Translate this page into:
Spike lavender essential oil attenuates hyperuricemia and induced renal injury by modulating the TLR4/NF-κB/NLRP3 signalling pathway
⁎Corresponding authors. ph.175@163.com (Jing Sun), 2051028@sntcm.edu.cn (Xiaofei Zhang)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Abstract
Hyperuricemia (HUA), recognized as the fourth “high” condition following hypertension, hyperlipidemia, and hyperglycemia, is a metabolic disorder that severely impairs renal function. Spike lavender essential oil (SLEO) exhibits substantial anti-inflammatory and antioxidant activities and can inhibit xanthine oxidase (XOD), suggesting its potential therapeutic potential against HUA. This of this study aimed to analyze the chemical constituents of SLEO with potential efficacy in treating HUA and to explore their mechanisms of action. Gas chromatography-mass spectrometry (GC–MS) combined with a greedy algorithm identified linalool, β-pinene, and β-caryophyllene as key active components of SLEO. Through network pharmacology “weight coefficient” method, the NOD-like signaling pathway emerged as a significant mechanism for SLEO in treating HUA. Investigating an in vitro uric acid (UA)-induced HK-2 cell model revealed that SLEO effectively inhibited the production of IL-1β and IL-18 in the supernatant of HK-2 cells compared to the NLRP3 (antagonist MCC950). Additionally, a HUA rat model demonstrated that SLEO administration significantly reduced hyperuricemia pathological indicators, such as UA, XOD, and blood urea nitrogen (BUN) levels, with renal histopathological sections showing a marked reduction in following SLEO treatment. Metabolomics and transcriptomics analyses further highlighted significant changes in differential metabolites such as arachidonic acid and glycine, as well as the regulation of differential genes such as pycard and PTGS2 in rats.
Immunohistochemistry, Western blotting, molecular docking and in vitro XOD activity inhibition assays elucidated the active components mechanisms of SLEO in treating HUA and associated renal inflammation. The findings concluded that SLEO mitigates HUA and renal inflammation by modulating the arachidonic acid metabolic pathway and the NF-κB signaling pathway. Moreover, linalool, β-pinene and β-caryophyllene in SLEO were shown to reduce UA production and lower UA levels in vivo by inhibiting the TLR4/NF-κB/NLRP3 pathway, thereby alleviating HUA-induced renal injury.
Keywords
Spike lavender essential oil
Hyperuricemia
Essential oil composition analysis
Multi-omics analysis
Renal injury
- AA
-
arachidonic acid
- BUN
-
blood urea nitrogen
- BCA
-
bicinchoninic acid
- CTD
-
Comparative Toxicogenomics Database
- CRE
-
creatinine
- CC
-
cellular component
- ELISA
-
enzyme-linked immunosorbent assay
- GC-MS
-
Gas chromatography coupled with mass spectrometry
- GO
-
Gene Ontology
- HE
-
hematoxylin and eosin
- HK-2
-
human proximaltubular epithelial cell line
- IL-1β
-
Interleukin-1 beta
- IL-18
-
Interleukin-18
- KEGG
-
Kyoto Encyclopedia of Genes and Genomes
- MDA
-
malonaldehyde
- MF
-
molecular function
- NF-ĸB
-
nuclear factor-kappa B
- NIST20
-
National Institute of Standards and Technology database
- OPLS-DA
-
Orthogonal partial least squares discriminant analysis
- OB
-
oral bioavailability
- PCA
-
principal component analysis
- PPI
-
protein-protein interaction
- PGE2
-
prostaglandin E2
- ROS
-
reactive oxygen species
- SD
-
Sprague Dawley
- SLEO
-
spike lavender essential oil
- S-plot
-
Scatter Plot
- TCA
-
tricarboxylic acid
- TNF-α
-
tumor necrosis factor-α
- TNF-α
-
tumor necrosis factor
- UA
-
uric acid
- XOD
-
xanthine oxidase
- XDH
-
Xanthine Dehydrogenase
- VIP
-
variable importance in prediction
Abbreviations
1 Introduction
Hyperuricemia (HUA) occurs when the body produces excessive uric acid (UA) or excretes insufficient amounts (Kimura et al., 2021). Diagnosis is confirmed by a fasting blood UA level exceeding 420 μmol/L in men and 360 μmol/L in women on two separate occasions. Approximately 98 % of UA in the body is present as sodium salts (Jalal 2016), and prolonged high levels of UA can result in severe inflammatory kidney damage and potential kidney failure. Current HUA treatments, such as allopurinol, which inhibits UA production, and benzbromarone, which promotes UA excretion, often cause adverse effects, including liver and kidney toxicity, anemia, and bone marrow suppression (Lu et al., 2019). Moreover, these drugs do not alleviate UA-induced renal inflammation. Therefore, there is an urgent need for a natural drug with fewer side effects, greater safety, and precise efficacy in treating HUA. HUA-induced renal disease frequently involves inflammatory responses and oxidative stress. Chronic exposure to high UA levels activates the classical nuclear factor-kappa B (NF-κB) inflammatory signaling pathway (Liu et al., 2017). NF-κB activation occurs through urate crystals interacting with Toll-like receptors and the stimulation of NOD, LRR, and pyrin domain-containing protein 3 (NLRP3) inflammasomes (Jo et al., 2016). NLRP3, a vital inflammatory response component, works alongside NLRP1, an apoptosis-associated speck-like protein containing a CARD (ASC), and cysteine protease-74 precursor (Mittal et al., 2014). Activated NLRP3 inflammasomes promote the production of mature interleukin-1β (IL-1β), triggering an inflammatory response (Liu and Liu 2020). Therefore, suppressing the NF-κB/NLRP3 pathway to mitigate renal inflammation and oxidative stress presents a promising novel therapy for HUA.
Spike lavender (Lavandula latifolia Med.), a Mediterranean plant from the Labiatae family, is one of the primary varieties of Provencal lavender. Its essential oil possesses notable aromatic medicinal properties. Due to its specific chemical composition, spike lavender essential oil (SLEO) demonstrates superior anti-inflammatory activity compared to essential oils from the other two varieties and true lavender (Karaca et al., 2023). SLEO effectively reduces the release of cellular inflammatory factors in rheumatic diseases caused by UA accumulation (Carrasco et al., 2016). Additionally, SLEO inhibits UA production by suppressing xanthine oxidase (XOD) activity. Abdelhakim Bouyahya et al. evaluated SLEO's in vitro antioxidant activity by measuring its inhibition of 2,2-diphenyl-1-picrylhydrazyl (DPPH), H2O2, and XOD. For XOD, the semi-inhibitory concentration of SLEO was 8.89 μg/mL, compared to 1.24 μg/mL for allopurinol, the standard for comparison (Hamad Al-Mijalli et al., 2022). Although SLEO's potency approximately 8 times that of allopurinol, it surpasses most natural products. Consequently, SLEO holds potential as a natural dual-action drug for treating HUA: reducing UA production by inhibiting XOD activity and effectively suppressing severe renal inflammation induced by excessive UA accumulation. However, further investigation into its active ingredients and mechanisms of action is necessary.
The traditional unidimensional transcriptomics approach of studying differential genes to understand mechanisms of action is inadequate for elucidating the dynamic changes during disease treatment. To address this, we integrated transcriptomics data with metabolomics to explore the correlations between changes in gene expression and metabolite accumulation (Ritchie et al., 2015). In vivo differential metabolomic data can aid in analyzing the “co-expression” of genes over time, constructing a core regulatory network, identifying key candidate genes, and uncovering the precise mechanisms of drug action during disease treatment. Current network pharmacology often fails to incorporate the most critical quantitative relationships regarding drug efficacy, resulting in inaccurate predictions of pathways and mechanisms (Li et al., 2024). This study proposes using the relative absorption of all drug components, determined by their relative content and oral bioavailability or percutaneous absorption rate, as the weighting value. Using these weight values, pathways in the network pharmacology were annotated according to biological process, cellular component, molecular function, and KEGG enrichment analysis, and then the pathways were reordered by their importance to identify reliable mechanisms of drug action (Wang et al., 2022b).
This research investigated the therapeutic effects of SLEO on HUA and its associated renal injury through in vivo and in vitro analyses. The study included identifying active components in SLEO, establishing a rat HUA model, and utilizing UA-induced HK-2 cells. Additionally, metabolomics, transcriptomics and “weighted” network pharmacology were employed to screen and predict the potential molecular mechanisms of SLEO on HUA. The ultimate goal was to provide a novel therapeutic approach for the clinical treatment of HUA.
2 Methods
2.1 Materials and reagents
SLEO with a purity of over 95 %, was sourced from Shanghai Polaris Aroma Pharmaceutical Technology Co., Ltd,. Assay kits for UA, blood urea nitrogen (BUN), and creatinine (CRE) were obtained from Nanjing Jianjian Biotechnology Co., Ltd., while ELISA kits for determining tumor necrosis factor-α(TNF-α), interleukin-1β (IL-1β), and interleukin-6(IL-6), were acquired from Jiangsu Meimian Industrial Co., Ltd.; Primary antibodies NF-κB-p65, p-NF-κB-p65, and NLRP3 were provided by Abcam Shanghai Trading Co., Ltd. (Shanghai, China). Antibodies for IKK-β, p-IKK-β, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) came from Proteintech Group, Inc. (Wuhan, China). Caspase-1 and XDH antibodies were sourced from Hangzhou HuaAn Biotechnology Co., Ltd. (Hubei, China). Horseradish peroxidase-conjugated secondary antibodies were obtained from Beyotime Biological Technology Co., Ltd. (Wuhan, China). The gas chromatography–-mass spectrometry (GC–MS) system (7890GC/5977MS) was purchased from Agilent Technologies Inc.
2.2 Preliminary evaluation of the efficacy of SLEO in the treatment of HUA
2.2.1 Replication of the HUA model
Forty-eight specific-pathogen-free Sprague Dawley male rats, weighing 180 ± 20 g, (Provided by Chengdu Dashuo Laboratory Animal Co. Ltd (Animal License No.: SCXK (Chuan) 2022-080). This study was approved by the Animal Ethics Committee of Shaanxi University of Traditional Chinese Medicine (SUCMDL20220703002). The rats were randomly assigned to six groups of eight rats each, following five days of acclimatization: blank control, model, positive drug, SLEO low-dose (20 mg/kg), SLEO medium-dose (40 mg/kg), and SLEO high-dose (80 mg/kg) (Honda et al., 2014). Rats in all groups were gavaged with adenine (25 mg/kg) and potassium salt of oxonic acid (300 mg/kg) diluted in 0.5 % sodium carboxymethylcellulose solution for 21 days in a row, with the exception of the blank control group. This was done to create a model of HUA (Qin et al., 2021). All the groups were administered their corresponding treatments 3 h after the induction of HUA. The blank and model groups received an aqueous solution of 1 % Tween 80 daily, the positive drug group received an aqueous solution of 1 % Tween 80 containing allopurinol (−), and the SLEO groups received the respective dose of SLEO for 21 consecutive days. Each dose of SLEO administered was configured with 1 % Tween 80 as the solvent. (Fig. 1A).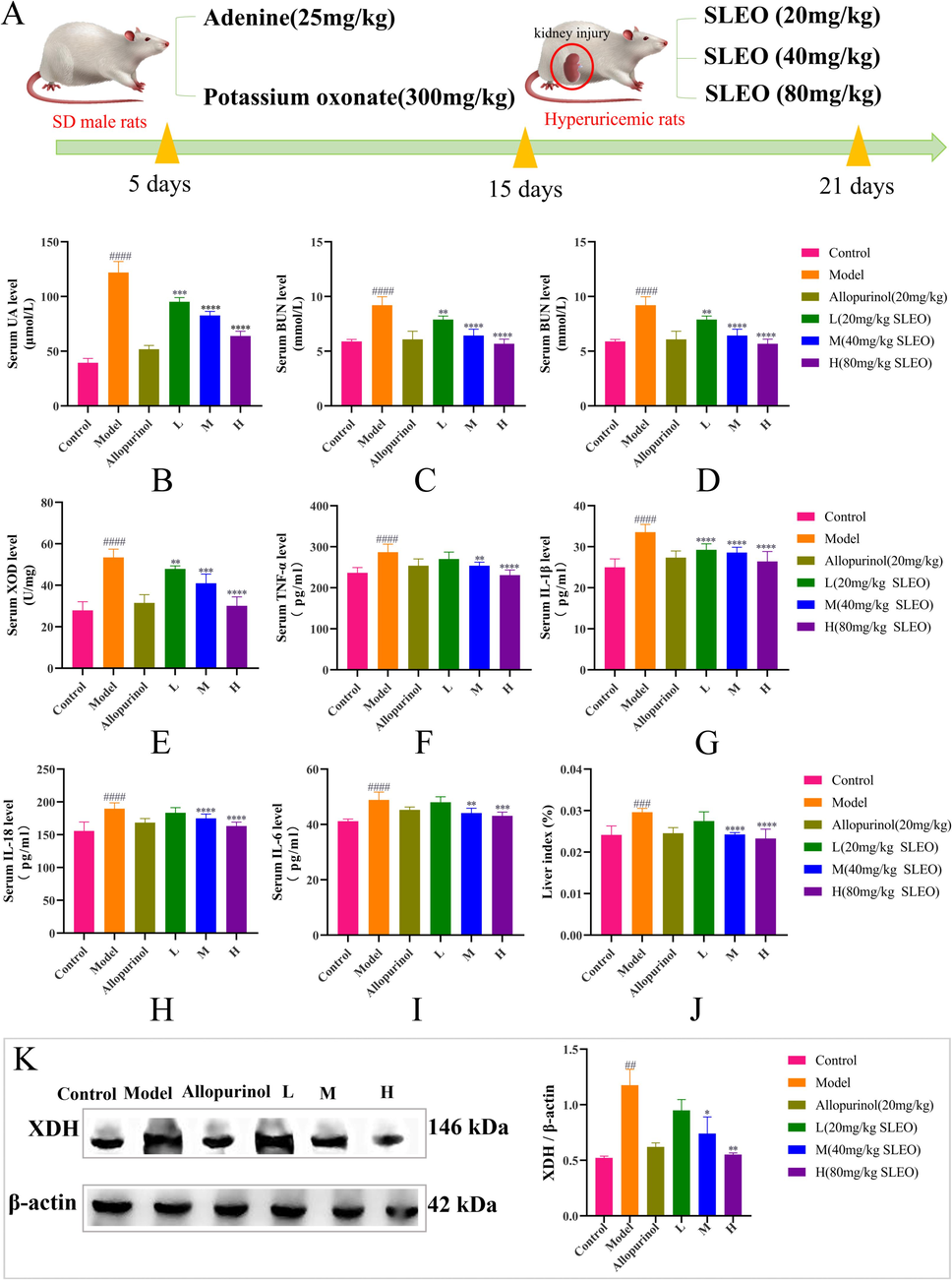
The serum levels of A. Hyperuricemia Modeling Flowchart, B. UA, C. BUN, D. CRE, E. XOD, F. TNF-α, G. IL-1β, H.IL-18 and I. IL-6 in each group of rats J. Liver index for each group of rats K. Levels of XDH protein expression levels in liver tissues of rats in each group (Standard deviation ± mean (n = 6), *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 vs. Model group., #P<0.05, ##P<0.01, ###P<0.001, ####P<0.0001 vs. Control group.).
