
-H
1119.5610、1073.5487、603.3922
Prototype
M1
12.73
CLD-Rha-Glc-Glc
C35H56O8
603.3907
603.3902
0.79
-H
603.3907
Metabolites
M2
9.31
CLD + SO3-O + 2H2
C54H92O26S
1187.5477
1187.5513
−3.03
+HCOO
1187.5477、671.3821、603.3922
Metabolites
M3
12.74
CLD + SO3-O + 2H2-Rha-Glc-Glc
C35H60O10S
671.3821
671.3823
−0.29
+HCOO
671.3821
Metabolites
M4
9.32
CLD-CH2 + HPO3-O + H2
C52H87O23P
1109.5322
1109.5302
1.8
-H
1109.5322、603.3920
Metabolites
Cauloside H
M0
9.17
Cauloside H
C59H96O28
1251.6039
1251.6012
2.16
-H
1251.6039、1135.5561、619.3865
Prototype
M1
8.79
CLH-Glc
C53H86O23
1089.5466
1089.5487
−1.93
-H
1135.5561、1089.5466、619.3865
Metabolites
M2
11.47
CLH-Glc-Rha-Glc-Glc
C35H56O9
619.3865
619.3851
2.26
-H
619.3865、573.3821
Metabolites
M3
29.2
CLH-Glc-Rha-Glc-Glc-HCOOH
C34H54O7
573.3821
573.3796
4.96
-H
573.3821
Metabolites
M4
8.8
CLH-Glc + SO3-O + 2H2
C54H92O27S
1203.5427
1203.5462
−2.91
+HCOO
1203.5427、619.3868、573.3821
Metabolites
M5
8.8
CLH-Glc-CH2 + HPO3-2O + H2
C52H87O24P
1125.5271
1125.5252
1.69
-H
1125.5271、619.3868
Metabolites
M6
18.65
CLH-Glc-Rha-Glc-Glc-Ara + C2H2O
C32H50O6
529.3555
529.3534
3.97
-H
529.3555
Metabolites
Leonticin D
M0
8.78
Leonticin D
C53H86O23
1089.5466
1089.5487
1.99
-H
1135.5566、1089.5466、619.3875
Prototype
M1
11.48
LD-Glc-Rha-Glc-Glc
C35H56O9
619.3873
619.3851
3.55
-H
619.3873
Metabolites
M2
20.33
LD-Glc-Rha-Glc-Glc-Ara
C30H48O5
487.3427
487.3429
−0.41
-H
487.3427
Metabolites
M3
8.8
LD + SO3-O + 2H2
C54H92O27S
1203.5443
1203.5462
−1.58
+HCOO
1203.5443
Metabolites
M4
8.8
LD-CH2 + HPO3-2O + H2
C52H87O24P
1125.5272
1125.5252
1.78
-H
1125.5272
Metabolites
3.2.1 Metabolites of Cauloside G
Eight metabolites (CLG-M (1–8)) of Cauloside G were identified (Fig. 4). These metabolites indicate that deglycosylation was the main metabolic pathway. Detailed information on metabolites is described in Table 1. CLG-M0 was eluted at 9.04 min with [M − H]− ion at m/z 1235.6068 and [M + HCOO − H]−ion at m/z 1281.6143, which produced fragment ions at m/z 765.4422, 603.3908, 471.3482, and 423.3273; the ion at m/z 765.4422 corresponded to a loss of –Rha-Glc-Glc from the ion at m/z 1235.6068. The ions at m/z 603.3908 and 471.3482 resulted from the loss of Glc and Ara, respectively. The ions at m/z 471.3482 corresponded to hederagenin. The ion at m/z 423.3233 is generated by the loss of 2H and HCOOH from the ion at m/z 471.3282.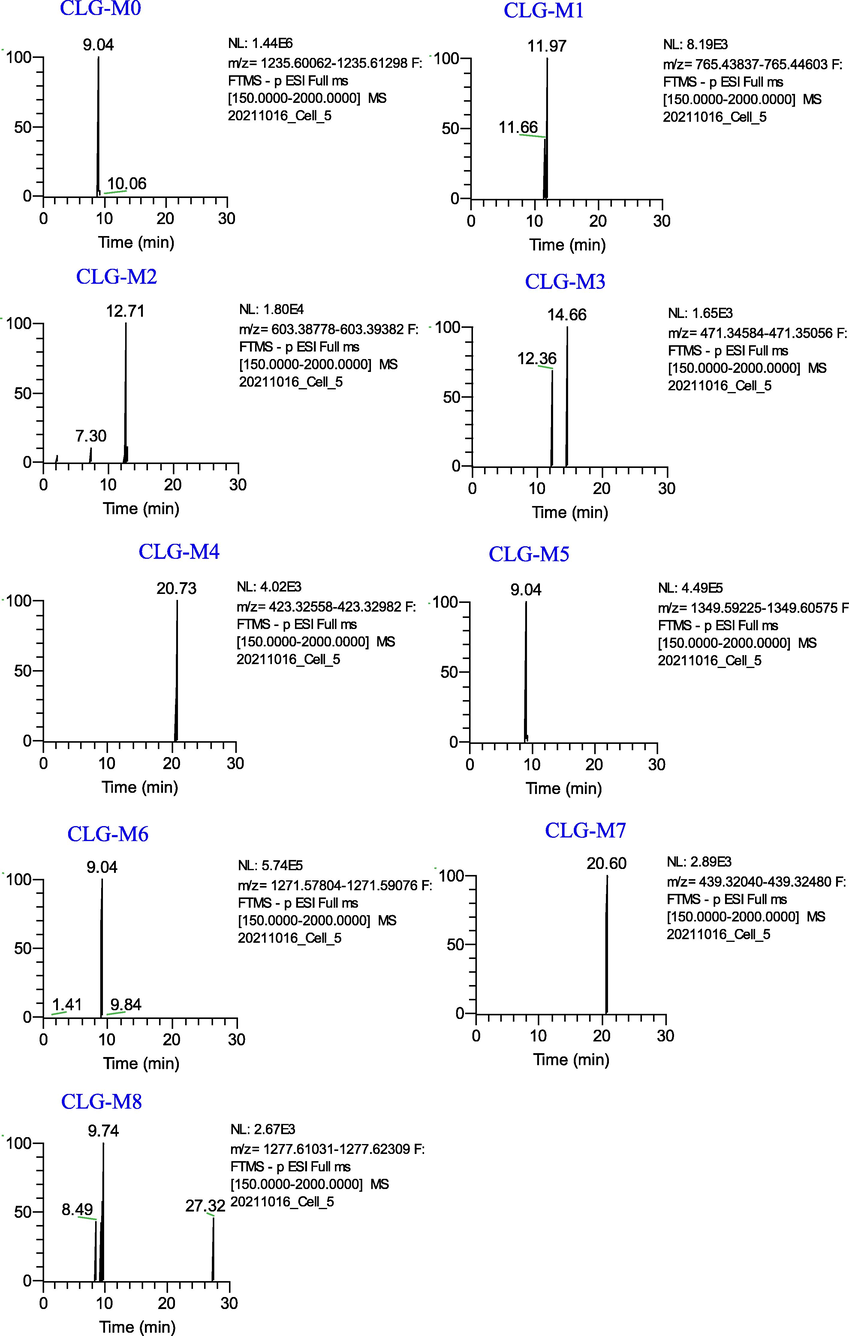
Extracted ion chromatograms of the metabolites of Cauloside G in the negative ion mode.
