Translate this page into:
A high-efficiency integrated strategy for the targeted discovery of novel barrigenol-type triterpenoid saponins from the shell of Xanthoceras sorbifolium Bunge by offline two-dimensional chromatography-Orbitrap mass spectrometry, neutral loss data acquisition, and predicted natural product screening
⁎Corresponding authors. zhangyi@cqidc.org.cn (Yi Zhang), mengdali@syphu.edu.cn (Dali Meng)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Natural products are considered as potential sources of leading compounds and play an important role in drug discovery. The liquid chromatography-mass spectrometry (LC-MS) technique is a powerful tool for compound-guided isolation from natural products. In this study, a high-efficiency integrated strategy was adopted to improve the new leading compounds discovery, including offline two-dimensional (2D) LC to extend the peak capacities, target neutral loss (NL) data-dependent acquisition (DDA) for barrigenol-type triterpenoids saponins and automatic screening through predicted natural product screening (PNPS) in TraceFinder. To validate the integrated strategy, the shell of Xanthoceras sorbifolium Bunge (XSB) was taken as a case. An offline 2D-LC system was constructed with hydrophilic interaction chromatography (HILIC) and reversed phase (RP) C18 column, and orthogonality of 0.66 and peak capacity of 3494. The 2D-LC system improved chromatographic baseline separation and peak resolution. Full MS/all ion fragmentation (AIF)/NL dd-MS2 DDA was employed for the detection of the barrigenol saponins. PNPS strategy was adopted and markedly extended the screening coverage. The combined strategy showed about 5 times improvement in the screening capability. The PNPS screening process, using TraceFinder software, discovered a total of 752 barrigenol saponins from the shell of XSB, including 707 potentially new barrigenol saponins, accounting for 94.02%. The feasibility of the strategy was also confirmed by the isolation of two novel barrigenol saponins, the structures of which were unambiguously identified using nuclear magnetic resonance (NMR). Furthermore, this strategy could also be applied to rapidly discover new bioactive constituents from other herbal medicines or other natural sources, especially the barrigenol saponins constituents.
Keywords
Offline two-dimensional liquid chromatography
MS-oriented targeted isolation
Predicted natural product screening
Xanthoceras sorbifolium Bunge
Barrigenol-type triterpenoids saponins
- AIF
-
all ion fragmentation
- DDA
-
data-dependent acquisition
- DPI
-
diagnostic product ions
- HILIC
-
hydrophilic interaction chromatography
- IT
-
ion trap
- LC
-
liquid chromatography
- LT
-
linear ion trap quadrupole
- NMR
-
nuclear magnetic resonance
- NC
-
normalized collision energies
- N
-
neutral loss
- PNP
-
predicted natural product screening
- Ppm
-
parts per million
- qTOF
-
quadrupole-time of flight
- RP
-
reversed phase
- TCM
-
traditional Chinese medicine
- 2D L
-
two-dimensional liquid chromatography
- XSB
-
Xanthoceras sorbifolium Bunge
Abbreviations
1 Introduction
Natural products have long been regarded as potential sources for the discovery and development of new drugs(Atanasov et al., 2015, Shen, 2015). More than 64.9 % of new drugs with small molecules are derived from natural products over nearly four decades from 01/1981 to 09/2019(Newman and Cragg, 2020). However, the complexity of natural products is one of the biggest challenges for discovering new drugs from natural products(Cieśla and Moaddel, 2016). A large number of natural products, such as traditional Chinese medicine (TCM), have the characteristics of complicated chemical composition, low content, wide polarity coverage and prevalent isomers(Qiao et al., 2015, Yan et al., 2017). Furthermore, the exposure and characterization of unknown components are usually interfered with the coexistence of known major compounds. There is a great need for sufficient separation, sensitive detection, and potent identification approaches to discover new compounds. In recent years, there has been significant promotion of these technologies due to the rapid development of column technology and instrumentation(Guo et al., 2020).
Mass spectrometry (MS), particularly coupled with chromatography, has become the workhorse in the discovery of novel compounds(Jarmusch et al., 2021). The various types of tandem high-resolution mass spectrometry with liquid chromatography (LC), including LC/quadrupole-time of flight (qTOF)-MS, LC/Orbitrap-MS, LC/ion trap (IT)-TOF and LC/linear ion trap quadrupole (LTQ)-Orbitrap allow rapid structural characterization of unknown compounds in complex natural products extracts(Qiao et al., 2015, Pan et al., 2017, Yao et al., 2018). However, the presence of known major compounds often hampers the discovery of low-content new compounds in natural product extracts. To address this issue, two-dimensional liquid chromatography (2D LC) systems, based on two independent separation mechanisms, have been extensively explored to increase chromatographic peak capacity and resolving power. Among them, offline 2D LC systems by combining orthogonal phase chemistries, such as hydrophilic interaction chromatography (HILIC) and reversed phase (RP) C18 (HILIC × RPLC or RPLC × HILIC) has been proven powerful in the separation of complex natural product extracts (Wang et al., 2022, Wang et al., 2022, Wu et al., 2022). Additionally, 2D LC systems provide significant advantages in the separation and identification of co-eluted compounds and isomers, such as fatty acid isomers, glycolytic phosphorylated carbohydrate metabolites isomers and other (Deáková et al., 2015, Olfert et al., 2022, Su et al., 2023). To further improve the efficiency of discovering new compounds, researchers have made considerable efforts, including MS data acquisition, data post-processing and annotation(Naz et al., 2017). Targeted screening strategies have been developed for specific chemical constituents in different medicinal plants, such as precursor ion list scanning(Qiao et al., 2015), neutral loss (NL) scanning(Sun et al., 2019), multiple reaction monitoring scanning (Navarro-Reig et al., 2017), and mass defect filter scanning(Pan et al., 2018), which can improve selectivity and sensitivity. However, after obtaining high-throughput datasets, data post-processing and annotation become critical and time-consuming steps in medicinal plants, especially in the rapid identification of known compounds and the discovery of previously unreported compounds. TraceFinder, an automated data processing platform, has been successfully applied in the metabolites identification of TCMs with direct database searching methods(Zhang et al., 2019). Metabolic prediction analysis has emerged as an effective solution for rapidly identifying unknown compounds. By utilizing metabolic prediction screening combination with offline two-dimensional liquid chromatography-mass spectrometry, 945 ginsenoside compounds, including 662 previously unreported ginsenosides in the leaves of Panax notoginseng were identified(Yao et al., 2018). Therefore, based on the unique structural characteristics of barrigenol saponins existed in the target medicinal materials, a metabolic prediction screening strategy was employed in the current study to expand the scope of metabolite screening.
Xanthoceras sorbifolium Bunge (XSB), a monotypic species belonging to the family Sapindaceae and the genus Xanthoceras, is widely distributed throughout China. XSB has considerable value as a biofuel feedstock and as a source of novel drugs(Shen et al., 2019, Zang et al., 2021). Phytochemical investigations have revealed that barrigenol-type triterpenoids are the main bioactive constituents of XSB(Zang et al., 2021). Barrigenol saponins are responsible for a wide range of pharmacological activities, including anti-inflammation(Qi et al., 2013), anti-tumor(Chan et al., 2008), anti-HIV(Jiao et al., 2014)and anti-Alzheimer's disease effects(Li et al., 2016, Ji et al., 2017, Jin et al., 2019, Zhou et al., 2019, Zhou et al., 2022). Given the promising therapeutic potential of barrigenol saponins for the above-mentioned diseases, it is crucial to comprehensively profile XSB and identify novel barrigenol saponins structures as a matter of urgency.
