Translate this page into:
A novel fluorescent sensor based on aptamer recognition and DNA walker amplification strategy and its determination of 17β-estradiol
⁎Corresponding author at: School of Public Health, Hebei Medical University, Shijiazhuang 050017, PR China. lingmei622@hotmail.com (LingMei Niu)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
17β-Estradiol (E2) is a common environmental endocrine and excessive estrogen will disturb the endocrine balance of human body and lead to diseases because of its accumulation in human body. The main aim of this study is to determine the trace amount of E2 in various foods. Based on aptamer specific recognition, DNA walker molecular machine and Exonuclease III-assisted dual cycle signal amplification technology, a fluorescent aptamer sensor was constructed and applied to the detection of 17β-estradiol. Atomic force microscopy (AFM) and polyacrylamide gel electrophoresis (PAGE) were used to verify the feasibility of DNA walker participating in the reaction. Under the optimal conditions, 17β-estradiol concentration showed a good linear relationship with fluorescence in the range of 5.0 × 10−10–1.8 × 10−8 M, and the detection limit was 5.6 × 10−11 M. The fluorescent aptamer sensor established in this study has good selectivity, simple operation and good specificity, and the results of its application in the determination of milk samples are satisfactory. Research findings revealed that the E2 concentration in water and milk is less than the safe limits, indicating the foods in the market are safe and can be consumed. Though the foods are safe in present content, the health risk still exists because of continuous intake of trace E2.
Keywords
Aptamer
Fluorescence sensor
DNA walker
Exonuclease III
17β-estradiol
1 Introduction
The use of various water resources for agricultural purposes has caused a serious problem in developing countries. For example, Professor Zafar Iqbal Khan evaluated the nickel in agricultural irrigation water that shows toxicity and negative health impacts on people. (Iqbal Khan et al., 2023) According to the statistics, the emission of hormones in China, such as 17β-estradiol, progesterone, estriol, and estrone, greatly exceeds that of the European Union (Tian et al., 2022), which indicated the serious pollution in some natural water in China. 17β-estradiol (E2) is a common environmental endocrine. Since it is widely used in animal husbandry to promote the growth of livestock and increase milk production, E2 may exist in surface water from animal excrement and industrial wastewater (Nameghi et al., 2019). On the other hand, E2 is also a natural estrogen, which has a strong biological activity and can regulate the human reproductive system and cardiovascular system (Yang and Xu 2022). Studies have shown that excessive estrogen will disturb the endocrine balance of human body and leads to diseases, such as male infertility and breast cancer (Zhang et al., 2021). E2 is easily absorbed by the human body through the food chain, and even low concentration of E2 can have a big effect on the human body due to the bio-accumulation. Recently, E2 has been detected in animal-derived foods such as pork, chicken, grass carp, eggs, milk and other dairy products (Wang et al., 2021). Therefore, it is very necessary to develop various detection methods suitable for lower concentrations of E2.
At present, the main detection methods of E2 include gas/liquid chromatography (GC/LC) (Shi et al., 2011), high performance liquid chromatography-mass spectrometer (HPLC-MS) (Tai and Welch 2005, Matějíček and Kubáň 2008), electrochemical sensor method (ECS) (Hao et al., 2022) and surface-enhanced Raman spectroscopy (SERS) (Pu et al., 2019a, 2019b), etc. Among of them, GC/LC and HPLC-MS cost more and require complex sample pretreatments (Pu et al., 2019a, 2019b). The sample preparation and detection process of ECS and SERS is time-consuming, and some studies have shown that E2 has weak electrochemical activity and high oxidation potential, which may cause electrode passivation, resulting in poor specificity and repeatability (Hu et al., 2020). However, fluorescence analysis has attracted more attention due to its advantages of high sensitivity, environmental friendliness and simple operation, and some studies have combined fluorescence method with aptamer-based nucleic acid amplification method, showing higher sensitivity (Yang et al., 2020, Huang et al., 2021).
Aptamer is a single-stranded oligonucleotide isolated from random sequence nucleic acid library by exponential enrichment method (SELEX), which has high affinity and specificity for a variety of target molecules, such as ions, small molecules, peptides and cells(Li et al., 2022). Aptamer can bind specifically to the target through hydrogen bonding, electrostatic interaction, van der Waals force or shape complementation, similar to antibody-antigen reactions, so they are often referred to as “chemical antibodies”(Zhao et al., 2021). Compared with antibodies, aptamers have the advantages of short SELEX process, small difference between production batches, and easy preservation and transportation (Cai et al., 2021), so they are often used to construct various biosensors. In order to further improve the sensitivity of the fluorescent aptamer sensor, researchers combined it with various cyclic amplification methods, such as polymerase chain reaction(Wang et al., 2011), “rolling ring” amplification reaction (RCA) (Tang et al., 2016), hybrid chain reaction (HCR) (Wang et al., 2016), and chain replacement amplification (SDA) (Zhou et al., 2017), etc. However, these methods still have some limitations. For example, the electron transfer rate of amplified DNA structure is slow, leading to non-specific enzyme digestion reaction and low catalytic cycle turnover (Ji et al., 2017).
In recent years, various DNA nanomachine have been developed, including DNA tweezers, DNA motors, and DNA walker (Zhao et al., 2019). Due to the programmable nature of its structure and function, DNA walker can achieve precise mechanical movement on the nanoscale along the specified two-dimensional or three-dimensional orbit (Lv et al., 2020). It is accompanied by changes in DNA conformation (Yin et al., 2020). Typically, DNA walker is initiated externally and activated by a bridge-burning mechanism through a series of energy transformations, such as enzyme cycle reactions or chain replacement reactions (SDA), which introduce a small number of targets and trigger a large number of signaling probes to achieve a cascade of amplification reactions. DNA walker is mainly composed of three parts: DNA walking strand, DNA trace and the energy that drives the walking chain to move (Jiang et al., 2021). The fluorescent aptamer sensor based on DNA walker is characterized by good flexibility, high controllability, good mobility (Huang et al., 2020), and sequence predictability and programmability (Zhang et al., 2019). Therefore, it has attracted wide attention, providing a new way to improve the detection sensitivity. Currently, researchers have used DNA origami and electrodes as the running track of DNA walker, which has the characteristics of flexibility and high addressability. However, nanomachines made from these materials are expensive, complex in design, and take a long time to prepare. Therefore, a low-cost and easily available material is needed to prepare nanomachines.