CLG-M1, CLG-M2 and CLG-M3 are formed via deglycosylation. CLG-M1 eluted at 11.96 min and yielded a deprotonated molecule [M − H]− ion at m/z 765.4422(C41H66O13), which was 469 (Rha-Glc-Glc) Da lower than the deprotonated ion of CLG-M0. CLG-M2 was observed at a retention time of 12.71 min and exhibited an [M − H]− ion at m/z 603.3908, which indicated 162 (C6H10O5) Da cleavage at C-3. CLG-M3 (14.66 min, C30H48O4) was formed by the cleavage of -Rha-Glc-Glc, -Glc, and -Ara of CLG-M0.
CLG-M4 (20.72 min) showed an [M − H]− ion at m/z 423.3273(C29H44O2), which is considered to be the deacidification and dehydrogenation product of CLG-M0.
CLG-M5 showed an [M + HCOO]− ion at m/z 1349.5990 (9.05 min, C60H102O31S), which was 68 Da (+SO3-O + 2H2) higher than the deprotonated ion of CLG-M0. Further fragment ions at 765.4425[M-H-C18H34O16S]−,603.3918[M-H-C18H34O16S-Glc]−, 585.3819[M-H-C18H34O16S-Glc-H2O]−and 471.3477[M-H-C18H34O16S-Glc-Ara]−. The metabolites showed that sulfation, hydrogenation, and deoxygenation centers were glycosyl substituents rather than the hederagenin skeleton.
CLG-M6 (9.05 min) exhibited a deprotonated ion at m/z 1271.5844 (C58H97O28P), which was 36 Da greater than that of CLG-M0. According to previous studies, it is speculated that it is obtained by phosphorylation, hydrogenation, and deoxygenation after the loss of –CH2O on the C-3 glucose group of CLG-M0. The product ions at m/z 765.4419,603.3920 and 471.3487 were also produced by the cleavage of -Rha-Glc-Glc, -Glc, and -Ara of CLG-M0, suggesting that the phosphorylation, hydrogenation, and deoxygenation sites were at the glycosyl substituents instead of the hederagenin skeleton.
CLG-M7 (20.60 min, C29H44O3) showed an [M − H]−ion at 439.3226, which was 32 Da (–CH2-H2O) less than CLG-M3, indicating that it was CLG-M3 dehydration and demethylation product.
CLG-M8 (9.75 min, 1277.6167) was considered to be the acetylation product of CLG-M0(C61H98O28).
3.2.2 Metabolites of Cauloside C
Seven metabolites (CLC-M (1–7)) of Cauloside C were identified (Fig. 5). Detailed information on these metabolites is provided in Table 1. CLC-M0 eluted at 11.97 min and yielded a deprotonated molecule [M − H]− ion at m/z 765.4417. In the negative ion mode, the product ions at m/z 603.3907 [M − H − Glc]− and 471.3491 [M − H − Glc − Ara]− were produced by the continuous loss of a sugar moiety from C-3 of Cauloside C,423.3285[M-H-Glc-Ara-2H-HCOOH]−.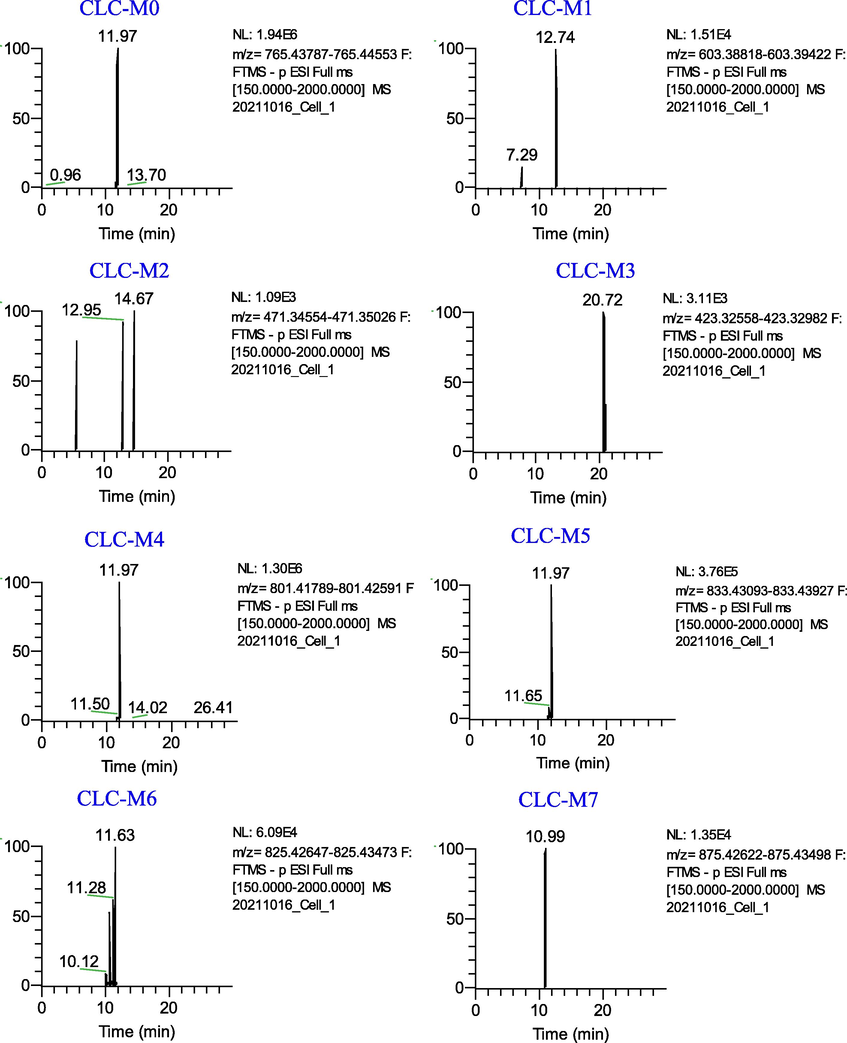
Extracted ion chromatograms of the metabolites of Cauloside C in the negative ion mode.
CLC-M1 and CLC-M2 were produced by cleavage of -Glc and -Ara at C-3 of CLC-M0, respectively. CLC-M1 showed the precursor ion [M − H]− at m/z 603.3912 (C35H56O8) and CLG-M2 (Hederagenin,14.67 min, C30H48O4) showed the [M − H]−ion at 471.3494.
