Translate this page into:
Insights on in-silico approaches for identifying potential bioactive inhibitors for TNF-α and IL-6 proteins associated with rheumatoid arthritis
⁎Corresponding author. frenlysara@gmail.com (Summya Rashid)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Rheumatoid arthritis (RA) is a severe autoimmune-mediated inflammatory disorder that primarily affects joints thereby leading to synovial inflammation, pain, stiffness, stress, and affecting the quality of life. Owing to the significant role of TNF-α and IL-6 in the inflammatory process, the inhibition of their activities as a potential target for drug development studies has gained much attention. Currently, steroidal and non-steroidal anti-inflammatory drugs are used in the treatment of RA, however, adverse side effects are associated with these medications. Bioactive compounds obtained from plants have great therapeutic potential and can be developed as drug entities using computational approaches.
Abstract
Rheumatoid arthritis (RA) is a chronic immunocompromised disorder that primarily affects joints thereby leading to synovial inflammation, pain, stiffness, stress, and affecting the quality of life. The present study aimed to determine and identify bioactive compounds that can be developed as potential inhibitors to target human TNF-α and IL-6 proteins associated with RA. The present study also aimed to determine physiochemical properties and toxicity of compounds by ADMET, identify binding pockets via CASTp server, stereochemical quality of protein by PROCHECK, MMGB/SA analysis by fastDRH and molecular dynamics (WebGro). The findings of the present study revealed staurosporine was the most effective compound against TNF-α with the highest binding affinity of −10.8 kcal/mol. Similarly, withanoside L showed the highest binding affinity of −8.2 kcal/mol when docked against IL-6. In the present study, rutin, withanoside II, and withanoside IV did not follow Lipinski’s rule of five whereas staurosporine, somnifericin, and withanolide L followed RO5. In the present study, the top binding pocket of TNF-α had highest area (SA) of 2743.25 Å2 and volume (SA) 4514.06 Å3 whereas IL-6 had highest area (SA) of 63.54 Å2 and volume (SA) of 38.58 Å3 respectively. In this study, Ramachandran plot analysis revealed that 90.2% and 95.2% residues for TNF-α and IL-6 proteins occupied the most favored region. In the present study, TNF-α rutin complex and IL-6 rutin complex showed the highest PB1 score of 13.69 kcal/mol and 12.30 kcal/mol respectively. WebGro server estimated molecular dynamic properties (RMSD, Rg, SASA, RMSF and hydrogen bonds) of the proteins. Bio2Byte tools predicted the biophysical behaviour of the proteins. The findings of the present study revealed that staurosporine and withanolide L could be developed as potential inhibitors of TNF- α and IL-6, however further in-vitro and in-vivo studies are required to validate these results.
Keywords
Rheumatoid arthritis
TNF-α
IL-6
Molecular Docking
MM/PB(GB) SA
Molecular dynamics simulation
- RA
-
Rheumatoid Arthritis
- TNF-α
-
Tumor Necrosis Factor
- IL-6
-
Interleukin −6
- ADMET
-
absorption, distribution, metabolism, excretion, and toxicity
- MM/GBSA
-
Molecular Mechanics with Generalised Born and Surface Area Solvation
- Simple Data Format
-
SDF
- 2D
-
Two-Dimensional
- 3D
-
Three Dimensional
- CASTp
-
Computed atlas of surface topography of proteins
- NMA
-
Normal Mode Analysis
- logS
-
Water solubility
- BBB
-
Blood-Brain Barrier
Abbreviations
1 Introduction
Rheumatoid arthritis (RA) is a severe autoimmune-mediated inflammatory disorder that approximately affects 1% of the global population (Hansildaar et al., 2021). RA primarily affects the synovial joints and is characterized by the accumulation of inflammatory cells, synovial hyperplasia, systemic complications, and progressive destruction of joints (Chen et al., 2019; Mcinnes and Schett, 2011). The outcome of the inflammatory process involves the host immune system, inflammatory mediator molecules, and the vascular compartments (Khalid et al., 2018). The inflammatory process involves the infiltration of various immune cells such as neutrophils, macrophages, and their products (cytokines and chemokines) (Ullah et al., 2020). Subsequently, these inflammatory mediators are released into the systemic circulation and generate systemic effects like fever, pain, redness, heat, and discomfort in the body (Mikuls et al., 2016). In the onset of inflammatory reaction, several mediator molecules are involved including tumor necrosis factor-α (TNF-α), interleukin-6 (IL-6), and interleukin-1β (IL-1β).
TNF-α is an important cytokine molecule that plays a pivotal role in apoptosis, inflammation, cell metabolism, proliferation, differentiation, and immune regulation (Montgomery and Bowers, 2012; Wijbrandts et al., 2008). TNF-α is a homotrimeric protein with a molecular weight of 17 kDa and consists of 157 amino acids. Activated lymphocytes, macrophages, and natural killer (NK) cells mainly synthesize this pleiotropic cytokine molecule. The dysfunction of TNF-α is associated with various diseases, such as RA, cancer, ankylosing spondylitis, psoriasis, neurological disorders, and inflammatory diseases (Hyrich et al., 2007; Schulz et al., 2014; Feldmann and Maini, 2003). Similarly, IL-6 is a soluble pleiotropic inflammatory mediator and an important member of the IL-6 family produced in response to infections and tissue damage (Tanaka et al., 2014). The abnormal IL-6 synthesis has been found in various diseased conditions including RA, multiple myeloma, autoimmune diseases, postmenopausal osteoporosis, and viral infections (Kamimura et al., 2014; Zheng et al., 2017; Angulo et al., 2017; Ergönül et al., 2017; Ulhaq and Soraya, 2020). In recent years, owing to the significant role of TNF-α and IL-6 in the inflammatory process, the inhibition of their activities as a potential target for drug development studies has gained much attention. Currently, steroidal and non-steroidal anti-inflammatory drugs are used in the treatment of RA (Wei et al., 2016; Inman et al., 2016). However, adverse side effects are associated with these medications that can affect the liver, kidney, heart, gastrointestinal system, and bone marrow (Felson et al., 2016; Solomon et al., 2018). In such a scenario, natural products obtained from plants have great therapeutic potential and can be developed as drug entities and in recent years, the use of bioactive molecules has gained much attention (Zhang and Wei, 2020).
Plant-derived bioactive molecules serve as a cheap and rich source in the development of novel drug molecules for use in clinical practice. Plant compounds could provide a plethora of potential phytochemicals for targeted therapies and have been used since folklore times. Medicinal plants serve an as important source for traditional folklore medicine, pharmaceutical intermediates, synthetic drugs, food supplements and nutraceuticals. These medicinal plants contain various active bioactives like phenolic compounds, alkaloids, flavonoids and tannins that possess the biological activity to be used for therapeutic purposes. Plant compounds like Curcumin, Zerumbone, Zingerone, Isosilychristin, Naringenin, Polydatin, Gallic Acid, Gingerenone A, Quercetin, Rutin, Staurosporine, Withanoside II, Somnifericin, Syringic Acid, Withanoside IV, Weldolactone, Chlorogenic acid, Genstein, Withaferin A, Tocotrienol, Rosamaric acid, Kaempferol,Withanolide L and Withanolide M are known to possess antioxidant, anti-inflammatory, anticancer, immunomodulatory, neuroprotective and antidepressant properties (Kou et al., 2023; Nosran et al., 2021; Chia et al., 2021; Viktorova et al., 2019; Gai et al., 2022; Malla et al., 2022; Rehman et al., 2022; Ali et al., 2023).
An emerging class of new compounds can be obtained from natural and semi-synthetic derived compounds (Ain et al., 2020; Sinha et al., 2018). Moreover, the development of computational approaches can serve as a rapid and reliable process in the screening of a large number of natural compounds. Molecular docking methods determine the binding affinity of ligands with the target protein and the mechanism by which interaction occurs (Sinha et al., 2018).
In the present study, an attempt was made to use computational approaches to evaluate the anti-inflammatory properties of various bioactives against human TNF-α and IL-6 proteins and to reveal possible interaction mechanisms. Furthermore, the present aimed at revealing the ADMET (absorption, distribution, metabolism, excretion, and toxicity), docking features, topological properties, molecular dynamics, rescoring profiles, and calculation of free binding energy via MM/GBSA (molecular mechanics with generalized born and surface area solvation) approach.
2 Materials and methods
2.1 Selection of ligands
In the present study, 24 bioactive compounds (Curcumin, Zerumbone, Zingerone, Isosilychristin, Naringenin, Polydatin, Gallic Acid, Gingerenone A, Quercetin, Rutin, Staurosporine, Withanoside II, Somnifericin, Syringic Acid, Withanoside IV, Weldolactone, Chlorogenic acid, Genstein, Withaferin A, Tocotrienol,Rosamaric acid, Kaempferol,Withanolide L and Withanolide M) were selected based on their anti-inflammatory properties. The phytocompounds used in the present study were selected based on previous literature. PubChem was used to obtain 2D structures of natural compounds in simple data format (SDF) (https://pubchem.ncbi.nlm.nih.gov/compound/ accessed on 25 July 2022). The ligands in the SDF were converted into Protein data bank (PDB) format by using Online Smiles Translator (https://cactus.nci.nih.gov/translate/ accessed on 25 July 2022).
3 Molecular docking
3.1 Preparation of protein/receptor
In the present study, the 3D structure of TNF-α and IL-6 were retrieved from a protein data bank with PDB ID of 2AZ5 and 1ALU respectively (https://www.rcsb.org/structure/ accessed on 25 July 2022). In the present study, AutoDock Vina (UCSF-chimera©, version 4.2.6) was used to perform docking analysis as per the procedure described by (Trott and Olson, 2010). In AutoDock Vina, both proteins were prepared separately by following a suite of steps. Initially from the proteins, molecules of water were removed followed by polar hydrogen addition. After this, Gasteiger charges were added to both the proteins. Similarly, the ligands were prepared by adding polar hydrogen, Gasteiger charges and performing energy minimization. Before grid preparation, the proteins were selected as macromolecule and the phytocompounds as ligands. After preparatory phase, both the proteins and ligand molecules were saved in pdbqt format.
