Translate this page into:
Identification of antidiabetic constituents in Polygonatum odoratum (Mill.) Druce by UPLC-Orbitrap-MS, network pharmacology and molecular docking
⁎Corresponding authors. wzhang@must.edu.mo (Wei Zhang), wangliang@gdph.org.cn (Liang Wang)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Abstract
Advanced UPLC-Orbitrap-MS method was developed to profile the composition of the methanol extract of P. odoratum. Core components of P. odoratum and key T2DM targets were identified through network pharmacology. Molecular docking was conducted to verity the genuine interactions between P. odoratum components and T2DM targets.
Abstract
Polygonatum odoratum has been known as having therapeutic effects on diabetes. However, due to its complex composition, it is difficult to elucidate the molecular mechanisms of its functions. In this study, we investigated the material basis and molecular mechanisms underlying the antidiabetic activities of methanol extract of P. odoratum (Mill.) Druce by ultra-performance liquid chromatography orbitrap mass spectrometry (UPLC-Orbitrap-MS) and network pharmacology. In specificity, we first constructed a compound-protein interaction network for type 2 diabetes mellitus (T2DM) to identify potential drug targets. Disease Ontology (DO), Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were then conducted to discover the pathways involved in the antidiabetic activities of P. odoratum. Among the 334 compounds detected in P. odoratum, 277 active constituents and 897 corresponding targets were identified to be associated with antidiabetic activities. Five compounds and five targets were consequently obtained by analyzing the compound-target-pathway network. The five compounds were 9-Aminocamptothecin, 9-Methoxycamptothecin, 5-Hydroxy-1-tetralone, 5,7,3′-Trihydroxy-6,4′,5′-trimethoxyflavone, and Diacerein, while the five targets were ACTB, JUN, STAT3, HIF1A, and MMP9. In addition, 30 antidiabetic-related pathways were recognized by using KEGG pathway analysis. Moreover, molecular docking was performed with the five essential compounds and the top five targets based on degree ranking. Our study confirms that active components in P. odoratum can exert antidiabetic activities via multi-component, multi-target, and multi-pathway mechanisms of action, which serves as a practical guidance for further experimental investigations into the biological functions of the methanol extract of P. odoratum in the treatment of T2DM.
Keywords
T2DM
Polygonatum odoratum
Network pharmacology
Mass spectrometry
Molecular docking
1 Introduction
Type 2 diabetes mellitus (T2DM) is one of the most common types of chronic disease (Taggart et al., 2018). It is often accompanied by metabolic syndromes and multisystem complications, leading to the malfunction of multiple organs, and causing severe diseases such as nephropathy, retinopathy, and neuropathy (Stefanidis et al., 2018; [1]; Kasznicki et al., 2012). T2DM can also affect mental health and life quality, which imposes tremendous economic and public health burdens on society. Recently, the 10th edition of the International Diabetes Federation (IDF) Diabetes Atlas reports that more than 1 in 10 adults now have diabetes globally in 2021, and the number of people with diabetes will continue to expand rapidly (Sun et al., 2022). Therefore, it is necessary to identify effective therapeutic methods and medicinal drugs to treat this chronic and refractory disease. Traditional Chinese medicine (TCM), based on the principles of syndrome differentiation and holistic treatment, has a long history of treating diabetes and its complications. Studies have described TCM as effective method in the treatment of T2DM, which is vastly different from the forced hypoglycemic and lipid-lowering effects of drugs commonly used in clinic, thereby providing a therapeutic strategy with all-round, multi-faceted, and multi-targeted advantages (Xiao and Luo, 2018).
Polygonatum odoratum (Mill.) Druce is a representative of the Liliaceae family, which is a perennial herbaceous plant widely distributed in East Asia and Europe (Zhang et al., 2020). In China, P. odoratum is known as Yu Zhu and has a long history as a traditional Chinese medicine (Wujisguleng et al., 2012). So far, various bioactive pharmacological compounds have been isolated from the plant, such as polysaccharides, saponins, glycosides, flavonoids, and alkaloids, etc. (Wang et al., 2018; Quan et al., 2015; Ngamsurach and Praipipat, 2021). Previous studies showed that P. odoratum possessed many pharmacological effects (Wang et al., 2013; Zhao et al., 2019; Chansiw et al., 2018; Jiang et al., 2018). For example, the aqueous extract of P. odoratum could prevent hyperglycemia and hypertriglyceridemia, and alleviate the symptoms of diabetes (Jiang et al., 2018). The neutral polysaccharides named POP-1 were purified from P. odoratum might affect the immunoregulatory activity (Zhao et al., 2019). The methanol extract of P. odoratum exhibits anti-hemolytic, antibacterial and anti-cancer activities, e.g., inhibition of Gram-positive bacteria growth and treatment of HL-60 cancer cell by causing decreased viability, increased G1-phase accumulation, and apoptosis induction (Chansiw et al., 2018).
Due to the compositional complexity of P. odoratum, traditional analytic methods are limited to dissecting the functions of components in P. odoratum. Ultra-performance liquid chromatography orbitrap mass spectrometry (UPLC-Orbitrap-MS) is a new analytical tool with good resolution, excellent sensitivity, and structural solid characterization capability (Xin et al., 2018). Moreover, it can provide a good separation effect and enable rapid and efficient analysis of complex components (Xin et al., 2018). Therefore, UPLC-Orbitrap-MS is an efficient and advanced tool for the compositional analysis of TCMs. For example, Wang et al. established a rapid and precise method based on UPLC-Orbitrap-MS/MS in both positive- and negative-ion modes to rapidly identify various chemical components in the tubers of Gymnadenia conopsea for the first time, which identified a total of 91 compounds based on the accurate mass within the error of 3 ppm (Wang et al., 2020). Therefore, we applied this advanced method for the compositional analysis of methanol extract of P. odoratum in this study.
The concept of network pharmacology was first proposed by Andrew Hopkins in 2007 (Hopkins, 2007). As a systematic analysis method, network pharmacology is efficient in studying disease and drug action mechanisms in the context of significant biological networks, which provides a comprehensive approach for analyzing complex drug mechanisms of action and potential disease interventions by identifying core targets common to drugs and diseases (Li et al., 2022). Through network pharmacology, we can not only explore the complex active molecular components and potential molecular targets in TCM compounds, but also understand the relationships between different components in the compound, and the molecular mechanism between these components and complex diseases. In recent years, network pharmacology has been widely used in elucidating the functions of TCMs for cardiovascular diseases, cancers, arthritis, diabetes, and other diseases due to its systematic and holistic advantages (Yu et al., 2019; Taha et al., 2020; Xie et al., 2021; Qin et al., 2019). In addition, the validation of the results of the network pharmacology method is critical (Jiao et al., 2021). Molecular docking, as a computer-aided drug design technology, can simulate the geometric structure and intermolecular interaction force of molecules through the stoichiometric calculation method to find the best binding mode of small molecule drugs and macromolecules of known structure. Therefore, the combination of network pharmacology and molecular docking can efficiently dissect the interactions between chemical substances and disease targets, providing a high-throughput approach to understanding the working mechanisms of TCMs ({Herrington et al., 2008).