CLC-M3 eluted at 20.73 min with an [M − H]−ion at m/z 423.3277(C29H44O2), which was 48 Da less than CLC-M2. The ion at m/z 423.3277 was generated by the loss of 2H and HCOOH from CLC-M2.
CLC-M4 exhibited a precursor ion [M − H]− at m/z 833.4311 (C41H70O15S), which was 68 Da higher than that of CLC-M0. Product ions at m/z 603.3883 and 471.3467 were obtained in the same manner as described above.
CLC-M5 eluted at 11.98 min with an [M − H]− ion at m/z 801.4219, which was presumed to have an elemental composition of C40H67O14P, 36 Da less than that of CLC-M0. Combined with the literature, CLC-M5 was tentatively speculated to be obtained by phosphorylation, hydrogenation, and deoxygenation of the glucose group at the C-3 position of CLC-M0 after the loss of CH2O. The major secondary fragment ions m/z 603.3930 and 471.3492 were generated by CLC-M5 through the loss of the C5H11O6P group and Ara, respectively, indicating that the phosphorylation, hydrogenation, and deoxygenation sites occurred at the glycosyl substituents of CLC-M0.
In addition, CLC-M6 (11.27 min) showed a deprotonated ion at m/z 825.4305 (C42H66O16), which was 14 Da (-O + H2) more than CLC-M0. CLC-M7 (10.98 min, 875.4306, C42H68O19) was identified as the oxidation product of CLC-M0.
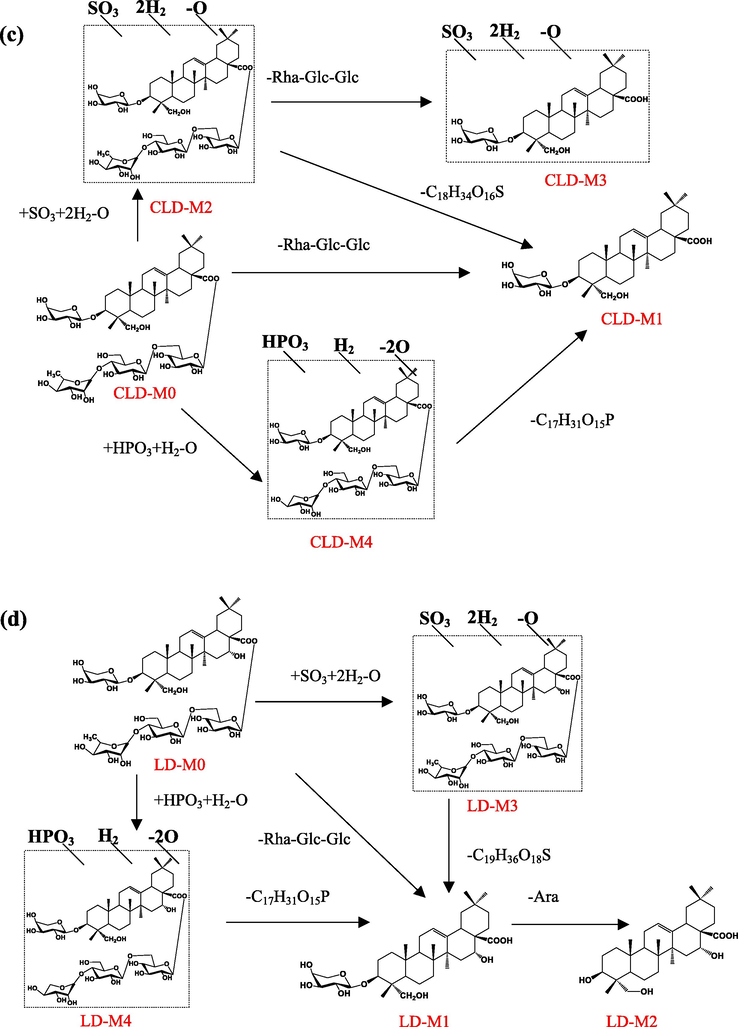
3.2.3 Metabolites of Cauloside D
Four metabolites (CLD-M (1–4)) of Cauloside D were identified (Fig. 6). Detailed information on metabolites is shown in Table 1. CLD-M0 was eluted at 9.31 min with an [M − H]− ion at m/z 1073.5487 and fragment ions at m/z 603.3922 [M − H − Rha − Glc − Glc]−.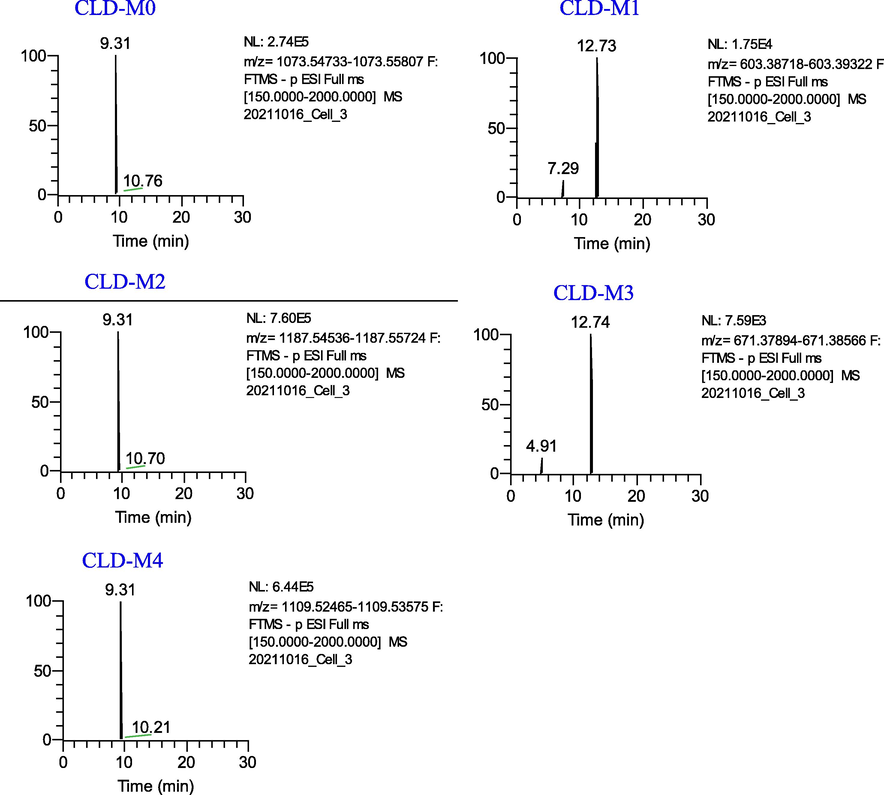
Extracted ion chromatograms of the metabolites of Cauloside D in the negative ion mode.
CLD-M1 and CLD-M3 are formed via deglycosylation. CLD-M1 (12.73 min, 603.3922, C35H56O8) was produced by the cleavage of sugar moieties at C-28 (-Rha-Glc-Glc) of CLD-M0. CLD-M3 was observed at a retention time of 12.74 min and the deprotonated molecule [M − H]− ion at m/z 671.3821, which was 469 Da (-Rha-Glc-Glc) lower than CLD-M2.