Graphene oxide (GO) is a two-dimensional nanomaterial, which is a 2D lattice composed of graphitized carbon atoms with a dense array of honeycomb hexagonal patterns (Adegoke et al., 2020). It has advantages such as low price, large specific surface area, good bio-compatibility, good dispersion in water and strong fluorescence quenching ability (Kou et al., 2021). It can interact with aptamer labeled with fluorescent groups by π-π stacking, thus transferring fluorescence resonance energy (FRET) to GO in the absence of the target. In the presence of the target, fluorescence is recovered, and GO has been widely used in the development of biosensors due to this advantage(Chen et al., 2022, Zhang et al., 2022). In addition, studies have shown that using GO as a DNA carrier and an effective energy receptor is conducive to the construction of a DNA walker based on energy transfer with low cost, short running time and low background signal (Liang et al., 2022).
Exonuclease III (Exo III) -assisted cycling is often used for nucleic acid signal amplification. It can progressively catalyze the cleavage of the concave or blunt 3′-hydroxyl terminus of double-stranded DNA, but has limited activity against the 3′-protruding ends of single-stranded DNA and double-stranded DNA. Compared with incision exonucleases, Exo III does not need to recognize specific nucleotide sequences, which makes Exo III-based DNA walker easier to design. Moreover, Exo III auxiliary signal amplification technology has universality and good repeatability (Qiang et al., 2015), which can be combined with DNA walker to improve the sensitivity of the sensor.
Considering the problems that most of the existing methods are single cycle amplification or rolling loop amplification, which showed long reaction time, low detection efficiency, and poor sensitivity, a new strategy of double amplification was innovated in this study. Therefore an ultra-sensitive and efficient fluorescence detection method for E2 was established based on aptamer, DNA walker and Exo III auxiliary signal amplification method. This method possesses the following four advantages: First, in the presence of the target object, the system can release two cycle initiation factors and start two signal cycle amplification reactions at the same time, which greatly improves the detection efficiency. The experimental results show that compared with the single cycle detection, the time used to determine a sample is reduced from the original 3 h to 1 h, and the efficiency is increased by 66.7 %. Secondly, the introduction of DNA walker solves the problems of unpredictable cycle process and disorderly hybridization of primers, which reduces the probability of autonomous mismatching of primers in the reaction. Thirdly, if DNA origami technology is used, the construction of such nanomachines requires hybridization of at least one long single-stranded deoxynucleotide scaffold and hundreds of short oligonucleotides (Liang et al., 2022), and the reaction cost is high. In this experiment, the running platform of DNA walker prepared by GO and dsDNA can reduce the cost by at least 50 %. Fourthly, due to the presence of quenching groups black hole quencher1 (BHQ1) and GO, the fluorescence signal of FAM group was doubly quenched, that is, FAM group was the only energy donor and both GO and BHQ1 were energy receptors, forming a dual energy transfer system to achieve fluorescence resonance energy transfer and greatly reducing the background signal value. As can be seen from the above, this method has shown great advantages in terms of detection cost, detection efficiency, detection sensitivity and reduction of background value. Compared with the reported methods, this system has the advantages of low detection cost, short detection time, high detection sensitivity and strong specificity, and shows good accuracy in the determination of E2 in actual samples. The above results show that the system will have important value and broad application prospect.
2 Material and methods
2.1 Chemical and material
17β-estradiol was purchased from Aladdin Biochemical Technology (Shanghai) Co., Ltd., Exonuclease III (Exo III) was purchased from Sangon Biotechnology (Shanghai) Co., Ltd., enzyme-free water (DEPC water) was purchased from Labgic (Beijing) Co., Ltd., and single-layer graphene oxide sheet was purchased from Xianfeng Nanomaterials Technology (Jiangsu) Co., Ltd. 10 × TBE buffer (500 mL) was purchased from Solaibio Technology (Beijing) Co., Ltd. The DNA non-denatured PAGE electrophoresis kit, nucleic acid rapid silver staining kit and 20 bp DNA ladder were purchased from Real-Times (Beijing) Biotechnology Co., Ltd. The reagents used in this experiment are all analytical grade reagents. All the synthesized oligonucleotides were from Sangon Biotechnology (Shanghai) Co., Ltd., and the sequences were as follows:
HP1:
5′-GGGTGGCTTCCAGCTTATTGAATTACACGCAGAGGGTAGCGGCTCTGCGCATTCAATTGCTGCGCGCTGAAGCGCGGAAGCCACCCTTCCGTTTT-3′ (The bold part in italics is the aptamer sequence of E2).
HP2:
5′-CCTGGTTTCAGTTATATGGATTTTTTTAACTGAAACCAGGCGGAAGGGTG-3′(the underline of HP1 and HP2 is the complementary bases).
BHQ1-DNA:
5′-BHQ1 -TTTCAGTTATATGGACTTCAAAAAAAAAAAAAAAAAAAA-3′ (The bold part complements with that of the FAM-DNA).
FAM-DNA:
5′-AGAGAAGTCCATATAACTGAAACCAGG- FAM-3′ (the curve part complements with that of HP2).
2.2 Apparatus
Fluorescence spectrophotometer (Hitachi F-7000, Japan), sequencing electrophoresis instrument (Eco-Mini, Biometra), high voltage power supply (P25T, Biometra), laboratory ultra-pure water preparation system (Q-POD), electric thermostatic incubator (ZXDP-B2120, ZXDP-B2120, Shanghai Zhicheng Analytical Instrument Manufacturing Co., Ltd.), Combined Full temperature Oscillating incubator (ZQPZ-228A, Tianjin Leibo terry Equipment Co., Ltd.), composite rotor centrifuge (Haimen Kylin-Bell Lab Instruments Co., Ltd.), Discoloring Shaker (TS-300 T, Hangzhou Miulab Instrument Co., Ltd.).