2.2.2 Serum and tissue sample collection
After the last drug administration for 3 h in each group, the rats were anesthetized with 1 % pentobarbital sodium solution before being sacrificed. Blood was collected from each group of rats, and the serum was centrifuged and stored frozen at −80 °C. Both the kidneys and liver tissues were extracted and weighed.
2.2.3 Measurement of serum indicators of HUA
Serum samples were thawed at room temperature, and the levels of UA, CRE, BUN, XOD, and inflammatory TNF-α, IL-1β, and IL-6 were measured using enzyme-linked immunosorbent assay (ELISA) kits. Additionally, XDH protein expression in rat livers of rats was assessed.
2.3 Determination of the active ingredients of SLEO using gas chromatography–mass spectrometry (GC–MS)
The components of SLEO (Shanghai Polaris Aroma Pharmaceutical Technology Co.; purity > 95 %) were analyzed and identified using GC–MS. An HP-5MS capillary column (30 m × 250 μm × 0.25 μm) was used for chromatography. It was equipped with a high-purity helium flow rate of 1 mL/min, a 1.5 μL injection volume, and a controlled heating mode. The temperature was increased from 65 °C to 90 °C at 5 °C/min, kept for 1 min, then elevated to 210 °C at 5 °C/min and maintained for 1 min. The following parameters applied to the mass spectrometer: full scan mode, 20–350 atomic mass units, ion source, electron ionization, electron energy, 70 V, MS quadrupole temperature, 150 °C, solvent delay, 4 min (Lebanov et al., 2021).
First, the GC–MS results were analyzed and retention indices of the components in SLEO were calculated against the standard spectral database of the National Institute of Standards and Technology (NIST). The components were screened based on the match, retention index and relevant literature. The retention indices of the components of SLEO were calculated based on the retention indices of the n-alkanes (Beale et al., 2017). See formula 1, n-hexadecane served as an internal standard, and a standard solution (0.05 g/mL) was prepared. The relative content of each component in SLEO was determined using the internal standard method.
2.4 Network pharmacological predictions
2.4.1 Identification of the targets of SLEO active ingredients and HUA target
The CAS numbers of the active ingredients in SLEO were entered into PubChem (https://pubchem.ncbi.nlm.nih.gov/) to obtain their simplified molecular-input line-entry system notations. The relevant targets of the active ingredients in SLEO were obtained through the SwissTargetPrediction (https://www.swisstargetprediction.ch/) and TargetNet (https://targetnet.scbdd.com/) databases (Li et al., 2023). HUA targets were retrieved from GeneCards (https://www.genecards.org/), Online Mendelian Inheritance in Man (https://www.omim.org/, OMIM), and Comparative Toxicogenomics Database (https://ctdbase.org/, CTD).
2.4.2 Constructing a “component–disease–target” interaction network
The overlap between the targets of SLEO active ingredients and HUA disease targets was obtained using Venny 2.1.0 and imported into the STRING database (https://cn.string-db.org/) to construct protein–protein interaction (PPI) networks and to hide relationship line segments with very low interaction scores. The degrees of PPI were evaluated and ranked, and the screened target proteins were imported into Cytoscape 3.9.1 to construct a component–disease–target visualization network (Zhou et al., 2023b).
2.4.3 Establishment of a weighting factor for introducing “ weight coefficient ”
Considering the route of administration of SLEO, weighting coefficients were assigned to network pharmacological predictions to accurately reflect the significance of each key pharmacodynamic component within the pharmacological mechanism. The weighting coefficient for each active ingredient was calculated by multiplying the probability of its oral bioavailability by its relative content in SLEO (Wang et al., 2023). The established relationships were as follows:
2.4.4 Reordering of enriched pathways based on the “quantitative efficiency” weighting coefficients
To accurately elucidate the mechanism of action of SLEO in treating HUA, the enriched signaling pathways identified through KEGG and Gene Ontology analysis were reordered based on their weighting coefficients, calculated using formula (2), (3), and (4).
2.5 Establishment of a model of uric acid-induced HK-2 cells
2.5.1 Cell culture
Human renal tubular epithelial cells (HK-2), obtained from Procell Life Science & Technology Co., Ltd., were cultured in DMEM with 10 % fetal bovine serum at 37 °C and 5 % CO2. In subsequent experiments, HK-2 cells were pretreated with SLEO or 10 μM NLRP3 inhibitor (MCC950, HY-12815, MedChem Express) for 24 h, followed by stimulation with appropriate amounts of UA (Zhou et al., 2023a).
2.5.2 Cell viability assay
The viability of HK-2 cells assessed at varying concentrations of UA (400, 300, 200, 100, 50, and 25 μg/mL) and SLEO (1, 0.5, 0.25, 0.125, 0.0625 and 0.0375 μL/mL) according to the MTT kit instructions, with absorbance was measured at 490 nm (Cui et al., 2020). Uric acid sodium salt (UA, 1198–77-2, MedChem Express) was prepared to a concentration of 200 μg/mL of soluble UA at the time of prophylaxis, and 10 μM MCC950 was dissolved in 0.9 % sodium chloride solution.
2.5.3 ELISA assay
Determination of important inflammatory indicators in the supernatant of each cell group was performed using standard ELISA kits.
2.6 Metabolomics analysis
2.6.1 Preparation and analysis of metabolomic samples
Thaw serum stored at −80 °C at room temperature. To 50 μL of serum, the internal standard seventeen carbonic acid and methanol were added. The mixture is homogenized and allowed to stand at −20 °C for 10 min, then centrifuged to remove the proteins. The supernatant was aspirated and freeze-dried. Its lyophilised powder was collected and dissolved in methoxyaminopyridine solution (15 mg/mL) and then reacted with methoxyamine hydrochloride for 2 h at 60 °C with stirring. The reaction was sealed again by adding N,O-bis(trimethylsilyl)trifluoroacetamide, cooled at 4 °C, and centrifuged at 12,000 rpm for 10 min. The supernatant was analyzed by GC–MS. Repeat all the above steps to prepare quality control samples (Shan et al., 2021).
HP-5MS capillary column (30 m × 250 μm × 0.25 μm) was used for chromatography, and it was filled with 1.5 μL of high-purity helium and flowed at a rate of 1 mL/min. The temperature was ramped up to 260 °C at 10 °C/min, then to 300 °C at 2 °C/min in the planned ramping mode, without shunt, after being held at 80 °C for two minutes. Mass spectrometer conditions were as follows: full scan mode in the range of 20–450 atomic mass units; ion source, electron ionization; electron energy, 70 V; MS quadrupole temperature, 150 °C; solvent delay, 5 min (Chen et al., 2016).
2.6.2 Metabolomics analysis
Serum metabolomics data were analyzed using specialized software, and standard mass spectra from the NIST 20 database employed to match the purified mass spectra of the identified metabolites. Compounds with less than 80 matches in the serum were removed, and the peak area of each sample was used as a variable and normalized for all samples. The collated data were imported into MetaboAnalyst 5.0 (https://www.metaboanalyst.ca/) for principal component analysis (PCA) to reveal the distribution of metabolites in the rat serum samples. An orthogonal partial least squares discriminant analysis (OPLS-DA) model was constructed using the SIMCA software to visualize the metabolic changes after SLEO treatment. Screening criteria were set with a variable importance in projection (VIP) score > 1 and P value < 0.05 were levied as the screening criteria, and the screened metabolites were considered as differential metabolites, which were imported into MetaboAnalyst 5.0 for metabolic pathway analysis (Zhu et al., 2023).
2.7 Transcriptomics analysis
2.7.1 RNA-seq sample preparation
Fifteen specific-pathogen-free, adult, male Sprague Dawley rats, each weighing approximately 200 g, were acclimatized to standard conditions of temperature (25 ± 2 °C) and humidity (60 ± 5 %) for 1 week before being randomly assigned to either the blank, model, or SLEO groups (five rats per group). The HUA model was established as described earlier, and the SLEO group received 40 mg/kg of SLEO for 21 days. At the end of the treatment, the rats were anesthetized with 1 % pentobarbital sodium before being sacrificed. Collect their kidney tissue and freeze it for preservation (Shen et al., 2023).
2.7.2 RNA-seq analysis
RNA-seq libraries were constructed using the Illumina NovaSeq Reagent Kit. Total RNA was extracted from rat kidney tissues using Trizol reagent. Subsequently, mRNA was isolated, fragmented, and primed, followed by synthesis of the first and second strands of cDNA were synthesized in a thermal cycler to construct a transcriptome library. RNA quality was assessed by the RNA integrity number (RIN) using the Agilent 2100 detector, where higher RIN values indicate better and more complete RNA. Finally, RNA-seq analysis was conducted using the free Majorbio Cloud platform (https://www.majorbio.com) (Wang et al., 2022a).
2.7.3 Weighted gene co-expression network analysis (WGCNA)
WGCNA can identifies highly synergistic gene sets and candidate biomarkers or therapeutic targets by examining gene set end-conjugation and gene set-phenotype associations. This method yields precise gene function enrichment results from transcriptome sequencing, revealing key hub genes (Kakati et al., 2019).
Differential gene and HUA trait files from transcriptome sequencing were analyzed using the Majorbio Cloud Platform (https://www.majorbio.com) WGCNA panel, excluding genes with expression levels ≤ 2 were screened out. Initially, the weighting coefficient β\betaβ gene correlations within of each sample group of samples were determined according to the scale-free network principle to construct the gene clustering tree and divide the gene modules. Subsequently, the gene module most associated with the trait was identified, based on module-trait associations and gene enrichment within the module. This approach established the gene regulatory network within the module uncovering the regulatory core genes (hub genes) and the regulatory relationships among individual genes (Cheng et al., 2022).
2.8 Integration of transcriptomics and metabolomics data
To elucidate the signaling and metabolic changes associated with SLEO treatment, the differential metabolites identified through metabolomics and the differential genes from transcriptomics were analyzed using the OmicShare tool (https://www.omicshare.com/tools). The correlations between the two sets of data were analyzed using Spearman's coefficient and a dynamic correlation network diagram were employed to analyze correlations between the two data sets. Additionally, key targets and differential metabolites identified from transcriptome sequencing were imported into MetScape, a Cytoscape 3.9.1 plug-in, to construct the compound–reaction–enzyme–gene network (Huang et al., 2023).
2.9 Validation of the mechanism of action of SLEO against HUA
2.9.1 Greedy algorithm to screen possible key components of SLEO for HUA treatment
For precise identification and screening of key active ingredients in SLEO for molecular docking studies, the greedy algorithm (Doi and Imai 1999, Gu et al., 2024), was introduced as an innovative tool for drug ingredient screening. Initially, intersection targets of SLEO and ingredient-disease relationships were merged and analyzed to map their correspondence between SLEO and intersection targets. Subsequently, the greedy algorithm, as detailed in the supplementary material, was used to identify the set of SLEO bioactive ingredients that cover the possible targets for HUA treatment. The final screened active ingredients represent the core components of SLEO for treating HUA, enhancing the accuracy of molecular docking predictions.
2.9.2 Molecular docking
The pdb structure files of possible target proteins for HUA treatment, identified through “weighted” network pharmacology and multi-omics analysis, were downloaded. Based on GC–MS analysis, “weighted” network pharmacology predictions, and greedy algorithm screening 2D structure files of the component compounds in sdf format were also downloaded. These the structure files were then imported into Discovery Studio 4.0 and PyMOL software for molecular docking and binding energy calculations.
2.9.3 Hematoxylin and eosin (HE) staining of renal tissue
Rat left kidney tissues were embedded in paraffin, dehydrated using an ethanol gradient, fixed in 4 % paraformaldehyde, sliced to a thickness of 5 μm, stained with HE following standard protocol, and examined under a light microscope (Nikon (Japan) ECLIPSE 80I) (Li et al., 2022).
2.9.4 Immunohistochemistry
Protein detection in rat kidney tissues was performed using immunohistochemistry. Paraffin-embedded tissues were sectioned into 5-μm slices, hydrated, incubated in 3 % H2O2 at room temperature for antigen retrieval, blocked 30 min, and probed overnight at 4 °C in a humid chamber with the corresponding primary antibodies targeting IL-1β, p-NF-κB-p65, and ASC, left at room temperature for 1 h on the following day, rinsed thrice in phosphate-buffered saline for 3 min each time, incubated for 60 min at 37 °C with horseradish peroxidase-labeled secondary antibodies developed for color and sealed for microscopic observation (Ren et al., 2020).
2.9.5 Western blotting
Proteins extracted from kidney and liver tissues were stored at −80 °C until analysis. Proteins were separated by electrophoresis and transferred to a membrane, which was blocked at room temperature for 70 min. The membrane was then probed overnight at 4 °C with primary antibodies NF-κB-p65, p-NF-κB-p65, NLRP3 (1:1000; Abcam, ab214185, Shanghai, China), caspase-1 (1:1000; HUAbio, ET1608-69, Hangzhou, Zhejiang, China), IKK-β, p-IKK-β, glyceraldehyde 3-phosphate dehydrogenase (1:1000; Proteintech, 60004–1-Ig, Wuhan, Hubei, China), and XDH(1:1000; HUAbio, 55156–1-AP, Wuhan, Hubei, China). After three washes at room temperature with Tris-buffered saline containing Tween 20 for 5 min each the membrane was incubated for 50 min at room temperature with horseradish peroxidase-conjugated secondary antibody (1:10,000; Beyotime, P0258, Beijing, China). Finally, the membrane was washed three times with Tris-buffered saline containing Tween 20 for 5 min each and developed using an enhanced chemiluminescence reagent.
2.10 Statistical analysis
All the experimental data were statistically analyzed using GraphPad Prism 9.4.1 and expressed as the mean ± standard deviation. One-way analysis of variance determined the statistical significance of intergroup differences, with P<0.05 and P<0.01 considered significant.
3 Results
3.1 Validation of the anti-HUA efficacy of SLEO
3.1.1 UA, CRE, BUN, and XOD levels after SLEO treatment
ELISA results indicated that serum levels of UA, CRE, BUN, and XOD were significantly elevated in the model group compared with the blank group (P<0.05), confirming the successful replication of the HUA model. Compared with the model group, the levels of UA, CRE, BUN, and XOD were significantly reduced compared to the model group (Fig. 1B–E). SLEO alleviated HUA by lowering UA levels and inhibiting XOD activity.