In the MS spectra, the deprotonated ion of CLD-M2 was observed at m/z 1187.5477 (C54H92O26S), which was 68 Da higher than the ions at m/z 1119.5616 (CLD-M0 + HCOO-H). Moreover, characteristic fragment ions at m/z 603.3921,671.3821 were found. Thus, it was identified as the sulfation, hydrogenation, and deoxygenation product of CLD-M0.
CLD-M4 was observed at the deprotonated molecule [M − H]− ion at m/z 1109.5322(C52H87O23P), which was 36 Da more than CLD-M0; therefore, it is speculated that it was obtained by phosphorylation, hydrogenation, and deoxygenation after the loss of –CH2 on the C-28 rhamnosyl of CLD-M0 and the further fragment ions were similar to CLD-M0, which indicated the phosphorylation, hydrogenation, and deoxygenation products of CLD-M0.
3.2.4 Metabolites of Cauloside H
Five metabolites (CLH-M(1–6)) were detected and identified in the Cauloside H group (Fig. 7). Detailed information on metabolites is described in Table 1. CLH-M0, which was characterized by an accurate [M − H]¯ at m/z 1251.604, And the MS2 spectrum showed a fragmented ion at m/z 619.3865 [M − H − Rha − Glc − Glc − Glc]¯.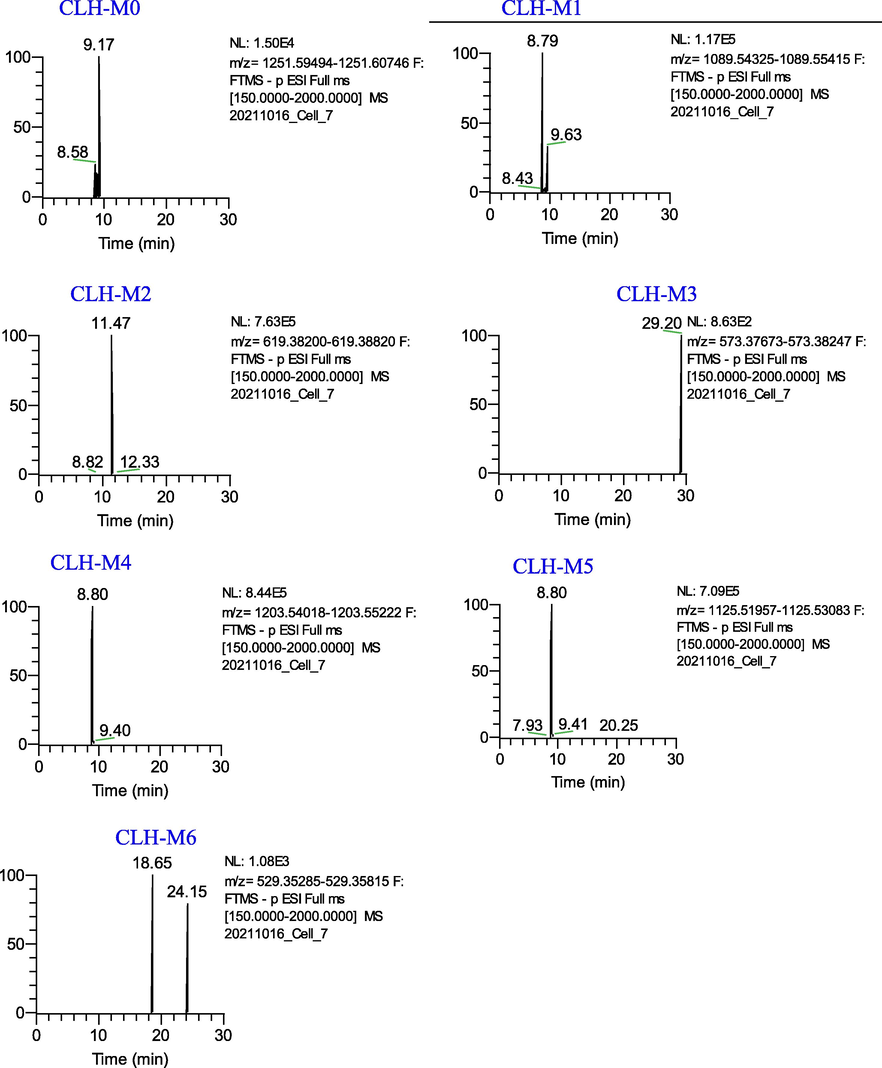
Extracted ion chromatograms of the metabolites of Cauloside H in the negative ion mode.
CLH-M1 was eluted at 8.80 min, which showed an accurate[M-H]¯ ion at m/z at 1089.5466 and [M + HCOO]¯ ion at m/z at 1135.5560, which was 162 Da (-C6H10O5) less than CLH-M0. CLH-M2 had an accurate [M − H]¯ ion at m/z 619.3865 (C35H56O9) and 469 Da (-Rha-Glc-Glc), less than CLH-M1.
CLH-M3(tR = 29.18 min) showed a protonated ion at m/z 573.3818 (C34H54O7), which was 46 Da (HCOOH) less than CLH-M2, indicating that it was tentatively identified as the deacidification product of CLH-M0.
CLH-M4 exhibited a protonated ion at m/z 1203.5427 (C54H92O27S) and a fragment ion at m/z 619.3868 [M − H − Rha − Glc − Glc − Glc]¯ and 573.3752[M-H-Rha-Glc-Glc-Glc-HCOOH]¯, which indicated sulfation, hydrogenation, and deoxygenation sites at glycosyl substituents.
CLH-M5 (8.80 min) exhibited a deprotonated ion at m/z 1125.5271 (C52H87O24P), which was 36 Da greater than that of CLH-M1. Based on the literature, CLH-M5 was tentatively speculated to be obtained by phosphorylation, hydrogenation, and deoxygenation after the loss of –CH2 on the C-28 rhamnosyl of CLH-M1 and the fragment ions at m/z 619.3866 [M − H − Rha − Glc − Glc − Glc]−. The further fragment ions were similar to CLH-M1, which indicated that the phosphorylation, hydrogenation, and deoxygenation sites occurred at the glycosyl substituents of CLH-M1.
In MS spectra, the deprotonated ion of CLH-M6 was at m/z 529.3555 (18.65 min, C32H50O6), which was 42 Da higher than the sapogenin (caulophyllogenin) of CLH-M0.