In this study, a high-efficiency integrated strategy based on offline comprehensive 2D LC coupled with high-resolution Orbitrap MS was explored and applied to automatically screen barrigenol saponins and discover novel compounds from XSB shell. The workflow was presented in Fig. 1. Firstly, an orthogonal HILIC × RPLC Orbitrap-MS system was utilized to characterize the barrigenol saponins present in XSB. Secondly, in-house data libraries containing barrigenol saponins from XSB were constructed. Thirdly, the barrigenol saponins were automatically screened out using predicted natural product screening (PNPS) in TraceFinder with the predicted library. The interpretations were based on fragmentation behavior studies of reference standard barrigenol saponins. Finally, two compounds were purified through LC-MS guided isolation, and their structures were unambiguously identified by nuclear magnetic resonance (NMR) to validate the strategy.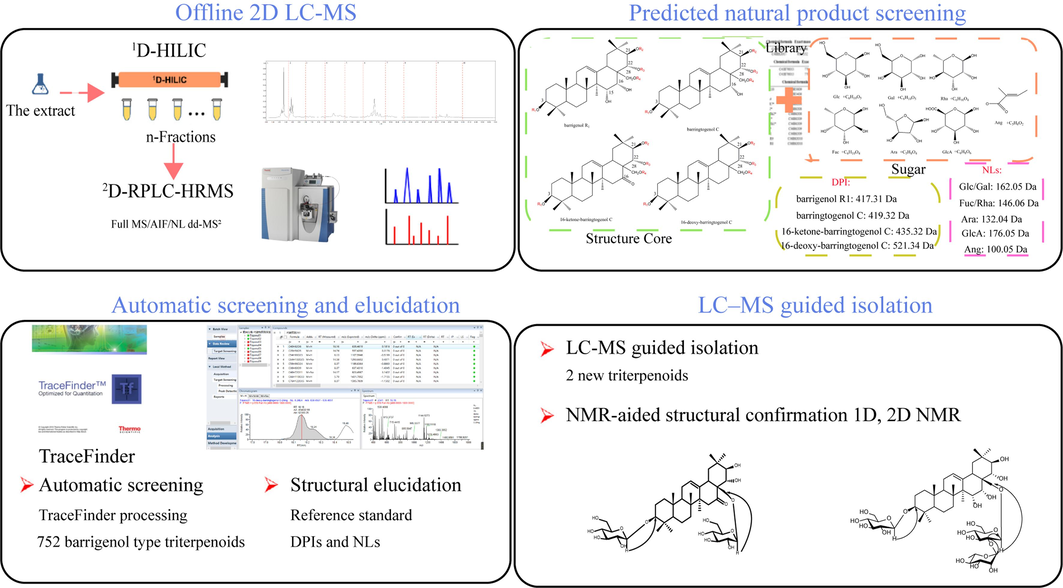
The workflow PNPS based offline 2D LC–MS guided isolation of barrigenol saponins from the shell of XSB.
2 Material and methods
2.1 Chemicals and reagents
A total of 21 barrigenol type triterpenoids were used for MS/MS fragmentation behavior studies and reference standards (Table S1). All triterpenoids were isolated from the shell of XSB. HPLC-grade acetonitrile and methanol were purchased from Fisher Chemical (Thermo Fisher Scientific, CA, USA). MS-grade formic acid was obtained from Fisher Chemical (Thermo Fisher Scientific, CA, USA). Deionized water was prepared by a Millipore Alpha-Q water purification system (Millipore, Bedford, USA).
2.2 Sample preparation
The dry XSB shell powder of 1.0 g was accurately weighed and ultrasonically extracted in 250 mL methanol (w/v) on a water bath (45 kHz) at room temperature for 1 h. The supernatant was filtered through a 0.22 μm membrane filter before analysis.
2.3 Offline comprehensive 2D LC separation of barrigenol saponins in XSB
Combining HILIC with RPLC was employed to separate the barrigenol saponins in XBS extract. The 1D HILIC separation was conducted Waters Xbridge Amide (4.6 × 100 mm, 3.5 µm) on a Waters e2695-2998 PDA (Waters Technologies, MA, USA). The mobile phase consisted of acetonitrile (A) and water (B) at a flow rate of 0.8 mL/min, and the optimized separation was achieved using the following gradient: 0–1 min, 95 % (A); 1–15 min, 95 %-85 % (A); 15–20 min, 85 % (A); 20–28 min, 85 %-50 % (A); 28–30 min, 50 % (A); 30–31 min, 50 %-95 % (A); 31–35 min, 95 % (A). The column temperature was set at 30℃, and the DAD detector was set at 210 nm, and the injection volume was 20 µL. The eluent was collected as a fraction according to peak and ten fractions were acquired (Fr. 1-Fr. 10). To prepare for 2D RPLC analysis, the fractions were evaporated to dryness using nitrogen gas. Next, each of the 10 fractions dried residues was dissolved in 100 µL of 50 % aqueous methanol (v/v) and centrifuged at 14000 rpm for 10 min.
The 2D LC separation was conducted on a Thermo Ultimate 3000 UHPLC system (Thermo Fisher Scientific, CA, USA) equipped with a binary solvent manager, a sample manager and a column manager. A Waters ACQUITY HSS T3 column (100 mm × 2.1 mm, 1.8 μm) maintained at 40℃ was applied for chromatographic separation with the mobile phase consisting of acetonitrile (A) and 0.1 % formic acid water (B) using the following gradient: 0–1 min, 10 % (A); 1–4 min, 10 %-30 % (A); 4–20 min, 30 %-65 % (A); 20–21 min, 65 %-99 % (A); 21–23 min: 99 % (A). The flow rate was set at 0.3 mL/min, and the injection volume was 2 µL.
2.4 Full MS/all ion fragmentation (AIF)/NL dd-MS2 data-dependent acquisition (DDA)
A Thermo Q Exactive MS (Thermo Fisher Scientific, CA, USA) operated in positive mode was coupled to UHPLC system via ESI interface for saponins data acquisition. ESI source parameters were set as follows: evaporation temperature, 350 °C; capillary temperature, 320 °C; spray voltage, 3.5 kV for positive ion mode; aux gas flow rate (arb), 10; sheath gas rate (arb), 35. NL scan parameters were set according to the characteristic MS/MS fragmentations of barrigenol saponins reference compounds, and 5 NL fragments were set: 142.05 Da (Rha, Fuc), 176.04 Da (GlcA), 162.05 Da (Glc, Gal), 132.04 Da (Ara), 100.06 Da (Ang). Full MS/AIF/NL dd-MS2 scan mode over m/z 400–1500 with a resolution of 70,000 was performed in profile format. AGC target and maximum injection time were set as 3e6 and 200 ms, respectively. The stepped normalized collision energies (NCE) were separately set as 30 %, 40 %, 50 %. AGC target and maximum injection time for dd-MS2 scan were separately 2 e5 and 100 ms, respectively. The isolation window was 2.0 (m/z). The resolution for MS2 scan was 35,000 and the spectra were recorded in profile format. Data acquisition and processing were conducted with Xcalibur 4.2 software (Thermo Fisher Scientific, CA, USA).
2.5 Targeted isolation and identification of two new barrigenol saponins
For verification of the PNPS strategy, two potential new barrigenol saponins (Comp 1: tR 6.06 min with molecular formula C42H68O15; Comp 2: tR 5.96 min with molecular formula C48H80O20) were selected and isolated under the guide of LC-MS analysis. After 75 % aqueous ethanol solvent extraction, this target new compound was enriched in the 95 % aqueous ethanol fraction on the microporous absorptive resin (D100), which was further separated on silica gel (200–300 mesh), eluted with a gradient system of CH2Cl2–MeOH (10:1, 5:1, 3:1, 1:1, 0:1) to afford Fr. 1–5. Fr. 3 (108.0 g) was further fractionated by an ODS column using MeOH–H2O (0:100–100:0) as the elution system to give five subfractions. The third subfraction was further isolated by reversed-phase C18 silica gel (Nacalai tesque, Japan) column chromatography using a gradient mixture of acetonitrile–water (from 15 % to 30 %, v/v) to afford target compounds (Comp 1, 6.3 mg; Comp 2, 8.5 mg, respectively). 1D and 2D NMR spectra were recorded on Bruker DRX 600 MHz (Bruker, Ettlingen, Germany) spectrometers, and the chemical shifts (δ) were expressed in parts per million (ppm) with reference to the solvent signals.
3-O-β-D-glucopyranosyl-28-O-β-D-glucopyranosyl-3β,21β,22α-trihydroxy-12-ene-16-ketone (Comp 1). White, amorphous powder (MeOH); HRESIMS [M + H]+: m/z 813.45947 (calcd. for C42H69O15, 813.4631); 1H (600 MHz, C5D5N) and 13C (150 MHz, C5D5N) NMR data, see Table 2.