3.2 Grid dimensions
In the present study, the grid box size dimensions (x, y, z) were 40 A° each with a spacing of 40 A° for both proteins and the centre dimensions (x = -13.690, y = 71.605, z = 27.00 A°) for TNF-α and IL-6 (x = 2.523, y = -19.960, z = 8.684 A°) respectively.
3.3 ADMET analysis
The drug-like properties of top six natural compounds that showed highest binding affinity scores via molecular docking with TNF-α and IL-6 were further assessed by admetSTAR (https://Immd.ecust.edu.cn/admetstar2/ accessed on 25 July 2022). In drug discovery and assessment of environment hazards, ADMET analysis plays a significant role in describing the properties of drug molecules and environmental chemicals. The foremost pharmacokinetic properties that can be evaluated by ADMET analysis include the blood–brain barrier, human intestinal absorption, CYP4502C9, and Caco-2 permeability (Cheng et al., 2012). The Lipinski’s rule of five parameters (RO5) i.e. molecular weight (<500 g/mol, topological polar surface area (TPSA) < 140 Å2, AlogP < 5 and number of hydrogen bond acceptors and donors<10 and 5) (Yang et al., 2019; Daina et al., 2017). The toxicity (hepatotoxicity and carcinogenicity) of the compounds was also determined via this tool.
3.4 Computed atlas of surface topography of proteins (CASTp)
The topological and geometric properties of the human TNF-α and IL-6 were determined by the CASTp server (https://sts.bioe.uic.edu/castp/ accessed on 25 July 2022). The pockets and cavities in the protein structures were located, delineated, and measured by this tool. In computational geometry, CASTp is based on principle of alpha shape and pocket algorithm. Using the molecular surface model (Connolly’s surface) and solvent accessible surface model (Richard’s surface), this web-based server measures area and volume of every pocket and void.
3.5 PROCHECK
This tool (https://saves.mbi.ucla.edu/ accessed on 25 July 2022) evaluates a protein structure's overall and residue-by-residue geometry to determine the stereochemical quality of the structure. For evaluating the calibre of structures solved by NMR, it includes PROCHECK-NMR. Based on the Ramachandran plot, this server provides geometric calculation that depends upon visualization of the dihedral angles Phi (ϕ) and Psi (ψ) of amino acids.
3.6 fastDRH
An easily available online web server (https://cadd.zju.edu.cn/fastdrh/ accessed on 25 July 2022) that determines the rescoring of docking poses for protein–protein interaction (PPI) inhibitors by using the Molecular Mechanics (MM) of Poisson-Boltzmann surface area (PB) and Generalized Born Surface Area (GBSA) (MM/PB(GB) SA) approaches. The binding affinity of the selected complexes is estimated by these methods.
3.7 Molecular dynamic simulation
WebGro is a completely automated online server for simulating the molecular dynamics of macromolecules (proteins) alone or in complex with ligands (small molecules) (https://simlab.uams.edu/ProteinInWater/index.html/ accessed on 25 July 2022). For completely solvated molecular dynamics simulations, WebGro uses the GROMACS simulation program. Molecular Dynamics simulation was utilized to investigate the binding stability of the final complexes using WEBGRO for macromolecular simulations. The sys-tem was solvated in water, neutralized, and 0.15 M NaCl salt was added using GRO-MOS96 43a1 force field parameters. The steepest descent method was used to reduce energy in 5000 stages. The types of constant quantity, volume, temperature (NVT/NPT), and pressure equilibration were utilized. The temperature was set to 300 K and the pressure was set to 1.0 bar for a simulation time of 60 ns and 1000 frames per simulation. Root Mean Square Deviation (RMSD), Root Mean Square Fluctuation (RMSF), Radius of Gyration (Rg), intermolecular H-bonding (H-bonds), and Solvent Accessible Surface Area (SASA) were the simulation parameters sought.
3.8 Bio2Byte (b2b) tools
This tool's objective is to identify the biophysical characteristics or behaviours of proteins that are difficult for structural biology and/or molecular dynamics approaches to adequately capture (https://bio2byte.be/b2tools/ accessed on 25 July 2022). Integrated predictions from DynaMine backbone and side-chain dynamics, conformational propensities, and derived EFoldMine early folding, DisoMine disorder, are provided by uploading a FASTA file or text-entry of a sequence.
4 Results and discussion
4.1 Molecular docking
The docking approach helps to model the interaction between a ligand and protein molecule at the atomic level. In the present study, staurosporine showed the highest binding affinity of −10.8 kcal/mol against TNF-α. The least binding affinity was shown by chlorogenic acid (-5.7 kcal/mol) against TNF-α. Similarly, against IL-6, the highest binding affinity was depicted by withanoside IV (-8.6 kcal/mol) followed by withanolide L (-8.2 kcal/mol) and lowest by zingerone (-5.0 kcal/mol). The top most five compounds that showed good binding affinities against both proteins were further analyzed in this study. The findings of docking analysis obtained in present studies are represented in Table 1.
Compounds
Binding energy (kcal/mol)
Compounds
TNF-α
IL-6
Curcumin
−6.2
−5.3
Zerumbone
−7.6
−6.3
Zingerone
−5.9
−5.0
Isosilychristin
−7.6
−6.9
Naringenin
−8.3
−6.4
Polydatin
−8.5
−6.8
Gallic Acid
−6.1
−5.5
Gingerenone A
−7.4
−5.4
Quecertin
−8.7
−6.3
Rutin
−9.0
−8.1
Staurosporine
−10.8
−7.6
Withanoside II
−9.2
−7.5
Somnifericin
−8.8
−7.3
Syringic Acid
−8.4
−6.4
Withanoside IV
−9.5
−8.6
Weldolactone
−7.9
−6.6
Chlorogenic acid
−5.7
−5.4
Genstein
−6.9
−6.3
Withaferin A
−7.8
−7.1
Tocotrienol
−7.4
−6.8
Withanolide L
−8.7
−8.2
Withanolide M
−8.6
−7.8
Rosamaric acid
−6.9
−6.2
Kaempferol
−6.8
−6.3
Fig. 1 graphically illustrates the 2D interaction of TNF-α protein with top most five compounds. In present study, the various types of interaction formed by proteins with compounds is shown in Table 2 respectively.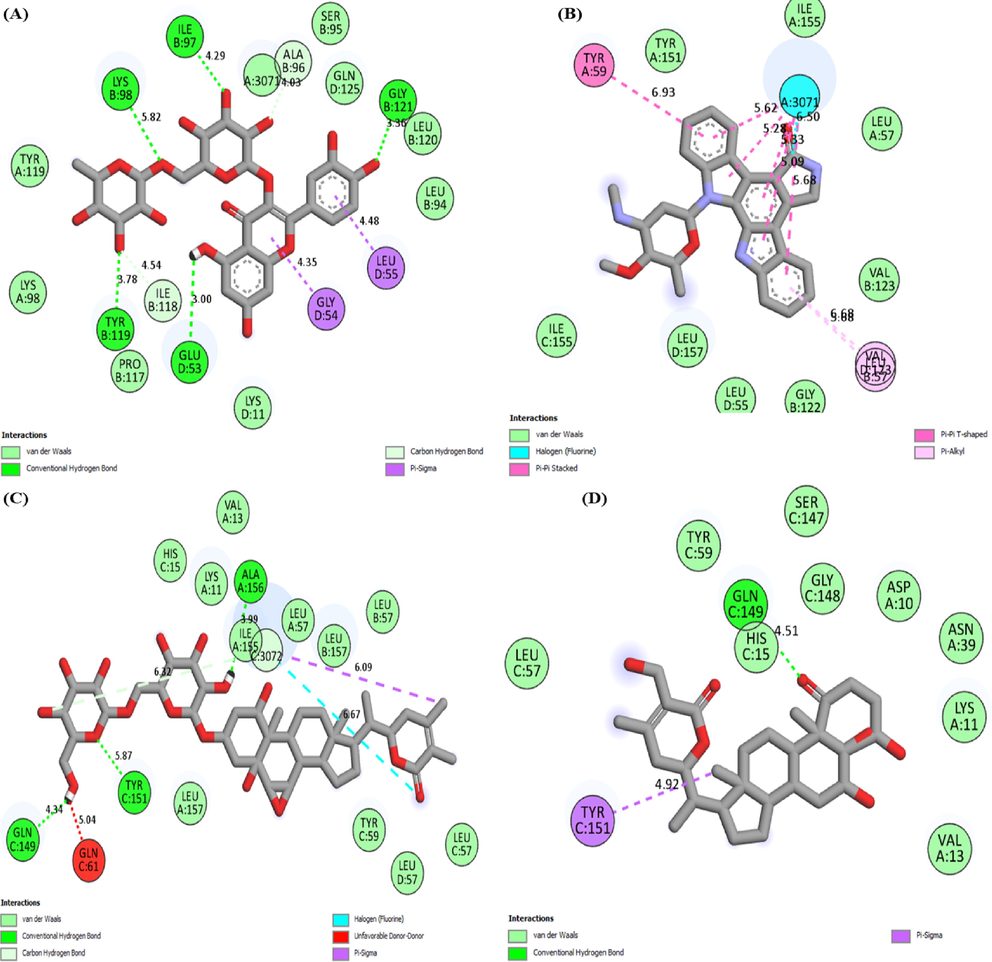
2D molecular interaction of TNF-α With (A) Rutin (B) Staurosporine (C) Withanoside II (D) Somnifericin (E) Withanoside IV.
2D molecular interaction of TNF-α With (A) Rutin (B) Staurosporine (C) Withanoside II (D) Somnifericin (E) Withanoside IV.