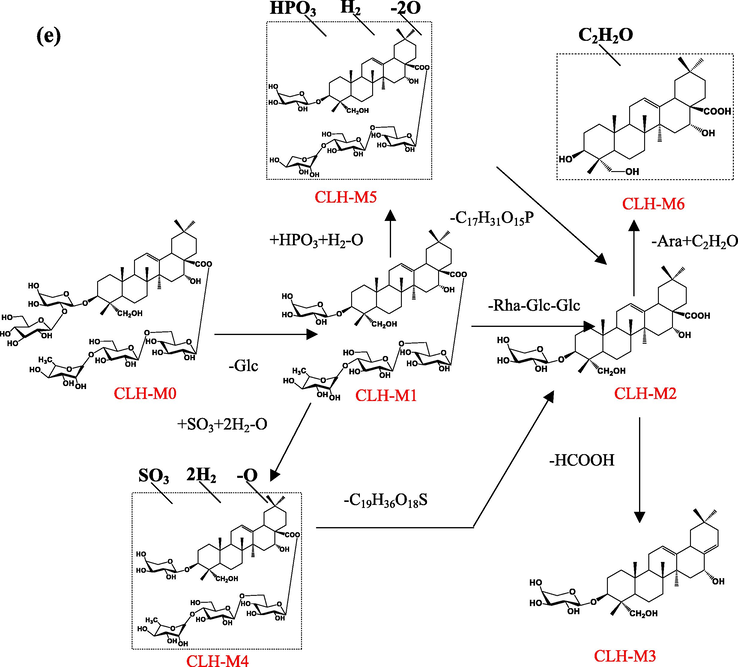
3.2.5 Metabolites of Leonticin D
A total of three metabolites (LD-M(1–4)) from the Leonticin D group were identified (Fig. 8). Detailed information on metabolites is shown in Table 1. LD-M0 was eluted at 8.77 min with [M − H]− ion at m/z 1089.5466, [M + HCOO − H]− ion at m/z 1135.5560, and fragment ions at m/z 619.3875 [M − H − Rha − Glc − Glc − Glc]−.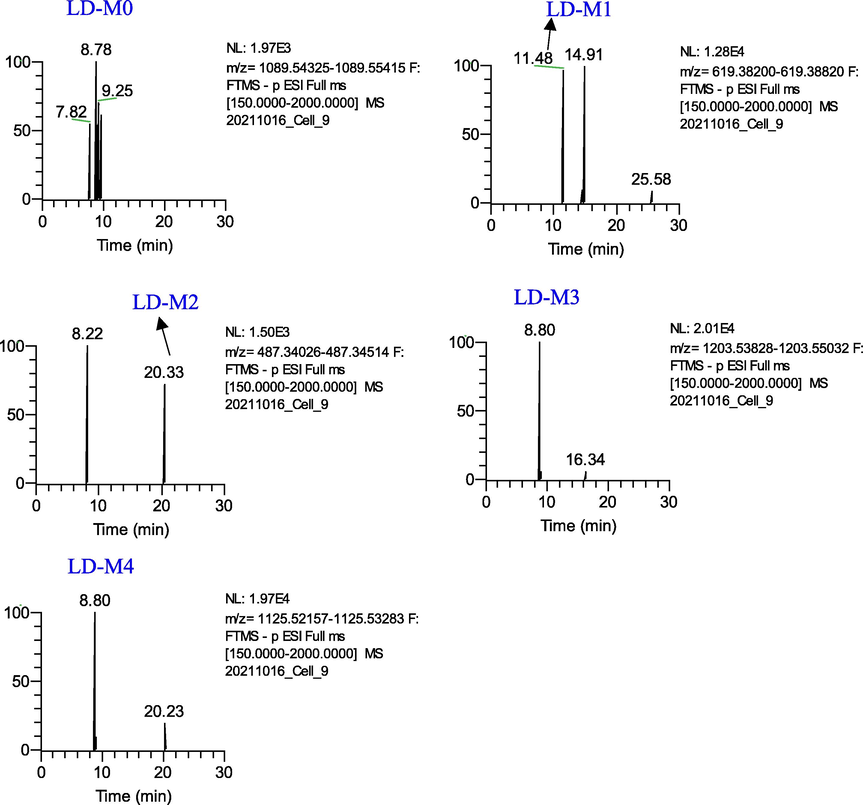
Extracted ion chromatograms of the metabolites of Leonticin D in the negative ion mode.
LD-M1(tR=11.48 min, C35H56O9), which was characterized by an accurate [M − H]¯ at m/z 619.3873, was identified as a deglycosylation product of LD-M0.
LD-M2 showed an [M − H]− ion at m/z 487.3427 (20.32 min, C30H48O5), which was 132 Da (-Ara) lower than the deprotonated ion of LD-M1. The ion at m/z 478.3427 corresponded to caulophyllogenin.
LD-M3 exhibited a protonated ion at m/z 1203.5443 (C54H92O27S), which was 68 Da (+SO3-O + 2H2) higher than the deprotonated ion of the [M + HCOO-H]− ion at m/z 1135.5560, which was tentatively considered to be the sulfation, hydrogenation, and deoxygenation products of LD-M0.
LD-M4 (8.80 min, 1125.5272, C52H87O24P) were tentatively identified as the phosphorylation, hydrogenation, and deoxygenation products of LD-M0. LD-M4 was 36 Da greater than LD-M0, and further fragment ions at m/z 603.3873 were produced. LD-M4 was tentatively speculated to have been obtained by phosphorylation, hydrogenation, and deoxygenation after the loss of –CH2 on the C-28 rhamnosyl of LD-M0.
In conclusion, by comparing the number and types of metabolites detected in FLS, we found that the metabolic pathways of the five saponin components of C. robustum were diversified, and the biological samples of the five saponins had the same metabolites. Among them, the deglycosylation products (m/z 603.3902, C35H56O8; m/z 471.3479, C30H48O4) and deacidification products (m/z 423.3268, C29H44O2) were the common metabolites of CLC and CLG. CLC, CLD, and CLG have the same diagnostic and characteristic fragment ion at 603.3902 (C35H56O8), and the same metabolite of CLH and LD is the monoglycoside (m/z 619.3851, C35H56O9). All five saponins produced phosphorylation and sulfation products (CLG-M5, M6; CLC-M4, M5; CLD-M2, M4; CLH-M4, M5; LD-M3, M4), and it is speculated that they may be metabolically active substances that exert their medicinal effects.