3-O-β-D-glucopyranosyl-28-O-[α-L-rhamnopyranosyl(1 → 2)]-β-D-glucopyranosyl-barrigenol R1 (Comp 2). White, amorphous powder (MeOH); HRESIMS [M + H]+: m/z 977.52722 (calcd. for C48H81O20, 977.5316); 1H (600 MHz, C5D5N) and 13C (150 MHz, C5D5N) NMR data, see Table 2.
2.6 Data mining
TraceFinder 5.1 software(Thermo Fisher Scientific, CA, USA), LC-MS data analysis, allows target screening and quantitative analysis detected. TraceFinder can also be used for automatic target screening based on our self-established barrigenol saponins database. And we can also establish a screening database for unknown targets by predicting the structure of metabolites. The theoretical peak capacity of the offline 2D LC system was carried out according to Guo, et al(Pan et al., 2018, Yao et al., 2018).
3 Results and discussion
3.1 Construction and evaluation of 2D LC-Q-Orbitrap-MS system
Sufficient chromatographic separation is a fundamental requirement in order to achieve the objective of comprehensive chemical profiling of low-content compounds in extracts of natural products extracts. Natural product extracts exhibit diverse characteristics, including a wide range of molecular mass and polarity, similar physicochemical properties, and significant differences in composition. Relying solely on a single separation mechanism, such as RP separation, may be insufficient for conducting a comprehensive chemical analysis. Therefore, our objective is to develop a 2D-LC approach to effectively resolve the multi components of XSB. The performance of chromatography in each dimension is influenced by key parameters, including the stationary phase, mobile phase, column temperature, and gradient eluting program. These parameters were systematically optimized through experiments.
The construction of 2D LC system mainly includes 1D chromatography construction, 2D chromatography construction, mass spectrometry data acquisition condition optimization, orthogonality and peak capacity evaluation of 2D LC system, etc. Firstly, the selection of columns for the 2D separation was operated. Three conventional RP columns, including Waters HSS T3 (2.1 × 100 mm, 1.8 µm), BEH C18 (2.1 × 100 mm, 1.7 µm) and Thermo Hypersil gold (2.1 × 100 mm, 3 µm), were tested. As a result, besides comparable resolution and similar selectivity, the HSS T3 column could resolve more peaks and showed shorter analysis time (Fig. S1). To render abalance between peak shape and resolving power, the HSS T3 column was finally chosen in the 2D separation. Consistently, the RP mode HSS T3 column was also utilized in UHPLC/QTOF-MS analysis of XSB in a recent report(Chen et al., 2023). Subsequently, under the same elution procedure, different mobile phase systems were investigated, including acetonitrile-5 mM ammonium formate aqueous solution, acetonitrile–water and acetonitrile-0.1 % formate water, respectively. The results showed that acetonitrile-0.1 % FA mobile phase system showed higher column efficiency and better peak shape, besides comparable resolution. (Fig. S2). Therefore, acetonitrile-0.1 % FA was finally selected as the mobile phase system. In this experiment, the effects of three different column temperatures on the separation of barrigenol saponins were investigated, including 25 ℃, 30 ℃ and 40 ℃, respectively. As a result, we selected a column temperature of 40 ℃ as it enabled advanced retention time of chromatographic peaks and improved separation of peaks occurring between 5 and 7 min and 22–24 min (Fig. S3).
Selection of the 1D stationary phase was based on the selectivity difference with HSS T3 column used for 2D separation. Thus, we choose Waters Xbridge Amide (4.6 × 100 mm, 3.5 µm) for 1D HILIC chromatography analysis. Consistently, the Xbridge Amide column was also utilized in 1D HILIC analysis of ginsenosides in a recent report (Yao et al., 2018). For 1D HILIC chromatography, acetonitrile–water was used as the mobile phase to facilitate the preparation of fractions. In the UV absorption wavelength selection, three different wavelengths were investigated, including 210 nm, 230 nm, and 254 nm, respectively (Fig. S4). As a result, the wavelength of 210 nm showed better peak shape and higher response. Therefore, 210 nm was finally selected as the detection wavelength. Finally, the optimized 2D LC separation conditions were used for sample preparation and data acquisition.
24 of barrigenol saponins components (containing 21 reference standards) were used as orthogonality evaluation index components (Table S1 and S2). The separation efficiency of the 2D LC/orbitrap-MS system was evaluated in terms of orthogonality and peakcapacity. The orthogonality (Ao) of the developed 2D LC system was calculated with a series of asterisk equations(Camenzuli and Schoenmakers, 2014). Orthogonality evaluation results showed that the values of Z−, Z+, Z1, Z2 were separately calculated as 0.84, 0.68, 0.92 and 0.83, and the orthogonality Ao was determined as 0.66. All results reflected the good distribution of components in 2D LC system. Compared to the previously reported offline HILIC × RP 2D LC method for ginsenosides with higher orthogonality value of 0.69, but this system required a greater number of fractions in the 1D LC, consequently leading to a longer analysis time in the 2D LC -MS(Yao et al., 2018). Peak capacity was compared according to the number of extracted ions, and unknown compound screening methods were established by using TraceFinder. The number of ions extracted from 10 fractions were Fr.1 (1 8 5), Fr.2 (1 0 6), Fr.3 (1 1 9), Fr.4 (1 2 4), Fr.5 (2 1 1), Fr.6 (1328), Fr.7 (3 9 4), Fr.8 (3 1 8), Fr.9 (6 3 7) and Fr.10 (72), respectively (Fig. 2B). 2D LC system significantly increases in peak capacity, approximately doubling it from 1558 to 3494. From the above results, it was clear that the combination of HILC and RPLC 2D LC chromatography separation could greatly improve the detection of barrigenol saponins in the XSB.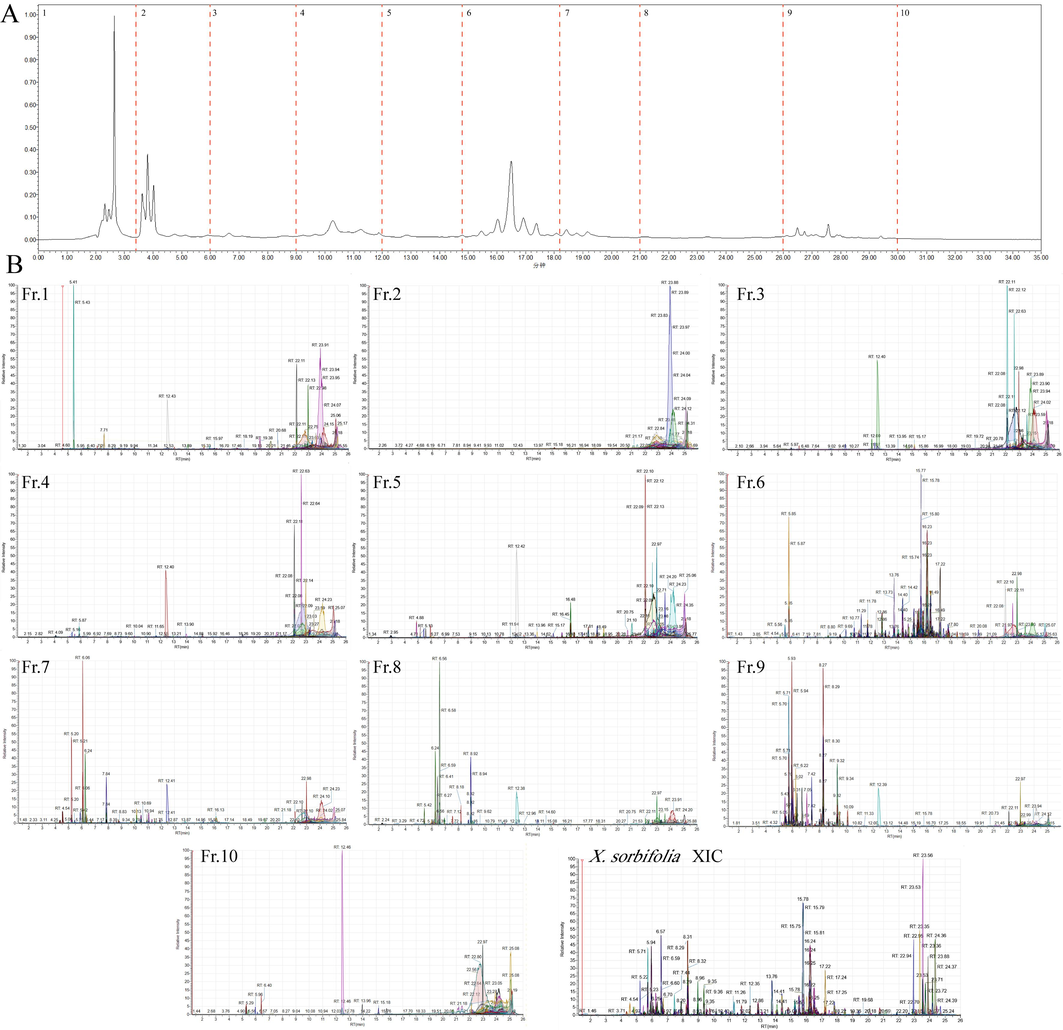
Chromatographic separation of XSB by the developed HILIC × RPLC 2D-LC/MS system. (A) 1D UV spectrum of the total extract by HILIC; (B) 2D extracted ion chromatogram of ten fraction samples and XSB total extract.