Protein
Bioactive
Interaction
Residue and Position
TNF-α
Rutin
Hydrogen bond
GLU53, ILE97, LYS98, TYR119, GLY121
Van der Waals
LYS11, LYS98, SER95, LEU94, TYR119, PRO117, LEU120, GLN125
Pi-sigma
GLY54, LEU55
Staurosporine
Vander waals
LEU155, TYR151, LEU57, ILE155, LEU157, VAL123, LEU55, GLY122
Pi-alkyl
VAL123, LEU57
Pi-stacked
TYR59
Withanoside II
Hydrogen bond
GLN149, TYR151, ALA156
Van der Waals
LYS11, VAL13, HIS15, TYR59, LEU57, LEU157, ILE155,
Somnifericin
Hydrogen bond
GLN149
Van der Waals
LEU57, TYR59, SER147, HIS15, GLY148, ASP10, ASN39, LYS11, VAL13
Pi-Sigma
TYR151
Hydrogen bond
GLY148, ALA156, PRO12, ASP10,
Withanoside IV
Van der Waals
HIS15, TYR59, TYR151, LEU57, LEU36, HIS15, GLN149, LEU55, LEU157, VAL123, GLY122, ASN39, VAL13,
Pi-alkyl
TYR59, ILE155
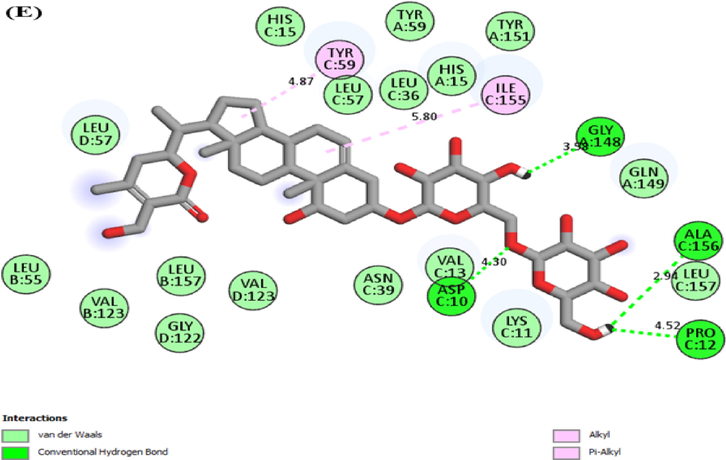
Fig. 2 graphically illustrates the 2D interaction of IL-6 protein with the top five compounds. In the present study, the various types of interaction formed by proteins with compounds are shown in Table 3 respectively.
2D molecular interaction of IL-6 with (A) Rutin (B) Staurosporine (C) Withanoside II (D) Withanolide L (E) Withanoside IV.

2D molecular interaction of IL-6 with (A) Rutin (B) Staurosporine (C) Withanoside II (D) Withanolide L (E) Withanoside IV.
Protein
Bioactives
Interaction
Residue and Position
IL-6
Rutin
Hydrogen bond
ASN103, GLU106, SER107, ARG104, GLN159, GLN156
Van der Waals
TYR100, SER108, PHE105, GLU42, SER42
Pi-Anion
ASP160
Pi-Alkyl
LYS46
Staurosporine
Van der waals
THR43, GLU42, THR163, SER107, GLU106, GLN156
Pi-Sigma
PHE105
Pi-Alkyl
LYS46
Pi-Cation
ASP160
Pi-Anion
ARG104
Withanoside II
Hydrogen bond
SER37
Van der Waals
GLN175, ILE32, GLY35, LEU33, LEU178, ARG30, LYS41, ALA38, ALA114,
Pi-Sigma
TYR31
Withanolide L
Hydrogen bond
LYS86
Van der Waals
ASN63, LEU64, PRO65, LYS66, ALA68, ASP71, GLU93, VAL96, TYR97,THR143, ASN144, LEU147,
Withanoside IV
Hydrogen bond
GLN116, ASN103
Van der Waals
VAL96, ASN144, THR149, GLN152, LEU148, GLN127, LEU92, ILE123, GLN124, LYS120, PRO141, THR142, ALA145, MET117, GLN102, ARG113
4.2 ADMET
In the present study, rutin, withanoside II, and withanoside IV did not follow Lipinski’s rule as molecular weight was greater than 500 g/mol, TPSA greater than 140 Å, number of hydrogen bond acceptors and donors were greater than 10 and 5 respectively whereas staurosporine and somnifericin followed the RO5. (Table 4). Moreover the lipophilicity (logP) of the compounds ranged from −0.19 to 4.35 and hence was within the acceptable range (<5) that indicates better absorption. In the present study, the TPSA scores of rutin, staurosporine and somnifericin was within range whereas withanoside II and withanoside IV had higher TPSA values. In drug development, water solubility (logS) is an important parameter and all the selected compounds have values < 5 respectively. The molar refractivity (MR) of the selected compounds were not within the acceptable range of 40–130. In transdermal drug development, skin permeability (log Kp) is a vital parameter and values greater than 2.5 represent lower skin permeability. In the present study, staurosporine and somnifericin had good bioavailability/GI absorption whereas rest of compounds had lower values. The absorption of an oral administered drug molecule is described by epithelial colorectal adenocarcinoma cell line (Caco-2) with scores greater than 0.90 predicting higher permeability. In this study, only rutin had a higher Caco-2 score of 0.90 whereas other compounds had lesser values. Staurosporine was the only compound found to cross the blood–brain barrier (BBB) whereas the rest of compounds showed poor distribution to the brain. The cytochrome p450 (CYP450) inhibitors assess the ability of a molecule whether it is going to inhibit CYP450 and its isoforms. In this study, rutin, staurosporine and soomnifericin showed active activity for hepatotoxicity. Ames toxicity is used to determine whether a compound is mutagenic or not and in this study, rutin and staurosporine were mutagenic. Moreover, none of the selected compounds showed active carcinogenicity profile and were inactive. The + sign in brackets indicates active for toxicity whereas the (-) sign represents inactive for respective toxicity. The roman numerals in brackets represent a class of toxicity.
ADMET
Natural compounds
Rutin
Staurosporine
Withanoside II
Somnifericin
Withanoside IV
Withanolide L
Molecular weight
610.52
466.54
798.92
490.64
782.92
452.58
AlogP
−1.69
4.35
−0.98
2.53
−0.19
4.34
Number of H-bond acceptors
16
6
16
7
15
5
Number of H-bond donors
10
2
9
4
9
2
Water solubility (logS)
−2.77
−2.85
−3.93
−3.72
−4.21
−4.35
TPSA (Å2)
92.68
69.45
257.82
124.29
245.29
83.83
Molar refractivity (MR)
141.38
194.21
193.69
131.32
194.21
127.61
Log Kp (skin permeation) cm/s
−10.26
−6.85
−11.09
−7.28
−10.37
−6.99
Bioavailability score
0.17
0.55
0.17
0.55
0.17
0.55
GI absorption
Low
High
Low
High
Low
High
Lipinski rule
No
Yes
No
Yes
No
Yes
Caco-2
0.92
0.58
0.88
0.74
0.88
0.86
Blood-Brain Barrier (BBB)
No
Yes
No
No
No
No
CYP3A4 inhibition
0.92
0.56
0.93
0.82
0.94
0.75
CYP2C9 inhibition
0.90
0.67
0.80
0.91
0.93
0.65
CYP2D6 inhibition
0.95
0.82
0.93
0.95
0.94
0.80
CYP2C19 inhibition
0.90
0.68
0.86
0.93
0.93
0.70
CYP1A2 inhibition
0.86
0.56
0.87
0.89
0.92
0.69
Hepatotoxicity
0.66 (+)
0.63 (+)
0.68 (-)
0.52 (+)
0.71 (-)
0.83 (-)
Ames mutagenesis
0.80 (+)
0.75 (+)
0.74 (-)
0.73 (-)
0.70 (-)
0.88(-)
Carcinogenicity (Binary)
1.00 (-)
0.89 (-)
0.99 (-)
1.00 (-)
0.99 (-)
0.80(-)
In the present study, 24 bioactive compounds were used for in silico studies and the top five compounds were further analyzed for docking analysis. In the present study, staurosporine was found to be the most significant compound that showed the highest binding affinity (-10.8 kcal/mol) against TNF-α. Staurosporine is a broad-spectrum competitive inhibitor of protein kinases and competes with ATP molecules by binding to surface pockets of target kinases (Gani and Engh, 2010). Staurosporine has been reported to be an effective antitumor compound and has shown potential activity in breast, oral, colon, and cervical cancer cell lines (del Solar et al., 2015; Yadav et al., 2015; Vasaturo et al., 2005). In many cancer cell lines, staurosporine is known to induce apoptosis (Stepczynska et al., 2001).
Similarly, withanoside IV showed the highest binding of −8.6 kcal/mol when docked against human IL-6 protein, however it did not follow RO5 that is considered to be important for drug development. As a result, withanolide L that simultaneously followed RO5 and showed second best binding affinity after it was considered. Withanoside L is an important constituent of Ashwagandha (Withania somnifera). Numerous research studies have reported Withania somnifera is quite effective in the treatment of asthma, cancer, arthritis, hypertension, insomnia, stress, thyroid gland function, anxiety, anti-inflammatory, and in the recovery of male and female sexual functions (Nasimi Doost Azgomi et al., 2018; Devkar et al., 2016; Langade et al., 2019; Gannon et al., 2019; Raut et al., 2012). In traditional medicine, Withania somnifera has been used in the treatment and management of polyarthritis, swellings, uterine infections, general debility, premature ejaculation, oligospermia, impotency, leucorrhoea, and hemorrhoid (Uddin et al., 2012; Imtiaz et al., 2013). Other research studies have reported that Withania somnifera increases the levels of luteinizing hormone, testosterone, and follicle-stimulating hormone in addition to enhancing folliculogenesis and spermatogenesis (Nirupama and Yajurvedi, 2015; Kumar et al., 2015).