2.3 The proposed scheme and detection of E2
The efficient fluorescent sensor was designed to amplify signals by two cycles. Cycle I was composed of hairpin DNA (HP1), hairpin DNA (HP2) and Exo III. When E2 existed in detection solutions, the hairpin will be opened after aptamer of which catching the objects. Then, the other bottom of HP1 hybridized with HP2. Consequently, the hybridization of HP1 and HP2 led Exo III to cut down some strand of HP2, releasing strand S1 and the opened HP1 then went back to hybridized with HP2 again. This was the first cycle of this strategy. The second cycle was that the generated S1 in cycle I can competitively combined with double-strand DNA immobilized on graphene oxide (GO) and the double-strand DNA was composed of two single-strand DNA containing BHQ1 quencher group and fluorescent group, respectively. Then, the fluorescent DNA was cut down after the competitive combination between strand S1 and fluorescent DNA. The fluorescent probe was consequently released. The more E2 will produce more strand DNA S1 and releasing much more fluorescent probe. Based on the above, an ultrasensitive E2 biosensor was constructed.
The detection of E2 was as follows: First, dsDNA was prepared by mixing 10.0 μL FAM-DNA (8.0 μM) with 10.0 μL BHQ1-DNA (8.0 μM) and heating at 95 °C for 5 min (Liang et al., 2022). DNA walker was prepared by incubating 20.0 μL dsDNA with 20.0 μL GO (2.0 mg/mL) at 37 °C for 2 h. The operation should be carried in dark place. The methods to detect E2 were as follows: 10.0 μL HP1 solution, 100 μL E2 with different concentrations and 1.0 μL 10 × buffer enzyme buffer solution was incubated at 37 °C for 30 min; Then, HP2 solution of 10.0 μL, Exo III solution of 10.0 μL (1.5 U/μL) and the prepared DNA walker running track were added. The mixture was then incubated at 37 °C for 1 h (Ning et al., 2021) and tested by fluorescence. The detection parameters of the fluorescence spectrophotometer were 5 nm for the excitation slit and 487 nm for the excitation wavelength, 500–600 nm for the wavelength range of the fluorescence emission spectrum, 700 V for the photomultiplier light (PMT) and 25 °C for the temperature.
2.4 Polyacrylamide gel electrophoresis
In this experiment, 15 % polyacrylamide gel electrophoresis was used to characterize the reaction process of this strategy. The volume ratio of the sample and 6 × loading buffer was 1:5 to prove that the recognition of E2 and HP1 containing aptamer sequence triggered exonuclease to play the enzyme digestion role, and started the DNA walker cycle reaction process. First clean the glass plate with ultra-pure water, and then fix it with a vertical rubber fixing frame after drying. Pour the configured 15 % polyacrylamide gel into the glass plate and add 95 % ethanol liquid seal, stand for about 30 min, gel solidification, pour away the ethanol; Then pour in 4 % polyacrylamide gel, insert comb, let stand for 40 min waiting for gel. The prepared gel was fixed in the electrophoresis tank and 1 × TBE buffer was added. Then, the prepared sample and DNA marker were added to the point sampling port successively, and the parameters of the electrophoresis instrument were set as follows: voltage was 200 V, current was 25 mA, and time was 90 min. After electrophoresis, the gel was stripped and silver dyed: rinsed twice with 100 mL ultra-pure water, then added 100 mL silver dye after discarding the water, and shook for 5 min on the shaking bed; Then discard the silver dye solution and wash it quickly with ultra-pure water, add 100 mL silver dye color rendering solution, and shake it in the shaking bed for 3–10 min until the ideal strip appears; The color developing solution was discarded and 100 mL ultra-pure water was added to stop the dyeing. The experimental results were observed after the strip became stable.
2.5 Optimization of detection conditions of E2
To achieve the best signals of E2, some conditions were optimized, such as HP1 concentration, HP2 concentration, reaction temperature, DNA walker running time, amount of Exo III, the ratio of fluorescence probe to GO, the reaction of temperature, time and amount of Exo, etc.
2.6 Analysis of E2 in actual sample
In this experiment, the recovery experiment of milk and water was carried out. The milk was added into the plugged centrifuge tube, the same volume of acetonitrile was added and the solution was mixed for 5 min and centrifuged at 10,000 r/min for 10 min (4 °C). Then, the supernatant was transferred to the clean centrifuge tube and the residue was extracted once again with acetonitrile, combined with the supernatant in the centrifuge tube. The supernatant was diluted 20 times, and the milk was stored in the refrigerator at 4 °C overnight (Qiao et al., 2021).
