Translate this page into:
Mechanistic and kinetic aspects of Natamycin interaction with serum albumin using spectroscopic and molecular docking methods
⁎Corresponding authors. ezzatij@tbzmed.ac.ir (Jafar Ezzati Nazhad Dolatabadi), torbatim@tbzmed.ac.ir (Mohammadali Torbati)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Natamycin (NT) is a polyene natural antimycotic, which has an antimicrobial effect against yeasts and molds and is used as a preservative in the food industry. In the present study, we evaluated NT interaction with bovine serum albumin (BSA) through surface plasmon resonance (SPR) and several spectroscopic techniques, which are accompanied by a molecular docking study. According to the results, the intensity of BSA fluorescence decreased by adding different concentrations of NT. The fluorescence quenching results showed that NT reduces the intensity of BSA fluorescence by forming a complex with BSA through a hybrid quenching. Binding constant decreases from 18.73 to 2.13 (102 M−1) with increasing temperature, which indicates a decrease in complex formation owing to the interaction of NT with BSA. Negative values of ΔH° and ΔS° confirmed that van der Waals forces and hydrogen bonds are the basic forces in the interaction of NT with BSA. Moreover, increasing the equilibrium constants values with increasing temperature indicated that BSA binding to NT decreased. Finally, BSA interaction occurring with NT through Ser 109, Asp 111, Lys 114, Leu 115, Glu 424, and Arg 458 have been verified via molecular docking analysis. Attained results via SPR and fluorimetry showed that the binding constant between BSA and NT decreased when the temperature was raised.
Keywords
Natamycin
Serum albumin
Spectroscopic technique
Surface plasmon resonance
Docking simulation
1 Introduction
Food additives are synthetic or natural substances that are added to food in small amounts to prolong the shelf life of the product and increase or change its attributes, including its structure, flavor, or appearance without reducing their nutritional value (Silva and Lidon, 2016). According to the EU rules, preservatives are food additives that protect food against microorganisms (fungi and/or bacteria) (Shim et al., 2011; Silva and Lidon, 2016). Moreover, the side effects of food additives on public health have become an argued subject in recent years (Javaheri-Ghezeldizaj et al., 2020). Natamycin (NT) (pimafucin, pimaricin) is a polyene natural antimycotic that is produced in immersed culture by different strains of actinomycetes, like Streptomyces. Chattanoogans, Streptomyces. gilvosporeus, and Streptomyces. natalensis (Pintado et al., 2010; Elsayed et al., 2013). NT has been used in the food industry to preserve cheese, sausages (the surface treatment), yogurt, and in some countries juices, and wine (Dalhoff and Levy, 2015). The acceptable daily intake (ADI) for NT is defined by JECFA as 0.03 mg/kg BW/day (Martínez et al., 2013). Due to its low toxicity, NT is classified by the FDA as a safe additive and in the EU as a natural preservative (da Silva et al., 2012). NT is additionally prescribed for the topical therapy of fungal infections of the eyes, throat, skin, and vaginal in humans (Levy, 2007). Since NT is a white to creamy powder, crystalline, odorless, and colorless, it causes no taste aversion in the consumer (Brik, 1981; Stark, 1999). This antifungal substance contains a large 22-carbon lactone ring that is attached to an amino sugar, a mycosamine moiety, by a glycosidic bond. NT is a polyene macrolide antibiotic and owing to its four conjugated double bonds assessed specifically as a tetraene antibiotic. NT has a low aqueous solubility (20–50 mg/L in water) and it must be used at high concentrations (Zunszain et al., 2008; Additives and Food, 2009). Most of the food available in the market contains diverse types of preservatives. These chemicals can cause immediate adverse effects including headaches, changes in energy levels, and changes in mental focus, behavior, or immune response. Or it may cause long-term effects, including an increased risk of cancer, cardiovascular disease, and other degenerative conditions. Thus, even very few doses of these additives, when consumed continuously, can lead to an irreversible toxic effect that ultimately leads to cancer and chromosomal and embryonic damage. So in recent years, researchers have paid much attention to the interaction between chemicals, especially food additives, and various biological molecules in the body (Li et al., 2007; Inetianbor et al., 2015).
Serum albumin (SA) is the most plenty biological molecule in the circulatory system of many living organisms. It is responsible for several physiological functions such as the transfer of exogenous and endogenous compounds (greasy acids, drugs, steroids, metal particles, and metabolites) (Mohammadzadeh-Aghdash et al., 2017; Szkudlarek et al., 2019). SA can increment the solubility of hydrophobic compounds and therefore, has a critical role in the absorption, metabolism, and distribution of chemicals from cells. The interaction of these chemicals with SA changes its primary and secondary structure as well as its physiological functions. So, in recent years, researchers have paid much attention to the interaction between chemicals, especially food additives, and various biological molecules in the body. Human serum albumin (HSA) consists of 585 amino acid residues in the monomer via 17 disulfide bridges that help maintain the heart-like shape. It is the foremost abundant bearer protein within the blood plasma. HSA binds to various drugs through several hydrophobic binding sites in its structure (Koch-Weser and Sellers, 1976; Petitpas et al., 2001). Another major soluble protein is bovine serum albumin (BSA) which has 80% structural similarity to HSA (583 amino acids similar to HSA). The difference is in the number of tryptophans, in that BSA has two tryptophan amino acids, while HSA has only one tryptophan. Because of its low cost, therapeutic care, accessibility, and binding characteristics, BSA is commonly chosen as a pattern for investigating drug-protein interactions (Xu et al., 2013; Rajendiran and Thulasidhasan, 2016; Szkudlarek et al., 2019). In 2022, Khaskhashi Moghadam et al. studied the interaction behavior of cyanidin with the albumin-holo transferrin complex of human serum with a new perspective, and the results showed a moderate binding affinity between cyanidin and both proteins throughout binary and ternary systems (Khashkhashi-Moghadam et al., 2022). (Basu and Suresh Kumar, 2015) stated that the tartrazine food additive can change the chemical structure of BSA and disrupt its physiological functions (Basu and Suresh Kumar, 2015). In 2023, Zaheri et al. investigated the kinetic and thermodynamic aspects of serum albumin interaction with sodium hydrosulfite and found that sodium hydrosulfite, by forming a complex with BSA, reduces the fluorescence intensity of BSA and changes the structure of BSA (Zaheri et al., 2023). Shahabadi et al. showed that the structure of BSA changes significantly after interacting with the TBHQ (Shahabadi et al., 2011). In 2017, Asaran Darban et al. investigated the interaction between two different angiotensins I converting enzyme inhibitory peptides from gluten hydrolysate and human serum albumin and concluded that both peptides quench the fluorescence intensity of HSA through a static mechanism (Assaran Darban et al., 2017).