RA is a severe autoimmune disorder marked by inflammatory changes in synovial joints, bones, and cartilage causing disability and death (Scherer et al., 2020). Presently, RA has a high incidence rate and it seriously affects the health/quality of life (Safiri et al., 2019; Myasoedova et al., 2020). In the drug discovery process, ADMET studies play a significant role with many drugs failing in the last stage of clinical trials due to undesirable efficacy, toxicity, poor absorption, metabolism distribution, and excretion (Caldwell et al., 2009). In the present study, staurosporine, somnifericin, withanolide L followed RO5, however, rutin, withanoside IV, and withanoside II did not follow. The drug-like properties of the bioactive compounds are determined by Lipinski’s rule (Lipinski, 2004; Aldossari et al., 2023). Our study identified that among the natural compounds used in the present study, staurosporine and withanolide L showed good results through in silico analysis and could be used to develop potential inhibitors against TNF- and IL-6 proteins involved in the pathogenesis of RA. The failure rate of preclinical drugs that are in clinical trials can be reduced by pharmacokinetics and toxicity (Yang et al., 2019; Ferreira and Andricopulo, 2019). The use of ADMET prediction on natural compounds has gained much attention in recent years owing to their cost-saving, easy availability, and alternative to experimental methods (Patel et al., 2020; Ali et al., 2023). Numerous in silico approaches have been effectively used for in vitro ADMET prediction of compounds and successful in silico models to replace in vivo models have been developed to predict the toxicity and pharmacokinetic properties of molecules (Wang et al., 2015; Alqahtani, 2017; Alsaffar et al., 2023). The progress of in silico prediction approaches has increased with the continuous growth of the cheminformatics field and has entered into the age of big data (Ferreira and Andricopulo, 2019).
4.3 CASTp
The topmost four binding pockets in the TNF-α and IL-6 proteins were identified by the CASTp server and are illustrated in Fig. 3 and Fig. 4. Pocket panels present several residues located in the pocket and they may serve as a target for drugs. Different pocket imprints showing various amino acid residues in TNF-α and IL-6 proteins are represented in Fig. 3(A-D) and Fig. 4 (A-D). The amino acid sequence of human TNF- α and IL-6 is described in Fig. 3E and Fig. 4E. The CASTp statistics showing surface accessible (SA) area pocket ID (Poc ID), and surface accessible (AS) volume is represented by Table 5 and Table 6 respectively. In the present study, the top binding pocket of TNF-α had highest area (SA) of 2743.25 Å2 and volume (SA) 4514.06 Å3 respectively.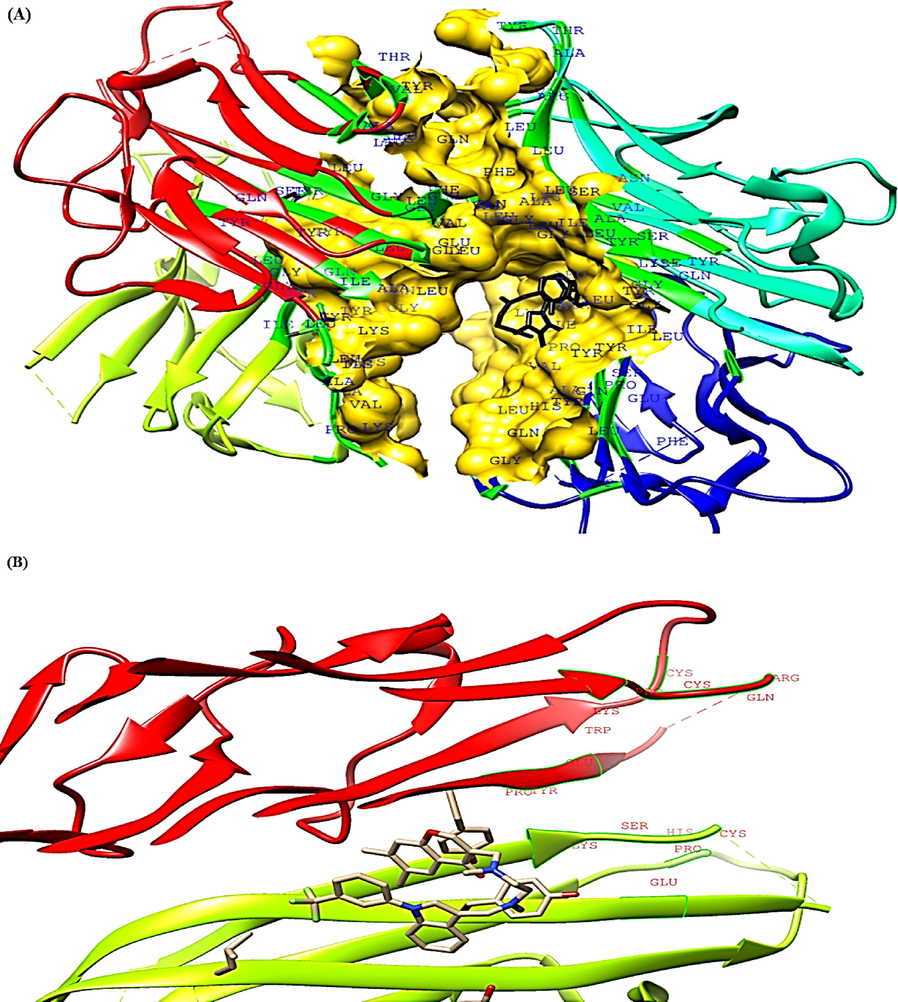
CASTp analysis of TNF-α (A-D) Top four binding pockets (E) Pocket sequence.
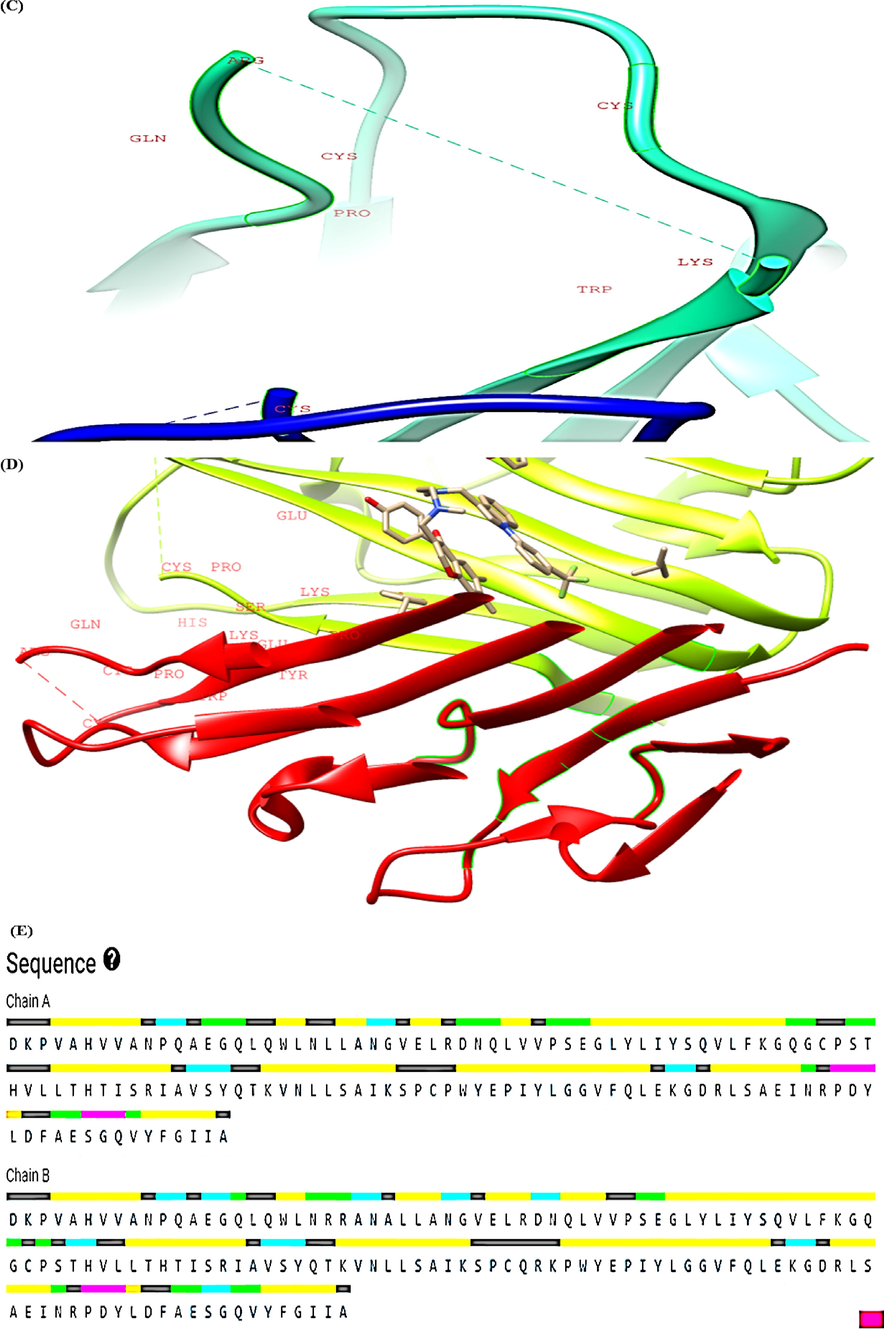
CASTp analysis of TNF-α (A-D) Top four binding pockets (E) Pocket sequence.
