Translate this page into:
Magnetic ligand fishing protocol combined with HPLC-FT-ICR-MS for screening potential α-Glucosidase inhibitors from UCG and in silico analysis
⁎Corresponding authors at: School of Pharmacy, Shenyang Pharmaceutical University, No. 103 Wenhua Road, Shenyang 110016, Liaoning, China. lab433@163.com (Chunjie Zhao), zm19871224@sina.com (Min Zhao)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Uremic Clearance Granule (UCG) is a traditional Chinese medicines (TCMs) that has been recognized as potentially effective treatment for diabetic nephropathy (DN). However, the chemical composition and the hypoglycemic ingredients of UCG remain unclear. In this experiment, a magnetic ligand fishing protocol combined with HPLC coupled Fourier transform ion cyclotron resonance mass spectrometry (HPLC-FT-ICR-MS) technology was developed to screen and identify α-Glucosidase inhibitors from UCG. First, constituents of UCG was identified by HPLC-FT-ICR-MS. Next, α-Glucosidase coated Fe3O4@SiO2@NH2 (Fe3O4@SiO2@NH2@α-Glucosidase) nano composites was synthesized. The essential parameters (concentration of Glutaraldehyde (GA), ratio of Fe3O4@SiO2@NH2 to enzyme, crosslinking time and immobilization time) were optimized to obtain maximum enzyme activity and the performance of immobilized enzyme was compared with that of free enzyme. The α-Glucosidase inhibitory activities were then evaluated using autodock and the 4-Nitrophenyl-α-D-glucopyranoside (pNPG) method. As a result, 142 compounds were identified were identified by comparing the retention time, molecular ions and fragmentation behaviors with the reference compounds or the in-house database. 20 possible α-Glucosidase activity inhibitory components were identified using HPLC-FT-ICR-MS. Molecular docking and inhibition studies showed that 4 compuunds including Limonexic acid, Sanggenol A, Glabrone, Matrine were more likely to stronger α-Glucosidase inhibitory activities than acarbose. In conclusion, the proposed approach, which combined highly specific screening with HPLC-FT-ICR-MS, provided a powerful platform for discovering bioactive components from multi-component and multi-target traditional Chinese medicines (TCMs).
Keywords
Diabetic kidney disease
HPLC-FT-ICR-MS
Ligand fishing
α-Glucosidase inhibitors
1 Introduction
Diabetes mellitus, a serious global public health problem associated with numerous complications that can damage many systems of the human body, is characterized by the disorder of glucose metabolism (Deng et al., 2022). The disorder of glucose metabolism is mainly caused by the absence or inhibition of α-Glucosidase in the human body (Rojo-Poveda et al., 2019). α-Glucosidase is a key enzyme that catalyzes the final step in the carbohydrate digestion process and is required for the synthesis of oligosaccharide chains in sugar metabolism (Wu et al., 2023). The low inhibitory activity of α-Glucosidase in organisms can lead to the disorder of glucose metabolism, which can lead to chronic diseases such as type 2 diabetes (Hu et al., 2024; Li et al., 2024). A study has highlighted that α-Glucosidase is an important drug target in the treatment of type 2 diabetes, and inhibition of α-Glucosidase can reduce the absorption of glucose (Wang et al., 2022). At present, hypoglycemic drug research strategies targeting α-Glucosidase has become a research hotspot, such as α-Glucosidase inhibitors which is a class of hypoglycemic drugs (Zhang et al., 2022; Min et al., 2024). However, prolonged utilization of synthetic α-Glucosidase inhibitors may result in adverse reactions, including diarrhea, vomiting, flatulence and severe stomach pain (Sekhon-Loodu and Rupasinghe, 2019; Xiao et al., 2023). Therefore, the development of new α-Glucosidase inhibitors with guaranteed efficacy and low side effects has become an urgent need. In recent years, many studies have shown that natural products have lower toxicity as inhibitory ligands of α-Glucosidase (Shen et al., 2020). Traditional Chinese medicine compounds are composed of several Chinese herbs, and α-Glucosidase inhibitors from different herbs may cooperate to achieve the effect of treating diseases (Lin et al., 2022). Therefore, it is significant to discover new classes of α −glucosidase inhibitors from traditional Chinese medicine compounds.
Many studies have reported that many traditional Chinese medicine compounds have α-Glucosidase inhibitory effects in vitro (Guo et al., 2020). Uremic Clearance Granule (UCG) is a traditional Chinese medicine compounds consisting of Radix Rhei Et Rhizome, Hedysarum Multijugum Maxim, Mori Cortex, Sophorae Flavescentis Radix, Atractylodes Macrocephala Koidz, Poria Cocos (Schw.) Wolf, Paeoniae Radix Alba, Radix Salviae, and licorice, which can invigorate spleen and Qi, remove dampness and activate blood (Wang et al., 2021a). Therefore, it is often used in the clinic to treat diseases such as intestinal dysfunction, inflammation, hypertension and diabetic nephropathy (Bai et al., 2021; Zhao et al., 2022). However, studies on the hypoglycemic active ingredients of UCG have yet to found. Meanwhile, systematic identification of UCG has not been report, so it is necessary to identify its chemical compositions and screen out the hypoglycemic active ingredients of UCG.
At present, there are various methods to screen out active ingredients, such as ultrafiltration (Dehghankar et al., 2023), affinity chromatography (Chi et al., 2023), membrane affinity chromatography (Chen et al., 2023). However, there are some disadvantages to these methods, such as low efficiency, high labor intensity, long time which is not suitable for the screening of traditional Chinese medicine compounds with complexity (He et al., 2022). A ligand-based target-fishing approach (ligand fishing) has been developed and effectively applied in the effective identification of biologically active compounds from traditional Chinese medicine compounds in order to improve the efficacy of screening active component(Guo et al., 2021). Ligand fishing is achieved by functionalizing the carrier surface with a targeted protein because the immobilized enzyme has high stability and utilization which can both retain the enzyme catalytic function and be easily separated from the reaction product (Xie et al., 2009). Novel carrier surface, including nanoparticle, microreactors and metal–organic frameworks, have gained extensive utilization in the assessment of active components from natural products (Wan et al., 2021a). There is a significant trend in developing biosensors using nanoparticle-based materials such as magnetic nanoparticle (Hooshiar et al., 2024a; Hooshiar et al., 2024b; Mirjalili et al., 2022). Magnetic nanoparticle provides excellent support for enzyme immobilization. Their interaction with magnetic forces enables the rapid separation of immobilized enzyme from solution, thereby reducing sample manipulation and simplifying the enzymatic reuse process (He et al., 2014). Fe3O4 magnetic nanoparticle owns high saturation magnetization, superparamagnetism and large specific surface area, and it has been widely used in the field of enzyme immobilization (Li et al., 2014a). In addition, targeted enzymes immobilized on Fe3O4 showed higher stability and acid resistance compared to free targeting enzymes (Li et al., 2014c). However, Fe3O4 magnetic nanoparticle is easily oxidized when exposed to air, so it is essential to modify the surface of Fe3O4 prior to immobilization in order to enhance its functionality. The surface coating of Fe3O4 with organic or inorganic materials can effectively solve the problem of easy oxidation (Liu et al., 2020). Among them, SiO2 coating is relatively stable, and the silanol groups formed are difficult to aggregate and can be connected with other functional groups (Reddy and Yun, 2016). Then active NH2 could be introduced by (3-Aminopropyl) triethoxysilane (APTES), and the synthesized Fe3O4@SiO2 can be combined with α-Glucosidase by Schiff base reaction (Amirjani and Rahbarimehr, 2021). Various reaction parameters including reaction temperature, reaction time, pH, concentration of crosslinker and enzyme affecting synthesis were investigated (Shao et al., 2015). To accurately identify α-Glucosidase inhibitors of UCG, it is of importance to utilize instruments that exhibit high sensitivity, precision and accuracy. HPLC-FT-ICR-MS can effectively identify the chemical composition of Chinese herbal medicine because of its high resolution and mass accuracy. Remarkably, many studies have successfully built ligand fishing in combination with mass spectrometry to screen bioactive ingredients in herbal extracts rapidly (Guo et al., 2019; Li et al., 2020a). However, α-Glucosidase immobilized on Fe3O4 magnetic nanoparticle has not been used as a tool to screen for active compounds in traditional Chinese medicine compounds extract, and its feasibility is our research point.
In this study, HPLC-FT-ICR-MS was applied to identify the chemical components. Next, immobilized enzyme was synthesized successfully. Based on the pharmacological substance basis, immobilized enzyme was used to screen α-Glucosidase inhibitors from UCG extracts. The potential α-Glucosidase inhibitors screened out were validated by molecular docking and α-Glucosidase inhibition assay.