3.3 Analysis of metabolic pathways of five representative saponins in FLS
Traditional Chinese medicine (TCM) has the characteristics of “multi-component, multi-pathway, and multi-target” and it is difficult to explore the metabolites and metabolic pathways of TCM as a whole in cells. Therefore, studying the metabolic processes of one or more monomeric components of TCM with significant pharmacological activity in cells can reveal its metabolic transformation more accurately. Metabolites of the five representative saponins of C. robustum showed that Deglycosylation is the main metabolic pathway in FLS. Deacidification, deoxygenation, oxidation, sulfation, phosphorylation, hydrogenation, demethylation, acetylation, and de-CH2O can also be detected by UHPLC-Q-Exactive-Plus-Orbitrap-MS. In this study, combined with previous ones (Lü et al.,2019; Xia et al.,2014; Wan et al.,2013; Fu et al.,2019; Feng et al.,2018; Zhang et al.,2016), the metabolic profiles of five saponins were analyzed. The possible biotransformation of the five saponins by FLS is shown in Fig. 9a, 9b, 9c, 9d, and 9e.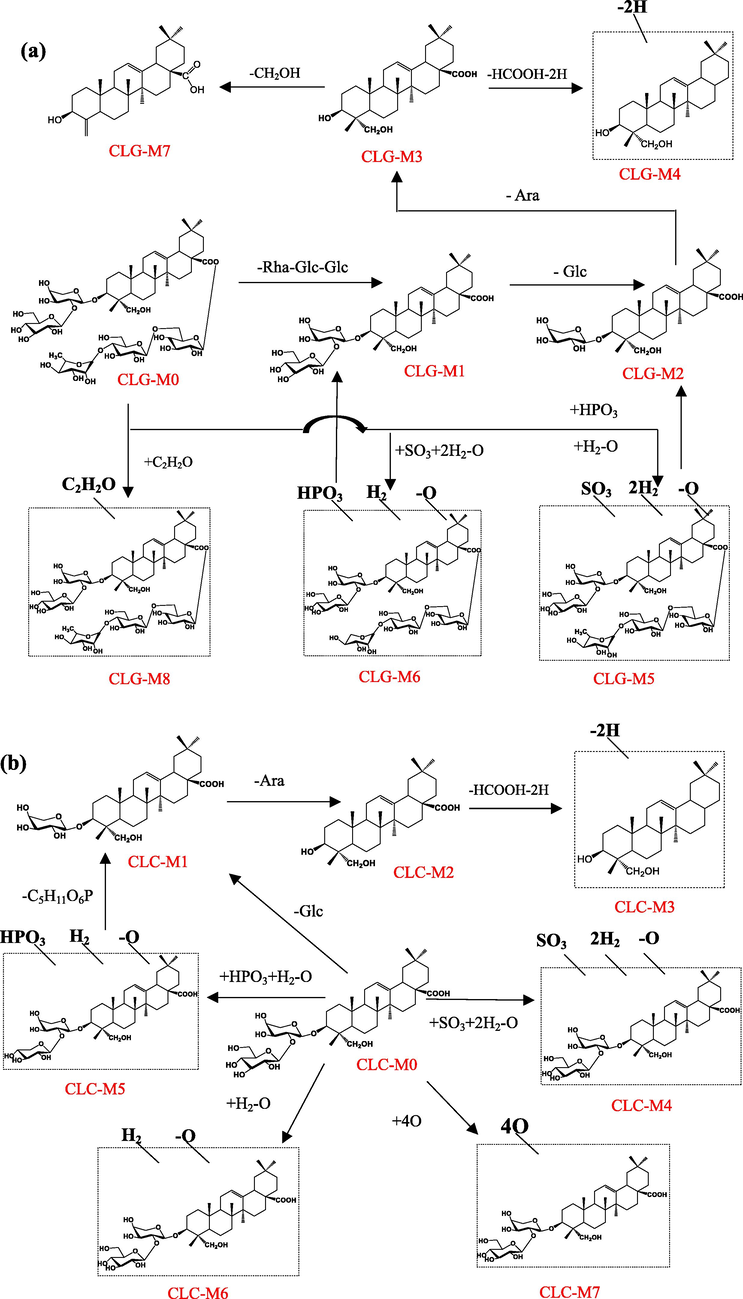
The proposed metabolic pathways of Cauloside G (a), Cauloside C (b), Cauloside D (c), Leonticin D(d), Cauloside H(e) by Fibroblast-like synoviocytes.
The proposed metabolic pathways of Cauloside G (a), Cauloside C (b), Cauloside D (c), Leonticin D(d), Cauloside H(e) by Fibroblast-like synoviocytes.
The proposed metabolic pathways of Cauloside G (a), Cauloside C (b), Cauloside D (c), Leonticin D(d), Cauloside H(e) by Fibroblast-like synoviocytes.
3.4 Comparison of five representative saponins metabolites and proposed metabolic pathways between normal and inflammatory states
RA highly expresses β-glucosidase (Zhao et al.,2021), which has catalytic activity and can catalyze the cleavage of non-reducing β-D-glycosidic bonds at the tail of compounds with sugar groups, releasing β-D-glucose or the corresponding monosaccharides, oligosaccharides, or complex sugars are released (Ketudat Cairns et al.,2015). However, whether the highly expressed enzyme affects the metabolism of saponins in FLS is also a matter of concern. In this study, β-glucosidase was added to FLS to simulate the inflammatory environment for the first time, and a comparative study of the degradation and transformation of C. robustum saponins in inflammatory and normal FLS was carried out. The results showed no significant differences in the metabolites and metabolic pathways produced in the two environments. Metabolite information of the inflammatory environment is presented in Fig.S1 and Table S1. However, the addition of β-glucosidase to FLS cannot fully mimic the real inflammatory environment. Therefore, the metabolism of C. robustum saponins in inflammatory FLS requires further study.
4 Discussion
Saponins are one of the most active and fastest-growing areas of traditional Chinese medicine research, with various important biological activities and extensive pharmacological effects. Although it shows biological effects such as cytotoxicity or changes in the cell membrane secretion process, activation/inhibition of ion channels, etc. (Aminin et al.,1999), the molecular mechanism of saponin metabolism and transformation in cells has rarely been reported. In recent years, the continuous development of microphysics and LC/MS technology has made it possible to study saponin absorption and degradation processes in a microscopic environment (Shen et al.,2019). The study found that the possible metabolic pathways of saponins in FLS are mainly the phase I reactions of deglycosylation, oxidation, demethylation, deoxygenation, and deacidification, and the phase II reactions of sulfation, phosphorylation, and acetylation. Among these, deglycosylation is the most important metabolic pathway. It has been reported that saponins are usually poorly bioavailable, and the deglycosylated metabolites formed by bacterial transformation in the gut have more potent biological activity (Lee et al.,2006; Wang et al.,2015; Tao et al.,2015).
In the deglycosylation reaction, we found that saponins with different sugar chain structures of the same type can be converted into each other in the cell, mainly as macromolecules CLH and CLG, which can be converted into LD and CLC, respectively, after removing the 162 Da glucose group. In addition, compared to the metabolites of total saponins of C.robustum in vivo (Lü et al.,2019), acetylated metabolites (CLG-M8), phosphorylated metabolites (CLG-M7, CLC-M7, CLD-M4, CLH-M6), oxidative metabolites (CLC-M9), and deacidified metabolites (CLG-M5, CLC-M4, CLH-M4) of C. robustum saponins were first discovered. Among them, the secondary fragmentation ions of the phosphorylated metabolites were consistent with the fragmentation information of the original compound, indicating that phosphorylation reactions may have occurred in the glycosyl chain. Liu et al. found that phosphorylation was present during the in vivo transformation of RT5 and PF11 with glycosyl chains, whereas ocotillol without glycosyl chains did not produce phosphorylation products (Liu et al.,2020). Usually, the binding reaction of the drug (phase II) converts it into a metabolite with increased polarity and inactivity, so that the drug can be excreted from the body. Among them, glucuronide binding, sulfation, and acetylation reactions are the most common. However, metabolites of the glucuronidation product of andrographolide have been reported to exhibit significant anti-inflammatory activity (Wang et al.,2023). In our study, all five saponins produced phosphorylation products, which could be potential active substances that deserve further investigation. In conclusion, the results showed that saponins of C. robustum might be transformed into various metabolites at the cellular level, which further clarified the pathway of action of C. robustum saponins.