Given the high importance of NT consumption in the food industry, investigating its effect on the structure and function of body macromolecules such as SA can be very valuable in nutrition, food science, food chemistry, and food safety. Therefore, to understand the type of structural changes of SA and the mechanisms of SA-NT complex formation, we decided to investigate the interaction of NT with BSA using surface plasmon resonance (SPR), fluorescence and UV–Vis spectroscopies, and molecular docking analysis.
2 Material and methods
2.1 Material
N-ethyl-N-(3-dimethyl aminopropyl) carbodiimide (EDC), N-hydroxysuccinimide (NHS), phosphate-buffered saline (PBS) solution, and ethanolamine-HCl was bought from Merck (Darmstadt, Germany). Acetate buffer, NT, and BSA was bought from Sigma-Aldrich. Carboxymethyl dextran (CMD) low-density-modified gold surface chips were bought from BioNavis Company (Tampere region, Finland). Other materials utilized were of the highest quality.
2.2 Stock preparation
Phosphate buffer was prepared by dissolving a tablet of saline phosphate buffer in deionized water. The stock solution of BSA (1.4 10−5 M, according to its molecular weight of 66,000D) was dissolved in a 0.01 M phosphate buffer solution (pH 7.40). NT stock solution at a concentration of 2 mM was prepared by dissolving 1.4 mg NT in PBS buffer solution. All ready solutions were stored in the refrigerator at 4 °C.
2.3 Fluorescence experiments
Fluorescence spectroscopy evaluations were performed at five distinct temperatures with a Jasco FP-750 fluorescence spectrometer (Jasco International Co, Kyoto, Japan) equipped with a thermostat. The measurements were done in a 10 mm quartz cell. To observe the fluorescence of Trp residues, a wavelength of 290 nm was used for excitation. The slit width for excitation and emission was 5.0 nm. The scanning speed of the device was set to 8000 nm/min. 1 mM BSA solution was titrated with concentrations of 1−6 × 10−5 and its fluorescence spectrum was recorded at a wavelength of 300–450 nm. All measurements were performed with PBS buffer at 288, 293, 398, 303, and 310 K. To eliminate the inner filter effects of protein and ligands, absorbance measurements were performed at the excitation and emission wavelengths of the fluorescence measurements. The fluorescence intensity was corrected using Eq. (1):
2.4 UV–Vis spectrophotometry
To record Absorption spectra of BSA without and with different concentrations of NT at a temperature of 25 °C, a UV–Vis spectrometer model T70 + manufactured by PG Instrument Company (Leicestershire, UK) was used. Titration of 1 mM BSA solution with distinct concentrations of NT (0–70 μM) was performed in a 10 mm quartz cuvette. Finally, the solution was thoroughly blended and incubated for up to 3 min to let NT interact with BSA.
2.5 SPR measurements
To investigate the interaction of BSA with NT and calculate the kinetic parameters, an SPR device (MP-SPR navi210 A, Bionavis Ltd, Tampere, Finland) with a dual-stream channel (using Kerchman's prism configuration) and a continuous peristaltic pump was used (Sharifi et al., 2017).
2.6 Immobilization of BSA on CMD sensor chip
The CMD sensor chip was settled within the SPR instrument. Acetate buffer (10 mM) with pH 4.5 was utilized to wash the entire machine. After 15 min, the sensor diagram reached the baseline, then NaCl (2 M) and NaOH with concentration (0.1 M) were used to clear the SPR channels for 3 min at a speed of 30 μl/min. In the next step to activate the carboxyl groups on the CMD sensor chip, a stream of solution (NHS 0.05 M + EDC 0.2 M) EDC/NHS in equal ratios of 1:1 was injected into the SPR instrument at a speed of 30 microliters per min for 7 min. Then, to stabilize BSA on the CMD sensor chip, a solution of BSA at a concentration of 1 mg/ml was infused into channel 1 for 7 min and acetate buffer was infused simultaneously into channel 2. Finally, a stream of Ethanolamine-HCl solution (1 M, pH 8.5) was injected at an amount of 30 μl/min to inactivate and block the active carboxyl groups on the surface of the CMD chip (Sharifi et al., 2017; Fathi et al., 2019).
2.7 Kinetic parameters analysis of BSA interaction with NT
To calculate the kinetic parameters including association constant (Ka), dissociation constant (Kd), and equilibrium constants (KD), which are calculated via , different concentrations of NT were prepared in PBS buffer with pH 7.4 and stream of the prepared solution at a speed of 30 μl/min for 2 min at 293, 298, 303, and 310 K was injected into both flow cells. Since BSA was immobilized in only one channel, both stream cells were utilized for sample injection and the stream cell without BSA was utilized as a reference. The need for the regeneration process is eliminated due to the rapid separation of NT from the BSA level. By measuring the SPR signal, the amount of binding of NT to BSA was determined. DrawerTM for SPR NaviTM was used to determine the kinetic parameters upon BSA interaction with NT (Maleki et al., 2020).
2.8 Study of molecular docking
To understand the possible interaction of NT with BSA as a potential target biomolecule, a molecular docking study was conducted via the AutoDock 4.2 package, which is validated by The Scripps Research Institute, La Jolla, CA, USA. The 3D molecular building of NT was achieved from the PubChem website (PubChem CID:5284447) (https://pubchem.ncbi.nlm.nih.gov/), and following that, its structure was optimized for our calculation through Gaussian 03 software (B3LYP method). Next, the BSA crystal structure (PDB code: 3 V03, 2.7 Å) was downloaded from Protein Data Bank Website (https://www.rcsb.org/). AutoDock Tools-1.5.6 program was used to apply Kollman/Gasteiger charges to the BSA structure (subunit A), and also eliminate all water molecules. To find the binding domain on BSA, a blind docking calculation was performed at a lace box of 126 × 126 × 126 Å (spacing parameter of 0.523 Å), and then the lowest binding score was considered for further analysis. To focus on the binding site, another docking study was accomplished at a grid box of 70 × 70 × 70 Å and a spacing parameter of 0.380 Å. The Lamarckian genetic algorithm was applied in all calculation runs. The results were displayed and analyzed via UCSF Chimera software (version 1.14).