3.2 Systematic characterization of multi-type barrigenol saponins in XSB extract
Barrigenol saponins usually have hydroxyl groups at C-3, 15, 16, 28, 21 and 22 positions in the parent nucleus. According to the difference in hydroxyl substitution position, it can be divided into four types barrigenol saponins: barrigenol R1 (C15-OH, C16-OH, C21-OH, C22-OH), barringtogenol C (C16-OH, C21-OH, C22-OH), 16-keto-barringtogenol C (C16-OH, C21-OH, C22-OH) and 16-deoxy-barringtogenol C (C21-OH, C22-OH), respectively (Fig. 3A). On the basis of the structure of the aglycone, the substitution of angelica acyl (Ang) and sugar groups also enriched the structural diversity of the barrigenol-type saponins. In this study, high-resolution mass spectrometry analysis of 10 fractions collected by 1D was performed, and a total of 128 compounds were identified, including 47 16-deoxy-barringtogenol C-type compounds, 55 barrigenol R1-type ones, 23 barringtogenol C-type ones, and 3 16-ketone-barringtogenol C-type ones, respectively (Table S3).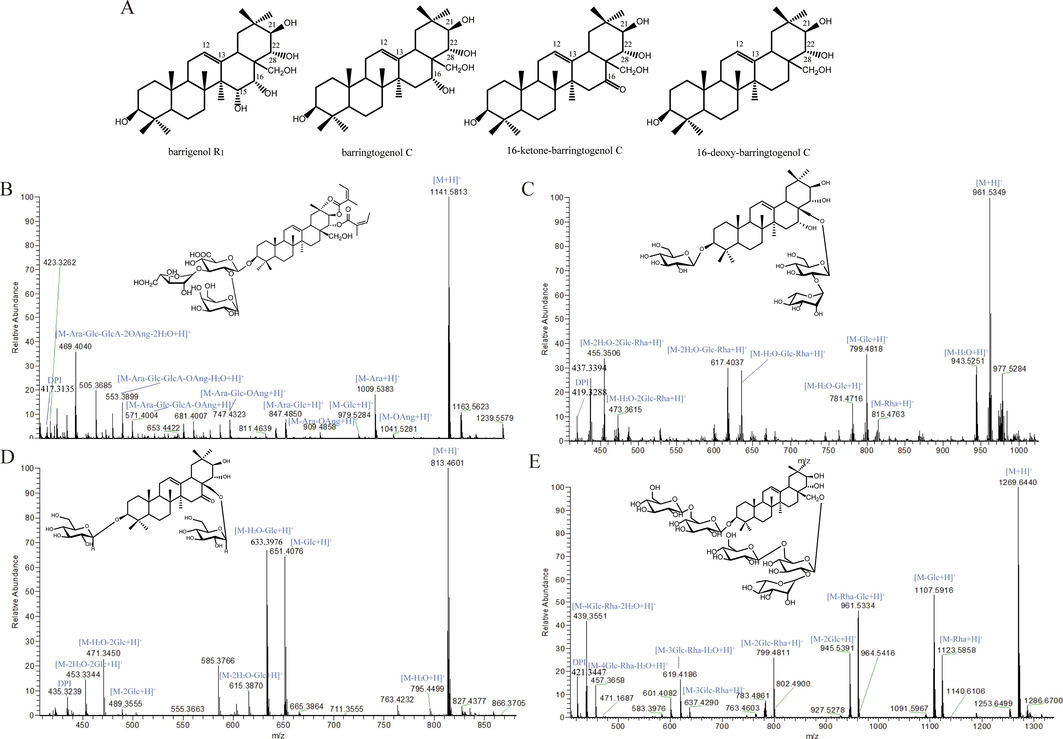
Systematic characterization of multi-type barrigenol saponins in XSB extract. (A) The types of four barrigenol type triterpenoids; (B) mass fragmentation pathway of barrigenol R1 type saponin; (C) mass fragmentation pathway of barringtogenol C; (D) mass fragmentation pathway of 16-ketone-barringtogenol C; (E) mass fragmentation pathway of 16-deoxy-barringtogenol C.
3.2.1 Barrigenol R1 type saponins
Xanthoceraside is a barrigenol R1 type saponin with 1 Glc, 1 Ara, 1 GlcA and 2 Ang groups, respectively. In the MS spectrum of xanthoceraside, molecular ions and sodium adduct ions were found at m/z 1141.5813 Da ([C57H89O23]+, calc. 1141.5789) and 1163.5623 Da ([C57H88O23Na]+, calc. 1163.5609), respectively (Fig. 3B). For the MS/MS spectrum, fragments at m/z 1041.53 Da corresponded to the [M−OAng + H]+; fragments at m/z 1009.54, 979.53, 847.49 and 671.44 Da corresponded to the [M−Ara + H]+, [M−Glc + H]+, [M−Ara−Glc + H]+ and [M−Ara−Glc−GlcA + H]+, respectively; fragments at m/z 471.34 Da corresponded to the [M−Ara−Glc−GlcA−2OAng + H]+. Therefore, the fragmentation pathway of barrigenol R1 was proposed in Fig. 3B, and a key fragment at m/z 417.3135 Da indicated the diagnostic product ions (DPI) of barrigenol R1.
3.2.2 Barringtogenol C type saponins
3-O-β-D-glucopyranosyl-28-O-α-L-rhamnopyranosyl(1 → 2)-β-D-glucopyranosyl −3β, 21β,22α-barringtogenol C is a barringtogenol C-type saponin with 2 Glc and 1 Rha moieties. In the MS spectrum, its molecular ion was found at m/z 961.1490 Da ([C48H81O19]+, calc. 961.5367) (Fig. 3C). For the MS/MS spectrum, fragments at m/z 943.52, 815.47, 799.48 and 635.41 Da corresponded to the [M−H2O + H]+, [M−Rha + H]+, [M−Glc + H]+ and [M−H2O−Glc−Rha + H]+, respectively; fragments at m/z 473.36, 455.35 and 437.36 corresponded to the [M−H2O−2Glc−Rha + H]+, [M−2H2O−2Glc−Rha + H]+ and [M−3H2O−2Glc−Rha + H]+, respectively. And a key fragment at m/z 419.3297 Da indicated the DPI of barringtogenol C.
3.2.3 16-ketone-barringtogenol C type saponins
3-O-β-D-glucopyranosyl-28-O-β-D-glucopyranosyl-3β,21β,22α-trihydroxy-12-ene-16-ketone is a 16-ketone-barringtogenol C-type saponin with 2 Glc substitution. In the MS spectrum, its molecular ion was found at m/z 813.4601 Da ([C42H69O15]+, calc. 813.4631) (Fig. 3D). For the MS/MS spectrum, fragments at 795.45, 651.41, 633.39, 615.39, 471.34 and 435.32 Da corresponded to the [M−H2O + H]+, [M−H2O−Glc + H]+, [M−2H2O−Glc + H]+, [M−H2O−2Glc + H]+ and [M−3H2O−2Glc + H]+, respectively. A key fragment at m/z 435.3239 Da indicated the DPI of 16-ketone-barringtogenol C.