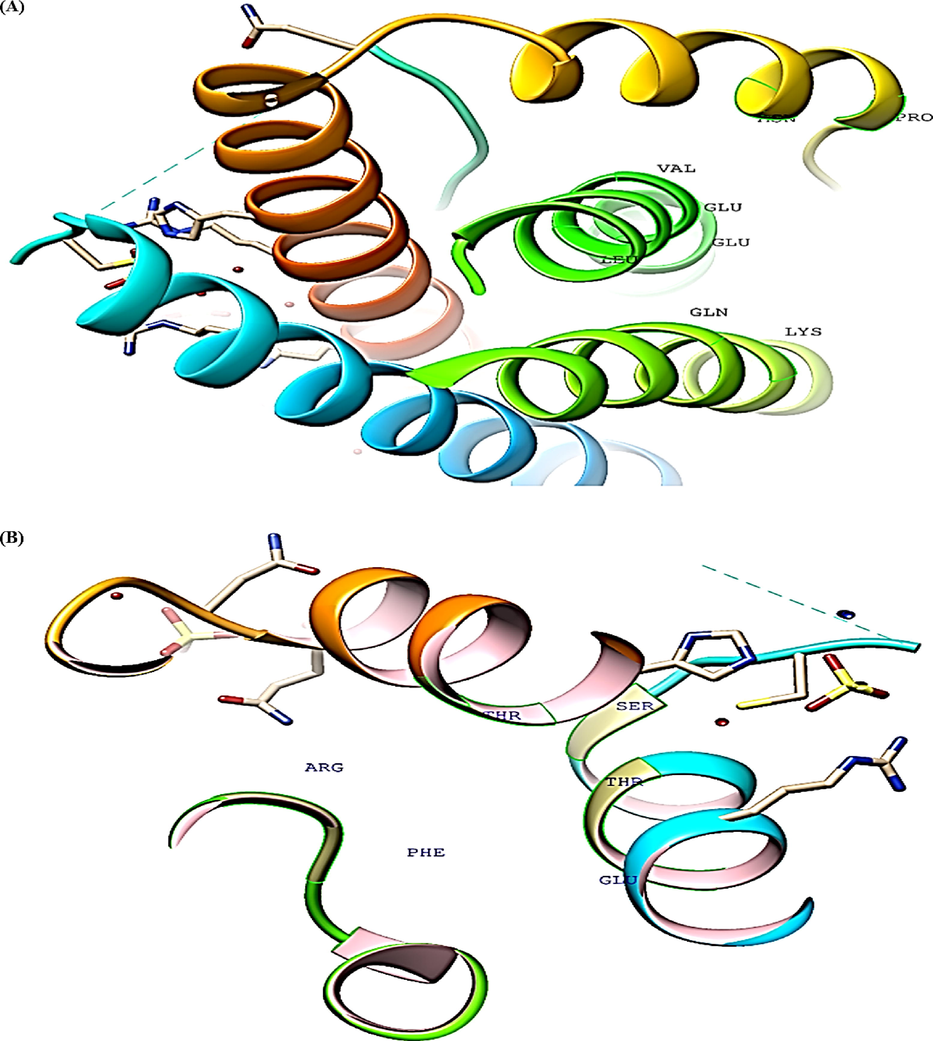
CASTp analysis of IL-6 protein (A-D) Top four binding pockets (E) Protein sequence.
CASTp analysis of IL-6 protein (A-D) Top four binding pockets (E) Protein sequence.
Poc ID
Area (SA) Å2
Volume (SA) Å3
1
2743.25
4514.06
2
109.88
36.10
3
104.50
35.04
4
100.43
84.38
Poc ID
Area (SA) Å2
Volume (SA) Å3
1
63.54
38.58
2
55.03
27.62
3
44.25
21.39
4
70.57
18.82
Similarly, the top binding pocket for IL-6 had highest area (SA) of 63.54 Å2 and volume (SA) of 38.58 Å3 respectively.
In the present study, the CASTp server identified all the surface pockets, cavities, and cross channels in TNF-α and IL-6 protein. This server also provided a detailed delineation of all atoms involved in their formation. The size of mouth openings, their exact volumes, and areas was also measured. These properties are analytically evaluated and calculated by using Connolly’s surface (molecular surface model) (Connolly, 1983) and Richard’s surface (solvent accessible surface model (Lee and Richards, 1971). The concave regions of a protein (surface pockets and interior voids) are primarily associated with binding events (Liang et al., 1998). This server measures mouth-size openings of separate pockets that help to reveal the accessibility of binding sites to different substrates and ligands. In many biological studies, CASTp computation analysis is quite useful (Paetzel and Strynadka, 1999; LiCata and Bernlohr, 1998; Ory and Banaszak, 1999; Li et al., 1999).
4.4 Homology modeling
The PROCHECK tool displayed the 3D geometry of the human TNF-α IL-6 proteins and highlighted residues with different colors, such as favored (red), allowed (yellow), generously allowed (pale yellow), disallowed (white) (Supplementary Fig. 1A-B).
In this study, TNF-α protein included 542 residues, 406 of which occupied the most favored region (A, B, L) (90.2%). There were 43 (9.6%) residues in the additionally allowed regions (a, b, I, p), 1 residue was detected in the generously allowed area (-a, -b, -I, -p), and none in the disallowed regions (Supplementary Fig. 1A). The number of non-glycine and non-proline residues in TNF-α were 450, while the number of end residues (excluding glycine and proline) was 20. Furthermore, there were 40 glycine residues (represented as triangles) and 32 proline residues. Similarly, in the IL-6 protein, there were 157 residues. In the most favored region (A, B, L) 139 residues (95.2%) were found. In additionally allowed regions (a, b, I, p) 7 (4.8%) residues were found. No residue was found in the generously allowed and disallowed regions. In the protein, about 146 non-glycine and non-proline residues were found. The number of end-residues (excluding glycine and proline) was 4. The number of glycine residues (represented as triangles) was 4 and the proline residues were 3 respectively. In this study, Supplementary Fig. 1B illustrates graphical representation of the Ramachandran plot for the IL-6 protein.
In this study. 93.4% residues occupied the favored region with almost all ϕ/ψ torsion angles found in the favored or additionally allowed regions. However, GLN88 residue was found in generously allowed region. The PROCHECK tool established the whole geometry of the protein using residue-by-residue analysis and offers information on the modeled protein's accuracy and stereochemical quality. Because 93.4% of the residues in the current study were in the preferred area, the modeled structure might be utilized for further research (Kleywegt and Jones, 1996). Ramachandran plot through PROCHECK analysis provided strong support for the stereochemical properties of the modeled protein, indicating the simulated structure may be adequate.
4.5 fastDRH
In the present study, the fastDRH tool was used to determine to decompose energy of TNF-α and IL-6 that included electrostatic energy (tele), total gas phase energy (TGAS), van der Waals contribution (VDW), non-polar and polar contribution to solvation (TGBSOL), and final estimated binding free energy (TGBTOT. In the present study, TNF-α rutin complex showed the highest binding score of PB1 score of 13.69 kcal/mol and GB1 score of −16.2 kcal/mol respectively. The profiles of free binding energies (kcal/mol) rescoring between the docked TNF-α and compounds are shown in Table 7.
Protein
Compound
Pose
Procedure and Score
1
PB1
PB3
PB4
GB1
GB2
GB5
GB6
TNF-α
Rutin
13.69
2.33
−2.7
−16.2
−9.81
−10.52
−3.12
Staurosporine
7.67
−2.88
−7.02
−14.67
−10.8
−10.29
−3.77
Withanoside II
7.17
−6.24
−7.51
−11.99
−8.06
−8.13
−4.10
Somnifericin
3.22
−7.62
−10.78
−12.97
−9.96
−10.17
−10.15
Withanoside IV
4.83
−4.59
−3.54
−10.27
−7.44
−7.4
−5.39
Similarly, the IL-6 rutin complex showed the highest PB1 score of 12.30 kcal/mol and GB1 score of −16.2 kcal/mol respectively. Moreover, the rescoring binding energy (kcal/mol) estimated between docked IL-6 and compounds is illustrated in Table 8.
Protein
Compound
Pose
Procedure and Score
1
PB1
PB3
PB4
GB1
GB2
GB5
GB6
IL-6
Rutin
12.30
1.90
−2.30
−15.5
−7.32
−7.91
−2.35
Staurosporine
5.80
−2.10
−6.38
−8.21
−7.90
−6.58
−2.19
Withanoside II
9.45
−10.38
−12.95
−13.19
−10.71
−10.45
−9.71
Withanolide L
6.40
−5.10
−8.40
−9.50
−6.90
−4.84
−2.20
Withanoside IV
6.92
−3.13
−2.37
−8.70
−5.36
−1.44
−2.54
In the present study, Table 9 shows the MM/PB(GB)SA findings of the TNF-α protein analyzed by the fast DRH server.
ELE
VDW
GAS
PBSOL
PBSUR/GBSUR
PBCAL/GB
PBSOL/GBSOL
PBELE/GBELE
PBTOT/GBTOT
−46.35
−17.39
−63.74
77.43
−14.5
64.85
77.43
18.5
13.69
Similarly, the MM/PB(GB)SA results of human IL-6 protein estimated via the fast DRH tool are represented in Table 10.
ELE
VDW
GAS
PBSOL
PBSUR/GBSUR
PBCAL/GB
PBSOL/GBSOL
PBELE/GBELE
PBTOT/GBTOT
−35.48
−10.92
−46.4
28.37
−10.02
41.12
50.68
5.64
12.30
fastDRH has the ability to predict correct binding conformations along with exact binding energy profile of complexes by using the MM/GBSA approach (Chen et al., 2016; Zhang et al., 2017). With per residue energy breakdown, MM/GBSA has been extremely effective in highlighting the important residues on the binding surface of protein–protein complexes (Chen et al., 2016; Corrada et al., 2016). Following docking analysis, MM/GBSA techniques have proven substantially more efficient in rescoring procedures (Genheden and Ryde, 2015; Sun et al., 2014). Because of their efficient performance in scoring methods, MM-PB (GB)SA techniques are considered an excellent choice for computing binding energies and revealing proper binding structures (Chen et al., 2016). The MM/PBSA approach was used to determine the binding energies of the protein complexes that included ELE, VDW, GAS, PBSOL, PBTOT, and GBSOL, corroborating the prediction of molecular docking. It is pertinent to mention that the total energies (GAS phase) and the VDW are the ones that contribute to most of the interaction energy of ligands with the protein. The least contribution to binding free energy is provided by entropy energies. The findings obtained confirm the binding free energy and ligands' tendency to bind to the protein structures depicting their capacity as potential targets for further studies.