3 Results and discussion
3.1 Study of BSA fluorescence quenching
The fluorescence quenching analysis was used for the investigation of NT interaction with BSA. It is a useful procedure for the study of protein interaction with various molecules and the evaluation of structural changes induced in the protein on the molecular level (Wani et al., 2018). Among the different types of molecular interactions, we can mention excited-state reactions, molecular rearrangements, energy transfer, ground-state complex formation, and quenching collisions that lead to quenching (Lakowicz, 2006). In general, the innate fluorescence of BSA is inferred from Trp, Tyr, and Phe. However, among them, the contribution of tryptophan is a maximum (Wu et al., 2019; Javaheri-Ghezeldizaj et al., 2021). Innate fluorescence of BSA is regularly utilized as a test to find the affinity of BSA to various food additives due to the Trp residues presence on the surface of BSA (Trp 134) and the interior of the hydrophobic pocket (Trp 213).
The fluorescent spectrum of BSA in the lack and existence of NT in the PBS buffer (pH = 7.4) is displayed in Fig. 1 (Javaheri-Ghezeldizaj et al., 2020). BSA has an intense fluorescence emission peak of about 350 nm below the excitation wavelength of 290 nm, and after adding NT from 0 to 60 μM, a decline in fluorescence spectra was recorded without any significant change in the emission wavelength. This trend indicates that the innate fluorescence of BSA is quenching owing to interaction with NT (Javaheri-Ghezeldizaj et al., 2020). Quenching mechanisms can be identified by three factors: temperature, viscosity, and excitation state life. In the static quenching method (by complex formation), increasing the temperature reduces the stability of the base state complex, followed by a reduction of the quenching constant (KSV), and therefore the formation of a ligand–protein complex with non-fluorescent properties (independent of diffusion) (Tantimongcolwat et al., 2019). In dynamic quenching, on the other hand, the absorption spectrum is not affected by extinction because it is effective only in the excitation mode of the quencher (Taheri et al., 2022). In dynamic quenching, high temperature causes quicker dissemination steady and progresses electron transfer, and therefore, KSV tends to increase (Wani et al., 2018; Banu et al., 2021). The Stern-Volmer steady-state Eq. (2) was utilized to confirm the nature of the BSA fluorescence quenching mechanism due to the formation of the complex with NT (Javaheri-Ghezeldizaj et al., 2020).
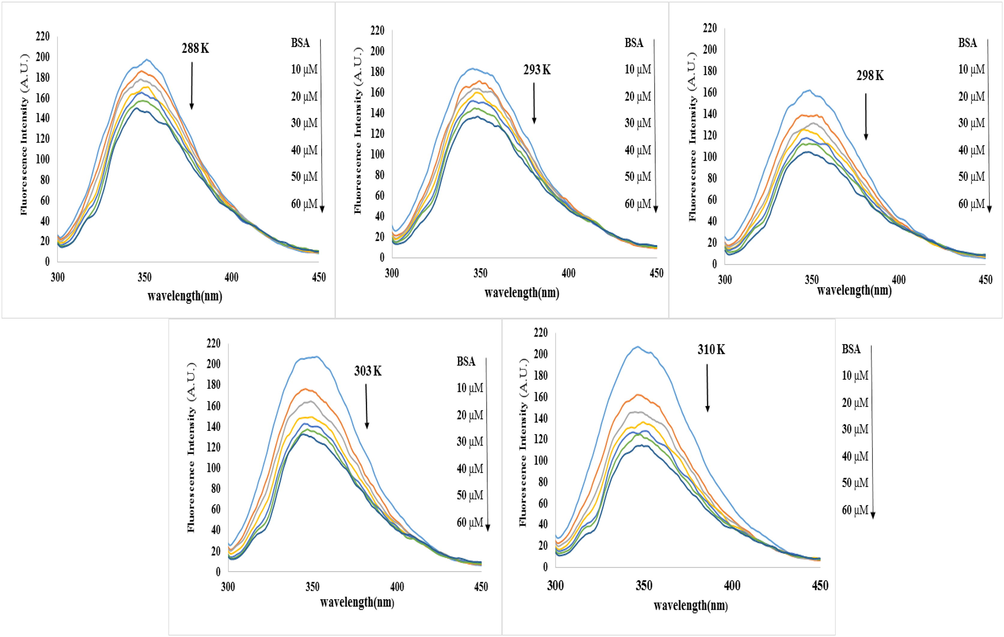
Inherent fluorescence spectrum of BSA (2 mM) in the absence and presence of various NT concentrations (0, 10, 20, 30, 40, 50 and 60 μM) at 288, 293, 298, 303, and 310 K temperature.