3.1.2 XDH expression and inflammatory factor levels in liver and serum after SLEO treatment
ELISA results, also showed that the serum levels of TNF-α, IL-1β, IL-6 and IL-18 in HUA rats were significantly different in HUA rats compared to the blank group (P<0.05). These levels were significantly reduced by SLEO treatment (P<0.05, Fig. 1F–I). In addition, the liver coefficients of HUA rats were significantly increased, whereas those of the SLEO high-dose group and the blank group did not significantly different (Fig. 1J). Western Blot analysis demonstrated that XDH protein expression was significantly reduced in the SLEO treatment group (P<0.01) (Fig. 1K).
3.2 Identification of the components of SLEO
Using GC–MS, the ion mass spectrum of SLEO was obtained, which was characterized in conjunction with retention indices by searching the NIST library. SLEO was found to contain 16 active ingredients, including eucalyptol, linalool, β-pinene, and β-caryophyllene (Fig. 2A and Table 1). SLEO was found to contain 16 active ingredients, including eucalyptol, linalool, β-pinene, and β-caryophyllene. Analyzing the content of active ingredients in SLEO, it was found that β-caryophyllene and linalool accounted for a large proportion of the content in SLEO (Fig. 2A and Table 1).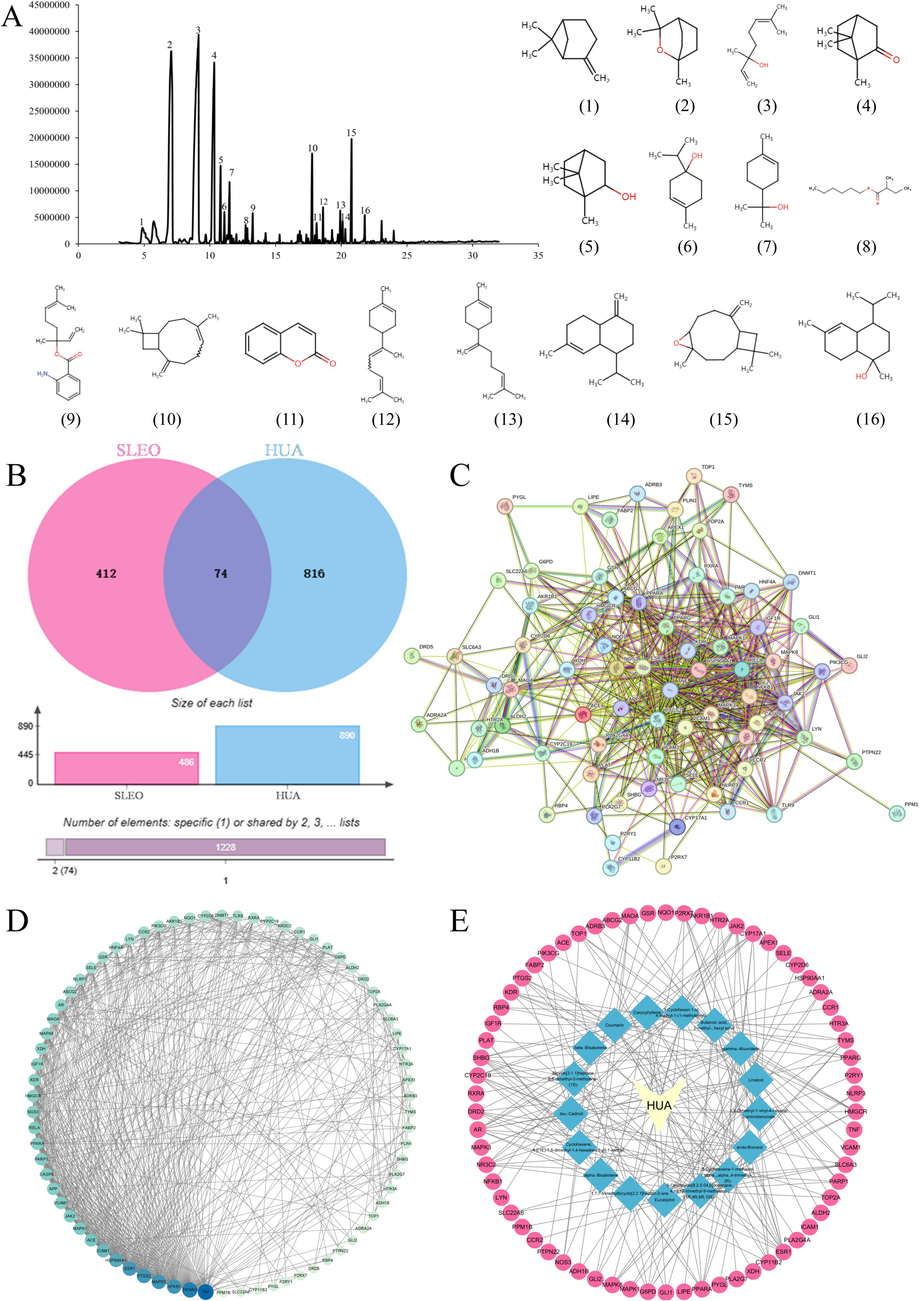
SLEO active ingredient identification A. Total ion flow diagram of SLEO (1–16 is the structure of active ingredient No. 1–16 in SLEO) B. Cross-targeting map of SLEO and hyperuricemia C. Protein interaction visualization network diagram D. Protein interaction degree value ranking E. “Target–disease” visualization network diagrams.
NO.
Compound name
CAS
RI
relative content
(mg/g)
1
Bicyclo[3.1.1]heptane,6,6-dimethyl-2-methylene-, (1S)-
018172-67-3
1079.386
0.52
2
Eucalyptol
000470-82-6
1128.31
2.61
3
Linalool
000078-70-6
1204.611
5.38
4
1,7,7-trimethylbicyclo[2.2.1]heptan-2-one
000076-22-2
1246.111
1.27
5
endo-Borneol
000507-70-0
1262.825
0.48
6
3-Cyclohexen-1-ol, 4-methyl-1-(1-methylethyl)-
000562-74-3
1272.912
1.54
7
3-Cyclohexene-1-methanol,.alpha.,.alpha.,4-trimethyl-, (R)-
007785-53-7
1286.166
2.10
8
Butanoic acid, 2-methyl-, hexyl ester
010032-15-2
1329.5
0.53
9
1,5-Dimethyl-1-vinyl-4-hexenyl 2-aminobenzoate
007149-26-0
1348.378
0.949
10
β-Caryophyllene
000087-44-5
1515.511
6.8
11
Coumarin
000091-64-5
1529.043
1.29
12
β-pinene
000495-61-4
1600.352
6.08
13
γ-Muurolene
030021-74-0
1608.044
3.34
14
Cyclohexene, 4-[(1E)-1,5-dimethyl-1,4-hexadien-1-yl]-1-methyl-
017627-44-0
1636.363
0.53
15
5-Oxatricyclo[8.2.0.04,6]dodecane, 4,12,12-trimethyl-9-methylene-, (1R,4R,6R,10S)-
001139-30-6
1678.321
1.08
16
T-Cadinol
005937-11-1
1734.441
0.05
3.3 Network pharmacological prediction
3.3.1 Key target prediction and interaction network construction
A total of 486 SLEO component-related targets and 890 disease targets were obtained through database queries and data deduplication, resulting in 74 were intersecting targets (Fig. 2B). The connection degrees between targets were obtained from the PPI network (Fig. 2C), and they were ranked by importing them into Cytoscape 3.9.1. The prediction results indicated that SLEO might treat HUA through four main targets, namely, TNF-α, NF-κB, peroxisome proliferator-activated receptor, and PTGS2 (Fig. 2D). Disease–component network interactions were visualized using Cytoscape 3.9.1 (Fig. 2E).
3.3.2 Functional enrichment of targets after introducing the “ weight coefficient ”
The 74 intersecting targets underwent enrichment analysis for cellular components and molecular functions using R. Weighting coefficients were applied to reorder the enriched biological processes and KEGG pathways. Among the 121 enriched KEGG pathways that were reordered, the nucleotide-binding oligomerization domain-like (NOD-like) receptor signaling pathway was promoted from the 27th to the 4th position after reordering, indicating that SLEO may alleviate HUA via the NOD-like receptor signaling pathway (Fig. 3A–B). Significant changes were also observed in the ranking of biological processes post-reordering (Fig. 3C–D). Notably, the NOD-like receptor signaling pathway is a classical inflammatory pathway, and HUA is closely linked to the activation of NLRP3 inflammasomes. The top 10 cellular components and molecular functions of the mechanism of action associated with SLEO treatment of HUA have been enriched using R, as shown in Fig. 3E–F.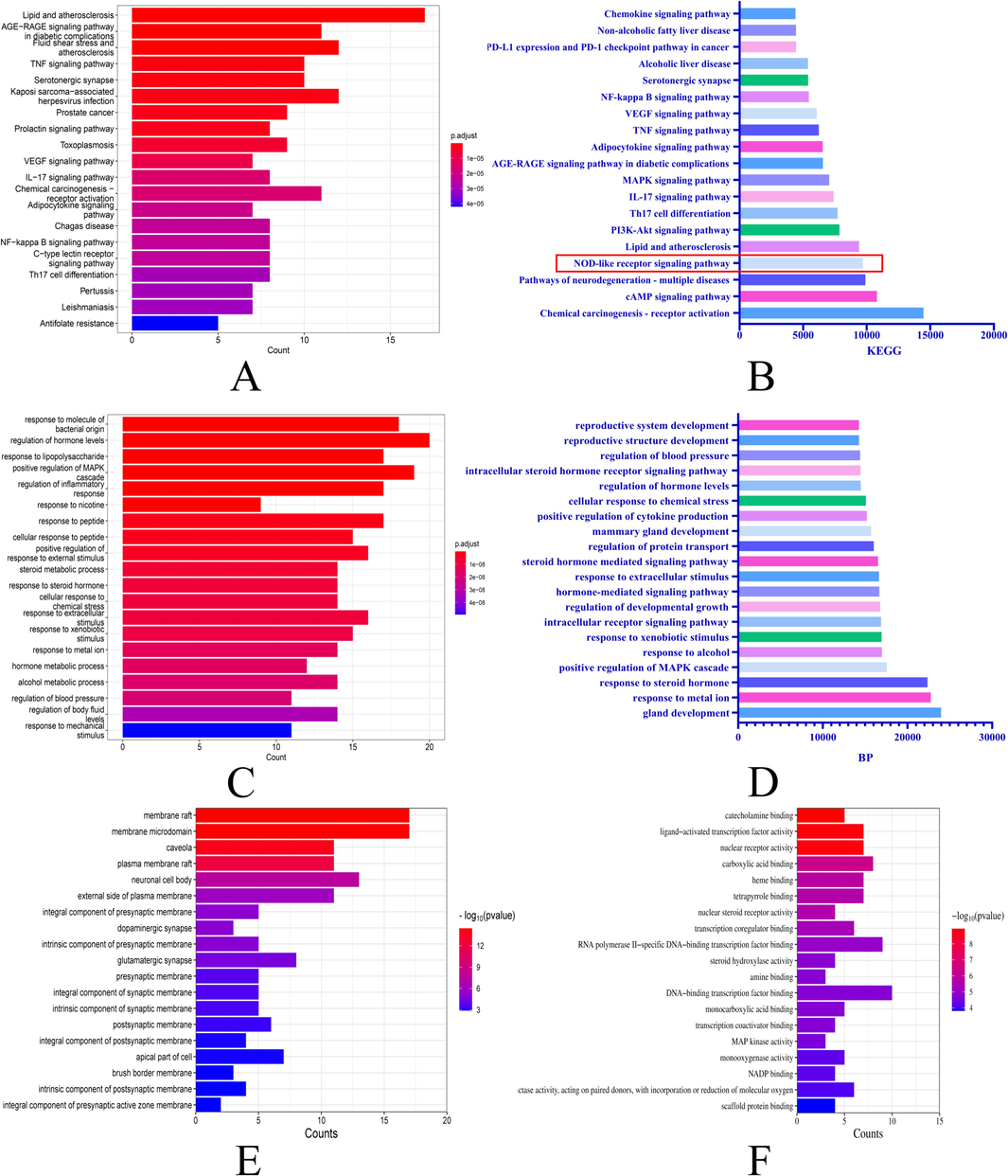
Changes in the ordering of KEGG pathways and BPs after introducing weighting coefficients A. KEGG pathways predicted by network pharmacology B. Reordering of KEGG pathways after introducing weighting factors C. BPs predicted by network pharmacology D. Reordering of BPs after introducing weighting factors E. Cellular components predicted by network pharmacology F. Molecular functions predicted by network pharmacology.
3.4 SLEO attenuates HUA-induced renal inflammation in vitro by inhibiting NLRP3 inflammasome activation
MTT results that cell viability remained above 90 % at a UA concentration of 200 μg/mL, and SLEO (0.125 μL/mL) had minimal impact on cell viability (Fig. 4A–B). Based on the above, subsequent in vitro experiments were performed using UA 200 μg/ mL and SLEO 0.125 μL/mL as the intervention concentrations for the cells. Elisa results showed that both SLEO and MCC950 had a significant inhibitory effect on inflammatory cytokines compared to the control (P<0.001) (Fig. 4C–E).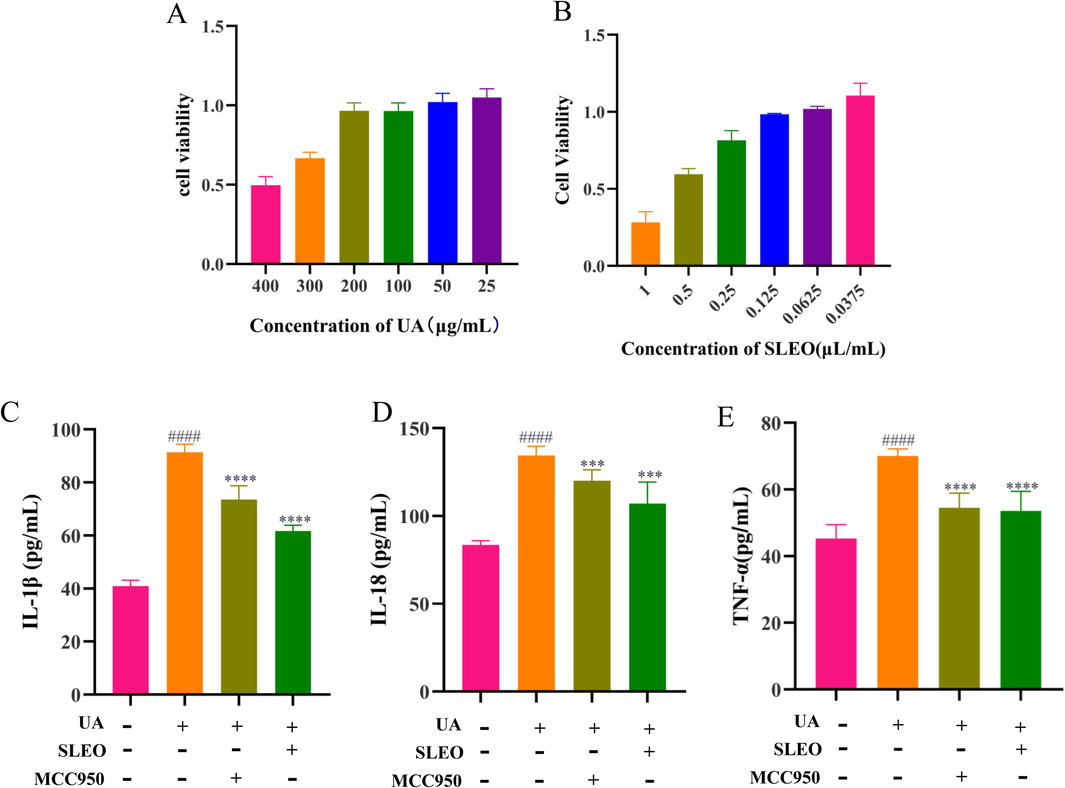
Effects of SLEO on UA-induced HK-2 cells A. Effect of UA on cell viability. B. Effect of SLEO on cell viability. C. Levels of IL-1β in cell supernatants. D. Levels of IL-18 in cell supernatants. E. Levels of TNF-α in cell supernatants (Standard deviation ± mean (n = 6), *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 vs. Model group., #P<0.05, ##P<0.01, ###P<0.001, ####P<0.0001 vs. Control group.).