3 Results and discussion
3.1 Detection mechanism of the fluorescent aptasensor
The fluorescent aptamer sensor designed in this study was mainly composed of DNA walker and the system of DNA walker contained graphene oxide (GO) with fluorescence quenching function, signal probe dsDNA, hairpin probe 1(HP1) containing E2 aptamer sequence, hairpin probe 2(HP2) and Exo III enzyme. First, two single-stranded DNA with fluorescence group FAM and quenching group BHQ1 respectively bound to form signal probe dsDNA, and then bound to GO through poly-A fragment (Xiao et al., 2016, Liang et al., 2022) at the 3′end of the BHQ1-DNA strand to form the running track of DNA walker. The designed hairpin DNA HP1 contained the E2 aptamer sequence and a separate designed sequence. In the presence of target E2, E2 and its aptamer can recognize and change the conformation of HP1. Sequences not involved in target recognition in HP1 can be combined with the unpaired part of the 3′end of hairpin HP2 to form a double-chain structure with a 3′end sag, which can be recognized and cut by Exo III until it reaches 4-T (Hu et al., 2012). This was the first phase of Exo III -assisted object-aptamer binding cycle amplification. The remaining part of HP2 was called S1, the walking strand of DNA walker. S1 can combine with dsDNA located on GO to form a double-stranded 3′terminal depression structure that can be cut by Exo III, so as to run DNA walker and release FAM group (Cai et al., 2023). With the increasing of the number of free fluorescent groups, the fluorescence signal gradually recovered, which was the second stage of signal amplification involving DNA walker. When E2 was absent, HP1 and HP2 existed in stable hairpin structure, and the fluorescence of FAM group was quenched by BHQ1 group and GO group. The fluorescent aptamer sensor achieved two cycles of amplification with good sensitivity and specificity, showing excellent sensitivity in real sample analysis (See Scheme 1).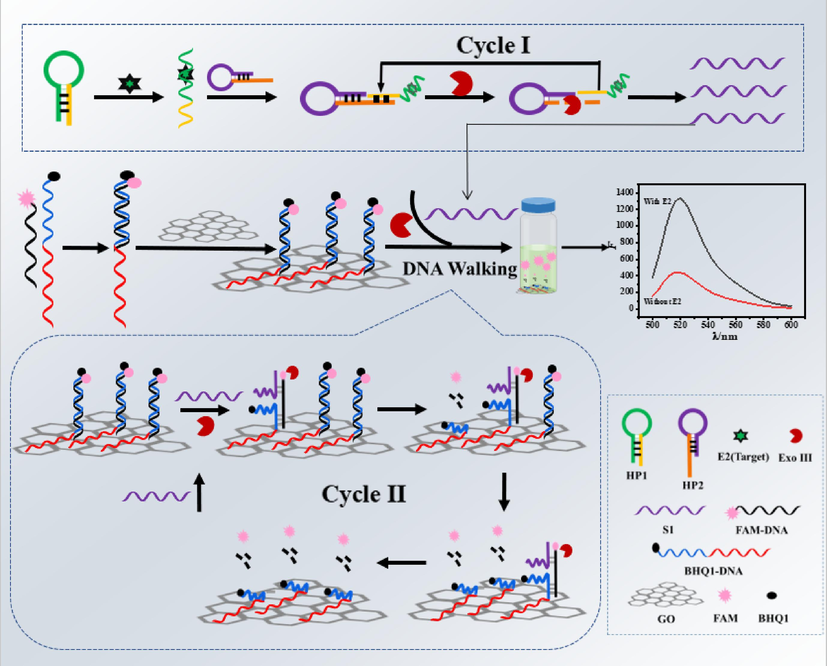
The strategy for the construction of fluorescent aptamer sensor.
3.2 Feasibility analysis
The prepared DNA walker was characterized by atomic force microscopy (AFM), and the successful assembly of DNA walker could be confirmed by comparing the thickness changes of GO before and after modification of dsDNA. Fig. 1(A) is the morphology of unmodified GO nanosheet, with a height of 1.0 nm. Fig. 1(B) is the thickness of GO surface fixed with dsDNA, reaching a height of 1.5 nm. Thus, the increased thickness indicated that dsDNA and GO has combined with each other. In addition, as shown in Fig. S1, the fluorescence spectra of FAM-DNA, dsDNA and dsDNA@GO showed that the fluorescence intensity decreased successively. In particular, the fluorescence signal dsDNA@GO complex could hardly be detected, which also proved that GO and dsDNA were bound each other. Based on this, whether GO participated in the reaction process was studied to explore the effect of double quenching strategy on the background values. The results showed that the fluorescence signal in presence of GO was 4 times higher than that in the absence of GO. Therefore, the double quenching efficiency was more effective than of single quenching group to reduce the background signal.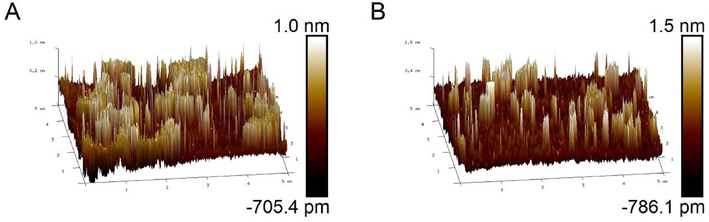
(A) Atomic Force microscopy before GO modification (B) Atomic force microscopy after GO modification of dsDNA.
In order to verify the feasibility of the aptamer sensor, the fluorescence intensity was compared with or without E2 in the reaction system. As shown in Fig. S2, when the target E2 was added to the system, the sequence specific recognition of the aptamer on E2 and HP1 changed the conformation of HP1, which was further combined with HP2 to be recognized and hydrolyzed by Exo III, starting the first round of cyclic reaction, and carrying out the second round of cyclic reaction with the addition of DNA walker. A large number of free FAM groups were obtained, and an obvious fluorescence signal was detected at 518 nm (curve a). When E2 was absent in the system, HP1 and HP2 existed in a stable hairpin structure. Under the action of BHQ1 and GO, the fluorescence signal of FAM was quenched, so only very low fluorescence intensity can be observed (curve b). Therefore, the comparison between curves a and b in the figure shows that the system has a good response to E2.
After the successful construction of DNA walker being confirmed, the feasibility of this proposed sensor was furtherly evaluated by the fluorescence in various circumstances. As shown in Fig. S3, when only dsDNA@GO existed, the system could not provide the DNA walking chain S1, so the fluorescence signal could hardly be detected (curve a). When HP1, HP2, E2 and dsDNA@GO existed in the system, the first cycle amplification could not be triggered due to the absence of Exo III in the system, consequently, S1 could not be produced either and the fluorescence signal could not be observed (curve b). Similarly, when HP1, E2, Exo III and dsDNA@GO existed in the system, the first cylce reaction could not be started because there is no HP2 to combine with HP1 and releasing S1. But because that Exo III in the system may perform non-specific cutting on the signal probe dsDNA, weak fluorescence signal could be detected (curve c). When HP2, E2, Exo III and dsDNA@GO were present in the system, a small amount of HP2 and dsDNA might react to form structures that could be cut by Exo III enzyme, thus releasing some FAM groups. Therefore, a certain fluorescence signal can be detected (curve d); Significant fluorescence signals were detected in the presence of HP1, HP2, E2, Exo III, and dsDNA@GO, indicating a cyclic amplification reaction with DNA walker running (curve e). In summary, all the substances were necessary to the whole reaction process.