3.2.4 16-deoxy-barringtogenol C type saponins
Xanifolia O5 is a 16-deoxy-barringtogenol C-type saponin with 4 Glc and 1 Rha moities. In the MS spectrum of xanifolia O5, the molecular ion was found at m/z 1269.6440 Da ([C60H101O28]+, calc. 1269.6474) (Fig. 3E). For the MS/MS spectrum, fragments at 1123.58, 1107.59, 961.53, 945.54, 799.49, 637.43, 619.42, 457.36 and 421.34 Da corresponded to the [M−Rha + H]+, [M−Glc + H]+, [M−Glc−Rha + H]+, [M−2Glc + H]+, [M−2Glc−Rha + H]+, [M−3Glc−Rha + H]+, [M−3Glc−Rha−H2O + H]+ and [M−4Glc−Rha−3H2O + H]+, respectively. A key fragment at m/z 421.3447 Da indicated the DPI of 16-deoxy-barringtogenol C.
The four types barrigenol saponins are more commonly reported in XSB. Of course, there are many other structural types of triterpenoid saponins, but due to the limited reports and lack of reference standards, their mass spectral fragmentation pathway have not been extensively explored. Therefore, the DPI of the four types of saponins were finally determined as barrigenol R1 (DPI: 417.3125), barringtogenol C (DPI: 419.3297), 16-keto-barringtogenol C (DPI: 435.3239), and 16-deoxy-barringtogenol C (DPI: 421.3447), respectively. By extracting the four diagnostic ions, the obtained base-peak ion extraction chromatograms (EIC) of the XSB extract were shown in Fig. S5.
3.3 Development of predicted natural product screening (PNPS) and confirmation methods
3.3.1 Construction PNPS strategy
Plant natural products are synthesized through biosynthesis pathways, so their structures are regular. In this study, PNPS in-house database was constructed by changing the composition and position of the substituents based on the reported structures of barrigenol saponins in XSB. Based on the literature reports(Zang et al., 2021, Chen et al., 2022), the structure characteristics of barrigenol saponins were summarized as follows: Based on the available literatures, the structural characteristics of barrigenol saponins could be summarized as follows: (1) Substituents were located at C-3, C-21, C-22, and C-28, respectively; (2) Sugar moieties usually substituted at C-3 and C-28, while Ang or acetyl groups were at the C-21 and C-22; (3) Typically, the sugar directly linked to C-3 and C-28 were Glc, while occasionally was GlcA at C-3; (4) The substituent at C-3′ of the Glc linking to the C-3 of aglycone was Glc, Ara, or occasionally Gal. The sugar directly linked to C-28 usually was Glc or Rha; (5) The C-6 of the Glc directly linked to both the C-3 and C-28 was generally substituted by Glc; (6) There were typically three sugar substituents at positions C-3 and C-28. Glc and Gal were isomers that were difficult to distinguish on the MS, so both of them were expressed as Glc. According to barrigenol saponins’ characteristics, the multi-step modification was applied to each barrigenol saponins, leading to 134 predicted metabolites for each barrigenol saponins (Table 1). A total of 536 possible barrigenol saponins, which was more than 5 times the number of saponins reported previously(compounds 1–124)(Zang et al., 2021).
C-3
Adducts
C-28
Adducts
C-21, C-22
Adducts
GlcA
+C6H9O6
Glc
+C6H11O5
Ang
+C5H7O1
Glc
+C6H11O5
Glc-Glc
+C12H21O10
GlcA-Glc
+C12H19O11
Glc-2Glc
+C18H31O15
GlcA-2Glc
+C18H29O16
Glc-Glc-Rha
+C18H31O14
GlcA-Glc-Ara
+C17H27O15
Glc-Glc
+C12H21O10
Glc-2Glc
+C18H31O15
Glc-Glc-Ara
+C17H29O15
No.
1
No.
2
δC
δH (J in Hz)
δC
δH (J in Hz)
1
39.2
1.16 m/0.91 m
1
39.5
1.50 m/0.96 m
2
27.0
2.28 m/1.87 m
2
27.2
2.27 m/1.86 m
3
89.2
3.41 dd (11.8, 4.1)
3
89.2
3.43 dd (11.7, 4.4)
4
40.0
4
40.0
5
56.1
0.76 d (11.9)
5
56.1
0.85 d (10.8)
6
18.8
1.46 m/1.28 m
6
19.3
1.43 m/1.33 m
7
33.1
1.39 m/1.12 m
7
37.0
2.16 m/2.05 m
8
40.9
8
42.1
9
47.2
1.55 m
9
47.4
1.77o
10
37.6
10
37.5
11
24.3
1.94 m
11
24.6
2.08 m/2.00 m
12
125.9
5.54 br s
12
125.5
5.60 br s
13
140.6
13
144.7
14
48.5
14
48.3
15
46.5
3.14o/2.02 d (14.4)
15
67.8
4.43 m
16
216.0
16
72.7
4.90 m
17
59.2
17
47.7
18
47.3
3.14o
18
42.4
2.74 dd (14.0, 4.0)
19
47.5
1.92 m/1.39 m
19
47.9
3.04 t (13.5)/1.37 m
20
37.2
20
36.8
21
77.5
4.28o
21
78.5
4.83 m
22
76.3
4.26o
22
77.0
4.49 d (10.1)
23
28.6
1.33 s
23
28.7
1.27 s
24
17.5
1.03 s
24
17.6
1.02 s
25
16.1
0.87 s
25
16.4
0.94 s
26
17.7
1.10 s
26
18.1
1.25 s
27
28.2
1.37 s
27
21.9
1.89 s
28
73.7
4.55 m/4.45 m
28
76.7
4.11o/4.07 m
29
30.7
1.23 s
29
31.1
1.31 s
30
19.1
1.20 s
30
20.4
1.28 s
3-Oglc
3-Oglc
1′
107.4
5.01 d (7.4)
1′
107.4
4.97 d (7.8)
2′
76.4
4.05 m
2′
76.4
4.05 m
3′
78.8
4.03 m
3′
78.9
4.06 m
4′
72.436
4.26 m
4′
72.3
4.25 m
5′
79.1
4.28 m
5′
79.3
4.28 m
6′
63.5
4.52 m
6′
63.5
4.62 dd (11.7, 2.5)/4.44 m
28-Oglc
28-Oglc
1′'
105.9
4.97 d (7.4)
1′'
103.9
4.78 d (7.8)
2′'
79.2
4.01 m
2′'
80.8
4.37 m
3′'
79.3
4.20 m
3′'
78.7
3.87 m
4′'
72.1
4.27 m
4′'
72.2
4.21 m
5′'
75.2
4.24 m
5′'
75.0
4.32 m
6′'
63.2
4.45 m
6′'
63.0
4.49 m
2′'-Orha
4.39 m
1′''
101.0
6.67 br s
2′''
73.1
4.73 m
3′''
73.1
4.66 dd (9.3, 3.4)
4′''
75.4
4.36 m
5′''
69.7
4.83 m
6′''
19.2
1.78 d (6.2)
In this study, the predicted combination of possible natural product structures was combined with isomers, resulting in a total of 261 possible barrigenol saponins. In the screening procedure, only the molecular formula information was taken into consideration. A in house database was established for the predicted and merged samples, and the information such as nomenclature, molecular formula, additive ion type and scan type were imported into the database (Fig. 4A). Finally, 752 kinds of barrigenol saponins were screened out by the established in-house library. In the screening results, 24, 12, 20, 19, 54, 163, 119, 123, 198 and 20 kinds of barrigenol saponins were screened from Fr.1–10, respectively (Fig. 4B), among which 45 ones had been reported, but there was still a large number of isomers that cannot be distinguished by MS. In addition, there were also 707 unreported saponins, accounting for 94.02 % of the total number of predicted screened saponins (Fig. 4C).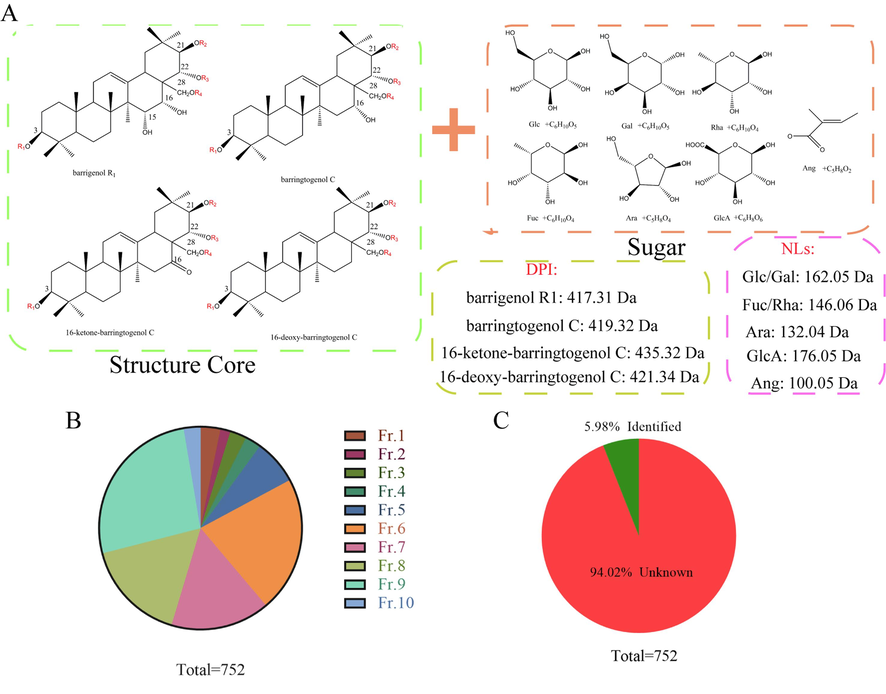
Schematic diagram for prediction of barrigenol type saponins. (A) Predicted metabolites screening strategy; (B-C) the screening results of barrigenol type saponins.