3.5 Metabolic differentials and metabolic pathways analysis
3.5.1 Serum metabolomics and the screening of differential metabolites
The differential serum metabolites were analyzed using MetaboAnalyst 5.0 and identified by subjecting the quality control samples, the control, model, positive drug, and SLEO treatment groups to PCA. The SLEO group was considerably separated from the model group in the PCA scatter plot, indicating that the metabolite content was significantly altered after treatment and metabolic processes might also be affected or reversed to some extent (Fig. 6A). We used SIMCA to analyze metabolic differences between the blank control, model and SLEO groups. The metabolomes of the blank control and the model groups were well-separated (R2X [cumulative] = 0.725, R2Y=0.999, Q2 [cumulative] = 0.956), corroborating the successful establishment of the HUA model (Fig. 5A). Analysis of metabolic data from model and SLEO groups (R2X [cumulative] = 0.337, R2Y=0.981, Q2 [cumulative] = 0.85), suggesting that SLEO significantly altered the metabolic content in the serum of HUA rats (Fig. 5D). In addition, these results were validated by S-plot and permutation tests (n = 200) (Fig. 5 B, E). A total of 200 permutation tests were performed to confirm the validity and applicability of the OPLS-DA model (Fig. 5C, F). Applying variable importance in projection score > 1 and P<0.05 in the OPLS-DA model as screening criteria, 12 differential metabolites were identified between the blank control and the model groups and 18 between the SLEO and the model groups (Tables 2 and 3).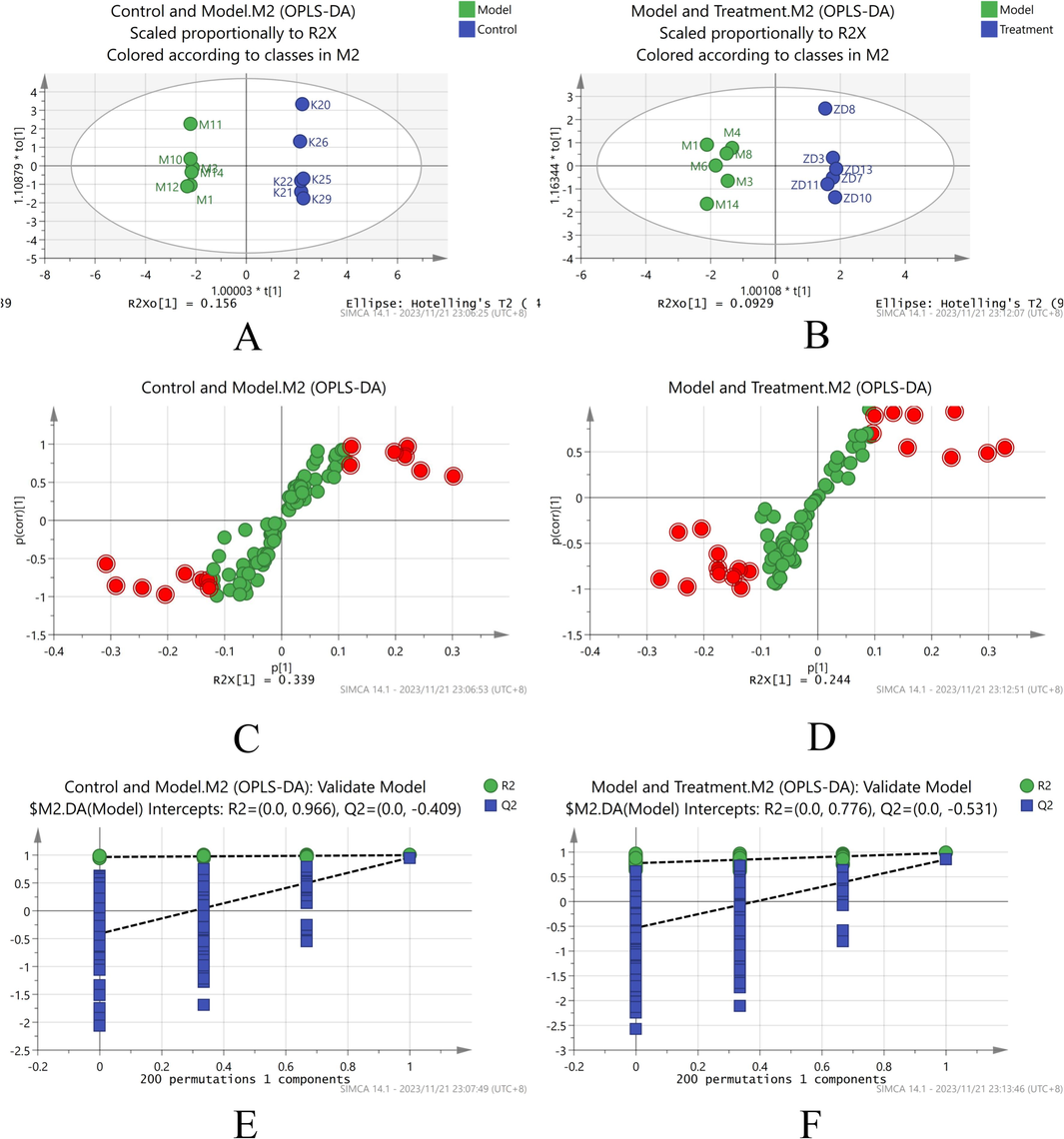
SIMCA Analysis of Metabolic Differentials. A. OPLS-DA of the blank control and model groups. B. OPLS-DA of the model and SLEO groups. C. S-plot of the control and model groups. D. S-plot of the model and SLEO groups. E. Stability experiments on the blank control and model groups. F. Stability experiments on the model and SLEO groups.
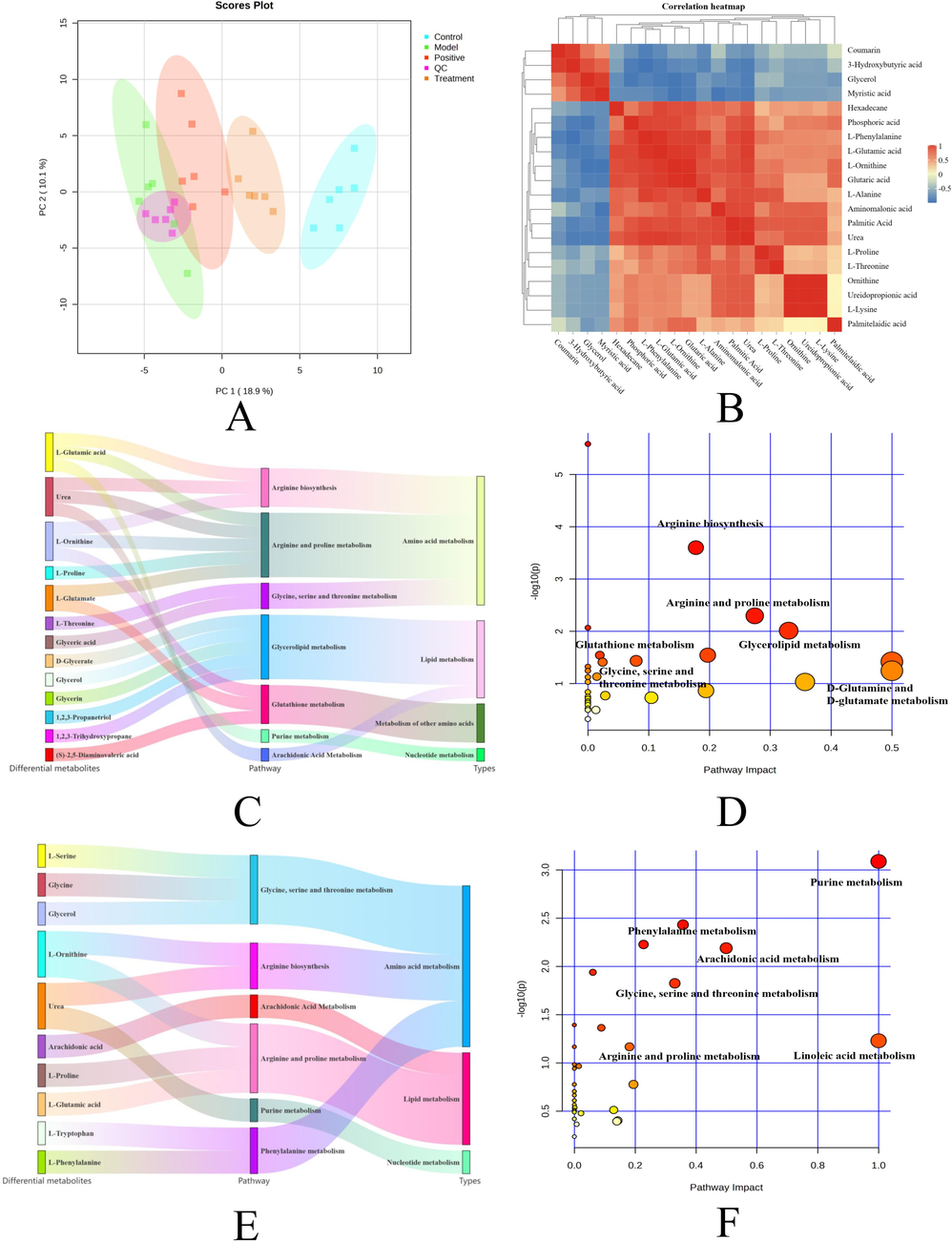
A. Plot of PCA scores for the five groups based on their serum metabolic profiles. Blue represents the control group, green represents the model group, red represents the positive group, orange represents the SLEO group, and purple represents the quality control group B. Heatmap of metabolic differences between the model and SLEO groups C. Sankey diagram depicting the functional enrichment of differential metabolites of the control and model groups. D. Functional enrichment of differential metabolites of the control and model groups. E. Sankey diagram depicting the functional enrichment of differential metabolites of the model and SLEO groups. F. Functional enrichment of differential metabolites of the model and SLEO groups.
CAS.
Metabolites
RT (min)
P. Value
VIP
Trend
002582-79-8
Myo-Inositol
18.2234
0.000829
1.32082
Down
005630-82-0
Glycine
9.1839
0.016574
1.10441
Down
006787-10-6
Glycerol
8.7319
1.45E-07
1.53657
Down
007364-47-8
L-Proline
9.0298
0.000226
1.38299
Up
007364-51-4
L-Phenylalanine
13.2613
0.042012
1.0086
Up
007536-83-6
Tyrosine
16.0597
0.003133
1.2806
Down
015985-07-6
L-Glutamic acid
13.1651
5.19E-05
1.4398
Up
017596-96-2
Lactic Acid
5.6066
0.00027
1.36949
UP
018297-63-7
Urea
8.3376
0.024103
1.01096
Down
024595-70-8
L-Ornithine
13.0783
1.08E-06
1.51554
UP
027844-07-1
L-Alanine
6.2124
0.00385
1.20967
UP
030274-77-2
Proline
12.0207
0.000687
1.33449
Down
031038-11-6
Phosphoric acid
14.8576
0.000222
1.36643
UP
038191-87-6
Glyceric acid
9.5397
0.005464
1.1673
Down
038191-88-7
Threonic acid
12.3477
0.001457
1.28665
Down
053925-65-8
Probucol
12.2419
0.000168
1.38953
Down
055133-94-3
3-Hydroxybutyric acid
7.049
9.82E-05
1.41549
Down
055429-07-7
L-Lysine
16.4441
0.002199
1.24191
UP
055520-89-3
Palmitic Acid
17.5022
0.00292
1.23373
UP
055520-95-1
Dodecanoic acid
13.4055
5.03E-05
1.46441
Down
055556-70-2
L-Ornithine
15.3865
0.000148
1.41828
UP
056259-07-5
Linoleic acid
19.0216
0.010085
1.1119
UP
060022-87-9
Glutaric acid
12.6265
0.002864
1.24869
UP
060615-84-1
Urea
7.395
2.85E-07
1.52964
UP
070125-39-2
L-Serine
8.4723
0.000805
1.31397
Down
113516-18-0
Arachidonic acid
19.0502
0.029044
1.12911
Up
1010333-14-1
L-Ornithine monochlorohydrate/ornithine
14.6557
0.000307
1.3631
Down
1033331-59-7
L-Tryptophan
19.1562
1.83E-05
1.47432
Down
1079394-06-1
Niacinamide
11.4438
1.02E-05
1.4743
Down
959048-60-3
L-Homoserine
13.7708
0.011169
1.1342
Up
959080-51-4
Aminomalonic acid
11.3765
3.42E-07
1.52448
Down
CAS.