So far, the chemical composition of the methanol extract of P. odoratum is still not clear and its functional mechanisms in the treatment of T2DM are still unclear. Moreover, the interactions between compounds of P. odoratum methanol extract and corresponding disease targets in T2DM have not been elucidated yet, though P. odoratum has been reported to have positive effects in T2DM treatment (Jiang et al., 2018). Therefore, an in-depth investigation into the molecular effects of methanol extract of P. odoratum on T2DM is in time and important. In this study, we first used UPLC-Orbitrap-MS to elucidate the chemical compositions of the ethanol extract of P. odoratum, based on which the T2DM targets were then collected via drug database analysis. The interactive network among P. odoratum components and T2DM disease targets were then constructed, while protein–protein interactions (PPI) among T2DM disease targets were also analyzed in order to identify core disease targets. Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were performed to study the biological function and metabolic pathways in terms of the therapeutical effects of P. odoratum. Finally, molecular docking was conducted to verify the binding abilities of the core active components in the methanol extract of P. odoratum to core disease targets for T2DM. Taken together, this study provided a comprehensively theoretical understanding in the molecular mechanisms of methanol extract of P. odoratum for the effective treatment of T2DM, which paved the way for subsequently experimental verifications and potential applications of P. odoratum in real-world settings.
2 Methods and materials
2.1 Sample preparation
Dried P. odoratum rhizomes were purchased from Beijing Tong Ren Tang Co., Ltd (Lot Number: 20210802). 2.0 g of P. odoratum rhizome powder was extracted by ultrasonic with methanol (1:25, w/v) for 30 min. The extraction solution was forced to pass through 0.22 μm filter membrane (Millipore, Merck). The filtered methanol extraction solution was ready for injection.
2.2 UPLC-Orbitrap-MS analysis of methanol extract of P. Odoratum
UPLC-Orbitrap-MS analysis was conducted on the methanol extract of P. odoratum via the Thermo Fisher Scientific UPLC system (Dionex UltiMate 3000) coupled with an LTQ Orbitrap Velos mass spectrometer. XCalibur software (version 2.0.7) was used for instrument control, data acquisition, and data analysis. The chromatographic separation was performed on an Agilent ZORBAX SB C18 column (2.1 mm × 150 mm, 3.5 μm). 0.1 % formic acid in water (solvent A) and methanol (solvent B) as mobile phase were used at a flow rate of 0.3 mL/min. The gradient program was set as follows: 0–5 min, 5–20% B; 5–15 min, 20–45% B; 15–25 min, 45–80% B; 20–30 min, 80–100% B; equilibration time of 5 min at 5% B. A total of running time was 35 min. The column temperature was set at 35 °C, and the injection volume was 10 μL. The MS data were acquired using electrospray ionization (ESI) in the negative ionization modes, spray voltage, 4 kV (-4 kV in ESI-); sheath gas (N2, >95%), 35 bar; auxiliary gas (N2, >95%), 15 bar; heater temperature, 275 °C; capillary temperature, 275 °C. MS Scanning mode: full MS scan ranging from 50 m/z to 1500 m/z and the resolution was 35,000.
2.3 Disease targets of the compounds in methanol extract of P. odoratum
Traditional Chinese Medicine Systems Pharmacology Database (TCMSP, https://tcmspw.com/tcmsp.php) was initially searched by using chemical names, InChIKeys, and CAS numbers of all the compounds identified in the methanol extract of P. odoratum via UPLC-Orbitrap-MS (Ru et al., 2014). A total of 183 components were identified; which corresponded to 313 targets. For the compound that cannot be found in TCMSP, we queried the Simplified Molecular Input Line Entry System (SMILES) numbers of the corresponding components in the PubChem database https://pubchem.ncbi.nlm.nih.gov/ and import them into Swiss Target Prediction https://swisstargetprediction.ch/ platform with screening threshold > 0.1. A total of 94 components were obtained, which corresponded to 673 targets. Taken together, we obtained 277 chemical components and 897 drug targets. The names of drug targets were then converted into gene symbols through the UniProt database https://www.uniprot.org/ (Consortium, 2020).
2.4 Disease targets of T2DM
To obtain the target genes related to diabetes among all identified targets, we used “type 2 diabetes mellitus” as the keyword to obtain 15,776 gene targets related to T2DM from DrugBank https://go.drugbank.com/, Online Mendelian Inheritance in Man (OMIM) https://www.omim.org/ and Gene Cards https://www.genecards.org databases (Hamosh et al., 2005, Safran et al., 2021, Wishart et al., 2018). All obtained targets were validated and calibrated against the UniProt database (Consortium, 2020). In order to obtain T2DM-related drug targets in terms of P. odoratum methanol extract, we used the online analysis tool Venny (https://bioinfogp.cnb.csic.es/tools/venny/) to get the intersection of identified targets of P. odoratum and T2DM. A total of 703 common targets were collected in this study.
2.5 Construction of the compounds-targets network
703 targets for T2DM and the corresponding 277 components in the methanol extract of P. odoratum were inputted into CytoScape (version 3.7.1) for the construction of the “component-target” network (Killcoyne et al., 2009). In order to identify core bioactive components, we used the “Analyze Network” function in CytoScape to perform a topology analysis of the network and calculate the degree value of each node in the network (Chin et al., 2014). Based on the degree numbers, the top five core active ingredients were selected. To further demonstrate the key “ingredient-target” network, only T2DM targets associated with the top five active ingredients were kept drawing a sub-network of the interaction between the core active ingredients and disease targets.
2.6 Construction and analysis of protein–protein interaction network
In order to understand the relationships among T2DM targets, we imported the targets corresponding to the active components of methanol extract of P. odoratum into the STRING database https://cn.string-db.org/ in order to obtain the protein–protein interaction (PPI) network. “Multiple proteins” were selected for the specific input method and “Homo sapiens” for species to remove the independent nodes in the network. TSV files were downloaded in the analysis results, which were then imported into CytoScape (version 3.7.1) for data visualization and network analysis. The Maximal clique centrality (MCC) algorithm in the Cytohubba plugin was used as the screening tool for the core target (Chin et al., 2014). All parameter settings were in default, and the nodes with the top five scores were selected as the hub nodes of the network. The first neighbors of each hub node in the network were grouped into one category. In the PPI network, nodes represented proteins and edges represented interactions between proteins.