The effect of the structure on the degradation of saponins in FLS is discussed. Generally, saponins with similar chemical structures are affected by slight differences in the number and position of the sugar chains, resulting in different cellular physiological responses. Therefore, structural differences may also affect the degradation of saponins. Liu et al. used UPLC/QTOF-MS to identify the metabolites and metabolic pathways of ocotilol, RT5, and PF11 in vivo and found that RT5 was the easiest to transform into metabolites, and ocotilol was more difficult to biotransform, mainly related to glycosyl number(Liu et al.,2020). Comparing the metabolites and biotransformation pathways of CLC, CLG, and CLD with different structures of the same type in FLS, the number of metabolites was CLG > CLC > CLD, showing that CLD was the most difficult to transform in FLS. Comparing the metabolites and biotransformation pathways of CLH and LD with different structures of the same type in FLS, the number of metabolites from CLH was greater than that of LD, showing that LD was the most difficult to transform in FLS. This result indicates that the easy or difficult degradation was related to the glycosyl at the C-3 position. The glycosyl chains of CLD and LD were shorter than those of CLC, CLG, and CLH, resulting in smaller steric hindrances when degradation occurred.
The metabolism of the five saponins into sapogenin products in the FLS was studied. It has been scientifically proven that the biological activity of most sapogenins is greater than that of the original saponins. For example, the antitumor activity of ginsenosides is in the order of aglycone > monoglycoside > diglycoside > triglycoside > tetraglycoside (Joaquín Navarro et al.,2018). Many sapogenins exhibit good anti-RA and immunosuppressive effects. Choi et al. extracted hederagenin from the stem of Lardizabalaceae to improve RA (Choi et al.,2005), and other studies found that the anti-RA-active aglycone (oleanolic acid) of Kochia scoparia fruits is better than saponin(Choi et al.,2002). However, sapogenin has poor solubility, low permeability, low absorption in vivo mediated by passive diffusion, and low bioavailability (Baky et al.,2022). It is a molecule that is difficult to enter cells. Generally, active secondary saponins or sapogenins are obtained in vivo through enzymatic catalysis, the transformation of microorganisms, and intestinal flora (Luo et al.,2020; He et al.,2019). Interestingly, we found that CLG, CLC, and LD can be degraded to sapogenin in FLS; no sapogenin product was found in the CLH cell samples, but we found an acetylated product (CLH-M6) of caulophyllogenin. Therefore, we speculate that it may have undergone biotransformation. The CLD is mainly degraded to monoglycosides. Why does CLD, the same nucleus as CLG, only degrade into monosaccharide glycosides? We suspect that the glycosyl chain at the C-3 position of CLD was shorter than that of CLG, which caused it to possess more minor steric hindrance when degradation occurred. Therefore, it is difficult to convert it to sapogenin. In summary, structural differences in saponins may affect their conversion to sapogenin products.
5 Conclusion
The metabolic behavior of saponins of Caulophyllum robustum Maxim in target cells was studied for the first time, and the metabolites and metabolic pathways were qualitatively characterized by the efficient and rapid UHPLC-Q-Exactive-Plus-Orbitrap-MS analysis method. The results showed that saponins of C. robustum can be converted into a variety of metabolites by FLS, and that deglycosylation, deacidification, deoxygenation, demethylation, oxidation, sulfation, phosphorylation, hydrogenation, and acetylation reactions might be involved in the transformation of C. robustum saponins in target cells. Notably, saponins with different sugar chain structures of the same type can be interconverted within target cells. Based on the structural information of the metabolites, structural differences may affect the degradation of saponins in cells. The results of this study are important for further clarifying the pathway of action of saponins in C. robustum.
6 Authors’ contributions
Writing, manuscript preparation, and overall conception: M.Z.; reviews and revisions: S.L.; Project administration, funding acquisition: Q.W. and H.K.; resources and analysis: Y.S., G.L., and B.Y. All authors read and approved the final manuscript.
Acknowledgments
This work was supported by the National Nature Science Foundation of China (grant number: 81373929), the State Key Creative New Drug Project of the 12th Five-Year Plan of China (2013ZX09102019), Chief Scientist of Qi-Huang Project of National Traditional Chinese Medicine Inheritance and Innovation “One Hundred Million” Talent Project (Grant Number: [2021] No. 7). Qi-Huang Scholar of National Traditional Chinese Medicine Leading Talents Support Program (Grant Number: [2018] No. 284).National Famous Old Traditional Chinese Medicine Experts Inheritance Studio Construction Program of National Administration of TCM(Grant Number: [2022] No. 75).The Seventh Batch of National Famous Old Traditional Chinese Medicine Experts Experience Heritage Construction Program of National Administration of TCM (Grant Number: [2022] No.76).Heilongjiang Touyan Innovation Team Program (Grant Number: [2019] No. 5).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Saponin-rich extract of Tribulus terrestris alleviates systemic inflammation and insulin resistance in dietary obese female rats: Impact on adipokine/hormonal disturbances. Biomed. Pharmacother.. 2022;2022(147):112639
- [CrossRef] [Google Scholar]
- The effect of pH on biological activity of plant cytotoxin cauloside C. Compar. Biochem. Physiol. Part A, Mol. Integr. Physiol.. 1999;122(1):45-51.
- [CrossRef] [Google Scholar]
- Phytochemical and biological diversity of triterpenoid saponins from family Sapotaceae: a comprehensive review. Phytochemistry. 2022;202:113345
- [CrossRef] [Google Scholar]
- Anti-rheumatoid arthritis effect of the Kochia scoparia fruits and activity comparison of momordin lc, its prosapogenin and sapogenin. Arch. Pharm. Res.. 2002;25(3):336-342.
- [CrossRef] [Google Scholar]
- Antinociceptive and anti-inflammatory effects of the saponin and sapogenins obtained from the stem of Akebia quinata. J. Med. Food. 2005;8(1):78-85.
- [CrossRef] [Google Scholar]
- Saponins regulate intestinal inflammation in colon cancer and IBD. Pharmacol. Res.. 2019;144:66-72.
- [CrossRef] [Google Scholar]
- Studies on the chemical and intestinal metabolic profiles of Polygalae Radix by using UHPLC-IT-MS n and UHPLC-Q-TOF-MS method coupled with intestinal bacteria incubation model in vitro. J. Pharm. Biomed. Anal.. 2018;148:298-306.