2 Material and methods
2.1 Chemicals and reagents
UCG was obtained from Consun Pharmaceutical (Horgos) Co., Ltd (Neimenggu, China). α-Glucosidase (EC3.2.1.20, from yeast, lyophilized powder) and tetraethoxysilane (TEOS) were purchased from Adamas Reagent Co., Ltd (Shanghai, China). 4-Nitrophenyl-α-D-gluco pyranoside (pNPG), p-nitrophenol (pNP), glutaraldehyde (50 %), (3-Aminopropyl) triethoxysilane (APTES) was obtained from Sigma-Aldrich (St. Louis, MO, USA). Acarbose (purity > 98 %) and Brilliant Blue G-250 were supplied by Macklin Biochemical Co., Ltd (Shanghai, China). Ferric chloride hexahydrate (FeCl3·6H2O), ferrous sulphateheptahydrate (FeSO4·7H2O), sodium hydroxide (NaOH), phosphate-buffered saline (PBS, pH 6.8, 0.1 M) and sodium carbonate (Na2CO3) were acquired from Ailan Chemical Technology Co., Ltd (Shanghai, China). Reference compounds (purity > 98 %) of Protocatechuic acid, Danshensu, Caffeic acid, Paeoniflorin, Liquiritin, Liquiritigenin, Luteolin, Aloe-emodin, 10,15-Octadecadienoic acid, Rhein, Limonexic acid and Matrine used in this study were from Macklin Biochemical Co., Ltd (Shanghai, China). Reference compound (purity = 99.8 %) of Sanggenol A used in this study was from Chengdu Push Bio-technology Co., Ltd. Reference compound (purity = 98 %) of Glabrone used in this study was from Shanghai yuanye Bio-Technology Co.,Ltd. Purified water was purchased from Wahaha (Hangzhou, China). Formic acid (LC/MS grade) was obtained from Fisher Scientific (Fair Lawn, NJ, USA). Ethanol absolute (HPLC grade), acetonitrile (HPLC grade) and ammonium hydroxide were from Concord Technology Co., Ltd (Tianjin, China).
2.2 Preparation of UCG solution and mixed standard solution
UCG were extracted by ultrasonic extraction method. Details can be seen in S 1.1.
2.3 Instrument and analytical conditions
HPLC-FT-ICR-MS was used for qualitative analysis of UCG extract, HPLC was used for quantitative analysis, and the conditions of the instrument can be found in S 1.2.
2.4 Synthesis of Fe3O4@SiO2@NH2@α-Glucosidase
Synthesis of Fe3O4@SiO2@NH2@α-Glucosidase was based on relevent literature with some modifications (Qing et al., 2012). As shown in Fig. 1, The FeCl3·6H2O (4.32 g) and FeSO4·7H2O (1.6 g) were dissolved in 20 mL of water, followed by the addition of 18 mL of 0.1 g/mL NaOH solution under nitrogen atmosphere. The reaction mixture was under stirring at 70 °C for 1 h. After the reaction, the product was washed with water and phosphate buffer solution (PBS) (0.1 M, pH 6.8) to neutral pH and dried at 60 °C in a vacuum oven to obtain Fe3O4.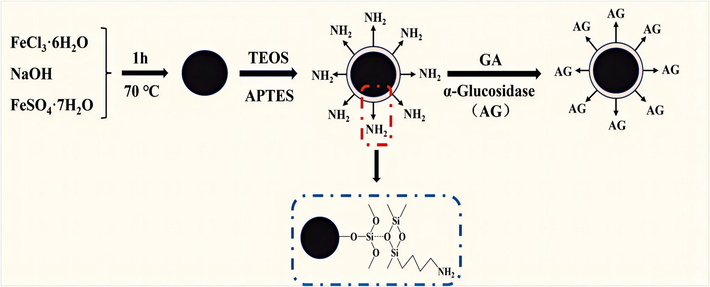
Synthesis of Fe3O4@SiO2@NH2@α-Glucosidase.
1.5 g of Fe3O4 was weighed and dispersed in an 120 mL ethanol solution consisting of water (40 mL) and ethanol (80 mL). Subsequently, 4 mL of tetraethoxysilane (TEOS) and 4.5 mL of ammonia were fully mixed and vigorously stirred in an ice bath for 12 h. At the end of the reaction, the product was washed with water and PBS (0.1 M, pH 6.8) to neutral pH and dried at 60 °C in a vacuum oven to obtain Fe3O4@SiO2.
Accurately weigh 1.0 g of Fe3O4@SiO2 into a three-necked flask, followed by adding 100 mL of ethanol. Then 6 mL of APTES was added and mechanically stirred at 70 °C for 8 h under nitrogen atmosphere. At the end of the reaction, the product was washed three times with water and PBS (0.1 M, pH 6.8) and dried at 60 °C in a vacuum drying oven to obtain Fe3O4@SiO2@NH2.
The Fe3O4@SiO2@NH2 nanoparticles (10 mg) were combined with 2 mL of 10 % glutaraldehyde solution in a centrifuge tube. After centrifugation at 5000 rpm for 1 h, the samples were subjected to disintegration via ultrasonication for 10 min. The supernatant was discarded, and the magnetic material was washed three times with PBS (0.1 M, pH 6.8). Subsequently, the Fe3O4@SiO2@NH2 was crosslinked with the α-Glucosidase solution and incubated for 4 h. After magnetic cleaning, the magnetically immobilized enzyme was stored in PBS (0.1 M, pH 6.8) at 4 °C.
2.5 Characterization of Fe3O4@SiO2@NH2@α-Glucosidase
2.5.1 Scanning electron microscopy with energy dispersive X-ray spectrometry (SEM-EDX)
The surface of samples were investigated using a Carl Zeiss Sigma 300 scanning electron microscope (SEM) (Carl Zeiss, Germany), while energy-disperse X-ray (EDX) scan was used to determine the elements present in the sample. The combined SEM-EDX technique can be used to analyze the elemental composition of the sample.
2.5.2 Fourier transform − infrared (FT-IR)
Fourier transform infrared (FT-IR) spectrum was collected from a Nicolet 10 (Thermo Nicolet Corporation, Waltham, MA, USA) IR spectrometer in the range of 4000–400 cm−1.
2.6 Optimization of Fe3O4@SiO2@NH2@α-Glucosidase
The optimization of four crucial immobilization parameters, including concentration of Glutaraldehyde (GA) (6 %, 8 %, 10 %, 12 %, 14 %, 16 %), ratio of Fe3O4@SiO2@NH2 to enzyme (10: 1, 20: 1, 30: 1, 40: 1, 50: 1), crosslinking time (0.5 h, 1.0 h, 1.5 h, 2.0 h, 2.5 h, 3.0 h) and the immobilization time (2 h, 4 h, 6 h, 8 h, 10 h, 12 h) were conducted to achieve maximum enzyme activity.
2.7 Performance study
The performance of immobilized enzyme was studied, including pH, temperature and reusability. Details can be seen in S 1.3.
2.8 Enzyme loading of Fe3O4@SiO2@NH2@α-Glucosidase
The study of α-Glucosidase loading was based on relevent literature with some modifications (Yi et al., 2022). Detailed steps can be seen in S 1.4.
2.9 Free and immobilized α-Glucosidase activity
In the study of enzyme activity, a few modifications was made according to the methodology reported in the literature (Miao et al., 2018). 4-Nitrophenyl-α-D-gluco pyranoside (pNPG) was used as a substrate, and its hydrolysis by α-Glucosidase resulted in the production of p-nitrophenol (pNP). The detailed procedure for examining enzyme activity can be seen in S 1.5.
2.10 Kinetic parameters of free and immobilized α-Glucosidase
The concentrated pNPG solution was diluted using PBS (pH 6.8, 0.1 M) in order to generate a series of solutions with concentrations ranging from 0.2 mM to 2 mM. Subsequently, kinetic studies were conducted on free α-Glucosidase and immobilized α-Glucosidase at 37 °C. The values for the enzymes' kinetic parameters were determined using Eq. (1):
The initial and maximum reaction rates are represented by v and vmax, respectively. [s] represents the concentration of the substrate, while km represents the Michaelis-Menten constant which signifies the enzyme's affinity for the substrate.