According to Eq. (2), F0 and F show the intensity of BSA fluorescence in the lack and existence of NT (quencher). KSV (Stern-Volmer constant), [Q] indicates the initial concentration of the quencher, kq is the quenching rate of biomolecules of a protein (BSA) and
0 is the lifespan of the exciting mood of the protein in the lack of any quencher (Maheri et al., 2022). KSV values can be attained by the slope of the F0/F plotting versus [Q]. The Ksv values are important from the viewpoint of fluorescence quenching. As revealed in Fig. 2A and Table 1, there is a powerful linear connection between F0/F and [Q] for the Stern-Volmer diagram. Static and dynamic quenching can result in a Stern-Volmer linear diagram. The difference is that in hybrid mechanisms the diagram is nonlinear and this diagram shows an upward curvature. In general, with a raising temperature, the KSV values decrease in the static quenching process but in the dynamic quenching process, temperature rising leads to a constant increase in diffusion and promotes electron transmission, so KSV tends to increase (Shi et al., 2017; Mahmoudpour et al., 2020; Javaheri-Ghezeldizaj et al., 2021). KSV and kq values are shown in Table 1. Considering the KSV values increase from 288 to 310 K (5.32 × 103 to 10.0 × 103) indicates that fluorescence quenching is more compatible with dynamic quenching instead of static quenching. However, at different temperatures, the kq values are more than the maximum collision quenching constant (2.0 × 1010 Lmol−1s−1), signifying the occurrence of static quenching. Therefore, it can be inferred that NT reduced the fluorescence of BSA via a hybrid quenching mechanism (Sandhya et al., 2011; Shi et al., 2017; Nan et al., 2019; Javaheri-Ghezeldizaj et al., 2021).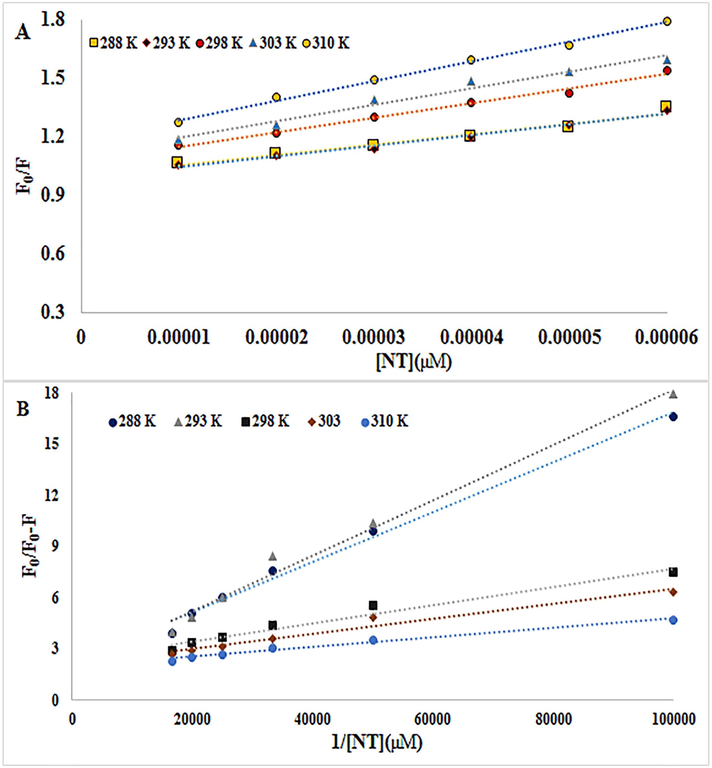
A) The Stern–Volmer diagrams of the quenching of BSA fluorescence by NT at five different temperatures B) Modified Stern-Volmer plots for quenching of BSA upon interaction with NT at five different temperatures.
T (K)
KSV (103 M−1)
Kq (1012 M−1S−1)
Ka (103m−1)
Kb (102 M−1)
ΔH°(kJ mol−1)
ΔS° (J mol−1 K−1)
ΔG°(kJ mol−1)
R2
288
2935.32
5.480.88
0.9121.696
9.65018.73
31.49
−18.73
−17.54
0.97
0.98
298
7.47
1.24
47.334
3.77
−87.16
−237.6
−16.35
0.98
303
3108.37
10.011.39
1.6651.995
65.7604.07
2.13
−15.16
−13.50
0.97
0.99
Fluorescence quenching data were additionally evaluated by a modified Stern-Volmer Eq. (3) (Ali et al., 2023):
3.2 Binding constant and number of binding locations
If small molecules such as NT interact separately with a set of equivalent sites on biomacromolecules such as BSA, the possible binding stoichiometry (n) and the binding constant (Kb) of NT-BSA can be analyzed by the slop and intercept double logarithm regression curve utilizing the following equation (Bhojwani et al., 2023):
3.3 Analysis of thermodynamic parameters
Intramolecular interaction forces consist of electrostatic bonding, hydrogen bonding, van der Waals forces, and hydrophobic interactions that play a significant function in protein binding to various ligands. Enthalpy change (ΔH°), Gibbs free energy (ΔG°), and entropy change (ΔS°) are among the thermodynamic parameters, which help to identify the type of forces influencing the interaction between two molecules. Positive values of ΔH° and ΔS° represent the presence of hydrophobic interaction between two molecules and negative values of ΔH° and ΔS° are signs of van der Waals forces or hydrogen bonds occurring. A value of ΔG° less than zero indicates that a spontaneous interaction between NT and BSA occurs at the appropriate temperature. Finally, a negative sign for Δ ° indicates that the interaction of NT with BSA is enthalpy-driven. Thermodynamic parameters can be calculated by the following van’t Hoff and Gibbs free energy Eqs. (5) and (6):
According to Eq. (5), R is the universal gas constant (8.314 J mol−1 K−1) and T describes the experiential temperature. Van't Hoff diagrams for the interaction of BSA and NT are demonstrated in Fig. 3A. The values for ΔH°, ΔG°, and ΔS° at five temperatures are given in Table 1. The results revealed that the values of ΔS° and ΔH° are −237.6 J mol−1 K−1and −87.16 kJ mol−1, respectively. Negative values of ΔH° and ΔS° confirmed that van der Waals forces, or hydrogen bonds, are the basic forces in the interaction of NT with BSA (Ross and Subramanian, 1981; Zhang et al., 2012; Wani et al., 2017; Wani et al., 2018; Wu et al., 2019; Javaheri-Ghezeldizaj et al., 2020).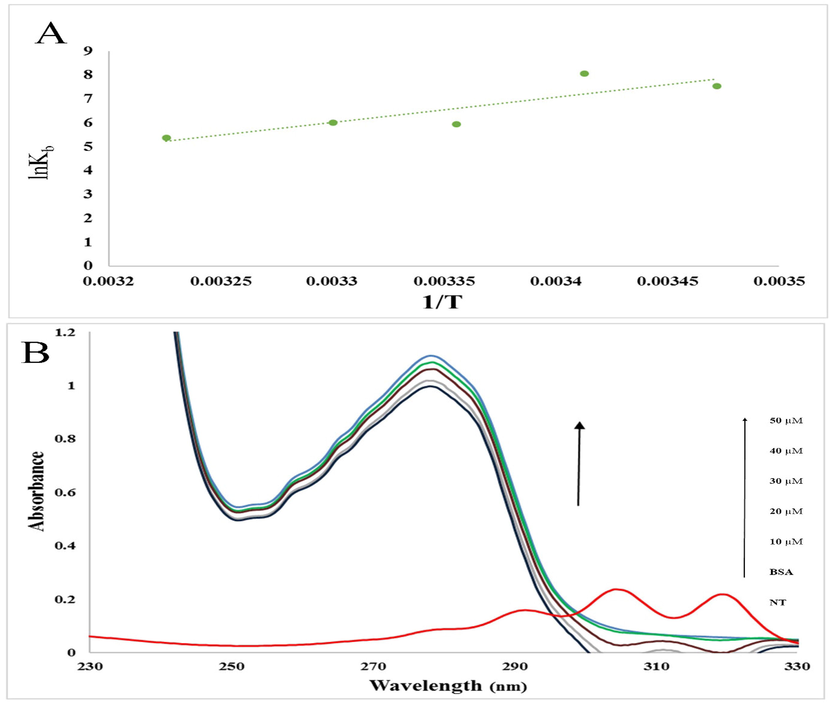
A) The van't Hoff diagrams for the interaction of BSA with NT B) Absorption spectra of BSA (2 mM) before and after the addition of NT (0, 10, 20, 30, 40, and 50 μM). The red line showed the absorption spectra of NT.