Thirdly, the feasibility of the method was verified by polyacrylamide gel electrophoresis. As shown in Fig. 2, lanes1, 2, 3 and 4 are bands of HP1, HP2, FAM-DNA, and BHQ1-DNA, respectively. Lane 5 is a double-stranded DNA which was formed by annealing of FAM-DNA and BHQ1-DNA, resulting in a new band. Lane 6 is the mixture of HP1, HP2, E2, Exo III and DNA walker. After the completion of the first round of cyclic reaction, HP2 will release a large amount of S1 under the action of Exo III, and then S1 will start the second cycle of amplification. Therefore, it can be seen that as soon as the short S1 band was produced, and the DNA fragment band was generated. Compared with lane 6, target E2was not added in lane 7 and could not be specifically identified by HP1, then the first cyclic amplification could not be started, consequently S1 band was not generated. In conclusion, the feasibility of this method was proved.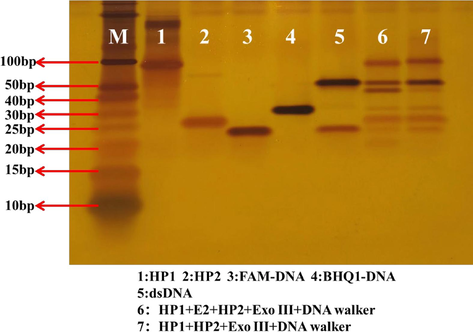
Graph of the results of polyacrylamide gel electrophoresis experiments. (1)HP1(4.0 μM);(2)HP2(4.0 μM);(3)FAM-DNA(2.0 μM);(4)BHQ1-DNA(2.0 μM);(5)dsDNA(1.0 μM);(6) HP1(4.0 μM) + E2(1.0 nM) + HP2(4.0 μM) + Exo III (20.0 U) + DNA walker;(7) HP1(4.0 μM) + HP2(4.0 μM) + Exo III (20.0 U) + DNA walker.
3.3 Optimization of reaction conditions for aptasensor
In order to obtain better analytical performance, the important influencing factors were optimized, including HP1 concentration, HP2 concentration, reaction temperature, DNA walker running time, amount of Exo III, and the ratio of fluorescence probe to GO.
The influence of HP1 concentration on the determination of E2 was investigated. As shown in Fig. S4, with the increase of HP1 concentration, the fluorescence intensity reached a peak when it reached 4.0 μM, and then stabilized. It might be that at 4.0 μM, the recognition of HP1 and E2 in the system reached saturation, and thus showed a stable trend. From above, it could be arrived at the conclusion that appropriate HP1 concentration was the guarantee of sensitivity for this method. If the concentration of HP1 was too low, the target could not be fully identified by HP1 in the system, and the HP1 participating in the cyclic amplification reaction was reduced, which reduced the sensitivity of the sensor. However, high concentration of HP1 will not increase the sensitivity of E2 detection. Further, It was not beneficial for saving the cost. Therefore, 4.0 μM was selected as the optimal HP1 concentration for further experiments.
After specific recognition of E2 and HP1, the complex bound to HP2 and initiated the first round of cyclic amplification, so HP2 was a key substance to provide the walking DNA. As shown in Fig. S5, with the increasing of HP2 concentration, the fluorescence intensity tended to be stable until it reached 10.0 μM, indicating that the first cycle reaction was fully carried out under this concentration condition. That may be due to the reason that when the concentration of HP2 was too low, the first round of cyclic amplification reaction could not be fully carried out, so that the solution containing a large amount of S1 could not be obtained. The reduction of DNA walker chain directly affected the subsequent reaction and reduced the sensitivity of the sensor. When the concentration of HP2 was too high, excess HP2 might react with the fluorescent probe on GO, so higher fluorescence intensity could be detected even when the target was not present. Therefore, 10.0 μM was selected as the optimal reaction concentration of HP2.
3.4 Effect of Exo III
Exo III not only participated in the first round of cyclic amplification, hydrolyzed HP2 to obtain a large amount of S1, but also participated in the second round of DNA walker cyclic amplification as energy supply material. Therefore, enzyme activity is an important factor affecting the sensitivity of the sensor (Ma et al., 2023). The main factors affecting enzyme activity are temperature, time and amount of Exo. As shown in Fig. S6, four temperature conditions were selected for optimization in this experiment. With the increase of temperature, the change of fluorescence intensity gradually increased, reaching the peak at 37 °C, and then the change of fluorescence intensity suddenly decreased as the temperature continued to rise.
As shown in Fig. S7, with the extension of reaction time, the change of fluorescence intensity gradually increased until it reached the peak value at 60 min, and then the change of fluorescence intensity gradually decreased, which probably because the E2 content in the system gradually decreased with the extension of reaction time. Therefore, 60 min was selected as the best reaction time. The amount of enzyme is also one of the important factors that affect the activity of enzyme. As shown in Fig. S8, with the amount of Exo III increasing, the change of fluorescence intensity gradually increased, indicating that the target object triggered more cyclic reactions, and gradually met the energy supply of DNA walker operation. When the dosage of Exo III was 15.0 U, it reached the peak and stabilized, indicating that the two-round cyclic amplification reaction had been completed. Therefore, 15.0 U was selected as the best reaction condition of Exo III.
3.5 Optimization of the ratio of fluorescence probe to GO
In this study, five DNA walkers were prepared by changing the volume ratio of fluorescence probe and GO, so as to determine the optimal detection conditions. As shown in Fig. S9, when the volume ratio between the fluorescent probe and GO was 1:1, the change in fluorescence intensity was the most obvious. It might be due to the reason of that if GO content in the system was too low, only a small number of fluorescent probes could be combined with GO, which meant that the expected fluorescence quenching effect could not be achieved. In this case, a large background value will appear in the detection of blank samples, affecting the sensitivity of the sensor. If there was too much GO content in the system, even if the two-round cyclic amplification reaction was fully carried out and a large number of free FAM groups were obtained, obvious fluorescence signals might still not be detected due to the strong fluorescence quenching ability of GO, which also affects the sensitivity of the sensor. So 1:1 was chosen as the optimal reaction condition.