2.11 Screening of inhibitors in UCG
The UCG solution was denoted as L0. Firstly, 400 μL of L0 was incubated at 37 °C for 5 min, then 10 mg of immobilized enzyme and Fe3O4@SiO2@NH2 was added respectively, and mixed at 1000 rpm for 15 min. After separating the solid using a magnet, the magnetic material was eluted with acetonitrile–water (90:10, v/v). The eluent of immobilized enzyme was recorded as L1, and the eluent of Fe3O4@SiO2@NH2 was recorded as blank. The chemical composition in both L0, L1 and blank were compared by HPLC-FT-ICR-MS to identify the active ingredient.
2.12 Molecular docking
The interaction between α-Glucosidase and the active ingredients was elucidated, docking simulations performed using AutoDock 4.2. The crystal structure of α-Glucosidase (PDB ID: 3A4A, Resolution: 1.60 Å) in pdb format was downloaded from the PDB database (https://www.rcsb.org/). The water and the original ligands (GLC and Ca2+) were then removed from the target protein through Pymol 2.5.5 and stored in PDBQT format. The mol2 format of the active ingredient was downloaded from the PubChem database (https://www.pubchem.com/) and stored in PDBQT format. A total number of fifty independent docking runs were performed for each active ingredient. Receptor-ligand interaction analysis and figure rending were performed using Discovery Studio.
2.13 Inhibition assay
The α-Glucosidase inhibition activity of the inhibitor extracted from UCG extract was verified by pNPG method, but with a slight modification (Miao et al., 2018). IC50 is determined by varying the concentration of inhibitors while maintaining a constant amount of pNPG (2 mM). Briefly, three groups were set for the assay: blank group, sample group and control group (n = 3). The blank group contained no sample or enzyme, while the control group had no sample. In the sample group, α-Glucosidase (50 μL) was mixed with the sample solution (50 μL) and substrate (2 mM pNPG, 50 μL). The mixture was then incubated for 30 min at 37 °C. Finally, 0.2 M Na2CO3 (50 μL) was added into the system and immediately removed 100 μL solution from 96-well plate. The absorbance at 405 nm was determined using a microplate reader. The optical density (OD) value was calculated as the percentage of inhibition. The calculation of the inhibition rate was performed using Eq. (2):
The absorbance values of the sample group, blank group and control group are represented by As, Ab and Ac, respectively.
2.14 Data analysis strategy
According to the exact molecular weights measured, the possible molecular formulas were calculated by Data Analysis 4.4 software (Bruker Corp., Karlsruhe, Germany) within the specified mass error range (within ± 3 ppm) and purity score range (more than 85 %). The chemical composition of UCG was finally determined by matching each possible molecular formula and its fragment information of secondary mass spectrometry with the in-house database or the reference compounds. An in-house database of chemical compounds which includes names, molecular formulas, theoretical mass, chemical structures and the MS/MS fragmentation profiles was established according to Chemspider (https://www.chemspider. com/), SciFinder (https://scifinder.cas.org/), PubMed (https://www.ncbi.nlm.nih.gov/pubmed), Massbank (https://massbank.eu/), CNKI (https://www.cnki.net/) and Web of Science (https://www.webofknowledge.com) (Liu et al., 2022; Zhang et al., 2021a; Ma et al., 2014; Wang et al., 2019; Li et al., 2022; Wang et al., 2015; Gong et al., 2020; Yan et al., 2016; Li et al., 2014b; Jiang et al., 2019; Zhang et al., 2021b; Sheng et al., 2021; Zhong et al., 2015).
3 Results
3.1 Qualitative analysis of components in UCG
Fig. 2 reveals the base peak ion chromatograms (BPCs) of UCG solution in positive and negative ion modes, together with the BPC of the mixed standard solution in negative ion mode. On the basis of its specific molecular weight, retention duration and in-house database, the chemical structure and cleavage patterns were inferred. As a consequence, 142 components in all were found in UCG, including 57 flavonoids, 21 terpenoids, 24 phenylpropanoids, 5 phenols, 8 alkaloids, 8 anthraquinones, 10 organic acids and 9 other ingredients. 9 components, including Danshensu, Caffeic acid, Paeoniflorin, Liquiritin, Liquiritigenin, Luteolin, Aloe-emodin, 10, 15-Octadecadienoic acid and Rhein, were identified by matching molecular weights, retention time and MS/MS fragment data to the references. Molecular weight-specific extracted ion chromatograms (EICs) were shown in the supplementary material Fig. S1. The results of each chemical structure are shown in Fig. S2. The fragmentation pathway of typical compounds is shown in Fig. S3. The formula, retention time, measured mass, theoretical mass, ion mode, error value and MS/MS are shown in Table S1. UCG component analysis of studies can be found in S 2.1.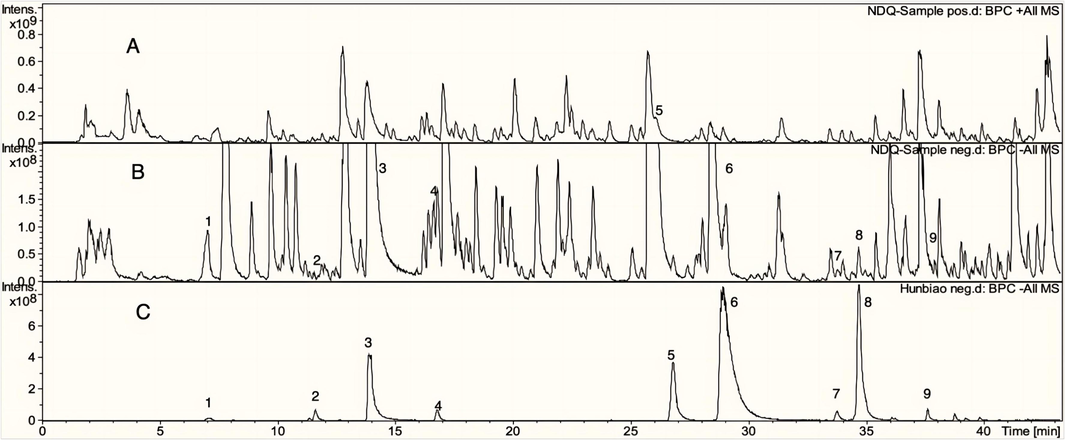
UCG's positive (A) and negative (B) ion base peak chromatograms (BPC), and the reference compounds in positive ion mode (C). 1-Danshensu, 2-Caffeic acid, 3-Paeoniforin, 4-Liquiritin, 5-Liquiritigenin, 6-Luteolin, 7-Aloe-emodin, 8–10,15-Oct adecadienoic, 9-Rhein.