3.3.2 Validation of PNPS
Two components were selected to evaluate the PNPS method, including 3-O-β-D-glucopyranosyl-28-O-α-L-rhamnopyranosyl(1 → 2)-β-D-glucopyranosyl-3β,21β,22α-barringtogenol C (barringtogenol C type saponin) and xanifolia O5 (16-deoxy-barringtogenol C type saponin).
The MS spectrum of barringtogenol C-type saponin showed molecular ions at m/z 961.1490 Da ([C48H81O19]+, calculated 961.5367). It had a retention time of 6.15 min and corresponded to compound No. 112 (Fr.9) in the screening results (Table s4), which could be matched in the three dimensions of molecular ions, retention time and MS/MS fragment (Fig. 5 A-B), respectively.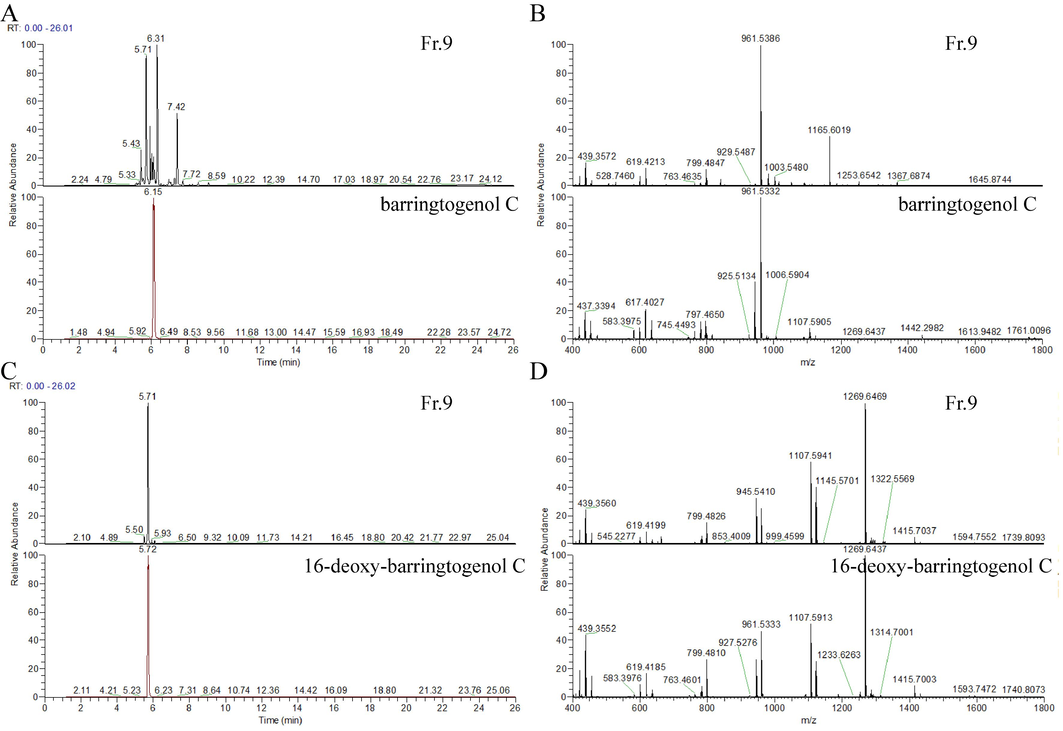
The validation of PNPS. (A) The extracted ion chromatogram of Fr.9 and reference (m/z 961.1490); (B) The MS/MS chromatogram of Fr.9 and reference; (C) The extracted ion chromatogram of Fr.9 and reference (m/z 1269.6440); (D) The MS/MS chromatogram of Fr.9 and reference.
The MS spectrum of 16-deoxy-barringtogenol C-type saponin showed molecular ions at m/z 1269.6440 Da ([C60H101O28]+, calc. 1269.6474). It had a retention time of 5.71 min and corresponded to compound No. 19 (Fr.9) in the screening results (Table s4), which could be matched in the three dimensions of molecular ions, retention time and MS/MS fragment (Fig. 5 C-D), respectively.