3.4 UV–Vis absorption spectroscopy
UV–Vis spectrum measurement is a procedure for analyzing structural changes and recognizing the formation of protein complexes upon interaction with small molecules, which is used in protein–ligand research. In this study, we evaluated the interaction of BSA with different amounts of NT using UV–Vis spectroscopy. Owing to the presence of aromatic amino acid residues (tryptophan, phenylalanine, and tyrosine residues) in the BSA structure, an innate absorption peak of about 210 (sharp peak) and 280 (weak peak) nm can be seen in Fig. 3B. By adding different amounts of NT (10 to 60 μM) to the BSA solution, the BSA adsorption spectra at 280 nm gradually increased. In general, abnormalities in the albumin absorption spectrum in the formation of the albumin ligand complex are evident. Based on this, it can be concluded that the change in the structure and microenvironment of BSA was owing to the interaction with NT and the formation of the BSA-NT complex (Hu et al., 2006; Banu et al., 2021).
3.5 Immobilization of BSA on CMD sensor chip
A schematic illustration of the BSA immobilization on the surface of a gold chip via amide linkage is shown in Fig. 4A (Fathi et al., 2019). As can be seen in the figure, the carboxylic groups on the surface of the CMD chip were activated using the EDC/NHS solution injected into the device. And in the next step, by injecting BSA into the device, a association was established between the active carboxyl groups on the surface of the CMD chip and BSA, and thus BSA was stabilized on the surface of the chip. In the last step, ethanolamine solution was used to block unreacted sites (Maleki et al., 2020). The sensorgram of these steps is shown in Fig. 4B. Measurements showed that at pH 7.4 the immobilization level of BSA on the CMD chip surface was acceptable (Taghipour et al., 2018).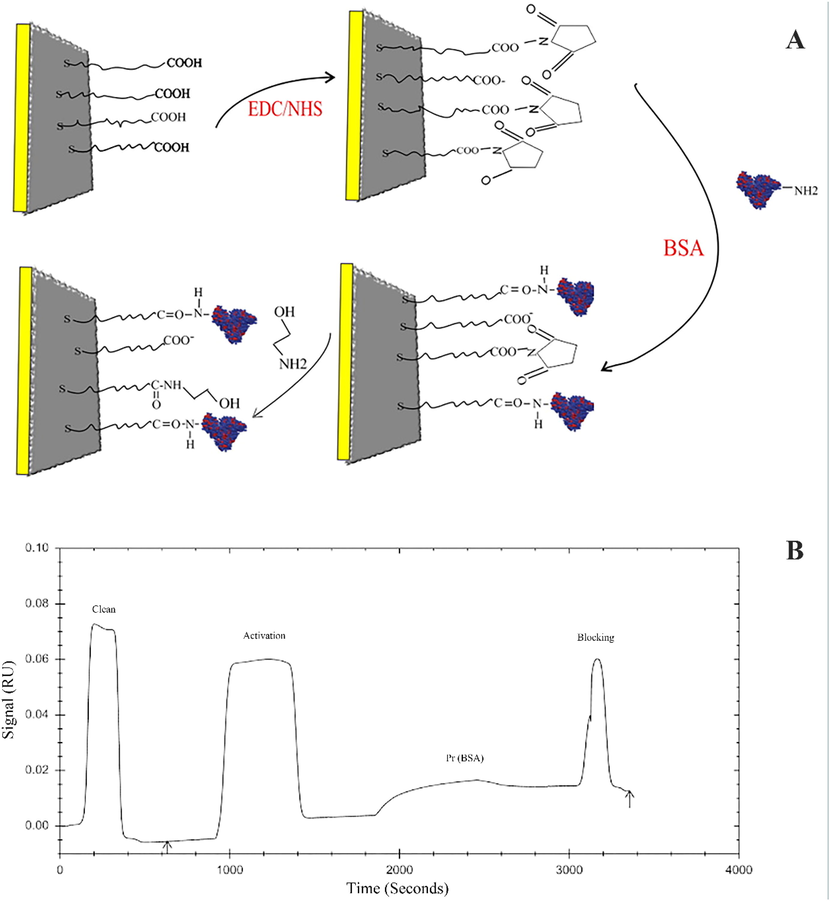
A) Schematic plot of BSA immobilization by amine coupling. B) SPR sensorgram of BSA immobilization processes on a CMD chip.
3.6 Kinetic parameters of BSA interaction with Natamycin
The reversible reaction of BSA-NT complex formation and separation can be shown by a simple Langmuir reaction:
To calculate the kinetic properties, it is necessary to calculate the ka, kd, and KD for the binding of BSA to NT. The association constant is denoted by ka, while the dissociation is denoted by kd.