3.2 Characterization of Fe3O4@SiO2@NH2@α-Glucosidase
3.2.1 SEM-EDX analysis
The SEM images of the Fe3O4, Fe3O4@SiO2, Fe3O4@SiO2@NH2 and Fe3O4@SiO2@NH2@α-Glucosidase were shown in Fig. 3. Their particle diameters were 20 nm, 66 nm, 77 nm and 83 nm, respectively. As shown in Fig. 3b, an increase in particle size accompanied by a more pronounced manifestation of the spherical and smooth structure was observed after the modification with TEOS. These findings prove the successful coating of SiO2 layer onto the surface of Fe3O4 (Cheng et al., 2019). As shown in Fig. 3c, after the modification with APTES, Fe3O4 magnetic nanoparticles become more agglomerate. In addition, As shown in Fig. 3d, in Fe3O4@SiO2@NH2@α-Glucosidase, a large number of APTES and α-Glucosidase act as a “bridge”, and a membrane was found on the surface of nanomaterials. These results indicated that the surface modification of every step was successful. The result of EDX is shown in Fig. 3e. The coexistence of Fe (pink) and O (green) in Fe3O4@SiO2@NH2@α-Glucosidase indicated the successful synthesis of Fe3O4. The existence of Si (cyan) was due to the successful modification of TEOS. The existence of N (yellow) indicated the successful modification of APTES and the active NH2 was successfully introduced. The SEM-EDX analysis reveals the successful synthesis of Fe3O4@SiO2@NH2.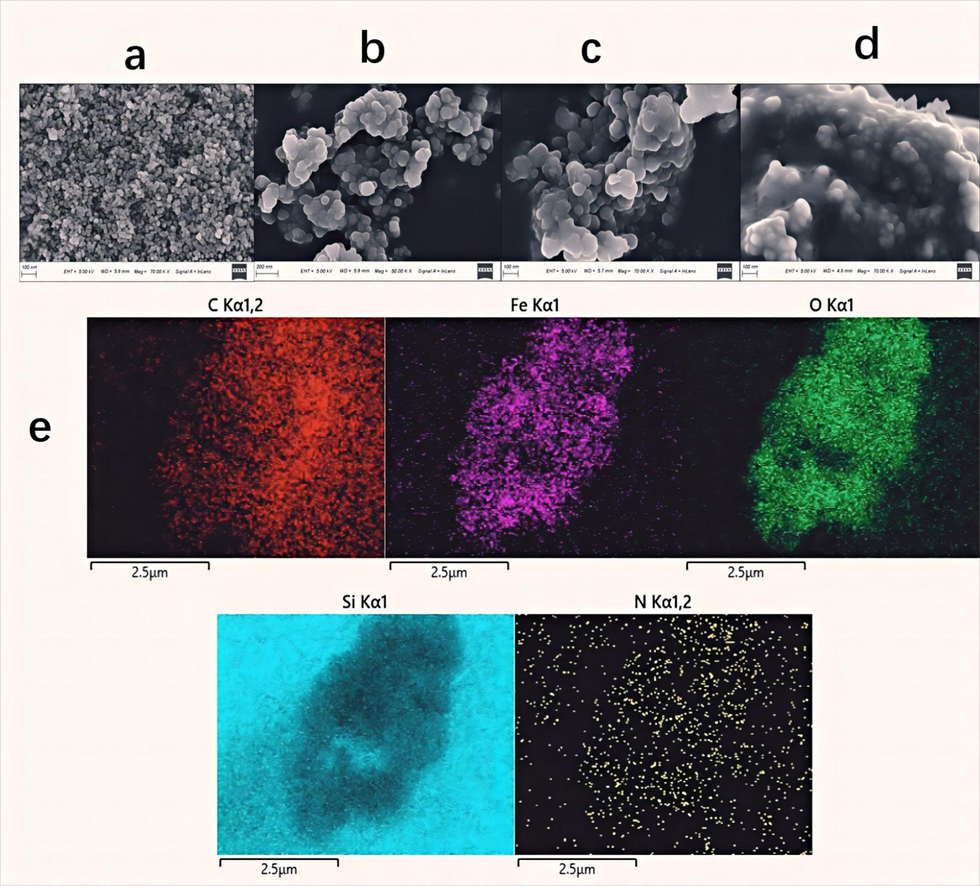
SEM images of Fe3O4 (a), Fe3O4@SiO2 (b), Fe3O4@SiO2@NH2 (c), Fe3O4@SiO2@NH2@ α-Glucosidase (d), and EDX mapping of different elements in the Fe3O4@SiO2@NH2@α-Glucosidase (e).
3.2.2 FT-IR analysis
FT-IR spectra of Fe3O4, Fe3O4@SiO2 and Fe3O4@SiO2@NH2@α-Glucosidase are shown in Fig. 4. The peak at about 560 cm−1 was ascribed to the vibrational absorption of Fe-O (Yang et al., 2019). The peaks observed at about 460 cm−1, about 789 cm−1 and about 1086 cm−1 were assigned to the bending vibration of O-Si-O, symmetric stretching vibration of Si-O-Si and stretching vibration of Si-O-Fe; and the decrease in the intensity of the Fe-O adsorption vibration both indicated the generation of Fe3O4@SiO2 (Shi et al., 2018). The presence of a new peak at 2155 cm−1 was observed in the spectrum of Fe3O4@SiO2@NH2@α-Glucosidase represents C⚌N stretching vibration of Schiff base, which confirmed the linking of the –NH2 group with the aldehyde group (–HCO) of GA (Agrawal et al., 2016). The overlapping of –NH and –OH stretching vibrations was assigned to a broad and strong band observed at about 3424 cm−1 (Shi et al., 2018, Amirjani et al., 2022, Fan et al., 2022). The absorption peak observed at about about 1628 cm−1 was assigned to the bending mode of absorbed molecule water on Fe3O4 (Wang et al., 2021b). In summary, the FT-IR analysis comfirmed the effective synthesis of Fe3O4@SiO2@NH2.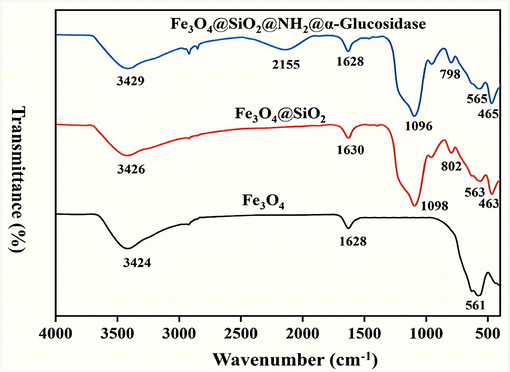
FT-IR spectra of sample: Fe3O4 (a); Fe3O4@SiO2 (b); Fe3O4@SiO2@NH2@α-Glucosidase (c).
3.3 Enzyme loading of Fe3O4@SiO2@NH2
We optimized the four key parameters and studied the performance of immobilized enzyme, and the result was presented in S 2.1 and S 2.2. Then, the efficiency of immobilization was initially examined. The quantification of enzyme loading on Fe3O4@SiO2@NH2 was determined to be 240.1 μg/mg, indicating successful immobilization of α-Glucosidase on Fe3O4@SiO2@NH2. The relation curve between the concentration and absorbance of the pNP solution was shown in Fig. 5a, and the activities of both immobilized and free enzyme were quantified. It was observed that the activity of the immobilized enzyme accounted for 33 % of the free enzyme, which was consistent with the literature (Guo et al., 2023). The decrease in enzyme activity can be attributed to the absence of the unbound form of the N-terminal region of α-Glucosidase.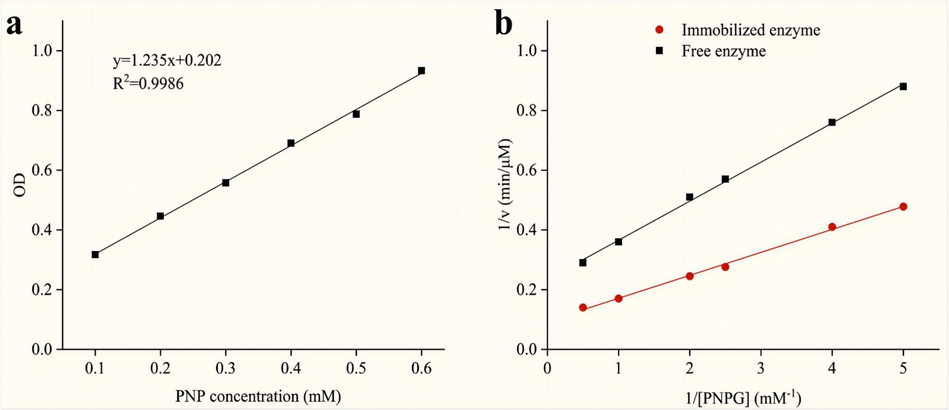
The standard curve of p-nitrophenol (pNP) (a), and Lineweaver-Burk plots of the free and immobilized α-Glucosidase (b).
3.4 Kinetic study
The Km is often used to evaluate the affinity of the enzyme for a specific substrate. The lower Km suggested higher level of affinity. The Lineweaver-Burk plots for Fe3O4@SiO2@NH2@α-Glucosidase and free α-Glucosidase (Fig. 5b) were as follows: 1/v = (0.13007 ± 0. 00262) (1/[S]) + (0. 21727 ± 0.00776) with R2 = 0.9980 and 1/v = (0.07633 ± 0.00171) (1/[S]) + (0.09559 ± 0.00507) with R2 = 0.9975, individually. And the values of Km were determined as 0.59 mM and 0.80 mM, respectively. The results showed that the affinity of α-Glucosidase was enhanced after immobilization. The reason is that the immobilized enzyme active site exhibited enhanced substrate accessibility and a reduced diffusion barrier effect compared with the free enzyme (Arana-Peña et al., 2021). The affinity changes obtained were consistent with some literatures (Li et al., 2020b; Wan et al., 2021b).