2.7 Functional classification of target proteins in T2DM
In addition to studying interactions among targets in T2DM, we also used the online classification system PANTHER https://pantherdb.org/to classify T2DM-related proteins for their functions (Mi et al., 2021). First of all, we imported the proteins into PANTHER, selected “ID List” for List Type, “Homo sapiens” for Organism, and “Functional classification in gene list” for analysis mode to display protein classification in pie chart. Based on the number of proteins (genes) covered, the top five protein categories were selected and the list information was downloaded for each protein category. In order to visualize the relationship between the active components of P. odoratum and protein classification, we used the online drawing platform HIGHCHARTS (https://www.highcharts.com.cn/) to draw a Sankey diagram so as to show the relationship between the five core active components of P. odoratum and the top five protein classes (PC).
2.8 Enrichment analysis
Disease Ontology (DO) function in the DOSE package (version 3.22.0) was used to enrich disease-related target proteins into pathways (Yu et al., 2014). In order to screen the enriched pathways with statistical difference, the specific parameters of DO were set to ont = “DO”, P-value Cutoff = 1 and Q-value Cutoff = 1. According to the size of Gene Ratio, a bubble chart was drawn for the result. For Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses, UniProtKB IDs obtained from the UniProt database for all the target proteins were imported into the database DAVID https://david.ncifcrf.gov/summary.jsp with the identifier selected as “UNIPROT_ACCESSION” to perform GO and KEGG enrichment analysis, the results of which were downloaded for further analysis (Huang da et al., 2009; Sherman et al., 2022). For the GO enrichment analysis based on the number of differentially expressed genes, the top 10 GO terms based on P-values in Biological Process (BP), Molecular Function (MF), and Cellular Component (CC) were selected to draw a bubble chart. For KEGG pathway enrichment analysis, the top 30 terms with based on P-values were selected to draw a bubble chart. The smaller the P-value, the more significant the enrichment. Redder nodes and larger node sizes in bubble chart indicated that the pathways were more important in the entire network.
2.9 Molecular docking
Molecular docking is a convenient method for screening the genuine interactions between active ingredients and target proteins, which also facilitated the understanding of the binding mechanism (Morris and Lim-Wilby, 2008). In this study, we conducted molecular docking to test whether the core active components in the ethanol extract of P. odoratum identified by network pharmacology were genuinely combined with the core target proteins. First of all, we downloaded the 3D Conformer small molecule files in sdf format corresponding to the core components from the PubChem (https://pubchem.ncbi.nlm.nih.gov) database and used Open Babel (version 3.1.1) software to convert sdf files into mol2 file. For the 3D structures of top 5 target proteins, their pdb files were sourced from the RCSB https://www.rcsb.org database, that is, actin Cytoplasmic 1 (ACTB) (PDB ID: 2OAN), transcription factor activator protein AP-1 (JUN) (PDB ID: 5VPC), signal transducer and activator of transcription 3 (STAT3) (PDB ID: 6NJS), hypoxia-inducible factor 1 alpha (HIF1A) (PDB ID: 1LQB), and matrix metalloproteinase 9 (MMP9) (PDB ID: 5UE3). PyMol (version 2.4.0) software was used to remove water molecules and redundant ligands for pre-docking of small molecule components to target proteins. AutoDock4 (version 1.5.7) was then used to hydrogenate proteins that were set as acceptors, the results of which were generated as pdbqt files. Small molecules were also conducted for hydrogenation operation, and torsion trees were automatically configured in the software, saving as a ligand file in pdbqt format. The pdbqt structure files of receptor and ligand were imported into AutoDock4 to define the molecular docking range. The target protein was taken as the center of the grid, while the parameters of the center coordinates (center X/Y/Z) and box size (size X/Y/Z) were adjusted to ensure that the protein macromolecules were completely covered by the docking box before molecular docking was conducted.
In AutoDock4, molecular docking was accomplished by detecting protein macromolecules and inserting tiny drug molecules, as well as configuring operating methods and docking parameters. The pdbqt format was used to compute the minimal binding energy. We converted the pdbqt file to PDB format to demonstrate hydrogen bonds between ligands and proteins, calculated the distance between the protein residue stick structure and the hydrogen bond. The final generated receptor and ligand complex PDB files were imported into LigPlot++ (version 2.2.5) software to visualize and analyze the two-dimensional interaction diagram of ligands and target proteins. In addition, we used the “pheatmap” package to draw a heat map of docking binding energy to visualize the binding energy between different receptors and ligands, and used the binding energy of these active components and target proteins as the evaluation criteria for the possibility of molecular docking. The smaller the binding energy, the greater the possibility of docking. In this study, the binding energy threshold was set to −4 kcal/mol. If the docking binding energy was less than −4 kcal/mol, it was considered that the binding ability between the ligand and the receptor was in good condition.
3 Results
3.1 Chemical composition
The total ion chromatogram (TIC) of UPLC-Orbitrap-MS in the negative ion mode is shown in Fig. 1. A total of 334 compounds belonging to five categories were identified, which were flavonoids, alkaloids, terpenoids, saponins, and organic acids. Supplementary Table S1 contains the retention time, molecular formula, molecular ion peak, and adduct ion of all the identified compounds. The results showed a relatively good mass spectrometry response in the negative ion mode.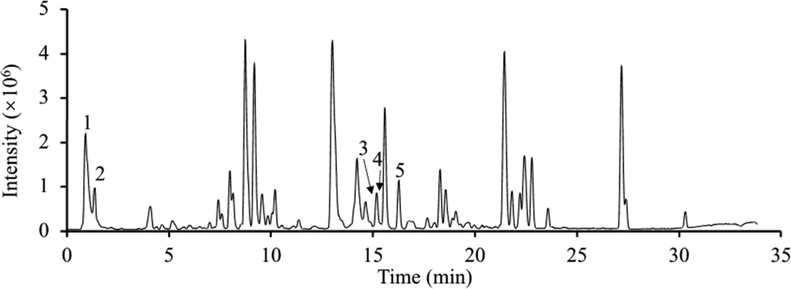
UPLC-Orbitrap-MS total ion current chromatogram of the methanol extract of P. odoratum in the negative ion mode. Representative components were labelled by numbers in the chromatogram. 1: 5,7,3′-Trihydroxy-6,4′,5′-trimethoxyflavone; 2: 9-Methoxycamptothecin; 3: 9-Aminocamptothecin; 4: Diacerein; 5: 5-Hydroxy-1-tetralone.