BSA sensorgrams in the existence and lack of various concentrations of NT (0.1–1.2 µM) at four temperatures (298, 303, 308, and 310 K) have been shown in Fig. 5 and its related data has been demonstrated in Table 2. The ka of the BSA interaction with NT decreased when temperature increased while the kd of the interaction was constant and therefore the values of KD have been increased. The attained results demonstrated that the values of ka play a major role in the variation of KD values. Besides, increasing KD values upon raising temperature indicated that BSA binding to NT decreased, which verified the attained data from the calculation of binding constants through a fluorimetry study.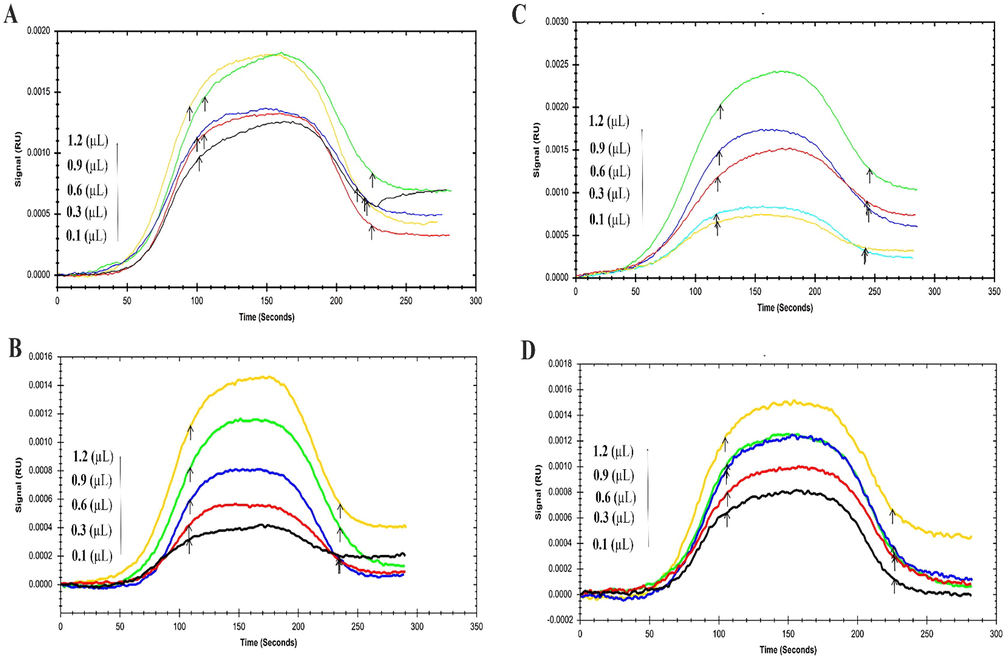
The SPR Sensorgram of immobilized BSA interaction with various concentrations of NT as a food additive at four temperatures (A) 293 °K, (B) 298 °K, (C) 303 °K, and (D) 310 °K.
T (K)
ka(1/M × s)
kd(1/s)
KD(M)
293
2.15 × 104
1.00 × 10−3
4.65 × 10−8
298
1.04 × 104
1.00 × 10−3
9.87 × 10−8
303
1.01 × 104
1.00 × 10−3
9.94 × 10−8
310
9.98 × 103
1.00 × 10−3
1.00 × 10−7
3.7 Molecular docking results
One of the most widely used methods for studying the interaction of small molecules with biomolecules is the study of molecular binding. In the present study, the interaction of the NT and BSA biomolecule was investigated by applying of Auto Dock 4.2 package. The tertiary structure of BSA is classified into three binding domains (homologous), which are nominated as I, II, and III with subdomains A and B. The major ligand-binding region is located in some hydrophobic pockets in subdomain IIIA (Szymaszek et al., 2022).
In the present study, the blind docking study displayed that NT could bind to a pit that is located among IIIA (Sudlow I) and IB domains (Fig. 6A). Sudlow I is an important binding site on BSA that several big molecules enable to bind there due to the extended entrance gate. The importance of Sudlow I lies in its ability to bind a wide range of drugs, which makes it a useful element for drug delivery and development. Many drugs, including non-steroidal anti-inflammatory drugs (NSAIDs), antibiotics, and anti-cancer agents, bind to Sudlow I with high affinity, leading to their sequestration in the plasma and reduced toxicity. Moreover, the binding of drugs to the Sudlow I site in BSA alters the pharmacokinetics and pharmacodynamics of the drugs. It affects the duration of drug action, the volume of distribution, the metabolism, and the elimination of the drug (Solanki et al., 2021).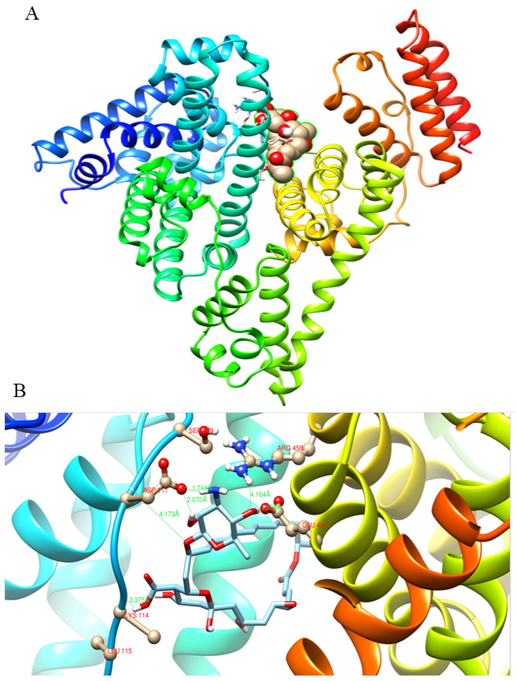
A) Represents the binding site of NT on BSA in a cavity that is located among IIIA and IB. B) The detailed interaction between NT molecule and BSA on IIA and IB binding domains.
To provide more details, another docking calculation was carried out with a particular focus on Site II. This site is known as an important position for the binding of many heterocyclic and aromatic compounds. Some studies also reported that Site II is an essential position for binding some fatty acids onto serum albumin (Tayeh et al., 2009; Belatik et al., 2012).
As indicated in Fig. 6B, Ser 109, Asp 111, Lys 114, Leu 115, Glu 424, and Arg 458 have an important role in NT-BSA interaction. The sign and magnitude of ΔS° and ΔH° indicated that the main significant interaction force for the binding between NT and BSA was van der Waals forces or hydrogen bond (Cheng, 2012). The binding was predicted to be −36.526 kJ mol−1, which confirms the spontaneous binding of this ligand onto BSA without any requirement for external energy. As shown in Table 1, the thermodynamic study found the binding free energy (ΔG°C) of the NT-BSA complex to be −16.35 kJ mol−1 at room temperature, but this varied from the molecular docking calculation. It is assumed that the theoretical research did not consider interstitial water molecules and buffer salts in the structure of BSA, which could have contributed to the association between NT and BSA molecules via the hydrogen bonding network (Diana et al., 1989; Abou-Zied and Al-Lawatia, 2011).