3.5 Identification of α-Glucosidase inhibitors
The base peak ion chromatograms (BPCs) of L0, L1 and blank are shown in Fig. 6. On the basis of their specific molecular weight, retention duration and in-house database, the chemical structures of inhibitors were inferred, which was shown in Table 1. As a consequence, 20 compounds were identified, including 9 flavonoids, 3 phenylpropanoids, 3 anthraquinones, 2 organic acids, 1 alkaloid, 1 terpenoid and 1 other class.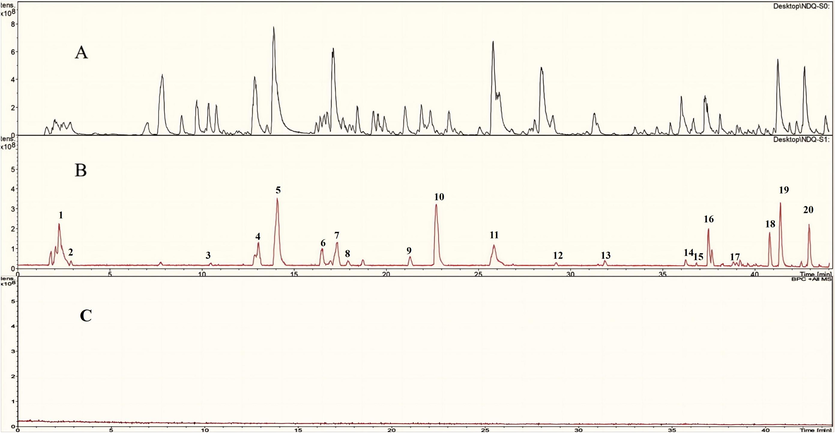
The base peak ion chromatograms (BPCs) of L0 (A), L1 (B) and Blank (C).
Peak No.
Identification
Formula
Theoretical mass (m/z)
Measured mass (m/z)
Error
(ppm)MS/MS (m/z)
Structural class
1
Limonexic acid
C6H8O7
192.0270
191.0197
−0.21
173.00, 129.01, 111.00, 87.01, 85.03
organic acids
2
Matrine
C15H24N2O
248.3661
249.1961
0.27
247.18, 230.15, 150.12, 148.11, 112.07
alkaloids
3
Chlorogenic acid
C16H18O9
354.0867
353.0878
−0.48
309.09, 295.08, 191.05, 179.03, 143.04, 127.03
phenylpropanoids
4
Mudanpioside E
C24H30O13
526.1614
525.1613
−0.43
449.14, 327.10, 165.05, 121.02
terpenoids
5
Cirsimarin
C23H24O11
476.4302
477.1391
0.20
313.74, 299.65, 281.07, 197.53, 169.47
flavonoids
6
Liquiritin
C21H20O9
418.3942
417.1191
0.09
255.07, 135.01, 119.05
flavonoids
7
Quercetin-3-rhamnoside
C21H20O11
448.3774
447.0933
−0.29
447.00, 405.00
flavonoids
8
Plantamajoside
C29H36O16
640.5863
639.193
0.45
178.95, 160.92, 133.39
phenylpropanoids
9
Kaempferol-7-O-α-L-rhamnoside
C21H20O10
432.3841
431.1129
−0.63
473.43, 447.32, 429.45, 284.78
flavonoids
10
Rosmarinic acid
C18H16O8
360.3148
359.0772
−0.65
197.04, 179.03, 161.02, 135.04, 133.02, 72.99
phenylpropanoids
11
Liquiritigenin
C15H12O4
256.2634
257.0808
−0.23
135.01, 119.05
flavonoids
12
Physcion
C16H12O5
284.2635
283.0612
−0.43
269.04, 240.04, 225.05
anthraquinones
13
Senkyunolide D
C12H14O4
222.0819
221.0819
−0.1
221.11, 177.43
other
14
Pinellic acid
C18H34O5
330.2330
329.2333
−0.56
229.14, 211.13, 183.43, 171.10
organic acids
15
Formononetin
C16H12O4
268.2643
267.0662
−0.35
254.05, 237.05, 213.09, 107.04
flavonoids
16
Rhein
C15H8O6
284.2204
283.0248
−0.35
255.06, 227.17, 183.26
anthraquinones
17
Maackiain
C16H12O5
284.0601
283.0612
−0.29
283. 06, 255. 06, 117. 05, 211. 07, 137. 02, 145. 02, 240. 04, 227. 06
flavonoids
18
Sanggenol A
C25H28O6
424.4862
423.1813
−0.66
298.13, 245,12, 151.55, 126.87
flavonoids
19
Emodin
C15H10O5
270.2374
269.0455
0.16
241.05, 225.05, 197.06
anthraquinones
20
Glabrone
C20H16O5
336.3382
335.0925
−0.58
335.09, 291.10, 231.08
flavonoids
3.6 Molecular docking
To further verify whether the 20 compounds had α-Glucosidase inhibitory, Molecular docking technique was employed to examine the affinity of the candidate compounds with α-Glucosidase. The docking results are shown in Table 2, To improve the screening accuracy, the affinity < -8.0 kcal/mol was used as the screening condition for potential inhibitors in this study.
Compound name
Affinity energy
(kcal/mol)
Limonexic acid
−9.69
Sanggenol A
−8.57
Glabrone
−8.06
Matrine
−8.04
Liquiritigenin
−7.62
Maackiain
−7.53
Kaempferol-7-O-α-L-rhamnoside
−7.41
Quercetin-3-rhamnoside
−7.25
Liquiritin
−7.18
Formononetin
−6.89
Rosmarinic acid
−6.85
Emodin
−6.75
Cirsimarin
−6.73
Mudanpioside E
−6.08
Senkyunolide D
−6.06
Physcion
−6.02
Acarbose
−6
Rhein
−5.9
Plantamajoside
−5.67
Chlorogenic acid
−5.55
Pinellic acid
−4.03
Limonexic acid, Sanggenol A, Glabrone, Matrine exhibited the high affinity with α-Glucosidase and, thus, displayed potential inhibitory activity against α-Glucosidase. The protein–ligand interactions of the 4 compounds were presented in Fig. 7 and illustrated in Table 3. As shown in Fig. 7a, strong hydrophobic interactions between Limonexic acid and active-site amino acid residues play an important role in their binding. The following residues, namely Lys 156, Tyr 158, Phe 303, Leu 313, Arg 315 and Arg 442, were found to be involved in both bonding and non-bonding interactions with Limonexic acid. The main characteristic of Limonexic acid is that it is composed of oxygen-containing functional groups, including ester group, ether group, carbonyl group, oxhydryl group. The ester group was bound with Lys 156 and Arg 442 via conventional hydrogen bonds, while the oxhydryl group was also bound with Leu 313 via conventional hydrogen bonds. These hydrogen bonds play a important role in enhancing the stability of the protein–ligand complex. Furthermore, the oxhydryl group played a crucial role in stabilizing the complex between the protein and ligand by generating an electrostatic attraction