3.4 Targeted isolation and structural elucidation of two potential new barrigenol saponins
To partially validate the MS-oriented targeted isolation, we selected and isolated two potentially new compounds from the extract of XSB. Comp.1 (corresponding to No. 93 in Table S4; tR 6.06 min, Fr. 9) had the molecular formula C42H68O15. Based on the transitions (m/z 813.45 → 651.41 → 489.36 → 435.32) and the aglycone ion species (Fig. 6B), it was primarily characterized as 16-ketone-barringtogenol C-type saponin substituted with 2 Glc moieties. The DPI ion (m/z 435.32) could help identify the aglycone as 16-ketone-barringtogenol C. By searching the molecular formula in SciFinder, this compound should be a new one. 13C and 1H NMR were further utilized to confirm the structure (Table 2). Comp.1 was obtained as a white amorphous powder, which gave positive results for the Liebermann–Burchard reaction and Molish reagent. In addition, we detected the optical rotation value of Comp.1 and found that the optical rotation value was negative ([α]20 D-95.29 (c 0.17 mg/ml, MeOH)). The 13C NMR spectrum of 1 displayed 42 carbon signals, of which 30 were assigned to the aglycon. The 1H and 13C NMR data of 1 indicated a pentacyclic triterpenoid saponin, and assignments were confirmed with the help of HMBC experiments. The 1H NMR spectrum exhibited the characteristic methyl singlets of triterpenoid in the higher field (δH 1.33, 1.03, 0.87, 1.10, 1.37, 1.23, 1.20), and the corresponding carbon signals in 13C NMR spectrum were at δC 28.6, 17.5, 16.1, 17.7, 28.2, 30.7, 19.1, respectively. In addition, one trisubstituted olefinic proton signal at δH 5.62 (br s, H-12), coupled with typical 13C NMR resonances at δC 125.9 and 140.6, as well as a hydroxymethyl signal at δC 73.7 and 216.0 in the 13C NMR spectrum, indicated the aglycon of 16-carbonyl-28-hydroxy-olean-12-ene. The 13C NMR spectrum of compound 1 showed one downfield shift at δC 215.2, which was attributed to carbonyl at C-16. Acid hydrolysis of 1 with 2 M HCl afforded the aglycon and D-glucose, respectively. The anomeric proton’s coupling constant and the chemical shift of C-3/5 suggesting the relative configurations of D-glucose was β(Zhao et al., 2022). Furthermore, four oxygenated methine protons [δH 3.41 (1H, dd, J = 11.8, 4.1 Hz), 4.28 (1H, o), 4.26 (1H, o), 4.55 (1H, o), 4.45 (1H, o)] and the corresponding carbons (δC 89.2, 77.5, 76.3 and 73.7) were given in the NMR spectra, suggesting the hydroxylation of C-3, C-21, C-22 and C-28, respectively. This presumption was corroborated by the HMBC long-range correlations of two methyl groups at δH 1.19 (s, Me-29) and 1.15 (s, Me-30) with the oxygenated methine carbon signal at δC 77.5 (C-21), as well as two methyls at δH 1.32 (s, Me-23) and 1.03 (s, Me-24) with the oxygenated methine carbon at δC 89.2 (C-3), and δH 4.55 (H-28) with δC 216.0 (C-16) and δH 4.55 (H-28) with δC 76.3 (C-22), respectively. In the HMBC spectrum, correlation peaks were observed from the signal at δH 4.95 (GlcI-H-1) to δC 89.2 (C-3), and δH 4.98 (GlcII-H-1) to δC 73.7 (C-28), which indicated that the sugar moiety were located at C-3 and C-28, respectively. The long-range HMBC correlations of H-28 (δH 4.55), H-22 (δH 4.26) and H-18 (δH 3.14) with C-16 confirmed the conclusion (Fig. 6A). From all the above given data, the structure of compound 1 was elucidated to be 3-O-β-D-glucopyranosyl-28-O-β-D- glucopyranosyl-3β,21β,22α-trihydroxy-12-ene-16-ketone.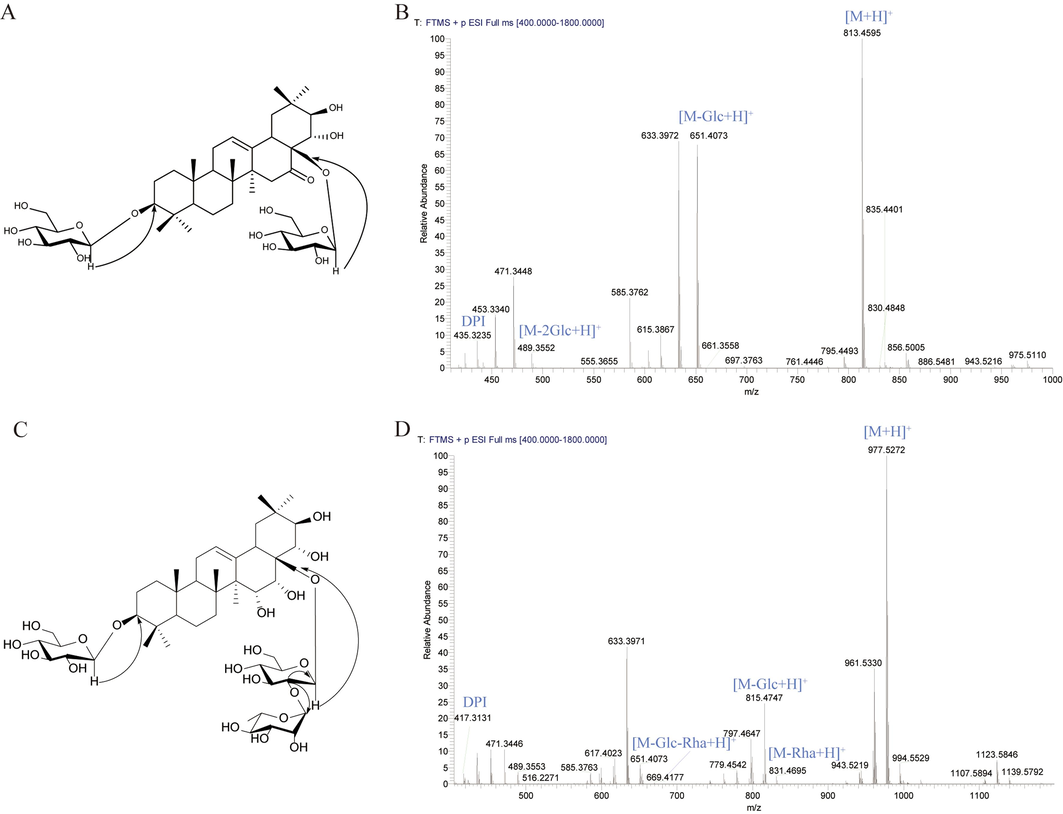
Targeted isolation and structural elucidation of two potential new barrigenol saponins. (A) The key HMBC correlations of Comp.1; (B) the proposed fragmentation pathways of Comp.1; (C) The key HMBC correlations of Comp.2; (D) the proposed fragmentation pathways of Comp.2.
Comp.2 (corresponding to No. 87 in Table S4; tR 5.96 min, Fr. 8) had the molecular formula of C48H80O20. Based on the transitions (m/z 977.53 → 831.47 → 815.47 → 669.42 → 417.31) and the aglycone ion species (Fig. 6D), it was primarily characterized as barrigenol R1-type saponin substituted with 2 Glc and 1 Rha moieties. The DPI ion (m/z 417.31) could help identify the aglycone as barrigenol R1. By searching the molecular formula in SciFinder, this compound also should be a new one. The 13C and 1H NMR were further utilized to confirm constructure (Table 2). Acid hydrolysis of 2 with 2 M HCl afforded the aglycon, D-glucose and L-rhamnose, respectively. The optical rotation data of 2 is [α]20 D-130.00 (c 1.00 mg/ml, MeOH). The detailed NMR analysis showed an additional hydroxy group at C-15 (δC 67.8), which was further confirmed by HMBC experiments from δH 1.89 (H-27) to δC 67.8 (C-15). In the HMBC spectrum, the correlations observed between δH 4.90 (H-16) and δC 67.8 (C-15), together with the HMBC cross-peak of δH 4.90 (H-16) of δC 42.4 (C-18) were also noted. The β-anomeric configurations for the glucose were determined from their 3JH1, H2 coupling constants (7.1–7.9 Hz) and α-configuration for the rhamnose unit based on the 13C NMR chemical shifts(Zhao et al., 2022). The linkage of the sugar units at C-3 was established by the HMBC correlations from δH 4.96 (GlcI-H-1) to δC 89.2 (C-3). Similarly, the sugar chain at C-28 was deduced from the HMBC correlations from δH 4.78 (GlcII-H-1) to δC 76.7 (C-28), as well as of δH 4.37 (GlcII-H-2) to δC 103.9 (GlcII-C-1) and δC 101.0 (RhaIII-C-1) (Fig. 6C). Thus, 2 was assigned as 3-O-β-D-glucopyranosyl-28-O-[α-L-rhamnopyranosyl(1 → 2)]-β-D-glucopyranosyl-barrigenol R1.
4 Conclusions
To improve the LC-MS guided isolation of new compounds from herbal medicines, an integrated strategy was developed. This strategy includes the use of offline 2D-LC-Orbitrap MS, multiple NL scanning, DPIs filtering and PNPS method, respectively. The approach was successfully applied to identify barrigenol saponins in XSB. The utilization of an offline 2D-LC system (HILIC × RPLC) exhibited an adequately separate chromatographic peak and significantly increased in peak capacity, approximately doubling it from 1558 to 3494. The DPIs filtering and NL scanning were employed to targeted identify and discover new barrigenol saponins in XSB. The PNPS method, utilizing TraceFinder software, enhanced the screening process by approximately 2 times, resulting in the identification of 752 barrigenol saponins, including 707 potentially new compounds, accounting for 94.02 % of the total predicted screened saponins. It was also confirmed by the separation of two novel barrigenol saponins, whose structures were unambiguously identified by NMR. Furthermore, this strategy could also be applied to rapidly discover new bioactive compounds from other herbal medicines or other natural sources, particularly the sequential compounds.
Acknowledgments
The authors acknowledge the financial support from National Natural Science Foundation of China (Grant No. 81573694), the Science Foundation of the Educational Department of Liaoning Province in 2021 (Grant No. LJKZ0920), the China Postdoctoral Science Foundation (Grant No. 2023M730444) and China Pharmaceutical Regulatory Science Action Plan of National Institutes for Food and Drug Control (Grant No. NMPAJGKX-2023-090).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnol Adv.. 2015;33:1582-1614.