4.6 Molecular dynamic simulations
In the present study, the root mean square deviation (RMSD) Cα backbone of TNF-α was found to be 0.30. ± 0.03A° (Fig. 5A). Similarly, the radius of gyration (Rg) was found to be 2.93 ± 0.01A° (Fig. 5B). In this study, solvent accessible surface area (SASA) was 318.7 ± 8.02 Å2 (Fig. 5C). Fig. 5D shows the relative number of hydrogen bonds with mean concentration of 612.9 ± 16.76. Moreover, the first 5 eigenvalues for Cα atoms of TNF-α protein was found to be 13.90 Å2 and the total variance found was 143.31 Å2 respectively (Fig. 5E). Fig. 5F illustrates the RMSF of the TNF-α protein with peaks in the protein representing the fluctuations of the residues.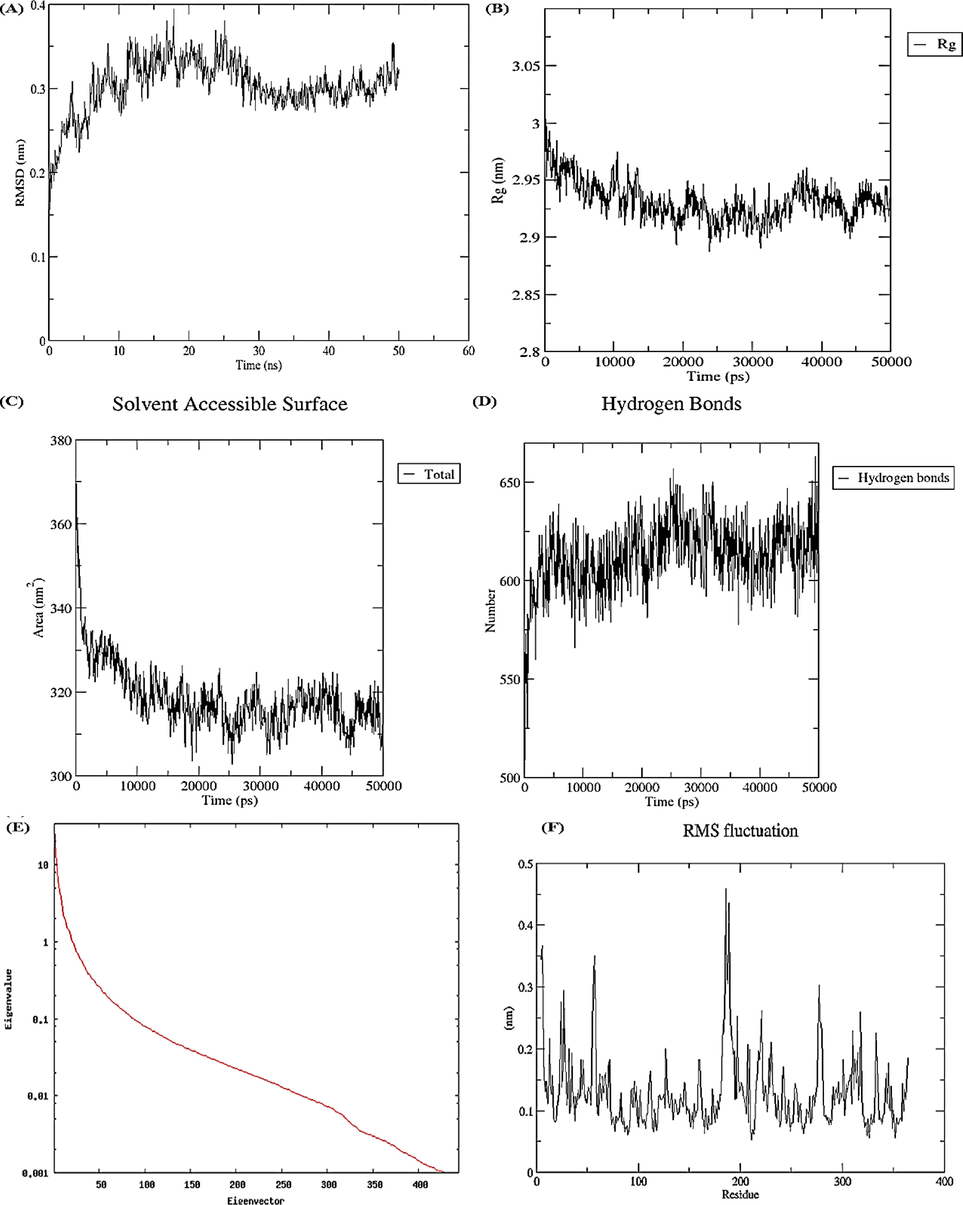
Molecular dynamic simulations via WebGro (A) RMSD (B) Rg (C) SASA (D) Hydrogen bonds (E) Eigenvalues (F) RMSF.
In this study, the Cα backbone of IL-6 showed an RMSD value of 0.31 ± 0.02 Å (Fig. 6A) and Rg was 1.64 ± 0.01 Å (Fig. 6B) respectively. In this study, the SASA (Å2) value was 92.39 ± 2.90 (Fig. 6C). In this study, the relative number of hydrogen bonds were 136.1 ± 6.94 as shown in Fig. 6D. The first eigenvalues of IL-6 protein showed the value of 91.23 Å2 whereas total variance was 970.13 Å2 (Fig. 6E). The RMSD of IL-6 protein is represented by Fig. 6F with fluctuating residues shown by the peaks.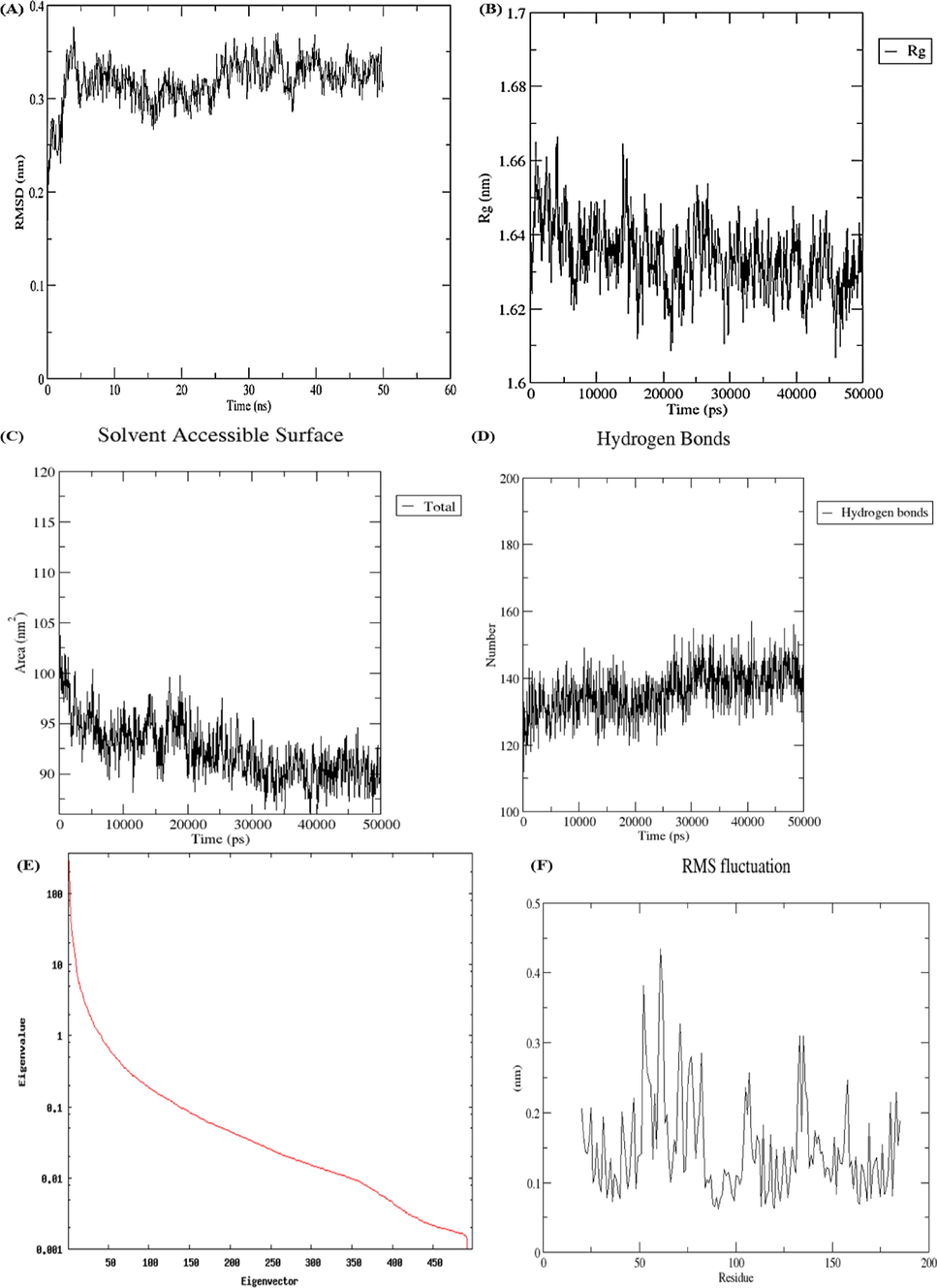
Molecular dynamic simulations via WebGro (A) RMSD (B) Rg (C) SASA (D) Hydrogen bonds (E) Eigenvalues (F) RMSF.
In the near physiological conditions, the insights on the dynamics of TNF-α and IL-6 proteins on a multi-nanosecond scale were provided by WebGro server. The information obtained from these studies could be used for many purposes including drug-design processes, and evolutionary and biophysical studies. RMSD values give insights into protein stability and flexibility by comparing reference protein molecules concerning corresponding structures (Schreiner et al., 2012). RMSD measures the difference between the protein backbones from their initial conformational structure to their final position. During the dynamic simulation process, information about protein stability concerning its relative conformation is provided by deviations. The smaller the deviations, the more stable will be the protein conformation whereas higher values depict more instability. The compactness of protein molecules can be assessed by Rg values with higher values depicting stable conformations. SASA measures the protein area that is sufficiently visible to make interactions with the neighbouring molecules of solvent. In determining the stability of proteins and folding analysis, SASA is considered an important factor (Miller et al., 1987). During simulation analysis, increased SASA values in the protein molecule represent structural relaxation and therefore decreased protein stability (Damjanovic et al., 2021). In the present study, TNF-α formed higher number of hydrogen bonds as compared to IL-6 protein. In the process of drug development, hydrogen bonding has a significant impact on drug specificity as it provides vital insights about protein–ligand interaction. The eigenvalues indicate the motion stiffness and its value is directly proportional to the required energy to deform protein conformation. Smaller eigenvalues represent easier deformation whereas higher values indicate more variance. In addition, RMSF scores of TNF-α and IL-6 proteins plotted against each residue did not reveal substantial residual fluctuations. Moreover, the peaks in the plots represent the areas of the proteins that fluctuated during the simulation process.