effect. The ether group was bound with Arg 442 via carbon hydrogen bond and Arg 442 constitutes a crucial part of the protein's residues responsible for regulating its active conformation. And six-membered ring and its substituted methyl groups were bound with Tyr 158, Phe 303 and Arg 315 via pi-alkyl bonds and alkyl bonds. The pi-alkyl interaction plays a crucial role in enhancing the stability of the protein–ligand complexes. As shown in Fig. 7b, the docked conformation of the Sannggenol A depicting the possible interactions with various amino acids of 3A4A: Lys 156, Tyr 158, Gly 161, Leu 177, Leu 313, Ala 418, Iso 419 and Glu 422. The methyl group showed a number of hydrophobic interactions with Lys 156, Tyr 158 and Leu 177. Furthermore, the benzene ring was bound with Lys 156, Ala 418 and Iso 419 via hydrophobic interactions, pi-alkyl interactions and alkyl interactions. In addition, the electropositive hydrogen atom of the oxhydryl group and the carbonyl group segment were bound with Lys 156, Tyr 158, Gly 161, Leu 313 and Glu 422 via strong hydrogen bonding. The presence of hydrogen bonding plays a crucial role in preserving the stability of the conformation. These interactions allow Sannggenol A and 3A4A to bind firmly together. As shown in Fig. 7c, Glabrone exhibited interactions with the subsequent amino acids via both bonding and non-bonding interactions: Lys 156, Tyr 158, Val 232, Pro 312, Leu 313, Phe 314 and Asn 415. The methyl group was involved in hydrophobic interactions with Val 232. The benzene ring was involved in important hydrophobic interactions:pi-pi T-shaped interaction with Phe 314, pi-alkyl and alkyl interactions with Val 232 and Leu 313, respectively. Moreover, carbon hydrogen bond was formed between carbon–carbon double bond and Pro 312. The electropositive hydrogen atom and electronegative oxygen atom of oxhydryl group and electronegative oxygen atom of carbonyl group were bound with with Pro 312, Tyr 158, Asn 415 and Lys 156. These high-affinity interactions allow Glabrone to be firmly bound to Glabrone. As shown in Fig. 7d, the conformation of Matrine, when docked with 3A4A, displayed strong interactions with several amino acids: Lys 156, Tyr 158, Arg 315 and Asp 242. The pyrimidine ring was involved in important hydrophobic interactions: pi-alkyl and alkyl interactions with Tyr 158 and alkyl interactions with Lys 156. Moreover, ortho-position of pyridine N part was bound to Asp 242 via Carbon hydrogen bond. The electronegative oxygen atom of carbonyl group was bound with Arg 315 via hydrogen bond. These interactions allow Matrine and 3A4A to bind firmly together. It can be seen that the hydrogen bond and hydrophobic interaction between ligand and enzyme may be beneficial to enhance the stability of inhibitor-enzyme complex, and thus play an important role in the inhibitory activity of α-Glucosidase.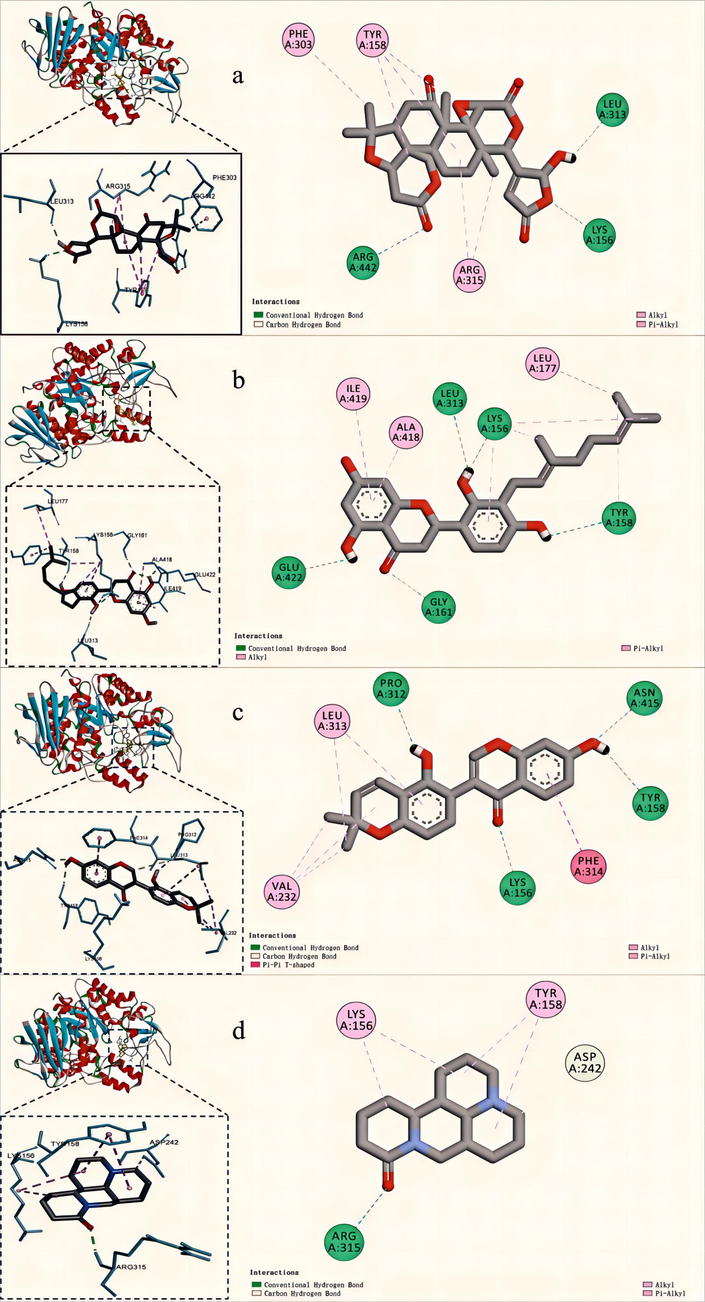
2D, 3D molecular docking graphs of Limonexic acid (a), Sanggenol A (b), Glabrone (c), Matrine (d) with 3A4A.
Compound
Protein-ligand interaction
Conventional Hydrogen Bond
Carbon Hydrogen Bond
Alkyl
Pi-Alkyl
Pi-Pi T-shaped
Limonexic acid
Lys 156, Arg 442
Arg 442
Arg 315
Tyr 158, Phe 303, Leu 313
−
Sanggenol
Lys 156, Tyr 158, Gly 161, Leu 313,
−
Lys 156, Tyr 158, Leu 177
Lys 156, Tyr 158, Ala 418, Ile 419, Glu 422
−
Glabrone
Lys 156, Tyr 158, Pro 312, Asn 415
Pro 312, Asn 415
Val 232, Leu 313
Leu 313
Phe 314
Matrine
Arg 315
Asp 242
Lys 156, Tyr 158
Tyr 158
−
3.7 Inhibitory activity of the potential inhibitors
The dose–response curves in Fig. 8 illustrate the inhibitory effects of the ligands and acarbose on α-Glucosidase activity. It was determined that the IC50 values of Acarbose, Limonexic acid, Sanggenol A, Glabrone and Matrine were measured at 16.57 μM, 5.91 μM, 8.52 μM, 10.91 μM, 11.51 μM, respectively, which showed that the inhibitory of Limonexic acid, Sanggenol A, Glabrone and Matrine surpasses that of Acarbose. Results of molecular docking and inhibition assays show that these 4 compounds exhibit promising prospects as α-Glucosidase inhibitors in field drug.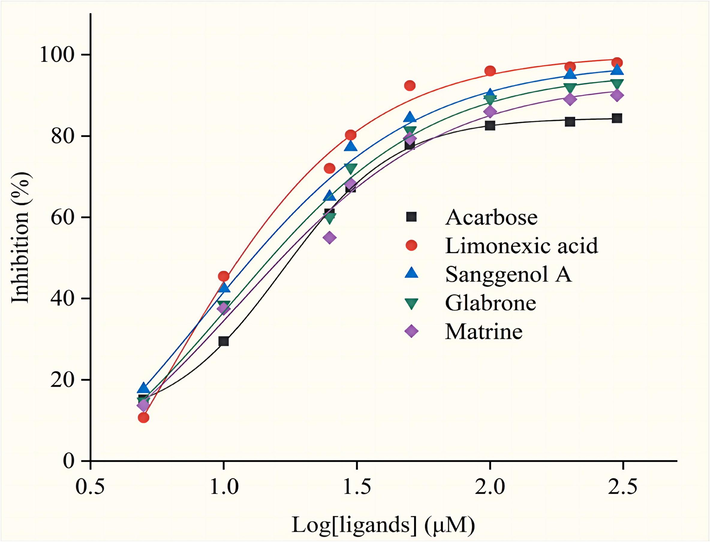
The inhibition curves of Limonexic acid, Sanggenol A, Glabrone, Matrine, and Acarbose.
3.8 Quantitative analysis
Quantitative analysis of the 4 compounds screened were carried out by HPLC. We determined the presence of 4 four compounds by comparison to the references, as shown in Fig. 9.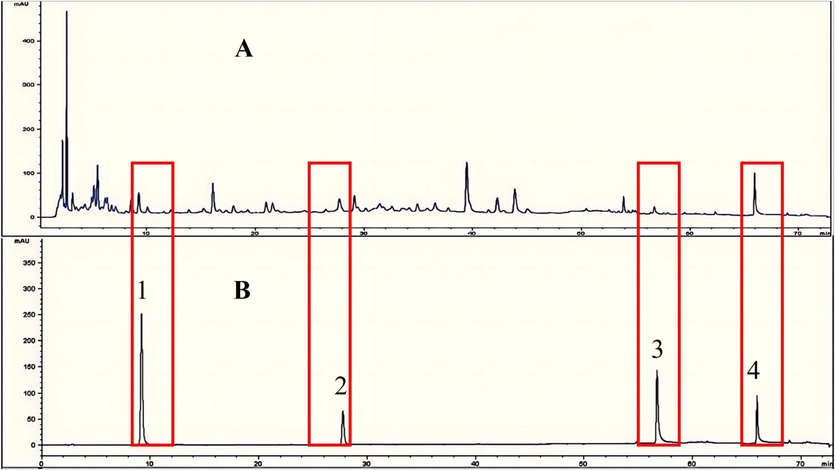
Chromatograms of sample (A), reference substances (B). 1-Limonexic acid; 2-Matrine; 3-Sanggenol A; 4-Glabrone.