3.2 Analysis of compound-target network
Due to the multi-component and multi-target characteristics of TCM functions, we constructed a component-target network for the methanol extract of P. odoratum in Fig. 2. The degree value of the component was calculated in the network according to the number of targets linked to each component. According to the degree values, the top 5 components were highlighted in the center as the five hub components with unique protein targets clustered in a circle and connected individually (see Table 1), while the shared protein targets formed the outermost circles; in addition, the remaining active components formed the purple circles, and the green circles represented the remaining targets (Fig. 2A). To highlight the hub components of P. odoratum, we also plotted a sub-network of hub component-target protein interaction (Fig. 2B), in which the shared components were displayed in the central area and separated the shared nodes of adjacent hub components to show their association.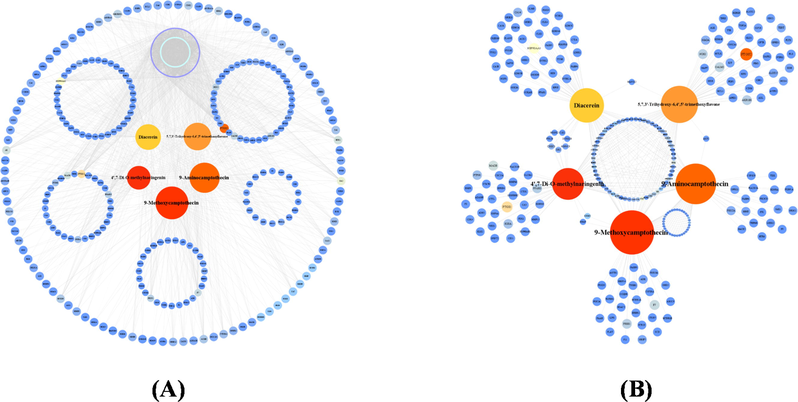
Network visualization of multi-components and multi-targets relationships for the methanol extract of P. odoratum. (A) Interactional network of active components in methanol extract of P. odoratum and T2DM disease target proteins. (B) Network of hub components and corresponding T2DM target proteins. Node sizes in the network indicated the importance of the nodes in the network.
No.
Compound Name
Chemical Structure
1
5,7,3′-Trihydroxy-6,4′,5′-trimethoxyflavone
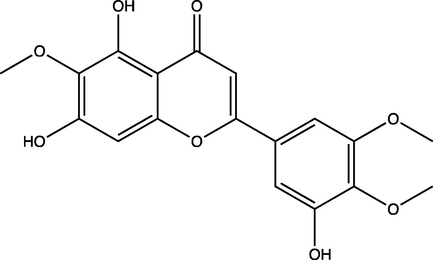
2
9-Methoxycamptothecin
3
9-Aminocamptothecin
4
Diacerein
5
5-Hydroxy-1-tetralone

3.3 PPI network construction and analysis
A PPI network was created from the obtained 703 common targets via STRING database, which was then imported into CytoScape for further visualization. The sizes represent the node degree in the network (Fig. 3A). In specificity, the PPI network contains 703 nodes and 16,494 edges, the average node degree is 46.9, and the expected number of edges is 7580, which means that these proteins have more functional and physical interactions with each other and can be biologically linked as a group. We used the size and color of the node to represent the interaction between proteins, where the larger the size and the redder the color, the more the node is associated with other nodes. In addition, Five core targets including Actin Cytoplasmic 1 (ACTB), Transcription factor AP-1 (JUN), Signal transducer and activator of transcription 3 (STAT3), Hypoxia-inducible factor 1 alpha (HIF1A), and Matrix metalloproteinase 9 (MMP9) and their directly connected targets in Fig. 3B. The common targets of 5 core targets were clustered in the center, and the associated targets between two of the core targets were shown in the adjacent area, and the targets associated with a single core target were shown in the outermost layer.
PPI network of active components in methanol extract of P. odoratum and T2DM disease targets. (A) PPI network of intersection targets of active components of P. odoratum. (B) Core target network. The size of a node in the network indicates how important the node is in the network.
3.4 Classification of core target proteins
Functional classification of all the target proteins were performed and the results were shown in the pie chart in Fig. 4A, according to which the top five subclasses were Protein Modifying Enzyme (PC00260), Metabolite Interconversion Enzyme (PC00262), Transmembrane Signal Receptor (PC00197), Transporter (PC00227) and Gene-Specific Transcriptional Regulator (PC00264) (Mi et al., 2021). In order to check which protein subclasses of the potential key active components of P. odoratum played a role, we plotted the top five protein classes Sankey diagram in Fig. 4B. The associations between components and proteins were shown according to the connections among hub components-core targets-protein classes, and it could be seen in Fig. 4B that Protein Modifying Enzyme (PC00260) was associated with 80 core targets and Metabolite Interconversion Enzyme (PC00262) was associated with 56 core targets. The results indicated that these two protein subclasses were enriched with more genes (proteins) related to T2DM and might play key roles in the treatment with P. odoratum active ingredients.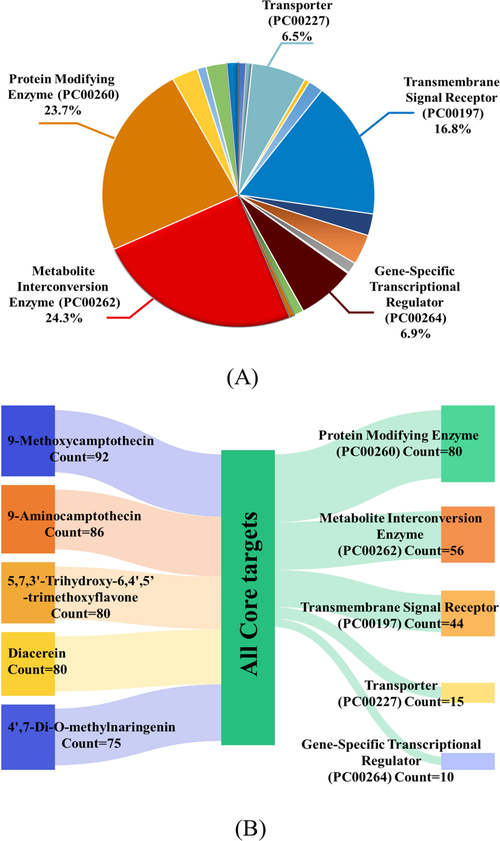
Pie chart of protein classification and Sankey diagram for top five protein classes. (A) Pie chart of PANTHER protein classes. (B) Sankey diagram of top five protein classes containing the most amount of core targets. The five hub components were shown on the left, while the top five protein classes with gene hits were displayed on the right, which were linked by all core targets of the five hub components.