3.6 Analytical performance of the E2 aptasensor
In order to verify the performance of the fluorescent aptamer sensor better, the sensor was used to measure E2 solution of different concentrations under optimal conditions, and all samples were measured in parallel for three times. As shown in Fig. 3, with the increase of E2 concentration, the change in fluorescence intensity gradually increased, which indicated that FAM groups were gradually accumulated in the reaction system, and it also proved that the sensor could be used to detect E2. In the range of 5.0 × 10−10–1.8 × 10−8 M, E2 concentration had a good linear relationship with the change of fluorescence intensity. The regression equation is ΔF = 188.6C + 1246.1 (C: nM), the correlation coefficient is 0.997, the detection limit is 5.6 × 10−11 mol/L. The performance of the fluorescent aptamer sensor in this study was compared with that of the E2 aptamer sensor reported in Table 1. Compared with electrochemical and HPLC methods, the detection limit was decreased by 100 times. Even compared with similar fluorescence analysis methods, the sensitivity was obviously improved. It can be seen that this method showed better sensitivity and lower detection limit.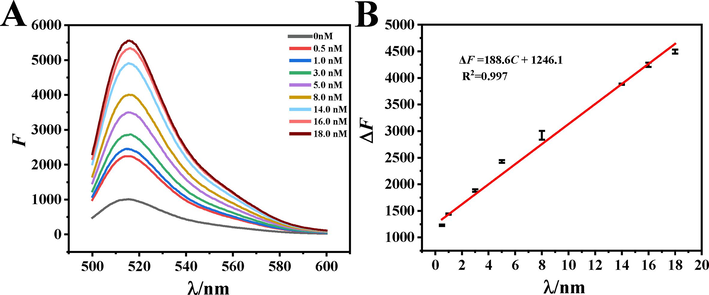
(A) Fluorescence spectra of aptamer sensors with different concentrations of E2 (B) Linear curve of E2 determination. The concentrations of HP1, HP2, Exo III and the volume ratio of dsDNA to GO were 4.0 μM, 10.0 μΜ,15.0 U and 1:1. The capture time of target is 30 min and enzyme cycle amplification temperature and reaction time were 37 °C and 60 min.
Methods
Materials
Linear range(nM)
LOD (nM)
Ref
Colorimetry
AuNP/aptamer
0.2–5
0.2
(Alsager et al., 2015)
HPLC
——
1.84–367
1.47
(Lu and Xu 2015)
Photocatalytic fuel cell (PFC) platform
ITO/SnS2/aptamer
1–500
0.12
(Yao et al., 2020)
Fluorescence
AuNP/aptamer
2–80
2
(Lin et al., 2012)
Fluorescence
FRET-based turn-on fluorescent aptasensor
0.37–36.71
0.35
(Zhang et al., 2018)
Electrochemistry
CuPc/P6LC/ Nafion film modified screen-printed electrode
80–7300
5
(Wong et al., 2019)
Fluorescence
Aptaer/quantum dot-bioconjugate
0.82–20.50
0.22
(Long et al., 2014)
Fluorescence
Aptamer/Exo III GO@dsDNA
0.5–18
0.056
This
work
3.7 Selectivity, stability and Reproducibility of fluorescent aptasensor for E2 detection
Selectivity is an important parameter to evaluate aptamer sensing platform. Multiple estrogens usually coexist in the body. Estriol (E3) and ethinylestradiol (EE) not only have similar physical and chemical properties, but also are very similar in structure to E2. Bisphenol A (BPA) and diethylstilbestrol (DES) can affect the endocrine function of human body. The change values of fluorescence intensity of various interfering agents at 10 times concentration of E2 were detected, such as E3, EE, BPA and DES. It showed that the selectivity of the proposed fluorescence aptamer sensor for E2 detection was excellent. The experimental results are shown in Fig. 4(A). The fluorescence intensity of E2 was much higher than that of other substances, which indicated that the disruptor could hardly bind specifically to HP1 and trigger subsequent amplification. The results indicated that the aptamer sensor in this study had good selectivity for E2 detection. Reproducibility is an effective method to test the precision of fluorescent aptamer sensors. As shown in Fig. 4(B), the same sample was measured in parallel using five constructed sensors, and the relative standard deviation (RSD) was1.82 %.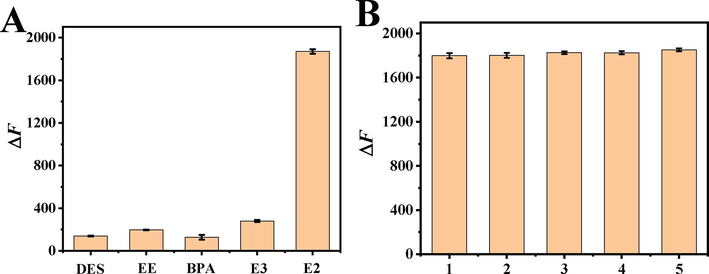
(A) Effect of different interferences on the variation value of fluorescence intensity of aptamer sensor. The concentrations of EE, BPA, DES and E3 were 20 nM. The concentrations of E2, HP1, HP2, Exo III and the volume ratio of dsDNA to GO were 2.0 nM,4.0 μM, 10.0 μΜ,15.0 U and 1:1. The capture time of target is 30 min and enzyme cycle amplification temperature and reaction time were 37 °C and 60 min.(B)Reproducibility of fluorescent aptamer sensors. The concentrations of E2, HP1, HP2, Exo III and the volume ratio of dsDNA to GO were 2.0 nM,4.0 μM, 10.0 μΜ,15.0 U and 1:1. The capture time of target is 30 min and enzyme cycle amplification temperature and reaction time were 37 °C and 60 min.
3.8 Analysis of E2 in real samples
The detection of actual samples is an important considering factor to evaluate the practical application of this proposed sensor and it was tested by standard addition method. The results were shown in Table 2. The recoveries and relative standard deviations of tap water samples were 106.81–112.00 % and 0.18–0.86 %, respectively. The recoveries and relative standard deviations of milk samples are 83.33–112.50 % and 0.10–0.85 %, respectively. It can be seen from the above that the prepared aptamer fluorescence sensor can be applied in the determination of actual samples.