3.8.1 Method validation
Methodological validation is performed before quantitative analysis of these two components. Table 4. displays the linear regression, precision, repeatability, stability and recovery for 4 substances in UCG. There is good linearity in a relatively wide concentration range. To evaluate the precision the quantitative method, the mixed standard solution was injected 6 times consecutively within one day and the RSD values of 4 components were 0.48 %, 1.05 %, 0.75 %, and 0.84 %, respectively (n = 6), which shows that precision of the instrumen is good. Six consecutive injections of sample solutions were used to verify repeatability. The RSD values of 4 compounts were 1.47 %, 1.98 %, 1.64 % and 1.29 %, respectively, indicating good repeatability. Stability was evaluated by testing the sample solutions six times within 24 h at room temperature and the RSD values of 4 compounts were 1.97 %, 1.83 %, 2.46 % and 1.58 %, respectively, indicating that the stability of the sample solutions was maintained for over 24 h when stored at room temperature. Moreover, the recovery of analytes were 98.33 %, 96.74 %, 102.5 % and 98.45 %, and the RSD values were 0.83 %, 0.96 %, 1.32 %, 1.53 %, repectively. Therefore, the precision, repeatability, stability and recovery of the method were validated as reliable and applicable for the quantitation of UCG.
Compound
Regression equation
r
linear range/(µg/mL)
Precision
RSD%
(n=6)Repeatability
RSD%
(n=6)Stability
RSD%Recovery
mean
RSD%
(n=6)
Limonexic acid
y = 14.63x + 436.6
0.9998
10.00–500.0
0.48
1.47
1.97
98.33
0.83
Matrine
y = 11.24x + 283.4
0.9997
10.00–500.0
1.05
1.98
1.83
96.74
0.96
Sanggenol A
y = 9.37x + 559.6
0.9992
10.00–500.0
0.75
1.64
2.46
102.5
1.32
Glabrone
y = 5.265x + 823.5
0.99994
10.00–500.0
0.84
1.29
1.58
98.45
1.53
3.8.2 Sample content determination
The chromatographic analysis of 3 batches of UCGwas recorded according to the chromatographic conditions S 1.2.2, and the contents of Limonexic acid, Sanggenol A, Glabrone, and Matrine were calculated by external standard method, and the results were shown in Table 5.
No.
W/mg·g−1
Limonexic acid
Matrine
Sanggenol A
Glabrone
1
0.1484
0.09362
0.05847
0.2418
2
0.1524
0.08647
0.05639
0.2371
3
0.1482
0.09284
0.05485
0.2262
4 Discussion
4.1 Selection of sample solution preparation
During the preparation of the sample solution, different extraction solvents (methanol, 50 % methanol, ethanol and 50 % ethanol), different extraction methods (reflux, ultrasound) and different extraction times (20, 30 and 40 min) were used to investigate the preparation method of UCG sample solution. The results showed that various spectral peaks was significant, and the overall peak shape was well separated with 50 % ethanol. After comprehensive consideration, it was determined to use 50 % ethanol solution as the extraction solvent, and the ultrasonic time was 30 min.
4.2 Selection of chromatographic conditions
This study investigated the mobile phase by gradient elution using methanol–water, acetonitrile-0.1 % formic acid and acetonitrile-0.1 % phosphoric acid, respectively. The results showed that when acetonitrile-0.1 % formic acid was used as the mobile phase, the separation effect of each chromatographic peak was better, and the baseline was stable. Finally, acetonitrile-0.1 % formic acid was determined as the mobile phase system for this experiment. Then, the column temperatures (20, 25, 30 and 35 °C) were investigated. The results showed that the column temperatures were 35 °C, and the chromatographic peaks were well separated. Therefore, the above chromatographic conditions were used in the experiment to ensure the method's accuracy.
5 Conclusion
In summary, a ligand fishing strategy based on Fe3O4@SiO2@NH2@α-Glucosidase combined with HPLC-FT-ICR-MS analysis was developed for the aim of identifying chemical compositions, and efficiently screening α-Glucosidase inhibitors from UCG extract. On basis of this approach, a total of 142 compounds were identified. 20 potential inhibitors were identified by HPLC-FT-ICR-MS. Through molecular docking and inhibitory assay, 4 inhibitors with high inhibitory activity were further validated. Moreover, these 4 compounds were identified by comparing them with the references and quantitatively analysized. This study provides important information for the study of hypoglycemic mechanism of UCG. In addition, this study also provides a reference for finding active ingredients of other TCMs. However, the current ligand fishing techniques do not allow biomedical imaging to further reveal the results of screening, which is of great significance for accurately assessing the bioactivity of screening ligands. It is easier to assess bioactivity if biomedical imaging and inhibition assays can be combined.
CRediT authorship contribution statement
Bo Yuan: Writing – original draft, Formal analysis, Conceptualization. Yumeng Zhang: Formal analysis. Xinting Man: Investigation. Chunjie Zhao: Writing – review & editing, Supervision, Project administration. Min Zhao: Writing – review & editing, Conceptualization.
Acknowledgements
This work was supported by the Career development support plan for young and middle-aged teachers of Shenyang Pharmaceutical University (ZQN2019014); the Applied Basic Research Program of the Department of Science and Technology of Liaoning Province (Youth Special Project, 2023JH2/101600013).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Application of nanoparticle-immobilized thermostable β-glucosidase for improving the sugarcane juice properties. Innov. Food. Sci. Emerg.. 2016;33:472-482.
- [Google Scholar]
- SPR-based assay kit for rapid determination of Pb2+. Anal. Chim. Acta. 2022;1220:340030
- [Google Scholar]
- Recent advances in functionalization of plasmonic nanostructures for optical sensing. Microchim. Acta.. 2021;188:1-17.
- [Google Scholar]
- Enzyme co-imm obilization : Always the biocatalyst designers' choice…or not? Biotechnol. Adv.. 2021;51:107584
- [Google Scholar]
- The clinical effectiveness and safety of traditional Chinese medicine uremic clearance granule combined with high-flux hemodialysis in the treatment of uremic pruritus: A protocol for systematic review and meta analysis. Medicine. 2021;100(25):e26423.
- [Google Scholar]
- Recent development and application of membrane chromatography. Anal. Bioanal. Chem.. 2023;415(1):45-65.
- [Google Scholar]
- α-Glucosidase immobilization on functionalized Fe3O4 magnetic nanoparticles for screening of enzyme inhibitors. Chin. Chem. Lett.. 2019;30(3):656-659.
- [Google Scholar]
- Construction of lipid raft-coupled agarose gels as bioaffinity chromatography materials and validation with tropomyosin-related kinase A-targeted drugs. J. Chromatogr. a.. 2023;1691:463803
- [Google Scholar]
- Synthesis and modification methods of metal-organic frameworks and their application in modification of polymeric ultrafiltration membranes: a review. J. Environ Chem. Eng. 2023109954
- [Google Scholar]
- Synthesis and bioactivities evaluation of oleanolic acid oxime ester derivatives as α-glucosidase and α-amylase inhibitors. J. Enzym. Inhib. Med.. 2022;37(1):451-461.
- [Google Scholar]
- Luminescent binuclear Zinc (II) organic framework as bifunctional water-stable chemosensor for efficient detection of antibiotics and Cr (VI) anions in water. Spectrochim. Acta. A. 2022;264:120232
- [Google Scholar]
- Identification of the metabolites of isochlorogenic acid A in rats by UHPLC-Q-Exactive Orbitrap MS. Pharm. Biol.. 2020;58(1):992-998.
- [Google Scholar]
- Screening of lipase inhibitors from bamboo leaves based on the magnetic ligand fishing combined with HPLC/MS. Microchem. J.. 2020;153:104497
- [Google Scholar]
- Comprehensive screening and identification of natural inducible nitric oxide synthase inhibitors from Radix Ophiopogonis by off-line multi-hyphenated analyses. J. Chromatogr. A. 2019;1592:55-63.
- [Google Scholar]
- Immobilized enzyme for screening and identification of anti-diabetic components from natural products by ligand fishing. Crit. Rev. Biotechnol.. 2021;43(2):242-257.
- [Google Scholar]
- Preparation of magnetic microcapsules of α-amylase and α-glucosidase for dual-target affinity screening of active components from Toona sinensis. Process. Biochem.. 2023;124:92-99.
- [Google Scholar]
- Magnetic separation techniques in sample preparation for biological analysis: a review. J. Pharm. Biomed. Anal.. 2014;101:84-101.
- [Google Scholar]
- Cell membrane-coated biomimetic magnetic nanoparticles for the bio-specific extraction of components from Gualou Guizhi decoction exhibiting activities against oxygen-glucose deprivation/reperfusion injury. J. Pharm. Biom. Anal.. 2022;209:114528
- [Google Scholar]
- Recent advances in nanomaterial-based biosensor for periodontitis detection. J. Biol. Eng.. 2024;18(1):28.
- [Google Scholar]
- The potential use of nanozymes as an antibacterial agents in oral infection, periodontitis, and peri-implantitis. J. Nanobiotechnol.. 2024;22(1):207.