3.5 Enrichment analysis
As for the enrichment analysis, a total of 703 target proteins were first analyzed via DO enrichment, according to which 706 DO terms were significantly enriched (P-value < 0.05). In the DO enrichment (Fig. 5A), the most significant disease pathways included urinary system disease, over-nutrition, kidney disease, and obesity. Furthermore, DO enrichment analysis also revealed multiple cancer-related disease signals. Since DO enrichment only suggested diseases associated with the input target proteins, we also performed GO functional enrichment and KEGG pathway enrichment analyses to further investigate the mechanisms of these core target proteins. For BP, CC, and MF, the core targets for the treatment of T2DM were analyzed using the DAVID database. The top 10 items of GO with statistical significance (P-value < 0.05) for each category were shown in Fig. 5B. The plasma membrane, an integral component of the plasma membrane, response to drugs, protein phosphorylation, protein kinase activity and enzyme binding and other metabolic aspects were most relevant to GO functional enrichment. Similarly, for the KEGG enrichment analysis, P-value < 0.05 was used as the threshold to obtain the signaling pathways in Fig. 5C. The top 30 items of KEGG results were mainly composed of signaling pathways such as pathways in cancer, neuroactive ligand-receptor interaction, lipid and atherosclerosis and age-rage signaling pathway in diabetic complications.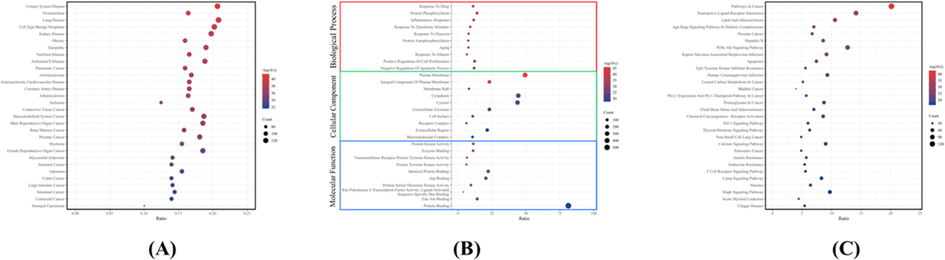
Enrichment analysis of target proteins. (A) DO disease enrichment analysis. (B) GO functional enrichment analysis. (C) KEGG pathway enrichment analysis. The abscissa ratio in the figure represented the percentage of the number of proteins enriched in each term or pathway to the total core target proteins, and the ordinate was the enriched term or pathway.
3.6 Molecular docking study
In order to validate the core components that have potential roles in the treatment of T2DM, we calculated the binding affinities of the top five bioactive components in P. odoratum and target proteins of T2DM. According to the heat map of the binding energy (Fig. 6), it could be found that the binding energies between active components and target proteins were less than 0 kcal/mol, indicating that the top 5 active components had the potential to bind to the top 5 target proteins. In particular, the binding energy between JUN and CID 5496475 was the highest at −0.44 kcal/mol, which suggested that the binding between the protein and the compound was very weak. As for other interactions, STAT3 and CID 1236175 had the binding energy of −4.12 kcal/mol, HIF1A and CID 1236175 at −4.02 kcal/mol, and MMP9 and CID 72402 at −4.56 kcal/mol. In particular, MMP9 and CID 1236175 had the smallest value of the binding energy value at −4.69 kcal/mol, indicating that a strong binding occurred between the two molecules.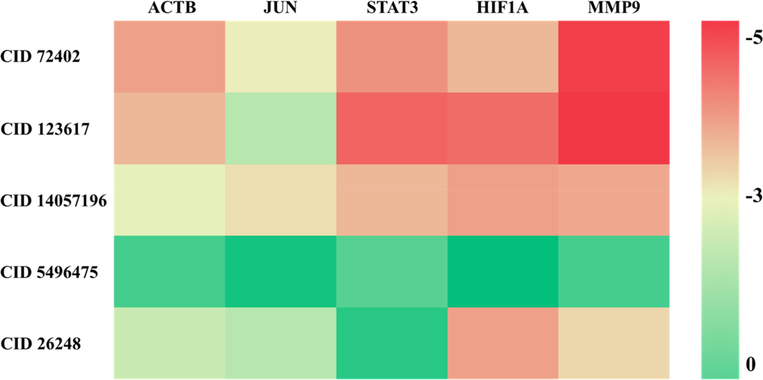
Heatmap of binding energies between potential active ingredients and core disease targets. The left side of the figure is the ID number of the active ingredient in PubChem, and the top of the figure is the core target name. The darker the color (red), the lower the binding energy and the easier the binding between the receptor and the ligand. CID 72402: 9-Aminocamptothecin, CID 123617: 9-Methoxycamptothecin, CID 14057196: 5-Hydroxy-1-tetralone, CID 5496475: 5,7,3′-Trihydroxy-6,4′,5′-trimethoxyflavone, CID 26248: Diacerein.
The optimal molecular docking diagrams of receptors and ligands were exhibited at Fig. 7. It could be seen that 9-Methoxycamptothecin formed two hydrogen bonds with GLN in STAT3 with bond lengths of 2.1 and 3.5, respectively (Fig. 7A). In addition, 9-Methoxycamptothecin formed a hydrogen bond with both PRO-496 and ILE-498 in HIF1A with bond lengths of 2.8 and 2.2, respectively (Fig. 7B). 9-Aminocamptothecin formed two hydrogen bonds with LYS-92 in MMP9 with bond lengths of 2.1 and 2.5 and formed one hydrogen bond with both ARG-95 and ASP-185 with bond lengths of 3.1 and 2.4, respectively (Fig. 7C). 9-Methoxycamptothecin formed one hydrogen bond with GLY-215 in MMP9 with a bond length of 2.0 and interacted with PHE-250 via two hydrogen bonds with bond lengths of 1.9 and 3.6 (Fig. 7D). For each receptor and ligand combination, we also mapped the protein–ligand interaction. For molecular docking diagrams between the remaining target proteins and active components, please see Supplementary Figure S1.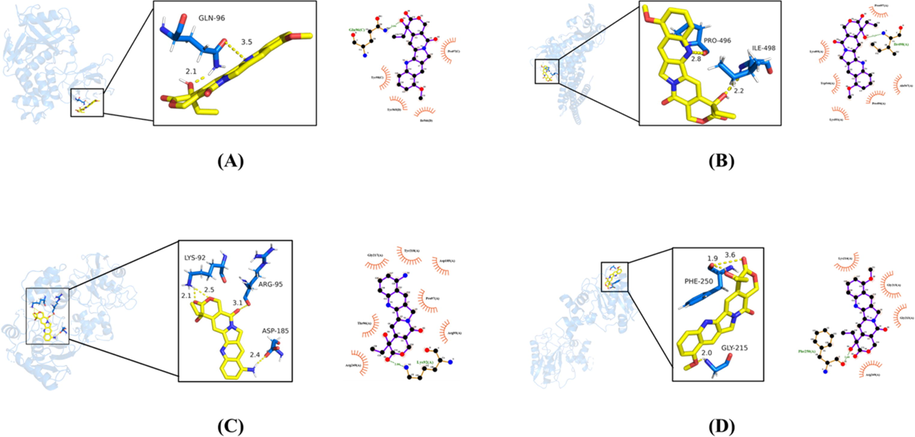
Molecular docking between active components in P. odoratum and core target proteins of T2DM disease. (A) 9-Methoxycamptothecin and STAT3. (B) 9-Methoxycamptothecin and HIF1A. (C) 9-Aminocamptothecin and MMP9. (D) 9-Methoxycamptothecin and MMP9. Yellow dotted lines indicated hydrogen bonds, while the numbers indicated bond lengths.