Sample
Spiked amount(nM)
Found level(nM)
Recovery (%)
RSD (%)
Water
0
——
——
——
0.60
0.65
108.33
0.86
6.00
6.72
112.00
0.58
16.00
17.09
106.81
0.18
Milk
0
——
——
——
0.60
0.50
83.33
0.85
6.00
6.75
112.50
0.10
16.00
17.37
108.56
0.55
4 Conclusions
An aptamer sensor for E2 fluorescence detection was designed based on DNA walker and Exo III assisted amplification. Research findings reveal that two signal cycle amplification reactions can obviously reduce the determination time. Compared DNA origami technology, less amount of oligonucleotides cut down the cost of sample determination significantly. Further, the double quenching effect of BHQ1 and GO, decreased the background signal value and greatly improved the sensitivity. The method has good specificity, low cost, high sensitivity, simple operation and detection limit of 5.6 × 10-11 M. The sensor is used for the determination of water samples and milk samples, and the results are accurate and reliable. In addition, the sensor designed in this study can provide a platform for the detection of other targets by changing the aptamer sequence on the hair clip DNA HP1, which provides a new idea and theoretical background for the analysis and determination of targets. Therefore, the constructed biosensor was expected to have a broad development prospect in the future.
CRediT authorship contribution statement
Yajun Zhang: Conceptualization, Data curation, Writing – review & editing. Licong Jia: Methodology, Data curation, Investigation. Wei Wang: Methodology, Data curation. Meng Jiang: Methodology, Data curation. Hongying Zhang: Data curation, Formal analysis. LingMei Niu: Supervision, Writing – review & editing.
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China under grant No. 82073601.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Aptamer-based cocaine assay using a nanohybrid composed of ZnS/Ag2Se quantum dots, graphene oxide and gold nanoparticles as a fluorescent probe. Mikrochim. Acta. 2020;187:104.
- [CrossRef] [Google Scholar]
- Self-Assembled DNA Nanoflowers triggered by a DNA walker for highly sensitive electrochemical detection of Staphylococcus aureus. ACS Appl. Mater. Interfaces. 2021;13:4905-4914.
- [CrossRef] [Google Scholar]
- Navigable DNA walker coupled with exonucleases-assisted signal amplification for sensitive fluorescent detection of Staphylococcus aureus. Sens. Actuat. B: Chem.. 2023;394:134475
- [CrossRef] [Google Scholar]
- Graphene oxide and fluorescent-aptamer-based novel aptasensors for detection of metastatic colorectal cancer cells. Polymers (Basel, Switz.). 2022;14:3040.
- [CrossRef] [Google Scholar]
- Ultrasensitive detection and application of estradiol based on nucleic acid aptamer and circulating amplification technology. J. Electroanal. Chem.. 2022;913:116284
- [CrossRef] [Google Scholar]
- An ultrasensitive fluorescent aptasensor for adenosine detection based on exonuclease III assisted signal amplification. Biosens. Bioelectron.. 2012;34:83-87.
- [CrossRef] [Google Scholar]
- Rapid nondestructive detection of mixed pesticides residues on fruit surface using SERS combined with self-modeling mixture analysis method. Talanta. 2020;217:120998
- [CrossRef] [Google Scholar]
- Double-site DNA walker based ternary electrochemiluminescent biosensor. Talanta. 2020;219:121274
- [CrossRef] [Google Scholar]
- Primer-template conversion-based cascade signal amplification strategy for sensitive and accurate detection of polynucleotide kinase activity. Anal. Chim. Acta. 2021;1187:339139
- [CrossRef] [Google Scholar]
- Evaluation of nickel toxicity and potential health implications of agriculturally diversely irrigated wheat crop varieties. Arab. J. Chem.. 2023;16:104934
- [CrossRef] [Google Scholar]
- Binding-induced DNA walker for signal amplification in highly selective electrochemical detection of protein. Biosens. Bioelectron.. 2017;96:201-205.
- [CrossRef] [Google Scholar]
- A three-dimensional dynamic DNA walker-mediated branching hybridization chain reaction for the ultrasensitive fluorescence sensing of ampicillin. Analyst. 2021;146:5413-5420.
- [CrossRef] [Google Scholar]
- Selection and truncation of aptamers for ultrasensitive detection of sulfamethazine using a fluorescent biosensor based on graphene oxide. Anal. Bioanal. Chem.. 2021;413:901-909.
- [CrossRef] [Google Scholar]
- Sensitive fluorescent aptasensing of tobramycin on graphene oxide coupling strand displacement amplification and hybridization chain reaction. Int. J. Biol. Macromol.. 2022;1:1287-1293.
- [CrossRef] [Google Scholar]
- Rational fabrication of a DNA walking nanomachine on graphene oxide surface for fluorescent bioassay. Biosens. Bioelectron.. 2022;211:114349
- [CrossRef] [Google Scholar]
- Label-free aptamer-based electrochemical impedance biosensor for 17beta-estradiol. Analyst.. 2012;137:819-822.
- [CrossRef] [Google Scholar]
- Fluorescence resonance energy transfer based aptasensor for the sensitive and selective detection of 17b-estradiol using a quantum dot-bioconjugate as a nano-bioprobe. Rsc Advances.. 2014;4:2935-2941.
- [CrossRef] [Google Scholar]
- Lu, H. and Xu, S., 2015. Mesoporous structured estrone imprinted Fe3O4@SiO2@mSiO2 for highly sensitive and selective detection of estrogens from water samples by HPLC. Talanta. 144, 303-311. https://doi.org/10.1016/j.talanta.2015.06.017.
- ZIF-8-Assisted NaYF4:Yb, Tm@ZnO converter with Exonuclease III-Powered DNA walker for near-infrared light responsive biosensor. Anal. Chem.. 2020;92:1470-1476.