- [CrossRef] [Google Scholar]
- Chemical and metabolic analysis of Achyranthes bidentate saponins with intestinal microflora-mediated biotransformation by ultra-performance liquid chromatography-quadrupole time-of-flight mass spectrometry coupled with metabolism platform. J. Pharm. Biomed. Anal.. 2019;170:305-320.
- [CrossRef] [Google Scholar]
- HPLC-MS/MS method for the determination and pharmacokinetic study of six compounds against rheumatoid arthritis in rat plasma after oral administration of the extract of Caulophyllum robustum Maxim. J. Pharm. Biomed. Anal.. 2020;181:112923
- [CrossRef] [Google Scholar]
- He, Y.,Hu, Z.,Li, A.,Zhu, Z.,Yang, N.,Ying, Z.,He, J.,Wang, C.,Yin, S., & Cheng, S. (2019). Recent Advances in Biotransformation of Saponins. Molecules (Basel, Switzerland), 24(13), null. https://doi.org/10.3390/molecules24132365.
- Joaquín Navarro del Hierro,Teresa Herrera,Tiziana Fornari...Diana Martin.(2018).The gastrointestinal behavior of saponins and its significance for their bioavailability and bioactivities. Journal of Functional Foods. DOI:10.1016/j.jff.2017.11.032.
- β-Glucosidases: multitasking, moonlighting or simply misunderstood? Plant Sci.: Int. J. Experimental Plant Biol.. 2015;241:246-259.
- [CrossRef] [Google Scholar]
- Physicochemical characteristics and bioavailability of a novel intestinal metabolite of ginseng saponin (IH901) complexed with beta-cyclodextrin. Int. J. Pharm.. 2006;316(1–2):29-36.
- [CrossRef] [Google Scholar]
- The metabolites and biotransformation pathways in vivo after oral administration of ocotillol, RT5, and PF11. Biomed. Chromatogr.: BMC. 2020;34(8):e4856.
- [Google Scholar]
- The treatment of rheumatoid arthritis using Chinese medicinal plants: from pharmacology to potential molecular mechanisms. J. Ethnopharmacol.. 2015;176:177-206.
- [CrossRef] [Google Scholar]
- Systematic screening and characterization of prototype constituents and metabolites of triterpenoid saponins of Caulopphyllum robustum Maxim using UPLC-LTQ Orbitrap MS after oral administration in rats. J. Pharm. Biomed. Anal.. 2019;168:75-82.
- [CrossRef] [Google Scholar]
- Mechanism of Caulophyllum robustum Maxim against rheumatoid arthritis using LncRNA-mRNA chip analysis. Gene. 2020;722:144105
- [CrossRef] [Google Scholar]
- A review of saponin intervention in metabolic syndrome suggests further study on intestinal microbiota. Pharmacol. Res.. 2020;160:105088
- [CrossRef] [Google Scholar]
- The fibroblast-like synovial cell in rheumatoid arthritis: a key player in inflammation and joint destruction. Clin. Immunol. (Orlando, Fla.). 2005;115(2):118-128.
- [CrossRef] [Google Scholar]
- Synovial fibroblasts as potential drug targets in rheumatoid arthritis, where do we stand and where shall we go? Ann. Rheum. Dis. 2022
- [CrossRef] [Google Scholar]
- Saponins as cytotoxic agents: a review. Phytochem. Rev.: Proc. Phytochem. Soc. Europe. 2010;9(3):425-474.
- [CrossRef] [Google Scholar]
- Saponins and their derivatives: potential candidates to alleviate anthracycline-induced cardiotoxicity and multidrug resistance. Pharmacol. Res.. 2022;182:106352
- [CrossRef] [Google Scholar]
- Identification of a novel triterpene saponin from Panax ginseng seeds, pseudoginsenoside RT8, and its antiinflammatory activity. J. Ginseng Res.. 2020;44(1):145-153.
- [CrossRef] [Google Scholar]
- Micro-PET imaging demonstrates 3-O-β-D-glucopyranosyl platycodigenin as an effective metabolite affects permeability of cell membrane and improves dosimetry of [18F]- Phillygenin in lung tissue. Front. Pharmacol.. 2019;10:1020.
- [CrossRef] [Google Scholar]
- Biotransformation of luteoloside by a newly isolated human intestinal bacterium using UHPLC-Q-TOF/MS. J. Chromatogr. B, Anal. Technol. Biomed. Life Sci.. 2015;991:1-8.
- [CrossRef] [Google Scholar]
- Optimization of ultrasound extraction of Tribulus terrestris L. leaves saponins and their HPLC-DAD-ESI-MS n profiling, anti-inflammatory activity and mechanism in vitro and in vivo. J. Ethnopharmacol.. 2021;278:114225
- [CrossRef] [Google Scholar]
- Biotransformation and metabolic profile of American ginseng saponins with human intestinal microflora by liquid chromatography quadrupole time-of-flight mass spectrometry. J. Chromatogr. A. 2013;1286:83-92.
- [CrossRef] [Google Scholar]
- Pharmacological effect of Caulophyllum robustum on collagen-induced arthritis and regulation of nitric oxide, NF-κB, and proinflammatory cytokines in vivo and in vitro. Evid. Based Complement. Alternat. Med.. 2017;2017:8134321.
- [CrossRef] [Google Scholar]
- A comparative study of Liandan Xiaoyan Formula metabolic profiles in control and colitis rats by UPLC-Q-TOF-MS combined with chemometrics. J. Pharm. Biomed. Anal.. 2023;223:115115
- [CrossRef] [Google Scholar]
- Intestinal interleukin-10 mobilization as a contributor to the anti-arthritis effect of orally administered madecassoside: a unique action mode of saponincompounds with poor bioavailability. Biochem. Pharmacol.. 2015;94(1):30-38.
- [CrossRef] [Google Scholar]
- Fibroblast-like synoviocytes in rheumatoid arthritis: surface markers and phenotypes. Int. Immunopharmacol.. 2021;93:107392
- [CrossRef] [Google Scholar]
- A strategy for characterization of triterpene saponins in Caulophyllum robustum hairy roots by liquid chromatography with electrospray ionization quadrupole time-of-flight mass spectrometry. J. Pharm. Biomed. Anal.. 2014;100:109-122.
- [CrossRef] [Google Scholar]
- Screening of intestinal bacterial metabolites of Platycodin D using ultra-performance liquid chromatography/quadrupole time-of-flight mass spectrometry. Am. J. Chin. Med.. 2016;44(4):817-833.
- [CrossRef] [Google Scholar]
- Acid Sphingomyelinase and acid β-Glucosidase 1 exert opposite effects on interleukin-1β-induced interleukin 6 production in rheumatoid arthritis fibroblast-like synoviocytes. Inflammation. 2021;44(4):1592-1606.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary material
The supplementary material shows the structures and mass spectra of each metabolite identified in the fibroblast-like synoviocytes. Metabolite information tables and mass spectra of five saponins in simulated inflammatory environments. Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2023.104757.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary data 1
Supplementary data 1