4 Discussion
Traditional Chinese medicine has a long history in Asian countries for treating many chronic and acute diseases, which has the advantages of being multi-component and multi-target system that is significantly distinct from western medicine (Wang et al., 2020). However, due to its complex chemical compositions, the molecular mechanisms of TCM are normally hard to explain (Zhang et al., 2019) because it is difficult to determine the specific active ingredients and disease targets. Therefore, it is necessary to combine information technology to make TCM more effective. With the development of the times, various network technologies are booming. Based on bioinformatics, systems biology, and high-throughput histology, network pharmacology that integrates pharmacology and informatics brings a new turning point for comprehending the system of TCM (Hopkins, 2007). Through the method of network pharmacology, we can directly identify active components and disease targets from a large amount of data and understand the molecular mechanisms and important pathways of TCM (Jiao et al., 2021).
P. odoratum is a well-known TCM and has been widely used for the treatment of T2DM. An earlier study showed that a steroidal glycoside from P. odoratum improved insulin resistance in 90% of pancreatectomized rats without adverse effect on insulin secretion (Choi and Park, 2014). Another study also found that aqueous extract from P. odoratum significantly reduced hyperglycemia caused by starch fed to normal and diabetic mice (Chen et al., 2001). However, up to now, there are no studies that clarified the mechanisms of action of the active substances in P. odoratum during the treatment of diabetes. Therefore, UPLC-Orbitrap-MS was employed in this study to analyze the components of P. odoratum with higher sensitivity and high resolution (Fig. 1). 334 compounds were found by comparing chemical database (Supplementary Table S1). Through searches of various public databases, potential active ingredients, disease targets, and related metabolic pathways for T2DM were identified.
In this study, we constructed a multi-layer network, which provided insights into compound-target relationships (Fig. 2). According to the results, five compounds including 9-Aminocamptothecin, 9-Methoxycampothecin, 5,7,3′-Trihydroxy-6,4′,5′-trimethoxyflavone, Diacerein, and 5-Hydroxy-1-tetralone were identified, all of which were interacted with multiple targets related with T2DM. The 5,7,3′-Trihydroxy-6,4′,5′-trimethoxyflavone was a derivation belonging to flavonoids. Flavonoids are potent antioxidants and exibit free radical scavenging abilities, leading to functions in anti-hyperglycemia, anti-inflammatory, anti-cancer and anti-hepatotoxic effects (Gul et al., 2022; Chen et al., 2019; Chen et al., 2021). For example, a recent study by Yazdiniapour et al. showed that the 5,7,3′-Trihydroxy-6,4′,5′-trimethoxyflavone isolated from Artemisia Kermanensis had apparent cytotoxic and apoptotic effects on breast cancer cell lines (Yazdiniapour et al., 2021). Another research conducted by Xia et al. proved that flavonoids (including 5,7,3′-Trihydroxy-6,4′,5′-trimethoxyflavone) from Artemisiae Argyi Folium exhibited antioxidant effect (Xia et al., 2019). Although there is no definite research on the anti-diabetic activity of 5,7,3′-Trihydroxy-6,4′,5′-trimethoxyflavone, flavonoids have been proven to have a beneficial effect on T2DM and hyperlipidemia. 9-Methoxycamptothecin and 9-Aminocamptothecin are modified by methoxyl group and amino acids at the 9-site of camptothecin, respectively. 20-(S)-Camptothecin is a natural alkaloid that was first extracted from the bark of camptotheca acuminate, which acts as a DNA topoisomerase 1 inhibitor with antitumor activities. Since the late 20th century, it has been widely used in cancer therapy (Martino et al., 2017). There were studies showing that small molecule Camptothecin was the inducer of a growth differentiation factor inducer 15 (GDF15). Therefore, in line with anorectic effect of GDF15, camptothecin suppresses food intake, thereby reducing body weight, blood glucose, and hepatic fat content in obese mice, which indicated that camptothecin could be a promising anti-obesity agent through activation of GDF15-GFRAL pathway (Lu et al., 2022). Diacerein is an anthraquinone derivative, a non-steroidal anti-inflammatory drug, which has demonstrated great efficacy with safety profile and symptomatic slow-acting treatment of osteoarthritis. Depending on its pharmacological efficacy in the mediation of anti-inflammation, antioxidation and anti-apoptosis, diacerein has been shown to have a potential role in moderating the risk of many diseases such as kidney injury, diabetes mellites, cancer, ulcerative colitis, testicular toxicity, cervical hyperkeratosis, and pain, etc. (Almezgagi et al., 2020). These studies further confirm that these essential compounds are mechanistically related to T2DM.
The results of PPIs revealed that five target genes were highly inter-connected, which were ACTB, JUN, STAT3, HIF1A, and MMP9. ACTB provides instructions for making a protein called β-actin, which is part of the actin protein family (Zhu et al., 2018). Proteins in this family are organized into a network of fibers called the actin cytoskeleton, which makes up the structural framework inside cells (Zhu et al., 2018). JUN is a transcription factor that regulates the expression of the genes encoding a number of proinflammatory mediators found in atheroma, including interleukin-1β (IL-1β) and tumor necrosis factor-α (TNF-α) (R□sen et al., 2001). STAT3 is recognized as an important survival signaling protein in β-cells, mediating the pro-survival effects of various growth factors and cytokines (García-Ocaña et al., 2001; Garcia-Ocaña et al., 2000). Enhanced STAT3 signaling protects β-cells from destruction induced by genotoxic stress (streptozotocin) and is a mechanism by which islet cell autoantigen 512/IA-2, an intrinsic tyrosine phosphatase-like protein of the secretory granules, activates a pathway for β-cell proliferation (Paula et al., 2018). HIF1A is the key regulator of oxygen homeostasis in response to hypoxia (Samanta et al., 2017). In diabetes, multiple tissues are hypoxic but adaptive responses to hypoxia are impaired due to insufficient activation of HIF signaling, which results from inhibition of HIF1A stability and function owing to hyperglycemia and elevated fatty acid levels (Catrina and Zheng, 2021). MMP9 regulates pathological remodeling processes that involve inflammation and fibrosis in cardiovascular disease and is considered important for diabetic retinopathy onset and progression (Yabluchanskiy et al., 2013). These suggest that P. odoratum can treat T2DM by modulating a complex network of multiple pathologies associated with T2DM.