Metabolites
RT (min)
P. Value
VIP
Trend
000473-08-5
Coumarin
16.6367
0.020515
1.05398
Up
000544-76-3
Hexadecane
11.0878
0.020189
1.05541
Down
006787-10-6
Glycerol
8.7319
1.96E-06
1.5033
Up
007364-47-8
L-Proline
9.0204
0.003614
1.23805
Down
007364-51-4
L-Phenylalanine
13.2613
2.01E-06
1.51441
Down
007537-02-2
L-Threonine
10.2994
0.001793
1.28998
Down
015985-07-6
L-Glutamic acid
13.1651
4.87E-06
1.5071
Down
018603-17-3
Myristic acid
15.5404
0.007628
1.23577
Up
024595-70-8
L-Ornithine
13.0784
0.001307
1.36257
Down
027844-07-1
L-Alanine
6.2124
0.001863
1.28225
Down
031038-11-6
Phosphoric acid
14.8576
0.004212
1.23107
Down
038191-87-6
Glyceric acid
9.5397
0.010126
1.16263
Down
038191-88-7
Threonic acid
12.3477
2.39E-07
1.54739
Up
055133-94-3
3-Hydroxybutyric acid
7.049
4.70E-05
1.46577
Up
055255-77-1
Ureidopropionic acid
10.7704
0.010418
1.46577
Down
055429-07-7
L-Lysine
16.4443
0.005543
1.20996
Down
055520-89-3
Palmitic Acid
17.5022
0.001545
1.3343
Down
055556-70-2
L-Ornithine
15.3865
7.51E-05
1.44497
Down
060022-87-9
Glutaric acid
12.6266
4.02E-05
1.47224
Down
060615-84-1
Urea
7.395
2.74E-07
1.53615
Down
1079394-06-1
Niacinamide
11.4438
5.20E-06
1.50691
Up
1206693-35-7
Palmitelaidic acid
17.3098
0.011256
1.11349
Down
959080-51-4
Aminomalonic acid
11.3765
0.009303
1.12768
Down
3.5.2 Differential metabolite screening and metabolic pathway analysis
The results indicated significant alterations in serum metabolite content of the rats changed after SLEO treatment (Fig. 6B). The differential metabolites identified between the model and SLEO groups were imported into MetaboAnalyst 5.0 to determine the metabolic pathways possibly altered. We discovered that 31 metabolic pathways were potentially enriched in the SLEO group relative to the model group, among which arginine biosynthesis, glutathione metabolism, arginine and proline metabolism, D-glutamine and D-glutamate metabolism, and the metabolism of glycine, serine, and threonine may be the key pathways involved in mediating the therapeutic effects of SLEO against HUA (Fig. 6D). Sankey diagrams illustrated the linkage of individual metabolites to the enriched pathways and to denote the type of metabolism (Fig. 6C). In addition, we also studied and analysed the metabolic pathways of the blank and model groups as a secondary reference for the administration group (Fig. 6E-F).
3.6 Differential gene screening, functional enrichment analysis and hub genes mining
The analysis of 30,560 differential genes was analysed utilized a false discovery rate < 0.05 and |log2 (fold- change) | > 2 as criteria. (Fig. 7A) Cluster analysis was used to visualize the similarity between samples (Fig. 7B). A heatmap comparing the model and the treatment groups revealed a large difference in gene expression between the two groups (Fig. 7C). KEGG enrichment analysis identified 85 pathways involving differential genes, such as including NOD-like receptor signaling, arachidonic acid metabolism, and NF-κB signaling (Fig. 7F). The molecular functions mainly enriched in the set of differential genes included intracellular calcium-activated chloride channel activity, intracellular chloride channel activity, and kringle domain-binding. The cellular components mainly enriched in the set of differential genes included protein-containing complex, extracellular region part and membrane part. The biological processes mainly enriched in the set of differential genes included cellular component organization or biogenesis, multicellular organismal process, biological regulation (Fig. 7D). Gene Set Enrichment Analysis linked closely associated genes and signaling pathways and, visualizing the expression trends of key target proteins in the NOD-like receptor signaling pathway, which ranked first in the functional enrichment analysis (Fig. 7G). Sankey diagrams illustrated the chief enriched pathways and key targets (Fig. 7E).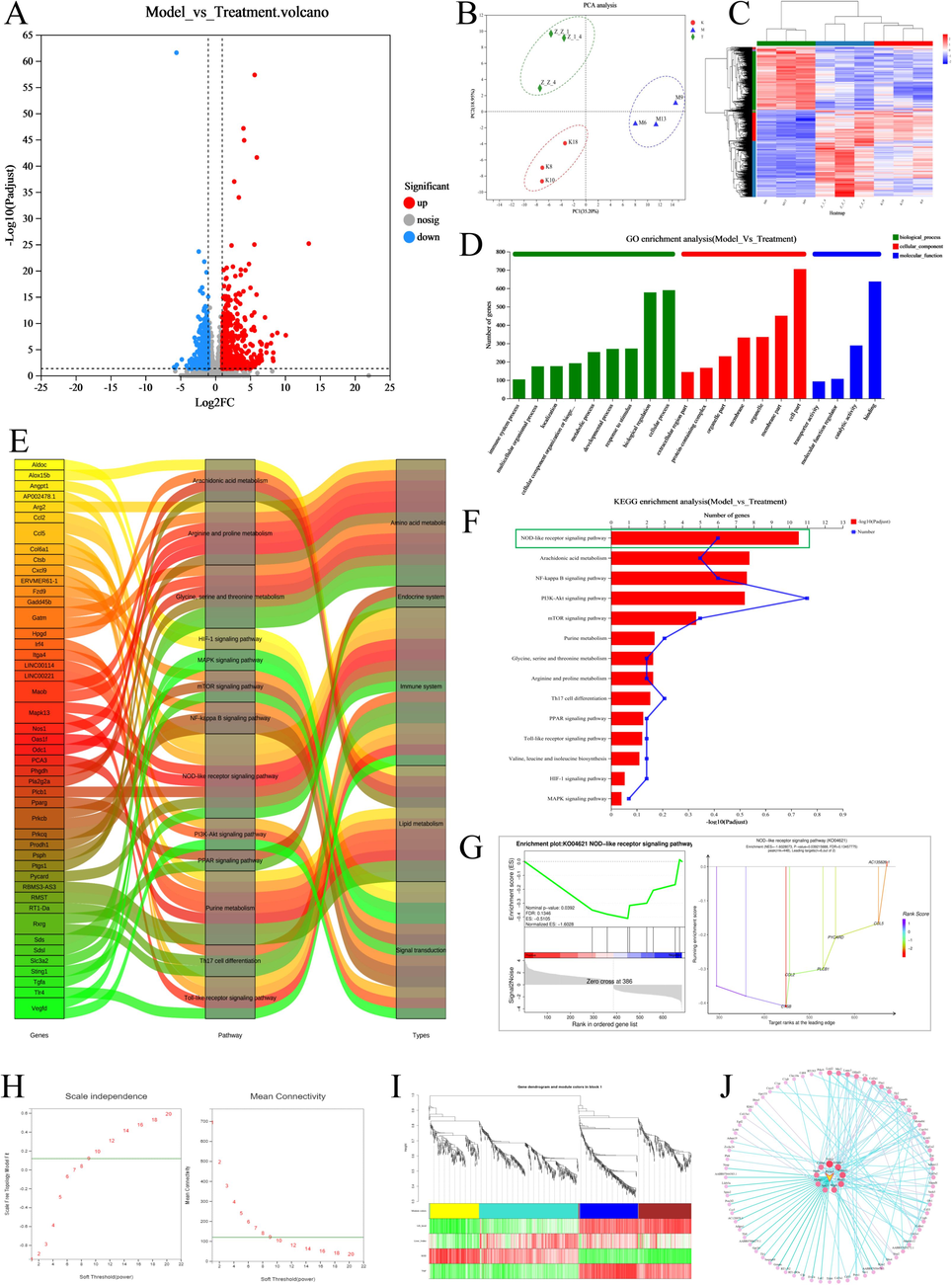
Transcriptome and WGCNA sequencing results A. Volcano plots of differential genes between the model and blank groups, showing both up- and downregulated genes B. PCA clustering plot of transcriptome sequencing samples demonstrates correlation between the samples C. Heatmap of differential gene clustering across groups. Analysis of gene clustering in each group of samples D. GO enrichment of BP, CC, and MF in the set of differential genes after the administration of SLEO E. Sankey diagrams show the major pathways through which SLEO might act and the key targets within them. F. The main KEGG pathways enriched in the set of differential genes between the model and the SLEO groups. G. GSEA of the NOD-like receptor signaling pathway. H. The soft threshold was set at 9, where the green horizontal line indicates the correlation coefficient of 0.8 I. Correlation analysis between traits and modules J. Mining of hub genes in the turquoise module visualization network.
A total of 1,553 differential genes were imported for WGCNA and 1,092 were retained based on TOM value screening. A power value of 9 was determined so that the correlation coefficient between genes was greater than 0.8 (Fig. 7H). Module hierarchical classification and module identification revealed that most genes were enriched in the turquoise module (Fig. 7I). We constructed a visual network to identify hub genes in the turquoise module. WGCNA indicated that SLEO may likely treat HUA via the ASC-encoding gene in the turquoise module (Fig. 7J).
3.7 Integrating transcriptomics and metabolomics to uncover key mechanisms of action of SLEO against HUA
The interactions between genes and metabolites were elucidated to explore the mechanism of action of SLEO against HUA by integrating metabolomics and transcriptomics data. Correlations between differential genes and metabolites and filtered out the “target–metabolite” combinations with the closest possible interaction (Fig. 8A). Studying the dynamic network relationships between differential genes and differential metabolites revealed that the targets nitric oxide synthase 1 (NOS1), XOD, and ASC were closely related to the differential metabolites in the SLEO group, suggesting that SLEO may impact metabolism through these targets (Fig. 8B).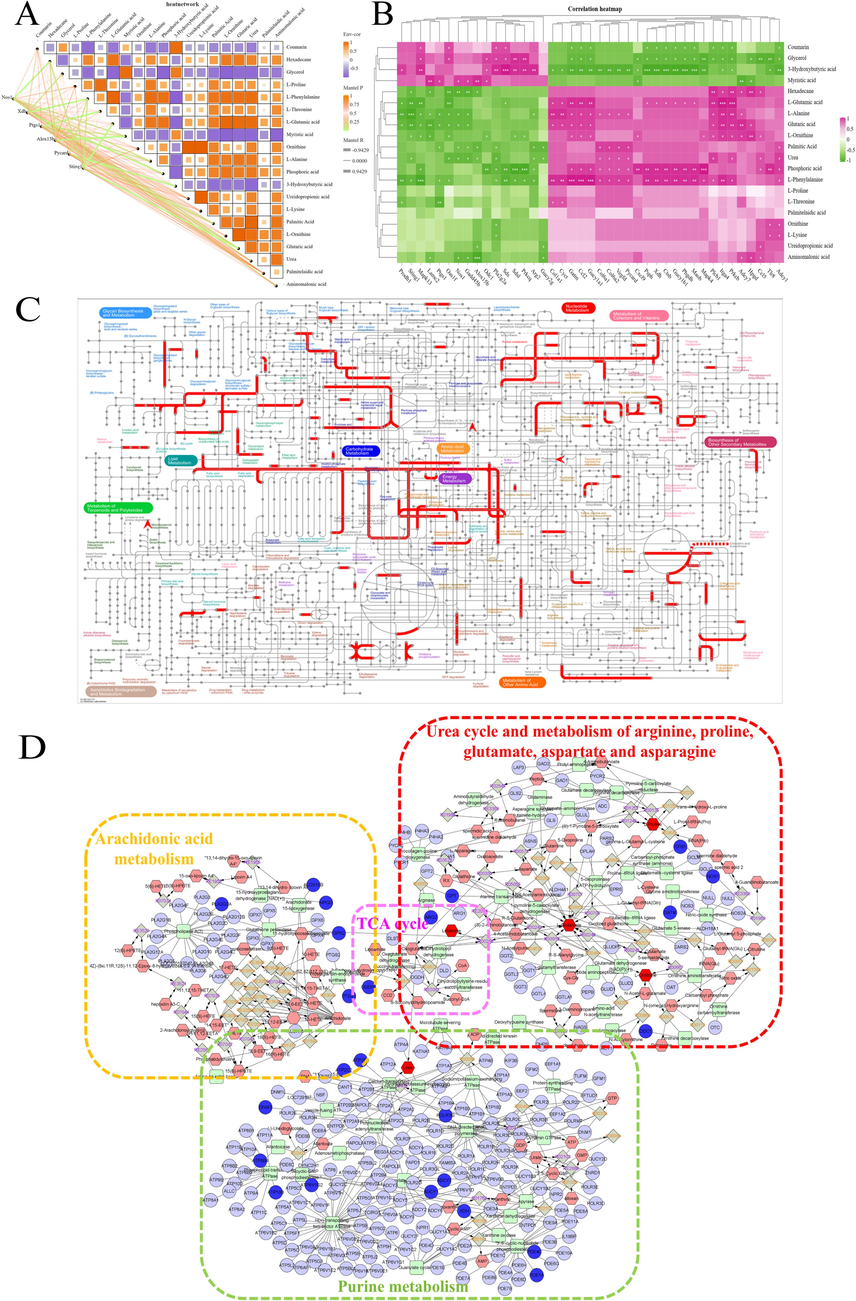
A. Dynamic correlation network map for differential metabolites and differential genes. B. Heatmap depicting the correlations between differential metabolites and differential genes. C. Integrated transcriptome and metabolome analysis in iPath 3.0. D. Compound–reaction–enzyme–gene network diagram.
An analysis of the compound–response–enzyme–gene network indicated that SLEO may affect the AA pathway in HUA rats, which is highly correlated with the inflammatory response, through PTGS1 and arachidonate 15 lipoxygenase. Additionally, changes in NOS1 expression after treatment may also be related to the urea synthesis pathway (Fig. 8D). An in-depth comprehensive analysis of metabolic differential-free and differential genes using iPath 3.0 (https://pathways.embl.de) revealed that amino acid metabolism, lipid metabolism, and nucleotide metabolism pathways were significantly enriched after SLEO treatment of hyperuricemia, and that SLEO influences hyperuricemia by modulating amino acid metabolic pathways and participating in arachidonic acid metabolic processes (Fig. 8C).