4 Conclusion
In the current study, the interaction between NT and BSA was explored using different spectroscopic techniques, SPR techniques, and molecular docking modeling. The results of fluorimetry measurements showed that NT reduces the intensity of BSA fluorescence by forming a complex with BSA through a hybrid quenching mechanism. Negative values of both ΔH° and ΔS° confirm that van der Waals forces or hydrogen bonds are the basic forces in the interaction of NT with BSA. The negative ΔG° value confirmed the spontaneous binding of this ligand onto BSA without any requirement for external energy. The results of the blind docking at molecular docking study displayed that NT could bind to a pit that is located among IIIA (Sudlow I) and IB domains. In addition, Ser 109, Asp 111, Lys 114, Leu 115, Glu 424, and Arg 458 also play important roles in NT-BSA interaction.
Acknowledgments
The authors would like to thank Tabriz University of Medical Sciences for supporting this project (grant No: 69708), which was a part of my M.Sc thesis.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Exploring the drug-binding site Sudlow I of human serum albumin: the role of water and Trp214 in molecular recognition and ligand binding. ChemPhysChem. 2011;12:270-274.
- [CrossRef] [Google Scholar]
- Additives, E. P. o. F., N. S. a. t. Food, 2009. Scientific Opinion on the use of natamycin (E 235) as a food additive. EFSA J. 7, 1412.
- Study of the binding of cuminaldehyde with bovine serum albumin by spectroscopic and molecular modeling methods. J. Spectroscopy. 2023;2023
- [Google Scholar]
- Multi-spectroscopic and molecular modeling studies of interaction between two different angiotensin I converting enzyme inhibitory peptides from gluten hydrolysate and human serum albumin. J. Biomol. Struct. Dyn.. 2017;35:3648-3662.
- [CrossRef] [Google Scholar]
- Multispectroscopic and computational studies of interaction of bovine serum albumin, human serum albumin and bovine hemoglobin with bisacodyl. J. Mol. Struct.. 2021;1249:131550
- [CrossRef] [Google Scholar]
- Thermodynamics of the interaction of the food additive tartrazine with serum albumins: A microcalorimetric investigation. Food Chem.. 2015;175:137-142.
- [CrossRef] [Google Scholar]
- Locating the binding sites of Pb(II) Ion with human and bovine serum albumins. PLoS One. 2012;7:e36723
- [Google Scholar]
- Synthesis and biological evaluation of benzamide-chalcone hybrids as potential c-Met kinase and COX-2 inhibitors. Arch. Pharm. 2023e2200405
- [Google Scholar]
- Studies on the interaction between scopoletin and two serum albumins by spectroscopic methods. J. Lumin.. 2012;132:2719-2729.
- [Google Scholar]
- Modelling natamycin release from alginate/chitosan active films. Int. J. Food Sci. Technol.. 2012;47:740-746.
- [Google Scholar]
- Does use of the polyene natamycin as a food preservative jeopardise the clinical efficacy of amphotericin B? A word of concern. Int. J. Antimicrob. Agents. 2015;45:564-567.
- [Google Scholar]
- Binding of nonsteroidal anti-inflammatory agents and their effect on binding of racemic warfarin and its enantiomers to human serum albumin. J. Pharm. Sci.. 1989;78:195-199.
- [CrossRef] [Google Scholar]
- Improvement in natamycin production by Streptomyces natalensis with the addition of short-chain carboxylic acids. Process Biochem.. 2013;48:1831-1838.
- [Google Scholar]
- Kinetic and thermodynamic insights into interaction of albumin with piperacillin: Spectroscopic and molecular modeling approaches. J. Mol. Liq.. 2019;296:111770
- [Google Scholar]
- Spectroscopic studies on the interaction between methylene blue and bovine serum albumin. J. Photochem. Photobiol. A Chem.. 2006;179:324-329.
- [Google Scholar]
- Effects of food additives and preservatives on man-a review. Asian J. Sci. Technol.. 2015;6:18.
- [Google Scholar]
- Albumin binding study to sodium lactate food additive using spectroscopic and molecular docking approaches. J. Mol. Liq.. 2020;310:113259
- [Google Scholar]
- Multi-spectroscopic, thermodynamic and molecular dockimg insights into interaction of bovine serum albumin with calcium lactate. Microchem. J.. 2020;154:104580
- [Google Scholar]
- Binding process evaluation of bovine serum albumin and Lawsonia inermis (henna) through spectroscopic and molecular docking approaches. J. Mol. Liq.. 2021;331:115792
- [Google Scholar]
- Novel perspective into the interaction behavior study of the cyanidin with human serum albumin-holo transferrin complex: Spectroscopic, calorimetric and molecular modeling approaches. J. Mol. Liq.. 2022;356:119042
- [CrossRef] [Google Scholar]
- Plasmonics in biology and plasmon-controlled fluorescence. Plasmonics. 2006;1:5-33.
- [Google Scholar]
- Does use of the polyene natamycin as a food. Adv. Drug Deliv. Rev.. 2007;59:645-666.
- [Google Scholar]
- Study on the interaction between clozapine and bovine serum albumin. J. Mol. Struct.. 2007;833:184-188.
- [Google Scholar]
- Glucokinase activity enhancement by cellulose nanocrystals isolated from jujube seed: A novel perspective for type II diabetes mellitus treatment (In vitro) J. Mol. Struct.. 2022;1269:133803
- [CrossRef] [Google Scholar]
- Thermodynamic analysis of albumin interaction with monosodium glutamate food additive: Insights from multi-spectroscopic and molecular docking approaches. J. Mol. Struct.. 2020;1221:128785
- [Google Scholar]
- Surface plasmon resonance, fluorescence, and molecular docking studies of bovine serum albumin interactions with natural coumarin diversin. Spectrochim. Acta A Mol. Biomol. Spectrosc.. 2020;230:118063
- [Google Scholar]
- Study on effect of lomefloxacin on human holo-transferrin in the presence of essential and nonessential amino acids: Spectroscopic and molecular modeling approaches. Int. J. Biol. Macromol.. 2017;97:688-699.
- [CrossRef] [Google Scholar]
- Effect of natamycin on cytochrome P450 enzymes in rats. Food Chem. Toxicol.. 2013;62:281-284.