- [CrossRef] [Google Scholar]
- A novel CRISPR/Cas14a1-Exo III aptasensor for melamine detection coupled with systematically studied binding mechanism of truncated aptamer. Sens. Actuat. B: Chem.. 2023;374:132847
- [CrossRef] [Google Scholar]
- Enhancing sensitivity of liquid chromatographic/ion-trap tandem mass spectrometric determination of estrogens by on-line pre-column derivatization. J. Chromatogr. A. 2008;1192:248-253.
- [CrossRef] [Google Scholar]
- An ultrasensitive electrochemical sensor for 17beta-estradiol using split aptamers. Anal. Chim. Acta. 2019;1065:107-112.
- [CrossRef] [Google Scholar]
- Graphene-based fluorometric determination of agrD gene transcription in methicillin-resistant Staphylococcus aureus using exonuclease III-aided target recycling and DNA walker cascade amplification. Microchim. Acta. 2021;188
- [CrossRef] [Google Scholar]
- Recent advances in the detection of 17β-estradiol in food matrices: A review. Crit. Rev. Food Sci. Nutr.. 2019;59:2144-2157.
- [CrossRef] [Google Scholar]
- Double strand DNA functionalized Au@Ag Nps for ultrasensitive detection of 17beta-estradiol using surface-enhanced raman spectroscopy. Talanta. 2019;195:419-425.
- [CrossRef] [Google Scholar]
- A fluorescent biosensing platform based on the polydopamine nanospheres intergrating with Exonuclease III-assisted target recycling amplification. Biosens. Bioelectron.. 2015;71:143-149.
- [CrossRef] [Google Scholar]
- Truncated affinity-improved aptamers for 17β-estradiol determination by AuNPs-based colorimetric aptasensor. Food Chem.. 2021;340
- [CrossRef] [Google Scholar]
- Selective determination of trace 17beta-estradiol in dairy and meat samples by molecularly imprinted solid-phase extraction and HPLC. Food Chem.. 2011;126:1916-1925.
- [CrossRef] [Google Scholar]
- Development and evaluation of a reference measurement procedure for the determination of Estradiol-17β in human serum using isotope-dilution liquid chromatography−tandem mass spectrometry. Anal. Chem.. 2005;77:6359-6363.
- [CrossRef] [Google Scholar]
- Electrochemical strategy for ultrasensitive detection of microRNA based on MNAzyme-mediated rolling circle amplification on a gold electrode. Mikrochim. Acta. 2016;183:3061-3067.
- [CrossRef] [Google Scholar]
- Mechanism of 17β-estradiol degradation by Rhodococcus equi via the 4,5-seco pathway and its key genes. Environ. Pollut.. 2022;312:120021
- [CrossRef] [Google Scholar]
- Simultaneous detection of Salmonella enterica, Escherichia coli O157:H7, and Listeria monocytogenes using oscillatory-flow multiplex PCR. Microchim. Acta. 2011;173:503-512.
- [CrossRef] [Google Scholar]
- A colorimetric assay for Hg(II) based on the use of a magnetic aptamer and a hybridization chain reaction. Mikrochimica Acta 2016
- [CrossRef] [Google Scholar]
- A magnetic relaxation switch sensor for determination of 17beta-estradiol in milk and eggs based on aptamer-functionalized Fe3O4 @Au nanoparticles. J. Sci. Food Agric.. 2021;101:5697-5706.
- [CrossRef] [Google Scholar]
- Voltammetric determination of 17ß-estradiol in different matrices using a screen-printed sensor modified with CuPc, Printex 6L carbon and Nafion film. Microchem. J.. 2019;147:365-373.
- [CrossRef] [Google Scholar]
- Exonuclease III-assisted graphene oxide amplified fluorescence anisotropy strategy for ricin detection. Biosens. Bioelectron.. 2016;85:822-827.
- [CrossRef] [Google Scholar]
- Highly-sensitive and simple fluorescent aptasensor for 17 β-estradiol detection coupled with HCR-HRP structure. Talanta. 2022;240:123094
- [CrossRef] [Google Scholar]
- DNAzyme-powered DNA walking machine for ultrasensitive fluorescence aptasensing of kanamycin. Mikrochim. Acta. 2020;187:678.
- [CrossRef] [Google Scholar]
- Ratiometric Self-Powered Sensor for 17beta-Estradiol Detection Based on a Dual-Channel Photocatalytic Fuel Cell. Anal. Chem.. 2020;92:8026-8030.
- [CrossRef] [Google Scholar]
- DNAzyme-powered three-dimensional DNA walker nanoprobe for detection amyloid beta-peptide oligomer in living cells and in vivo. Anal. Chem.. 2020;92:9247-9256.
- [CrossRef] [Google Scholar]
- A simple FRET-based turn-on fluorescent aptasensor for 17ß-estradiol determination in environmental water, urine and milk samples. Sens. Actuators.. 2018;B.273:1648-1653.
- [CrossRef] [Google Scholar]
- Exonuclease III-powered DNA walking machine for label-free and ultrasensitive electrochemical sensing of antibiotic. Sens. Actuators B. 2019;297:126771
- [CrossRef] [Google Scholar]
- Visual-afterglow dual-mode immunochromatographic strip for 17beta-estradiol detection in milk. Talanta. 2021;232:122427
- [CrossRef] [Google Scholar]
- Development of a graphene oxide nanosheet and double-stranded DNA structure based fluorescent “signal off” aptasensor for ochratoxin A detection in malt. Food Chem.. 2022;X. 14:100308
- [CrossRef] [Google Scholar]
- Ultrasensitive detection of exosomes by target-triggered three-dimensional DNA walking machine and Exonuclease III-assisted electrochemical ratiometric biosensing. Anal. Chem.. 2019;91:14773-14779.
- [CrossRef] [Google Scholar]
- Aptamer-based fluorescent sensors for the detection of cancer biomarkers. Spectrochim. Acta A Mol. Biomol. Spectrosc.. 2021;247:119038
- [CrossRef] [Google Scholar]
- Fluorometric determination of microRNA based on strand displacement amplification and rolling circle amplification. Microchim. Acta. 2017;184:4359-4365.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2023.105340.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary data 1
Supplementary data 1