As for the enrichment analysis, DO enrichment showed that urinary system disease, overnutrition, kidney disease, and obesity were the most significant disease pathways. In addition, DO enrichment analysis also focused on multiple cancer-related disease signals. Previous studies identified an association between elevated serum uric acid and components of metabolic syndrome, such as cardiovascular events in obese people (Chu et al., 2017; Reschke et al., 2015). In addition, the top 30 items of GO biological function analysis results were screened (Fig. 5B), among which the plasma membrane, an integral component of the plasma membrane, response to drugs, protein phosphorylation, protein kinase activity, enzyme binding, and other metabolic aspects were most relevant to GO functional enrichment. As for the top 30 items of KEGG, the results were mainly composed of signaling pathways such as pathways in cancer, neuroactive ligand-receptor interaction, lipid and atherosclerosis, and age-rage signaling pathways in diabetic complications. Based on the KEGG analysis results, we also found that these core targets were related to signaling pathways such as insulin resistance and endocrine resistance.
The results of molecular docking discovered the binding properties of five key compounds, that is, 9-Aminocamptothecin, 9-Methoxycampothecinm, 5,7,3′-Trihydroxy-6,4′,5′-trimethoxyflavone, Diacerein, and 5-Hydroxy-1-tetralone, to five essential proteins of ACTB, JUN, STAT3, HIF1A, and MMP9. The results showed that these proteins had the binding solid ability with small molecular compound including STAT3 and 9-Methoxycamptothecin, HIF1A and 9-Methoxycamptothecin, MMP9 and 9-Aminocamptothecin, MMP9 and 9-Methoxycamptothecin, respectively. The visualization diagram indicated that 9-Methoxycamptothecin, 9-Aminocamptothecin, START3, HIF1A, and MMP9 had a better combination effect. These results predicted that these active ingredients played an anti-T2DM role by regulating the related collaterals.
5 Conclusion
In summary, we used network pharmacology and molecular docking to identify compounds from the methanol extract of P. odoratum in ameliorating T2DM. Based on the results in docking studies, the main ingredients, 9-Aminocamptothecin, 9-Methoxycampothecin, 5,7,3′-Trihydroxy-6,4′,5′-trimethoxyflavone, Diacerein, and 5-Hydroxy-1-tetralone were expected to bind with and regulate the function of ACTB, JUN, STAT3, HIF1A, and MMP9, which could be important in the treatment of T2DM. Taken together, this study provided new insights into the complex molecular mechanisms underlying the functions of the methanol extract of P. odoratum in the treatment of T2DM, which shall be further validated via experimental investigations.
Acknowledgments
We greatly appreciate the reviewers whose comments significantly improved the quality of the manuscript. LW was financially supported by the National Natural Science Foundation of China [Grant No. 32171281] and Xuzhou Key R&D Plan Social Development Project [Grant No. KC22300]. WZ was funded by the Science and Technology Development Fund, Macau SAR [File No. 0040/2021/AGJ].
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Diacerein: recent insight into pharmacological activities and molecular pathways. Biomed. Pharmacother.. 2020;131:110594
- [Google Scholar]
- Hypoxia and hypoxia-inducible factors in diabetes and its complications. Diabetologia. 2021;64(4):709-716.
- [Google Scholar]
- Anti-hemolytic, antibacterial and anti-cancer activities of methanolic extracts from leaves and stems of Polygonum odoratum. Asian Pac. J. Trop. Biomed.. 2018;8(12)
- [Google Scholar]
- Antioxidant and anti-inflammatory properties of flavonoids from lotus plumule. Food Chem.. 2019;277:706-712.
- [Google Scholar]
- Hypoglycemic effects of aqueous extract of Rhizoma Polygonati Odorati in mice and rats. J. Ethnopharmacol.. 2001;74(3):225-229.
- [Google Scholar]
- Garcinia linii extracts exert the mediation of anti-diabetic molecular targets on anti-hyperglycemia. Biomed. Pharmacother.. 2021;134:111151
- [Google Scholar]
- cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol.. 2014;8(4):S11.
- [Google Scholar]
- cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014:8(S4).
- [Google Scholar]
- A Steroidal Glycoside fromPolygonatum odoratum(Mill.) Druce. Improves Insulin Resistance but does not Alter Insulin Secretion in 90% Pancreatectomized Rats. Biosci. Biotech. Bioch.. 2014;66(10):2036-2043.
- [Google Scholar]
- The association of uric acid calculi with obesity, prediabetes, Type 2 diabetes mellitus, and hypertension. Biomed Res. Int.. 2017;2017:1-6.
- [Google Scholar]
- UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res.. 2020;49(D1):D480-D489.
- [Google Scholar]
- Hepatocyte growth factor overexpression in the islet of transgenic mice increases beta cell proliferation, enhances islet mass, and induces mild hypoglycemia. J. Biol. Chem.. 2000;275(2):1226-1232.
- [Google Scholar]
- Transgenic overexpression of hepatocyte growth factor in the β-cell markedly improves islet function and islet transplant outcomes in mice. Diabetes. 2001;50(12):2752-2762.
- [Google Scholar]
- Alpinetin: a dietary flavonoid with diverse anticancer effects. Appl. Biochem. Biotechnol.. 2022;194(9):4220-4243.
- [Google Scholar]
- Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res.. 2005;33(Database issue):D514-D517.
- [Google Scholar]
- Molecular Docking. Molecular modeling of proteins. Methods Mol. Biol. 2008:365-382.
- [Google Scholar]
- Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc.. 2009;4(1):44-57.
- [Google Scholar]
- Physicochemical properties and antidiabetic effects of a polysaccharide obtained from Polygonatum odoratum. Int. J. Food Sci. Technol.. 2018;53(12):2810-2822.
- [Google Scholar]
- A comprehensive application: Molecular docking and network pharmacology for the prediction of bioactive constituents and elucidation of mechanisms of action in component-based Chinese medicine. Comput. Biol. Chem.. 2021;90
- [Google Scholar]
- Evaluation of oxidative stress markers in pathogenesis of diabetic neuropathy. Mol. Biol. Rep.. 2012;39(9):8669-8678.