- [Google Scholar]
- Spectroscopic studies on the interaction of cinnamic acid and its hydroxyl derivatives with human serum albumin. J. Mol. Struct.. 2004;692:71-80.
- [Google Scholar]
- Multi-spectroscopic and molecular modeling studies of bovine serum albumin interaction with sodium acetate food additive. Food Chem.. 2017;228:265-269.
- [Google Scholar]
- Interaction of graphene oxide with bovine serum albumin: A fluorescence quenching study. Spectrochim. Acta A Mol. Biomol. Spectrosc.. 2019;210:348-354.
- [CrossRef] [Google Scholar]
- Crystal structures of human serum albumin complexed with monounsaturated and polyunsaturated fatty acids. J Mol Biol.. 2001;314:955-960.
- [CrossRef] [Google Scholar]
- Control of pathogenic and spoilage microorganisms from cheese surface by whey protein films containing malic acid, nisin and natamycin. Food Control. 2010;21:240-246.
- [Google Scholar]
- Spectroscopic, electrochemical and molecular docking studies of dothiepin and doxepin with bovine serum albumin and DNA base. Luminescence. 2016;31:1438-1447.
- [CrossRef] [Google Scholar]
- Thermodynamics of protein association reactions: forces contributing to stability. Biochemistry. 1981;20:3096-3102.
- [Google Scholar]
- Interaction of triprolidine hydrochloride with serum albumins: Thermodynamic and binding characteristics, and influence of site probes. J. Pharm. Biomed. Anal.. 2011;54:1180-1186.
- [CrossRef] [Google Scholar]
- Changes in binding affinity between ofloxacin and calf thymus DNA in the presence of histone H1: Spectroscopic and molecular modeling investigations. J. Lumin.. 2018;203:599-608.
- [Google Scholar]
- Molecular interactions of danofloxacin with bovine serum albumin: An experimental and theoretical investigation. Iranian. J. Anal. Chem. 2022
- [Google Scholar]
- Multispectroscopic studies on the interaction of 2-tert-butylhydroquinone (TBHQ), a food additive, with bovine serum albumin. Food Chem.. 2011;124:1063-1068.
- [CrossRef] [Google Scholar]
- Kinetic and thermodynamic study of bovine serum albumin interaction with rifampicin using surface plasmon resonance and molecular docking methods. J. Biomed. Opt.. 2017;22:037002
- [Google Scholar]
- In vitro study on binding interaction of quinapril with bovine serum albumin (BSA) using multi-spectroscopic and molecular docking methods. J. Biomol. Struct. Dyn.. 2017;35:2211-2223.
- [Google Scholar]
- Characterization of interactions of simvastatin, pravastatin, fluvastatin, and pitavastatin with bovine serum albumin: multiple spectroscopic and molecular docking. J. Biomol. Struct. Dyn.. 2017;35:1529-1546.
- [Google Scholar]
- Consumers’ knowledge and safety perceptions of food additives: Evaluation on the effectiveness of transmitting information on preservatives. Food Control. 2011;22:1054-1060.
- [Google Scholar]
- Food preservatives–An overview on applications and side effects. Emirates J. Food Agric. 2016:366-373.
- [Google Scholar]
- Anticancer nano-delivery systems based on bovine serum albumin nanoparticles: A critical review. Int J Biol Macromol.. 2021;193:528-540.
- [CrossRef] [Google Scholar]
- Stark, J., 1999. PRESERVATIVES| Permitted Preservatives–Natamycin.
- Influence of piracetam on gliclazide—glycated human serum albumin interaction. A spectrofluorometric study. Molecules.. 2019;24:111.
- [Google Scholar]
- Molecular interactions of bovine serum albumin (BSA) with pyridine derivatives as candidates for non-covalent protein probes: aspectroscopic investigation. J. Mol. Liq.. 2022;347:118262
- [CrossRef] [Google Scholar]
- Bovine serum albumin binding study to erlotinib using surface plasmon resonance and molecular docking methods. J. Photochem. Photobiol. B Biol.. 2018;183:11-15.
- [Google Scholar]
- Exploring the HSA/DNA/lung cancer cells binding behavior of p-Synephrine, a naturally occurring phenyl ethanol amine with anti-adipogenic activity: multi spectroscopic, molecular dynamic and cellular approaches. J. Mol. Liq.. 2022;368:120826
- [CrossRef] [Google Scholar]
- Unravelling the interaction mechanism between clioquinol and bovine serum albumin by multi-spectroscopic and molecular docking approaches. Spectrochim. Acta A Mol. Biomol. Spectrosc.. 2019;216:25-34.
- [CrossRef] [Google Scholar]
- Fluorescence spectral resolution of tryptophan residues in bovine and human serum albumins. J. Pharm. Biomed. Anal.. 2009;50:107-116.
- [CrossRef] [Google Scholar]
- Study of binding interaction of rivaroxaban with bovine serum albumin using multi-spectroscopic and molecular docking approach. Chem. Cent. J.. 2017;11:1-9.
- [Google Scholar]
- Spectroscopic and molecular modeling studies of binding interaction between bovine serum albumin and roflumilast. Drug Des. Devel. Ther.. 2018;12:2627.
- [Google Scholar]
- Study on the interaction of fisetholz with BSA/HSA by multi-spectroscopic, cyclic voltammetric, and molecular docking technique. J. Biomol. Struct. Dyn.. 2019;37:3496-3505.
- [Google Scholar]
- Characterization of the interaction between eupatorin and bovine serum albumin by spectroscopic and molecular modeling methods. Int. J. Mol. Sci.. 2013;14:14185-14203.
- [Google Scholar]
- Kinetic and thermodynamic aspects on the interaction of serum albumin with sodium hydrosulfite: Spectroscopic and molecular docking methods. J. Photochem. Photobiol. A Chem.. 2023;442:114804
- [CrossRef] [Google Scholar]
- Multispectroscopic studies on the interaction of maltol, a food additive, with bovine serum albumin. Food Chem.. 2012;133:264-270.
- [Google Scholar]
- Crystallographic analysis of human serum albumin complexed with 4Z, 15E-bilirubin-IXα. J. Mol. Biol.. 2008;381:394-406.
- [Google Scholar]