3.8 Identification of key active ingredients in SLEO and their corresponding binding targets
3.8.1 Molecular docking results
Sixteen active ingredients of SLEO identified through GC–MS were analyzed. Using the greedy algorithm, these ingredients that could cover the 75 intersecting targets by the correspondence between the 16 ingredients and the 75 intersecting targets of the ingredients and diseases predicted by network pharmacology. The active ingredient set includes 11 key active ingredients from SLEO. Among them, linalool, caryophyllene and β-pinene play important roles in the treatment of HUA by SLEO (Fig. 9F and Table S1).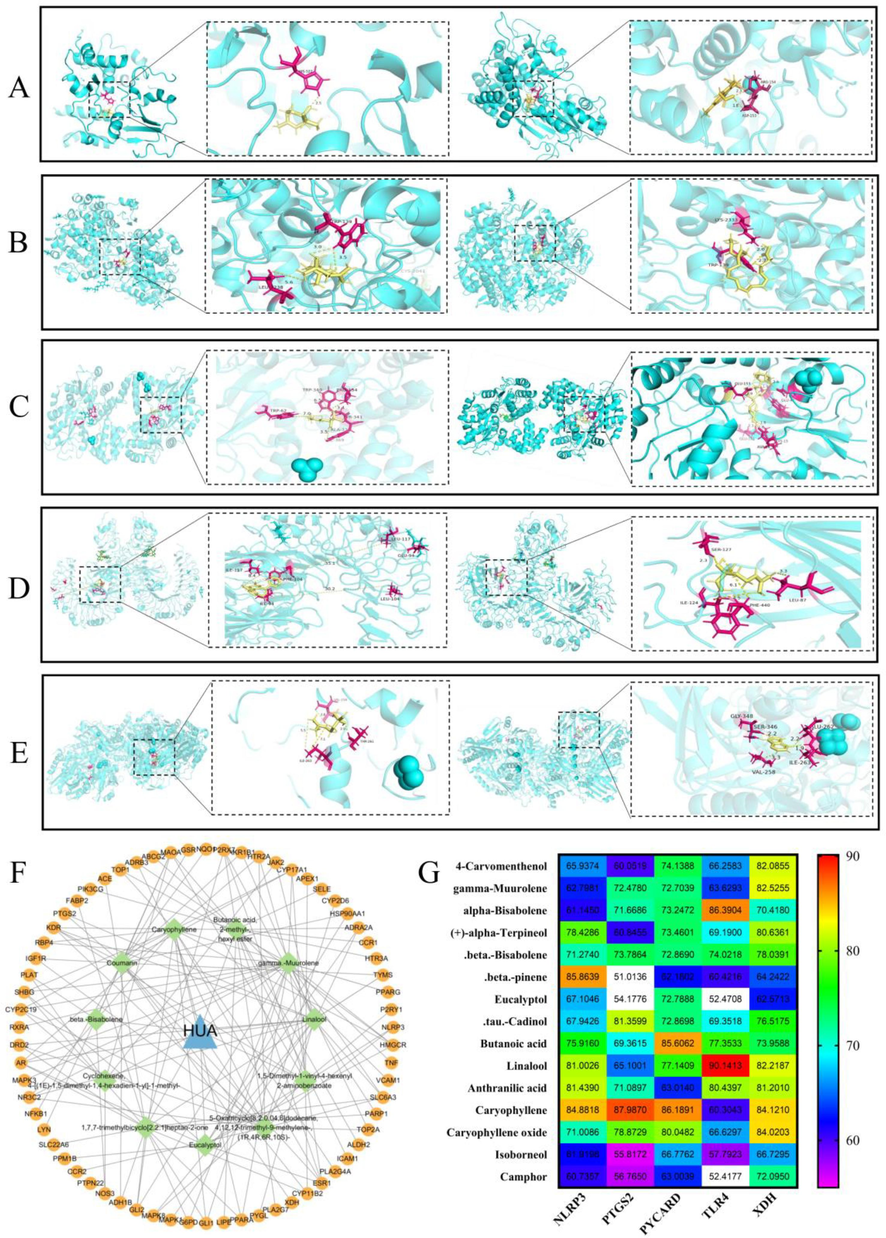
Molecular docking results. A. Docking results of β-pinene and NLRP3 (left); positive drug indomethacin and NLRP3(right) B. Docking results of β-caryophyllen and COX-2(left); positive drug celecoxib and COX-2 (right) C. Docking results of β-caryophyllen and ASC (left); positive drug melphalan(right) and ASC D. Docking results of linalool and TLR4(left); positive drug alprazolam and TLR4 (right) E. Docking results of β-caryophyllen and XOD (left); positive drug allopurinol and XOD (right) F. Plot of key components in 11 SLEO screened by the greedy algorithm in relation to the target network. G The binding scores of the key active ingredients to the targets.
Key target proteins such as TLR4, NLRP3, XOD, ASC, and COX-2 were selected as the key target proteins based on their PPI degree ranking and the core components of SLEO screened by greedy algorithm. The active ingredients of SLEO constituted the ligands, while fostamatinib, celecoxib, alprazolam, allopurinol, and melphalan were selected as positive control drugs. The binding scores of linalool to TLR4, β-caryophyllen to XOD, and β-pinene to NLRP3 were comparable to those of the positive control drugs to their protein targets, suggesting that linalool, β-caryophyllene, and β-pinene maybe the key active components of SLEO for the treatment of HUA and kidney injury (Fig. 9A-E). Binding fractions of active ingredients to proteins and 2D binding structural formulae (Fig. S1). The binding scores of the key active ingredients to the targets are shown in Fig. 9G.
3.8.2 Preliminary validation of molecular docking results
The molecular docking results were preliminarily verified by in vitro determination of the contents of the key factors in the relevant mechanism of action (Khalaf et al., 2022). First, the maximum administered dose of the three active ingredients was determined by the MTT, which yielded a cell survival rate of > 90 % at linalool (80 μM), β-pinene (10 μM) and β-stigmasterol (80 μM), shown in Fig. 10A-C. Two active components of SLEO, linalool and β-caryophyllene, exhibit strong binding properties with TLR4 and COX-2 on the NF-κB signaling pathway. To preliminarily verify this finding, classical inflammatory factors TNF-α and IL-6, downstream of NF-κB (Fu et al., 2023), were measured in cell supernatant after in vitro UA-induced HK-2 modeling and administration of linalool and β-caryophyllene (Wei et al., 2023). In vitro experiments with linalool and β-caryophyllene significantly reduced TNF-α and IL-6 levels (p < 0.05) are shown in Fig. 10G-H, suggesting that these components in SLEO may block the NF-κB pathway by binding to TLR4 and COX-2 targets. Additionally, β-pinene and β-caryophyllene were closely linked to the activation of NLRP3 inflammasomes. The activation of NLRP3 inflammasomes leads to increased levels of inflammatory factors, such as IL-1β and IL-18, exacerbating kidney injury (Özenver and Efferth 2021). In vitro experiments demonstrated that the administration of β-pinene and β-caryophyllene significantly reduced IL-1β and IL-18 levels (p < 0.05), shown in Fig. 10E-F. The study also provides preliminary validation of the molecular docking results through XOD in vitro activity inhibition assays, shown in Fig. 10D.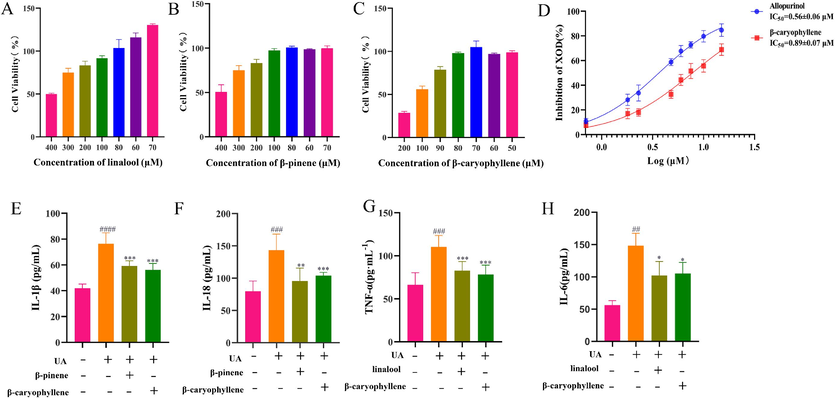
A Determination of the maximum administered dose of linalool by the MTT. B Determination of the maximum administered dose of β-pinene by the MTT. C Determination of the maximum administered dose of β-caryophyllene by the MTT. D Experimental results of XOD inhibition by β-caryophyllene. E Effects of β-caryophyllene and β-pinene on IL-1β in NLRP3 inflammasome activation. F Effects of β-caryophyllene and β-pinene on IL-18 in NLRP3 inflammasome activation. G Effect of linalool and β-caryophyllene on TNF-α content on the NF-κB pathway. H Effect of linalool and β-caryophyllene on IL-6 content on the NF-κB pathway. (*P<0.05, **P<0.01, ***P<0.001vs. Model group., ##P<0.01, ###P<0.001, ####P<0.0001 vs. Control group.).
3.9 Validation of key target proteins and the therapeutic mechanism of SLEO against HUA
3.9.1 HE staining of renal tissues
HE-stained sections of renal tissues revealed prominent infiltration of inflammatory cells (green arrows) in the tissues of the model group, while a small number of renal tubular epithelial cells appeared to be sparsely edematous and detached (yellow arrows). In contrast, the renal tissues of rats in the SLEO group showed a significant reduction in inflammatory cell infiltration, including a significant improvement in pathological features in the renal tissues of the high-dose group (Fig. 11).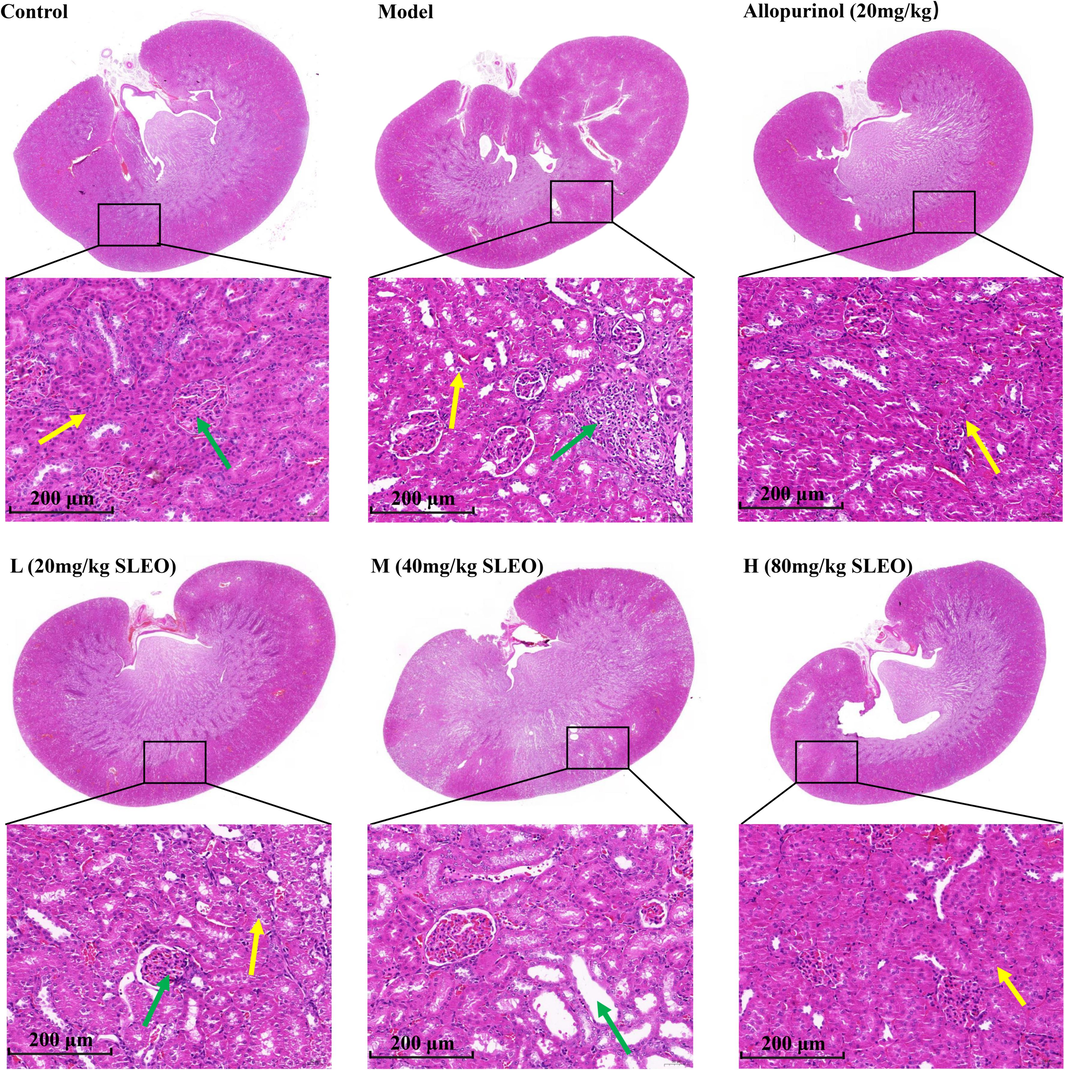
HE staining of rat kidney tissues after treatment of hyperuricemia with SLEO. Renal tissues in the model group had more severe infiltration of inflammatory factors compared with those in the control group, while the renal histopathology of rats in the high-dose group was significantly improved by SLEO treatment.
3.9.2 Immunohistochemistry
The expression of the inflammation-related proteins ASC, IL-1β, and p-NF-κB-p65 was higher in the renal tissues of rats in the model group compared with those in the blank group (P<0.05), but it was lower in the renal tissues of rats in the SLEO group compared with those in the model group (P<0.01) (Fig. 12A-D).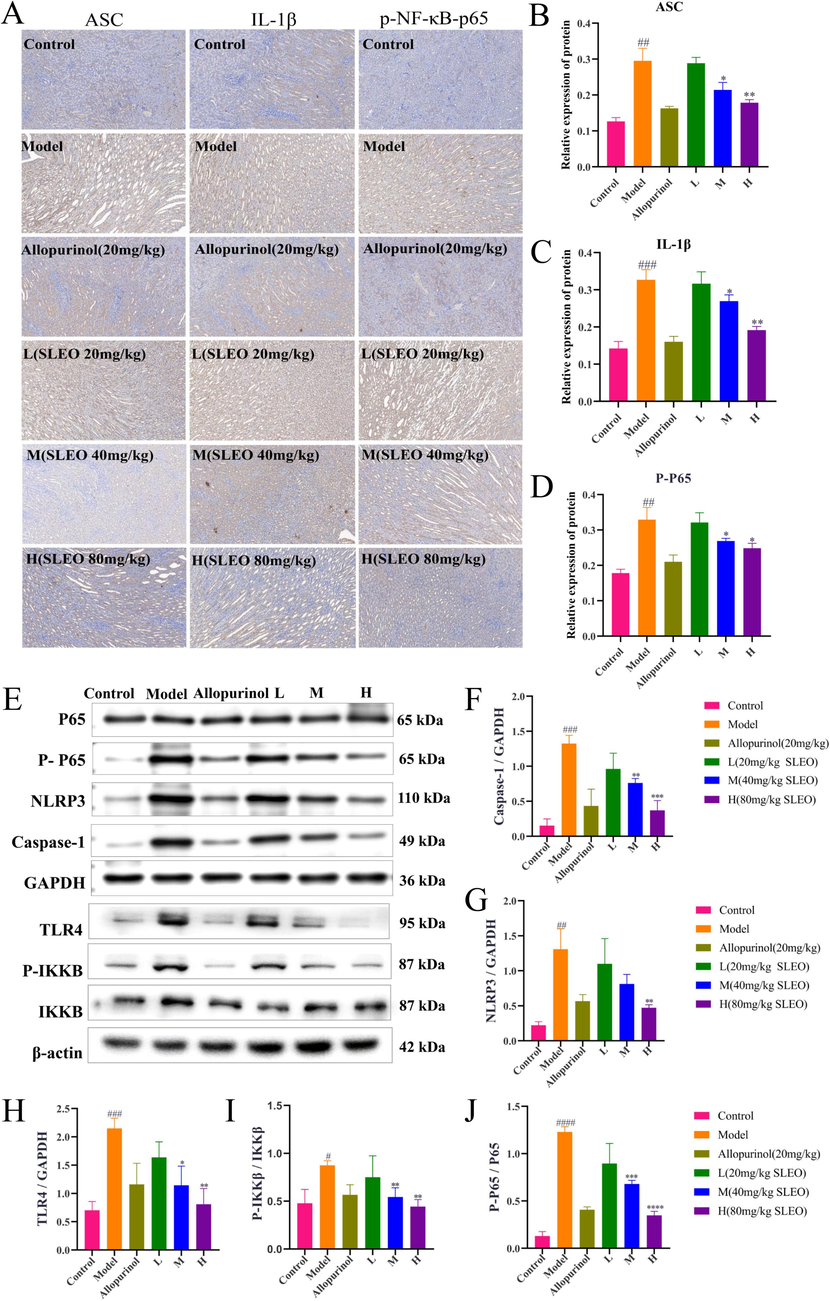
A-D The immunohistochemical detection of protein expression in the renal tissues of rats in various group. E-J The detection of protein expression in the kidney tissues of rats in each group by Western blotting. (Standard deviation ± mean (n = 3), *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 vs. Model group., #P<0.05, ##P<0.01, ###P<0.001, ####P<0.0001 vs. Control group.).