4.7 b2b analysis
In this study, the backbone DynaMine values greater than 0.8 suggest rigid conformations, scores below 0.69 depict flexible areas and values greater than 1 represent membrane-spanning regions. The scores ranging between 0.69 and 0.80 are content dependent and proficient enough to be either flexible or rigid. The higher sidechain DynaMine dynamics values indicate the rigid structure and these values are highly dependent on the type of amino acid. The higher DynaMine conformational (helix, coil, sheet, ppII) scores indicate higher propensities.
In the present study, EFoldMine early folding propensity gives insights into protein folding with residue values greater than 0.169 and predicts the start of the folding process depending upon indigenous interactions with other amino acids. In the present study, the scores obtained from Disomine disorder analysis determine that values greater than 0.5 predict disordered residues.
In this study, PSP (Phase Separating Protein) determines whether the protein is likely to be phase-separated with a specific mechanism that involves interaction with RNA. This analysis provides scores that highlight protein regions that are involved mechanistically. In this study, both TNF-α and IL-6 had a PSP score of 0.27 and 0.23 indicating the proteins were likely not a phase separating protein.
In the present study, Supplementary Fig. S2 and Supplementary Fig. S3 represent the findings of TNF-α and IL-6 protein obtained from different servers of b2b tools. Supplementary Fig. 2A and Supplementary Fig. 3A illustrate the structure prediction of TNF-α and IL-6 estimated via multiple online tools. Similarly, Supplementary Fig. 2B and Supplementary Fig. 3B represent the findings of TNF-α and IL-6 obtained via the PSP server.
5 Conclusion
Rheumatoid arthritis is a severe immune-mediated disorder in which the immune system of the host destroys the healthy tissues in the body. The findings of our study suggest that staurosporine and withanoside L could be developed as potential inhibitors that can target TNF-α and IL-6 proteins. Lipinski’s rule is important to determine the ADMET properties of bioactives and in our study rutin, withanoside II and withanoside IV did not obey RO5. CASTp revealed the top binding pocket in TNF-α and IL-6 protein along with their accessible surface area and volume. Homology modelling predicted 90.2% residues found in the most favoured regions. fastDRH calculated the rescoring profiles of the protein–ligand complexes with TNF-α rutin and IL-6 rutin complex with highest affinity profile scores. The insights about MM/PB(GB)SA were provided by fast DRH. WebGro determined the molecular dynamics of the proteins and revealed the behaviour of protein molecules. b2b studies provided insights about the rigidity, flexibility, protein folding and phase-separating properties of protein molecules. Through these in silico studies, significant insights were gained that could prove useful for the further development of novel inhibitors for rheumatoid arthritis.
CRediT authorship contribution statement
Rana M Aldossari: Writing – original draft. Aarif Ali: Writing – review & editing, Conceptualization, Writing – original draft, Methodology, Software, Formal analysis, Visualization, Supervision. Summya Rashid: Writing – review & editing. Muneeb U Rehman: Writing – review & editing, Formal analysis, Supervision. Sheikh Bilal Ahmad: Writing – review & editing, Supervision. Bashir Ahmad Malla: Writing – review & editing.
Acknowledgement
The authors acknowledge the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number (IF2/PSAU/2022/03/22951).
Funding
The authors extend their appreciation to the Deputyship for Research and Innovation, “Ministry of Education” in Saudi Arabia for funding this research (IF2/PSAU/2022/03/22951).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- TLR4-targeting therapeutics: structural basis and computer-aided drug discovery approaches. Molecules. 2020;25(3):627.
- [Google Scholar]
- Computational approaches for identification of potential plant bioactives as novel G6PD inhibitors using advanced tools and databases. Molecules. 2023;28(7):3018.
- [Google Scholar]
- In silico analysis and molecular docking studies of natural compounds of Withania somnifera against bovine NLRP9. J. Mol. Model.. 2023;29:171.
- [Google Scholar]
- Biochemical and computational assessment of acute phase proteins in dairy cows affected with subclinical mastitis. Curr. Issues Mol. Biol.. 2023;45(7):5317-5346.
- [Google Scholar]
- Alqahtani S (2017). In silico ADME-Tox modeling: progress and prospects Expert Opin Drug Metab Toxicol 13 1147–1158 doi: 101080/1742525520171389897.
- Alsaffar, R. M., Ali, A., Rashid, S. M., Ahmad, S. B., Alkholifi, F. K., Kawoosa, M. S.,. & Rehman, M. U. (2023). Zerumbone Protects Rats from Collagen-Induced Arthritis by Inhibiting Oxidative Outbursts and Inflammatory Cytokine Levels. ACS Omega.
- Serum levels of interleukin-6 are linked to the severity of the disease caused by Andes Virus. PLoS Negl. Trop. Dis.. 2017;11(7):e0005757.
- [Google Scholar]
- Caldwell G W, Yan Z, Tang W, Dasgupta M, and Hasting B (2009). ADME optimization and toxicity assessment in early- and late-phase drug discovery Curr Top Med Chem 9 965–980 doi: 102174/156802609789630929,
- Chen Z, Bozec A, Ramming A, and Schett G (2019). Anti-inflammatory and Immune-Regulatory Cytokines in Rheumatoid Arthritis Nat Rev Rheumatol 159–17 doi:101038/s41584-018-0109-2.
- Assessing the performance of the MM/PBSA and MM/GBSA methods 6 Capability to predict protein–protein binding free energies and re-rank binding poses generated by protein–protein docking. PCCP. 2016;18(32):22129-22139.
- [Google Scholar]
- Cheng F, Li W, Zhou Y, Shen J, Wu Z, Liu G, & Tang Y (2012). admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties.
- Zerumbone ameliorates neuropathic pain symptoms via cannabinoid and ppar receptors using in vivo and in silico models. Molecules. 2021;26(13):3849.
- [Google Scholar]
- Deciphering dimerization modes of PAS domains: computational and experimental analyses of the AhR: ARNT complex reveal new insights into the mechanisms of AhR transformation. PLoS Comput. Biol.. 2016;12(6):e1004981.
- [Google Scholar]
- Daina A, Michielin O, & Zoete V (2017). SwissADME: a free web tool to evaluate pharmacokinetics drug-likeness and medicinal chemistry friendliness of small molecules Scientific reports 7(1) 1-13.
- Elucidating solution structures of cyclic peptides using molecular dynamics simulations. Chem. Rev.. 2021;121(4):2292-2324.
- [Google Scholar]
- del Solar V, Lizardo D Y, Li N, Hurst J J, Brais C J, & Atilla-Gokcumen G E (2015). Differential regulation of specific sphingolipids in colon cancer cells during staurosporine-induced apoptosis Chemistry & biology 22(12) 1662-1670.
- Hepatoprotective effect of withanolide-rich fraction in acetaminophen-intoxicated rat: decisive role of TNF-α IL-1β COX-II and iNOS. Pharm. Biol.. 2016;54(11):2394-2403.
- [Google Scholar]
- Cytokine response in Crimean-Congo hemorrhagic fever virus infection. J. Med. Virol.. 2017;89(10):1707-1713.
- [Google Scholar]
- TNF defined as a therapeutic target for rheumatoid arthritis and other autoimmune diseases. Nat. Med.. 2003;9(10):1245-1250.
- [Google Scholar]
- Felson D T, Niu J, Neogi T, Goggins J, Nevitt M C, Roemer F, & Group M I (2016). Synovitis and the risk of knee osteoarthritis: the MOST Study Osteoarthritis and cartilage 24(3) 458-464.
- Ferreira L L G, and Andricopulo A D (2019). ADMET modeling approaches in drug discovery Drug Discov Today 24 1157–1165 doi: 101016/jdrudis201903015.
- Exploration of potential targets and mechanisms of Naringenin in treating autism spectrum disorder via network pharmacology and molecular docking. Medicine. 2022;101(46)
- [Google Scholar]
- Protein kinase inhibition of clinically important staurosporine analogues. Nat. Prod. Rep.. 2010;27(4):489-498.
- [Google Scholar]
- Effects of a standardized extract of Withania somnifera (Ashwagandha) on depression and anxiety symptoms in persons with schizophrenia participating in a randomized placebo-controlled clinical trial. Ann. Clin. Psychiatry. 2019;31(2):123-129.
- [Google Scholar]
- The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov.. 2015;10(5):449-461.
- [Google Scholar]
- Cardiovascular risk in inflammatory arthritis: rheumatoid arthritis and gout. Lancet Rheumatol.. 2021;3(1):e58-e70.
- [Google Scholar]
- Hyrich K L, Lunt M, Watson K D, Symmons D P M, and Silman A J (2007). Outcomes after Switching from One Anti-tumor Necrosis Factor α Agent to a Second Anti-tumor Necrosis Factor α Agent in Patients with Rheumatoid Arthritis: Results from a Large UK National Cohort study Arthritis Rheum 56 13–20 doi:101002/art22331.
- Medicinal plants used by traditional healers of Punjab Pakistan. Canad. J. Appl. Sci.. 2013;3:113-132.
- [Google Scholar]
- Serum biomarkers and changes in clinical/MRI evidence of golimumab-treated patients with ankylosing spondylitis: results of the randomized placebo-controlled GO-RAISE study. Arthrit. Res. Ther.. 2016;18(1):1-7.