- [Google Scholar]
- Cytoscape: a community-based framework for network modeling. protein networks and pathway analysis. Methods Mol. Biol. 2009:219-239.
- [Google Scholar]
- Network pharmacology prediction and molecular docking-based strategy to explore the potential mechanism of Huanglian Jiedu Decoction against sepsis. Comput. Biol. Med.. 2022;144
- [Google Scholar]
- Camptothecin effectively treats obesity in mice through GDF15 induction. PLoS Biol.. 2022;20(2):e3001517.
- [Google Scholar]
- The long story of camptothecin: From traditional medicine to drugs. Bioorg. Med. Chem. Lett.. 2017;27(4):701-707.
- [Google Scholar]
- PANTHER version 16: a revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res.. 2021;49(D1):D394-D403.
- [Google Scholar]
- Modified alginate beads with ethanol extraction of cratoxylum formosum and polygonum odoratum for antibacterial activities. ACS Omega. 2021;6(47):32215-32230.
- [Google Scholar]
- Exercise training protects human and rodent β cells against endoplasmic reticulum stress and apoptosis. FASEB J.. 2018;32(3):1524-1536.
- [Google Scholar]
- Systematic investigation of the mechanism of Cichorium glandulosum on type 2 diabetes mellitus accompanied with non-alcoholic fatty liver rats. Food Funct.. 2019;10(5):2450-2460.
- [Google Scholar]
- Chemical constituents from Polygonatum odoratum. Biochem. Syst. Ecol.. 2015;58:281-284.
- [Google Scholar]
- Elevated uric acid and obesity-related cardiovascular disease risk factors among hypertensive youth. Pediatr. Nephrol.. 2015;30(12):2169-2176.
- [Google Scholar]
- The role of oxidative stress in the onset and progression of diabetes and its complications: asummary of a Congress Series sponsored byUNESCO-MCBN, the American Diabetes Association and the German Diabetes Society. Diabetes Metab. Res. Rev.. 2001;17(3):189-212.
- [Google Scholar]
- TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminf.. 2014;6(1):13.
- [Google Scholar]
- DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update) Nucleic Acids Res.. 2022;50(W1):W216-W221.
- [Google Scholar]
- The contribution of genetic variants of SLC2A1 gene in T2DM and T2DM-nephropathy: association study and meta-analysis. Ren. Fail.. 2018;40(1):561-576.
- [Google Scholar]
- IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract.. 2022;183
- [Google Scholar]
- Health promotion and wellness initiatives targeting chronic disease prevention and management for adults with intellectual and developmental disabilities: recent advancements in Type 2 Diabetes. Curr. Dev. Disord. Rep.. 2018;5(3):132-142.
- [Google Scholar]
- Identifying cancer-related molecular targets of Nandina domestica Thunb. by network pharmacology-based analysis in combination with chemical profiling and molecular docking studies. J. Ethnopharmacol. 2020:249.
- [Google Scholar]
- Polygonatum odoratum Polysaccharides Modulate Gut Microbiota and Mitigate experimentally Induced Obesity in Rats. Int. J. Mol. Sci.. 2018;19(11)
- [Google Scholar]
- Antioxidant activities of different extracts and homoisoflavanones isolated from thePolygonatum odoratum. Nat. Prod. Res.. 2013;27(12):1111-1114.
- [Google Scholar]
- Distribution of traditional Chinese medicine syndromes in type 2 diabetes mellitus with chronic heart failure. Medicine. 2020;99(30)
- [Google Scholar]
- Rapid characterizaiton of chemical constituents of the tubers of gymnadenia conopsea by UPLC–Orbitrap–MS/MS Analysis. Molecules. 2020;25(4)
- [Google Scholar]
- DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res.. 2018;46(D1):D1074-D1082.
- [Google Scholar]
- Ethnobotanical review of food uses of Polygonatum (Convallariaceae) in China. Acta Soc. Bot. Pol.. 2012;81(4):239-244.
- [Google Scholar]
- Simultaneous determination of phenolic acids and flavonoids in Artemisiae Argyi Folium by HPLC-MS/MS and discovery of antioxidant ingredients based on relevance analysis. J. Pharm. Biomed. Anal.. 2019;175:112734
- [Google Scholar]
- Alternative therapies for diabetes: a comparison of western and traditional Chinese Medicine (TCM) approaches. Curr. Diabetes Rev.. 2018;14(6):487-496.
- [Google Scholar]
- Targets of hydroxychloroquine in the treatment of rheumatoid arthritis. A network pharmacology study. Joint Bone Spine. 2021;88(2)
- [Google Scholar]
- UPLC–Orbitrap–MS/MS combined with chemometrics establishes variations in chemical components in green tea from Yunnan and Hunan origins. Food Chem.. 2018;266:534-544.
- [Google Scholar]
- Matrix Metalloproteinase-9: many shades of function in cardiovascular disease. Physiology. 2013;28(6):391-403.
- [Google Scholar]
- Prevalence of diabetic retinopathy, proliferative diabetic retinopathy and non-proliferative diabetic retinopathy in Asian T2DM patients: a systematic review and Meta-analysis. Int. J. Ophthalmol. 2019
- [Google Scholar]
- Isolation and characterization of methylated flavones from Artemisia kermanensis. Adv. Biomed. Res. 2021:10.
- [Google Scholar]
- DOSE: an R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics. 2014;31(4):608-609.
- [Google Scholar]
- Uncovering the pharmacological mechanism of Carthamus tinctorius L. on cardiovascular disease by a systems pharmacology approach. Biomed. Pharmacother. 2019:117.
- [Google Scholar]
- Systems pharmacology for investigation of the mechanisms of action of traditional Chinese medicine in drug discovery. Front. Pharmacol.. 2019;10
- [Google Scholar]
- Comparative transcriptomic analysis of rhizomes, stems, and leaves of Polygonatum odoratum (Mill.) Druce reveals candidate genes associated with polysaccharide synthesis. Gene 2020:744.
- [Google Scholar]
- Purification, characterization and immunomodulatory activity of fructans from Polygonatum odoratum and P. cyrtonema. Carbohydr. Polym.. 2019;214:44-52.
- [Google Scholar]
- Diagnosis of known sarcoma fusions and novel fusion partners by targeted RNA sequencing with identification of a recurrent ACTB-FOSB fusion in pseudomyogenic hemangioendothelioma. Mod. Pathol.. 2018;32(5):609-620.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2023.105032.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary data 1
Supplementary data 1
Supplementary data 2
Supplementary data 2