- [CrossRef] [Google Scholar]
- A new measure of orthogonality for multi-dimensional chromatography. Anal Chim Acta.. 2014;838:93-101.
- [CrossRef] [Google Scholar]
- Cytotoxic acylated triterpene saponins from the husks of Xanthoceras sorbifolia. J Nat Prod.. 2008;71:1247-1250.
- [CrossRef] [Google Scholar]
- Traditional uses, phytochemistry, pharmacology and current uses of underutilized Xanthoceras sorbifolium bunge: A review. J Ethnopharmacol.. 2022;283:114747
- [CrossRef] [Google Scholar]
- Extraction, purification of saponins components from Xanthoceras sorbifolium Bunge leaves: potential additives in the food industry. J Food Meas Charact.. 2023;17:916-932.
- [Google Scholar]
- Comparison of analytical techniques for the identification of bioactive compounds from natural products. Nat Prod Rep.. 2016;33:1131-1145.
- [CrossRef] [Google Scholar]
- Two-dimensional high performance liquid chromatography for determination of homocysteine, methionine and cysteine enantiomers in human serum. J Chromatogr a.. 2015;1408:118-124.
- [CrossRef] [Google Scholar]
- Recent advances in non-targeted screening analysis using liquid chromatography - high resolution mass spectrometry to explore new biomarkers for human exposure. Talanta.. 2020;219:121339
- [CrossRef] [Google Scholar]
- Advancements in capturing and mining mass spectrometry data are transforming natural products research. Nat Prod Rep.. 2021;38:2066-2082.
- [CrossRef] [Google Scholar]
- The total triterpenoid saponins of Xanthoceras sorbifolia improve learning and memory impairments through against oxidative stress and synaptic damage. Phytomedicine.. 2017;25:15-24.
- [CrossRef] [Google Scholar]
- Xanthoceraside induces apoptosis in melanoma cells through the activation of caspases and the suppression of the IGF-1R/Raf/MEK/ERK signaling pathway. J Med Food.. 2014;17:1070-1078.
- [CrossRef] [Google Scholar]
- Xanthoceraside prevented synaptic loss and reversed learning-memory deficits in APP/PS1 transgenic mice. J Physiol Sci.. 2019;69:477-488.
- [CrossRef] [Google Scholar]
- Xanthoceras sorbifolia extracts ameliorate dendritic spine deficiency and cognitive decline via upregulation of BDNF expression in a rat model of Alzheimer's disease. Neurosci Lett.. 2016;629:208-214.
- [CrossRef] [Google Scholar]
- Untargeted comprehensive two-Dimensional liquid chromatography coupled with high-resolution mass spectrometry analysis of rice metabolome using multivariate curve resolution. Anal Chem.. 2017;89:7675-7683.
- [CrossRef] [Google Scholar]
- Development of a liquid chromatography–high resolution mass spectrometry metabolomics method with high specificity for metabolite identification using all ion fragmentation acquisition. Anal Chem.. 2017;89:7933-7942.
- [CrossRef] [Google Scholar]
- Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod.. 2020;83:770-803.
- [CrossRef] [Google Scholar]
- Comprehensive profiling of conjugated fatty acid isomers and their lipid oxidation products by two-dimensional chiral RP×RP liquid chromatography hyphenated to UV- and SWATH-MS-detection. Anal Chim Acta.. 2022;1202:339667
- [CrossRef] [Google Scholar]
- Mass defect filtering-oriented classification and precursor ions list-triggered high-resolution mass spectrometry analysis for the discovery of indole alkaloids from Uncaria sinensis. J Chromatogr a.. 2017;1516:102-113.
- [CrossRef] [Google Scholar]
- An enhanced strategy integrating offline two-dimensional separation and step-wise precursor ion list-based raster-mass defect filter: Characterization of indole alkaloids in five botanical origins of Uncariae Ramulus Cum Unicis as an exemplary application. J Chromatogr a.. 2018;1563:124-134.
- [CrossRef] [Google Scholar]
- Xanthoceraside inhibits pro-inflammatory cytokine expression in Aβ25–35/IFN-γ–stimulated microglia through the TLR2 receptor, myD88, nuclear factor-κB, and mitogen-activated protein kinase signaling pathways. J Pharmacol Sci.. 2013;122:305-317.
- [CrossRef] [Google Scholar]
- Global profiling and novel structure discovery using multiple neutral loss/precursor ion scanning combined with substructure recognition and statistical analysis (MNPSS): characterization of terpene-conjugated curcuminoids in Curcuma longa as a case study. Anal Chem.. 2015;88:703-710.
- [CrossRef] [Google Scholar]
- A new golden age of natural products drug discovery. Cell.. 2015;163:1297-1300.
- [CrossRef] [Google Scholar]
- Evaluation of biodiesel from Xanthoceras sorbifolia Bunge seed kernel oil from 13 areas in China. J for Res (harbin).. 2019;30:869-877.
- [Google Scholar]
- Isomer selectivity of one- and two-dimensional approaches of mixed-mode and hydrophilic interaction liquid chromatography coupled to tandem mass spectrometry for sugar phosphates of glycolysis and pentose phosphate pathways. J Chromatogr a.. 2023;1688:463727
- [CrossRef] [Google Scholar]
- High throughput identification of pentacyclic triterpenes in Hippophae rhamnoides using multiple neutral loss markers scanning combined with substructure recognition (MNLSR) Talanta.. 2019;205:120011
- [CrossRef] [Google Scholar]
- A novel hybrid scan approach enabling the ion-mobility separation and the alternate data-dependent and data-independent acquisitions (HDDIDDA): Its combination with off-line two-dimensional liquid chromatography for comprehensively characterizing the multicomponents from Compound Danshen Dripping Pill. Anal Chim Acta.. 2022;1193:339320
- [CrossRef] [Google Scholar]
- A multi-dimensional liquid chromatography/high-resolution mass spectrometry approach combined with computational data processing for the comprehensive characterization of the multicomponents from Cuscuta chinensis. J Chromatogr a.. 2022;1675:463162
- [CrossRef] [Google Scholar]
- In-depth exploration and comparison of chemical constituents from two Lilium species through offline two-dimensional liquid chromatography combined with multimode acquisition of high-resolution mass spectrometry. J Chromatogr a.. 2022;1670:462980
- [CrossRef] [Google Scholar]
- Simultaneous determination of components with wide polarity and content ranges in Cistanche tubulosa using serially coupled reverse phase-hydrophilic interaction chromatography-tandem mass spectrometry. J Chromatogr a.. 2017;1501:39-50.
- [CrossRef] [Google Scholar]
- Global profiling combined with predicted metabolites screening for discovery of natural compounds: Characterization of ginsenosides in the leaves of Panax notoginseng as a case study. J Chromatogr a.. 2018;1538:34-44.
- [CrossRef] [Google Scholar]
- Xanthoceras sorbifolium Bunge: A Review on Botany, Phytochemistry, Pharmacology, and Applications. Front Pharmacol.. 2021;12:708549
- [CrossRef] [Google Scholar]
- A high-throughput metabolomics approach for the comprehensive differentiation of four Pulsatilla Adans herbs combined with a nontargeted bidirectional screen for rapid identification of triterpenoid saponins. Anal Bioanal Chem.. 2019;411:2071-2088.
- [CrossRef] [Google Scholar]
- Barrigenol-like triterpenoid saponins from the husks of Xanthoceras sorbifolia bunge and their anti-inflammatory activity by inhibiting COX-2 and iNOS expression. Phytochemistry.. 2022;204:113430
- [CrossRef] [Google Scholar]
- Xanthoceraside Could Ameliorate Alzheimer’s Disease Symptoms of Rats by Affecting the Gut Microbiota Composition and Modulating the Endogenous Metabolite Levels. Front Pharmacol.. 2019;10:1035.
- [CrossRef] [Google Scholar]
- Xanthoceraside exerts anti-Alzheimer's disease effect by remodeling gut microbiota and modulating microbial-derived metabolites level in rats. Phytomedicine.. 2022;98:153937
- [CrossRef] [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2023.105445.
Appendix A
Supplementary data
The following are the Supplementary data to this article:Supplementary data 1
Supplementary data 1