- [Google Scholar]
- Kamimura D, Arima Y, Hirano T, Ogura H, & Murakami M (2014). IL-6 and inflammatory diseases In Cytokine Frontiers (pp 53-78) Springer Tokyo.
- Khalid S, Ullah MZ, Khan AU, Afridi R, Rasheed H, Khan A, & Khan S (2018). Antihyperalgesic properties of honokiol in inflammatory pain models by targeting of NF-κB and Nrf2 signaling Frontiers in pharmacology 9 140.
- Kleywegt G J, & Jones T A (1996). Phi/psi-chology: Ramachandran revisited Structure 4(12) 1395-1400.
- Effect of curcumin on rheumatoid arthritis: a systematic review and meta-analysis. Front. Immunol.. 2023;14:1121655.
- [Google Scholar]
- Phytoremedial effect of Withania somnifera against arsenic-induced testicular toxicity in Charles Foster rats. Avicenna J. Phytomed.. 2015;5(4):355.zx.
- [Google Scholar]
- Langade D, Kanchi S, Salve J, Debnath K, & Ambegaokar D (2019). Efficacy and safety of Ashwagandha (Withania somnifera) root extract in insomnia and anxiety: a double-blind randomized placebo-controlled study Cureus 11(9).
- The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol.. 1971;55(3):379-IN4.
- [Google Scholar]
- Crystal structures of zinc-free and -bound heme domain of human inducible nitric-oxide synthase. J. Biol. Chem.. 1999;274:21276-21284.
- [Google Scholar]
- Anatomy of protein pockets and cavities: measurement of binding site geometry and implications for ligand design. Protein Sci.. 1998;7(9):1884-1897.
- [Google Scholar]
- Surface properties of adipocyte lipid-binding protein: Response to lipid binding and comparison with homologous proteins. Proteins: Struct. Funct. Bioinform.. 1998;33(4):577-589.
- [Google Scholar]
- Lead-and drug-like compounds: the rule-of-five revolution. Drug Discovery Today: Technol.. 2004;1(4):337-341.
- [Google Scholar]
- Insights into molecular docking and dynamics to reveal therapeutic potential of natural compounds against P53 protein. J. Biomol. Struct. Dyn. 2022:1-20.
- [Google Scholar]
- Mcinnes IB, and Schett G (2011). The Pathogenesis of Rheumatoid Arthritis N Engl J Med 365 2205–2219 doi:101056/nejmra1004965.
- Autoimmunity of the lung and oral mucosa in a multisystem inflammatory disease: The spark that lights the fire in rheumatoid arthritis? J. Allergy Clin. Immunol.. 2016;137(1):28-34.
- [Google Scholar]
- Tumor necrosis factor-alpha and the roles it plays in homeostatic and degenerative processes within the central nervous system. J. Neuroimmune Pharmacol.. 2012;7(1):42-59.
- [Google Scholar]
- Myasoedova E, Davis J, Matteson E L, and Crowson C S (2020). Is the Epidemiology of Rheumatoid Arthritis Changing? Results from a PopulationBased Incidence Study 1985-2014 Ann Rheum Dis 79 440–444.
- Nasimi Doost Azgomi R, Zomorrodi A, Nazemyieh H, Fazljou S M B, Sadeghi Bazargani H, Nejatbakhsh F, & Ahmadi AsrBadr Y (2018). Effects of Withania somnifera on reproductive system: a systematic review of the available evidence BioMed Research International 2018.
- Efficacy of Ashwagandha (Withania somnifera L) root extracts in preventing stress induced testicular damage in rat. Eur. J. Biomed. Pharma. Sci.. 2015;2(7):413-424.
- [Google Scholar]
- Design, synthesis, molecular docking, anti-quorum sensing, and anti-biofilm activity of pyochelin-zingerone conjugate. Drug Dev. Res.. 2021;82(4):605-615.
- [Google Scholar]
- Studies of the ligand binding reaction of adipocyte lipid binding protein using the fluorescent probe 1 8-anilinonaphthalene-8-sulfonate. Biophys. J.. 1999;77(2):1107-1116.
- [Google Scholar]
- Common protein architecture and binding sites in proteases utilizing a Ser/Lys dyad mechanism. Protein Sci.. 1999;8(11):2533-2536.
- [Google Scholar]
- Patel C N, Kumar S P, Rawal R M, Patel D P, Gonzalez F J, and Pandya H A (2020). A multiparametric organ toxicity predictor for drug discovery Toxicol Mech Methods 30 159–166 doi: 101080/153765162019 1681044.
- Exploratory study to evaluate tolerability safety and activity of Ashwagandha (Withania somnifera) in healthy volunteers. J. Ayurveda Integr. Med.. 2012;3(3):111.
- [Google Scholar]
- In Silico molecular docking and dynamic analysis of natural compounds against major non-structural proteins of SARS-COV-2. J. Biomol. Struct. Dyn. 2022:1-17.
- [Google Scholar]
- Safiri S, Kolahi A A, Hoy D, Smith E, Bettampadi D, Mansournia M A, et al (2019). Global Regional and National burden of Rheumatoid Arthritis 1990- 2017: a Systematic Analysis of the Global Burden of Disease Study 2017 Ann Rheum Dis 78 1463–1471.
- Scherer H U, Häupl T, and Burmester G R (2020). The Etiology of Rheumatoid Arthritis J Autoimmun 110 102400 doi:101016/jjaut2019102400.
- Schreiner W, Karch R, Knapp B, & Ilieva N (2012). Relaxation estimation of RMSD in molecular dynamics immunosimulations Computational and mathematical methods in medicine 2012.
- Schulz M, Dotzlaw H, and Neeck G (2014). Ankylosing Spondylitis and Rheumatoid Arthritis: Serum Levels of TNF-Alpha and its Soluble Receptors during the Course of Therapy with Etanercept and Infliximab Biomed Res Int 2014 675108 doi:101155/2014/675108.
- Sinha S, Doble M, & Manju S L (2018). Design synthesis and identification of novel substituted 2-amino thiazole analogues as potential anti-inflammatory agents targeting 5-lipoxygenase European journal of medicinal chemistry 158 34-50.
- Lorlatinib in patients with ALK-positive non-small-cell lung cancer: results from a global phase 2 study. Lancet Oncol.. 2018;19(12):1654-1667.
- [Google Scholar]
- Staurosporine and conventional anticancer drugs induce overlapping yet distinct pathways of apoptosis and caspase activation. Oncogene. 2001;20(10):1193-1202.
- [Google Scholar]
- Assessing the performance of MM/PBSA and MM/GBSA methods. 4. Accuracies of MM/PBSA and MM/GBSA methodologies evaluated by various simulation protocols using PDBbind data set. PCCP. 2014;16(31):16719-16729.
- [Google Scholar]
- Tanaka T, Narazaki M, & Kishimoto T (2014). IL-6 in inflammation immunity and disease Cold Spring Harbor perspectives in biology 6(10) a016295.
- AutoDock Vina: improving the speed and accuracy of docking with a new scoring function efficient optimization and multithreading. J. Comput. Chem.. 2010;31(2):455-461.
- [Google Scholar]
- Uddin Q, Samiulla L, Singh V K, & Jamil S S (2012). Phytochemical and pharmacological profile of Withania somnifera Dunal: a review Journal of Applied Pharmaceutical Science (Issue) 170-175.
- Interleukin-6 as a potential biomarker of COVID-19 progression. Med. Maladies Infect.. 2020;50(4):382.
- [Google Scholar]
- Poncirin attenuates CCL4-induced liver injury through inhibition of oxidative stress and inflammatory cytokines in mice. BMC Complement. Med. Ther.. 2020;20(1):1-14.
- [Google Scholar]
- Role of extracellular matrix in regulation of staurosporine-induced apoptosis in breast cancer cells. Oncol. Rep.. 2005;13(4):745-750.
- [Google Scholar]
- Antioxidant, anti-inflammatory, and multidrug resistance modulation activity of silychristin derivatives. Antioxidants. 2019;8(8):303.
- [Google Scholar]
- Wang Y, Xing J, Xu Y, Zhou N, Peng J, Xiong Z, et al (2015). In silico ADME/T modelling for rational drug design Q Rev Biophys 48 488–515 doi: 101017/S0033583515000190.
- The effects of dopamine receptor 2 expression on B cells on bone metabolism and TNF-α levels in rheumatoid arthritis. BMC Musculoskelet. Disord.. 2016;17(1):1-7.
- [Google Scholar]
- Wijbrandts C A, Dijkgraaf M G W, Kraan M C, Vinkenoog M, Smeets T J, Dinant H, et al (2008). The Clinical Response to Infliximab in Rheumatoid Arthritis Is in Part Dependent on Pretreatment Tumour Necrosis Factor Expression in the Synovium Ann Rheum Dis 67 1139–1144 doi:101136/ ard2007080440.
- Anti-tumor activity of staurosporine in the tumor microenvironment of cervical cancer: An in vitro study. Life Sci.. 2015;133:21-28.
- [Google Scholar]
- Yang Z Y, Yang Z J, Dong J, Wang L L, Zhang L X, Ding J J, et al (2019). Structural analysis and identification of colloidal aggregators in drug discovery J Chem Inf Model 59 3714–3726 doi: 101021/acsjcim9b00541.
- admetSAR 20: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics. 2019;35(6):1067-1069.
- [Google Scholar]
- Zhang X, Perez-Sanchez H, & C Lightstone F (2017). A comprehensive docking and MM/GBSA rescoring study of ligand recognition upon binding antithrombin Current topics in medicinal chemistry 17(14) 1631-1639.
- Anti-inflammatory and immunoregulatory effects of paeoniflorin and total glucosides of paeony. Pharmacol. Ther.. 2020;207:107452
- [Google Scholar]
- Zheng J, Shi Y, Xiong L, Zhang W, Li Y, Gibson P G, & Wang F (2017). The expression of IL-6 TNF-α and MCP-1 in respiratory viral infection in acute exacerbations of chronic obstructive pulmonary disease Journal of immunology research 2017.
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2023.105200.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary data 1
Supplementary data 1