- [Google Scholar]
- Synthesis and biological evaluation of indole derivatives containing thiazolidine-2,4-dione as α-glucosidase inhibitors with antidiabetic activity. Eur. J. Med. Chem.. 2024;264:115957
- [Google Scholar]
- Chemometrics coupled with UPLC-MS/MS for simultaneous analysis of markers in the raw and processed Fructus Xanthii, and application to optimization of processing method by BBD design. Phytomedicine.. 2019;57:191-202.
- [Google Scholar]
- A rapid strategy for screening high-efficiency PCSK9 inhibitors from Ginkgo biloba leaves by ligand fishing, HPLC-Q-TOF-MS and interdisciplinary assay. J. Food. Drug. Anal.. 2020;28(2):273.
- [Google Scholar]
- Pharmacokinetics of plantamajoside and acteoside from Plantago asiatica in rats by liquid chromatography–mass spectrometry. J. Pharm. Biomed. Anal.. 2014;89:251-256.
- [Google Scholar]
- Oriented covalent immobilization of esterase BioH on hydrophilic-modified Fe3O4 nanoparticles. Biotechnol. Appl. Biochem.. 2014;61(5):603-610.
- [Google Scholar]
- α-Glucosidase immobilization on polydopamine-coated cellulose filter paper and enzyme inhibitor screening. Anal. Biochem.. 2020;605:113832
- [Google Scholar]
- Bioactivity-guided isolation of compounds from Sophora flavescens with antibacterial activity against Acinetobacter baumannii. Nat. Prod. Res.. 2022;36(17):4334-4342.
- [Google Scholar]
- Thiazolidine-2, 4-dione derivatives as potential α-glucosidase inhibitors: Synthesis, inhibitory activity, binding interaction and hypoglycemic activity. Bioorg. Chem.. 2024;144:107177
- [Google Scholar]
- Molecular imprinting and immobilization of cellulase on to magnetic Fe3O4@SiO2 nanoparticles. J. Nanosci. Nanotechnol.. 2014;14(4):2931-2936.
- [Google Scholar]
- Synthesis and Biological Evaluation of 5-Fluoro-2-Oxindole Derivatives as Potential α-Glucosidase Inhibitors. Front. Chem.. 2022;10:928295
- [Google Scholar]
- Preparation, surface functionalization and application of Fe3O4 magnetic nanoparticles. Adv. Colloid. Interface. Sci.. 2020;281:102165
- [Google Scholar]
- Rapid characterization of the chemical constituents of Wangbi Capsule by UPLC coupled with Fourier transform ion cyclotron resonance mass spectrometry. Microchem. J.. 2022;180:107603
- [Google Scholar]
- Identification and determination of the chemical constituents in a herbal preparation, Compound Kushen injection, by HPLC and LC-DAD-MS/MS. J. Liq. Chromatogr. Relat. Technol.. 2014;37(2):207-220.
- [Google Scholar]
- Active compounds, antioxidant activity and α-glucosidase inhibitory activity of different varieties of Chaenomeles fruits. Food. Chem.. 2018;248:330-339.
- [Google Scholar]
- Investigation on the inhibition mechanism and binding behavior of cryptolepine to α-glucosidase and its hypoglycemic activity by multi-spectroscopic method. J. Lumin.. 2024;269:120437
- [Google Scholar]
- Facile and rapid detection of microalbuminuria by antibody-functionalized gold nanorods. Plasmonics. 2022;17(3):1269-1277.
- [Google Scholar]
- Identification of enzyme inhibitors using therapeutic target protein–magnetic nanoparticle conjugates. Anal. Methods. 2012;4(6):1612-1615.
- [Google Scholar]
- Spinel ferrite magnetic adsorbents: alternative future materials for water purification? Chem. Soc. Rev.. 2016;315:90-111.
- [Google Scholar]
- Effects of particle size and extraction methods on cocoa bean shell functional beverage. Nutrients. 2019;11(4):867.
- [Google Scholar]
- Evaluation of antioxidant, antidiabetic and antiobesity potential of selected traditional medicinal plants. Front. Nutr.. 2019;6:53.
- [Google Scholar]
- Improvement of antioxidant and moisture -preserving activities of Sargassum horneri polysaccharide enzymatic hydrolyzates. Int. J. Biol. Macromol.. 2015;74:420-427.
- [Google Scholar]
- Construction of Fe3O4@ α-glucosidase magnetic nanoparticles for ligand fishing of α-glucosidase inhibitors from a natural tonic Epimedii Folium. Int. J. Biol. Macromol.. 2020;165:1361-1372.
- [Google Scholar]
- UHPLC–MS/MS analysis on flavonoids composition in Astragalus membranaceus and their antioxidant activity. Antioxidants. 2021;10(11):1852.
- [Google Scholar]
- Enhanced Cr (VI) removal from acidic solutions using biochar modified by Fe3O4@SiO2@NH2 particles. Sci. Total. Environ.. 2018;628:499-508.
- [Google Scholar]
- α-glucosidase immobilization on magnetic core-shell metal-organic frameworks for inhibitor screening from traditional Chinese medicines. Colloids Surf. B: Biointerfaces. 2021;205:111847
- [Google Scholar]
- Photocatalytic C-C coupling from carbon dioxide reduction on copper oxide with mixed-valence copper (I)/copper (II) J. Am. Chem. Soc.. 2021;143(7):2984-2993.
- [Google Scholar]
- Traditional Chinese Medicine in the treatment of chronic kidney diseases: theories, applications, and mechanisms. Front Pharmacol.. 2021;13:917975
- [Google Scholar]
- Effects of salvianolic acid A on intestinal microbiota and lipid metabolism disorders in Zucker diabetic fatty rats. Diabetol. Metab. Syndr.. 2022;14(1):135.
- [Google Scholar]
- Phytochemical information and biological activities of quinolizidine alkaloids in Sophora: a comprehensive review. Curr. Drug. Targets. 2019;20(15):1572-1586.
- [Google Scholar]
- Characterization and rapid identification of chemical constituents of NaoXinTong capsules by UHPLC-linear ion trap/Orbitrap mass spectrometry. J. Pharm. Biomed. Anal.. 2015;111:104-118.
- [Google Scholar]
- Synthesis and anti-α-glucosidase activity evaluation of betulinic acid derivative. Arab. J. Chem.. 2023;16(5):104659
- [Google Scholar]
- Identification of 1, 3, 4-oxadiazolyl-containing β-carboline derivatives as novel α-glucosidase inhibitors with antidiabetic activity. Europ. J. Med. Chem.. 2023;261:115795
- [Google Scholar]
- Recent advance in the support and technology used in enzyme immobilization. Afr. J. Biotechnol.. 2009;8
- [Google Scholar]
- HPLC-TOF-MS and HPLC-MS/MS combined with multivariate analysis for the characterization and discrimination of phenolic profiles in nonfumigated and sulfur-fumigated rhubarb. J. Sep. Sci.. 2016;39(14):2667-2677.
- [Google Scholar]
- β-Glucosidase from tartary buckwheat immobilization on bifunctionalized nano-magnetic iron oxide and its application in tea soup for aroma and flavonoid aglycone enhancement. Food. Funct.. 2019;10(9):5461-5472.
- [Google Scholar]
- A preparation strategy for protein-oriented immobilized silica magnetic beads with Spy chemistry for ligand fishing. J. Pharm. Anal.. 2022;12(3):415-423.
- [Google Scholar]
- Development and validation of a UPLC-MS/MS method for the quantitative determination and pharmacokinetic analysis of cirsimarin in rat plasma. Biomed. Res. Int.. 2021;2021:1-7.
- [Google Scholar]
- Rapid characterization of the chemical constituents of Yinchen Wuling Powder by UPLC coupled with Fourier transform ion cyclotron resonance mass spectrometry. J. Pharm. Biomed. Anal.. 2021;198:114022
- [Google Scholar]
- Synthesis and biological evaluation of coumarin derivatives containing oxime ester as α-glucosidase inhibitors. Arab. J. Chem.. 2022;15(9):104072
- [Google Scholar]
- Comparative efficacy of seven Chinese patent medicines for early diabetic kidney disease: a bayesian network meta-analysis. Complement. Ther. Med.. 2022;67:102831
- [Google Scholar]
- Identification of anti-inflammatory constituents from Kalimeris indica with UHPLC-ESI-Q-TOF-MS/MS and GC-MS. J. Ethnopharmacol.. 2015;165:39-45.
- [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2024.105899.
Appendix A
Supplementary data
The following are the Supplementary data to this article:Supplementary Data 1
Supplementary Data 1