3.9.3 Western blotting
Since we have established that SLEO alleviates HUA by modulating the NF-κB/NLRP3 pathway, we investigated the changes in protein expression in this pathway after SLEO administration. The protein expression of TLR4, p-IKKB/IKKB, p-P65/P65, NLRP3, and caspase-1 was significantly increased in the renal tissues of the model group compared with those of the control group. SLEO treatment decreased the expression of these proteins in a dose-dependent manner (Fig. 12E-J). The results of Western blotting indicated that SLEO could inhibit the release of inflammatory factors mediated by NF-κB/NLRP3, thereby mitigating renal inflammation.
4 Discussion
During the development of HUA, XOD promotes the overproduction of UA in the body, leading to the formation of urate crystals (Pan et al., 2020), These crystals are highly pro-inflammatory, inducing local inflammation that can eventually cause rheumatoid arthritis (Zamudio-Cuevas et al., 2015).Besides the joints, urate crystals were shown to be deposited in large numbers in other tissues (Ahmad et al., 2021), with the collecting ducts of the kidneys being the most susceptible to damage (Khanna et al., 2020). The crystals can adhere to the renal tubular epithelium and stimulate the release of inflammatory mediators from inflammatory vesicles within the epithelial cells (Su et al., 2020), leading to the rupture of the tubular wall and the escape of the crystals into the interstitium, which results in chronic interstitial inflammation and aggravates kidney damage (Cameron and Simmonds 1981). The anti-inflammatory and antioxidant active components in SLEO as determined by GC–MS, such as linalool, β-pinene, and caryophyllene, may play a key antagonistic role in the pathogenesis of HUA.
The NF-κB/NLRP3 axis exacerbates HUA and promotes renal failure. In this study, “weight coefficient ” network pharmacology predicted that SLEO ameliorated HUA via the NOD-like receptor signaling pathway. RNA-seq analysis further revealed that the expression of NOD-like receptor signaling and NF-κB signaling was significantly altered after SLEO treatment. We successfully established an HUA model and found that the levels of IL-1β, IL-6, and TNF-α were elevated in the sera of HUA rats. The expression of NLRP3, ASC, and caspase-1 was also enhanced in the renal tissues of HUA rats. The classical NLRP3 inflammasome can be activated through two pathways (Aachoui et al., 2013). One mode is NF-κB-mediated activation, which mainly relies on the binding of pathogen-associated and damage-associated molecular patterns to pattern recognition receptors (Boaru et al., 2015) and the consequent upregulation of NLRP3 and pro-IL-1β genes, eventually priming the NLRP3 inflammasome into a state where it can be activated (Zhong et al., 2016). IL-1β is a cytokine associated with inflammatory immune responses (Joosten et al., 2013). During NLRP3 inflammasome activation, caspase-1 will prompt the conversion of pro-IL-1β into mature IL-1β for extracellular secretion (McGettrick and O'Neill 2013). The other mode of NLRP3 inflammasome activation is transcription-independent. NF-κB can be activated via TLR, inducing NLRP3 phosphorylation on S194, which triggers the assembly of inflammatory vesicles (Tannahill and O'Neill 2011). TLR4 is a pattern recognition receptor (Zhang et al., 2022) that binds TIR-domain-containing adaptor protein and recruits myeloid differentiation primary response protein 88 to trigger a signaling cascade that includes IκB phosphorylation and NF-κB-mediated inflammation when the organism is injured (Li et al., 2021). NF-κB enters the nucleus and stimulates the expression of many proinflammatory mediators, such as cyclooxygenase-2 (COX-2), IL-1β, IL-6, inducible NOS, and TNF-α (van Loo and Beyaert 2011). However, in the present study, in vitro uric acid induced HK-2 cells, and I found that both the SLEO administration group and the NLRP inhibitor (MCC950) group significantly inhibited the production of IL-1β and IL-18.SLEO had the same effect as MCC950. In addition, the serum levels of IL-1β, IL-6, TNF-α and IL-18 as well as the expression levels of NLRP3, ASC, caspase-1 and TLR4 in renal tissues were significantly reduced in the rats administered with SLEO.
The AA pathway is closely related to the therapeutic mechanism of SLEO against HUA. AA, a major component of cell membrane lipids, is mainly metabolized via three enzymatic pathways: COX, lipoxygenase (LOX), and cytochrome P450 (CYP450) (Zhang et al., 2023). Through these enzymatic pathways, AA can trigger severe inflammatory responses. In the kidney, the main metabolites produced by AA are prostaglandins, thromboxanes, leukotrienes, and hydroxyeicosatetraenoic acids (HETEs) (Wang et al., 2019), all of which contribute to impairing renal function. In the late stage of HUA, crystals deposited in the renal tubules increase intratubular pressure and intrarenal vascular resistance while decreasing renal blood flow, which ultimately reduces the glomerular filtration rate and causes acute renal failure (Ejaz et al., 2012). LOX and CYP450 in the AA metabolic pathway were reported to be closely related to renal blood flow and glomerular filtration rate (Imig and Khan 2015). 20-HETE is generated through the CYP450-catalyzed ω-hydroxylation of AA, which is also mediated by CYP1A, CYP4A11, and CYP4F in P450 metabolism (Hoff et al., 2011). LOXs metabolize AA into leukotrienes, HETEs, and lipotoxins. Leukotrienes generated by 15-LOX promote renal inflammation. Leukotrienes B4 and D4 cause a decrease in glomerular filtration rate and aggravate tubulointerstitial injury (Surh et al., 2001). AA is released from cell membranes when they are subjected to external stimuli, especially inflammatory responses, and COX2 converts free arachidonic acid in vivo into PGE2 (Breyer and Harris 2001), which stimulates the release of inflammatory factors from renal tubular epithelial cells and promotes the production of ROS by acting on E-prostanoids, thereby enhancing inflammation as well as oxidative stress in renal tissues (Wu 2005). In addition, COX2 is an important proinflammatory factor downstream of the NF-κB signaling pathway. According to genomic and lipidomic analyses, COX1 and COX2 function in a coordinated manner during the inflammatory response, and the two COX isoforms exhibit differential reciprocity in maintaining the ability to synthesize prostaglandins in the context of inflammation (Li et al., 2018). In the present study, SLEO alleviated the renal inflammatory response during HUA mostly by inhibiting the transcriptional activity of NF-κB, downregulating COX2, and modulating the AA metabolic pathway.
Amino acid metabolism changed significantly following SLEO treatment. The integration of RNA-seq and serum metabolomic data revealed significant changes in arginine and proline metabolism. L-arginine is a substrate for inducible NOS and is also essential for the biosynthesis of proteins and other amino acids. By converting arginine to ornithine and urea in the urea cycle, arginase competes with inducible NOS for the same substrate (Cinelli et al., 2020). Inducible NOS is an important mediator of immune activation and inflammation (Anavi and Tirosh 2020). Recent studies have implicated NF-κB in the regulation of iNOS expression. L-glutamate, a major metabolite in the tricarboxylic acid cycle of energy metabolism and a precursor for urate synthesis, inhibits the glutamate–cystine reverse transport system (Shen et al., 2021), which significantly attenuates the intracellular glutathione content, stimulates ROS production, and induces oxidative stress in the organism. In the present study, L-glutamate levels were higher in the serum of HUA rats compared with the blank control rats.
Molecular docking results indicated high binding activities for linalool with TLR4, β-caryophyllene with XOD, and β-pinene with NLRP3. Moreover, the expression levels of key proteins in the NOD-like receptor signaling and NF-κB signaling pathways were reduced in the renal tissues of SLEO-administered rats, and the serum levels of XOD, UA, CRE, and BUN were also attenuated. Therefore, we concluded that SLEO treats HUA and alleviates renal inflammation through multitarget regulation of the TLR4/NF-κB/NLRP3 signaling axis (Fig. 13).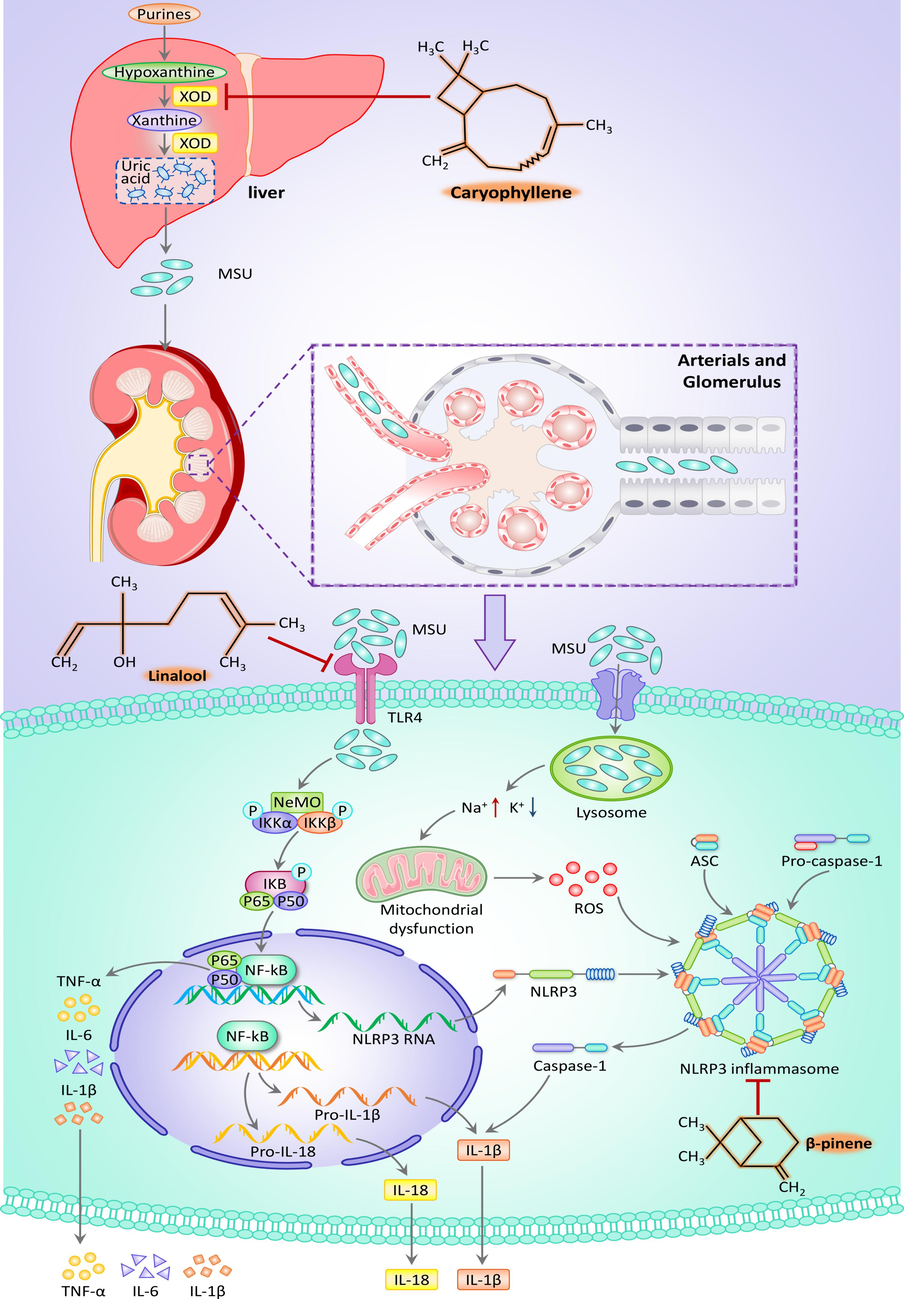
Diagram of the mechanism of action of SLEO in the treatment of HUA.
5 Conclusion
This study revealed that SLEO significantly inhibited XOD activity in HUA rats and uric acid-induced HK-2 cells, reduced uric acid production, lowered the levels of inflammatory factors such as IL-1β, TNF-α and IL-6, and alleviated the HUA-induced renal inflammatory injury. The mechanism of action may be as follows: the three active components of SLEO, β-pinene, linalool and β-caryophyllene, inhibit the expression of proteins such as NLRP3, ASC, IKKB, TLR4, NF-κB-p65, and caspase-1, etc., and inhibit the expression of these proteins through the TLR4/NF-κB/NLRP3 signaling axis, purine metabolism pathway, and purine metabolism pathway. axis, purine metabolism pathway, and arachidonic acid pathway to treat hyperuricemia and alleviate its induced renal inflammation. The study concluded that SLEO can be used as a safe and efficacious natural medicine for the treatment of hyperuricemia.
Funding
This work was supported by the National Key Research and Development Program of China ( 2021YFD1601004, 2023YFD1600402, 2023YFD1600403); Shaanxi Provincial Traditional Chinese Medicine Administration (ZYIXG-Y23005); Xi'an Science and Technology Plan Project(20231H-JSJ0-0007); Inner Mongolia Autonomous Region Science and Technology Program (2022YFSH0001); Shaanxi Provincial Department of Science and Technology Project (2023-YBSF-474); Key R&D Program of Shaanxi Province (2024SF-ZDCYL-03-11); Project of Shaanxi Provincial Administration of Traditional Chinese Medicine (2021-PY-005); Scientific Research Program Funded by Education Department of Shaanxi Provincial Government (23JP034); Shaanxi Provincial University Youth Innovation Team of Aromatic Chinese Medicine Industrialization Key Technology; Shaanxi Provincial University Engineering Research Center of Chinese Medicine Aromatic Industry; Key Discipline of High Level Traditional Chinese Medicine in Shaanxi Province, Traditional Chinese Medicine Processing; Shaanxi Provincial Engineering Research Center for Traditional Chinese Medicine Pieces.
CRediT authorship contribution statement
Peijie Zhou: Writing – original draft. Biao Zhang: Software. Xuan Wang: Methodology. Jiawei Duan: Data curation. Jinkai Li: Investigation. Jie Wang: Software. Ning Xia: Data curation. Shihao Zhang: Methodology. Jinghui Wang: Software. Dongyan Guo: Methodology. Chongbo Zhao: Data curation. Huanxian Shi: Software. Jiangxue Cheng: Software. Yundong Xie: Investigation. Jing Sun: Writing – review & editing, Funding acquisition. Xiaofei Zhang: Writing – review & editing, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Inflammasome-mediated pyroptotic and apoptotic cell death, and defense against infection. Curr. Opin. Microbiol.. 2013;16:319-326.
- [CrossRef] [Google Scholar]
- iNOS as a metabolic enzyme under stress conditions. Free Radic. Biol. Med.. 2020;146:16-35.
- [CrossRef] [Google Scholar]
- Chemometric analysis of lavender essential oils using targeted and untargeted GC-MS acquired data for the rapid identification and characterization of oil quality. Molecules (Basel, Switzerland).. 2017;22
- [CrossRef] [Google Scholar]
- NLRP3 inflammasome expression is driven by NF-κB in cultured hepatocytes. Biochem. Biophys. Res. Commun.. 2015;458:700-706.
- [CrossRef] [Google Scholar]
- Cyclooxygenase 2 and the kidney. Curr. Opin. Nephrol. Hypertens.. 2001;10:89-98.
- [CrossRef] [Google Scholar]
- Lavandula angustifolia and Lavandula latifolia essential oils from Spain: aromatic profile and bioactivities. Planta Med.. 2016;82:163-170.
- [CrossRef] [Google Scholar]
- Effect of a traditional Chinese medicine prescription Quzhuotongbi decoction on hyperuricemia model rats studied by using serum metabolomics based on gas chromatography-mass spectrometry. J. Chromatogr. B, Anal. Technol. Biomed. Life Sci.. 2016;1026:272-278.
- [CrossRef] [Google Scholar]
- An immune-related gene signature for the prognosis of human bladder cancer based on WGCNA. Comput. Biol. Med.. 2022;151:106186
- [CrossRef] [Google Scholar]
- Inducible nitric oxide synthase: regulation, structure, and inhibition. Med. Res. Rev.. 2020;40:158-189.
- [CrossRef] [Google Scholar]
- Phloretin ameliorates hyperuricemia-induced chronic renal dysfunction through inhibiting NLRP3 inflammasome and uric acid reabsorption. Phytomed.: Int. J. Phytother. Phytopharmacol.. 2020;66:153111
- [CrossRef] [Google Scholar]
- A greedy algorithm for minimizing the number of primers in multiple PCR experiments. Genome Informat. Workshop Genome Informat.. 1999;10:73-82.
- [Google Scholar]
- Lowering serum uric acid to prevent acute kidney injury. Med. Hypotheses. 2012;78:796-799.
- [CrossRef] [Google Scholar]
- Klotho alleviates contrast-induced acute kidney injury by suppressing oxidative stress, inflammation, and NF-KappaB/NLRP3-mediated pyroptosis. Int. Immunopharmacol.. 2023;118:110105
- [CrossRef] [Google Scholar]
- Deciphering the antifungal mechanism and functional components of cinnamomum cassia essential oil against Candida albicans through integration of network-based metabolomics and pharmacology, the greedy algorithm, and molecular docking. J. Ethnopharmacol.. 2024;319:117156
- [CrossRef] [Google Scholar]
- Hamad Al-Mijalli, S., E. L. ER, E. M. Abdallah, et al., 2022. Determination of Volatile Compounds of Mentha piperita and Lavandula multifida and Investigation of Their Antibacterial, Antioxidant, and Antidiabetic Properties. Evidence-based complementary and alternative medicine : eCAM. 2022, 9306251. DOI: 10.1155/2022/9306251.
- Inhibition of 20-HETE synthesis and action protects the kidney from ischemia/reperfusion injury. Kidney Int.. 2011;79:57-65.
- [CrossRef] [Google Scholar]
- Administered chrysanthemum flower oil attenuates hyperuricemia: mechanism of action as revealed by DNA microarray analysis. Biosci. Biotechnol. Biochem.. 2014;78:655-661.
- [CrossRef] [Google Scholar]
- Isovitexin alleviates hepatic fibrosis by regulating miR-21-mediated PI3K/Akt signaling and glutathione metabolic pathway: based on transcriptomics and metabolomics. Phytomed.: Int. J. Phytotherapy Phytopharmacol.. 2023;121:155117
- [CrossRef] [Google Scholar]
- Cytochrome P450 and lipoxygenase metabolites on renal function. Compr. Physiol.. 2015;6:423-441.
- [CrossRef] [Google Scholar]
- Hyperuricemia, the kidneys, and the spectrum of associated diseases: a narrative review. Curr. Med. Res. Opin.. 2016;32:1863-1869.
- [CrossRef] [Google Scholar]
- Molecular mechanisms regulating NLRP3 inflammasome activation. Cell. Mol. Immunol.. 2016;13:148-159.
- [CrossRef] [Google Scholar]
- Interleukin-1β in innate inflammation, autophagy and immunity. Semin. Immunol.. 2013;25:416-424.
- [CrossRef] [Google Scholar]
- Comparison of methods for differential co-expression analysis for disease biomarker prediction. Comput. Biol. Med.. 2019;113:103380
- [CrossRef] [Google Scholar]
- Enhanced bioactivity of rosemary, sage, lavender, and chamomile essential oils by fractionation, combination, and emulsification. ACS Omega. 2023;8:10941-10953.
- [CrossRef] [Google Scholar]
- Ameliorate impacts of scopoletin against vancomycin-induced intoxication in rat model through modulation of Keap1-Nrf2/HO-1 and IκBα-P65 NF-κB/P38 MAPK signaling pathways: molecular study, molecular docking evidence and network pharmacology analysis. Int. Immunopharmacol.. 2022;102:108382
- [CrossRef] [Google Scholar]
- Systemic urate deposition: an unrecognized complication of gout? J. Clin. Med.. 2020;9
- [CrossRef] [Google Scholar]
- Uric acid in inflammation and the pathogenesis of atherosclerosis. Int. J. Mol. Sci.. 2021;22
- [CrossRef] [Google Scholar]
- Data handling and data analysis in metabolomic studies of essential oils using GC-MS. J. Chromatogr. A. 2021;1640:461896
- [CrossRef] [Google Scholar]
- Integrating network pharmacology and pharmacological validation to explore the effect of Shi Wei Ru Xiang powder on suppressing hyperuricemia. J. Ethnopharmacol.. 2022;298:115679
- [CrossRef] [Google Scholar]
- Genomic and lipidomic analyses differentiate the compensatory roles of two COX isoforms during systemic inflammation in mice. J. Lipid Res.. 2018;59:102-112.
- [CrossRef] [Google Scholar]
- Rosemary (Rosmarinus officinalis L.) hydrosol based on serotonergic synapse for insomnia. J. Ethnopharmacol.. 2024;318:116984
- [CrossRef] [Google Scholar]
- Targeting microglial α-synuclein/TLRs/NF-kappaB/NLRP3 inflammasome axis in Parkinson's disease. Front. Immunol.. 2021;12:719807
- [CrossRef] [Google Scholar]
- The mechanism of action of paeoniae radix rubra-angelicae sinensis radix drug pair in the treatment of rheumatoid arthritis through PI3K/AKT/NF-κB signaling pathway. Front. Pharmacol.. 2023;14:1113810.
- [CrossRef] [Google Scholar]
- Liu, Y. and Z. Liu, 2020. Jianpi Huazhuo Tiaozhi granules reduce oxidative stress injury in macrophages by inhibiting the nicotinamide adenine dinucleotide phosphate oxidase/reactive oxygen species-nuclear transcription factor kappa B pathway. Journal of traditional Chinese medicine = Chung i tsa chih ying wen pan. 40, 922-927. DOI: 10.19852/j.cnki.jtcm.2020.06.004.
- High uric acid-induced epithelial-mesenchymal transition of renal tubular epithelial cells via the TLR4/NF-kB signaling pathway. Am. J. Nephrol.. 2017;46:333-342.
- [CrossRef] [Google Scholar]
- CPUY192018, a potent inhibitor of the Keap1-Nrf2 protein-protein interaction, alleviates renal inflammation in mice by restricting oxidative stress and NF-κB activation. Redox Biol.. 2019;26:101266
- [CrossRef] [Google Scholar]
- NLRP3 and IL-1β in macrophages as critical regulators of metabolic diseases. Diabetes Obes. Metab.. 2013;15(Suppl 3):19-25.
- [CrossRef] [Google Scholar]
- Reactive oxygen species in inflammation and tissue injury. Antioxid. Redox Signal.. 2014;20:1126-1167.
- [CrossRef] [Google Scholar]
- Phytochemical inhibitors of the NLRP3 inflammasome for the treatment of inflammatory diseases. Pharmacol. Res.. 2021;170:105710
- [CrossRef] [Google Scholar]
- Mechanistic insights of soluble uric acid-related kidney disease. Curr. Med. Chem.. 2020;27:5056-5066.
- [CrossRef] [Google Scholar]
- Ameliorative effect and mechanism of Yi-Suan-Cha against hyperuricemia in rats. J. Clin. Lab. Anal.. 2021;35:e23859.
- [Google Scholar]
- Flavonoid fisetin alleviates kidney inflammation and apoptosis via inhibiting Src-mediated NF-κB p65 and MAPK signaling pathways in septic AKI mice. Biomed. Pharmacotherapy = Biomedecine & Pharmacotherapie.. 2020;122:109772
- [CrossRef] [Google Scholar]
- Methods of integrating data to uncover genotype-phenotype interactions. Nat. Rev. Genet.. 2015;16:85-97.
- [CrossRef] [Google Scholar]
- Untargeted metabolomics reveal the therapeutic effects of Ermiao wan categorized formulas on rats with hyperuricemia. J. Ethnopharmacol.. 2021;281:114545
- [CrossRef] [Google Scholar]
- Integrated network pharmacology, transcriptomics, and metabolomics analysis to reveal the mechanism of salt Eucommiae cortex in the treatment of chronic kidney disease mineral bone disorders via the PPARG/AMPK signaling pathway. J. Ethnopharmacol.. 2023;314:116590
- [CrossRef] [Google Scholar]
- Serum metabolomics identifies dysregulated pathways and potential metabolic biomarkers for hyperuricemia and gout. Arthritis Rheumatol. (Hoboken, N.J.). 2021;73:1738-1748.
- [CrossRef] [Google Scholar]
- Research advances in the mechanisms of hyperuricemia-induced renal injury. Biomed Res. Int.. 2020;2020:5817348.
- [CrossRef] [Google Scholar]
- Molecular mechanisms underlying chemopreventive activities of anti-inflammatory phytochemicals: down-regulation of COX-2 and iNOS through suppression of NF-kappa B activation. Mutat. Res.. 2001;480–481:243-268.
- [CrossRef] [Google Scholar]
- The emerging role of metabolic regulation in the functioning of Toll-like receptors and the NOD-like receptor Nlrp3. FEBS Lett.. 2011;585:1568-1572.
- [CrossRef] [Google Scholar]
- Negative regulation of NF-κB and its involvement in rheumatoid arthritis. Arthritis Res. Ther.. 2011;13:221.
- [CrossRef] [Google Scholar]
- Arachidonic acid metabolism and kidney inflammation. Int. J. Mol. Sci.. 2019;20
- [CrossRef] [Google Scholar]
- Valerian essential oil for treating insomnia via the serotonergic synapse pathway. Front. Nutr.. 2022;9:927434
- [CrossRef] [Google Scholar]
- Aberrant human ClpP activation disturbs mitochondrial proteome homeostasis to suppress pancreatic ductal adenocarcinoma. Cell Chem. Biol.. 2022;29:1396-1408.e1398.
- [CrossRef] [Google Scholar]
- Cinnamon essential oil based on NLRP3 inflammasome and renal uric acid transporters for hyperuricemia. Food Bioscience. 2023;56:103285.
- [CrossRef] [Google Scholar]
- Mechanism investigation of Shi-Xiao-San in treating blood stasis syndrome based on network pharmacology, molecular docking and in vitro/vivo pharmacological validation. J. Ethnopharmacol.. 2023;301:115746
- [CrossRef] [Google Scholar]
- Control of cyclooxygenase-2 transcriptional activation by pro-inflammatory mediators. Prostaglandins Leukot. Essent. Fat. Acids. 2005;72:89-93.
- [CrossRef] [Google Scholar]
- Molecular basis of oxidative stress in gouty arthropathy. Clin. Rheumatol.. 2015;34:1667-1672.
- [CrossRef] [Google Scholar]
- Toll-like receptor 4 (TLR4) inhibitors: current research and prospective. Eur. J. Med. Chem.. 2022;235:114291
- [CrossRef] [Google Scholar]
- NF-κB restricts inflammasome activation via elimination of damaged mitochondria. Cell. 2016;164:896-910.
- [CrossRef] [Google Scholar]
- Shizhifang ameliorates pyroptosis of renal tubular epithelial cells in hyperuricemia through inhibiting NLRP3 inflammasome. J. Ethnopharmacol.. 2023;317:116777
- [CrossRef] [Google Scholar]
- Evaluation of the mechanism of action of rosemary volatile oil in the treatment of alzheimer's disease using gas chromatography -mass spectrometry analysis and network pharmacology. Combinatorial Chem. High Throughput Screening.. 2023;26:2321-2332.
- [CrossRef] [Google Scholar]
- Study on the mechanism of Orthosiphon aristatus (Blume) Miq. in the treatment of hyperuricemia by microbiome combined with metabonomics. J. Ethnopharmacol.. 2023;317:116805
- [CrossRef] [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2024.105897.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary Data 1
Supplementary Data 1







